Abstract
Glioblastoma multiforme (GBM), a lethal brain tumor developing in the white matter of the adult brain, contains a small population of GBM stem cells (GSCs), which potentially cause chemotherapeutic resistance and tumor recurrence. However, the mechanisms underlying the pathogenesis and maintenance of GSCs remain largely unknown. A recent study reported that incorporation of ribosomes and ribosomal proteins into somatic cells promoted lineage trans‐differentiation toward multipotency. This study aimed to investigate the mechanism underlying stemness acquisition in GBM cells by focusing on 40S ribosomal protein S6 (RPS6). RPS6 was significantly upregulated in high‐grade glioma and localized at perivascular, perinecrotic, and border niches in GBM tissues. siRNA‐mediated RPS6 knock‐down significantly suppressed the characteristics of GSCs, including their tumorsphere potential and GSC marker expression; STAT3 was downregulated in GBM cells. RPS6 overexpression enhanced the tumorsphere potential of GSCs and these effects were attenuated by STAT3 inhibitor (AG490). Moreover, RPS6 expression was significantly correlated with SOX2 expression in different glioma grades. Immunohistochemistry data herein indicated that RPS6 was predominant in GSC niches, concurrent with the data from IVY GAP databases. Furthermore, RPS6 and other ribosomal proteins were upregulated in GSC‐predominant areas in this database. The present results indicate that, in GSC niches, ribosomal proteins play crucial roles in the development and maintenance of GSCs and are clinically associated with chemoradioresistance and GBM recurrence.
Keywords: cancer stem cell, glioblastoma, GBM stem‐like cells, ribosomal protein S6, ribosome
We evaluated ribosomal protein S6 (RPS6) that has important roles in developing glioblastoma stem‐like cells. RPS6‐positive cells were highly expressed in glioblastoma stem‐like cell dominant areas. Also, we tested the RPS6 regulated stem cell properties of glioblastoma stem‐like cells. These findings are a break‐through in targeting therapy for ribosomal proteins to downregulate stem cell properties in glioblastoma.
Abbreviations
- CT
cellular tumor
- GBM
glioblastoma multiforme
- GSC
GBM stem cell
- HBV
Hyperplastic blood vessels in cellular tumor
- IT
infiltrating tumor
- LE
leading edge
- MVP
microvascular proliferation
- NSC
neural stem cell
- PA
pilocytic astrocytomas
- PAN
pseudopalisading cells around necrosis
- PNZ
perinecrotic zone
- RPS6
ribosomal protein 6
1. INTRODUCTION
Glioblastoma multiforme is the most aggressive type of brain tumor, 1 with a mean survival of 14.6 mo. 2 , 3 Standard therapies for GBM include surgical resection and chemoradiotherapy. 3 Despite several therapeutic strategies, the prognosis for patients has not improved sufficiently over the past 3 decades. GBM usually recurs around the cavity of the resected tumor, even upon complete surgical resection of enhanced mass lesions detected by magnetic resonance imaging. Part of the reason for its chemoradioresistance is the presence of a small population of cells with stem cell‐like characters, so called GBM stem cells (GSC) in GBM. 4 GSCs are generally present in a special microenvironment, the GSC niche, and retain their potential for self‐renewal, chemoradioresistance, and tumorigenicity. 5 Despite different types of GSC niches, such as perivascular, perinecrotic, and border niches, 6 , 7 the mechanisms underlying the promotion of stem cell‐like features in glioma cells by these niches remain largely unknown. Therefore, to improve survival of GBM patients, it is important to understand the detailed mechanism underlying GSC development and to develop new therapeutic strategies targeting GSCs.
Ribosomal proteins participate in numerous biological phenomena, primarily including protein synthesis. 8 Ribosomes are comprised of two subunits, small and large, which synergistically function and translate mRNA into a polypeptide chain during protein synthesis. 8 , 9 Ribosomes contain approximately 80 ribosomal proteins, which contribute to tumorigenesis, immune signaling, development, and several diseases. 10 Ribosomal incorporation into somatic cells reportedly promoted lineage trans‐differentiation toward multipotency, and these cells differentiated into different germ layer‐derived cells upon induction with respective media, such as adipocytes, osteocytes, and chondrocytes. 11 , 12 , 13 These reports suggest that levels of ribosomal proteins influence the induction and maintenance of stem cell‐like features.
Ribosomal protein S6, a component of the 40S ribosomal subunit, 14 plays important roles in cell proliferation and DNA repair. 15 shRNA‐mediated knock‐down of RPS6 reportedly suppressed cancer cell proliferation and metastasis in lung cancer, 16 and RPS6 phosphorylation is important for pancreatic cancer pathogenesis. 17 However, the mechanism of action of RPS6 in GBM and whether it induces malignant characters in GBM cells remain unclear. This study aimed to investigate the effect of RPS6 on stemness induction in GBM.
2. MATERIALS AND METHODS
2.1. Antibodies and reagents
Rabbit polyclonal anti‐RPS6 antibody was obtained from Abcam. Mouse polyclonal anti‐β‐actin antibody, rabbit polyclonal anti‐STAT3, rabbit polyclonal anti‐p‐STAT3(Tyr705), and rabbit polyclonal anti‐SOX2 were purchased from CST. Rabbit polyclonal anti‐glial fiblirally acidic protein (GFAP) antibody was obtained from DAKO. Rabbit polyclonal anti‐Olig2 antibody was obtained from IBL. Mouse polyclonal anti‐MAP2 was obtained from Sigma and mouse monoclonal anti‐Nestin antibody was obtained from MERCK. AG490 was obtained from Sigma and dissolved in 50 µmol/L DMSO (Sigma) and then diluted in culture medium.
