ABSTRACT
The human parasite Trypanosoma brucei transitions from the trypomastigote form to the epimastigote form in the insect vector by repositioning its mitochondrial genome and flagellum-associated cytoskeleton. The molecular mechanisms underlying such changes in cell morphology remain elusive, but recent works demonstrated the involvement of three flagellar proteins, FLAM3, ClpGM6 and KIN-E, in this process by controlling the elongation of the flagellum attachment zone (FAZ). In this report, we identified a FAZ flagellum domain-localizing protein named FAZ27 and characterized its role in cell morphogenesis. Depletion of FAZ27 in the trypomastigote form caused major morphological changes and repositioning of the mitochondrial genome and flagellum-associated cytoskeleton, generating epimastigote-like cells. Furthermore, proximity biotinylation and co-immunoprecipitation identified FLAM3 and ClpGM6 as FAZ27-interacting proteins, and analyses of their functional interplay revealed an interdependency for assembly into the FAZ flagellum domain. Finally, we showed that assembly of FAZ27 occurred proximally, identical to the assembly pattern of other FAZ sub-domain proteins. Taken together, these results demonstrate a crucial role for the FAZ flagellum domain in controlling cell morphogenesis and suggest a coordinated assembly of all the FAZ sub-domains at the proximal end of the FAZ.
KEY WORDS: Trypanosoma brucei, Flagellum attachment zone, FLAM3, ClpGM6, Epimastigote, Trypomastigote
Summary: The flagellum attachment zone protein FAZ27 interacts with two flagellar proteins to maintain trypanosome cells in the trypomastigote form. Depletion of FAZ27 results in the formation of epimastigote-like cells.
INTRODUCTION
Trypanosoma brucei, an early-branching protozoan parasite that is the causative agent of sleeping sickness in humans and nagana in cattle in sub-Saharan Africa, possesses a motile flagellum that is also required for maintaining cell morphology, defining the cell division plane and mediating cell–cell communications (Gull, 1999). The flagellum is nucleated from the basal body (BB), a centriole-like microtubule-organizing center located at the posterior portion of the cell, exits the cell body through the flagellar pocket and attaches, along most of its length, to the cell body via a proteinaceous cytoskeletal structure termed the flagellum attachment zone (FAZ) (Gull, 1999). The FAZ consists of multiple sub-domains at the cell body–flagellum junction, including the FAZ flagellum domain, the FAZ intracellular domain and the FAZ filament domain, and is required for flagellum attachment and cell morphogenesis (Sunter and Gull, 2016). At the proximal portion of the flagellum, near the BB, another cytoskeletal structure termed the hook complex (HC), which is marked by the TbMORN1 protein (Esson et al., 2012), sits atop of the TbBILBO1-marked flagellar pocket collar (FPC) (Bonhivers et al., 2008) and plays essential roles in the regulation of flagellum positioning and attachment (Zhou et al., 2010). A centrin protein-marked structure termed the centrin arm (CA), which was originally named the bilobe structure (He et al., 2005), associates with the shank part of the HC and the proximal end of the intracellular FAZ filament (Esson et al., 2012). The biogenesis and segregation of these flagellum-associated cytoskeletal structures are well coordinated with the elongation of the flagellum during the cell cycle.
T. brucei has a complex life cycle that involves alternating between the insect vector tsetse fly and the mammalian hosts. Inside the insect vector, the parasite transitions to different developmental forms during their life cycle. These life cycle developmental forms differ morphologically due to changes in the position of the mitochondrial genome or kinetoplast relative to the nucleus, the position of the flagellum relative to the posterior cell end, as well as the length of the free, unattached flagellum (Hoare and Wallace, 1966). T. brucei develops from the trypomastigote form to the epimastigote form in the proventriculus of the tsetse fly, and in the salivary gland the epimastigote cells further develop into the mammal-infective metacyclic form (Hoare and Wallace, 1966). The molecular mechanisms underlying the transition from the trypomastigote form to the epimastigote form remain poorly understood. Recent works have demonstrated the involvement of three flagellar proteins, ClpGM6 (encoded by Tb927.11.1090), FLAM3 (encoded by Tb927.8.4780) and KIN-E (encoded by Tb927.5.2410), in regulating the morphology changes (An and Li, 2018; Hayes et al., 2014; Sunter et al., 2015a). Although these three proteins all localize to the flagellum, they have distinct sub-cellular localizations. ClpGM6 localizes to the FAZ flagellum domain (Hayes et al., 2014), whereas FLAM3 is enriched in the FAZ flagellum domain and is detected, at a lower level, in the unattached (free) flagellum, except the flagellar tip (Rotureau et al., 2014; Sunter et al., 2015a). Moreover, FLAM3 is additionally localized to the flagella connector or the distal tip of the newly formed flagellum (Sunter et al., 2015a). ClpGM6 and FLAM3 form a complex and are interdependent for assembly into the flagellum (Sunter et al., 2015a), demonstrating that they function together to control cell morphogenesis. KIN-E is a kinetoplastid-specific orphan kinesin and localizes to the entire flagellum with an enrichment at the distal tips of both old and new flagella (An and Li, 2018). KIN-E interacts with FLAM3 and is required for FLAM3 localization to the flagellum (An and Li, 2018). In addition to the abovementioned flagellar proteins, two FAZ filament proteins, FAZ9 (encoded by Tb927.10.14320) (McAllaster et al., 2015) and TbSAS-4 (encoded by Tb927.11.330) (Hu et al., 2015), appear to also be involved in cell morphogenesis. These findings suggest that morphology transitions from the trypomastigote form to the epimastigote form in T. brucei require the modulation of the abundance of FAZ-associated proteins.
In this report, we identify another FAZ flagellum domain protein named FAZ27, which interacts with FLAM3 and ClpGM6 and controls cell morphogenesis in T. brucei. We also demonstrate the interdependence for assembly into the FAZ between FAZ27 and its interacting partners, providing evidence to support their functional interplay in maintaining cell morphology. Finally, we show that the assembly of the FAZ flagellum domain occurs at the proximal end of the FAZ, in accordance with the assembly of other FAZ sub-domains. These findings provided insights into the roles of FAZ flagellum-domain proteins in cell morphogenesis.
RESULTS
FAZ27 is a new component of the FAZ flagellum domain in T. brucei
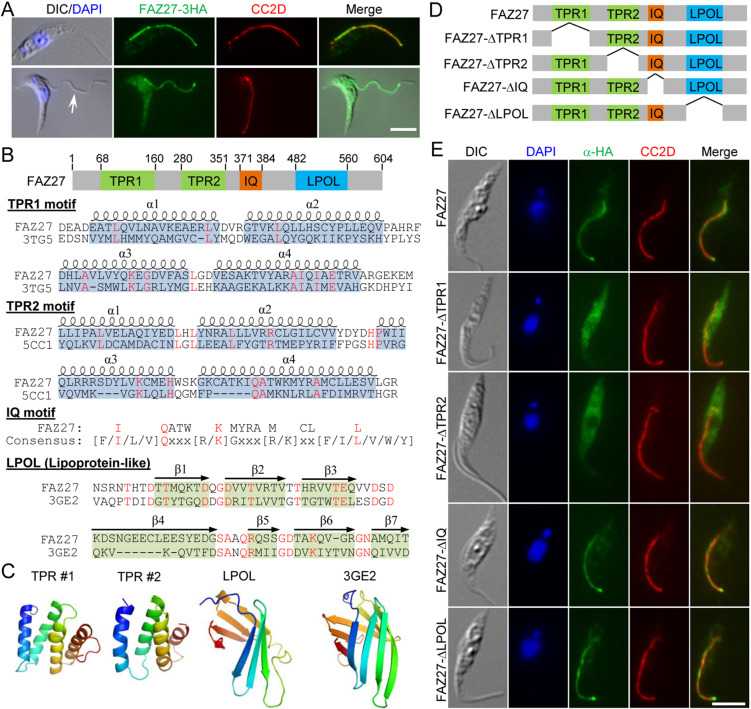
During the course of work that aimed to identify novel BB-associated proteins by epitope tagging of numerous hypothetical proteins whose expression is highly enriched during the S phase of the cell cycle (Dang et al., 2017), one hypothetical protein (Tb927.9.8350) was found to localize to the FAZ by immunofluorescence microscopy (Fig. 1A) and thus was named FAZ27. The protein is conserved in all Kinetoplastida parasites, but not outside of the kinetoplastids, suggesting that FAZ27 is a kinetoplastid-specific protein. Because the FAZ consists of multiple sub-domains in the junction between the flagellum and the cell body (Sunter and Gull, 2016), we attempted to determine the FAZ sub-domain that FAZ27 localizes to. To this end, we searched for cells in which the flagellum was detached during sample preparation, so as to distinguish between the FAZ sub-domains between the flagellum and the cell body. In cells with a detached flagellum, we found that FAZ27 associated with the flagellum, but not the FAZ filament domain marked by the anti-CC2D antibody (Fig. 1A), suggesting that FAZ27 resides on the FAZ sub-domains inside the flagellum, but not the FAZ sub-domains inside the cell. By conserved domain analysis and structural modeling, four structural motifs, including two tetratricopeptide repeat (TPR) domains, an IQ calmodulin-binding motif and a lipoprotein-like domain (herein abbreviated as LPOL), were identified in FAZ27 (Fig. 1B,C). The TPR domain consists of a series of antiparallel amphipathic α-helices and is involved in protein–protein interactions (Zeytuni and Zarivach, 2012). Thus, the two TPR domains in FAZ27 might be involved in protein–protein interactions. The IQ motif is a basic unit of ∼25 amino acids comprising a core consensus sequence ([F/I/L/V]QxxxRGxxx[R/K], where x represents any residue) and forms an amphiphilic seven-turn α-helix capable of binding to calmodulin in a calcium-dependent manner (Bähler and Rhoads, 2002). It is therefore possible that the IQ motif in FAZ27 might be able to bind to calmodulin and might be involved in calcium signaling. A lipoprotein consists of multiple anti-parallel β-sheets (Fig. 1B,C) and is primarily involved in transporting hydrophobic lipids. Hence, the lipoprotein-like domain in FAZ27 might be capable of binding to the flagellum membrane. However, FAZ27 does not contain any transmembrane domain; therefore, it is unlikely to localize on the flagellar membrane.
Fig. 1.
