Significance
The survival of many malignancies critically depends on l-methionine (l-Met) needed for S-adenosyl-methionine formation, protein synthesis, cysteine production, and polyamine synthesis. Partial dietary l-Met restriction or the administration of bacterial l-Met–degrading enzymes displays potent antitumor effects in multiple murine cancer models. Bacterial l-Met–degrading enzymes are problematic due to immunogenicity, and dietary restriction is not practical or desirable in some therpaeutic settings. Here we describe the development of an engineered human enzyme (hMGL-4.0) that reduces systemic l-Met levels and significantly inhibits the growth of multiple prostate cancer allografts/xenografts without weight loss or toxicity, even when administered for over a month. Our data suggests hMGL-4.0 represents a drug candidate targeting the l-Met metabolic vulnerability in prostate and other cancers.
Keywords: prostate cancer, l-methionine depletion, hMGL
Abstract
Extensive studies in prostate cancer and other malignancies have revealed that l-methionine (l-Met) and its metabolites play a critical role in tumorigenesis. Preclinical and clinical studies have demonstrated that systemic restriction of serum l-Met, either via partial dietary restriction or with bacterial l-Met–degrading enzymes exerts potent antitumor effects. However, administration of bacterial l-Met–degrading enzymes has not proven practical for human therapy because of problems with immunogenicity. As the human genome does not encode l-Met–degrading enzymes, we engineered the human cystathionine-γ-lyase (hMGL-4.0) to catalyze the selective degradation of l-Met. At therapeutically relevant dosing, hMGL-4.0 reduces serum l-Met levels to >75% for >72 h and significantly inhibits the growth of multiple prostate cancer allografts/xenografts without weight loss or toxicity. We demonstrate that in vitro, hMGL-4.0 causes tumor cell death, associated with increased reactive oxygen species, S-adenosyl-methionine depletion, global hypomethylation, induction of autophagy, and robust poly(ADP-ribose) polymerase (PARP) cleavage indicative of DNA damage and apoptosis.
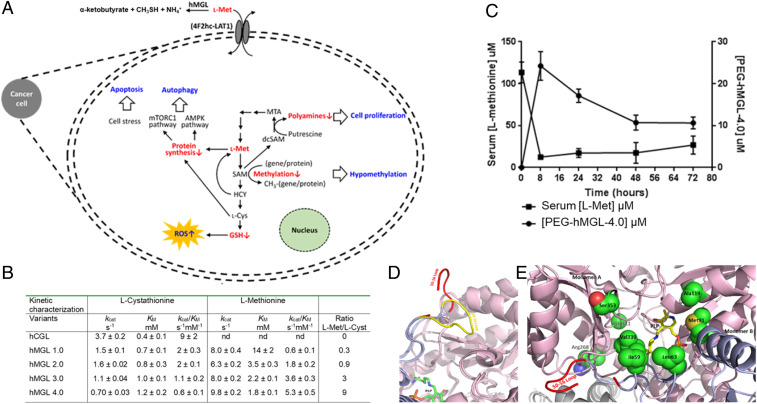
Many tumors have such a high demand for l-methionine (l-Met) that C11-methionine positron emission tomography can be used for imaging, particularly in glioblastomas (GBM), astrocytomas, prostate carcinomas (PCa), and certain lymphomas (1–6). Extensive studies have revealed that cancer cells are much more sensitive to the availability of l-Met compared to healthy tissues and these findings have generated significant interest in therapeutic strategies aimed at systemically reducing l-Met levels (7–12). The high l-Met demand of tumor cells is partly related to their increased proliferation rate, as l-Met is required for l-cysteine (l-Cys) production for protein synthesis and also for glutathione (GSH) and antioxidant responses. l-Met is also required for polyamine synthesis and for S-adenosyl-methionine (SAM) formation, which serves as the methyl donor for DNA and protein methylation (Fig. 1A). Aberrant methylation and increased polyamine synthesis are hallmarks of numerous tumors and known contributors to tumorigenesis (13–16) (Fig. 1A). In addition to the elevated l-Met demand associated with the rapid proliferation of cancer cells, the requirement for high rates of l-Met uptake is exacerbated by genetic defects in salvage pathways that recycle l-homocysteine (l-Hcy) back to l-Met. For example, genetic alterations that impact methionine salvage pathways arise in a large fraction of tumors and include mutations that impact methionine synthase (encoded by the MTR gene, occurring at a frequency of about 8% across all tumor types) and methylthioadenosine phosphorylase (MTAP deletions, 15% of all tumors), which cleaves 5-methylthioadenosine (5-MTA, a byproduct of polyamine synthesis) to 5-methylthioribose-1-phosphate that is further metabolized back to l-Met (7, 17–25) (Fig. 1A).
Fig. 1.
Engineered human methioninase for cancer treatment. (A) Overview of the role of methionine in cell metabolism and the key metabolic and physiological effects arising from its depletion. (B) Michaelis–Menten parameters for engineered hMGL variants and for the parent enzymes, hCGL. (C) Serum l-Met concentration and pharmacokinetics of PEG/hMGL-4.0 following a single-dose intraperitoneal administration (50 mg/kg) in C57BL/6J mice. (D and E) Structural features of hMGL-4.0. (D) Comparison of the 50–56 loop in the “open” state in hMGL-4.0 (shown in red) and in the “closed” state in wild-type hCGL (shown in yellow). The secondary structures of CGL are shown in ribbon diagram with one monomer colored pink and the neighboring one light blue, with the exception the 50–56 loop colored in yellow for the wild-type and for hMGL-4.0 in red. PLP is shown in stick with carbon atom colored green. (E) The location of the mutated residues in hMGL-4.0. Residues that were engineered to create hMGL-4.0 that have been shown in space-filling mode. Coloring scheme for the secondary structures of CGL is identical with D. The PLP cofactor is shown in stick with carbon atoms colored yellow.
Collectively, these observations have provided the impetus for the development of therapeutic approaches for cancer treatment by limiting systemic availability of l-Met. The most straightforward way to achieve a reduction in l-Met levels is via dietary restriction and in several small clinical trials, 1- to 3-d cycles of l-Met fasting in combination with chemotherapy were shown to result in improved outcomes in GBM, melanoma, gastric cancer, and colorectal cancers (26–29). l-Met fasting results in a decrease in the concentration of l-Met in circulation by ∼40 to 50% (27, 29), which is reversed very quickly upon return to a complete diet. However, patients can only be maintained on a complete l-Met–free diet for relatively short intervals (≤4 d) due to rapid weight loss and other toxicities. Alternatively, adverse effects of complete restriction may be avoided by partial l-Met reduction and several recent studies have demonstrated the potential for this approach, either alone or in combination with chemotherapeutic agents or radiation for several cancers using preclinical models, with the clinical validation of this appeach awaiting further studies (30–32).
Hoffman and coworkers demonstrated that the intravenous administration of bacterial methionine-γ-lyase from Pseudomonas putida (pMGL), which degrades l-Met to α-ketobutyrate, methane thiol, and NH3, enables a drastic depletion of serum l-Met to a level <20% of the physiological serum concentration, in turn resulting in dramatic reduction in the growth of numerous tumors, including GBM, neuroblastoma, colon cancer, and PCa in murine xenograft and patient-derived (PDX) xenograft models (33–39). Treatment with pMGL was shown to cause G2 arrest and thus to synergize with S/G2-dependent chemotherapeutic agents. However, preclinical studies in the macaque model subsequently showed that pMGL rapidly loses activity in serum with a t1/2 of ∼2 h and, equally importantly, was shown to trigger severe immune responses that can result in anaphylactic shock and death (40).
Immunogenicity is almost always a major concern with the clinical use of nonhuman proteins. Unfortunately, neither humans nor other mammals encode l-Met–degrading enzymes. Red blood cell encapsulation of pMGL is being evaluated as a means of preventing immune recognition of the enzyme; however, this approach does not overcome the rapid deactivation of pMGL, which occurs due to loss of its cofactor, pyridoxal phosphate (PLP, or vitamin B6) (41). The closest human homolog to pMGL (61% amino acid identity) is cystathionine-γ-lyase (hCGL), the last enzyme of the mammalian transsulfuration pathway that catalyzes the α,γ-elimination of l-cystathionine to l-Cys, α-ketobutyrate, and NH3. Although pMGL and hCGL both belong to the γ-family of PLP enzymes and are structural homologs performing a related reaction chemistry, the latter enzyme shows no catalytic activity with l-Met. Previously, we used structure-guided scanning saturation mutagenesis of key residues in the active site of hCGL to engineer a prototypical human methionine-γ-lyase (hMGL-1.0 variant: hCGL-E59N/R119L/E339V) (42). We showed that enzyme administration in animals maintained on dietary l-Met restriction led to a >20-fold reduction in the serum concentration of l-Met relative to that in control animals. Only diet + enzyme administration (but not diet alone) resulted in a dramatic decrease in the growth of neuroblastoma xenografts in nude mice. However, this treatment strategy suffered from three key limitations: First, it required daily dosing of hMGL-1.0 at a level that is impractical for humans; second, it was accompanied by significant weight loss in the animals (in both the diet and diet + enzyme groups) that, third, necessitated intermittent treatment consisting of 3 d of severe l-Met deprivation (diet + enzyme administration) followed by a 4-d recovery period (42).
Here we report the development of a catalytically optimized hMGL capable of lowering the plasma l-Met concentration by >75% for more than 3 d after single intraperitoneal administration without the need for any dietary interventions. We show that the sustained low concentration of serum l-Met achieved by enzyme treatment is well tolerated and results in no weight loss, liver enzyme abnormalities, or other toxicities but importantly, it leads to significant retardation in the growth of multiple murine prostate tumor allograft and xenograft models. Mechanistic studies revealed that limiting l-Met availability depletes SAM and induces global hypomethylation, causing a G2 cell cycle arrest and robust PARP cleavage indicative of DNA damage and apoptosis.