2.2. Patients, tissue specimens, and immunohistochemistry
For western blot analysis, resected glioma tissues were harvested from 12 patients (seven males, five females, low‐grade: six cases, high‐grade: six cases) at Kumamoto University Hospital, Japan, after having obtained informed consent from the patients and in accordance with the guidelines of the Research Ethics Committee (Kumamoto University Hospital No. 1470). All samples were rapidly frozen after surgery and stored at − 80°C. Demographic data of the patients are provided in Table 1.
TABLE 1.
Demographic data of the patients
| Patient | Grade | Age | Sex | Diagnosis |
|---|---|---|---|---|
| 1 | 1 | 29 y | M | Pilocytic astrocytoma |
| 2 | 1 | 6 y | M | Pilocytic astrocytoma |
| 3 | 1 | 9 y | F | Pilocytic astrocytoma |
| 4 | 2 | 60 y | F | Astrocytoma |
| 5 | 2 | 48 y | M | Astrocytoma |
| 6 | 2 | 10 mo | F | Astrocytoma |
| 7 | 3 | 49 y | F | Anaplastic astrocytoma |
| 8 | 3 | 35 y | M | Anaplastic oligoastrocytoma |
| 9 | 3 | 58 y | M | Anaplastic astrocytoma |
| 10 | 4 | 74 y | M | GBM |
| 11 | 4 | 58 y | M | GBM |
| 12 | 4 | 69 y | F | GBM |
For immunohistochemical analysis, tumor tissues were fixed in formalin overnight, embedded in paraffin, and then cut into 3‐μm‐thick sections. The following antibodies were used to detect antigens: rabbit anti‐RPS6 (1:100; Abcam), rabbit anti‐GFAP (1:4000; DAKO), rabbit anti‐SOX2 (1:100; CST), rabbit anti‐Nestin (1:200; Merck), rabbit anti‐Olig2 (1:100; IBL), and mouse anti‐MAP2 (1:5000; Sigma) antibodies. Reactions were visualized using a diaminobenzidine substrate system (Nichirei).
2.3. Cell lines and cell culture
The human GBM cell line U251MG was obtained from the Japanese Collection of Research Bio Resources Cell Bank. Cells were cultured in Dulbecco's modified Eagle's medium and Ham's medium (GIBCO) with 10% FBS in an atmosphere containing 5% CO2 in air at 37°C.
2.4. Sphere‐formation assay
U251MG cells (104 cells/100 µL/well) were cultured in NSC culturing condition (serum‐free medium supplemented with growth factors, as recently reported 18 , 19 ) using a 96‐well plate (Corning) for 3 d. The spheres (size >50 µm) were enumerated using a microscope. Upon transfection of plasmid‐DNA or siRNA, U251MG cells were cultured in NSC medium.
2.5. Western blot analysis
Cells were lysed in ice‐cold lysis buffer containing phosphatase inhibitor (CST). Whole patient tissues and proteins from cultured cells were extracted using cell lysis buffer in accordance with the manufacturer's instructions (CST). Proteins were separated through SDS‐PAGE in a 10% resolving gel and electrotransferred onto polyvinylidene difluoride membranes (Merck).
Membranes were blocked in 3% skim milk (GE Healthcare) with TBS‐T at ambient temperature for 1 h with agitation and incubated with the following primary antibodies overnight at 4°C: rabbit polyclonal anti‐RPS6 antibody (1:1000), mouse polyclonal anti‐β‐actin antibody (1:1000), rabbit polyclonal anti‐STAT3 (1:1000), rabbit polyclonal anti‐p‐STAT3 (Tyr705) (1:1000), rabbit polyclonal anti‐SOX2 (1:1000), and mouse monoclonal anti‐Nestin (1:1000) antibodies. After three washes, the membranes were incubated with horseradish peroxidase‐conjugated secondary antibody (rabbit: GE Healthcare, mouse: GE Healthcare) with 3% skim milk for 1 h. Finally, immunoreactive protein bands were visualized using ECL select detection reagents (GE Healthcare) and detected using LAS4000EPUVmini (GE Healthcare).
2.6. siRNA transfection
U251MG cells were incubated in 6‐well plates (Corning) (1.0 × 105 cells/2 mL/well) for 24 h and transiently transfected with RPS6‐specific siRNA (10 nmol/L), using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s protocol. After transfection and incubation for 48 h, experiments were performed. Silencer Negative Control siRNA (Applied Biosystems, Life Technologies) was used as the control. The RPS6‐specific siRNA sequences were 5‐GAAGCGUAUGGCCACAGAAtt for the top strand and 5‐UUCUGUGGCCAUACGCUUCtc for the bottom strand (Thermo Fisher Scientific).
2.7. Transfection with plasmid‐DNA
U251MG cells were incubated in 12‐well plates (Corning) (1.6 × 105 cells/mL/well) for 24 h and transiently transfected with plasmid‐DNA (pcDNA3.0‐RPS6; Addgene) and negative control (pcDNA3.0) at 1 µg/mL each, using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer’s protocol.