FAZ27 is a new FAZ flagellum domain-localizing protein. (A) Subcellular localization of FAZ27. Cells expressing FAZ27–3HA from its endogenous locus were stained with FITC-conjugated anti-HA antibody to label FAZ27–3HA, anti-CC2D antibody to label the FAZ and DAPI to stain DNA. The arrow indicates the detached flagellum in a 1N1K cell during sample preparation. (B) Schematic illustration of the structural motifs in FAZ27. TPR, tetratricopeptide repeat; IQ, isoleucine-glutamine motif; LPOL, lipoprotein-like motif. Alignment of the TPR domains in FAZ27 with those of the human SMYD2 protein (PBD: 3TG5) and the human S425G Glucocorticoid receptor DNA-binding domain (PBD: 5CC1), alignment of the lipoprotein-like domain in FAZ27 with the putative lipoprotein SP_0198 from Steptococcus pneumoniae (PBD: 3GE2) and alignment of the FAZ27 IQ motif with the IQ motif consensus sequence are shown. Identical residues are highlighted in red and the positions of α-helices and β-sheets are indicated. (C) Homology modeling of the TPR domains and the LPOL domain in FAZ27 using the structures mentioned in B as templates. (D) Schematic illustration of wild-type FAZ27 and its various deletion mutants, with domains indicated as in B. (E) Subcellular localization of ectopically expressed wild-type FAZ27 and its various deletion mutants. Cells harboring the overexpression constructs of wild-type FAZ27 and its various deletion mutants, which were tagged with a triple HA epitope at their C-terminus, were incubated with 1.0 µg/ml tetracycline for 24 h and then co-immunostained with FITC-conjugated anti-HA antibody and anti-CC2D antibody and counterstained with DAPI for nuclear and kinetoplast DNA. Scale bars: 5 µm. Images in A and E are representative of three experiments.
To examine the potential requirement of these structural motifs for FAZ27 localization, four FAZ27 mutants, in which each of the four structural motifs was deleted (Fig. 1D), were tagged with a C-terminal triple HA epitope and ectopically expressed in procyclic trypanosome cells. When overexpressed from the ectopic locus, wild-type FAZ27 correctly localized to the FAZ (Fig. 1E). Both the TPR1- and TPR2-deletion mutants were found to mislocalize to the cytosol, whereas the IQ-deletion mutant and the LPOL-deletion mutant were correctly localized to the FAZ (Fig. 1E). These results demonstrated that FAZ27 localization required each of the two TPR domains.
Depletion of FAZ27 causes major changes in cell morphology
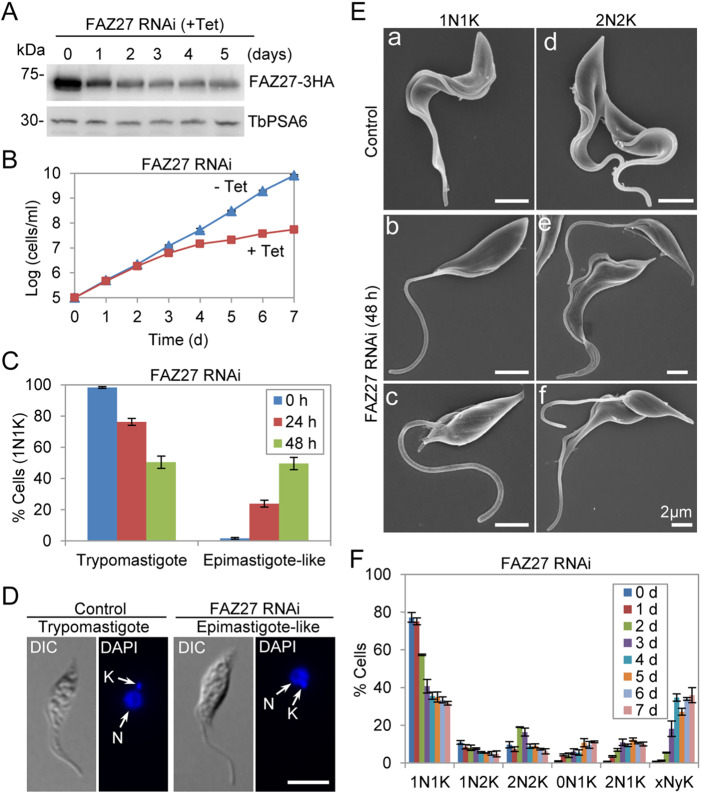
We investigated the function of FAZ27 in the procyclic (trypomastigote) form of T. brucei using inducible RNAi-mediated gene ablation. Tetracycline-induced RNAi resulted in a gradual decrease of the protein level of FAZ27, which was endogenously tagged with a triple HA epitope at its C-terminus, but did not result in the complete depletion of FAZ27 protein after 5 d of RNAi induction (Fig. 2A), likely due to low RNAi efficiency. This downregulation of FAZ27 caused growth defects (Fig. 2B), indicating that FAZ27 is required for cell proliferation in the procyclic form. Strikingly, RNAi of FAZ27 caused substantial changes in cell morphology, resulting in the production of epimastigote-like cells among ∼24% and ∼50% of cells with one nucleus and one kinetoplast (1N1K) after RNAi induction for 24 and 48 h, respectively (Fig. 2C). These epimastigote-like cells contained a repositioned kinetoplast, which was either anterior or juxtaposed to the nucleus, and a long, unattached (free) flagellum (Fig. 2D). Scanning electron microscopy showed that the free flagellum of the FAZ27 RNAi-deficient one-flagellum cells was much longer than that of the control one-flagellum cells (Fig. 2E, panels a–c), confirming the epimastigote-like morphology of these one-flagellum cells. The flagellum in these FAZ27 RNAi cells either attached to a small part of the cell body (Fig. 2E, panel b) or did not attach to the cell body (Fig. 2E, panel c). In the control two-flagellum cells, both flagella attached to the cell body for most of its length (Fig. 2E, panel d), whereas in the FAZ27 RNAi-induced two-flagellum cells, most (∼80%) of the cells possessed a long, unattached new flagellum and a normal, attached old flagellum (Fig. 2E, panel e), which, after cell division, appeared to generate an epimastigote-like cell and a normal-shaped cell. The rest (∼20%) of the two-flagellum cells possessed long, unattached new and old flagella (Fig. 2E, panel f). These cells appeared to have developed from the one-flagellum cells that had a long, unattached flagellum following cell cycle progression and, after cell division, appeared to produce two epimastigote-like cells. We also noted that FAZ27 RNAi induction for shorter times of up to 2 d caused major cell morphological changes, and RNAi induction for longer times after 3 d produced multi-nucleated (xNyK, x>2, y≥1) cells (Fig. 2F). These multi-nucleated cells all contained multiple long, unattached flagella, suggesting that they were derived from the epimastigote-like 1N1K cells.
Fig. 2.
Knockdown of FAZ27 by RNAi in the trypomastigote form produced epimastigote-like cells. (A) Western blotting to monitor the efficiency of RNAi against FAZ27. FAZ27 was endogenously tagged with a triple HA epitope in cells containing the FAZ27 RNAi construct, and was detected using an anti-HA antibody. TbPSA6 served as the loading control. Blots shown are representative of three experiments. (B) Effect of FAZ27 RNAi on cell proliferation, comparing RNAi-induced (+ Tet) and uninduced (− Tet) cells. Data are mean±s.d. from three replicates. (C) Percentages of 1N1K cells of trypomastigote and epimastigote-like morphology from control and FAZ27 RNAi cells. A total of 200 1N1K cells were counted for each time point. Data are mean±s.d. from three replicates. (D) Morphology of control and FAZ27 RNAi cells examined under a light microscope, with nuclear and kinetoplast DNA stained using DAPI. N, nucleus; K, kinetoplast. Scale bar: 5 μm. (E) Morphology of control and FAZ27 RNAi cells examined by scanning electron microscopy. Scale bars: 2 μm. (F) Effect of FAZ27 RNAi on cell cycle progression, examined by quantitation of cells with different numbers of nuclei (N) and kinetoplasts (K). xNyK indicates cells with more than two nuclei and more than one kinetoplast. A total of 350 cells were counted for each time point. Data are mean±s.d. from three independent experiments.
FAZ27 is required for elongation of the FAZ and positioning flagellum-associated cytoskeletal structures
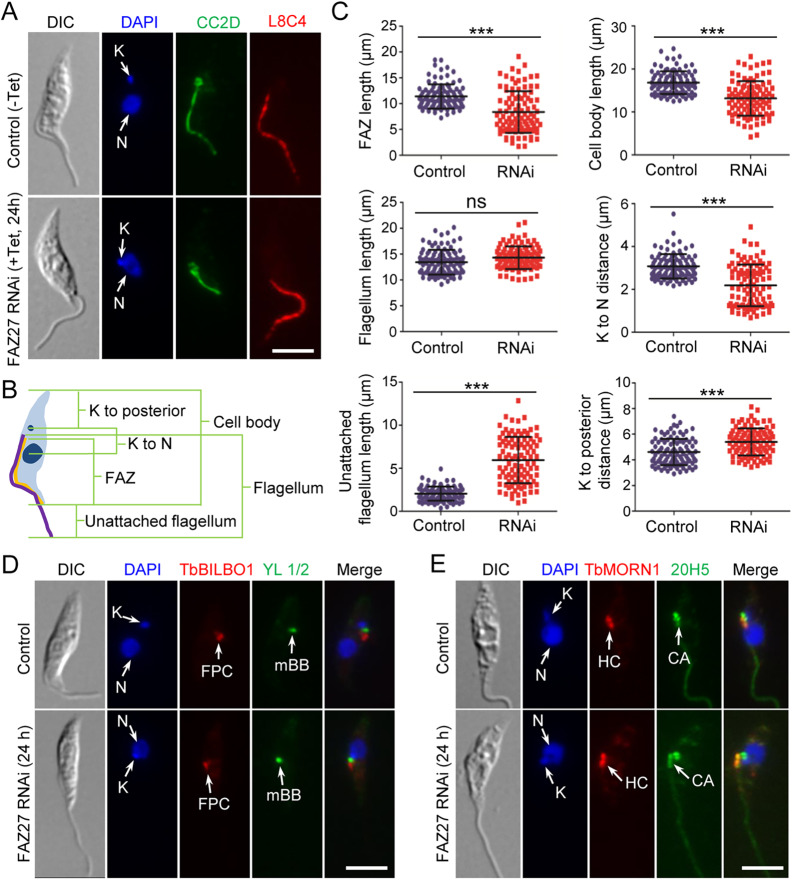
We further characterized the epimastigote-like cells to examine the length of the FAZ and the positioning of flagellum-associated cytoskeletal structures by immunofluorescence microscopy. Cells were immunostained with anti-CC2D antibody (Zhou et al., 2011) and anti-PFR2 antibody to label the FAZ filament inside the cell and the paraflagellar rod (PFR) inside the flagellum, respectively (Fig. 3A). We then measured the length of the FAZ, the flagellum, the unattached flagellum and the cell body, as well as the distance between the kinetoplast and the nucleus and the distance between the kinetoplast and the posterior tip of the cell (Fig. 3B). The results showed that those 1N1K cells produced by FAZ27 RNAi treatment possessed a shorter FAZ than the control 1N1K cells (an average length of 8.3 µm versus 11.4 µm) (Fig. 3C). Although the length of the flagellum was not significantly changed in FAZ27 RNAi cells (an average length of 14.3 µm versus 13.4 µm) (Fig. 3C), the length of the unattached flagellum of the FAZ27-deficient 1N1K cells was significantly longer than the unattached flagellum of the control 1N1K cells (an average length of 5.9 µm versus 2.0 µm) (Fig. 3C), which correlated with the reduced length of the FAZ after RNAi induction (Fig. 3C). Moreover, the length of the cell body of the FAZ27-deficient 1N1K cells was significantly shorter than that of the control 1N1K cells (an average cell body length of 13.2 µm versus 16.8 µm) (Fig. 3C). Furthermore, the kinetoplast–nucleus distance was reduced from 3.1 µm in control cells to 2.2 µm in FAZ27 RNAi cells, whereas the distance between the kinetoplast and the posterior tip was increased from 4.6 µm in control cells to 5.4 µm in FAZ27 RNAi cells (Fig. 3C). Finally, all of the flagellum-associated cytoskeletal structures, such as the FPC, the flagellar BB, and the HC, were repositioned toward the nucleus in the FAZ27-deficient 1N1K cells, as compared with that in the control 1N1K cells (Fig. 3D,E). Altogether, these results demonstrated that FAZ27 RNAi caused the change from the trypomastigote morphology to an epimastigote-like morphology.
