Abstract
Mitochondrial double-stranded RNA (mtdsRNA) and its innate immune responses have been reported previously; however, mtdsRNA generation and its effects on alcoholic liver disease (ALD) remain unclear. Here, we report that hepatic mtdsRNA stimulates toll-like receptor 3 (TLR3) in Kupffer cells through the exosome to enhance IL-17A production in ALD. Following binge ethanol drinking, IL-17A production primarily increased in γδ T cells of wild type (WT) mice, whereas the production of IL-17A was mainly facilitated by CD4+ T cells in acute on chronic ethanol consumption. These were not observed in TLR3 KO or Kupffer cell-depleted WT mice. The expression of PNPase, an mtdsRNA restricting enzyme, was significantly decreased in ethanol-exposed livers and hepatocytes of WT mice. Immunostaining revealed that mtdsRNA co-localized with the mitochondria in ethanol-treated hepatocytes from WT mice and healthy humans. Bioanalyzer analysis revealed that small-sized RNAs were enriched in ethanol-treated exosomes (EtOH-Exo) rather than EtOH-treated microvesicles in hepatocytes of WT mice and humans. qPCR and RNA sequencing analyses indicated that mRNA expression of mitochondrial genes encoded by heavy and light strands was robustly increased in EtOH-Exo from mice and humans. After direct treatment with EtOH-Exo, IL-1β expression was significantly increased in WT Kupffer cells but not in TLR3 KO Kupffer cells, augmenting IL-17A production of γδ T cells in mice and humans.
Conclusion:
Ethanol-mediated generation of mtdsRNA contributes to TLR3 activation in Kupffer cells through exosomal delivery. Consequently, increased IL-1β expression in Kupffer cells triggers IL-17A production in γδ T cells at the early stage that may accelerate IL-17A expression in CD4+ T cells in the later stage of ALD. Therefore, mtdsRNA and TLR3 may function as novel therapeutic targets in ALD.
Keywords: extracellular vesicle, γδ T cell, interleukin-1β, Kupffer cell, microvesicle
Alcoholic liver disease (ALD) results from multiple inflammatory responses mediated by self or non-self ligands (e.g., damage- and pathogen-associated molecular patterns; DAMPs and PAMPs) through toll-like receptors (TLRs) or the inflammasome, in which damaged and stimulated hepatocytes, non-parenchymal cells, and immune cells release various inflammatory mediators such as cytokines, chemokines, and extracellular vesicles (EVs).(1, 2)
EVs play important roles in intercellular communication by delivering diverse cargos including mRNA, microRNA (miRNA), DNA, proteins, and lipids.(3, 4) Depending on their size and biogenesis, they are classified into three types that include exosome (50-100 nm), microvesicle (100-1000 nm) or apoptotic body (500-2000 nm). EVs transfer their cargos to neighboring or target cells via receptor-ligand interactions, fusion, phagocytosis, or endocytosis. Recently, lines of evidence indicate that EVs released from hepatocytes after ethanol exposure play a substantial role in the inflammatory responses of ALD.(5–8) Following acute binge or chronic ethanol exposure in mice, rats and humans, damaged hepatocytes release EVs by cytochrome P450 2E1 (CYP2E1)-mediated reactive oxygen species (ROS) or activated caspase-3-depedent mechanisms.(5, 6) Their cargos including miRNA, mitochondrial DNA (mtDNA), CD40 ligand (CD40L), and heat shock protein 90, increase the production of inflammatory cytokines such as interleukin (IL)-1β in macrophages/Kupffer cells and neutrophils.(5–10) In addition to the IL-1β production by macrophages that is important in the context of ALD, EVs may also stimulate or augment IL-17 production of hepatic γδ T cells, as these cells produce innate IL-17 in response to IL-1β and IL-23 stimulation without T cell receptor engagement.(11, 12) It has not been reported if EV-mediated IL-1β production by macrophage/Kupffer cells stimulates innate IL-17 production by γδ T cells. Additionally, the mechanism underlying this process remain unknown.
Ethanol metabolism increases ROS production primarily in the mitochondria of hepatocytes, where mtDNA may be a major target of ethanol-induced ROS, ultimately promoting depletion of mtDNA and cell death by apoptosis.(13, 14) Interesting studies have suggested that mtDNA-enriched EVs, such as mitochondrial DAMPs, induced by ethanol increase inflammation through TLR9-mediated neutrophilia(10) and that oxidized mtDNA plays critical roles in mediating NLRP3 inflammasome activation in macrophages.(15) TLR9-deficient mice reduce chronic and binge ethanol feeding-mediated liver injury by decreasing neutrophil infiltration in a manner that is dependent on TLR9 expression of hepatocytes and hepatic stellate cells (HSCs), but not Kupffer cells, indicating a partial involvement of Kupffer cells in neutrophil recruitment.(16) In addition to mtDNA, recent studies have reported that the process of bidirectional transcription of mtDNA generates mitochondrial double-stranded RNA (mtdsRNA), but it undergoes rapid decay by the RNA degradosome to prevent deleterious function of mtdsRNA through immunogenic receptors including TLRs.(17, 18). The key enzymes of the degradosome are mitochondrial RNA helicase SUV3 and polynucleotide phosphorylase (PNPase) that function to restrict the levels of mtdsRNA,(17) and the loss of either enzyme results in a large accumulation of mtdsRNA that escapes into the cytoplasm in a PNPase-dependent manner.(18) However, whether ethanol metabolic processes within the mitochondria could affect the expression and cytosolic appearance of mtdsRNA have not been investigated, leading to the transfer of mtdsRNA into EVs.
Previously, dsRNA of natural origin (viral, nuclear and ribosomal) has been indicated as a potential inhibitor of protein synthesis and a modest inducers of interferons;(19) however, polyinosinic-polycytidylic acid (poly I:C), a synthetic dsRNA, efficiently induces interferons and does not inhibit protein synthesis,(20) suggesting differential interactions and effects of various dsRNA at specific subcellular locations. Currently, three types of pattern recognition receptors including retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5) and TLR3, are thought to play key roles in the host defense against dsRNA viral infection.(21, 22 Extracellular dsRNA are internalized by cells through clathrin-dependent endocytosis, in which TLR3 can recognizes dsRNA (>40 bp) in the endosome or internalized dsRNA exits from the endosome to the cytosol via a mammalian dsRNA transporter systemic RNA interference defective protein-1 transmembrane family member 2, SIDT2.(22) In the cytoplasm, RIG-I recognizes short dsRNA with 5’-triphosphate end generated by viral polymerase, while MDA5 detects long dsRNA (>1,000 bp).(21, 22) Although the activation of RNA sensors is strictly regulated to avoid autoimmunity, various self-RNAs including mtdsRNA, mRNA, and non-coding RNA activate MDA5 and TLR3.(12, 18, 23, 24) Here, we investigated ethanol-mediated mtdsRNA generation, enrichment of mtdsRNA within EVs, mtdsRNA-mediated TLR3 activation in Kupffer cells, and the impacts of these processes on innate IL-17 production by γδ T cells in ALD.
Materials and Methods
ANIMALS
Male C57BL/6 and TLR3 knockout (TLR3 KO) mice (C57BL/6 background) were purchased from The Jackson Laboratory (Bar Harbor, ME), and maintained in a specific pathogen-free facility (Bio Model System Park; KAIST, Daejeon, Korea). Chimeric mice were generated by reciprocal bone marrow transplantation as described previously.(12) All animals received humane care in accordance with the “Guide for the Care and Use of Laboratory Animals”, published by the National Institutes of Health, and all animal experiments were approved by the KAIST Institutional Animal Care and Use Committee.
INDUCTION OF ETHANOL-INDUCED LIVER INJURY
For singe binge ethanol drinking model, 4 g/kg ethanol was administered to mice by oral gavage and the mice were sacrificed at several time points. As previously reported,(25) to induce acute-on-chronic liver injury, mice were fed a 4.5% Lieber-DeCarli ethanol liquid diet (Dyets Inc., Bethlehem, PA) for 10 days and 4 g/kg ethanol was administered to mice by oral gavage and the mice were sacrificed at 6 hour after last oral gavage. For depletion of Kupffer cells, 10 μl/g clodronate liposomes (Haarlem, Netherlands) were injected intraperitoneally in each mouse. Depletion was confirmed at 2-day after the injection by FACS analyses.
ISOLATION OF PRIMARY HEPATIC CELLS
HSCs, Kupffer cells and hepatocytes of mice were isolated by collagenase perfusion followed by differential centrifugation on an Opti-Prep (Sigma, St. Louis, MO) density gradient as described previously.(26) For human cells, we obtained small sized-healthy liver during liver resection from the department of surgery and the department of internal medicine, Chungnam National University Hospital (Daejeon, South Korea) and collected human primary cells through liver perfusion. More detailed procedures were described in the supporting documents. Authorization for the use of these tissues for research purposes was obtained from the Institutional Review Board of Chungnam National University Hospital (IRB number: 2016-03-02-003), Daejeon, Korea. Informed consents were received from the entire patients who had provided the tissue.
ISOLATION OF EXTRACELLULAR VESICLES
To collect EVs from ethanol-treated hepatocytes (100 mM, 24 hours), we established the differential centrifugation methods. Briefly, collected supernatants were centrifuged at 300 g for 10 minutes and carefully collect the supernatant except the pellet and it was further centrifuge at 2,000 g for 12 minutes to remove the apoptotic bodies. Then the supernatant was carefully centrifuge at 15,000 g for 30 minutes to collect microvesicles. Finally, exosomes were isolated from the collected supernatants using ExoQuick-TC™ exosome precipitation solution (System Biosciences Inc., Mountain View, CA). The fractions of microvesicles and exosomes were confirmed and counted using a Dynamic Light Scattering System and a Nanosight Nanoparticle Tracking Analysis System (Malvern Instruments Ltd, Worcestershire, UK), respectively.