Results
Engineering of a Therapeutically Relevant Human Methionine-γ-Lyase.
Enzymes in the same superfamily derive from a common ancestor with subsequent evolution to differential catalytic functions occurring by fixing particular amino acids at certain positions key for dictating substrate specificity and catalytic mechanism (43–45). We searched the National Center for Biotechnology Information database for CGL and MGL enzymes that share an average of 30 to 40% amino acid sequence identity. Using a phylogenetic tree comprising eukaryotic and prokaryotic CGL sequences, as well as prokaryotic MGL sequences, we identified ∼67 amino acid differentially conserved positions classified as heterotactic/type I or type II (43–45). Type I amino acid positions are conserved in MGL sequences but variable in CGL sequences, and type II positions are those that have a conserved amino acid in MGL and a different conserved amino acid in CGL sequences. We selected residues for mutagenesis both based on phylogenetic conservation patterns and on their positions in the hCGL structure.
First, we constructed a binary positional library using hMGL-1.0 as a template at 11 positions within the C-terminal domain such that each position could either be wild-type or changed to the consensus amino acid identified on the MGL branch of the phylogenetic tree, with a theoretical diversity of 211 = 2,048 variants. Approximately 4,000 Escherichia coli transformants (∼2× of the theoretical diversity of this library) were screened using a microtiter plate assay (42). Ten variants with the highest activity were purified and their kinetics analyzed, in turn leading to the identification of hMGL-2.0 containing three additional mutations (K268R/T311G/I353S) and having a threefold improved kcat/KM for l-Met degradation over the parent (Fig. 1B). In parallel, 20 residues identified from the phylogenetic analysis and located within the first and second shell of the active site were individually mutated to the respective conserved MGL amino acid. Two of these amino acid substitutions, S63L and L91M, resulted in higher catalytic activity and when introduced into the hMGL-2.0 scaffold increased the kcat/KM for l-Met by twofold, yielding hMGL-3.0. A further increase in kcat/KM was achieved following mutagenesis of the E59 and R119 positions.
In summary, the phylogeny-based protein engineering campaign led to the creation of hMGL-4.0 with eight substitutions (hCGL-E59I/S63L/L91M/R119A/K268R/T311G/E339V/I353S) having ninefold higher kcat/KM for the degradation of l-Met relative to hMGL-1.0. Additionally, hMGL-4.0 had 15-fold lower catalytic activity for l-cystathionine, the physiological substrate of CGL relative to the wild-type enzyme and 3-fold lower than hMGL-1.0. Importantly, unlike the bacterial pMGL, the hCGL-4.0 enzyme was extremely stable in serum (t1/2 = 83 ± 2 h) (SI Appendix, Fig. S1). Single-dose intraperitoneal administration of hMGL-4.0, PEGylated to prevent renal clearance (by conjugation to methoxy PEG succinimidyl carboxymethyl ester, molecular mass 5,000 Da; PEG/hMGL-4.0) (SI Appendix, Fig. S2) in C57BL/6 mice at 50-mg/kg weight resulted in sustained reduction of serum l-Met for >72 h (Fig. 1C). At 2 and 3 d following treatment, the serum l-Met concentration was reduced to 17 and 26 μM, respectively, compared to 110 μM at t = 0, before enzyme administration. Allometric scaling suggests that a dose of 50 mg/kg in mice translates to ∼7 mg/kg in humans, which is comparable to the typical dosing of several antibody therapeutics (46). Utilizing an anti-PEG sandwich enzyme-linked immunosorbent assay (ELISA) to detect circulating levels of drug, we further determined that PEG/hMGL-4.0 had favorable pharmacokinetic properties with a clearance t1/2 of ∼44 h (Fig. 1C).
hMGL 4.0 Structure.
The X-ray crystal structure of hMGL-4.0 with a PLP internal aldimine was solved at a resolution of 2.73 Å (SI Appendix, Table S1). The enzyme has a near identical structure to hCGL (rmsd = 0.60 Å at the Cα atoms), with a dimer of dimers architecture wherein the four active sites are composed of residues at the interface of adjacent monomers (47) (SI Appendix, Fig. S3A). However, subtle differences are observed involving, or close to, the residues mutated in hMGL-4.0. In wild-type CGL, a loop region comprising amino acids 50 to 56, which is located at the entrance of the active site and controls access of the substrate assumes a “closed” conformation, placing P52 in contact with I353 (SI Appendix, Fig. S3B). In contrast, in hMGL-4.0 the hydrophilic I353S substitution greatly destabilizes the closed conformation of the 50 to 56 loop which, as a result, is rotated by ∼100° and becomes completely solvent-exposed (Fig. 1D). The loop seems to be highly flexible with some of the residues showing no traceable electron density (Fig. 1D). Six of the eight mutations in hMGL-4.0 are located close to the active site (Fig. 1E), including residues E59, S63, and E339 that in CGL act to coordinate the binding of l-cystathionine. In hMGL-4.0 these are mutated to apolar amino acids (E59I, S63L, and E339V), forming a hydrophobic patch that provides effective l-Met binding within the active site (hydrophobicity of l-Met >> l-cystathionine) (SI Appendix, Fig. S3C). The R119A substitution in hMGL-4.0 removes the hydrogen bond of the R119 side chain to N241, which serves to anchor the S63 hydroxyl group in the wild-type enzyme. As a result, in hMGL-4.0, N241 assumes an alternative isomeric state that accommodates the hydrophobic replacement of S63 with leucine. Overall, the X-ray crystal structure of hMGL-4.0 suggests that increased loop mobility near the entrance to the active site plays a significant role in the markedly improved enzymatic activity for degradation of l-Met.
Enzyme-Mediated Systemic Depletion of l-Met Inhibits Tumor Growth in Mice.
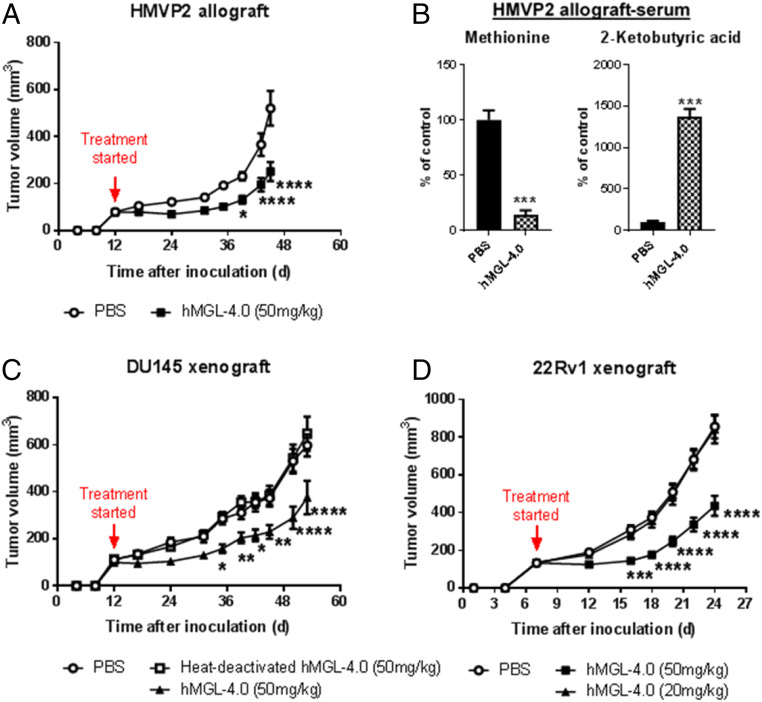
Tumors formed by the HMVP2 mouse cell line constitute a relevant model to human disease, including possessing characteristics of cancer stem cells, expressing stem cell markers such as LINneg/Sca-1high/CD49fhigh, CK14, and CD29, and having the capability of forming spheroids under appropriate culture conditions (48). Syngeneic male FVB/N mice bearing tumors (80 ± 7 mm3) were treated with PEG/hMGL-4.0 and either vehicle alone (phosphate buffered saline [PBS]) or heat-deactivated enzyme in PBS as controls. Heat-deactivated, PEGylated protein, rather than mutated inactive enzyme, was used as a control to properly account for the effects of protein batch-to-batch variation and for the presence of trace levels of expression host-derived impurities. Enzyme treatment had a significant inhibitory effect on the growth of allogeneic HMVP2 tumors associated with a reduction in the serum l-Met by 86% and a concomitant increase in the concentration of the reaction product 2-ketobutyrate (Fig. 2 A and B). Similarly, we observed a comparable degree of retardation of tumor growth in athymic nude mice bearing xenograft tumors formed by either DU145 cells or the more aggressive 22Rv1 human PCa cell line (tumor volumes at start of treatment = 110 ± 20 mm3 and 134 ± 4.4 mm3, respectively) (Fig. 2 C and D). Importantly, and in striking contrast to the severe weight loss and toxicities we and others have reported for mice maintained on a complete l-Met–free diet, treatment with PEG/hMGL-4.0, and the ensuing sustained reduction in serum l-Met for well over 30 d, was very well tolerated with no differences in body weight, food consumption, blood counts, or markers of liver (serum alanine aminotransferase [ALT] levels) and kidney (serum urea levels) toxicity relative to control mice (SI Appendix, Figs. S4 and S5). Additionally, histological analysis at necropsy revealed no gross abnormalities in any major organs of mice subjected to enzyme-mediated systemic l-Met depletion. Finally, analyses of H&E-stained liver sections from mice treated with the enzyme (SI Appendix, Fig. S6) showed a normal appearance and lack of any treatment related abnormalities consistent with the lack of any changes in serum toxicity markers.
Fig. 2.
