Abstract
Purpose
Machine learning-based approaches now outperform competing methods in most disciplines relevant to diagnostic radiology. Image-guided procedures, however, have not yet benefited substantially from the advent of deep learning, in particular because images for procedural guidance are not archived and thus unavailable for learning, and even if they were available, annotations would be a severe challenge due to the vast amounts of data. In silico simulation of X-ray images from 3D CT is an interesting alternative to using true clinical radiographs since labeling is comparably easy and potentially readily available.
Methods
We extend our framework for fast and realistic simulation of fluoroscopy from high-resolution CT, called DeepDRR, with tool modeling capabilities. The framework is publicly available, open source, and tightly integrated with the software platforms native to deep learning, i.e., Python, PyTorch, and PyCuda. DeepDRR relies on machine learning for material decomposition and scatter estimation in 3D and 2D, respectively, but uses analytic forward projection and noise injection to ensure acceptable computation times. On two X-ray image analysis tasks, namely (1) anatomical landmark detection and (2) segmentation and localization of robot end-effectors, we demonstrate that convolutional neural networks (ConvNets) trained on DeepDRRs generalize well to real data without re-training or domain adaptation. To this end, we use the exact same training protocol to train ConvNets on naïve and DeepDRRs and compare their performance on data of cadaveric specimens acquired using a clinical C-arm X-ray system.
Results
Our findings are consistent across both considered tasks. All ConvNets performed similarly well when evaluated on the respective synthetic testing set. However, when applied to real radiographs of cadaveric anatomy, ConvNets trained on DeepDRRs significantly outperformed ConvNets trained on naïve DRRs (p < 0.01).
Conclusion
Our findings for both tasks are positive and promising. Combined with complementary approaches, such as image style transfer, the proposed framework for fast and realistic simulation of fluoroscopy from CT contributes to promoting the implementation of machine learning in X-ray-guided procedures. This paradigm shift has the potential to revolutionize intra-operative image analysis to simplify surgical workflows.
Keywords: Monte Carlo simulation, Artificial intelligence, Computer assisted surgery, Robotic surgery, Segmentation, Image guidance
Introduction
Clinical context
Machine learning-based approaches, and in particular deep convolutional neural networks (ConvNets), now define the state of the art across a variety of well-studied problems in computer vision, and more recently, medical imaging. It is not surprising that the most impressive improvements until now were achieved in the sub-discipline of medical image computing [16,29]. This field is dominated by diagnostic imaging tasks where traditionally:
All imaging data are archived,
Expert annotations that define potential learning targets are well established (e.g., for mammographic lesion detection [13] or brain lesion segmentation [20]) or can be approximated efficiently in a self-supervisory manner (e.g., using automatic tools that yield either imperfect [24] or sparse [17] signals), and
Comparably simple augmentation strategies, such as rigid and non-rigid displacements [21,23], ease the limited data problem.
One may be tempted to conclude from the above reasoning that diagnostic image analysis provides a comfortable environment for machine learning, but this is not generally the case. This is because, compared to general computer vision, datasets in diagnostic image analysis are still small considering the dimensionality of the data and variability of its presentation. However, in interventional imaging, and particularly in X-ray-guided procedures, the situation is even more complicated. First, while hundreds of X-ray images are acquired for procedural guidance [4], only very few radiographs are archived suggesting a severe lack of meaningful data. Second, learning targets are not well established or defined. Third, interventional images are acquired from multiple view points onto the anatomy, the exact poses of which cannot accurately be reproduced during surgery [9,33]. Finally, the overall variability in the data is further amplified by surgical changes in anatomy and the presence of tools. Overall, the archived data are heavily unstructured and exhibit enormous variation, which challenges meaningful augmentation. As a consequence, in order to enable machine learning for fluoroscopy-guided procedures the dataset curation and annotation problem must be addressed first. Successfully overcoming this hurdle seems challenging and is reflected in the observation that, despite clear opportunities, only very little work has considered learning in this context.
Contribution
In this manuscript, we describe a hybrid pipeline for fast, physics-based simulation of digitally reconstructed radiographs (DRRs) with corresponding annotations from labeled 3D CT volumes. We extend our DeepDRR framework introduced previously [34] with the capability of simulating tools made from arbitrary materials that may move relative to anatomy defined by the CT volume. The code will be integrated into our open-source repository1 upon publication of this manuscript. Compared to ConvNets trained on naïve DRRs and not considering re-training or domain adaptation, ConvNets trained on DeepDRRs generalize better to unseen clinical data. We demonstrate this on two use cases: (1) viewpoint-invariant detection of anatomical landmark of the pelvis and (2) concurrent segmentation and localization of dexterous robotic end-effectors. In both scenarios, we train the same ConvNet on naïve DRRs and DeepDRRs, respectively, to reveal performance changes resulting solely from realistic modeling of image formation.
Related work
Physics-based simulation of image formation from volumetric data is at the core of the proposed methodology. Traditionally, realistic simulation is achieved using Monte Carlo (MC) simulation during which photons emitted in a specified imaging geometry are traced as they propagate through the medium [2]. Consequently, the distribution of materials within the volume to be imaged must be precisely known, since photon–tissue interaction is highly material dependent. The concept of MC simulation is often used for radiation treatment planning since MC simulation not only yields an image, but also provides highly accurate estimates of tissue dose deposited upon irradiation [27,28]. In addition, it has recently regained some attention for generating medical images in the context of virtual clinical trials [3]. These two examples suggest that MC simulation is capable of producing highly realistic results, but comes at the cost of long computation times that are prohibitive when trying to generate training datasets with hundreds–thousands of images. Our own experiments revealed that accelerated MC simulation [2] on an NVIDIA Titan Xp takes ≈ 4hours for a single X-ray image with 1010 photons. The medical physics community provides strategies for acceleration if prior knowledge on the problem exists. A well-studied example is variance reduction for scatter correction in cone-beam CT, since scatter is of low frequency [30]. Due to these limitations, the use of MC simulation of X-ray images to train machine learning algorithms is not a particularly tractable solution and has, to the best of our knowledge, only been explored by highly specialized groups for scatter estimation [19,38], a task for which no other approach is possible.
