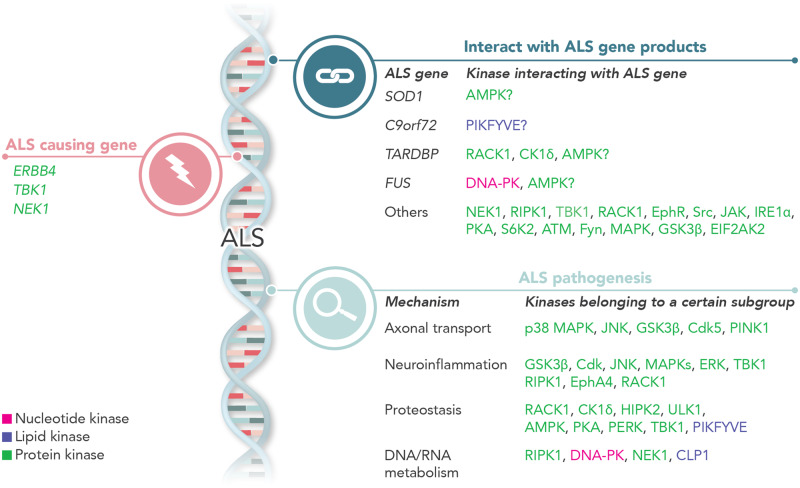
By reviewing recent findings on kinase function in ALS, Guo et al. show that kinases are implicated in ALS disease progression on three levels: a) genes encoding kinases are causal or risk genes in ALS; b) kinases interact with proteins encoded by ALS genes; and c) kinases participate in major ALS disease mechanisms.
Keywords: kinase, amyotrophic lateral sclerosis, motor neuron, phosphorylation
Abstract
Amyotrophic lateral sclerosis is the most common degenerative disorder of motor neurons in adults. As there is no cure, thousands of individuals who are alive at present will succumb to the disease. In recent years, numerous causative genes and risk factors for amyotrophic lateral sclerosis have been identified. Several of the recently identified genes encode kinases. In addition, the hypothesis that (de)phosphorylation processes drive the disease process resulting in selective motor neuron degeneration in different disease variants has been postulated. We re-evaluate the evidence for this hypothesis based on recent findings and discuss the multiple roles of kinases in amyotrophic lateral sclerosis pathogenesis. We propose that kinases could represent promising therapeutic targets. Mainly due to the comprehensive regulation of kinases, however, a better understanding of the disturbances in the kinome network in amyotrophic lateral sclerosis is needed to properly target specific kinases in the clinic.
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective and progressive degeneration of motor neurons in the brain, brainstem and spinal cord. Motor neuron deterioration leads to muscle weakness and results in death of the patient due to respiratory failure typically within 3 to 5 years after diagnosis. The life time risk of developing ALS is estimated to be ∼1 in 400 (Alonso et al., 2009).
In most patients, ALS begins at ∼50–60 years of age with asymmetric, painless weakness in one limb. Initially, abnormalities can be focal, with the disease spreading relentlessly over time. Nevertheless, ALS is a heterogeneous disorder. Time of disease onset varies between the first and the seventh decade of life and there are clear differences in location of symptom onset, rate of disease progression, and degree of cognitive impairments (reviewed in Swinnen and Robberecht, 2014). Besides variations in clinical presentation, there is also considerable variation in disease aetiology. Approximately 10% of patients report a family member also diagnosed with ALS (familial ALS), strongly supporting a direct genetic cause of the disease. However, 90% of patients suffer from the sporadic form of the disease (sporadic ALS), as no family member was ever diagnosed with ALS. Therefore, considerable efforts to identify environmental factors causing ALS were made, but these were unsuccessful until now. This supports the idea that for most ALS cases, including sporadic ALS, there might be an important genetic contribution (Simpson and Al-Chalabi, 2006).
A cascade of different processes could result in motor neuron death, independent of the exact cause of each ALS case. One of the processes that could drive selective motor neuron degeneration is the aberrant regulation of kinases (Hu et al., 2003; Krieger et al., 2003). The beneficial effects of inhibiting the Src/c-Abl pathway in multiple models of ALS suggests that kinases can closely regulate shared downstream processes in ALS pathogenesis (Katsumata et al., 2012; Wenqiang et al., 2014; Imamura et al., 2017). A compound screening based on a survival assay of induced pluripotent stem cell (iPSC) derived motor neurons from ALS patients (Imamura et al., 2017) also identified the Src/c-Abl as a major pathway related to motor neuron survival (Imamura et al., 2017). In addition, genetic knockdown and pharmacological inhibition of Src/c-Abl rescued the degeneration of iPSC-derived motor neurons from patients with familial ALS mutations in SOD1, C9orf72, TARDBP, and from sporadic ALS patients (Imamura et al., 2017). In line with these in vitro observations, Src/c-Abl inhibitors also attenuated ALS phenotypes both in mutant SOD1 and in TDP-43 transgenic mice (Katsumata et al., 2012; Wenqiang et al., 2014). Moreover, phosphorylation of Src/c-Abl was increased in post-mortem spinal cord tissue from ALS patients (Katsumata et al., 2012) indicating that this pathway could also play a role in motor neurons from ALS patients. Taken together, the beneficial effects obtained by inhibiting the Src/c-Abl pathway indicate that a shared downstream kinome pathway could be involved in the selective death of motor neurons in ALS.
Aberrant phosphorylation of various ALS-related proteins (e.g. SOD1 TDP-43 and FUS) by kinases could affect the localization and function of these proteins. Furthermore, kinases play a pivotal role in biochemical reactions involved in protein, lipid and nucleotide metabolism (Box 1). However, the exact role these alterations play in motor neuron degeneration remains elusive. In addition, multiple studies showed that mutations in genes encoding different kinases can cause or confer susceptibility to ALS, suggesting that alterations in the function of specific kinases and/or their downstream targets are vital to (motor) neuron survival (Freischmidt et al., 2015; Higelin et al., 2018). In this review, we provide a focused update on the potential role of kinases in ALS genetics and pathophysiology in view of some recent data.
Box 1.
Classification of kinases
Kinases are transferases catalysing the addition of a phosphate group () to hydroxyl groups of various substrates including lipids, nucleic acids, and amino acids. The phosphate normally originates from adenosine triphosphate (ATP). Phosphorylation is involved in nearly all signal transduction processes, and thus kinases play a pivotal role in regulating cellular metabolism, cell cycle, transport, secretory processes, and many other pathways (for a review see Rask-Andersen et al., 2014).
The total number of kinases is very large; more than 900 genes in the human genome encode kinases. Kinases are classified based on their substrates and functions. Based on the substrate, kinases can be classified as protein kinases, lipid kinases and nucleotide kinases (Rask-Andersen et al., 2014).
Protein kinases are the largest category of kinases with more than 500 encoding genes identified. These kinases are responsible for phosphorylating amino acids. Based on its specificity, protein kinases can be categorized into four classes: (i) protein-histidine kinases, which phosphorylate histidine residue; (ii) protein-tyrosine kinases, which phosphorylate tyrosine residue; (iii) protein-serine/threonine kinases, which phosphorylate serine and/or threonine residues; and (iv) dual-specificity kinases, which phosphorylate both tyrosine and serine/threonine residues. Protein tyrosine kinases can be classified into two additional subtypes: the receptor-type and the non-receptor-type based on the function of the substrates (Rask-Andersen et al., 2014).
Lipid kinases are a group of kinases that are responsible for phosphorylating lipid molecules. These lipids comprise membrane structures including the plasma membrane, as well as the membranes of the organelles. Inositol is phosphorylated by lipid kinases to generate phosphoinositol and phosphoinositide lipids. The lipid phosphorylation process is involved in the membrane signal transmission throughout the endomembrane system.
Nucleotide kinases are responsible for the phosphorylation of nucleic acids that are the basic units of RNA and DNA. In RNA and DNA polymers, the backbone is composed of repeating phospho-ribose units. Kinases transfer the phosphate to the nucleoside, creating a nucleotide monophosphate. This process is also involved in regulating the synthesis of nucleotides.
ALS genetics: the emerging role of kinases
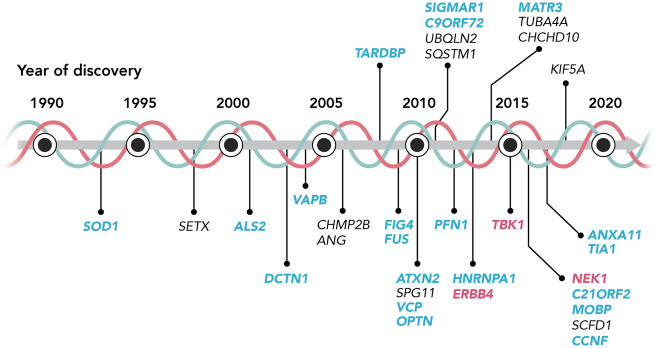
Since the discovery that mutations in the superoxide dismutase 1 (SOD1) gene cause ALS (Rosen et al., 1993), a multitude of genes have been linked to ALS (reviewed in Volk et al., 2018) Remarkably, the gene products can be grouped into a few biological processes: proteostasis (UBQLN2, VCP, OPTN, TBK1, C9orf72, VAPB), RNA metabolism (FUS, TARDBP, HNRNPA1, MATR3), and cytoskeletal dynamics (PFN1, TUBA4A, KIF5A, DCTN1). This indicates the potential importance of these processes in ALS pathobiology, especially when considering loss-of-function mutations in these genes (Taylor et al., 2016). While mutations in several genes are associated with ALS, point mutations in SOD1, TARDBP, FUS, or a hexanucleotide repeat expansion in C9orf72 explain more than half of the familial ALS cases (Fig. 1 and Box 2) (Taylor et al., 2016). Recently developed sequencing techniques resulted in multiple new discoveries, and suggest also polygenetic modes of inheritance (Oskarsson et al., 2018). Several of these newly discovered ALS-causing or ALS risk genes encode protein kinases.
Figure 1.
ALS genetics, the emerging role of kinases. Chronological overview of the discovery of gene mutations involved in ALS. Genes in blue are ALS genes of which the gene product could interact with kinases pathologically. Genes in pink are encoding kinases. Each of these gene mutations is reported in more than one ALS-affected family, or in multiple, unrelated cases of sporadic ALS.
Box 2.
Main ALS genes and pathology
At present, mutations in four genes are linked to classical ALS and explain up to 80% of familial ALS cases. More than 25 years ago, linkage analysis identified dominant missense mutations in the SOD1 gene as the first genetic cause for ALS (Rosen et al., 1993). This Cu-Zn superoxide dismutase is an abundant, ubiquitously expressed, cytoplasmic enzyme that catalyses the conversion of highly reactive superoxide into hydrogen peroxide. More than 170 SOD1 mutations were reported occurring in ∼12% of familial ALS and ∼2% of sporadic ALS cases (for a review see Renton et al., 2014). The most commonly used mouse model for ALS is the mutant SOD1G93A mouse (Gurney et al., 1994). This transgenic model develops adult-onset neurodegeneration of spinal motor neurons and progressive motor deficits leading to paralysis (Gurney et al., 1994).
An important discovery in the context of ALS was the identification of the TDP-43 protein as the major constituent of ubiquitin-positive neuronal inclusions (Neumann et al., 2006). This was followed by the discovery of mutations in the TARDBP gene encoding TDP-43 (Gitcho et al., 2008; Sreedharan et al., 2008). TARDBP mutations are relatively rare and it is estimated that ∼4% of familial ALS patients and only a small percentage of sporadic ALS cases is caused by these mutations (Renton et al., 2014). As TDP-43 plays an important role in DNA/RNA metabolism, the discovery of TDP-43 in ALS pathogenesis highlighted the importance of RNA processing in ALS (for a review see Ling et al., 2013).
Soon after the discovery of the TARDBP mutations, mutations in FUS, another gene encoding a DNA/RNA-binding protein, were discovered (Kwiatkowski et al., 2009; Vance et al., 2009). FUS mutations are also responsible for a small subset of ALS cases. It is estimated that they account in the Western world for 4% and 1% of familial ALS and sporadic ALS, respectively (Kwiatkowski et al., 2009; Vance et al., 2009; Van Damme et al., 2010). ALS mutations in FUS are mainly in the N-terminal low-complexity domain and in the highly-conserved C-terminal nuclear localization signal (NLS) (Ling et al., 2013). They lead to the mislocalization of FUS to the cytoplasm and this results in the formation of cytoplasmic FUS inclusions (Dormann et al., 2010).
More recently, an intronic hexanucleotide (GGGGCC) tandem repeat expansion was identified in the C9orf72 gene (DeJesus-Hernandez et al., 2011; Renton et al., 2011), which is the most common cause of ALS explaining ∼40% of familial ALS and ∼7% of sporadic ALS cases in the Western world (for a review see Renton et al., 2014). While control individuals usually harbour two to eight repeats, patients have more than 30, and up to hundreds or even thousands (Renton et al., 2014). The exact underlying pathological mechanism remains elusive, although a combination of loss-of-function and gain-of-function was suggested (Shi et al., 2018).
TBK1
TBK1 encodes TANK-binding kinase, a serine/threonine kinase interacting with proteins involved in the innate immune response and autophagy (Pottier et al., 2015). Genetic variants in TBK1 are associated with glaucoma (Traynis et al., 2014) and herpes (Herman et al., 2012). In addition, two independent whole exosome and whole genome sequencing studies linked different mutations in TBK1 to ALS (Fig. 1) (Cirulli et al., 2015; Freischmidt et al., 2015). Mutations in TBK1, as identified in ALS, caused a loss of kinase function (de Majo et al., 2018) and TBK1 knockout mice showed dendritic swellings, abnormally shaped astrocytes, and p62- and ubiquitin-positive aggregates in the cerebellum (Duan et al., 2019). Interestingly, families with TBK1 mutations showed an increased risk to develop cognitive defects in addition to their motor symptoms (Oakes et al., 2017), and ALS patients with TBK1 mutations displayed a bulbar onset more frequently (van der Zee et al., 2017). Post-mortem neuropathological analysis of TBK1 mutation carriers showed massive TDP-43-positive perinuclear inclusions in temporal lobe neurons, but not in the spinal cord, and showed p62/sequestosome 1 (SQSTM1)-positive perinuclear inclusion in the right para-hippocampal gyrus (van der Zee et al., 2017). p62/SQSTM1 is encoded by the ALS gene SQSTM1 (Fecto et al., 2011), and acts as a major autophagy receptor (Aparicio et al., 2019). Interestingly, TBK1 phosphorylates p62/SQSTM1 to ensure its binding to polyubiquitinated proteins and to efficiently target these proteins for degradation in autophagosomes (Matsumoto et al., 2015). As most patients with ALS displayed p62/SQSTM1 positive cytoplasmic inclusions (Teyssou et al., 2013), and as TBK1 was an inducer of type-1 interferons and affected autophagy and mitophagy (de Majo et al., 2018), it is possible that reduced TBK1-mediated p62/SQSTM1 phosphorylation disrupts cellular proteostasis (Vinet and Zhedanov, 2011). As a consequence, disturbed autophagy could (partially) explain the TBK1-related pathophysiology in ALS. An additional ALS-related protein that is also a target of TBK1 is optineurin. This protein is encoded by the OPTN gene, is highly abundant, and is involved in the inflammatory response, autophagy, Golgi maintenance, and vesicular transport. Recessive mutations in OPTN are considered as a rare genetic cause of ALS (Richter et al., 2016). In view of the loss of kinase function from TBK1 mutations identified in ALS (de Majo et al., 2018), it is likely that this also affects the ubiquitin-directed breakdown of aggregates through decreased optineurin targeting (Li et al., 2018).
