Abstract
The ribonuclease A family of proteins is well studied from the biochemical and biophysical points of view, but its evolutionary origins are obscure, as no sequences homologous to this family have been reported outside of vertebrates. Recently, the spatial structure of the ribonuclease domain from a bacterial polymorphic toxin was shown to be closely similar to the structure of vertebrate ribonuclease A. The absence of sequence similarity between the two structures prompted a speculation of convergent evolution of bacterial and vertebrate ribonuclease A-like enzymes. We show that bacterial and homologous archaeal polymorphic toxin ribonucleases with a known or predicted ribonuclease A-like fold are distant homologs of the ribonucleases from the EndoU family, found in all domains of cellular life and in viruses. We also detected a homolog of vertebrate ribonucleases A in the transcriptome assembly of the sea urchin Mesocentrotus franciscanus. These observations argue for the common ancestry of prokaryotic ribonuclease A-like and ubiquitous EndoU-like ribonucleases, and suggest a better-grounded scenario for the origin of animal ribonucleases A, which could have emerged in the deuterostome lineage, either by an extensive modification of a copy of an EndoU gene, or, more likely, by a horizontal acquisition of a prokaryotic immunity-mediating ribonuclease gene.
Keywords: ribonuclease A, ribonuclease EndoU, protein folding
INTRODUCTION
Bovine pancreatic ribonuclease (RNase A; EC 3.1.27.5) is an enzyme that is secreted in the pancreas of ruminants and is able to cleave RNA at the 3′ side of pyrimidine residues, using a catalytic mechanism that involves a nucleoside 2′,3′-cyclic phosphodiester intermediate (Cuchillo et al. 2011). The ribonuclease activity in pig pancreas was discovered in 1920 by W. Jones (Jones 1920), and M. Kunitz developed a protocol for purification of the bovine pancreatic enzyme in the 1930s (Kunitz 1939). In the early 1950s, the Armour company, most famous for its meatpacking business, antibacterial Dial soap, and Dale Carnegie, produced a kilogram of the crystalline enzyme from bovine pancreatic tissue and offered free samples to any interested research laboratory (Richards 1992).
Early availability of pure RNase A in bulk, together with a small size of the molecule (124 amino acids in the secreted form of the protein), its exceptional stability, and high activity that did not require any prosthetic groups, made RNase A the favorite model protein for many enzymologists interested in RNA biology and general principles of protein structure and function; it has been called “the most studied enzyme of the 20th century” (Raines 1998). RNase A was the first enzyme (and third protein altogether, after insulin and hemoglobin) for which an amino acid sequence was determined by direct chemical sequencing, long before DNA sequence determination became available (Smyth et al. 1963); the first enzyme for which a mechanism of catalysis was proposed based on the experimental evidence, even without the benefit of knowing the complete sequence or the spatial structure of the protein (Findlay et al. 1962); the fourth protein for which a three-dimensional structure was determined (Avey et al. 1967; Wyckoff et al. 1967); and the first protein recovered in the active form, albeit slowly and at low yield, after total chemical synthesis and in vitro refolding (Gutte and Merrifield 1969; Hirschmann et al. 1969). Famously, Anfinsen's studies of experimental disruption and restoration of biophysical properties and biochemical activity of RNase A have been viewed as the evidence that protein folding is a spontaneous process occurring under thermodynamic control (Anfinsen 1973).
From the physiological point of view, the RNase A sequence family is of interest because many lineages of vertebrates have multiple homologs of this enzyme, which play diverse biological roles. Despite the initial suggestion of the predominantly nutritional role of pancreatic RNase A (i.e., the facilitation of hydrolysis in the gut of stray RNA, presumably from dead bacteria), the functions of RNases A expressed in pancreas and especially in other tissues appear not to be related to the digestion of bulk RNA, but likely have to do with intracellular signaling during the development and with responding to bacterial and viral infection, in what could be one of the branches of RNA-triggered innate immunity response (Dyer and Rosenberg 2006; Cho and Zhang 2007; Ardelt et al. 2008; Pulido et al. 2013; Koczera et al. 2016; Ferguson et al. 2019). Curiously, some of these biological functions do not correlate with the rate or specificity of RNA hydrolysis displayed by the proteins in the in vitro assays (Domachowske et al. 1998; Dyer and Rosenberg 2006; Pizzo et al. 2006).
In the field of protein engineering, the clues obtained from the structure-function analysis of the RNase A family have allowed researchers to improve the therapeutic/oncolytic potential of some family members, or test the limits of functional change, such as reaction rate or specificity toward defined oligonucleotide substrates, that can be obtained by modifying a natural protein scaffold (Smith and Raines 2006; Pizzo and D'Alessio 2007; Vacca et al. 2008; Pulido et al. 2013; Prats-Ejarque et al. 2019). The RNase A family also has been the subject of extensive molecular evolutionary analyses, which addressed questions such as the lineage-specific evolution rates in proteins (Singhania et al. 1999; Zhang et al. 2000; Rosenberg et al. 2001; Cho et al. 2005; Cho and Zhang 2006), signatures of drift and selection in biological sequences (Zhang et al. 1998; Osorio et al. 2007; Premzl 2014), and the possibility of reconstructing and resurrecting ancestral sequences on the basis of the known sequences of their present-day descendants, with some surprising conclusions about the preference of the ancestral enzyme to double-strand RNA (Stackhouse et al. 1990; Jermann et al. 1995).
In contrast to this wealth of knowledge about structural determinants of protein stability and RNA degradation within the RNase A family, there is no clarity about its evolutionary origin. Until very recently, proteins with sequence or structural similarities to pancreatic RNase A were detected only in vertebrates. In the mammalian genomes, the RNase A gene family typically consists of 13 members, most of which are expressed narrowly in one or a few tissues in a secreted form (Cho and Zhang 2006; Premzl 2014), though the size of the family may be expanded in some lineages, to as many as 21 members in marsupials (Cho and Zhang 2006). The families in other vertebrates are smaller; for example, only three RNase A homologs have been found in chicken and four in zebrafish (Pizzo et al. 2006; Cho and Zhang 2007; Kazakou et al. 2008). These gene products, easily detected by sequence database searches, mostly share the conserved hallmarks of the family, such as the conserved trio of the catalytic residues, two histidines and a lysine, and between two and four pairs of disulfide bonds. The absence of RNase A sequence homologs outside of vertebrates received a special mention in the paper describing the initial sequencing and analysis of the human genome, where its status as the only vertebrate-specific enzyme was noted as an evolutionary oddity (Lander et al. 2001).