The general idea of boosting dataset size by simulating digital radiographs from volumetric CT is fairly obvious and not new, but the challenge is achieving scenarios in which results obtained on synthetic data generalize to clinical cases. Most of the previous works [1,14,15,32] use straightforward ray casting for image formation, which is very fast and allows for the generation of arbitrarily large datasets. Ray casting mimics mono-energetic X-ray sources passing through objects that consist of a single material, an assumption that undoubtedly is violated in clinical X-ray imaging. As a consequence, the synthetic images do not exhibit the characteristic artifacts encountered in practice, such as beam hardening and complex noise characteristics. This shortcoming raises questions regarding performance on real data, with several manuscripts limiting evaluation to synthetic images generated using the same tools [14,32,37] or reporting poor generalizability when applied to clinical data [34,42].
In addressing this problem, unpaired conditional image-to-image style transfer networks [43] constitute the largest competitor to realistic simulation. For digital radiography-based procedures, Zhang et al. show that ConvNets trained on naïvely generated DRRs can be applied to clinical data if the clinically acquired image is first processed with such ConvNet to explicitly mimic the appearance of DRRs [42]. This approach is currently studied across various disciplines [18,25,35] with convincing results; however, it is unclear how well this approach handles new target image styles as this may require a large amount of new target images to retrain the style transfer network.
Methodology
We describe a pipeline for realistic DRR generation from CT volumes that is implemented in Python, PyCUDA, and PyTorch to seamlessly integrate with the tools native to machine learning, and enables fast data generation due to its hybrid design that relies on analytic formulations of projection, attenuation, and noise injection while approximating statistic and heuristic processes via state-of-the-art ConvNets. The pipeline consists of four major modules: (1) decomposition of CT volumes into multiple materials using a deep segmentation ConvNet; (2) material and X-ray source spectrum-aware forward projection using GPU-parallelized ray casting; (3) neural network-based Rayleigh scatter estimation; and (4) quantum and electronic readout noise injection. A schematic overview of the pipeline is provided in Fig. 1 and explained in greater detail in the remainder of this section.
Fig. 1.
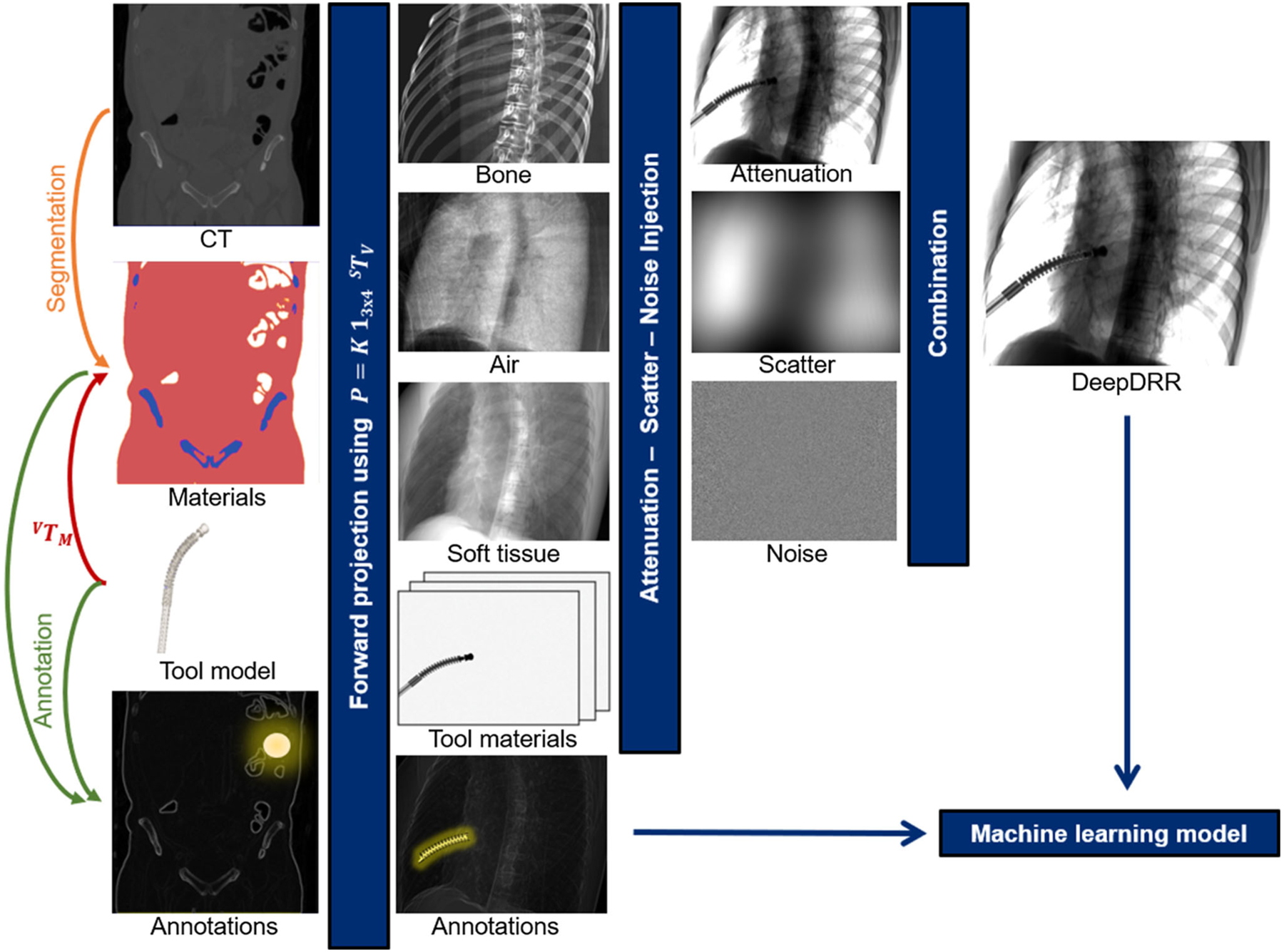
Schematic overview of our realistic simulation pipeline
Physics-based simulation pipeline: DeepDRR
Material decomposition and tool integration
X-ray absorption heavily depends on material properties and photon energy, giving rise to several physical effects such as beam hardening that must be modeled accurately. To this end, we need to derive the 3D distribution of materials from the grayscale CT volume. Traditionally, the task of material decomposition is accomplished by simple thresholding of the CT intensity values. This becomes possible since CT intensities, measured in Hounsfield units (HU), are related to the average linear attenuation coefficient of a particular material and can thus be used to define material characteristic HU ranges [26]. Thresholding works reasonably well for large HU discrepancies, e.g., separating air ([−1000] HU) and bone ([200, 3000] HU), but is prone to failure otherwise, particularly between soft tissue ([−150, 300] HU) and bone in the presence of low mineral density. This is problematic since, despite similar HU, and thus, average linear attenuation, the energy-resolved attenuation characteristic of bone is substantially different from soft tissue [11]. To overcome the limitations of thresholding-based material decomposition, we use a deep volumetric ConvNet adapted from [21] to automatically decompose air, soft tissue, and bone in CT volumes. The ConvNet is of encoder–decoder structure with skip-ahead connections to retain information of high spatial resolution while enabling large receptive fields. For the experiments described here, we use the ConvNet described in [34] that accepts patches with 1283 voxels with voxel sizes of 0.86 × 0.86 × 1.0 mm3 yielding a material map M(x) that assigns a candidate material to each 3D point x. The material decomposition ConvNet was trained (validated) on 10 (2) whole-body CT data from the NIH Cancer Imaging Archive [8] that were manually annotated using ITK Snap [39] using the multi-class Dice loss [21,31] as optimization target for prediction of M = 3 materials, i.e., air, soft tissue, and bone. The network achieved a mis-classification rate of (2.03 ± 3.63)%, a performance that is in agreement with previous studies using similar architectures [12,21,40]. During application, patches of 1283 voxels are fed-forward with stride of 64 since only labels for the central 643 voxels are accepted. After application of the ConvNet to a CT volume of size Nx, Ny, Nz voxels, we have the material map and corresponding material density .
