Abstract
Recognition of highly degenerate mammalian splice sites by the core spliceosomal machinery is regulated by several protein factors that predominantly bind exonic splicing motifs. These are postulated to be single-stranded in order to be functional, yet knowledge of secondary structural features that regulate the exposure of exonic splicing motifs across the transcriptome is not currently available. Using transcriptome-wide RNA structural information we show that retained introns in mouse are commonly flanked by a short (≲70 nucleotide), highly base-paired segment upstream and a predominantly single-stranded exonic segment downstream. Splicing assays with select pre-mRNA substrates demonstrate that loops immediately upstream of the introns contain pre-mRNA-specific splicing enhancers, the substitution or hybridization of which impedes splicing. Additionally, the exonic segments flanking the retained introns appeared to be more enriched in a previously identified set of hexameric exonic splicing enhancer (ESE) sequences compared to their spliced counterparts, suggesting that base-pairing in the exonic segments upstream of retained introns could be a means for occlusion of ESEs. The upstream exonic loops of the test substrate promoted recruitment of splicing factors and consequent pre-mRNA structural remodeling, leading up to assembly of the early spliceosome. These results suggest that disruption of exonic stem–loop structures immediately upstream (but not downstream) of the introns regulate alternative splicing events, likely through modulating accessibility of splicing factors.
INTRODUCTION
To remove introns, mammalian splicing machinery recognizes 5′ (donor) and 3′ (acceptor) splice sites (SS) flanking the intron, as well as the intervening branchpoint site (BS) and the polypyrimidine tract (PPT) near the 3′-SS, yet the precise mechanisms of recognition remain unclear (1). The functionality of these ‘splice signals’ is dependent not only on their sequences but also on the context in which they are present within the pre-mRNA (2). Exonic segments flanking an intron contain a variety of splicing regulatory elements (3) including exonic splicing enhancers (ESEs) and exonic splicing silencers (ESSs) (4), both of which contain loosely conserved sequences for engagement of different RNA binding proteins and are important for regulated assembly of the spliceosome (5). One of the most studied regulators of ESE-dependent splicing activation is the serine–arginine-rich (SR) family of RNA-binding proteins that contain an N-terminal RNA-binding domain (RBD) and an Arg-Ser-rich (RS) domain at their C-terminus (6). SR-proteins promote constitutive splicing and regulate alternative splicing in vivo by binding to ESEs present within a short distance (∼50 nucleotides) upstream and downstream of 5′- and 3′-SS, respectively (7–11). Other trans-acting splicing factors have also been implicated in ESE-dependent splicing activation (12).
Recent studies indicate that RNA transcripts can adopt multiple secondary and tertiary structures in vivo (13–17), and that these structures modulate binding to partner protein complexes and regulate pre-mRNA processing (18–20). Co-transcriptional RNA folding is also known to be regulated by the rate of transcription elongation in eukaryotes (21), suggesting a coordinated regulation of splicing and transcription elongation (22–26). Although it is postulated that functional ESE and ESS sequences are present on single-stranded RNA (27), little information on the correlation of secondary structure of pre-mRNAs with transcriptome-wide distribution of ESE/ESS sequences for splicing regulation is available.
In this study, we investigate the secondary structure of intron proximal exonic regions. We observe that exonic segments immediately upstream of the 5′-SS display increased base-pairing in retained introns relative to spliced introns of the mouse embryonic stem cell transcriptome. A detailed investigation of these exonic segments to correlate secondary structure, the presence of enhancer/silencer elements, and SR protein binding indicates that the degree of base-pairing within exonic segments immediately upstream of the 5′-SS regulates assembly of the early spliceosome.
MATERIALS AND METHODS
Cloning and protein expression
cDNAs of SR proteins were purchased from Open Biosystems and cloned in a T7 promoter-based Escherichia coli expression vector (pET24d; EMD Millipore). Proteins contained a hexa-histidine (His6) -tag that is removable by TEV protease. Proteins were expressed in E. coli BL21 (DE3) cells with isopropyl β-d-1-thiogalactopyranoside induction and purified by Ni2+-nitrilotriacetate (Ni-NTA) affinity chromatography. The His6-tags were removed from all proteins used in our experiments by treatment with His6-TEV protease overnight at room temperature, and residual uncleaved proteins and His6-tagged TEV protease were removed by subsequent passage through Ni-NTA resin. Tag-removed proteins were further purified by size-exclusion chromatography (Superdex 75; GE Healthcare Lifesciences). Two functional variants of SR proteins used in our experiments were the RNA binding domain (RBD) of SRSF1 (amino acids 1–203) (28) and RBD of SRSF2 (amino acids 1–127) in chimera with fully phosphomimetic RS domain of SRSF1 (amino acids 197–246) in which all serine residues were replaced with glutamate (29). All purified proteins were confirmed to be RNase-free by incubating a small aliquot of the purified protein with a long RNA (e.g. β-globin RNA) overnight at room temperature and analyzing the RNA quality by urea-PAGE after phenol extraction.
Pre-mRNA constructs
The mouse pre-mRNA constructs with retained introns used in this study are 2610507B11Rik IVS17 (192+127+203) (chr11:78272778-78273299, mm10) and Amt IVS1 (104+102+427) (chr9:108296922-108297295, mm10). The model pre-mRNA substrates used are human β-globin IVS1 (109+130+204) and Adenovirus 2 major later transcript IVS1 (AdML) (65+123+50). The lengths of exon–intron–exon segments of pre-mRNA constructs are provided within parenthesis.
Electrophoretic mobility shift assay (EMSA)
Uniformly radiolabeled pre-mRNA was synthesized with [α-P32]UTP (3000 Ci/mmol; 10 μCi/μl) by in vitro (run off) transcription driven by T7 RNA polymerase (New England Biolabs), Next, the DNA template was digested with 2 units of DNase I (New England Biolabs) for 1 h at 37°C and the RNA was desalted twice by Illustra Microspin G-25 columns (GE Healthcare Life Sciences). Method for EMSA was modified from a previously published protocol (30). ∼10 pM radiolabeled pre-mRNA was incubated with an SR protein for 20 min at 30°C in 20 mM HEPES–NaOH (pH 7.5), 250 mM NaCl, 1 mM DTT, 2 mM MgCl2, 1 M urea, 20% glycerol and 0.3% polyvinyl alcohol (P-8136, Sigma) in 15 μl volume. Reaction products were resolved on 4% (89:1) polyacrylamide gels containing 2.5% glycerol and 50 mM Tris–glycine buffer. Gels were run at 250 V for 90 min at 4°C, dried, and analyzed by phosphorimaging.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)
The SHAPE assay was performed broadly following a protocol published before (31). Denatured and renatured β-globin pre-mRNA (25 nM) was incubated with SRSF1 (750 nM), SRSF2 (250 nM), or an equal volume SR protein storage buffer in the abovementioned EMSA condition (without glycerol) for 15 min. Similarly, denatured and renatured AdML pre-mRNA (25 nM) was incubated with SRSF1 (250 nM). Freshly prepared N-methylisatoic anhydride (NMIA, Aldrich) in dimethyl sulfoxide (DMSO, Sigma) was added to the test reactions at 2 mM (final concentration), and the reaction was allowed to proceed for 15 min at 30°C and then 15 min at room temperature. Equal volume of DMSO was added to the control reactions. Reaction mixtures were treated with proteinase K at 37°C for 10 min (New England Biolabs) and the RNA samples were purified by RNeasy Mini Kit (Qiagen). Reverse transcription reaction was carried out using MuLV Super-RT reverse transcriptase (Biobharati Life Science, India) using manufacturer's protocol at 50°C. cDNAs were precipitated with ethanol, resuspended in deionized formamide containing bromophenol blue, xylene cyanol and 0.5 mM EDTA, and separated in 12% and 8% 7 M urea sequencing gels of 0.35 mm thickness. In 8% gels, the same samples were loaded a second time into empty lanes when xylene cyanol of the first load migrated 12.5 cm. Electrophoresis was continued till xylene cyanol from the second load migrated 28 cm. We could resolve up to 125 nucleotides with each primer by this method. The individual bands were identified and quantified using Fiji (32). Where the lanes did not run straight, they were divided into several segments with overlapping regions, with each individual segment being straight. The quantified band intensities within the overlapping regions were used to normalize the values between consecutive segments. The independent values of a primer series from different gel runs (i.e. 12% and 8%) were also normalized. The band intensity values of the DMSO-treated control RNA were subtracted from those of NMIA-treated pre-mRNAs to obtain the raw SHAPE reactivity profiles. The raw reactivity data were processed as described in the earlier work (31). Briefly, all raw SHAPE reactivity values for a primer series were divided by the average of the top 10% values excluding the high-value outliers to obtain the processed values. Raw SHAPE reactivity values that were ≥1.5 times the median of all values for a primer series—capped at top 5% of all band intensity values in each primer series as the number of bands obtained with each primer was small, about 100–150—were considered outliers. All negative values were considered to be zero. Maximum values were capped at 3. For β-globin, we also processed the raw SHAPE-reactivity by 90% Winsorization for comparison to the trimming-based method described above. The processed values were analyzed with the ‘RNAstructure’ (33) software, which considered these values as pseudo-free energy of folding to generate the secondary structure models. Two variable parameters of RNA folding, the slope and the intercept, were set to 2.6 and –0.8, respectively. The RNA secondary structure models were drawn using VARNA (34). ΔSHAPE-reactivity for SR protein-bound RNA was calculated by subtracting the SHAPE reactivity of each nucleotide of the protein-free RNA from that of the corresponding nucleotide of the SR protein-bound RNA.
