Significance
The innovation of stomatal pores in plant evolution enables the success and diversification of land plants over the past 400 million years. The production and patterning of stomata are regulated by a group of deeply conserved bHLH transcription factors in land plants, with the founding member SPEECHLESS (SPCH) playing a pivotal role in the initiation of stomatal lineage. The protein stability and function of SPCH are tightly regulated by protein phosphorylation mediated by multiple kinases in Arabidopsis. Here, we establish the highly conserved PP2A protein phosphatases as positive regulators of SPCH and thus stomatal production, underscoring a key function of PP2A in fine-tuning stomatal development, a highly plastic biological process that influences many plant responses to changing environmental conditions.
Keywords: Arabidopsis thaliana, stomatal development, PP2A protein phosphatases, bHLH SPCH, protein phosphoregulation
Abstract
Stomatal guard cells control gas exchange that allows plant photosynthesis but limits water loss from plants to the environment. In Arabidopsis, stomatal development is mainly controlled by a signaling pathway comprising peptide ligands, membrane receptors, a mitogen-activated protein kinase (MAPK) cascade, and a set of transcription factors. The initiation of the stomatal lineage requires the activity of the bHLH transcription factor SPEECHLESS (SPCH) with its partners. Multiple kinases were found to regulate SPCH protein stability and function through phosphorylation, yet no antagonistic protein phosphatase activities have been identified. Here, we identify the conserved PP2A phosphatases as positive regulators of Arabidopsis stomatal development. We show that mutations in genes encoding PP2A subunits result in lowered stomatal production in Arabidopsis. Genetic analyses place the PP2A function upstream of SPCH. Pharmacological treatments support a role for PP2A in promoting SPCH protein stability. We further find that SPCH directly binds to the PP2A-A subunits in vitro. In plants, nonphosphorylatable SPCH proteins are less affected by PP2A activity levels. Thus, our research suggests that PP2A may function to regulate the phosphorylation status of the master transcription factor SPCH in stomatal development.
Stomata are epidermal pores, each of which is girdled by a pair of guard cells (GCs) that control stomatal opening and closing to mediate gas exchange between plants and the atmosphere. The formation of stomata involves stereotypic cell divisions and cell fate differentiation of the stomatal lineage cells, all of which are tightly controlled by a suite of closely related and sequentially expressed basic helix–loop–helix (bHLH) transcriptional factors in the model plant Arabidopsis thaliana. The bHLH SPEECHLESS (SPCH) acts earliest and is mainly expressed in meristemoid mother cells (MMCs) that divide to produce meristemoids (Ms) and stomatal lineage ground cells (SLGCs). SPCH is required to initiate the MMCs, and thus the formation of stomata, in the epidermis of a plant (1). The differential developmental trajectories of the Ms and the SLGCs coordinate the production of stomatal GCs and pavement cells in a leaf. The closely related bHLH MUTE is mainly expressed in late Ms to promote cell fate transition to guard mother cells (GMCs) (2). Another related bHLH, FAMA, is expressed in GMCs and young GCs, driving GMC terminal differentiation into GCs (3). In addition, two bHLH-leucine zipper (bHLH-LZ) proteins, INDUCER OF CBF EXPRESSION 1 (ICE1, also known as SCREAM) and SCREAM2 (SCRM2), function to promote the three distinct sequential phases of differentiation in stomatal development by acting as partners of SPCH, MUTE, and FAMA (4). Among these bHLH transcription factors, SPCH appears to be a master regulator that directly drives the expression of other bHLH factors, for example, MUTE, SCRM/ICE1, and SCRM2 (5).
Upstream of the core bHLH functional module, both environmental and developmental signals regulate stomatal development, most, if not all, of which eventually feed into the regulation of SPCH expression. In a well-established linear signaling pathway, the extracellular peptide ligands, EPIDERMAL PATTERNING FACTORs, are perceived by receptors at the plasma membrane, for example, the receptor-like protein TOO MANY MOUTHS (TMM), the receptor-like kinase ERECTA and the SOMATIC EMBRYOGENESIS RELATED KINASE (SERK) families (6–8). The ligand−receptor signaling is then transduced to a mitogen-activated protein kinase (MAPK) cascade, including MAPKKK YODA (YDA), MKK4/5, and MAPK3/6 (9, 10), to trigger SPCH phosphorylation and protein degradation (11). The plant hormone brassinosteroid (BR) may promote stomatal production in the hypocotyl by suppressing a negative regulator of SPCH, the serine/threonine glycogen synthase kinase 3 (GSK3)/SHAGGY-like BRASSINOSTEROID insensitive 2 (BIN2), which phosphorylates SPCH for degradation (12). The cell-cycle regulator CYCLIN-DEPENDENT KINASE A;1 (CDKA;1) also phosphorylates SPCH but positively affects SPCH function via an unknown mechanism (13). Protein phosphorylation is highly dynamic, and rapid phosphorylation−dephosphorylation cycles underlie many critical biological responses. The identity of the phosphatase(s) that counteracts these kinases in the regulation of SPCH stability and function in stomatal development remains a long-standing question.
The conserved Protein Phosphatase 2A (PP2A) family of ubiquitously expressed Ser/Thr protein phosphatases in eukaryotes are heterotrimeric complexes that comprise a catalytic subunit C, a regulatory subunit B, and a scaffolding subunit A (14). The Arabidopsis genome encodes multiple isoforms for each subunit (3 As, 17 Bs, and 5 Cs) (15) (Fig. 1A), and differentially assembled PP2A complexes regulate plant growth, development, metabolism, and stress responses (15, 16). Functions of PP2A have been linked to the regulation of blue light-triggered stomatal movement by inhibiting the kinase activity of phototropin PHOT2 (17). Also, by associating with the SnRK2-type kinases, PP2A participates in ABA-mediated stomatal closure (18). In addition, the determination of cell division orientation in maize stomatal complexes and in Arabidopsis embryos and seedlings requires PP2A activities in the formation of the preprophase band (19, 20). Here, we establish a new function of PP2A in stomatal development: PP2A phosphatases positively regulate SPCH protein stability to promote stomatal production. Evidence is presented for a physical association of PP2A with SPCH mediated by direct interactions between SPCH and the A subunits. Thus, our research revealed a missing regulator of SPCH homeostasis in stomatal development. In addition, the PP2A components, as newly identified signaling molecules in the regulation of stomatal production, will be candidates to be manipulated in the future to fine tune stomatal development for plants to better adjust to unfavorable environmental conditions.
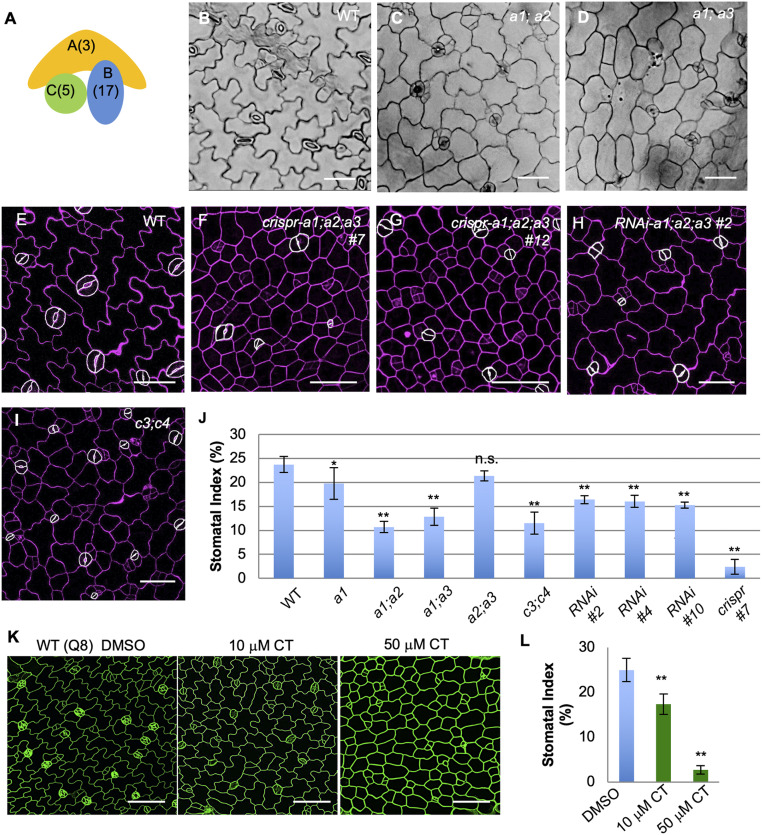
Fig. 1.
