SUMMARY
Acute physical activity leads to several changes in metabolic, vascular and immune pathways. While studies have examined selected changes in these pathways, the system-wide molecular response to an acute bout of exercise has not been fully characterized. We performed longitudinal multi-omic profiling of plasma and peripheral blood mononuclear cells including metabolome, lipidome, immunome, proteome and transcriptome from 36 well-characterized volunteers, before and after a controlled bout of symptom-limited exercise. Time-series analysis revealed thousands of molecular changes and an orchestrated choreography of biological processes involving energy metabolism, oxidative stress, inflammation, tissue repair, growth factor response as well as regulatory pathways. Most of these processes were dampened and some were reversed in insulin resistant participants. Finally, we discovered biological pathways involved in cardiopulmonary exercise response and developed prediction models revealing potential resting blood-based biomarkers of peak oxygen consumption.
Keywords: cardiopulmonary exercise, fitness, peak VO2, multi-omics, integrative omics, systems biology, insulin resistance, predictive analytics, time-series analysis, outlier analysis
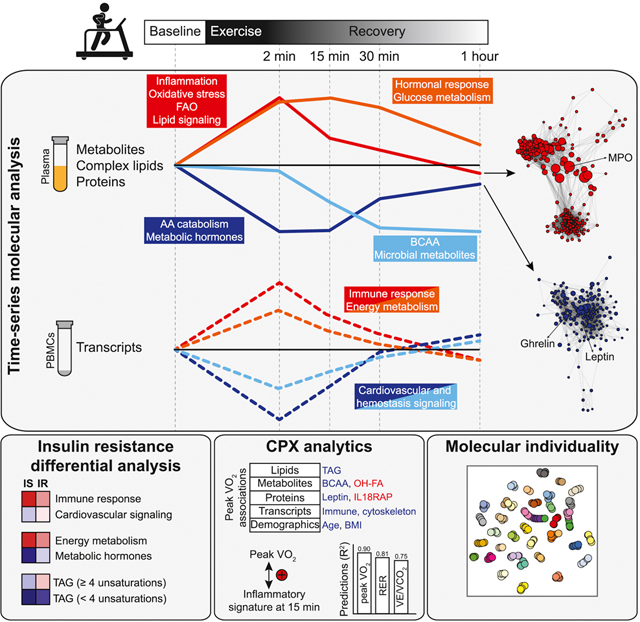
Graphical Abstract
INTRODUCTION
Physical activity is one of the pillars of cardiovascular, immune and cognitive health (Warburton et al., 2006). In addition to its role in promoting health, acute exercise testing can unmask early signs of cardiopulmonary disease and provide prognostic markers of outcome (e.g. peak oxygen consumption and ventilatory efficiency) as well as predictive markers of response to therapy (e.g. contractile reserve) (Arena and Sietsema, 2011; Force et al., 2007).
Acute physical activity triggers complex molecular responses including changes in acute inflammatory markers (e.g. interleukin-6) (Fischer, 2006) and metabolic pathways (e.g. glycolysis and fatty acid oxidation) (Goodwin et al., 2007). However, previous studies have been limited by the breadth of molecules measured and biological processes covered. A better understanding of these processes through detailed analyses can improve our knowledge of exercise physiology and guide the development of exercise analytics in clinical practice.
In this context, we performed longitudinal multi-omic profiling of blood components (i.e. plasma and peripheral blood mononuclear cells) before and after a controlled bout of acute cardiopulmonary exercise (CPX) in participants with a wide spectrum of insulin resistance. The goal was to characterize the detailed series of events that occur in response to exercise and understand how they relate to fitness measures, and how they are affected by insulin resistance. Several ventilatory parameters are measured during CPX including peak oxygen consumption (peak VO2), a marker of fitness and minute ventilation/carbon dioxide production slope (VE/VCO2), a marker of respiratory drive and ventilatory dead space (Arena et al., 2004). Molecules and pathways associated with peak VO2 have not been systematically investigated but are expected to be valuable in understanding cardiovascular fitness and exercise limitations as well as identifying novel biomarkers of disease (Shah and Hunter, 2017).
Assessing the molecular response to exercise not only in healthy individuals but also in individuals at risk for cardiometabolic disease may be particularly relevant with the current epidemic of obesity and diabetes mellitus (Schussler-Fiorenza Rose et al., 2019; Tabak et al., 2012). Insulin resistance is one of the central pathophysiological processes involved in obesity and diabetes mellitus that contributes to end-organ dysfunction. The effects of physical activity on insulin sensitivity have been previously investigated (Bird and Hawley, 2016); however, the molecular response to exercise across the continuum of insulin resistant individuals has not been thoroughly studied.
Our deep longitudinal profiling revealed an orchestrated molecular choreography in response to acute exercise illustrating the complex interplay between biological processes across various organ systems. We also defined healthy molecular profiles of peak VO2 and demonstrated the ability of baseline multi-omic analytes to predict key CPX parameters. We showed that insulin resistance is associated with altered molecular response to exercise in several major biological processes, and we explored the clinical relevance of molecular outlier analysis for exercise response at an individual level. Altogether, this study illustrates the value of deep longitudinal profiling to decipher complex physiological processes in humans and provides a valuable open access resource for the integrated study of multi-level molecular response to acute exercise.
RESULTS
Cohort characteristics and research design.