BIOANALYZER ANALYSIS
Total RNA was extracted by TRIzol reagent (ThermoFisher Scientific, Waltham, MA, USA) from extracellular vesicles. After quantification of extracted total RNA by using NANODROP LITE spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA), RNA Nano Chips for use with the Agilent 2100 Bioanalyzer System was used for the analysis of RNA (Agilent Technologies) according to the manufacturer’s instruction.
DOUBLE-STRANDED RNA STAINING
Fixed cells with 4 % paraformaldehyde were washed and added with 0.5% Triton X-100 for 15 minutes at room temperature (RT). After 3 washes with 0.05% phosphate buffered saline Tween-20, blocking was performed with 5% goat serum at RT for 1 hour. Primary antibodies anti-dsRNA (#10010500, J2 monoclonal antibody, mouse, IgG2a, kappa chain, SCICONS) were incubated for 3 hours at 37 °C. Cells were washed three times and then incubated with secondary donkey anti-mouse IgG conjugated with Alexa Fluor 488 (#ab150109, Abcam, Cambridge, UK) at (1:300) concentration at RT for 1 hour. Primary hepatocytes were incubated with MitoTracker Deep Red (M22426, Invitrogen, 100 nM) for 30 minutes at 37 °C before fixing. More detailed procedures were described in the supporting documents.
EXOSOMAL RNA ANALYSIS
Total exosomal RNA was isolated using TRIzol according to manufacturer’s instructions. cDNA was generated from 0.05 μg of RNA using random hexamers and reverse transcriptase (Maxima, Thermo Scientific). For amplifying transcripts originated from mouse and human mitochondrial genome, the primers are designed and provided in Supporting Tables S1 and S2. The relative quantities of mitochondrial RNAs were calculated using the ΔΔCt method without normalization. For high throughput RNA-sequencing, we followed standard protocol of Illumina sequencing using TruSeq Stranded Total RNA kit with Ribo-Zero (Part#15031048 Rev. E) and NovaSeq6000 S4 (150bp PE). Analyses were performed on two paired-ends samples with 3.1 Gb Raw data/2.6 Gb Trimmed data for Control sample and 3.4 Gb/2.8 Gb for EtOH-treated sample. The Q30 percentage (% of bases with quality over phred score 30) of raw and trimmed data are 93%/95% and 92%/95% respectively. Trimmed reads are mapped to reference genome (HG19 with chrM transcripts)(27) with HISAT2 resulting in 27% and 44% mapping radio. After the read mapping, Stringtie was utilized for transcript assembly. Expression profile was calculated for each sample and transcript/gene as read count and FPKM (Fragment per Kilobase of transcript per Million mapped reads). (Supporting Table S3).
STATISTICAL ANALYSIS
Data are presented as the means ± SEM. To compare values obtained from two groups, Student’s t test or one-way analysis of variance was performed and one way ANOVA with Dunnett’s test for multiple comparison vs control. A value of P < 0.001, 0.01 or 0.05 was considered statistically significant.
Other detailed materials and methods are described in the supporting information.
Results
KUPFFER CELLS CONTRIBUTE TO IL-17A PRODUCTION IN γδ T CELLS IN ACUTE ALCOHOLIC LIVER INJURY
Serum and FACS analyses revealed that liver injury and the frequency of IL-17A producing lymphocytes peaked at 6 hour after single binge ethanol feeding in wild type (WT) mice (Fig. 1A,B). Intracellular cytokine staining indicated that hepatic γδ T cells were the major producers of IL-17A, although the population of these cells was relatively small compared to that of CD4+ and CD8+ T cells (Fig. 1B). In qRT-PCR analysis, Il1b and Il23a mRNA expression significantly increased only in freshly isolated Kupffer cells from binge ethanol-fed mouse liver compared to that of control mouse liver; however, this increase was not observed in isolated HSCs (Fig. 1C). Ccl20 mRNA expression was slightly decreased but still higher in Kupffer cells than that of HSCs (Fig. 1C). Additionally, Il17 mRNA expression was not increased in freshly isolated Kupffer cells and HSCs after binge ethanol consumption (Supporting Fig. 1A). These data suggest that Kupffer cells may be the major activator of IL-17A production in γδ T cells. To confirm this hypothesis, WT mice were administered a single binge ethanol dose after Kupffer cells were depleted using clodronate (Supporting Fig. 1B,C). Although liver injury was not different after binge ethanol drinking, IL-17A producing lymphocytes were significantly decreased in mice treated with clodronate compared to mice without clodronate treatment (Fig. 1D; Supporting Fig. 1D). Interestingly, Kupffer cell depletion induced a remarkable decrease of IL-17A production in γδ T cells (Fig. 1E,F and Supporting Fig. 1E). Collectively, these data suggested that Kupffer cells contributed to IL-17A production in hepatic γδ T cells by increasing IL-1β expression in response to acute alcoholic liver injury.
Fig 1. Kupffer cells contribute to increased IL-17A production of γδ T cells in acute alcoholic liver injury.
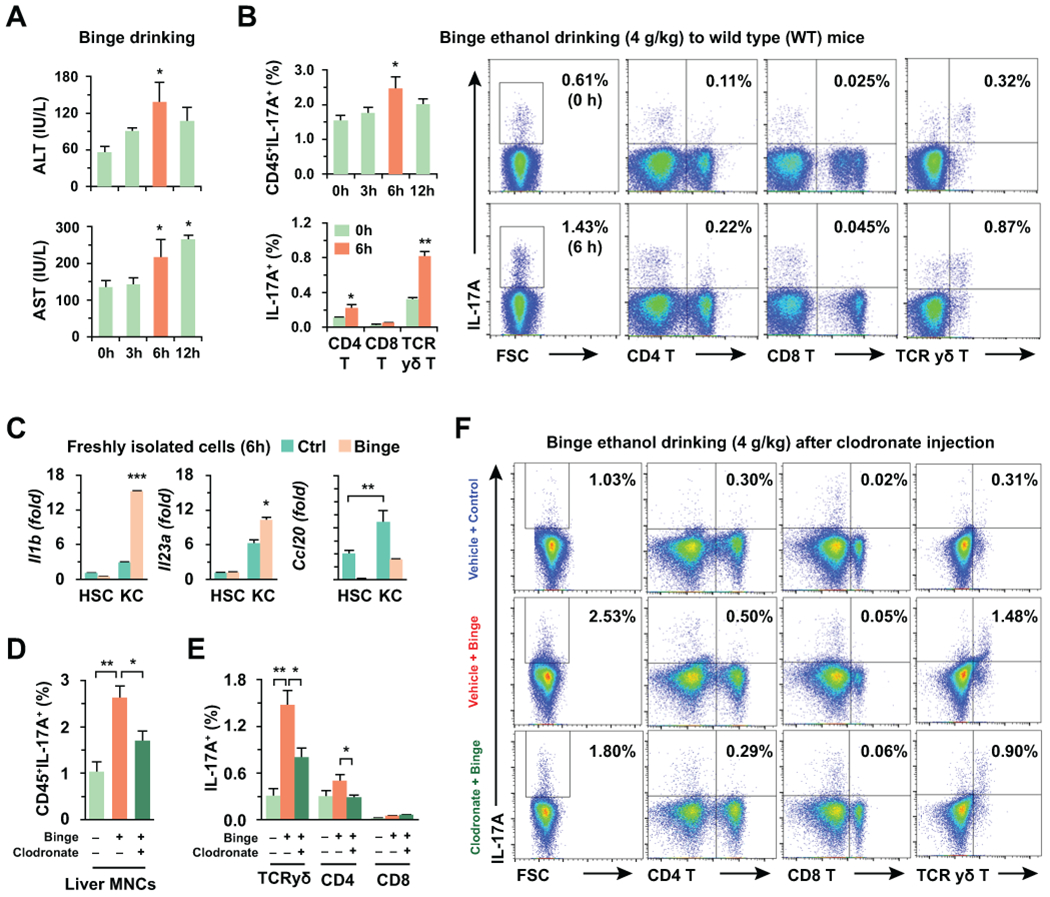
Wild type (WT) mice were fed with 4 g/kg of ethanol (binge drinking) for various time points (n = 5/group). (A) Serum levels of ALT and AST were measured. (B) IL-17A expressing liver MNCs were analyzed by flow cytometry after binge drinking. (C) Freshly isolated hepatic stellate cells (HSCs) and Kupffer cells were subjected to quantitative real-time PCR (qRT-PCR) analyses (3 replicates). (D-F) WT mice were sacrificed at 6 hour after binge drinking with or without 2-day pretreatment of clodronate (10 μl/g) (n = 5/group). Total frequency (D) and each cell type (E) of IL-17A producing liver MNCs were analyzed by flow cytometry (F). Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
ETHANOL-DERIVED HEPATIC EXTRACELLULAR VESICLES STIMULATE IL-1β PRODUCTION IN KUPFFER CELLS
Next, we tested whether ethanol-derived EVs from hepatocytes stimulate IL-1β production in Kupffer cells. Total EVs were isolated from media of cultured hepatocytes with or without ethanol treatment, and hepatic microvesicles (MV) and exosomes (Exo) were then separated by differential isolation techniques, and confirmed using a dynamic light scattering system (Fig. 2A; Supporting Fig. 2A). The shapes and the increased numbers of MV and Exo from EtOH-treated hepatocytes (EtOH-MV and EtOH-Exo) were identified and compared to those of control hepatocytes using the electron microscopy and the nanoparticle tracking analysis, respectively (Fig. 2B; Supporting Fig. 2B). Upon direct treatments of isolated WT Kupffer cells with EVs (designated as isolated MV and Exo in this study), EtOH-EV treatment significantly increased the IL-1β concentration in media compared to those observed in the untreated and vehicle-EV (Veh-EV) treatment controls (Fig. 2C). Based on the above findings, we next investigated whether increased IL-1β production by Kupffer cells could influence IL-17A expression in γδ T cells. In the experiments using a co-culture system, EtOH-EV treatment significantly increased the mRNA expression of Il1b, Il23a and Ccl20 in Kupffer cells and Il17a mRNA expression in γδ T cells compared to the expression levels observed in Veh-EV treatment (Fig. 2D and Supporting Fig. 2C). Similar with previous reports,(11, 12) IL-17A production and its expression in γδ T cells were significantly increased by the treatment of IL-1β and IL-23 (Supporting Fig. 2D). Upon separate treatments with EtOH-MV or EtOH-Exo, the expression of Il1b, Nlrp3, and Ccl20 mRNA was significantly increased in Kupffer cells by both EtOH-MV and EtOH-Exo (Fig. 2E). Consistent with these findings, both treatments substantially increased the protein levels of NLRP3, pro-IL-1β and pro-caspase-1 along with caspase-1 activation in Kupffer cells compared to the levels observed in Veh-treated Kupffer cells (Fig. 2F). However, Tlr3 mRNA expression was significantly increased only in EtOH-Exo-treated Kupffer cells, while Aim2 and Tlr9 mRNA expression, encoding receptors for dsDNA and CpG sequences of DNA, were unchanged or markedly decreased (Fig. 2E). Based on above data, we speculated that dsRNA and TLR3 may be involved in EtOH-Exo-mediated IL-1β production in Kupffer cells.