Recently, the spatial structure of a type of the carboxy-terminal effector domain of bacterial polymorphic toxin, CdiAYkris from Yersinia kristensenii, has been determined, and its high structural similarity to RNase A came to light (Batot et al. 2017). CdiAYkris is a part of the system for self/non-self recognition in bacteria (Zhang et al. 2011, 2012), which posesses an RNase activity, but there is no sequence similarity between CdiAYkris and vertebrate RNases A. The spatial structures of RNase A and CdiAYkris, on the other hand, can be superimposed readily, with the root mean square deviation of 3.6 Å over 89 aligned α-carbon atoms, even though only one catalytic residue in both molecules, the N-proximal histidine (His 12 in the pancreatic RNase A), was located in the same position in both structures. This structural fit in the absence of discernible sequence similarity (including the absense of cysteines in CdiAYkris sequence) prompted the hypothesis that the RNase A-like and CdiAYkris-like families have evolved convergently toward the same fold (Batot et al. 2017).
Although the RNase A spatial structure can no longer be viewed as unique to vertebrates, its evolutionary origins, and the relationship between two sequence families within it, remain unclear. In particular, there was a question of whether the distinct RNase A-like and CdiAYkris-like families indeed have evolved convergently—and if so, what were the unrelated sequences and structures that have attained the same fold in the course of convergent evolution. Alternatively, the RNase A-like and CdiAYkris-like families could have evolved divergently from a common ancestral sequence family—and if so, there is a question of what that family was and which fold did it have.
In this study, we present the evidence of similarities between prokaryotic CdiAYkris-like RNases and another family of ribonucleases, called the EndoU family after the best-studied representative, a ribonuclease EndoU that has been first characterized as the factor involved in biogenesis of a particular small nucleolar RNA encoded by an intron of a ribosomal protein in X.laevis (Gioia et al. 2005). EndoU-like enzymatic domains are also found within large proteins encoded by Nidovirales, an order of animal viruses with large RNA genomes (Ricagno et al. 2006), whereas the prokaryotic homologs of EndoU have been detected in a distinct group of bacterial and archaeal polymorphic toxins (Zhang et al. 2011). In the following, we describe the sequence-level and structure-level connections between EndoU and RNase A families and discuss various evolutionary and functional implications of these connections.
RESULTS AND DISCUSSION
Recently, a significant expansion of the repertoire of polymorphic toxins in archaea has been reported, and a subset of effector domains in archaeal toxins was shown to be related to CdiAYkris and its bacterial homologs (Makarova et al. 2019). In particular, a match between a putative toxin from a partially sequenced archaeon Thermococcus sp. EXT12c (GenBank ID WP_099209516) and the model of CdiAYkris build on the sequence of the PDB entry 5e3e has been observed in the HHPred analysis (K Makarova, pers. comm.). We performed the same search in October 2019, using as a query the WP_099209516 sequence except for a putative signal peptide in amino acids 1–22. The model for CdiAYkris/5e3e was the only match, with the P-value 1.8 × 10−5, when the entire conserved domain database (NCBI CDD) was used as the search space. The HHboost results also showed that WP_099209516 was a true match to bacterial CdiA-CT RNase A (Pfam ID PF18431, an alignment and the model built on CdiAYkris sequence) with probability 0.678, but an even better match to a protein annotated as a bacterial EndoU homolog (Pfam ID PF14436; probability 0.999).
Connections between CdiAYkris-like/RNase A-like toxins (which we will call prokRNases A, to distinguish them from the vertebrate RNases A) and prokaryotic EndoU-like toxins were also revealed by iterative searches of the NCBI NR database using the psi-blast program (Altschul et al. 1997). For example, when the search was initiated by WP_099209516 sequence, only archaeal homologs were seen at the first pass, but a homolog from a firmicute bacterium Lysinibacillus varians was detected at the iteration 2 (WP_025219942, E-value 10−6), followed by many other bacterial and archaeal matches; at the iteration 7, a sequence annotated as an EndoU nuclease, SNY81002 from an actinomycete Nocardia amikacinitolerans, was detected (E-value 2 × 10−4). Hundreds of other sequences were retrieved in this search, which converged after 19 iterations; we then clustered the homologs by pairwise sequence similarity and used representative sequences to identify additional homologs. Altogether, more than 1000 non-identical sequences were retrieved, many of them already annotated as toxins, nucleases, ribonucleases or EndoU-like ribonucleases; many of them are large multidomain proteins, with the effector nuclease typically occupying the extreme carboxy-terminal region of the polyprotein, while others are reported as stand-alone ORFs. The searches did not appear to be exhaustive; it is likely that many thousands of representatives of the prokRNase A/EndoU superfamily are already in the sequence databases.
A few trends in the distribution of these domains are noticeable: In archaea, they are found mostly in mesophiles and only a few thermophiles, and in bacteria, they are encoded mostly by the free-living species with relatively large genomes, representing multiple divisions of proteobacteria, firmicutes, actinomycetes, high GC Gram-positive bacteria, cyanobacteria, the FCB group, Verrucomicrobia, and a few other lineages known mostly from the environmental samples. Despite broad presence of these protein domains in many clades of bacteria and archaea, all such clades also have some species that lack recognizable homologs of those proteins. On the other hand, sometimes evolutionarily distant species of prokaryotes encode prokRNase A/EndoU homologs with very similar sequences, supporting the notion of an important role of gene gain by horizontal gene transfer and gene loss in the evolutionary dissemination of the polymorphic toxin systems (Zhang et al. 2012).
Sequence similarity searches as well as HMM comparisons also support direct evolutionary relationship of the prokaryotic EndoU-like proteins with their eukaryotic counterparts; for example, the psi-blast search seeded by the sequence of EndoU catalytic domain from Xenopus laevis (pdb 2c1w) recovers at the 11th iteration an ortholog from an oomycete Phytophthora parasitica P1976 (ETO85362). When the latter protein is used to start a new search, an annotated EndoU-like nuclease from a cyanobacterium Calothrix sp. NIES-4071 (WP_096729112) is seen at the 4th iteration with E-value 10−4, followed by many bacterial homologs intermingled with eukaryotic EndoU-like proteins. Curiously, two of the prokRNase A/EndoU-like homologs recovered in our searches, purportedly encoded by the draft genome assemblies of a mollusk Lottia gigantea (XP_009060737) and a sea anemone Exaiptasia pallida (KXJ13895), match only bacterial sequences in the database scans. We suspect that these gene products represent bacterial consorts inadvertently sequenced together with the invertebrate genomes, rather than genes encoded by the genomes of the two hosts; on the other hand, real eukaryotic-type EndoU sequences are encoded by these species, for example, Lottia XP_009056913 and XP_009047139.
It is notable that, despite apparently high sensitivity of our analysis, we did not detect any vertebrate RNases A when the database searches were initiated with prokRNAse A or EndoU sequences and sequence family models. Similarly, the reverse database scans initiated using the vertebrate RNase A sequences or family models did not find any of the prokRNase A/EndoU proteins. Noticing that some of the searches initiated with vertebrate RNases A were reporting low-scoring spurious matches to unrelated cysteine-rich proteins, we wondered whether the models of vertebrate RNases A were biased by the conservation of multiple cysteine residues, which could preclude detection of relevant similarities in other residues. To account for this possibility, we produced modified alignments of RNases A, in which each conserved cysteine in every sequence was replaced by sampling from the distribution of non-cysteine residues within the same secondary structure element (Supplemental Additional File 1). Such engineered alignments also could not be matched to the prokRNase A/EndoU family models, suggesting evolutionary remoteness of vertebrate RNases A from the prokaryotic proteins.