Efficacy of hMGL-4.0 administration in murine PCa models. (A) Quantitation of tumor volume in male FVB/N mice bearing allograft tumors of HMVP2 PCa spheroids following treatment with hMGL-4.0 or controls (PBS, n = 17; hMGL-4.0 [50 mg/kg], n = 18). (B) Relative concentrations of l-Met and 2-ketobutyric acid in serum determined by MS following termination of the HMVP2 allograft studies (n = 5 per group). (C) Quantitation of tumor volume in male nude mice bearing xenograft tumors of DU145 PCa cells following treatment with hMGL-4.0 or controls (PBS, n = 9; heat-deactivated hMGL-4.0 [50 mg/kg], n = 15; hMGL-4.0 [50 mg/kg], n = 18). (D) Quantitation of tumor volume in male nude mice bearing xenograft tumors of 22Rv1 PCa cells following treatment with PBS, n = 12; hMGL-4.0 (50 mg/kg), n = 12; hMGL-4.0 (20 mg/kg), n = 12. Throughout, data are expressed as mean ± SEM. (A, C, and D) Repeated-measures two-way ANOVA followed by Bonferroni’s multiple-comparison test; (B) two-tailed Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Metabolomic and Signaling Effects Induced by Enzyme-Mediated Depletion of l-Met.
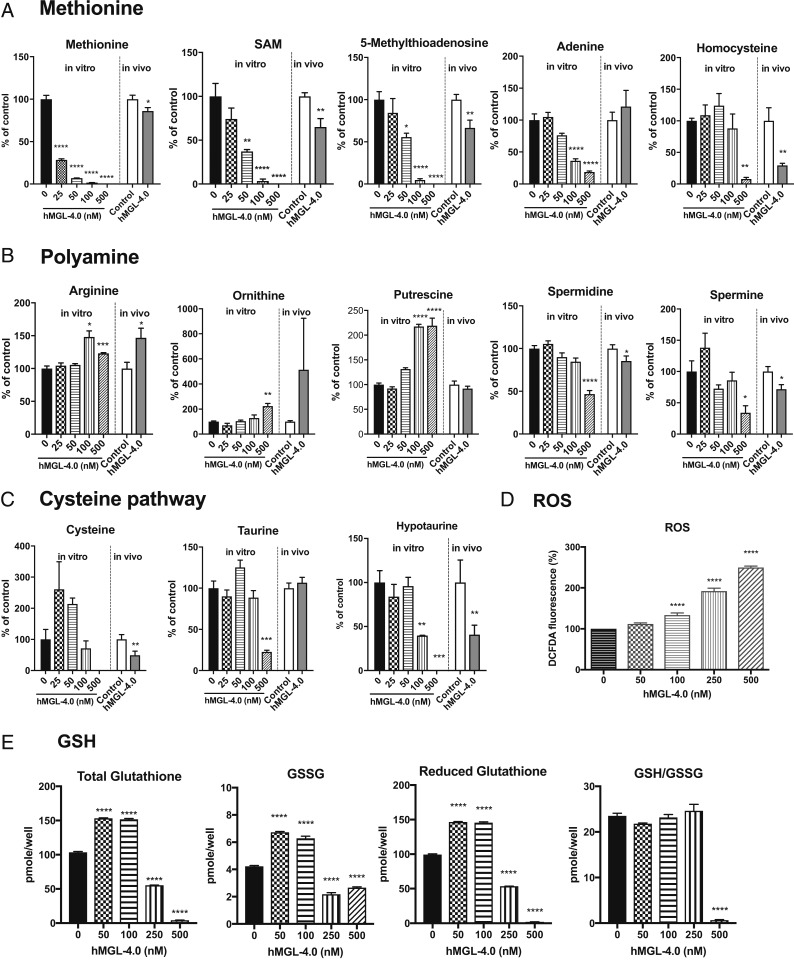
In vitro treatment of HMVP2 cells incubated with hMGL-4.0 resulted in dose-dependent and—given the enzyme kinetic parameters—time-dependent decreases in the levels of intracellular l-Met and SAM with complete elimination of both metabolites at higher concentrations (Fig. 3A). Consistent with a loss of SAM needed for spermidine/spermine synthesis, the polyamine precursors ornithine and putrescine were elevated approximately twofold concomitant with dose-dependent decreases in the levels of 5-MTA and its adenosine salvage pathway product, adenine (Fig. 3A). A significant reduction in spermidine/spermine levels was only evident with the highest enzyme concentration used and was concurrent with complete SAM depletion (Fig. 3B). Significant reductions in l-Hcy, l-Cys, and total and reduced GSH, as well as hypotaurine and taurine, were detected at higher concentrations of enzyme consistent with the near complete depletion of cellular l-Met (Fig. 3 A, C, and E). As expected, depletion of GSH was accompanied by a significant increase in intracellular reactive oxygen species (ROS) (Fig. 3D). Addition of N-Acetyl-l-Cysteine (NAC) partially rescued the survival of HMVP2 cells in the presence of hMGL-4.0 (SI Appendix, Fig. S7). Metabolite levels from tumor tissues isolated from HMVP2 allografts treated with hMGL-4.0 showed very similar changes to those observed in tumor cells treated in vitro (Fig. 3 A–C; see also SI Appendix, Fig. S8 for absolute l-Met levels). Specifically, tumor tissues from hMGL-4.0–treated mice showed a significant decrease in l-Met, SAM, 5-MTA, and l-Hcy (Fig. 3A). SAM and l-Hcy levels were more depressed relative to the observed l-Met levels in tumor tissues in comparison to the in vitro results; however, it should be kept in mind that the in vivo metabolite data account for the contribution of stromal cell contents that reduce tumor purity and, furthermore, reflect a much longer treatment period relative to the in vitro experiment. Overall, the metabolomic analysis indicates that the in vitro results are recapitulated in vivo and underscore the significant decreases in spermidine/spermine levels along with significant reductions in l-Cys and hypotaurine mediated by treatment (Fig. 3 B and C).
Fig. 3.
Metabolomic analyses of HMVP2 PCa cells in treating with hMGL-4.0. (A–D) Relative concentrations of methionine and nonmethionine intracellular metabolites in HMVP2 PCa cells as a function of increasing concentration hMGL-4.0 after 24-h treatment (bars left of the dividing lines) and in HMVP2 allograft tumor tissues after treatment with control or hMGL-4.0 50 mg/kg (bars right of the dividing lines) as determined by MS (n = 3 cell culture replicates and n = 8 or 9 for control and hMGL-4.0 50-mg/kg treated tumor tissues). Select metabolites in (A) methionine pathway, (B) polyamine pathway, (C) cysteine pathway, and (D) relative ROS levels in HMVP2 PCa cells as a function of increasing concentration hMGL-4.0. Cellular ROS levels were measured by DCFDA fluorescence 4-h posttreatment (data are from four independent experiments; for each experiment n = 3 cell culture replicates at each dose). (E) Total, oxidized, and reduced GSH levels were measured at 24-h time point by the spectrophotometric method (n = 3 cell culture replicates at each dose). All data are expressed as mean ± SEM. One-way ANOVA followed by Bonferroni’s multiple comparison test (for in vitro cell culture) and Student t test (for in vivo tumor tissues). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
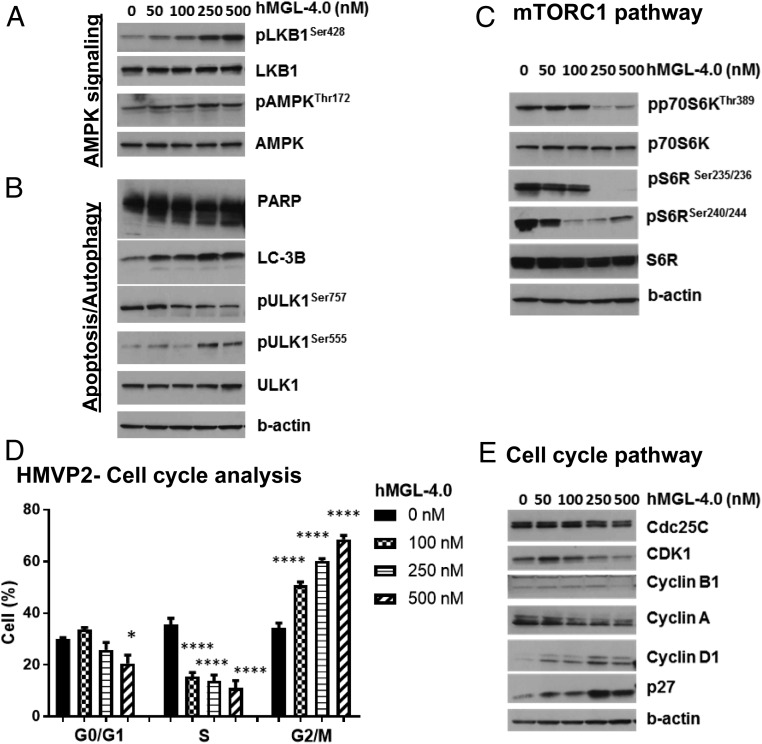
Corresponding with a nutrient starvation response, we observed a slight increase (∼1.3-fold) in AMPK phosphorylation (pAMPKThr172) and robust phosphorylation of its upstream regulator LKB1 (pLKB1) (Fig. 4A). This was accompanied by decreased phosphorylation of ULK1 (pULK1Ser757), along with increases in phosphorylated ULK1Ser555 (pULK1Ser555) (Fig. 4B). A significant reduction of mammalian target of rapamycin (mTOR)C1 signaling was evidenced by marked reduction in phosphorylation of p70S6K, its downstream target pS6R (Fig. 4C), and increased levels of the autophagosome marker LC-3 II (Fig. 4B). Although autophagy is often utilized as a survival mechanism, we found significant PARP cleavage indicative of apoptosis occurring in hMGL-4.0–treated cells (Fig. 4B), suggesting that l-Met depletion causes unique cellular responses beyond nutrient deprivation. Furthermore, treatment with hMGL-4.0 caused a significant concentration-dependent arrest in G2/M phase with a corresponding decrease in the proportion of cells primarily in the S-phase population (Fig. 4D and SI Appendix, Fig. S9). We confirmed that the arrest in G2/M was correlated at the protein level with concentration-dependent decreases of cdc25c, CDK1, cyclin B1, and cyclin A and an increase of the cyclin-dependent kinase inhibitor p27 (Fig. 4E). As DNA methyltransferase (DNMT) inhibitors, such as adenosine-2,3-dialdehyde, 5-aza-2′-deoxycytidine, and 5-azacytidine have been shown to cause a G2 cell cycle arrest in a variety of tumors (49–52), we hypothesized that a methylation deficiency driven by l-Met depletion may be responsible for arrest in G2/M. Indeed, corresponding with the observed loss of SAM, an assessment of the global methylation status of these cells revealed a significant decrease in DNA methylation approaching a near 50% reduction in cells treated with the highest enzyme concentrations (SI Appendix, Fig. S10).