In order to enable the integration of tools, a second volume is defined that fully contains the tool model, e.g., by voxelizing CAD surface models, yielding a map of materials MM(x) with associated material densities ρM(x). The spatial relation of the tool to anatomy is defined by a rigid body transform VTM. When the tool is positioned within anatomy as defined by the CT volume V, CT densities in the overlapping area must be omitted. However, this cannot be achieved by simply setting ρ(x) to zero at appropriate positions, since Mm, ρm can be of different resolutions than the CT volume ρ(x) and arbitrarily oriented relative to the CT coordinate system. Rather, we allocate a third volume MO with densities ρO that is defined by
| (1) |
This will allow compensating for the excess contributions of the overlapping area in projection domain described as follows.
Primary computation via ray casting
Once material maps of the considered materials in CT {air, soft tissue, bone} and materials of tools, e.g., {Nitinol, steel}, are available, the contribution of each material to the total attenuation density at detector position u can be computed. We project in predetermined cone-beam geometry (pin-hole camera model) described by projection matrix , that is defined by a rigid transformation XTV that maps CT volume coordinates to the X-ray source coordinate frame followed by projection onto a detector described by intrinsic parameter matrix K:
| (2) |
Then, using the X-ray spectral density p0(E), the total attenuation is obtained by ray-tracing as
| (3) |
| (4) |
where δ (·, ·) is the Kronecker delta, lu is the 3D ray connecting the source position and 3D location of detector pixel u determined by P, is the material- and energy-dependent linear attenuation coefficient [11], and ρ(x) is the material density at position x that can either be derived from HU values or defined explicitly (e.g., for tools). For a single volume, Eq. 4 can be evaluated in a straightforward manner. For multiple volumes as considered here, however, it is beneficial to first compute the line integral L(u, m, M, ρ) of every volume and material first in order to merge the volumes of different sizes and resolutions in projection domain as discussed above. Then and using the total set of materials and line-integral images of every material , we compute the total contribution per material as
| (5) |
Then, Eq. 3 is evaluated using the overlap-adjusted line integrals
| (6) |
The projection domain image p(u) s then used as basis for scatter prediction.
Learning-based scatter estimation
Traditional scatter estimation relies on variance-reduced MC simulations [30], which requires a complete MC setup. In clinical applications, kernel-based methods are widely used due to their simplicity and have proven to work reasonably well since scatter is of low frequency. Recent approaches to scatter estimation via ConvNets outperform these methods [19] while retaining a low computational demand. In addition, they inherently integrate with deep learning software environments. Unfortunately, our task fundamentally differs from most previous scatter estimation scenarios: In clinical applications, we are interested in separating signal from scatter in an already acquired image. This is in contrast to our image synthesis pipeline that must only rely on clean, high-frequency data to estimate scatter. Considering the very low frequency nature of the scatter signal, kernel-based methods are well justified, as they basically perform low-pass filtering of the joint signal and thus result in a well-separated scatter signal. Solely relying on kernel-based methods in our case would impute that the scatter signal not only is of low frequency, but linearly depends on the frequency content of anatomy, as this corresponds to the information that is extracted via kernel-based low-pass filtering. The success of MC simulations from uncorrected 3D reconstructions [30] affirms that the above does not hold, rendering the direct utilization of kernel-based methods unsuitable in our application.
Nevertheless, the ten-layer ConvNet architecture as presented in [34] is heavily inspired by kernel-based methods but introduces modeling of a nonlinear relationship between the anatomy and scatter signal. The first six layers consist of 11 × 11 kernels arranged in multiple channels to extract basic image features, required for the generation of Rayleigh scatter estimates and the last four layers, with 31×31 kernels and a single channel, ensure smoothness. The network was trained on 330 images generated via MC simulation [2] from whole-body CT volumes, and then augmented by random rotations and reflections. As per [34], the trained scatter estimation network achieved a normalized mean squared error of 9.96%.