2.8. Analysis of the GBM database
To analyze the publicly available GBM database, we analyzed IVY GAP databases. 20 Patient clinical data were obtained from a previous study. 20 RNA sequencing (RNA‐seq) data were obtained from the Ivy GAP database 20 made publicly available by the Allen Institute (©2015 Allen Institute for Brain Science. Ivy Glioblastoma Atlas Project2). We compared the expression levels of ribosomal proteins including RPS6 within the tumor mass (CT zone) with those in zones predominantly containing GSC‐like cells (PAN, MVP, LE, and IT identified through hematoxylin and eosin staining). In total, 122 RNA samples were generated from 10 tumors. Samples from the anatomic structures were harvested through laser microdissection and sequenced to a depth of ~30 million reads (~15 million mapped reads). Sets of genes specific to or enriched in CT, MVP, and PAN were identified. In particular, genes meeting the following statistical criteria were included in the initial list of potential marker genes (https://ivygap.swedish.org/home). RNA‐seq data for anatomic structures and putative cancer stem cell clusters were obtained via laser microdissection. Tumorigenesis was assessed using the IVY GAP database (http://glioblastoma.alleninstitute.org/ish). Fragments per kilobase of exon per million mapped fragments (FPKM) values were averaged across replicates within regions to generate single values for each sample within a particular region.
2.9. Statistical analysis
Data are expressed as mean ± standard deviation values. Student t test was used to assess differences between the two groups. P‐values <.05, **P mean <.01, and ***P mean <.001 were considered statistically significant.
3. RESULTS
3.1. RPS6 expression in glioma tissues
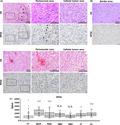
RPS6 is upregulated and contributes to malignancy in several cancers such as lung cancer and renal cell carcinoma 16 , 21 ; however, its role in glioma remains unclear. To assess the role of RPS6 in GBM cells, we evaluated RPS6 expression in GBM tissues. We performed immunohistochemical staining of GBM tissue samples with RPS6 polyclonal antibody. Strong immunostaining of RPS6 was detected in the tumor area; however, weak immunostaining was detected in areas containing lesser tumor cells (Figure 1A). Furthermore, to evaluate RPS6 expression in gliomas of different grades, tissues of GBM and PA were used. RPS6 immunoreactivity in PA was weaker than that in GBM (Figure 1B). Moreover, we assessed the expression of SOX2 and Nestin, GSC markers; GFAP, an astrocyte marker; MAP2, a neuronal marker; and Olig2, an oligodendrocyte progenitor cell marker. Both SOX2 and Nestin were upregulated in the area where RPS6 was detected. GFAP and Olig2 but not MAP2 were detected in the tumor area (Figure 1C). Immunofluorescent double staining of GBM tissues showed that the majority of RPS6‐positive cells were positive for Nestin (99.3%), Olig2 (89.5%), and SOX2 (86.0%) (Figure S1). These immunohistochemistry results indicate that RPS6 expression is strongly associated with the primitive feature and malignant phenotype of gliomas.
FIGURE 1.
Ribosomal protein S6 (RPS6) expression in glioma tissues. Representative images of hematoxylin and eosin (HE) and immunohistochemical staining. A, Images showing positive RPS6 immunoreactivity in tumor areas and lesser tumor cells in glioblastoma multiforme (GBM). B, Images showing RPS6 expression in GBM and pilocytic astrocytoma. C, Images showing immunoreactivity of GFAP, MAP2. RPS6, SOX2, Nestin, and Olig2, which are markers of stem and progenitor cells, in GBM tissue containing tumor cells. Scale bars, 100 µm
3.2. RPS6 knock‐down suppressed the sphere‐forming potential and GSC marker expression in GBM cells
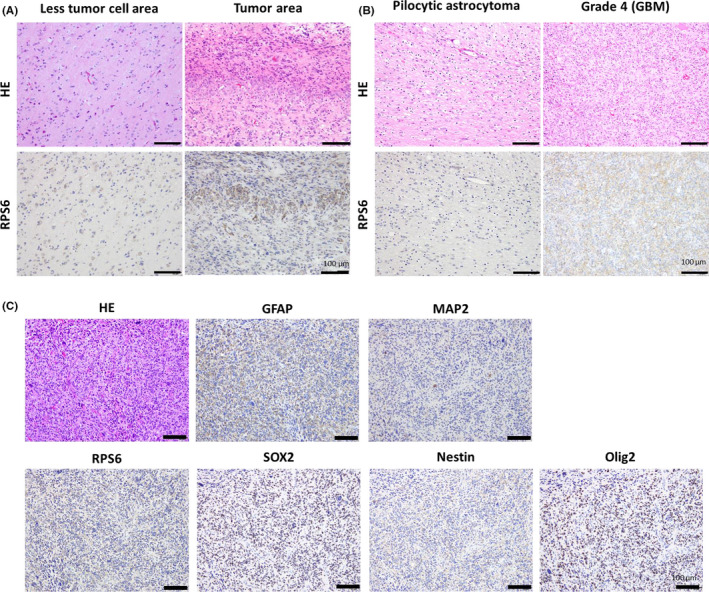
To investigate whether RPS6 expression is associated with stemness properties in glioma cells, the sphere formation assay was performed using the GBM cell line U251MG. RPS6 was strongly inhibited in U251MG cells upon transfection of RPS6‐specific siRNA (siRPS6) compared with that in cells transfected with control siRNA (siCont) (decreased by 54%) (Figure 2A). Cells transfected with both siRPS6 and siCont were cultured in NSC medium for 3 d and their sphere‐forming potential was assessed by enumerating spheres larger than 50 μm in diameter. The sphere‐forming potential of siRPS6 was significantly lower than that of siCont (decreased by 6%, P < .05) (Figure 2B), and the average size of spheres decreased (Figure S2A). To confirm this result, we performed the same experiment using U87MG cells and obtained similar results (Figure S4A). Furthermore, stemness‐related proteins Nestin and SOX2 were strongly suppressed in cells transfected with siRPS6 rather than with siCont (Nestin: decreased by 8%, P < .05, SOX2: decreased by 10%, P < .05), as revealed through western blotting (Figure 2C). These results were confirmed in experiments using another RPS6‐specific siRNA (siRPS6#2) (Figure S3). These results suggest that RPS6 downregulation by siRPS6 clearly diminished the stemness properties in GBM cells.
