Abstract
Ten years ago we reviewed how the cellular DNA damage response (DDR) is controlled by changes in the functional and structural properties of nuclear proteins, resulting in a timely coordinated control of gene expression that allows DNA repair. Expression of genes that play a role in DDR is regulated not only at transcriptional level during mRNA biosynthesis but also by changing steady-state levels due to turnover of the transcripts. The 3′ end processing machinery, which is important in the regulation of mRNA stability, is involved in these gene-specific responses to DNA damage. Here, we review the latest mechanistic connections described between 3′ end processing and DDR, with a special emphasis on alternative polyadenylation, microRNA and RNA binding proteins-mediated deadenylation, and discuss the implications of deregulation of these steps in DDR and human disease.
Keywords: 3′ end processing, deadenylation, DNA damage response, polyadenylation
1 |. INTRODUCTION
DNA damage occurs as the result of exogenous and endogenous sources of stress, either in the form of energetically driven chemical alterations to the DNA, such as UV-mediated intra-strand crosslinks, or agents that promote genome instability, such as inhibition of torsional stress relief by topoisomerases (Hande, 1998; Mahadevan, Bowerman, & Luger, 2019; Mao & Wyrick, 2019; Merchut-Maya, Bartek, & Maya-Mendoza, 2019; Setlow, Swenson, & Carrier, 1963; T. E. Wilson & Sunder, 2019). Both forms disrupt cellular homeostasis and increase the risk of mutation passing to daughter cells during division. Because of the importance to the organism of minimizing mutational inheritance, considerable cellular resources have evolved to detect and repair DNA damage in both a general and lesion-specific manner. As the damage to DNA can occur as quickly as electron-transfer, the DNA damage response (DDR) arms the cell with mechanisms for dynamic changes in gene expression beyond the time consuming processes of transcription, nuclear export and translation of mRNAs. A paradigm of the DDR is that transcripts involved in the response are expressed before the DNA repair process begins, suggesting that gene-specific compensatory mechanisms are needed on these genes to ensure their expression, allowing the cell to react to genotoxic stresses and maintain genomic integrity. For a better understanding of DDR and its regulation in mammalian biology, we suggest the reading of the following reviews (Jachimowicz, Goergens, & Reinhardt, 2019; Jackson & Bartek, 2009; O’Connor, 2015). Posttranscriptional processing of transcripts represents an essential mechanism for dynamic and proper control of gene expression at different time-points during DDR. For example, mRNA 3′ end processing modulates the length of the poly(A) tail of an mRNA providing a widespread strategy used to control mRNA stability and protein production.
3′ end processing of most eukaryotic mRNAs is a multistep maturation involving co-transcriptional recognition of pre-mRNA cis-elements, following by cleavage of the nascent RNA and non-templated addition of ∼200 adenosine residues in human cells. These mRNA 3′ end modifications confer stability and translational efficiency to the transcripts (Bentley, 2014; Jalkanen, Coleman, & Wilusz, 2014; Proudfoot, 2011; Y. Shi & Manley, 2015). Conversely, removal of the poly(A) tail by deadenylation can signal mRNA decay and/or translational repression of poly(A) + transcripts (Aström, Aström, & Virtanen, 1991; Mayya & Duchaine, 2019; Nicholson & Pasquinelli, 2019; Webster et al., 2018; Yi et al., 2018; X. Zhang, Kleiman, & Devany, 2014).These modifications in the 3′ end are controlled by cis-acting elements present in the mRNA and trans-acting regulatory factors, such as RNA binding proteins (RBPs) and RNAs with complementary base-pairing, such as, but not restricted to, microRNAs (miRNAs). For better understanding of basic aspects of 3′ end formation and its regulation, we suggest the reading of comprehensive reviews that have covered this topic (Hollerer, Grund, Hentze, & Kulozik, 2014; Neve, Patel, Wang, Louey, & Furger, 2017; Y. Shi & Manley, 2015). An additional level of 3′ end formation control is by alternative polyadenylation (APA), where the conserved poly(A) signal AAUAAA, AUUAAA, and less efficacious derivatives can occur in multiple incidences in a single transcriptional unit generating templated mRNAs that differ by the 3′ end (Tian & Manley, 2017). Finally, non-adenosine nucleotide transfers to the 3′ end of the mRNA are emerging as potential global regulators of DDR (Lackey, Welch, & Marzluff, 2016). Here, we present an update of our previous review (Cevher & Kleiman, 2010) on how undergoing research has shown new and updated links between mRNA 3′ end processing and DDR (Figure 1).
FIGURE 1.
Categories of events and pathways involved at the intersection of mRNA 3′ end processing and DNA damage response, as discussed in this review
2 |. CLEAVAGE AND POLYADENYLATION
2.1 |. CstF-50 and RNA polymerase II
One of the first functional connections described between the cleavage and polyadenylation (CpA) machinery and DDR was the interaction of the tumor suppressors BRCA1/BARD1 (breast and ovarian cancer type 1 susceptibility protein 1)/(BRCA1 associated RING domain 1) with not only DNA repair proteins PCNA and Rad51 (Scully et al., 1997) but also the cleavage factor CstF-50 and RNA Polymerase II (RNAP II) (Kleiman & Manley, 1999, 2001). After stress, BRCA1/BARD1/CstF-50 inhibits the cleavage step of polyadenylation (Kleiman & Manley, 1999, 2001) and induces the ubiquitination and proteasomal degradation of RNAP II, an activator of pre-mRNA 3′ cleavage (Figure 2) (Hirose & Manley, 1998; Mirkin et al., 2008). Interestingly, recent studies show that when RNAP II is inhibited by depletion of CpA machinery some protein-coding genes exhibit read through transcription and backsplicing, leading to circular RNAs (Liang et al., 2017; J. E. Wilusz, 2017). Consequently, Serine 2 phosphorylation of RNAP II C-terminal domain (CTD) is necessary for CpA factors recruitment (Ahn, Kim, & Buratowski, 2004). Consistent with this, RNAP II degradation was also inhibited in cleavage factor I (CFI) mutants in yeast (Gaillard & Aguilera, 2014). The cleavage factor CstF-50 regulates the chromatin remodeling, including histone and RNAP II ubiquitination, of differently expressed genes after DNA damage (Fonseca et al., 2018). In fact, DDR-responsive gene loci show similar patterns for RNAP II CTD phosphorylation at Serine 2, a marker associated with transcription termination, and stress-induced phosphorylation of histone 3 (H3) by AKT (J. H. Lee et al., 2015). Interestingly, abolishing the AKTmediated phosphorylation of H3 results in the dysregulation of 3′ end processing of DNA damage-activated genes, reducing RNA decay downstream of the cleavage site and the release of RNAP II from the chromatin. In addition, the CstF complex subunit CstF-64 and its analogue τCstF-64 (Dass et al., 2001; Wallace et al., 1999) are serine phosphorylated after ionizing radiation treatment, possibly signaling a role in DNA repair (Matsuoka et al., 2007).
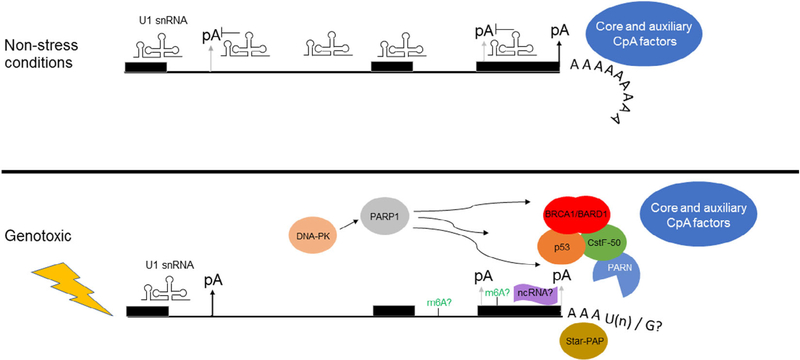
FIGURE 2.
Factor flux before and after the introduction of genotoxic stress. Top: In nonstress conditions, promoter-proximal poly(A) signals are suppressed by the “telescripting” effect of U1 snRNA in a 5′→3′ direction, including both intronic and proximal 3′ UTR poly(A) signals (Berg et al., 2012). The CpA machinery operates unhindered as a multitude of core and auxiliary factors function to process the precursor mRNA, including PAP and CstF-50 (Y. Shi et al., 2009). Bottom: Upon occurrence and recognition of genotoxic stress, the ribonucleoprotein components of CpA function are altered, as U1 snRNA levels are decreased, allowing for recognition of specific promoter-proximal poly(A) signals (Devany et al., 2016). Recruitment of BRCA1/BARD1/p53 complex(es) takes place, inhibiting CpA via CstF-50 (Kleiman & Manley, 2001), and activating deadenylation through factors such as PARN (Devany, Zhang, Park, Tian, & Kleiman, 2013). A number of CpA factors undergo DNA-PK- and/or PARP1-mediated posttranslational modifications, affecting their activity (Jungmichel et al., 2013). Star-PAP is also recruited to certain stress-response genes (W. Li et al., 2012) as well as poly(U) polymerases in the cytoplasm during apoptosis (Thomas et al., 2015). There are also potential mRNA methylation events and guanines added to the mRNA 3′ ends (Lim et al., 2018), though these are yet to be described in DDR context. The role of noncoding RNAs in this regulation is also a nascent field (Huang et al., 2017). CpA, cleavage and polyadenylation; DDR, DNA damage response; PAP, poly(A) polymerase; PARN, poly(A)-specific ribonuclease
The UV-induced depletion of RNAP II, in part by BRCA1/BARD1/CstF-50-mediated ubiquitination (Kleiman et al., 2005), might also be involved in the induction of APA. Recent studies showed that depletion of RNAP II promotes the usages of APA sites for several transcripts (Yu, Rege, Peterson, & Volkert, 2016), such as Retinol Binding Protein 2 (RBP2), peptide transporter PTR2, and Discoidin Domain Receptor Tyrosine Kinase 2 (DDR2). Indeed, while under nonstress conditions RPB2 mRNA terminates at a proximal poly(A) site, after UV treatment a distal poly(A) site is preferentially used, resulting in a longer transcript. As transcription and CpA are globally inhibited upon DNA damage, an important question arises: how are DDR-responsive genes able to circumvent such inhibition? Increased transcription for some DDR-responsive genes has been described (reviewed in Christmann & Kaina, 2013) and it is permitted because transcription inhibition factors are engage everywhere else in the genome (Rockx et al., 2000). As transcription is tightly coupled with 3′ end RNA processing, it is possible that the increase in the number of transcripts of DDR-responsive genes compensate the inhibition of their processing resulting in an increase in their expression despite a number of collateral unprocessed transcripts. Another possibility is that the overall strength of the poly(A) signals, including upstream (USEs) and downstream (DSEs) sequence elements, is higher in DDR-responsive genes, requiring less active CpA complex in the vicinity for effective 3′ end processing (Proudfoot, 2011; Takagaki, Seipelt, Peterson, & Manley, 1996; Tian & Manley, 2017). Third, studies have shown that certain RNA secondary structures enhance gene-specific CpA. For example, TP53 pre-mRNA possesses G-quadruplex structure downstream of poly(A) site, which was found to be necessary for processing during DDR (Decorsière, Cayrel, Vagner, & Millevoi, 2011). Subsequent work elucidated that the RNA helicase DHX36 is required for G-quadruplex mediated maintenance of TP53 pre-mRNA processing (Figure 3) (Newman et al., 2017).
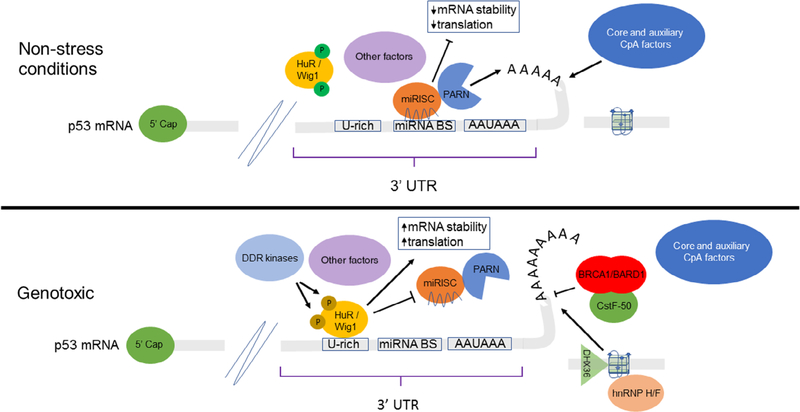
FIGURE 3.