Based on the targets of TBK1 (e.g. p62/SQSTM1 and optineurin) and because ALS-causing TBK1 mutations result in a loss of kinase function, we hypothesize that the impaired kinase function of TBK1 induces impairments in the clearance of proteins by autophagy or by the ubiquitin proteasome system, thereby contributing to the motor neuron degeneration. These mechanisms may act alone or in combination with other affected processes. Therapeutically stimulating the kinase function of TBK1 may be beneficial. However, more studies are needed to find out the exact therapeutic potential of TBK1 modulation in ALS, eventually also in those ALS patients without TBK1 mutations.
NEK1
Another kinase associated with ALS is NIMA related kinase 1 (NEK1) (Brenner et al., 2016; Kenna et al., 2016). Recently, different NEK1 variants have been identified in both familial and sporadic ALS (Kenna et al., 2016; Gratten et al., 2017; Nguyen et al., 2018; Shu et al., 2018; Tripolszki et al., 2019). NEK1 risk variants occur in ∼3 to 5% of ALS cases, although no ALS pedigrees have been identified with a clear segregation of NEK1 mutations with the disease (Nguyen et al., 2018). While most of the variants are missense variants, a large proportion of NEK1 variants lead to a loss-of-function (Nguyen et al., 2018). NEK1 variants are either heterozygous or homozygous in ALS patients (Shu et al., 2018; Goldstein et al., 2019), and often occur in ALS patients with a mutation in another ALS gene, such as SOD1, C9orf72, TUBA4A, or TARDBP (Nguyen et al., 2018; Shu et al., 2018).
The NEK1 protein contains an N-terminal kinase domain and an extended C-terminal domain with several predicted coiled-coil regions interacting with other proteins (Melo-Hanchuk et al., 2017). Although NEK1 contains two classical nuclear localization signals (NLS), the full length protein exclusively localizes to the cytoplasm, as the cytoplasmic localization signal originates from the extended C-terminal domain (Feige et al., 2006). Interestingly, a short protein fragments with its C-terminal ending at the first NLS has also been identified and this truncated version of NEK1 enters the nucleus (Feige et al., 2006). Overexpression of these nuclear NEK1 isoform caused abnormal chromatin condensation and dispersal of the nuclear pore complex (Feige et al., 2006). Therefore, a (genetic) modification of NEK1 might affect chromatin modifications and DNA stability, as well as several other cellular functions, including cilia formation, DNA-damage response, microtubule stability, neuronal morphology and axonal polarity (reviewed in Nguyen et al., 2018). This is supported by the phenotype of the NEK1 null mice suffering from developmental abnormalities including pleiotropic malfunctions, including facial dysmorphism, male sterility, dwarfism and anaemia, although no neurodegeneration was reported (Upadhya et al., 2000). Human fibroblasts from ALS patients with homozygous NEK1 truncations showed abnormalities in cilia number, cilia structure and microtubule stability (Kenna et al., 2016). Moreover, in vitro silencing of NEK1 led to distorted neuronal morphology with disturbed polarity and deacetylation of microtubules via histone deacetylase 6 (HDAC6) and to disrupted microtubule stability and growth (Chang et al., 2009; Cohen et al., 2013). Besides neuronal morphology and axonal polarity, NEK1 also regulates cellular viability and the permeability of the mitochondrial membrane through phosphorylation of the voltage-dependent anion channel 1 (VDAC1) (Chen et al., 2009). In addition, compromised NEK1 expression in patient-derived cells showed increased DNA damage which was accompanied by the deregulation of the cell cycle (Higelin et al., 2018). Interestingly, NEK1 can also interact with multiple ALS-related gene products, including alsin, VAPB and C21ORF2 (Nguyen, et al., 2018). One example is the interaction of NEK1 with C21ORF2, which is needed for efficient DNA damage repair responses (Fang et al., 2015).
While the above information highlights potential mechanisms by which variants in NEK1 might affect motor neuron viability in ALS, it is currently unclear which of these processes is involved in motor neuron degeneration and/or whether these are viable therapeutic targets. The generation of NEK1-ALS patient-derived iPSCs and subsequent motor neuron studies could aid in gaining a better understanding of this.
ERBB4
Mutations in ERBB4 have been identified in ALS patients (Takahashi et al., 2013), although these findings have not (yet) been replicated. However, the modifying role of ERBB4 and neuregulin 1 in ALS has been extensively investigated (Takahashi et al., 2013; Mancuso et al., 2016; Mòdol-Caballero et al., 2017). Erb-B2 receptor tyrosine kinase 4 (ERBB4) is a tyrosine kinase receptor that is able to activate multiple signal transduction cascades including the mitogen-activated protein kinase (MAPK), Agrin/MuSK, mTORC1, and STAT pathways (Trinidad et al., 2000; Eto et al., 2010; Sundvall et al., 2012; Nie et al., 2018). ERBB4 plays a role in various biological processes, including neurodevelopment. It belongs to the epidermal growth factor (EGF) subfamily of receptor tyrosine kinases (RTKs) and can be activated upon binding of neuregulins (NRGs) to the extracellular ligand-binding domain (Takahashi et al., 2013). In primate brain, expression of both full length and shorter fragments of ERBB4 was widely found in neuronal soma and nucleus throughout the brain of juvenile and adult primates, which could indicate a regulatory role for the ERBB4/NRG pathway in the CNS (Thompson et al., 2007). ERBB4 mutations identified in ALS patients decreased the auto-phosphorylation of ERBB4 upon neuregulin 1 stimulation in vitro (Takahashi et al., 2013). As a transmembrane receptor tyrosine kinase, ERBB4 binds to neuregulin 1 activating its signalling, and an impaired neuregulin ERBB4 pathway is involved in the pathogenesis of ALS (Takahashi et al., 2013; Mancuso et al., 2016; Mòdol-Caballero et al., 2017). The shortest ectodomain fragments of ERBB4 are generated in the presence of neuregulin 1 (Lopez-Font et al., 2019). Interestingly, ERBB4 ectodomain fragments were decreased in the CSF from ALS patients, as well as in the plasma of SOD1G93A and TDP-43A315T mice, indicating an involvement of ERBB4 in different ALS subtypes (Lopez-Font et al., 2019).
ALS is considered as a ‘dying back’ or ‘distal axonopathy’, in which the first pathological changes occur at the neuromuscular junction (NMJ) prior to motor neuron degeneration and the onset of clinical symptoms (Campanari et al., 2016). NRG1 is mainly produced by neurons and muscles and it mediates the crosstalk between terminal Schwann cells and the peripheral motor axon at the end-plate. This process is tightly regulated by the ERBB receptor family including ERBB3, ERBB4 and their co-receptors ERBB1 and ERBB2 (Morano et al., 2018). ERBB4 is especially enriched in the neuromuscular junctions and inhibition of ERBB4 impaired neuromuscular development in zebrafish embryos (Paatero et al., 2019). This is in line with results showing that loss-of-function mutations in ERBB4 could likely be the cause of autosomal-dominant ALS (Takahashi et al., 2013). Furthermore, the expression of NRG1 type III isoform was reduced in both ALS patients and SOD1G93A mice in parallel with motor neuron loss (Lasiene et al., 2016). In addition, the expression of NRG1 type I isoform was increased and associated with neuroinflammation and glial activation in spinal cord of ALS patients and SOD1G93A mice (Song et al., 2012). Moreover, overexpression of NRG1 in skeletal muscle promoted NMJ maintenance in the SOD1G93A mouse model of ALS (Mancuso et al., 2016). This suggests a potential cell type specific effect, which should be taken into consideration in the development of potential therapeutic strategies targeting this kinase.
As abnormal expression and activation of ERBBs is associated with many human cancers (Hynes and Lane, 2005), modulation of the activity of these kinases has already been studied (Qiu et al., 2008), and could facilitate the development of ERBB4 modulation as a potential ALS therapeutic strategy. However, more work is needed to evaluate how exclusive the interaction between ERBB4 and neuregulin 1 is, and whether other ERBB4 interactors play a pathogenic role in ALS.
The multiple roles of kinases in ALS pathophysiology
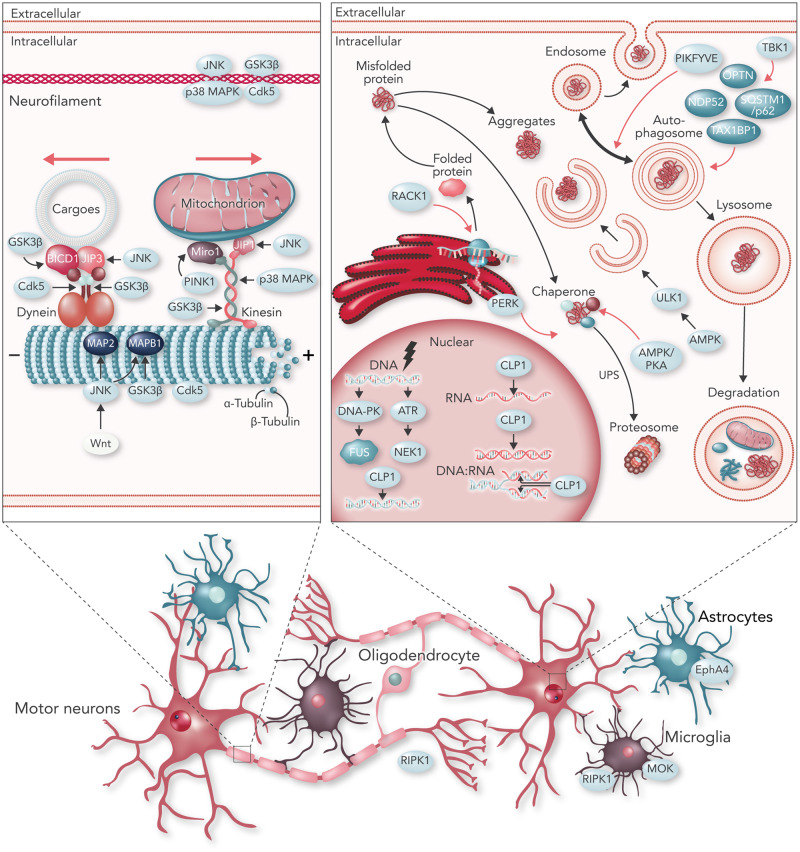
While some kinases are associated with ALS as they are encoded by (potential) ALS-related genes, other kinases are involved in processes linked to selective motor neuron death (Fig. 2). p38 mitogen-activated protein kinase (p38 MAPK), c-Jun N-terminal kinase (JNK), TBK1, and DNA-dependent protein kinase (DNA-PK) are kinases that have been associated with different ALS-related pathophysiological changes (Deng et al., 2014; Oakes et al., 2017; Naumann et al., 2018).
Figure 2.
Kinases in neurodegenerative processes involved in ALS. Schematic overview of kinases involved in different ALS pathophysiological processes. Names identified in light blue ovals indicate kinases. Names in dark blue ovals indicate adaptors that interact with kinases. Top left: Examples of kinases regulating axonal transport by interacting with subunits of the transport process including dynein (retrograde transport), kinesin (anterograde transport) and their adaptors, microtubules, neurofilaments, and cargo specific-adaptors. Bottom: Examples of kinases activated in non-neuronal cells including astrocytes, microglia and oligodendrocytes. Top right: Examples of kinases regulating proteostasis including protein synthesis through ribosomes on the endoplasmic reticulum (ER), the ubiquitin-proteasome system (UPS) and autophagy by direct or indirect phosphorylation. Examples of kinases activated by DNA damage and involved in RNA-related processes are also shown.
p38 MAPK belongs to the class of mitogen-activated protein kinases (MAPKs), which participate in signalling responses to cytokines and stress. Increased levels of active p38 MAPK were detected in post-mortem spinal cord and brain tissue of both familial and sporadic ALS patients (Bendotti et al., 2004). Moreover, we and others observed an upregulation of activated p38 MAPK in the SOD1G93A mice during disease progression (Tortarolo et al., 2003; Dewil et al., 2007), and inhibition of p38 MAP kinase had a moderate positive effect on the survival of the SOD1G93A mice (Dewil et al., 2007). More recently, it was shown that p38 MAPK regulates axonal transport and neuroinflammation in different ALS models (Sama et al., 2017; Gibbs et al., 2018).
JNKs, another subgroup of the MAPK family, function as stress-activated protein kinases. Activation of JNK/c-jun signalling occurs in motor neurons of SOD1G93A mice and is involved in the phosphorylation of TDP-43. Phosphorylated TDP-43 is a major component of the inclusions observed in neurons and glial cells in many cases of frontotemporal dementia (FTD) and/or ALS (Arai et al., 2006; Neumann et al., 2006; Hasegawa et al., 2008; Sreedharan et al., 2008; Mackenzie et al., 2010; Gitler and Shorter, 2011; Suzuki and Matsuoka, 2013; Ratti and Buratti, 2016). Multiple phosphorylation epitopes (pS379, pS403/404, pS409, pS410 and pS409/410) were identified in aggregated TDP-43 via phosphorylation-specific anti-TDP-43 antibodies. This abnormal phosphorylation and accumulation of TDP-43 is regulated by casein kinase-1 (CK1) (Hasegawa et al., 2008). Besides JNK and CK1, glycogen synthase kinase 3β (GSK3β) was shown to be a suppressor of ALS and FTD pathogenesis (Sreedharan et al., 2008; Suzuki and Matsuoka, 2013).
As described above, TBK1 is a serine/threonine-protein kinase implicated in both neuroinflammation and autophagy in ALS (Oakes et al., 2017). DNA-PKs are nuclear protein serine/threonine kinases, which sense DNA damage and participate in the ligation step of the non-homologous end joining (NHEJ) pathway of DNA double strand break repair. Combined with cytoplasmic FUS accumulations, DNA-PKs are activated upon DNA damage in ALS (Naumann et al., 2018). Below, we summarize the regulation of some of the main kinases in the pathophysiology of ALS (Fig. 2).
Kinases and axonal transport
Intracellular transport of cargoes is essential to maintain the structure and function of motor neurons because of their extreme morphology and polarization. Axonal transport mediates the distribution of cargoes, such as mRNAs, protein and organelles, mainly synthesized in the cell body across the cell (Box 3). In addition, axonal transport maintains the essential long-distance communication between the cell body and the synaptic terminals which allows neurons to react to their surroundings via trafficking of signalling endosomes (for a review see De Vos and Hafezparast, 2017). Axonal transport defects were observed in many different ALS models, and mutations in components of the axonal transport machinery were linked to ALS (López et al., 2015; Wloga et al., 2017). Moreover, axonal transport defects are an early, presymptomatic phenotype, which has also been observed in iPSC-derived motor neurons carrying a wide variety of ALS-causing mutations (Sivadasan et al., 2016; Guo et al., 2017; Kreiter et al., 2018; Naumann et al., 2018; Pal et al., 2018). There is increasing support for the hypothesis that (de)phosphorylation processes are key regulators of axonal transport (Box 3) (reviewed in Brady and Morfini, 2017). Kinases regulate axonal transport by directly phosphorylating molecular motors, adaptors, cargoes and/or the microtubular network, and this affects the interaction between motor proteins and their cargoes or between motor proteins and microtubules, hence affecting axonal transport (Brady and Morfini, 2017; De Vos and Hafezparast, 2017). In addition, modifying the microtubule network affects axonal transport by altering the microtubule stability. Below, we discuss the major kinases described to impact these interactions (Fig. 2A).
Box 3.