Fig 2. Ethanol-derived hepatic extracellular vesicles stimulate IL-1β production in Kupffer cells.
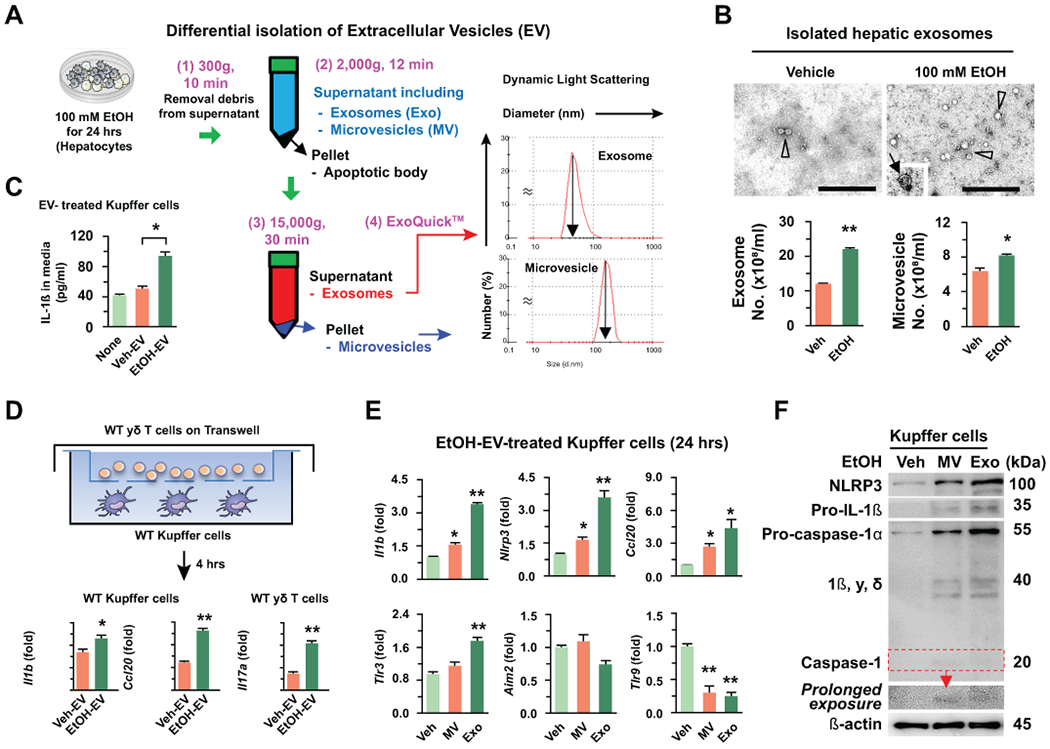
(A) Differential isolation of hepatic extracellular vesicles (EV). Microvesicles (MV) and exosomes (Exo) were collected by step-wise centrifugation and ExoQuick-TC™. Each size was analyzed by dynamic light scattering. (B) Shapes and numbers of MV (arrow in an insert) and Exo (arrow head) were assessed by electron microscope and nanoparticle tracking analysis. Bar = 500 nm. (C) Freshly isolated hepatocytes were treated with or without 100 mM EtOH for 24 hours. Next, the IL-1β concentration was measured in the media after treatments of Kupffer cells with vehicle-induced EV (Veh-EV) and ethanol-induced EV (EtOH-EV) for 24 hours. (D) Schematic diagram of the co-culturing system using Transwell with 3 μm pore. After pretreatment of WT Kupffer cells with Veh- or EtOH-Exo for 12 hours, the media were changed and then they were co-cultured with WT γδ T cells for an additional 4 hours. Kupffer cells and migrated γδ T cells were subjected to qRT-PCR. (E, F) After treatments of Kupffer cells with vehicle (medium), EtOH-induced MV and Exo, the expression changes of genes and proteins were analyzed by qRT-PCR and immunoblotting, respectively. Values and images represent the results from three experimental replicates. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
MITOCHONDRIAL dsRNA AND DNA ARE DELIVERED TO KUPFFER CELLS THROUGH EXOSOMES AND MICROVESICLES
To test the hypothesis mentioned above, we investigated RNA contents within isolated EVs. RNA bioanalyzer analysis revealed that small and diverse-sized RNAs (approximately 25 ~ 500 nucleotides) were mainly identified in isolated Exo compared to MV, and their contents were significantly increased in EtOH-Exo compared to Veh-Exo (Fig. 3A). These findings suggested that, in addition to microRNA (about 22 nucleotides), hepatic exosomal delivery of small-sized RNA might affect Kupffer cells. In immunostaining analyses using J2 antibody, dsRNA co-localized with DiI-stained EtOH-Exo in Kupffer cells (Fig. 3B). In qRT-PCR analyses, mitochondrial mRNAs coded from heavy- and light-strand mtDNA were mostly observed and their expression was significantly increased in EtOH-Exo compared to Veh-Exo (Fig. 3C).
Fig. 3. Ethanol-derived mitochondrial dsRNA is delivered to Kupffer cells through exosomes.
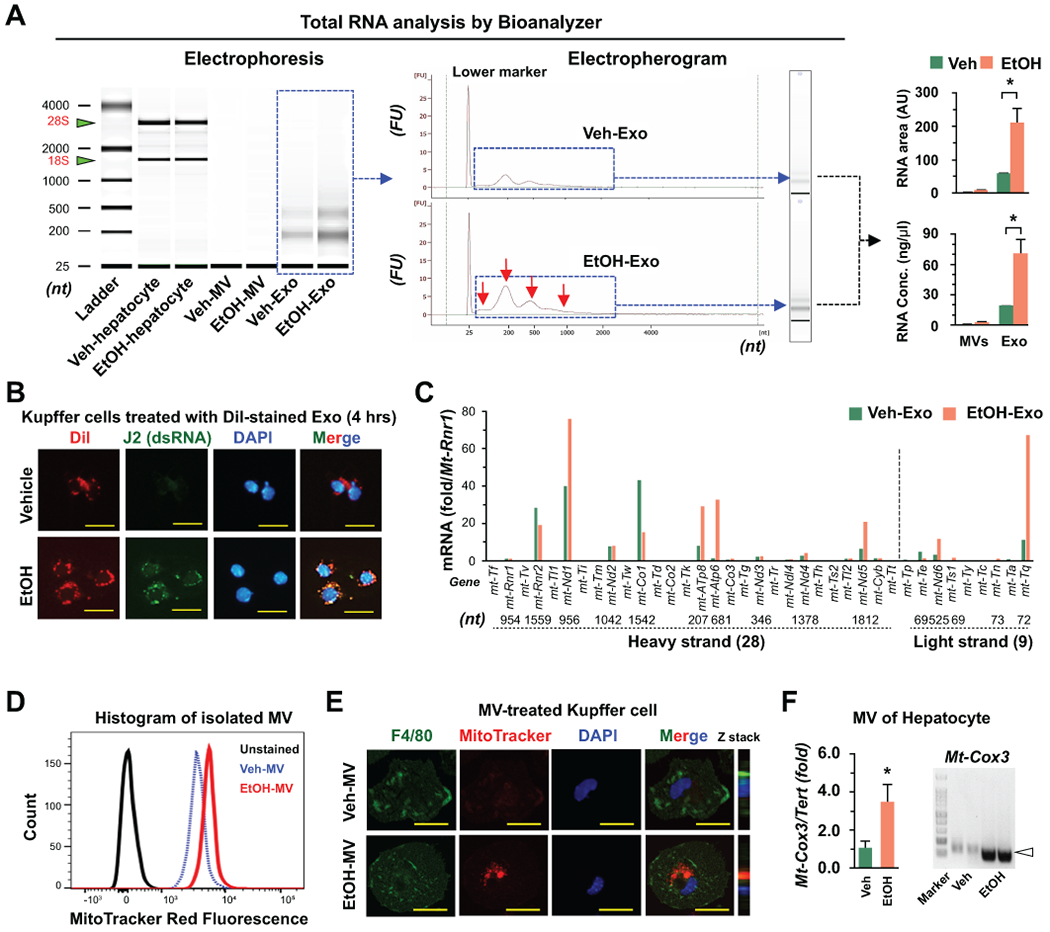
(A) Size distribution and contents of total RNA isolated from hepatocyte, MV and Exo were analyzed using Bioanalyzer. (B) Representative immunostaining of J2 antibody in Kupffer cells after incubation with DiI-stained Veh- and EtOH-Exo (4 hour). Bar = 20 μm. (C) Representative mitochondrial mRNA expression in isolated Veh- and EtOH-Exo was assessed by qRT-PCR (Relative expression vs mitochondrially encoded 12S RNA, Rnr1). (D,E) MitoTracker-stained mitochondrial components within MVs of Veh- and EtOH-treated hepatocytes were assessed using by flow cytometry (D) and immunocytochemistry (E). Bar = 10 μm. (F) Presence of mitochondria DNA (mtDNA) in MVs was assessed by PCR analysis. Values and images represent the results from three experimental replicates. Data are expressed as the mean ± SEM. *P < 0.05 compared to the corresponding control.