Fig. 3.
Morphological analysis of the epimastigote-like cells generated by FAZ27 RNAi. (A) Morphology of the 1N1K cells from control and FAZ27 RNAi cells. Cells were co-immunostained with anti-CC2D antibody to label the FAZ and anti-PFR2 (clone L8C4) antibody to label the flagellum, and then counterstained with DAPI for nuclear (N) and kinetoplast (K) DNA. Scale bar: 5 μm. (B) Diagram depicting a 1N1K cell used for measuring the length of cell body, flagellum, unattached flagellum and FAZ, and for measuring the distance from kinetoplast (K) to the posterior cell tip and from kinetoplast to nucleus (N). (C) Morphometric measurements of non-induced control cells and FAZ27 RNAi-induced (24 h) cells. Cells were co-immunostained with anti-CC2D and anti-PFR2 antibodies. The length of the cell body, FAZ, flagellum and unattached flagellum, and the distance from kinetoplast to nucleus and from kinetoplast to the cell posterior were measured and plotted (n=100). Mean±s.d. values are indicated. ***P<0.001; ns, no statistical significance (Student's t-test). (D) Position of the flagellar basal body and flagellar pocket collar (FPC) in control and FAZ27 RNAi cells. Cells were co-immunostained with YL 1/2 antibody to label the mature basal body (mBB) and anti-TbBILBO1 antibody to label the FPC, and counterstained with DAPI for nuclear (N) and kinetoplast (K) DNA. Scale bar: 5 μm. (E) Position of the hook complex (HC) and the centrin arm (CA) in control and FAZ27 RNAi cells. Cells were co-immunostained with anti-TbMORN1 antibody to label the HC and 20H5 antibody to label the CA, and counterstained with DAPI for nuclear (N) and kinetoplast (K) DNA. Scale bar: 5 μm. Images in D,E are representative of three experiments.
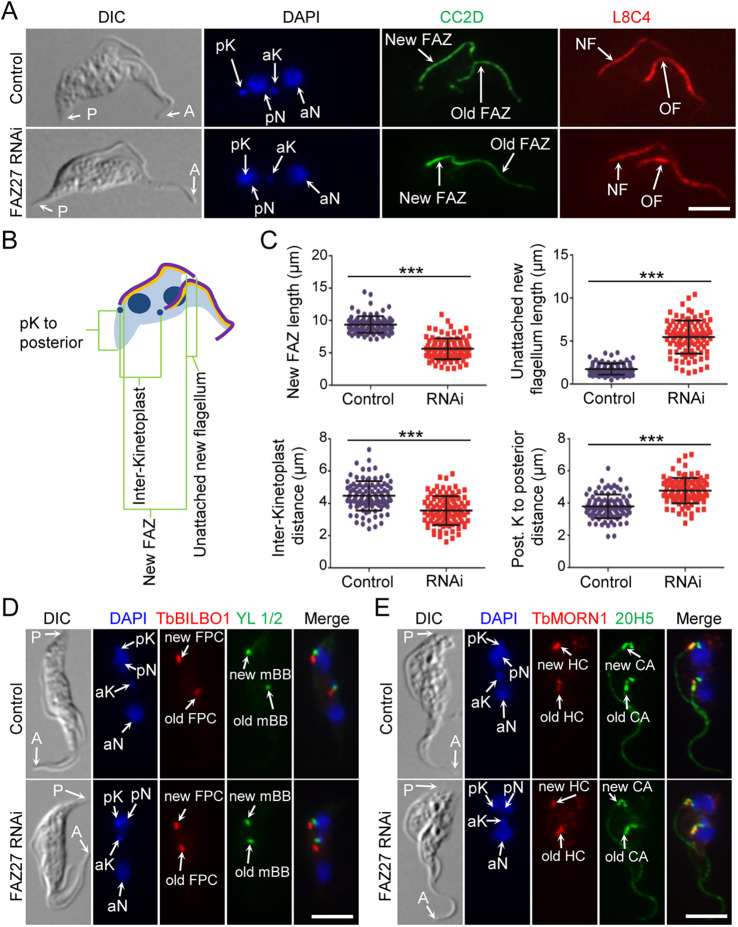
It is unlikely that FAZ27 RNAi affected the elongation of the existing (old) FAZ and the positioning of the existing (old) kinetoplast and flagellum-associated cytoskeletal structures, since these structures had already been assembled or positioned prior to RNAi induction. Instead, FAZ27 RNAi very likely affected the elongation of the new FAZ and the positioning of the new flagellum-associated structures. Therefore, we examined the cells with two nuclei and two kinetoplasts (2N2K), which are at later stages of the cell cycle and have segregated their duplicated organelles and cytoskeletal structures (Fig. 4A). We focused on the new FAZ and the new flagellum, and found that the new FAZ of the RNAi cells was significantly shorter than that of the control cells (an average length of 5.6 µm versus 9.4 µm) (Fig. 4A–C). The unattached portion of the new flagellum of the RNAi cells was significantly longer than that of the control cells (an average length of 5.5 µm versus 1.7 µm) (Fig. 4A–C). We also found that the new, posterior kinetoplast (pK) was repositioned towards the new, posterior nucleus (pN), resulting in an increase of the distance between the new kinetoplast to the cell posterior (an average length of 4.8 µm in RNAi cells versus 3.8 µm in control cells) and a decrease in inter-kinetoplast distance (an average length of 3.5 µm in RNAi cells versus 4.5 µm in control cells) (Fig. 4A–C). Moreover, all of the newly formed flagellum-associated cytoskeletal structures, such as the new BB, the new HC, and the new FPC, were repositioned toward the new nucleus (Fig. 4D,E). These results demonstrated that RNAi of FAZ27 disrupted the elongation of the new FAZ and abolished the migration of the newly formed kinetoplast and flagellum-associated structures, thus causing the newly formed flagellum to be unable to attach to the cell body.
Fig. 4.
FAZ27 RNAi disrupted the elongation of the new FAZ and positioning of flagellum-associated cytoskeletal structures. (A) Morphology of 2N2K cells from control and FAZ27 RNAi cells. 2N2K cells were immunostained with anti-CC2D and anti-PFR2 (L8C4) antibodies to label the FAZ and the flagellum, respectively. Cells were counterstained with DAPI to stain nuclear (N) and kinetoplast (K) DNA. pK, posterior kinetoplast; aK, anterior kinetoplast; pN, posterior nucleus; aN, anterior nucleus; NF, new flagellum; OF, old flagellum; P, posterior; A, anterior. Scale bar: 5 μm. (B) Diagram depicting a 2N2K cell used for measuring the length of the new FAZ and the unattached new flagellum, the distance from the posterior kinetoplast (pK) to the posterior cell tip and the distance between the two kinetoplasts. (C) Morphometric measurements of control and FAZ27-deficient 2N2K cells. 2N2K cells from control and FAZ27 RNAi (24 h) were immunostained with anti-CC2D and anti-PFR2 antibodies. Shown are the length of the new FAZ, the length of the unattached new flagellum, the inter-kinetoplast distance and the posterior kinetoplast to cell posterior distance for control (n=100) and FAZ27 RNAi cells (n=100). Mean±s.d. values are indicated. ***P<0.001 (Student's t-test). (D) Position of the flagellar pocket collar (FPC) and the basal body in control and FAZ27 RNAi cells. Shown are 2N2K cells that were co-immunostained with YL 1/2 antibody to label the mature basal body (mBB) and anti-TbBILBO1 antibody to label the FPC. Cells were counterstained with DAPI to stain nuclear (N) and kinetoplast (K) DNA. Abbreviations are as in A. Scale bar: 5 μm. (E) Positions of the hook complex (HC) and the centrin arm (CA) in control and FAZ27 RNAi cells. Shown are 2N2K cells that were co-immunostained with anti-TbMORN1 antibody and 20H5 antibody to label the HC and the CA, respectively, and counterstained with DAPI for nuclear and kinetoplast DNA. Abbreviations are as in A. Scale bar: 5 μm. Images in D,E are representative of three experiments.