Fig. 4.
Cell cycle and signaling pathway analyses of HMVP2 PCa cells in treatment with hMGL-4.0. (A–E) HMVP2 cells were treated with indicated concentrations of hMGL-4.0 for 24 h. Metabolic stress markers and cell cycle regulatory proteins were measured by immunoblot. Immunoblots were performed at least three times with β-actin controls for each experiment. (D) Cell-cycle phase distribution was measured by guava-based flow cytometry at 24-h time point (n = 5 independent experiments); one-way ANOVA followed by Bonferroni’s multiple-comparison test. *P < 0.05, ****P < 0.0001.
As noted with the mouse PCa HMVP2 cells above, treatment of the human PCa cell line 22Rv1 with hMGL-4.0 elicited a near identical concentration-dependent depletion of intracellular l-Met and similar overall changes in downstream metabolites. SAM, 5-MTA, and l-Cys were eliminated at the highest concentration of enzyme accompanied by significant increases in ornithine and putrescine, decreases in GSH, and elevated levels of ROS (SI Appendix, Fig. S11). We again observed elevated levels of cleaved PARP, indicating cell killing via an apoptotic mechanism. Autophagic responses were also evidenced by pAMPK accumulation, including reduction of pp70S6KThr389, pS6Ser235/236, and pS6Ser240/244, as well as LC-3 II formation (SI Appendix, Fig. S12). Concentration-dependent decreases of cdc25c, CDK1, cyclin B1, and cyclin A and an increase of the cyclin-dependent kinase inhibitor p27 were also observed in the 22Rv1 cell line.
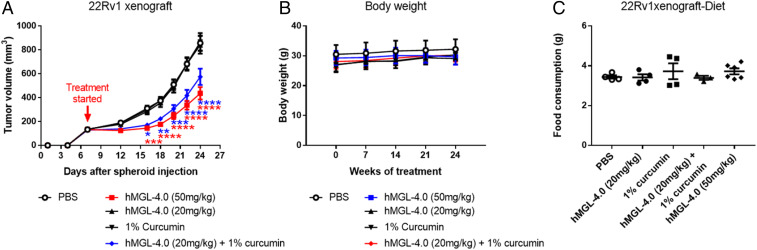
Enzyme-Mediated l-Met Depletion Synergizes with Curcumin for Inhibition of Tumor Growth.
As the treatment of hMGL-4.0 in PCa cells caused increased ROS and decreased GSH levels, we examined the efficacy of hMGL-4.0 in combination with agents that affect other major antioxidant pathways, such as the natural product curcumin, which has been reported to irreversibly inhibit thioredoxin reductase (TXNR) (53). As shown in (SI Appendix, Fig. S13), in vitro combination of curcumin with PEG/hMGL-4.0 produced a synergistic reduction in cell survival in both mouse (HMVP2) and human PCa (DU145 and 22Rv1) cells. To determine if these effects could be translated to an in vivo model, we treated male nude mice bearing 22Rv1 tumors with either curcumin (1% in diet), a low dose of PEG/hMGL-4.0 (20 mg/kg) or the combination of the two. At this dosing level, neither curcumin nor PEG/hMGL-4.0 as single agents showed any effect upon tumor growth compared to control; however, coadministration of PEG/hMGL-4.0 and curcumin together significantly inhibited tumor growth. In this experiment, the lower dose of hMGL-4.0 (20 mg/kg) produced a 63% reduction in serum l-Met level compared to the control group. Treatment with the higher dose of hMGL-4.0 (50 mg/kg) produced >90% reduction in l-Met serum level. Combination treatment was not found to be associated with any major toxicity (Fig. 5 and SI Appendix, Fig. S14). The synergistic effect of hMGL-4.0 in combination with curcumin may be due to inhibition of TXNR by curcumin in combination with a reduction in GSH pathway by hMGL-4.0, leading to further induction of ROS. In support of this hypothesis, the combination of curcumin and hMGL-4.0 produced further increases of intracellular ROS (SI Appendix, Fig. S15) compared to either agent alone. Since curcumin has multiple targets in addition to TXNR, additional experiments were performed using a combination of hMGL-4.0 together with a specific TXNR inhibitor, Auranofin (54). The combination of hMGL-4.0 and Auranofin showed strong synergistic inhibition of cell survival and increased ROS levels similar that seen with the combination of curcumin and hMGL-4.0 (SI Appendix, Figs. S16 and S17).
Fig. 5.
Efficacy of hMGL-4.0 and curcumin. (A) Quantitation of tumor volume in male nude mice bearing xenograft tumors of 22Rv1 PCa cells following treatment with PBS, n = 12; hMGL-4.0 (50 mg/kg), n = 12; hMGL-4.0 (20 mg/kg), n = 12; 1% curcumin in diet, n = 10; or hMGL-4.0 (20 mg/kg) and curcumin in combination, n = 15. (B and C) Quantitation of (B) body weight (n = 7 to 8) and average (C) food consumption (n = 3 to 6) for each treatment group. Throughout, data are expressed as mean ± SEM. Repeated-measures two-way ANOVA (A and B) or one-way ANOVA (C) followed by Bonferroni’s multiple comparison test was used for statistical analyses. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
We demonstrated that administration of PEG/hMGL-4.0 as a single agent caused a sustained decrease (75%) of l-Met in mice that remarkably did not cause any weight loss. In contrast, long-term dietary l-Met restriction results in substantial weight loss despite only decreasing serum l-Met by 40 to 50% (27, 29, 42, 55, 56). It is likely that the toxicities and weight loss associated with dietary l-Met restriction result from its effect on the gastrointestinal tract. In earlier studies, maintenance of animals on l-Met– and l-Met/choline-deficient diets, which cause nonalcoholic steatohepatitis, was reported to result in intestinal barrier deterioration, macroscopic shortening of the intestine and intestinal villi, and in gut microbial dysbiosis (57, 58). Hepatocellular injury, oxidative stress, and high ALT levels are caused by a significant decrease in liver SAM and GSH in these models (59), which is not surprising given that >75% of the blood supplied to the liver is via the portal vein that collects blood from nearly the entire gastrointestinal tract (60). In contrast, systemic administration of PEG/hMGL-4.0 not only allowed deeper and more precise control of serum l-Met levels compared to what may be achieved by dietary intervention but, even more importantly, suggests that it does not directly perturb normal gastrointestinal and liver metabolism as there were no observations of weight loss nor changes in liver and kidney function biomarkers. In fact, repeated administration of PEG/hMGL-4.0 for over a month in the presentent study was not associated with any significant adverse effects.
There is considerable ongoing interest in capitalizing upon the benefits of l-Met restriction for cancer treatment. For example, there are three open clinical oncology trials exploring the effects of dietary l-Met restriction: 1) NCT03186937, for enhancing tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor-2 expression in triple-negative breast cancer, and in turn enhanced sensitization to TRAIL receptor agonists (61); 2) NCT03733119, in which l-Met restriction in combination with an Akt/ERK inhibitor is being evaluated in metastatic or inoperable triple-negative breast cancer; and 3) NCT03574194, for sensitizing nonsmall-cell lung carcinoma (NSCLC) to stereotactic body radiation therapy. Additionally, radiation in combination with dietary l-Met restriction was found to be highly effective in treating mouse tumor models (62), as have other studies combining radiation with epigenetic modifiers, such as DNMT inhibitors (63). Since complete l-Met restriction is associated with severe weight loss and toxicity, recent studies have utilized partial l-Met restriction (30–32). For example, Gao et al. (30) reported inhibition of a chemotherapy-resistant RAS-driven patient-derived xenograft model of colorectal cancer with 0.12% l-Met diet compared to control diet (0.86% l-Met). They also showed a synergistic effect of this partial l-Met restriction in combination with low-dose 5-FU in this cancer model. These authors also showed that l-Met retriction alone did not alter the tumor growth in an aggressive autochthonus mouse model of soft tissue sarcoma; however, in combination with radiation it produced significant reduction of tumor growth in this radiation-resistant mouse model of cancer. However, the clinical impact of partial dietary l-Met restriction for cancer therapy has not yet been evaluated.
In the present study, depletion of l-Met with treatment of hMGL-4.0 produced significant decreases in the level of several metabolites. Notably, l-Met, SAM, 5-MTA, l-Hcy, l-Cys, hypotaurine, and spermine/spermidine levels were reduced in both cultured PCa cells and in tumor tissue following treatment with hMGL-4.0 (Fig. 3). The results are consistent with recently published report by Gao et al. (30), showing that mice maintained on partial l-Met–restricted diet had significant decreases in l-Met, l-Hcy, and hypotaurine in tumor tissues. l-Met restriction in primary colorectal cancer cells also similarly decreased levels of l-Met, SAM, 5-MTA, l-Hcy, and hypotaurine (30). We found that hMGL-4.0 not only mediates l-Met/SAM depletion resulting in global hypomethylation in PCa cells, but also inhibited the mTOR signaling pathway, which other studies have cotargeted by using DNA methyltransferase inhibitors in combination with mTOR inhibitors to achieve synergistic proapoptotic anticancer effects in colorectal, thyroid, and nasopharyngeal carcinoma models (64–66).