Axonal transport and the ‘dying back’ hypothesis in ALS
The most particular morphological feature of neuronal cells in comparison to other cell types is their extreme polarity and very long axons. Axons allow for the efficient communication between soma and axonal terminals. Axonal communication is especially important for motor neurons, as they not only need to connect with each other but also reach over a long distance to muscles to properly control muscle contraction (reviewed in De Vos and Hafezparast, 2017). Axonal transport maintains the efficient supply of cargoes including proteins, RNAs, lipids and organelles and it is also responsible to clear or recycle misfolded proteins or aggregates during cellular stress (reviewed in Prior et al., 2017). Axonal transport uses microtubules as tracks and α-tubulin and β-tubulin are the building blocks of these microtubules. The growing ‘plus’ end of the microtubules is at the axonal distal part, and the ‘minus’ end is located at the side of the soma. Two types of ATP-dependent motors, including kinesins and dyneins, are responsible for moving cargoes (including organelles, proteins, RNAs etc.) along the microtubules (reviewed in Guo et al., 2019). Kinesins are a superfamily of proteins that are mainly responsible for anterograde axonal transport (from ‘minus’ to ‘plus’ end of the microtubules) (Guo et al., 2019), while some kinesins move towards the ‘minus’ end. This enables the kinesins to transport cargo in both directions (Endow, 1999). Dyneins are also a family of proteins that can be further divided into two isoforms including cytoplasmic dynein and axonemal dynein. Cytoplasmic dynein is responsible for retrograde axonal transport (from ‘plus’ to ‘minus’ end of the microtubules) (for a review see Reck-Peterson et al., 2018). Adaptor proteins connect motor proteins to their cargo (Guo et al., 2019).
The ‘dying-back’ hypothesis explains the sequence of events during motor neuron degeneration in ALS (Dadon-Nachum et al., 2011; Baker 2014). The underlying idea is that motor neurons lose their connection with muscle fibres and that the axon retracts towards the soma. This ultimately results in motor neuron death. This hypothesis is supported by the observation that motor neuron pathology begins at the terminal part of the axon and proceeds in a ‘dying back’ pattern (Dadon-Nachum et al., 2011). In addition, the longest and largest neurites with the highest metabolic demand seem to be the most susceptible to this “dying back” phenomenon (Dadon-Nachum et al., 2011).
p38 MAPK
Similar to patients, SOD1G93A transgenic mice and transgenic squid axoplasm carrying ALS-linked FUS mutations (G230C, R521G and R495X) showed activation of p38 MAPK and impaired axonal transport (Tortarolo et al., 2003; Bendotti et al., 2004; Sama et al., 2017). The mechanism underlying the toxic function of p38 MAPK overactivation is the phosphorylation of kinesin-1 on its serines 175 and 176 by p38 MAPK. This inhibits translocation of kinesin-1 along axonal microtubules and will cause transport defects (Morfini et al., 2013). Inhibition of p38 MAPK showed multiple beneficial effects in rodent ALS models, including protection of mutant SOD1-induced motor neuron degeneration, prolongation of survival of SOD1G93A mice, restoration of axonal transport defects in both SOD1G93A mice and in murine motor neurons transfected with mutant FUS (Dewil et al., 2007; Sama et al., 2017; Gibbs et al., 2018) (Table 1). p38 MAPK can also phosphorylate neurofilament medium and heavy chain (NF-M and NF-H) sidearms, which hampered axonal transport (Ackerley et al., 2004). This is in line with the observation that increased co-localization of p38 MAPK with phosphorylated neurofilaments was present in neurons of SOD1G93A mice (Bendotti et al., 2004). This implies that inhibition of p38 MAPK could be therapeutically relevant in ALS, as we have previously shown by using semapimod in SOD1G93A mice, although the beneficial therapeutic effect was moderate (Dewil et al., 2007). We assume that this moderate beneficial effect in vivo was because of the fact that semapimod could not restore the function of the neuromuscular junctions (Dewil et al., 2007). Therefore, we propose a combined treatment regimen targeting the entire motor unit in vivo.
Table 1.
Compounds targeting kinases in ALS
| Kinase | Possible target/mechanism | Inhibitor | Preclinical model or clinical trial phase | Effect | BBB permeability | FDA approval (ALS/other diseases) | Ref /clinical trial registration number a |
|---|---|---|---|---|---|---|---|
| AMPK | A2A adenosine receptor (A2AR) | JMF1907 | NSC34 cells TDP-43 transgenic mouse | Normalized the mislocalization of TDP-43 in vitro Improved the motor function (rotarod performance, forelimb grip strength) in vivo | NE | NE | Liu et al., 2015 |
| ASK1 | Stress-responsive | K811, K812 NQDI-1 | SOD1G93A transgenic mouse SOD1G85R transgenic squid axoplasm | Increased survival of motor neurons Inhibited the activation of glial cells Extended survival of SOD1G93A transgenic mouse Rescued anterograde axonal transport in SOD1G85R transgenic squid axoplasm | NE | NE | Song et al., 2013; Fujisawa et al., 2016 |
| CK1 | Phosphorylate TDP-43 directly | Inhibitor20 | TDP-43 transgenic fly | Extended fly lifespan | Yes | NE | Salado et al., 2014 |
| DLK | JNK pathway | GNE-3511 | SOD1G93A transgenic mouse | Delayed neuromuscular junction denervation | Yes | NE | Le Pichon et al., 2017 |
| ERK | EGFR pathway signalling | Erlotinib | SOD1G93A transgenic mouse | Delays disease progression; no extend survival | Yes | Yes | Le Pichon et al., 2013 |
| EphA4 | EphA4-LBD | 123C4 | SOD1G93A transgenic mouse | Extended survival | NE | NE | Wu et al., 2017 |
| EphA4 | EPHA4 | EphA4-ASO | SOD1G93A and PFN1G118V transgenic mouse | No improvement of motor function or survival | NE | NE | Ling et al., 2018 |
| GSK3β | COX-2 | GSK-3 inhibitor VIII | SOD1G93A transgenic mouse | Increased motor neuron survival; delayed disease onset and extended survival | NE | NE | Koh et al., 2007 |
| GSK3β | Changes of transcription factors | Lithium plus valproate | SOD1G93A transgenic mouse | Delayed the onset of motor dysfunction Extended survival and reducing neurological deficits | Yes | Yes | Feng et al., 2008 |
| GSK3β | NE | JGK-263 | SOD1G93A transgenic mouse | Increased motor neuron survival Improved motor function and delayed the onset of motor dysfunction, rotarod failure, and survival | Yes | NE | Ahn et al., 2014 |
| JAK3 | NE | WHI-P131 | SOD1G93A transgenic mouse | Increased survival | NE | NE | Trieu et al., 2000 |
| p38 MAPK | Kinesin1 | SB239063 | SOD1G93A transgenic mouse | Restored the rate of axonal retrograde transport in vivo | Yes | NE | Gibbs et al., 2018 |
| p38 MAPK | TNFs | Semapimod | SOD1G93A transgenic mouse | Increased motor neuron survival Delayed disease onset and extended survival | NE | NE | Dewil et al., 2007 |
| p38 MAPK | Stress response | MW069 | SOD1G85R transgenic squid axoplasm | Rescue anterograde axonal transport | NE | NE | Song et al., 2013 |
| PERK | eIF2α | GSK2606414 | Primary rat cortical neurons TDP-43 transgenic fly | Increased survival of neurons; mitigation of TDP-43-induced climbing dysfunction in fly | Yes | NE | Kim et al., 2014 |
| PIKFYVE | RAB5 | YM201636 Apilimod | C9orf72 patient iMNs | Increased EEA1+ endosome size Increased patient iMN survival | NE | NE | Shi et al., 2018 |
| RIPK1 | Inflammation | DNL747 | Phase I in ALS | Not yet available | Yes | NE | NCT03757351 |
| ROCK | Actin cytoskeleton and neuronal survival | Fasudil | Phase II in ALS | Not yet available | Yes | NE | Lingor et al., 2019; NCT03792490 |
| Src/c-Abl | Autophagy | Bosutinib | iPSC-MNs (sporadic, TDP-43, C9orf72, SOD1) SOD1G93A transgenic mouse | Increased survival of iPSC-derived MNs Delayed disease onset and extended survival | Yes | Yes | Imamura et al., 2017 |
| Tyrosine kinase (pan) | Inflammation (add-on treatment with riluzole) | Masitinib | Phase II / III in ALS | Phase II: improvement of life quality, respiratory function and delay of death | Yes | NE | Mora et al., 2019; NCT02588677 |
As registered on clinicaltrials.gov.
BBB = blood–brain barrier; iMN = induced motor neuron; NE = no evidence.
JNK
Increased phosphorylation of c-Jun occurred in motor neurons of SOD1G93A mice and JNK signalling also seemed to be involved in TDP-43-related toxicity (Vlug et al., 2005; Suzuki and Matsuoka, 2013). Limited TDP-43 overexpression in NSC34 motor neuronal cells and primary cortical neurons induced neuronal cell death through the upregulation of Bim and CHOP expression and downregulation of Bcl-xL expression (Suzuki and Matsuoka, 2013). Furthermore, TDP-43 overexpression increased the phosphorylation of JNK and inhibition or downregulation of JNK inhibited TDP-43-induced cell death, suggesting a link between the JNK/c-Jun signalling and TDP-43 induced cell death (Suzuki and Matsuoka, 2013). JNK directly phosphorylated the motor domain of kinesin-1 (DeBerg et al., 2013). Moreover, JNK could disrupt binding between kinesin-1 and JIP1, which is a cargo adaptor of kinesin, via activation of MAP kinase kinase kinase (MAPKKK), Wallenda (a homologue of dual leucine zipper-bearing kinase) and MAPKK Hemipterous (a homologue of MKK7) in Drosophila (Horiuchi et al., 2007). Furthermore, JNK regulated retrograde axonal transport by affecting the binding of JIP3 to p150 Glued and dynein light chain (DLIC) (Horiuchi et al., 2007). Similar to p38 MAPK, Jun N-terminal kinase-1 and -3 (JNK1 and JNK3) could also phosphorylate NF-M and NF-H side-arm domains, interrupting axonal transport (Ackerley et al., 2004). JNK signalling can also be activated by dual leucine zipper kinase (DLK). Genetic deletion and pharmacological inhibition of DLK protected against axon degeneration, neuronal loss, and functional decline in SOD1G93A mice by reducing phosphorylated c-Jun (Le Pichon et al., 2017). In addition, JNK can be activated by the Wnt pathway to phosphorylate microtubule-associated proteins, such as microtubule-associated protein 2 (MAP2) and microtubule-associated protein 1B (MAP1B). This results in changes in the microtubular dynamics and eventually disrupts neuronal axonal transport in cooperation with GSK3β (Ciani and Salinas, 2007). Dramatic upregulation of the expression of several members of the Wnt family in astrocytes as a function of disease progression has been shown in spinal cord of SOD1G93A mice, as well as in post-mortem tissue of ALS patients (Chen et al., 2012; Yu et al., 2013; González-Fernández et al., 2019). This suggests that glial proliferation-related neurodegeneration activates the Wnt signalling pathway. Subsequently, JNKs are activated by Wnt and eventually disturb the stability of the microtubules. The interaction between different kinases indicates that a series of kinome events could be involved in ALS pathogenesis. As a consequence, targeting a single kinase may also influence the function of other related kinases. On the other hand, targeting only one single kinase could be insufficient to result in positive therapeutic effects.
GSK3β
GSK3β is a multifunctional serine/threonine kinase that was originally identified as a regulator of glycogen synthase (Du et al., 2010). GSK3β modulated transport of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA)-containing vesicles by mediating phosphorylation of kinesin light chain 2 (KLC2) in kinesin-dependent axonal transport (Du et al., 2010). In addition, GSK3β can also regulate dynein-dependent axonal transport. Two conserved residues from two different dynein intermediate chain (IC) isoforms, S87/T88 in IC-1B and S88/T89 in IC-2C, are targeted by GSK3β. Although these isoforms are fairly ubiquitous, the IC-1B isoform is described to play a prominent role in axonal transport in neurons (Gao et al., 2015). The phosphorylated residues are within an Ndel1-binding domain, which is responsible for the interaction between ICs and Ndel1 (Gao et al., 2015). Moreover, the phosphorylation of a dynein adaptor called BICD1 was GSK3β dependent (Fumoto et al., 2006). Pharmacological and genetic inhibition of GSK3β could increase dynein motility (Gao et al., 2015) (Table 1). GSK3β can also mediate semaphorin3A-induced bidirectional axonal transport through phosphorylation of the axis inhibitor-1 (Hida et al., 2015). Interestingly, semaphorin 3A signalling through neuropilin-1 was an early trigger for the distal axonopathy in SOD1G93A mice (Venkova et al., 2014), implying that GSK3β signalling plays a pivotal role in distal axonal degeneration in ALS. The activation of GSK3β was also observed in ALS-associated defects in endoplasmic reticulum (ER)/mitochondrial communication (Stoica et al., 2016). Therefore, GSK3β may also influence axonal transport indirectly by affecting ER–mitochondria interactions (Stoica et al., 2014, 2016; Guo et al., 2017). Moreover, GSK3β could influence axonal transport by fostering NF-NF associations that compete with transport by phosphorylating the C-terminal tail of the NF-H (Vohnoutka et al., 2017), and GSK3β regulated the release of tau from microtubules by phosphorylation which influenced microtubule stability (Rankin et al., 2007). GSK3β is also the main kinase responsible for TDP-43 phosphorylation and deletion of the GSK3β gene protected against TDP-43-induced toxicity (Sreedharan et al., 2008). Although extensive research identified a direct and indirect involvement of GSK3β in ALS pathology, the real therapeutic potential in ALS patients is not yet clear. Until now, different GSK3β inhibitors with blood–brain barrier (BBB) permeability have been developed and investigated in SOD1G93A mice (Table 1) showing that lithium had a neuroprotective effects in SOD1G93A mice (Fornai et al., 2008). As most of these inhibitors showed beneficial effects including delayed disease onset, increased motor neuron survival, and improved motor function (Feng et al., 2008; Ahn et al., 2014), clinical trials targeting GSK3β have been conducted. A completed Phase III trial administering lithium to ALS patients did not show a benefit on survival (Al-Chalabi et al., 2013), although a meta-analysis identified a beneficial effect on mutant UNC13A carriers but not on mutant C9orf72 carriers (van Eijk et al., 2017). Further research using other GSK3β inhibitors in different ALS model systems should be performed to validate the potential of GSK3β as a therapeutic target.
CDK5
CDK5 is a serine/threonine kinase that belongs to the mitotic cyclin-dependent kinases family, and activation of CDK5 occurred in the spinal cord of SOD1G37R mice (Nguyen et al., 2001). Similar to GSK3β, CDK5 phosphorylates Ndel1, inducing the formation of the Lis1 and Ndel1 complex. Binding of this complex to dynein inhibited the dynein-mediated transport through blockage of the ATP-dependent release of dynein from microtubules (Klinman and Holzbaur, 2015). This process was validated by monitoring the transport of lysosomes, autophagosomes, mitochondria, and endosomes (Pandey and Smith, 2011; Klinman and Holzbaur, 2015). This was further confirmed by a restoration of axonal transport defects through inhibition of CDK5 in dorsal root ganglion (DRG) neurons from SOD1G93A mice (Klinman and Holzbaur, 2015). However, it is unclear whether this also affected motor neuron survival. Furthermore, activation of CDK5 was also required to maintain initial segment integrity of the axon, which was responsible for both microtubular organization and polarized dynein-dependent sorting of axodendritic cargos in neurons (Klinman et al., 2017). In addition, CDK5 could regulate axonal transport in neuronal cells by phosphorylating the NF-H subunit (Shea et al., 2004). Through sharing both up and downstream pathways with GSK3β (Engmann and Giese, 2009), CDK5 might also participate in a kinase network that regulates axonal transport, which might contribute to motor neuron survival.