TP53 mRNA post-transcriptional regulation before and after DNA damage. Under non-stress conditions, multiple different miRNAs as part of Ago2-containing miRISC complex possess binding sites for TP53 3′ UTR, recruiting deadenylases, such as PARN (Cevher et al., 2010), leading to decay or miRNA-mediated translational repression (Devany et al., 2013). HuR and other RBPs are phosphorylated by cellular kinases (Kim, Abdelmohsen, & Gorospe, 2010), preventing binding to mRNA. Under stress conditions, HuR and Wig1 are dephosphorylated by DDR kinases allowing their binding to ARE-sequences present in mRNA targets (Mukherjee et al., 2011; Vilborg et al., 2009). Steric overlap of miRNA seed sequence with ARE-sequence prevents Ago2/PARN association, leading to increase mRNA stability and translational capacity (Mukherjee et al., 2011; X. Zhang et al., 2015). Concomitantly, global inhibition of CpA by CstF-50/BRCA1/BARD1 (Kleiman & Manley, 1999, 2001) is overcome in part through functional complementation of RNA helicase DHX36 and factor hnRNP H/F binding to G-quadruplex structure downstream of TP53 cleavage site (Newman et al., 2017). ARE, AU-rich element; CpA, cleavage and polyadenylation; DDR, DNA damage response; PARN, PARN, poly(A)-specific ribonuclease; RBPs, RNA binding proteins
2.2 |. DDR factors functionally associated with the 3′ end processing complex
Given the complexity of the CpA machinery, both in size and component abundance, it is not surprising that studies have described DDR-factors associated with core CpA factors such as protein kinase complex DNA-PKcs/Ku70–86 and PARP1 (Y. Shi et al., 2009). Although the nuclear serine/threonine kinase DNA-PKcs/Ku70–86 is involved in nonhomologous end joining and double-strand break (DSB) repair (Davis, Chen, & Chen, 2014; Ryan & Bauer, 2008), a direct role of DNA-PK in phosphorylating 3′ end processing factors has not yet been described. However, DNA-PK can phosphorylate PARP1 (Ariumi et al., 1999), which is associated with the 3′ complex (Y. Shi et al., 2009) and is involved in DNA damage detection and repair (reviewed in Ji & Tulin, 2010). PARP1 can also associate with and PARylate poly(A) polymerase (PAP) under stress conditions, resulting in inhibition of polyadenylation as modified PAP is unable to bind substrate mRNA (Di Giammartino, Shi, & Manley, 2013). Other 3′ end processing factors, such as the FIP1L1 component of the cleavage and polyadenylation specificity factor (CPSF) and poly(A) binding protein PABPN1, can be modified by PARP-1 (Jungmichel et al., 2013; Y. Zhen, Zhang, & Yu, 2017), suggesting that PARP1 might regulate 3′ processing under different conditions, including DDR. Consistent with this, recent studies have shown that PARP1-mediated chromatin modifications affect not only RNAP II elongation but also co-transcriptional modifications (Matveeva, Al-Tinawi, Rouchka, & Fondufe-Mittendorf, 2019) and that PARP1 is an mRNA-binding protein (Melikishvili, Chariker, Rouchka, & Fondufe-Mittendorf, 2017). Besides, PARP1 binds to and PAR-ylates embryonic lethal abnormal vision-like 1 (Elavl1)/human antigen R (HuR), allowing its nucleocytoplasmic shuttling and binding to its mRNA targets 3′ ends, increasing their stability (Y. Ke et al., 2017). Y. Shi et al. (2009) also described RBBP6, an E3 ubiquitin ligase originally described to interact with p53 (Simons et al., 1997), as part of the 3′ end processing molecular architecture. Some RBBP6 isoforms generated by alternative mRNA processing led to increased use of distal poly(A) sites genome-wide (Di Giammartino et al., 2014). In fact, efficient mRNA 3′ processing requires a zinc knuckle, ubiquitin-like (UBL) and RING finger domains of Mpe1, the yeast RBBP6 homolog (S. D. Lee & Moore, 2014). The zinc knuckle and RING domains bind RNA with no sequence-specificity (Di Giammartino et al., 2014; S. D. Lee & Moore, 2014). Mpe1 can ubiquitinate Pap1, indicating a functional association between ubiquitin pathway and CpA machinery.
RNA:DNA hybrid-mediated R-loops are associated with genomic instability (reviewed in Crossley, Bocek, & Cimprich, 2019). In yeast, a screening for factors involved in ameliorating R-loop mediated genome instability identified seven essential protein components of 3′ end processing machinery, including the human homologues for two of the components of cleavage factor IIm (CFIIm), CstF-50, CstF-64, CPSF100, Fip1, and WDR33 (Stirling et al., 2012). Trf4, a gene encoding a noncanonical PAP involved in RNA surveillance, also participates in maintenance of genome integrity (Gavaldá, Gallardo, Luna, & Aguilera, 2013). While future studies will be required to elucidate the contribution of each factor, it is possible that these mRNA processing factors allow the transcripts to dissociate from chromatin playing a major role in DNA homeostasis. In that scenario, we could speculate that the presence of multiple poly(A) signals in a single transcriptional unit might play a role not only in generating different mRNAs and proteins (Tian, Hu, Zhang, & Lutz, 2005) but also in ensuring genomic stability. As the spliceosomal U1 snRNP binds the nascent RNA to inhibit the usage of intronic poly(A) sites within first introns (Almada, Wu, Kriz, Burge, & Sharp, 2013; Berg et al., 2012; Kaida et al., 2010), U1 snRNP might also participate in genome integrity maintenance, possibly through currently undescribed complex(es) with genome integrity and DDR factors. In fact, under DNA damaging conditions, the levels of U1 snRNA decrease inducing intronic APA at the promoter-proximal side of the genes (Devany et al., 2016), potentially allowing greater access to DNA repair factors and dissociation of nascent transcripts.
2.3 |. Integrator complex and 3′ end processing
Eight subunits of the integrator complex are part of the molecular architecture of the pre-mRNA 3′ processing complex (Y. Shi et al., 2009). The integrator complex binds to RNAP II (Baillat et al., 2005) and is involved in homeostatic transcription termination of specific genes, including non-polyadenylated snRNAs (reviewed in J. Chen & Wagner, 2010; Rienzo & Casamassimi, 2016) and histone mRNAs (Skaar et al., 2015). These genes undergo a specialized processing of the 3′ ends of their RNAs (J. Chen et al., 2012; Ezzeddine et al., 2011; Marzluff, Wagner, & Duronio, 2008), and the disruption of mechanisms controlling snRNA transcription termination results into extended snRNAs with poly(A) tails in human cells (O’Reilly et al., 2014; Yamamoto et al., 2014). Whether this disruption occurs physiologically during DDR is to our knowledge currently unknown. Interestingly, proteins involved in polyadenylation can participate in the termination of snRNA transcription (Egloff, Al-Rawaf, O’Reilly, & Murphy, 2009; O’Reilly et al., 2014). These results indicate that the integrator complex functionally overlaps with the mRNA CpA machinery to promote cleavage and couple snRNA 3′ end processing with termination. Transcription termination also plays a role in 3′ processing of replication-dependent histone genes, which also undergo a specialized processing of the 3′ ends of their RNAs (reviewed in Marzluff et al., 2008). Like with snRNA, when the termination machinery is disrupted, replication-dependent histone genes are polyadenylated and lose the regulation of their stability and expression during cell-cycle progression (Sullivan, Steiniger, & Marzluff, 2009). Transcription pausing at promoter proximal sites could function as a decision point for transcription elongation or termination/3′ processing (Brannan et al., 2012), and this mechanism plays a role in controlling the expression of highly inducible genes, such as stress response genes (Adelman & Lis, 2012). In fact, in addition to the integrator’s noted role in termination of snRNA and histone transcription, integrator binds at promoter-proximal sites of polyadenylated stress-response genes, highlighting integrator’s role in DDR (Rienzo & Casamassimi, 2016; Skaar et al., 2015). Additionally, some subunits of the integrator complex interact with proteins involved in DDR, such as DNA-binding protein 1 (hSSB1), and regulate the accumulation of RAD51 and BRCA1 at DNA damage sites and the correlated homologous recombination (Vidhyasagar et al., 2018; F. Zhang, Ma, & Yu, 2013; J. Zhang, Sun, etal., 2013).
2.4 |. Poly(A) polymerases
Canonical PAP are responsible for the co-transcriptional addition of an adenosine tail at the 3′ end of mRNAs in the nucleus. There are two ubiquitously expressed forms; PAPα and PAPγ (Q. Yang, Nausch, Martin, Keller, & Doublié, 2014), and the testis-specific PAPα (Y. J. Lee, Lee, & Chung, 2000). However, noncanonical Star-PAP (Speckle Targeted PIPKIα Regulated Poly(A) Polymerase) differs from canonical PAPs through its ability to bind to pre-mRNA directly (Laishram & Anderson, 2010). During cellular stress elicitation, Star-PAP is capable of regulating 3′ end processing in a gene-specific and condition-dependent manner (Figure 2) (W. Li et al., 2012). In etoposide treatment, Star-PAP is recruited to pro-apoptotic gene Bcl-2 interacting killer (BlK) mRNA (W. Li et al., 2012). On the other hand, nuclear phosphoinositide stress signaling (Barlow, Laishram, & Anderson, 2010) during oxidative stress leads to Star-PAP-mediated regulation of cytoprotective enzymes HO-1 (heme oxygenase-1, Mellman et al., 2008) and NQO-1 (NAD[P]H:Quinone Oxidoreductase; Gonzales, Mellman, & Anderson, 2008). Interestingly, Star-PAP induces the use of APA of tumor suppressor PTEN (phosphatase and tensin homolog deleted on chromosome 10) gene and this is essential for DNA damage-induced increase of PTEN protein levels (W. Li, Li, et al., 2017; X. Li, Xiong, et al., 2017), indicating that Star-PAP regulates APA in a signaling- and target gene-specific manner. Genome/functional analysis of Star-PAP and PIPKIα depleted cells indicated that mRNAs encoding anti-invasive factors, such as CDH1, CDH13, FEZ1, KISS1R, NME1, WIF1, are also targets of this polyadenylation activity (Laishram & Anderson, 2010). Outside of nuclear regulation, mitochondrial mRNA poly(A) polymerase (mtPAP) was observed to be associated with increased radiosensitivity, concurrent with increased reactive oxygen species (ROS) and DSBs (Martin et al., 2014).
As several RNA processing factors are known to localize to sites of DNA damage and to interact with DNA repair proteins (Gaillard & Aguilera, 2014) suggesting that they may play a direct role in the DDR, it is not surprising that several proteomic and functional screens have identified RNA 3′ end processing factors as part of DDR (Jungmichel et al., 2013; Montecucco & Biamonti, 2013; Paulsen et al., 2009). Remarkably, structural analysis of the recognition of the AAUAAA polyadenylation signal identified that the scaffold complex CPSF160-WDR33 possesses structural similarity to the DNA repair complex DDB1-DDB2, suggesting recent evolutionary divergence and/or some functional redundancy (Q. Sun, Hao, & Prasanth, 2018; Y. Sun, Zhang, et al., 2018).
3 |. ALTERNATIVE POLYADENYLATION
3.1 |. APA factors involved in DDR
As genes sometimes consist of tens of thousands of base pairs, the statistical likelihood to find the conserved poly(A) signal AAUAAA, AUUAAA, and less efficacious derivatives is very high. Approximately 70% of human genes are characterized by multiple poly(A) sites that result in different transcript isoforms with variable 3′ ends (Derti et al., 2012) and this significantly contributes to transcriptome diversity (Tian et al., 2005). A large scale analysis elucidated the impact of different CpA factors on APA and how APA is regulated by the location of the poly(A) signals relative to the transcription start site, distance between competing poly(A) signals, cis elements near poly(A) signal and concentrations of core CpA factors (W. Li et al., 2015). In keeping with some of the earliest data on APA (Takagaki et al., 1996), CstF-64 was identified as a key effector (W. Li et al., 2015). It is currently unknown whether the levels of available CpA effectors or the usage of different poly(A) signals are affected under various genotoxic agents, however, it has been shown that UV-treatment does not change the levels CstF-50 but its availability is altered through differential complex formation (Mirkin et al., 2008). Fip1, a polymerase-regulating component of the CpA complex, not only regulates APA (Lackford et al., 2014; W. Li et al., 2015) but also maintains genome integrity by suppressing R-loop formation, suggesting that Fip1 function may be relevant to certain human cancers (Stirling et al., 2012). RBBP6 is part of the CpA complex (Y. Shi et al., 2009) that plays a role in genome stability and DNA replication (Miotto et al., 2014; Motadi, Lekganyane, & Moela, 2018). An RBBP6 isoform generated by intronic APA is able to compete with full-length RBBP6 for association with the remainder of the polyadenylation machinery, thereby inhibiting polyadenylation and regulating APA, with an enrichment for affecting 3′ end processing of AU-rich RNAs (Di Giammartino et al., 2014). RBBP6 also appears to be important for regulating the stability of mRNAs with AU-rich elements in their 3′ UTRs.
Factors not considered part of the core CpA machinery have been implicated in alternative 3′ end processing after DNA damage. Cyclin-dependent kinase CDK12 is a regulatory kinase that pairs with Cyclin K and phosphorylates the RNAP II CTD to maintain processive elongation (Bartkowiak et al., 2010; Blazek et al., 2011; Ekumi et al., 2015; Fusby et al., 2015). CDK12 depletion in cell-based (Blazek et al., 2011) or xenograft models (Johnson et al., 2016) is associated with genome instability and DNA repair defects. CycK/Cdk12 regulates the expression of predominantly long genes with high numbers of exons, including DDR genes such as BRCA1, ATR (ataxia telangiectasia and Rad3-related), FANCI, and FANCD2. In fact, CDK12 has been implicated in 3′ end processing of c-MYC (Davidson, Muniz, & West, 2014) and c-FOS (Eifler et al., 2015), suggesting that CDK12 regulates CpA through RNAP II phosphorylation at the CTD (Ahn et al., 2004). Comparing normal cells and breast cancer cells containing a CDK12 amplification, it was shown that CDK12 regulates alternate last exon (ALE) splicing, a subset of splicing events that involves APA (Elkon, Ugalde, & Agami, 2013; Tien et al., 2017). It remains to be determined whether this CDK12-mediated effect on ALE splicing in breast cancer is dependent on RNAP II CTD or another undescribed pathway of CDK12. Recent studies showed that CDK12 not only play a role in ALE regulation but also in protecting the cells from premature CpA and allowing expression of long genes, a sizable portion of which include DDR genes (Krajewska et al., 2019). p38 kinase (Danckwardt et al., 2011) and protein kinase C (PKC)δ (W. Li et al., 2012) are involved in the modulation of poly(A) signal usage of prothrombin pre-mRNA in response to the stress-inducing agent anisomycin and BIK pre-mRNA in response to DNA damage, respectively.