As previously reported,(10, 15) MitoTracker-stained mitochondrial components and their delivery to Kupffer cells were significantly increased in EtOH-MV compared to those of Veh-MV (Fig. 3D,E). Additionally, mtDNA sequences, such as cytochrome c oxidase subunit 3 (Cox3 gene), were increased in EtOH-MV compared to Veh-MV (Fig. 3F). These data suggested that mtdsRNA and mtDNA were enriched in EtOH-induced EVs and that they could be delivered to Kupffer cells through hepatic Exo and MV, respectively.
ETHANOL-INDUCED EXOSOMAL mtdsRNA STIMULATES IL-1β EXPRESSION IN KUPFFER CELLS
In contrast to mtDNA, the effects of mtdsRNA have not been clearly investigated. Previously, an interesting study demonstrated that mtdsRNA in mammalian cells can be generated by downregulating the expression of mitochondrial RNA helicase SUV3 (Supv3l1) and polynucleotide phosphorylase PNPase (Pnpt1).(18) qRT-PCR and Western blot analyses revealed that Pnpt1 gene expression and protein levels of PNPase were significantly decreased in liver tissues of mice after binge ethanol feeding and hepatocytes treated with various doses of ethanol in vitro, whereas Supv3l1 expression and SUV3 levels were unchanged by ethanol (Fig. 4A–C). The EtOH-mediated decrease in Pnpt1 expression in hepatocytes was also verified using an in situ closed perfusion system (Fig. 4D). Immunostaining revealed that increased dsRNA generation (J2 foci) was observed in the mitochondria at 12 hour after ethanol treatment, and the number of J2 foci (dsRNA) was decreased and primarily observed outside of the mitochondria at 24 hour (Fig. 4E). These findings suggested a translocation of dsRNA from mitochondria to cytosol and subsequent release of dsRNA through the exosome. Next, we investigated the effects of exosomal mtdsRNA on Kupffer cells using RNase III, a ribonuclease of dsRNA. In immunostaining, RNase III significantly decreased J2 foci in EtOH-treated hepatocyte (Fig. 4F). In parallel with these results, EtOH-Exo-mediated Il1b expression by Kupffer cells was completely suppressed after RNase III treatment (Fig. 4F), suggesting mtdsRNA-mediated Il1b expression in Kupffer cells.
Fig. 4. Ethanol treatment decreases PNPase expression and generates mitochondrial dsRNA in hepatocytes.
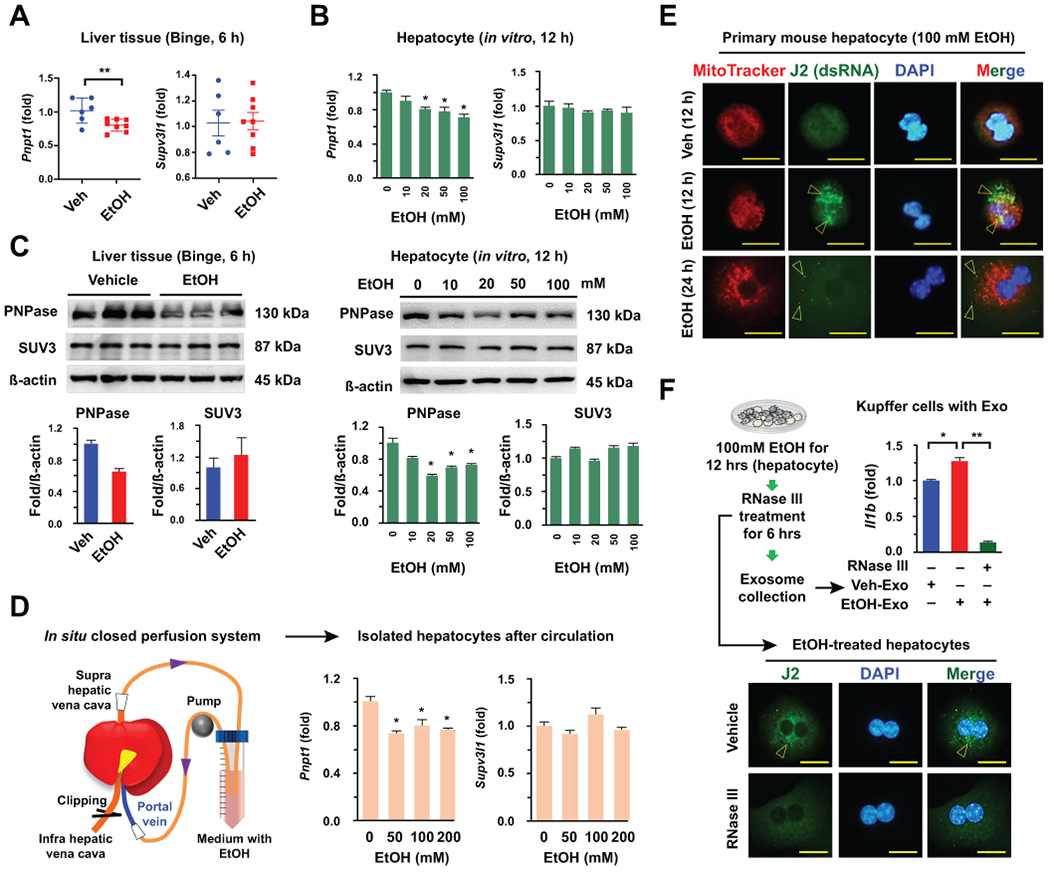
(A-C) Expression levels of genes and proteins were assessed by qRT-PCR and Western blotting in mouse livers (n = 6 ~ 8) and isolated hepatocytes with or without EtOH treatments, respectively. (D) After circulation of EtOH for 2 hours, hepatocytes were isolated and subjected to qRT-PCR. (E) Representative immunostaining of J2 antibody (arrowhead) in Veh- and 100 mM EtOH-treated mouse hepatocyte (12 and 24 hour). Bar = 20 μm. (F) After treatment with 100 mM EtOH for 12 hours, RNase III was added to the medium for an additional 6 hours. Then, hepatocytes were fixed and immunostained with J2 antibody (Bar = 20 μm), and collected exosomes from the medium were co-incubated with freshly isolated mouse Kupffer cells for 24 hours. Kupffer cells were subjected to qRT-PCR. Values and images represent the results from three experimental replicates. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
BINGE ETHANOL DRINKING STIMULATES IL-17A EXPRESSION BY γδ T CELLS IN TLR3- AND KUPFFER CELL-DEPENDENT MANNERS
Next, we investigated whether exosomal mtdsRNA-mediated TLR3 activation in Kupffer cells could stimulate IL-17A production by γδ T cells in acute alcoholic liver injury. After binge ethanol drinking to WT, TLR3 KO, or TLR3 KO mice with or without clodronate treatment, there were no differences in liver injury and frequencies of F4/80+CD11b+ cells (infiltrating macrophages) and Ly6G+CD11b+ cells (neutrophils) between WT and TLR3 KO mice or between TLR3 KO mice with and without Kupffer cell deletion (Fig. 5A; Supporting Fig. 3A,B). The frequency of IL-17A producing lymphocytes, however, was significantly decreased in γδ T cells of TLR3 KO mice compared to that of WT mice, while TLR3 KO mice with Kupffer cell depletion did not increase IL-17A production of γδ T cells in response to acute alcoholic liver injury (Fig. 5B). In qRT-PCR analysis, the expression of Il1b, Il23a and Ccl20 mRNA was significantly decreased in isolated Kupffer cells of TLR3 KO mice compared to that of WT mice, while Kupffer cell-depleted TLR3 KO mice possessed similar or lower expression of Il1b, Ccl20 and Nlrp3 mRNA than that of TLR3 KO mice (Fig. 5C; Supporting Fig. 3C,D). Consistently, the expression of Il17a, and Il1b was decreased in liver MNCs of WT mice compared to that of TLR3 KO mice (Supporting Fig. 3C). The expression levels of Aim2 and Tlr9 mRNA were not significantly affected by acute alcohol liver injury (Supporting Fig. 3D,E). These data suggested that TLR3 may be responsible for the expression of Il1b and Ccl20 in Kupffer cells and for inducing migration and IL-17A production in γδ T cells. To confirm these findings, we co-cultured EtOH-Exo-treated WT and TLR3 KO Kupffer cells with WT γδ T cells using a Transwell system. Migration of WT γδ T cells was significantly decreased in TLR3 KO Kupffer cells compared to that of WT Kupffer cells (Fig. 5D). Additionally, IL-17A-expression was also decreased in WT γδ T cells that were co-cultured with TLR3 KO Kupffer cells compared to that of WT Kupffer cells (Fig. 5E). In vitro poly I:C treatment, a synthetic TLR3 ligand, of WT Kupffer cells, remarkably increased the expression of Il1b and Nlrp3 at 3 hour and then this expression gradually decreased, but the expression of Aim2 and Tlr9 was unchanged and significantly decreased (Fig. 5F). Collectively, TLR3-mediated IL-1β expression in Kupffer cells stimulates innate IL-17A production by γδ T cells at the early stage of ALD.
