Abstract
The cell nucleus is a remarkably well-organized organelle with membraneless but distinct compartments of various functions. The largest of them, euchromatin and heterochromatin, are spatially segregated in such a way that the transcriptionally active genome occupies the nuclear interior, whereas silent genomic loci are preferentially associated with the nuclear envelope. This rule is broken by rod photoreceptor cells of nocturnal mammals, in which the two major compartments have inverted positions. The inversion and dense compaction of heterochromatin converts these nuclei into microlenses that focus light and facilitate nocturnal vision. As often the case in biology, when a mutation helps to understand normal processes and structures, inverted nuclei have served as a tool to unravel general principles of nuclear organization, including mechanisms of heterochromatin tethering to the nuclear envelope, autonomous behavior of small genomic segments and euchromatin heterochromatin segregation.
Introduction
The nuclei of practically all animal cells have a common general plan of genomic organization: transcriptionally active euchromatin is positioned within the nuclear interior, whilst silent heterochromatin resides at the periphery [1, 2]. A unique exception from this nuclear plan is found in rod photoreceptors of nocturnal mammals, in which eu-and heterochromatin have exchanged positions [2, 3]. Regardless of the intranuclear positioning of eu-and heterochromatin, their segregation appears to be central to the functional organization of the genome and has been widely accepted as one of the crucial factors in the regulation of transcription [4–6]. In the past decade, substantial progress has been made in understanding genome folding within the nucleus, as well as the mechanisms of eu-and heterochromatin compartmentalization [7–10]. Just as a mutation helps us to understand universal processes and structures, the abnormal organization of rod photoreceptor nuclei in nocturnal mammals can be exploited to understand the basic principles of nuclear organization. In particular, mouse rod nuclei with their clearly segregated and concentrically arranged compartments serve as an excellent model to study molecular tethers that hold heterochromatin at the nuclear periphery, mechanisms that drive nuclear compartmentalization and the major players in these processes.
In the first half of this review we outline the current knowledge about the two types of nuclear organization, conventional and inverted, and discuss why the inverted nuclear structure may be disadvantageous. The second part of the review focuses on aspects of nuclear organization that were elucidated through studies of inverted rod nuclei.
Two strategies to build the cell nucleus
The universal conventional and the unique inverted nuclear designs
Our current knowledge about the organization of the animal genome in the cell nucleus is based, on the one hand, on various types of microscopy and, on the other hand, on genome wide approaches, such as DamID and Chromosome Conformation Capture (3C)-techniques (Box 1). Both approaches provide growing evidence that the genome is spatially segregated into large chromatin domains with different structural and functional characteristics.
Text box 1. Genome-wide approaches to study genome spatial organization.
Microscopy detects overall spatial arrangements of genomes, chromosomes, chromosomal segments or single genes in the nucleus. Genome-wide methods such as Hi-C, mostly detect relative spatial genome arrangements. Other genome-wide approaches, such as Dip-C [16, 102], TSA-Seq [103], SPRITE [104], assess both relative and overall distributions of chromosomal loci. Another genome-wide technique DamID [105] allows one to define genomic loci residing in proximity to a structure of interest. Below we describe only a selection of genome-wide methods.
Chromosome conformation capture (3C) technology.
3C-based methods are a series of methods that use chromatin crosslinking, restriction enzyme digestion, ligation and sequencing to detect pairwise DNA interactions, e.g. between two genomic loci (3C) or genome-wide (HiC). In the latter case, contact frequencies between genomic regions are used to generate global 3D maps of the genome either in a population of cells (bulk HiC, [13] or in single cells (single-cell-HiC, [106]). In contrast to microscopy, bulk Hi-C detects the relative spatial arrangement of chromosomes and compartments, rather than their overall position. For instance, enrichment of interchromosomal contacts between small chromosomes and depletion of contacts between small and large chromosomes, detected in human lymphoblastoid cells [13], only hint at the more central position of small gene-rich chromosomes (chr 17, 19–22) and the more peripheral location of the larger (chr 1–8) and gene-poor (chr 18) chromosomes, a finding made by microscopists 10 years earlier [107, 108].
DipC.
DipC is a single-cell chromatin Hi-C that uses a high-coverage whole-genome amplification with multiplex end-tagging amplification (META) [102]. With DipC the two homologous chromosomes can be distinguished from each other in diploid cells. Remarkably, the inverted organization of mouse rod nuclei was deduced from DipC contact maps [102], but not from bulk Hi-C maps, which suggests that DipC is a powerful technique to study the radial distribution of genomic loci. Mouse chromocenters consist of merged blocks of subcentromeric major satellite repeats from different chromosomes and thus represent typical constitutive heterochromatin, which cannot be “seen” by analyses based on sequencing. However, single cell Dip-C allows “to outline” the position of chromocenters in mouse nuclei, at least in cases when there are only 1 or 2 chromocenters [16].
SPRITE, split-pool recognition of interactions by tag extension.
SPRITE is a powerful new technique that enables genome-wide mapping of multiple DNA interactions [104]. SPRITE, likewise Hi-C, relies on chromatin crosslinking and digestion but does not use restriction enzymes allowing for multiple regions of interacting DNA to stay together in a cluster that receives a unique barcode enabling its identification and the mapping of the interacting sequences. Another advantage of the method is that it identifies interactions at further nuclear distances and with nuclear bodies such as the nucleolus or speckles. It can also be used to create more complex maps of combined DNA, RNA and protein interactions.
TSA-seq, tyramide signal amplification sequencing.
TSA-seq is a method used for measuring cytological distances genome-wide and relative to a particular nuclear compartment [103]. It is based on the ability of TSA to label DNA directly and uniformly by producing biotin-tyramide free radicals using a HRP-conjugated antibody to a particular target. Upon TSA the concentration of free radicals is highest at the target and spreads away from it, thus creating a concentration gradient, which provides distance-dependent biotin labeling.
DamID, DNA adenine methyltransferase identification.
One of the best established genome-wide “spatial” technique is DamID, the approach that withstood the test of time in the last 15 years. The method utilizes the bacterial N6-adenine methyl-transferase Dam fused to a protein of interest to detect genomic regions that are in close proximity to the Dam fusion partner and thus undergo methylation by Dam [105]. In the nuclear field, it was initially used to identify genomic loci associated with the nuclear lamina, so called lamina-associated domains, or LADs [46]. Fusion of DNA adenine methyltransferase with nuclear lamina proteins allows to correlate the dynamics of lamina-association with the transcriptional activity of genes during differentiation and development [109], as well as can predict the average nuclear position (internal/peripheral) and shape (voluminous/flat) of a particular chromosome [2].
Microscopy after immunostaining or fluorescence in situ hybridization (FISH), allows one to distinguish three major types of chromatin occupying different nuclear positions (Figure 1A): euchromatin (EC), heterochromatin (HC) and constitutive heterochromatin (cHC) [2]. EC is gene-rich, SINE-rich and transcriptionally active, marked with transcription-associated histone modifications, such as H3K4me3, H3K27ac, H3K36me3, H3/H4ac, and resides in the nuclear interior. HC is gene-poor, LINE/LTR-rich, mostly transcriptionally inactive and found at the nuclear and nucleolar peripheries. Despite their different functions for chromatin regulation, the two histone modifications, H3K27me3 and H3K9me2/3, both mark HC, although H3K27me3 is also frequently found within EC as a mark of a special type of short Polycomb-silenced regions [11]. The third type of chromatin, cHC, consists of highly repetitive sequences (satellites), typically accumulated in subcentromeric regions, lacks expression of coding RNA, although considered not completely silent [12], is marked with H3K9me2,3 modifications, and has the same nuclear locations as the other heterochromatin (Figure 1A).
Figure 1. Comparison of conventional and inverted nuclear organization as seen by microscopy (A,B) and Hi-C (C).