3.2 |. Global analyses of APA after cellular stress
A number of global studies have been published on APA regulation under DNA damaging conditions in both single celled and multicellular eukaryotes. In yeast, treatment with a UV-mimetic led to a global lengthening of transcripts due to the inhibition of CpA and the decrease in the levels of several mRNA 3′ end processing factors (Graber et al., 2013). In mammalian cells, DSBs created by topoisomerase inhibitor treatment led to induction of ALE through APA site usage (Dutertre et al., 2014). Those studies identified HuR as a regulator of ALE maturation in response to doxorubicin, a topoisomerase inhibitor used in breast cancer chemotherapy, and implicate doxorubicin-regulated ALEs in DDR and cell cycle regulation. Work by the Kulozik group described that cells treated with anisomycin, an inhibitor of the peptidyl transferase activity of the ribosome causing ribotoxic stress, show an overall lengthening of transcripts with a decrease in the utilization of promoter-proximal poly(A) sites in introns and increase in the utilization of promoter-distal poly(A) sites in intergenic regions (Hollerer et al., 2016). Interestingly, work from the Kleiman and Tian labs showed that treatment of cells with UV induced a ∼twofold increase in intronic polyadenylation and promoter-proximal poly(A) site usage, affecting gene groups with important functions in DDR and cancer (Figure 2) (Devany et al., 2016). UV-induced intronic APA activation correlates with the previously described decrease in U1 snRNA levels after UV treatment (Morra, Lawler, Eliceiri, & Eliceiri, 1986) and the global suppression of promoter-proximal sites by U1 (Berg et al., 2012; Gunderson, Polycarpou-Schwarz, & Mattaj, 1998). Consistent with this, U1 snRNA overexpression reverses these effects on APA and mitigates UV-induced apoptosis. A possible explanation for the discrepancies observed on the effect of anisomycin and UV treatments on APA may be due to how these stress-treatments affect U1 levels. The effect of anisomycin on the U1 snRNA was not determined in those studies. Treatment of cells with arsenite also led to overall transcript shortening by preferential usage of proximal poly(A) sites and enhanced degradation of transcripts with long 3′ UTR during recovery (D. Zheng et al., 2018). T cell-restricted intracellular antigen-1 (TIA1), an RBP involved in the recruitment of RNAs into stress-granules, preferentially interacts with the long 3′ UTR isoforms through U-rich motifs inducing their decay and allowing mRNAs with short 3′ UTRs generated by stress-induced APA to evade degradation. These APA changes induced by arsenite stress represent a mechanism to regulate the transcriptome in proliferating and differentiated cells.
3.3 |. Non-coding RNAs in 3′ end processing
The role of noncoding RNAs in both 3′ end processing and DDR has not been extensively studied. Recently, a small nucleolar RNA (snoRNA), normally associated with rRNA maturation, was shown to compete with CpA component Fip1 for binding to AAUAAA signal (Huang et al., 2017; J. Shi, Huang, Huang, & Yao, 2018). SNORD50A inhibits mRNA 3′ processing by blocking the Fip1-poly(A) site interaction, and SNORD50A depletion changed APA profiles genome-wide. Interestingly, many long noncoding RNA (lncRNAs) undergo APA in poly(A) sites upstream of the most 3′ exon (Hoque, Li, & Tian, 2014). LncRNA functions might be regulated by APA, as most of the conserved sequences are located upstream of the first poly(A) site. In fact, genome-wide analysis showed that lncRNA expression is controlled by PABPN1 resulting in the regulation of their stability (Beaulieu, Kleinman, Landry-Voyer, Majewski, & Bachand, 2012). Different pool of snoRNAs derived from lncRNA is induced by DNA damage and lncRNA are involved in the regulation of DDR (Dianatpour & Ghafouri-Fard, 2017; Krell et al., 2014; Michelini et al., 2017), suggesting a possible unexplored relationship between regulatory RNAs and control of RNA processing during DDR.
4 |. DEADENYLATION: RBPS AND MICRORNAS
4.1 |. Regulation of deadenylation during DDR
A great number of studies in the last 10 years have identified different aspects of an intermediate branch of the DDR that works on posttranscriptional regulatory pathways that include control of mRNA stability. In these pathways, RBPs and miRNAs can serve not only as targets of DNA damage signaling but also as transducers of signals to other gene expression pathways. Given that cellular stress cannot be predicted, when genotoxic stress occurs some level of transcripts of many DDR genes are capable of increasing very quickly in their stability and translational capacity (Young & Wek, 2016; Zander et al., 2016). Additionally, the median mRNA half-life under steady-state conditions is over 7 hr (Sharova et al., 2009), which reduces for housekeeping genes upon stress introduction (Cevher et al., 2010). Thus, removal of the poly(A) tail offers a convenient mechanism to regulate mRNA stability, and hence gene expression, while the cell assesses the extent of damage and initiates DNA repair. Deadenylation is under certain conditions the first step in mRNA decay (C. Y. Chen & Shyu, 2003). However, deadenylation is not a DNA damage-specific cellular response, as it is necessary to precisely regulate intracellular mRNA homeostasis in other physiological pathways (Y. B. Yan, 2014), such as removal of stem cell mRNAs in differentiating cells (Solana et al., 2013) and oocyte meiotic maturation (Vieux & Clarke, 2018).
4.2 |. Poly(A)-specific ribonuclease
During stress, expanding the central role of CstF-50 in promoting DDR-mediated 3′ end processing inhibition, it was shown that CstF-50 can exist in (a) nuclear complex(es) with the deadenylase poly(A)-specific ribonuclease (PARN) (Cevher et al., 2010). The CstF-50/PARN complex activated deadenylation of nuclear mRNA targets, including housekeeping genes, under DNA damaging conditions (Figure 2) (Cevher et al., 2010). Those studies also showed that CstF/PARN complex has a role in decreasing the levels of mRNAs involved in DDR, control of cell growth and differentiation, keeping their expression levels low under nonstress conditions. Expression of the tumor suppressors p53 and Gadd45α, key core components of DDR, are affected by PARN deadenylase (Devany et al., 2013; Reinhardt et al., 2010). PARN keeps p53 expression levels low in non-stress conditions by controlling TP53 mRNA stability, and the UV-induced increase in p53 activates PARN deadenylase, regulating gene expression during DDR in a transactivation-independent manner (Figure 2). This mechanism represents a feedback loop between p53 and PARN (Devany et al., 2013; X. Zhang et al., 2015). It is important to highlight that p53 can also form complex(es) with CstF-50/BRCA1/BARD1, promoting the inhibition of CpA reaction (Nazeer et al., 2011), suggesting that p53 might play an important role in the decision of either polyadenylate or deadenylate a target mRNA. While PARN deadenylase phosphorylation is necessary for Gadd45α mRNA stabilization after DNA damage, PARN deadenylase activity is not needed (Reinhardt et al., 2010). L. N. Zhang and Yan (2015) showed that the cellular functions of PARN are dependent on cell-type and stress-specific protein expression profile. It is important to highlight that CCR4-NOT has been described as the main cellular deadenylase, and that PARN and PAN2/PAN3 deadenylases do not have substantial effect on the poly(A) length and abundance of most mRNAs (Yi et al., 2018), suggesting that these deadenylases might have a more specific role in controlling mRNA decay. PARN hereditary mutations affects telomere biology and causes dyskeratosis congenita and related syndromes, a group of bone marrow failure disorders (Dhanraj et al., 2015; Mason & Bessler, 2015; Tummala et al., 2015). In fact, PARN mediates the biogenesis of the telomerase RNA component by removing a post-transcriptionally acquired poly(A) tail allowing telomere maintenance (Moon et al., 2015). In concert with nuclear regulation, control of cytoplasmic deadenylation of ARE-containing mRNA during DDR has also been observed (Blattner et al., 2000; Bollig et al., 2002; Gowrishankar et al., 2005; W. Wang et al., 2000). Ccr4-Not complex, also the major cytoplasmic deadenylase, has been shown to contribute to DDR by affecting expression of checkpoint genes after hydroxyurea (HU) and methylmethane sulfonate (MMS) treatments (Traven, Hammet, Tenis, Denis, & Heierhorst, 2005). Besides, Ccr4-Not complex plays a role in controlling the expression of the ribonucleotide reductase enzymatic complex involved during DNA damage and replication stress (Mulder, Winkler, & Timmers, 2005; Woolstencroft et al., 2006) and in maintaining genomic integrity by controlling the ubiquitination and degradation of RNAP II (H. Jiang, Wolgast, Beebe, & Reese, 2019).
4.3 |. Sequence specific RBPs
Among the enormous population of proteins present in the cell, a portion have evolved the capacity to bind directly to RNA species. This particularly true of polyadenylated mRNAs, which are highly regulated co- and post-transcriptionally. Indeed, the “mRNA-bound proteome” has been investigated in the last decade, elucidating further the many pathways and factors revolving RNA biology (Baltz et al., 2012; Castello et al., 2012; Conrad et al., 2016). Through both nonspecific and sequence specific binding, RBPs contribute to gene expression control in the early detection of DNA damage by relaying the signals generated by DNA damage sensors to the effectors of RNA metabolism, for example deadenylases. RBPs can themselves act as sensors by sending signals to downstream factors involved in DNA repair and chromatin modifications at DNA-damage sites. In that way, RBPs play a role in maintaining genomic integrity. RBPs can also determine cell fate by regulating cell cycle and transcription. For a comprehensive outline of general RBP function during DDR see Kai (2016) and Dutertre and Vagner (2017).
Phosphorylation of nucleolin, an abundant stress-responsive RBP, can regulate PARN deadenylase activity during cellular stress response (X. Zhang et al., 2018). Under nonstress conditions, nucleolin forms (a) complex(es) with factors that regulate deadenylation, such as p53 and the AU-rich (ARE)-binding protein HuR; and these interactions are favored by hypophosphorylated nucleolin after UV treatment. Nucleolin activates PARN activity and binds PARN substrates, such as TP53 and BCL2 mRNAs, playing a role in their downregulation under nonstress conditions. The RBP Wig-1, a p53 target protein, binds to TP53 mRNA and stabilizes it by protecting it from deadenylation (Vilborg et al., 2009), representing a mechanism to enhance p53 expression in a positive feedback loop (Figure 3). On the other hand, Wig1 promotes p53-target FAS mRNA degradation via binding to deadenylase CNOT6 in cytoplasmic stress granules (Bersani, Xu, Vilborg, Lui, & Wiman, 2014), decreasing cell death and reducing cell cycle arrest upon DNA damage, providing further opportunity for down-regulation of mRNAs that have escaped nuclear degradation or already are cytoplasmically localized upon stress.
4.4 |. Human antigen R
Another well-characterized ARE-binding protein is HuR (Grammatikakis, Abdelmohsen, & Gorospe, 2017; Lόpez de Silanes, Zhan, Lal, Yang, & Gorospe, 2004). HuR is known to affect multiple posttranscriptional processes, including mRNA stability and translation (reviewed in García-Mauriño et al., 2017; Grammatikakis et al., 2017). HuR has been shown to increase the stability of transcripts involved in carcinogenesis, cell proliferation and survival, and oxidative and genotoxic cellular response (Fan et al., 2011; Y. Li, Estep, & Karginov, 2018). Some of those transcripts include proto-oncogenes c-Fos (C. Y. Chen, Xu, & Shyu, 2002) and c-Myc (Gunzburg et al., 2015); cyclooxygenase-2 (COX-2; Doller et al., 2007); iNOS (Z. Guo & Geller, 2014); tumor suppressors TP53 (Abdelmohsen et al., 2014) and von Hippel–Lindau (Galbán et al., 2003). Interestingly, HuR can also regulate APA of target genes dependent on U-rich sequences proximal to poly(A) signal (Barnhart, Moon, Emch, Wilusz, & Wilusz, 2013; Berkovits & Mayr, 2015; Zhu, Zhou, Hasman, & Lou, 2007), of genes involved in stress responses (Kraynik et al., 2015), and of its own gene through reduction of CstF-64 recruitment (Dai, Zhang, & Makeyev, 2012).