After overnight fasting, 36 highly characterized participants underwent symptom-limited CPX testing and serial blood collection (Figure 1a). The cohort was composed of individuals with an age range of 40–75 (mean (± SD) of 59 ± 8 years old), a body mass index (BMI) of 28.4 ± 5.1 kg/m2 and 58% were male. Participants were selected to span a wide range of peripheral insulin resistance with a steady-state plasma glucose (SSPG) of 153 ± 67 mg/dl as determined by the modified insulin suppression test (Schussler-Fiorenza Rose et al., 2019). Resting and stress echocardiography, as well as vascular ultrasound were performed to exclude heart failure, stress-induced ischemia or atherosclerotic disease. The majority of the participants (86%) reached a respiratory exchange ratio (RER) > 1.05 at peak exercise and the remaining individuals reached > 95% maximal predicted heart rate for their age. Table S1.1 provides detailed information about participant’s baseline demographics, echocardiographic and CPX characteristics. Intravenous blood specimens were collected before exercise (baseline) as well as 2, 15, 30 min and 1-hour in recovery. Fifteen participants also provided a fasted blood sample the next morning to assess inter-day variability and a subset of the cohort (n = 14) participated in a control experiment to evaluate the natural deviation of analytes in the absence of exercise.
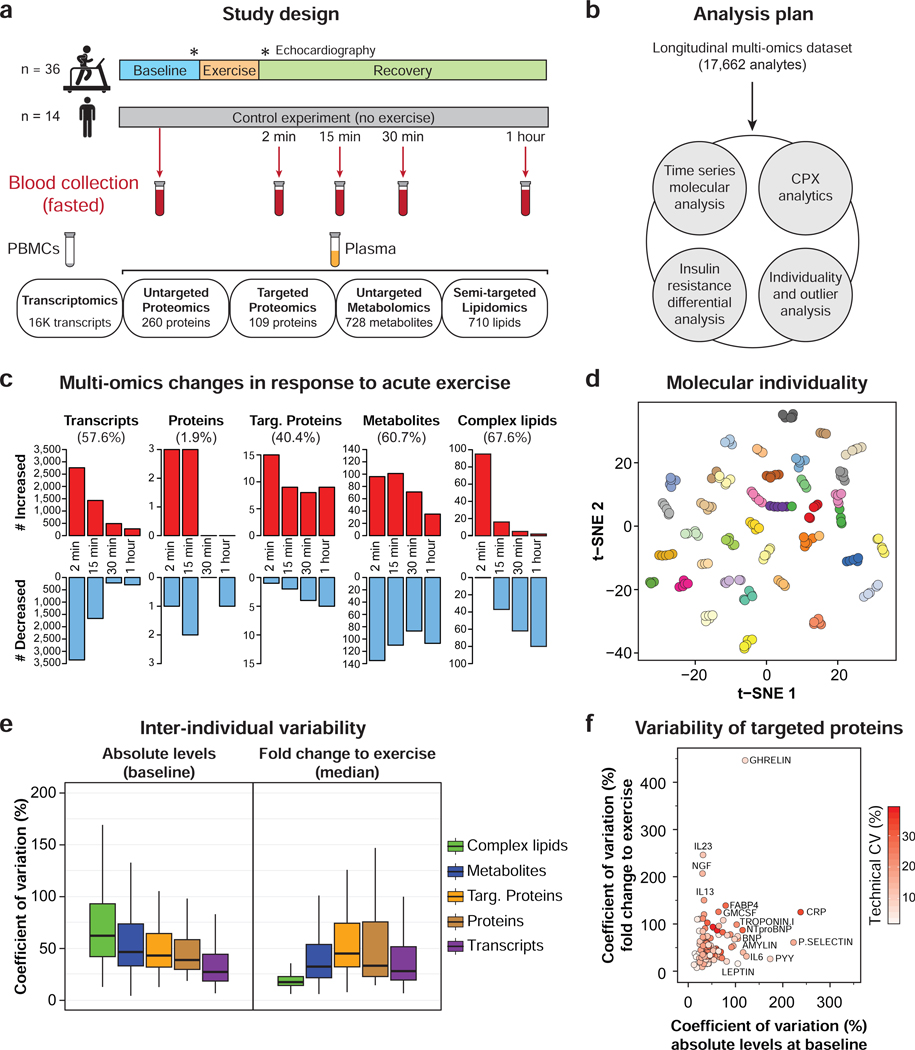
Figure 1. Study design, molecular response to exercise and inter-individual variability.
(a) Overview of the study design including an acute bout of exercise (symptom-limited cardiopulmonary exercise, CPX), cardiovascular phenotyping and longitudinal multi-omic profiling from blood specimens. PBMCs: peripheral blood mononuclear cells. (b) Analysis plan. (c) Multi-omic changes in response to acute exercise. (d) 2D visualization of all multi-omic analytes using t-distributed stochastic neighbor embedding (tSNE) technique. Each dot represents a single sample colored by participants. (e) Inter-individual variability at baseline (absolute levels) and in response to exercise (median of fold change to exercise) across molecule types. (f) Inter- individual variability of targeted proteins (technical, at baseline and in response to exercise). See also Figures S1–S3 and Tables S1–S3.