The primers used for reverse transcription of β-globin were 5′ACGTGCAGCTTGTCACAGTG (βgRT1), 5′TTTCTTGCCATGAGCCTTC (βgRT2), 5′AGTGGACAGATCCCCAAAG (βgRT3), 5′GGAAAATAGACCAATAGGC (βgRT4), 5′TTCTCTGTCTCCACATGCC (βgRT5), and 5′AACTTCATCCACGTTCACC (βgRT6). For reverse transcription of the EH3+4 mutant of β-globin, βgRT6 was replaced with EH-RT6 (5′CCAACTTCATGCTGGTGACG). Reverse transcription of AdML was performed with primers: 5′AAGAGTACTGGAAAGACCGC (AdRT1), 5′GGACAGGGTCAGCACGAAAG (AdRT2), and 5′CAGCGATGCGGAAGAGAGTG (AdRT3).
In vivo splicing assay
In vivo splicing assays with minigene constructs were performed in HeLa cells grown in a six-well plate (9 cm2 surface area) transfected with 1 μg of the purified plasmid. RNA was harvested 36 h post transfection using Trizol reagents (Thermoscientific) following manufacturer's instructions, treated with RNase-free DNase I (New England Biolabs), and purified by phenol:chloroform extraction and ethanol precipitation. cDNA was synthesized in a 20 μl volume using 100 ng RNA with MuLV Super RT reverse transcriptase (Biobharati Life Science, India) at 50°C following manufacturer's instructions. 1 μl of this cDNA was used in a 25 μl reaction volume for PCR. Non-saturating PCR products were analyzed in 2% agarose gels. Densitometric estimation was carried out by Fiji (32). The splicing efficiency (% spliced) was calculated using the formula: (mRNA/(mRNA + pre-mRNA + aberrant product))/(WT mRNA/(WT mRNA + WT pre-mRNA)) × 100. Each splicing assay was repeated three times. All substrates were expressed in vivo under the control of a CMV promoter (pcDNA3.1) since it represents a strong promoter (transcription rate ∼6–7 mRNAs per hour) near the median strength of endogenous mammalian promoters (35,36). New England Biolabs (Ipswich, MA, USA) DNA ladder 100 bp is used throughout the study and all size markers are annotated in order to indicate the size of the RT PCR products.
In vitro spliceosome assembly assay
Nuclear extract was prepared from HeLa cells as published (37). Spliceosomal complex assembly assay was carried out as described before (38).
RNA-seq data analysis
icSHAPE data was generated from publicly available raw data (15). The datasets polyA(+) icSHAPE DMSO biological replicates (Gene Expression Omnibus accession # GSM1464037, GSM1464038), polyA(+) icSHAPE in vitro NAI-N3 biological replicates (GSM1464039, GSM1464040), and polyA(+) icSHAPE in vivo NAI-N3 biological replicates (GSM1464041, GSM1464042) were used. The icSHAPE enrichment score was calculated as in (15) using previously published icSHAPE pipeline (https://github.com/qczhang/icSHAPE). Briefly, the difference between the reverse transcription stops in the treated sample and the control sample was divided by base density in the control sample. Outliers were removed by 90% Winsorization and the scores were then scaled from 0 to 1. This method tacitly integrates the biological variation between replicates into the icSHAPE enrichment score but does not generate separate icSHAPE enrichment score for individual replicates. Since NAI-N3 adducts formed at flexible 2′-OH group of the RNA is quickly hydrolyzed, only reasonably stable RNA structural features are likely captured by NAI-N3.
All exon–exon and exon–intron boundaries were identified using annotations in the GENCODE release M12. Only transcripts annotated to contain retained introns were searched for exon-retained intron boundaries. To identify the position of the retained intron, exon start and stop positions from retained intron transcripts were compared to those of coding transcripts originating from the same gene. Exons from retained intron transcripts that spanned two sequential exons in a related coding transcript were considered to contain introns, and the exon-retained intron boundaries were defined as the 3′ end of the upstream exon and the 5′ end of the downstream exon in the coding sequence. A 70-nt long region upstream of the 5′ boundary and downstream of the 3′ boundary was extracted for analysis. For comparison, 70-nt upstream and downstream of 10 000 randomly selected annotated exon-exon junctions in coding transcripts were analyzed. For both sets, transcripts with another exon-exon or exon–intron boundary, or a transcript end, within the 70 bp were discarded. The same method was used to isolate the exons downstream of the introns. Custom scripts sjshapePC.py and sjshapePC_3prime.py were used to collect icSHAPE enrichment score upstream and downstream of exon–exon junctions, respectively. intronHunterPC.py and intronHunterPC_3prime.py were used to collect the same for the exonic segments upstream and downstream of retained introns. The scripts are made available at https://github.com/englandwe/NAR_splicing and in Supplementary File 9. For standard error of the mean (SEM), we calculated the mean of all enrichment scores at the given position relative to the splice junction of a set of transcripts, with or without retained introns.
Counting ESE and ESS sequences
Hexamers assigned as ESE, ESS, or neutral sequences in (4) within target (70-nt) and non-target (80-nt) regions were counted using Jellyfish (39). ‘Splice score’ was calculated by summing up the individual scores of ESE and ESS as described before (4).
In vivo DMS-seq
HeLa cells transfected with the plasmids were treated with dimethyl sulfate as described before (40). For isolation of nuclei (41), transfected and DMS-treated cells were washed with ice-cold phosphate-buffered saline (PBS), scraped cells were centrifuged for 30 s at 1000 g, the supernatant was removed, cells were resuspended in 10 mM HEPES 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and 0.05% Nonidet P-40 and kept on ice for 10 min with occasional flicking. Then the suspension was centrifuged for 10 min at 730 g at 4°C. The pellet was resuspended in Trizol (Thermoscientific) and RNA was isolated following manufacturer's protocol. Reverse transcription of β-globin constructs was performed with βgRT5 (nested in the intron ensuring production of the cDNA synthesis from the pre-mRNA and not the mRNA). Reverse transcription and densitometric analyses were carried out as in SHAPE. For densitometric measurements, background of the images was subtracted.
Statistical analyses
The statistical significance of splicing assay results was calculated using one-sample t-test. P-values for icSHAPE and ESE/ESS analyses are from Welch's two-sample t-tests, with Bonferroni correction; the R-scripts are uploaded onto github (https://github.com/englandwe/NAR_splicing) and available in Supplementary File 9.
Calculation of ‘probability of being unpaired’ (PU) values by MEMERIS, RNAplfold and LocalFold
‘Probability of being unpaired’ or PU values for each hexamer within the given sequences were calculated using MEMERIS (27), RNAplfold (42), as well as LocalFold (43). MEMERIS uses RNAfold algorithm (44) to fold the input sequence. For each motif, we iterated MEMERIS calculations 20 times by varying the maximum lengths of the regions flanking the hexamer of interest from 11 to 30 nt (input sequence = 11–30 + hexamer + 11–30) and finally generated an average PU value for each motif. Thus, MEMERIS uses RNAfold to determine local RNA structure. We have made two corrections in the ‘Getsecondarystructurevalues.pl’ script of MEMERIS: We changed the regular expression to allow for multiple spaces preceding the free energy value in the RNAfold output to accommodate current RNAfold output formatting (lines 145 and 200), and modified output text formatting to handle non-numerical return values if RNAfold output is not properly recognized (at line 377). We have uploaded the revised script on github (https://github.com/englandwe/NAR_splicing) and provided in Supplementary File 9.
On the other hand, RNAplfold (42) also determines local structure in sliding windows (W) by imposing a maximum base-pair span (L; L≤ W) and calculates the probability of occurrence of a specific base-pair over all folding windows containing the sequence interval with the base-pair. RNAplfold was used with two sets of parameter for calculating PU values of individual hexamers: with L = W = 70 and with L = 70, W = 120 (W = L might cause unusually high base pairing probabilities of long-range base pairs, which could be resolved by setting W = L+50 (43)).
LocalFold uses RNAplfold algorithm to fold the RNA but corrects a prediction bias at each artificial border of the sliding window introduced by RNAplfold by ignoring the outlier values of 10-nt at the window borders but including them in the structure. We used L = 70 and W = 120 for LocalFold calculations.
For transcriptome-wide analysis, we kept up to a 30-nt sequence flanking the 70-nt ‘target region’ (see Results). The flanking 30-nt region from the transcript was collected using custom scripts flankFetch_2way*.py, which are available at https://github.com/englandwe/NAR_splicing and in Supplementary File 9.