As HuR functions are regulated by myriad DDR kinases (Kim et al., 2008, 2010), elucidating the full extent of the role of HuR posttranslational modifications in 3′ end processing during stress is of great interest. For example, CHK2 that gets activated after DNA damage and phosphorylates HuR after genotoxic stress, influencing HuR binding to mRNA and survival to genotoxic stress (Abdelmohsen et al., 2007; Lebedeva et al., 2011; Mukherjee et al., 2011; C. J. Wilusz & Wilusz, 2007). Additionally, following exposure to H2O2, HuR phosphorylation results in its dissociation from the stress-response SIRT1 transcript reducing its half-life. Transcriptome wide analysis of samples from cells exposed to IR radiation revealed a decrease in HuR binding to its target mRNAs due to activated CHK2-mediated phosphorylation (Masuda et al., 2011). Upon genotoxic stress, HuR can be PARylated by poly(ADP-ribose) polymerase 1 (PARP1) facilitating its cytoplasmic translocation and regulation of its binding to target mRNAs (Chand et al., 2017; Gagné et al., 2008; Y. Ke et al., 2017). dePARylation of HuR facilitates its release from target mRNAs and its shuttling back into the nucleus.
4.5 |. Competition between RBPs in controlling gene expression during DDR
The stability of ARE-containing mRNAs, and thus the level of expression of their protein products, is regulated by the antagonistic behavior of different RBPs. Binding by HuR generally leads to mRNA stabilization by inhibiting deadenylase recruitment (X. Zhang et al., 2015). In contrast, binding by AUF1, tristetraprolin (TTP; Clement, Scheckel, Stoecklin, & Lykke-Andersen, 2011) and KH-type splicing regulatory protein (KSRP; Winzen et al., 2007) generally lead to rapid degradation of the mRNA by recruitment of deadenylases (Cevher & Kleiman, 2010) or recruitment of 3′→5′ exosome as in the case for AUF1 (C. Y. Chen et al., 2002). Analysis of the binding site(s) for HuR and AUF1 present in androgen receptor MTA1 mRNA show that the same sequence is contacted by both proteins (Barker et al., 2012), consistent with the idea that both RBPs compete for binding to their cognate recognition sequences. While CDKN1A transcripts are destabilized by AUF1 and RBPs from the poly(C)-binding family under nonstress conditions (Barreau, Paillard, & Osborne, 2005; Scoumanne, Cho, Zhang, & Chen, 2011; Waggoner, Johannes, & Liebhaber, 2009), HuR and heterogeneous nuclear ribonucleoprotein C1 (hnRNP C1) interaction with CDKN1A mRNA stabilizes the transcript via binding to its ARE following UV, gamma radiation, and other stress causing treatments (Cho, Zhang, & Chen, 2010; W. Wang et al., 2000).
A similar functional competition between HuR and AUF1 was described for transcripts of c-fos, granulocyte macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and cyclin D under different stress condition (Barreau et al., 2005). The DNA damage-inducible Gadd45α transcript, which is up-regulated in response to stress stimuli, is downregulated by AUF1 and T-cell-restricted intracellular antigen (TIA) 1-related protein (TIAR), which prevents the mRNA association with translating polysomes (Lal et al., 2006). After UV or MMS treatment, MAPKAP kinase-2 (p38/MK2) increases Gadd45α mRNA stability through phosphorylation of the RBPs hnRNPA0, TIAR, and PARN deadenylase (Reinhardt et al., 2010). In contrast, the PCBP (poly(C)-binding protein) family of RBPs, composed of five major members hnRNP K, PCBP1, PCBP2, PCBP3, and PCBP4, binds CU-rich elements in the 3′ UTR to negatively regulate p21 expression (Scoumanne et al., 2011; Waggoner et al., 2009).
4.6 |. Competition between RBPs and miRNAs in controlling gene expression during DDR
RBPs interacting with the 3′ UTRs of target genes can affect the potential of miRNAs to regulate gene expression (Boucas et al., 2012; Meisner & Filipowicz, 2011; Vos, Leedman, Filipovska, & Rackham, 2019). In fact, deadenylation is a major component of miRNA-mediated repression (Eulalio et al., 2009) and miRNAs play a large role in DDR (reviewed in Wan, Mathur, Hu, Zhang, & Lu, 2011; Y. Wang & Taniguchi, 2013). For example, HuR relieves miRNA-mediated translational repression of cationic acid transporter-1 (CAT-1) only under stress conditions (Bhattacharyya, Habermacher, Martine, Closs, & Filipowicz, 2006). HuR has also been shown to compete with miRNA-induced silencing complex (miRISC) loaded with miR125b for binding TP53 mRNA, resulting in the inhibition of PARN-mediated deadenylation and stabilization of TP53 under stress conditions (X. Zhang et al., 2015). A computational study described a specific group of miRNA recognition sites enriched within 50 nucleotides from the RBP recognition sites for Pumilio and UAUUUAU, suggesting a potential competitive or cooperative regulation to maintain mRNA homeostasis (P. Jiang, Singh, & Coller, 2013). For example, it has been reported that over 75% of mRNAs with Ago binding sites in the 3′ UTR also have HuR binding sites (Figure 3), and most of these Ago and HuR binding sites overlap or are adjacent, with a distance of less than 10 nt of one another (Mukherjee et al., 2011). While some studies have shown that ARE-mediated decay can occur independent of miRNA functions (Helfer, Schott, Stoecklin, & Förstemann, 2012), other publications have shown that miRNAs machinery can functionally interact with ARE-BPs regulating ARE-mediated decay. For example, binding of HuR to AREs present in c-Myc 3′ UTR facilitates the targeting of let-7-loaded miRISC resulting in downregulation of c-Myc mRNA levels (Kim et al., 2009). Furthermore, miR-130a and mir-301a cooperate with nucleolin in the deadenylation of CSF-1 mRNA (Woo, Baker, Laszlo, & Chambers, 2013), and binding of HuR to adjacent mi125b seed sequence in TP53 3′ UTR inhibits miRNA-dependent translation repression (Ahuja, Goyal, & Ray, 2016). Additionally, cooperation of the ARE-BP tristetraprolin and miRISC that results in the recruitment of the deadenylase for tumor necrosis factor-α mRNA degradation has been described (Jing et al., 2005). Thus, a dynamic and complex flux exists between RBP and miRNA regulation of mRNAs during disruption of homeostasis. As the role of miRNAs-mediated regulation of mRNA stability during DDR has been extensively reported (reviewed in Han, Wan, Langley, Zhang, & Lu, 2012; He, Zhou, Li, & Guo, 2016; H. Hu & Gatti, 2011; Wan et al., 2011), here we will only discuss few examples of this regulatory mechanism. In the earlier steps of detection of DNA damage, miRNAs are involved in the regulation of key regulators of DDR, such as Ataxia-telangiectasia-mutated (ATM) (Maréchal & Zou, 2013). miR-18a (Cao et al., 2018; Song et al., 2011), miR-421 (Mansour et al., 2013), miR-101 (D. Yan et al., 2010), and miR-181 (Y. Wang, Huang, et al., 2011; Y. Wang, Yu, et al., 2011) have been shown to suppress ATM expression and in some cases the formation of nuclear foci by its downstream substrates H2AX and 53BP1. ATM phosphorylates H2AX, a required step for the assembly of DNA repair proteins at the sites of damaged chromatin and activation of checkpoints proteins (Turinetto & Giachino, 2015). Overexpression of miR-24 (Lal et al., 2009) and miRNA-138 (Y. Wang, Huang, et al., 2011; Y. Wang, Yu, et al., 2011; H. Yang, Luo, Liu, Zhou, & Luo, 2015) results in cells hypersensitive to DNA-damaging agents and decrease in foci formation of phosphorylated H2AX. On the effectors side, BRCA1, a critical tumor suppressor recruited to DNA damage lesions to facilitate DNA repair, is regulated by miR-182 (Moskwa et al., 2011), miR-146a, and 146b-5p (Garcia et al., 2011). The expression of the tumor suppressor p53 is downregulated by miR-125b (Le et al., 2011) and miR-504 (W. Hu et al., 2010) in several types of human cells. Interestingly, p53 can increase the cellular levels of miRNAs that target Mdm2 expression, such as miR-605 (Xiao, Lin, Luo, Luo, & Wang, 2011) and miR-143/miR-145 (F. Zhang, Ma, et al., 2013; J. Zhang, Sun, et al., 2013), allowing rapid accumulation of p53 by a positive feedback loop that ensures rapid activation of DDR. As mentioned above, p53 can promote the inhibition of CpA reaction (Nazeer et al., 2011) and PARN-mediated activation of deadenylation of non-DDR genes (Devany et al., 2013).
4.7 |. RNA structure as 3′ end processing regulatory element
The binding of proteins and/or noncoding RNAs to specific recognition sequences is not the only mechanism to regulate 3′ end processing, as secondary structures within mRNAs can also dictate mRNA stability by recruiting specific RBPs and enzyme complexes (Wu & Brewer, 2012). For example, ribonucleoprotein hnRNP H/F plays a key role in binding to G-quadruplex structure in TP53 mRNA maintaining 3′ end processing during global CpA inhibition (Decorsière et al., 2011). Additionally, the stem-loop structure formed by histone pre-mRNAs is recognized by stem-loop binding protein (SLBP) and is necessary for the 3′ end processing of histone mRNAs (Battle & Doudna, 2001; Dominski & Marzluff, 1999; Lampert, Brodersen, & Peter, 2017; Marzluff & Koreski, 2017). More recent research has uncovered that the ubiquitin ligase CRL4, a protein complex notable for its role in genome stability (Abbas & Dutta, 2011) and UV-mediated proteolysis of cell cycle regulator p21 (Abbas et al., 2008), also monoubiquitinates SLBP, permitting proper histone mRNA 3′ end maturation (Brodersen et al., 2016; Lampert et al., 2017). During G2 stress, cyclin-F, the substrate recognition subunit of the Skp1-Cul1-F-box E3 ligase complex, targets SLBP to proteasomal degradation and limits H2A.X signaling and apoptosis following DNA damage (Dankert et al., 2016).
5 |. NON-POLYADENYLATE 3′ END MODIFICATIONS AND EPITRANSCRIPTOMICS
5.1 |. Polyuridylation
In addition to PAP-mediated ribonucleotidyl transferase activity and deadenylases, a number of poly(U) polymerases that add poly(U) rather than poly(A) to their RNA substrates have been described (Kwak & Wickens, 2007; Rissland & Norbury, 2008). While uridylation has been shown to play a role in promoting decay of mature non-coding RNAs, it plays a role in regulating mRNA deadenylation, translation, and possibly storage (De Almeida, Scheer, Zuber, & Gagliardi, 2018). A global analysis identified that most mRNAs in mammalian cells undergo 3′ end uridylation with a negative correlation with mRNA stability (Chang, Lim, Ha, & Kim, 2014), suggesting an overall role in gene expression dynamics during conditions requiring low mRNA half-life. Histone mRNAs are the only known mRNAs canonically known not to be polyadenylated upon RNA cleavage, instead these mRNAs are stabilized by a stem loop structure tens of nucleotides downstream from the stop codon (Pandey & Marzluff, 1987). Due to the lack of a poly(A) tail, it was proposed that histone mRNA stability was only regulated at the translational level (Kaygun & Marzluff, 2005). However, inhibition of DNA synthesis by HU treatment was shown to lead to uridylation of 3′ end of histone mRNAs, which involved recruitment of both 3′→5′ exonucleases and decapping enzymes (Mullen & Marzluff, 2008). The oligo(U) tail serves as a cis-element for the exoRnase Eri1 to process the stem loop structure (Hoefig et al., 2013). Interestingly, arsenic treatment led to polyadenylation of histone mRNAs by downregulation of SLBP expression, and these poly(A) + mRNAs were not susceptible to normal degradation resulting in increased canonical histones levels (Brocato et al., 2014), which is associated with cellular sensitivity to DNA damaging agents (Arita & Costa, 2009; Brocato & Costa, 2013; Kurat et al., 2014).
In addition to histone mRNA regulation, global poly(A) + mRNAs are regulated by 3′ end uridylation during DDR. When cellular stress has reached a critical threshold, the global apoptotic program is activated, leading to cell death. Preexisting mRNAs, but not some ncRNAs, are rapidly and markedly degraded early after apoptosis induction by diverse classical apoptotic stimuli before other apoptotic events occur such as membrane lipid scrambling, DNA fragmentation, and inactivation of translation (Thomas et al., 2015). Genotoxic agents that induce mitochondrial outer membrane permeabilization (MOMP) and caspase activation, but not caspase-independent cell death or oxidative stress, led to widespread poly(A) + mRNA decay (Thomas et al., 2015). Interestingly, the RNA decay intermediates first undergo a non-templated 3′ end uridylation by uridylyl transferases TUT4 and TUT7 and then are recognized by the 3′→5′ exoribonuclease DIS3L2, which preferentially recognizes oligouridylated 3′ ends (Figure 2). The recognition and decay of which promote apoptotic pathway (Thomas et al., 2015). Interestingly, many ncRNAs are protected from this degradation by structures such as stem loops (X. Liu et al., 2018). These studies suggest that mRNA decay is important for making sure the death program passes the “point of no return.” In non- apoptotic cells, this pathway of TUTases and DIS3L2 act in quality control pathways of mRNAs and ncRNAs (Pirouz, Du, Munafò, & Gregory, 2016; Ustianenko et al., 2016). In contrast to the destabilizing effect of terminal uridyl, a more recent report has identified promiscuous nucleotidyl transferase activity leading to guanylation of the poly(A) tail that protects mRNAs from degradation (Chang et al., 2014; Y. Lim et al., 2018). TENT4A (PAPD7) and TENT4B (PAPD5) are noncanonical poly(A) polymerases with terminal nucleotidyltransferase activity that catalyze preferentially the transfer of ATP and GTP on mRNA 3′ poly(A) tail (Y. Lim et al., 2018). Importantly, a single guanosine residue is sufficient to impede the mRNA decay-mediated by deadenylase CCR4-NOT complex. While the role of guanylation in DDR is not known, these guanidyl transferases are also a catalytic subunit of the Trf4/Air2/Mtr4p polyadenylation (TRAMP)-like complex which has a poly(A) RNA polymerase activity and is involved in a post-transcriptional quality control mechanism (Ogami, Chen, & Manley, 2018). Given their similar mRNA abundances (D. Zheng & Tian, 2014), in future studies, it will be necessary to determine whether this modification is involved in DDR as uridylation.