In-depth multi-omic profiling was performed on each sample including plasma proteomics (targeted and untargeted), metabolomics (untargeted), and lipidomics (semi-targeted) and gene expression (transcriptomics) from peripheral blood mononuclear cells (PBMCs). Complex lipids refer to glycerolipids, glycerophospholipids, sphingolipids and sterol lipids, and targeted proteins were selected given their relevance to exercise physiology focusing on metabolic, cardiovascular and immune proteins (i.e. immunome) (Table S1.2). After data curation and annotation, the final dataset contained a total of 17,662 analytes which included 15,855 transcripts, 260 proteins from the untargeted analysis, 109 targeted proteins, 728 metabolites and 710 complex lipids. A list containing all the detected analytes can be found in Table S1.3. The longitudinal multi-omic dataset was used to i) characterize the dynamic molecular response to acute exercise, ii) determine molecular associations with peak VO2 and predict key measurements of exercise physiology, iii) analyze the differential response to exercise in insulin resistant participants and iv) examine the clinical relevance of outlier analysis at an individual level (Figure 1b).
Multi-omic changes in response to acute exercise.
The quality of each omic dataset was first examined to ensure technical reproducibility and the absence of batch effect (Figure S1a and Figure S2a). One participant underwent CPX testing twice, 10 months apart at the beginning and at the end of the study. Samples from both sessions clustered together indicating that the exercise protocol and sample collection were reproducible (Figure S1b). Molecules significantly affected by exercise were identified using linear models adjusted for personal baselines, age, sex, race/ethnicity and BMI. Acute exercise induced extensive changes in 9,815 analytes spanning all omic layers (56.9% of the detected analytes, FDR < 0.05) indicating large system-wide changes (Figure 1c and Table S2.1). Different patterns of changes were observed across the molecule types; transcripts (n = 9,132) exhibited a very rapid response reaching a maximum/minimum level early post-exercise and returning to baseline within 60 min (93.7%) whereas metabolites (n = 442) and complex lipids (n = 192) were altered across all time points and a large proportion (19.5% and 28.9%, respectively) remained significantly different at 60 min in recovery (Figure S1c). Despite the global molecular impact of exercise, samples from the same participant tended to cluster together (Figure 1d) indicating greater individual similarity even in the presence of the exercise perturbation.
In order to demonstrate that the changes reported are induced by exercise and not timing or fasting, 14 individuals from the cohort participated in a control experiment that followed the same protocol (i.e. sample collection and processing) but without exercise. Linear models revealed that < 2% of the analytes that changed with exercise (targeted proteins, metabolites and complex lipids) also varied naturally within a 1-hour time window without exercise (FDR < 0.10, Figure S3a). These molecules all presented a slow decrease (except hydroxybutyrate) whereas their trajectories with exercise were different either in amplitude or direction (Figure S3b,c). These results demonstrate that the changes reported in this study are due to exercise and not explained by the natural variation of the analytes or fasting. In addition, we found that the level of molecular response to exercise was at least two-fold larger than the individual inter-day variation at 24 hours (n = 15, Figure S3d,e).
Baseline versus exercise-induced individual omic variability.
The inter-individual variability both at baseline and in response to exercise has not previously been compared across the omic layers. At baseline, complex lipids presented the highest coefficient of variation (CV) followed by metabolites, proteins and transcripts (62.0%, 46.2%, 38.9% and 26.9%, respectively; Figures 1e, S1d, S2b and Table S3.1). Among lipids, triacylglycerol (TAG) and diacylglycerol (DAG) species were the most variable (Figure S2c), consistent with the total TAG content measured by a clinic test (CV = 57.1%). Similarly, xenobiotics – small molecules acquired from the environment or generated by the microbiome – were the most variable metabolites (e.g. secondary bile acids and indoles, Figure S2d). Enrichment analysis using variable transcripts (CV > 100%, n = 412) highlighted inflammation as the most variable biological process in the cohort with pathways such as ‘communication between innate and adaptive immune cells’ (FDR = 3.0E-07) (Table S3.2). This was further supported by the variability in C-reactive protein (CRP), interleukin 6 (IL-6) and serum amyloid A1 (SAA1) and A2 (SAA2) (Figure 1f and Table S3.1).
The inter-individual variability in response to exercise differed from baseline with proteins varying the most (CV = 36.8%), followed by metabolites, transcripts, and complex lipids (32.1%, 27.7%, 17.0%, respectively; Figures 1e, S2e and Table S3.3). Some proteins demonstrated high variability in response to exercise despite low baseline and technical variability (e.g. interleukins (IL-13 and IL-23) as well as nerve growth factor (NGF)) (Figure 1f). Among metabolic hormones, acylated ghrelin and leptin differed with high and low variability, respectively, despite similar technical and baseline variability. Finally, the most variable transcripts in response to exercise (CV > 100%, n = 1,334) were enriched for ‘osteoarthritis pathway’ (FDR = 5.6E-05) and ‘hepatic fibrosis/hepatic stellate cell activation’ (FDR = 1.6E-04), indicating differential regulation of these pathways in response to exercise (Table S3.2).
Time-series system-wide molecular analysis.
We took advantage of the high sampling density post- exercise to i) define longitudinal clusters (FDR < 0.05) and ii) calculate pairwise correlations between molecules within each cluster. Using c-means clustering, four main clusters of longitudinal trajectories were identified that delineated key biological processes encompassing early (i.e. energy metabolism, oxidative stress and immune response) and late events (i.e. energy homeostasis, tissue repair and remodeling) (Figure 2a and Table S2.1). Some molecules increased following exercise and quickly returned to baseline (cluster 1) whereas others presented a delayed increase post-exercise before returning to baseline (cluster 2). The remaining analytes decreased in response to exercise with some returning to baseline within one hour (cluster 3) and others continuing to decrease in recovery (cluster 4). Correlation networks were generated for each cluster highlighting potential regulators of biological processes and novel molecular functions through unexpected connections.