Fig. 5. Ablation of TLR3 decreases IL-17A production in γδ T cells in acute alcoholic liver injury.
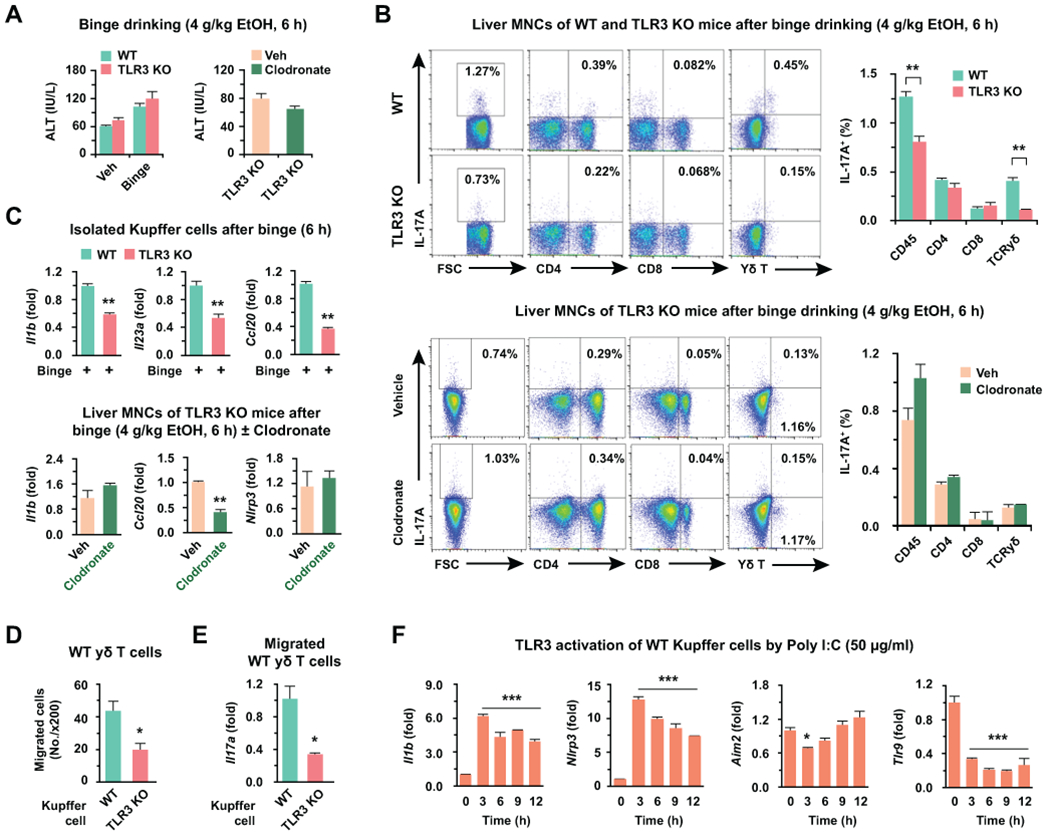
Wild type (WT) and toll-like receptor 3 (TLR3) knockout (KO) mice were fed with 4 g/kg of ethanol (binge drinking) for 6 hours with or without 2-day pretreatment with clodronate (10 μl/g) (n = 5/group). (A) Serum level of ALT were measured. (B) IL-17A expressing liver MNCs were analyzed by flow cytometry in WT and TLR3 KO mice or in TLR3 KO mice with or without clodronate treatment. (C) After binge drinking, gene expression was assessed by qRT-PCR in freshly isolated Kupffer cells from WT and TLR3 KO mice, or TLR3 KO mice with or without clodronate. (D, E) WT γδ T cells were co-cultured with WT and TLR3 KO Kupffer cells for 12 hours after pretreatment with EtOH-Exo (4 hours) using 3 μm pore-sized Transwell systems. Then, migrated γδ T cells were counted under the microscope (D; x200 magnification) and subjected to qRT-PCR analyses (E). (F) Gene expressions were analyzed after poly I:C stimulation in primary WT Kupffer cells (3 replicates). Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
KUPFFER CELLS WITH TLR3 DEPLETION AMELIORATES LIVER INJURY IN CHRONIC AND ACUTE ALCOHOL INGESTION
To investigate the additional effects of TLR3 on Kupffer cells and the involvement of IL-17A production at the late stage of ALD, we first fed a liquid ethanol diet to WT and TLR3 KO mice for 8 weeks (alcoholic steatosis model). Although liver injury was decreased in TLR3 KO mice compared to WT mice, there were no differences in the frequencies of IL-17A producing cells in liver MNCs between WT and TLR3 KO mice (Supporting Fig. 4). Next, we generated chimeric mice possessing TLR3-deficient Kupffer cells by transplantation of WT bone marrow (BM) into WT mice (WTWT) and TLR3 KO (TLR3 KOWT) mice. After confirming TLR3 chimerism in Kupffer cells (Fig. 6A), the mice were fed a liquid EtOH diet for 10 days, and this was followed by one binge drinking to induce alcoholic hepatitis as previously reported.(25) At sacrifice, the levels of serum ALT and liver TG were both significantly attenuated in TLR3 KOWT mice compared to those of WTWT mice (Fig. 6B, C). The numbers of total liver MNCs and IL-17A producing cells were significantly decreased in TLR3 KOWT mice compared to WTWT mice (Fig. 6D). In contrast to observations after single binge drinking, the major populations of IL-17A producing cells were CD4+ T cells in chronic and binge ethanol ingestion (Fig. 6D). The frequencies of CD11b+Ly6G+ and CD11b+F4/80+ cells were similar, but their numbers were decreased in TLR3 KOWT mice compared to that of WTWT mice (Fig. 6E). Based on the qRT-PCR analysis, the expression of Il1b, Il23a, 1l17a and Rorc mRNA was significantly decreased in TLR3 KOWT mice compared to that of WTWT mice (Fig. 6F). Indeed, the expression levels of Cxcl1 and Cxcr2 mRNAs for the recruitments of neutrophils and macrophages were decreased in TLR3 KOWT mice (Fig. 6F). In contrast, the expression of CCl20 and Fasl mRNAs, critical factors for the migration and activation of γδ T cells in liver fibrosis,(28) was not changed in TLR3 KOWT mice compared to that of WTWT mice (Fig. 6F). Based on these findings, hepatic exosome-mediated TLR3 activation of Kupffer cells stimulated innate IL-17A production by γδ T cells at early alcoholic liver injury. Meanwhile, it could enhance the production of IL-1β and IL-23 in macrophages, which subsequently leads to the acceleration of steatosis and IL-17A production in CD4+ T cells at the later stage of alcoholic liver injury.
Fig. 6. Ablation of TLR3 in Kupffer cells ameliorates liver injury in chronic and acute alcohol consumption.