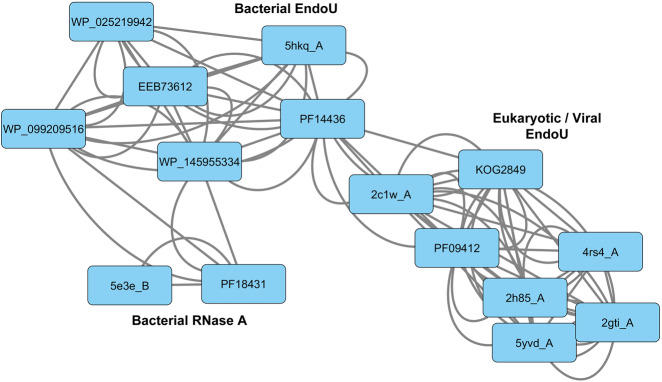
To build a graphical representation of the relationships between various prokRNase A/EndoU-related proteins, we used HHboost probabilities as a measure of sequence similarity between them and constructed a weighted similarity network (Fig. 1). While there are no direct connections linking prokRNase A and eukaryotic/viral EndoU proteins, a number of bacterial EndoU proteins serve as the network hubs connecting the two groups into a large and diverse sequence superfamily.
FIGURE 1.
Network of prokRNase A-related and EndoU-related conserved PFAM domains and sequence models based on select queries from the NR and PDB databases. The arc between domains/models indicates a statistically supported match obtained by the HHPred or HHBoost searches.
We collected a representative sample of sequences from various clusters and produced their multiple alignment. The structures examined in the course of the analysis were: CdiAYkris (5e3e); an EndoU-like toxin from Escherichia coli STEC031 (5hkq; coincidentally also named CdiA, but before this study thought to be unrelated to CdiAYkris); and two EndoU enzymes, one from X.laevis (2ciw) and the other from Murine hepatitis coronavirus (2gti). For comparison, the structures of animal RNases A were also overlaid on the alignment, using the superimposition of the pancreatic ribonuclease 1u1b with 5e3e.
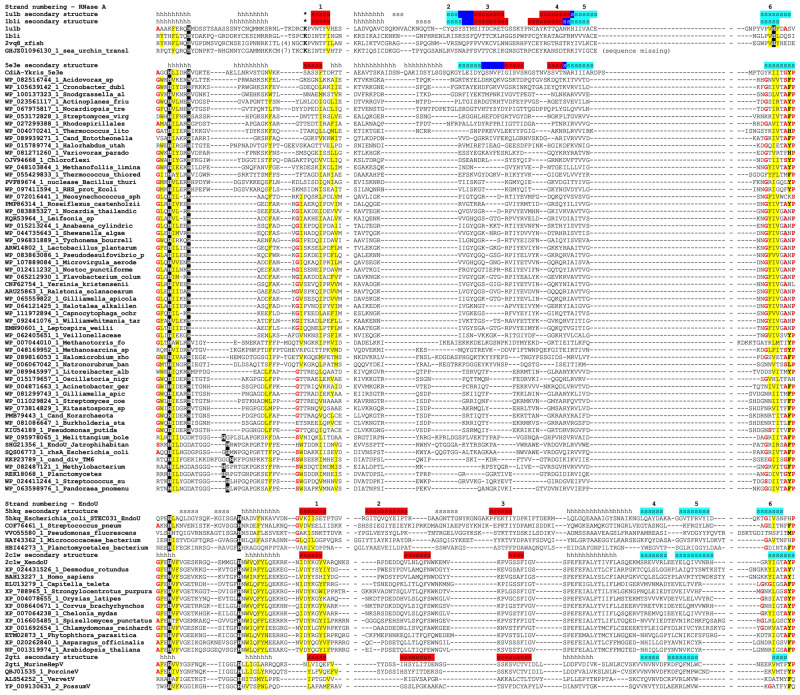
The analysis of the multiple alignment, shown in Figure 2, reveals several common sequence and structure features in all RNase A-like and EndoU-like proteins. The conservation is relatively high in the regions corresponding to the terminal segments within the prokRNase A and prokaryotic EndoU-like domains (in the following, “amino-terminal” and “carboxy-terminal” refer to the regions within the RNase A and EndoU catalytic domains themselves, even though many of them are embedded within longer proteins). In the amino-terminal part of the alignment, there is a pattern of a long loop, sometimes partially structured into a short helix or strand, followed by a conserved helix-turn-strand arrangement. This region includes two conserved histidines in almost all prokaryotic family members. One of those histidines is also preserved in animal RNases A, where it is crucially important for the catalysis, serving as the general base initiating the hydrolysis of the phosphoester bond (Cuchillo et al. 2011). In the carboxy-terminal region, there is a β-strand with a motif consisting of a proline next to an aliphatic or more commonly aromatic residue; in animal RNases A, this strand contains the second catalytic histidine that serves as a general acid in the catalysis. Despite the relatively short lengths of these discontinuous sequence segments, both of them were observed as the main high-scoring segment in at least some of the PSI-BLAST or HHPred/Hhboost searches, suggesting that both these segments retain a strong evolutionary signal. At the same time, the middle portions of the bacterial, archaeal and eukaryotic homologs are less well conserved, and shared sequence motifs there could not be identified with confidence.
FIGURE 2.
Multiple alignment of RNase A-like and EndoU-like protein sequences. Unique identifiers in the NR or PDB databases are shown before each sequence. In the secondary structure lines, h stands for a helical structure and s stands for an elongated structure (a strand). Vertebrate and sea urchin RNase A sequences are superimposed on the alignment to maximize the overlap of the structurally equivalent secondary structure elements. Conserved catalytic histidines are shown in white-on-black type, conserved hydrophobic residues (I, F, L, M, V, W, Y) are indicated by yellow shading, conserved residues with the propensity to make turns or kinks in the main chain (A, G, S, P) are indicated by bold red type, and catalytic lysine in animal RNases A is shown by asterisks in the secondary structure line. The strands that belong to the same β-sheet in EndoU family, or to the same wing of a sheet in RNases A, are shaded with the same color, and dark blue shading indicates the crease connecting the two wings in RNases A. The numbering of the β-strands is modified to consider only structurally equivalent strands, accounting for the contributions of the long strand to both wings in RNases A (see text for details).