Noise injection
After adding scatter, p(u) expresses the energy deposited by a photon in detector pixel u. The number of photons is estimated as:
| (7) |
to obtain the number of registered photons N(u) and perform realistic noise injection. In Eq. 7, N0 (potentially location dependent N0(u), e.g., due to bow-tie filters) is the emitted number of photons per pixel. Noise in X-ray images is a composite of multiple sources, mainly uncorrelated quantum noise due to photon statistics that becomes correlated due to pixel crosstalk, and correlated readout noise [41]. Due to beam hardening, the spectrum arriving at any detector pixel differs. This effect is most prominent at locations with highly attenuating objects in the beam path. To account for this fact in the Poisson noise model, we compute a mean photon energy for each pixel by and estimate quantum noise as , where pPoisson is the Poisson generating function. Since real flat panel detectors suffer from pixel crosstalk, we correlate the quantum noise of neighboring pixels by convolving the noise signal with a blurring kernel [41]. The second major noise component is electronic readout noise. While the magnitude of crosstalk-correlated quantum noise depends on the signal, i.e., the number of photons incident on the detector pixel, electronic noise is signal independent and can be modeled as additive Gaussian noise is correlated along the rows due to the sequential readout of current detectors [41]. After noise injection, we obtain a realistically simulated DRR that we refer to as DeepDRR.
Experimental setup, results, and discussion
Since the DeepDRR pipeline as such was previously evaluated in [34], here we focus on task-based evaluation of the proposed realistic simulation pipeline. To this end, we consider two applications in X-ray-guided surgery: Anatomical landmark detection and concurrent segmentation and localization of snake-like robots that will be described in greater detail in Sects. 3.1 and 3.2, respectively. For both applications, we train ConvNets on synthetic data and then investigate how well the learned models generalize to unseen clinical data without any re-training or domain adaptation. The synthetic data we train on will be generated (1) using DeepDRR the physics-driven approach described here, or (2) using the naïve approach to DRR generation:
| (8) |
a special case of Eq. 6, where p0 represents the total spectral density and is the mean linear attenuation coefficient averaged over all materials and energies. In other words, naïve DRR generation considers a mono-energetic source, objects made from a single material, and neglects image corruption, e.g., due to noise. In addition to these extremal cases, we perform an ablation study for the application of anatomical landmark detection to determine the contributions of ConvNet-based material decomposition and scatter estimation, respectively. For DeepDRR simulation, we used the spectrum of a tungsten anode operated at 120 kV with 4.3 mm aluminum filtration and assumed a high-dose acquisition with N0 = 5 · 105 photons per pixel. While our simulation yields intensity domain images, all ConvNets are trained on line-integral domain data, i.e., after logarithmic transform, to decrease the dynamic range. For images acquired with a real C-arm X-ray system, N0 is not known, and therefore, approximated by the highest intensity value in that image. All real X-ray images of cadaveric specimens used in the experiments were acquired in the International Center of Orthopedic Advancement at Johns Hopkins Bayview Medical Center using a Siemens Cios Fusion C-arm X-ray system (Siemens Healthineers, Forchheim, Germany) equipped with a flat panel detector.
Anatomical landmark detection
Experimental setup
This experiment focuses on detecting anatomical landmarks of the pelvis in radiographs from arbitrary views [6]. In 20 CT scans of the pelvis (split 18-1-1 into training, validation, and testing), we annotate 23 anatomical landmarks (Fig. 2, last column contains all landmarks) and generate naïve and DeepDRRs with corresponding annotations on a spherical segment centered on the anterior–posterior pelvis covering 120° and 90° in RAO/LAO and CRAN/CAUD, respectively. A total of 20,000 DRRs were generated per method, i.e., 1000 per CT volume. Then, a sequential prediction framework [6,36] is trained and, upon convergence, used to predict the 23 anatomical landmarks in the respective testing set and in unseen, real X-ray images of cadaveric lower torso specimens. We trained the network using Adam over 30 epochs with a learning rate of 10−6. We then evaluate the ConvNet on the respective synthetic test set, where exact landmark locations are known, and on 78 real X-ray images of two cadaveric lower torso specimens. To establish reference anatomical landmark positions in real radiographs, we rely on metal beads (BBs) injected into the pelvis prior to CT and radiographic imaging. The BBs were annotated and matched semi-automatically, so that correspondences between BB locations in 3D CT and all X-rays were known. Using this information and for every X-ray image i, we recover projection matrices Pi that map 3D CT points onto the detector. Finally, we manually annotate the 23 target landmarks in the CT scans of both cadaveric specimens and project them via Pi to establish our X-ray image-domain reference for quantitative evaluation on real data. For synthetic data generation, we consider the following ablations:
Fig. 2.
