Abstract
Carbapenems are last‐resort β‐lactam antibiotics used in healthcare facilities to treat multidrug‐resistant infections. Thus, most studies on identifying and characterizing carbapenem‐resistant bacteria (CRB) have focused on clinical settings. Relatively, little is still known about the distribution and characteristics of CRBs in the environment, and the role of soil as a potential reservoir of CRB in the United States remains unknown. Here, we have surveyed 11 soil samples from 9 different urban or agricultural locations in the Los Angeles–Southern California area to determine the prevalence and characteristics of CRB in these soils. All samples tested contained CRB with a frequency of <10 to 1.3 × 104 cfu per gram of soil, with most agricultural soil samples having a much higher relative frequency of CRB than urban soil samples. Identification and characterization of 40 CRB from these soil samples revealed that most of them were members of the genera Cupriavidus, Pseudomonas, and Stenotrophomonas. Other less prevalent genera identified among our isolated CRB, especially from agricultural soils, included the genera Enterococcus, Bradyrhizobium, Achromobacter, and Planomicrobium. Interestingly, all of these carbapenem‐resistant isolates were also intermediate or resistant to at least 1 noncarbapenem antibiotic. Further characterization of our isolated CRB revealed that 11 Stenotrophomonas, 3 Pseudomonas, 1 Enterococcus, and 1 Bradyrhizobium isolates were carbapenemase producers. Our findings show for the first time that both urban and agricultural soils in Southern California are an underappreciated reservoir of bacteria resistant to carbapenems and other antibiotics, including carbapenemase‐producing CRB.
Keywords: Achromobacter, Bradyrhizobium, carbapenemase, carbapenem‐resistant bacteria, Cupriavidus, Enterococcus, Planomicrobium, Pseudomonas, soil, Stenotrophomonas
The study reported here is the first specific study about the distribution and characteristics of carbapenem‐resistant bacteria (CRB) in urban and agricultural environmental soils in the United States. We have found that the soils from Los Angeles–Southern California area are an underappreciated reservoir of bacteria resistant to carbapenems and other antibiotics, including carbapenemase‐producing CRB. Our findings also show a much higher relative frequency of CRB on soils from locations adjacent to farms compared to soils from urban locations.
1. INTRODUCTION
Carbapenems are broad‐spectrum β‐lactam antibiotics that act as potent inhibitors of bacterial cell wall synthesis because of their high affinity for penicillin‐binding proteins (Papp‐Wallace, Endimiani, Taracila, & Bonomo, 2011). While most β‐lactams have a cishydroxyethyl side chain, carbapenems have a transhydroxyethyl side chain. This unique feature confers carbapenems increased resistance to hydrolysis by most β‐lactamases, including extended‐spectrum β‐lactamases, and thus has led to their use as last‐resort drugs to treat multidrug‐resistant infections (Papp‐Wallace et al., 2011; Vardakas, Tansarli, Rafailidis, & Falagas, 2012).
Carbapenem‐resistant bacteria (CRB), especially Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumanii, have been designated by the Centers for Disease Control and Prevention (CDC) and other health organizations as a major public health threat because the infections they cause are difficult to treat, their high associated mortality rates, and their rising prevalence in healthcare settings (Centers for Disease Control & Prevention, 2013a, 2013b; Cuzon et al., 2011; Guh et al., 2015).
Resistance to carbapenems can occur through three major mechanisms: decreased outer membrane permeability (Livermore, Mushtaq, & Warner, 2005; Shin et al., 2012; Warner et al., 2013), increased efflux (Livermore et al., 2005; Papp‐Wallace et al., 2011; Rodríguez‐Martínez, Poirel, & Nordmann, 2009; Warner et al., 2013), and production of carbapenemases, which are unique β‐lactamases capable of degrading carbapenems (Marsik & Nambiar, 2011; Queenan & Bush, 2007). Carbapenemase‐producing CRB (CP‐CRB) are especially concerning because carbapenemase genes are often located on transmissible genetic elements that can quickly spread to other bacteria (Mathers et al., 2011; Walsh, 2010).
Because the use of carbapenems is restricted to healthcare facilities (Bradley et al., 1999; Paterson, 2000), most studies on isolating and characterizing CRB have also focused on these and immediately related settings (Gupta, Limbago, Patel, & Kallen, 2011; Kallen, Hidron, Patel, & Srinivasan, 2010; Khuntayaporn, Montakantikul, Mootsikapun, Thamlikitkul, & Chomnawang, 2012; Rhomberg & Jones, 2009; Ssekatawa, Byarugaba, Wampande, & Ejobi, 2018). However, other β‐lactams including extended‐spectrum penicillins and cephalosporins are used to treat patients outside healthcare facilities and are used in agriculture as well. For example, in the United States, penicillins account for 12% of antibiotics used in food‐producing animals (United States Food & Drug Administration Center for Veterinary Medicine, 2017). Even though there is no established relationship between the broad use of β‐lactams or extended‐spectrum β‐lactams and resistance to carbapenems, the use of these and other drugs is predicted to cause selection favoring carbapenem resistance in the environment (Meletis, 2016; Mollenkopf et al., 2017). Recent findings of CRB in environmental samples from Europe, Africa, Asia, and North America (Adelowo, Vollmers, Mäusezahl, Kaster, & Müller, 2018; Ash, Mauck, & Morgan, 2002; Aubron, Poirel, Ash, & Nordmann, 2005; Di, Jang, Unno, & Hur, 2017; Girlich, Poirel, & Nordmann, 2010; Harmon et al., 2019; Henriques et al., 2012; Hrenovic et al., 2019; Isozumi et al., 2012; Mills & Lee, 2019; Poirel et al., 2012; Potron, Poirel, Bussy, & Nordmann, 2011; Sivalingam, Pote, & Prabakar, 2019; Tacão, Correia, & Henriques, 2015; Zou et al., 2020; Zurfluh, Hachler, Nuesch‐Inderbinen, & Stephan, 2013) seem to support this hypothesis. However, further studies are needed to fully understand the role of the environment as a reservoir of CRB and carbapenem resistance genes.
Knowledge about the environmental distribution and characteristics of CRB is especially lacking in the United States. For example, there have only been three studies about CRB in freshwater environments in the United States (Ash et al., 2002; Aubron et al., 2005; Harmon et al., 2019) and no specific studies about the prevalence or characteristics of CRB in U.S. soils. However, recent studies in soil and related environmental samples from Africa and Europe suggest that soil may be an underappreciated reservoir of CRB. For example, CRB including CP‐CRB have been isolated from agricultural and nonagricultural soil samples from Algeria, Spain, England, Germany, Denmark, and Norway (Gudeta et al., 2016) and Croatia (Hrenovic et al., 2019), as well as from swine and poultry farms from Germany (Borowiak et al., 2017; Fischer et al., 2013), and natural soil samples from Algeria (Djenadi, Zhang, Murray, & Gaze, 2018), among other locations.
Although there are no specific studies about the prevalence or characteristics of CRB in U.S. soils, a few studies suggest that CRB may also be prevalent in U.S. soils. For example, a study on soil samples from the Midwestern United States that used penicillins as selective agents identified three isolates that were carbapenem‐resistant (Crofts et al., 2018). CRB and CP‐CRB have also been isolated from fecal samples from dairy farms in New Mexico and Texas (Webb et al., 2016), as well as from fecal and environmental samples recovered from a swine nursery in Ohio (Mollenkopf et al., 2017). These findings are very significant because farm animal feces are routinely used as manure, which may lead to the spread of CRB and carbapenemase genes to the soil, water, and other environments.
To contribute to addressing the information gap about the role of U.S. soils as potential sinks and sources of CRB, we report here the first study specifically aimed at determining the prevalence and characteristics of CRB in soil from the West Coast of the United States. Our findings indicate that both urban and agricultural soils from the highly populated Los Angeles–Southern California area are a significant reservoir of CRB and CP‐CRB, which we found to be also resistant to other classes of antibiotics as well.
2. MATERIALS AND METHODS
2.1. Collection of soil samples and isolation of carbapenem‐resistant bacteria
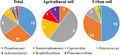
We collected 11 different soil samples from 9 different locations in the Los Angeles (California) area between June 2016 and January 2019. The location (Figure 1) and characteristics of sampling sites are summarized in Table 1. For each sample, we collected surface soil in 50‐ml sterile conical tubes and immediately transported the sample to the laboratory. We then weighed 4 g of the soil sample into a sterile 15‐ml conical tube, added 10 ml of sterile saline (0.85% NaCl), and vortexed the mixture continuously for 5 min to homogenize the sample and extract the bacteria present in the soil. Soil debris was then removed by centrifugation for 10 min at 1,000 × g, and the supernatant containing the extracted soil bacteria collected for subsequent analyses.
FIGURE 1.
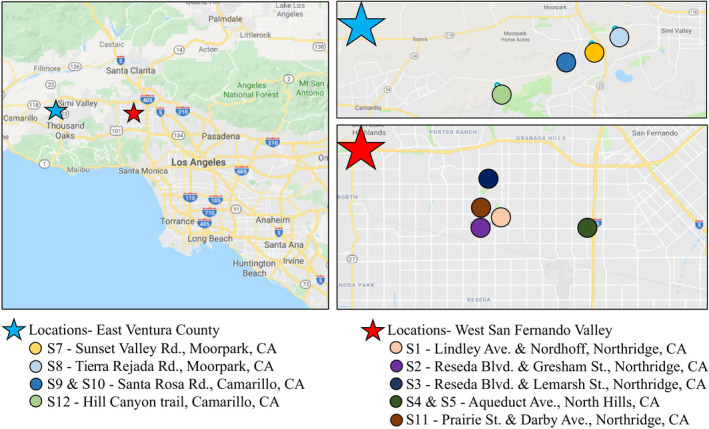
Map of the location of the soil samples analyzed in this study. Left panel: A general map of the Southern California region with the two major areas sampled in the East Ventura County (labeled with a blue star) and the West San Fernando Valley County (labeled with a red star). Top right panel: Detailed map of the soil locations sampled in the East Ventura County. Bottom right panel: Detailed map of the soil locations sampled in the West San Fernando Valley County
TABLE 1.