As another approach to reducing methionine cycle metabolites, pharmacologic inhibition of methionine adenosyltransferase 2A (MAT2A) with FIDAS-5 was shown to significantly decrease lung tumor growth without showing any toxicity or loss of body weight while significantly reducing SAM and SAH levels (67). However, as expected, in that study l-Met levels were not reduced by treatment with the drug. In contrast, treatment with hMGL-4.0 led to decreased levels of SAM, l-Hcy, and 5-MTA, in addition to significantly decreased levels of l-Met, as noted above. However, in preliminary experiments with FIDAS-5 we observed only modest reductions in PCa cell survival at higher concentrations and therefore we did not pursue further studies with this compound.
Our data also indicate that increased ROS and reduced GSH levels resulting from hMGL-4.0 administration contribute an additional mechanism to SAM depletion that could enhance ROS/DNA damaging effects, and accordingly we showed that using dietary curcumin as a ROS enhancer in combination with hMGL-4.0 synergistically inhibited growth of 22Rv1 PCa xenografts. Further in vitro studies with the specific TXNR inhibitor, Auranofin, led to the same conclusion. The ability of hMGL-4.0 to potently control l-Met levels without the need for dietary restriction and inhibit the growth of multiple PCa allograft/xenograft models bodes well for its further development as a drug candidate for treating patients with PCa and other malignancies.
Materials and Methods
Data Availability Statement.
All data are shown in the main manuscript and SI Appendix, except for the atomic coordinates and structure factors of hMGL-4.0, which have been deposited in the Protein Data Bank (PDB ID code 6OVG).
Site-Directed Mutagenesis.
We used the QuikChange Site-Directed Mutagenesis kit (Agilent Technology) to generate single site-mutated enzyme variants. All used oligonucleotides were designed based on the requirement of the kit protocol and ordered from Integrated DNA Technologies. Mutant strand synthesis reaction (Thermal cycling) was as follows: 50-µL reaction included 250 ng oligonucleotide pair (2 × 125 ng), 50 to 100 ng template plasmid (hCGL was cloned into pET28a vector), 10 nmol dNTP, 10-fold concentrated PFU turbo buffer (Agilent Technology), and 2.5 U PFU turbo DNA polymerase (Agilent Technology). Cycling parameter was: One cycle of 95 °C for 30 s and 16 cycles of 95 °C for 30 s, 55 °C for 1 min, 68 °C for 2 min. After thermal cycling, reactions were treated with 20 units of DpnI (New England BioLabs) at 37 °C for 1 h. Finally, 4 µL of the resulting reaction was used for heat-shock transformation on BL21 (DE3)-competent cells.
Combinatorial Library Construction.
Overlap extension PCR was used to generate a combinatorial library comprised of 11 positions selected from phylogenetic analysis for altering substrate specificity. All oligonucleotides were ordered from Integrated DNA Technologies and were further purified by gel chromatography. Sixteen oligonucleotides were designed to cover 336 base pairs of the C terminal of hCGL. Thermal cycling condition for fragment assembly was as follows: 50-µL reaction included 0.1 μM oligonucleotides (final concentration), 20 nmol dNTP, KOD hot start DNA polymerase buffer, 1 unit of KOD hot start DNA polymerase (EMD4Biosciences). The following program was used for thermal cycling: One cycle of 94 °C for 2 min, followed by 24 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min; 2.5 µL of each of the assembled reactions were used for further amplification and the amplified product was gel extracted for cloning into a pET28a plasmid.
Enzyme Kinetics.
Kinetic analyses were performed as described previously to measure α-ketobutyrate production by reaction with 3-methyl-2-benzothiazolone hydrazone hydrochloride (MBTH) to generate a chromophore with a λmax of ∼320 nm (42). Briefly, Eppendorf tubes (1.5 mL) containing 220 μL of substrate in a 100 mM sodium phosphate buffer, 1 mM EDTA (pH 7.3), and 10 µM PLP were incubated at 37 °C in a heat block. Reactions were started by adding 20.3 μL of enzyme solution and quenched with 26.7 μL of 50% trichloroacetic acid after a set length of time. Reactions and blanks were then mixed with 733 μL of MBTH solution (2.2 mL: 1.6 mL of 1 M acetate buffer pH 5.0, and 0.6 mL of 0.1% MBTH in same) and incubated at 50 °C for 40 min. After cooling, the samples were transferred to cuvettes and the A320 nm was determined. The resulting data were directly fit to the Michaelis–Menton equation using the software program KaleidaGraph to determine kcat and KM.
96-Well Plate Screening.
This screening method is based on the previous paper. Single colonies of E. coli BL21 (DE3), containing plasmids encoding either hMGL-V3.1 or variant hCGL enzymes, were packed into 96-well culture plates containing 75 µL of TB media per well and 50 µg/mL kanamycin. Cells were grown at 37 °C on a plate shaker to an OD600 of ∼0.8 to 1, cooled to 25 °C, whereupon an additional 75 µL of media containing 50 µg/mL kanamycin, and 2 mM isfopropyl-β-d-thiogalactopyranoside (IPTG) were added and incubation with shaking was continued for 2 h at 25 °C. Subsequently, 100 µL of culture per well were transferred to a fresh 96-well plate (assay plate). The assay plates were centrifuged (10 min, 3,500 × g) to pellet the cells, the media was removed, and the cells were chemically lysed by addition of 50 µL per well of B-PER protein extraction reagent (Pierce) and mixing for 25 min on a plate shaker. Then 20 µL of 5 mM l-Met at pH 7.3 was added to the lysates and incubated at 37 °C for ∼12 h. The α-keto butyrate acid reaction product is then derivatized by addition of 146 µL of MBTH solution to 54 µL of reaction and heated for 40 min at 50 °C. The absorbance at 320 nm was determined spectrophotometrically using a microtiter plate reader. Variants exhibiting high absorbance values indicative of product generation can thus be identified and selected for further characterization.
Pharmacokinetic Analyses.
The amount of PEG/hMGL-4.0 was assayed on sera following injection of 50 mg/kg of enzyme in C57BL/6J mice. Serum samples were obtained at 0, 8, 24, 48, and 72 h after injection. Diluted serum samples incubated with ELISA plate coating with anti-PEG antibodies (Abcam PEG-B-47) and then washed with PBST (PBS with 0.05% Tween 20) for five times. Finally, the captured PEG/hMGL-4.0 was detected by HRP-labeled anti-PEG antibodies (Abcam PEG-B-47 HRP). Half-lives of enzyme was calculated by KaleidaGraph using the pharmacokinetics equation y = k × (e(−ke × x) −e(−ka × x)), where ke is the elimination rate constant and ka is the absorption rate constant; t1/2 = ln (2)/ke.
Protein Purification.
E. coli BL21 (DE3) cells harboring plasmids with a given enzyme gene were grown in TB media containing 50 μg/mL kanamycin at 37 °C to an OD600 of ∼0.6, upon which time IPTG was added to a concentration of 1 mM. After an additional ∼12 h of incubation at 25 °C, cells were collected by centrifugation, resuspended in immobilized metal affinity chromatography (IMAC) buffer (10 mM NaPO4/10 mM imidazole/300 mM NaCl, pH 8), and lysed by a French pressure cell. The lysates were centrifuged at 14,000 × g for 30 min at 4 °C, the resulting supernatant applied to a nickel IMAC column, washed with 10 to 20 column volumes of IMAC buffer, and eluted with IMAC elution buffer (50 mM NaPO4/250 mM imidazole/300 mM NaCl, pH 8). All five chromatography procedures was performed at 4 °C. Eluted enzyme was then incubated with 10 mM pyridoxal-5′-phosphate (Sigma) for 30 min to 1 h at 25 °C. Using a 10,000 molecular weight cutoff (MWCO) centrifugal filter device (Amicon), protein was then buffer exchanged into a solution comprised of 1× PBS, 10% glycerol, pH 7.4. Aliquots of purified enzyme were then flash frozen in liquid nitrogen and stored at −80 °C. To determine protein concentrations, extinction coefficients were calculated based on amino acid sequence (ε280 = 29,870 M−1cm−1 for hCGL, 22,330 M−1cm−1 for pMGL) (4). All protein concentrations were determined from the absorption at 280 nm (A280) in 6 M guanidinium hydrochloride, 20 mM phosphate buffer, pH 6.5.
Pharmacological Optimization of hMGL-4.0 Variants.
Human methioninase variants were conjugated to lysyl residues using methoxy PEG succinimidyl carboxymethyl ester of molecular weight 5,000 (PEG-5K) (JenKem). Purification was performed as described above, with inclusion of an additional on column wash step using 100 column volumes of IMAC buffer containing 0.1% Triton X-114 (Sigma) in order to remove endogenous endotoxin. Purified proteins were then buffer exchanged into 1× PBS using a 10,000 MWCO filtration device (Amicon). PEG-5K was added at a 100:1 molar ratio to enzyme and allowed to react for 1 h at 25 °C in glass vials with gentle stirring. PEGylated hMGL-4.0 variants were extensively buffer exchanged using a 100,000 MWCO centrifugal filter device (Amicon) into storage buffer (PBS made 10% [vol/vol] with glycerol). Aliquots of PEGylated enzyme were flash-frozen in liquid nitrogen and stored at -80 °C. All PEGylated enzymes were analyzed for lipopolysaccharide (LPS) content using a Limulus Amebocyte Lysate kit (Cape Cod Inc.).
Serum Stability Analysis.
Stability analyses were performed to measure methanethiol production by reaction with 5,5′-dithio-bis-(2-nitrobenzoic acid) (DTNB, Thermo Fisher Scientific) to generate a chromophore with a λmax of ∼412 nm. Briefly, the purified hMGL-4.0 protein was incubated with 100% pooled human serum to generate enzyme-serum mixture (90% serum) at 37 °C. Each 15-µL aliquot was then taken from an enzyme–serum mixture at different time points to measure its residual activity by incubating with a mixture of 3.5 µL of 100 mM sodium phosphate buffer, 31.5 µL of 100% pooled human serum, 25 µL of 2mM of substrate, and 25 µL of 2mM DNTB. The absorbance changes at different time points were then determined and plotted to determine the half-life of hMGL-4.0.