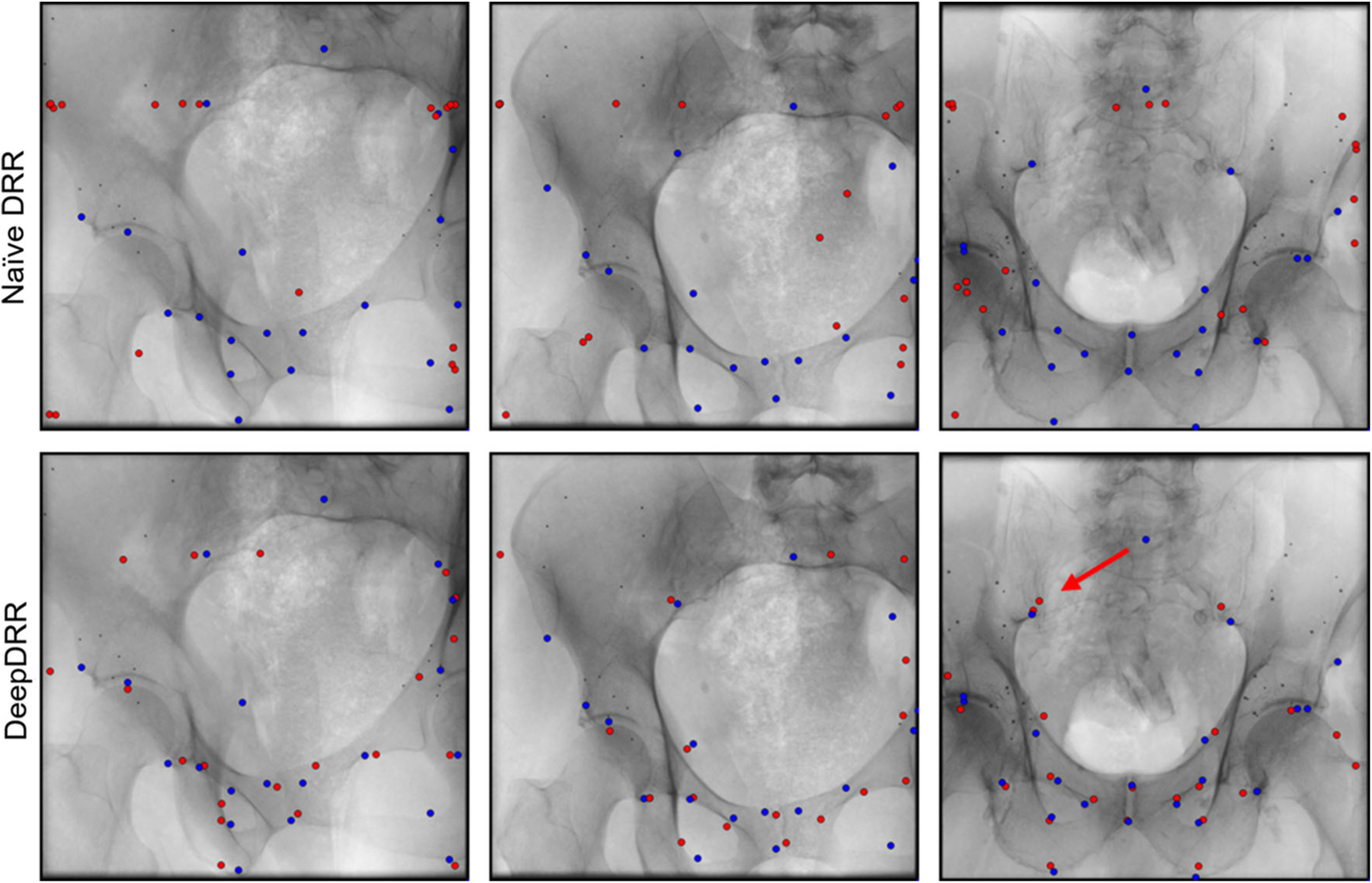
Anatomical landmark predictions on real X-ray images of cadaveric specimen. Reference landmark positions are shown in blue, while ConvNet predictions are shown in red. We highlight a representative outlier using a red arrow that connects a target landmark with the corresponding prediction
Ablation 1 DRRs generated as per Eq. 8: Mono-energetic, single material, no noise, no scatter.
Ablation 2 DRRs generated as per Eq. 6: Poly-chromatic, multi-material, noise injection, no scatter. Material decomposition via thresholding (air < −700 HU, soft tissue ∈ [−700, 250]HU, bone > 250 HU [26])
Ablation 3 DRRs generated as per Eq. 6: Poly-chromatic, multi-material, noise injection, no scatter. Material decomposition via ConvNet (Sect. 2.1)
Ablation 4 DRRs generated as per Eq. 6: Poly-chromatic, multi-material, noise injection, scatter. Material decomposition and scatter estimation via ConvNets (Sect. 2.1)
Results and discussion
The quantitative results of the ablation study are summarized in Table 1. In every ablation, ConvNets perform similarly well when applied to the respective synthetic test sets. However, when applied to real data we observe substantial improvements in landmark detection accuracy as the DRR simulation pipeline more accurately models X-ray image formation. From Table 1, it becomes apparent that the improvement is most notable when transitioning from Ablation 1 to Ablation 2, i.e., when considering the material dependence and energy dependence of photon attenuation. This is likely due to the fact that Ablation 2 introduces beam hardening, which increases bone to soft tissue contrast, substantially altering image appearance. Accuracy is further improved slightly as threshold-based material decomposition is replaced by the proposed ConvNet-based decomposition (Ablation 3). This improvement can partially be attributed to the capability of the ConvNet to use structural information to disambiguate soft tissue in the [−150, 300] HU interval and trabecular bone > 200 HU, thus yielding more realistic X-ray image appearance. However, we noticed that the poor performance in Ablation 2 is affected by catastrophic failure cases in 8 of the 78 real X-ray images. Omitting these from analysis would reduce this error to 27.3±27.0 mm. However, while the addition of scatter (Ablation 4, the full DeepDRR pipeline) further improves detection accuracy, this improvement is not significant. This observation is further discussed in Sect. 3.3. Our analysis suggests that, while learning-based material decomposition and scatter estimation can further improve detection accuracy, the significant boost in performance (p < 0.01) is to be attributed to poly-energetic and multi-material-based modeling of image formation, and is less sensitive to the method used for material decomposition. In light of the results of our ablation study, we limit all subsequent analysis to the two extreme cases Ablation 1 and Ablation 4, i.e., naïe DRRs and DeepDRRs, respectively. A qualitative analysis of landmark predictions versus true reference locations is presented in Fig. 2. For landmark prediction on real data, we observe both, compromised performance and a very high standard deviation that is partly due to outliers. Considering this decrease in performance, it is unclear whether directly interpreting the detected landmarks is clinically practical. However, the amount of outliers can, in principle, be reduced substantially by only considering landmarks that are detected with a certain belief [5,6], yielding landmark detection accuracy that is appropriate for initializing 2D/3D registration. While this approach proved feasible for the DeepDRR-based ConvNet, using a threshold above 20% would reduce the accepted detections in the naïve DRR-based ConvNet close to zero, impeding meaningful validation. It is worth mentioning that reference targets used for quantitative assessment are likely imperfect since (1) anatomical landmarks in 3D CT are defined manually, and (2) Pi are estimated from data. While this observation challenges the overall magnitude of error reported on real data, comparisons remain valid since both methods are affected in the same way.
Table 1.
