Abstract
DNA-encoded libraries of small molecules are being explored extensively for the identification of binders in early drug-discovery efforts. Combinatorial syntheses of such libraries require water- and DNA-compatible reactions, and the paucity of these reactions currently limit the chemical features of resulting barcoded products. The present work introduces strain-promoted cycloadditions of cyclic allenes under mild conditions to DNA-encoded library synthesis. Owing to distinct cycloaddition modes of these reactive intermediates with activated olefins, 1,3-dipoles, and dienes, the process generates diverse molecular architectures from a single precursor. The resulting DNA-barcoded compounds exhibit unprecedented ring and topographic features, related to elements found to be powerful in phenotypic screening.
Graphical Abstract
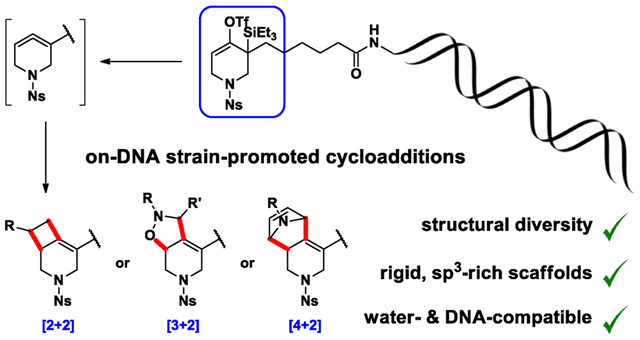
INTRODUCTION
The identification of small molecules that bind biological macromolecules is a key step in early drug discovery. Target-directed, binding-based approaches include high-throughput, fragment, and in silico screening. While each of these techniques has had a significant impact on successful drug development programs, they also have shortcomings. An additional approach uses DNA-encoded libraries (DELs), which comprise collections of compounds individually barcoded with DNA sequences that report on the synthetic reactions leading to their formation.1 DELs are typically prepared by split-and-pool synthesis from central scaffolds and readily available building blocks as appendages. DEL screens are commonly performed using immobilized proteins, and barcode enrichment, as a surrogate for binding, is determined by next-generation sequencing of PCR-amplified DNA. DELs are a promising source of hit compounds with several examples having advanced to clinical candidates.2 Beyond affinity-based screens, recent work relying on spatial separation of individual library members by microfluidics hint that DELs may become amenable to activity-based screens.3,4 As for all hit-finding approaches, compound libraries spanning diverse chemical space with features well-suited for binding are considered most promising,5 especially in the absence of known target binders.
Diversity-oriented synthesis (DOS) has been particularly successful in generating structure-diverse, stereochemistry-rich libraries that include many distinct, rigid ring skeletons that can reduce the entropic cost of binding and effectively display appendages in three-dimensional space.6 Often fueled by advancements in reaction methodology, the application of DOS principles has delivered numerous chemical probes and clinical candidates.7 Merging the logic of DOS with DNA-barcoding holds great promise for the identification of protein binders that can function by novel mechanisms of action.8
The conventional solution-phase synthesis of DELs relies on reactions that tolerate water (to keep DNA in solution) and maintain barcode integrity.9,10 These constraints have limited the range of transformations applicable to DEL construction and led to an enrichment of sp2-rich structures and peptidomimetics in published libraries. In response, recent work has expanded the toolbox of DEL chemists by identifying DNA-compatible conditions for established off-DNA reactions. Examples for such efforts include decarboxylative radical additions to Michael acceptors,11,12 Ullmann-type N-arylations,13 maleimide Diels–Alder reactions14a and intramolecular nitrone cycloadditions,14b and Ni/Ir dual catalytic alkylations of aryl halides.15 The Brunschweiger group has reported on DNA-compatible micellar catalysis16 and introduced an approach in the solid phase toward hexathymidine-conjugated heterocycles.17,18 In an alternative approach, two recent reports describe DNA-immobilization on quaternary ammonium resins to enable reactions under near anhydrous conditions including decarboxylative sp2-sp3 cross couplings, electrochemical aminations of aryl iodides, reductive aminations of ketones, as well as copper-mediated formation of heterocycles (tin amine protocol, SnAP).19,20
Strain-promoted reactions have found widespread application in chemical biology. Prominent examples are the copperfree [3 + 2] cycloaddition of cyclooctyne-derivatives with organic azides21,22 and the inverse-electron-demand Diels–Alder reaction of trans-cyclooctene with tetrazines (Figure 1A).23,24 Having significantly advanced the field of bioconjugation chemistry, these and related reactions are compatible with water and biopolymers by necessity.25 With only one recent report on inverse-electron-demand Diels–Alder reactions26 and reactions of cyclooctyne-DNA conjugates in the solid phase,27,28 strain-promoted reactivity has not yet found general application in the context of DELs.
Figure 1.