PP2A promotes stomatal production in Arabidopsis. (A) Diagram depicting a heterotrimeric PP2A complex. Numbers in parentheses indicate the number of genes in the Arabidopsis genome encoding the corresponding subunit type. (B–D) DIC images of 5-dpg cotyledons in (B) Col, (C) a1;a2, and (D) a1;a3 seedlings. (E–I) Confocal images of 3-dpg adaxial epidermis of cotyledons in the (E) WT, (F) crispr-a1;a2;a3 #7, (G) crispr-a1;a2;a3 #12, (H) RNAi-a1;a2;a3 #2, and (I) c3;c4. (Scale bars in B−I, 50 μm.) (J) Quantification of SI in 5-dpg adaxial cotyledon epidermis. Data are mean ± SD n = 930 to 3,334 total epidermal cells of 10 to 12 individual plants; asterisk denotes significantly different compared with the WT values (Student’s t test, *P < 0.05, **P < 0.001); n.s., not significant. GCs are manually traced with white highlights to improve visibility. Cell walls were stained with PI, shown in magenta. (K) Confocal images of 3-dpg WT seedlings (Q8 GFP expression to show cell outlines in green) grown with DMSO, 10 μM CT (the PP2A inhibitor), and 50 μM CT, respectively. (Scale bars, 50 μm.) (L) Quantification of SI in 3-dpg adaxial cotyledons shown in K. Data are mean ± SD. n = 1,103 to 1,996 cells. Asterisk denotes significantly different from the WT values (Student’s t test, **P < 0.001).
Results
The PP2A-A Mutants Are Defective in Stomatal Production.
It has been previously reported that PP2A plays pivotal roles in plant development and growth patterning. Mutations in PP2A components led to defects in many developmental processes in Arabidopsis, such as inflorescence development (A subunits) (21), root elongation (A subunits) (21, 22), pavement cell morphogenesis (C subunits) (23), and cell division patterns (B subunits) (20). The Arabidopsis genome encodes three A subunits that are highly similar in their sequences (A2/PDF1 and A3/PDF2 are 94% identical, and A2/PDF1 is 86% identical to A1/RCN1, shortened to A1, A2, and A3 throughout this manuscript) (21), and we observed all three A subunit genes to be ubiquitously expressed in leaves (SI Appendix, Fig. S1 H–J). We examined the developmentally defective PP2A-A transfer DNA (T-DNA) insertion mutants (21) for possible defects in stomatal development. Indeed, both a1;a2 and a1;a3 mutants produced fewer stomata, measured by Stomatal Index (SI; number of stomata as a percentage of the total number of epidermal cells in a given area). The SI of a1;a2 and a1;a3 mutant plants were reduced to 10.7% and 12.9% (n = 930 and 1,844 total number of cells), respectively, compared to 23.7% in wild-type (WT) plants (Fig. 1 B–D and quantification in Fig. 1J). The other double mutant, a2;a3, did not show overall growth defects and produced normal leaves with typical stomatal patterning (Fig. 1J and SI Appendix, Fig. S1D), suggesting the important contribution of A1 in stomatal development. In fact, mutating A1 alone already reduced stomatal production in plants (Fig. 1J and SI Appendix, Fig. S1 B and C; 19.8%, n = 2,445 vs. 23.7% in WT, n = 1,628), while neither a2 nor a3 mutations alone obviously changed stomatal and other developmental processes. We also crossed the double mutants a1;a2 and a1;a3, but failed to obtain homozygous triple mutants, suggesting that some A subunit function is essential for plant early development.
To further investigate the collective roles of the three A subunits in the regulation of stomatal development, we deployed CRISPR-Cas9−mediated mutagenesis (24) to simultaneously knock them out. Based on the high sequence similarity among the three PP2A-A subunits (21), we designed two guide RNAs (sgRNA1 and sgRNA2) that specifically target A1 and A2/A3, respectively (SI Appendix, Fig. S1A). Simultaneously introducing the two sgRNAs with Cas9 into Arabidopsis plants was anticipated to give rise to mutations in all three A genes (SI Appendix, Fig. S1A). Indeed, we noted that the first generation of transgenic plants (T1s) already showed growth defects, including dwarfed stature (SI Appendix, Fig. S1K) and inhibited root growth, as previously reported (21). Two independent lines (crispr-a1;a2;a3 #7, and #12) were selected for thorough genotype and phenotype analyses. We found that the mutants show greatly reduced SI (Fig. 1 E–G, and quantified in Fig. 1J; 2.4% in crispr-a1;a2;a3 #7, n = 2,183 total epidermal cells vs. 23.7% in WT, n = 1,628). Genotyping data suggested that these individual mutant plants were not homogenous knockouts, and instead carry various somatic mutations. Specifically, the plant crispr-a1;a2;a3 #12 (SI Appendix, Fig. S1K) carried 1-base pair (bp) insertions in A1 and A3 and a 6-bp deletion in A2 (SI Appendix, Fig. S1L). On the other hand, the crispr-a1;a2;a3 #7 plant is chimeric, harboring a 1-bp deletion in A3 and multiple mutations in A1 and A2. We were unable to obtain any Cas9-free mutants from the plants that showed strong stomatal phenotypes, hinting that the loss-of-function mutations in all three PP2A-A genes might be detrimental to gametophyte and/or early embryo development.
To further verify the stomatal phenotype we observed, we generated knockdown plants using the micro-RNA (miRNA)-induced gene silencing strategy (25). Specifically, a highly conserved coding region in PP2A-A2 (93% identical to A3, and 81% identical to A1) was cloned into a destination vector together with a miR173 target site, which can trigger the production of transacting small interfering RNAs, so that the expression of all three endogenous PP2A-A genes can be suppressed simultaneously. Among the RNA interference (RNAi) progenies, three independent transgenic RNAi-a1;a2;a3 lines with reduced expression level of PP2A-A genes were isolated (SI Appendix, Fig. S1G). T3 plants showed abnormal pavement cell shape (Fig. 1H and SI Appendix, Fig. S1 E and F) and reduced SI (RNAi-a1;a2;a3 #2 = 16.4%, n = 2,655; #4 = 16.1%, n = 3,117; #10 = 15.2%, n = 3,334 vs. WT = 23.7%, n = 1,628) (Fig. 1J). Phenotypic quantification showed that the stomatal defects of the RNAi plants were weaker than those of crispr-a1;a2;a3 #7 lines, but were comparable to the double mutants a1;a2 and a1;a3 (Fig. 1J). Taken together, we collected multiple lines of genetic evidence that demonstrate a positive role of the PP2A phosphatases in stomatal production.
PP2A Functions to Promote Stomatal Production in Arabidopsis.
As the PP2A protein phosphatases are heterotrimeric, disruptions of the catalytic PP2A-C subunits were anticipated to lead to developmental defects similar to those caused by the A mutations. The c3;c4 double mutants, as previously reported, show a dwarfed growth phenotype, mirroring that of a1;a2 and a1;a3 mutants (20) as well as defects in pavement cell morphogenesis (23). We also found dramatically reduced stomatal density (Fig. 1 I and J) in the leaf epidermis of the c3;c4 mutants, consistently demonstrating the positive role of PP2A in stomatal production.
Besides genetic strategies, we deployed pharmacological treatment to define PP2A’s function in stomatal development. Cantharidin (CT) is a widely used PP2A inhibitor that preferentially inhibits protein phosphatases PP2A and PP1 (26). WT seeds expressing the cell outline marker Q8 (GFP-tagged plasma membrane channel PIP2A) (27) were germinated on half-strength Murashige and Skoog (MS) medium supplemented with 10 μM or 50 μM CT, while the corresponding amount of dimethyl sulfoxide (DMSO) solvent was used as control. In 3-d postgermination (3-dpg) seedlings, we found that CT treatment suppressed stomatal production (SI reduced to 17.3% by 10 μM CT n = 1,752, and to 2.7% by 50 μM CT n = 1,103, compared to 25.0% in the DMSO control experiments, n = 1,996) (Fig. 1 K and L).