Crystallization and Data Collection.
The hMGL-4.0 protein was found to crystallize under conditions containing 30% isopropanol, 150 mM sodium citrate, and 100 mM sodium cocadylate pH 6.5 as was reported for the hCGL-E59N, R119L, E339V variant (68). Sheet-like crystals were visualized after 2 d by mixing protein and mother liquid in a ratio of 2:1 sealed at 4 °C using a sitting-drop vapor diffusion technique and grew to their final size within 7 d. The crystals were then cryoprotected in 20% 2-methyl-2,4-pentanediol before being vitrified in liquid nitrogen for data collection. Crystal diffraction data were collected at the Advanced Photon Source Beamline BL23-ID-B (Argonne, IL). The resulting data were processed using the program HKL2000 (69). The statistics for data collection are summarized in SI Appendix, Table S1.
Structure Determination and Refinement.
The structure of hMGL-4.0 was determined by molecular replacement. A significant off-origin peak in the Patterson map with over 15% height of original peak was observed using the FFT program in CCP4 suite, which implicates the existence of pseudotranslational symmetry (tNCS) in crystal. Using Phaser in the CCP4 suite, this pseudotranslational vector was refined to be (0.5, 0.38, 0.5) (70–72). The application of this vector to correct the intensity of the diffraction spots allowed us to identify the correct solution for this crystal form using Phenix (73–75). A model of wild-type CGL (PDB ID code 2NMP) was used by Phaser from the CCP4 program suite to find the initial solution. The fitted model with mutated residues was rebuilt using COOT (76) and several iterative cycles of optimization was carried out to improve the quality of the model. The atomic models were refined using Phenix.refine, reserving 5% of diffraction as an unbiased test set for cross validation (Rfree) (73–75). The final Rwork is 18% with an Rfree of 23%. Checking the model using Molprobity (77) showed a clashscore of 6.3 (99th percentile) and Molprobity score for 1.58 (100th percentile). Detailed refinement statistics are summarized in SI Appendix, Table S1. Figures were prepared with PyMol (78).
Cell Culture.
Human PCa cell lines DU145, and 22Rv1 were purchased from the American Type Culture Collection. Cells were cultured in RPMI-1640 medium (Life Technologies) supplemented with FBS (10%; Life Technologies). The murine prostate tumor cell line, HMVP2, was derived from the ventral prostate of 1-y-old HiMyc transgenic mice (79) and cultured in RPMI-1640 medium containing 10% FBS. All cells were cultured at 37 °C in 95% air and 5% CO2.
Cell Survival Assay.
Cell survival was measured by Crystal violet assay (80). Briefly, cells (1 to 15 × 103/mL) in 96-well plates were treated with varying concentrations of hMGL-4.0. At indicated time points, the cells were fixed with 10% formalin and stained with 0.05% Crystal violet. After washing with water once, dye was extracted with 10% acetic acid solution. The absorbance was measured at 595 nm using a microplate reader (Tecan Group). Cell survival was further confirmed by 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma) or Trypan blue dye exclusion test (81, 82).
Measurement of ROS.
The intracellular ROS concentration was measured using 2′,7′-dichlorofluorescin diacetate (DCFDA) fluorescence. HMVP2 cells in a 96-well plate were stained with DCFDA (20 µmol/L; Sigma) at 37 °C for 30 min and then treated with indicated concentrations of hMGL-4.0 for 4 h and fluorescence intensity was measured at the respective excitation and emission wavelengths of 485 nm and 535 nm using a fluorescent plate reader (Tecan Group).
Measurement of Intracellular GSH Levels of HMVP2 Cells.
HMVP2 cells were treated with indicated concentrations of hMGL-4.0 for 24 h, the cells were then washed with PBS, pelleted, and mixed with 3 volumes of 5% metaphosphoric acid (R&D Systems Inc.). Following cell lysis (via two cycles of freeze–thaw), the resulting mixture was centrifuged and the GSH level in the supernatant was measured using a commercially available GSH detection kit (R&D Systems Inc.).
Cell Cycle Analysis.
HMVP2 cells were treated with various concentrations of hMGL-4.0 for 24 h after which cells were harvested, washed twice with PBS, and fixed in 70% ethanol overnight at −20 °C. Following fixation, cells were washed, resuspended in PBS containing RNase (250 mg/mL; Sigma), and incubated at 37 °C for 30 min. Cells were then treated with propidium iodide (50 μg/mL; Sigma) solution, incubated in the dark for 30 min, and the distribution of the various cell cycle phases were analyzed by Guava-based flow cytometry software (Millipore).
Western Blotting.
Antibodies against LKB1, pLKB1S428, AMPK, pAMPKThr172, p70S6K, pp70S6KThr389, S6 Ribo, pS6 RiboS235/236, pS6 RiboS240/244, ULK1, pULK1S555, pULKS757, LC-3B, cyclin A, cyclin D1, and PARP were purchased from Cell Signaling. Antibodies for p27, cyclin B1, and CDC25C were from Santa Cruz Biotechnology, and β-actin from Sigma-Aldrich. Secondary antibodies were purchased from GE Healthcare. Cells were treated with indicated concentrations of hMGL-4.0 for the specified time. After incubation, cells were washed with ice-cold PBS and lysed in radioimmunoprecipitation assay buffer. Proteins were separated by using 4 to 15% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. After blocking in 5% BSA for 1 h, the membranes were probed with specific primary antibodies (listed above) overnight at 4 °C. Following secondary antibody incubation, the membranes were visualized using a commercially available chemiluminescent detection kit (Pierce Biotechnology).
Mouse Studies.
All protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas at Austin. All mice were allowed to acclimate for at least 1 wk prior to use in experiments. For all tumor studies, tumor size was measured two to three times weekly using a digital caliper. Tumor volume was calculated by the formula: 0.5236 D1(D2)2, where D1 and D2 are the long and short diameter, respectively. Food consumption and body weight of the mice were measured weekly.
Deactivation of PEG/hMGL-4.0 as a Control for Mouse Studies.
Control PEG/hMGL-4.0 samples were deactivated by incubation at 100 °C for 10 min followed by centrifugation (16,000 × g for 5 min) to remove any precipitates. The samples (both before and after boiling) were subsequently tested using the aforementioned kinetic assay to ensure there was no remaining activity in the heat-inactivated enzyme control.
Mouse Allograft Tumor Studies.
The HMVP2 allograft tumor study was performed in syngeneic FVB/N male mice. Mice were fed a semipurified diet (AIN76A, 10 kcal%; Research Diets) and water ad libitum. To generate spheroids, HMVP2 cells were plated in ultralow attaching tissue culture dishes. After 3 d, spheroids were harvested, mixed 1:1 with matrigel (BD Biosciences), and injected subcutaneously into the flank. After the tumors were palpable, mice were divided into groups such that the average tumor volumes in all of the groups were approximately equal. Each group was treated by intraperitoneal injection (once every 2 d) with one of the following: PBS, or 50 mg/kg PEG/hMGL-4.0.
22Rv1 PCa Xenograft Studies.
The 22Rv1 cells (2 × 106) were mixed with matrigel (1:1) and injected subcutaneously into both flanks of male 6- to 7-wk-old athymic nude mice (JAX). Treatment was initiated when tumor volume reached 120 mm3 (day 8) with one of the following: PBS (intraperitoneal injections every 2 d), 20 mg/kg hMGL/4.0 (intraperitoneal injections every 2 d), 50 mg/kg PEG/hMGL-4.0 (intraperitoneal injections every 2 d), curcumin (1% [wt/wt] in diet fed ad libitum), or combinations of 20 mg/kg PEG/hMGL-4.0 and curcumin (n = 7 per group).
DU145 PCa Xenograft Studies.
Male athymic nude mice (JAX) were injected subcutaneously with 2.5 × 106 DU145 cells mixed with matrigel (1:1) into both flanks. The animals were fed a semipurified diet (AIN76A, 10 Kcal%; Research Diets) and water ad libitum. After the tumors were palpable, mice were divided into two groups such that the average tumor volumes in all of the groups were approximately equal. Each group was treated by intraperitoneal injection once every 2 d with either PBS, heat inactivated hMGL/4.0 (50 mg/kg), or 50 mg/kg PEG/hMGL-4.0.
Analysis of Blood and Serum Toxicity Profile.
Blood was collected by cardiac puncture following killing. Immediately after cardiac puncture, 90 μL of blood was mixed with 10 μL of potassium EDTA (K2EDTA) solution (18 mg/mL; Fisher Scientific) and red blood cells number and size were counted with an automated cell counter (Nexcelom Bioscience). For white blood cells the K2EDTA-containing blood was mixed with acridine orange/propidium iodide (1:1) solution (Nexcelom Bioscience) and number and size of white blood cells were counted with the same cell counter. Plasma samples were isolated from the remaining blood according to the established procedure (83) The biochemical parameters of hepatotoxicity:ALT, as well as urea level (as an index of renal function) were measured in these samples using commercially available kits (Sigma).
Metabolomics
Sample Preparation.
Cell samples.
The extraction of metabolites from human (22Rv1) and murine (HMVP2) cells was performed using a modified Bligh-Dyer procedure (84) by adding water, methanol, and chloroform in equal volumes (1:1:1). The resulting solution was vortexed vigorously and stirred at 1,500 rpm for 10 min, then centrifuged at 4,750 rpm for 20 min at 4 °C. The polar fraction was collected in Eppendorf tubes, evaporated to dryness in a CentriVap refrigerated vacuum concentrator (Labconco), at 4 °C and resuspended in 180 µL ultrapure water and 20 µL of internal standards solution, as previously described (85, 86). The 200-µL solution was filtered through Nanosep 3K ultracentrifugal device (Pall Co.) at 8,000 rpm for 20 min at 4 °C (87). The resultant filtrate was poured into the LC vial and stored at −20 °C until LC-MS analysis.