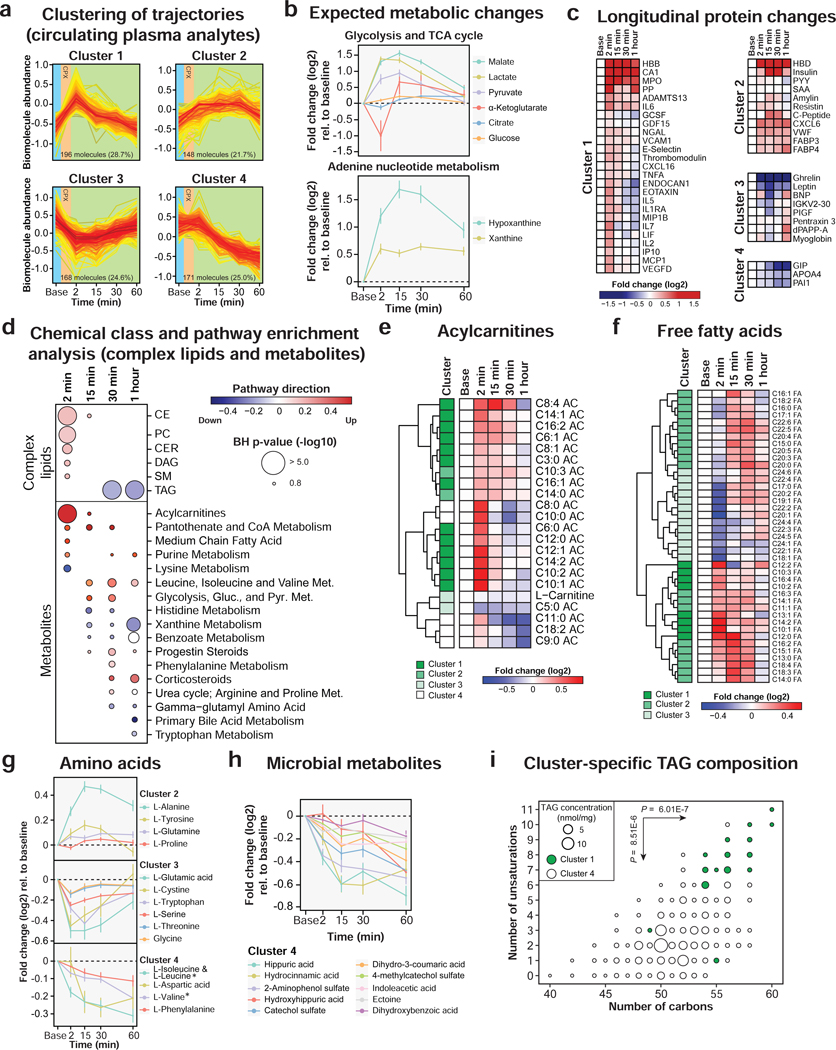
Figure 2. Multi-omic changes in response to acute exercise.
(a) Clustering of longitudinal trajectories using significant circulating plasma analytes (FDR < 0.05). (b) Expected metabolic changes in response to exercise including glycolysis, TCA cycle and adenine nucleotide metabolism. The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). (c) Heatmap of significant proteins representing the median log2 fold change relative to baseline in the cohort. Proteins were grouped by clusters. (d) Pathway/chemical class enrichment analysis of circulating plasma metabolites and complex lipids. Pathway direction is the median log2 fold change relative to baseline of significant molecules in each pathway (blue: downregulated, red: upregulated). The dot size represents pathway significance. Heatmaps representing the median log2 fold change relative to baseline for acylcarnitines (e) and free fatty acids (f). The clusters are indicated on the left side of the heatmaps. Longitudinal trajectories of significant amino acids (g) and microbial metabolites (h) in response to exercise. The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). * Branched chain amino acids. (i) Triacylglycerol (TAG) fatty acid composition in clusters 1 and 4. Two-sided Welsh t-tests were used to calculate differential enrichment in TAG composition. See also Figures S4–S5 and Table S2.