PP2A Functions Upstream of SPCH.
To study the genetic relationship of PP2A with the other key components in the regulation of stomatal development (depicted in Fig. 2A), we introduced crispr-a1;a2;a3 mutations in a number of stomatal mutants. ERECTA (ER) is a receptor-like protein kinase, and the loss-of-function mutant er105 produces excessive asymmetric cell divisions and meristemoids (6). After the crispr-a1;a2;a3 construct was introduced into the er105 mutant, the SI of er105 (15.0%, n = 1,362) was greatly reduced to 2.4% (n = 1,254) (Fig. 2 B and C and SI Appendix, Fig. S2A), suggesting crispr-a1;a2;a3 is epistatic to er105 in stomatal production. The success of generating crispr-a1;a2;a3 mutations in these transgenic plants was manifested by a number of phenotypic features, including small and dark green leaves, short roots, and abnormal pavement cell shapes, etc.
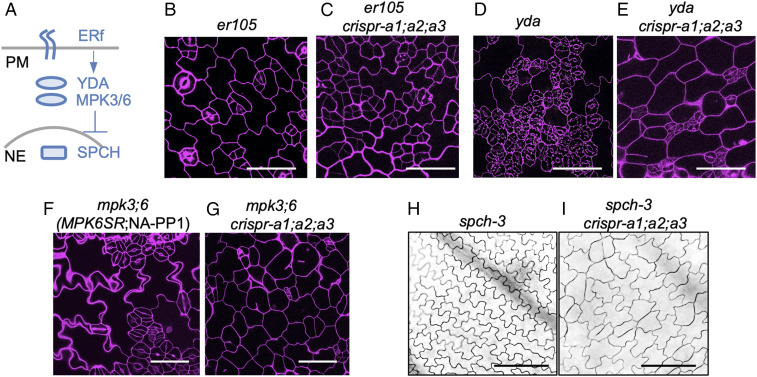
Fig. 2.
PP2A functions upstream of SPCH. (A) Simplified diagram to describe key signaling molecules in stomatal development. ERf, the ERECTA receptor-like kinase family; YDA, the MAPKK Kinase; MPK3/6, MAPK 3 and MAPK 6; SPCH, the bHLH transcription factor. PM, plasma membrane; NE, nuclear envelope. (B–G) Confocal images of 3-dpg seedlings of (B) er105, (C) er105;crispr-a1;a2;a3, (D) yda, (E) yda;crispr-a1;a2;a3, (F) mpk3;6, and (G) mpk3;6;crispr-a1;a2;a3. PI staining marks the cell outlines (magenta). (Scale bars, 50 μm) (H and I) DIC images of 5-dpg seedlings of (H) spch-3 and (I) spch-3;crispr-a1;a2;a3. (Scale bars, 100 μm.)
Downstream of the cell-surface ER receptors, the MAPKKK YODA (YDA) functions as a molecular switch of stomatal identity in the epidermis (9). The null yda-3 mutation leads to excessive GC formation and severe stomatal clustering (28). After the crispr-a1;a2;a3 construct was introduced into yda-3 mutant, the stomatal overproduction and clustering phenotype was greatly suppressed in the progeny, with SI reduced to 16.0% (n = 3,858) compared with 42.3% (n = 1,287) in yda-3 (Fig. 2 D and E and SI Appendix, Fig. S2A), suggesting that crispr-a1;a2;a3 is partially epistatic and PP2A functions in parallel or downstream of YDA.
MAPK3 and MAPK6 function redundantly downstream of the MAPKKK YDA in stomatal development (10). We examined a chemically inducible loss-of-function double mutant of mpk3;mpk6, named as MPK6SR (29), which harbors the double loss-of-function T-DNA insertional mutations that are functionally complemented by an MPK6 variant (MPK6YG). In the presence of the chemical inhibitor NA-PP1 (4-amino-1-tert-butyl-3-(1′-naphthyl) pyrazolo [3,4-d] pyrimidine), the complementation of MPK6YG can be disrupted, resulting in the loss-of-function, severely clustered stomatal phenotype (29). When crispr-a1;a2;a3 was introduced into MPK6SR, the NA-PP1 induction of stomatal overproliferation was greatly impaired (Fig. 2 F and G; SI of 61.1%, n = 546 reduced to 7.3%, n = 1,553; quantification in SI Appendix, Fig. S2A). These results indicate that crispr-a1;a2;a3 stomatal phenotype is partially epistatic to mpk3;6. The above genetic results were further confirmed by the CT treatment of two dominant negative mutants of the pathway, DNyda (9) and DNmpk6 (30). In both cases, the severe stomatal clustering phenotype was alleviated by CT treatment (SI Appendix, Fig. S3 A and B), consistent with PP2A acting in parallel or downstream of the YDA MAPK cascade.
In the nucleus, the bHLH transcription factor SPCH is the master regulator that initiates the stomatal lineage in Arabidopsis (1) and is directly targeted by the YDA MAPK cascade for degradation (11). The null spch-3 mutation eliminates stomatal production in the epidermis (1). When we introduced crispr-a1;a2;a3 mutations into spch-3, the progeny produced the stomataless epidermis of spch-3 with round pavement cells, a typical feature of crispr-a1;a2;a3 (Fig. 2 H and I), suggesting that SPCH is downstream of PP2A and that PP2A functions somewhere between MAPKs and SPCH.
Inhibition of PP2A Suppresses Protein Expression of Key Transcription Factors.
Stomatal development in Arabidopsis involves sequential events of 1) lineage initiation controlled by SPCH with partners SCRM/ICE1 and SCRM2 (4), 2) stomatal fate transition mediated by MUTE (2), and 3) terminal fate differentiation switched by FAMA (3). To determine which step in stomatal development is regulated by PP2A, we took advantage of pharmacological treatment, the PP2A inhibitor CT, to examine the responses of the corresponding marker expression. The Arabidopsis marker lines expressing SPCH-CFP (5), MUTE-GFP (2), and GFP-FAMA (3), all driven by their endogenous promoters, were germinated and grown in the presence of 50 μM CT, and the adaxial sides of 3-dpg cotyledon epidermis were examined for protein accumulation. Because all these markers are cell type specific and expressed transiently, the detection of such low-abundance proteins in the whole-plant tissue by biochemical strategies, for example, immunoblotting, was not trivial. We therefore relied on counting the frequency of marker-expressing cells to indicate protein expression level. We observed that the long-duration CT treatment (plants grown for 3 d) greatly reduced the percentage of cells expressing each of the three transcription factors, for example, SPCH-CFP (dropped down to 3.0%, n = 2,048 CFP-positive cells in the CT treatment, compared to 18.5%, n = 8,112 in the control), MUTE-GFP (0.0% in CT, n = 10,776 vs. 9.0% in DMSO, n = 6,080), and GFP-FAMA (1.8% in CT, n = 1,512 vs. 20.5% in DMSO, n = 774) (Fig. 3 A–C; quantification in SI Appendix, Fig. S2B). Interestingly, the expression of the SPCH partner, SCRM/ICE1-YFP, was not significantly affected by CT (Fig. 3D; quantification in SI Appendix, Fig. S2B), suggesting the specific regulation of PP2A on SPCH/MUTE/FAMA.