PINK1
PTEN-induced kinase 1 (PINK1) is a mitochondrial serine/threonine-protein kinase that localizes at the outer membrane of mitochondria, but can also be found throughout the cytosol. Mitochondria are one of the most important cargoes transported along the axon to maintain proper energy supply and normal synaptic function of motor neurons (Saxton and Hollenbeck, 2012; Vandoorne et al., 2018, 2019). The efficient clearance of damaged mitochondria largely relies on proper axonal transport (Saxton and Hollenbeck, 2012). PINK1 regulates axonal transport by phosphorylating mitochondrial Rho GTPase 1 (Miro1) (Wang et al., 2011). Miro1 serves as a component of the primary motor/adaptor complex that anchors kinesin to the mitochondrial surface (MacAskill et al., 2009; López-Doménech et al., 2018). It plays a pivotal role in regulating mitochondrial transport in response to Ca2+ levels within and outside of mitochondria by interacting with kinesin (MacAskill et al., 2009; López-Doménech et al., 2018) (Fig. 2A). When mitochondria are damaged, the depolarized mitochondria stabilize PINK1 on their surface, promoting the interaction between PINK1 and Miro, and causing PINK1 to phosphorylate Ser156 of Miro. Subsequently, the interaction of parkin with Miro removes Miro from the mitochondrial membrane and it is subsequently degraded by the proteasome. Kinesin is released from the mitochondria and this eventually stops mitochondrial transport (Wang et al., 2011). The pivotal role of PINK1, often through Miro1, in ALS pathogenesis has been suggested in different models as well as in ALS patients. In spinal cord tissue of ALS patients as well as in the SOD1G93A and TDP-43M337V mouse models, it was shown that Miro1 expression was reduced (Zhang et al., 2015). In addition, mutations in Miro genes caused anterograde mitochondrial transport defects in distal synaptic terminals of flies (Guo et al., 2005; Russo et al., 2009). Moreover, PINK1 can be stimulated by mitochondrial damage, can induce Miro degradation and can halt mitochondrial transport (Wang et al., 2011). Interestingly, downregulation of PINK1 suppressed the TDP-43-induced degenerative phenotypes in a Drosophila model, and PINK1 also functioned as a genetic modifier of FUS-induced neurodegeneration (Chen et al., 2016; Sun et al., 2018). This implies that PINK1 might serve as a potential therapeutic target for ALS through regulating mitochondrial transport.
Kinases and neuroinflammation
Neuroinflammation is the inflammatory response within the brain or spinal cord. It is mediated by the production of cytokines, chemokines and reactive oxygen species produced by resident microglia, astrocytes and endothelial cells, and/or by peripherally derived immune cells (reviewed in DiSabato et al., 2016). It is a common denominator in neurodegeneration (Hammond et al., 2019). In the CNS of ALS patients and of ALS animal models, a chronic activation of microglia, astrocytes and T lymphocytes was a prominent pathological observation (Liu and Wang, 2017). A genome-wide transcriptional study on motor cortex samples from 31 patients with sporadic ALS and 10 healthy control subjects identified aberrant expression of 1573 of 2637 inflammatory-related genes in ALS patient tissues (Morello et al., 2017). Several critical mediators of neuroinflammation, including GSK3β, Cdk, JNK, MAPKs, and ERK, are kinases (Wang et al., 2012; Chen et al., 2016). However, it remains to be determined whether neuroinflammation is driving the ALS disease process or represents a mere consequence of the neurodegeneration. This is especially difficult to assess in ALS patients as diagnostic certainty is mostly reached in an advanced stage of the disease. However, the discovery of TBK1 as an ALS-causing gene suggests a direct link between neuroinflammation and kinases in ALS (Cirulli et al., 2015; Freischmidt et al., 2015). In addition, decreasing microgliosis by administration of masitinib, a tyrosine kinase inhibitor, in symptomatic SOD1G93A rats prolonged survival by 40% and offered motor neuron protection in the spinal cord (Trias et al., 2016). Despite this and many other findings that suggest that inflammation is important in ALS, one should also keep in mind that many therapeutic strategies counteracting neuroinflammation failed in clinical trials despite showing promising therapeutic effects in animal models (reviewed in McCauley and Baloh, 2019). Below, we discuss the role of various neuroinflammation-related kinases in the context of ALS (Fig. 2B).
TBK1
As described above, different genetic studies identified TBK1 as an ALS-causing gene and/or a disease modifier (Cirulli et al., 2015; Freischmidt et al., 2015). TBK1 belongs to the IKK-kinase family, which is involved in immune signalling pathways by regulating the production of interferon (IFN)α and IFNβ, and activation of TBK1 was observed in microglia (Moore and Holzbaur, 2016; Oakes et al., 2017). TBK1 also functions as an important regulator of dendritic cells, which are crucial for mediating immune responses. Dendritic cell-specific deletion of TBK1 caused T-lymphocyte activation and autoimmune symptoms (Xiao et al., 2017). In T cell-specific TBK1 knockout mice, the migration of T lymphocytes from the lymph nodes was impaired and resulted in reduction of the number of T lymphocytes in the CNS, eventually causing TBK1-dependent neurotoxicity (Yu et al., 2015). Studies in SOD1G93A mice showed that neuroprotective anti-inflammatory mediators were present during the early stage of the disease, and these changed into cytotoxic pro-inflammatory mediators at later stages of the disease (Beers et al., 2011). As a recent phase I clinic trial showed that administration of T lymphocytes to ALS patients was safe and well tolerated (Thonhoff et al., 2018), it will be interesting to investigate whether this strategy could rescue ALS-related phenotypes in TBK1 conditional knockout mice. In addition, further experiments evaluating the effect of TBK1 expression on neuroinflammation would be helpful to pinpoint which of the canonical functions of TBK1 are essential for its link to ALS.
RIPK1
In post-mortem tissue from ALS patients, multiple biochemical hallmarks of necroptosis—a form of programmed necrosis caused by inflammation—were observed (Ito et al., 2016). Although there is no causal evidence linking receptor-interacting kinase 1 (RIPK1) mutations to ALS, this gene is considered to be associated with ALS by its close association to the ALS gene OPTN and it is a critical regulator of cell death and inflammation (Humphries et al., 2015; Ito et al., 2016). In SOD1G93A mice, loss-of-function mutations in the OPTN gene resulted in progressive demyelination and axonal degeneration through engagement of the necroptotic machinery in the CNS (Ito et al., 2016). RIPK1 regulates necroptosis via the sequential activation of two downstream targets: RIPK3 and mixed lineage kinase domain-like protein (MLKL) (Wang et al., 2019). In ALS, OPTN significantly suppressed RIPK1-dependent signalling by regulating its turnover (Ito et al., 2016). Moreover, elevated levels of RIPK1, RIPK3 and MLKL were observed in the SOD1G93A mouse model, which could contribute to the axonal pathology and motor dysfunction in these mice (Ito et al., 2016). As the inhibition of RIPK1 prevented progressive axonal degeneration, RIPK1 could also play an important role in mediating axonal degeneration (Ito et al., 2016). In addition, through interaction with TBK1, RIPK1 promoted ageing-related inflammation that has been suggested to contribute to ALS disease progression (Xu et al., 2018). Recently, a phase I clinical ALS trial, which will test the safety, tolerability, pharmacokinetics, and pharmacodynamics of the RIPK1 inhibitor, DNL747, was announced (NCT03757351) (Table 1). Overall, RIPK1 is an interesting target to provide axonal protection in ALS and eventually in other human degenerative disorders characterized by axonal degeneration.
MOK
MAPK/MAK/MRK overlapping kinase (MOK) belongs to the MAP kinase superfamily. It is localized in the cytoplasm and phosphorylates several exogenous substrates or undergoes autophosphorylation. Cytoplasmic TDP-43 aggregation is a pathological hallmark in both familial and sporadic ALS patients (reviewed in Hergesheimer et al., 2019). TDP-43 aggregates purified from the brains of ALS patients were hyperphosphorylated and/or ubiquitinated (Hergesheimer et al., 2019). These intracellular aggregates were cytotoxic and caused extracellular TDP-43 accumulation that could be internalized and modified by microglia (Leal-Lasarte et al., 2017). MOK seems to play a role as an important mediator in TDP-43 aggregates exposed to microglia. MOK co-localized with TDP-43 aggregates and reduced phosphorylation after exposure of microglia to TDP-43 aggregates (Leal-Lasarte et al., 2017). By targeting MOK, extracellular TDP-43 aggregates stimulated inflammasome dependent IL-1β and IL-18 secretion, and neuroinflammation-linked caspase 3 activation that eventually could cause neurodegeneration (Leal-Lasarte et al., 2017). This may be a direct consequence of abnormal engagement of MOK into TDP-43 aggregates, and, as a consequence, modulation of MOK activation status may affect cellular responses that are modulated by downstream signalling pathways. In addition, TDP-43 also triggered inflammasome activation of the NF-κB pathway through p38 MAPK, which led to secretion of IL-1β and IL-18 (Huang et al., 2019). As both MOK and p38 MAPK belong to the MAPK superfamily, the interaction between MOK and p38 MAPK still needs to be investigated in more detail to obtain a better picture of the kinase network in TDP-43-related neuroinflammation.
EPHA4
EPHA4 receptor tyrosine kinase belongs to the Ephrin receptor subfamily of the protein-tyrosine kinase family and was initially identified as an ALS modifier in a morpholino-based zebrafish screen (Van Hoecke et al., 2012). EPHA4 receptor tyrosine kinase regulates axonal guidance in the corticospinal tract, but also functions as a mediator of inflammation in spinal cord injury (Goldshmit et al., 2004; Zhao et al., 2018). In mice, EPHA4 expression was also upregulated in activated astrocytes after spinal cord injury (Goldshmit et al., 2004). In our previous study, we showed that EPHA4 modulated motor neuron degeneration and disease progression in ALS (Van Hoecke et al., 2012). Another recent study showed that decreased signalling of EPHA4 improved functional performance and motor neuron survival in the SOD1G93A mice (Zhao et al., 2018). In addition, very rare loss-of-function variants in the EPHA4 gene were associated with prolonged survival in ALS patients (Van Hoecke et al., 2012). We obtained EPHA4-specific nanobodies (Schoonaert et al., 2017) and selective EPHA4 agonists were also developed to treat ALS (Wu et al., 2017) with promising results in delaying the progression of disease in the SOD1G93A mouse model (Wu et al., 2017) (Table 1). Despite the fact that these results indicate that EPHA4 receptor tyrosine kinase may serve as a therapeutic target for ALS, downregulation of EPHA4 using antisense oligonucleotides had no protective effect (Ling et al., 2018). In addition, it becomes clearer that the ephrin system is complicated and that interfering with this signalling system in the context of ALS does not always result in the expected outcome (Rué et al., 2019).
RACK1
Receptor of activated protein C kinase 1 (RACK1) functions as an intracellular protein receptor for protein kinase C, and has been associated to ALS as it mislocalized in spinal cord sections of ALS patients (Russo et al., 2017). RACK1 modulated microglial resistance against LPS-induced inflammatory injury (Yin et al., 2017). Moreover, TDP-43 inhibited the inflammatory response by modulating expression of RACK1 in human osteoarthritis (Huang et al., 2017). Intriguingly, mislocalization of RACK1 to TDP-43-positive cytoplasmic inclusions in motor neurons was observed in ALS patients (Russo et al., 2017). Although there is no clear mechanism underlying the role of RACK1 in ALS yet, the current findings suggest that RACK1 might mediate ALS pathogenesis through its effects on neuroinflammation.
Kinases and disrupted proteostasis
Proteostasis is dependent on a complex regulatory network that maintains protein homeostasis (Box 4). It is vital for cell health and survival and consists of several pathways controlling protein biosynthesis, folding, trafficking, and degradation (reviewed in Webster et al., 2017). It also includes specific protein stress pathways such as the unfolded protein response (UPR) in the ER, the mitochondrial UPR and the cytosolic heat shock response (Webster et al., 2017). Ubiquitin-positive inclusions are a hallmark of ALS. Hyperphosphorylated, ubiquitinated and/or cleaved TDP-43 are major constituent of these inclusions (Neumann et al., 2006). This, in combination with preclinical studies showing altered autophagy and proteasomal pathways (reviewed in Ramesh and Pandey, 2017), and genetic studies linking mutations in genes encoding proteins involved in these pathways to ALS (Webster et al., 2017; Alexander et al., 2018), suggests that proteostasis could be a key player in ALS pathogenesis. More recently, defects in protein synthesis have also been implicated in ALS (López-Erauskin et al., 2018; Koskimäki et al., 2019). Kinases actively participate in several steps of protein synthesis and degradation (Box 4). Therefore, we discuss the main kinases impacting proteostasis that could be involved in ALS (Fig. 2C).
Box 4.
Proteostasis in ALS
Proteostasis is the concept that there are cellular mechanisms that control the synthesis, folding, trafficking and degradation of proteins inside and outside the cell (Powers et al., 2009). In humans, approximately one-third of all proteins are synthesized in the ER and then transit to membrane compartments (Hetz and Mollereau, 2014). The quality control of protein folding happens in the ER (Hetz and Mollereau, 2014). In pathological conditions, as well as during ageing, the efficient protein folding process is hampered with misfolded proteins accumulating and clumping together as aggregates (for a review see Boeynaems and Gitler, 2018). Under normal physiological conditions, protein aggregation could play a protective and beneficial role to minimize the toxic effects of misfolded proteins. Some proteins can switch between liquid and solid phases, a process called phase transition (Boeynaems et al., 2018; Wang and Zhang, 2019). If this process is impaired, toxicity can result in disease-relevant pathological changes (Boeynaems et al., 2018). It has been shown that the phosphorylation of the prion-like domain can play a pivotal role in phase transition of proteins (Boeynaems and Gitler, 2018). In addition, disrupted proteostasis can cause ER stress (Hetz and Saxena, 2017). ER stress induces the ubiquitin proteasome system (UPS), a signalling network that transduces information about the protein folding status from the ER to the cytosol and nucleus to conjugate ubiquitin to substrates. This results in the degradation of these proteins by the proteasome (Hetz and Saxena, 2017). Besides the UPS, autophagy is another mechanism that contributes to protein degradation, a process in which autophagosomes and lysosome are involved (reviewed in Kwon and Ciechanover, 2017).
Abnormal protein aggregation is one of the earliest pathological features observed in spinal cord and brain tissue of ALS patients (Blokhuis et al., 2013). Different ALS-related gene mutations in SOD1, TARDBP or FUS lead to protein misfolding and aggregation (Blokhuis et al., 2013). Activation of autophagy delays disease onset, reduces neurological deficits and prolongs survival in SOD1G93A mice (Kim et al., 2013; Staats et al., 2013). As a consequence, proper synthesis, protein folding, and degradation are important processes to maintain efficient proteostasis in motor neurons.
AMPK
AMP activated kinase (AMPK) is a serine/threonine kinase that can be activated in response to energetic stress (Garcia and Shaw, 2017). The activity of AMPK is induced by a high ratio of intracellular AMP to ATP levels. It is a member of the protein kinase A (PKA) family, which comprises enzymes that are dependent for their activity on cellular levels of cyclic AMP (cAMP). Levels of cAMP are affected by the ubiquitin-proteasome system (UPS) to further modulate phosphorylation-dependent downstream signalling. The AMPK/PKA pathway regulates multiple aspects of cell survival (Rinaldi et al., 2015). Motor neurons of ALS patients exhibiting cytoplasmic TDP-43 mislocalization showed an increased level of AMPK activation (Liu et al., 2015). Moreover, AMPK-mediated phosphorylation was significantly upregulated in spinal cords of SOD1G93A mice upon disease onset (Liu et al., 2015). C9ORF72 interacts with Rab1a and the Unc‐51‐like kinase 1 (ULK1) autophagy initiation complex, which might undergo a close regulation by AMPK (Yang et al., 2016; Nwadike et al., 2018). In contrast to the data obtained in SOD1G93A mice, AMPK activity was drastically diminished in spinal cords and brains of presymptomatic and symptomatic TDP-43A315T mice (Perera et al., 2014). Eventually, this could be explained by the milder motor neuron loss in the spinal cord of these mutant TDP-43A315T mice (Perera et al., 2014). In the brain cortex of the TDP-43A315T mice, pronounced neuronal loss and ubiquitin pathology was observed, which could be consistent with a lower AMPK activity in the brain (Wegorzewska et al., 2009; Perera et al., 2014). Furthermore, the TDP-43A315T mice showed a progressive weight gain, increased body fat and adipocyte hypertrophy leading to AMPK inactivation (Stallings et al., 2013; Perera et al., 2014). Although the presence of activated AMPK in SOD1G93A mice at symptom onset argues against a role for AMPK signalling in disease initiation, it could be a determinant of disease progression (Perera et al., 2014). However, the role of elevated AMPK signalling in SOD1G93A mice should be further investigated, as the AMPK-mediated transcription of its downstream targets was unaltered (Perera et al., 2014). Altogether, these data suggest a different effect of AMPK activation in mutant SOD1 compared to mutant TDP-43 transgenic mice (Liu et al., 2015). In addition, the activation of AMPK in mouse spinal cords induced the mislocalization of TDP-43, recapitulating a key neuropathological characteristic of ALS. Inhibition of AMPK activity rescued the mislocalization of TDP-43 and delayed disease progression in TDP-43 transgenic mice (Liu et al., 2015) (Table 1). In addition, cAMP-induced phosphorylation by PKA of the 26S proteasome on Rpn6/PSMD11 at Ser14 enhanced its activity to hydrolyse ubiquitinated proteins, ATP and small peptides (Lokireddy et al., 2015). It also stimulated the degradation of protein aggregates, including SOD1, TDP-43, and FUS in motor neurons, as well as in cultured cortical neurons (Lokireddy et al., 2015).