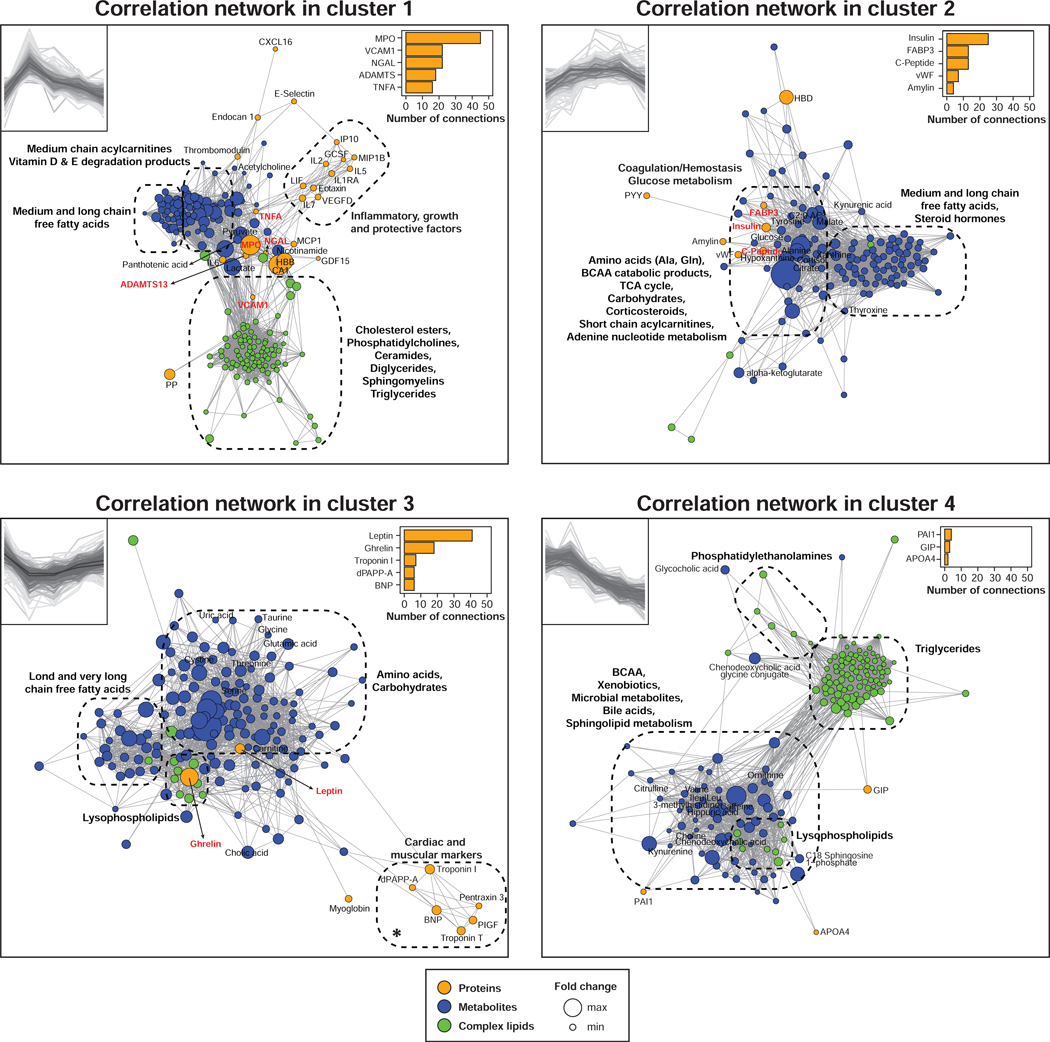
Figure 3. Integrative multi-omic analysis of circulating analytes.
Pairwise spearman correlation networks of multi-omic measures belonging to each cluster as defined in Figure 2a. Nodes were color-coded by molecule type and their size represent the median fold change relative to baseline. The top 5 proteins with the greatest number of first order connections in each correlation network were displayed. Proteins with more than 10 connections are in bold and red. * Cardiac and muscular markers belong to cluster 3 but the decrease is not significant. See also Figures S4 and Table S2.
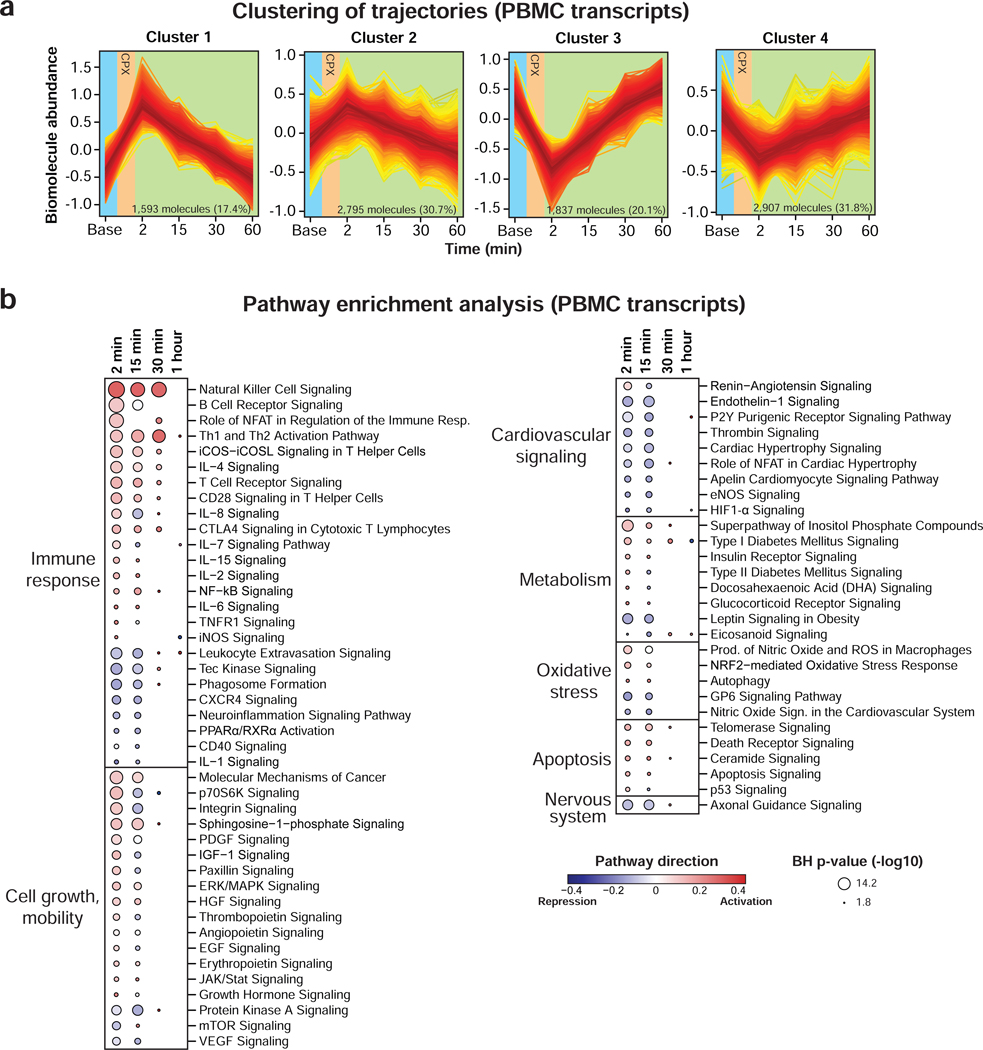
Figure 4. PBMC gene expression changes in response to acute exercise.