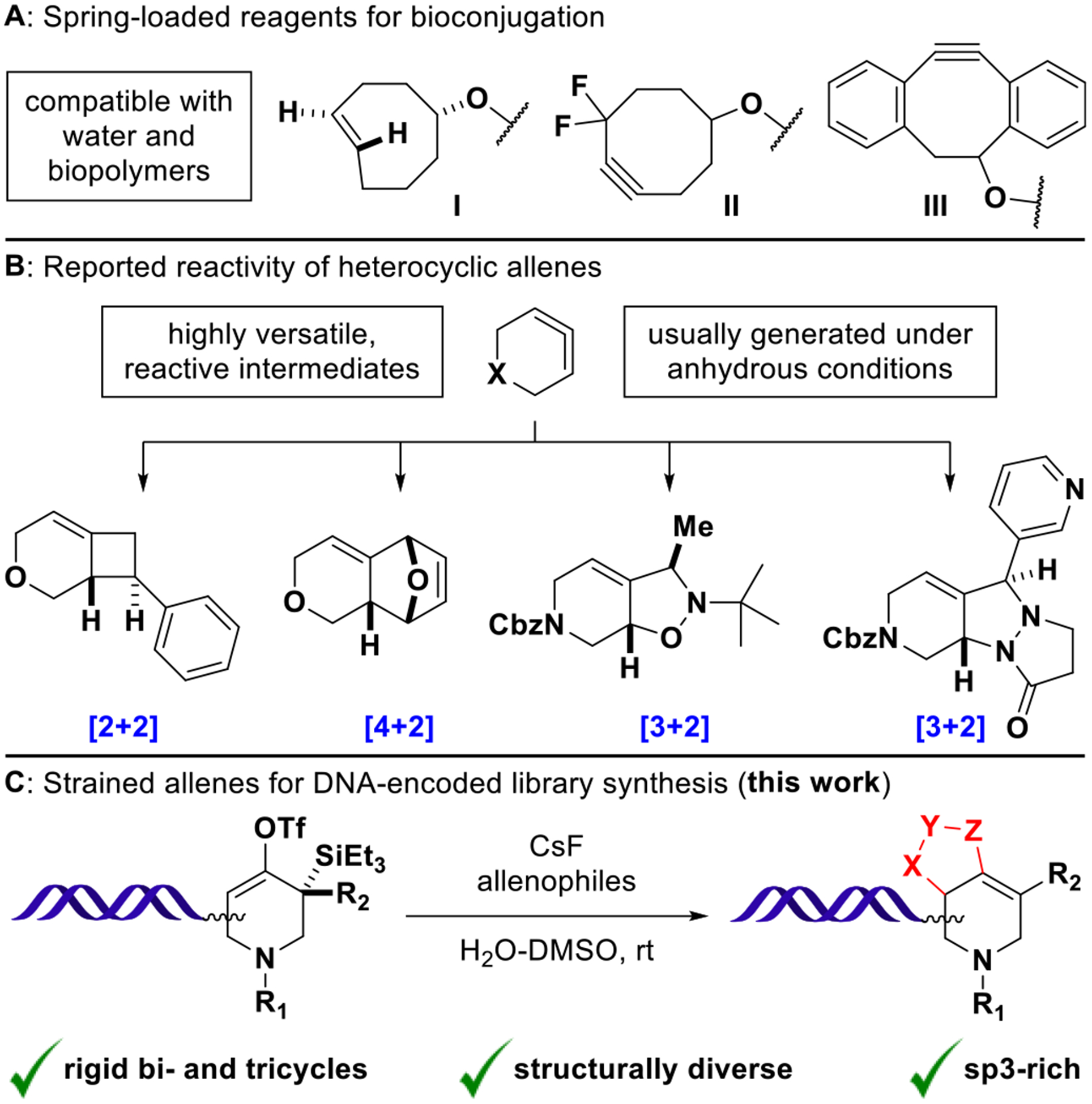
(A) Water-compatible reagents for bioorthogonal chemistry: trans-Cyclooctene (I) rapidly undergoes inverse-electron-demand Diels–Alder reactions with tetrazines. Cyclooctyne derivatives (II, III) undergo strain-promoted azide–alkyne cycloadditions. (B) In situ generated heterocyclic allenes exhibit distinct cycloaddition modes with activated olefins, dienes, and 1,3-dipoles (major diastereomers shown). (C) Fluoride-induced formation of DNA- conjugated heterocyclic allenes and trapping with various allenophiles affords structurally diverse cycloaddition products.
When contained within 8-membered rings and smaller, cyclic allenes show increased reactivity and readily undergo strain-releasing reactions.29,30 Both carbo- and heterocyclic allenes have been prepared under anhydrous conditions by the action of alkyllithiums on dihalocyclopropane precursors (Doering–Moore–Skattebøl rearrangement),31,32 by base-induced elimination of vinyl bromides33 or more recently, by fluoride-induced β-elimination of silyl-vinyl-triflates.34–36 Importantly, strained allenes exhibit distinct cycloaddition modes with olefins, 1,3-dipoles, and furans/N-substituted pyrroles to undergo [2 + 2], [3 + 2], and [4 + 2] reactions, respectively.36–38 The possibility to generate diverse molecular architectures from a common precursor (Figure 1B) renders these intermediates particularly interesting in the context of diversity-oriented synthesis. Motivated by a recent report on azacyclic allene reactivity by the Garg group,37 we heredocument the successful implementation of strain-promoted cycloadditions for DNA-encoded library synthesis (Figure 1C).
RESULTS AND DISCUSSION
Our efforts commenced with the preparation of DNA- conjugated allene precursor 3 designed to minimally perturb steric and electronic factors of piperidine-derivative 4 used in Garg’s off-DNA study.37 Racemic benzoic acid-derivative rac-2 was prepared in four steps from 1 closely resembling the published synthetic strategy toward 4 (see SI, Section 4c); rac- 2 was elaborated into 3 by amide conjugation to a double stranded DNA-headpiece (DNA-HP) developed by research- ers at GlaxoSmithKline (see Figure 2).39
Figure 2.
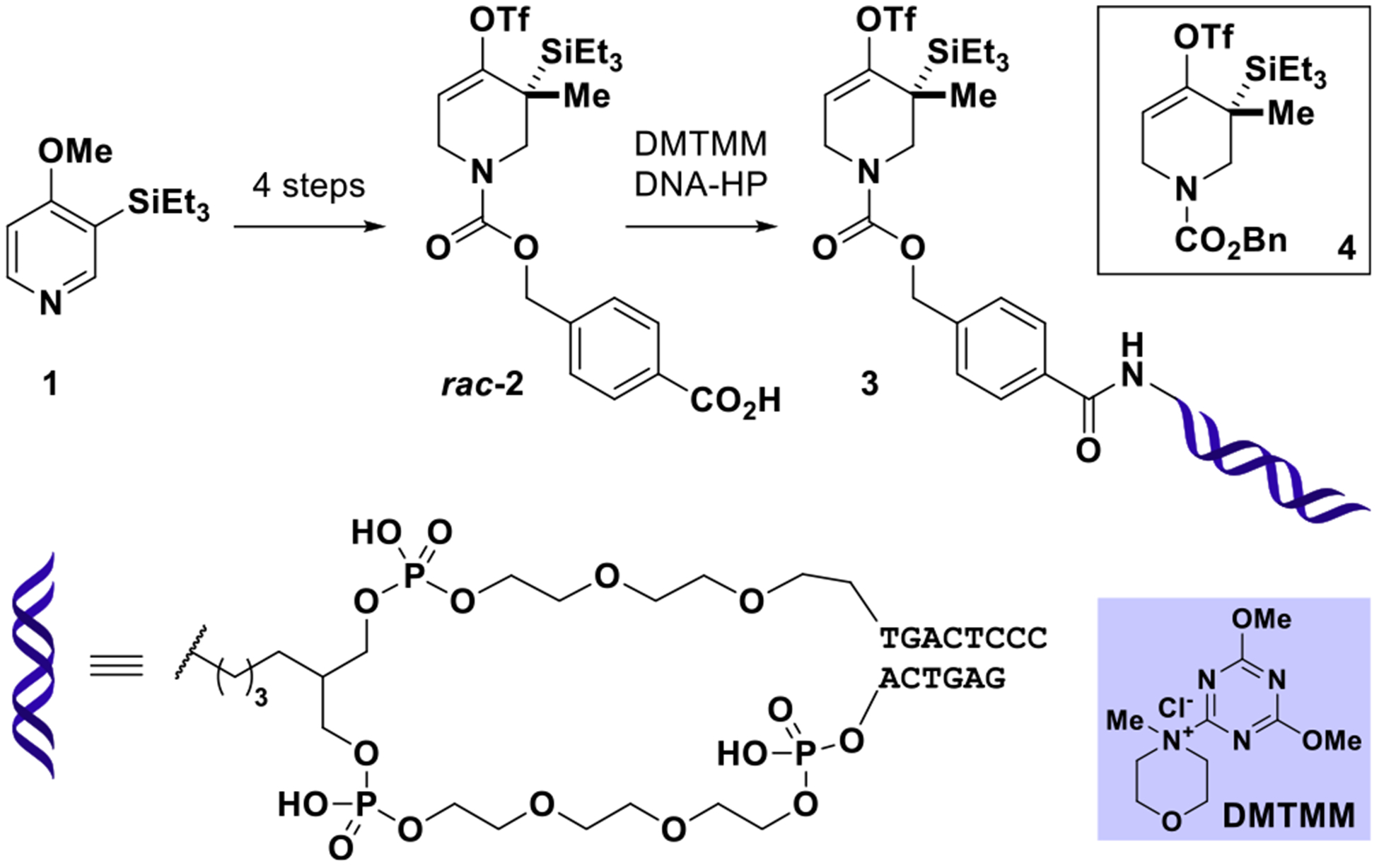
Preparation of on-DNA strained allene precursor 3 by conjugation of rac-2 to DNA-headpiece (DNA-HP). For experimental details, see SI, Section 4c.