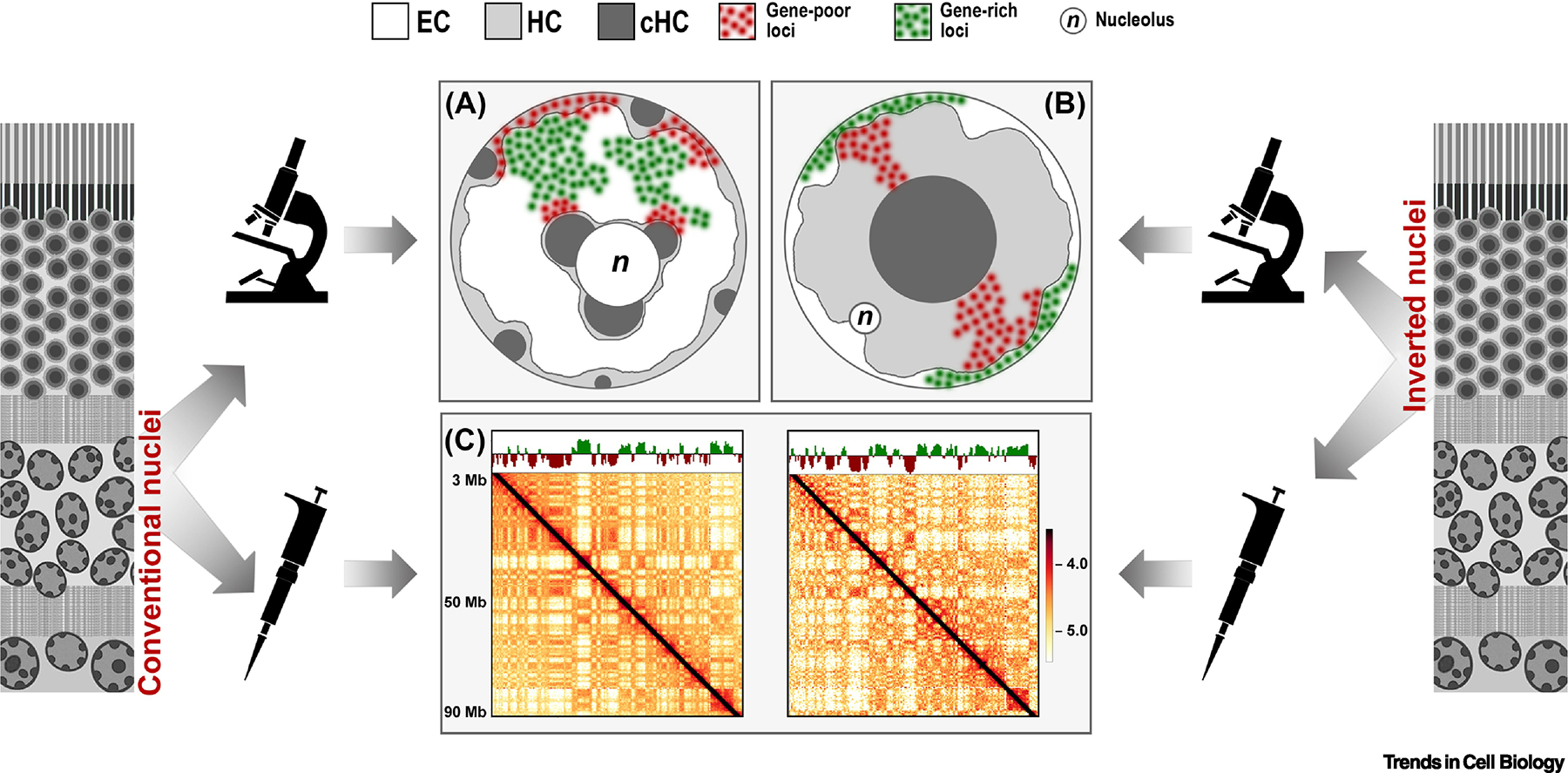
Microscopy reveals a striking difference in the nuclear organization of conventional nuclei, e.g., nuclei of non-photoreceptor retinal neurons (A), and inverted nuclei of rod photoreceptors (B) from mouse retina. In conventional nuclei, both types of heterochromatin, HC and cHC, underlie the nuclear envelope and surround the nucleolus (n), whereas EC occupies the nuclear interior. In inverted nuclei, HC and EC exchange their locations: HC is densely packed in the nuclear interior and EC is relocated to the very nuclear periphery. Correspondingly, if genic EC loci (green), easily establish contacts in the nuclear interior of conventional nuclei, contacts between EC loci in inverted nuclei are hampered by the practically two-dimensional organization of the thin layer of peripheral chromatin. In difference to microscopy, bulk Hi-C analysis does not sense the dramatic difference between the two spatial chromatin organizations and in both cases reveals qualitatively similar features, such as chromosome territories, compartments and TADs (C). Shown are Hi-C maps of chromosome 1 in conventional (left) and inverted (right) nuclei.
3C techniques, in particular Hi-C, measure contact frequencies among genomic regions, with the exception to highly repetitive segments formed by cHC (Box1), and thus detect two distinct compartments alternating along chromosomes historically called A and B (Figure 1C). A correlation of A and B compartments with epigenetic signatures and gene expression profiles allowed to identify the two compartments to be EC and HC, respectively [2, 8, 13].
The conventional nuclear arrangement described above is characteristic of all animal cell types with one known exception, the rod photoreceptor cells of nocturnal mammals. In rod nuclei, the positions of EC and HC are inverted: heterochromatin, including both cHC and HC, occupies the nuclear center, while euchromatin is displaced to the periphery [3] (Figure 1B). Remarkably, nuclear inversion, albeit not currently known to hinder any nuclear processes, is likely to be strongly disadvantageous for nuclear function (see the next chapter) but, as it turned out, confers improved vision under low-light conditions [3, 14].
Since Hi-C measures contacts between pairs of genomic regions, it is insensitive to the spatial location of these regions in the nucleus, as long as a pair stays together (Box 1) and, therefore, Hi-C maps do not show changes in the global spatial arrangement if contact frequencies stay the same. This is why, in contrast to those using microscopy and seeing striking morphological differences between inverted and conventional nuclei (Figure 1A,B), researchers utilizing 3C technology are not particularly excited by the fairly similar Hi-C maps of the two nuclear types (Figure 1C). Indeed, the major nuclear features that can be deduced from Hi-C experiments compartments, chromosome territories, topologically associating domains, or TADs, are present in rods as in other cell types [15, 16]. This finding is in agreement with the fact that in both inverted and conventional nuclei the three chromatin classes, EC, HC and cHC, have the same epigenetic marks and are enriched in similar chromatin-associated proteins [17, 18].
Some quantitative differences between nuclei of rods and other cells, however, can be detected in Hi-C data. Chromosomes in rods exhibit a high trans/cis contact ratio, pointing at a weak chromosome territoriality, which has also been confirmed by microscopy [15]. Since the chromosome territory is apparently a reflection of packaging chromatin into a compact mitotic chromosome for cell division but not for nuclear function, the longer a chromosome remains in a postmitotic state, the less territorial it becomes as a result of various types of chromatin movements [19–22]. This explains why in postmitotic neurons territoriality is weaker than in cycling lymphocytes [15, 23]. What is more, weak territoriality in rods is caused not only by “long life” but also by “hard life”. Nuclei of postmitotic rod precursor cells are primarily conventional and in the process of inversion, they undergo global spatial reorganization, involving dissociation of chromocenters from the nuclear periphery and their merging in the nuclear center [3]. Such dynamics, reinforced by gradual loss of tethering to the nuclear lamina, leads to chromosome refolding and stretching within nuclei of differentiating rods [15]. A weak territoriality has also been reported for olfactory sensory neurons, which too undergo global nuclear rearrangements during differentiation [24, 25]. The number of interchromosomal contacts increases from 17.9% in multipotent olfactory progenitors to 35.6% in mature neurons [26]. The complicated chromosome dynamics during the differentiation of these two types of neurons ultimately results in a partial chromosome mixing and hence less discrete chromosome territoriality.