5.2 |. 3′ end processing and epitranscriptomics
While epigenomic studies on consequences that chromatin marks have in cellular functions have elucidated much detail (reviewed in Stricker, Köferle, & Beck, 2017), it is still challenging to determine the causative roles that these marks actually have on gene expression. On the other hand, much less is known about epitranscriptomics, basically how dynamic RNA modification affects RNA fate and affects gene expression. The major mRNA modifications in the transcriptome of eukaryotic cells are N6-methyladenosine (m6A), N6, 2′-O-dimethyladenosine, 5-methylcytidine, 5-hydroxylmethylcytidine, inosine, pseudouridine, and N1-methyladenosine (Roundtree, Evans, Pan, & He, 2017). Most of our understanding in this relatively new field is based on developing technologies, so the cellular functions of each modification are in question (Boulias et al., 2019; Sendinc et al., 2019), and how many of these modifications found on mRNAs and ncRNAs are legitimate (Khoddami et al., 2019; W. Li, Li, et al., 2017; X. Li, Xiong, et al., 2017; Safra et al., 2017). However, m6A has been repeatedly demonstrated to be involved in post-transcriptional regulation of gene expression (Yue et al., 2018; Yue, Liu, & He, 2015). Interestingly, the majority of m6A modifications have been detected in the last exon and 3′ UTRs (Figure 2), and the presence of this modification correlated with increased usage of distal poly(A) sites in human (S. Ke et al., 2015) and regulation of APA during mouse oocyte development (Kasowitz et al., 2018). Although UV-mediated regulation of DNA repair using m6A as molecular beacon has been described (Nishida, Kuwano, Nishikawa, Masuda, & Rokutan, 2017; Xiang et al., 2017), usage of epitranscriptomic marks during DDR is grossly understudied. It will be of interest to determine whether additional RNA modifications, such as m1A, play a role in the regulation of polyadenylation/deadenylation. RNA modifications could also play a role in other non-polyadenylate mechanism directly or through ncRNA modifications.
6 |. CONCLUSIONS
In the last 10 years extensive studies have been undertaken to understand the role of mRNA 3′ end processing in DDR. While the direct connection between DDR and mRNA 3′ end processing machinery, particularly for tumor suppressors, provides an obvious link to clinical situations in diseases such as cancer, other clinical connections have been described (Curinha, Braz, Pereira-Castro, Cruz, & Moreira, 2014). For instance, several diseases, including fragile X syndrome, myotonic dystrophy, Huntington’s disease, arise, and are exacerbated by increased microsatellite repeat expansion, such as (CAG)n, in coding and non-coding regions of mRNAs (López Castel, Cleary, & Pearson, 2010; McMurray, 2010). Repeat instability is facilitated by DNA repair enzymes, such as Cockayne Syndrome B (Lin & Wilson, 2007), and factors involved in the mRNA 3′ processing machinery, such as the endoribonuclease CpA specificity factor 73 (CPSF73) (McGinty et al., 2017). CPSF73 suppresses (GAA)n expansion in actively transcribed regions as a result of enhanced transcription elongation rate, highlighting the interplay of DNA repair and CpA processes. Affecting mRNA length and composition by APA or disrupted CpA is also involved in variety of disease models such as α- and β-thalassemia (Harteveld et al., 1994; Higgs et al., 1983; Orkin, Cheng, Antonarakis, & Kazazian, 1985; Rund et al., 1992), cancer (Elkon et al., 2012; Mayr & Bartel, 2009; Sandberg, Neilson, Sarma, Sharp, & Burge, 2008; Singh et al., 2009; Xia et al., 2014), diabetes (Garin et al., 2010), thrombosis (Gehring et al., 2001; Lane & Grant, 2000), and oculopharyngeal muscular dystrophy (Brais et al., 1998). To illustrate, some of the notable diseases with known causative or associative roles that function at the intersection between DDR and mRNA 3′ end processing are highlighted in Table 1.
TABLE 1.
Factors or complexes with roles in DDR and mRNA 3′ end processing found to have a causative or associative link with various non-tumorigenic human diseases
| CpA/DDR factors attributed to non-cancer related pathologies | ||
|---|---|---|
| Factor/complex | Disease(s) | Reference |
| PARN | Dyskeratosis congenita | Tummala et al., 2015 |
| Integrator | Neurodevelopmental delay | Oegema et al., 2017 |
| DIS3L2 | Perlman syndrome | Astuti et al., 2012 |
| SLBP | Osteoarthritis (SNP association) | Castaño-Betancourt et al., 2016 |
| Ago2 | Addiction (SNP association) | Barragán et al., 2016 |
| PABPN1 | Oculopharyngeal muscular dystrophy (OMPD) | Schreuder, de Die-Smulders, Herbergs, & Koehler, 2006 |
| FIP1L1 | Hypereosinophilic syndrome | Cools, Stover, & Gilliland, 2006 |
| Rad51 | Fanconi anemia-like | A. T. Wang etal., 2015 |
| DNA-PKcs | Severe combined immunodeficiency | van der Burg et al., 2009 |
| PARP1 | Parkinson’s disease | Kam et al., 2018 |
| hnRNP K | Au-Kline syndrome | Au et al., 2018 |
| POLH | Xeroderma Pigmentosum variant V | J. Guo, Zhou, Zhang, Song, & Bian, 2013 |
| mtPAP | Spastic ataxia with optic atrophy | W. C. Wilson et al., 2014 |
| CstF-64 | Intellectual disability (SNP association) | Grozeva et al., 2015 |
| Clp1 (CFIIm) | Pontocerebellar hypoplasia | Karaca et al., 2014 |
Note: Unless specified, the gene-disease relationship was experimentally determined to play a direct role. Single nucleotide polymorphismss (SNP) identified near DDR/3′ processing genes by genome-wide association studies (GWAS) are indicated as “SNP association.” SNP associations were cataloged and searchable by gene or disease on DisGeNET (Piñero et al., 2017).
Abbreviations: CpA, cleavage and polyadenylation; DDR, DNA damage response; PARN, poly(A)-specific ribonuclease.
Sometimes, 3′ end processing acts as the signal rather than the effector for DDR. For instance, recent work uncovered the fascinating link between transcription and replication, whereby deregulation of mRNA cleavage impaired replication fork speed and excessive origin activity, activating ATR-dependent DDR (Teloni et al., 2019). These studies indicate that the CpA machinery protects cells from replication-stress-associated DNA damage, suggesting that pre-mRNA cleavage allows an efficient release of nascent transcripts and prevents genomic instability. Aside from transcriptional effects, genotoxic-stress mediated DNA translesion synthesis by PolH is itself regulated by APA. Specifically, APA generates a PolH transcript with shortened 3′ UTR that results in enhanced PolH expression, increasing cancer cell resistance to genotoxic stress (J. Zhang et al., 2019).
Ultimately, the pathways identified above represent only a fraction of the actual occurrences in how mRNA 3′ end processing and DDR are intimately coupled. It will be of great interest to see how the further elucidation of roles of ncRNAs in this functional connection, as well as the continuation of sequencing from patient samples uncovering epistatic interactions between DDR and mRNA processing components hitherto unexplored. We look forward to the next 10 years of research at the exciting intersection of these fields.
ACKNOWLEDGMENTS
We apologize to those authors whose work was not cited because of space limitations.
Funding information
National Cancer Institute, National Institutes of Health (NIH), Grant/Award Numbers: 1U54CA221704-01A, R21 CA204610-01
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIRES ARTICLES
Connections between 3′-end processing and DNA damage response
FURTHER READING
- Abdelmohsen K, & Gorospe M (2010). Posttranscriptional regulation of cancer traits by HuR. Wiley Interdisciplinary Reviews. RNA, 1, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P, & Ross J (1989). Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends in Biochemical Sciences, 14, 373–377. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, … Karin M (2001). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell, 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Colgan DF, & Manley JL (1997). Mechanism and regulation of mRNA polyadenylation. Genes & Development, 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, & Prives C (1992). Wild-type p53 activates transcription in vitro. Nature, 358, 83–86. [DOI] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, & Keller W (1996). Sequence similarity between the 73-Kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- Kim HH, & Gorospe M (2008). Phosphorylated HuR shuttles in cycles. Cell Cycle, 7, 3124–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, & Oren M (2009). The first 30 years of p53: Growing ever more complex. Nature Reviews. Cancer, 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, & Petrenko O (2003). The MDM2-p53 interaction. Molecular Cancer Research: MCR, 14, 1001–1008. [PubMed] [Google Scholar]
- Richard P, Trollet C, Stojkovic T, de Becdelievre A, Perie S, Pouget J, … Neurologists of French Neuromuscular Reference Centers CORN-EMUS and FILNEMUS. (2017). Correlation between PABPN1 genotype and disease severity in oculopharyngeal muscular dystrophy. Neurology, 88, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong JC, Zhang J, & Chen X (2010). RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Research, 38, 2256–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szcześniak MW, & Makałowska I (2016). lncRNA-RNA interactions across the human transcriptome. PLoS One, 11, e0150353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, & Volkert MR (2013). UV damage regulates alternative polyadenylation of the RPB2 gene in yeast. Nucleic Acids Research, 41, 3104–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- Abbas T, & Dutta A (2011). CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle, 10, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, & Dutta A (2008). PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes & Development, 22, 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, … Gorospe M (2014). 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Research, 42, 10099–10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Pullmann R Jr., Lal A, Kim HH, Galban S, Yang X, … Gorospe M (2007). Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Molecular Cell, 25, 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, & Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nature Reviews Genetics, 13, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Kim M, & Buratowski S (2004). Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Molecular Cell, 13, 67–76. [DOI] [PubMed] [Google Scholar]
- Ahuja D, Goyal A, & Ray PS (2016). Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biology, 13, 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, & Sharp PA (2013). Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature, 499, 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita A, & Costa M (2009). Epigenetics in metal carcinogenesis: Nickel, arsenic, chromium and cadmium. Metallomics: Integrated Biometal Science, 1, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Masutani M, Copeland TD, Mimori T, Sugimura T, Shimotohno K, … Noda M (1999). Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene, 18, 4616–4625. [DOI] [PubMed] [Google Scholar]
- Aström J, Aström A, & Virtanen A (1991). In vitro deadenylation of mammalian mRNA by a HeLa cell 3′ exonuclease. The EMBO Journal, 10, 3067–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Morris MR, Cooper WN, Staals RHJ, Wake NC, Fews GA, … Maher ER (2012). Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nature Genetics, 44, 277–284. [DOI] [PubMed] [Google Scholar]
- Au PYB, Goedhart C, Ferguson M, Breckpot J, Devriendt K, Wierenga K, … Care for Rare Canada Consortium. (2018). Phenotypic spectrum of Au-Kline syndrome: A report of six new cases and review of the literature. European Journal of Human Genetics: EJHG, 26, 1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, & Shiekhattar R (2005). Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell, 123, 265–276. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, … Landthaler M (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Molecular Cell, 46, 674–690. [DOI] [PubMed] [Google Scholar]
- Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MCJ, Wilce JA, … Leedman PJ (2012). Sequence requirements for RNA binding by HuR and AUF1. Journal of Biochemistry, 151, 423–437. [DOI] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, & Anderson RA (2010). Nuclear phosphoinositides: A signaling enigma wrapped in a compartmental conundrum. Trends in Cell Biology, 20, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MD, Moon SL, Emch AW, Wilusz CJ, & Wilusz J (2013). Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Reports, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán R, Coltell O, Asensio EM, Francés F, Sorlí JV, Estruch R, … Corella D (2016). MicroRNAs and drinking: Association between the pre-miR-27a rs895819 polymorphism and alcohol consumption in a Mediterranean population. International Journal of Molecular Science, 16, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, & Osborne HB (2005). AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Research, 33, 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkowiak B, Liu P, Phatnani HP, Fuda NJ, Cooper JJ, Price DH, … Greenleaf AL (2010). CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes & Development, 24, 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, & Doudna JA (2001). The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA, 7, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu YB, Kleinman CL, Landry-Voyer AM, Majewski J, & Bachand F (2012). Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genetics, 8, e1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014). Coupling mRNA processing with transcription in time and space. Nature Reviews Genetics, 15, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, … Dreyfuss G (2012). U1 snRNP determines mRNA length and regulates isoform expression. Cell, 150, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, & Mayr C (2015). Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature, 522, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersani C, Xu LD, Vilborg A, Lui WO, & Wiman KG (2014). Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14–3-3σ. Oncogene, 33, 4407–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, & Filipowicz W (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell, 125, 1111–1124. [DOI] [PubMed] [Google Scholar]
- Blattner C, Kannouche P, Litfin M, Bender K, Rahmsdorf HJ, J, H., … Herrlich, P. (2000). UV-induced stabilization of c-Fos and other short-lived mRNAs. Molecular and Cellular Biology, 20, 3616–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek D, Kohoutek J, Bartholomeeusen K, Johansen E, Hulinkova P, Luo Z, … Peterlin BM (2011). The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes and Development, 25, 2158–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig F, Winzen R, Kracht M, Ghebremedhin B, Ritter B, Wilhelm A, … Holtmann H (2002). Evidence for general stabilization of mRNAs in response to UV light. European Journal of Biochemistry, FEBS, 269, 5830–5839. [DOI] [PubMed] [Google Scholar]
- Boucas J, Riabinska A, Jokic M, Herter-Sprie GS, Chen S, Höpker K, & Reinhardt HC (2012). Posttranscriptional regulation of gene expression-adding another layer of complexity to the DNA damage response. Frontiers in Genetics, 3, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, … Greer L (2019). Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Molecular Cell, 75, 631–643. 10.1016/j.molcel.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chrétien N, Tomé FM, … Rouleau GA (1998). Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nature Genetics, 18, 164–167. [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, … Bentley DL (2012). mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Molecular Cell, 46, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato J, & Costa M (2013). Basic mechanics of DNA methylation and the unique landscape of the DNA Methylome in metal-induced carcinogenesis. Critical Reviews in Toxicology, 43, 493–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocato J, Fang L, Chervona Y, Chen D, Kiok K, Sun H, … Costa M (2014). Arsenic induces polyadenylation of canonical histone mRNA by down-regulating stem-loop-binding protein gene expression. The Journal of Biological Chemistry, 289, 31751–31764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen MML, Lampert F, Barnes CA, Soste M, Piwko W, & Peter M (2016). CRL4(WDR23)-mediated SLBP ubiquitylation ensures histone supply during DNA replication. Molecular Cell, 62, 627–635. [DOI] [PubMed] [Google Scholar]