PERK
Protein kinase R-like endoplasmic reticulum kinase (PERK), also known as eukaryotic translation initiation factor 2-alpha kinase 3, is a transmembrane protein kinase of the pancreatic eukaryotic initiation factor-2alpha kinase (PEK) family located in the ER membrane. ER stress evokes the activation of the UPS which is followed by activation of three major ER transmembrane proteins known as inositol-requiring enzyme 1 (IRE1), PERK, and activating transcription factor 6 (ATF6) (Kametaka et al., 2007). The PERK pathway is the only one modulating protein synthesis as an adaptive response (Bell et al., 2016). Typical ER stress markers were observed in soluble extracts of spinal cord tissue of sporadic ALS patients (Medinas et al., 2017). Modulation of the eIF2α phosphorylation, involved in the PERK signalling, was protective against TDP-43 toxicity in flies and mammalian neurons (Kim et al., 2014). Mutant TDP-43 transgenic flies showed increased eIF2α phosphorylation and an impaired motor function (Kim et al., 2014). In line with this, knockdown of the fly homologue of PERK improved the motor function in these flies (Casci and Pandey, 2015). In addition, treatment of rat neurons and flies with a PERK inhibitor rescued the toxicity induced by TDP-43 overexpression (Kim et al., 2014) (Table 1). Phosphorylation of eIF2α by PERK led to increased expression of ATF4 particularly in spinal cord of SOD1G93A mice (Kikuchi et al., 2006). In addition, SOD1G93A mice showed more ATF4 expression at symptomatic stage compared to earlier stages, which suggests that PERK is activated during the disease progression (Kikuchi et al., 2006). Furthermore, the assembly of stress granules induced by poly-PR encoded from the hexanucleotide repeat sequence in the C9orf72 gene by repeat-associated non-ATG (RAN) translation was dependent on eIF2α phosphorylation (Boeynaems et al., 2017). This is indicative for a potential toxic function of eIF2α phosphorylation by PERK in C9ORF72-related ALS. Considering the fact that TDP-43, FUS, as well as the arginine-containing dipeptide repeat proteins (DPRs) derived from the hexanucleotide repeats in C9ORF72, are all involved in stress granule dynamics, it could be that there is a more general role for PERK signalling in ALS (Smith and Mallucci, 2016). Phosphorylation of eIF2α and subsequent reduction of protein translation increased stress granules formation (Aulas et al., 2017). Moreover, PERK-mediated phosphorylation of eIF2α initiated stress granule formation (Walker et al., 2013) and a disturbance in stress granule dynamics might eventually cause neuronal dysfunction (Chitiprolu et al., 2018; Fernandes et al., 2018). Whether inhibition of PERK could have a beneficial effect for different ALS subtypes needs to be investigated further.
TBK1
Besides its role in inflammation, TBK1 also plays an important role in autophagy through phosphorylation of different autophagy adaptors including p62/SQSTM1, OPTN, nuclear dot protein 52 kDa (NDP52) and Tax1 binding protein 1 (TAX1BP1) (Richter et al., 2016). Disrupted autophagy of mutant SOD1 and TDP-43 seems to be involved in ALS patients carrying mutations in p62/SQSTM1 (Teyssou et al., 2013; Hadano et al., 2016). In addition, TBK1 expression was reduced in SOD1G93A mice and deletion of TBK1 disrupted autophagy reproducing behavioural and locomotor symptoms in mice (Duan et al., 2019). Moreover, ALS-associated mutations in OPTN downregulated autophagy and protein clearance (Markovinovic et al., 2017). The assembly of ubiquitin chains triggered autophagy adaptor recruitment resulting in the activation of TBK1. In this process, TBK1 interacted with autophagy-relevant sites including ubiquitin- and LC3-binding domains of OPTN and p62/SQSTM1, as well as with the SKICH domains of NDP52 and TAX1BP1 (Richter et al., 2016). Impairment of autophagy could be the result of defective recognition of LC3 binding domain by mutant p62/SQSTM1, and could lead to neurodegeneration in vivo (Goode et al., 2016). TBK1 phosphorylates OPTN to promote mitophagy and it phosphorylates p62/SQSTM1 to promote autophagosome maturation (Vinet and Zhedanov, 2011). Mutations in TBK1 decreased the clearance of dysfunctional mitochondria by reducing the binding of TBK1 to OPTN (Weil et al., 2016), suggesting that TBK1 could indeed play a role in ALS pathogenesis. As different mutations in the TBK1 gene cause different diseases, it was proposed that gain-of-function mutations in TBK1 are associated with normal tension glaucoma, while loss-of-function mutations result in ALS/FTD or in herpes encephalitis (Ahmad et al., 2016). So far, there are at least six different small molecules that are known to inhibit TBK1 that showed beneficial results improving diet-induced metabolic dysfunctions in mice (Cruz and Brekken, 2018). Considering the role of metabolic alterations in ALS patients and models (for a review see Vandoorne et al., 2018), targeting TBK1 could also be considered as a therapeutic approach for ALS, although the effect of TBK1 inhibition on autophagy and mitophagy should be first clarified in more detail.
PIKFYVE
FYVE finger-containing phosphoinositide kinase (PIKFYVE) belongs to a large family of evolutionarily-conserved lipid kinases, and inhibiting its activity increased C9orf72 motor neuron survival (Shi et al., 2018). PIKFYVE phosphorylates phosphatidylinositol 3-phosphate (PtdIns3P) to form phosphatidylinositol (3,5)-bisphosphate [PtdIns(3,5)P2]. The FYVE finger domain of PIKFYVE plays a vital role in localizing the protein to the cytosolic leaflet of endosomes through directly binding to membrane PtdIns3P, and is thereby involved in multiple processes of endosome dynamics (reviewed in Hasegawa et al., 2017). Ubiquitous knockout of this protein was embryonic lethal in mice, while one active allele was sufficient for normal embryonic development and survival (Ikonomov et al., 2011). C9ORF72 co-localized with Rab proteins, which are implicated in autophagy and endocytic transport in human spinal motor neurons, indicating a role in Rab-mediated endosomal trafficking in ALS (Farg et al., 2014). In a recent study, induced motor neurons were generated from human iPSCs derived from patients carrying the hexanucleotide repeat expansion in C9orf72. The patient-derived induced motor neurons showed a lower survival after stress compared to control induced motor neurons (Shi et al., 2018). In a subsequent small molecule screen, a PIKFYVE inhibitor, YM201636, significantly increased the survival of C9ORF72 patient-derived motor neurons by converting PtdIns3P into PtdIns(3,5)P2, which enhanced the fusion of lysosomes with both endosomes and autophagosomes under cell stress (Shi et al., 2018) (Table 1). These results were confirmed using a different PIKFYVE inhibitor, apilimod (Shi et al., 2018). In addition, brief treatment of apilimod directly into the hippocampal area rescued neurons from excitotoxic injury in vivo (Shi et al., 2018), and dose-dependently reduced N-methyl d-aspartate (NMDA)-induced neurodegeneration in the hippocampus (Staats et al., 2019). These results suggest that inhibition of PIKFYVE might be a viable therapeutic target in C9-ALS.
Kinases and DNA/RNA metabolism
DNA/RNA metabolism is the process in which nucleic acids are synthesized and degraded (for a review see: Voet et al., 2016). The discovery that mutations in the genes encoding the DNA/RNA binding proteins FUS and TDP-43 can cause ALS highlights the fact that DNA/RNA metabolism is an important player in ALS (Sreedharan et al., 2008; Kwiatkowski et al., 2009; Vance et al., 2009). Different in vitro and in vivo experiments concerning FUS and TDP-43 confirmed that both proteins play a key role in various aspects of DNA/RNA metabolism (reviewed in Lagier-Tourenne et al., 2010). Mutations in the angiogenin gene (ANG) have also been linked to familial and sporadic ALS (Wu et al., 2007). Mechanistically, ANG-induced tRNA fragmentation was proposed to play a key role in the stress granule formation and neuronal loss (Emara et al., 2010; Hanada et al., 2013). Moreover, interfering with DNA damage represents another therapeutic avenue given the post-mitotic nature of motor neurons. In addition to familial ALS caused by mutations in SOD1, where protein misfolding is proposed to lead to a reduced protection against oxidative DNA damage (Barbosa et al., 2010), two recent studies using FUS-ALS patient iPSC-derived motor neurons showed increased levels of DNA damage and an impaired DNA repair response (Naumann et al., 2018; Wang et al., 2018). Moreover, recent studies showed that different kinases are actively involved in DNA repair and tRNA processing in the context of ALS (Hanada et al., 2013; Deng et al., 2014; Kenna et al., 2016; Havali-Shahriari et al., 2017). Below, we focus on the main kinases currently described to play a role in these different processes (Fig. 2C).
DNA-PK
DNA-PK is a serine/threonine protein kinase that belongs to the phosphatidylinositol 3-kinase-related kinase protein family. It is required for the NHEJ pathway of DNA repair, which repairs DNA double-strand breaks (Yin et al., 2017). This kinase could play a role in ALS as one of its substrates is FUS (Rhoads et al., 2018). Upon DNA damage, DNA-PK phosphorylates the N-terminal serine/threonine residues of FUS, resulting in the cytoplasmic translocation of FUS (Deng et al., 2014). This implies that the proper function of FUS relies on phosphorylation. Recent studies showed that the prion-like low complexity domain (PrLD) of FUS in combination with an RGG domain are responsible for condensing FUS from a soluble condition into a gel-like phase, which could be the stepping stone for macroscopic aggregate formation (Patel et al., 2015; Shorter, 2017; Bogaert et al., 2018). Phosphorylation of the PrLD by DNA‐PK inhibited the transition of FUS from a liquid to a solid phase and influenced the formation of FUS aggregates (Deng et al., 2014; Patel et al., 2015; Shang and Huang, 2016). Similar to FUS, TDP-43 also contains a prion-like domain and showed a similar ability to phase separate (Li et al., 2018). It is not a stretch to predict that DNA-PK might also regulates TDP-43 by a similar mechanism.
NEK1
The NEK kinases are a family of serine/threonine kinases that play an important role in the regulation of the disjunction of the centrosome, the assembly of the mitotic spindle and the DNA damage response (Fry et al., 2012). NEK1 is the only member of the NEK kinase family required for the activation of the DNA damage response through ataxia telangiectasia and Rad3-related (ATR) kinase (Melo-Hanchuk et al., 2017). As described above, variants in NEK1 are associated with ALS (Brenner et al., 2016;Kenna et al., 2016), and the C21ORF2 protein can interact with NEK1 during DNA repair (Fang et al., 2015). IPSC-derived motor neurons carrying a heterozygous nonsense mutation in NEK1 showed increased DNA damage and impaired DNA damage response after induction of DNA damage (Higelin et al., 2018). This indicates that loss-of-function of NEK1 can induce deficits in DNA damage repair that might contribute to neurodegeneration in ALS. Therefore, promoting the DNA damage response could be a potential approach to compensate for the loss-of-function of NEK1. As described above, expression of nuclear NEK1 isoform affected chromatin stability and induced nuclear pore complex dispersal (Feige et al., 2006). Recently, different studies suggested that defects in nucleocytoplasmic transport serve as a shared downstream consequence in different ALS subtypes (reviewed in Kim and Taylor, 2017). These include the identification of cytoplasmic protein aggregates of TDP-43, FUS, OPTN, UBQLN2 and recognition of impaired nucleocytoplasmic transport of C9ORF72 (Kim and Taylor, 2017). In line with the fact that NEK1 mutants co-occur with different other ALS genes and as it might play a role as an ALS disease modifier, mutations in NEK1 could eventually affect the nuclear pore complex interfering with nucleocytoplasmic transport of proteins and RNAs (White and Sreedharan, 2016; Nguyen et al., 2018; Shu et al., 2018). Targeting NEK1 might contribute to reverse nucleocytoplasmic defects (Walker and El-Khamisy, 2018). Therefore, clarification of the nuclear function and substrates of NEK1 is crucial to understand its role in ALS.
CLP1
Polyribonucleotide 5′-hydroxyl-kinase Clp1 (CLP1) is a polynucleotide kinase and a component of the tRNA splicing endonuclease (TSEN) complex (Schaffer et al., 2014). As the first discovered mammalian RNA kinase, it phosphorylates the 5′-hydroxyl groups of double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), double-stranded DNA (dsDNA) and double-stranded DNA/RNA hybrids (Hanada et al., 2013). CLP1 knockout mice showed progressive loss of spinal motor neurons, axonal degeneration in peripheral nerves, denervation of neuromuscular junctions resulting in impaired motor function, muscle weakness, paralysis and eventually respiratory failure leading to a reduced life span (Hanada et al., 2013). This indicates that there is a link between CLP1 and the tRNA process which can lead to motor neuron degeneration in mice. Despite being indicative for a link between CLP1 and motor neuron degeneration, more data are required to propose CLP1 as a potential target for treating motor neuron diseases, including ALS.
Modulating kinase activity: a potential therapeutic avenue for ALS?
As receptor tyrosine kinase signalling pathways have been successfully targeted to inhibit proliferation and angiogenesis in the context of cancer therapies (Bhullar et al., 2018) and as kinase deregulation has been shown to play a role in ALS, kinases could potentially play a pivotal role in novel drug developments for ALS. Interestingly, kinase inhibitor drug discovery programs have recently broadened their focus, including an expanded range of kinase targets (Ferguson and Gray, 2018).
The discoveries related to the potential role of different kinases in ALS resulted in testing different compounds, mainly kinase inhibitors, in different preclinical ALS models and in clinical trials (Table 1). Except some major kinases mentioned above, inhibiting some other individual kinases (e.g. apoptosis signal-regulating kinase 1 (ASK1), casein kinase 1δ (CK1δ), Janus kinase 3 (JAK3), extracellular signal-regulated kinase (ERK), Rho-associated, coiled-coil containing protein kinase (ROCK) also showed beneficial results in both ALS mouse models and clinical trials (Trieu et al., 2000; Le Pichon et al., 2013; Song et al., 2013; Salado et al., 2014; Fujisawa et al., 2016; Lingor et al., 2019) (Table 1). As a loss-of-function of different kinases could be involved in ALS, it is also possible that activating specific kinases could be a new approach to treat ALS. Interestingly, the most effective FDA-approved ALS drug, riluzole, was reported to function as an antagonist of protein kinase C (PKC) (Krieger et al., 2003), despite the fact that the therapeutic effect of riluzole is mainly connected to an effect on excitotoxicity (Cheah et al., 2010). Based on the significant results in SOD1G93A rats (Trias et al., 2016), a recent phase II clinical trial (NCT02588677) in ALS patients showed a benefit of masitinib as an add-on therapy to riluzole resulting in a 25% delay in disease progression (Mora et al., 2019). It is important that this positive effect will be confirmed in a phase III study (NCT02588677). Masitinib seems to modulate inflammatory processes by targeting a limited number of kinases at a safe dosage (Mora et al., 2019). This is in line with the idea that a combined kinase-based treatment might be necessary to be effective. Given the widespread involvement of different kinases in various biological processes, a ‘kinase targeting cocktail’ treatment could be an even better option to achieve optimal clinical benefits for ALS patients. Yet, given the widespread physiological roles of kinases in various cell types extensive preclinical testing will be needed to minimize the potential side effects before kinase modulating drugs can be used in clinical practice. Whether this will be possible, given the pleiotropic role of most kinases in different pathways, is an open question and it might be more complicated than anticipated to answer this question.