FIGURE 2.
Ribosomal protein S6 (RPS6) knock‐down suppressed the sphere‐forming potential and expression of glioblastoma multiforme (GBM) stem cell (GSC) markers in glioma cells. A, GBM cells (U251MG) were transfected with control siRNA (siCont) or siRNA specific for RPS6 (siRPS6). RPS6 expression was confirmed at the protein level upon western blotting. B, RPS6 knock‐down experiments; sphere‐forming potential in GBM cell line (U251MG) cultured in 10% FCS and NSC medium. Graph showing the number of spheres (>50 µm). C, Western blot data showing that RPS6 knock‐down affected the expression of GSC markers Nestin and SOX2 in GBM cell line (U251MG) cultured in neural stem cell (NSC) medium. A graph showing the expression of Nestin and SOX2 in siRPS6‐transfected cells or siCont‐transfected cells. D, Empty control vector (Control) or RPS6‐expressing plasmid vector (OE‐RPS6) was transfected into GBM cell line (U251MG). Images displaying sphere formation by OE‐RPS6. Graph showing the number of spheres (>50 µm) in NSC medium (values are presented as means ± SD of triplicate samples, *P < .05 siCont vs siRPS6 and Cont vs OE‐RPS6). Scale bar, 100 µm
Conversely, to investigate the effect of RPS6 in GBM cells, a plasmid vector expressing RPS6 was transfected into U251MG cells (OE‐RPS6). RPS6 was consequently upregulated by 1.7‐fold in OE‐RPS6 in western blots, the sphere‐forming potential was significantly increased by 1.5‐fold (P < .05) (Figure 2D), and the average size of spheres was increased (Figure S2B). To confirm these results, we performed the same experiments using U87MG cells and obtained similar results (Figure S4B). These data indicate that RPS6 plays important roles in the acquisition of stem cell‐like characters in GBM cells in vitro.
3.3. RPS6 regulates the stemness of GBM cells via STAT3 signaling
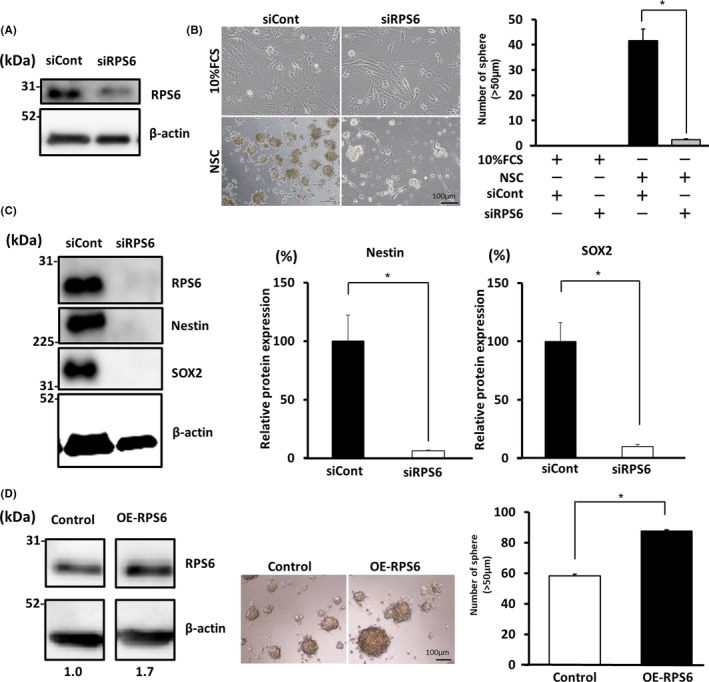
To determine the pathway involved in the acquisition of stemness properties, we assessed phosphorylated STAT3 (pSTAT3) levels because pSTAT3 is critical for chemoradioresistance, anti‐apoptosis, stemness, and tumorigenesis. 5 , 22 , 23 STAT3 is reportedly upregulated in several cancers including glioma. 24 , 25 , 26 Furthermore, this signal is increased in high‐grade glioma. 25 First, STAT3 and pSTAT3 expression was evaluated in GBM cells. However, pSTAT3 was downregulated in siRPS6 rather than in siCont conditions (Figure 3A). This result was confirmed in the experiment using another RPS6‐specific siRNA (siRPS6#2) (Figure S3C). Relative protein expression of pSTAT3/STAT3 in the siRPS6 condition was significantly decreased by 22% in comparison with that in the siCont condition (P < .05) (Figure 3A). To confirm the actual effects of STAT3, we performed a sphere formation assay using GBM cells treated with JAK‐STAT inhibitor (AG490) in NSC culture medium. The sphere‐forming potential of GBM cells was suppressed through treatment with 10 µmol/L (P < .05) and 25 µmol/L (P < .01) AG490 (Figure 3B). Furthermore, the sphere‐forming potential was suppressed by 67% upon AG490 treatment (25 µmol/L) compared with that of mock‐OE‐RPS6 upon RPS6 overexpression (Figure 3C and Figure S2C). To confirm these results, we performed the same experiments using U87MG cells and obtained similar results (Figure S4C). Thus, the sphere‐forming potential of GBM cells was enhanced upon RPS6 overexpression, however it was attenuated upon treatment with AG490. Expression of Nestin and SOX2 was also decreased by treatment with AG490 (Figure S5B). These results suggest that RPS6 is partially involved in STAT3 signaling to regulate stem cell‐like characters in GBM cells.