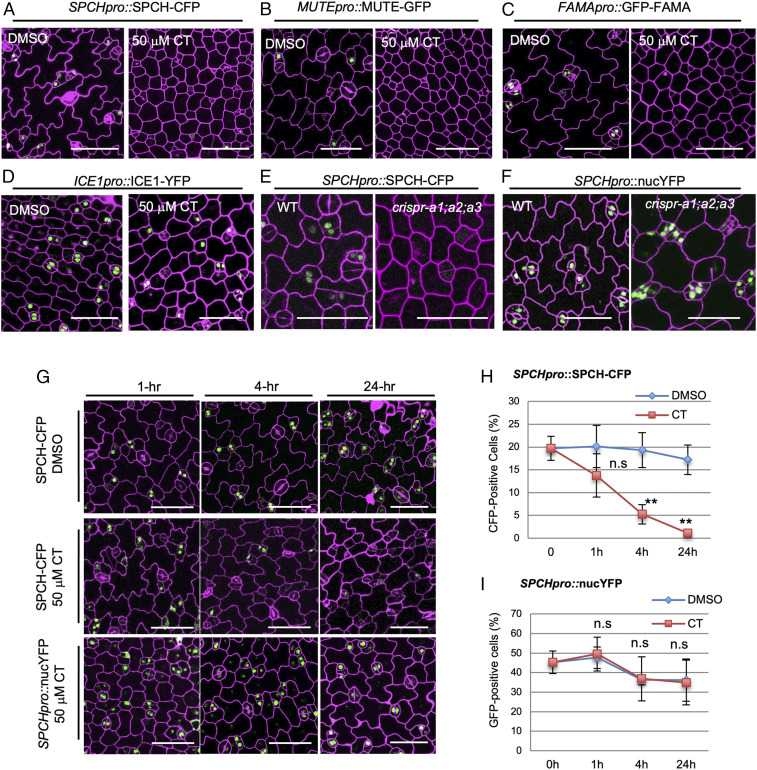
Fig. 3.
PP2A promotes SPCH protein stability. (A–D) Confocal images of 3-dpg seedlings expressing (A) SPCH-CFP, (B) MUTE-GFP (C), GFP-FAMA, and (D) SCRM/ICE1-YFP grown with DMSO and 50 μM CT. (E and F) Confocal images of 3-dpg cotyledons of the (E) translational fusion SPCHpro::SPCH-CFP vs. (F) the transcriptional fusion SPCHpro::nucYFP seedlings in the WT background (Left) vs. in crispr-a1;a2;a3 (Right). (G) Confocal images of 3-dpg adaxial cotyledons of SPCH-CFP and SPCHpro::nucYFP (green) treated with DMSO or 50 μM CT for (Left to Right) 1, 4, and 24 h. Cell outlines are visualized by PI staining (magenta). (Scale bars in A–G, 50 μm.) (H and I) Quantification of CFP- or YFP-positive cells in 3-dpg (H) SPCHpro::SPCH-CFP, n = 884 to 2,865 cells and (I) SPCHpro::nucYFP, n = 650 to 1,482 cells. Data are mean ± SD. Student’s t test, **P < 0.001, n.s., not significant.
We also tested whether PP2A may modulate protein expression levels of other stomatal factors, including the receptor-like protein TMM (7) (TMM-GFP driven by the endogenous promoter), the MAPKKK YDA (YDA-YFP driven by its native promoter and the dominant-negative DNyda-YFP, equivalent to the kinase inactive version, driven by the 35S promoter), MPK6 (the kinase inactive DNmpk6-YFP driven by the 35S promoter), the polarity factor BASL (GFP-BASL driven by its own promoter), and SCRM/ICE1 (SCRM/ICE1-YFP driven by its own promoter). We found that none of these markers were discernably changed by CT-mediated disruption of PP2A functions or in the crispr-a1;a2;a3 lines (SI Appendix, Fig. S3), consistent with the genetic position of PP2A between the plasma membrane receptors and the nuclear SPCH function.
PP2A Promotes SPCH Protein Stability.
Because the early stomatal gene SPCH appeared to directly regulate key factors in cell division and cell fate determination and the chromatin immunoprecipitation assays supported the physical binding of SPCH with the promoter of MUTE, but not that of FAMA (5), we suspected that the lowered expression of MUTE and FAMA in response to CT (Fig. 3 A–C) could result from lowered levels of SPCH protein. To test this possibility, we shortened the CT treatment duration to examine the immediate consequences of protein accumulation. The three translational marker lines (SPCH-CFP, MUTE-GFP, and GFP-FAMA) were grown for 3 d, followed by short-term CT treatment (50 μM for 1, 4, and 24 h, respectively). We found that SPCH-CFP was very sensitive to CT treatment: After 1 h of incubation, the percentage of cells expressing SPCH-CFP was reduced by 30% (13.8% in CT, n = 2,016 vs. 19.7%, n = 884 in DMSO); after 4 h, 70% of SPCH-CFP−positive cells were lost (5.2%, n = 1,670 vs. 19.7% in DMSO); and, after 24 h, SPCH-CFP were almost absent (only 1.1% CFP-positive, n = 2,865). In the DMSO control experiment, the number of cells expressing SPCH-CFP was maintained stably at all three time points (Fig. 3G; quantification in Fig. 3H). Interestingly, MUTE-GFP showed similar, though milder, responses to 50 μM CT: After 1 h of incubation, the percentage of cells expressing MUTE-GFP was reduced by 21% (from 6.0%, n = 965 dropped to 4.7%, n = 2,309); after 4 h, 46% of MUTE-GFP−positive cells were lost (3.2%, n = 2,888); and, after 24 h, 76% of MUTE-GFP were lost (1.3%, n = 2,249). Again, in the control experiment, MUTE-GFP expression remained stable in 24-h DMSO (SI Appendix, Fig. S5A; quantification in SI Appendix, Fig. S5B). Strikingly, however, the expression levels of GFP-FAMA remained almost unaffected at all of the time points within 24 h of 50 μM CT treatment (SI Appendix, Fig. S5D; quantification in SI Appendix, Fig. S5E). These results clearly demonstrated that the protein accumulation of SPCH and MUTE is more sensitive to short-term CT-triggered protein phosphoregulation, but FAMA did not appear to be regulated by this mechanism. As both GFP- and CFP-tagged proteins were assayed in our experiments, we evaluated possible influences of different fluorescent protein (FP) tags to protein stability by testing the responsiveness of GFP-tagged SPCH (1) versus the CFP-tagged SPCH under the same treatment. Quantification data showed that SPCH-GFP responded similarly to SPCH-CFP in CT treatment with regards to their protein instability (SI Appendix, Fig. S4B). Thus, different FP tags (GFP/YFP/CFP) were interchangeably used in our study.
We further evaluated whether the short-term CT-triggered lowered expression of SPCH-CFP and MUTE-GFP occur at the transcriptional level or at the posttranslational level, by comparing the transcriptional reporters (SPCHpro::nucYFP and MUTEpro::nucYFP) vs. the translational reporter lines (SPCHpro::SPCH-CFP and MUTEpro::MUTE-GFP). The data showed that, unlike the protein fusion, the expression of SPCHpro::nucYFP was not discernably sensitive to CT and remained at equivalently high levels after 1, 4, and 24 h of treatment (Fig. 3G; quantification in Fig. 3I). Consistently, the transcriptional fusion SPCHpro::nucYFP was not affected by crispr-a1;a2;a3 (Fig. 3F). Also, quantitative PCR results demonstrated that comparable transcript levels of SPCH-CFP were found in both WT and crispr-a1;a2;a3 (SI Appendix, Fig. S4A), although SPCH-CFP proteins were greatly reduced in the mutants (Fig. 3E). Thus, our results strongly suggest that the positive role of PP2A in the regulation of SPCH expression occurs at the protein level but not at the transcription level.
On the other hand, we found that, in contrast to the stable expression of SPCHpro::nucYFP, the transcription fusion MUTEpro::nucYFP was sensitive to CT treatment, with the reduction rates of 38% (from 17.1%, n = 995 to 10.3%, n = 1,923) at 4-h and 73% (from 17.1%, n = 995 to 4.65%, n = 2,129) at 24-h 50 μM CT treatment (SI Appendix, Fig. S5A; quantification in SI Appendix, Fig. S5C). In the same set of experiments, the DMSO controls showed negligible changes (SI Appendix, Fig. S5C). Considering that the SPCH protein could bind to the MUTE promoter to activate gene expression, it is probably not surprising that MUTEpro::nucYFP is sensitive to CT, because the indirect effects resulted from the reduced SPCH abundance. However, we cannot attribute all of the observed reduction in MUTE-GFP to the reduction in MUTE transcription, since MUTE protein abundance declined earlier than the promoter activity in responding to 1-h CT treatment (SI Appendix, Fig. S5B vs. SI Appendix, Fig. S5C). Thus, these results indicated the impacts of the PP2A inhibitor CT on the expression of different stomatal genes: CT may inhibit the protein accumulation of SPCH, while the suppression of MUTE most likely occurs at the transcriptional level, which may result from lowered SPCH abundance.