RESULTS
Exonic segments immediately upstream of retained introns are highly base-paired
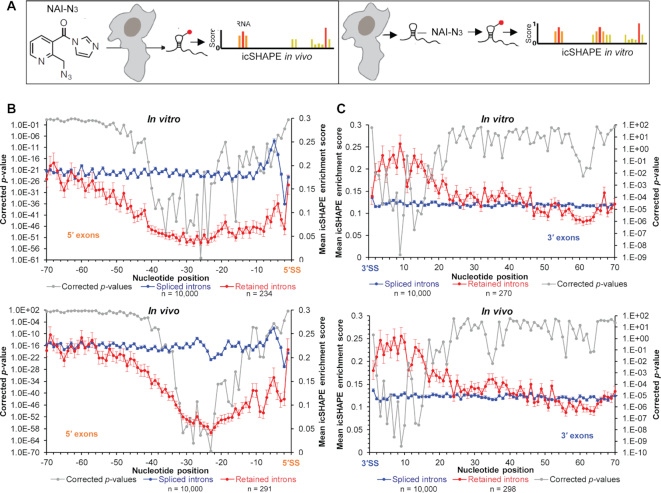
To assess structural flexibility of exons flanking mammalian splice sites, we analyzed structural profiles of mRNA using ‘in vivo click selective 2′-hydroxyl acylation and profiling experiment’ (icSHAPE) data for the mouse embryonic stem cell transcriptome (15). icSHAPE identifies single-stranded nucleotides by measuring 2′-hydroxyl flexibility. mRNAs used in the study are poly-A purified and thus have undergone processing. A schematic of the in vivo (reaction performed in cell) and in vitro (reaction performed with purified RNA) icSHAPE experiments is shown in Figure 1A. In vivo the ribonucleotide flexibility is governed by both base-pairing and by interactions with proteins; in vitro only the degree of base-pairing influences the readout. Both in vitro and in vivo icSHAPE data indicate low average reactivities of nucleotides in exonic regions immediately upstream of the 5′-SS preceding retained introns, suggesting that these segments exhibit a higher degree of base-pairing (Figure 1B). In contrast, equivalent exonic segments that precede spliced-out introns display a higher average icSHAPE reactivity. icSHAPE reactivities and sequences of the exons preceding retained introns are provided in Supplementary File 1. A similar study of the exonic regions (20–50 nt long) immediately downstream of the retained introns reveal higher icSHAPE reactivities compared to similar regions downstream of spliced introns (Figure 1C). The raw icSHAPE enrichment score of the in vivo dataset are provided in Supplementary File 10.
Figure 1.
icSHAPE analyses of mRNA from mouse embryonic stem cells. (A) Schematic of in vivo and in vitro icSHAPE analyses. (B) In vitro (top panel) and in vivo (bottom panel) icSHAPE enrichment score for 70-nt region upstream of 5′-SS in 10 000 exon–exon junctions with spliced introns (blue line) and 234/291 (in vitro/in vivo dataset with 219 shared) exon–intron junctions with retained introns in polyadenylated mRNAs (red line); error bars indicate standard error of the mean indicating variation in icSHAPE enrichment score at each position among the transcripts in each group (retained or spliced); P-values (corrected) for icSHAPE reactivity at individual nucleotide positions are indicated with a grey line. (C) In vitro (top panel) and in vivo (bottom panel) icSHAPE data for 70-nt region downstream of 3′-SS in 10 000 exon–exon junctions with spliced introns (blue line) and 270/298 (in vitro/in vivo dataset with 257 shared) intron–exon junctions with retained introns in polyadenylated mRNAs (red line), corrected P-values (gray line).
We estimated the G/C content of exonic segments and observed an enrichment in exons upstream of retained introns compared to those upstream of spliced introns (Supplementary Figure S1A). However, no statistically significant difference was observed in the GC content of exonic segments downstream of the retained introns compared to the spliced introns (Supplementary Figure S1B).
Then we calculated PU (probability of being unpaired) values of the exonic segments flanking the retained and spliced introns (described in the Method section). Plots showing the mean PU values per hexamer obtained by MEMERIS, RNAplfold (L = W = 70), RNAplfold (L = 70, W = 120), and LocalFold (L = 70, W = 120) of the spliced (Supplementary Figure S1C and D) and retained (Supplementary Figure S1E and F) introns across the transcriptome (in vivo dataset) are provided. The mean PU value per nucleotide was obtained by averaging the PU values of each hexamer overlapping an individual nucleotide. The six nucleotides of each hexamer were identified based on the fact that the RNAplfold output provides hexamer end-positions and MEMERIS output hexamer start positions. Plots representing the mean PU values per nucleotide are provided in Supplementary Figure S1G, H, I and J. The PU values obtained with RNAplfold (L = 70, W = 120) and LocalFold (L = 70, W = 120) appeared to be almost identical. A moderate correlation (r ≈ 0.7) was observed between mean icSHAPE enrichment score and mean PU values of the 70-nt long ‘target region’ upstream of the retained introns obtained by RNAplfold and LocalFold (Supplementary Table S1). The PU values of the other segments obtained with any method did not exhibit moderate or better (r ≥ 0.7) correlation with the mean icSHAPE enrichment score (Supplementary Table S1). The mean PU values are provided in Supplementary File 2 and raw PU values in Supplementary File 10.
We next investigated variations in size, distribution, and types of the retained introns. Intron size ranges between 67 and 15 574 nucleotides and distribution (Supplementary File 1) also varies widely. We looked for the presence of ‘exitrons’ (a small and unique class of retained introns present within protein-coding exons (45), the splicing of which inhibits functional protein synthesis), and observed four human exitron homologues in the in vitro mouse retained intron dataset (Supplementary File 1). We asked if a subset of the retained introns could be ‘detained introns’ (i.e. retained introns with longer half-lives undergoing post-transcriptional splicing (46)), and found that about one-fifth of the retained introns are also reported to be detained introns (Supplementary File 1).
Exonic loops immediately upstream of the 5′-SS are important for splicing
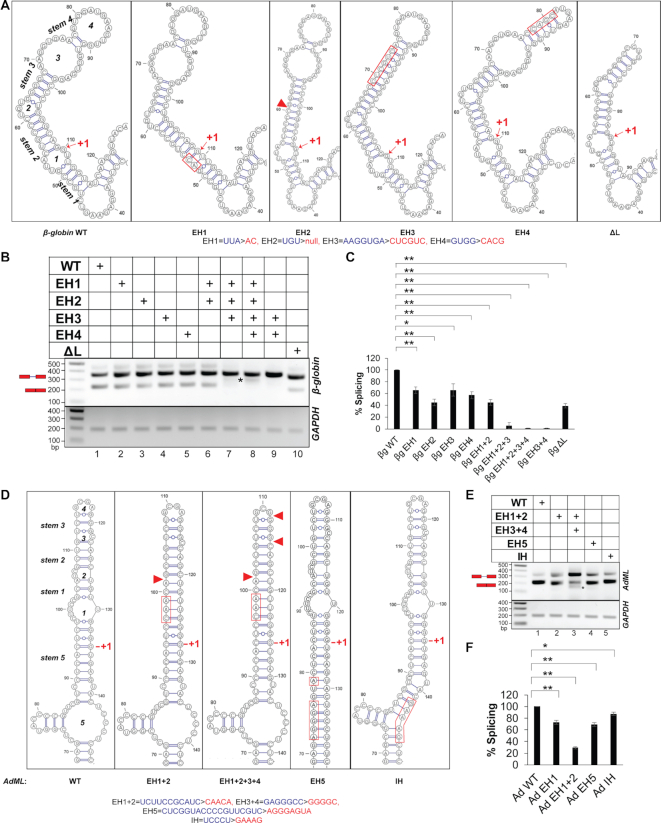
Since the retained introns are preceded by hybridized exonic segments, we hypothesized that loops in this region might be important for splicing activation. We tested this possibility by studying the structures and splicing activities of two well-studied, constitutively spliced substrates, human β-globin IVS1 (47) and Adenovirus 2 major late transcription unit IVS1 (AdML) (48). First, we determined secondary structural models of these substrates using in vitro SHAPE reactivity, which revealed that both substrates have four loops (loops 1–4) in the exonic segment immediately upstream of the 5′-SS (see Supplementary Figure S2A and B for SHAPE-derived secondary structure models and Supplementary Files 4 and 5 for representative examples of raw data). We then generated mutants with select loops eliminated (by replacing nucleotides on one strand, allowing it to hybridize to the opposite strand) and named them exon hybridization (EH) mutants. We also generated intron hybridization (IH) mutants with loops downstream of the 5′-SS (within introns) hybridized (Figure 2A and D, Supplementary Figure S2A and B). The 9-nt of 5′-SS and its three flanking nucleotides were unaltered.
Figure 2.