FIGURE 3.
Ribosomal protein S6 (RPS6) regulates stemness in glioblastoma multiforme (GBM) cells via STAT3 signaling. A, Western blot showing the expression of pSTAT3 and STAT3 in GBM cell line (U251MG) transfected with siRPS6 or siCont in neural stem cell (NSC) medium. Graph showing relative protein expression of pSTAT3/STAT3 (B) Treatment with 10 and 25 µmol/L of AG490 (JAK‐STAT inhibitor) affected the sphere‐forming potential of GBM cell line (U251MG) relative to the control. C, Images and graphs showing the sphere‐forming potential of GBM cell line (U251MG) and the number of spheres (>50 µm). Values are presented as mean ± SD of triplicate samples; *P < .05, **P < .01. Scale bar, 500 µm
3.4. RPS6 expression was correlated with SOX2 expression in glioma tissue
In vitro experiments revealed the roles of RPS6 in inducing stem cell‐like features in GBM cells. To determine whether the expression pattern of RPS6 was correlated with that of SOX2, a stemness marker, western blotting was performed using glioma tissues at different grades. We assessed tissue samples from 12 glioma patients (seven males, five females; low‐grade: six cases, high‐grade: six cases, Table 1). Consequently, RPS6 expression in high‐grade glioma was 2.65‐fold that in low‐grade glioma (P < .05). Similarly, SOX2 expression in high‐grade glioma was 5.37‐fold that in low‐grade gliomas (P < .05) (Figure 4A). RPS6 and SOX2 expression was strongly correlated (y = 0.8262x + 0.1464, *P < .05) (Figure 4B). These data suggest that RPS6 expression modulates stemness in high‐grade gliomas; in other words, RPS6 expression induces malignant characters in GSCs.
FIGURE 4.
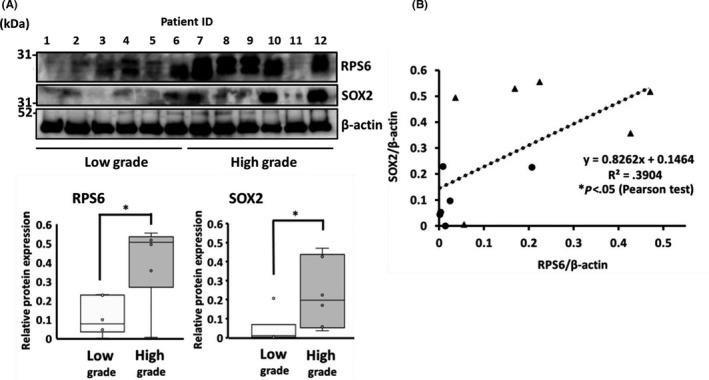
Ribosomal protein S6 (RPS6) expression was correlated with SOX2 expression in glioma tissue. A, RPS6 and SOX2 expression in human glioma tissues was analyzed via western blotting. B, Graph showing the positive correlation between RPS6 and SOX2 expression. (▲: high‐grade glioma sample, ●: low‐grade sample). Values are presented as means ± SD of six samples; *P < .05
3.5. RPS6 was detected in areas predominantly containing GSCs
Glioblastoma multiforme tissue displayed heterogeneous pathological findings, including areas displaying higher to lower cellularity, MVP, necrosis, and infiltration of glioma cells into the brain. To assess the detailed expression patterns of RPS6 at different characteristic sites in GBM tissue, we focused on perivascular, perinecrotic, and border areas because GSCs exist predominantly in these areas, which are considered GSC niches. RPS6 was predominantly expressed in these areas as a GSC niche compared with that in other areas (Figure 5A,B).
FIGURE 5.
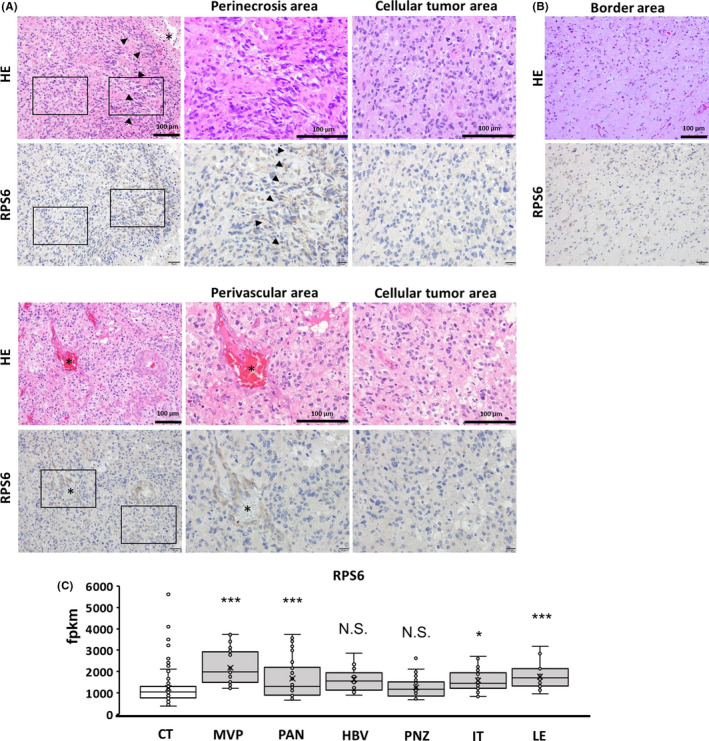
Ribosomal protein S6 (RPS6) expression was detected at areas predominantly containing GBM stem cells (GSCs). A, Pictures displaying hematoxylin and eosin staining and immunohistochemical staining in perivascular and perinecrotic areas. B, RPS6‐positive cells in the border area. C, Graph showing RPS6 expression in the GSC‐dominant area in accordance with the IVY GAP database. RPS6 expression in areas of microvascular proliferation, pseudopalisading cells around necrosis, infiltrating tumors, and leading edge. Asterisk indicates blood vessel cavities, black arrowhead indicates necrotic areas. Scale bars, 100 µm
Moreover, to confirm these results, we used the IVY GAP database, wherein GBM tissues were divided into several areas through microdissection, and RNA expression was analyzed. 20 We compared RPS6 expression in the CT, MVP, PAN, HBV, PNZ, IT, and LE.