As SPCH was postulated to be phosphorylated and then degraded by the 26S proteasome system (12), we tested the hypothesis that PP2A might be the phosphatase counteracting this process. We combined the 26S proteasome inhibitor MG132 with CT in the treatment, anticipating that MG132 would abolish the CT-triggered protein instability of SPCH. Our data showed that 24-h 50 μM MG132 treatment almost fully recovered robust SPCH-CFP expression in CT-treated 3-dpg seedlings (Fig. 4 A and B). Thus we propose that PP2A, likely through protein dephosphorylation, promotes the stability of SPCH protein which, in turn, promotes stomatal production in Arabidopsis.
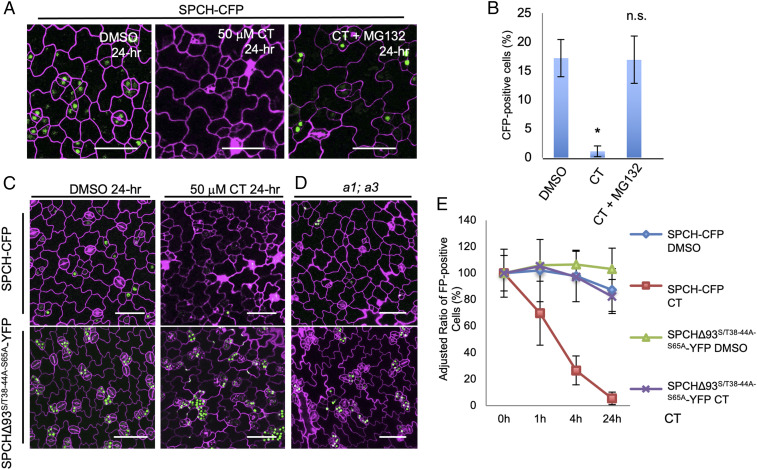
Fig. 4.
Phosphodeficient SPCH is less affected by defective PP2A. (A) Confocal images of SPCH-CFP seedlings (3-dpg) after 24-h incubation with DMSO, 50 μM CT, or 50 μM CT mixed with 50 μM MG132. (B) Quantification of CFP-positive cells in total epidermal cells shown in A, n = 798 to 2,865 cells from 10 to 12 seedlings. (C and D) Confocal images of 3-dpg SPCH-CFP (Upper) and SPCHΔ93S/T38−44A−S65A-YFP (Lower) seedlings after 24-h incubation with (C) DMSO (Left) and 50 μM CT (Right), and (D) in a1;a3. (E) Quantification of the adjusted ratios of fluorescence-positive cells in SPCH-CFP and SPCHΔ93S/T38−44A−S65A-YFP in 3-dpg cotyledons in responding to DMSO and 50 μM CT at different time points (0, 1, 4, and 24 h after treatment), n = 1,915 to 2,865 cells. The initial ratios of FP-positive cells were defined as 100%, and the relative ratios (adjusted) were calculated at each time point. In A and C, the expression GFP/YFP/CFP was artificially colored with green, and cell outlines were visualized by PI staining (magenta). (Scale bars, 50 μm.) Data are mean ± SD. Asterisk denotes significantly different from the DMSO control values (Student’s t test, *P < 0.05).
Differential Sensitivity of SPCH Phosphovariants to Lowered PP2A Activities.
The SPCH sequence contains multiple possible phosphorylation sites for MPK3/6, CDKA;1, and BIN2 (11–13); many of these sites are distributed in the unique MAPK targeting domain (MPKTD), with a few additional ones in the N-terminal domain (SI Appendix, Fig. S5F). It was reported that overexpression of the SPCH variant with the MPKTD deleted (SPCHΔ93) generated large clusters of stomata (11), presumably due to low phosphorylation levels, and thus elevated protein stability and activity of SPCH. On top of SPCHΔ93, we further mutated five putative Ser/Thr phosphorylation sites (S38, T40, S43, T44, and S65) to Alanines to produce an enhanced phosphodeficient version, SPCHΔ93S/T38−44A−S65A. If PP2A indeed functions to lower SPCH phosphorylation level, we anticipate that the phosphodeficient SPCHΔ93S/T38−44A−S65A would be less sensitive to PP2A activity in plants. To test this hypothesis, we introduced SPCHΔ93S/T38−44A−65A-YFP driven by the SPCH promoter into plants and found that, compared to the WT SPCH-CFP that is very sensitive to CT, the expression of SPCHΔ93S/T38−44A−65A-YFP responded to CT less sensitively (Fig. 4C). The quantification data showed that, compared to the nearly 100% reduction of SPCH-CFP expression by 24-h CT treatment (n = 2,865; Fig. 4E), CT-treated SPCHΔ93S/T38−44A−65A-YFP plants showed only a 20% reduction (n = 1,915) (Fig. 4E). Similarly, while the SPCH-CFP expression was greatly reduced in a1;a3 mutants, SPCHΔ93S/T38−44A−65A-YFP maintained high expression levels (Fig. 4D). These results collectively demonstrate that the phosphodeficient SPCH variant is less affected by defective PP2A activity, suggesting that PP2A may contribute to stabilizing SPCH through protein de-phosphorylation.
The PP2A-A Subunits Physically Interact with SPCH.
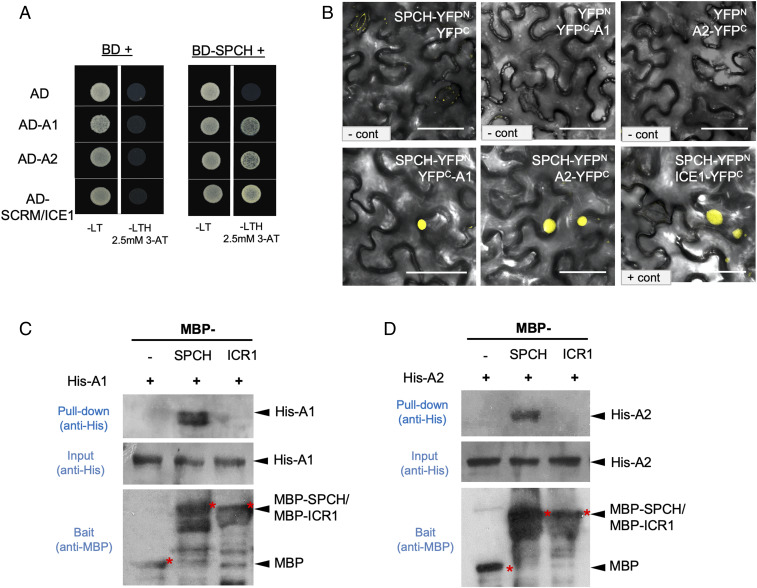
Supported by the above experimental evidence, we asked whether PP2A directly targets the SPCH protein to regulate its phosphorylation status. Canonical PP2A complexes are composed of one A (scaffolding), one B (regulatory), and one C (catalytic) subunit, and, in Arabidopsis, each subunit type is encoded by multiple genes (Fig. 1A) (14). A noncanonical complex containing a C subunit bound by the TAP42/alpha4/TAP46 regulatory protein has also been identified in plants and other eukaryotes (31). To determine which subunit(s) may directly interact with SPCH, based on their expression profiles (32) and sequence similarity analysis, we selected PP2A subunits (A1, A2, A3, B’β/B4, B”α/B13, FASS/TON2/B12, and C1) and one regulatory protein TAP46 to test their possible interactions with SPCH in planta, in yeast, and in vitro. The yeast two-hybrid assay detected the interaction of SPCH with A1 and A2 (Fig. 5A; because A2 and A3 are highly similar, A3 was not further tested), but not with the B and C subunits (SI Appendix, Fig. S6A). Bimolecular fluorescence complementation (BiFC) assays in tobacco epidermal cells indicated that SPCH associates with A1, A2, and A3 (Fig. 5B and SI Appendix, Fig. S6C). We quantified the BiFC fluorescence intensity values obtained from the same confocal settings and found that the absolute fluorescence intensity levels (compared with the positive control SPCH-ICE1) supported physical association of SPCH with the A1/A2 subunits (SI Appendix, Fig. S6E). It is worth noting that, although the PP2A-A subunits were ubiquitously distributed/localized at the subcellular level in tobacco epidermal cells (SI Appendix, Fig. S6B), their interaction with SPCH occurred only in the nucleus (Fig. 5B and SI Appendix, Fig. S6C). As predicted, the SPCH homolog, MUTE, did not show interactions with A1/A2 in the BiFC (SI Appendix, Fig. S6 D and E). The physical association between SPCH and A1/A2 was further confirmed by in vitro pull-down assays with Escherichia coli-made recombinant proteins (Fig. 5 C and D). As a further negative control for the pull-down assays, we included the Interactor of Constitutive active ROPs 1 (ICR1) (33) protein that has molecular weight and electric charge (38 kDa and immunoprecipitation [IP] 4.99) comparable to those of SPCH (33 kDa and IP 4.5). The results showed that no obvious interactions were detected between ICR1 and A1/A2 (Fig. 5 C and D).