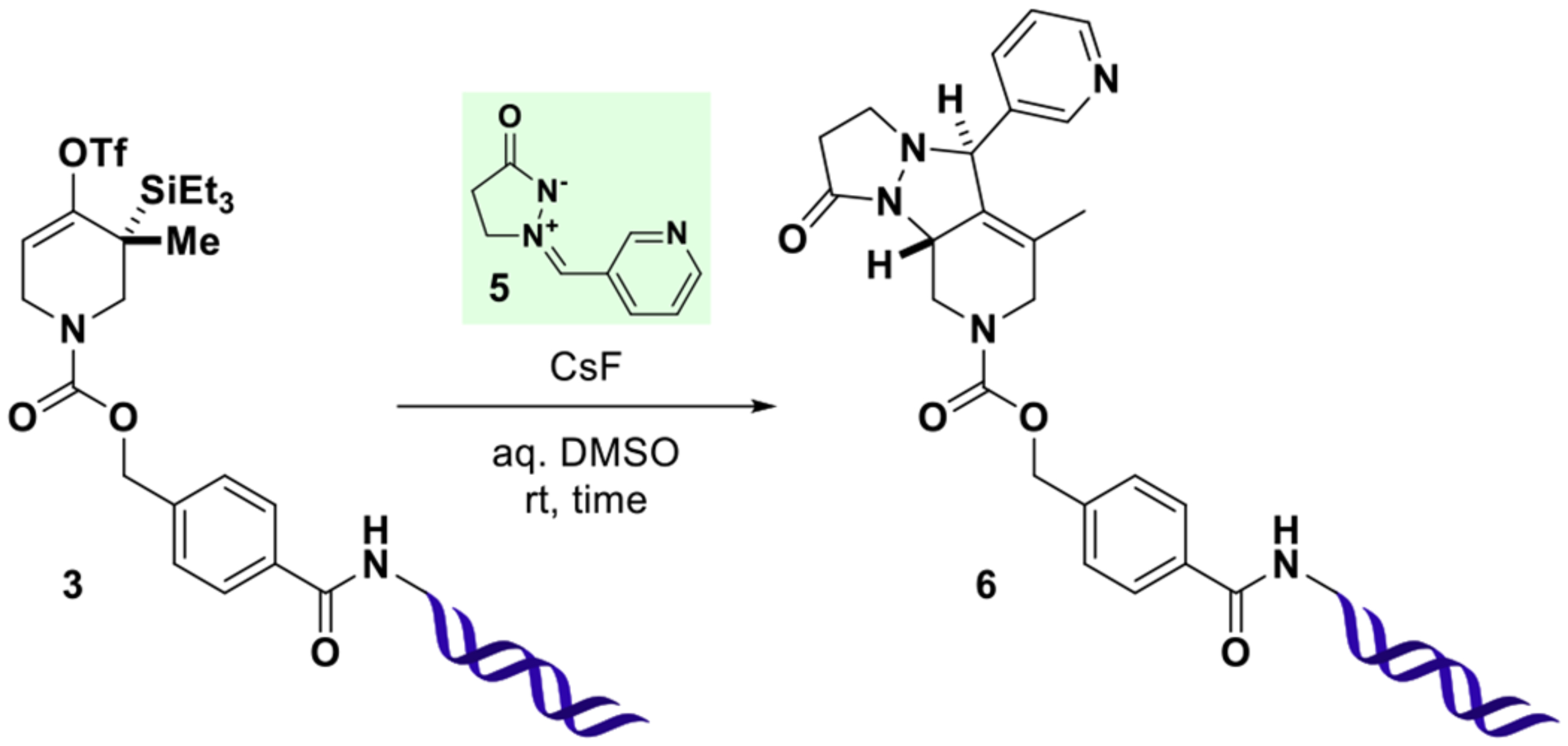
With this material in hand, on-DNA strained allene generation was investigated. To this end, 3 was combined with varying amounts of cesium fluoride and azomethine imine 5 in DMSO-water mixtures (10 μL total volume) for defined time intervals (Table 1). Following ethanol precipitation, reaction outcomes were analyzed by UPLC-MS and quantified by integration of UV absorption (260 nm), neglecting non- DNA species as judged by the absence of signal in the total ion chromatogram. In these experiments, water content emerged as a critical parameter inversely correlating with the consumption of 3 (entries 1–3). After 24 h, significant conversion was observed only for the reaction with the lowest water content (75% DMSO), resulting in efficient formation of a new species, the deconvoluted mass of which agreed with cycloaddition product 6. The slow conversion in the presence of water, presumably originating from fluoride ion hydration, could be overcome by increasing the concentration of activating agent (entries 4–7). With further reduced water content (90% DMSO), full consumption of 3 was observed with as little as 125 equiv of cesium fluoride (ca. 6 mM final concentration) within 1 h (entries 8 and 9). Importantly, throughout the series, the only detected DNA-species were 3 and 6, provided the concentration of trapping agent 5 was sufficiently high (entries 10–12), indicating the desired transformation to be highly selective.
Table 1.