Ablation results of anatomical landmark detection on synthetic and real data
| Ablation 1 | Ablation 2 | |||
|---|---|---|---|---|
| Synthetic | Real | Synthetic | Real | |
| Mean L2 | 6.70 ± 4.32 | 78.2 ± 26.5 | 6.09 ± 4.19 | 34.4 ± 41.9 |
| Ablation 3 | Ablation 4 | |||
| Synthetic | Real | Synthetic | Real | |
| Mean L2 | 6.60 ± 5.44 | 26.9 ± 15.1 | 6.35 ± 5.02 | 24.7 ± 21.3 |
All results are stated in mm on the detector. Ablation 4 corresponds to the full DeepDRR pipeline
Robotic end-effector localization
Experimental setup
This experiment builds upon our previous work on detecting continuous dexterous manipulators in radiographs [10]. An encoder–decoder-like architecture with skip connections is trained to concurrently segment and localize a snake-like robot end-effector in X-ray images. Localization in this context refers to predicting the location of two keypoints on the end-effector, namely the start and end point of its centerline. In the decoding stage, the final feature layer is shared across the segmentation and localization branch, and the final segmentation mask is backward concatenated to this shared feature to boost keypoint detection. The robot end-effector consists of body and base made from Nitinol, and a shaft and inserted drilling tool made from stainless steel. In our simulation, the shape of the snake-like body is controlled by adjusting the control points of a cubic spline that describes the snake’s centerline. Once a body shape is defined, the tool model is voxelized with high resolution to preserve details. We use a total of 5 CT volumes (4 training, 1 test) and, for each of the 10 femurs, manually define VTM such that the robot is enclosed in bone. Then, fluoroscopic images are generated by randomly sampling the snake shape by adjusting the control point angles ∈ [−7.9°, 7.9°], and the C-arm pose relative to scene by adjusting the source-to-isocenter distance ∈ [400 mm, 500 mm], the source rotation (LAO/RAO ∈ [0°, 360°] and CRAN/CAUD ∈ [75°, 105°]), and translation along all three axes ∈ [−20, 20]mm. For every femur, 1000 configurations were sampled randomly and for each configuration a DeepDRR, a naïve DRR, and the corresponding segmentation mask and keypoint locations were generated. The ConvNet was then trained using Dice loss for the segmentation task and the standard L2 loss for the localization task. Learning rate started at 10−3 and decayed by 10−1 every 10 epochs. For quantitative evaluation on real data, we manually annotated 87 X-ray images of a dexterous robot [22] drilling in femoral bone specimens.
Results and discussion
The results are summarized in Table 2. ConvNets trained on naïve and DeepDRRs performed similarly well on their respective synthetic testing data. However, when considering cadaveric data from a real C-arm system, the ConvNet trained on DeepDRRs substantially outperformed the network trained on naïve DRRs in the segmentation task (p < 0.01) and, particularly, in the keypoint detection task (p ≪ 0.01). Representative results on synthetic and real data are shown in Figs. 3 and 4, respectively. While the naïve DRR ConvNet performed comparably on some real images (cf. Fig. 4, bottom row), it was prone to over-segmentation reducing the overall Dice score (cf. Fig. 4, middle row). In addition, we observed that, when applied to real data, the network would often detect the same landmark twice giving rise to the large L2 error for this task (cf. Fig. 4, top row). The cadaveric dataset contains scenarios not observed during training, e.g., the end-effector being fully in air before insertion into femoral bone, causing deteriorated performance which is reflected in the high standard deviation for both, ConvNets trained on naïve and DeepDRRs.
Table 2.
Segmentation and localization accuracy in %-dice and in mm, respectively
| Naïve DRR | DeepDRR | |||
|---|---|---|---|---|
| Synthetic | Real | Synthetic | Real | |
| Segmentation | ||||
| Mean dice | 99.3 ± 0.3 | 88.7 ± 7.7 | 99.6 ± 0.1 | 91.5 ± 6.3 |
| Median dice | 99.3 | 91.7 | 99.6 | 93.8 |
| Localization | ||||
| Mean L2 | 0.43 ± 0.36 | 26 ± 19 | 0.37 ± 0.35 | 2.5 ± 1.0 |
| Median L2 | 0.37 | 32 | 0.31 | 2.8 |
Fig. 3.

Representative results of concurrent segmentation and keypoint detection [10] on synthetic X-ray images generated by the same method the ConvNets were trained on. Segmentations are shown in green, keypoints as red heatmaps, and magnifications of the region of interest are shown in the top right corner of each image, delimited by an orange border
Fig. 4.

Representative results of concurrent segmentation and keypoint detection [10] on real X-ray images of cadaveric anatomy of ConvNets trained on naïe DRRs and DeepDRRs, respectively. We also show the input X-ray image after logarithm transform and reference annotations is obtained manually. Segmentation results are shown as green overlay, while keypoint predictions are shown as red heatmaps. Magnifications of a region of interest around the snake end-effector are provided in the top right corner of every image, marked-off by an orange border
Challenges and limitations
While DeepDRR-based ConvNets significantly outperformed their counterpart trained on naïve DRRs in all studied cases, we observed compromised performance on real data compared to the synthetic testing data. This degradation results from multiple factors: (1) The reference annotations for evaluation on real data are obtained using semi-automatic techniques, suggesting that the targets used for quantitative assessment may not be the true targets. While this shortcoming slightly compromises synthetic–real comparisons, they do not affect comparisons of real data performance between naïve- and DeepDRR-based ConvNets since for such comparison the above error is systematic. (2) The images provided by the clinical C-arm X-ray system are not, in fact, raw images but are pre-processed by a vendor-specific, proprietary filtering pipeline (including but not limited to noise reduction and scatter correction) that can substantially alter image characteristics. As a consequence, the DeepDRR framework cannot accurately mimic image formation since part of the process is unknown. (3) DRRs are generated from 3D CT volumes that exhibit finite resolution. This shortcoming is emphasized during DRR generation due to magnification in cone-beam geometry and detectors with very high spatial resolution down to . An interesting next step would be to investigate whether super-resolution strategies, as they have become popular in MRI [7], could mitigate this problem. It is worth mentioning that the proposed DeepDRR pipeline could be formulated as an end-to-end neural network, since forward projection can be easily incorporated as a layer [37]. This is appealing, since it would allow incorporation of all ConvNet-based processing, e.g., for 3D segmentation and super-resolution, or 2D pre-processing, and enable end-to-end training of these networks by maximizing the resulting DRRs’ similarity with real X-ray images of the target application via adversarial training.