Fig. 5.
PP2A-A subunits may directly bind to SPCH. (A) Yeast two-hybrid assay for BD-SPCH with PP2A-A (AD-A1 and AD-A2). The BD and AD empty vectors were used as negative control, and SCRM/ICE1 was used as positive control. Yeast growth controls are shown on the Left (-Leu-Trp). Interaction tests are shown on the Right (-Leu-Trp-His with 2.5 mM 3-AT). (B) Confocal images of BiFC assays to test interactions between SPCH and PP2A-A subunits. The expression of half YFPs (YFPN and YFPC) were used as negative controls. SCRM/ICE1 was used as positive control. Yellow shows complemented expression of YFP signal. (Scale bars, 50 μm.) (C and D) In vitro pull-down assays by using E. coli-made recombinant proteins. Amylose resins-bound MBP, MBP-SPCH, or MBP-ICR1 were incubated with (C) His-A1 or (D) His-A2 proteins. MBP and MBP-ICR1 proteins were used as negative controls. Immunoblots were visualized by anti-His antibody. Input His-tagged proteins and MBP-tagged proteins were visualized by anti-His and anti-MBP antibodies, respectively. Red asterisks indicate the positions of related proteins.
Taken together, we collected multiple lines of evidence supporting that SPCH may directly interact with the A subunits of PP2A, allowing the dephosphorylation of SPCH (SI Appendix, Fig. S7A), but we do not exclude the possibility that PP2A may regulate other stomatal factors, such as MUTE, or the regulators of SPCH in stomatal development.
Discussion
Stomatal production is not only vital for plant growth and development, it is also highly related to the ecosystem water balance on Earth. Strong genetic evidence supports the crucial role of SPCH in the initiation of the stomatal lineage cells in both monocots (34, 35) and dicots (1). Stomatal production is highly plastic in responding to environmental changes that necessitate SPCH expression levels to be highly adjustable. For example, the SPCH promoter is directly targeted and transcription is suppressed by the bHLH transcription factor PIF4, a core regulator in high-temperature signaling (36). The C2H2 zinc finger IDD16 transcription factor that functions in plant organ morphogenesis also directly suppresses SPCH expression (37). At the protein level, SPCH stability and function both are tightly related to its phosphorylation status. In Arabidopsis, both MPK3/6 and the GSK3-like kinase BIN2 target SPCH for phosphorylation and degradation (SI Appendix, Fig. S5F) (11, 12). On the other hand, the CDKA;1 kinase phosphorylates SPCH on one specific amino acid that seemed to promote SPCH’s activity in stomatal divisions (13). Here, through pharmacological treatment (CT, preferentially inhibiting PP2A and PP1) and genetic strategies (CRISPR and RNAi), we found that PP2A may stabilize SPCH and promote its abundance. In our experiments, overexpression of PP2A-A driven by the stomatal lineage BASL promoter does not give discernable phenotypes in stomatal development (SI Appendix, Fig. S7B). We suspect that the overexpression of an A subunit alone, without sufficient amount of the other two subunits, would not give rise to obvious biological consequences, including the regulation of SPCH phosphorylation levels. With the limitation of SPCH being destabilized in the PP2A mutant backgrounds, mapping out the specific PP2A-mediated dephosphorylation sites in SPCH protein is highly desired, but technically restricted. A previous study showed that PP2A dephosphorylates the bHLH factor HAND1 to regulate its dimerization and functional specificity in limb development (38). In stomatal development, the bHLH SCRM/ICE1 scaffolds the physical interaction between MAPKs and SPCH (39), and SCRM protein stability is also regulated by protein phosphorylation (40). Although our research did not suggest that PP2A dephosphorylates SCRM/ICE1, future research should address the question of whether heterodimerization of SPCH and SCRM/ICE1 is regulated by SPCH phosphorylation status.
Posttranslational modifications, including protein phosphorylation, are dynamically modulated by opposing regulators, such as kinases and phosphatases. With several kinases identified to phosphorylate SPCH, how SPCH dephosphorylation is regulated has been unknown before our study. While the Arabidopsis genome encodes more than 150 protein phosphatase catalytic subunit genes (36, 41), PP2A family phosphatases constitute a major group of protein Ser/Thr phosphatases and are implicated in many biological processes (15). It is intriguing to note that PP2A appeared to be functionally coupled with a few highly conserved kinases, such as MAPKs and the GSK3-like kinases, in multiple pathways. For example, the BR-responsive transcription factor BZR1 is phosphorylated and inactivated by BIN2 in the absence of BR, but, in the presence of BR, BZR1 is activated by PP2A-mediated dephosphorylation, which relieves the BIN2-mediated inhibition of BR responses (42, 43). The key ethylene biosynthetic enzymes, 1-aminocyclopropane 1-carboxylate synthases 2 and 6 (ACS2/6), are phosphorylated (and stabilized) by MPK6 (44) and dephosphorylated (and destabilized) by PP2A (45). In mammals, the p70 ribosomal protein S6 kinase (p70S6K) regulates cell growth, proliferation, and cell differentiation, and its phosphorylation status is tightly controlled by the antagonistic actions of the GSK3β kinase and PP2A (46). Here, our study suggested the opposing functions of PP2A with MAPKs and GSK3-like kinases, providing another example of kinase−phosphatase antagonism in the control of stomatal development, and underscoring the sophisticated signaling network that shapes this highly plastic plant developmental program.
Stomatal development and functional behavior are highly responsive to environmental cues (47). Genetic evidence showed that light, temperature, and carbon dioxide are influential to stomatal production, and the corresponding key regulators have been identified. PP2A function has been linked to plant responses to environmental changes/stresses and developmental cues (17, 22, 43, 48). Therefore, it is tempting to speculate that PP2A transduces environmental and/or developmental cues to optimize the SPCH protein level, and thus stomatal production. In the past, phosphatases were often considered as housekeeping dephosphorylation enzymes, without much specificity; however, they are now known to be both highly specific and subject to sophisticated regulatory mechanisms (15, 49). In our study, the in vitro data supported direct physical binding between SPCH and the scaffolding A subunits of PP2A, differing from many PP2A−substrate interactions that are largely controlled by the B regulatory subunits (50). Interestingly, it was recently found that small Rab GTPases, prominent regulators of vesicular trafficking, interact with the A subunits and may compete with the catalytic subunit for the formation of a functional PP2A complex (51). Whether SPCH may, by binding to the A subunits in vivo, regulate PP2A’s function in the nucleus will be an intriguing research direction in the future. On the other hand, the microarray dataset indicated that several genes encoding PP2A subunits, particularly the B and some C subunits, are responsive to environmental factors, for example, abiotic and biotic stresses, chemical treatments, and light (16). Future study will focus on investigating under what conditions PP2A phosphatases are activated or inactivated, and how this regulation feeds into the PP2A−SPCH connection in stomatal development, and on defining how functional specificity is achieved by these convoluted signaling networks. A systematic investigation of expression pattern, subcellular localization, and genetic redundancy of the B and C subunits will be helpful to further characterize the functional specificity of PP2A in different developmental contexts.
Methods
Plant Materials and Growth Conditions.