Summary of the origin, count of total gram‐negative bacteria, and count of carbapenem‐resistant bacteria (CRB) obtained for the 11 soil samples from the Los Angeles–Southern California area tested in this study
| Sample | Date | Location (Type) | Urban/agricultural | GPS location | Total bacteria (cfu/g) | CRB (cfu/g) |
|---|---|---|---|---|---|---|
| S1 | 10/3/2016 | Lindley Ave. and Nordhoff St., Northridge (adjacent to CSUN Pond) | Urban | 34.235587–118.5274932 | 1.2·105 | 8.5·102 |
| S2 | 1/9/2017 | Reseda Blvd. and Gresham St., Northridge (highly transited intersection) | Urban | 34.2307707–118.5382339 | 9.6·104 | <10 |
| S3 | 2/4/2017 | Reseda Blvd. and Lemarsh St., Northridge (Northridge Recreation Center park) | Urban | 34.2543024–118.5345628 | TNTC | <10 |
| S4 | 4/16/2017 | Aqueduct Ave., North Hills (private chicken coop, sample A) | Urban | 34.2308032–118.4751102 | 3.0·104 | 1.3·104 |
| S5 | 4/16/2017 | Aqueduct Ave., North Hills (private chicken coop, sample B) | Urban | 34.2308032–118.4751102 | 3.0·104 | 1.3·104 |
| S7 | 8/6/2017 | Sunset Valley Rd., Moorpark (adjacent to produce farm) | Agricultural | 34.2558565–118.8558643 | 1.9·103 | 1.6·103 |
| S8 | 8/6/2017 | Tierra Rejada Rd., Moorpark (adjacent to horse farm) | Agricultural | 34.26555732–118.8345638 | 2.7·103 | 5.6·102 |
| S9 | 8/6/2017 | Santa Rosa Rd. and Moorpark Rd., Camarillo (adjacent to an avocado orchard, sample A) | Agricultural | 34.2461891–118.8708311 | 1.1·103 | 2.8·102 |
| S10 | 8/6/2017 | Santa Rosa Rd. and Moorpark Rd., Camarillo (adjacent to an avocado orchard, sample B) | Agricultural | 34.2461891–118.8708311 | 2.4·103 | 4.4·102 |
| S11 | 12/26/2018 | Prairie Rd. and Darby Ave., Northridge (grass area recently fertilized) | Urban | 34.2391393–118.5360182 | 1.3·105 | 2.5·102 |
| S12 | 1/7/2019 | Hill Canyon trail, Camarillo (hiking trail near a strawberry farm) | Agricultural | 34.2281655–118.9322636 | 9.3·104 | 3.0·102 |
Abbreviation: TNTC, too numerous to count.
The total count of bacteria was determined using MacConkey medium (Fisher Scientific) as a primary selection for enteric bacteria and gram‐negatives, which were the main target in our study. The bacterial count was determined by direct plating of 100 µl of soil supernatant as well as by spot plating of 10 µl of a 100 to 10–4 dilution bank of soil supernatants in sterile saline on MacConkey agar plates, followed by incubation for 24 hr at 37°C. The count of carbapenem‐resistant bacteria (CRB) was determined by the same procedure except for using MacConkey agar plates containing 4 µg/ml of meropenem (Ark Pharm, Inc.), which is the Clinical Laboratory Standards Institute (CLSI) minimum inhibitory concentration (MIC) clinical breakpoint for this antibiotic in Enterobacteriaceae (Clinical & Laboratory Standards Institute, 2018). We selected meropenem because it is the most commonly prescribed carbapenem in the United States and is highly active against a broad spectrum of gram‐negative bacteria (Papp‐Wallace et al., 2011). Because of the low concentration of CRB in samples S2 and S3, all 10 ml of supernatant containing the extracted soil bacteria were concentrated by filtration using 0.45‐µm filters (Merck Millipore). The filters were then placed onto MacConkey‐meropenem plates as described above to obtain CRB colonies.
For each sample, we patched up to 50 distinct meropenem‐resistant colonies on Mueller‐Hinton (Fisher Scientific) agar plates supplemented with meropenem at 4 μg/ml (Enterobacteriaceae breakpoint) and 16 μg/ml (CLSI meropenem MIC breakpoint for other non‐Enterobacteriaceae gram‐negatives; Clinical & Laboratory Standards Institute, 2018). Growth in at least 4 μg/ml of meropenem was confirmed for nearly all patched colonies. In total, we selected 40 CRB isolates—up to 8 distinct CRB isolates per sample, prioritizing those that grew in 16 μg/ml of meropenem—for culturing, long‐term storage at −80°C, and preparation of cell suspension templates for PCR, as previously described (Harmon et al., 2019).
2.2. Identification of CRB by PCR and sequencing of the 16S rRNA gene, and oxidase test
The 40 selected soil CRB isolates were identified following the procedures described in Harmon et al. (2019). Briefly, we used PCR amplification of the 16S rRNA gene of each selected isolate, followed by Sanger sequencing, BLAST analysis (Altschul et al., 1997) of the obtained sequences, and oxidase test analysis. The oxidase test was used to further distinguish between closely related S. maltophilia, which is oxidase negative, and Pseudomonas species, most of which are oxidase‐positive (Bergey & Holt, 1994).
Besides, we constructed a phylogenetic tree for each genus isolated in our study (Achromobacter, Bradyrhizobium, Cupriavidus, Enterococcus, Planomicrobium, Pseudomonas, and Stenotrophomonas) to further characterize the taxonomic relationship between our soil isolates across different locations, as well as between our isolates and isolates from previous studies. We used MEGA X 10.1 software (Hall, 2013) to align the 16S rRNA genes and construct phylogenetic trees based on the Jukes–Cantor model and the neighbor joining method.
2.3. Determination of the antibiotic susceptibility profile of the isolated CRB
Determination of the antibiotic susceptibility profile of the 40 selected carbapenem‐resistant isolates was performed using the CLSI disk diffusion method (Clinical & Laboratory Standards Institute, 2018) and the reference strain Escherichia coli ATCC 25922 as quality control, as previously described (Harmon et al., 2019). The meropenem, imipenem, cefotaxime, ciprofloxacin, gentamicin, and tetracycline antibiotic disks were purchased from Becton Dickinson. To determine whether an isolate was susceptible, intermediate, or resistant to an antibiotic, we used CLSI zone diameter breakpoint values (Clinical & Laboratory Standards Institute, 2018). Unless otherwise indicated, for taxa in which the CLSI zone diameter breakpoints are not provided, we used the CLSI Enterobacteriaceae breakpoint values (Clinical & Laboratory Standards Institute, 2018).
2.4. Identification of carbapenemase‐producing isolates by the CarbaNP and mCIM assays, and detection of the L1 carbapenemase gene in Stenotrophomonas isolates
We identified carbapenemase‐producing CRB isolates using the CarbaNP assay (Dortet, Poirel, & Nordmann, 2012a, 2012b; Nordmann, Poirel, & Dortet, 2012). The assay was performed as described by CLSI (Clinical & Laboratory Standards Institute, 2018) using 6 mg/ml or either meropenem or imipenem. For each CRB isolate, colonies were grown overnight on plain Mueller‐Hinton agar (to detect constitutively expressed carbapenemases) and Mueller‐Hinton agar with the highest concentration of meropenem with growth (to detect inducible carbapenemases). Isolates that turned yellow at 37°C within 2 hr in the presence of meropenem or imipenem were considered carbapenemase‐positive. Isolates that were positive for carbapenemase production when grown on Mueller‐Hinton agar with the antibiotic but negative when grown on plain Mueller‐Hinton were considered to have an inducible carbapenemase.
For CarbaNP‐positive isolates, we confirmed that they produce carbapenemases by the modified Carbapenem Inactivation Method (mCIM; Pierce et al., 2017). This assay was performed as described by CLSI (Clinical & Laboratory Standards Institute, 2018). A zone of inhibition between 6 and 15 mm for E. coli ATCC 25922 when grown in the presence of a meropenem disk previously incubated in the presence of the isolate to be tested was a confirmed carbapenemase‐positive isolate.
PCR amplification to confirm the presence of the L1 carbapenemase gene (bla L1) in carbapenemase‐producing Stenotrophomonas isolates was performed using the primers and program described by Henriques et al. (2012) to amplify bla L1 as previously described (Harmon et al., 2019).
3. RESULTS
3.1. Distribution, frequency, and identification of carbapenem‐resistant bacteria in soil samples from the Los Angeles–Southern California area
We analyzed 11 different soil samples from 9 different urban and agricultural locations in the Los Angeles–Southern California area (United States; Figure 1; Table 1). Using meropenem as a selective agent, we found that all soil samples analyzed contained CRB. The frequency of CRB in these samples was between <10 and 1.3 × 104 cfu per gram of soil (Table 1). Interestingly, S4 and S5, the two samples with the most abundance of CRB, were obtained from the soil of a private urban chicken coop, which suggests that animal feces might be an important contributor to soil CRB. Overall, samples could be classified into those with a low relative frequency of CRB (<1%) compared to the total bacterial counts obtained (S1–S3 and S11–S12; mostly urban soils) and those with a high relative frequency of CRB (18%–80%, urban chicken coop, and most agricultural soil samples) compared to the total bacterial count obtained (S4–S10; Table 2).