- Cao P, Zhang M, Wang L, Sai B, Tang J, … Xiang J (2018). miR-18a reactivates the Epstein-Barr virus through defective DNA damage response and promotes genomic instability in EBV-associated lymphomas. BMC Cancer, 18, 1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Betancourt MC, Evans DS, Ramos YFM, Boer CG, Metrustry S, Liu Y, … van Meurs JBJ (2016). Novel genetic variants for cartilage thickness and hip osteoarthritis. PLoS Genetics, 12, e1006260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, … Hentze MW (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406. [DOI] [PubMed] [Google Scholar]
- Cevher MA, & Kleiman FE (2010). Connections between 3′-end processing and DNA damage response. Wiley Interdisciplinary Reviews, RNA, 1, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, … Kleiman FE (2010). Nuclear Deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. The EMBO Journal, 29, 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S, … Brody JR (2017). Posttranscriptional regulation of PARG mRNA by HuR facilitates DNA repair and resistance to PARP inhibitors. Cancer Research, 77, 5011–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, & Kim VN (2014). TAIL-Seq: Genome-wide determination of poly(A) tail length and 3– end modifications. Molecular Cell, 53, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chen CY, & Shyu AB (2003). Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Molecular and Cellular Biology, 23, 4805–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Xu N, & Shyu AB (2002). Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Molecular and Cellular Biology, 22, 7268–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ezzeddine N, Waltenspiel B, Albrecht TR, Warren WD, Marzluff WF, & Wagner EJ (2012). An RNAi screen identifies additional members of the Drosophila integrator complex and a requirement for Cyclin C/Cdk8 in snRNA 3′-end formation. RNA, 18, 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, & Wagner EJ (2010). snRNA 3′ end formation: The dawn of the integrator complex. Biochemical Society Transactions, 38, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Zhang J, & Chen X (2010). RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Research, 38, 2256–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann M, & Kaina B (2013). Transcriptional regulation of human DNA repair genes following genotoxic stress: Trigger mechanisms, inducible responses and genotoxic adaptation. Nucleic Acids Research, 41, 8403–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement SL, Scheckel C, Stoecklin G, & Lykke-Andersen J (2011). Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Molecular and Cellular Biology, 31, 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Albrecht AS, de Melo Costa VR, Sauer S, Meierhofer D, & Ørom UA (2016). Serial interactome capture of the human cell nucleus. Nature Communications, 7, 11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools J, Stover EH, & Gilliland DG (2006). Detection of the FIP1L1-PDGFRA fusion in idiopathic hypereosinophilic syndrome and chronic eosinophilic leukemia. Methods in Molecular Medicine, 125, 177–187. [DOI] [PubMed] [Google Scholar]
- Crossley MP, Bocek M, & Cimprich KA (2019). R-loops as cellular regulators and genomic threats. Molecular Cell, 73, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curinha A, Braz SO, Pereira-Castro I, Cruz A, & Moreira A (2014). Implications of polyadenylation in health and disease. Nucleus, 5, 508–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Zhang G, & Makeyev EV (2012). RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Research, 40, 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, … Kulozik AE (2011). p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Molecular Cell, 41, 298–310. [DOI] [PubMed] [Google Scholar]
- Dankert JF, Rona G, Clijsters L, Geter P, Skaar JR, … Pagano M (2016). Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2. Molecular Cell, 64, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Muniz L, & West S (2014). 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes & Development, 28, 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass B, McMahon KW, Jenkins NA, Gilbert DJ, Copeland NG, & MacDonald CC (2001).The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome19 and is expressed in pachytene spermatocytes in mice. The Journal of Biological Chemistry,276, 8044–8050. [DOI] [PubMed] [Google Scholar]
- Davis AJ, Chen BPC, & Chen DJ (2014). DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair, 17, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida C, Scheer H, Zuber H, & Gagliardi D (2018). RNA uridylation: A key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdisciplinary Reviews. RNA, 9, e1440 10.1002/wrna.1440 [DOI] [PubMed] [Google Scholar]
- Decorsière A, Cayrel A, Vagner S, & Millevoi S (2011). Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes & Development, 25, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, … Babak T (2012). A quantitative atlas of polyadenylation in five mammals. Genome Research, 22, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devany E, Park JY, Murphy MR, Zakusilo G, Baquero J, Zhang X, … Kleiman FE (2016). Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discovery, 2, 16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devany E, Zhang X, Park JY, Tian B, & Kleiman FE (2013). Positive and negative feedback loops in the p53 and mRNA 3′ processing pathways. PNAS, 110, 3351–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanraj S, Gunja SMR, Deveau AP, Nissbeck M, Boonyawat B, Coombs AJ, … Dror Y (2015). Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN). Journal of Medical Genetics, 52, 738–748. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Li W, Ogami K, Yashinskie JJ, Hoque M, Tian B, & Manley JL (2014). RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3′ UTRs. Genes & Development, 28, 2248–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, & Manley JL (2013). PARP1 represses PAP and inhibits polyadenylation during heat shock. Molecular Cell, 49, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianatpour A, & Ghafouri-Fard S (2017). The role of long non coding RNAs in the repair of DNA double strand breaks. International Journal of Molecular Medicine, 6, 1–12. [PMC free article] [PubMed] [Google Scholar]
- Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, & Eberhardt W (2007). Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: Implications for posttranscriptional regulation of cyclooxygenase-2. Molecular Biology of the Cell, 18, 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, & Marzluff WF (1999). Formation of the 3′ end of histone mRNA. Gene, 239, 1–14. [DOI] [PubMed] [Google Scholar]
- Dutertre M, Chakrama FZ, Combe E, Desmet FO, Mortada H, Espinoza MP, … Auboeuf DA (2014). Recently evolved class of alternative 3′-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nature Communications, 5, 3395. [DOI] [PubMed] [Google Scholar]
- Dutertre M, & Vagner S (2017). DNA-damage response RNA-binding proteins (DDRBPs): Perspectives from a new class of proteins and their RNA targets. Journal of Molecular Biology, 429, 3139–3145. [DOI] [PubMed] [Google Scholar]
- Egloff S, Al-Rawaf H, O’Reilly D, & Murphy S (2009). Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Molecular and Cellular Biology, 29, 4002–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eifler TT, Shao W, Bartholomeeusen K, Fujinaga K, Jäger S, Johnson JR, … Peterlin BM (2015). Cyclin-dependent kinase 12 increases 3′ end processing of growth factor-induced c-FOS transcripts. Molecular and Cellular Biology, 35, 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekumi KM, Paculova H, Lenasi T, Pospichalova V, Bosken CA, Rybarikova J, … Barboric M (2015). Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic Acids Research, 43, 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, & Agami R (2012). E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biology, 13, R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, & Agami R (2013). Alternative cleavage and polyadenylation: Extent, regulation and function. Nature Reviews. Genetics, 14, 496–506. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, & Izaurralde E (2009). Deadenylation is a widespread effect of miRNA regulation. RNA, 15, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, … Wagner E (2011). A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and Spliceosomal snRNA 3′-end formation. Molecular and Cellular Biology, 31, 328–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ishmael FT, Fang X, Myers A, Cheadle C, Huang SK, … Stellato C (2011). Chemokine transcripts as targets of the RNA-binding protein HuR in human airway epithelium. Journal of Immunology, 186, 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca D, Baquero J, Murphy MR, Aruggoda G, Varriano S, Sapienza C, … Kleiman FE (2018). mRNA processing factor CstF-50 and ubiquitin escort factor p97 are BRCA1/BARD1 cofactors involved in chromatin remodeling during the DNA damage response. Molecular and Cellular Biology, 38, 364–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusby B, Kim S, Erickson B, Kim H, Peterson ML, & Bentley DL (2015). Coordination of RNA polymerase II pausing and 3′ end processing factor recruitment with alternative polyadenylation. Molecular and Cellular Biology, 36, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, … Poirier GG (2008). Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Research, 36, 6959–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, & Aguilera A (2014). Cleavage factor I links transcription termination to DNA damage response and genome integrity maintenance in Saccharomyces cerevisiae. PLoS Genetics, 10, e1004203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbán S, Martindale JL, Mazan-Mamczarz K, Lopez de Silanes I, Fan J, Wang W, … Gorospe M (2003). Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Molecular and Cellular Biology, 23, 7083–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, … Mazoyer S (2011). Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Molecular Medicine, 3, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mauriño SM, Rivero-Rodríguez F, Velázquez-Cruz A, Hernández-Vellisca M, Díaz-Quintana A, De la Rosa MA, & Díaz-Moreno I (2017). RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Frontiers in Molecular Biosciences, 4, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin I, Edghill EL, Akerman I, Rubio-Cabezas O, Rica I, Locke JM, … Hattersley AT (2010). Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. PNAS, 107, 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavaldá S, Gallardo M, Luna R, & Aguilera A (2013). R-loop mediated transcription-associated recombination in trf4Δ mutants reveals new links between RNA surveillance and genome integrity. PLoS One, 8, e65541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, & Kulozik AE (2001). Increased efficiency of mRNA 3′ end formation: A new genetic mechanism contributing to hereditary thrombophilia. Nature Genetics, 28, 389–392. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, Mellman DL, & Anderson RA (2008). CKIα is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. The Journal of Biological Chemistry, 283, 12665–12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar G, Winzen R, Bollig F, Ghebremedhin B, Redich N, Ritter B, … Holtmann H (2005). Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biological Chemistry, 386, 1287–1293. [DOI] [PubMed] [Google Scholar]
- Graber JH, Nazeer FI, Yeh PC, Kuehner JN, Borikar S, … Moore CL (2013). DNA damage induces targeted, genome-wide variation of poly(A) sites in budding yeast. Genome Research, 23, 1690–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis I, Abdelmohsen K, & Gorospe M (2017). Posttranslational control of HuR function. Wiley Interdisciplinary Reviews. RNA, 8, e1372 10.1002/wrna.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D, Carss K, Spasic-Boskovic O, Tejada M-I, Gecz J, Shaw M, … Raymond FL (2015). Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Human Mutation, 36, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, & Mattaj IW (1998). U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Molecular Cell, 1, 255–264. [DOI] [PubMed] [Google Scholar]
- Gunzburg MJ, Sivakumaran A, Pendini NR, Yoon JH, Gorospe M, Wilce MCJ, & Wilce JA (2015). Cooperative interplay of Let-7 mimic and HuR with MYC RNA. Cell Cycle, 14, 2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhou G, Zhang W, Song Y, & Bian Z (2013). A novel POLH mutation causes XP-V disease and XP-V tumor proneness may involve imbalance of numerous DNA polymerases. Oncology Letters, 6(6), 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, & Geller DA (2014). microRNA and human inducible nitric oxide synthase. Vitamins and Hormones, 96, 19–27. [DOI] [PubMed] [Google Scholar]
- Han C, Wan G, Langley RR, Zhang X, & Lu X (2012). Crosstalk between the DNA damage response pathway and microRNAs. Cellular and Molecular Life Sciences, 69, 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hande KR (1998). Etoposide: Four decades of development of a topoisomerase II inhibitor. European Journal of Cancer, 34, 1514–1521. [DOI] [PubMed] [Google Scholar]
- Harteveld CL, Losekoot M, Haak H, Heister GA, Giordano PC, & Bernini LF (1994). A novel polyadenylation signal mutation in the alpha 2-globin gene causing alpha thalassaemia. British Journal of Haematology, 87, 139–143. [DOI] [PubMed] [Google Scholar]
- He M, Zhou W, Li C, & Guo M (2016). MicroRNAs, DNA damage response, and cancer treatment. International Journal of Molecular Sciences, 17, 2087–2101. 10.3390/ijms17122087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer S, Schott J, Stoecklin G, & Förstemann K (2012). AU-rich element-mediated mRNA decay can occur independently of the miRNA machinery in mouse embryonic fibroblasts and Drosophila S2-cells. PLoS One, 7, e28907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SEY, Lamb J, Clegg JB, Weatherall DJ, & Proudfoot NJ (1983). Alpha-thalassemia caused by a polyadenylation signal mutation. Nature, 306, 398–400. [DOI] [PubMed] [Google Scholar]
- Hirose Y, & Manley JL (1998). RNA polymerase II is an essential mRNA polyadenylation factor. Nature, 395, 93–96. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, … Heissmeyer V (2013). Eril degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nature Structural & Molecular Biology, 20, 73–81. [DOI] [PubMed] [Google Scholar]
- Hollerer I, Curk T, Haase B, Benes V, Hauer C, Neu-Yilik G, … Kulozik AE (2016). The differential expression of alternatively polyadenylated transcripts is a common stress-induced response mechanism that modulates mammalian mRNA expression in a quantitative and qualitative fashion. RNA, 22, 1441–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollerer I, Grund K, Hentze MW, & Kulozik AE (2014). mRNA 3′ end processing: A tale of the tail reaches the clinic. EMBO Molecular Medicine, 6, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Li W, & Tian B (2014). Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods in Molecular Biology, 1125, 119–129. [DOI] [PubMed] [Google Scholar]