Serum samples.
For metabolite extraction, 100 µL of serum was filtered through a Nanosep 3K ultra centrifugal device at 8,000 rpm for 2 h at 4 °C (87), resultant filtrate (80 µL) was diluted with 8 µL of the internal standards solution, poured into the LC vial, and stored at −20 °C until LC-MS analysis.
Tumor tissue samples.
Tumors were excised, combined with 0.5 mL cold methanol/H2O (50:50) and homogenized in a Precellys24 cooled tissue homogenizer equipped with Cryolys system for liquid-nitrogen cooling (Bertin Corp.), operating at 5,000 rpm × 2 cycles × 30 s for three times. Calculated volumes of homogenates with 20 mg of tissue for each sample were transferred into glass vials with 0.5 mL cold chloroform, and methanol/H2O (50/50) was added to bring the volume up to 1 mL. Samples were vigorously vortexed for 30 s and then at 2,500 rpm for 10 min. The samples were then centrifuged at 4,750 rpm at 4 °C for 20 min, the polar fraction was carefully collected, and dried using a CentriVap refrigerated vacuum concentrator. Dried samples were resuspended in 200 µL LC-grade water spiked with 10% internal standards, and filtered through Nanosep 3K ultra centrifugal device at 10,000 rpm for 4 h at 4 °C. The filtrates were transferred to LC vials and analyzed by LC-MS.
Metabolomic Analyses.
Metabolomic analyses of polar fractions was performed on a Hybrid quadrupole-Orbitrap mass spectrometer (Q Exactive, Thermo Scientific) hyphenated with a Thermo Scientific Accela 1250 UHPLC system via electrospray ionization source, simultaneously operating in positive/negative polarity switching ionization mode. Chromatographic separation of metabolites was achieved on a Kinetex C18 150 × 2.1 mm (2.6 μm, 100 Å) column (Phenomenex) with gradient elution of 0.2% FA in water (A) and methanol (B) at a flow rate of 150 µL min−1 within 30 min. The gradient elution was programmed as follows: 0 to 4 min, 2% B; 4 to 14 min, 2 to 80% B; 14 to 15 min, 80 to 98% B; 15 to 20 min, 98 to 98% B; 20 to 25 min, 98 to 2% B, equilibration time 5 min. Detection of metabolites was performed in full MS scan under the following conditions: Spray voltage, 4.0 kV; capillary temperature, 300 °C; sheath gas, 50 (arbitrary units); auxiliary gas, 10 (arbitrary units); microscans, 1; AGC target, 3e6; maximum injection time, 200 ms; mass resolution, 70,000 full-width half-maximum; m/z range, 50 to 750. To ensure mass accuracy below 5 ppm, the MS detector was calibrated prior to analysis using commercial calibration solutions. The LC-MS platform of analysis was controlled by the XCalibur 2.2 software (Thermo Scientific).
A quality control (QC) sample, representing the equivalent concentration of all samples, was prepared to control possible instrumental error (drift) in data acquisition and run once every five samples (88). All raw MS datasets were processed using Sieve 2.2 (Thermo Fisher Scientific) and features with coefficient of variation lower than 25% in the QC samples were considered for further analysis. Peaks were scaled according to PQN (89) and features were then mined against an in-house database of accurate masses and retention times generated in our laboratory using the IROA 300 (85), Mass Spectrometry Metabolite Library of Standards (MSMLS; IROA Technologies). In addition, databases of accurate masses taken from the Kyoto Encyclopedia of Genes and Genomes (90) and the Human Metabolome database (91) were also mined.
Absolute Quantification of Methionine.
For absolute quantification of methionine, 5 ppm (in the concentration range of all samples) 1-13C-methionine (Sigma-Aldrich) was spiked in the QC sample as an internal standard, followed by running the mixture in LC-MS. Thus, the methionine concentrations in samples were determined based on the peak area ratios of 13C-/12C-methionine and the amount of the 13C-standard spiked to the QC sample.
Statistical Analyses.
Data are representative of at least three independent experiments unless otherwise indicated. Data are reported as mean values ± SEM. Statistical analyses were performed using Student’s t test, one-way ANOVA followed by Bonferroni’s multiple comparison test and repeated measure two-way ANOVA followed by Bonferroni’s multiple comparison test. Significance was set at P ≤ 0.05.
Accession Codes.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank: PDB ID code 6OVG for hMGL-4.0.
Supplementary Material
Acknowledgments
Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at The University of Texas at Austin. We acknowledge the support of the Advanced Light Source and the Advanced Photon Source (operated for the US Department of Energy Office of Science by Argonne National Laboratory and supported by the US Department of Energy under Contract DE-AC02-06CH11357) for X-ray crystallographic data collection. This work was supported by grants funded by the National Institutes of Health R01 GM104896 and GM125882 (to Y.J.Z.), CA154754 (to G.G. and E.S.), and CA189623 (to J.D., G.G., and E.S.); the Cancer Prevention and Research Institute of Texas RP180590 (to E.S. and J.D.); and the Welch Foundation F-1778 (to Y.J.Z.).
Footnotes
Competing interest statement: W.-C.L., G.G., and E.S. are inventors on intellectual property related to this work, and G.G. and E.S. have an equity interest in Aeglea Biotherapeutics, a company that has licensed the commercial development of hMGL-4.0.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 6OVG).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917362117/-/DCSupplemental.
References
- 1.Aki T. et al., Evaluation of brain tumors using dynamic 11C-methionine-PET. J. Neurooncol. 109, 115–122 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Tang B. N. et al., Three-dimensional Gaussian model to define brain metastasis limits on 11C-methionine PET. Radiother. Oncol. 89, 270–277 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Galldiks N. et al., [11C]-L-methionine positron emission tomography in the management of children and young adults with brain tumors. J. Neurooncol. 96, 231–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tóth G., et al. , Detection of prostate cancer with 11C-methionine positron emission tomography. J. Urol. 173, 66–69, discussion 69 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Kaste S. C. et al., Comparison of 11C-methionine and 18F-FDG PET/CT for staging and follow-up of pediatric lymphoma. J. Nucl. Med. 58, 419–424 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Palanichamy K., Chakravarti A., Diagnostic and prognostic significance of methionine uptake and methionine positron emission tomography imaging in Gliomas. Front. Oncol. 7, 257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halpern B. C., Clark B. R., Hardy D. N., Halpern R. M., Smith R. A., The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc. Natl. Acad. Sci. U.S.A. 71, 1133–1136 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreis W., Goodenow M., Methionine requirement and replacement by homocysteine in tissue cultures of selected rodent and human malignant and normal cells. Cancer Res. 38, 2259–2262 (1978). [PubMed] [Google Scholar]
- 9.Breillout F., Antoine E., Poupon M. F., Methionine dependency of malignant tumors: A possible approach for therapy. sJ. Natl. Cancer Inst. 82, 1628–1632 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Kreis W., Baker A., Ryan V., Bertasso A., Effect of nutritional and enzymatic methionine deprivation upon human normal and malignant cells in tissue culture. Cancer Res. 40, 634–641 (1980). [PubMed] [Google Scholar]
- 11.Kreis W., Tumor therapy by deprivation of L-methionine: Rationale and results. Cancer Treat. Rep. 63, 1069–1072 (1979). [PubMed] [Google Scholar]
- 12.Chaturvedi S., Hoffman R. M., Bertino J. R., Exploiting methionine restriction for cancer treatment. Biochem. Pharmacol. 154, 170–173 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Thomas T., Thomas T. J., Polyamine metabolism and cancer. J. Cell. Mol. Med. 7, 113–126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Megosh L. et al., Increased frequency of spontaneous skin tumors in transgenic mice which overexpress ornithine decarboxylase. Cancer Res. 55, 4205–4209 (1995). [PubMed] [Google Scholar]
- 15.Auvinen M., Paasinen A., Andersson L. C., Hölttä E., Ornithine decarboxylase activity is critical for cell transformation. Nature 360, 355–358 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Casero R. A. Jr., Murray Stewart T., Pegg A. E., Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman R. M., Jacobsen S. J., Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 77, 7306–7310 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman R. M., Jacobsen S. J., Erbe R. W., Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc. Natl. Acad. Sci. U.S.A. 76, 1313–1317 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman R. M., Erbe R. W., High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl. Acad. Sci. U.S.A. 73, 1523–1527 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stern P. H., Mecham J. O., Wallace C. D., Hoffman R. M., Reduced free-methionine in methionine-dependent SV40-transformed human fibroblasts synthesizing apparently normal amounts of methionine. J. Cell. Physiol. 117, 9–14 (1983). [DOI] [PubMed] [Google Scholar]
- 21.Stern P. H., Wallace C. D., Hoffman R. M., Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J. Cell. Physiol. 119, 29–34 (1984). [DOI] [PubMed] [Google Scholar]
- 22.Stern P. H., Hoffman R. M., Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro 20, 663–670 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Backlund P. S. Jr., Chang C. P., Smith R. A., Identification of 2-keto-4-methylthiobutyrate as an intermediate compound in methionine synthesis from 5′-methylthioadenosine. J. Biol. Chem. 257, 4196–4202 (1982). [PubMed] [Google Scholar]
- 24.Backlund P. S. Jr., Smith R. A., Methionine synthesis from 5′-methylthioadenosine in rat liver. J. Biol. Chem. 256, 1533–1535 (1981). [PubMed] [Google Scholar]
- 25.Ashe H. et al., N5-methyltetrahydrofolate: Homocysteine methyltransferase activity in extracts from normal, malignant and embryonic tissue culture cells. Biochem. Biophys. Res. Commun. 57, 417–425 (1974). [DOI] [PubMed] [Google Scholar]
- 26.Goseki N. et al., Synergistic effect of methionine-depleting total parenteral nutrition with 5-fluorouracil on human gastric cancer: A randomized, prospective clinical trial. Jpn. J. Cancer Res. 86, 484–489 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durando X. et al., Optimal methionine-free diet duration for nitrourea treatment: A phase I clinical trial. Nutr. Cancer 60, 23–30 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Thivat E. et al., Phase II trial of the association of a methionine-free diet with cystemustine therapy in melanoma and glioma. Anticancer Res. 29, 5235–5240 (2009). [PubMed] [Google Scholar]