Heterochromatin in rods builds a lens
In mouse rods, all the cHC regions are tightly packed in one central chromocenter of ca. 2 μm in diameter that is surrounded by a HC shell with a thickness of ca. 1 μm, which is in turn surrounded by a not more than 0.5 μm thick EC shell (Figure 1B). Thus, the volume of the rod nucleus, which is ca. 5 μm in diameter, is divided approximately in half between EC and HC. While the central rod chromocenter (cHC) occupies about 6% of nuclear volume, it constitutes about 10% of the entire mouse genome [27, 28]. This suggests about 1.8-fold higher volume density in the chromocenter than in the rest of the chromatin, and is consistent with an approximately 2-fold higher density in the densest region in the conventional interphase nucleus and mitotic chromosomes measured by ChromEMT [29]. Nonetheless, the physical properties of the densely packed heterochromatic core, formed by cHC and HC, are pretty unusual for instance, it is not penetrable to free GFP molecules in rods, whereas in conventional nuclei GFP molecules freely go through heterochromatin of both types [14]. Moreover, the core is highly refractive and shows physical properties of a lens focusing light [3, 30]. Given that the difference in DNA concentration between the heterochromatic core and EC is not high, one can speculate that the unusual density of the rod core structure is caused by a high accumulation of proteins. Since the exact molecular components of the rod nuclear core are not known, we can only speculate in favor of this notion based on the comparable refractivity of the rod nuclear core and the nucleoli, the most protein-rich structures in the nucleus.
Since rod perikarya are almost entirely occupied by nuclei, regular columns of rod perikarya in nocturnal retinas (up to 10–12 perikarya in a column) are practically composed of lenses stacked upon each other (Figure 2A,B). Due to this arrangement, light travelling through the columns of lenses is significantly less scattered, facilitating light sensitivity in low-light (scotopic) conditions [3, 14, 30]. In agreement with this, the inverted nuclear arrangement is found only in nocturnal mammals, whereas rod nuclei of diurnal mammals (living in photopic conditions) have conventional architecture [3, 31, 32] (Figure 2B,C). Of note, rod nuclei of nocturnal species that do not possess such big cHC blocks as mice, e.g. rat and ringtail possum, still have a dense central HC core (Figure 2B), in this case, consisting presumably of interspersed repeats.
Figure 2. Rod nuclear inversion facilitates nocturnal vision but is disadvantageous for nuclear functions.
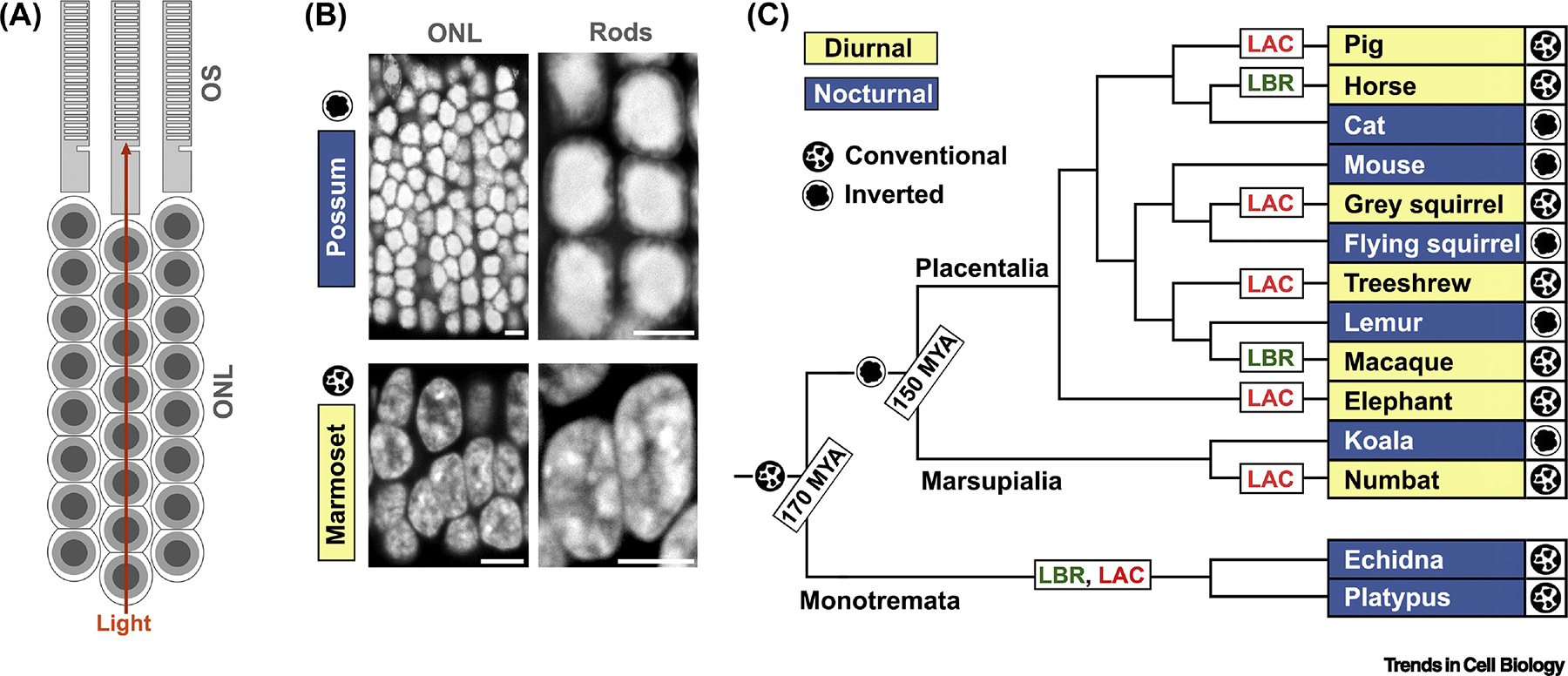
(A) Schematic drawings of an outer nuclear layer (ONL) of nocturnal retina showing columns of rod cell perikarya and rod outer segments (OS), in which photoreception occurs. Rod columns facilitate light propagation by reducing light scattering in the ONL. The path of light propagation through the ONL is indicated by an orange arrow. (B) Structure of ONL and rod nuclei in nocturnal and diurnal mammals, exemplified by possum and marmoset retinas, respectively. Note the difference between nuclei of nocturnal rods with densely packed single heterochromatic globules and diurnal rods with multiple granules of heterochromatin distributed throughout the nucleoplasm. DAPI staining. Scale bars, 5 μm. (C) Phylogenetic tree of mammals with representative taxa showing the structure of rod nuclei. In mammals, nocturnal species (white on blue fields) have inverted nuclei, whereas diurnal species (black on yellow fields) have conventional nuclei. Nuclear inversion occurred before the splitting of Placentalia and Marsupialia but after the separation of therian mammals from Monotremata, because only therian species have rods with inverted nuclei. Since mammalian ancestors were nocturnal animals, acquisition of a diurnal lifestyle by several taxa occurred independently. Acquisition of diurnality is accompanied by restoration of either LBR- or LAC-dependent tether expression in rod cells and, consequently, by reorganization of inverted nuclei to conventional ones, suggesting that the inverted chromatin arrangement is strongly disadvantageous.
Inverted see better, conventional work better
Despite the major spatial reorganization, the inverted architecture does not hamper nuclear functions or, at least, does not interfere with high transcriptional activity in rods. Does it mean that the spatial arrangement of chromatin is not important for nuclear functions? Before answering this question, we need to understand some evolutionary aspects of inversion.
Inverted rod nuclei are present in Theria but not Monotremata taxa (our unpublished data), indicating that inversion has probably evolved as a specific adaptation to nocturnal vision before the splitting of Theria to Placentalia and Marsupialia (ca. 150 MYA), but after the divergence of Theria and Monotremata (ca. 170 MYA) [3] (Figure 2C), which corresponds to data on the evolution of cone visual pigments in mammals [33–35]. Since vision is such an important trait for survival, it is tempting to speculate that it could have driven vertebrate genome evolution. Thus, an attractive hypothesis is that, since nocturnal rods required more heterochromatin to build up microlenses, repetitive non-coding DNA became abundant in Theria genomes as an adaptation to vision. Indeed, on average, the genomes of Placentalia and Marsupialia are larger in comparison to Monotremata and other diurnal taxa, such as birds and reptiles, and enriched in highly repetitive sequences, as well as in interspersed AT rich LINE1 and LTR repeats [36–38].