Analysis of the representative three-dimensional structures of RNases A, EndoU, and their bacterial homologs provides more insight into the discontinuous pattern of their sequence conservation. The RNase A fold has been described as an anti-parallel β-sheet, consisting of three long strands and elaborated by additional sheets or helices in some family members. On the other hand, the structure of EndoU catalytic domain has been described as the duplication of a unit comprising an α-helix and three β-strands, so that the entirety of the EndoU fold consists of two β-sheets, each with three main strands. We have performed a structural comparison of RNase A-like and EndoU-like enzymes, confirming a close similarity of two folds and explaining the similarities and differences of their topology (Fig. 3).
FIGURE 3.
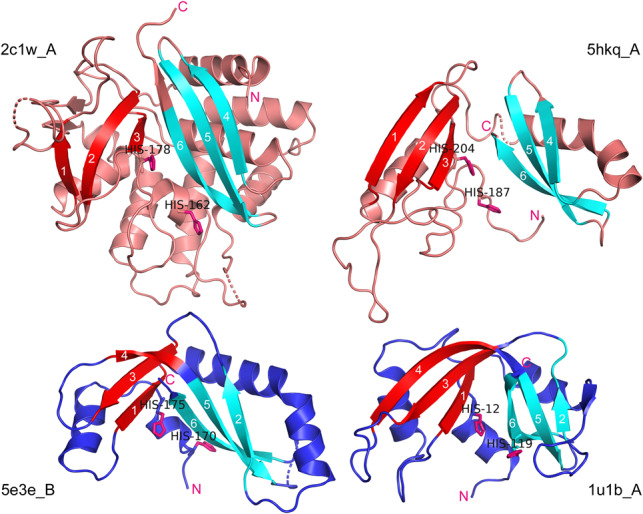
Spatial structure and topology of EndoU, prokRNase A, and animal RNase A proteins. (Upper left) X. laevis EndoU (2c1w_A); (upper right) E.coli EndoU-like toxin (5hkq_A); (lower left) Y. kristensenii RNase A-like toxin (5e3e_B); (lower right) bovine pancreatic ribonuclease (1u1b_A). Known or putative catalytic histidines are shown in all structures. The β-sheets or “wings” of a creased sheet are colored in cyan and red, to match the shading of the strands in Figure 2. The amino-termini in all chains are closely followed by two conserved histidines (or by single His-12 in 1u1b_A), and the carboxy-termini in all chains are located immediately downstream from the conserved Strand 6.
A striking feature of the three-stranded anti-parallel β-sheet forming the core of the RNase A fold is that it is sharply bent in the middle, along the line perpendicular to the main direction of the strands’ backbones, forcing many amino acids in the middle of two strands to kink out of the canonical β-strand-like conformation. The resulting bent sheet in RNase A may be described as consisting of two “wings,” each formed by three strands that do not follow each other along the sequence. The first of the wings includes the conserved strands 1, 3 and the amino-terminal portion of the long conserved strand 4, whereas the other sheet comprises strand 2, the carboxy-terminal part of the conserved strand 4, and strand 5. The loop between strands 2 and 3 together with the kink in the middle of strand 4 form a crease, which dissects the β-sheet into two distinct smaller β-sheets (“wings”), positioned at an angle to each other. Thus, ignoring the peripheral strands and helices, and splitting the longest strand 4 into two strands (new strands 4 and 5), we obtain the new strand numbering 1 through 6 for RNase A-like enzymes, and the strand composition of the first β-sheet is then 1 + 3+ 4 (red-shaded in the secondary structure lines in Fig. 2, and red-colored in Fig. 3), and that of the second sheet is 2 + 5+ 6 (cyan shading or coloring in the figures). In contrast to this mixed topology, in EndoU homologs the three strands comprising each sheet are contiguous in sequence (except for occasional insertions that do not affect the main core topology), so that the strand compositions of the β-sheets are 1 + 2+ 3 and 4 + 5+ 6; the two sheets in EndoU are separated by loops, which set the sheets at a slightly acuter angle than in RNase A and its structural relatives (Fig. 3).
The rewiring of strands in EndoU compared to RNase A cannot be achieved by a one-step circular permutation of the gene and protein sequences—a process that is better studied and is amenable to automated detection (Lo and Lyu 2008)—and would require a relatively complex sequence of genetic events for converting one to another. This structural difference is the likely reason why the relationship between RNase A and EndoU ribonucleases has eluded detection until recently.
Despite the rearrangements described above, the two most conserved sequence elements shared by RNase A and EndoU families, that is, strand 6 and the assemblage of helices and loops preceding strand 1 (Fig. 2), are found close to each other in all available spatial structures, often with a potential for van der Waals interactions or hydrogen bonding. Taking strand 6 as a reference and focusing on the close contacts (within 3.8 Å), in the pancreatic ribonuclease 1u1b_A, the backbone of residues P117 and F120 from strand 6 interacts with the side chain of the F8 residue from the amino-terminal conserved region; this interaction brings the two catalytic residues H12 and H119, which are remote in the sequence, into close spatial proximity. In human angiogenin (1b1i), the side chains of residues P112 and L115 interact with the side chains of Y6 and F9, also likely contributing to proper positioning of the two catalytic histidines. In the CdiAYkris, the key structural role within strand 6 appears to be played by the side chain of T276, which makes contacts with L171 and with both of the group-specific histidine residues, H170 and H175. In the bacterial EndoU-like ribonuclease (5hkq_A), a similar role is played by the tripeptide Phe-His-Pro that is located just after the carboxyl terminus of strand 6 (a histidine residue in this position is not highly conserved and is likely to play a structural rather than catalytic role), which interacts with a structurally conserved leucine in the extra strand upstream of strand 1, as well as two family-specific conserved N-proximal histidines. In some cases, residues within these conserved elements also directly contact the substrate oligonucleotides.
The persistence of paired histidines in the amino-terminal regions of the RNase superfamily defined here, as well as the conservation of a network of interactions between the neighbors of those histidines and the distal, carboxy-terminal residues, may have implications for the catalytic mechanism of the prokRNase A and EndoU enzymes. In the case of CdiAYkris/5e3e_B, the histidine-175 is thought to be functionally equivalent to catalytic H12 in RNase A, and it has been speculated that the role of the purportedly missing second catalytic histidine is fulfilled by T276, which is postulated to participate in general acid-base catalysis through a hydroxyl group in its side chain (Batot et al. 2017). Our sequence and structure analyses argue that the catalytic center of CdiAYkris and other prokaryotic RNase A-like enzymes in fact includes two histidine residues, conserved H170 and H175, and that T276 may play another role.
The essential catalytic roles of the two N-proximal His residues in eukaryotic and viral EndoU enzymes have been documented using the in vitro assays, and some of the developmental phenotypes of XendoU homologs are dependent on the intactness of those histidines as well (Gioia et al. 2005; Posthuma et al. 2006; Schwarz and Blower 2014). Although the two histidines in EndoU homologs is found at a wider separation along the primary structure than in the case of CdiAYkris (usually 12–18 amino acids apart in the former vs. 5–6 amino acids in the latter), these pairs are found at similar spatial distances of 3.5–4 Å. In the published structure of CdiAYkris/5e3e_B, the side chain of H170 points away from the putative substrate-binding cleft, but, given high sequence conservation of this position over a long evolutionary span and the functional importance of both histidines in the EndoU family, we predict that further studies of CdiAYkris and its homologs will refine the orientation of this histidine residue and will implicate it more directly in the catalysis.