Conclusions and future perspectives
Some time ago, alterations of several protein kinases and their pathways were reported in SOD1G93A mice by Krieger et al. (2003). Since then, research findings expanded the scope beyond mutant SOD1 and illustrated that there could be a multi-faceted role for kinases in ALS pathogenesis (Fig. 3). Evidence supporting the importance of kinases in ALS stems from many different sources. First, recent genetic studies showed that mutations in kinase-encoding genes can be a risk factor or can even be a cause of ALS. Second, kinases interact with various ALS-related genes/gene products, and affect disease progression in many cases. Even without being a causal gene, different kinases directly regulate ALS-causing gene products and influence their function. Last but not least, kinases actively participate in regulating major pathological processes in ALS. However, it remains unclear how and to what extent these observations are interconnected. Therefore, investigating ALS-related alterations in kinase pathways, as well as their functional consequences is crucial. With the rapidly developing genetic and compound screening platforms, it is expected to be only a matter of time to obtain a clear kinome-wide picture of ALS. This, in combination with the availability of efficient kinase-targeting therapeutics, offers the intriguing opportunity to target multiple ALS-related processes simultaneously, which hopefully could lead to a new treatment strategy for ALS.
Figure 3.
The multifaceted role of kinases in ALS. Overview of the different types of kinases involved in ALS genetics and pathophysiology. These include protein kinases (green), lipid kinases (purple) and nucleotide kinases (pink).
Funding
The work of the authors is supported by VIB, KU Leuven (C1 and ‘Opening the Future’ Fund), the ‘Fund for Scientific Research Flanders’ (FWO-Vlaanderen), the Thierry Latran Foundation, the ‘Association Belge contre les Maladies neuro-Musculaires’ (ABMM), the Muscular Dystrophy Association (MDA), the ALS Liga België (A Cure for ALS) and the ALS Association (ALSA). W.G. is supported by a postdoctoral mandate (PDM fellowship) from KU Leuven (PDM/18/213). T.V. is supported by a strategic basic research PhD fellowship awarded by the FWO (1S60116N). J.S. is supported by a PhD fellowship of the Agency for Innovation by Science and Technology in Flanders (IWT131535).
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- ALS =
amyotrophic lateral sclerosis
- AMPK =
AMP activated kinase
- DNA-PK =
DNA-dependent protein kinase
- ER =
endoplasmic reticulum
- GSK3β =
glycogen synthase kinase 3β
- iPSC =
induced pluripotent stem cell
- JNK =
c-Jun N-terminal kinases
- Miro1 =
mitochondrial Rho GTPase 1
- MOK =
MAPK/MAK/MRK overlapping kinase
- NF-M/H =
neurofilament medium/heavy chain
- p38 MAPK =
p38 mitogen-activated protein kinases
- PERK =
protein kinase R-like endoplasmic reticulum kinase
- PtdIns(3,5)P2 =
phosphatidylinositol (3,5)-bisphosphate
- TDP-43 =
TAR DNA-binding protein 43
References
- Ackerley S, Grierson AJ, Banner S, Perkinton MS, Brownlees J, Byers HL, et al. p38alpha stress-activated protein kinase phosphorylates neurofilaments and is associated with neurofilament pathology in amyotrophic lateral sclerosis. Mol Cell Neurosci 2004; 26: 354–64. [DOI] [PubMed] [Google Scholar]
- Ahmad L, Zhang S-Y, Casanova J-L, Sancho-Shimizu V.. Human TBK1: a Gatekeeper of Neuroinflammation. Trends Mol. Med 2016; 22: 511–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S-W, Jeon GS, Kim M-J, Shon J-H, Kim J-E, Shin J-Y, et al. Neuroprotective effects of JGK-263 in transgenic SOD1-G93A mice of amyotrophic lateral sclerosis. J Neurol Sci 2014; 340: 112–6. [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, Allen C, Counsell C, Farrin A, Dickie B, Kelly J, et al. Lithium in patients with amyotrophic lateral sclerosis (LiCALS): a phase 3 multicentre, randomised, double-blind, placebo-controlled trial. Lancet Neurol 2013; 12: 339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander EJ, Ghanbari Niaki A, Zhang T, Sarkar J, Liu Y, Nirujogi RS, et al. Ubiquilin 2 modulates ALS/FTD-linked FUS–RNA complex dynamics and stress granule formation. Proc Natl Acad Sci U S A 2018; 115: E11485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Logroscino G, Jick SS, Hernán MA.. Incidence and lifetime risk of motor neuron disease in the United Kingdom: a population-based study. Eur J Neurol 2009; 16: 745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio R, Rana A, Walker DW.. Upregulation of the autophagy adaptor p62/SQSTM1 prolongs health and lifespan in middle-aged Drosophila. Cell Rep 2019; 28: 1029–40.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 2006; 351: 602–11. [DOI] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 2017; 130: 927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MR. ALS–dying forward, backward or outward? Nat Rev Neurol 2014; 10: 660.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa LF, Cerqueira FM, Macedo AFA, Garcia CCM, Angeli JPF, Schumacher RI, et al. Increased SOD1 association with chromatin, DNA damage, p53 activation, and apoptosis in a cellular model of SOD1-linked ALS. Biochim Biophys Acta 2010; 1802: 462–71. [DOI] [PubMed] [Google Scholar]
- Beers DR, Henkel JS, Zhao W, Wang J, Huang A, Wen S, et al. Endogenous regulatory T lymphocytes ameliorate amyotrophic lateral sclerosis in mice and correlate with disease progression in patients with amyotrophic lateral sclerosis. Brain 2011; 134: 1293–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendotti C, Atzori C, Piva R, Tortarolo M, Strong MJ, DeBiasi S, et al. Activated p38MAPK is a novel component of the intracellular inclusions found in human amyotrophic lateral sclerosis and mutant SOD1 transgenic mice. J Neuropathol Exp Neurol 2004; 63: 113–9. [DOI] [PubMed] [Google Scholar]
- Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer 2018; 17: 48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis AM, Groen EJN, Koppers M, van den Berg LH, Pasterkamp RJ.. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol 2013; 125: 777–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol 2018; 28: 420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 2017; 65: 1044–55.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Gitler AD.. Phosphorylation leads the way for protein aggregate disassembly. Dev Cell 2018; 45: 279–81. [DOI] [PubMed] [Google Scholar]
- Bogaert E, Boeynaems S, Kato M, Guo L, Caulfield TR, Steyaert J, et al. Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep 2018; 24: 529–37.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady ST, Morfini GA.. Regulation of motor proteins, axonal transport deficits and adult-onset neurodegenerative diseases. Neurobiol Dis 2017; 105: 273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Muller K, Wieland T, Weydt P, Bohm S, Lule D, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain 2016; 139: e28.. [DOI] [PubMed] [Google Scholar]
- Bell MC, Meier E S, Ingram AL, Abisambra JF.. PERK-opathies: an endoplasmic reticulum stress mechanism underlying neurodegeneration. Car 2016; 13: 150–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanari M-L, García-Ayllón M-S, Ciura S, Sáez-Valero J, Kabashi E.. Neuromuscular junction impairment in amyotrophic lateral sclerosis: reassessing the role of acetylcholinesterase. Front Mol Neurosci 2016; 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci I, Pandey UB.. A fruitful endeavor: modeling ALS in the fruit fly. Brain Res 2015; 1607: 47–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Baloh RH, Milbrandt J.. The NIMA-family kinase Nek3 regulates microtubule acetylation in neurons. J Cell Sci 2009; 122: 2274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah BC, Vucic S, Krishnan A, Kiernan MC.. Riluzole, neuroprotection and amyotrophic lateral sclerosis. Curr Med Chem 2010; 17: 1942–59. [DOI] [PubMed] [Google Scholar]
- Chen N-F, Chen W-F, Sung C-S, Lu C-H, Chen C-L, Hung H-C, et al. Contributions of p38 and ERK to the antinociceptive effects of TGF-β1 in chronic constriction injury-induced neuropathic rats. J Headache Pain 2016; 17: 72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Craigen WJ, Riley DJ.. Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle 2009; 8: 257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Deng J, Wang P, Yang M, Chen X, Zhu L, et al. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet 2016; 25: 5059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guan Y, Liu H, Wu X, Yu L, Wang S, et al. Activation of the Wnt/β-catenin signaling pathway is associated with glial proliferation in the adult spinal cord of ALS transgenic mice. Biochem Biophys Res Commun 2012; 420: 397–403. [DOI] [PubMed] [Google Scholar]
- Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 2018; 9: 2794.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L, Salinas PC.. c-Jun N-terminal kinase (JNK) cooperates with Gsk3beta to regulate dishevelled-mediated microtubule stability. BMC Cell Biol 2007; 8: 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015; 347: 1436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Aizer A, Shav-Tal Y, Yanai A, Motro B.. Nek7 kinase accelerates microtubule dynamic instability. Biochim Biophys Acta-Mol Cell Res 2013; 1833: 1104–13. [DOI] [PubMed] [Google Scholar]
- Cruz VH, Brekken RA.. Assessment of TANK-binding kinase 1 as a therapeutic target in cancer. J Cell Commun Signal 2018; 12: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadon-Nachum M, Melamed E, Offen D.. The ‘dying-back’ phenomenon of motor neurons in ALS. J Mol Neurosci 2011; 43: 470–7. [DOI] [PubMed] [Google Scholar]
- DeBerg HA, Blehm BH, Sheung J, Thompson AR, Bookwalter CS, Torabi SF, et al. Motor domain phosphorylation modulates kinesin-1 transport. J Biol Chem 2013; 288: 32612–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Majo M, Topp SD, Smith BN, Nishimura AL, Chen H-J, Gkazi AS, et al. ALS-associated missense and nonsense TBK1 mutations can both cause loss of kinase function. Neurobiol Aging 2018; 71: 266.e1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, et al. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci 2014; 34: 7802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Hafezparast M.. Neurobiology of axonal transport defects in motor neuron diseases: opportunities for translational research? Neurobiol Dis 2017; 105: 283–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewil M, Dela Cruz VF, Van Den Bosch L, Robberecht W.. Inhibition of p38 mitogen activated protein kinase activation and mutant SOD1(G93A)-induced motor neuron death. Neurobiol Dis 2007; 26: 332–41. [DOI] [PubMed] [Google Scholar]
- DiSabato DJ, Quan N, Godbout JP.. Neuroinflammation: the devil is in the details. J Neurochem 2016; 139: 136–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J 2010; 29: 2841–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Wei Y, Liu L, Wang Y, Khairova R, Blumenthal R, et al. A kinesin signaling complex mediates the ability of GSK-3 to affect mood-associated behaviors. Proc Natl Acad Sci U S A 2010; 107: 11573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Guo M, Yi L, Zhang J, Bi Y, Liu Y, et al. Deletion of Tbk1 disrupts autophagy and reproduces behavioral and locomotor symptoms of FTD‐ALS in mice. Aging 2019; 11: 2457–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 2010; 285: 10959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA. Determinants of molecular motor directionality. Nat Cell Biol 1999; 1: E163–7. [DOI] [PubMed] [Google Scholar]
- Engmann O, Giese KP.. Crosstalk between Cdk5 and GSK3beta: implications for Alzheimer’s disease. Front Mol Neurosci 2009; 2: 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K, Hommyo A, Yonemitsu R, Abe S.. ErbB4 signals Neuregulin1-stimulated cell proliferation and c-fos gene expression through phosphorylation of serum response factor by mitogen-activated protein kinase cascade. Mol Cell Biochem 2010; 339: 119–25. [DOI] [PubMed] [Google Scholar]
- Fang X, Lin H, Wang X, Zuo Q, Qin J, Zhang P.. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim Biophys Sin (Shanghai) 2015; 47: 834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, Levina V, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 2014; 23: 3579–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol 2011; 68: 1440–6. [DOI] [PubMed] [Google Scholar]
- Feige E, Shalom O, Tsuriel S, Yissachar N, Motro B.. Nek1 shares structural and functional similarities with NIMA kinase. Biochim Biophys Acta-Mol Cell Res 2006; 1763: 272–81. [DOI] [PubMed] [Google Scholar]
- Feng H-L, Leng Y, Ma C-H, Zhang J, Ren M, Chuang D-M.. Combined lithium and valproate treatment delays disease onset, reduces neurological deficits and prolongs survival in an amyotrophic lateral sclerosis mouse model. Neuroscience 2008; 155: 567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson FM, Gray NS.. Kinase inhibitors: the road ahead. Nat Rev Drug Discov 2018; 17: 353–77. [DOI] [PubMed] [Google Scholar]
- Fernandes N, Eshleman N, Buchan JR.. Stress granules and ALS: a case of causation or correlation? Adv Neurobiol 2018; 173–212. [DOI] [PubMed] [Google Scholar]
- Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 2008; 105: 2052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci 2015; 18: 631–6. [DOI] [PubMed] [Google Scholar]
- Fry AM, O’Regan L, Sabir SR, Bayliss R.. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci 2012; 125: 4423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Takahashi M, Tsukamoto Y, Yamaguchi N, Nakoji M, Endo M, et al. The ASK1-specific inhibitors K811 and K812 prolong survival in a mouse model of amyotrophic lateral sclerosis. Hum Mol Genet 2016; 25: 245–53. [DOI] [PubMed] [Google Scholar]
- Fumoto K, Hoogenraad CC, Kikuchi A.. GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome. EMBO J 2006; 25: 5670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FJ, Hebbar S, Gao XA, Alexander M, Pandey JP, Walla MD, et al. GSK-3β phosphorylation of cytoplasmic dynein reduces ndel1 binding to intermediate chains and alters dynein motility. Traffic 2015; 16: 941–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Shaw RJ.. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell 2017; 66: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs KL, Kalmar B, Rhymes ER, Fellows AD, Ahmed M, Whiting P, et al. Inhibiting p38 MAPK alpha rescues axonal retrograde transport defects in a mouse model of ALS. Cell Death Dis 2018; 9: 596.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol 2008; 63: 535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Shorter J.. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion 2011; 5: 179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmit Y, Galea MP, Wise G, Bartlett PF, Turnley AM.. Axonal regeneration and lack of astrocytic gliosis in EphA4-deficient mice. J Neurosci 2004; 24: 10064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein O, Kedmi M, Gana-Weisz M, Twito S, Nefussy B, Vainer B, et al. Rare homozygosity in amyotrophic lateral sclerosis suggests the contribution of recessive variants to disease genetics. J Neurol Sci 2019; 402: 62–8. [DOI] [PubMed] [Google Scholar]
- González-Fernández C, Gonzalez P, Andres-Benito P, Ferrer I, Rodríguez FJ.. Wnt signaling alterations in the human spinal cord of amyotrophic lateral sclerosis cases: spotlight on Fz2 and Wnt5a. Mol Neurobiol 2019; 56: 6777–91. [DOI] [PubMed] [Google Scholar]
- Goode A, Butler K, Long J, Cavey J, Scott D, Shaw B, et al. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy 2016; 12: 1094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Zhao Q, Benyamin B, Garton F, He J, Leo PJ, et al. Whole-exome sequencing in amyotrophic lateral sclerosis suggests NEK1 is a risk gene in Chinese. Genome Med 2017; 9: 97.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 2005; 47: 379–93. [DOI] [PubMed] [Google Scholar]
- Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 2017; 8: 861.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Stoklund Dittlau K, Van Den Bosch L.. Axonal transport defects and neurodegeneration: Molecular mechanisms and therapeutic implications. Semin Cell Dev Biol 2019. doi: 10.1016/j.semcdb.2019.07.010. [DOI] [PubMed] [Google Scholar]
- Gurney M, Pu H, Chiu A.. Motor neuron degeneration in mice that express a human Cu, Zn superoxide dismutase mutation. Science 1994; 264: 1772–5. [DOI] [PubMed] [Google Scholar]
- Hadano S, Mitsui S, Pan L, Otomo A, Kubo M, Sato K, et al. Functional links between SQSTM1 and ALS2 in the pathogenesis of ALS: cumulative impact on the protection against mutant SOD1-mediated motor dysfunction in mice. Hum Mol Genet 2016; 25: 3321–40. [DOI] [PubMed] [Google Scholar]