Later in evolution, some mammalian groups independently re-acquired a diurnal lifestyle (Figure 2C). These events were accompanied by re-arrangement of rod nuclear structure from inverted to conventional. In other words, as soon as the evolutionary pressure for nocturnal adaptations is removed and retinas do not need a microlens system, rod nuclei return to the conventional shape [3, 31], suggesting that there should be strongly disadvantageous aspects in the inverted chromatin arrangement. Development of inversion or re-inversion is apparently a long process that requires millions of years. This is why in secondary nocturnal mammals, such as the primates Aotus and Tarsius, conventional rod nuclei do not undergo a new round of inversion but the groups develop other anatomical adaptations to nocturnal vision [32].
We do not know why inversion is disadvantageous for nuclear functions (see Outstanding Questions). One hypothesis could be that the inverted chromatin arrangement hampers the intra and inter-chromosomal interactions necessary for proper regulation of transcription or efficient DNA repair. Indeed, squeezing genic regions of chromosomes into a narrow peripheral shell converts rod euchromatin into a practically two-dimensional structure (Figure 1B), compared to the three-dimensional euchromatin in the nuclear interior of other cell types (Figure 1A). This may limit the options for spatial regulation of transcription [39–41], or efficient DNA repair of double-strand breaks [42], e.g. by reducing the efficiency of interactions between homologous chromosomes for DSB repair. Moreover, the lack in inverted nuclei of HC tethering to the nuclear lamina, that is widely accepted as a major mechanism of gene silencing and suppression of mobile elements [43], might be another strong drawback rendering inverted nuclei disadvantaged.
Outstanding Questions Box.
What is the price that nocturnal mammals pay for gaining a nuclear microlens system? Why are inverted nuclei disadvantageous?
What are other tethers of peripheral heterochromatin?
What are other components of the lamin A/C-depended tether?
What are the molecular players that mediate attractions among heterochromatic regions?
Is increased density of heterochromatin critical for its silencing function?
What is the functional relevance of segregating euchromatin from heterochromatin and placing heterochromatin at the nuclear periphery?
Another known example of nuclei without intra and inter-chromosomal interactions and without a broad contact with the nuclear envelope is found in Diptera somatic cells containing polytene chromosomes [44, 45]. As the case with rods, the cells with polytene chromosomes are postmitotic and highly specialized for massive expression of certain genes. In both discussed examples the basic rules of genome regulation are apparently modified to satisfy other cell type-specific physiological needs.
In conclusion, although inverted rod nuclei improve vision and are capable of high gene expression, they are selected against evolutionarily. This confirms the current paradigm of nuclear biology that spatial chromatin arrangement is highly important for nuclear function.
What inverted nuclei tell us about conventional nuclei
Chromatin positioning by two major peripheral heterochromatin tethers
Mechanisms that position chromatin within the nucleus should be clearly distinguished from mechanisms that lead to segregation of different types of chromatin. The main and the best studied mechanism of chromatin positioning is anchoring of HC to the nuclear lamina, resulting in sequestration of silenced loci to the nuclear periphery and formation of genomic regions named lamina-associated domains (LADs) [46–49]. LADs represent one third of the mouse and human genomes and predominantly reside in the heterochromatic compartment. Correspondingly, LADs are enriched in LINEs/LTR and silencing histone modifications (H3K9me2, H3K9me3), and are either depleted of genes or contain mostly lowly expressed or silenced genes [49]. Maintenance of lamina-proximal positioning is attributed to H3K9 di and trimethylation and Lamin B1 methylation [50–53].
The term “lamina-associated domain” is somewhat misleading because LADs can be found in both silencing compartments-near the nuclear lamina and around the nucleoli [48]. LADs are tethered to the nuclear lamina via transmembrane proteins (see below), but what attracts LADs to nucleoli remains unknown [2, 48, 54]. The nucleolus is a highly structured nuclear organelle formed around ribosomal repeats and surrounded by so called perinucleolar chromatin [55, 56]. Most of the perinucleolar chromatin, but not all [57], originates from HC arrays flanking nucleolar organizing regions and excluded from the nucleolar interior. One can speculate that the layer of perinucleolar HC attracts other HC regions via a phase separation mechanism (see below) that is possibly mediated by the Ki-67 protein [58], and thus initiates formation of the perinucleolar HC rim. Of note, similarly to the layer of peripheral HC, the thickness and composition of the perinucleolar HC rim strongly vary between species, cell types, and differentiation stages [57], indicating a highly dynamic nature of these nuclear subcompartments.
How are LADs released from the nuclear lamina in inverted nuclei and what anchors them to it in conventional nuclei? In contrast to other cell types, inverted rods lack two ubiquitous proteins of the nuclear envelope, Lamin B Receptor (LBR) and Lamin A/C (LAC) (Figure 3B1). An extensive study of LBR and LAC expression in multiple mouse tissues at different developmental stages showed that in various cell types at least one, if not both proteins are present (Figure 3A1). Moreover, expression of LBR and LAC is temporarily coordinated during development and cell differentiation with LBR preceding LAC. Thus, during embryogenesis and the first postnatal days, all neurons in the retina express only LBR, which is then gradually substituted by LAC [31]. In contrast to all other retinal cells, rods strongly downregulate LBR expression to an apparently negligible level [59], but never turn on expression of LAC. Importantly, LBR downregulation temporally coincides with the reorganization of the initially conventional rod nuclei into inverted ones, which is manifested firstly, by the dissociation of chromocenters from the nuclear periphery and their merging into a single central chromocenter and, secondly, by the formation of a HC shell around the chromocenter [3]. Comparable merging of chromocenters was also observed in mouse olfactory neurons, which similarly to rods downregulate LBR expression during differentiation [25, 60], but maintain lamin A/C expression and thus remain conventional with HC distributed throughout the nucleoplasm and several chromocenters adjacent to the nuclear envelope. These observations led to the hypothesis that the lack of or low level of both LBR and LAC causes nuclear inversion [31] (Figure 3).
Figure 3. Two major tethers of peripheral heterochromatin.