FAZ27 forms a complex with FLAM3 and ClpGM6
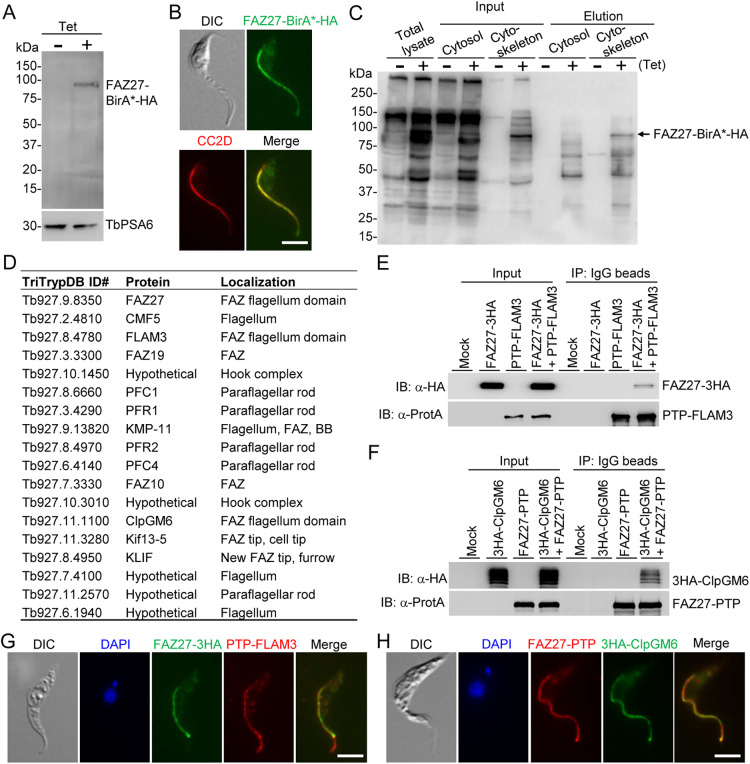
Our goal was to identify FAZ27-interacting proteins and understand their functional relationship. To this end, we planned to carry out proximity-dependent biotin identification (BioID) by ectopically expressing FAZ27–BirA*–HA in trypanosomes in a tetracycline-inducible manner to identify potential FAZ27-interacting proteins, and then verify them by co-immunoprecipitation. Western blotting and immunofluorescence microscopy using an anti-HA antibody confirmed the expression and correct localization of FAZ27–BirA*–HA (Fig. 5A,B). Affinity purification of biotinylated proteins (Fig. 5C) was performed for both the non-induced control and tetracycline-induced cells, and subsequent mass spectrometry identified FAZ27 and 86 other proteins from the tetracycline-induced cells (Table S1). We excluded 26 proteins that were either also identified from the non-induced control cells or were known contaminants (Table S2). Among the 60 putative FAZ27 proximal proteins (Table S3), 17 proteins localized to the vicinity of FAZ27 (Fig. 5D), including FLAM3 and ClpGM6, both of which have been previously reported to be involved in cell morphogenesis (Hayes et al., 2014; Sunter et al., 2015a). We noted that KIN-E, another known regulator of cell morphogenesis in T. brucei (An and Li, 2018), was not identified by FAZ27 BioID, likely because KIN-E is enriched at the flagellar tip and thus does not colocalize with FAZ27. Because FAZ27, FLAM3 and ClpGM6 are all involved in controlling cell morphogenesis, we tested whether FAZ27 interacts with FLAM3 and/or ClpGM6 by co-immunoprecipitation. Cells co-expressing FAZ27–3HA and PTP–FLAM3 or co-expressing FAZ27–PTP and 3HA–ClpGM6 from their respective endogenous loci were generated and used for co-immunoprecipitation. We found that immunoprecipitation of PTP–FLAM3 was able to pull down FAZ27–3HA (Fig. 5E) and immunoprecipitation of FAZ27–PTP was able to pull down 3HA–ClpGM6 (Fig. 5F), demonstrating that FAZ27 interacts with both FLAM3 and ClpGM6 in vivo in trypanosomes. Immunofluorescence microscopy showed that FAZ27 colocalized with FLAM3 and ClpGM6 along the entire length of the FAZ flagellum domain, but not in the unattached flagellum where FLAM3 was additionally localized (Fig. 5G,H). Because previous studies demonstrated that FLAM3 and ClpGM6 formed a complex in the FAZ flagellum domain (Sunter et al., 2015a), it is possible that FAZ27, FLAM3 and ClpGM6 reside in the same protein complex in the flagellum.
Fig. 5.
Proximity-dependent biotin identification and co-immunoprecipitation identified FLAM3 and ClpGM6 as FAZ27-interacting proteins. (A) Western blotting to detect the expression of FAZ27–BirA*–HA in procyclic trypanosomes. FAZ27–BirA*–HA was detected with anti-HA antibody, and TbPSA6 served as the loading control. (B) Immunofluorescence microscopy to detect the subcellular localization of FAZ27–BirA*–HA. Cells were immunostained with FITC-conjugated anti-HA antibody and anti-CC2D antibody. Scale bar: 5 μm. (C) Western blotting with anti-streptavidin antibody to monitor the purification of biotinylated proteins in close proximity to FAZ27–BirA*–HA. (D) List of proteins identified by FAZ27 BioID that localize to the vicinity of FAZ27. (E) Co-immunoprecipitation to test the interaction between FAZ27 and FLAM3. PTP–FLAM3 and FAZ27–3HA were co-expressed from their respective endogenous loci. Cells expressing FAZ27–3HA or PTP–FLAM3 alone served as controls. PTP–FLAM3 was detected by anti-Protein A (α-ProtA) antibody, and FAZ27–3HA was detected by anti-HA antibody. (F) Co-immunoprecipitation to test the interaction between FAZ27 and ClpGM6. FAZ27–PTP and 3HA–ClpGM6 were co-expressed from their respective endogenous loci. Cells expressing FAZ27–PTP or 3HA–ClpGM6 alone served as controls. FAZ27–PTP was detected by anti-Protein A (α-ProtA) antibody, and 3HA–ClpGM6 was detected by anti-HA antibody. (G) Co-immunofluorescence microscopy to detect the colocalization between FAZ27 and FLAM3. FAZ27–3HA was detected by FITC-conjugated anti-HA antibody, and PTP–FLAM3 was detected by anti-Protein A antibody. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. Scale bar: 5 μm. (H) Co-immunofluorescence microscopy to detect the colocalization between FAZ27 and ClpGM6. FAZ27–PTP was detected by anti-Protein A antibody, and 3HA–ClpGM6 was detected by FITC-conjugated anti-HA antibody. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. Scale bar: 5 μm. Blots shown in A,C,E,F are representative of three experiments, and images in B,G,H are representative of three experiments.
FAZ27 and FLAM3 are interdependent for assembly into the FAZ
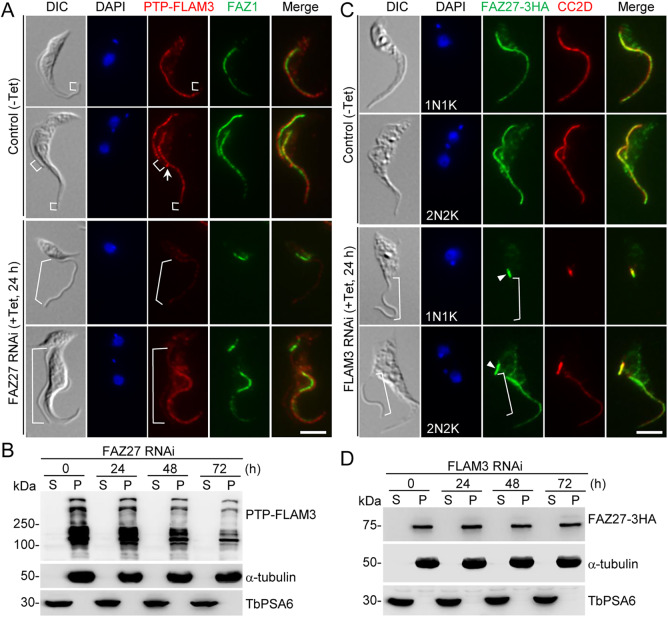
We investigated the functional relationship between FAZ27 and FLAM3 by examining the effect of knocking down one protein on the localization and stability of the other. FLAM3 was endogenously tagged with an N-terminal PTP epitope in cells harboring the FAZ27 RNAi construct. Cells were immunostained with anti-Protein A antibody to detect PTP–FLAM3 and anti-FAZ1 (clone L3B2) antibody to label the FAZ. In the non-induced control one-flagellum (1N1K) cells, FLAM3 was detected in the flagellum, with the exception of the distal portion (Fig. 6A, brackets). In the epimastigote-like one-flagellum (1N1K) cells derived from depletion of FAZ27, however, the FLAM3 fluorescence signal was significantly reduced in the entire flagellum (Fig. 6A, brackets). In the non-induced control bi-flagellated (1N2K and 2N2K) cells, FLAM3 was detected in the old and the new flagella, except for the distal portion of both flagella (Fig. 6A, brackets), and additionally localized to the distal tip of the new flagellum (Fig. 6A, arrow). In the FAZ27-deficient bi-flagellated (1N2K and 2N2K) cells, however, the FLAM3 signal in the old flagellum was not changed, but it was significantly reduced in the entire new flagellum (Fig. 6A, brackets). Western blotting showed that FLAM3 protein was only detectable in the cytoskeletal fraction, and after FAZ27 RNAi for up to 72 h the level of FLAM3 protein gradually decreased (Fig. 6B). These results suggest that FAZ27 RNAi disrupted the assembly of FLAM3 into the flagellum and destabilized the FLAM3 protein.
Fig. 6.
Interdependence between FAZ27 and FLAM3 for assembly into the FAZ. (A) Effect of FAZ27 RNAi on the localization of FLAM3 to the flagellum. FLAM3 was tagged with a PTP tag at its N-terminus in the FAZ27 RNAi cell line. RNAi-induced and control cells were co-immunostained with anti-Protein A antibody and anti-FAZ1 (clone L3B2) antibody. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. The white arrows indicate the PTP–FLAM3 signal at the flagella connector, and the white brackets indicate the regions without PTP–FLAM3 signal. Scale bar: 5 μm. (B) Effect of FAZ27 RNAi on the distribution of FLAM3 in cytosolic and cytoskeletal fractions. Cell lysate from Control (0 h) and FAZ27 RNAi cells expressing PTP–FLAM3 was spun down to separate the soluble cytosolic (S) and cytoskeletal pellet (P) fractions for western blotting with anti-Protein A (α-ProtA) antibody to detect PTP–FLAM3 in the two fractions. The same blot was probed with anti-α-tubulin antibody and anti-PSA6 antibody to serve as cytoskeleton and cytosol markers, respectively. (C) Effect of FLAM3 depletion on FAZ27 localization. FAZ27 was endogenously tagged with a triple HA epitope in the FLAM3 RNAi cell line. Cells were co-immunostained with FITC-conjugated anti-HA antibody to detect FAZ27–3HA and anti-CC2D antibody to label the FAZ. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. The white brackets indicate the regions without FAZ27–3HA signal. Arrowheads indicate the FAZ27 protein located at the proximal end of the new FAZ. Scale bar: 5 μm. (D) Effect of FLAM3 RNAi on the distribution of FAZ27 in cytosolic and cytoskeletal fractions. The soluble cytosolic (S) fraction and cytoskeletal pellet (P) fraction were prepared as described in B. FAZ27–3HA was detected by anti-HA antibody. The same blot was probed with anti-α-tubulin antibody and anti-TbPSA6 antibody to serve as cytoskeleton and cytosol markers, respectively. Images in A and C, and blots in B and D are representative of three experiments.