RPS6 mRNA was upregulated in the MVP, PAN, IT, and LE (Figure 5C). RPS6 mRNA expression levels in the MVP and PNZ were slightly, but not significantly, higher than those in the CT. The present data from immunohistochemistry and the IVY GAP database indicate that RPS6 is predominantly present at sites containing GSC niches. These results strongly suggest that RPS6 upregulation in GSC niches is associated with the induction and maintenance of stem cell‐like characters in GBM cells.
3.6. Upregulation of ribosomal proteins in GSC‐dominant areas
Finally, we focused on the expression patterns of ribosomal proteins in the IVY GAP database. Similar to the expression patterns of RPS6, most ribosomal proteins were upregulated in GSC‐dominant areas including the PAN, MVP, LE, and IT in comparison with the CT. Furthermore, RPS6 expression levels at both the PNZ and HBV were not as high as those in other areas (Figure 6A). These data suggest that upregulation of ribosomal proteins including RPS6 in GSC‐dominant areas is universal in GBM. Moreover, not only RPS6 but also many other ribosomal proteins might promote stem cell‐like characters in GBM cells at perivascular, perinecrotic, and border niches (Figure 6B).
FIGURE 6.
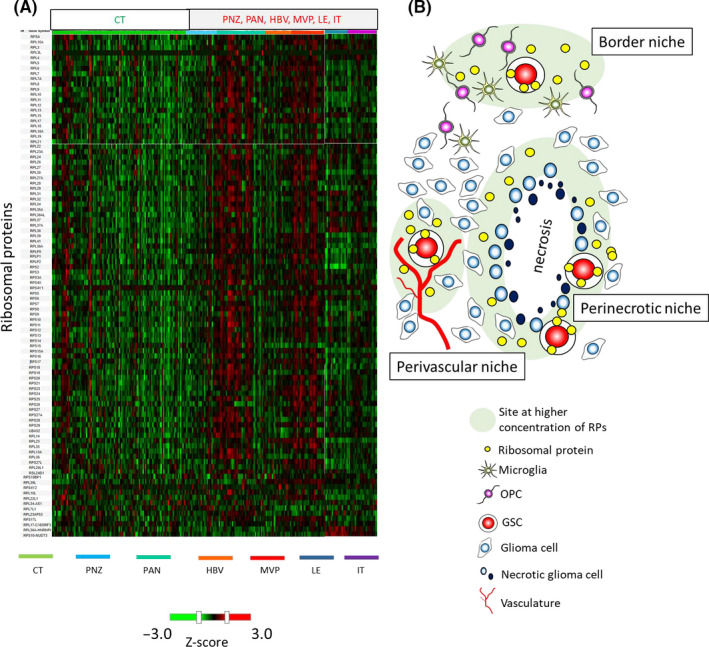
Upregulation of ribosomal proteins and a hypothetical scheme of incorporation of ribosomal proteins into glioblastoma multiforme (GBM) cells. A, Heatmap showing RNA expression of 96 ribosomal proteins including ribosomal protein S6 (RPS6) in cellular tumor and GBM stem cell (GSC)‐dominant areas (perinecrotic zone, pseudopalisading cells around necrosis, hyperplastic blood vessels in cellular tumor, microvascular proliferation, leading edge, and infiltrating tumor) from the IVY GAP database. The value in the heat map is the Z‐score. Red indicates RNA upregulation, and green indicates RNA downregulation. B, Scheme showing the concepts of GSC niches and ribosomal proteins. GBM cells were affected by inflammation and stress in perivascular, perinecrotic, and border niches, where GBM cells expressed RPS6 and other ribosomal proteins. GBM cells generate microvesicles containing RNAs and proteins including ribosomal proteins, which function as mediators for intercellular communication. We hypothesized that upregulated intrinsic ribosomal proteins and their incorporation from other GBM cells may promote the induction of stem cell properties in GBM cells and from GSC niches
4. DISCUSSION
Malignant GSCs are clinically chemoradioresistant and cause recurrence. To decrease the mortality rates of GBM patients, it is important to understand the mechanism underlying the formation of GSCs and their maintenance in tissues. This study shows that RPS6 and other ribosomal proteins play important roles in promoting malignant features in GBM cells.
Ribosomes are associated with bone marrow failure, skeletal and other developmental abnormalities, immune signaling, and cancer predisposition, 27 , 28 , 30 and are overexpressed in several cancers such as hepatocellular cancer and gastric cancer. 31 , 32 Moreover, downregulation of ribosomal proteins reportedly suppresses tumorigenesis and progression, 33 suggesting that ribosomes constitute a potential target for cancer treatment. 34 , 35
Interestingly, Ito et al 11 reported that the incorporation of ribosomes or small ribosomal proteins induces stem cell‐like characters and multipotency in somatic cells. Originally, multipotency was induced by bacterial ribosomes 11 , 12 , however incorporation of human ribosomes induced dedifferentiation and multipotency in human skin fibroblasts. 11 Increased levels of ribosomal proteins are reportedly associated with tumorigenesis, however we investigated ribosomal proteins from the viewpoint that they induce stemness through chemoradioresistance and recurrence of GBM.