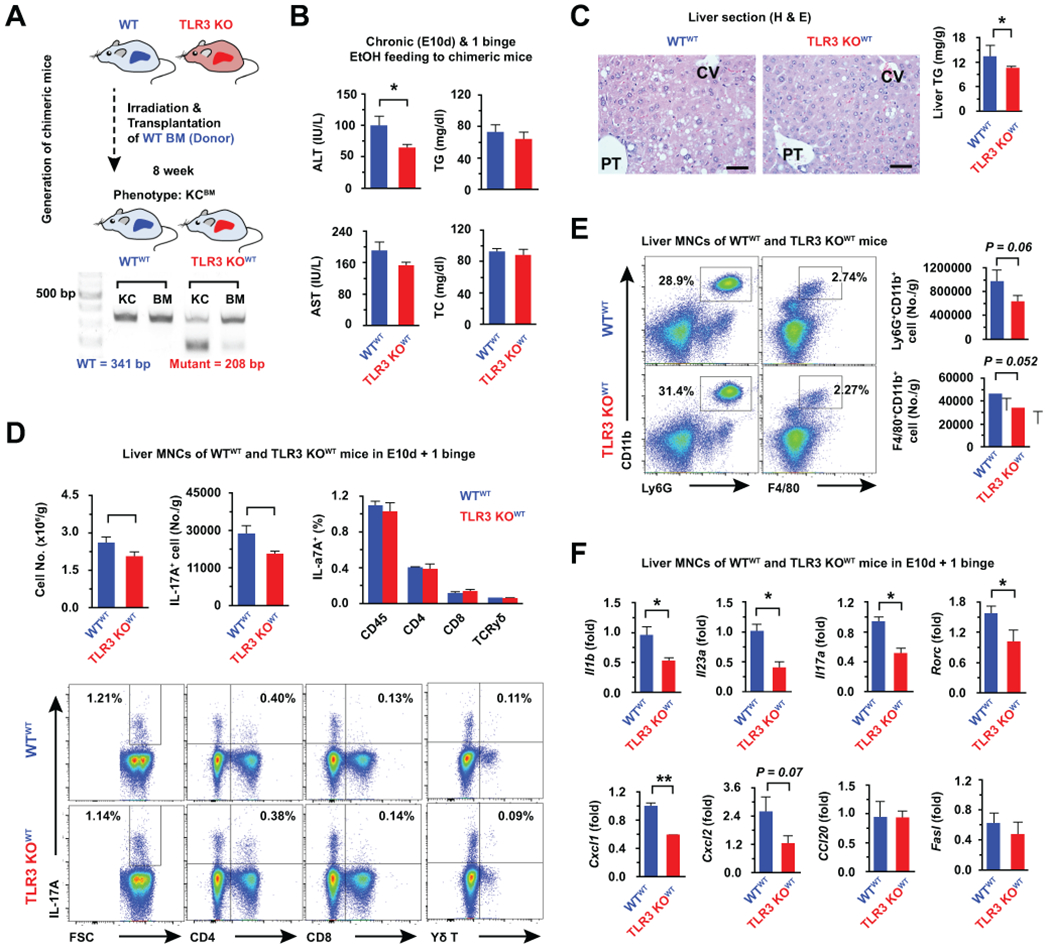
(A) After 8 weeks of WT bone marrow (BM) transplantation to WT mice and TLR3 KO mice, chimerism of Kupffer cells, BMs, or PBMCs was assessed by PCR. After confirmation of chimerism, the mice were fed a liquid EtOH diet for 10 days (E10d), and 1 binge drinking (4 g/kg) (n = 5/group). (B) Serum levels of ALT, AST, TG and TC were measured. (C) Liver sections were stained with H & E. Bar = 50 μm. (D, E) IL-17A expressing T, Ly6G+CD11b+ and F4/80+CD11b+ cells of liver MNCs were analyzed by flow cytometry after chronic and binge ethanol consumption. (F) Liver MNCs were subjected to qRT-PCR analyses (3 replicates). Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
HUMAN KUPFFER CELLS STIMULATED BY EXOSOMAL mtdsRNA AUGMENT IL-17A PRODUCTION OF HUMAN γδ T CELLS
To confirm our findings in human cells, we freshly isolated primary human hepatocytes, Kupffer cells and γδ T cells from healthy livers for use in vitro experiments. Ethanol treatment decreased PNPT1 expression and the protein levels of PNPase, responsible for degradation of mitochondrial light-strand transcripts,(17) in primary hepatocytes at 24 hour, suggesting the occurrence of mtdsRNA accumulation (Fig. 7A). Consistent with the above findings, immunostaining revealed that J2 foci were significantly increased in human hepatocytes after EtOH treatment (Fig. 7B). Based on qRT-PCR analysis, the expression levels of IL1B, IL23A, CCL20, TLR3, and IRF3 were significantly increased in EtOH-Exo-treated human Kupffer cells (hKCs) compared to those in Veh-Exo-treated hKCs (Fig. 7C). In parallel with gene expression, EtOH-Exo-treated hKCs recruited and interacted with co-cultured human γδ T cells, leading to increased expression of IL17A in γδ T cells compared to that in co-cultured γδ T cells with Veh-Exo-treated hKCs (Fig. 7D). To test whether mtdsRNA might induce IL1B expression, hKCs were treated with EtOH-Exo in the presence or absence of RNase III. Increased IL1B expression by EtOH-Exo was decreased in hKCs after RNase III-mediated degradation of mtdsRNA (Fig. 7E). In direct treatment with poly I:C, the expression of IL1B was remarkably increased in hKCs along with others genes such as IL23A, CCL20, and NLRP3 (Fig. 7F). These findings suggested that EtOH-Exo stimulated IL1B expression of hKCs and their interaction with γδ T cells through the mtdsRNA-TLR3 signaling pathway. Collectively, these data suggested that ethanol-induced hepatic exosomes contain dsRNA and can be delivered to Kupffer cells by endocytosis, which augments IL-17A production of γδ T cells through dsRNA/TLR3-mediated IL-1β production of Kupffer cells.
Fig. 7. Mitochondrial double-stranded RNA in exosome stimulates IL-1β production of human primary Kupffer cells.
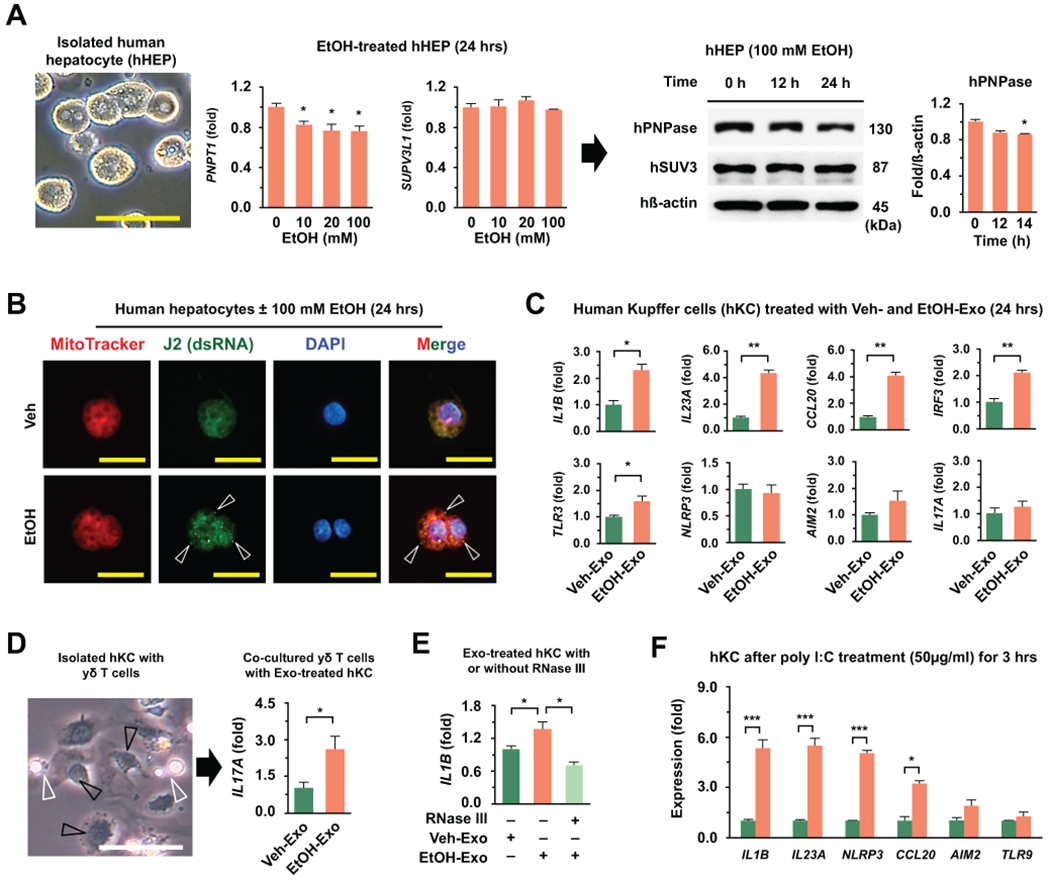
(A) In EtOH-treated human hepatocytes, expression of genes (PNPT1 and SUPV3L1) and proteins (PNPase and SUV3) were assessed by qRT-PCR and Western blotting, respectively (12, 24 hour). Bar = 20 μm. (B) Representative immunostaining of J2 antibody in Veh- and EtOH (100 mM)-treated human primary hepatocyte (24 hour). dsRNA (Arrowhead), Bar = 10 μm. (C) Freshly isolated EtOH-EVs from primary human hepatocytes were treated to primary human Kupffer cells (hKC) for 24 hours and then gene expression was analyzed. (D) hKCs were co-incubated with WT γδ T cells (white arrowhead) for an additional 4 hours after treatments with Veh- and EtOH-Exo. (E) Primary hKC (black arrowhead) were treated with Veh- and EtOH-Exo in the absence and presence of RNase III (24 hour). (F) Primary hKC were treated with poly I:C (50 μg/g) for 3 hours and they were subjected to qRT-PCR. Values and images represent the results from three experimental replicates. Data are expressed as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 compared to the corresponding control, based on unpaired t-test between two groups and one way ANOVA with Dunnett’s test for multiple comparison vs control.
ENRICHED TRANSCRIPT EXPRESSION OF MITOCHONDRIAL GENES ENCODED BY HEAVY AND LIGHT STRANDS IN EXOSOMES OF ETHANOL-TREATED HUMAN HEPATOCYTES
Next, we investigated RNA contents in human hepatic EVs after EtOH treatment. Similar to observations in mice, RNA content was enriched in hepatic exosomes compared to that of microvesicles, and RNA content was also significantly increased in EtOH-Exo compared to that of Veh-Exo (Fig. 8A; Supporting Fig. 5A). Based on RNA sequencing analysis, mitochondria-originated (chrM) RNA was mainly increased in the total RNA reads of EtOH-Exo compared to that of Veh-Exo (Fig. 8B). In the top 50 RNA reads, 84% of total reads (14 mitochondrial genes) was mitochondria-originated RNA in EtOH-Exo (Fig. 8C). In addition, RNA sequencing and qRT-PCR analyses revealed that nearly all of the mitochondrial RNA and transcripts including NADH dehydrogenase (MT-ND1, 3, 4, 4L, 5 and 6), cytochrome c oxidase (MT-CO1, MT-CO3), coenzyme Q-cytochrome c reductase/cytochrome b (MT-CYB), ATP synthase (MT-ATP6) and transfer RNA were increased in EtOH-Exo compared to their levels in Veh-Exo (Fig. 8D; Supporting Fig. 5B). These transcripts originated from both heavy- and light-stranded mtDNA, reflecting the presence of mtdsRNA within EtOH-Exo of human hepatocytes.