TABLE 2.
Summary of the number and characteristics of soil carbapenem‐resistant bacteria isolated from samples described in Table 1
| Genus | Sample of origin | Number of isolates | Number of CP a isolates | Antibiotic resistant/intermediate (number of isolates) b |
|---|---|---|---|---|
| Achromobacter | S10 | 1 | 0 | MP (1), CF (1) |
| Bradyrhizobium | S11 | 1 | 1 | MP (1), IM (1), CF (1), CI (1), GE (1), TE (1) |
| Cupriavidus | S2, 7, 8, 9 | 8 | 0 | MP (8), IM (2), CF (2), GE (4) |
| Enterococcus | S7, 11, 12 | 3 | 1 | MP (3), IM (2), CF (3), GE (1) |
| Planomicrobium | S7 | 1 | 0 | MP (1), IM (1), CF (1), GE (1), TE (1) |
| Pseudomonas | S2, 3, 4, 5, 11 | 15 | 3 | MP (15), IM (5), CF (14), GE (1), TE (1) |
| Stenotrophomonas | S1, 7, 11 | 11 | 11 | MP (11), IM (11), CF (3), GE (8), TE (8) |
| Total | 40 | 16 | MP (40), IM (22), CF (33), CI (1), GE (17), TE (11) |
CP = carbapenemase‐producing isolates as determined by the CarbaNP test and confirmed using the mCIM method.
The number of isolates that were resistant or intermediate to meropenem (MP), imipenem (IM), cefotaxime (CF), ciprofloxacin (CI), gentamicin (GE), and tetracycline (TE) is shown in parentheses. The detailed antibiotic susceptibility profile and carbapenemase production result for each isolate are provided in Table 3.
We selected a total of 40 CRB isolates for further identification and characterization. We identified them using their 16S rRNA gene sequence as well as phylogenetic analyses (Figure 2 and Figures A1, A2, A3, A4, A5, A6, A7). We also used the oxidase test to distinguish between members of the Stenotrophomonas genus and closely related members of the genus Pseudomonas. We preliminarily identified our isolates as 1 Achromobacter marplatensis, 1 Bradyrhizobium elkanii, 8 Cupriavidus (3 C. alkaliphilus and 5 C. respiraculi), 3 Enterococcus (1 E. durans and 2 E. gallinarum), 1 Planomicrobium glaciei, 15 Pseudomonas (1 P. alkylphenolica, 1 P. putida, 10 P. stuzeri, and 4 P. vranovensis), and 11 Stenotrophomonas maltophilia isolates (Figure 2; Tables 2 and 3).
FIGURE 2.
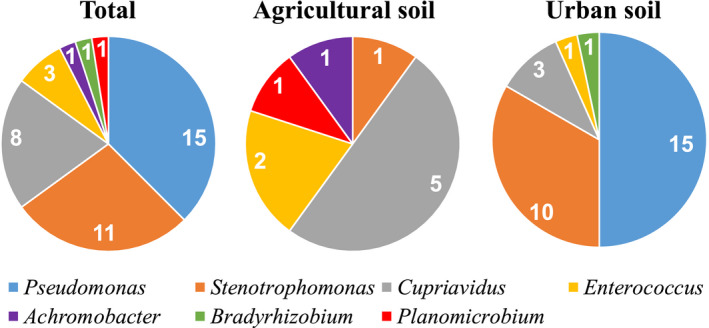
The abundance of the seven genera of carbapenem‐resistant isolates from soil identified in this study: Total abundance is shown on the left chart, abundance in agricultural soils is shown in the center chart, and abundance in urban soils is shown on the right chart
TABLE 3.
Carbapenem‐resistant soil isolates identified and characterized in this study
| Closest species identified by BLAST using 16S rRNA gene a | Isolate # | Inhibition zone (diameter in mm) b | Carbapenemase c | |||||
|---|---|---|---|---|---|---|---|---|
| MP | IM | CF | CI | GE | TE | |||
| Achromobacter marplatensis | S10‐1 | 14 | 33 | 14 | 30 | 21 | 27 | − |
| Bradyrhizobium elkanii | S11‐1 | 0 | 0 | 0 | 0 | 0 | 0 | + |
| Cupriavidus alkaliphilus | S2‐2 | 0 | 37 | 47 | 43 | 13 | 33 | − |
| Cupriavidus alkaliphilus | S2‐3 | 0 | 37 | 47 | 40 | 13 | 34 | − |
| Cupriavidus alkaliphilus | S2‐4 | 0 | 38 | 45 | 39 | 14 | 32 | − |
| Cupriavidus respiraculi | S7‐6 | 10 | 18 | 37 | 41 | 14 | 33 | − |
| Cupriavidus respiraculi | S8‐1 | 8 | 26 | 25 | 39 | 23 | 31 | − |
| Cupriavidus respiraculi | S8‐2 | 10 | 26 | 29 | 37 | 27 | 29 | − |
| Cupriavidus respiraculi | S9‐1 | 12 | 17 | 37 | 39 | 20 | 30 | − |
| Cupriavidus respiraculi | S9‐2 | 13 | 22 | 19 | 39 | 21 | 31 | − |
| Enterococcus durans | S12‐1 | 11 | 20 | 0 | 24 | 14 | 35 | − |
| Enterococcus gallinarum | S7‐2 | 0 | 51 | 17 | 31 | 27 | 35 | + |
| Enterococcus gallinarum | S11‐3 | 15 | 21 | 0 | 21 | 18 | 19 | − |
| Planomicrobium glaciei | S7‐3 | 0 | 0 | 15 | 34 | 0 | 14 | − |
| Pseudomonas alkylphenolica | S2‐1 | 0 | 36 | 45 | 40 | 14 | 33 | − |
| Pseudomonas putida | S11‐2 | 12 | 35 | 16 | 38 | 0 | 0 | + |
| Pseudomonas stutzeri | S4‐1 | 14 | 23 | 13 | 41 | 28 | 34 | − |
| Pseudomonas stutzeri | S4‐2 | 16 | 29 | 17 | 40 | 28 | 32 | − |
| Pseudomonas stutzeri | S4‐3 | 10 | 24 | 13 | 42 | 34 | 29 | − |
| Pseudomonas stutzeri | S5‐1 | 16 | 21 | 20 | 39 | 27 | 30 | − |
| Pseudomonas stutzeri | S5‐2 | 15 | 21 | 19 | 42 | 31 | 31 | − |
| Pseudomonas stutzeri | S5‐3 | 17 | 20 | 17 | 41 | 33 | 32 | − |
| Pseudomonas stutzeri | S5‐4 | 14 | 21 | 17 | 39 | 30 | 32 | − |
| Pseudomonas stutzeri | S5‐5 | 16 | 22 | 17 | 41 | 32 | 32 | − |
| Pseudomonas stutzeri | S5‐6 | 17 | 23 | 19 | 43 | 32 | 19 | − |
| Pseudomonas vranovensis | S3‐1 | 10 | 29 | 0 | 26 | 26 | 22 | + |
| Pseudomonas vranovensis | S3‐2 | 9 | 26 | 21 | 29 | 27 | 19 | + |
| Pseudomonas vranovensis | S3‐3 | 11 | 30 | 0 | 28 | 26 | 16 | − |
| Pseudomonas vranovensis | S3‐4 | 11 | 27 | 0 | 35 | 23 | 24 | − |
| Stenotrophomonas maltophilia | S1‐1 | 0 | 0 | 13 | 26 | 11 | 13 | + |
| Stenotrophomonas maltophilia | S1‐2 | 0 | 0 | 12 | 28 | 12 | 14 | + |
| Stenotrophomonas maltophilia | S1‐3‐1 | 0 | 0 | 20 | 23 | 10 | 14 | + |
| Stenotrophomonas maltophilia | S1‐3‐2 | 0 | 0 | 17 | 24 | 11 | 15 | + |
| Stenotrophomonas maltophilia | S1‐4 | 0 | 0 | 18 | 26 | 0 | 13 | + |
| Stenotrophomonas maltophilia | S1‐5 | 0 | 0 | 12 | 25 | 10 | 12 | + |
| Stenotrophomonas maltophilia | S1‐6 | 0 | 0 | 12 | 26 | 10 | 13 | + |
| Stenotrophomonas maltophilia | S1‐7 | 0 | 0 | 13 | 27 | 15 | 15 | + |
| Stenotrophomonas maltophilia | S7‐1 | 0 | 0 | 9 | 27 | 30 | 20 | + |
| Stenotrophomonas maltophilia | S11‐4 | 0 | 0 | 0 | 24 | 0 | 13 | + |
| Stenotrophomonas maltophilia | S11‐5 | 0 | 0 | 0 | 23 | 16 | 11 | + |
Abbreviations: CF, cefotaxime; CI, ciprofloxacin; GE, gentamicin; IM, imipenem; MP, meropenem; TE, tetracycline.
For each isolate, we obtained their 16S rRNA gene sequence and used BLAST (Altschul et al., 1997) to determine the closest known strain. In all cases, the DNA identity between our isolate and the top BLAST known strain hit was ≥98% (≥99% for 34 out of 40 isolates).