Formation and Trapping of a DNA-Conjugated Strained Allenesa
 | ||||||
|---|---|---|---|---|---|---|
| # | 5 [mM] | CsF [mM] | %DMSO | t [h] | 3 [%] | 6 [%] |
| 1 | 15 | 50 | 25 | 24 | 100 | 0 |
| 2 | 15 | 50 | 50 | 24 | 97 | 3 |
| 3 | 15 | 50 | 75 | 24 | 0 | 100 |
| 4 | 100 | 1000 | 50 | 1 | 2 | 98 |
| 5 | 100 | 500 | 50 | 1 | 28 | 72 |
| 6 | 100 | 750 | 75 | 1 | 1 | 99 |
| 7 | 100 | 375 | 75 | 1 | 28 | 72 |
| 8 | 15 | ~6 | 90 | 1 | 0 | 100 |
| 9 | 15 | ~3 | 90 | 1 | 27 | 73 |
| 10 | 2 | 50 | 90 | 12 | 0 | 100 |
| 11 | 1 | 50 | 90 | 12 | 0 | 95 |
| 12 | 0.5 | 50 | 90 | 12 | 0 | 84 |
Reactions were performed at room temperature with 3(0.5 nmol) in a total volume of 10 μL for the indicated time. Following ethanol precipitation, residual 3and newly formed 6were detected by UPLC-MS and quantified (%AUC) by integration of UV chromatograms (260 nm) considering DNA-species only. The relative configuration of the two newly formed stereogenic centers in 6indicates the expected major product as observed in off-DNA precedence.37 For additional data, see SI, Table S1.
These initial experimental results allow for the following conclusions: (1) DNA-conjugated strained allenes can be formed in aqueous solution and exhibit considerable lifetime in the medium; (2) cycloadditions with azomethine imine 5 take place efficiently and rapidly in aqueous mixtures of DMSO (for other solvents, see SI, Table S2); (3) water content critically influences the rate of conversion (higher water content slows the reaction); (4) higher fluoride concentrations increase the rate of conversion. Encouraged by these results, the scope of the reaction was evaluated with a variety of 1,3-dipoles, olefins, and N-substituted pyrroles (Figure 3). In order to attenuate potential reactivity differences, building blocks were used in excess (180 mM final concentration). Nitrones derived from both aromatic and aliphatic aldehydes (7a–7d) participated in the transformation to give the expected [3 + 2] cycloaddition products of type 7 in high purity (>95%AUC). Ketone-derived nitrone 7e resulted in a sluggish reaction profile, presumably due to higher steric hindrance of the dipole. Pivaldehyde derived nitrone 7f did not afford the expected product. Instead, a species with an apparently missing tert-butyl group was detected. This result could be explained by fast cycloaddition of a competing nitrone formed from small quantities of formaldehyde present in DMSO (see SI, Figure S1). [4 + 2] reactions of N-substituted pyrroles (8a–8h) efficiently afforded bridged tricyclic compounds of type 8 and tolerated the presence of numerous functional groups (nitro, aniline, phenol, aldehyde, ester). The presence of carboxylic acid 8k slowed the consumption of starting material and resulted in formation of a species exhibiting a mass in agreement with either a ketone originating from triflate hydrolysis and desilylation or an allylic alcohol resulting from hydration of the intermediate allene (req m/z 5208 Da, found 5209 Da). A species with the same retention time and mass spectral properties was formed in the presence of excess primary amine 8l. The slowed consumption of starting material in the presence of acidic protons as in 8k appears to be a general phenomenon (see SI, Table S3). The supposed fluoride sequestration might be explained by hydrogen bond formation and could be overcome by lowering the concentration of the acidic building block, thereby increasing the ratio of fluoride to acid (see SI, Table S4). Alternatively, fluoride sequestration may also be overcome by the addition of basic buffer (see SI, Figure S2).
Figure 3.
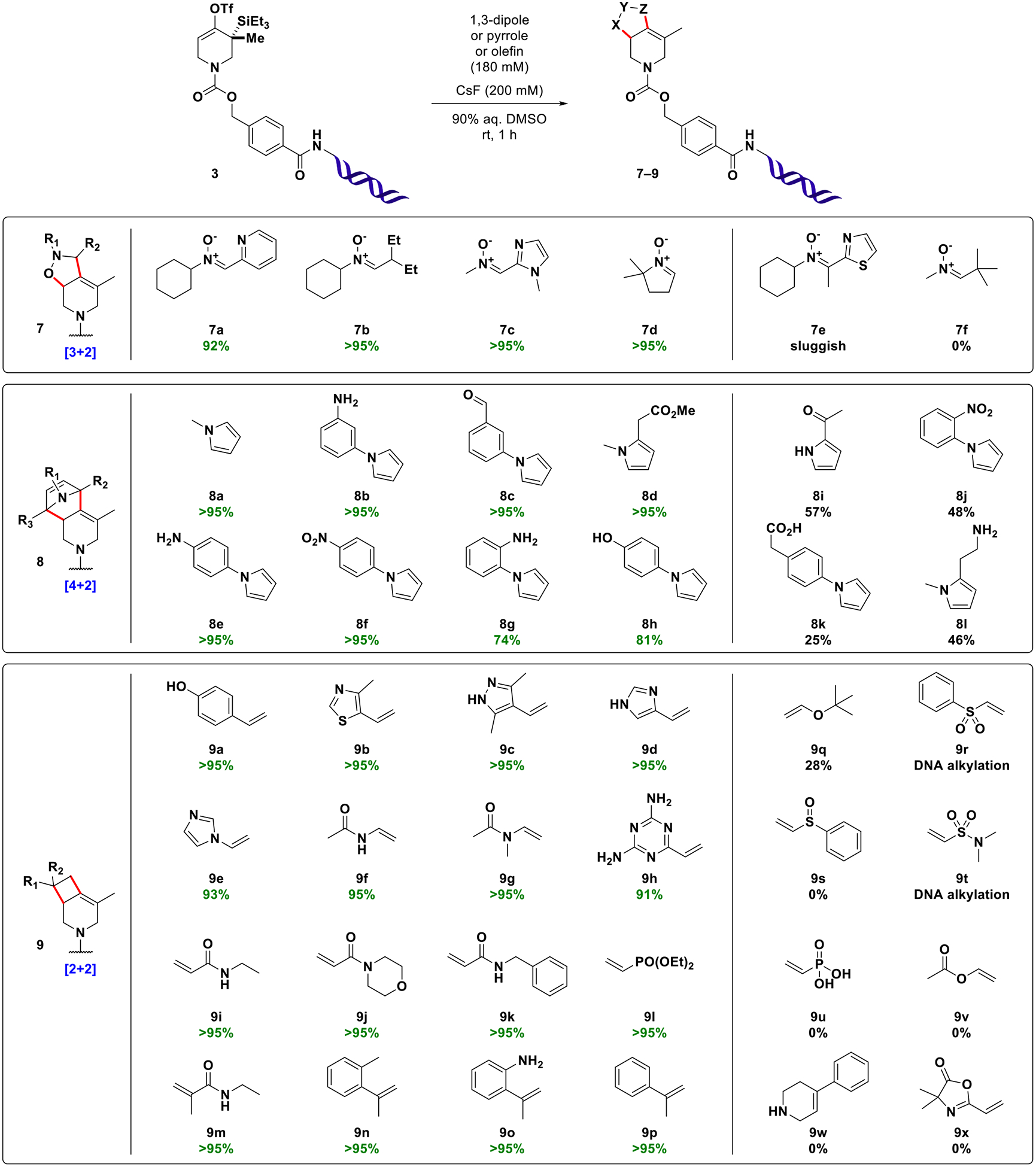
Selected examples of building blocks used in strain-promoted cycloaddition reactions of DNA-conjugated allenes derived in situ from 3. Product display (%AUC) was determined by integration of UV signals (260 nm) considering DNA-species only. For additional examples of [2 + 2] reactions, see SI, Figure S5. For titration experiments with each type of cycloaddition partner, see SI, Figure S6.