(a) Clustering of longitudinal gene expression trajectories (FDR < 0.05). (b) Pathway analysis using PBMC transcripts significantly changing in response to exercise. Pathway direction is the median log2 fold change relative to baseline of significant transcripts in each pathway (blue: downregulated, red: upregulated). The dot size represents pathway significance. See also Figures S5 and Table S2.
Cluster 1.
Cluster 1 was enriched in molecules (n = 196) associated with anaerobic metabolism, immune response, oxidative stress, fatty acid oxidation and complex lipid metabolism (Figure S4a). As expected, we observed a sharp increase in plasma concentrations of glycolysis products (i.e. lactate, pyruvate) and tricarboxylic acid (TCA) cycle intermediates (i.e. malate) presumably reflecting heightened anaerobic metabolism (Figure 2b) (Lewis et al., 2010).
Oxidative stress signaling was detected through the accumulation of myeloperoxidase (MPO). MPO was among the most responsive molecules (in fold change) and emerged as a centrally connected proteomic feature with the greatest number of connections (n = 45) (Figure 3). MPO is predominantly released from neutrophils via degranulation and is believed to signal skeletal muscle damage or stress and recruit macrophages to damaged sites (Morozov et al., 2006; Reihmane et al., 2012). It was connected to all omic layers bridging inflammatory and growth/protective factors (n = 5) to acylcarnitines (n = 12) and complex lipids (n = 9) suggesting potential novel roles for MPO in regulating aspects of inflammation and lipid metabolism. For example, MPO was strongly associated with the inflammatory marker neutrophil gelatinase-associated lipocalin (NGAL) (FDR < 1.0E-13) (Otto et al., 2015), and it can potentiate inflammation by activating endothelial cells to release more cytokines (Odobasic et al., 2016).
Inflammatory response post-exercise was evident via the secretion of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) (Golbidi and Laher, 2014; Kinugawa et al., 2003; Vijayaraghava et al., 2015) (Figure 2c). These changes were concomitant with an increase of IL-1 receptor antagonist (IL-1RA) and vascular endothelial growth factor D (VEGF-D) which provide a compensatory anti-inflammatory response. Pro-inflammatory properties of TNF-α were confirmed by its association with many other cytokines (i.e. IL-2, IL- 6, IL-7, granulocyte-colony stimulating factor (G-CSF), monocyte chemoattractant protein 1 (MCP-1), leukemia inhibitory factor (LIF) and eotaxin) (Figure 3). Interestingly, TNF-α also correlated with pantothenic acid (vitamin B5), an essential nutrient required for cellular energy metabolism (Tahiliani and Beinlich, 1991), confirming its role as a potent metabolic regulator (Chen et al., 2009; Sethi and Hotamisligil, 1999).
Endothelial markers and vascular adhesion molecules also constituted key molecules of cluster 1 with vascular cell adhesion molecule-1 (VCAM-1), E-selectin and endothelial cell-specific molecule 1 (endocan 1). VCAM-1 was connected to a variety of molecules (n = 22) including neutrophil inflammatory and oxidative stress markers (MPO and NGAL), glycolytic products (lactate and pyruvate), pantothenic acid and many complex lipids (n = 14). VCAM-1 has been previously shown to increase with exercise in untrained individuals as well as in patients with peripheral vascular disease (Brevetti et al., 2001; Jilma et al., 1997) and thus may be a marker for cardiovascular function.
Fatty acid oxidation (FAO) was activated by exercise as indicated by the early accumulation of many acylcarnitines (n = 18) and free fatty acids (n = 30) (Figure 2d and Table S2.1, S2.2). Distinct trajectories were evident depending on fatty acid composition. Medium-chain acylcarnitines (i.e. C6:0, C8:0, C10:0, C12:0) accumulated the most following exercise and returned to baseline by 15–30 min in recovery whereas others (i.e. C6:1, C8:1, C14:0, C16:1) accumulated to a lesser extent and returned to baseline more slowly (30 min to 1 hour) (Figure 2e). The increased abundance of circulating medium-chain acylcarnitines reflects partial FAO in skeletal muscle (Lehmann et al., 2010; Zhang et al., 2017). The level of circulating free carnitine demonstrated an inverse trajectory (cluster 3) suggesting that it is used to form acylcarnitines from free fatty acid molecules. Free fatty acids exhibited three main trajectories with some reaching a maximum at 2 min post-exercise (10–12 carbons, cluster 1), others at 15 min (14–18 carbons, cluster 2) and the remainder decreasing at 2 min in recovery (20–24 carbons, cluster 3) (Figure 2f). These observations were confirmed by differential expression analysis between each consecutive time points (FDR < 0.05, Figure S5a and Table S2.3) and suggests that long-chain fatty acids, in particular the ones with 20–22 carbons (C20:1, C20:2, C22:1, C22:2 and C22:3), are preferentially oxidized during exercise while partial FAO results in an increased abundance of medium-chain fatty acids.