- 29.Durando X. et al., Dietary methionine restriction with FOLFOX regimen as first line therapy of metastatic colorectal cancer: A feasibility study. Oncology 78, 205–209 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Gao X. et al., Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon H. et al., Methionine deprivation suppresses triple-negative breast cancer metastasis in vitro and in vivo. Oncotarget 7, 67223–67234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha R. et al., Dietary methionine restriction inhibits prostatic intraepithelial neoplasia in TRAMP mice. Prostate 74, 1663–1673 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Poirson-Bichat F., Gonçalves R. A., Miccoli L., Dutrillaux B., Poupon M. F., Methionine depletion enhances the antitumoral efficacy of cytotoxic agents in drug-resistant human tumor xenografts. Clin. Cancer Res. 6, 643–653 (2000). [PubMed] [Google Scholar]
- 34.Kokkinakis D. M. et al., Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Cancer Res. 61, 4017–4023 (2001). [PubMed] [Google Scholar]
- 35.Tan Y. et al., Efficacy of recombinant methioninase in combination with cisplatin on human colon tumors in nude mice. Clin. Cancer Res. 5, 2157–2163 (1999). [PubMed] [Google Scholar]
- 36.Hu J., Cheung N. K. V., Methionine depletion with recombinant methioninase: In vitro and in vivo efficacy against neuroblastoma and its synergism with chemotherapeutic drugs. Int. J. Cancer 124, 1700–1706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igarashi K. et al., Recombinant methioninase in combination with doxorubicin (DOX) overcomes first-line DOX resistance in a patient-derived orthotopic xenograft nude-mouse model of undifferentiated spindle-cell sarcoma. Cancer Lett. 417, 168–173 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Murakami T. et al., Recombinant methioninase effectively targets a Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget 8, 35630–35638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaguchi K. et al., Combination treatment with recombinant methioninase enables temozolomide to arrest a BRAF V600E melanoma in a patient-derived orthotopic xenograft (PDOX) mouse model. Oncotarget 8, 85516–85525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z. et al., Pharmacokinetics, methionine depletion, and antigenicity of recombinant methioninase in primates. Clin. Cancer Res. 10, 2131–2138 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Gay F. et al., Methionine tumor starvation by erythrocyte‐encapsulated methionine gamma‐lyase activity controlled with per os vitamin B6. Cancer Med. 6, 1437–1452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone E. et al., De novo engineering of a human cystathionine-γ-lyase for systemic (L)-Methionine depletion cancer therapy. ACS Chem. Biol. 7, 1822–1829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole M. F., Gaucher E. A., Exploiting models of molecular evolution to efficiently direct protein engineering. J. Mol. Evol. 72, 193–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cacan E., Kratzer J. T., Cole M. F., Gaucher E. A., Interchanging functionality among homologous elongation factors using signatures of heterotachy. J. Mol. Evol. 76, 4–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cole M. F., Gaucher E. A., Utilizing natural diversity to evolve protein function: Applications towards thermostability. Curr. Opin. Chem. Biol. 15, 399–406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrikx J. J. M. A. et al., Fixed dosing of monoclonal antibodies in oncology. Oncologist 22, 1212–1221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Q. et al., Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S. J. Biol. Chem. 284, 3076–3085 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Saha A., Blando J., Fernandez I., Kiguchi K., DiGiovanni J., Linneg Sca-1high CD49fhigh prostate cancer cells derived from the Hi-Myc mouse model are tumor-initiating cells with basal-epithelial characteristics and differentiation potential in vitro and in vivo. Oncotarget 7, 25194–25207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong H. et al., Inhibition of DNA methyltransferase induces G2 cell cycle arrest and apoptosis in human colorectal cancer cells via inhibition of JAK2/STAT3/STAT5 signalling. J. Cell. Mol. Med. 13, 3668–3679 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desjobert C. et al., Combined analysis of DNA methylation and cell cycle in cancer cells. Epigenetics 10, 82–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiemjit A. et al., p21(WAF1/CIP1) induction by 5-azacytosine nucleosides requires DNA damage. Oncogene 27, 3615–3623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fialova B. et al., Epigenetic modulation of AR gene expression in prostate cancer DU145 cells with the combination of sodium butyrate and 5′-Aza-2′-deoxycytidine. Oncol. Rep. 36, 2365–2374 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Fang J., Lu J., Holmgren A., Thioredoxin reductase is irreversibly modified by curcumin: A novel molecular mechanism for its anticancer activity. J. Biol. Chem. 280, 25284–25290 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Roder C., Thomson M. J., Auranofin: Repurposing an old drug for a golden new age. Drugs R D. 15, 13–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Epner D. E., Morrow S., Wilcox M., Houghton J. L., Nutrient intake and nutritional indexes in adults with metastatic cancer on a phase I clinical trial of dietary methionine restriction. Nutr. Cancer 42, 158–166 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Forney L. A., Wanders D., Stone K. P., Pierse A., Gettys T. W., Concentration-dependent linkage of dietary methionine restriction to the components of its metabolic phenotype. Obesity (Silver Spring) 25, 730–738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider K. M. et al., Intestinal microbiota protects against MCD diet-induced steatohepatitis. Int. J. Mol. Sci. 20, 308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye J.-Z. et al., Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J. Gastroenterol. 24, 2468–2481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caballero F. et al., Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: Impact on mitochondrial S-adenosyl-L-methionine and glutathione. J. Biol. Chem. 285, 18528–18536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eipel C., Abshagen K., Vollmar B., Regulation of hepatic blood flow: The hepatic arterial buffer response revisited. World J. Gastroenterol. 16, 6046–6057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strekalova E., Malin D., Good D. M., Cryns V. L., Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin. Cancer Res. 21, 2780–2791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miousse I. R. et al., Modulation of dietary methionine intake elicits potent, yet distinct, anticancer effects on primary versus metastatic tumors. Carcinogenesis 39, 1117–1126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwon H.-M. et al., Combinatorial effects of an epigenetic inhibitor and ionizing radiation contribute to targeted elimination of pancreatic cancer stem cell. Oncotarget 8, 89005–89020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian X. J. et al., Inhibition of DNA methyltransferase as a novel therapeutic strategy to overcome acquired resistance to dual PI3K/mTOR inhibitors. Oncotarget 6, 5134–5146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y.-J. et al., Combined inhibition of Dnmt and mTOR signaling inhibits formation and growth of colorectal cancer. Int. J. Colorectal Dis. 24, 629–639 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Vitale G. et al., Synergistic activity of everolimus and 5-aza-2′-deoxycytidine in medullary thyroid carcinoma cell lines. Mol. Oncol. 11, 1007–1022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z. et al., Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 25, 825–837 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Yan W., Stone E., Zhang Y. J., Structural snapshots of an engineered cystathionine-γ-lyase reveal the critical role of electrostatic interactions in the active site. Biochemistry 56, 876–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otwinowski Z., Minor W., [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Winn M. D. et al., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ten Eyck L. F., Crystallographic fast Fourier transforms. Acta Crystallogr. A 29, 183–191 (1973). [Google Scholar]
- 72.Read R. J., Schierbeek A. J., A phased translation function. J. Appl. Cryst. 21, 490–495 (1988). [Google Scholar]
- 73.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Afonine P., Grosse-Kunstleve R., Adams P., The Phenix refinement framework. CCP4 Newsletter 42 (2005). [Google Scholar]
- 75.Headd J. J. et al., Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr. D Biol. Crystallogr. 68, 381–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis I. W. et al., MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35 (suppl. 2), W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeLano W. L., Unraveling hot spots in binding interfaces: Progress and challenges. Curr. Opin. Struct. Biol. 12, 14–20 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Ellwood-Yen K. et al., Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Yang S. et al., Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saha A. et al., 6-Shogaol from dried ginger inhibits growth of prostate cancer cells both in vitro and in vivo through inhibition of STAT3 and NF-κB signaling. Cancer Prev. Res. (Phila.) 7, 627–638 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Saha A., Kuzuhara T., Echigo N., Suganuma M., Fujiki H., New role of (-)-epicatechin in enhancing the induction of growth inhibition and apoptosis in human lung cancer cells by curcumin. Cancer Prev. Res. (Phila.) 3, 953–962 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Kleiner H. E., Vulimiri S. V., Starost M. F., Reed M. J., DiGiovanni J., Oral administration of the citrus coumarin, isopimpinellin, blocks DNA adduct formation and skin tumor initiation by 7,12-dimethylbenz[a]anthracene in SENCAR mice. Carcinogenesis 23, 1667–1675 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Bligh E. G., Dyer W. J., A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 85.Lu X. et al., The early metabolomic response of adipose tissue during acute cold exposure in mice. Sci. Rep. 7, 3455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lodi A. et al., Combinatorial treatment with natural compounds in prostate cancer inhibits prostate tumor growth and leads to key modulations of cancer cell metabolism. NPJ Precis. Oncol. 1, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tiziani S. et al., Optimized metabolite extraction from blood serum for 1H nuclear magnetic resonance spectroscopy. Anal. Biochem. 377, 16–23 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Sumner L. W. et al., Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dieterle F., Ross A., Schlotterbeck G., Senn H., Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Okuda S. et al., KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 36 (suppl. 2), W423–W426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wishart D. S. et al., HMDB: The human metabolome database. Nucleic Acids Res. 35 (suppl. 1), D521–D526 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are shown in the main manuscript and SI Appendix, except for the atomic coordinates and structure factors of hMGL-4.0, which have been deposited in the Protein Data Bank (PDB ID code 6OVG).