To determine whether our isolates were resistant (highlighted in red), intermediate (highlighted in yellow) or sensitive (no highlight) to the antibiotics tested, we used the CSLI zone diameter clinical breakpoint values (Clinical & Laboratory Standards Institute, 2018). For taxa in which the CLSI zone diameter breakpoint values were not available, we used the Enterobacteriaceae values. Enterococci are considered clinically resistant to aminoglycosides even if they test as susceptible in vitro (Clinical & Laboratory Standards Institute, 2018).
All carbapenemase‐producing isolates were carbapenemase‐positive when the CarbaNP test was performed measuring the hydrolysis of both meropenem and imipenem, and all were confirmed as positives using the mCIM test. Carbapenemase production was inducible on all carbapenemase‐producing isolates except for S. maltophilia isolates S1‐2 and S1‐3‐2.
Interestingly, the majority of the urban soil isolates belonged to the genera Pseudomonas and Stenotrophomonas, whereas the most represented agricultural soil isolates belonged to the genus Cupriavidus (Figure 2). Overall, we identified carbapenem‐resistant (CR) Pseudomonas in 5 (all urban soils) out the 11 samples analyzed; CR Stenotrophomonas maltophilia in 3 samples (2 urban and 1 agricultural soil); CR Cupriavidus in 1 urban and 3 agricultural soil samples; and CR Enterococcus in 3 samples (2 agricultural and 1 urban soil), whereas CR Achromobacter marplatensis, Bradyrhizobium elkanii, and Planomicrobium glaciei were identified only in one agricultural, urban, and agricultural soil samples, respectively (Figure 2; Table 2).
3.2. Characterization of the antibiotic susceptibility profile of CRB isolates
We next characterized the antibiotic susceptibility profile of the 40 identified CRB isolates using disk diffusion experiments with the two most clinically used carbapenems (meropenem and imipenem) and 4 noncarbapenem antibiotics (cefotaxime, ciprofloxacin, gentamicin, and tetracycline; Tables 2 and 3; and Figure 3). All 40 isolates were resistant to meropenem, confirming them as CRB. Moreover, most of the isolates were also resistant or intermediate to imipenem (55% of the isolates) and cefotaxime (83% of isolates), which although not a carbapenem, it is also a β‐lactam (third‐generation cephalosporin; Figure 3; Table 3). In contrast, the number of isolates that were resistant to the three different classes of non‐β‐lactam antibiotics tested was much lower. Overall, 43% and 28% of the CRB isolates characterized were resistant or intermediate to aminoglycoside gentamicin and tetracycline, respectively (Figure 3; Table 3). Furthermore, only one CRB isolate, identified as Bradyrhizobium elkanii, was resistant to the fluoroquinolone ciprofloxacin (Figure 3; Table 3). These findings highlight the importance of Southern California soils as reservoirs of CRB, including CRB that are also resistant to other antibiotics.
FIGURE 3.
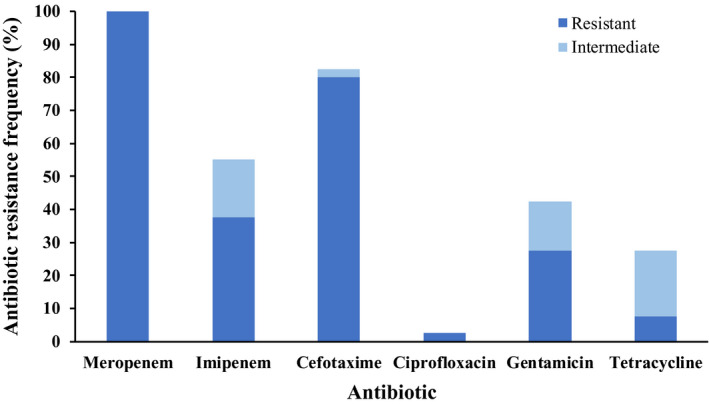
Antibiotic resistance frequency of the soil isolates characterized in this study for carbapenem (meropenem and imipenem) and noncarbapenem (cefotaxime, ciprofloxacin, gentamicin, and tetracycline) antibiotics. For each antibiotic tested, the percentage of resistant isolates is shown in dark blue, and the percentage of intermediate isolates is shown in light blue
3.3. Identification of CRB isolates that produce carbapenemases
Given the importance of carbapenemase genes in spreading resistance to carbapenems, we next used the CarbaNP test to determine which CRB isolates produce carbapenemases. Interestingly, 16 out of the 40 CRB isolates tested (40%) were positive for carbapenemase production when tested by the CarbaNP using both meropenem and imipenem, and as confirmed by the mCIM test (Tables 2 and 3). These carbapenemase‐positive isolates were 1 Bradyrhizobium elkanii, 1 E. gallinarum, 1 P. putida, 2 P. vranovensis, and all 11 S. maltophilia (Table 3). To our knowledge, this is the first report of carbapenemase production for E. gallinarum and P. vranovensis as well as in the genus Bradyrhizobium.
4. DISCUSSION
Carbapenem‐resistant bacteria are a major public health threat all over the world (Centers for Disease Control & Prevention, 2013a, 2013b; Cuzon et al., 2011; Guh et al., 2015). However, little is still known about the distribution and characteristics of CRB outside health care or immediately related settings (Gupta et al., 2011; Kallen et al., 2010; Khuntayaporn et al., 2012; Rhomberg & Jones, 2009; Ssekatawa et al., 2018). This gap in knowledge is especially significant in the United States, where only three specific studies about the prevalence of CRB in the environment, all three in freshwater, have been performed (Ash et al., 2002; Aubron et al., 2005; Harmon et al., 2019). CRB in the United States have also been found in fecal samples from dairy farms in New Mexico and Texas and a swine nursery in Ohio (Mollenkopf et al., 2017; Webb et al., 2016). Thus, not only clinical facilities but also farms may contribute to spread CRB to the environment. Recent findings in other parts of the world, especially in Europe, have found CRB in agricultural and nonagricultural soil samples (Borowiak et al., 2017; Djenadi et al., 2018; Fischer et al., 2013; Gudeta et al., 2016; Hrenovic et al., 2019) and suggest that soil may be an underrecognized reservoir of CRB. In the United States, 3 CRB isolates were identified among a collection of penicillin‐resistant isolates obtained from soil samples from the Midwestern United States (Crofts et al., 2018). However, studies that specifically address the distribution and characteristics of CRB in United States soils are still lacking.
The study reported here is significant for several reasons. To our knowledge, this is the first specific study about the distribution and characteristics of CRB in urban and agricultural environmental soils in the United States. Moreover, this is only the second study about the environmental distribution of CRB in the West Coast of the United States, the first one being our previous report on CRB in freshwater (Harmon et al., 2019). Here, we have found that CRB were present in all soil samples analyzed from both from urban and agricultural‐related locations in the Los Angeles–Southern California area. Furthermore, 40% of the CRB isolates characterized were carbapenemase producers (CR‐CRB), and all CRB isolates characterized were resistant or intermediate to at least one of the noncarbapenem antibiotics tested. For most urban soil samples as well as S12 (a hiking trail sample), the relative frequency of CRB compared to the total bacterial counts obtained was less than 1%, which is similar to the relative frequencies of CRB we had previously observed in freshwater environments from the Los Angeles–Southern California area (Harmon et al., 2019). In contrast, most agricultural soil samples (and the urban chicken coop soil samples) had a much higher relative frequency of CRB to the total bacterial count (from 18% up to 80% in soil S7, which was obtained adjacent to a produce farm). Although further studies comparing soil samples from locations at different proximities from farms are necessary, our results support the hypothesis that the use of antibiotics (or the use of manure from antibiotic‐treated animals) in farms might contribute to the spread of CRB to the environment (Mollenkopf et al., 2017; Webb et al., 2016), including CP‐CRB and CRB also resistant to other antibiotics.
In a previous study, Hrenovic et al. (2019) used a similar approach than the one we used in our study, but a different growth medium (CHROMagar™ Acinetobacter medium with CR102 supplement in their study, compared to MacConkey agar medium supplement with meropenem in our study) and temperature (37°C and 42°C in their study, compared to 37°C in our study) to determine the presence of CRB in different soils samples from Croatia. Hrenovic et al. (2019) found that at 37°C, most soil isolates were S. maltophilia, except for two soil samples in which they were absent. As is further discussed below, S. maltophilia are widespread in soil and other environments, and are intrinsically resistant to carbapenems (Brooke, 2012; Harmon et al., 2019; Tacão et al., 2015; Youenou et al., 2015). They also found that isolating CRB at 42°C, which suppresses the growth of S. maltophilia, increased the diversity of CRB recovered from their samples, including CRB of potential anthropogenic origin (Hrenovic et al., 2019). In the future, as we expand our studies to additional soil samples and locations, it will be interesting to analyze our samples at both 37°C and 42°C to compare the abundance and diversity of CRB obtained at both temperatures. However, of the 40 CRB isolates identified and characterized in the present study, only 11 of them (from 3 different soil samples) were S. maltophilia (Tables 2 and 3), and we were able to isolate, among other CRB, carbapenem‐resistant (CR) Cupriavidus, and Pseudomonas strains, as reported by Hrenovic et al. (2019) at 42°C. These findings suggest that, although S. maltophilia may be an important contributor to the abundance and wide distribution of CRB found in the soils we analyzed, other CRB were also an important factor. Moreover, although different soil locations were tested in both studies, our findings, as well as those from Djenadi et al. (2018), suggest that at 37°C, using MacConkey medium instead of CHROMagar might contribute to isolating more diverse CRB, even without using 42°C to suppress the growth of S. maltophilia. Also, CR Pseudomonas were the most abundant (15 out of 40 CRB identified in our study) CRB we found, compared to only one CR Pseudomonas isolate identified by Hrenovic et al. (2019) at 42°C. Although further studies analyzing the same soil samples with both growth media and temperatures are necessary, this finding suggests that isolation of CRB at 42°C may not only suppress the growth of S. maltophilia, but also of closely related Pseudomonas.