Exercise was also accompanied by a transient accumulation of diverse complex lipids including cholesteryl esters (CE, n = 20), phosphatidylcholines (PC, n = 23), diacylglycerols (DAG, n = 10), ceramides (CER, n = 9) and sphingomyelins (SM, n = 8) (Figure 2d). Sphingolipids, and in particular ceramides, may be involved in signaling inflammation in response to exercise similarly to that described for inflammatory diseases (Maceyka and Spiegel, 2014).
Cluster 2.
Molecules in cluster 2 (n = 148) presented a delayed increase post-exercise and a large proportion of these molecules were associated with carbohydrate metabolism. Exercise triggered the secretion of numerous hormones including steroid and thyroid hormones as well as corticosteroids to restore homeostatic balance (Figure 2d and 3). Correlation networks provided insights into hormonal responses of exercise. We detected an increase of cortisol which can stimulate gluconeogenesis in the liver leading to a rise of circulating glucose (Kjaer, 1998). We also observed a significant positive correlation between glucose and insulin levels (rho = 0.44, FDR = 1.30E-05); insulin secretion enables cellular glucose absorption to meet tissue energy demand. These changes were concomitant with an accumulation of fatty acid binding proteins 3 and 4 (FABP3 and FABP4) that facilitate glucose and free fatty acid uptake in skeletal muscle and heart tissue (Kusudo et al., 2011). Insulin was the most connected proteomic feature in the cluster (n = 25) and was highly correlated with the proinsulin C-peptide and amylin (co-secreted with insulin). Increased glucose metabolism correlated with TCA cycle constituents (malate, citrate, α-ketoglutarate) and resulted in a marked increase of products of adenine nucleotide catabolism (i.e. hypoxanthine and xanthine) that are markers of ATP turnover (Lewis et al., 2010) (Figure 2b). In addition, we detected a delayed increase of the purine end-product uric acid, presumably due to increased synthesis and decreased renal excretion (Sutton et al., 1980) (Figure S4). Finally, we also detected an increase of coagulation and hemostasis factors, such as von Willebrand factor (vWF) and A Disintegrin and Metalloprotease with ThromboSpondin motif repeats 13 (ADAMTS-13), likely in response to the shear stress induced by treadmill exercise (Stakiw et al., 2008).
Cluster 3.
Cluster 3 contained molecules (n = 168) that decreased in response to exercise and returned to baseline within 1 hour. The correlation network was centered on two metabolic hormones leptin and ghrelin, suggesting a role in regulation of appetite by exercise (Figure 3). Leptin is predominantly secreted by adipose tissue whereas ghrelin is produced in the stomach (Klok et al., 2007) and both have been reported to decrease following intense exercise and suppress hunger (King et al., 1994). The levels of many (15 of 20) amino acids changed upon exercise (FDR < 0.05) reflecting a central role in exercise physiology (Figures 2e and S4a). Six amino acids (i.e. glutamic acid, cystine, tryptophan, serine, threonine and glycine) belonged to cluster 3 suggesting that they were catabolized presumably by skeletal muscle cells to produce energy (Henriksson, 1991) and re-synthesized in the recovery phase. Four amino acids (i.e. alanine, tyrosine, glutamine and proline) presented an opposite trajectory (cluster 2) accumulating as a product of increased cellular metabolism. Alanine and glutamine released in plasma is expected due to muscle ammonia detoxification (Lewis et al., 2010).
Cluster 4.
Molecules in cluster 4 (n = 171) were metabolized in response to exercise but did not return to baseline within the 1-hour recovery phase (Figure 3). Some amino acids presented this trajectory and included branched-chain amino acids (BCAA) leucine, isoleucine and valine (Figure 2g). BCAA are essential amino acids that cannot be synthesized by the body and are preferentially catabolized by skeletal muscle (Henriksson, 1991) and used to repair damaged skeletal muscle fibers (Negro et al., 2008). BCAA catabolism was evident with a marked increase in branched-chain ketoacids (Figure S4). Many other metabolites had the same trajectory including microbial metabolites (Figure 2h), xenobiotics (caffeine metabolism) and bile acids (Figure S4)
Cluster 4 also contained many TAG species reflecting hydrolysis to release fatty acids necessary for energy production. While most TAG belonged to this cluster, we identified a subset of TAG that increased transiently following exercise (cluster 1). A close examination of their fatty acid composition revealed that TAG in cluster 1 contained fatty acids with more carbons (P = 6.01E-07) and unsaturations (P = 8.51E-06) than TAG enriched in cluster 4 (Figure 2i). TAG in cluster 1 contained fatty acids with signaling properties including arachidonic acid (AA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) suggesting that the transient burst might play a role in signaling (AA) or compensating for inflammation (EPA, DHA) (Calder, 2013). In contrast, TAG with shorter and saturated fatty acids may be preferentially used for energy production (Ranallo and Rhodes, 1998). Altogether, our dense sampling revealed dynamically and functionally distinct subclasses of TAG.