A. thaliana Col-0 was used as the WT unless otherwise noted. The mutants and marker lines used in this study were er105 (CS89504), yda-3 (Salk_105078), MPK6SR (29), spch-3 (1), 35S::DNyda-YFP (28), and 35S::DNmpk6-YFP (28). Other lines also included BASLpro::GFP-BASL (52), YDApro::YDA-YFP (28), ICE1pro::ICE1-YFP (40), and TMMpro::TMM-GFP (7). The PP2A-A mutant lines a1 (rcn1-6, SALK_055903) (48), a1;a2, and a1;a3 lines were constructed by crossing rcn1-6 with the a2-1 (SALK_042724) and a3-1 (SALK_014113) mutants (21). The c3;c4 seeds (20) were the generous gift of Martine Pastuglia, Institut Jean-Pierre Bourgin, UMR1318 INRA-AgroParisTech-ERL3559 CNRS, Versailles, France.
Unless otherwise noted, Arabidopsis seeds were germinated on half-strength MS medium at 22 °C in constant light for 6 d to 10 d. Seedlings were then transferred to potting mix for growth in 22 °C growth chambers with 16-h light/8-h dark cycles.
The Arabidopsis Genome Initiative numbers under investigation in the study are PP2A-A1 (RCN1, AT1G25490), PP2A-A2 (PDF1, AT3G25800), PP2A-A3 (PDF2, AT1G13320), PP2A-C1 (AT1G59830), PP2A-C3 (AT2G42500), PP2A-C4 (AT3G58500), PP2A-B”α/B13 (AT5G44090), PP2A-B' 'β/B4 (AT3G09880), TAP46 (AT5G53000), and FASS/TON2/B12 (AT5G18580).
CT Treatment.
The PP2A inhibitor CT (MilliporeSigma) was dissolved in DMSO for storage and usage. For long-duration CT treatment, Arabidopsis seeds were germinated and grown on half-strength MS/Agar medium supplemented with 10 μM or 50 μM CT, or with the corresponding amount of the DMSO solvent (mock). Seedlings were grown at 22 °C in constant light for 3 d to 5 d before images were captured. For short-term treatment, Arabidopsis seeds were grown on half-strength MS medium at 22 °C in constant light for 3 d, followed by submerging into sterile water with the specified amount of CT and/or DMSO for treatment. Plant samples were kept in 22 °C growth chambers for 1, 4, or 24 h before images were captured.
Plasmid Construction and Primer Designs.
The LR Clonase II-based gateway cloning technology (Invitrogen) was used to generate most constructs, unless otherwise noted. The promoter fragments (∼2 kb upstream of start codon), the coding regions of PP2A-A genes, were first cloned into the pENTR/D-TOPO vector (Invitrogen) and subsequently subcloned into binary destination vectors. Primers used to amplify these sequences are listed in SI Appendix, Table S1. To introduce amino acid mutations into SPCH, the plasmid pENTR/D-TOPO carrying SPCH coding sequence without the stop codon was used as template, and specific mutations were introduced by QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). The binary vector pMDC43 (53), with the 35S promoter replaced by an endogenous promoter, was used for PP2A-A overexpression. To generate the BiFC constructs for tobacco transient expression, pENTR/D-TOPO vectors containing the coding sequences of SPCH, MUTE, PP2A-A1, PP2A-A2, PP2A-A3, and SCRM/ICE1 were recombined to destination vectors pH35YG, pH35GY, pXNGW, pNXGW, pXCGW, and/or pCXGW (54). The pENTR/D-TOPO carrying PP2A-A promoters were recombined to pBGYN backbone (55) to generate PP2A-Apro::nucYFP. Plasmids were transformed into Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation and tobacco leaf infiltration.
To generate RNAi-PP2A-A mutants, the protocol described in ref. 25 was followed. Specifically, a 481-bp highly conserved region among the three PP2A-A genes was amplified from the A2 gene by RNAi-a1;a2;a3 primers and cloned into pCAMBIA3300 together with the miRNA173 target site by overlapping PCR. To generate crispr-a1;a2;a3 mutants, we followed the protocol described in ref. 24. Specifically, two pairs of oligos 100% matching PP2A-A1 and PP2A-A2/3, respectively, were designed. The sgRNA oligos were phosphorylated by T4 PNK (NEB) and annealed in thermocycler, followed by ligation with pMD18-T containing pAtU6-pAtUBQ-Cas9 cascade into the BbsI site. Then the chimeric AtU6-crispr-a1-pAtUBQ-Cas9 cassette was cloned into pCAMBIA1300 or pCAMBIA2300 through HindIII and EcoRI restriction sites to obtain p1300-crispr-a1 and p2300-crispr-a1, respectively. The AtU6-crispr-a2/3-pAtUBQ-Cas9 cascade was generated similarly, except that the pAtU6-crispr-a2/3 cassette was inserted into p1300-cripsr-a1 or p2300-crispr-a1 through KpnI and EcoRI sites to obtain p1300-crispr-a1;a2;a3 and p2300-crispr-a1;a2;a3, respectively. To genotype the crispr-a1;a2;a3 mutants, primers flanking the gRNA-targeting sites were designed to amplify the mutagenized regions of the three A genes. The amplicons were first assayed by gel electrophoresis. Single DNA bands were sent for Sanger sequencing. Heterogeneous amplicons were cloned in a TA cloning vector, and individual clones (10 to 20 for each PCR) were sent for Sanger sequencing. To introduce the crispr mutations into different genetic backgrounds (Fig. 2 and SI Appendix, Fig. S3), p1300-crispr-a1;a2;a3 or p2300-crispr-a1;a2;a3 was transformed into corresponding plant materials and screened for resistant transformants. The success of generation of crispr-a1;a2;a3 mutations was first evaluated by plant phenotypes, followed by PCR and sequencing. Primer sequences used in this study are listed in SI Appendix, Table S1.
Confocal Imaging and Image Processing.
All confocal images were captured with a Leica TCS SP5 II confocal microscope. Unless otherwise noted, adaxial epidermis from cotyledons of 3-dpg Arabidopsis seedlings were used. Cells outlines were visualized by the propidium iodide (PI; Invitrogen) staining. FPs were excited at 458 nm (CFP), 488 nm (GFP), 514 nm (YFP), 543 nm (mCherry), and 594 nm (PI). Emissions were collected at 480 nm to 500 nm (CFP), 501 nm to 528 nm (GFP), 520 nm to 540 nm (YFP), 600 nm to 620 nm (mCherry), and 591 nm to 636 nm (PI). Confocal images were false colored and brightness/contrast was adjusted with Fiji ImageJ software.
Quantitative Analysis of Stomatal Phenotype and SPCH Protein Level.
For quantification analysis of stomatal phenotype, 5-dpg seedlings were stained with PI (Invitrogen), and adaxial cotyledon images were captured with a Carl Zeiss Axio Scope A1 fluorescence microscope equipped with a ProgRes camera (Jenoptik). SI was calculated as the percentage of stomatal GC number relative to the total number of epidermal cells. The Student’s t test was used to determine whether difference is significant between lines.
To quantify SPCH protein expression levels, frequencies of CFP/YFP-positive cells were counted for comparison. Seedlings expressing SPCH-CFP and other variants were stained with PI, and confocal images were captured from adaxial cotyledon epidermis. The number of CFP/YFP-positive cells and total epidermal cells were counted with Fiji ImageJ software, and the ratio of CFP/YFP-positive cells among total epidermal cells were calculated. The Student’s t test was used to determine whether the difference is significant between treatments.
RNA Extraction and Real-Time PCR.
Total RNAs were extracted from 5-dpg Arabidopsis seedlings using RNeasy Plant Mini Kit (Qiagen). The first-strand cDNAs were synthesized by the qScript cDNA SuperMix (Quantabio), with 500 ng of total RNAs as template.
For real-time PCR assay, reactions were prepared by a SYBR Green Master Mix kit (ThermoFisher) and conducted by the StepOnePlus Real-Time PCR System (ThermoFisher). Three technical replicates were performed for each of three biological replicates. Expression values were normalized to the reference gene ACTIN2 using the ΔCT method. Data are presented as mean ± SD. Primers used in this study are listed in SI Appendix, Table S1. The CFP primers used in this study to evaluate the expression level of SPCH-CFP are as described in SI Appendix, Table S1.
Yeast Two-Hybrid Assay.