Conversely, the effect of FLAM3 depletion on FAZ27 localization and protein stability was also examined. FAZ27 was endogenously tagged with a triple HA epitope in cells harboring the FLAM3 RNAi construct, and cells were co-immunostained with anti-HA antibody to detect FAZ27–3HA and anti-CC2D antibody to label the FAZ. Similar to the effect of FAZ27 RNAi on FLAM3, knockdown of FLAM3 disrupted the localization of FAZ27 to most of the FAZ of the epimastigote-like one-flagellum (1N1K) cells and to most of the new FAZ of the bi-flagellated (1N2K and 2N2K) cells (Fig. 6C, brackets), with the exception of the proximal end of the FAZ (Fig. 6C, arrowheads). Western blotting showed that FAZ27 was only detectable in the cytoskeletal fraction, and knockdown of FLAM3 did not affect the levels of FAZ27 protein (Fig. 6D). Taken together, these results demonstrated that knockdown of FLAM3 disrupted the assembly of FAZ27 into the FAZ flagellum domain, but unlike the effect of FAZ27 knockdown on the stability of FLAM3, RNAi of FLAM3 did not cause destabilization of FAZ27, indicating distinct mechanisms of regulation.
FAZ27 and ClpGM6 are interdependent for assembly into the FAZ
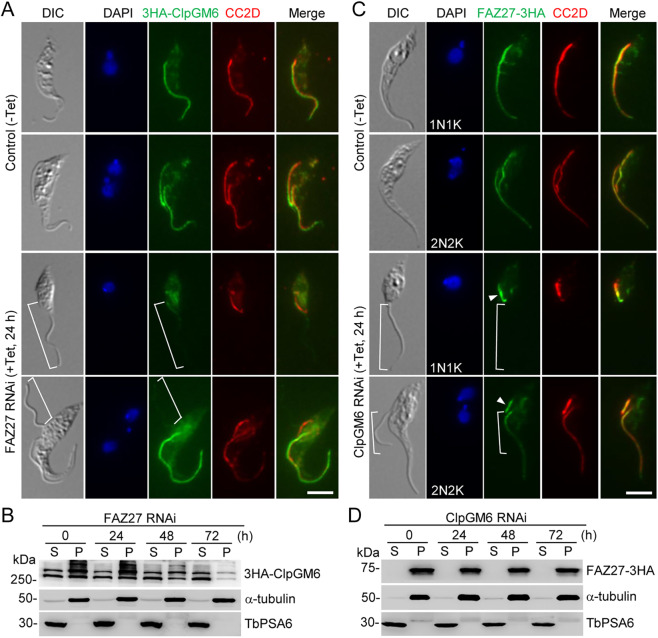
The functional relationship between FAZ27 and ClpGM6 was also investigated. We first examined the effect of FAZ27 RNAi on the localization and protein stability of ClpGM6. To this end, ClpGM6 was endogenously tagged with a triple HA epitope at its C-terminus in cells containing the FAZ27 RNAi construct, and cells were then immunostained with an anti-HA antibody to detect ClpGM6–3HA and an anti-CC2D antibody to label the FAZ. In the non-induced control one-flagellum (1N1K) cells, ClpGM6 was detected in the FAZ flagellum domain (Fig. 7A). However, in the FAZ27-deficient, epimastigote-like one-flagellum (1N1K) cells, the ClpGM6 fluorescence signal was reduced to a very low level in the flagellum (Fig. 7A, brackets). In the control bi-flagellated (1N2K and 2N2K) cells, ClpGM6 was detected in both the new flagellum and the old flagellum (Fig. 7A), but in the FAZ27 RNAi-induced bi-flagellated (1N2K and 2N2K) cells, the ClpGM6 fluorescence signal in the new flagellum, but not in the old flagellum, was significantly reduced (Fig. 7A, brackets). Western blotting showed that the ClpGM6 protein was present in both the cytosolic (soluble) fraction and the cytoskeletal (pellet) fraction and its level gradually decreased in the cytoskeletal fraction after FAZ27 RNAi induction for 48 h and longer (Fig. 7B). These results suggest that FAZ27 knockdown disrupted the assembly of ClpGM6 into the flagellum and destabilized ClpGM6.
Fig. 7.
Interdependence between FAZ27 and ClpGM6 for assembly into the FAZ. (A) Effect of FAZ27 RNAi on the localization of ClpGM6 to the flagellum. ClpGM6 was tagged with an N-terminal triple HA epitope in the FAZ27 RNAi cell line. Cells were co-immunostained with FITC-conjugated anti-HA antibody and anti-CC2D antibody. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. The white brackets indicate the regions without 3HA–ClpGM6 signal. Scale bar: 5 μm. (B) Effect of FAZ27 RNAi on the distribution of 3HA–ClpGM6 in cytosolic and cytoskeletal fractions. The soluble cytosolic (S) fraction and cytoskeletal pellet (P) fraction were prepared as described in Fig. 6B. 3HA–ClpGM6 was detected by anti-HA antibody. α-tubulin and TbPSA6 served as cytoskeleton and cytosol markers, respectively. (C) Effect of ClpGM6 depletion on FAZ27 localization. FAZ27 was endogenously tagged with a triple HA epitope in the FLAM3 RNAi cell line. Cells were immunostained with FITC-conjugated anti-HA antibody to detect FAZ27-3HA and anti-CC2D antibody to label the FAZ. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. The white brackets indicate the regions without FAZ27–3HA signal. Arrowheads indicate the FAZ27 protein located at the proximal end of the new FAZ. Scale bar: 5 μm. (D) Effect of ClpGM6 RNAi on the distribution of FAZ27 in cytosolic and cytoskeletal fractions. The soluble cytosolic (S) fraction and cytoskeletal pellet (P) fraction were prepared as described in Fig. 6B. FAZ27–3HA was detected by anti-HA antibody. α-tubulin and TbPSA6 served as cytoskeleton and cytosol markers, respectively. Images in A,C and blots in B,D are representative of three experiments.
We next investigated the effect of ClpGM6 knockdown on the localization and stability of FAZ27 by immunofluorescence microscopy and western blotting. FAZ27 was endogenously tagged with a triple HA epitope in the ClpGM6 RNAi cell line, and cells were then immunostained with an anti-HA antibody to detect FAZ27–3HA and an anti-CC2D antibody to label the FAZ. The results showed that knockdown of ClpGM6 disrupted the localization of FAZ27 to most of the FAZ except the proximal end in the epimastigote-like one-flagellum (1N1K) cells and to most of the new FAZ except the proximal end in bi-flagellated (1N2K and 2N2K) cells (Fig. 7C, brackets). Western blotting showed that knockdown of ClpGM6 did not affect the level of FAZ27 protein (Fig. 7D). These results demonstrated that ClpGM6 is required for assembly of FAZ27 into the FAZ flagellum domain.
FAZ27 is assembled at the proximal end of the FAZ
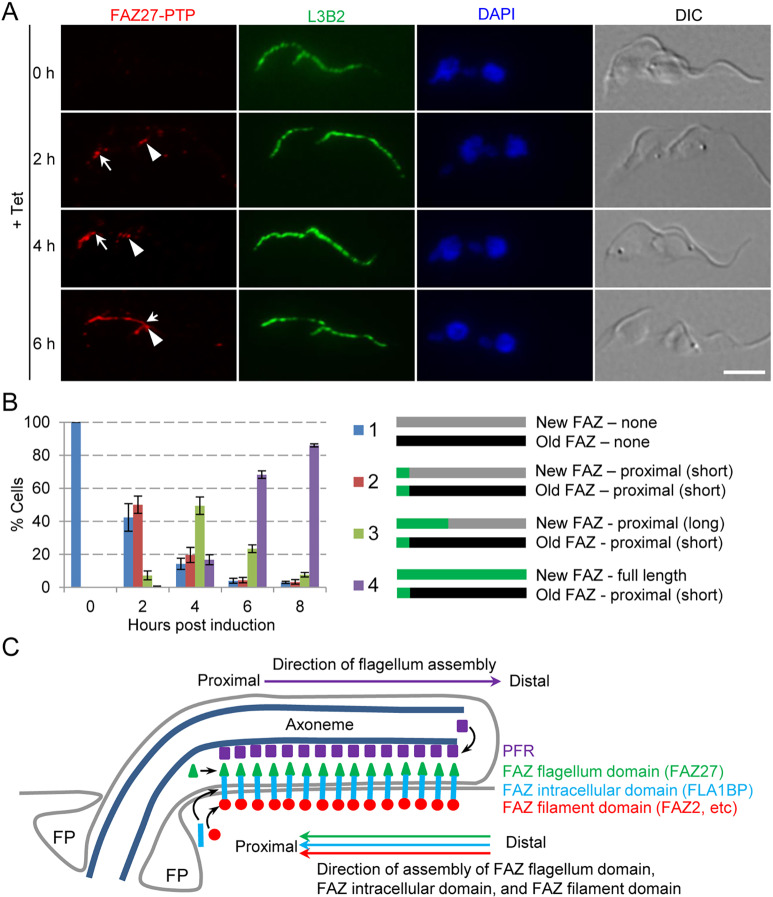
Previously, we and others have independently demonstrated that the assembly of the FAZ filament proteins (Sunter et al., 2015b; Zhou et al., 2015) and the FAZ intracellular-domain protein FLA1BP (Sunter et al., 2015b) occurs at the proximal end of the FAZ, opposite to the assembly of the flagellar components, which occurs at the distal tip of the flagellum (Bastin et al., 1999). It remains unclear whether the assembly of the FAZ flagellum domain follows the same pattern of assembly as other FAZ sub-domains (i.e. proximal assembly) or as the flagellar components (i.e. distal assembly). We chose FAZ27 as a representative of the FAZ flagellum-domain proteins due to its small size, which allowed easy cloning of its gene into the ectopic expression vector. We monitored the incorporation of FAZ27 into the elongating FAZ using the same approach that had been used previously (Sunter et al., 2015b; Zhou et al., 2015). We ectopically expressed FAZ27–PTP and examined its localization at different time points of induction of FAZ27–PTP expression by immunofluorescence microscopy (Fig. 8A). After 2 h of induction, FAZ27–PTP fluorescence signal was detected at the proximal end of both the new FAZ and the old FAZ (Fig. 8A,B). After 4 h and 6 h of induction, the FAZ27–PTP signal at the new FAZ was gradually elongated towards the anterior end of the new FAZ, whereas the FAZ27–PTP signal at the old FAZ was no more elongated than after 2 h of induction (Fig. 8A,B). These results suggest that FAZ27 was assembled at the proximal end of the FAZ flagellum domain during the assembly of the new FAZ flagellum domain. Given that the FAZ filament-domain proteins and the FAZ intracellular-domain proteins are similarly assembled at the proximal end (Sunter et al., 2015b; Zhou et al., 2015), it suggests that assembly of the FAZ flagellum domain is coordinated with that of other FAZ sub-domain proteins at the proximal end of the FAZ during the assembly of the new FAZ (Fig. 8C).