To further characterize the diversity of CRB present in the soils we studied, we identified 40 CRB soil isolates. Identification of these isolates revealed a diversity of species that included Achromobacter marplatensis, Bradyrhizobium elkanii, Cupriavidus alkaliphilus, Cupriavidus respiraculi, Enterococcus durans, Enterococcus gallinarum, Planomicrobium glaciei, Pseudomonas alkylphenolica, Pseudomonas putida, Pseudomonas stuzeri, Pseudomonas vranovensis, and Stenotrophomonas maltophilia (Table 3). Of the soil CRB characterized, Cupriavidus, Pseudomonas, and S. maltophilia isolates were the most abundant and widely distributed in soils from the Los Angeles area. Carbapenem‐resistant (CR) Pseudomonas and S. maltophilia isolates were also the most abundant CRB in freshwater samples from the same area (Harmon et al., 2019) and have been found before both in clinical settings and in soil, freshwater, animal feces, and other environments (Aubron et al., 2005; Brooke, 2012; Centers for Disease Control & Prevention, 2013b; Djenadi et al., 2018; Gudeta et al., 2016; Hrenovic et al., 2019; Tacão et al., 2015; Webb et al., 2016). However, this is to our knowledge the first report of carbapenem‐resistant P. alkylphenolica and P. vranovensis isolates. Resistance to carbapenems in Pseudomonas can occur by different mechanisms such as the production of different carbapenemases, overexpression of efflux pumps, and decreased outer membrane permeability (Papp‐Wallace et al., 2011; Rizek et al., 2014; Rodríguez‐Martínez et al., 2009). Interestingly, only 1 P. putida and 2 P. vranovensis out of the 15 Pseudomonas isolates characterized produced carbapenemases. In contrast, all S. maltophila isolates were carbapenemase producers. It is well‐documented that carbapenem resistance in S. maltophilia is predominantly caused by the bla L1 gene, which encodes for the intrinsic L1 carbapenemase in both clinical and environmental isolates (Brooke, 2012; Harmon et al., 2019; Tacão et al., 2015; Youenou et al., 2015). Using PCR, we could confirm that this carbapenemase gene was also present in all our S. maltophilia isolates (data not shown).
The third most abundant CR soil isolates obtained belonged to the genus Cupriavidus, which we identified in four different samples. Members of this genus are usually found in soil or water environments and occasionally as opportunistic pathogens (Coenye et al., 1999; Coenye, Goris, Spilker, Vandamme, & LiPuma, 2002; Harmon et al., 2019; Henriques et al., 2012; Hrenovic et al., 2019; Karafin et al., 2010; Kobayashi et al., 2016; Wang et al., 2015). However, carbapenem‐resistant C. alkaliphilus isolates have not been reported before in either clinical or environmental samples. None of the soil CR Cupriavidus isolates characterized in this study produced carbapenemases. A related species, C. gilardii, is intrinsically resistant to carbapenems despite also not producing carbapenemases likely because of its large array of multidrug efflux pumps (Ruiz, McCarley, Espejo, Cooper, & Harmon, 2019).
Other notable but less abundant CR soil isolates included Enterococcus gallinarum, which is associated with nosocomial‐ and community‐acquired bacteremia and other infections (Narciso‐Schiavon et al., 2015; Quinones, Goni, Rubio, Duran, & Gomez‐Lus, 2005; Reid, Cockerill, & Patel, 2001; Schouten, Voss, & Hoogkamp‐Korstanje, 1999); Enterococcus durans, an infrequent human pathogen mostly associated with diarrhea in piglets and calves (Cheon & Chae, 1996; Quinones et al., 2005; Rogers, Zeman, & Erickson, 1992; Schouten et al., 1999); Achromobacter marplatensis, a soil microbe that has also been found in cystic fibrosis patients (Gomila et al., 2011; Papalia et al., 2019); Bradyrhizobium elkanii, a soil bacterium and legume symbiont used commercially as an inoculant to improve the growth of legume plants (Crovadore et al., 2016; Faruque et al., 2015; Hungria, Delamuta, Ribeiro, & Nogueira, 2019); and Planomicrobium glaciei, an infrequently isolated bacterium, first found in a glacier and later in food (Tshipamba, Lubanza, Adetunji, & Mwanza, 2018; Zhang et al., 2009). Finding these isolates is significant for several reasons. First, neither resistance to carbapenems in these species nor the production of carbapenemases found in one E. gallinarum isolate and the B. elkanii isolate has been reported before. Given that E. gallinarum can cause infections in humans, including bacteremia, resistance to carbapenems and carbapenemase production in this species may impact therapy directly or by the transmission of the carbapenemase gene to other pathogens. In the case of B. elkanii, although it is not known to infect humans or animals, the fact that this isolate was completely resistant (0 mm inhibition zone diameters) to all carbapenem and noncarbapenem antibiotics tested makes this bacterium a potential reservoir of multiple antibiotic resistance genes. Further genomic studies are necessary to fully characterize this isolate and determine whether its antibiotic resistance determinants are conserved among other B. elkanii isolates and whether these determinants are located in mobile elements that may facilitate their transmission to other bacteria. However, the role of this species as a potential reservoir of resistance genes should be taken into account when considering its commercial use in crops.
5. CONCLUSIONS
In conclusion, our findings show for the first time that soils from the Los Angeles–Southern California area are a previously underappreciated reservoir of different species of CRB that are also resistant to other antibiotics, including carbapenemase‐producing CRB. Our study also shows a much higher relative frequency of CRB on most soils from locations adjacent to farms, compared to most soils from urban locations, which suggest a potential role of farms in spreading bacteria resistant to carbapenems and other antibiotics.
CONFLICTS OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Nicolas V. Lopez: Conceptualization (supporting); Data curation (lead); Formal analysis (equal); Investigation (lead); Methodology (equal). Cameron J. Farsar: Data curation (supporting); Formal analysis (supporting); Investigation (supporting). Dana E. Harmon: Conceptualization (supporting); Data curation (supporting); Formal analysis (supporting); Investigation (supporting); Methodology (supporting); Supervision (supporting). Cristian Ruiz: Conceptualization (lead); Data curation (supporting); Formal analysis (equal); Funding acquisition (lead); Investigation (supporting); Methodology (equal); Project administration (lead); Supervision (lead); Writing‐original draft (equal); Writing‐review & editing (lead).
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
N.V. Lopez was supported by the National Institutes of Health Research Initiative for Scientific Enhancement (NIH‐RISE, grant number 5R25GM063787‐16) program at California State University Northridge. This work was supported by the California State University Northridge (CSUN) start‐up funds and the CSUN Research, Scholarship and Creative Activity Award grant to C. Ruiz.
APPENDIX 1.
FIGURE A1.
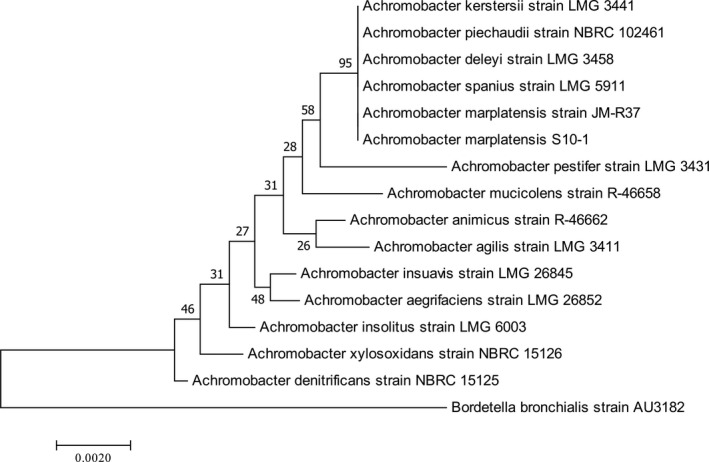
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between the Achromobacter isolate from this study and Achromobacter isolates from previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A2.
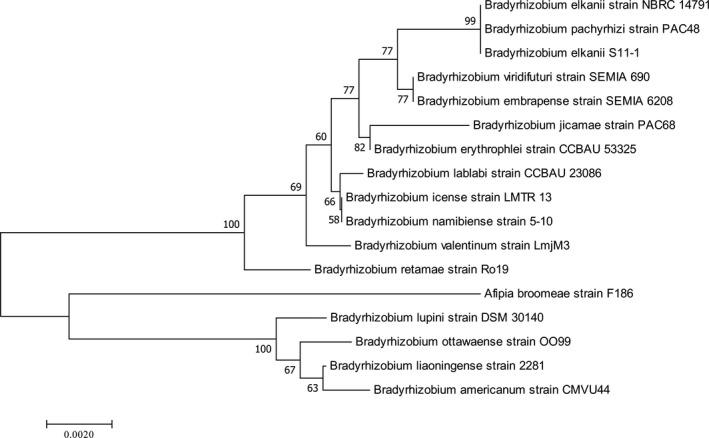
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between the Bradyrhizobium isolate from this study and Bradyrhizobium isolates from previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A3.