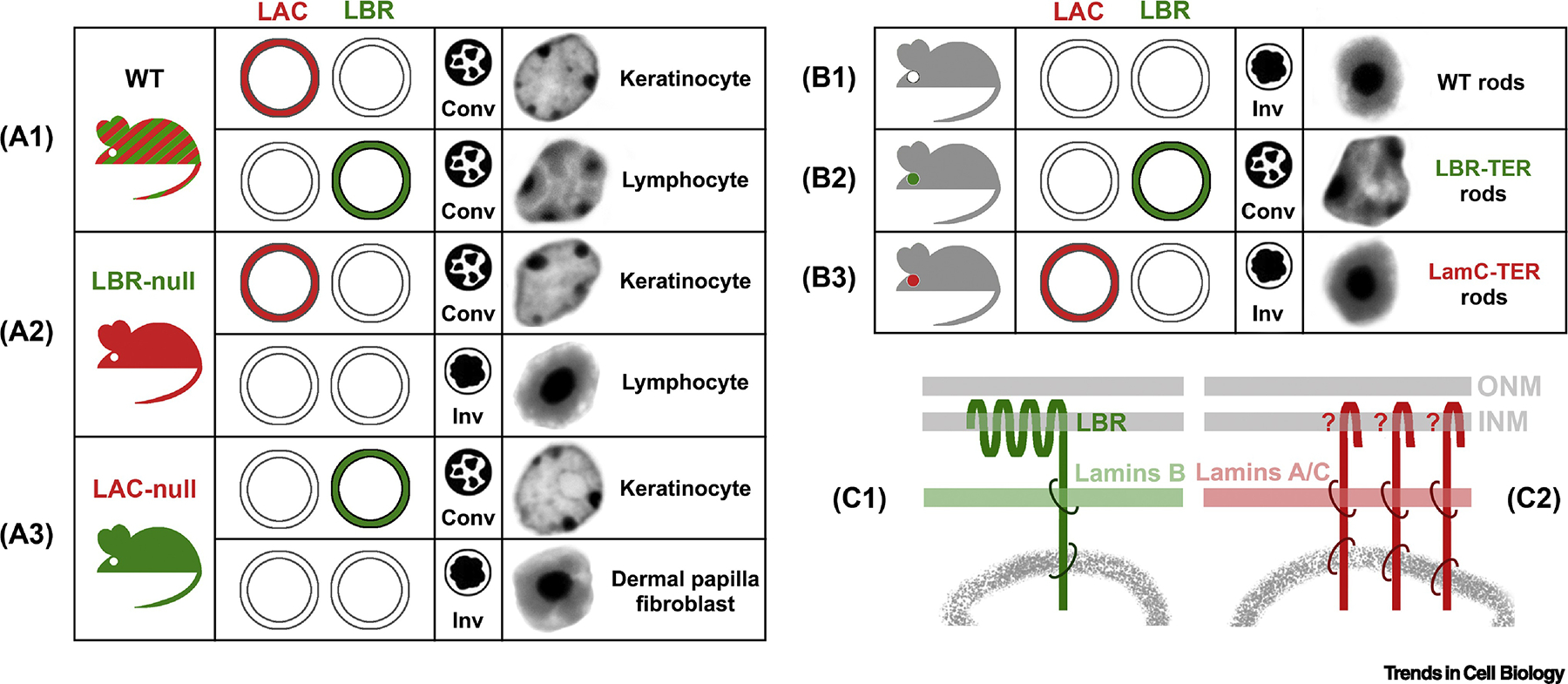
(A1) LBR and lamin A/C (LAC) are ubiquitously expressed proteins of the nuclear lamina. WT cells either express both proteins or at least one of them and thus have conventional nuclei. (A2) In LBR-null mice, nuclei of those cells that do not normally express LAC, become inverted, e.g., lymphocytes. (A3) In LAC-null mice, nuclei of cells that do not express LBR in WT, become either inverted (e.g, fibroblasts of the dermal papilla) or remain conventional as a result of abnormally upregulated LBR expression (e.g., keratinocytes). (B1) Rods are the only cell type naturally lacking both proteins. (B2) Transgenic expression of LBR in mouse rods effectively counteracts nuclear inversion and restores the conventional architecture in these cells. (B3) Transgenic expression of lamin C is not sufficient for restoration of the conventional architecture, suggesting that LAC needs partners for heterochromatin binding, which are not expressed in mouse rods. (C) Schematics of the two major types of peripheral heterochromatin tethers. LBR-dependent tether (C1) binds B-lamins and peripheral heterochromatin. LAC-dependant tether (C2) includes transmembrane proteins (marked with “?”) binding both LAC and heterochromatin. conv, conventional nuclei; inv, inverted nuclei; LBR-TER, transgenic rods expressing ectopic LBR; LamC-TER, transgenic rods expressing ectopic lamin C; nuclei of exemplified cells on (A) and (B) are shown as inverted greyscale images of DAPI staining; ONM, outer nuclear membrane; INM, inner nuclear membrane.
Work with knockout mice deficient in LBR, LAC, or both, revealed that these proteins are parts of the two different tethers of peripheral heterochromatin (Figure 3C1,2). In LBR-null mice, nuclei of those cells that do not normally express LAC, become inverted, e.g. lymphocyte nuclei (Figure 3A2). In LAC-null mice, nuclei of those cells that do not normally express LBR, become either inverted, e.g., fibroblasts of the dermal papilla, or remain conventional as a result of upregulation and prolongation of LBR expression, e.g., keratinocytes (Figure 3A3). Transgenic expression of LBR in mouse rods is sufficient to localize HC and cHC to the nuclear periphery and thus effectively counteracts nuclear inversion, restoring a conventional architecture (Figure 3B2). At the same time, expression of only lamin C does not change the inverted nuclear arrangement (Figure 3B3), pointing to lamins A and C being scaffold proteins that bind chromatin primarily through their partners, which might be not expressed in mouse rods.
The two proteins are sequentially used during cellular differentiation and development by exerting differential gene regulation [31, 61, 62]. Based on the developmental dynamics of the two tethers, one can speculate that the LBR-dependent tether is more ancient than the LAC-dependent tether, with the latter evolving as an additional layer of gene regulation during cell differentiation. This consideration agrees with the existence of many transmembrane proteins that bind both LAC and chromatin and are differentially expressed during development and differentiation in various cell types [63–66].
As expected, rods of various nocturnal mammals do not express LBR or LAC, in contrast to rods of diurnal mammals [31, 32], where just one of these tethers is present and sufficient to maintain the conventional nuclear architecture in these cells (Figure 2C). In support of the concept of an independent origin of diurnality in mammalian groups, different mammalian groups switch on different peripheral tether (Figure 2C).
The discovery of the two major peripheral tethers does not exclude the existence of other mechanisms for HC binding to the nuclear periphery. In this respect, cultured human cells with oncogene-induced senescence draw special attention. Upon induction of senescence, cells significantly downregulate the levels of nuclear membrane proteins, such as lamins A/C and B1, LBR, MAN1, which in turn causes partial dissociation of heterochromatin from the nuclear periphery and formation of so-called senescence-associated heterochromatin foci [67–69]. However, it is still unknown what causes eviction of peripheral HC and what the mechanisms of foci formation are. Interestingly, both phenomena have been recently attributed to an increased number of nuclear pores in senescent cells and, more specifically, to a high concentration of the nucleoporin TPR [70].
In summary, studies of inverted nuclei led to the identification of two major tethers of heterochromatin to the nuclear periphery (Figure 3C). Whether the tethers act on the same genomic regions and recognize the same histone marks and types of heterochromatin (HC or cHC) and what the components of the A/C-depended tether are, remains to be investigated. Further work is needed to uncover other tethers that anchor heterochromatin to the nuclear lamina (see Outstanding Questions).
Chromatin compartmentalization: attraction drives segregation
Maintenance of spatial segregation of HC and EC in inverted nuclei shows that the segregation is driven by a process that does not involve tethering to the lamina. From the physics point of view, spatial separation of EC and HC is reminiscent of the classical picture of microphase separation in a mixture of different types of polymers, or in polymers made of monomers of different types (block copolymers). Higher affinity (i.e. attraction) between monomers of the same type drives formation of areas predominantly occupied by monomers of one type, even if these regions remain liquid in nature. This well-studied phenomenon in polymer physics has recently attracted a great deal of attention in biology [8], with proposals that formation of membraneless compartments in both the cytoplasm [71–73] and nucleoplasm [74, 75] constitute such liquid-liquid phase separation. In the context of the nucleus, separation of chromatin can be driven by higher affinity of EC regions to each other, HC regions to each other, or both.
EC-centered models of compartmentalization suggest that clustering of active genomic regions around the transcription [39, 41, 76] and splicing [77–79] machineries mediates their mutual affinity and generates separation. The plausibility of such a scenario was supported by computational modeling [80, 81]. Moreover, super-resolution microscopy of zebrafish embryos showed that cluster aggregation within EC is driven by the exclusion of inactive chromatin from RNA-enriched microenvironments containing expressed genes [82]. Furthermore, transcription factor activation domains can form phase-separated condensates with the Mediator coactivator at super-enhancers and thus compartmentalize the transcription apparatus [39, 83–86]. Hi-C studies also observed a particularly high contact frequency between transcribed long multi-exonic genes, suggesting the role of splicing in these interactions [87, 88]. The spatial segregation of HC and EC, however, seems to be independent of transcription because the intra-and inter-chromosomal contacts persist after transcription inhibition [89] or in largely transcriptionally inert pronuclei [90] and sperm cells [91, 92]. Together, these studies suggest that although transcription is a major organizing force within the euchromatic compartment, it is not what drives large-scale segregation between EC and HC. A HC-centered model of compartmentalization (see below) is based on the observation of high affinity between HC regions marked by silencing histone modifications and associated proteins. It remains to be seen whether such affinity is driven by direct interactions between histone tails [75], or mediated by other proteins, e.g. HP1α that can be induced to form droplets [93–95].
Compartments are marked by repeats
Mouse rod nuclei are a convenient model to study the formation of compartments, since they exhibit a remarkable degree of segregation between EC and HC, which is evident from both microscopy [17] and compartment strength detected by Hi-C analysis [15]. All three types of chromatin are marked by a characteristic repeat repertoire: cHC is formed by satellites, HC is enriched in interspersed repeats LINEs and LTRs, EC is populated by SINEs [2, 3]. Such conspicuous differential enrichment led to the suggestion that EC and HC segregation in general can be based on mutual recognition and attraction of homotypic repeats [2, 96–98]. Moreover, this hypothesis predicts that building of the interphase nucleus is a self-organizing and largely predetermined process, because homotypic repetitive sequences find each other during early interphase, and as such can serve as a minimal model for the separation of EC from HC.