The [2 + 2] cycloadditions of commercial activated and nonactivated olefins were investigated next. Vinyl (hetero) aromatics and acrylamides including 1,1-disubstituted congeners performed very reliably in the reaction (9a--9p). Interestingly, N-vinyl amide 9g efficiently afforded the expected species while the analogous process with enol ether derivative 9v resulted in no significant product formation. This finding is in agreement with relative estimated radical stabilization energies (RSE) of initial diradical species, which are the presumed intermediates in this type of [2 + 2] cycloaddition.30,40 The difference in RSE of relevant O- and N- stabilized radicals was calculated to favor the latter by ca. 25 kJ/mol (see SI, Figure S3).41
The reaction with vinyl sulfone 9r afforded several species that could be only partially resolved chromatographically. Inspection of mass spectra indicated formation of multiple adducts, most likely arising from the desired [2 + 2] cycloaddition and additional DNA alkylation reactions (up to seven events by mass spectrometry (see SI, Figure S4). This finding prompted us to examine a variety of acceptor substituted olefins of varied electrophilicity on the Mayr reactivity scale42 (see SI, Table S5). Qualitative correlation of the reaction outcomes with the corresponding electrophilicity parameters of building blocks revealed that electrophiles with EDMSO > −19 tend to undergo DNA alkylation reactions under the chosen conditions (90% aq. DMSO, 180 mM building block, 200 mM CsF, 1 h, rt). Less electrophilic building blocks did not show signs of DNA alkylation. Although observed in 90% aq. DMSO, these findings might have implications not only in the context of DNA-encoded library synthesis, but for the field of bioconjugate chemistry in general, where maleimides (EDMSO ≈ −14) are often used, as well as for toxicological assessment of electrophilic drugs.
Unlike reagent classes such as boronic acids, aldehydes or amines, the number of commercially available 1,3-dipoles is limited. Thus, a combinatorial synthesis of such building blocks ideally avoiding tedious purification would be highly desirable. We therefore attempted the synthesis of a test set of azomethine imines by simply combining 3-pyrazolidinone with various aldehydes in ethanol. Incubation of the resulting mixtures at room temperature overnight, removal of the volatiles and reconstitution of residual material in DMSO afforded solutions for immediate use in on-DNA reactions, notably without purification. Assuming quantitative azome- thine imine formation, on-DNA precursor 3 was combined with 1,3-dipoles (15 mM final concentration) and cesium fluoride (50 mM final concentration) in 90% aqueous DMSO. In this nonoptimized procedure, about half of the building block solutions thus prepared validated (>80%AUC) to afford the expected DNA-species as identified by UPLC-MS analysis following ethanol precipitation (see SI, Figure S7).
While DNA-conjugate 3 was shown to undergo fluoride- induced elimination readily to form a highly reactive strained allene that could be trapped even in aqueous media, it is amenable to one-step diversification only, provided no additional functionalities are introduced by virtue of the cycloaddition partners. To highlight the promise of the described transformation in terms of combinatorial DEL synthesis, a second-generation substrate (11) was prepared by conjugating rac-10 to a PEG-linker extended version of DNA-HP (AOP-HP) via amide bond formation. Substrate 11 exhibits a nosyl group on nitrogen, the on-DNA deprotection of which has been described,43 and as such offers two points of diversification (strained allene cycloaddition, N-capping). In addition, library synthesis should benefit from spatial separation of the nucleophilic nitrogen and the cycloaddition product appendages, providing more uniform reactivity during N-capping reactions. Figure 4 shows four examples of a synthetic sequence involving strain-promoted cycloadditions/Ns-deprotection to afford intermediate piperidines, which were subjected to N-sulfonylations using 2-methylbenzenesulfonyl chloride as a representative N-capping reaction (Figure 4A). Final products 12–15 were derived in 79–96%AUC as evident from the UV chromatograms (Figure 4B).
Figure 4.