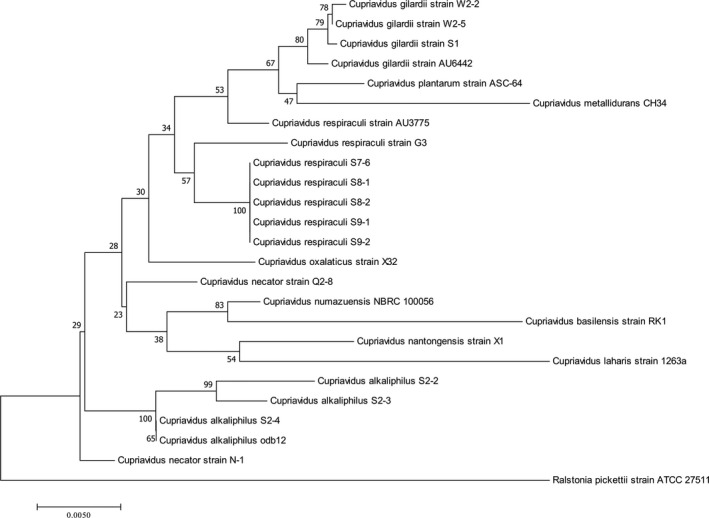
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between Cupriavidus isolates from this and previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A4.
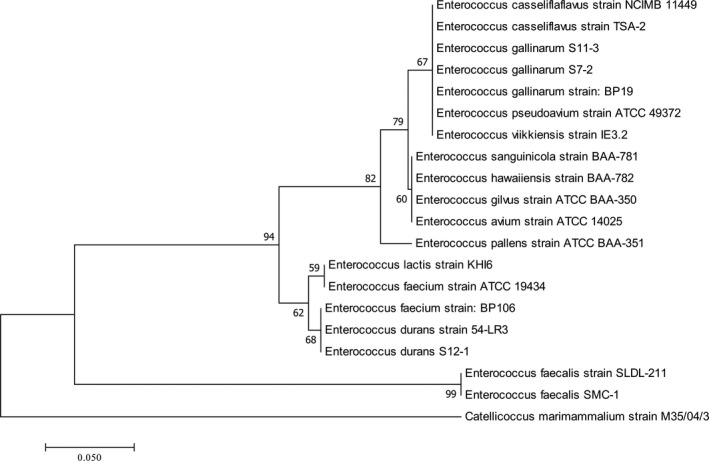
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between Enterococcus isolates from this and previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A5.
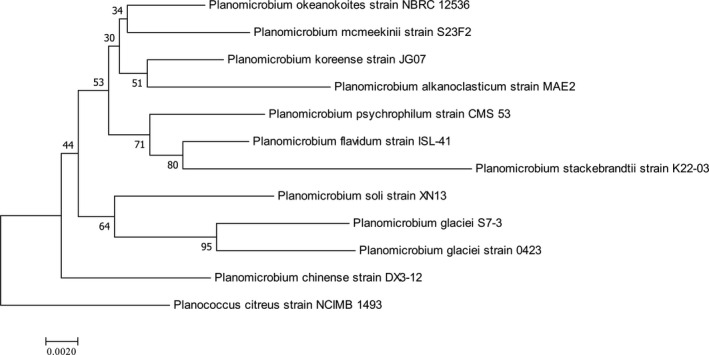
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between the Planomicrobium isolate from this study and Planomicrobium isolates from previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A6.
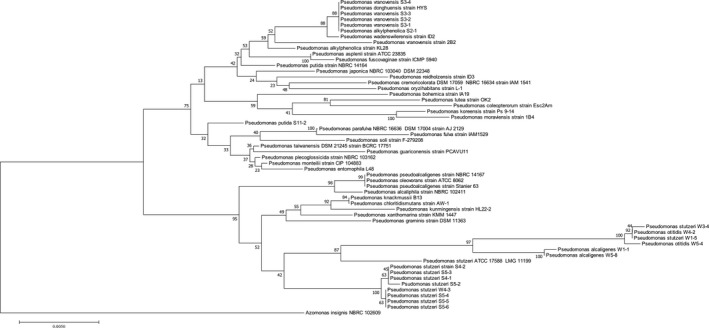
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between Pseudomonas isolates from this and previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
FIGURE A7.
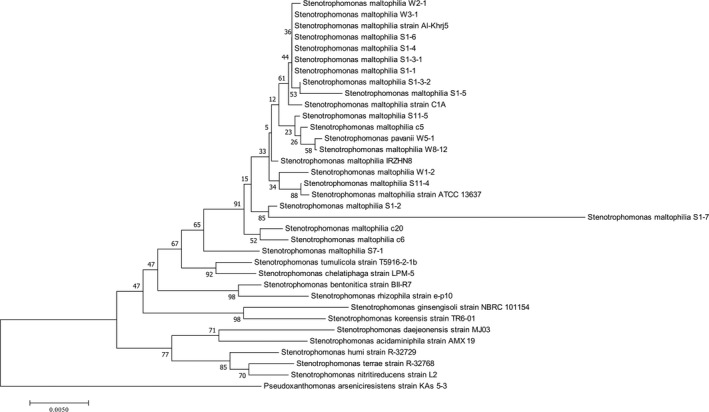
Phylogenetic tree constructed using 16S rRNA gene sequences showing the relatedness between Stenotrophomonas isolates from this and previous studies. The scale bar at the bottom represents the number of nucleotide substitutions per site
Lopez NV, Farsar CJ, Harmon DE, Ruiz C. Urban and agricultural soils in Southern California are a reservoir of carbapenem‐resistant bacteria. MicrobiologyOpen. 2020;9:e1034 10.1002/mbo3.1034
DATA AVAILABILITY STATEMENT
All 16S rRNA gene sequences obtained in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: MN732973–MN733008, MN810328–MN810330, and MN813762.
REFERENCES
- Adelowo, O. O. , Vollmers, J. , Mäusezahl, I. , Kaster, A.‐K. , & Müller, J. A. (2018). Detection of the carbapenemase gene blaVIM‐5 in members of the Pseudomonas putida group isolated from polluted Nigerian wetlands. Scientific Reports, 8(1), 15116 10.1038/s41598-018-33535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F. , Madden, T. L. , Schäffer, A. A. , Zhang, J. , Zhang, Z. , Miller, W. , & Lipman, D. J. (1997). Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash, R. J. , Mauck, B. , & Morgan, M. (2002). Antibiotic resistance of gram‐negative bacteria in rivers, United States. Emerging Infectious Diseases, 8(7), 713–716. 10.3201/eid0807.010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubron, C. , Poirel, L. , Ash, R. J. , & Nordmann, P. (2005). Carbapenemase‐producing Enterobacteriaceae, U.S. rivers. Emerging Infectious Diseases, 11(2), 260–264. 10.3201/eid1102.030684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey, D. H. , & Holt, J. G. (1994). Bergey's manual of determinative bacteriology (9th ed.). Baltimore, MD:Williams & Wilkins. [Google Scholar]
- Borowiak, M. , Fischer, J. , Hammerl, J. A. , Hendriksen, R. S. , Szabo, I. , & Malorny, B. (2017). Identification of a novel transposon‐associated phosphoethanolamine transferase gene, mcr‐5, conferring colistin resistance in d‐tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. Journal of Antimicrobial Chemotherapy, 72(12), 3317–3324. 10.1093/jac/dkx327 [DOI] [PubMed] [Google Scholar]
- Bradley, J. S. , Garau, J. , Lode, H. , Rolston, K. V. , Wilson, S. E. , & Quinn, J. P. (1999). Carbapenems in clinical practice: a guide to their use in serious infection. International Journal of Antimicrobial Agents, 11(2), 93–100. [DOI] [PubMed] [Google Scholar]
- Brooke, J. S. (2012). Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clinical Microbiology Reviews, 25(1), 2–41. 10.1128/CMR.00019-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013a). Vital Signs: Carbapenem‐Resistant Enterobacteriaceae. Morbidity and Mortality Weekly Report, 62(9), 165–170. [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2013b). Antibiotic resistance threats in the United States. Retrieved from http://www.cdc.gov/drugresistance/threat‐report‐2013/pdf/ar‐threats‐2013‐508.pdf. [Google Scholar]
- Cheon, D. S. , & Chae, C. (1996). Outbreak of diarrhea associated with Enterococcus durans in piglets. Journal of Veterinary Diagnostic Investigation, 8(1), 123–124. 10.1177/104063879600800123 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (2018). Performance standards for antimicrobial susceptibility testing (28th ed.). CLSI Supplement M100, Wayne, PA. [Google Scholar]
- Coenye, T. , Falsen, E. , Vancanneyt, M. , Hoste, B. , Govan, J. R. W. , Kersters, K. , & Vandamme, P. (1999). Classification of Alcaligenes faecalis‐like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. International Journal of Systematic Bacteriology, 49(Pt 2), 405–413. 10.1099/00207713-49-2-405 [DOI] [PubMed] [Google Scholar]
- Coenye, T. , Goris, J. , Spilker, T. , Vandamme, P. , & LiPuma, J. J. (2002). Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. Journal of Clinical Microbiology, 40(6), 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts, T. S. , Wang, B. , Spivak, A. , Gianoulis, T. A. , Forsberg, K. J. , Gibson, M. K. , … Dantas, G. (2018). Shared strategies for beta‐lactam catabolism in the soil microbiome. Nature Chemical Biology, 14(6), 556–564. 10.1038/s41589-018-0052-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crovadore, J. , Calmin, G. , Chablais, R. , Cochard, B. , Schulz, T. , & Lefort, F. (2016). Whole‐Genome sequence of Bradyrhizobium elkanii Strain UASWS1016, a potential symbiotic biofertilizer for agriculture. Genome Announcements, 4(5), e01095‐16 10.1128/genomeA.01095-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzon, G. , Naas, T. , Villegas, M. V. , Correa, A. , Quinn, J. P. , & Nordmann, P. (2011). Wide dissemination of Pseudomonas aeruginosa producing beta‐lactamase bla KPC‐2 gene in Colombia. Antimicrobial Agents and Chemotherapy, 55(11), 5350–5353. 10.1128/AAC.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, D. Y. , Jang, J. , Unno, T. , & Hur, H. G. (2017). Emergence of Klebsiella variicola positive for NDM‐9, a variant of New Delhi metallo‐beta‐lactamase, in an urban river in South Korea. Journal of Antimicrobial Chemotherapy, 72(4), 1063–1067. 10.1093/jac/dkw547 [DOI] [PubMed] [Google Scholar]
- Djenadi, K. , Zhang, L. , Murray, A. K. , & Gaze, W. H. (2018). Carbapenem resistance in bacteria isolated from soil and water environments in Algeria. Journal of Global Antimicrobial Resistance, 15, 262–267. 10.1016/j.jgar.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Dortet, L. , Poirel, L. , & Nordmann, P. (2012a). Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrobial Agents and Chemotherapy, 56(12), 6437–6440. 10.1128/AAC.01395-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet, L. , Poirel, L. , & Nordmann, P. (2012b). Rapid detection of carbapenemase‐producing Pseudomonas spp. Journal of Clinical Microbiology, 50(11), 3773–3776. 10.1128/JCM.01597-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruque, O. M. , Miwa, H. , Yasuda, M. , Fujii, Y. , Kaneko, T. , Sato, S. , & Okazaki, S. (2015). Identification of Bradyrhizobium elkanii Genes Involved in Incompatibility with Soybean Plants Carrying the Rj4 Allele. Applied and Environment Microbiology, 81(19), 6710–6717. 10.1128/AEM.01942-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, J. , Rodríguez, I. , Schmoger, S. , Friese, A. , Roesler, U. , Helmuth, R. , & Guerra, B. (2013). Salmonella enterica subsp. enterica producing VIM‐1 carbapenemase isolated from livestock farms. Journal of Antimicrobial Chemotherapy, 68(2), 478–480. 10.1093/jac/dks393 [DOI] [PubMed] [Google Scholar]