The hypothesis that repeats are the driving force for compartmentalization was tested experimentally by using a human artificial chromosome introduced into mouse. The chromosome included 4 Mb of human chromosome 1 and consisted of 3 structurally different segments (Figure 4): centromeric cHC, gene-poor HC and gene-rich EC, enriched in the corresponding interspersed repeats [99]. Since in rod nuclei, chromatin types are clearly segregated and form regular concentric shells [3, 17] (Figure 4), they offer an opportunity to estimate the spatial distribution of human artificial chromosome segments between different chromatin types microscopically. Using retinas from mice carrying human chromosome, it has been shown that xenospecific segments are positioned strictly within the same mouse chromatin type and thus demonstrate autonomous behavior [100]. In particular, about 85% of human centromeres cluster with mouse centromeres in the proximity to chromocenters; about 90% of LINE/LTR-rich HC segments reside within the LINE/LTR-rich HC shell and more than 90% of SINE-rich EC segments are associated with the SINE-rich peripheral shell of rod nuclei [100]. These results on rods have been further confirmed by 4C analysis of mouse fibroblast nuclei carrying a human artificial chromosome, showing that xenospecific EC and HC segments share the corresponding nuclear compartments also in conventional nuclei [100]. A recent study on mouse ESCs provides further experimental support to the role of homotypic repeat clustering. It presents an evidence that L1 transcripts play a crucial role in the segregation of L1 and B1 (LINEs and SINEs) enriched compartments [101]. Thus, there is a clear indication that self-attraction of different types of chromatin can underlie the segregation, as opposed to intranuclear positioning of HC and EC. Relative contributions of HC/EC-specific histone marks and direct DNA sequences to compartmentalization are yet to be understood.
Figure 4. Probing rod nuclei with xenospecific sequences.
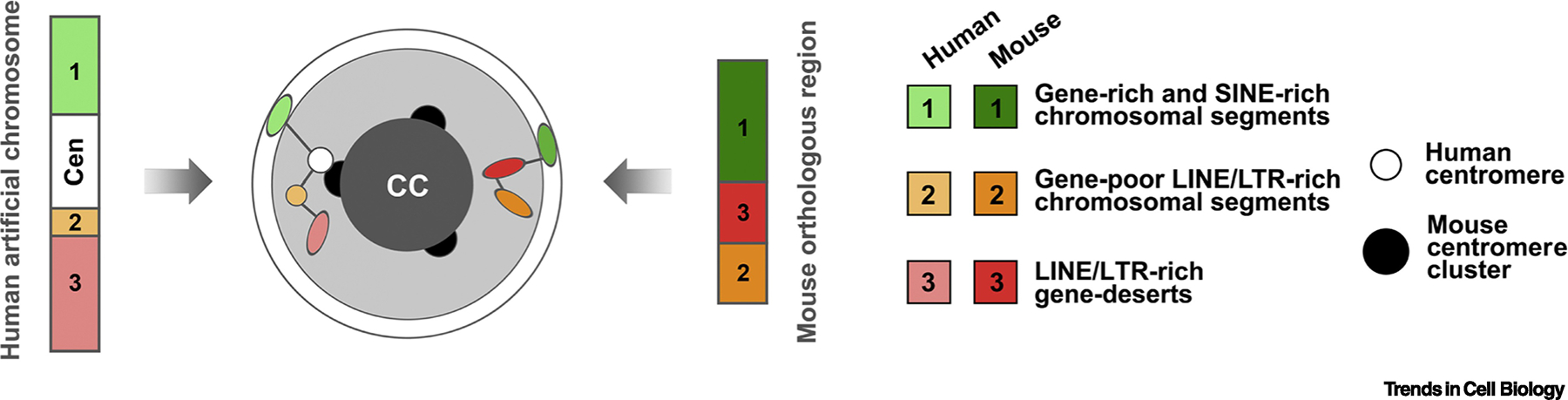
The mouse rod nucleus is built by three concentric layers of chromatin: EC enriched in SINEs (white), HC enriched in LINEs/LTRs (light grey) and cHC consisting of satellites (dark grey). This regular nuclear structure allows to microscopically track the positions of small xenospecific human (left schematics) and endogenous mouse (right schematics) chromosomal segments. In rods, the segments enriched in either SINEs or LINEs/LTRs, faithfully localize to the corresponding chromatin layer, marked by the same repeats. Human and mouse centromeres, both consisting of satellite sequences, also coalesce with each other.
Heterochromatin drives compartmentalization
In the context of many competing ideas for the mechanisms of HC and EC compartmentalization discussed above, inverted rod nuclei constitute an ideal system for testing proposed models. First, the absence of lamina tethers helps to isolate the roles of interactions between EC and between HC loci. Second, development of nuclear inversion upon loss of HC peripheral tethering [31] provides sufficient constraints to develop models that can reproduce separation of HC and EC in both conventional and inverted nuclei.
To examine the roles of EC, HC and cHC in compartmentalization, a polymer model of mouse chromosomes has been developed to reproduce both compartmentalization measured from Hi-C maps and spatial segregation seen in microscopy [15]. In the model, each chromatin type is given varying affinities for itself and for the other types (Figure 5). Depending on these affinities, chromosomes fold differently. The model demonstrates that in the absence of tethering to the lamina, the inverted architecture could be reproduced if cHC regions had the strongest affinity to themselves, followed by strong affinity among HC regions, whereas attractions between EC regions are dispensable (Figure 5). Moreover, even in models without cHC, reflecting the lack of prominent chromocenters in other mammalian rods, simultaneous agreement with Hi-C and microscopy data can still be achieved. By adding interactions between heterochromatin and the lamina, the same model can successfully recapitulate the conventional nuclear organization [15] (Figure 5). Importantly, cHC, HC and EC compartments formed due to this process remain liquid, and the relative density in HC is only 20% higher than in EC, and about 2-fold higher in cHC than in EC. Since the inverted nuclear morphology arises from the conventional one during terminal differentiation of rod photoreceptor cells after downregulation of LBR expression, it is particularly interesting to see that models of initially conventional nuclei invert in a manner closely mirroring the events observed in vivo during rod differentiation upon turning off lamina-interactions [15]. Furthermore, nuclear compartments in modeled inversion remain separated during the whole reorganization process similarly to observed separation of compartments in differentiating rods in vivo [3].
Figure 5. Polymer models aid in understanding the interactions that drive nuclear organization.
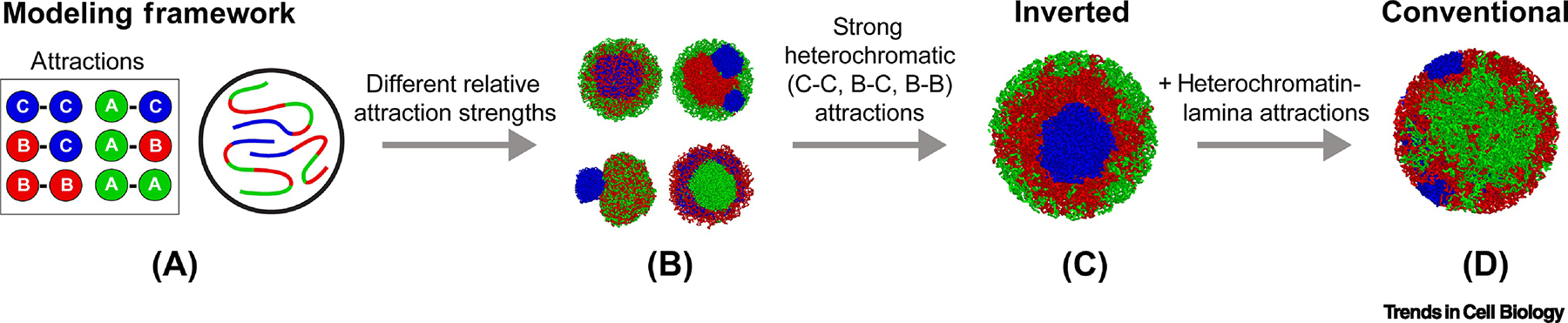
Models of chromosomes are built with confined polymers of three monomer types: A, B and C, representing EC, HC and cHC, respectively. A pair of monomers attract each other with different strengths depending on the monomer types involved (far left). Different relative strengths of the attractions result in a wide array of possible chromatin organizations (mid-left). Only certain relative strengths of attraction between monomers result in an inverted arrangement (mid-right). The same simulation parameters, but complemented with attraction of HC and cHC to the nuclear lamina, successfully recapitulate the conventional arrangement (far right).