Secondary structure models and splicing activities of pre-mRNAs. (A) SHAPE-derived secondary-structure model of the WT β-globin segment immediately upstream of the 5′-SS (leftmost panel); the designation of loops and stems is noted; anticipated secondary structure models (based on the WT structure) of the exonic segment upstream of the 5′-SS of the mutants corresponding to hybridization/deletion of loops (EH/ΔL) are shown (remaining panels); substituted and deleted native sequence in the mutants are indicated with red rectangle and red triangle, respectively; the original and mutated sequences are shown at the bottom (also see Supplementary Figure S2A); the first nucleotide of the intron is marked with ‘+1’. (B) A representative image showing splicing products of WT, EH and ΔL mutants of β-globin in transfection-based splicing assay; asterisks indicate aberrantly processed products; pre-mRNA and mRNA positions are marked with a red schematic; GAPDH amplification performed as an internal control shown at bottom. (C) Bar-graph showing changes in spliced mRNA products for mutants as a percentage of wild-type RNA and normalized with total RNA generated from the transfected plasmids (n = 3); error bars indicate standard deviation; statistical significance was tested to validate if EH/ΔL mutants of β-globin have lower splicing competence than the WT substrate; ‘*’0.005 < P≤ 0.05, ‘**’P≤ 0.005, N.S. = not significant; the background subtracted gel-images of two additional biological replicates are shown in Supplementary File 8. (D) SHAPE-derived secondary-structure model of WT AdML exonic segment immediately upstream of the 5′-SS (leftmost panel); anticipated secondary structure models of the exonic and intronic hybridization mutants (EH and IH) are shown; original and mutated sequences are shown at the bottom; also see Supplementary Figure S2B. (E) A representative image of splicing products of WT, EH and IH mutants of AdML obtained by transfection-based splicing assay; asterisk and red schematic as in (B). (F) Bar-graph showing quantified mRNA along with the statistical significance of the differences; statistical significance was tested to validate if EH/IH mutants of AdML have lower splicing competence than the WT substrate; the background subtracted gel-images of biological replicates are shown in Supplementary File 8.
A transfection-based assay indicated that splicing was significantly reduced in all EH mutants. While up to a 50% reduction was observed in individual EH mutants and the EH1+2 mutant, a complete or near complete abolition of splicing was observed in the other combinatorial mutants (EH1+2+3, EH1+2+3+4, EH3+4) (Figure 2B and C). We confirmed base-pairing between strands (i.e. elimination of the loops) in the β-globin EH3+4 mutant experimentally by in vitro SHAPE (Supplementary Figure S2C, see Supplementary Files 4E–H and 5 for representative examples of raw data). In the mutant with all four terminal loops hybridized (EH1+2+3+4) (Figure 2A), authentic splicing was abolished, and an aberrant product was detected (Figure 2B, lane 8, aberrant product marked with an asterisk). The aberrant product matched the β-globin EH1+2+3+4 pre-mRNA with a deletion of a 63-nt region within the 5′ exon (between the 32nd and 96th nucleotides, highlighted by red borders in Supplementary Figure S3A, middle panel). We also generated a β-globin ΔL mutant in which loops 3 and 4 were replaced by a tetraloop (Figure 2A); this displayed a ∼50% loss of splicing (Figure 2B, compare lanes 1 & 10, Figure 2C). In contrast to the EH mutants, the IH mutants of β-globin (Supplementary Figure S2A) showed up to an approximately two-fold enhancement of splicing (Supplementary Figure S2D and E).
We next examined if splicing was restored by reintroducing loop(s) into the 5′ exon of the β-globin EH1+2+3+4 mutant (Supplementary Figure S3A, middle panel). We added loops in the same general locations of Loops 1 and 2 in the WT substrate but composed of sequences different from WT, and called the new variant EH1+2+3+4+5L2 (Supplementary Figure S3A, right panel; the straight cyan line in the middle panel indicates the sequence of EH1+2+3+4 mutated to generate EH1+2+3+4+5L2). We observed that addition of the new loops eliminated the aberrant product of EH1+2+3+4 and slightly improved bona fide splicing. We also added a large loop in the general location of loops 3+4 of the WT substrate to generate a terminal loop (TL) in the EH1+2+3+4 mutant, and called it EH1+2+3+4+TL (Supplementary Figure S3A, left panel; the straight black line in the middle panel indicates the sequence of EH1+2+3+4 mutated to generate EH1+2+3+4+TL). Splicing in this variant was restored to ∼50% of WT levels (Supplementary Figure S3B and C).
Similar to the β-globin mutants, AdML EH mutants (Figure 2D) also displayed defects in splicing. The most severe defect was observed when all four loops upstream of the 5′-SS were hybridized (EH1+2+3+4); an aberrant product was also noticed (Figure 2E, compare lanes 1 & 3, 3F). Sequencing of this aberrantly processed product indicated that a GU dinucleotide (at positions 76 & 77, demarcated with red borders in Supplementary Figure S2B) can act as a cryptic 5′-SS and pair with the authentic 3′-SS. Hybridization-mutation at an intronic location of the 5′-SS-containing stem-loop (IH) (Figure 2D) produced a small (∼10%) defect in splicing (Figure 2E, compare lanes 1 & 5, Figure 2F). These results suggest that single-stranded regions immediately upstream of the 5′-SS may play essential roles in promoting splicing; however, these experiments have not clarified the role of specific nucleotide sequences within these exonic regions.
Single-stranded sequence immediately upstream of the 5′-SS promotes splicing in a sequence- and context-specific manner
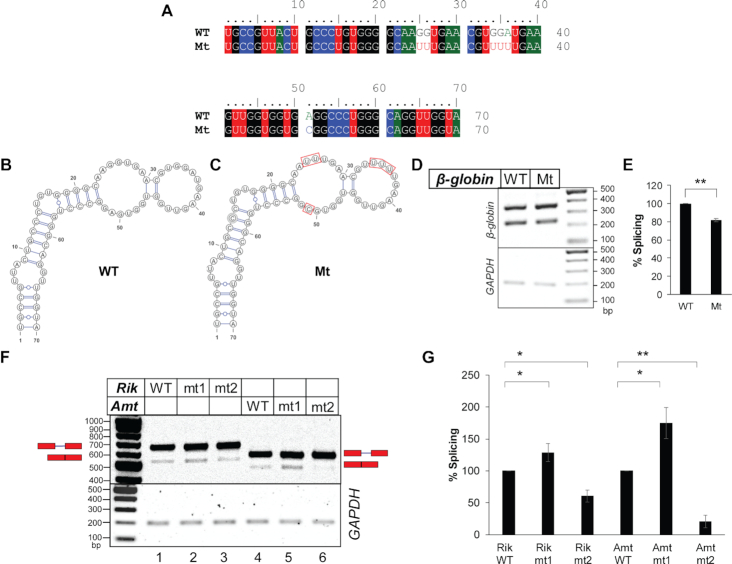
We assessed the importance of the nucleotide sequence within native exonic loops for splicing of constitutively spliced substrates. Here we focused on the loop regions of β-globin stem–loops 3 and 4. We made multiple purine-to-pyrimidine substitutions within the loops without disrupting the secondary structure of the stem–loop as predicted by ‘RNAstructure’ (33). Among these mutants, only mutant Mt (see Figure 3A for mutation, 3B and 3C for predicted secondary structure models of Mt and WT) exhibited a ∼20% reduction in splicing efficiency compared to the WT (Figure 3D and E). The remaining mutants did not exhibit any change in splicing. Due to high degeneracy in consensus sequence for binding of splicing factors and the presence of numerous splicing factors inside the nuclei with varied sequence specificity, mutations without an effect on splicing is not useful for evaluating the importance of an RNA sequence on splicing. Therefore, the results of the mutants other than that of Mt are not shown.
Figure 3.
Sequence- and context-specific effects of single-stranded nucleotide sequence in the exonic segments immediately upstream of the introns on splicing. (A) Sequence alignment of the 5′ exonic segment of WT and mutant β-globin variants. (B, C) Secondary structure models of the above exonic segment of WT β-globin (B) and the mutant β-globin (C) (both predicted by ‘RNAstructure’), which are identical to experimentally derived secondary structure of WT β-globin shown in Supplementary Figure S2A. (D) Transfection-based splicing assay of the WT and the mutant β-globin variants. (E) Plots of quantified band-intensities shown in (D) (n = 3); statistical significance was tested to validate if the splicing competence of Mt mutant of β-globin is less than that of WT β-globin; ‘*’0.005 < P≤ 0.05, ‘**’P≤ 0.005, N.S. = not significant; background-subtracted gel-images of two additional biological replicates are shown in Supplementary File 8. (F) Splicing products of mouse pre-mRNAs 2610507B11Rik IVS17 (Rik) and Amt IVS1 (Amt) and their mt1 and mt2 mutants (see Supplementary Figure S5) visualized in agarose gel (representative); positions of pre-mRNAs and mRNAs are marked and GAPDH amplification used as an internal control shown; the original ‘Image Lab’ image File is provided as Supplementary File 3. (G) A bar-graph showing comparison of spliced mRNAs normalized with total RNA generated from the transfected plasmids; error bars (n = 3) indicate standard deviation; statistical significance was tested to validate if mt1 mutants have higher and mt2 mutants have lower splicing competence than the WT substrate; ‘*’0.005 < P≤ 0.05, ‘**’P≤ 0.005; the background subtracted gel-images of two additional biological replicates used for densitometric analysis are shown in Supplementary File 8.