- Hammond TR, Marsh SE, Stevens B.. Immune signaling in neurodegeneration. Immunity 2019; 50: 955–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013; 495: 474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol 2008; 64: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Strunk BS, Weisman LS.. PI5P and PI(3,5)P2: minor, but essential phosphoinositides. Cell Struct Funct 2017; 42: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havali-Shahriari Z, Weinfeld M, Glover J.. Characterization of DNA substrate binding to the phosphatase domain of the DNA repair enzyme polynucleotide kinase/phosphatase. Biochemistry 2017; 56: 1737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergesheimer RC, Chami AA, de Assis DR, Vourc’h P, Andres CR, Corcia P, et al. The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight? Brain 2019; 142: 1176–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M, Ciancanelli M, Ou Y-H, Lorenzo L, Klaudel-Dreszler M, Pauwels E, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med 2012; 209: 1567–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Mollereau B.. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci 2014; 15: 233–49. [DOI] [PubMed] [Google Scholar]
- Hetz C, Saxena S.. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol 2017; 13: 477–91. [DOI] [PubMed] [Google Scholar]
- Hida T, Nakamura F, Usui H, Takeuchi K, Yamashita N, Goshima Y.. Semaphorin3A-induced axonal transport mediated through phosphorylation of Axin-1 by GSK3β. Brain Res 2015; 1598: 46–56. [DOI] [PubMed] [Google Scholar]
- Higelin J, Catanese A, Semelink-Sedlacek LL, Oeztuerk S, Lutz A-K, Bausinger J, et al. NEK1 loss-of-function mutation induces DNA damage accumulation in ALS patient-derived motoneurons. Stem Cell Res 2018; 30: 150–62. [DOI] [PubMed] [Google Scholar]
- Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM.. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol 2007; 17: 1313–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J-H, Zhang H, Wagey R, Krieger C, Pelech SL.. Protein kinase and protein phosphatase expression in amyotrophic lateral sclerosis spinal cord. J Neurochem 2003; 85: 432–42. [DOI] [PubMed] [Google Scholar]
- Huang Y, Huang Q, Su H, Mai X, Feng E, Cao Z, et al. TAR DNA-binding protein 43 inhibits inflammatory response and protects chondrocyte function by modulating RACK1 expression in osteoarthritis. Biomed Pharmacother 2017; 85: 362–71. [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang Z, Qin F, Tang W, Liu D, Wu P, et al. The mechanism of TDP‐43 gene expression on inflammatory factors and the JNK and p38 MAPK signalling pathways in ischaemic hypoxic stress dependence. Int Wound J 2019; 16: 724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries F, Yang S, Wang B, Moynagh PN.. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ 2015; 22: 225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, Lane HA.. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–54. [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin J-P, Rappolee D, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development. J Biol Chem 2011; 286: 13404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Izumi Y, Watanabe A, Tsukita K, Woltjen K, Yamamoto T, et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci Transl Med 2017; 9: eaaf3962.. [DOI] [PubMed] [Google Scholar]
- Ito Y, Ofengeim D, Najafov A, Das S, Saberi S, Li Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 2016; 353: 603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Moriyama K, Burgos PV, Eisenberg E, Greene LE, Mattera R, et al. Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. MBoC 2007; 18: 2991–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata R, Ishigaki S, Katsuno M, Kawai K, Sone J, Huang Z, et al. c-Abl inhibition delays motor neuron degeneration in the G93A mouse, an animal model of amyotrophic lateral sclerosis. PLoS One 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PTC, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet 2016; 48: 1037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi H, Almer G, Yamashita S, Guegan C, Nagai M, Xu Z, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci U S A 2006; 103: 6025–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim T-Y, Cho K-S, Kim HN, Koh J-Y.. Autophagy activation and neuroprotection by progesterone in the G93A-SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurobiol. Dis 2013; 59: 80–5. [DOI] [PubMed] [Google Scholar]
- Kim H-J, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 2014; 46: 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Taylor JP.. Lost in transportation: nucleocytoplasmic transport defects in ALS and other neurodegenerative diseases. Neuron 2017; 96: 285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, Holzbaur E.. Stress-induced CDK5 activation disrupts axonal transport via Lis1/Ndel1/Dynein. Cell Rep 2015; 12: 462–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, Tokito M, Holzbaur E.. CDK5-dependent activation of dynein in the axon initial segment regulates polarized cargo transport in neurons. Traffic 2017; 18: 808–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S-H, Kim Y, Kim HY, Hwang S, Lee CH, Kim SH.. Inhibition of glycogen synthase kinase-3 suppresses the onset of symptoms and disease progression of G93A-SOD1 mouse model of ALS. Exp Neurol 2007; 205: 336–46. [DOI] [PubMed] [Google Scholar]
- Koskimäki J, Zhang D, Li Y, Saadat L, Moore T, Lightle R, et al. Transcriptome clarifies mechanisms of lesion genesis versus progression in models of Ccm3 cerebral cavernous malformations. Acta Neuropathol Commun 2019; 7: 132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter N, Pal A, Lojewski X, Corcia P, Naujock M, Reinhardt P, et al. Age-dependent neurodegeneration and organelle transport deficiencies in mutant TDP43 patient-derived neurons are independent of TDP43 aggregation. Neurobiol Dis 2018; 115: 167–81. [DOI] [PubMed] [Google Scholar]
- Krieger C, Hu JH, Pelech S.. Aberrant protein kinases and phosphoproteins in amyotrophic lateral sclerosis. Trends Pharmacol Sci 2003; 24: 535–41. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco D. A, Leclerc A L, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009; 323: 1205–8. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Ciechanover A.. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci 2017; 42: 873–86. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW.. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet 2010; 19: R46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasiene J, Komine O, Fujimori-Tonou N, Powers B, Endo F, Watanabe S, et al. Neuregulin 1 confers neuroprotection in SOD1-linked amyotrophic lateral sclerosis mice via restoration of C-boutons of spinal motor neurons. Acta Neuropathol Commun 2016; 4: 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal-Lasarte MM, Franco JM, Labrador-Garrido A, Pozo D, Roodveldt C.. Extracellular TDP-43 aggregates target MAPK/MAK/MRK overlapping kinase (MOK) and trigger caspase-3/IL-18 signaling in microglia. FASEB J 2017; 31: 2797–816. [DOI] [PubMed] [Google Scholar]
- Le Pichon CE, Dominguez SL, Solanoy H, Ngu H, Lewin-Koh N, Chen M, et al. EGFR inhibitor erlotinib delays disease progression but does not extend survival in the SOD1 mouse model of ALS. PLoS One 2013; 8: e62342.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pichon CE, Meilandt WJ, Dominguez S, Solanoy H, Lin H, Ngu H, et al. Loss of dual leucine zipper kinase signaling is protective in animal models of neurodegenerative disease. Sci Transl Med 2017; 9: eaag0394.. [DOI] [PubMed] [Google Scholar]
- Li H-R, Chiang W-C, Chou P-C, Wang W-J, Huang J.. TAR DNA-binding protein 43 (TDP-43) liquid-liquid phase separation is mediated by just a few aromatic residues. J Biol Chem 2018; 293: 6090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xu D, Wang Y, Zhou Z, Liu J, Hu S, et al. Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation. Autophagy 2018; 14: 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KK, Jackson M, Alkam D, Liu D, Allaire N, Sun C, et al. Antisense-mediated reduction of EphA4 in the adult CNS does not improve the function of mice with amyotrophic lateral sclerosis. Neurobiol. Dis 2018; 114: 174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW.. Converging mechanisms in als and FTD: disrupted RNA and protein homeostasis. Neuron 2013; 79: 416–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingor P, Weber M, Camu W, Friede T, Hilgers R, Leha A, et al. ROCK-ALS: protocol for a randomized, placebo-controlled, double-blind phase IIa trial of safety, tolerability and efficacy of the rho kinase (ROCK) inhibitor fasudil in amyotrophic lateral sclerosis. Front Neurol 2019; 10: 293.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-J, Ju T-C, Chen H-M, Jang Y-S, Lee L-M, Lai H-L, et al. Activation of AMP-activated protein kinase α1 mediates mislocalization of TDP-43 in amyotrophic lateral sclerosis. Hum Mol Genet 2015; 24: 787–801. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang F.. Role of neuroinflammation in amyotrophic lateral sclerosis: cellular mechanisms and therapeutic implications. Front Immunol 2017; 8: 1005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, Kukushkin NV, Goldberg AL.. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A 2015; 112: E7176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López E, Casasnovas C, Giménez J, Santamaría R, Terrazas JM, Volpini V.. Identification of two novel KIF5A mutations in hereditary spastic paraplegia associated with mild peripheral neuropathy. J Neurol Sci 2015; 358: 422–7. [DOI] [PubMed] [Google Scholar]
- López-Doménech G, Covill-Cooke C, Ivankovic D, Halff EF, Sheehan DF, Norkett R, et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J 2018; 37: 321–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Erauskin J, Tadokoro T, Baughn MW, Myers B, McAlonis-Downes M, Chillon-Marinas C, et al. ALS/FTD-linked mutation in FUS suppresses intra-axonal protein synthesis and drives disease without nuclear loss-of-function of FUS. Neuron 2018; 100: 816–30.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Font I, Sogorb-Esteve A, Javier-Torrent M, Brinkmalm G, Herrando-Grabulosa M, García-Lareu B, et al. Decreased circulating ErbB4 ectodomain fragments as a read-out of impaired signaling function in amyotrophic lateral sclerosis. Neurobiol Dis 2019; 124: 428–38. [DOI] [PubMed] [Google Scholar]
- MacAskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, et al. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron 2009; 61: 541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Rademakers R, Neumann M.. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 2010; 9: 995–1007. [DOI] [PubMed] [Google Scholar]
- Mancuso R, Martínez-Muriana A, Leiva T, Gregorio D, Ariza L, Morell M, et al. Neuregulin-1 promotes functional improvement by enhancing collateral sprouting in SOD1G93A ALS mice and after partial muscle denervation. Neurobiol Dis 2016; 95: 168–78. [DOI] [PubMed] [Google Scholar]
- Markovinovic A, Cimbro R, Ljutic T, Kriz J, Rogelj B, Munitic I.. Optineurin in amyotrophic lateral sclerosis: multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog Neurobiol 2017; 154: 1–20. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Shimogori T, Hattori N, Nukina N.. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet 2015; 24: 4429–42. [DOI] [PubMed] [Google Scholar]
- McCauley ME, Baloh RH.. Inflammation in ALS/FTD pathogenesis. Acta Neuropathol 2019; 137: 715–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medinas DB, González JV, Falcon P, Hetz C.. Fine-tuning ER stress signal transducers to treat amyotrophic lateral sclerosis. Front Mol Neurosci 2017; 10: 216.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Hanchuk TD, Slepicka PF, Meirelles GV, Basei FL, Lovato DV, Granato DC, et al. NEK1 kinase domain structure and its dynamic protein interactome after exposure to. Sci Rep 2017; 7: 5445.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mòdol-Caballero G, Santos D, Navarro X, Herrando-Grabulosa M.. Neuregulin 1 reduces motoneuron cell death and promotes neurite growth in an in vitro model of motoneuron degeneration. Front Cell Neurosci 2017; 11: 431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, Holzbaur E.. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A 2016; 113: E3349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora JS, Genge A, Chio A, Estol CJ, Chaverri D, Hernández M, et al. Masitinib as an add-on therapy to riluzole in patients with amyotrophic lateral sclerosis: a randomized clinical trial. Amyotroph Lateral Scler Frontotemporal Degener 2019; 1–10. [DOI] [PubMed] [Google Scholar]
- Morano M, Ronchi G, Nicolò V, Fornasari BE, Crosio A, Perroteau I, et al. Modulation of the Neuregulin 1/ErbB system after skeletal muscle denervation and reinnervation. Sci Rep 2018; 8: 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello G, Spampinato AG, Cavallaro S.. Neuroinflammation and ALS: transcriptomic insights into molecular disease mechanisms and therapeutic targets. Mediators Inflamm 2017; 2017: 7070469.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfini GA, Bosco DA, Brown H, Gatto R, Kaminska A, Song Y, et al. Inhibition of fast axonal transport by pathogenic SOD1 involves activation of p38 MAP kinase. PLoS One 2013; 8: e65235.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, et al. Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun 2018; 9: 335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314: 130–3. [DOI] [PubMed] [Google Scholar]
- Nguyen MD, Larivière RC, Julien JP.. Deregulation of Cdk5 in a mouse model of ALS: toxicity alleviated by perikaryal neurofilament inclusions. Neuron 2001; 30: 135–47. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Van Broeckhoven C, van der Zee J.. ALS genes in the genomic era and their implications for FTD. Trends Genet 2018; 34: 404–23. [DOI] [PubMed] [Google Scholar]
- Nguyen HP, Van Mossevelde S, Dillen L, De Bleecker JL, Moisse M, Van Damme P, et al. NEK1 genetic variability in a Belgian cohort of ALS and ALS-FTD patients. Neurobiol Aging 2018; 61: 255.e1–7. [DOI] [PubMed] [Google Scholar]
- Nie S-D, Li X, Tang C-E, Min F-Y, Shi X-J, Wu L-Y, et al. High glucose forces a positive feedback loop connecting ErbB4 expression and mTOR/S6K pathway to aggravate the formation of tau hyperphosphorylation in differentiated SH-SY5Y cells. Neurobiol Aging 2018; 67: 171–80. [DOI] [PubMed] [Google Scholar]
- Nwadike C, Williamson LE, Gallagher LE, Guan J-L, Chan E.. AMPK inhibits ULK1-dependent autophagosome formation and lysosomal acidification via distinct mechanisms. Mol Cell Biol 2018; 38: e00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes JA, Davies MC, Collins MO.. TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain 2017; 10: 5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson B, Gendron TF, Staff NP.. Amyotrophic lateral sclerosis: an update for 2018. Mayo Clin Proc 2018; 93: 1617–28. [DOI] [PubMed] [Google Scholar]
- Paatero I, Veikkolainen V, Mäenpää M, Schmelzer E, Belting HG, Pelliniemi LJ, et al. ErbB4 tyrosine kinase inhibition impairs neuromuscular development in zebrafish embryos. Mol Biol Cell 2019; 30: 209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal A, Glaß H, Naumann M, Kreiter N, Japtok J, Sczech R, et al. High content organelle trafficking enables disease state profiling as powerful tool for disease modelling. Sci Data 2018; 5: 180241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JP, Smith DS.. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci 2011; 31: 17207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015; 162: 1066–77. [DOI] [PubMed] [Google Scholar]
- Perera ND, Sheean RK, Scott JW, Kemp BE, Horne MK, Turner BJ.. Mutant TDP-43 deregulates AMPK activation by PP2A in ALS models. PLoS One 2014; 9: e90449.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C, Bieniek KF, Finch N, van de Vorst M, Baker M, Perkersen R, et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol 2015; 130: 77–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE.. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 2009; 78: 959–91. [DOI] [PubMed] [Google Scholar]