Fig. 8.
Proximal assembly of FAZ27 in the elongating FAZ. (A) Incorporation of ectopically expressed FAZ27–PTP into the elongating FAZ. FAZ27–PTP was expressed ectopically and its localization to the new FAZ and the old FAZ was determined by immunofluorescence microscopy using an anti-Protein A antibody. Anti-FAZ1 (clone L3B2) was used as a marker for the FAZ filament. Cells were counterstained with DAPI to stain nuclear and kinetoplast DNA. Arrows indicate the distal tip of the FAZ27–PTP signal at the new (elongating) FAZ, and arrowheads indicate the distal tip of the FAZ27–PTP signal at the old FAZ. Scale bar: 5 μm. (B) Quantitation of cells with different patterns of FAZ27–PTP signal at the new FAZ and the old FAZ. A total of 100 2N2K cells were counted for each time point. Data are mean±s.d. from three independent replicates. (C) Model of FAZ27 assembly during FAZ elongation relative to the assembly of the components of the FAZ filament domain, the FAZ intracellular domain, and the paraflagellar rod (PFR). FP indicates the flagellar pocket.
DISCUSSION
Trypanosome life cycle development from trypomastigote to epimastigote appears to require the shortening of the length of the FAZ, which can be achieved through the modulation of the abundance of certain FAZ flagellum-domain proteins, such as ClpGM6 and FLAM3 (Hayes et al., 2014; Rotureau et al., 2014; Sunter et al., 2015a). Knockdown of certain FAZ filament proteins, such as FAZ9 and TbSAS-4 (Hu et al., 2015; McAllaster et al., 2015), also produces epimastigote-like cells, but epimastigote-like cells generated by FAZ9 RNAi only contain a repositioned kinetoplast but not a shortened FAZ (McAllaster et al., 2015) and the epimastigote-like cells produced by TbSAS-4 RNAi contain a shortened FAZ that is longer than the shortened FAZ found in the cells depleted of either ClpGM6 or FLAM3 (Hu et al., 2015). We reported here that another FAZ flagellum domain protein, FAZ27, also functions in regulating cell morphogenesis in T. brucei (Fig. 2). Morphologically, the trypomastigote form and the epimastigote form differ in the length of the FAZ and the relative position of kinetoplast, flagellum and flagellum-associated structures (Hoare and Wallace, 1966). Cells following FAZ27 RNAi treatment possessed characteristic epimastigote morphology, exhibiting a shorter FAZ and a repositioned kinetoplast, flagellum and flagellum-associated structures (Figs 2–4). FAZ27 RNAi in the trypomastigote cells produced cells with an epimastigote-like morphology (Fig. 2), suggesting that FAZ27 likely acts to maintain the morphology of the trypomastigote form by promoting the elongation of the FAZ flagellum domain, thereby promoting the attachment of the flagellum and the positioning of the flagellum and its associated structures.
The specialized FAZ cytoskeletal structure in T. brucei plays an essential role in determining cell length and organelle position, and it consists of multiple sub-domains in the junction between the flagellum and the cell body (Sunter and Gull, 2016). Studies from multiple research groups showed that RNAi-mediated ablation of the proteins from different FAZ sub-domains caused distinct phenotypic defects (Hayes et al., 2014; LaCount et al., 2002; Rotureau et al., 2014; Sun et al., 2013; Sunter et al., 2015a; Vaughan et al., 2008; Zhou et al., 2015, 2011). Knockdown of FAZ flagellum-domain proteins caused shortening of the FAZ and produced epimastigote-like morphology (Hayes et al., 2014; Rotureau et al., 2014; Sunter et al., 2015a), whereas knockdown of FAZ filament proteins mostly caused repositioning of the kinetoplast and flagellum detachment (Vaughan et al., 2008; Zhou et al., 2015, 2011). Although more than a dozen FAZ filament proteins have been identified (Hu et al., 2015; McAllaster et al., 2015; Sunter et al., 2015b; Vaughan et al., 2008; Zhou et al., 2015, 2011), only three proteins, ClpGM6 (Hayes et al., 2014), FLAM3 (Rotureau et al., 2014; Sunter et al., 2015a) and FAZ27 (this work), have been shown to localize to the FAZ flagellum domain, and they apparently function together to regulate cell morphogenesis. The identification of FAZ27 as a FAZ flagellum domain component required for cell morphogenesis further supports the notion that the FAZ flagellum domain is implicated in morphological transitions during trypanosome life cycle development.
Although knockdown of FAZ27, KIN-E, FLAM3 and ClpGM6 in the trypomastigote form all produced epimastigote-like cells, these RNAi cell lines exerted distinct effects on cell proliferation. RNAi of FAZ27 (Fig. 2), RNAi of KIN-E (An and Li, 2018) and RNAi of FLAM3 (Rotureau et al., 2014; Sunter et al., 2015a) all inhibited cell proliferation and produced multi-nucleated cells, whereas depletion of ClpGM6 had little effect on cell proliferation (Hayes et al., 2014). We postulate that the phenotypic distinctions among these RNAi cell lines are likely due to the different effects that they exerted on the assembly of the FAZ. Previously, Sunter et al. (2015a) suggested that the minimum length of the FAZ that supports trypanosome cytokinesis is 3 µm. Therefore, if the length of the new FAZ in any RNAi cell line is shorter than 3 µm, cell proliferation will be inhibited and multi-nucleated cells will be generated. Indeed, the length of the FAZ in the epimastigote-like cells generated by RNAi of ClpGM6 is between 5 and 10 µm (Hayes et al., 2014), but many epimastigote-like cells generated by RNAi of FLAM3 (Sunter et al., 2015a), RNAi of KIN-E (An and Li, 2018) and RNAi of FAZ27 (Fig. 3) contain a FAZ that is shorter than 3 µm. It appears that FAZ27 and its interacting partner proteins play distinct functions in controlling the assembly of the FAZ, despite forming parts of a protein complex.
FAZ27 and its interacting partner proteins FLAM3 and ClpGM6 were interdependent for assembly into the flagellum (Figs 6, 7); however, there appeared to be an additional effect of FAZ27 depletion on the stability of FLAM3 and ClpGM6, but not vice versa (Figs 6, 7). Previous results showed that FLAM3 and ClpGM6 were interdependent for maintaining their levels in the flagellum (Sunter et al., 2015a). Thus, it is possible that FAZ27 knockdown exerted effects on the stability of both, or one of, FLAM3 and ClpGM6. In the latter scenario, destabilization of either FLAM3 or ClpGM6 would cause the destabilization of the other partner proteins, as the two proteins are interdependent for stability (Sunter et al., 2015a). It should be noted that levels of both FLAM3 and ClpGM6 were significantly decreased in the flagellum of the epimastigote-like one-flagellum cells and in the new flagellum of the bi-flagellated cells generated by FAZ27 RNAi, whereas the level of FAZ27 was increased at the proximal end of the FAZ of the epimastigote-like one-flagellum cells and the proximal end of the new FAZ of the bi-flagellated cells generated by knockdown of FLAM3 or ClpGM6 (Figs 6, 7). As the level and the cytoskeletal association of FAZ27 protein were not changed in FLAM3 RNAi cells and ClpGM6 RNAi cells (Figs 6, 7), it is possible that the FAZ27 protein was accumulated at the proximal end of the FAZ in these RNAi cells. In contrast, the reduction in the protein levels of FLAM3 and ClpGM6 in FAZ27 RNAi cells (Figs 6, 7) suggests that these two proteins were degraded when the protein complex was partially disrupted after knockdown of FAZ27.
Although FAZ27 and ClpGM6 both localize in the FAZ flagellum domain (Fig. 5H), the localization of FLAM3 in the flagellum further extends from the FAZ flagellum domain to the unattached, free flagellum, where it does not colocalize with FAZ27 (Fig. 5G) and ClpGM6 (Sunter et al., 2015a). Knockdown of FAZ27 (Fig. 6A,B) and RNAi of ClpGM6 (Sunter et al., 2015a) both reduced, but did not completely remove, FLAM3 protein levels in the flagellum. It appeared that only a proportion of FLAM3 protein formed a complex with FAZ27 and ClpGM6 in the FAZ flagellum domain, and this proportion of FLAM3 protein became unstable when FAZ27 or ClpGM6 was depleted by RNAi. In this scenario, the remaining proportion of FLAM3 was not integrated into the protein complex and, hence, remained to be assembled along the length of the flagellum, with the exception of the distal tip (Fig. 6A). Previously, Sunter et al. (2015a) observed a reciprocal dependency between FLAM3 and ClpGM6 and suggested a hierarchy of assembly between FLAM3 and ClpGM6, in which FLAM3 maintains ClpGM6 localization and stability in the flagellum. We also observed a reciprocal dependency between FAZ27 and FLAM3, but we found that FAZ27 remained detectable at the proximal portion of the flagellum and was not destabilized by FLAM3 knockdown (Fig. 6), unlike ClpGM6, which disappeared from the entire FAZ flagellum domain and was degraded upon FLAM3 knockdown (Sunter et al., 2015a). These findings suggest that the functional interplay between FAZ27 and FLAM3 is likely to be different from that between ClpGM6 and FLAM3.
Using FAZ27 as a representative of the FAZ flagellum domain components, we investigated the site of assembly of the FAZ flagellum domain, and we provided experimental evidence to suggest that the FAZ flagellum-domain proteins are assembled at the proximal portion of the FAZ (Fig. 8). This proximal assembly is identical to that of those FAZ proteins located in the FAZ filament domain, such as FAZ2, FAZ8 and KMP-11 (Sunter et al., 2015b; Zhou et al., 2015), and the FAZ intracellular domain, such as FLA1BP (Sunter et al., 2015b), but is opposite to the distal assembly of PFR components located within the flagellum, such as PFR2 (Bastin et al., 1999). Based on the proximal assembly of FLA1BP, a transmembrane protein located on the flagellar membrane (Sun et al., 2013), it was proposed that this reflected the assembly of the FAZ flagellum domain (Sunter et al., 2015b). However, FLA1BP resides mostly outside of the flagellum membrane and interacts with the cell membrane-associated FLA1 protein (Sun et al., 2013). FAZ27 and its interacting partner proteins, however, likely reside inside the flagellum between the flagellar membrane and the PFR or in zone 1 of the FAZ, as designated by Sunter and Gull (2016). It should also be noted that although FLA1BP was primarily assembled into the proximal portion of the FAZ, it was additionally assembled into the distal end of the FAZ (Sunter et al., 2015b). Such a pattern of assembly was not observed for FAZ27 (Fig. 8), indicating that the assembly of FLA1BP cannot reflect the assembly of the FAZ flagellum-domain proteins. Nevertheless, these results suggest that the assembly of the different FAZ sub-domains is controlled in a coordinated manner at the proximal portion of the FAZ, where they maintain cell morphology and mediate cell–flagellum adhesion.