Fig. 8. RNA sequencing reveals increased mitochondrial RNA contents in exosomes of ethanol-treated human hepatocytes.
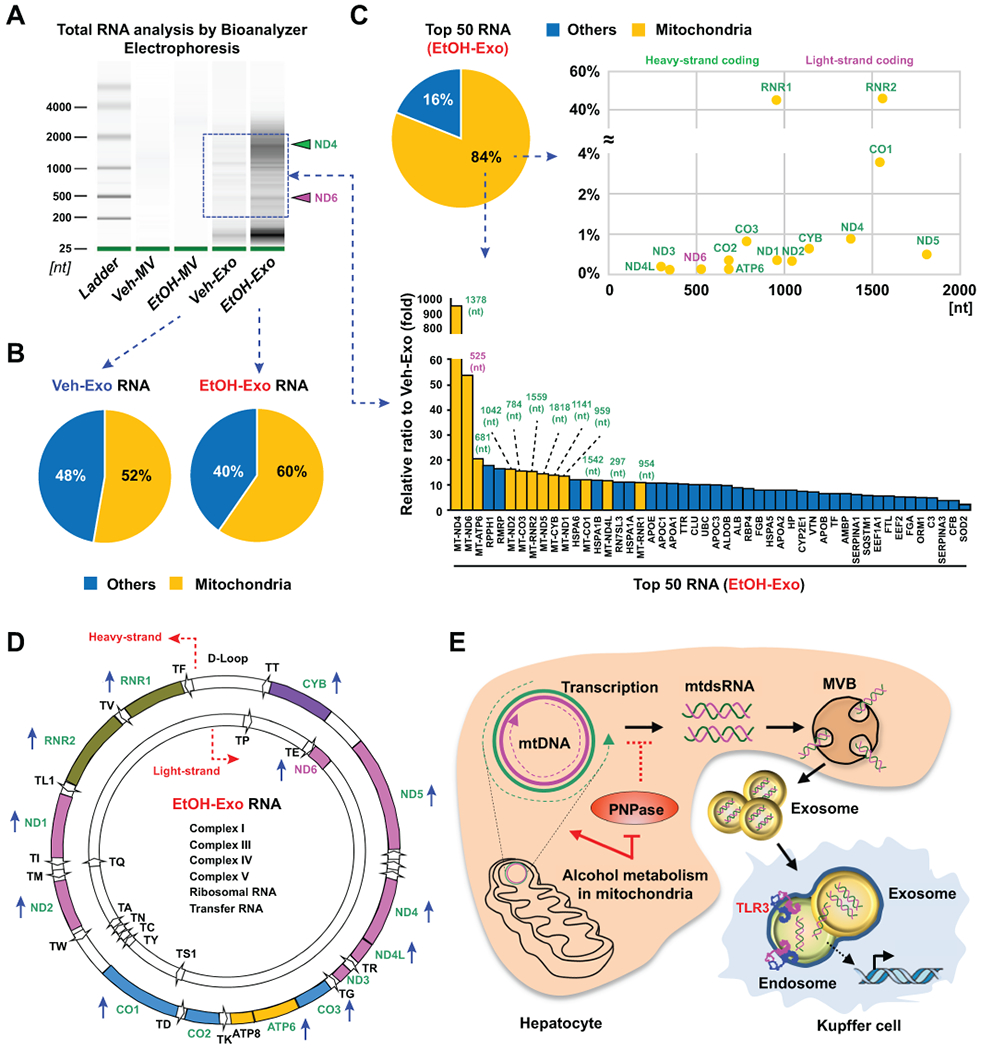
Human hepatocytes were treated with or without 100 mM ethanol for 24 hours, following which hepatic exosomes were isolated from media. (A) Size distribution and total RNA content isolated from hepatic exosomes were assessed by electrophoresis (3 replicates). (B) Distribution of the mitochondrial gene transcripts from Veh-Exo RNA (left) or EtOH-Exo RNA (right) sequencing. (C,D) Distribution of the mitochondrial gene transcripts (chrM) in the top 50 RNA from EtOH-Exo RNA sequencing, and their relative expression compared to those of Veh-Exo RNA. (D) Heavy (outer circle)- and light (inner circle)-stranded location of increased mitochondrial gene transcripts (blue-colored arrow) from EtOH-Exo RNA sequencing. (E) Interaction mechanisms between mtdsRNA of hepatocytes and TLR3 in Kupffer cells mediated by exosomes.
Discussion
In the present study, we clearly demonstrated that alcoholic stress in hepatocytes generates mtdsRNA, and exosomal delivery of mtdsRNA to TLR3 in Kupffer cells stimulates IL-1β production to augment innate IL-17A production of γδ T cells at the early stages of ALD, subsequently leading to IL-17A production in CD4+ T cells at the later stage of ALD. The mechanisms underlying the generation, delivery, and TLR3 activation of exosomal mtdsRNA are summarized in Fig. 8E.
Although EVs are known to deliver diverse cargos including mtDNA, miRNA, lipids and proteins,(7–10) the generation, delivery, and immunologic functions of dsRNA in EVs have not been clearly investigated in ALD. Ethanol metabolism by alcohol dehydrogenase generates cytosolic NADH that is further oxidized by mitochondrial electron transport complexes I, III, and IV, resulting in mitochondrial ROS formation, that in turn induces inappropriate functions of mitochondria through mtDNA strand breaks, oxidized mtDNA, and mtDNA depletion.(13, 14) In fact, oxidized mtDNA by alcohol or high-fat diet consumption is delivered to neutrophils and macrophages in mice and humans through EV-mediated delivery, contributing to the initiation of inflammatory responses through TLR9 and NLRP3 inflammasome activation.(10, 15, 29, 30) In contrast, in the present study, we demonstrated that ethanol-mediated metabolic stresses generated mtdsRNA, and exosomal delivery of mtdsRNA stimulated IL-1β expression in WT Kupffer cells in a TLR3-dependent manner. Uniquely, individual mitochondrial rRNAs and mRNAs can be released and matured after excising 22 interspersed tRNAs from two polycistronic transcripts of heavy and light-strands.(27) However, imbalanced turnover of polycistronic transcripts by loss of SUV3 and PNPase (encoded by SUPV3L1 and PNPT1 respectively) may cause accumulation of mtdsRNA.(17, 18). Accordingly, an interesting study examining the mitochondrial transcriptome reported that the ND5 3’UTR is antisense to the ND6 gene, suggesting stable antisense transcripts.(27) In our study, PNPase expression in hepatocytes was significantly decreased by ethanol treatment in mice and humans, resulting in the generation and accumulation of mtdsRNA. Moreover, RNA sequencing analysis revealed that the expression of MT-ND5 and ND6 mRNAs was enriched in EtOH-Exo, where dsRNA was also detected by J2 antibody; however, these findings were not observed in Veh-Exo. Therefore, further studies are needed to elucidate the formation of dsRNA between of MT-ND5 and ND6 mRNAs in exosomes.
Limited information regarding the detailed features of mtdsRNA delivery to target cells is available, and in particular, the sensing mechanisms of mtdsRNA by pattern recognition receptors such as RIG-I, MDA5, or TLR3 are poorly understood.(21, 22) A recent study demonstrated that mtdsRNA could engage cytosolic MDA5 to trigger Type I interferon production through mitochondrial antiviral signaling proteins within the same cytosol.(18) In our study, we could elucidate the contribution of mtdsRNA in hepatocytes to endosomal TLR3 of target Kupffer cells through exosomal delivery. Additionally, lines of evidence support our notion that diverse self-ligands including mRNA from necrotic cells, non-coding RNA from ultraviolet B-irradiated skin, and unknown-self ligands of injured hepatocytes stimulate TLR3 signaling in dendritic cells, keratinocytes and hepatic stellate cells.(12, 23, 24) These data also suggest an explanation for the underlying cause of certain autoimmune diseases; self-ligands such as mtdsRNA may contribute to TLR3-mediated inflammatory disease. In contrast, some exosomes limit the inappropriate activation of RIG-I-like receptors by endogenous RNA to prevent autoimmune disorders.(31) In fact, mitochondria and its circular DNA originated from an ancient α-proteobacterium and its components have recently been demonstrated to play important roles in host immune responses.(32) Given these conflicting data, it remains uncertain whether mitochondria-derived immune responses were derived from self- or non-self-ligands. Further investigations are required to clarify this notion, as the activation of RNA sensors is strictly regulated to avoid self-RNA-mediated autoimmunity. Next question is how captured mtdsRNA in exosomes could be delivered to TLR3 in the endosomes of Kupffer cells. A study provided evidence that circulating exosomes were efficiently internalized by Kupffer cells.(33) Diverse ligands present on the exosome surface are required to dock exosomes to the surface of target cells. CD40L/CD40 interaction may contribute to exosomal endocytosis in macrophages either directly or indirectly.(6, 34) Interestingly, endocytosed exosome-endosome fusion has been reported in dendritic cells and hepatocytes in regard to presentation of alloantigen peptides and efficient transfer of IFN-α-induced anti-HBV activity.(33, 35) Based on these findings, mtdsRNA might be transferred to TLR3 by fusion machinery. Regardless, further studies are necessary to unveil this mechanism.