- Girlich, D. , Poirel, L. , & Nordmann, P. (2010). Novel ambler class A carbapenem‐hydrolyzing beta‐lactamase from a Pseudomonas fluorescens isolate from the Seine River, Paris, France. Antimicrobial Agents and Chemotherapy, 54(1), 328–332. 10.1128/AAC.00961-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila, M. , Tvrzova, L. , Teshim, A. , Sedlacek, I. , Gonzalez‐Escalona, N. , Zdrahal, Z. , … Murialdo, S. E. (2011). Achromobacter marplatensis sp. nov., isolated from a pentachlorophenol‐contaminated soil. International Journal of Systematic and Evolutionary Microbiology, 61(Pt 9), 2231–2237. 10.1099/ijs.0.025304-0 [DOI] [PubMed] [Google Scholar]
- Gudeta, D. D. , Bortolaia, V. , Amos, G. , Wellington, E. M. , Brandt, K. K. , Poirel, L. , … Guardabassi, L. (2016). The soil microbiota harbors a diversity of carbapenem‐hydrolyzing beta‐lactamases of potential clinical relevance. Antimicrobial Agents and Chemotherapy, 60(1), 151–160. 10.1128/AAC.01424-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guh, A. Y. , Bulens, S. N. , Mu, Y. I. , Jacob, J. T. , Reno, J. , Scott, J. , … Kallen, A. J. (2015). Epidemiology of carbapenem‐resistant Enterobacteriaceae in 7 US Communities, 2012–2013. JAMA, 314(14), 1479–1487. 10.1001/jama.2015.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N. , Limbago, B. M. , Patel, J. B. , & Kallen, A. J. (2011). Carbapenem‐resistant Enterobacteriaceae: Epidemiology and prevention. Clinical Infectious Diseases, 53(1), 60–67. 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Molecular Biology and Evolution, 30(5), 1229–1235. 10.1093/molbev/mst012 [DOI] [PubMed] [Google Scholar]
- Harmon, D. E. , Miranda, O. A. , McCarley, A. , Eshaghian, M. , Carlson, N. , & Ruiz, C. (2019). Prevalence and characterization of carbapenem‐resistant bacteria in water bodies in the Los Angeles‐Southern California area. MicrobiologyOpen, 8(4), e00692 10.1002/mbo3.692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques, I. S. , Araújo, S. , Azevedo, J. S. N. , Alves, M. S. , Chouchani, C. , Pereira, A. , & Correia, A. (2012). Prevalence and diversity of carbapenem‐resistant bacteria in untreated drinking water in Portugal. Microbial Drug Resistance, 18(5), 531–537. 10.1089/mdr.2012.0029 [DOI] [PubMed] [Google Scholar]
- Hrenovic, J. , Ivankovic, T. , Durn, G. , Dekic, S. , Kazazic, S. , & Kisic, I. (2019). Presence of carbapenem‐resistant bacteria in soils affected by illegal waste dumps. International Journal of Environmental Health Research, 29(2), 154–163. 10.1080/09603123.2018.1522423 [DOI] [PubMed] [Google Scholar]
- Hungria, M. , Delamuta, J. R. M. , Ribeiro, R. A. , & Nogueira, M. A. (2019). Draft Genome Sequence of Bradyrhizobium elkanii Strain SEMIA 938, used in commercial inoculants for Lupinus spp. in Brazil. Microbiology Resource Announcements, 8(28), e00546‐19 10.1128/MRA.00546-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isozumi, R. , Yoshimatsu, K. , Yamashiro, T. , Hasebe, F. , Nguyen, B. M. , Ngo, T. C. , … Arikawa, J. (2012). bla (NDM‐1)‐positive Klebsiella pneumoniae from environment, Vietnam. Emerging Infectious Diseases, 18(8), 1383–1385. 10.3201/eid1808.111816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen, A. J. , Hidron, A. I. , Patel, J. , & Srinivasan, A. (2010). Multidrug resistance among gram‐negative pathogens that caused healthcare‐associated infections reported to the National Healthcare Safety Network, 2006–2008. Infection Control and Hospital Epidemiology, 31(5), 528–531. 10.1086/652152 [DOI] [PubMed] [Google Scholar]
- Karafin, M. , Romagnoli, M. , Fink, D. L. , Howard, T. , Rau, R. , Milstone, A. M. , & Carroll, K. C. (2010). Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. Journal of Clinical Microbiology, 48(3), 1005–1007. 10.1128/JCM.01482-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuntayaporn, P. , Montakantikul, P. , Mootsikapun, P. , Thamlikitkul, V. , & Chomnawang, M. T. (2012). Prevalence and genotypic relatedness of carbapenem resistance among multidrug‐resistant P. aeruginosa in tertiary hospitals across Thailand. Annals of Clinical Microbiology and Antimicrobials, 11, 25 10.1186/1476-0711-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T. , Nakamura, I. , Fujita, H. , Tsukimori, A. , Sato, A. , Fukushima, S. , … Matsumoto, T. (2016). First case report of infection due to Cupriavidus gilardii in a patient without immunodeficiency: A case report. BMC Infectious Diseases, 16, 493 10.1186/s12879-016-1838-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore, D. M. , Mushtaq, S. , & Warner, M. (2005). Selectivity of ertapenem for Pseudomonas aeruginosa mutants cross‐resistant to other carbapenems. Journal of Antimicrobial Chemotherapy, 55(3), 306–311. 10.1093/jac/dki009 [DOI] [PubMed] [Google Scholar]
- Marsik, F. J. , & Nambiar, S. (2011). Review of carbapenemases and AmpC‐beta lactamases. The Pediatric Infectious Disease Journal, 30(12), 1094–1095. 10.1097/INF.0b013e31823c0e47 [DOI] [PubMed] [Google Scholar]
- Mathers, A. J. , Cox, H. L. , Kitchel, B. , Bonatti, H. , Brassinga, A. K. C. , Carroll, J. , … Sifri, C. D. (2011). Molecular dissection of an outbreak of carbapenem‐resistant enterobacteriaceae reveals intergenus KPC carbapenemase transmission through a promiscuous plasmid. MBio, 2(6), e00204‐11 10.1128/mBio.00204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis, G. (2016). Carbapenem resistance: Overview of the problem and future perspectives. Therapeutic Advances in Infectious Disease, 3(1), 15–21. 10.1177/2049936115621709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, M. C. , & Lee, J. (2019). The threat of carbapenem‐resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environmental Pollution, 255(Pt 1), 113143 10.1016/j.envpol.2019.113143 [DOI] [PubMed] [Google Scholar]
- Mollenkopf, D. F. , Stull, J. W. , Mathys, D. A. , Bowman, A. S. , Feicht, S. M. , Grooters, S. V. , … Wittum, T. E. (2017). Carbapenemase‐producing Enterobacteriaceae Recovered from the environment of a swine Farrow‐to‐Finish operation in the United States. Antimicrobial Agents and Chemotherapy, 61(2), e01298‐16 10.1128/AAC.01298-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso‐Schiavon, J. L. , Borgonovo, A. , Marques, P. C. , Tonon, D. , Bansho, E. T. O. , Maggi, D. C. , … Schiavon, L. D. L. (2015). Enterococcus casseliflavus and Enterococcus gallinarum as causative agents of spontaneous bacterial peritonitis. Annals of Hepatology, 14(2), 270–272. [PubMed] [Google Scholar]
- Nordmann, P. , Poirel, L. , & Dortet, L. (2012). Rapid detection of carbapenemase‐producing Enterobacteriaceae. Emerging Infectious Diseases, 18(9), 1503–1507. 10.3201/eid1809.120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalia, M. , Steffanowski, C. , Traglia, G. , Almuzara, M. , Martina, P. , Galanternik, L. , … Radice, M. (2019). Diversity of Achromobacter species recovered from patients with cystic fibrosis, in Argentina. Revista Argentina De Microbiología, 52(1), 13–18. 10.1016/j.ram.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Papp‐Wallace, K. M. , Endimiani, A. , Taracila, M. A. , & Bonomo, R. A. (2011). Carbapenems: Past, present, and future. Antimicrobial Agents and Chemotherapy, 55(11), 4943–4960. 10.1128/AAC.00296-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, D. L. (2000). Recommendation for treatment of severe infections caused by Enterobacteriaceae producing extended‐spectrum beta‐lactamases (ESBLs). Clinical Microbiology and Infection, 6(9), 460–463. [DOI] [PubMed] [Google Scholar]