Like vertebrate RNases A, eukaryotic and viral EndoU enzymes cleave RNA producing 2′–3′-cyclic phosphates, but unlike RNases A, the catalytic activity of EndoUs in X.laevis and SARS coronavirus is thought to require the presence of Mn2+ ions (Laneve et al. 2003; Ricagno et al. 2006). The exact role of the magnesium ion in the catalysis remains unspecified, and the ion does not seem to be tightly associated with the enzyme during purification (see discussion in Gioia et al. 2005). The sequence and structure similarities between EndoU and metal-independent RNase A described in this study, as well as the inspection of the three-dimensional structures of the homologous enzymes that do not seem to retain a bivalent metal cation in catalytically relevant positions, may suggest that the role of Mn2+ ions in EndoU catalysis is more auxiliary than has been thought before.
Given a wide conservation of the arrangement of two N-proximal histidines in the RNases A and EndoU across many bacteria and archaea (as well as eukaryotes and their viruses in the case of EndoU), we speculate that this represents an ancestral configuration of the catalytic center, and the relocation of the second histidine to the carboxy-terminal conserved region in the structurally similar animal-specific RNases A is a derived state in the evolution of this enzymatic scaffold. An indirect support to this hypothesis is given by the protein engineering experiments, in which two histidines have been introduced into the amino-terminal phosphate-binding subsite next to the first catalytic histidine of bovine pancreatic ribonuclease (positions 7 and 10 of the mature chain), and the native catalytic histidines 12 and 119 were mutated (Moussaoui et al. 2007). The resulting enzyme still retained 9% of the wild-type activity, suggesting that animal RNases A may digest RNA in the absence of a histidine within the carboxy-terminal β-strand, and that the putative ancestral form with the amino-terminal pair of catalytic histidines may have had some utility to its owner.
In this work, we used probabilistic comparisons of protein sequences and sequence families to obtain the evidence of a specific sequence relationship between prokaryotic RNase A-like polymorphic toxins and another RNase family, EndoU, which is similarly found as a domain in bacterial and archaeal polymorphic toxins, but also is present in most eukaryotes sequenced to date, as well as in the RNA viruses from the order Nidovirales. The analysis of sequence and structure relationship of RNase A-like nucleases in prokaryotes and EndoU-like nucleases in all domains of Life suggests that they may have evolved from a common ancestor by sequence divergence, topological rearrangement of at least one strand, and relocation of some helices. A spatial similarity and tentative homology between RNase A and EndoU families has been noted in the latest releases of two databases of conserved structural domains, ECOD (Cheng et al. 2015) and SCOPe (Chandonia et al. 2019)—see entries for ECOD domain ID372 (http://prodata.swmed.edu/ecod/complete/tree?id=372) and SCOPe domain d.294.1—but, as far as we know, this automatically detected relationship has not been discussed in the literature until this study.
In eukaryotes, the genetic evidence has implicated EndoU domains in a variety of cellular and developmental phenotypes, including: clearance of ribonucleoprotein particles from the nuclear membrane surface with concomitant remodeling of the endoplasmic reticulum in frog oocytes (Schwarz and Blower 2014), the survival of peripheral B cell in mice (Poe et al. 2014), neuron survival in fruit fly (Laneve et al. 2017), and synaptic remodeling in the nematode (Ujisawa et al. 2018). In viruses, EndoU domains play a still incompletely understood role in virus genome replication, possibly by intervening with the host RNA surveillance systems (Posthuma et al. 2006; Wang et al. 2019). Finally, EndoU-like and related prokRNaseA-like polymorphic toxins in bacteria and archaea mediate the interspecific conflicts in these microbes by targeting the cells of the other species, as well as the intraspecific conflicts by secreting the factors that restrict the growth of non-self strains; such toxins also are involved, on all sides, in the conflicts between genomes and selfish replicons (Zhang et al. 2011, 2012). All this suggests an ancient association of the EndoU and RNase A ribonucleases with the RNA-level regulation of the events in the cell–cell and cell–parasite competition, as well as in cell differentiation in metazoa; on the other hand, a putative function of some vertebrate RNases A, such as pancreatic ribonuclease, in nutritional RNA salvage appears to be a side line in the superfamily.
The evolutionary scenario connecting the nuclease domain of eukaryotic EndoU enzymes to the bacterial EndoU-like polymorphic toxins with nuclease activity is relatively straightforward; the eukaryotic homologs most likely have evolved from their bacterial ancestors, acquiring an additional helical amino-terminal domain in the process. A shared sequence ancestry is also apparent between the prokaryotic EndoU and RNase A/CdiAYkris-like enzymes, though the high level of sequence divergence makes it difficult to say with confidence which one might be more ancient than the other. Earlier structural comparisons (Zhang et al. 2012) have suggested a link between the “duplicated half” of the EndoU catalytic domain and several other nuclease families, such as: a vast sequence family of homologous ribonucleases from prokaryotes and fungi, including barnase, RNases T1, U2, F1, Ms, and sarcin; a subset of bacterial colicin RNases, namely colicin E5 and colicin D; and the RNase domain of the RelE toxin. The entire group was named BECR fold, after Barnase, EndoU, Colicins, and RelE (Zhang et al. 2012). The structural theme uniting the fold has been described as a β-sheet formed by four strands, some of which may be long and meandering; the sheet is preceded by an α-helix and may be followed by the fifth short strand. It has been noted that there are no sequence-level connections between the four families within the BECR fold, that each of the four families uses a distinct set of catalytic residues, and that their spatial positions of these residues are different too (Zhang et al. 2012; Gucinski et al. 2019); we also note that the sequence-structure motifs that link eukaryotic and prokaryotic RNases A to EndoU are missing from the “B,” “C,” and “R” members of the BECR fold. These observations lend no extra support to the hypothesis of a common sequence ancestor shared by all families within the BECR fold. Noticing, however, that the BECR fold is found in its minimal form in the “B,” “C,” and “R” families, and as a duplication of that minimal form in the EndoU family, it can be tentatively suggested that the prokaryotic EndoU could have emerged first by duplication of a minimal BECR-like fold, and that prokRNase A-like enzymes have evolved by a more recent permutation of a prokaryotic EndoU-like enzyme.