- Prior R, Van Helleputte L, Benoy V, Van Den Bosch L.. Defective axonal transport: a common pathological mechanism in inherited and acquired peripheral neuropathies. Neurobiol Dis 2017; 105: 300–20. [DOI] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Choi SH, Sathyamurthy A, Bose R, Banjade S, et al. Mechanism of activation and inhibition of the HER4/ErbB4 Kinase. Structure 2008; 16: 460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N, Pandey UB.. Autophagy dysregulation in ALS: when protein aggregates get out of hand. Front Mol Neurosci 2017; 10: 263.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin CA, Sun Q, Gamblin TC.. Tau phosphorylation by GSK-3beta promotes tangle-like filament morphology. Mol Neurodegener 2007; 2: 12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M, Zhang J, Fabbro D, Schiöth HB.. Advances in kinase targeting: current clinical use and clinical trials. Trends Pharmacol. Sci 2014; 35: 604–20. [DOI] [PubMed] [Google Scholar]
- Ratti A, Buratti E.. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem 2016; 138: 95–111. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, Carter AP.. The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol 2018; 19: 382–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chiò A, Traynor BJ.. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 2014; 17: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads SN, Monahan ZT, Yee DS, Leung AY, Newcombe CG, O’Meally RN, et al. The prionlike domain of FUS is multiphosphorylated following DNA damage without altering nuclear localization. MBoC 2018; 29: 1786–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A 2016; 113: 4039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L, Sepe M, Donne RD, Feliciello A.. A dynamic interface between ubiquitylation and cAMP signaling. Front Pharmacol 2015; 6: 177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993; 362: 59–62. [DOI] [PubMed] [Google Scholar]
- Rué L, Oeckl P, Timmers M, Lenaerts A, van der Vos J, Smolders S, et al. Reduction of ephrin-A5 aggravates disease progression in amyotrophic lateral sclerosis. Acta Neuropathol Commun 2019; 7: 114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo GJ, Louie K, Wellington A, Macleod GT, Hu F, Panchumarthi S, et al. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J Neurosci 2009; 29: 5443–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Scardigli R, La Regina F, Murray ME, Romano N, Dickson DW, et al. Increased cytoplasmic TDP-43 reduces global protein synthesis by interacting with RACK1 on polyribosomes. Hum Mol Genet 2017; 26: 1407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salado IG, Redondo M, Bello ML, Perez C, Liachko NF, Kraemer BC, et al. Protein kinase CK-1 inhibitors as new potential drugs for amyotrophic lateral sclerosis. J Med Chem 2014; 57: 2755–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sama RRK, Fallini C, Gatto R, McKeon JE, Song Y, Rotunno MS, et al. ALS-linked FUS exerts a gain of toxic function involving aberrant p38 MAPK activation. Sci Rep 2017; 7: 115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Hollenbeck PJ.. The axonal transport of mitochondria. J Cell Sci 2012; 125: 2095–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AE, Eggens VRC, Caglayan AO, Reuter MS, Scott E, Coufal NG, et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014; 157: 651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonaert L, Rué L, Roucourt B, Timmers M, Little S, Chávez-Gutiérrez L, et al. Identification and characterization of Nanobodies targeting the EphA4 receptor. J Biol Chem 2017; 292: 11452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Huang EJ.. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res 2016; 1647: 65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea TB, Yabe JT, Ortiz D, Pimenta A, Loomis P, Goldman RD, et al. Cdk5 regulates axonal transport and phosphorylation of neurofilaments in cultured neurons. J Cell Sci 2004; 117: 933–41. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 2018; 24: 313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J. Liquidizing FUS via prion-like domain phosphorylation. EMBO J 2017; 36: 2925–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S, Lei X, Liu F, Cui B, Liu Q, Ding Q, et al. Mutation screening of NEK1 in Chinese ALS patients. Neurobiol Aging 2018; 71: 267.e1–4. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Al-Chalabi A.. Amyotrophic lateral sclerosis as a complex genetic disease. Biochim Biophys Acta-Mol Basis Dis 2006; 1762: 973–85. [DOI] [PubMed] [Google Scholar]
- Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci 2016; 19: 1610–8. [DOI] [PubMed] [Google Scholar]
- Smith HL, Mallucci GR.. The unfolded protein response: mechanisms and therapy of neurodegeneration. Brain 2016; 139: 2113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Chiang P, Wang J, Ravits J, Loeb JA.. Aberrant neuregulin 1 signaling in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 2012; 71: 104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Nagy M, Ni W, Tyagi NK, Fenton WA, López-Giráldez F, et al. Molecular chaperone Hsp110 rescues a vesicle transport defect produced by an ALS-associated mutant SOD1 protein in squid axoplasm. Proc Natl Acad Sci U S A 2013; 110: 5428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science (80-.) 2008; 319: 1668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats KA, Hernandez S, Schönefeldt S, Bento-Abreu A, Dooley J, Van Damme P, et al. Rapamycin increases survival in ALS mice lacking mature lymphocytes. Mol Neurodegener 2013; 8: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats KA, Seah C, Sahimi A, Wang Y, Koutsodendris N, Lin S, et al. Small molecule inhibition of PIKFYVE kinase rescues gain- and loss-of-function C9ORF72 ALS/FTD disease processes in vivo. bioRxiv 2019:685800.
- Stallings NR, Puttaparthi K, Dowling KJ, Luther CM, Burns DK, Davis K, et al. TDP-43, an ALS linked protein, regulates fat deposition and glucose homeostasis. PLoS One 2013; 8: e71793.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica R, Paillusson S, Gomez-Suaga P, Mitchell JC, Lau DHW, Gray EH, et al. ALS/FTD-associated FUS activates GSK-3β to disrupt the VAPB–PTPIP 51 interaction and ER—mitochondria associations. EMBO Rep 2016; 17: 1326–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica RD, Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau K-F, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 2014; 5: 3996.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Duan Y, Qin C, Li J-C, Duan G, Deng X, et al. Distinct multilevel misregulations of Parkin and PINK1 revealed in cell and animal models of TDP-43 proteinopathy. Cell Death Dis 2018; 9: 953.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Korhonen A, Vaparanta K, Anckar J, Halkilahti K, Salah Z, et al. Protein inhibitor of activated STAT3 (PIAS3) protein promotes SUMOylation and nuclear sequestration of the intracellular domain of ErbB4 protein. J Biol Chem 2012; 287: 23216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Matsuoka M.. The JNK/c-Jun signaling axis contributes to the TDP-43-induced cell death. Mol Cell Biochem 2013; 372: 241–8. [DOI] [PubMed] [Google Scholar]
- Swinnen B, Robberecht W.. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 2014; 10: 661–70. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Fukuda Y, Yoshimura J, Toyoda A, Kurppa K, Moritoyo H, et al. ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am J Hum Genet 2013; 93: 900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Cleveland DW.. Decoding ALS: from genes to mechanism. Nature 2016; 539: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssou E, Takeda T, Lebon V, Boillée S, Doukouré B, Bataillon G, et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol 2013; 125: 511–22. [DOI] [PubMed] [Google Scholar]
- Thompson M, Lauderdale S, Webster MJ, Chong VZ, McClintock B, Saunders R, et al. Widespread expression of ErbB2, ErbB3 and ErbB4 in non-human primate brain. Brain Res 2007; 1139: 95–109. [DOI] [PubMed] [Google Scholar]
- Thonhoff JR, Beers DR, Zhao W, Pleitez M, Simpson EP, Berry JD, et al. Expanded autologous regulatory T-lymphocyte infusions in ALS: a phase I, first-in-human study. Neurol Neuroimmunol Neuroinflamm 2018; 5: e465.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortarolo M, Veglianese P, Calvaresi N, Botturi A, Rossi C, Giorgini A, et al. Persistent activation of p38 mitogen-activated protein kinase in a mouse model of familial amyotrophic lateral sclerosis correlates with disease progression. Mol Cell Neurosci 2003; 23: 180–92. [DOI] [PubMed] [Google Scholar]
- Traynis I, De Moraes CG, Raza AS, Liebmann JM, Ritch R, Hood DC.. Prevalence and nature of early glaucomatous defects in the central 10° of the visual field. JAMA Ophthalmol 2014; 132: 291.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trias E, Ibarburu S, Barreto-Núñez R, Babdor J, Maciel TT, Guillo M, et al. Post-paralysis tyrosine kinase inhibition with masitinib abrogates neuroinflammation and slows disease progression in inherited amyotrophic lateral sclerosis. J Neuroinflamm 2016; 13: 177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu VN, Liu R, Liu XP, Uckun FM.. A specific inhibitor of janus kinase-3 increases survival in a transgenic mouse model of amyotrophic lateral sclerosis. Biochem Biophys Res Commun 2000; 267: 22–5. [DOI] [PubMed] [Google Scholar]
- Trinidad JC, Fischbach GD, Cohen JB.. The Agrin/MuSK signaling pathway is spatially segregated from the neuregulin/ErbB receptor signaling pathway at the neuromuscular junction. J Neurosci 2000; 20: 8762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripolszki K, Gampawar P, Schmidt H, Nagy ZF, Nagy D, Klivényi P, et al. Comprehensive genetic analysis of a Hungarian amyotrophic lateral sclerosis cohort. Front Genet 2019; 10: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya P, Birkenmeier EH, Birkenmeier CS, Barker JE.. Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc Natl Acad Sci U S A 2000; 97: 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009; 323: 1208–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Goris A, Race V, Hersmus N, Dubois B, Van Den Bosch L, et al. The occurrence of mutations in FUS in a Belgian cohort of patients with familial ALS. Eur J Neurol 2010; 17: 754–6. [DOI] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Van Mossevelde S, Perrone F, Dillen L, Heeman B, et al. TBK1 mutation spectrum in an extended european patient cohort with frontotemporal dementia and amyotrophic lateral sclerosis. Hum Mutat 2017; 38: 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandoorne T, De Bock K, Van Den Bosch L.. Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathol 2018; 135: 489–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandoorne T, Veys K, Guo W, Sicart A, Vints K, Swijsen A, et al. Differentiation but not ALS mutations in FUS rewires motor neuron metabolism. Nat Commun 2019; 10: 4147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk RPA, Jones AR, Sproviero W, Shatunov A, Shaw PJ, Leigh PN, et al. Meta-analysis of pharmacogenetic interactions in amyotrophic lateral sclerosis clinical trials. Neurology 2017; 89: 1915–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med 2012; 18: 1418–22. [DOI] [PubMed] [Google Scholar]
- Venkova K, Christov A, Kamaluddin Z, Kobalka P, Siddiqui S, Hensley K.. Semaphorin 3A signaling through neuropilin-1 is an early trigger for distal axonopathy in the SOD1 G93A mouse model of amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 2014; 73: 702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinet L, Zhedanov A.. A ‘missing’ family of classical orthogonal polynomials. J Phys A: Math Theor 2011; 44: 085201. [Google Scholar]
- Vlug AS, Teuling E, Haasdijk ED, French P, Hoogenraad CC, Jaarsma D.. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur J Neurosci 2005; 22: 1881–94. [DOI] [PubMed] [Google Scholar]
- Voet D, Voet JG, Pratt CW.. Fundamentals of biochemistry: Life at the molecular level. New York: John Wiley & Sons; 2016. [Google Scholar]
- Vohnoutka RB, Boumil EF, Liu Y, Uchida A, Pant HC, Shea TB.. Influence of a GSK3β phosphorylation site within the proximal C-terminus of Neurofilament-H on neurofilament dynamics. Biol Open 2017; 6: 1516–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk AE, Weishaupt JH, Andersen PM, Ludolph AC, Kubisch C.. Current knowledge and recent insights into the genetic basis of amyotrophic lateral sclerosis. Medgen 2018; 30: 252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C, El-Khamisy SF.. Perturbed autophagy and DNA repair converge to promote neurodegeneration in amyotrophic lateral sclerosis and dementia. Brain 2018; 141: 1247–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Soo KY, Sundaramoorthy V, Parakh S, Ma Y, Farg MA, et al. ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 2013; 8: e81170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Feng J, Yu J, Chen G.. FKBP12 mediates necroptosis by initiating RIPK1–RIPK3–MLKL signal transduction in response to TNF receptor 1 ligation. J Cell Sci 2019; 132: jcs227777.. [DOI] [PubMed] [Google Scholar]
- Wang H, Guo W, Mitra J, Hegde PM, Vandoorne T, Eckelmann BJ, et al. Mutant FUS causes DNA ligation defects to inhibit oxidative damage repair in Amyotrophic Lateral Sclerosis. Nat Commun 2018; 9: 3683.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Pan J, Chen S-D.. Kinases and kinase signaling pathways: potential therapeutic targets in Parkinson’s disease. Prog Neurobiol 2012; 98: 207–21. [DOI] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, et al. PINK1 and Parkin target miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 2011; 147: 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang HPS.. Transition and autophagic degradation of proteins in development and pathogenesis. Trends Cell Biol 2019; 29: 417–27. [DOI] [PubMed] [Google Scholar]
- Webster CP, Smith EF, Shaw PJ, De Vos KJ.. Protein homeostasis in amyotrophic lateral sclerosis: therapeutic opportunities? Front Mol Neurosci 2017; 10: 123.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH.. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A 2009; 106: 18809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil R, Laplantine E, Génin P.. Regulation of TBK1 activity by Optineurin contributes to cell cycle-dependent expression of the interferon pathway. Cytokine Growth Factor Rev 2016; 29: 23–33. [DOI] [PubMed] [Google Scholar]
- Wenqiang C, Lonskaya I, Hebron ML, Ibrahim Z, Olszewski RT, Neale JH, et al. Parkin-mediated reduction of nuclear and soluble TDP-43 reverses behavioral decline in symptomatic mice. Hum Mol Genet 2014; 23: 4960–9. [DOI] [PubMed] [Google Scholar]
- White MA, Sreedharan J. Amyotrophic lateral sclerosis: recent genetic highlights. Curr Opin Neurol 2016; 29: 557–64. [DOI] [PubMed] [Google Scholar]
- Wloga D, Joachimiak E, Fabczak H.. Tubulin post-translational modifications and microtubule dynamics. Int J Mol Sci 2017; 18: 2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, De SK, Kulinich A, Salem AF, Koeppen J, Wang R, et al. Potent and selective EphA4 agonists for the treatment of ALS. Cell Chem Biol 2017; 24: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Yu W, Kishikawa H, Folkerth RD, Iafrate AJ, Shen Y, et al. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol 2007; 62: 609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zou Q, Xie X, Liu T, Li HS, Jie Z, et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J Exp Med 2017; 214: 1493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jin T, Zhu H, Chen H, Ofengeim D, Zou C, et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 2018; 174: 1477–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Liang C, Swaminathan K, Herrlinger S, Lai F, Shiekhattar R, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv 2016; 2: e1601167–e1601167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Liu M, Tian Y, Wang J, Xu Y.. Cryo-EM structure of human DNA-PK holoenzyme. Cell Res 2017; 27: 1341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song S, Pan X.. Knockdown of miR-155 protects microglia against LPS-induced inflammatory injury via targeting RACK1: a novel research for intracranial infection. J Inflamm (Lond) 2017; 14: 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Guan Y, Wu X, Chen Y, Liu Z, Du H, et al. Wnt signaling is altered by spinal cord neuronal dysfunction in amyotrophic lateral sclerosis transgenic mice. Neurochem Res 2013; 38: 1904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhou X, Chang M, Nakaya M, Chang J-H, Xiao Y, et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun 2015; 6: 6074.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang W, Siedlak SL, Liu Y, Liu J, Jiang K, et al. Miro1 deficiency in amyotrophic lateral sclerosis. Front Aging Neurosci 2015; 7: 100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Cooper LT, Boyd AW, Bartlett PF.. Decreased signalling of EphA4 improves functional performance and motor neuron survival in the SOD1G93A ALS mouse model. Sci Rep 2018; 8: 11393.. [DOI] [PMC free article] [PubMed] [Google Scholar]