Abstract

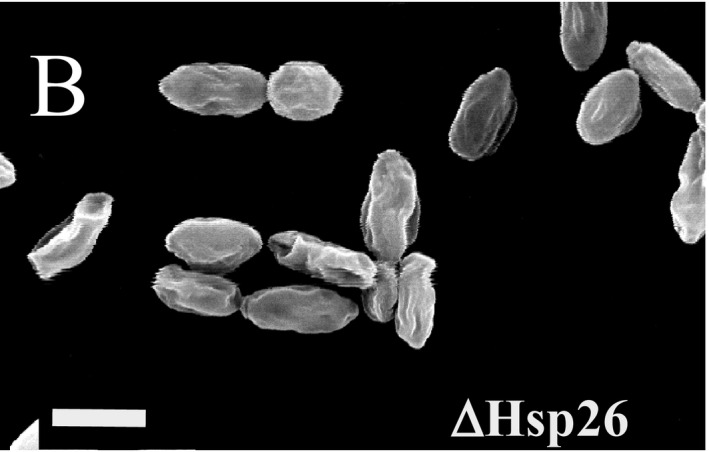
The authors of this report contacted The EMBO Journal to report a discrepancy in the assembly of Fig 9. The authors state that assembly of Fig 9B was mixed up in the process of resizing the images, with the microscopy image in the original Fig 9B representing a crop from the same micrograph also displayed in Fig 9F. In the corrected Fig 9, the phenotypic effects for ∆Hsp26 at physiological conditions (30°C until early stationary phase) are much less significant than those observed for ∆Hsp42, consistent with previous results by Susek and Lindquist (1989) as discussed in the original paragraph of the results section. Since the phenotypic differences between ∆Hsp26 and ∆Hsp42 at physiological temperatures had not been emphasized in the original result section, the following sentence on p. 645 of the article:
“At late logarithmic phase, consistent with the in vivo and in vitro results on the chaperone function of Hsp42 and Hsp26, both deletion strains showed a dramatic change in cell morphology when analyzed under heat shock conditions.”
is herewith being corrected to:
“Consistent with the in vivo and in vitro results on the chaperone function of Hsp42 and Hsp26, significant changes at late logarithmic phase were only visible for deletion of Hsp42. However, both deletion strains showed a dramatic change in cell morphology when analyzed under heat shock conditions.”
Despite these changes, the main conclusions of the article remain unchanged, and have since been verified and further refined by multiple studies, e.g., in Specht et al (2011), Ungelenk et al (2016), Mackenzie et al (2016), and Grousl et al (2018). Additional replicates for panel 9B are included as Source Data with this corrigendum.
Per journal policy, The EMBO Journal conducted additional image screening of all other figures in this article. These analyses identified further duplications of two panels for experimental controls of gene deletion strains (top middle panel, bottom left panel) within Fig 5B and 5C, respectively. The Journal subsequently received scans of Western blots from the original laboratory books, confirming the absence of bands in the respective experimental settings, and stress induction of small heat shock proteins in other panels. The authors note that duplication of the “empty” panels appears to have been an oversight during figure assembly and does not affect the conclusions from Fig 5. The original scans from laboratory book pages are published as Source Data with this corrigendum.
Faint lines in Figs 3 and 4C were judged to be image scanning/processing artifacts based on available source data.
Figure 9B. Original.
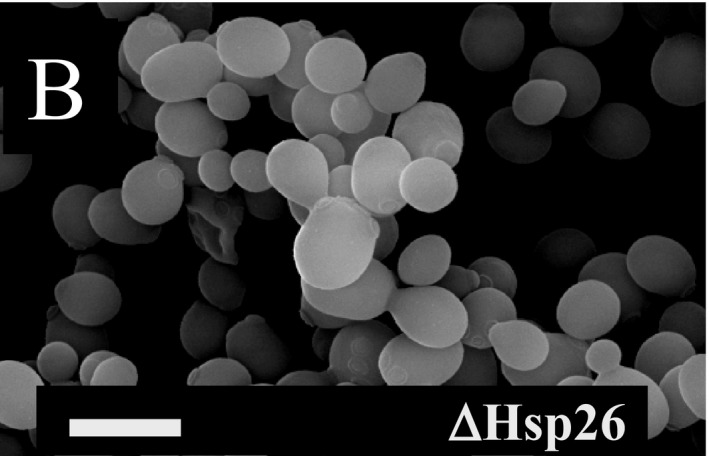
Figure 9B. Corrected.
Supporting information
Source Data for Figure 5
Source Data for Figure 9
Correction to: The EMBO Journal (2004) : 638–649. DOI 10.1038/sj.emboj.7600080 | Published online 29 January 2004
References
- Grousl T, Ungelenk S, Miller S, Ho CT, Khokhrina M, Mayer MP, Bukau B, Mogk A (2018) A prion‐like domain in Hsp42 drives chaperone‐facilitated aggregation of misfolded proteins. J Cell Biol 217: 1269–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie RJ, Lawless C, Holman SW, Lanthaler K, Beynon RJ, Grant CM, Hubbard SJ, Eyers CE (2016) Absolute protein quantification of the yeast chaperome under conditions of heat shock. Proteomics 16: 2128–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae . J Cell Biol 195: 617–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Lindquist SL (1989) hsp26 of Saccharomyces cerevisiae is related to the superfamily of small heat shock proteins but is without a demonstrable function. Mol Cell Biol 9: 5265–5271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungelenk S, Moayed F, Ho CT, Grousl T, Scharf A, Mashaghi A, Tans S, Mayer MP, Mogk A, Bukau B (2016) Small heat shock proteins sequester misfolding proteins in near‐native conformation for cellular protection and efficient refolding. Nat Commun 7: 13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Source Data for Figure 5
Source Data for Figure 9