In the course of this study, we also detected a bona fide homolog of vertebrate RNases A in the assembly of RNA transcriptome from a non-vertebrate metazoan animal, red sea urchin Mesocentrotus franciscanus (translation product of GHJZ01096130; Fig. 2). The sequence from Mesocentrotus is more distantly related to vertebrate RNases A than they are to each other, and resolves as an isolated long branch in a rootless phylogenetic tree (A Mushegian, unpubl.), suggesting that the evolutionary origin of this family may be more ancient than the common ancestor of all vertebrates, going at least as far back as the stem of the deuterostome lineage. This observation moves the evolutionary age of the animal RNase A back in time, at least to the common ancestor of the deuterostomes, and narrows the evolutionary gap between prokaryotic and animal RNases A.
Only a structural similarity (albeit a close one), but not a sequence-level similarity, can be discerned between the CdiAYkris-like prokaryotic RNases A and their animal counterparts. The evolutionary scenarios for the origin of the latter remain tentative for this reason, but two main hypotheses are worth considering. One possibility could be that animal RNases A have evolved in the deuterostome lineage by duplication of a gene that coded for the deuterostome version of pan-eukaryotic EndoU. This would have been followed by a loss of an extra all-helical domain observed in all eukaryotic EndoU and a complex rearrangement of strands in the catalytic domain in deuterostomes, leading to a convergent recreation of the prokRNase A-like domain topology. Such scenario would require a dramatic change in many other properties of the animal EndoU gene copy and of its product, such as massive loss of introns (eukaryotic EndoU genes are intron-rich, whereas animal RNase A genes lack introns in the coding region) and optimization of the formerly intracellular protein sequence for a new role as a secreted protein.
An alternative hypothesis is that RNases A have been acquired by a metazoan ancestor by a horizontal gene transfer of CdiAYkris-like RNase A domain from a prokaryote, for example, a bacterial or archaeal consort associated with the deuterostome host. Such an intronless open reading frame would have coded for a product already optimized to traverse at least some membranes, and could have been of more direct use to the host, for example, in mediating the interactions with its own microbiome. Each of these two scenarios must also account for the acceleration of sequence evolution in the animal RNase A lineage, which would place it beyond recognition by the current sequence comparison methods, and for rapid accrual of stabilizing pairs of disulfide bonds—a feature relatively uncommon in prokaryotic proteins but strongly associated with eukaryotic proteins (Wong et al. 2011). More extravagant scenarios, such as the convergent origin of animal RNase A and/or its transfer from metazoa to prokaryotes, cannot be formally excluded at this time, but appear less likely.
In the studies of pancreatic RNase A folding and stability, much attention has been given to the dynamics of experimental disruption and restoration of its tertiary structure, with the emphasis on the formation of the correct set of disulfide bonds (Anfinsen 1973; Wearne and Creighton 1988; Talluri et al. 1994; Neira and Rico 1997; Klink et al. 2000; Wedemeyer et al. 2000; Mamathambika and Bardwell 2008). The observation of a variety of homologous and structurally similar proteins which do not have equivalently positioned cysteine residues, and often have no cysteines at all, should bring to focus various other factors contributing to folding of RNase A, and perhaps even prompt a reevaluation of its folding mechanism. The interaction between the amino-terminal and carboxy-terminal regions of RNases A/EndoU nucleases is particularly interesting in that regard. The spatial proximity of the protein termini on the surfaces of protein molecules is a statistical trend in the entire space of the known protein structures, but the reasons for that tendency remain unknown (Krishna and Englander 2005; Jacob and Unger 2007). Recently, it has been proposed that such proximity may be a signature of the universal cotranslational protein folding mechanism, in which the ribosome-based protein folding machine holds the ends of the nascent chain throughout protein synthesis, releasing them only after the majority of the protein chain has been set into a defined conformation (Sorokina and Mushegian 2017, 2018). Under that model, the closeness of the two terminal regions is seen as a “scar” of the folding process, not adaptive by default. At least in some proteins, however, functionally important interactions between the termini could evolve, facilitated by their proximity. The conserved, and therefore likely functionally important, interaction between the amino-terminal and carboxy-terminal elements in the RNases A/EndoU family may be a factor in determining the efficiency of folding of these proteins both in vivo and in vitro. The results described here should provide a new guidance for the experimental studies of these possibilities.
MATERIALS AND METHODS
Sequence searches were performed in October 2019. Iterative protein sequence searches were done using the psi-blast program (Altschul et al. 1997), usually with inclusion cutoff 0.05 and composition-based statistics option, using the NCBI NR database as the search space. To build a hidden Markov model (HMM) of a protein family and compare it to all other HMMs in the NCBI CDD and PDB70 databases, the HHPred server (Zimmermann et al. 2018) was used with the alignment mode option set at realign-global. The HHPhed results were reanalyzed with HHboost software (M Dlakić, in prep.; http://hhboost.dlakiclab.org/). HHboost is a new implementation of HHsvm (Dlakić 2009), a program that utilizes a support vector machine (SVM) classifier trained to identify the homologs from HHPred results even in the absence of statistically significant E-values in the raw output of HHPred. In the HHboost implementation, the SVM classifier is replaced by gradient boosting machines, resulting in improved sensitivity and specificity (M Dlakić, in prep.). Multiple sequence alignments of proteins were done with MAFTT (Katoh and Standley 2013) and PROMALS-3D (Pei et al. 2008) algorithms, with iterative manual refinement aided by the information about secondary structure elements and topologies of several proteins for which the spatial structures are available. Structural comparisons were done using the MICAN program (Minami et al. 2013). The images of the protein spatial structures were rendered using the PyMOL software (The PyMOL Molecular Graphics System, Open-Source PyMOL v. 0.99rc6, Schrödinger, LLC).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Kira Makarova for communicating the results on archaeal prokRNAse A homologs prior to publication and for helpful discussions. This work was supported in part by the National Institutes of Health IDeA Program (COBRE Grant GM110732MD) to M.D. A.M. is a Program Director at the National Science Foundation (NSF), the agency of the U.S. Government; his work on this project was supported in part by the NSF Independent Research and Development Program, but the statements and opinions expressed herein are made in the personal capacity and do not constitute the endorsement by NSF or the government of the United States.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.074385.119.