Next, we examined if expansion of single-stranded segments (by disrupting base-pairing in the stems) immediately upstream of the 5′-SS of β-globin and AdML enhances splicing. We generated exonic de-hybridization (ED) mutants by removing base pairs from stems 1, 2, 3 or 4 of β-globin (Supplementary Figure S4A) and stems 1, 2, 3 or 5 of AdML (Supplementary Figure S4D). Splicing of these substrates revealed no clear trend. Only ED1 of β-globin exhibited splicing enhancement while the rest did not. On the other hand, all but one AdML ED mutant exhibited a reduction in splicing; only (ED1+5) exhibited no significant change. (Supplementary Figure S4B, C, E and F). Since the base substitutions to generate ED mutants were random (without any emphasis on specific sequence), the unpredictable splicing changes observed were likely due to random changes in the strength of available ESE and ESS elements of β-globin.
Next, we examined how replacing a single-stranded as well as a base-paired exonic sequence immediately upstream of the 5′-SS with a strong single-stranded heterologous hexameric ESE (4) altered the splicing activity of two mouse pre-mRNA substrates exhibiting intron retention. The mouse pre-mRNAs chosen have sufficient reads mapped to the retained introns in both in vivo and in vitro datasets (2610507B11Rik IVS17 and Amt IVS1, Supplementary Figure S5A). Secondary structure models based on their in vitro icSHAPE reactivities were generated using ‘RNAstructure’ (33) (Supplementary Figures S5B and C). Mutant ‘mt1’ was generated by replacing a base-paired native sequence with a single-stranded, high-ranking ESE (UCAUCG- ranked 35) (4) and ‘mt2’ was generated by replacing a loop sequence with another strong ESE (GAAGAA- ranked 17). Rik IVS17 mt1 exhibited a small increase in splicing efficiency (Figure 3F and G); on the other hand, Rik mt2 exhibited a decrease in splicing efficiency. mt1 of Amt IVS1 was created by replacing the base-paired native sequence with AAGAAA (ESE rank 25), and in mt2 of Amt IVS1 the native sequence of the loop was replaced with AAGAAC (ESE rank 102). Amt IVS1 mt1 exhibited an increase in splicing while mt2 exhibited a decrease (Figure 3F and G).
Overall, these results suggest that insufficient single-stranded ESE sequences within exons immediately upstream of the retained introns could be responsible for splicing defects, and that loops could display context-specific ESE which may not be replaceable with a heterologous ESE. Specific unwinding of the naturally hybridized exonic sequence immediately upstream of the retained introns and demonstration of its functional relevance in promoting splicing is difficult. However, the observation of enhanced splicing in the mt1 mutants of both mouse pre-mRNA substrates suggests that a single-stranded segment in place of the base-paired stem in the exonic region immediately upstream of the 5′-SS could promote splicing of the retained introns in a sequence/context-specific manner.
Exonic base-pairs immediately upstream of the retained introns appear to occlude ESEs transcriptome-wide
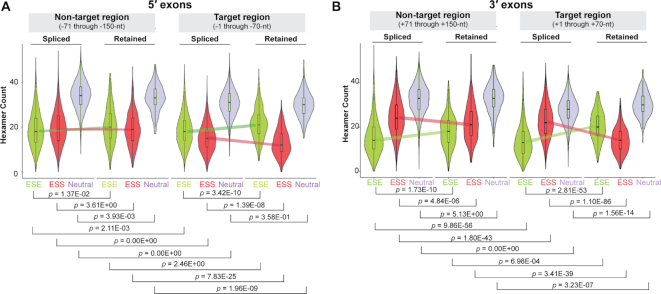
The results shown above suggest that pre-mRNA-specific ESEs are present in the single-stranded loop regions and may be occluded within the base-paired regions immediately upstream of the mouse retained introns. We therefore asked if this base-pairing occludes ESE sequences throughout the transcriptome. We chose the set of hexameric ESE, ESS, and neutral sequences identified by Ke et al. (4) and examined their occurrences in the exonic segments upstream and downstream of both retained and spliced introns across the transcriptome. Although the strength of ESE and ESS sequences is context-dependent, the observation of any distinct transcriptome-wide trend will likely provide a general idea regarding the distribution of ESE and ESS sequences with respect to the exonic secondary structure. A 70-nt stretch upstream or downstream of the intron was considered the ‘target region’, and the next 80-nt long exonic segment upstream or downstream was regarded as the ‘non-target region’. Figure 4A and B shows combination (violin + box) plots of distribution of recognized hexameric ESE and ESS, as well as neutral hexameric sequences (4) in both upstream (5′) and downstream (3′) exonic segments of retained and spliced introns in the in vivo dataset. Higher ESE and lower ESS sequence counts were observed in both 5′ and 3′ exonic ‘target regions’ flanking retained introns than were observed in similar regions flanking spliced introns (Figure 4A and B). ‘Non-target regions’ in the 3′ exons but not the 5′ exons showed a similar difference.
Figure 4.
Distribution of ESE/ESS counts in the exonic segments flanking the retained introns. (A) Counts of occurrences of hexameric ESE, ESS, and neutral sequences (green, red and blue, respectively) (4) in the –1 to –70-nt (target region) and –71 to –150-nt (non-target region) ranges upstream of the 5′-SS of retained and spliced introns. (B) Similar counts in the +1 to +70-nt (target region) and +71 to +150-nt (non-target region) ranges downstream of the 3′-SS of retained and spliced introns; P-values are indicated; colored lines indicate deviation of medians between spliced and retained introns.
In order to gain an idea of the average splicing potential for specific exonic segments (both target and non-target regions), we calculated their overall ‘splice score’ by summing up individual ESE and ESS scores. The values for hexameric ESE and ESS sequences (positive and negative, respectively) were obtained from Ke et al. (4) (and were therefore calculated based on the potential of these sequences to regulate splicing within the context of their experiments). We observed ‘splice scores’ of target regions flanking the retained introns to be higher than regions flanking the spliced introns (Supplementary Figure S6A and B). Also, in the case of retained introns, the splice scores for target regions were higher than those for non-target regions.
Since the presence of common motifs within the exonic segments flanking the retained introns might indicate similarity in their splicing regulation, we looked for de novo motifs in exonic segments upstream of retained introns in the in vivo dataset versus the control group (spliced introns) using MEME (49) and MAST (50). The sequence motif CTTCTG and its reverse complement (sequence logos shown in Supplementary Figure S6C) occurred 122 and 235 times respectively (Supplementary File 1) within the exonic segments upstream of 207 retained introns. The CTTCTG hexamer as well as its reverse complement were classified as ESEs in a previous study (4).
Since occlusion of ESE sequences by base-pairing potentially inhibits their ability to promote splicing (27,51), and the exonic segments flanking the retained introns are on average enriched in hexameric ESE sequences identified by Ke et al. (4), we hypothesized that occlusion of ESE or ESE-like sequences present in the 5′ exonic region by means of base-pairing could be one of the reasons for intron retention. However, comparison of icSHAPE reactivity (Figure 1B and C) and ESE count (Figure 4A and B) between retained and spliced introns indicated a more significant icSHAPE reactivity difference and a less significant ESE count difference in the ‘target region’ of upstream exons compared to those of downstream exons. These findings suggest that base-pairing of the ‘target regions’ may have additional purposes beyond regulating the exposure of ESEs.
WT β-globin exhibits greater degree of single-strandedness than its EH3+4 mutant in vivo
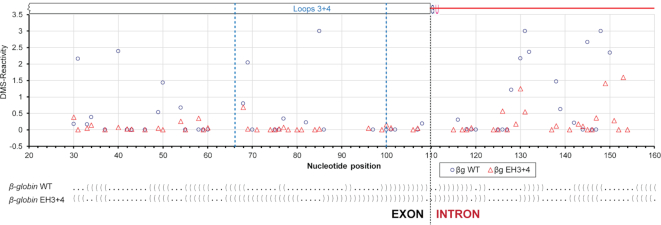
The hybridization mutations are planned based on ‘static’ in vitro models of full-length pre-mRNAs. Therefore, the questions remain whether the mutants acquire the expected structure upon mutagenesis in vivo. In order to explore possibilities of how the exonic loops in the target region promote splicing in vivo, we wanted to examine the secondary structural features of β-globin WT and the EH3+4 mutant in vivo. We carried out in vivo DMS-footprinting of the 5′ exon–intron junction region of β-globin variants expressed from the transfected minigene. In vivo DMS-reactivity is shown in Figure 5 and the raw data are provided as Supplementary Files 6A–D and 7. The DMS reactivity of A and C residues are mapped onto the SHAPE-derived secondary structure model of β-globin (Supplementary Figure S7). It is anticipated that an ensemble of structural states during the course of splicing are represented in vivo. The exonic region corresponding to stem–loops 3 and 4 in the splicing-competent WT substrate showed higher reactivity than that in the splicing-defective EH3+4 mutant substrate. Significantly, we also observed higher DMS-reactivity at a number of additional segments in both exons and introns outside of stem–loops 3 and 4 in the WT substrate compared to the EH3+4 mutant. In summary, in vivo WT β-globin exhibited more single-strandedness within as well as beyond the exonic segment immediately upstream of the 5′-SS compared to the EH3+4 mutant. Since protein-coding RNAs tend to be less structured in vivo than in vitro due to structural modulation predominantly caused by protein binding (15,52), we surmise that the exonic loops promote recruitment of splicing factors that in turn may induce structural modulation of the pre-mRNA.