Functions of RPS6 in GBM have not been discussed so far. Here, RPS6 was detected in perivascular, perinecrotic, and border niches, and these results were confirmed in relation to the IVY GAP database, indicating that RPS6 was upregulated at the MVP, PAN, IT, and LE (Figure 5A‐C). Together with the IVY GAP database, our results indicate that RPS6 is produced, secreted, and/or leaked at high levels at GSC‐dominant areas by stresses such as inflammation and cell damage. Considering the increase in the sphere‐forming potential and stemness marker expression owing to RPS6 upregulation, RPS6 upregulation was associated with the induction and maintenance of stem cell‐like characters in GBM cells. Additionally, pRPS6 (Ser235/236) was detected in glioma tissues and its expression was higher in high‐grade than in low‐grade gliomas (Figure S7). These data suggested that not only RPS6 but also its phosphorylation may be important for the regulation of the stem cell characters in glioma. Further investigation will also focus on the possible involvement of RPS6 phosphorylation in the regulation of GSC.
The involvement of RPS6 in inducing stemness may induce general effects in cancers because RPS6 was upregulated in not only GBMs but also several other cancers including lung cancer and renal cell carcinoma. 15 , 16 , 17 , 21 , 35 , 36 Moreover, the findings displayed in Figure 6A show that numerous ribosomal proteins are upregulated in the GSC niche, and uncontrolled high amounts of ribosomal proteins in GBM cells might be one of the key events resulting in malignant transformation. Moreover, these data indicate that not only RPS6 but also other ribosomal proteins possess a similar function of inducing stem cell characters.
Furthermore, ribosomes are assembled in the nucleolus via ribosomal RNA (rRNA). 37 The nucleolus is larger in malignant than in not‐malignant cancer cells. 33 A recent study reported that translation of rRNA and nucleolar regulation promote glioma tumorigenesis. 33 , 34 , 35 , 38
This study shows that RPS6 regulates STAT3 signals with regard to the sphere‐forming potential. Moreover, RPS6 expression was positively related to pSTAT3/STAT3 in high‐grade gliomas (Figure S8). In addition, our preliminary result showed that RPS6 knock‐down downregulated phospho‐JAK2 (Figure S5A). Furthermore, the sphere‐forming ability induced by IL‐6, a STAT3‐activating ligand, was abrogated by siRPS6 transfection (Figure S6). These findings suggest that RPS6 may regulate upstream of JAK2, such as IL‐6R or IL‐6 levels. Conversely, previous studies have also suggested that IL‐6 influences RPS6 expression and that IL‐6‐STAT3 signaling promotes GBM progression. 39
RPS6 expression in GBM is partially regulated by IL‐6, which in turn is associated with inflammation. 40 , 41 Furthermore, IL‐1β is secreted from macrophages at border niches, 7 and inflammation due to IL‐1β potentially influences RPS6 expression. Moreover, GBM tissues characteristically contain necrotic areas owing to the deficiency of vasculature, nutrition, and oxygen. 6 Around necrotic areas, inflammation is promoted by suppressing CYLD expression. 42 Therefore, inflammation at border niches and perinecrotic niches might induce RPS6 expression and the stem cell‐like features acquired by GBM cells (Figure 6B).
However, GBM cells reportedly generate microvesicles containing RNAs and proteins, which are used for intercellular communication, promote tumor growth, 43 and transmit the malignant phenotype. 44 GBM cells potentially incorporate not only RNA but also ribosomal proteins from neighboring cells through microvesicles and acquire stem cell‐like features in GSC niches.
In conclusion, this study shows that upregulation of ribosomal proteins in GBM cells and intercellular communication through microvesicles including ribosomal proteins in GSC niches are candidate treatment targets for GBM. Further studies are required to investigate the mechanism underlying RPS6 upregulation at GSC niches, RPS6‐mediated induction of stemness in GBM cells, and RPS6‐mediated intercellular communication. Studies on ribosomal proteins would reveal prominent methods to improve the prognosis of GBM patients by inhibiting chemoradioresistance and recurrence in cancer stem cell niches.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
ACKNOWLEDGMENTS
The authors thank Masayo Obata for excellent technical assistance with immunohistochemistry.
This work was supported by Grants in Aid for Scientific Research (B) 18H02591 (to Hirofumi Jono), and Grant in Aid for Young Scientists (A) 26713006 (Hirofumi Jono) from Ministry of Education, Culture, Sports, Science and Technology Japan Society for the Promotion of Science.
Shirakawa Y, Hide T, Yamaoka M, et al. Ribosomal protein S6 promotes stem‐like characters in glioma cells. Cancer Sci. 2020;111:2041–2051. 10.1111/cas.14399
Contributor Information
Takuichiro Hide, Email: thide@med.kitasato-u.ac.jp.
Hirofumi Jono, Email: hjono@kuh.kumamoto-u.ac.jp.