REFERENCES
- Altschul SF, Madden TI, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- Ardelt W, Shogen K, Darzynkiewicz Z. 2008. Onconase and amphinase, the antitumor ribonucleases from Rana pipiens oocytes. Curr Pharm Biotechnol 9: 215–225. 10.2174/138920108784567245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avey HP, Boles MO, Carlisle CH, Evans SA, Morris SJ, Palmer RA, Woolhouse BA, Shall S. 1967. Structure of ribonuclease. Nature 213: 557–562. 10.1038/213557a0 [DOI] [PubMed] [Google Scholar]
- Batot G, Michalska K, Ekberg G, Irimpan EM, Joachimiak G, Jedrzejczak R, Babnigg G, Hayes CS, Joachimiak A, Goulding CW. 2017. The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily. Nucleic Acids Res 45: 5013–5025. 10.1093/nar/gkx230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia J-M, Fox NK, Brenner SE. 2019. SCOPe: classification of large macromolecular structures in the structural classification of proteins—extended database. Nucleic Acids Res 47: D475–D481. 10.1093/nar/gky1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Liao Y, Schaeffer RD, Grishin NV. 2015. Manual classification strategies in the ECOD database: ECOD Manual Classification Strategies. Proteins 83: 1238–1251. 10.1002/prot.24818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Zhang J. 2006. Ancient expansion of the ribonuclease A superfamily revealed by genomic analysis of placental and marsupial mammals. Gene 373: 116–125. 10.1016/j.gene.2006.01.018 [DOI] [PubMed] [Google Scholar]
- Cho S, Zhang J. 2007. Zebrafish ribonucleases are bactericidal: implications for the origin of the vertebrate RNase A superfamily. Mol Biol Evol 24: 1259–1268. 10.1093/molbev/msm047 [DOI] [PubMed] [Google Scholar]
- Cho S, Beintema JJ, Zhang J. 2005. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics 85: 208–220. 10.1016/j.ygeno.2004.10.008 [DOI] [PubMed] [Google Scholar]
- Cuchillo CM, Nogués MV, Raines RT. 2011. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry 50: 7835–7841. 10.1021/bi201075b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakić M. 2009. HHsvm: fast and accurate classification of profile-profile matches identified by HHsearch. Bioinformatics 25: 3071–3076. 10.1093/bioinformatics/btp555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domachowske JB, Bonville CA, Dyer KD, Rosenberg HF. 1998. Evolution of antiviral activity in the ribonuclease A gene superfamily: evidence for a specific interaction between eosinophil-derived neurotoxin (EDN/RNase 2) and respiratory syncytial virus. Nucleic Acids Res 26: 5327–5332. 10.1093/nar/26.23.5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer KD, Rosenberg HF. 2006. The RNase a superfamily: generation of diversity and innate host defense. Mol Divers 10: 585–597. 10.1007/s11030-006-9028-2 [DOI] [PubMed] [Google Scholar]
- Ferguson R, Holloway DE, Chandrasekhar A, Acharya KR, Subramanian V. 2019. The catalytic activity and secretion of zebrafish RNases are essential for their in vivo function in motor neurons and vasculature. Sci Rep 9: 1107 10.1038/s41598-018-37140-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay D, Herries DG, Mathias AP, Rabin BR, Ross CA. 1962. The active site and mechanism of action of bovine pancreatic ribonuclease. 7. The catalytic mechanism. Biochem J 85: 152–153. 10.1042/bj0850152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia U, Laneve P, Dlakic M, Arceci M, Bozzoni I, Caffarelli E. 2005. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem 280: 18996–19002. 10.1074/jbc.M501160200 [DOI] [PubMed] [Google Scholar]
- Gucinski GC, Michalska K, Garza-Sánchez F, Eschenfeldt WH, Stols L, Nguyen JY, Goulding CW, Joachimiak A, Hayes CS. 2019. Convergent evolution of the Barnase/EndoU/Colicin/RelE (BECR) fold in antibacterial tRNase toxins. Structure 27: 1660–1674.e5. 10.1016/j.str.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutte B, Merrifield RB. 1969. The total synthesis of an enzyme with ribonuclease A activity. J Am Chem Soc 91: 501–502. 10.1021/ja01030a050 [DOI] [PubMed] [Google Scholar]
- Hirschmann R, Nutt RF, Veber DF, Vitali RA, Varga SL, Jacob TA, Holly FW, Denkewalter RG. 1969. Studies on the total synthesis of an enzyme. V. The preparation of enzymatically active material. J Am Chem Soc 91: 507–508. 10.1021/ja01030a055 [DOI] [PubMed] [Google Scholar]
- Jacob E, Unger R. 2007. A tale of two tails: Why are terminal residues of proteins exposed? Bioinformatics 23: e225–e230. 10.1093/bioinformatics/btl318 [DOI] [PubMed] [Google Scholar]
- Jermann TM, Opitz JG, Stackhouse J, Benner SA. 1995. Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily. Nature 374: 57–59. 10.1038/374057a0 [DOI] [PubMed] [Google Scholar]
- Jones W. 1920. The action of boiled pancreas extract on yeast nucleic acid. Am J Physiol 52: 203–207. 10.1152/ajplegacy.1920.52.1.203 [DOI] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazakou K, Holloway DE, Prior SH, Subramanian V, Acharya KR. 2008. Ribonuclease A homologues of the zebrafish: polymorphism, crystal structures of two representatives and their evolutionary implications. J Mol Biol 380: 206–222. 10.1016/j.jmb.2008.04.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink TA, Woycechowsky KJ, Taylor KM, Raines RT. 2000. Contribution of disulfide bonds to the conformational stability and catalytic activity of ribonuclease A. Eur J Biochem 267: 566–572. 10.1046/j.1432-1327.2000.01037.x [DOI] [PubMed] [Google Scholar]
- Koczera P, Martin L, Marx G, Schuerholz T. 2016. The ribonuclease A superfamily in humans: canonical RNases as the buttress of innate immunity. Int J Mol Sci 17: E1278 10.3390/ijms17081278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna MMG, Englander SW. 2005. The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci 102: 1053–1058. 10.1073/pnas.0409114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitz M. 1939. Isolation from beef pancreas of a crystalline protein possessing ribonuclease activity. Science 90: 112–113. 10.1126/science.90.2327.112 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E. 2003. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J Biol Chem 278: 13026–13032. 10.1074/jbc.M211937200 [DOI] [PubMed] [Google Scholar]
- Laneve P, Piacentini L, Casale AM, Capauto D, Gioia U, Cappucci U, Di Carlo V, Bozzoni I, Di Micco P, Morea V, et al. 2017. Drosophila CG3303 is an essential endoribonuclease linked to TDP-43-mediated neurodegeneration. Sci Rep 7: 41559 10.1038/srep41559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W-C, Lyu P-C. 2008. CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships. Genome Biol 9: R11 10.1186/gb-2008-9-1-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Karamycheva S, Zhang D, Aravind L, Koonin EV. 2019. Antimicrobial peptides, polymorphic toxins, and self-nonself recognition systems in archaea: an untapped armory for intermicrobial conflicts. MBio 10: e00715-19 10.1128/mBio.00715-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamathambika BS, Bardwell JC. 2008. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol 24: 211–235. 10.1146/annurev.cellbio.24.110707.175333 [DOI] [PubMed] [Google Scholar]