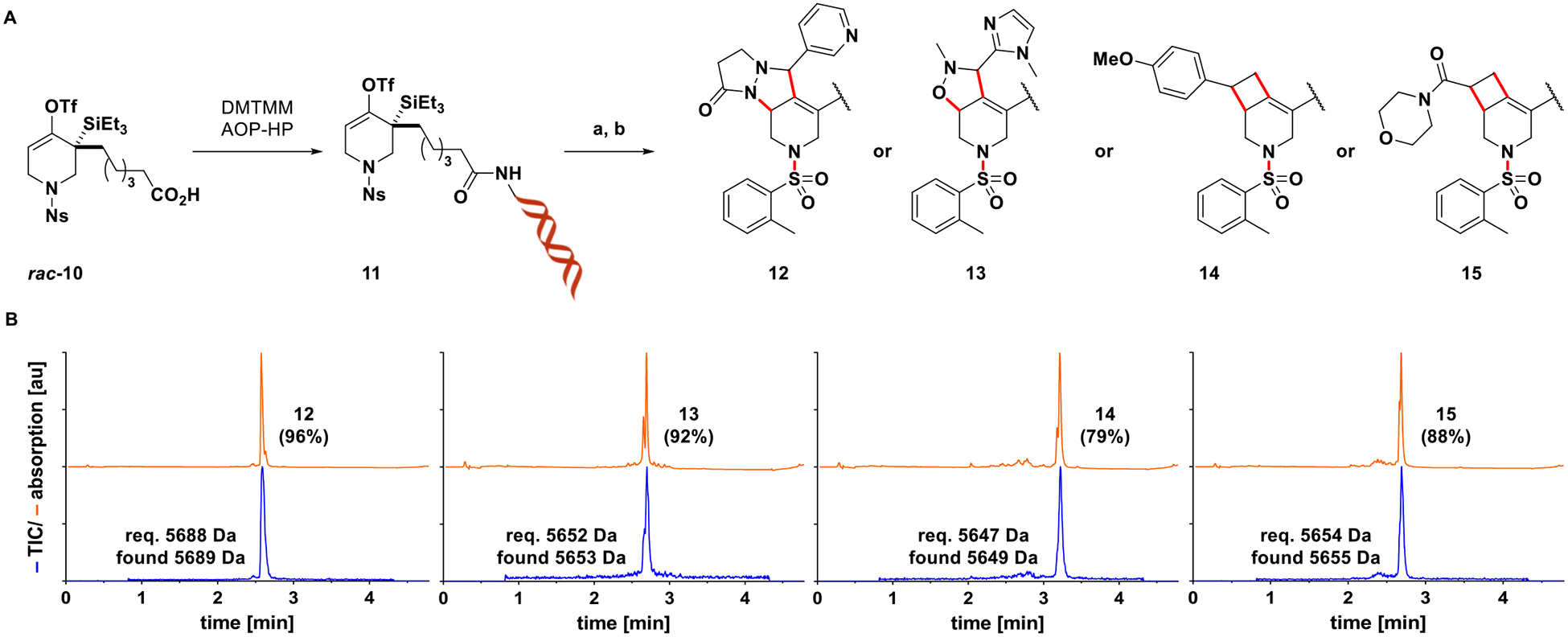
(A) Synthesis of DNA-conjugated strained allene precursor 11 enables two-step diversification. Reagents and conditions: (a) 11 (5 nmol), 1,3-dipole or styrene derivative (25 mM), CsF (23 mM), 90% aq. DMSO, rt, 1 h; then 4-methoxythiophenol (85 mM), carbonate buffer (pH10), 60% aq. DMSO, 80 °C, 1 h. For UPLC-MS analysis of intermediate piperidines, see SI, Figure S8; (b) 2-methylbenzenesulfonyl chloride (40 mM), phosphate buffer (pH8), 20% aq. MeCN, rt, 12 h. (B) UPLC-MS analysis of N-sulfonylation reactions following EtOH precipitation indicates efficient formation of compounds 12–15 (orange: UV (260 nm), blue: total ion chromatogram). Figures in parentheses refer to %AUC of the corresponding species.
In accord with off-DNA precedence,38,44,45 we note that the here-described on-DNA transformations are expected to form diastereoisomeric mixtures of varying ratios. Indeed, the corresponding UV chromatograms regularly show split peaks (see Figure 4B and SI) sharing the same mass spectral properties. We acknowledge that this property of the presented on-DNA process may complicate off-DNA hit validation following a library screening campaign but advocate for its widespread use given the unprecedented nature of the formed products. In this context, a substructure search of cores 7–9 in the ChEMBL database returned zero hits,46 supporting the notion that derivatives of these structures are indeed covering uncharted chemical space.
In an additional experiment, aliquots of the intermediate piperidine-DNA conjugates, isolated and purified by standard ethanol precipitation, were subjected to T4 DNA ligase-mediated ligation reactions with a 23-basepair primer sequence. The success of these ligations was confirmed by gel electrophoretic analysis (see SI, Figure S9). Furthermore, real-time polymerase chain reaction (qPCR) studies were performed to assess the extent of potential DNA degradation during the reaction.9 To this end, a full-length DNA-encoded library was subjected to conditions with varied DMSO and cesium fluoride content (see SI, Figure S10). Subsequent quantification of amplifiable material did not indicate any DNA degradation in samples treated with CsF (100 mM) in 85% aq. DMSO. Samples incubated with very high fluoride concentrations (4 M CsF in 50% aq. DMSO or 3 M CsF in 75% aq. DMSO) showed ca. 80% remaining amplifiable material. Notably, all test conditions in this degradation study used substantially more activating agent than that needed for efficient allene formation (see Table 1 and SI, Table S1). In conclusion, the successful outcome of enzymatic ligation reactions with samples that underwent strain-promoted cycloadditions along with negligible DNA degradation warrants a promising integration of the described transformation into existing DEL synthesis workflows.47
CONCLUSION
The present work describes a rare example of a DNA- compatible process allowing the synthesis of highly structurally diverse, rigid core structures of high sp3-content from a single precursor. Extension of the concept to other strained allene precursors including bicyclic and seven-membered systems may further expand the scope of unprecedented structures that can be incorporated into DNA-encoded libraries. Given the convenient reaction setup, the remarkably efficient and selective reaction profiles, and the number of commercially available or easily prepared building blocks, strain-promoted cycloaddition reactions should find widespread use in the field of DNA-encoded library synthesis.
Supplementary Material
ACKNOWLEDGMENTS
John Capece, Jennifer Poirier, Philip Michaels, Carmelina Rakiec, Thomas Dice, and Ritesh Tichkule are gratefully acknowledged for excellent technical and analytical support. Bruce Hua and Drs. Christopher Gerry, Wenyu Wang, Christian Gampe, Simone Bonazzi, Nichola Smith, and Frédéric Berst are gratefully acknowledged for general support, fruitful discussions and valuable feedback during preparation of this manuscript. The research was supported in part by the National Institute of General Medical Sciences (R35GM127045 awarded to S.L.S.) and by the NIBR Scholar’s Program.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.9b13186.
Experimental procedures (off and on-DNA work) and analytical data (PDF)
The authors declare the following competing financial interest(s): S.L.S. serves on the Board of Directors of the Genomics Institute of the Novartis Research Foundation (GNF); is a shareholder and serves on the Board of Directors of Jnana Therapeutics; is a shareholder of Forma Therapeutics; is a shareholder and advises Kojin Therapeutics, Kisbee Therapeutics, Decibel Therapeutics and Eikonizo Therapeutics; serves on the Scientific Advisory Boards of Eisai Co., Ltd., Ono Pharma Foundation, Exo Therapeutics, and F-Prime Capital Partners; and is a Novartis Faculty Scholar.
Contributor Information
Matthias V. Westphal, Chemical Biology and Therapeutics Science Program, Broad Institute, Cambridge, Massachusetts 02142, United States; Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States
Liam Hudson, Chemical Biology and Therapeutics Science Program, Broad Institute, Cambridge, Massachusetts 02142, United States; Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States.