The pENTR/D-TOPO vector containing coding sequences of SPCH, SCRM/ICE1, PP2A-A, PP2A-B, and PP2A-C genes was cloned into bait vector pGBKT7 or prey vector pGADT7 (Clontech). The EZ-Transformation Kit (MP Bio-medicals) was used for yeast transformation by following the manufacturer’s instructions. The bait and prey plasmids were cotransformed into the yeast strain AH109, and positive transformants were selected with SD/-Leu/-Trp. The interactions were tested on SD/-Leu/-Trp/-His medium supplemented with appropriate concentration of 3-amino-1,2,4-triazole (3-AT).
Transient Expression in Nicotiana benthamiana, and BiFC Assay.
A. tumefaciens strain GV3101 carrying binary constructs was cultured overnight in 10 mL of Luria−Bertani medium with appropriate antibiotics. Agrobacterial cells were collected at 3,000 rpm for 10 min and resuspended in 10 mL of 10 mM MgCl2, followed by washing with 10 mM MgCl2, then optical density at 600 nm (OD600) of cell cultures was measured. For single construct transient expression assay, cell cultures were diluted to final OD600 of 0.5. For the BiFC assay, cell cultures of the BiFC pairs were equally mixed with p19, followed by final dilution to OD600 at 0.5. In both assays, mixed cell cultures were then infiltrated to 4-wk-old tobacco leaves. Leaf disks were excited 2 d to 3 d after infiltration, and fluorescence was checked with the confocal microscope. Three independent experiments were performed to obtain the representative images. For quantification, absolute fluorescence intensity levels of YFP were obtained from the same confocal settings, for example, laser intensity and smart gains, applied to both positive/negative controls and testing samples. For each sample, n = 15 cells were collected from three independent experiments.
Recombinant Protein Production and Pull-Down Assay.
Recombinant protein expression and purification of His-A1, His-A2, MBP, MBP-SPCH, and MBP-ICR1 were performed using standard protocols. For the pull-down assays, 10 μg of His-A1/-A2 proteins were incubated with prewashed MBP or MBP-SPCH/MBP-ICR1 beads in 1 mL of tris-buffered saline with Tween 20 (TBST) buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 0.05% Tween 20] for 3 h at 4 °C with gentle shaking. Then, the beads were collected and washed with the TBST buffer three to five times to eliminate nonspecific bindings. The beads were then boiled with sodium dodecyl sulfate (SDS) sample buffer and loaded on a SDS/polyacrylamide gel electrophoresis gel, and the pull-down proteins were analyzed by Western Blot with the α-His antibody (Cell Signaling Technology). Inputs were analyzed by Western Blot with α-His antibody (Cell Signaling Technology) or α-MBP antibody (NEB). The pull-down assays were repeated at least three times to acquire the representative image.
Data Availability.
All data are included in the manuscript and supporting information. The materials described in the manuscript are freely available to readers from Waksman Institute of Microbiology.
Supplementary Material
Acknowledgments
We thank our laboratory members for critical readings of the manuscript. We appreciate Dr. Martine Pastuglia (French National Institute of Agricultural Research) and Dr. Haiyang Wang (Chinese Academy of Agricultural Sciences) for contributing genetic materials. We thank the Frommer Lab (previously at Carnegie, Stanford) for sharing the BiFC constructs. This work is supported by grants from NIH (Grants R01GM109080 and R35GM131827 to J.D.). C.B. is supported by the Chinese Scholar Council, Waksman Busch Pre-doctoral Fellowship, and Rutgers University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.G.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912075117/-/DCSupplemental.
References
- 1.MacAlister C. A., Ohashi-Ito K., Bergmann D. C., Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Pillitteri L. J., Sloan D. B., Bogenschutz N. L., Torii K. U., Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Ohashi-Ito K., Bergmann D. C., Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanaoka M. M. et al., SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau O. S. et al., Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345, 1605–1609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shpak E. D., McAbee J. M., Pillitteri L. J., Torii K. U., Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309, 290–293 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Nadeau J. A., Sack F. D., Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Meng X. et al., Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann D. C., Lukowitz W., Somerville C. R., Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Ngwenyama N., Liu Y., Walker J. C., Zhang S., Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lampard G. R., Macalister C. A., Bergmann D. C., Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Gudesblat G. E. et al., SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yang K. Z. et al., Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in Arabidopsis. Mol. Plant 8, 783–795 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Janssens V., Goris J., Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhrig R. G., Labandera A. M., Moorhead G. B., Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 18, 505–513 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Durian G., Rahikainen M., Alegre S., Brosché M., Kangasjärvi S., Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front. Plant Sci. 7, 812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng T. S., Briggs W. R., The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell 22, 392–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waadt R. et al., Identification of open stomata1-interacting proteins reveals interactions with sucrose non-fermenting1-related protein kinases2 and with type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol. 169, 760–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright A. J., Gallagher K., Smith L. G., discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21, 234–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spinner L. et al., A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 4, 1863 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Zhou H. W., Nussbaumer C., Chao Y., DeLong A., Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16, 709–722 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michniewicz M. et al., Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Li H. et al., Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res. 21, 970–978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao Y. et al., Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felippes F. F., Wang J. W., Weigel D., MIGS: miRNA-induced gene silencing. Plant J. 70, 541–547 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Honkanen R. E., Cantharidin, another natural toxin that inhibits the activity of serine/threonine protein phosphatases types 1 and 2A. FEBS Lett. 330, 283–286 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R., Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Wang P., Shao W., Zhu J.-K., Dong J., The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J. et al., A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bush S. M., Krysan P. J., Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 58, 2181–2191 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Harris D. M., Myrick T. L., Rundle S. J., The Arabidopsis homolog of yeast TAP42 and mammalian alpha4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol. 121, 609–617 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter D. et al., An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Gu Y., Yan A., Lord E., Yang Z. B., RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol. Plant 1, 1021–1035 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z. et al., Multiple transcriptional factors control stomata development in rice. New Phytol. 223, 220–232 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Raissig M. T., Abrash E., Bettadapur A., Vogel J. P., Bergmann D. C., Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U.S.A. 113, 8326–8331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau O. S. et al., Direct control of SPEECHLESS by PIF4 in the high-temperature response of stomatal development. Curr. Biol. 28, 1273–1280.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi S. L. et al., IDD16 negatively regulates stomatal initiation via trans-repression of SPCH in Arabidopsis. Plant Biotechnol. J. 17, 1446–1457 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Firulli B. A. et al., PKA, PKC, and the protein phosphatase 2A influence HAND factor function: A mechanism for tissue-specific transcriptional regulation. Mol. Cell 12, 1225–1237 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Putarjunan A. et al., Bipartite anchoring of SCREAM enforces stomatal initiation by coupling MAP kinases to SPEECHLESS. Nat. Plants 5, 742–754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C. et al., MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43, 618–629.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H., Chevalier D., Larue C., Ki Cho S., Walker J. C., The protein phosphatases and protein kinases of Arabidopsis thaliana. Arabidopsis Book 5, e0106 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He J. X., Gendron J. M., Yang Y., Li J., Wang Z. Y., The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang W. et al., PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo S., Liu Y., Lueth A., Zhang S., MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Skottke K. R., Yoon G. M., Kieber J. J., DeLong A., Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 7, e1001370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin S., Wolgamott L., Yu Y., Blenis J., Yoon S. O., Glycogen synthase kinase (GSK)-3 promotes p70 ribosomal protein S6 kinase (p70S6K) activity and cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 108, E1204–E1213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casson S. A., Hetherington A. M., Environmental regulation of stomatal development. Curr. Opin. Plant Biol. 13, 90–95 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Blakeslee J. J. et al., Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 146, 539–553 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLong A., Switching the flip: Protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 9, 470–477 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Shi Y., Serine/threonine phosphatases: Mechanism through structure. Cell 139, 468–484 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Sacco F. et al., A subset of RAB proteins modulates PP2A phosphatase activity. Sci. Rep. 6, 32857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong J., MacAlister C. A., Bergmann D. C., BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtis M. D., Grossniklaus U., A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rombolá-Caldentey B., Rueda-Romero P., Iglesias-Fernández R., Carbonero P., Oñate-Sánchez L., Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell 26, 2905–2919 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubo M. et al., Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and supporting information. The materials described in the manuscript are freely available to readers from Waksman Institute of Microbiology.