- Pierce, V. M. , Simner, P. J. , Lonsway, D. R. , Roe‐Carpenter, D. E. , Johnson, J. K. , Brasso, W. B. , … Das, S. (2017). Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. Journal of Clinical Microbiology, 55(8), 2321–2333. 10.1128/JCM.00193-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, L. , Barbosa‐Vasconcelos, A. , Simões, R. R. , Da Costa, P. M. , Liu, W. , & Nordmann, P. (2012). Environmental KPC‐producing Escherichia coli isolates in Portugal. Antimicrobial Agents and Chemotherapy, 56(3), 1662–1663. 10.1128/AAC.05850-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron, A. , Poirel, L. , Bussy, F. , & Nordmann, P. (2011). Occurrence of the carbapenem‐hydrolyzing beta‐lactamase gene bla OXA‐48 in the environment in Morocco. Antimicrobial Agents and Chemotherapy, 55(11), 5413–5414. 10.1128/AAC.05120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan, A. M. , & Bush, K. (2007). Carbapenemases: the versatile beta‐lactamases. Clinical Microbiology Reviews, 20(3), 440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones, D. , Goni, P. , Rubio, M. C. , Duran, E. , & Gomez‐Lus, R. (2005). Enterococci spp. isolated from Cuba: Species frequency of occurrence and antimicrobial susceptibility profile. Diagnostic Microbiology and Infectious Disease, 51(1), 63–67. 10.1016/j.diagmicrobio.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Reid, K. C. , Cockerill, I. F. , & Patel, R. (2001). Clinical and epidemiological features of Enterococcus casseliflavus/flavescens and Enterococcus gallinarum bacteremia: A report of 20 cases. Clinical Infectious Diseases, 32(11), 1540–1546. 10.1086/320542 [DOI] [PubMed] [Google Scholar]
- Rhomberg, P. R. , & Jones, R. N. (2009). Summary trends for the Meropenem yearly susceptibility test information collection program: A 10‐year experience in the United States (1999–2008). Diagnostic Microbiology and Infectious Disease, 65(4), 414–426. 10.1016/j.diagmicrobio.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Rizek, C. , Fu, L. , dos Santos, L. C. , Leite, G. , Ramos, J. , Rossi, F. , … Costa, S. F. (2014). Characterization of carbapenem‐resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Annals of Clinical Microbiology and Antimicrobials, 13, 43 10.1186/s12941-014-0043-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Martínez, J. M. , Poirel, L. , & Nordmann, P. (2009). Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa . Antimicrobial Agents and Chemotherapy, 53(11), 4783–4788. 10.1128/AAC.00574-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, D. G. , Zeman, D. H. , & Erickson, E. D. (1992). Diarrhea associated with Enterococcus durans in calves. Journal of Veterinary Diagnostic Investigation, 4(4), 471–472. 10.1177/104063879200400423 [DOI] [PubMed] [Google Scholar]
- Ruiz, C. , McCarley, A. , Espejo, M. L. , Cooper, K. K. , & Harmon, D. E. (2019). Comparative genomics reveals a well‐conserved intrinsic resistome in the emerging multidrug‐resistant pathogen Cupriavidus gilardii . mSphere, 4(5), e00631‐19 10.1128/mSphere.00631-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten, M. A. , Voss, A. , & Hoogkamp‐Korstanje, J. A. (1999). Antimicrobial susceptibility patterns of Enterococci causing infections in Europe. The European VRE Study Group. Antimicrobial Agents and Chemotherapy, 43(10), 2542–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S. Y. , Bae, I. K. , Kim, J. , Jeong, S. H. , Yong, D. , Kim, J. M. , & Lee, K. (2012). Resistance to carbapenems in sequence type 11 Klebsiella pneumoniae is related to DHA‐1 and loss of OmpK35 and/or OmpK36. Journal of Medical Microbiology, 61(Pt 2), 239–245. 10.1099/jmm.0.037036-0 [DOI] [PubMed] [Google Scholar]
- Sivalingam, P. , Pote, J. , & Prabakar, K. (2019). Environmental prevalence of carbapenem resistance Enterobacteriaceae (CRE) in a tropical ecosystem in India: Human health perspectives and future directives. Pathogens, 8(4), 174 10.3390/pathogens8040174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ssekatawa, K. , Byarugaba, D. K. , Wampande, E. , & Ejobi, F. (2018). A systematic review: the current status of carbapenem resistance in East Africa. BMC Research Notes, 11(1), 629 10.1186/s13104-018-3738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacão, M. , Correia, A. , & Henriques, I. S. (2015). Low prevalence of carbapenem‐resistant bacteria in river water: Resistance is mostly related to intrinsic mechanisms. Microbial Drug Resistance, 21(5), 497–506. 10.1089/mdr.2015.0072 [DOI] [PubMed] [Google Scholar]
- Tshipamba, M. E. , Lubanza, N. , Adetunji, M. C. , & Mwanza, M. (2018). Molecular characterization and antibiotic resistance of foodborne pathogens in street‐vended ready‐to‐eat meat sold in South Africa. Journal of Food Protection, 81(12), 1963–1972. 10.4315/0362-028X.JFP-18-069 [DOI] [PubMed] [Google Scholar]
- United States Food & Drug Administration Center for Veterinary Medicine (2017. ). 2017 Summary report on antimicrobials sold or distributed for use in food‐producing animals. Retrieved from https://www.fda.gov/media/119332/download [Google Scholar]
- Vardakas, K. Z. , Tansarli, G. S. , Rafailidis, P. I. , & Falagas, M. E. (2012). Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended‐spectrum β‐lactamases: A systematic review and meta‐analysis. Journal of Antimicrobial Chemotherapy, 67(12), 2793–2803. 10.1093/jac/dks301 [DOI] [PubMed] [Google Scholar]
- Walsh, T. R. (2010). Emerging carbapenemases: A global perspective. International Journal of Antimicrobial Agents, 36(Suppl. 3), S8–14. 10.1016/S0924-8579(10)70004-2 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Chen, M. , Xiao, J. , Hao, L. , Crowley, D. E. , Zhang, Z. , … Wu, J. (2015). Genome sequence analysis of the naphthenic acid degrading and metal resistant bacterium Cupriavidus gilardii CR3. PLoS ONE, 10(8), e0132881 10.1371/journal.pone.0132881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, D. M. , Yang, Q. , Duval, V. , Chen, M. , Xu, Y. , & Levy, S. B. (2013). Involvement of MarR and YedS in carbapenem resistance in a clinical isolate of Escherichia coli from China. Antimicrobial Agents and Chemotherapy, 57(4), 1935–1937. 10.1128/AAC.02445-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, H. E. , Bugarel, M. , den Bakker, H. C. , Nightingale, K. K. , Granier, S. A. , Scott, H. M. , & Loneragan, G. H. (2016). Carbapenem‐resistant bacteria recovered from faeces of dairy cattle in the high plains region of the USA. PLoS ONE, 11(1), e0147363 10.1371/journal.pone.0147363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youenou, B. , Favre‐Bonté, S. , Bodilis, J. , Brothier, E. , Dubost, A. , Muller, D. , & Nazaret, S. (2015). Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strains with different antibiotic resistance profiles. Genome Biology and Evolution, 7(9), 2484–2505. 10.1093/gbe/evv161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. C. , Liu, H. C. , Xin, Y. H. , Yu, Y. , Zhou, P. J. , & Zhou, Y. G. (2009). Planomicrobium glaciei sp. nov., a psychrotolerant bacterium isolated from a glacier. International Journal of Systematic and Evolutionary Microbiology, 59(Pt 6), 1387–1390. 10.1099/ijs.0.002592-0 [DOI] [PubMed] [Google Scholar]
- Zou, H. , Berglund, B. , Xu, H. , Chi, X. , Zhao, Q. , Zhou, Z. , … Zheng, B. (2020). Genetic characterization and virulence of a carbapenem‐resistant Raoultella ornithinolytica isolated from well water carrying a novel megaplasmid containing blaNDM‐1. Environmental Pollution, 260, 114041 10.1016/j.envpol.2020.114041 [DOI] [PubMed] [Google Scholar]
- Zurfluh, K. , Hachler, H. , Nuesch‐Inderbinen, M. , & Stephan, R. (2013). Characteristics of extended‐spectrum beta‐lactamase‐ and carbapenemase‐producing Enterobacteriaceae Isolates from rivers and lakes in Switzerland. Applied and Environment Microbiology, 79(9), 3021–3026. 10.1128/AEM.00054-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All 16S rRNA gene sequences obtained in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: MN732973–MN733008, MN810328–MN810330, and MN813762.