MATERIALS AND METHODS
Trypanosome cell culture and RNAi
The procyclic form of T. brucei strain 427 was cultivated in SDM-79 medium (Thermo Fisher Scientific/Gibco) containing 10% heat-inactivated fetal bovine serum (Sigma-Aldrich) at 27°C. The procyclic trypanosome strain 29-13 (Wirtz et al., 1999), which expresses the T7 RNA polymerase and the tetracycline repressor, was cultured at 27°C in SDM-79 medium supplemented with 10% heat-inactivated fetal bovine serum, 15 µg/ml G418 and 50 µg/ml hygromycin. Cells were diluted with fresh medium every 3 d or when the cell density reached 5×106/ml.
To generate the FAZ27 RNAi cell line, a 554 bp DNA fragment (nucleotides 223–776) corresponding to the N-terminal coding region of FAZ27 was cloned into the pZJM vector (Wang et al., 2000). To generate the ClpGM6 RNAi cell line, a 659 bp DNA fragment (nucleotides 4302–4960) of the ClpGM6 gene was cloned into the pZJM vector. The same DNA fragment of ClpGM6 gene was used for RNAi previously (Hayes et al., 2014). The resulting plasmids were each linearized by restriction digestion with NotI and transfected into the 29-13 strain by electroporation. Successful transfectants were selected with 2.5 µg/ml phleomycin and cloned by limiting dilution in a 96-well plate. The FLAM3 RNAi cell line generated in our lab has been described previously (An and Li, 2018). To induce RNAi, cells were incubated with 1.0 µg/ml tetracycline, and cell number was counted daily with a hemacytometer under a light microscope. Three independent clonal cell lines were selected and analyzed, and identical phenotypes were obtained. Only one out of the three clonal cell lines was used throughout the course of work for characterization.
In situ epitope tagging of proteins
Endogenous epitope tagging of FAZ27, FLAM3 (Rotureau et al., 2014), and ClpGM6 (Hayes et al., 2014) was carried out using the previously described one-step PCR-based method (Shen et al., 2001). For FAZ27 and FLAM3 colocalization, FAZ27 was tagged with a C-terminal triple HA epitope (neomycin resistance) and FLAM3 was tagged with an N-terminal PTP epitope (puromycin resistance) in the 427 cell line. For FAZ27 and ClpGM6 colocalization, FAZ27 was tagged with a PTP epitope (neomycin resistance) at its C-terminus and ClpGM6 was tagged with an N-terminal triple HA epitope (puromycin resistance) in the 427 cell line. Successful transfectants were selected under 40 µg/ml G418 and 1 µg/ml puromycin, and cloned by limiting dilution. FAZ27 was also tagged with a C-terminal triple HA epitope (puromycin resistance) in the FAZ27 RNAi cell line, the FLAM3 RNAi cell line and the ClpGM6 RNAi cell line. Conversely, FLAM3 was tagged with an N-terminal PTP epitope (puromycin resistance) in the FAZ27 RNAi cell line, and ClpGM6 was tagged with a C-terminal triple HA epitope in the FAZ27 RNAi cell line. Successful transfectants were selected in SDM-79 medium containing 1 µg/ml puromycin, 15 µg/ml G418, and 50 µg/ml hygromycin B, and then cloned by limiting dilution.
Ectopic overexpression of FAZ27 and its truncation mutants
To ectopically express FAZ27 and its truncation mutants, the full-length coding sequence and the truncated sequences of the FAZ27 gene were each cloned into the pLew100-3HA-BLE vector or pLew100-PTP-BLE vector (Wirtz et al., 1999). The resulting plasmids were each linearized with NotI and transfected into the 29-13 strain by electroporation. Successful transfectants were selected with 2.5 µg/ml phleomycin and cloned by limiting dilution. Expression of 3HA-tagged FAZ27 and its truncation mutants or PTP-tagged FAZ27 were induced with 0.5 µg/ml tetracycline and were verified by western blotting and immunofluorescence microscopy.
Proximity-dependent biotin identification and mass spectrometry
The full-length coding sequence of FAZ27 was amplified from genomic DNA by PCR and cloned into the pLew100-BirA*-HA vector (Hu et al., 2015). The resulting plasmid pLew100-FAZ27-BirA*-HA was linearized by restriction digestion with NotI and transfected into the 29-13 strain. Successful transfectants were selected with 2.5 µg/ml phleomycin and cloned by limiting dilution. Expression of FAZ27–BirA*–HA was induced with 0.5 µg/ml tetracycline and confirmed by western blotting and immunofluorescence microscopy.
Affinity purification of biotinylated proteins was performed according to our published procedures (Hu et al., 2015). Cells expressing FAZ27–BirA*–HA were incubated with 0.5 µg/ml tetracycline for 24 h and then incubated with 50 µM biotin for an additional 24 h. Cells treated with PEME buffer (100 mM PIPES pH 6.9, 2 mM EGTA, 0.1 mM EDTA and 1 mM MgSO4) containing 0.5% Nonidet P-40, and cytosolic (soluble) and cytoskeletal (pellet) fractions were separated by centrifugation. The cytoskeletal fraction was further extracted with lysis buffer (0.4% SDS, 500 mM NaCl, 5 mM EDTA, 1 mM DTT and 50 mM Tris-HCl, pH 7.4), and solubilized cytoskeletal materials were collected. The combined cytosolic extract and the cytoskeletal extract were incubated with 500 µl pre-washed streptavidin-coated Dynabeads (Invitrogen) at 4°C for 4 h.
The Dynabeads were washed five times with PBS, five times with 50 mM ammonium bicarbonate and finally re-suspended in 100 mM ammonium bicarbonate. Subsequently, 10% DTT was added to the suspension, followed by addition of 50% iodoacetamide. Finally, 5% DTT was added to the suspension, and proteins bound on the Dynabeads were digested with trypsin at 37°C overnight. Trypsin digestion was stopped by adding trifluoroacetic acid to the solution to make the pH ∼2.0. The trypsin-digested peptides were desalted and then analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) interfaced with an Eksigent nano-LC 2D plus chipLC system (Eksigent Technologies). Data analysis was performed according to our published methods (Wei et al., 2014; Zhou et al., 2015). Raw data files were searched against the T. brucei genome database using the Mascot search engine. The search conditions used peptide tolerance of 10 ppm and tandem mass spectrometry (MS/MS) tolerance of 0.8 Da with the enzyme set as trypsin and two missed cleavages permitted. The Mascot score for a protein is the logarithmic score for the individual peptides, e.g. peptide masses and peptide fragment ion masses, for all peptides matching a given protein.
Co-immunoprecipitation and western blotting
Co-immunoprecipitation was carried out according to our previous procedures (An and Li, 2018) with slight modifications. Cells (5×107) co-expressing FAZ27–3HA and PTP–FLAM3 or co-expressing FAZ27–PTP and 3HA–ClpGM6 were lysed by sonication in 1 ml immunoprecipitation buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1 mM DTT, 1% NP-40 and protease inhibitor cocktail). Cleared lysate was incubated with 50 µl settled IgG beads for 1 h at 4°C, and immunoprecipitates were washed six times with the immunoprecipitation buffer. Proteins bound to the IgG beads were eluted with 10% SDS, separated by SDS–PAGE, transferred onto a PVDF membrane and immunoblotted with anti-HA antibody (Sigma-Aldrich, Clone HA-7, H9658, 1:1000 dilution) to detect FAZ27–3HA or 3HA–ClpGM6 and with anti-Protein A antibody (Sigma-Aldrich, P3775, 1:2000 dilution) to detect PTP–FLAM3 or FAZ27–PTP. Cells expressing FAZ27–3HA alone, PTP–FLAM3 alone, FAZ27–PTP alone, and 3HA–ClpGM6 alone were included as negative controls.
Immunofluorescence microscopy
Trypanosome cells were adhered to glass coverslips for 30 min at room temperature, incubated with cold methanol (−20°C) for 30 min and then rehydrated with PBS for 5 min at room temperature. Cells on the coverslips were blocked with 3% BSA in PBS for 1 h at room temperature and then incubated with the primary antibody for 1 h at room temperature. The following primary antibodies were used: FITC-conjugated anti-HA monoclonal antibody for 3HA-tagged proteins (Sigma-Aldrich, Clone HA-7, H7411, 1:400 dilution), anti-Protein A polyclonal antibody for PTP-tagged proteins (Sigma-Aldrich, P3775, 1:400 dilution), anti-CC2D polyclonal antibody (Dr Cynthia Y. He, National University of Singapore, Singapore; 1:1000 dilution; Zhou et al., 2011) for the FAZ filament, 20H5 monoclonal antibody (1:400 dilution) for the bilobe structure (He et al., 2005), YL 1/2 monoclonal antibody (1:2000 dilution) for the mature basal body (Sherwin et al., 1987), anti-PFR2 (clone L8C4) monoclonal antibody (Dr Keith Gull, University of Oxford, UK; 1:50 dilution) for the flagellum (Kohl et al., 1999), anti-FAZ1 (clone L3B2) monoclonal antibody (Dr Keith Gull, University of Oxford, UK; 1:50 dilution) for the FAZ filament (Kohl et al., 1999), anti-TbBILBO1 polyclonal antibody (Dr Derrick Robinson, University of Bordeaux, France; 1:400 dilution) for the flagellar pocket collar (Bonhivers et al., 2008) and anti-TbMORN1 polyclonal antibody (Dr Brooke Morriswood, University of Wurzburg, Germany; 1:400 dilution) for the hook complex (Morriswood et al., 2009). Cells were washed three times with PBS and then incubated with FITC-conjugated anti-mouse IgG (Sigma-Aldrich, F0257, 1:400 dilution), Cy3-conjugated anti-rabbit IgG (Sigma-Aldrich, AP132C, 1:400 dilution) or FITC-conjugated anti-rat IgG (Sigma-Aldrich, F6258, 1:1000 dilution) for 1 h at room temperature. Cells on the coverslips were washed three times with PBS, mounted with DAPI-containing VectaShield mounting medium (Vector Labs) and imaged under an inverted fluorescence microscope (Olympus IX71) equipped with a cooled CCD camera (model Orca-ER, Hamamatsu) and a PlanApo N 60×1.42 NA lens. Images were acquired using the Slidebook 5 software.