Jeremy W. Mason, Chemical Biology and Therapeutics Science Program, Broad Institute, Cambridge, Massachusetts 02142, United States; Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States
Johan A. Pradeilles, Chemical Biology and Therapeutics Science Program, Broad Institute, Cambridge, Massachusetts 02142, United States; Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States
Frédéric J. Zécri, Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States
Karin Briner, Novartis Institutes for BioMedical Research, Cambridge, Massachusetts 02139, United States.
Stuart L. Schreiber, Chemical Biology and Therapeutics Science Program, Broad Institute, Cambridge, Massachusetts 02142, United States; Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138, United States.
REFERENCES
- (1).Neri D; Lerner RA DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem 2018, 87, 479–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Satz AL What Do You Get from DNA-Encoded Libraries? ACS Med. Chem. Lett 2018, 9, 408–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cochrane WG; Malone ML; Dang VQ; Cavett V; Satz AL; Paegel BM Activity-Based DNA-Encoded Library Screening. ACS Comb. Sci 2019, 21, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).MacConnell AB; Price AK; Paegel BM An Integrated Microfluidic Processor for DNA-Encoded Combinatorial Library Functional Screening. ACS Comb. Sci 2017, 19, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Galloway WRJD; Isidro-Llobet A; Spring DR Diversity- Oriented Synthesis as a Tool for the Discovery of Novel Biologically Active Small Molecules. Nat. Commun 2010, 1, 80 Nat. Commun. 2010, 1, 80,. [DOI] [PubMed] [Google Scholar]
- (6).Clemons PA; Wilson JA; Dancík V; Muller S; Carrinski HA; Wagner BK; Koehler AN; Schreiber SL Quantifying Structure and Performance Diversity for Sets of Small Molecules Comprising Small-Molecule Screening Collections. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 6817–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gerry CJ; Schreiber SL Chemical Probes and Drug Leads from Advances in Syn-thetic Planning and Methodology. Nat. Rev. Drug Discovery 2018, 17, 333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Schreiber SL A Chemical Biology View of Bioactive Small Molecules and a Binder-Based Approach to Connect Biology to Precision Medicines. Isr. J. Chem 2019, 59, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ratnayake AS; Flanagan ME; Foley TL; Smith JD; Johnson JG; Bellenger J; Montgomery JI; Paegel BM A Solution Phase Platform to Characterize Chemical Reaction Compatibility with DNA-Encoded Chemical Library Synthesis. ACS Comb. Sci 2019, 21, 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Malone ML; Paegel BM What Is a “DNA-Compatible” Reaction? ACS Comb. Sci 2016, 18, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang J; Lundberg H; Asai S; Martín-Acosta P; Chen JS; Brown S; Farrell W; Dushin RG; O’Donnell CJ; Ratnayake AS; Richardson P; Liu Z; Qin T; Blackmond DG; Baran PS Kinetically Guided Radical-Based Synthesis of C(Sp3)-C(Sp3) Linkages on DNA. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E6404–E6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kölmel DK; Loach RP; Knauber T; Flanagan ME Employing Photoredox Catalysis for DNA-Encoded Chemistry: Decarboxylative Alkylation of α-Amino Acids. ChemMedChem 2018, 13, 2159–2165. [DOI] [PubMed] [Google Scholar]
- (13).Ruff Y; Berst F Efficient Copper-Catalyzed Amination of DNA-Conjugated Aryl Iodides under Mild Aqueous Conditions. MedChemComm 2018, 9, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).(a) Buller F; Mannocci L; Zhang Y; Dumelin CE; Scheuermann J; Neri D Design and synthesis of a novel DNA- encoded chemical library using Diels-Alder cycloadditions. Bioorg. Med. Chem. Lett 2008, 18, 5926–5931. [DOI] [PubMed] [Google Scholar]; (b) Gerry CJ; Yang Z; Stasi M; Schreiber SL DNA-Compatible [3 + 2] Nitrone-Olefin Cycloaddition Suitable for DEL Syntheses. Org. Lett 2019, 21, 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Phelan JP; Lang SB; Sim J; Berritt S; Peat AJ; Billings K; Fan L; Molander GA Open-Air Alkylation Reactions in Photoredox-Catalyzed DNA-Encoded Library Synthesis. J. Am. Chem. Soc 2019, 141, 3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Škopić MK; Götte K; Gramse C; Dieter M; Pospich S; Raunser S; Weberskirch R; Brunschweiger A Micellar Brønsted Acid Mediated Synthesis of DNA-Tagged Heterocycles. J. Am. Chem. Soc 2019, 141, 10546–10555. [DOI] [PubMed] [Google Scholar]
- (17).Potowski M; Kunig VBK; Losch F; Brunschweiger A Synthesis of DNA-Coupled Isoquinolones and Pyrrolidines by Solid Phase Ytterbium- and Silver-Mediated Imine Chemistry. MedChem- Comm 2019, 10, 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Skopic MK; Salamon H; Bugain O; Jung K; Gohla A; Doetsch LJ; Santos D. d.; Bhat A; Wagner B; Brunschweiger A Acid- and Au(I)-Mediated Synthesis of Hexathymidine-DNA-Heterocycle Chimeras, an Efficient Entry to DNA-Encoded Libraries Inspired by Drug Structures. Chem. Sci 2017, 8, 3356–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ruff Y; Martinez R; Pellé X; Nimsgern P; Fille P; Ratnikov M; Berst F An Amphiphilic Polymer-Supported Strategy Enables Chemical Transformations Under Anhydrous Conditions for DNA-Encoded Library Synthesis. ACS Comb. Sci 2020, 22, 120. [DOI] [PubMed] [Google Scholar]