Collectively, microscopy, Hi-C analysis and polymer modeling showed that attractions between HC regions are central for the separation of the active from the inactive genome in inverted and conventional nuclei, whereas lamina-heterochromatin attraction is needed for the spatial organization of conventional nuclei. Strikingly, the building of compartments and their proper arrangement in the modeled nuclei do not require interactions among euchromatic regions. In light of these findings, we suggest that inversion is the default nuclear status, which is perturbed by lamina-association. The outstanding question is why heterochromatin tethering to the nuclear lamina that generates spatial positioning of HC and EC in conventional nuclei, is advantageous (see Outstanding Questions).
Concluding Remarks
During the last quarter of a century, the awareness about the spatial organization of the genome within the nucleus as a key component of genome function has become a textbook knowledge. For practical reasons, most of the accumulated knowledge is based on studies of permanent or primary cell cultures, that all have somewhat similar nuclear organization. In this review we tried to provide evidence for the importance of working with cells in their native tissue environment allowing to see and take advantage of the diversity of nuclear architectures there. To justify this notion, we explored rod photoreceptors in nocturnal mammals, whose nuclei turned out to be dramatically different from the nuclei of all other mammalian cells. The peculiar inverted structure of nocturnal rods is paradoxically not linked to nuclear function but plays an important role in retinal optics. Nonetheless, comparison of conventional and inverted nuclei allows studying fundamental aspects of the nuclear architecture. These aspects include general mechanisms of chromatin segregation and spatial positioning of chromatin compartments. The exact molecular players in these processes, however, remain largely unknown and we foresee that inverted nuclei of mammalian rods will be further harnessed to resolve other questions in the field, e.g. those listed in the Outstanding Questions Box.
Highlights.
The segregation of the eukaryotic genome into active euchromatic and silent heterochromatic nuclear compartments is crucial for the realization of nuclear processes. In most cells euchromatin occupies the nuclear interior, while heterochromatin is found at the nuclear and nucleolar peripheries
Nuclei of rod photoreceptor cells of nocturnal mammals represent the only known unique exception and have an inverted arrangement of the nuclear compartments with heterochromatin concentrated in the nuclear center and acting as a microlens focusing light and thus facilitating light propagation with reduced photon scattering
Inverted rod nuclei, with their perfectly segregated and concentrically arranged compartments, serve as a model to understand general principles of genome organization such as heterochromatin tethering at the nuclear periphery, compartment recognition, and mechanisms of genome segregation
Acknowledgements
YF has been supported by the Medical University of Plovdiv, grant DPDP - 10/2018, and the Bulgarian Ministry of Education and Science, National program “Young scientists and Postdoctoral candidates”. LM and MF are supported by NIH GM114190 and NIH Common Fund 4D Nucleome Program DK107980. IS has been supported by Deutsche Forschungsgemeinschaft grants SO1054/1-3, SO1054/2-1 and SFB1064.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van Steensel B and Belmont AS (2017) Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 169 (5), 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solovei I et al. (2016) How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol 40, 47–59. [DOI] [PubMed] [Google Scholar]
- 3.Solovei I et al. (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137 (2), 356–368. [DOI] [PubMed] [Google Scholar]
- 4.Zheng H and Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol 20 (9), 535–550. [DOI] [PubMed] [Google Scholar]
- 5.Kim S and Shendure J (2019) Mechanisms of Interplay between Transcription Factors and the 3D Genome. Mol. Cell 76 (2), 306–319. [DOI] [PubMed] [Google Scholar]
- 6.Jerkovic I et al. (2019) Higher-order chromosomal structures mediate genome function. J. Mol. Biol, pii: S0022–2836 (19), 30610–2. [DOI] [PubMed] [Google Scholar]
- 7.van Steensel B and Furlong EEM (2019) The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol 20 (6), 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirny LA et al. (2019) Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol 58, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eagen KP (2018) Principles of Chromosome Architecture Revealed by Hi-C. Trends Biochem. Sci 43 (6), 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowley MJ and Corces VG (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet 19 (12), 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Velazquez Camacho O et al. (2017) Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife 6, pii: e25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieux-Rochas M et al. (2015) Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc. Natl. Acad. Sci. U S A 112 (15), 4672–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman-Aiden E et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326 (5950), 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian K et al. Rod nuclear architecture determines contrast transmission of the retina and behavioral sensitivity in mice. bioRxiv 752444; doi: 10.1101/752444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falk M et al. (2019) Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570 (7761), 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan L et al. (2019) Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat. Struct. Mol. Biol 26 (4), 297–307. [DOI] [PubMed] [Google Scholar]
- 17.Eberhart A et al. (2013) Epigenetics of eu-and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 21 (5), 535–554. [DOI] [PubMed] [Google Scholar]
- 18.Mo A et al. (2016) Epigenomic landscapes of retinal rods and cones. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soutoglou E and Misteli T (2007) Mobility and immobility of chromatin in transcription and genome stability. Curr. Opin. Genet. Dev 17 (5), 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner MR and Spector DL (2010) Chromatin dynamics. Annu. Rev. Biophys 39, 471–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dion V and Gasser SM (2013) Chromatin movement in the maintenance of genome stability. Cell 152 (6), 1355–1364. [DOI] [PubMed] [Google Scholar]
- 22.Cuvier O and Fierz B (2017) Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat. Rev. Genet 18 (8), 457–472. [DOI] [PubMed] [Google Scholar]
- 23.Rosa A and Everaers R (2008) Structure and dynamics of interphase chromosomes. PLoS Comput. Biol 4 (8), e1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armelin-Correa LM et al. (2014) Nuclear compartmentalization of odorant receptor genes. Proc. Natl. Acad. Sci. U S A 111 (7), 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clowney EJ et al. (2012) Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151 (4), 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horta A et al. Cell type-specific interchromosomal interactions as a mechanism for transcriptional diversity. bioRxiv 287532; doi: 10.1101/287532 [DOI] [Google Scholar]
- 27.Kit S (1961) Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J. Mol. Biol 3, 711–716 [DOI] [PubMed] [Google Scholar]
- 28.Vissel B and Choo KH (1989) Mouse major (gamma) satellite DNA is highly conserved and organized into extremely long tandem arrays: implications for recombination between nonhomologous chromosomes. Genomics 5 (3), 407–414. [DOI] [PubMed] [Google Scholar]
- 29.Ou HD et al. (2017) ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357 (6349), pii: eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreysing M et al. (2010) Physical insight into light scattering by photoreceptor cell nuclei. Opt. Lett 35 (15), 2639–2641. [DOI] [PubMed] [Google Scholar]
- 31.Solovei I et al. (2013) LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598. [DOI] [PubMed] [Google Scholar]
- 32.Joffe B et al. (2014) Diurnality and Nocturnality in Primates: An Analysis from the Rod Photoreceptor Nuclei Perspective. Evol. Biol 41 (1), pp 1–11. [Google Scholar]
- 33.Bowmaker JK and Hunt DM (2006) Evolution of vertebrate visual pigments. Curr. Biol 16 (13), R484–489. [DOI] [PubMed] [Google Scholar]
- 34.Davies WL et al. (2007) Visual pigments of the platypus: a novel route to mammalian colour vision. Curr. Biol 17 (5), R161–163. [DOI] [PubMed] [Google Scholar]
- 35.Trezise AE and Collin SP (2005) Opsins: evolution in waiting. Curr. Biol 15 (19), R794–796. [DOI] [PubMed] [Google Scholar]
- 36.Canapa A et al. (2015) Transposons, Genome Size, and Evolutionary Insights in Animals. Cytogenet. Genome Res 147 (4), 217–239. [DOI] [PubMed] [Google Scholar]
- 37.Gregory TR (2005) Genome Size Evolution in Animals In The Evolution of the Genome, pp. 3–87, San Diego: Elsevier. [Google Scholar]
- 38.Kapusta A et al. (2017) Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci. U S A 114 (8), E1460–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hnisz D et al. (2017) A Phase Separation Model for Transcriptional Control. Cell 169 (1), 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J Jung I et al. (2019) A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet 5 (10), 1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenfelder S and Fraser P (2019) Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet 20 (8), 437–455. [DOI] [PubMed] [Google Scholar]
- 42.Frohns A et al. (2014) Inefficient double-strand break repair in murine rod photoreceptors with inverted heterochromatin organization. Curr. Biol 24 (10), 1080–1090. [DOI] [PubMed] [Google Scholar]