Scanning electron microscopy
Scanning electron microscopy was performed as described previously (Hu et al., 2019; Zhou et al., 2016a,b). Cells were directly fixed with 2.5% (v/v) glutaraldehyde in culture medium at room temperature for 2 h. The fixed cells were washed twice with PBS and then settled onto glass coverslips. Cells were washed three times with PBS and then dehydrated in alcohol. After critical point drying, cell samples were coated with an 8 nm metal film (Pt:Pd 80:20, Ted Pella Inc.) using a sputter coater (Cressington Sputter Coater 208 HR, Ted Pella Inc.) and imaged using Nova NanoSEM 230 (FEI). The scanning work distance was set at 5 mm, and the accelerating high voltage was set at 8 kV.
Data analysis and statistical analysis
ImageJ (National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/) was used to measure the length of the cytoskeletal structures and the distance between organelles, and data analysis was performed using GraphPad Prism5. Statistical analysis was conducted using the Student's t-test in Microsoft Excel and GraphPad Prism5. Error bars represent standard deviation (s.d.) from the mean of three independent biological replicates. For immunofluorescence microscopy experiments, images were taken randomly and all images were used for analysis.
Supplementary Material
Acknowledgements
We are grateful to Dr Cynthia Y. He of National University of Singapore, Dr Keith Gull of University of Oxford, Dr Brooke Morriswood of University of Wurzburg, and Dr Derrick Robinson of University of Bordeaux for providing the anti-CC2D antibody, anti-PFR2 (clone L8C4) and anti-FAZ1 (clone L3B2) antibodies, anti-TbMORN1 antibody, and anti-TbBILBO1 antibody, respectively. We also thank Dr Jianhua Gu for assistance with scanning electron microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.A., Q.Z., Z.L.; Methodology: T.A., Q.Z., H.H., H.C.; Validation: T.A., Q.Z., H.H., H.C.; Formal analysis: T.A., Q.Z.; Investigation: T.A., Q.Z., H.H., H.C., Z.L.; Writing - original draft: Q.Z., Z.L.; Writing - review & editing: T.A., Q.Z., Z.L.; Visualization: T.A., Q.Z., H.H., H.C.; Supervision: Z.L.; Project administration: Z.L.; Funding acquisition: Z.L. Deposited in PMC for release after 12 months.
Funding
This work was supported by the National Institutes of Health R01 grants AI118736 and AI101437 to Z.L. Deposited in PMC for release after 12 months.
Data availability
Raw mass spectrometry data can be accessed at the PeptideAtlas database under the following accession number: PASS01567.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.245258.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.245258.reviewer-comments.pdf
References
- An T. and Li Z. (2018). An orphan kinesin controls trypanosome morphology transitions by targeting FLAM3 to the flagellum. PLoS Pathog. 14, e1007101 10.1371/journal.ppat.1007101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler M. and Rhoads A. (2002). Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107-113. 10.1016/S0014-5793(01)03239-2 [DOI] [PubMed] [Google Scholar]
- Bastin P., MacRae T. H., Francis S. B., Matthews K. R. and Gull K. (1999). Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 19, 8191-8200. 10.1128/MCB.19.12.8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhivers M., Nowacki S., Landrein N. and Robinson D. R. (2008). Biogenesis of the trypanosome endo-exocytotic organelle is cytoskeleton mediated. PLoS Biol. 6, e105 10.1371/journal.pbio.0060105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H. Q., Zhou Q., Rowlett V. W., Hu H., Lee K. J., Margolin W. and Li Z. (2017). Proximity interactions among basal body components in Trypanosoma brucei identify novel regulators of basal body biogenesis and inheritance. MBio 8, e02120-16 10.1128/mBio.02120-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esson H. J., Morriswood B., Yavuz S., Vidilaseris K., Dong G. and Warren G. (2012). Morphology of the trypanosome bilobe, a novel cytoskeletal structure. Eukaryot. Cell 11, 761-772. 10.1128/EC.05287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull K. (1999). The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53, 629-655. 10.1146/annurev.micro.53.1.629 [DOI] [PubMed] [Google Scholar]
- Hayes P., Varga V., Olego-Fernandez S., Sunter J., Ginger M. L. and Gull K. (2014). Modulation of a cytoskeletal calpain-like protein induces major transitions in trypanosome morphology. J. Cell Biol. 206, 377-384. 10.1083/jcb.201312067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Y., Pypaert M. and Warren G. (2005). Golgi duplication in Trypanosoma brucei requires Centrin2. Science 310, 1196-1198. 10.1126/science.1119969 [DOI] [PubMed] [Google Scholar]
- Hoare C. A. and Wallace F. G. (1966). Developmental stages of trypanosomatid flagellates: a new terminology. Nature 212, 1385-1386. 10.1038/2121385a0 [DOI] [Google Scholar]
- Hu H., Zhou Q. and Li Z. (2015). SAS-4 protein in Trypanosoma brucei controls life cycle transitions by modulating the length of the flagellum attachment zone filament. J. Biol. Chem. 290, 30453-30463. 10.1074/jbc.M115.694109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., An T., Kurasawa Y., Zhou Q. and Li Z. (2019). The trypanosome-specific proteins FPRC and CIF4 regulate cytokinesis initiation by recruiting CIF1 to the cytokinesis initiation site. J. Biol. Chem. 294, 16672-16683. 10.1074/jbc.RA119.010538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L., Sherwin T. and Gull K. (1999). Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46, 105-109. 10.1111/j.1550-7408.1999.tb04592.x [DOI] [PubMed] [Google Scholar]
- LaCount D. J., Barrett B. and Donelson J. E. (2002). Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277, 17580-17588. 10.1074/jbc.M200873200 [DOI] [PubMed] [Google Scholar]
- McAllaster M. R., Ikeda K. N., Lozano-Núñez A., Anrather D., Unterwurzacher V., Gossenreiter T., Perry J. A., Crickley R., Mercadante C. J., Vaughan S. et al. (2015). Proteomic identification of novel cytoskeletal proteins associated with TbPLK, an essential regulator of cell morphogenesis in Trypanosoma brucei. Mol. Biol. Cell 26, 3013-3029. 10.1091/mbc.E15-04-0219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriswood B., He C. Y., Sealey-Cardona M., Yelinek J., Pypaert M. and Warren G. (2009). The bilobe structure of Trypanosoma brucei contains a MORN-repeat protein. Mol. Biochem. Parasitol. 167, 95-103. 10.1016/j.molbiopara.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Rotureau B., Blisnick T., Subota I., Julkowska D., Cayet N., Perrot S. and Bastin P. (2014). Flagellar adhesion in Trypanosoma brucei relies on interactions between different skeletal structures in the flagellum and cell body. J. Cell Sci. 127, 204-215. 10.1242/jcs.136424 [DOI] [PubMed] [Google Scholar]
- Shen S., Arhin G. K., Ullu E. and Tschudi C. (2001). In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113, 171-173. 10.1016/S0166-6851(00)00383-2 [DOI] [PubMed] [Google Scholar]
- Sherwin T., Schneider A., Sasse R., Seebeck T. and Gull K. (1987). Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol. 104, 439-446. 10.1083/jcb.104.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. Y., Wang C., Yuan Y. A. and He C. Y. (2013). An intracellular membrane junction consisting of flagellum adhesion glycoproteins links flagellum biogenesis to cell morphogenesis in Trypanosoma brucei. J. Cell Sci. 126, 520-531. 10.1242/jcs.113621 [DOI] [PubMed] [Google Scholar]
- Sunter J. D. and Gull K. (2016). The flagellum attachment zone: ‘the cellular ruler’ of trypanosome morphology. Trends Parasitol. 32, 309-324. 10.1016/j.pt.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter J. D., Benz C., Andre J., Whipple S., McKean P. G., Gull K., Ginger M. L. and Lukes J. (2015a). Modulation of flagellum attachment zone protein FLAM3 and regulation of the cell shape in Trypanosoma brucei life cycle transitions. J. Cell Sci. 128, 3117-3130. 10.1242/jcs.171645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter J. D., Varga V., Dean S. and Gull K. (2015b). A dynamic coordination of flagellum and cytoplasmic cytoskeleton assembly specifies cell morphogenesis in trypanosomes. J. Cell Sci. 128, 1580-1594. 10.1242/jcs.166447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan S., Kohl L., Ngai I., Wheeler R. J. and Gull K. (2008). A repetitive protein essential for the flagellum attachment zone filament structure and function in Trypanosoma brucei. Protist 159, 127-136. 10.1016/j.protis.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E. and Englund P. T. (2000). Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275, 40174-40179. 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- Wei Y., Hu H., Lun Z.-R. and Li Z. (2014). Centrin3 in trypanosomes maintains the stability of a flagellar inner-arm dynein for cell motility. Nat. Commun. 5, 4060 10.1038/ncomms5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C. and Cross G. A. M. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89-101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Zeytuni N. and Zarivach R. (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397-405. 10.1016/j.str.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Gheiratmand L., Chen Y., Lim T. K., Zhang J., Li S., Xia N., Liu B., Lin Q. and He C. Y. (2010). A comparative proteomic analysis reveals a new bi-lobe protein required for bi-lobe duplication and cell division in Trypanosoma brucei. PLoS ONE 5, e9660 10.1371/journal.pone.0009660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Liu B., Sun Y. and He C. Y. (2011). A coiled-coil- and C2-domain-containing protein is required for FAZ assembly and cell morphology in Trypanosoma brucei. J. Cell Sci. 124, 3848-3858. 10.1242/jcs.087676 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Hu H., He C. Y. and Li Z. (2015). Assembly and maintenance of the flagellum attachment zone filament in Trypanosoma brucei. J. Cell Sci. 128, 2361-2372. 10.1242/jcs.168377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Gu J., Lun Z.-R., Ayala F. J. and Li Z. (2016a). Two distinct cytokinesis pathways drive trypanosome cell division initiation from opposite cell ends. Proc. Natl. Acad. Sci. USA 113, 3287-3292. 10.1073/pnas.1601596113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Hu H. and Li Z. (2016b). An EF-hand-containing protein in Trypanosoma brucei regulates cytokinesis initiation by maintaining the stability of the cytokinesis initiation factor CIF1. J. Biol. Chem. 291, 14395-14409. 10.1074/jbc.M116.726133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.