Figure 5.
In vivo structural probing of β-globin WT and its EH3+4 mutant expressed from transfected minigenes. Quantified reverse transcription stops (DMS-reactivity) of individual nucleotides (A & C only) of β-globin WT (o) and EH3+4 mutant (Δ) are plotted against nucleotide positions; dot-bracket notations, which are aligned with the nucleotide positions in the plot, of the SHAPE-derived secondary structure models of protein-free WT β-globin and its EH3+4 mutant are shown at the bottom (dot = unpaired nucleotide, bracket = base-paired nucleotide).
Single-strandedness immediately upstream of the intron is important for structural remodeling of β-globin and AdML by SR proteins
The results described above led us to examine if binding of splicing factors causes structural modulations in vitro, and whether such remodeling is regulated by secondary structure of the exonic segment immediately upstream of the 5′-SS. We focused on SR protein-mediated structural modulations of the model substrates, β-globin and AdML since they are known to be activated for splicing by the SR proteins (53–55).
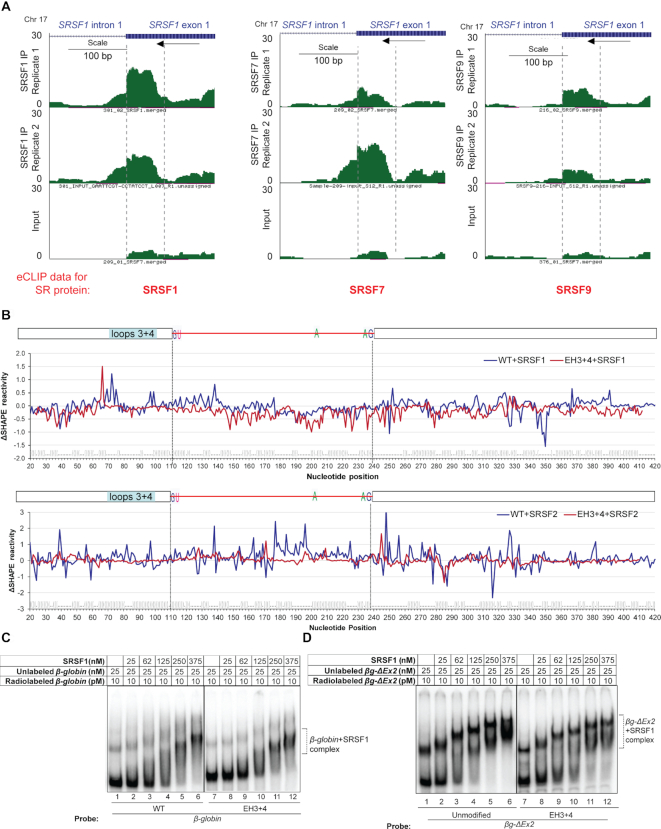
We examined data from multiple transcriptome-wide crosslinking and immunoprecipitation experiments followed by RNA-seq (CLIP-seq), which identified that SR proteins bind predominantly to exonic regions within ∼50-nt of the 5′-SS and 3′-SS (8–10). Enhanced CLIP (eCLIP) data (9) for binding of three different SR proteins (SRSF1, SRSF7 and SRSF9) to SRSF1 exon 1 are shown in Figure 6A (www.encodeproject.org), which shows the presence of eCLIP peaks for all three SR proteins within the ∼70-nt region immediately upstream of SRSF1 intron 1. We used EMSA to assess binding of SR protein SRSF1 to β-globin and AdML pre-mRNAs (500 nM unlabeled pre-mRNAs with radiolabeled tracer) (Supplementary Figure S8A, B and C). The stoichiometric binding assay revealed that ∼30 molecules of SRSF1 are needed to saturate a WT β-globin pre-mRNA molecule (Supplementary Figure S8A, lane 6) and ∼9 molecules for a WT AdML molecule (Supplementary Figure S8B, lane 6). This difference may be reflective of the much longer 3′ exon of β-globin (200-nt) compared to that of AdML (50-nt). A similar experiment with SRSF2 indicated that 10 molecules are required for saturation of a β-globin molecule (Supplementary Figure S8C, lane 5). Overall, these studies show binding of multiple molecules of SR proteins to a pre-mRNA molecule, which may be related to dose-dependent splicing regulation by SR proteins (56).
Figure 6.
Effect of exonic loops immediately upstream of the 5′-SS to functional SR protein binding and pre-mRNA structural modulation. (A) Genome browser views of 5′ exon of SRSF1 IVS1 with mapped SR protein (SRSF1, SRSF7 and SRSF9) eCLIP reads (9) showing the exonic region immediately upstream of the 5′-SS to overlap with high confidence SR protein binding sites; mapped SR protein read densities from two eCLIP biological replicate experiments performed in HepG2 cells and one size-matched input are plotted at the same y-axis scale; scale bar and the directions of transcription (by an arrow) are denoted (www.encodeproject.org); 70-nt exonic segment immediately upstream of the intron is marked with two vertical broken lines. (B) (Top) SHAPE reactivity differential (ΔSHAPE reactivity) of free and SRSF1-bound β-globin WT (blue line) and EH3+4 mutant (red line). (Bottom) SHAPE reactivity differential of free and SRSF2-bound β-globin WT (blue line) and EH3+4 mutant (red line); dot-bracket notation of WT β-globin secondary structure is shown at the bottom, which is aligned with nucleotide positions; position of 70-nt at the 3′ end of the upstream exon (−1 through −70-nt) is indicated. (C) Comparison of SRSF1 binding to full-length WT β-globin and its EH3+4 mutant by EMSA. (D) Comparison of SRSF1 binding to βg-ΔEx2 variants (truncated β-globin lacking the downstream exon) with and without EH3+4 mutation (compare lanes 4, 5, and 6 with 10, 11 and 12, respectively).
We next probed structural modulation of WT β-globin and its EH3+4 mutant upon binding of SRSF1 and SRSF2, and of WT AdML upon binding of SRSF1 by monitoring changes to in vitro SHAPE reactivity. WT β-globin and its EH3+4 mutant were saturated independently with SRSF1 and SRSF2, and WT AdML with SRSF1. The resulting SHAPE reactivity differentials (ΔSHAPE-reactivity) were plotted against individual nucleotide positions (see Figure 6B top panel for β-globin+SRSF1, Figure 6B bottom panel for β-globin+SRSF2, Supplementary Figure S8D for AdML+SRSF1, and Supplementary Files 4 and 5 for representative examples of raw data of β-globin WT+SRSF1 and β-globin EH3+4+SRSF1). Positive and negative ΔSHAPE-reactivity values indicate gain and loss of reactivity upon SR protein binding and imply an increase and decrease of nucleotide flexibility respectively. ΔSHAPE-reactivities (both loss and gain) of the β-globin EH3+4 mutant were diminished compared to the reactivities of the WT substrate. We also analyzed the raw SHAPE data of β-globin and β-globin+SRSF1 by 90% Winsorization and compared the resulting ΔSHAPE-reactivity with that described above obtained by ‘trimming’ (Supplementary File 5, sheet 3). Both methods yielded comparable results (P = 1.53E–02).
As performed for the retained and spliced introns, we calculated the PU values for hexamers of β-globin and AdML using MEMERIS, RNAplfold (L = 70, W = 120) and LocalFold (L = 70, W = 120). RNAplfold and LocalFold values were almost identical while MEMERIS values were somewhat different (Supplementary Figure S9). No obvious positive correlation between the SHAPE reactivity of protein-free pre-mRNAs as well as SRSF1-bound pre-mRNAs and the PU values for different segments of the model substrates were observed (Supplementary Table S2). Similarities in trend of the curves of SHAPE reactivity of protein-free as well as SRSF1-bound pre-mRNAs and the PU values in certain small patches were observed by visual inspection of the plots (Supplementary Figure S9). PU values obtained by both MEMERIS and LocalFold of the PPT region (215–239-nt) of β-globin are distinctly high but the protein-free pre-mRNA shows only eight unpaired nucleotides in this region (Supplementary Figure S2A). Interestingly, SR protein binding to the WT substrates appeared to maintain, enhance, or weakly suppress the flexibility of the PPT region while SR protein binding to the EH3+4 mutant appeared to strongly suppress the flexibility of this region (Figure 6B): the P-value of the difference in ΔSHAPE reactivity in the PPT and its flanking areas [150–250 nt] of β-globin WT+SRSF1 and β-globin EH3+4+SRSF1 mutant is 3.90E–08; and that of β-globin WT+SRSF2 and β-globin EH3+4+SRSF2 is 1.88E–09. This might be reflective of the requirement of a more single-stranded PPT region for splicing (57).