- 43.Guerreiro I and Kind J (2019) Spatial chromatin organization and gene regulation at the nuclear lamina. Curr. Opin. Genet. Dev 55, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eagen KP et al. (2015) Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell 163 (4), 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathog D et al. (1984) Characteristic folding pattern of polytene chromosomes in Drosophila salivary gland nuclei. Nature 308 (5958), 414–421. [DOI] [PubMed] [Google Scholar]
- 46.Guelen L et al. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453 (7197), 948–951. [DOI] [PubMed] [Google Scholar]
- 47.Kind J et al. (2015) Genome-wide Maps of Nuclear Lamina Interactions in Single Human Cells. Cell 163 (1), 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kind J et al. (2013) Single-cell dynamics of genome-nuclear lamina interactions. Cell 153 (1), 178–92. [DOI] [PubMed] [Google Scholar]
- 49.Meuleman W et al. (2013) Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23 (2), 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bian Q et al. (2013) beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol 203 (5), 767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harr JC et al. (2016) Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep. 17 (2), 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao RA et al. (2019) KMT1 family methyltransferases regulate heterochromatin-nuclear periphery tethering via histone and non-histone protein methylation. EMBO Rep. 20 (5), pii: e43260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towbin BD et al. (2012) Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150 (5), 934–947. [DOI] [PubMed] [Google Scholar]
- 54.Nemeth A et al. (2010) Initial genomics of the human nucleolus. PLoS Genet. 6 (3), e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pederson T (2011) The nucleolus. Cold Spring Harb. Perspect. Biol 3 (3), pii: a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smirnov E et al. (2016) Nucleolar DNA: the host and the guests. Histochem. Cell Biol 145 (4), 359–372. [DOI] [PubMed] [Google Scholar]
- 57.Vertii A et al. (2019) Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 29 (8), 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun X and Kaufman PD (2018) Ki-67: more than a proliferation marker. Chromosoma 127 (2), 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes AE et al. (2017) Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors. Sci. Rep 7, 43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bashkirova E and Lomvardas S (2019) Olfactory receptor genes make the case for inter chromosomal interactions. Curr. Opin. Genet. Dev 55, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mattar P et al. (2018) Casz1 controls higher-order nuclear organization in rod photoreceptors. Proc. Natl. Acad. Sci. U S A 115 (34), E7987–E7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thanisch K et al. (2017) Nuclear envelope localization of LEMD2 is developmentally dynamic and lamin A/C dependent yet insufficient for heterochromatin tethering. Differentiation 94, 58–70. [DOI] [PubMed] [Google Scholar]
- 63.Brachner A and Foisner R (2011) Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem. Soc. Trans 39 (6), 1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Las Heras JI et al. (2013) Tissue specificity in the nuclear envelope supports its functional complexity. Nucleus 4 (6), 460–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robson MI et al. (2016) Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol. Cell 62 (6), 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuleger N et al. (2013) Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 14 (2), R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandra T et al. (2015) Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 10 (4), 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lenain C et al. (2017) Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 27 (10), 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lenain C et al. (2015) Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis 36 (11), 1263–1274. [DOI] [PubMed] [Google Scholar]
- 70.Boumendil C et al. (2019) Nuclear pore density controls heterochromatin reorganization during senescence. Genes Dev. 33 (3–4), 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18 (5), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hyman AA et al. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol 30, 39–58. [DOI] [PubMed] [Google Scholar]
- 73.Ries RJ et al. (2019) m(6)A enhances the phase separation potential of mRNA. Nature 571 (7765), 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erdel F and Rippe K (2018) Formation of Chromatin Subcompartments by Phase Separation. Biophys. J 114 (10), 2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh HR and Ostwal YB (2019) Post-Translational Modification, Phase Separation, and Robust Gene Transcription. Trends Genet. 35 (2), 89–92. [DOI] [PubMed] [Google Scholar]
- 76.Papantonis A and Cook PR (2013) Transcription factories: genome organization and gene regulation. Chem. Rev 113 (11), 8683–8705. [DOI] [PubMed] [Google Scholar]
- 77.Herzel L et al. (2017) Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol 18 (10), 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu Y et al. (2009) Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol 185 (1), 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith KP et al. (1999) Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol 144 (4), 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brackley CA et al. (2016) Simulated binding of transcription factors to active and inactive regions folds human chromosomes into loops, rosettes and topological domains. Nucleic Acids Res. 44 (8), 3503–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shrinivas K et al. (2019) Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 75 (3), 549–561 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hilbert L et al. Transcription organizes euchromatin similar to an active microemulsion. bioRxiv 234112; doi: 10.1101/234112 [DOI] [Google Scholar]
- 83.Boija A et al. (2018) Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175 (7), 1842–1855 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cho WK et al. (2018) Mediator and RNA polymerase II clusters associate in transcription dependent condensates. Science 361 (6400), 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo YE et al. (2019) Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572 (7770), 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361 (6400), pii: eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonev B et al. (2017) Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171 (3), 557–572 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kerpedjiev P et al. (2018) HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19 (1), 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palstra RJ et al. (2008) Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One 3 (2), e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flyamer IM et al. (2017) Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte to-zygote transition. Nature 544 (7648), 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Du Z et al. (2017) Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547 (7662), 232–235. [DOI] [PubMed] [Google Scholar]
- 92.ung YH et al. (2017) Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep. 18 (6), 1366–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Larson AG et al. (2017) Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547 (7662), 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Larson AG and Narlikar GJ (2018) The role of phase-separation in heterochromatin formation, function and regulation. Biochemistry 57 (17), 2540–2548.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Strom AR et al. (2017) Phase separation drives heterochromatin domain formation. Nature 547 (7662), 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bouwman BA and de Laat W (2015) Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol. 16, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Krijger PH and de Laat W (2013) Identical cells with different 3D genomes; cause and consequences? Curr. Opin. Genet. Dev 23 (2), 191–196. [DOI] [PubMed] [Google Scholar]
- 98.Tang SJ (2011) Chromatin Organization by Repetitive Elements (CORE): A Genomic Principle for the Higher-Order Structure of Chromosomes. Genes (Basel) 2 (3), 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weuts A et al. (2012) Telomere length homeostasis and telomere position effect on a linear human artificial chromosome are dictated by the genetic background. Nucleic Acids Res. 40 (22), 11477–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van de Werken HJG et al. (2017) Small chromosomal regions position themselves autonomously according to their chromatin class. Genome Res. 27 (6), 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu JY et al. L1 and B1 repeats blueprint the spatial organization of chromatin. bioArxiv 802173; doi: 10.1101/802173 [DOI] [Google Scholar]
- 102.Tan L et al. (2018) Three-dimensional genome structures of single diploid human cells. Science 361 (6405), 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Y et al. (2018) Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol 217 (11), 4025–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Quinodoz SA et al. (2018) Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 174 (3), 744–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Steensel B and Henikoff S (2000) Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol 18 (4), 424–428. [DOI] [PubMed] [Google Scholar]
- 106.Nagano T et al. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502 (7469), 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cremer M et al. (2001) Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9 (7), 541–567. [DOI] [PubMed] [Google Scholar]
- 108.Croft JA et al. (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol 145 (6), 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peric-Hupkes D et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38 (4), 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]