- Hu H, & Gatti RA (2011). MicroRNAs: New players in the DNA damage response. Journal of Molecular Cell Biology, 3, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, … Feng Z (2010). Negative regulation of tumor suppressor p53 by microRNA miR-504. Molecular Cell, 38, 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Shi J, Guo Y, Huang W, Huang S, Ming S, … Yao C (2017). A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Research, 45, 8647–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachimowicz RD, Goergens J, & Reinhardt HC (2019). DNA double-strand break repair pathway choice - from basic biology to clinical exploitation. Cell Cycle, 18, 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, & Bartek J (2009). The DNA-damage response in human biology and disease. Nature, 461, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalkanen AL, Coleman SJ, & Wilusz J (2014). Determinants and implications of mRNA poly(A) tail size—does this protein make my tail look big? Seminars in Cell & Developmental Biology, 34, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, & Tulin AV (2010). The roles of PARP1 in gene control and cell differentiation. Current Opinion in Genetics & Development, 20, 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wolgast M, Beebe LM, & Reese JC (2019). Ccr4-not maintains genomic integrity by controlling the ubiquitylation and degradation of arrested RNAPII. Genes & Development, 33, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Singh M, & Coller HA (2013). Computational assessment of the cooperativity between RNA binding proteins and MicroRNAs in transcript decay. PLoS Computational Biology, 9, e1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, … Han J (2005). Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell, 120, 623–634. [DOI] [PubMed] [Google Scholar]
- Johnson SF, Cruz C, Greifenberg AK, Dust S, Stover DG, Chi D, … Shapiro GI (2016). CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Reports, 17, 2367–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, & Nielsen ML (2013). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Molecular Cell, 52, 272–285. [DOI] [PubMed] [Google Scholar]
- Kai M (2016). Roles of RNA-binding proteins in DNA damage response. International Journal of Molecular Sciences, 17, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, & Dreyfuss G (2010). U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature, 468, 664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam T-I, Mao X, Park H, Chou S-C, Karuppagounder SS, Umanah GE, … Dawson VL (2018). Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease. Science, 362, eaat8407 10.1126/science.aat8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, … Lupski JR (2014). Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell, 157, 636–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, … Wang PJ (2018). Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genetics, 14, e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H, & Marzluff WF (2005). Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Molecular and Cellular Biology, 25, 6879–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, … Darnell RB (2015). A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes & Development, 29, 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, … Zeng X (2017). PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nature Communications, 8, 14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, & Cairns BR (2019). Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. PNAS, 116, 6784–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr., Yang X, Galban S, … Gorospe M (2008). Nuclear HuR accumulation through phosphorylation by Cdk1. Genes & Development, 22, 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, & Gorospe M (2009). HuR recruits Let-7/RISC to repress c-Myc expression. Genes & Development, 23, 1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, & Gorospe M (2010). Regulation of HuR by DNA damage response kinases. Journal of Nucleic Acids, 2010, 981487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, & Manley JL (1999). Functional interaction of BRCAl-associated BARD1 with polyadenylation factor CstF-50. Science, 285, 1576–1579. [DOI] [PubMed] [Google Scholar]
- Kleiman FE, & Manley JL (2001). The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell, 104, 743–753. [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Wu-Baer F, Fonseca D, Kaneko S, Baer R, & Manley JL (2005). BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes and Development, 19, 1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Dries R, Grassetti AV, Dust S, Gao Y, Huang H, … George RE (2019). CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nature Communications, 10, 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynik SM, Gabanic A, Anthony SR, Kelley M, Paulding WR, Roessler A, … Tranter M (2015). The stress-induced heat shock protein 70.3 expression is regulated by a dual-component mechanism involving alternative polyadenylation and HuR. Biochimica et Biophysica Acta, 1849, 688–696. [DOI] [PubMed] [Google Scholar]
- Krell J, Frampton AE, Mirnezami R, Harding V, De Giorgio A, Roca Alonso L, … Castellano L (2014). Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One, 9, e98561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Recht J, Radovani E, Durbic T, Andrews B, & Fillingham J (2014). Regulation of histone gene transcription in yeast. Cellular and Molecular Life Sciences, 71, 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, & Wickens M (2007). A family of poly(U) polymerases. RNA, 13, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey PE, Welch JD, & Marzluff WF (2016). TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA, 22, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, … Shi Y (2014). Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. The EMBO Journal, 33, 878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, & Anderson RA (2010). The poly a polymerase star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. The EMBO Journal, 29, 4132–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, … Gorospe M (2006). Posttranscriptional derepression of GADD45alpha by genotoxic stress. Molecular Cell, 22, 117–128. [DOI] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, … Chowdhury D (2009). miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nature Structural & Molecular Biology, 16, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Brodersen MML, & Peter M (2017). Guard the guardian: A CRL4 ligase stands watch over histone production. Nucleus, 8, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DA, & Grant PJ (2000). Role of hemostatic gene polymorphisms in venous and arterial thrombotic disease. Blood, 95, 1517–1532. [PubMed] [Google Scholar]
- Le MTN, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J, … Lim B (2011). Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genetics, 7, e1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, & Rajewsky N (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Molecular Cell, 43, 340–352. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kang BH, Jang H, Kim TW, Choi J, Kwak S, … Youn HD (2015). AKT phosphorylates H3-threonine 45 to facilitate termination of gene transcription in response to DNA damage. Nucleic Acids Research, 43, 4505–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SD, & Moore CL (2014). Efficient mRNA polyadenylation requires a ubiquitin-like domain, a zinc knuckle, and a RING finger domain, all contained in the Mpe1 protein. Molecular and Cellular Biology, 34, 3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Lee Y, & Chung JH (2000). An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Letters, 487, 287–292. [DOI] [PubMed] [Google Scholar]
- Li W, Laishram RS, Ji Z, Barlow CA, Tian B, & Anderson RA (2012). Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCδ signaling. Molecular Cell, 45, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li W, Laishram RS, Hoque M, Ji Z, … Anderson RA (2017). Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases. Nucleic Acids Research, 45, 8930–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, … Tian B (2015). Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genetics, 11, e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, … Yi C (2017). Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Molecular Cell, 68, 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Estep JA, & Karginov FV (2018). Transcriptome-wide identification and validation of interactions between the miRNA machinery and HuR on mRNA targets. Journal of Molecular Biology, 430, 285–296. [DOI] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, … Wilusz JE (2017). The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Molecular Cell, 68, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Kim D, Lee YS, Ha M, Lee M, Yeo J, … Kim VN (2018). Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science, 361, 701–704. [DOI] [PubMed] [Google Scholar]
- Lin Y, & Wilson JH (2007). Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Molecular and Cellular Biology, 27, 6209–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu R, Pan Y, Meza-Sosa KF, Zhang Z, & Lieberman J (2018). PNPT1 release from mitochondria during apoptosis triggers decay of poly(A) RNAs. Cell, 174, 187–201. [DOI] [PubMed] [Google Scholar]
- López Castel A, Cleary JD, & Pearson CE (2010). Repeat instability as the basis for human diseases and as a potential target for therapy. Nature Reviews. Molecular Cell Biology, 11, 165–170. [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, & Gorospe M (2004). Identification of a target RNA motif for RNA-binding protein HuR. PNAS, 101, 2987–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan J, Bowerman S, & Luger K (2019). Quantitating repair protein accumulation at DNA lesions: Past, present, and future. DNA Repair (Amst), 81, 102650–102656. 10.1016/j.dnarep.2019.102650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour WY, Bogdanova NV, Kasten-Pisula U, Rieckmann T, Köcher S, Borgmann K, … Dahm-Daphi J (2013). Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiotherapy and Oncology: Journal of the European Society for Therapeutic Radiology and Oncology, 106, 147–154. [DOI] [PubMed] [Google Scholar]
- Mao P, & Wyrick JJ (2019). Organization of DNA damage, excision repair, and mutagenesis in chromatin: A genomic perspective. DNA Repair (Amst), 8, 102645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, & Zou L (2013). DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspectives in Biology, 5, 12716–12733. 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NT, Nakamura K, Paila U, Woo J, Brown C, Wright JA, … Concannon P (2014). Homozygous mutation of MTPAP causes cellular radiosensitivity and persistent DNA double-strand breaks. Cell Death & Disease, 5, e1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, & Koreski KP (2017). Birth and death of histone mRNAs. Trends in Genetics, 33, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, & Duronio RJ (2008). Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nature Reviews. Genetics, 9, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ, & Bessler M (2015). mRNA deadenylation and telomere disease. The Journal of Clinical Investigation, 125, 1796–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Abdelmohsen K, Kim MM, Srikantan S, Lee EK, Tominaga K, … Gorospe M (2011). Global dissociation of HuR–mRNA complexes promotes cell survival after ionizing radiation. The EMBO Journal, 30, 1040–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Al-Tinawi QMH, Rouchka EC, & Fondufe-Mittendorf YN (2019). Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics & Chromatin, 12, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, & Bartel DP (2009). Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in Cancer cells. Cell, 138, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayya VK, & Duchaine TF (2019). Ciphers and executioners: How 3′-untranslated regions determine the fate of messenger RNAs. Frontiers in Genetics, 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RJ, Puleo F, Aksenova AY, Hisey JA, Shishkin AA, Pearson EL, & Mirkin SM (2017). A defective mRNA cleavage and polyadenylation complex facilitates expansions of transcribed (GAA)n repeats associated with Friedreich’s Ataxia. Cell Reports, 20, 2490–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray CT (2010). Mechanisms of trinucleotide repeat instability during human development. Nature Reviews. Genetics, 11, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, & Filipowicz W (2011). Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Advances in Experimental Medicine and Biology, 700, 106–123. [DOI] [PubMed] [Google Scholar]
- Melikishvili M, Chariker JH, Rouchka EC, & Fondufe-Mittendorf YN (2017). Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discovery, 3, 17043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, & Anderson RA (2008). A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature, 451, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Merchut-Maya JM, Bartek J, & Maya-Mendoza A (2019). Regulation of replication fork speed: Mechanisms and impact on genomic stability. DNA Repair (Amst), 8, 102654. [DOI] [PubMed] [Google Scholar]
- Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, … Di Fagagna DA (2017). Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nature Cell Biology, 19, 1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Chibi M, Xie P, Koundrioukoff S, Moolman-Smook H, Pugh D, … Defossez PA (2014). The RBBP6/ZBTB38/MCM10 Axis regulates DNA replication and common fragile site stability. Cell Reports, 7, 575–587. [DOI] [PubMed] [Google Scholar]
- Mirkin N, Fonseca D, Mohammed S, Cevher MA, Manley JL, & Kleiman FE (2008). The 3′ processing factor CstF functions in the DNA repair response. Nucleic Acids Research, 36, 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, … Shiloh Y (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316, 1160–1166. [DOI] [PubMed] [Google Scholar]
- Montecucco A, & Biamonti G (2013). Pre-mRNA processing factors meet the DNA damage response. Frontiers in Genetics, 4, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, … Agarwal S (2015). Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nature Genetics, 47, 1482–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra DS, Lawler SH, Eliceiri BP, & Eliceiri GL (1986). Inhibition of small nuclear RNA synthesis by ultraviolet radiation. The Journal of Biological Chemistry, 261, 3142–3146. [PubMed] [Google Scholar]
- Mansour WY, Bogdanova NV, Kasten-Pisula U, Rieckmann T, Köcher S, Borgmann K, … Gatti RA (2013). Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiotherapy and Oncology, 106, 147–154. [DOI] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, … Chowdhury D (2011). miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Molecular Cell, 41, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motadi LR, Lekganyane MM, & Moela P (2018). RBBP6 expressional effects on cell proliferation and apoptosis in breast cancer cell lines with distinct p53 statuses. Cancer Management and Research, 10, 3357–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, … Keene JD (2011). Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Molecular Cell, 43, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder KW, Winkler GS, & Timmers HTM (2005). DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-not complex. Nucleic Acids Research, 33, 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, & Marzluff WF (2008). Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes & Development, 22, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazeer FI, Devany E, Mohammed S, Fonseca D, Akukwe B, Taveras C, & Kleiman FE (2011). p53 inhibits mRNA 3′ processing through its interaction with the CstF/BARDl complex. Oncogene, 30, 3073–3083. [DOI] [PubMed] [Google Scholar]
- Neve J, Patel R, Wang Z, Louey A, & Furger AM (2017). Cleavage and polyadenylation: Ending the message expands gene regulation. RNA Biology, 14, 865–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M, Sfaxi R, Saha A, Monchaud D, Teulade-Fichou MP, & Vagner S (2017). The G-Quadruplex-specific RNA helicase DHX36 regulates p53 pre-mRNA 3′-end processing following UV-induced DNA damage. Journal of Molecular Biology, 429, 3121–3131. [DOI] [PubMed] [Google Scholar]
- Nicholson AL, & Pasquinelli AE (2019). Tales of detailed poly(A) tails. Trends in Cell Biology, 29, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Kuwano Y, Nishikawa T, Masuda K, & Rokutan K (2017). RNA binding proteins and genome integrity. International Journal of Molecular Sciences, 18, 1341–1354. 10.3390/ijms18071341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor MJ (2015). Targeting the DNA damage response in cancer. Molecular Cell, 60, 547–560. [DOI] [PubMed] [Google Scholar]