Although the effects of IL-17 on ALD have been reported in mice and humans, the initial source of IL-17 and its production mechanism are not well understood in ALD, particularly in non-pathogen (e.g. DAMPs)-mediated liver injury. Exosomal miRNA delivery or ligand-mediated stimulation of macrophages induces inflammatory responses by producing a number of cytokines such as IL-1β that further contributes to severe inflammation and steatosis in mice and patients with ALD.(6–9) Our results strongly indicate that mtdsRNA-mediated IL-1β production might be involved in the IL-17A production of γδ T cells in TLR3- and Kupffer cell-dependent manners at the early stage of ALD, this subsequently affects IL-17A production by CD4+ T cells in chronic and acute ethanol ingestion. In response to IL-1β and IL-23, γδ T cells secrete innate IL-17 to promote Th17 responses in autoimmune inflammation and chronic ALD.(2, 11) Moreover, binge alcohol intake specifically induces the inhibition of TLR4 signaling by heat shock protein 70 and protein phosphatase 1, but not the TLR3-interferon regulatory factor-3 (IRF3) pathways in human monocytes.(36) Furthermore, chronic and acute ethanol ingestion increases liver injury through IRF3-induced apoptosis of restorative Ly6Clow macrophages.(37) However, the authors did not investigate which molecules activated TLR3 and IRF3 signaling in macrophages after binge ethanol feeding. Here, we clearly demonstrated that generated mtdsRNA in response to acute binge ethanol ingestion activated TLR3 in Kupffer cells through exosomal delivery.
In the current study, our findings demonstrate that ethanol exposure in hepatocytes induces the generation, enrichment, and release of mtdsRNA into exosomes, ultimately triggering IL-1β production of neighboring Kupffer cells through mtdsRNA-mediated TLR3 activation. Additionally, TLR3-mediated activation of Kupffer cells further leads to the recruitment of γδ T cells and their expression of IL-17A at the early stage of ALD. Therefore, our results suggest that mtdsRNA and TLR3 could serve as possible therapeutic targets to improve the treatment of ALD.
Supplementary Material
Acknowledgments:
We thank all study investigators and patients, and especially appreciate Prof. Ho-Min Kim and Young Seok Ju for technical supports of our study.
Financial Support
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIP) (2018R1A2A1A05077608), Korea Mouse Phenotyping Project (2014M3A9D5A01073556), and the Intelligent Synthetic Biology Center of Global Frontier Project (2011-0031955) and National Institutes of Health (R01AA027532).
List of Abbreviations
- ALD
alcoholic liver disease
- DAMPs
damage-associated molecular patterns
- PAMPs
pathogen-associated molecular patterns
- TLRs
toll-like receptors
- EVs
extracellular vesicles
- miRNA
microRNA
- CYP2E1
cytochrome P450 2E1
- ROS
reactive oxygen species
- mtDNA
mitochondrial DNA
- CD40L
CD40 ligand
- IL
interleukin
- HSCs
hepatic stellate cells
- mtdsRNA
mitochondrial double-stranded RNA
- PNPase
polynucleotide phosphorylase
- Poly I:C
polyinosinic-polycytidylic acid
- RIG-I
retinoic acid-inducible gene I
- MDA5
melanoma differentiation-associated gene 5
- MNCs
mononuclear cells
- RT
room temperature
- FACS
fluorescence-activated cell sorting
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- WT
wild type
- IRF3
interferon regulatory factor-3
References
- 1.Gao B, Ahmad MF, Nagy LE, Tsukamoto H. Inflammatory pathways in alcoholic steatohepatitis. J Hepatol 2019;70:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma HY, Xu J, Liu X, Zhu Y, Gao B, Karin M, et al. The role of IL-17 signaling in regulation of the liver-brain axis and intestinal permeability in Alcoholic Liver Disease. Curr Pathobiol Rep 2016;4:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S ELA, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov 2013;12:347–357. [DOI] [PubMed] [Google Scholar]
- 4.Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular Vesicles: Novel Mediators of Cell Communication In Metabolic Disease. Trends Endocrinol Metab 2017;28:3–18. [DOI] [PubMed] [Google Scholar]
- 5.Cho YE, Mezey E, Hardwick JP, Salem N Jr., Clemens DL, Song BJ. Increased ethanol-inducible cytochrome P450–2E1 and cytochrome P450 isoforms in exosomes of alcohol-exposed rodents and patients with alcoholism through oxidative and endoplasmic reticulum stress. Hepatol Commun 2017;1:675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verma VK, Li H, Wang R, Hirsova P, Mushref M, Liu Y, et al. Alcohol stimulates macrophage activation through caspase-dependent hepatocyte derived release of CD40L containing extracellular vesicles. J Hepatol 2016;64:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eguchi A, Lazaro RG, Wang J, Kim J, Povero D, Willliams B, et al. Extracellular vesicles released by hepatocytes from gastric infusion model of alcoholic liver disease contain a MicroRNA barcode that can be detected in blood. Hepatology 2017;65:475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep 2015;5:9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha B, Momen-Heravi F, Furi I, Kodys K, Catalano D, Gangopadhyay A, et al. Extracellular vesicles from mice with alcoholic liver disease carry a distinct protein cargo and induce macrophage activation through heat shock protein 90. Hepatology 2018;67:1986–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y, Xu MJ, Koritzinsky EH, Zhou Z, Wang W, Cao H, et al. Mitochondrial DNA-enriched microparticles promote acute-on-chronic alcoholic neutrophilia and hepatotoxicity. JCI Insight 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, Mills KH. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 2009;31:331–341. [DOI] [PubMed] [Google Scholar]
- 12.Seo W, Eun HS, Kim SY, Yi HS, Lee YS, Park SH, et al. Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by gammadelta T cells in liver fibrosis. Hepatology 2016;64:616–631. [DOI] [PubMed] [Google Scholar]
- 13.Hoek JB, Cahill A, Pastorino JG. Alcohol and mitochondria: a dysfunctional relationship. Gastroenterology 2002;122:2049–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansouri A, Gattolliat CH, Asselah T. Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology 2018;155:629–647. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Z, Liang S, Sanchez-Lopez E, He F, Shalapour S, Lin XJ, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 2018;560:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh YS, Zhang B, Loomba R, Seki E. TLR2 and TLR9 contribute to alcohol-mediated liver injury through induction of CXCL1 and neutrophil infiltration. Am J Physiol Gastrointest Liver Physiol 2015;309:G30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res 2013;41:1223–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018;560:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libonati M, Carsana A, Furia A. Double-stranded RNA. Mol Cell Biochem 1980;31:147–164. [DOI] [PubMed] [Google Scholar]
- 20.Content J, Lebleu B, De Clercq E. Differential effects of various double-stranded RNAs on protein synthesis in rabbit reticulocyte lysates. Biochemistry 1978;17:88–94. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol 2006;7:131–137. [DOI] [PubMed] [Google Scholar]
- 22.Tatematsu M, Funami K, Seya T, Matsumoto M. Extracellular RNA Sensing by Pattern Recognition Receptors. J Innate Immun 2018;10:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kariko K, Ni H, Capodici J, Lamphier M, Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem 2004;279:12542–12550. [DOI] [PubMed] [Google Scholar]
- 24.Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med 2012;18:1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc 2013;8:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh YG, Kim JK, Byun JS, Yi HS, Lee YS, Eun HS, et al. CD11b(+) Gr1(+) bone marrow cells ameliorate liver fibrosis by producing interleukin-10 in mice. Hepatology 2012;56:1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, et al. The human mitochondrial transcriptome. Cell 2011;146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerich L, Bangen JM, Govaere O, Zimmermann HW, Gassler N, Huss S, et al. Chemokine receptor CCR6-dependent accumulation of gammadelta T cells in injured liver restricts hepatic inflammation and fibrosis. Hepatology 2014;59:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, et al. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest 2016;126:859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoyt LR, Randall MJ, Ather JL, DePuccio DP, Landry CC, Qian X, et al. Mitochondrial ROS induced by chronic ethanol exposure promote hyper-activation of the NLRP3 inflammasome. Redox Biol 2017;12:883–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 2014;15:839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 2017;17:363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004;104:3257–3266. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Chen L, Wang G, Tang H. Cholesterol-dependent and -independent CD40 internalization and signaling activation in cardiovascular endothelial cells. Arterioscler Thromb Vasc Biol 2007;27:2005–2013. [DOI] [PubMed] [Google Scholar]
- 35.Yao Z, Qiao Y, Li X, Chen J, Ding J, Bai L, et al. Exosomes exploit the virus entry machinery and pathway to transmit IFN-alpha-induced antiviral activity. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muralidharan S, Lim A, Catalano D, Mandrekar P. Human Binge Alcohol Intake Inhibits TLR4-MyD88 and TLR4-TRIF Responses but Not the TLR3-TRIF Pathway: HspA1A and PP1 Play Selective Regulatory Roles. J Immunol 2018;200:2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz-Garcia C, Poulsen KL, Bellos D, Wang H, McMullen MR, Li X, et al. The non-transcriptional activity of IRF3 modulates hepatic immune cell populations in acute-on-chronic ethanol administration in mice. J Hepatol 2019;70:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