We further compared SRSF1 binding to WT and EH3+4 mutant of β-globin by EMSA (Figure 6C). No difference in SRSF1 binding efficiency between the WT and the mutant β-globin was clearly discernible, except that the band formed upon addition of 375 nM SRSF1 was slightly less compact in the case of β-globin EH3+4 than in WT β-globin. Since the presence of a long (200-nt) 3′ exon could suppress any binding difference, we generated a βg-ΔEx2 construct with the 3′ exon of β-globin removed. The band formed in the presence of 375 nM SRSF1 with 25 nM βg-ΔEx2 is clearly more compact than the one formed with 25 nM βg-ΔEx2-EH3+4 (Figure 6D, compare lanes 6 with 12). These results suggest that loops in the exonic segments immediately upstream of the 5′-SS govern the nature of SRSF1 binding to β-globin.
Single-strandedness immediately upstream of the intron is important for E-complex assembly
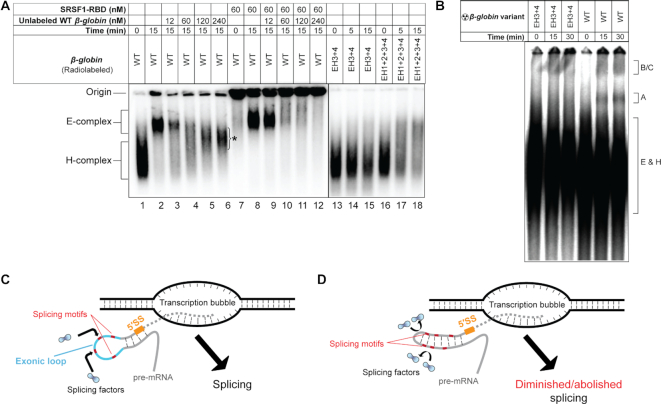
To assess the effect of exonic secondary structure on formation of the splicing complex, we examined assembly of the E-complex on β-globin WT and its EH3+4 and EH1+2+3+4 mutants in nuclear extract. Native sequence pre-mRNA formed the H-complex at 0 min (i.e., upon mixing) and E-complex in 15 min (Figure 7A, lanes 1 and 2). E-complex formation was compromised in both with EH3+4 and EH1+2+3+4 mutants (Figure 7A, lanes 13–18), in solid agreement with the results from the in vivo splicing assay results in Figure 2. Additionally, we observed that pre-incubation of native β-globin (performed with ∼10 pM radiolabeled pre-mRNA) with SRSF1 resulted in greater efficiency of E-complex formation (Figure 7A, compare lanes 2 and 8). Pre-incubated substrate was able to survive a challenge of ∼1000-fold excess (unlabeled) substrate while untreated substrate was not (compare lanes 3 and 9). As expected, β-globin of native sequence was able to form spliceosomal complexes A and B, but its EH3+4 mutant was not (Figure 7B). A schematic of this function of exonic secondary structure is presented in Figure 7C and D.
Figure 7.
Importance of exonic loops immediately upstream of the 5′-SS for assembly of the early spliceosome. (A) Comparison of E-complex assembly with 10 pM radiolabeled β-globin pre-incubated with SRSF1 in the presence or the absence of 12 nM untreated unlabeled competitor β-globin (compare lanes 3 and 9); E-complex assembly assay performed with β-globin EH3+4 and EH1+2+3+4 mutants (lanes 13–18); an asterisk indicates undefined pre-mRNA complexes; nuclear extract from the same preparation is used in all experiments. (B) Spliceosomal complex (A, B/C) assembly assay with β-globin WT and its EH3+4 mutant. (C, D) Schematics depicting the role of exonic secondary structure in spliceosome assembly; a pre-mRNA in the process of being released from the transcription bubble is shown; solid grey lines indicate exons and dotted grey lines intron; orange line indicates 5′-SS, cyan line the exonic loop(s) immediately upstream of the 5′-SS, and short red lines putative splicing motifs. In case of splicing competent introns, the exonic segment immediately upstream of the 5′-SS contain single-stranded segments, which promote recruitment of appropriate splicing factors for splicing. Stems in place of exonic loops either occlude the splicing motifs within or provide steric hindrance to recruitment of a splicing factor at nearby sites or both, thus diminishing splicing.
DISCUSSION
Exonic stem–loops are postulated to provide structural context to splice sites, predominantly by regulating exposure of ESE and ESS sequences (27,58) and influencing their ability to recruit splicing factors. We observed that exonic segments immediately upstream of retained introns transcriptome-wide are extensively base-paired and enriched in at least one set of previously identified hexameric ESE sequences compared to those upstream of the spliced introns. In contrast, the downstream exonic segments, while also enriched in ESE sequences compared to their spliced counterparts, appeared to be more single-stranded. With select pre-mRNA substrates, we observed that loops present in the upstream exonic segments promote splicing in a sequence- and context-dependent manner. In vitro, we observed that these loops facilitate recruitment of SR proteins and induce structural modulation of the pre-mRNA, perhaps exposing additional splice signals. In vivo, the pre-mRNA appeared more single-stranded in the presence of the loops than in their absence, suggesting involvement of splicing factors in structural modulation of the pre-mRNA. In addition to being enriched in ESE sequences, the exonic segments flanking the retained introns also exhibited a dearth of ESS sequences identified by Ke et al. (4) compared to the exonic segments flanking the spliced introns. This might suggest an apparent higher splicing potential of the exonic segments flanking the retained introns than those of the spliced introns. Based on these observations, we propose the following model for mammalian splicing regulation: Single-stranded ESEs immediately up- and down-stream of splice sites bind sequence-specific protein factors which modulate the pre-mRNA structure to sequentially recruit components of the spliceosome core. Extensive base-pairing immediately upstream of the retained introns reduces functional interactions between ESEs and splicing factors. Exposed ESEs immediately downstream of the retained introns cannot sufficiently substitute for the function of occluded ESEs in the 5′-exon. Promotion of splicing triggered by unwinding of the exonic segments upstream of the retained introns likely is not interfered by exposed ESS sequences due to their low counts. A previous study suggested that ∼40-nt exonic regions immediately upstream but not downstream of the retained introns are highly base-paired in Arabidopsis thaliana (14), suggesting that a similar mechanism of intron retention might also prevail in higher plants.
It will be intriguing to determine if the structural feature(s) associated with the exonic segments are correlated with alternative, biologically significant intron retention events. However, in the absence of structural data (e.g. icSHAPE reactivity) from multiple cell lines or cells under different conditions, we can only hypothesize about the possibility of exonic secondary structure contributing to alternative retention of an intron in different circumstances. Biologically, significant alternative intron retention regulated by exonic secondary structure might be effectuated by different RNA polymerase II elongation rates affecting co-transcriptional pre-mRNA folding, differential activities of helicases, or both either in different cell-types or in the same cell-type under different conditions. Differential folding of a pre-mRNA may not only change the accessibility of specific sequences within the exons but could also affect the efficiency of unwinding by helicases, which in turn may depend on the strength of base-pairing within the stem.
Observation of minor correlation between PU values obtained by various methods and SHAPE/icSHAPE reactivity in a few segments/patches could stem from our incomplete understanding of the principles of RNA folding. The prediction of local structure is dependent on fixed ‘motif-length’ and fixed L and/or W parameters and thus, could be limited. Furthermore, in vivo, RNA could fold differently in the presence of interacting proteins and RNA folding is often regulated by the transcription elongation rate; these possibilities cannot be easily considered for calculation of PU values. Overall, our data indicates a lack of a strong obvious correlation between PU values and SHAPE/icSHAPE reactivity and emphasize the need for experimentally determined RNA structure for a clearer understanding of RNA structure-function relationship.
There are many additional questions that need to be answered to better appreciate the impact of exonic stem–loops in splicing. Splicing efficiency of full-length pre-mRNAs with multiple exons and introns could depend upon a number of factors including cis-acting signals present at any distance from individual introns (59). It is not yet clear how these signals integrate with exonic stem-loop properties to regulate splicing. It is also not known if the binding of other splicing factors besides SR proteins is regulated by exonic secondary structure. Additionally, our data does not reflect how the extent of single-strandedness in the upstream exonic segment correlates with splicing efficiency. For example, AdML splices more efficiently than β-globin in spite of the fact that the former contains a less single-stranded region immediately upstream of the intron than the latter.