- Oegema R, Baillat D, Schot R, van Unen LM, Brooks A, Kia SK, … Mancini GMS (2017). Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genetics, 13, e1006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Chen Y, & Manley JL (2018). RNA surveillance by the nuclear RNA exosome: Mechanisms and significance. Non-Coding RNA, 4, 8–28. 10.3390/ncrna4010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D, Kuznetsova OV, Laitem C, JZaborowska J, Dienstbier M, & Murphy S (2014). Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Research, 42, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Cheng TC, Antonarakis SE, & Kazazian HH (1985). Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. The EMBO Journal, 4, 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, & Marzluff WF (1987). The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Molecular and Cellular Biology, 7, 4557–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, … Cimprich KA (2009). A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Molecular Cell, 35, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, … Furlong LI (2017). DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research, 45, D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz M, Du P, Munafò M, & Gregory RI (2016). Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Reports, 16, 1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2011). Ending the message: Poly(A) signals then and now. Genes & Development, 25, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MATM, Wang X, … Yaffe MB (2010). DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Molecular Cell, 40, 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienzo M, & Casamassimi A (2016). Integrator complex and transcription regulation: Recent findings and pathophysiology. Biochimica et Bio- physica Acta, 1859, 1269–1280. [DOI] [PubMed] [Google Scholar]
- Rissland OS, & Norbury CJ (2008). The Cid1 poly(U) polymerase. Biochimica et Biophysica Acta, 1779, 286–294. [DOI] [PubMed] [Google Scholar]
- Rockx DA, Mason R, van Hoffen A, Barton MC, Citterio E, Bregman DB, … Mullenders LH (2000). UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. PNAS, 97, 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, & He C (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund D, Dowling C, Najjar K, Rachmilewitz EA, Kazazian HH Jr., & Oppenheim A (1992). Two mutations in the beta-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. PNAS, 89, 4324–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, & Bauer DL (2008). Finishing touches: Post-translational modification of protein factors involved in mammalian pre-mRNA 3′ end formation. The International Journal of Biochemistry & Cell Biology, 40, 2384–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, … Schwartz S (2017). The m1A landscape on cytosolic and mitochondrial Mrna at single-base resolution. Nature, 551, 251–255. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, & Burge CB (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science, 320, 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder AH, de Die-Smulders CE, Herbergs J, & Koehler PJ (2006). From gene to disease; the PABN1 gene and oculopharyngeal muscular dystrophy. Ned Tijdschr Geneeskd, 150, 1124–6. [PubMed] [Google Scholar]
- Scoumanne A, Cho SJ, Zhang J, & Chen X (2011). The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Research, 39, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, & Livingston DM (1997). Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell, 90, 425–435. [DOI] [PubMed] [Google Scholar]
- Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, … Shi Y (2019). PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Molecular Cell, 75, 620–630. 10.1016/j.molcel.2019.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Swenson PA, & Carrier WL (1963). Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science, 142, 1464–1466. [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, & Ko MSH (2009). Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Research, 16, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Huang C, Huang S, & Yao C (2018). snoRNAs associate with mRNA 3′ processing complex: New wine in old bottles. RNA Biology, 15, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, … Manley JL (2009). Molecular architecture of the human pre-mRNA 3′ processing complex. Molecular Cell, 33, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, & Manley JL (2015). The end of the message: Multiple protein–RNA interactions define the mRNA polyadenylation site. Genes & Development, 29, 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, & Rotter V (1997). PACT: Cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene, 14, 145–155. [DOI] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, … Graber JH (2009). Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Research, 69, 9422–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, … Pagano M (2015). The integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Research, 25, 288–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Gamberi C, Mihaylova Y, Grosswendt S, Chen C, Lasko P, & Aboobaker AA (2013). The CCR4-NOT complex mediates deadenylation and degradation of stem cell mRNAs and promotes planarian stem cell differentiation. PLoS Genetics, 9, e1004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Lin C, Wu Z, Gong H, Zeng Y, Wu J, … Li J (2011). miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One, 6, e25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, … Hieter P (2012). R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes & Development, 26, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker SH, Köferle A, & Beck S (2017). From profiles to function in epigenomics. Nature Reviews. Genetics, 18, 51–66. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, & Marzluff WF (2009). A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Molecular Cell, 34, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Hao Q, & Prasanth KV (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends in Genetics: TIG, 34, 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, & Tong L (2018). Molecular basis for the recognition of the human AAUAAA polyadenylation signal. PNAS, 115, E1419–E1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, & Manley JL (1996). The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell, 87, 941–952. [DOI] [PubMed] [Google Scholar]
- Teloni F, Michelena J, Lezaja A, Kilic S, Ambrosi C, Menon S, … Altmeyer M (2019). Efficient pre-mRNA cleavage prevents replication-stress-associated genome instability. Molecular Cell, 73, 670–683 (e12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, … Lieberman J (2015). Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Reports, 11, 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, & Lutz CS (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Research, 33, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, & Manley JL (2017). Alternative polyadenylation of mRNA precursors. Nature Review Molecular Cell Biology, 18, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien JF, Mazloomian A, Cheng SWG, Hughes CS, Chow CCT, Canapi LT, … Morin GB (2017). CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Research, 45, 6698–6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Hammet A, Tenis N, Denis CL, & Heierhorst J (2005). Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics, 169, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, … Dokal I (2015). Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. The Journal of Clinical Investigation, 12, 2151–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V, & Giachino C (2015). Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Research, 43, 2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, … Vanacova S (2016). TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. The EMBO Journal, 35, 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg M, Ljspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, … van Gent DC (2009). A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. The Journal of Clinical Investigation, 119, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidhyasagar V, He Y, Guo M, Talwar T, Singh RS, Yadav M, … Wu Y (2018). Biochemical characterization of INTS3 and C9ORF80, two subunits of hNABP1/2 heterotrimeric complex in nucleic acid binding. The Biochemical Journal, 475, 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieux KF, & Clarke HJ (2018). CNOT6 regulates a novel pattern of mRNA deadenylation during oocyte meiotic maturation. Scientific Reports, 8, 6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, et al. (2009). The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. PNAS, 106, 15756–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos PD, Leedman PJ, Filipovska A, & Rackham O (2019). Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cellular and Molecular Life Sciences, 76, 3745–3752. 10.1007/s00018-019-03163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner SA, Johannes GJ, & Liebhaber SA (2009). Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of Cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. The Journal of Biological Chemistry, 284, 9039–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Mathur R, Hu X, Zhang X, & Lu X (2011). miRNA Response to DNA damage. Trends in Biochemical Sciences, 36, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, … Smogorzewska A (2015). A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Molecular Cell, 59(3), 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, … Gorospe M (2000). HuR regulates p21 mRNA stabilization by UV light. Molecular and Cellular Biology, 20, 760–769.10629032 [Google Scholar]
- Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, … Taniguchi T (2011). MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Molecular Cancer Research, 9, 1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, & Taniguchi T (2013). MicroRNAs and DNA damage response: Implications for cancer therapy. Cell Cycle, 12, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, … Wang SE (2011). Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene, 30, 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, … Passmore LA (2018). mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-not nucleases. Molecular Cell, 70, 1089–1100.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, … MacDonald CC (1999). Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. PNAS, 96, 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, & Sunder S (2019). Double-strand breaks in motion: Implications for chromosomal rearrangement. Current Genetics. 10.1007/s00294-019-01015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WC, Hornig-Do H-T, Bruni F, Chang JH, Jourdain AA, Martinou JC, … Lightowlers RN (2014). A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Human Molecular Genetics, 23, 6345–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, & Wilusz J (2007). HuR-SIRT: The hairy world of posttranscriptional control. Molecular Cell, 25, 485–487. [DOI] [PubMed] [Google Scholar]
- Wilusz JE (2017). Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biology, 14, 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, … Holtmann H (2007). Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Molecular and Cellular Biology, 27, 8388–8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HH, Baker T, Laszlo C, & Chambers SK (2013). Nucleolin mediates microRNA-directed CSF-1 mRNA deadenylation but increases translation of CSF-1 mRNA. Molecular & Cellular Proteomics: MCP, 12, 1661–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstencroft RN, Beilharz TH, Cook MA, Preiss T, Durocher D, & Tyers M (2006). Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. Journal of Cell Science, 119, 5178–5192. [DOI] [PubMed] [Google Scholar]
- Wu X, & Brewer G (2012). The regulation of mRNA stability in mammalian cells: 2.0. Gene, 500, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, & Li W (2014). Dynamic analyses of alternative polyadenylation from RNA-Seq reveal a 3′-UTR landscape across seven tumour types. Nature Communications, 5, 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, … Shi Y (2017). RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature, 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Lin H, Luo X, Luo X, & Wang Z (2011). miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. The EMBO Journal, 30, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, & Yamaguchi Y (2014). DSIF and NELF interact with integrator to specify the correct post-transcriptional fate of snRNA genes. Nature Communications, 5, 4263. [DOI] [PubMed] [Google Scholar]
- Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, … Wang Y (2010). Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One, 5, e11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan YB (2014). Deadenylation: Enzymes, regulation, and functional implications. Wiley Interdisciplinary Reviews. RNA, 5, 421–443. [DOI] [PubMed] [Google Scholar]
- Yang H, Luo J, Liu Z, Zhou R, & Luo H (2015). MicroRNA-138 regulates DNA damage response in small cell lung cancer cells by directly targeting H2AX. Cancer Investigation, 33, 126–136. [DOI] [PubMed] [Google Scholar]
- Yang Q, Nausch LWM, Martin G, Keller W, & Doublié S (2014). Crystal structure of human poly(A) polymerase gamma reveals a conserved catalytic core for canonical poly(A) polymerases. Journal of Molecular Biology, 426, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Park J, Ha M, Lim J, Chang H, & Kim VN (2018). PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Molecular Cell, 70, 1081–1088. [DOI] [PubMed] [Google Scholar]
- Young SK, & Wek RC (2016). Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. The Journal of Biological Chemistry, 291, 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Rege M, Peterson CL, & Volkert MR (2016). RNA polymerase II depletion promotes transcription of alternative mRNA species. BMC Molecular Biology, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, … Liu J (2018). VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discovery, 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, & He C (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes & Development, 29, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, & Krebber H (2016). mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature, 540, 593–596. [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma T, & Yu Y (2013). A core hSSB1-INTS complex participates in the DNA damage response. Journal of Cell Science, 126, 4850–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, & Chen WT (2013). Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene, 32, 61–69. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sun W, Ren C, Kong X, Yan W, & Chen X (2019). A PolH transcript with a short 3′UTR enhances PolH expression and mediates Cisplatin resistance. Cancer Research, 79, 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LN, & Yan YB (2015). Depletion of poly(A)-specific ribonuclease (PARN) inhibits proliferation of human gastric cancer cells by blocking cell cycle progression. Biochimica et Biophysica Acta, 1853, 522–534. [DOI] [PubMed] [Google Scholar]
- Zhang X, Devany E, Murphy MR, Glazman G, Persaud M, & Kleiman FE (2015). PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic Acids Research, 43, 10925–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kleiman FE, & Devany E (2014). Deadenylation and its regulation in eukaryotic cells. Methods in Molecular Biology, 1125, 289–296. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xiao S, Rameau RD, Devany E, Nadeem Z, Caglar E, … Saxena A (2018). Nucleolin phosphorylation regulates PARN deadenylase activity during cellular stress response. RNA Biology, 15, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Zhang Y, & Yu Y (2017). A cell-line specific atlas for PARP-mediated protein asp/Glu-ADP-ribosylation in breast cancer. Cell Reports, 21, 2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, & Tian B (2014). Sizing up the poly(A) tail: Insights from deep sequencing. Trends in Biochemical Sciences, 39, 255–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Wang R, Ding Q, Wang T, Xie B, Wei L, … Tian B (2018). Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nature Communications, 9, 2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou HL, Hasman RA, & Lou H (2007). Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. The Journal of Biological Chemistry, 282, 2203–2210. [DOI] [PubMed] [Google Scholar]