It is interesting to see that the degradation observed here occurs to a similar extent in the most similar work to ours that uses unpaired image-to-image style transfer to convert real X-ray images to a DRR-like counterpart [42]. On the example of anatomical landmark detection, where we now have a solid basis for evaluation, we plan on investigating how the approaches compare and whether combining both strategies can yield real data performance that is on a par with the performance on synthetic data. Doing so will require us to collect a sufficiently large database of clinical pelvic X-ray images, ideally acquired without tools in the image. While these images would not need to be annotated for style transfer, the dataset must still be representative with respect to anatomy and views thereof for style transfer to succeed. Since intra-operative X-rays are not commonly archived in pelvis surgery, curating this database requires a prospective study design which we will implement in future work.
Currently, simulation using DeepDRR is limited to flat panel detectors. Supporting image intensifiers is possible but would require adjustment of noise model and incorporation of distortions that occur in this detector technology.
In contrast to MC simulation where the rendering time is dependent on the number of photons injected, simulation using DeepDRR requires the same computation time irrespective of the required imaging parameters. It is worth mentioning, however, that material decomposition using the 3D ConvNet is comparably slow and is performed before every rendering. For practical application of the DeepDRR pipeline, this suggests that a large set of projections should be defined first and then submitted for rendering to reduce the per-image computation time. As an example, for 1000 images with 620 × 480 px, the simulation per image took 0.56s on an Nvidia Quadro P6000.
Conclusion
We proposed a fast and realistic framework for simulating intra-operative X-ray images with corresponding annotations from 3D CT volumes. The framework supports modeling of tools or surgical implants and is publicly available and open source. We evaluated our framework on two complementary tasks in X-ray-guided interventions, namely detecting anatomical landmark on the pelvis and segmenting and localizing robotic end-effectors during drilling in the femur. ConvNets trained on DeepDRRs, generated with our physics-based framework, significantly outperformed networks trained on naïve DRRs on real X-ray images, suggesting that realistic generation of X-ray image formation is a practicable way to establishing machine learning in fluoroscopic image analysis. While our results are promising, in future work we will investigate strategies to further improve the real data performance of ConvNets trained on synthetic data, e.g., via image style transfer for domain adaptation. Once introduced into clinical practice, learning-based algorithms that analyze intra-operative images to automatically extract semantic or contextual information, e.g., by measuring objects of interest, have the potential to radically improve the state-of-care by simplifying surgical workflows.
Acknowledgements
We gratefully acknowledge the support of R21 EB020113, R01 EB016703, R01 EB0223939, and the NVIDIA Corporation with the donation of the GPUs used for this research.
Footnotes
Publisher's Disclaimer: Disclaimer The concepts and information presented in this paper are based on research and are not commercially available.
Conflict of interest The authors have no conflict of interest to declare.
Informed consent This article does not contain patient data.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Available at https://github.com/mathiasunberath/DeepDRR.
References
- 1.Albarqouni S, Fotouhi J, Navab N (2017) X-ray in-depth decomposition: revealing the latent structures In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 444–452 [Google Scholar]
- 2.Badal A, Badano A (2009) Accelerating Monte Carlo simulations of photon transport in a voxelized geometry using a massively parallel graphics processing unit. Med Phys 36(11):4878–4880 [DOI] [PubMed] [Google Scholar]
- 3.Bakic PR, Myers KJ, Glick SJ, Maidment AD (2016) Virtual tools for the evaluation of breast imaging: state-of-the science and future directions In: International workshop on digital mammography. Springer, Berlin, pp 518–524 [Google Scholar]
- 4.Baumgartner R, Libuit K, Ren D, Bakr O, Singh N, Kandemir U, Marmor MT, Morshed S (2016) Reduction of radiation exposure from C-arm fluoroscopy during orthopaedic trauma operations with introduction of real-time dosimetry. J Orthop Trauma 3(2):e53–e58 [DOI] [PubMed] [Google Scholar]
- 5.Bier B, Goldmann F, Zaech JN, Fotouhi J, Hegeman R, Grupp R, Armand M, Osgood G, Navab N, Maier A, Unberath M (2019) Learning to detect anatomical landmarks of the pelvis in X-rays from arbitrary views. Int J Comput Assisted Radiol Surg 20:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bier B, Unberath M, Zaech JN, Fotouhi J, Armand M, Osgood G, Navab N, Maier A (2018) X-ray-transform invariant anatomical landmark detection for pelvic trauma surgery In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin [Google Scholar]
- 7.Chen Y, Shi F, Christodoulou AG, Xie Y, Zhou Z, Li D (2018) Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 91–99 [Google Scholar]
- 8.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L (2013) The cancer imaging archive (tcia): maintaining and operating a public information repository. J Dig Imaging 26(6):1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Silva T, Punnoose J, Uneri A, Goerres J, Jacobson M, Ketcha MD, Manbachi A, Vogt S, Kleinszig G, Khanna AJ, Wolinsky JP, Osgood G, Siewerdsen J (2017) C-arm positioning using virtual fluoroscopy for image-guided surgery In: Webster RJ III, Fei B (eds) Medical imaging 2017: image-guided procedures, robotic interventions, and modeling, vol 10135. International Society for Optics and Photonics, Bellingham, p 101352K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C, Unberath M, Taylor R, Armand M (2019) Localizing dexterous surgical tools in X-ray for image-based navigation. arXiv preprint [Google Scholar]
- 11.Hubbell JH, Seltzer SM (1995) Tables of X-ray mass attenuation coefficients and mass energy-absorption coefficients 1 keV to 20 MeV for elements Z = 1 to 92 and 48 additional substances of dosimetric interest. Technical report, National Institute of Standards and Technology [Google Scholar]