REFERENCES
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stupp R, Hegi ME, Gorlia T, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071–22072 study): a multicentre, randomised, open‐label, phase 3 trial. Lancet Oncol. 2014;15(10):1100‐1108. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Engl J Med. 2005;352:987‐996. [DOI] [PubMed] [Google Scholar]
- 4. Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6(6):425‐436. [DOI] [PubMed] [Google Scholar]
- 5. Leidgens V, Proske J, Rauer L, et al. Stattic and metformin inhibit brain tumor initiating cells by reducing STAT3‐phosphorylation. Oncotarget. 2017;8(5):8250‐8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends in Cancer. 2015;1(4):252‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hide T, Komohara Y, Miyasato Y, et al. Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine. 2018;30:94‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73(1):657‐704. [DOI] [PubMed] [Google Scholar]
- 9. Ito N, Anam MB, Ahmad SAI, Ohta K. Transdifferentiation of human somatic cells by ribosome. Dev Growth Differ. 2018;60(5):241‐247. [DOI] [PubMed] [Google Scholar]
- 10. Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7(2):92‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ito N, Katoh K, Kushige H, et al. Ribosome incorporation into somatic cells promotes lineage transdifferentiation towards multipotency. Sci Rep. 2018;8(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito N, Ohta K. Reprogramming of human somatic cells by bacteria. Dev Growth Differ. 2015;57(4):305‐312. [DOI] [PubMed] [Google Scholar]
- 13. Ohta K, Kawano R, Ito N. Lactic acid bacteria convert human fibroblasts to multipotent cells. PLoS ONE. 2012;7(12):e51866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520(7549):640‐645. [DOI] [PubMed] [Google Scholar]
- 15. Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31(6):342‐348. [DOI] [PubMed] [Google Scholar]
- 16. Chen B, Tan Z, Gao J, et al. Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non‐small cell lung cancer. J Exp Clin Cancer Res. 2015;34(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khalaileh A, Dreazen A, Khatib A, et al. Phosphorylation of ribosomal protein S6 attenuates DNA damage and tumor suppression during development of pancreatic cancer. Cancer Res. 2013;73(6):1811‐1820. [DOI] [PubMed] [Google Scholar]
- 18. Balenci L, Clarke ID, Dirks PB, et al. IQGAP1 protein specifies amplifying cancer cells in glioblastoma multiforme. Cancer Res. 2006;66(18):9074‐9082. [DOI] [PubMed] [Google Scholar]
- 19. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396‐401. [DOI] [PubMed] [Google Scholar]
- 20. Ecol M. An anatomic transcriptional atlas of human glioblastoma. Science. 2017;25(5):1032‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knoll M, Macher‐Goeppinger S, Kopitz J, et al. The ribosomal protein S6 in renal cell carcinoma: functional relevance and potential as biomarker. Oncotarget. 2016;7(1):418‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonnin DAA, Havrda MC, Lee MC, et al. Secretion‐mediated STAT3 activation promotes self‐renewal of glioma stem‐like cells during hypoxia. Oncogene. 2018;37(8):1107‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383‐2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spitzner M, Ebner R, Wolff HA, Michael Ghadimi B, Wienands J, Grade M. STAT3: a novel molecular mediator of resistance to chemoradiotherapy. Cancers. 2014;6(4):1986‐2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Birner P, Toumangelova‐Uzeir K, Natchev S, Guentchev M. STAT3 tyrosine phosphorylation influences survival in glioblastoma. J Neurooncol. 2010;100(3):339‐343. [DOI] [PubMed] [Google Scholar]
- 26. Bromberg J, Wang TC. Inflammation and Cancer: IL‐6 and STAT3 Complete the Link Jacqueline. Cancer Cell. 2009;15(2):79‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yelick PC, Trainor PA. Ribosomopathies: global process, tissue specific defects. Rare Dis. 2015;3(1):e1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulkarni S, Dolezal JM, Wang H, et al. Ribosomopathy‐like properties of murine and human cancers. PLoS ONE. 2017;12(8):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wegman‐Ostrosky T, Savage SA. The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol. 2017;177(4):526‐542. [DOI] [PubMed] [Google Scholar]
- 31. Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over‐expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39(1):129‐138. [DOI] [PubMed] [Google Scholar]
- 32. Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6;1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Derenzini M, Trerè D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol. 2000;191(2):181‐186. [DOI] [PubMed] [Google Scholar]
- 34. Catez F, Dalla Venezia N, Marcel V, Zorbas C, Lafontaine DLJ, Diaz JJ. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem Pharmacol. 2019;159:74‐81. [DOI] [PubMed] [Google Scholar]
- 35. Brighenti E, Treré D, Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: a real possibility? Oncotarget. 2015;6(36):38617‐38627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pallis M, Harvey T, Russell N. Phenotypically dormant and immature leukaemia cells display increased ribosomal protein S6 phosphorylation. PLoS ONE. 2016;11(3):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lam YW, Trinkle‐ L. The nucleolus. J Cell Sci. 2005;2005(1):1335‐1337. [DOI] [PubMed] [Google Scholar]
- 38. Kofuji S, Hirayama A, Eberhardt AO, et al. IMP dehydrogenase‐2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21(8):1003‐1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brighenti E, Giannone FA, Fornari F, et al. Therapeutic dosages of aspirin counteract the IL‐6 induced pro‐tumorigenic effects by slowing down the ribosome biogenesis rate. Oncotarget. 2016;7(39):63226‐63241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. West AJ, Tsui V, Stylli SS, et al. The role of interleukin‐6‐STAT3 signalling in glioblastoma. Oncol Lett. 2018;16(4):4095‐4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tanaka T, Narazaki M, Kishimoto T. IL‐6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect Biol. 2014;6:1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo J, Shinriki S, Su Y, et al. Hypoxia suppresses cylindromatosis (CYLD) expression to promote inflammation in glioblastoma: possible link to acquired resistance to anti‐VEGF therapy. Oncotarget. 2014;5(15):6353‐6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skog J, Würdinger T, van Rijn S , et al. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers Johan. Nat Cell Biol. 2008;10(12):1470‐1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Al‐Nedawi K, Meehan B, Micallef J, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619‐624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8