- Minami S, Sawada K, Chikenji G. 2013. MICAN: a protein structure alignment algorithm that can handle multiple-chains, inverse alignments, Cα only models, alternative alignments, and non-sequential alignments. BMC Bioinformatics 14: 24 10.1186/1471-2105-14-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaoui M, Cuchillo CM, Nogués MV. 2007. A phosphate-binding subsite in bovine pancreatic ribonuclease A can be converted into a very efficient catalytic site. Protein Sci 16: 99–109. 10.1110/ps.062251707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira JL, Rico M. 1997. Folding studies on ribonuclease A, a model protein. Fold Des 2: R1–R11. 10.1016/S1359-0278(97)00001-1 [DOI] [PubMed] [Google Scholar]
- Osorio DS, Antunes A, Ramos MJ. 2007. Structural and functional implications of positive selection at the primate angiogenin gene. BMC Evol Biol 7: 167 10.1186/1471-2148-7-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36: 2295–2300. 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzo E, D'Alessio G. 2007. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene 406: 8–12. 10.1016/j.gene.2007.05.006 [DOI] [PubMed] [Google Scholar]
- Pizzo E, Buonanno P, Di Maro A, Ponticelli S, De Falco S, Quarto N, Cubellis MV, D'Alessio G. 2006. Ribonucleases and angiogenins from fish. J Biol Chem 281: 27454–27460. 10.1074/jbc.M605505200 [DOI] [PubMed] [Google Scholar]
- Poe JC, Kountikov EI, Lykken JM, Natarajan A, Marchuk DA, Tedder TF. 2014. EndoU is a novel regulator of AICD during peripheral B cell selection. J Exp Med 211: 57–69. 10.1084/jem.20130648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma CC, Nedialkova DD, Zevenhoven-Dobbe JC, Blokhuis JH, Gorbalenya AE, Snijder EJ. 2006. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol 80: 1653–1661. 10.1128/JVI.80.4.1653-1661.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats-Ejarque G, Lu L, Salazar VA, Moussaoui M, Boix E. 2019. Evolutionary trends in RNA base selectivity within the RNase A superfamily. Front Pharmacol 10: 1170 10.3389/fphar.2019.01170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premzl M. 2014. Comparative genomic analysis of eutherian ribonuclease A genes. Mol Genet Genomics 289: 161–167. 10.1007/s00438-013-0801-5 [DOI] [PubMed] [Google Scholar]
- Pulido D, Moussaoui M, Nogués MV, Torrent M, Boix E. 2013. Towards the rational design of antimicrobial proteins: single point mutations can switch on bactericidal and agglutinating activities on the RNase A superfamily lineage. FEBS J 280: 5841–5852. 10.1111/febs.12506 [DOI] [PubMed] [Google Scholar]
- Raines RT. 1998. Ribonuclease A. Chem Rev 98: 1045–1066. 10.1021/cr960427h [DOI] [PubMed] [Google Scholar]
- Ricagno S, Egloff M-P, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci 103: 11892–11897. 10.1073/pnas.0601708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards FM. 1992. Linderstrøm-Lang and the Carlsberg laboratory: the view of a postdoctoral fellow in 1954. Protein Sci 1: 1721–1730. 10.1002/pro.5560011221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg HF, Zhang J, Liao YD, Dyer KD. 2001. Rapid diversification of RNase A superfamily ribonucleases from the bullfrog, Rana catesbeiana. J Mol Evol 53: 31–38. 10.1007/s002390010188 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD. 2014. The calcium-dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J Cell Biol 207: 41–57. 10.1083/jcb.201406037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania NA, Dyer KD, Zhang J, Deming MS, Bonville CA, Domachowske JB, Rosenberg HF. 1999. Rapid evolution of the ribonuclease A superfamily: adaptive expansion of independent gene clusters in rats and mice. J Mol Evol 49: 721–728. 10.1007/PL00006594 [DOI] [PubMed] [Google Scholar]
- Smith BD, Raines RT. 2006. Genetic selection for critical residues in ribonucleases. J Mol Biol 362: 459–478. 10.1016/j.jmb.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Smyth DG, Stein WH, Moore S. 1963. The sequence of amino acid residues in bovine pancreatic ribonuclease: revisions and confirmations. J Biol Chem 238: 227–234. [PubMed] [Google Scholar]
- Sorokina I, Mushegian A. 2017. Rotational restriction of nascent peptides as an essential element of co-translational protein folding: possible molecular players and structural consequences. Biol Direct 12: 14 10.1186/s13062-017-0186-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina I, Mushegian A. 2018. Modeling protein folding in vivo. Biol Direct 13: 13 10.1186/s13062-018-0217-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackhouse J, Presnell SR, McGeehan GM, Nambiar KP, Benner SA. 1990. The ribonuclease from an extinct bovid ruminant. FEBS Lett 262: 104–106. 10.1016/0014-5793(90)80164-E [DOI] [PubMed] [Google Scholar]
- Talluri S, Rothwarf DM, Scheraga HA. 1994. Structural characterization of a three-disulfide intermediate of ribonuclease A involved in both the folding and unfolding pathways. Biochemistry 33: 10437–10449. 10.1021/bi00200a027 [DOI] [PubMed] [Google Scholar]
- Ujisawa T, Ohta A, Ii T, Minakuchi Y, Toyoda A, Ii M, Kuhara A. 2018. Endoribonuclease ENDU-2 regulates multiple traits including cold tolerance via cell autonomous and nonautonomous controls in Caenorhabditis elegans. Proc Natl Acad Sci 115: 8823–8828. 10.1073/pnas.1808634115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca RA, Giannattasio S, Capitani G, Marra E, Christen P. 2008. Molecular evolution of B6 enzymes: binding of pyridoxal-5′-phosphate and Lys41Arg substitution turn ribonuclease A into a model B6 protoenzyme. BMC Biochem 9: 17 10.1186/1471-2091-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chen J, Yu C, Zhu X, Xu S, Fang L, Xiao S. 2019. Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J Virol 93: e00623-19 10.1128/JVI.00623-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SJ, Creighton TE. 1988. Further experimental studies of the disulfide folding transition of ribonuclease A. Proteins 4: 251–261. 10.1002/prot.340040404 [DOI] [PubMed] [Google Scholar]
- Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. 2000. Disulfide bonds and protein folding. Biochemistry 39: 4207–4216. 10.1021/bi992922o [DOI] [PubMed] [Google Scholar]
- Wong JWH, Ho SYW, Hogg PJ. 2011. Disulfide bond acquisition through eukaryotic protein evolution. Mol Biol Evol 28: 327–334. 10.1093/molbev/msq194 [DOI] [PubMed] [Google Scholar]
- Wyckoff HW, Hardman KD, Allewell NM, Inagami T, Johnson LN, Richards FM. 1967. The structure of ribonuclease-S at 3.5 A resolution. J Biol Chem 242: 3984–3988. [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci 95: 3708–3713. 10.1073/pnas.95.7.3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dyer KD, Rosenberg HF. 2000. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc Natl Acad Sci 97: 4701–4706. 10.1073/pnas.080071397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, Aravind L. 2011. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39: 4532–4552. 10.1093/nar/gkr036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, Aravind L. 2012. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7: 18 10.1186/1745-6150-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430: 2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.