In this report, we experimentally verified the effects of base-pairing and single-strandedness of the exonic segment on splicing using pre-mRNAs containing short introns. It is not clear if such a regulatory mechanism is universal for all pre-mRNAs. Further studies exploring a genome-wide secondary structural profile of pre-mRNAs in cells with arrested splicing will reveal the extent to which single-stranded segments upstream of the 5′-SS are required for splicing. Additionally, our data show that SR protein-mediated structural modulation of the pre-mRNA is important for splicing but it does not explain whether or how variation in intron retention (often achieved by varying the expression level of SR proteins (8)), is regulated by SR protein-mediated pre-mRNA structural remodeling. The mechanism behind the compensatory effects of multiple SR proteins on splicing (8) and the likely link to pre-mRNA structural modulation are also unclear and requires further investigation. Finally, as observed with exonic secondary structure, the intronic secondary structure may also have a regulatory role in splicing. For example, certain types of intronic hybridization have been proposed to be involved in the promotion of splicing (60). Our preliminary observations of intronic hybridization (IH) mutants of β-globin showing distinct enhancement in splicing and the IH mutant of AdML exhibiting a small defect in splicing suggest that intronic secondary structure could have contributed to splicing regulation. However, regulatory schemes mediated by intronic secondary structure is likely different and different sets of splicing factors could be involved. Further studies are required to unravel the role of intronic secondary structure on splicing.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Drs Stephan Leung and Suhyung Cho for experiments at the early stage of this work, Drs Gene Yeo and Stefan Aigner for providing expertise on eCLIP data analysis, Drs Matt Daugherty and Shankar Subramaniam for critical discussion on bioinformatic analyses, and Drs Simpson Joseph and Tom Huxford for critical reading of the manuscript.
Author contributions: K.S. designed and performed majority of the experiments, analyzed the data, and wrote the paper. T.B. analyzed data and wrote the paper. M.F. performed protein purification, cloning, and site-directed mutagenesis experiments, and helped write the paper. R.S. and W.E. performed and analyzed the icSHAPE data and wrote the paper. G.G. planned experiments, analyzed data, wrote the paper, and supervised the project.
Notes
Present address: Mike Minh Fernandez, Department of Systems Biology, Beckman Research Institute, City of Hope, 1500 E Duarte Rd, Duarte, CA 91010-3000, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM085490 and GM084277 to G.G., 5UM1HG009443 to R.C.S.]; R.C.S. is a Pew Biomedical Scholar; Helmsley Charitable Trust (Scholarship to K.S.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Will C.L., Luhrmann R.. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wong M.S., Kinney J.B., Krainer A.R.. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol. Cell. 2018; 71:1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goren A., Ram O., Amit M., Keren H., Lev-Maor G., Vig I., Pupko T., Ast G.. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol. Cell. 2006; 22:769–781. [DOI] [PubMed] [Google Scholar]
- 4. Ke S., Shang S., Kalachikov S.M., Morozova I., Yu L., Russo J.J., Ju J., Chasin L.A.. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res. 2011; 21:1360–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hollander D., Naftelberg S., Lev-Maor G., Kornblihtt A.R., Ast G.. How are short exons flanked by long introns defined and committed to splicing. Trends Genet. 2016; 32:596–606. [DOI] [PubMed] [Google Scholar]
- 6. Zhou Z., Fu X.-D.. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013; 122:191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Änkö M.L., Müller-McNicoll M., Brandl H., Curk T., Gorup C., Henry I., Ule J., Neugebauer K.M.. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012; 13:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Jr M.A., Fu X.-D.. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell. 2013; 50:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al.. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanford J.R., Wang X., Mort M., VanDuyn N., Cooper D.N., Mooney S.D., Edenberg H.J., Liu Y.. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009; 19:381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Screaton G.R., Cáceres J.F., Mayeda A., Bell M.V., Plebanski M., Jackson D.G., Bell J.I., Krainer A.R.. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995; 14:4336–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chasin L.A. Blencowe B.J., Graveley B.R.. Alternative Splicing in the Postgenomic Era. 2007; Austin: Landes Biosciences; 85–106. [Google Scholar]
- 13. Lu Z., Zhang Q.C., Lee B., Flynn R.A., Smith M.A., Robinson J.T., Davidovich C., Gooding A.R., Goodrich K.J., Mattick J.S. et al.. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016; 165:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M.. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014; 505:696–700. [DOI] [PubMed] [Google Scholar]
- 15. Spitale R.C., Flynn R.A., Zhang Q.C., Crisalli P., Lee B., Jung J.W., Kuchelmeister H.Y., Batista P.J., Torre E.A., Kool E.T. et al.. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015; 519:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corley M., Solem A., Phillips G., Lackey L., Ziehr B., Vincent H.A., Mustoe A.M., Ramos S.B.V., Weeks K.M., Moorman N.J. et al.. An RNA structure-mediated, posttranscriptional model of human α-1-antitrypsin expression. Proc. Natl Acad. Sci. U.S.A. 2017; 114:E10244–E10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahen E.M., Watson P.Y., Cottrell J.W., Fedor M.J.. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010; 8:e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C., Burge C.B.. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambert N., Robertson A., Jangi M., McGeary S., Sharp P.A., Burge C.B.. RNA bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014; 54:887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mustoe A.M., Busan S., Rice G.M., Hajdin C.E., Peterson B.K., Ruda V.M., Kubica N., Nutiu R., Baryza J.L., Weeks K.M.. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell. 2018; 173:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan T., Sosnick T.. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006; 35:161–175. [DOI] [PubMed] [Google Scholar]
- 22. Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014; 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jimeno-González S., Payán-Bravo L., Muñoz-Cabello A.M., Guijo M., Gutierrez G., Prado F., Reyes J.C.. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14840–14845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Voong L.N., Xi L., Sebeson A.C., Xiong B., Wang J.-P., Wang X.. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016; 167:1555–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X.-D., Bentley D.L.. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014; 28:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong J.J., Gao D., Nguyen T.V., Kwok C.T., van Geldermalsen M., Middleton R., Pinello N., Thoeng A., Nagarajah R., Holst J. et al.. Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 2017; 8:15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hiller M., Zhang Z., Backofen R., Stamm S.. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007; 3:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cho S., Hoang A., Sinha R., Zhong X.-Y., Fu X.-D., Krainer A.R., Ghosh G.. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandler S.D., Mayeda A., Yeakley J.M., Krainer A.R., Fu X.-D.. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:3596–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kohtz J.D., Jamison S.F., Will C.L., Zuo P., Lührmann R., Garcia-Blanco M.A., Manley J.L.. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994; 368:119–124. [DOI] [PubMed] [Google Scholar]
- 31. McGinnis J.L., Duncan C.D., Weeks K.M.. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009; 468:67–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathews D.H. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinformatics. 2014; doi:10.1002/0471250953.bi1206s13. [DOI] [PubMed] [Google Scholar]
- 34. Darty K., Denise A., Ponty Y.. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., Singer R.H.. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007; 14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 37. Mayeda A., Krainer A.R.. Haynes S.R. Methods in Molecular Biology (RNA-protein interaction protocols). 1999; 118:Humana Press (Springer)309–314. [Google Scholar]
- 38. Das R., Reed R.. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999; 5:1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marcais G., Kingsford C.. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011; 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zubradt M., Gupta P., Persad S., Lambowitz A.M., Weissman J.S., Rouskin S.. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods. 2017; 14:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nicolaides N.C., Stoeckert C.J. Jr. A simple, efficient method for the separate isolation of RNA and DNA from the same cells. BioTechniques. 1990; 8:154–156. [PubMed] [Google Scholar]
- 42. Bernhart S.H., Hofacker I.L., Stadler P.F.. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006; 22:614–615. [DOI] [PubMed] [Google Scholar]
- 43. Lange S.J., Maticzka D., Mohl M., Gagnon J.N., Brown C.M., Backofen R.. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012; 40:5215–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorith. Mol. Biol.: AMB. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marquez Y., Hopfler M., Ayatollahi Z., Barta A., Kalyna M.. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 2015; 25:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boutz P.L., Bhutkar A., Sharp P.A.. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015; 29:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krainer A.R., Maniatis T., Ruskin B., Green M.R.. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984; 36:993–1005. [DOI] [PubMed] [Google Scholar]
- 48. Bennett M., Michaud S., Kingston J., Reed R.. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992; 6:1986–2000. [DOI] [PubMed] [Google Scholar]
- 49. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 50. Bailey T.L., Gribskov M.. Combining evidence using P-values: application to sequence homology searches. Bioinformatics. 1998; 14:48–54. [DOI] [PubMed] [Google Scholar]
- 51. Buratti E., Baralle F.E.. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004; 24:10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S.. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014; 505:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhu J., Krainer A.R.. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000; 14:3166–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hertel K.J., Maniatis T.. Serine–arginine (SR)-rich splicing factors have an exon-independent function in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2651–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fu X.-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993; 365:82–85. [DOI] [PubMed] [Google Scholar]
- 56. Eperon I.C., Makarova O.V., Mayeda A., Munroe S.H., Caceres J.F., Hayward D.G.. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000; 20:8303–8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kralovicova J., Sevcikova I., Stejskalova E., Obuca M., Hiller M., Stanek D., Vorechovsky I.. PUF60-activated exons uncover altered 3′ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. 2018; 46:6166–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramanouskaya T.V., Grinev V.V.. The determinants of alternative RNA splicing in human cells. Mol. Genet. Genomics: MGG. 2017; 292:1175–1195. [DOI] [PubMed] [Google Scholar]
- 59. Kim S.W., Taggart A.J., Heintzelman C., Cygan K.J., Hull C.G., Wang J., Shrestha B., Fairbrother W.G.. Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res. 2017; 45:9503–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Soemedi R., Cygan K.J., Rhine C.L., Glidden D.T., Taggart A.J., Lin C.L., Fredericks A.M., Fairbrother W.G.. The effects of structure on pre-mRNA processing and stability. Methods. 2017; 125:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.