- 12.Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78 [DOI] [PubMed] [Google Scholar]
- 13.Kooi T, Litjens G, van Ginneken B, Gubern-Mérida A, Sánchez CI, Mann R, den Heeten A, Karssemeijer N (2017) Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 35:303–312 [DOI] [PubMed] [Google Scholar]
- 14.Kügler D, Stefanov A, Mukhopadhyay A (2018) i3posnet: instrument pose estimation from X-ray. arXiv preprint arXiv:1802.09575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Liang W, Zhang Y, An H, Tan J (2016) Automatic lumbar vertebrae detection based on feature fusion deep learning for partial occluded C-arm X-ray images In: 2016 IEEE 38th annual international conference of the engineering in medicine and biology society (EMBC). IEEE, pp 647–650 [DOI] [PubMed] [Google Scholar]
- 16.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88 [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Sinha A, Unberath M, Ishii M, Hager GD, Taylor RH, Reiter A (2018) Self-supervised learning for dense depth estimation in monocular endoscopy In: OR 2.0 context-aware operating theaters, computer assisted robotic endoscopy, clinical image-based procedures, and skin image analysis. Springer, Berlin, pp 128–138 [Google Scholar]
- 18.Mahmood F, Durr NJ (2018) Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med Image Anal 48:230. [DOI] [PubMed] [Google Scholar]
- 19.Maier J, Berker Y, Sawall S, Kachelrieß M (2018) Deep scatter estimation (DSE): feasibility of using a deep convolutional neural network for real-time X-ray scatter prediction in cone-beam CT In: Medical imaging 2018: physics of medical imaging, vol 10573. International Society for Optics and Photonics, p 105731L [Google Scholar]
- 20.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber M, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin H, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Leemput KV (2015) The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans Med Imaging 34(10):1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation In: 2016 fourth international conference on 3D vision (3DV). IEEE, pp 565–571 [Google Scholar]
- 22.Murphy RJ, Kutzer MD, Segreti SM, Lucas BC, Armand M (2014) Design and kinematic characterization of a surgical manipulator with a focus on treating osteolysis. Robotica 32(6):835–850 [Google Scholar]
- 23.Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 234–241 [Google Scholar]
- 24.Roy AG, Conjeti S, Sheet D, Katouzian A, Navab N, Wachinger C (2017) Error corrective boosting for learning fully convolutional networks with limited data In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 231–239 [Google Scholar]
- 25.Sankaranarayanan S, Balaji Y, Castillo CD, Chellappa R (2017) Generate to adapt: aligning domains using generative adversarial networks. ArXiv e-prints arXiv:1704.01705 [Google Scholar]
- 26.Schneider W, Bortfeld T, Schlegel W (2000) Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys Med Biol 45(2):459. [DOI] [PubMed] [Google Scholar]
- 27.Sempau J, Wilderman SJ, Bielajew AF (2000) DPM, a fast, accurate Monte Carlo code optimized for photon and electron radiotherapy treatment planning dose calculations. Phys Med Biol 45(8):2263. [DOI] [PubMed] [Google Scholar]
- 28.Sharma S, Kapadia A, Abadi E, Fu W, Segars WP, Samei E (2018) A rapid GPU-based Monte-Carlo simulation tool for individualized dose estimations in CT In: Medical imaging 2018: physics of medical imaging, vol 10573. International Society for Optics and Photonics, Bellingham, p 105733V [Google Scholar]
- 29.Shen D, Wu G, Suk HI (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sisniega A, Zbijewski W, Badal A, Kyprianou I, Stayman J, Vaquero JJ, Siewerdsen J (2013) Monte Carlo study of the effects of system geometry and antiscatter grids on cone-beam CT scatter distributions. Med Phys 40(5):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudre CH, Li W, Vercauteren T, Ourselin S, Cardoso MJ (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Berlin, pp 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terunuma T, Tokui A, Sakae T (2018) Novel real-time tumor-contouring method using deep learning to prevent mistracking in X-ray fluoroscopy. Radiol Phys Technol 11(1):43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unberath M, Fotouhi J, Hajek J, Maier A, Osgood G, Taylor R, Armand M, Navab N (2018) Augmented reality-based feedback for technician-in-the-loop C-arm repositioning. Healthc Technol Lett 5(5):143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unberath M, Zaech JN, Lee SC, Bier B, Fotouhi J, Armand M, Navab N (2018) Deepdrr—a catalyst for machine learning in fluoroscopy-guided procedures In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin [Google Scholar]
- 35.Visentini-Scarzanella M, Sugiura T, Kaneko T, Koto S (2017) Deep monocular 3D reconstruction for assisted navigation in bronchoscopy. Int J Comput Assisted Radiol Surg 12(7):1089–1099 [DOI] [PubMed] [Google Scholar]
- 36.Wei SE, Ramakrishna V, Kanade T, Sheikh Y (2016) Convolutional pose machines. In: CVPR, pp 4724–4732 [Google Scholar]
- 37.Würfl T, Hoffmann M, Christlein V, Breininger K, Huang Y, Unberath M, Maier AK (2018) Deep learning computed tomography: learning projection-domain weights from image domain in limited angle problems. IEEE Trans Med Imaging 37(6):1454–1463 [DOI] [PubMed] [Google Scholar]
- 38.Xu S, Prinsen P, Wiegert J, Manjeshwar R (2017) Deep residual learning in CT physics: scatter correction for spectral CT. arXiv preprint arXiv:1708.04151 [Google Scholar]
- 39.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128 [DOI] [PubMed] [Google Scholar]
- 40.Zeng G, Yang X, Li J, Yu L, Heng PA, Zheng G (2017) 3D U-net with multi-level deep supervision: fully automatic segmentation of proximal femur in 3D MR images In: International workshop on machine learning in medical imaging, pp 274–282. Springer, Berlin [Google Scholar]
- 41.Zhang H, Ouyang L, Ma J, Huang J, Chen W, Wang J (2014) Noise correlation in CBCT projection data and its application for noise reduction in low-dose CBCT. Med Phys 41(3):031906. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Miao S, Mansi T, Liao R (2018) Task driven generative modeling for unsupervised domain adaptation: application to X-ray image segmentation In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin [Google Scholar]
- 43.Zhu JY, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. arXiv preprint [Google Scholar]