- (20).Flood DT; Asai S; Zhang X; Wang J; Yoon L; Adams ZC; Dillingham BC; Sanchez BB; Vantourout JC; Flanagan ME; Piotrowski DW; Richardson P; Green SG; Shenvi RA; Chen JS; Baran PS; Dawson PE Expanding Reactivity in DNA- Encoded Library Synthesis via Reversible Binding of DNA to an Inert Quaternary Ammonium Support. J. Am. Chem. Soc 2019, 141, 9998–10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Agard NJ; Prescher JA; Bertozzi CR A Strain-Promoted [3 + 2] Azide-Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J. Am. Chem. Soc 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- (22).Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Lo A; Codelli JA; Bertozzi CR Copper-Free Click Chemistry for Dynamic in Vivo Imaging. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Blackman ML; Royzen M; Fox JM Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels-Alder Reactivity. J. Am. Chem. Soc 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lang K; Davis L; Wallace S; Mahesh M; Cox DJ; Blackman ML; Fox JM; Chin JW Genetic Encoding of Bicyclononynes and Trans-Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels-Alder Reactions. J. Am. Chem. Soc 2012, 134, 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Prescher JA; Bertozzi CR Chemistry in Living Systems. Nat. Chem. Biol 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- (26).Li H; Sun Z; Wu W; Wang X; Zhang M; Lu X; Zhong W; Dai D Inverse-Electron-Demand Diels-Alder Reactions for the Synthesis of Pyridazines on DNA. Org. Lett 2018, 20, 7186–7191. [DOI] [PubMed] [Google Scholar]
- (27).Singh I; Freeman C; Heaney F Efficient Synthesis of DNA Conjugates by Strain-Promoted Azide-Cyclooctyne Cycloaddition in the Solid Phase. Eur. J. Org. Chem 2011, 2011, 6739–6746. [Google Scholar]
- (28).Singh I; Heaney F Solid Phase Strain Promoted “Click” Modification of DNA via [3 + 2]-Nitrile Oxide-Cyclooctyne Cycloadditions. Chem. Commun 2011, 47, 2706–2708. [DOI] [PubMed] [Google Scholar]
- (29).Daoust KJ; Hernandez SM; Konrad KM; Mackie ID; Winstanley J; Johnson RP Strain Estimates for Small-Ring Cyclic Allenes and Buta-trienes. J. Org. Chem 2006, 71, 5708–5714. [DOI] [PubMed] [Google Scholar]
- (30).Christl M Allenes Up to Seven-Membered Rings In Modern Allene Chemistry; John Wiley & Sons, Ltd: 2008, 243–357. [Google Scholar]
- (31).Moore WR; Moser WR Reaction of 6,6- Dibromobicyclo[3.1.0]Hexane with Methyllithium. Evidence for the Generation of 1,2-Cyclohexadiene and 2,2’-Dicyclohexenylene. J. Am. Chem. Soc 1970, 92, 5469–5474. [Google Scholar]
- (32).Moore WR; Moser WR Reaction of 6,6- Dibromobicyclo[3.1.0]Hexane with Methyllithium. Efficient Trapping of 1,2-Cyclohexadiene by Styrene. J. Org. Chem 1970, 35, 908–912. [Google Scholar]
- (33).Wittig G; Fritze P On the Intermediate Occurrence of 1,2- Cyclohexadiene. Angew. Chem., Int. Ed. Engl 1966, 5, 846–846. [Google Scholar]
- (34).Lofstrand VA; West FG Efficient Trapping of 1,2- Cyclohexadienes with 1,3-Dipoles. Chem. - Eur. J 2016, 22, 10763–10767. [DOI] [PubMed] [Google Scholar]
- (35).Quintana I; Peña D; Pérez D; Guitiáń E Generation and Reactivity of 1,2-Cyclohexadiene under Mild Reaction Conditions. Eur. J. Org. Chem 2009, 2009, 5519–5524. [Google Scholar]
- (36).Barber JS; Styduhar ED; Pham HV; McMahon TC; Houk KN; Garg NK Nitrone Cycloadditions of 1,2-Cyclohexadiene. J. Am. Chem. Soc 2016, 138, 2512–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Barber JS; Yamano MM; Ramirez M; Darzi ER; Knapp RR; Liu F; Houk KN; Garg NK Diels-Alder Cycloadditions of Strained Azacyclic Allenes. Nat. Chem 2018, 10, 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Schreck M; Christl M Freisetzung Und Abfangreaktionen von 1-Oxa-3,4-Cyclohexadien. Angew. Chem 1987, 99, 720–721. [Google Scholar]
- (39).Clark MA; Acharya RA; Arico-Muendel CC; Belyanskaya SL; Benjamin DR; Carlson NR; Centrella PA; Chiu CH; Creaser SP; Cuozzo JW; Davie CP; Ding Y; Franklin GJ; Franzen KD; Gefter ML; Hale SP; Hansen NJV; Israel DI; Jiang J; Kavarana MJ; Kelley MS; Kollmann CS; Li F; Lind K; Mataruse S; Medeiros PF; Messer JA; Myers P; O’Keefe H; Oliff MC; Rise CE; Satz AL; Skinner SR; Svendsen JL; Tang L; van Vloten K; Wagner RW; Yao G; Zhao B; Morgan BA Design, Synthesis and Selection of DNA-Encoded Small-Molecule Libraries. Nat. Chem. Biol 2009, 5, 647–654. [DOI] [PubMed] [Google Scholar]
- (40).Boafo EA; Darko K; Afriyie BA; Tia R; Adei E Trapping of 1,2-cyclohexadiene: A DFT mechanistic study on the reaction of 1,2-cyclohexadiene with olefins and nitrones. J. Mol. Graphics Modell 2018, 81, 1–13. [DOI] [PubMed] [Google Scholar]
- (41).Hioe J; Zipse H Radical Stability and Its Role in Synthesis and Catalysis. Org. Biomol. Chem 2010, 8, 3609–3617. [DOI] [PubMed] [Google Scholar]
- (42).Allgäuer DS; Jangra H; Asahara H; Li Z; Chen Q; Zipse H; Ofial AR; Mayr H Quantification and Theoretical Analysis of the Electro-philicities of Michael Acceptors. J. Am. Chem. Soc 2017, 139, 13318–13329. [DOI] [PubMed] [Google Scholar]
- (43).Franch T; Lundorf MD; Jakobsen S; Olsen EK; Andersen AL; Holtmann A; Hansen AH; Sørensen AM; Goldbech A; De Leon D; Kaldor DK; Sløk FA; Husemoen GN; Dolberg J; Jensen KB; Petersen L; Nørregaard-Madsen M; Godskesen MA; Schrøder Glad S; Neve S; Thisted T; Kronborg TTA; Sams C; Felding J; Freskgaard P-O; Haahr Gouliaev A; Pedersen H (Nuevolution A/S), Enzymatic Encoding Methods for Efficient Synthesis of Large Libraries WO/2007/062664, 2007.
- (44).Christl M; Braun M Freisetzung und Abfangreaktionen von 1-Oxa-2,3-cyclohexadien. Chem. Ber 1989, 122, 1939–1946. [Google Scholar]
- (45).Yamano M; Knapp R; Ngamnithiporn A; Ramirez M; Houk K; Stoltz B; Garg NK Cycloadditions of Oxacyclic Allenes and a Catalytic Asymmetric Entryway to Enantioenriched Cyclic Allenes. Angew. Chem., Int. Ed 2019, 58, 5653–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46). [Nov. 9th];ChEMBL database. accessed on. under https://www.ebi.ac.uk/chembl/.
- (47).The synthesis of a pilot library is ongoing. For preliminary results, see Supporting Information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