Clinical correlates of time-series clusters.
We also explored associations of key changes described above with demographics and clinical parameters including body composition and CPX metrics (Table S2.4). Molecules involved in metabolic functions tended to associate with body composition and exercise physiological measures. As an example, the abundance of leptin at minimum intensity (15 min post-exercise) was strongly positively associated with estimated fat mass (P = 9.0E-06) and estimated percent fat (P = 8.2E-08) based on the National Health and Nutrition Examination Survey (NHANES) formulas (Lee et al., 2017). In contrast, oxidative stress and immune molecules as well as lactate did not associate strongly with any of the tested parameters (P > 0.01), indicating they are largely independent of body composition and exercise physiological measures over the tested ranges.
Time-series PBMC gene expression analysis.
We investigated PBMC gene expression in response to exercise both because immune cells play a critical role in muscle stress response and as a system-wide marker of alterations in gene expression (Gjevestad et al., 2015; Philippou et al., 2012; Radom-Aizik et al., 2009; Ulven et al., 2015). Transcript trajectories were categorized in 4 clusters with up- and down-regulated genes (high and low amplitudes) reaching a maximum response at 2 min and rapidly returning to baseline within 30–60 min (Figure 4a and Table S2.1). Pathway enrichment analysis using all significant transcripts at each time point (FDR < 0.05) revealed expected and novel pathways (Figure 4b and Table S2.5).
Exercise induced a robust inflammatory response with increased transcripts of ‘natural killer cell’, ‘Th1 and Th2 activation’, ‘B cell receptor’, ‘T cell receptor’, ‘NF-kB signaling’ and many interleukin signaling pathways as previously reported (Carlson et al., 2011; Connolly et al., 2004; Gjevestad et al., 2015). We also detected a transient overexpression of the human leukocyte antigen (HLA) class I genes (i.e. HLA-A, HLA-B and HLA-C) that participate in the immune response. Interestingly, these genes presented dynamic allele- specific expression (ASE) in response to exercise (FDR < 0.05) (Figure S5c,d) suggesting that genetic variation in cis-regulatory elements may be responsible for HLA class I genes dysregulation in the context of exercise. This observation is consistent with a recent study reporting differential ASE in many HLA genes during T-cell activation (Gutierrez-Arcelus et al., 2019). Pathways related to oxidative stress (e.g. ‘production of nitric oxide and reactive oxygen species in macrophages’) and apoptosis (e.g. ‘telomerase signaling’, ‘death receptor signaling’) were also altered in response to exercise. These results are consistent with the early release in circulation of pro- and anti-inflammatory proteins.
In addition to inflammatory and immune functions, we found a myriad of pathways associated with cell growth and mobility that are likely involved in muscle tissue repair and remodeling (Connolly et al., 2004). For instance, angiogenesis and wound healing pathways were upregulated in response to exercise (i.e. ‘PDGF signaling’, ‘HGF signaling’ and ‘EGF signaling’). We also detected many pathways associated with cardiovascular and hemostasis related signaling highlighting the interconnection between exercise and cardiovascular health. These pathways were mainly downregulated and included ‘endothelin-1 signaling’, ‘P2Y purigenic receptor signaling pathway’, ‘thrombin signaling’ and ‘cardiac hypertrophy signaling’. In addition, we observed dysregulated metabolic pathways including a repression of ‘leptin signaling pathway in obesity’ consistent with a decrease of circulating leptin abundance and activation of ‘superpathway of inositol phosphate compounds’ that may be involved in PBMC activation through synthesis of phosphoinositides (Huang et al., 2007).
Although most pathways returned to baseline 30 min post-exercise, some pathways persisted for more than 1 hour (‘Th1 and Th2 activation pathway’) suggesting longer lasting effects of some biological processes. Differential gene expression analysis on consecutive time points provided additional insights into pathway dynamics (Figure S5b and Table S2.6). In particular, most pathways were dysregulated immediately post- exercise and started reverting back 15 min later. It is known that exercise impacts immune cell count in a cell type-dependent fashion (Millard et al., 2013; Shinkai et al., 1992). Hence, changes in gene expression may occur by a combination of changes in cell population (most likely by mobilization) and cellular activation.
Multi-omic features of CPX parameters.
Several CPX parameters are strongly predictive of outcome. Maximum oxygen consumption (peak VO2) - a measure of aerobic fitness - is among the best predictors of longevity in the general population (Ladenvall et al., 2016) as well as survival in patients with heart failure (Sarullo et al., 2010). In our study, the range of peak VO2 (scaled to body weight, BW) was representative of a reference population (Myers et al., 2017) with a mean (± SD) of 30.6 ± 8.7 ml/kg/min (Figure S6a,b). Ventilatory efficiency measured as the slope of the minute ventilation to carbon dioxide production relationship (VE/VCO2 slope) is a strong prognostic marker in heart failure (Arena et al., 2004). The respiratory exchange ratio (RER) which is the ratio between the amount of carbon dioxide (CO2) produced in metabolism and oxygen (O2) used is a marker of maximum effort (Albouaini et al., 2007). We investigated key associations of omic measures with peak VO2 at baseline and in recovery and developed predictive models of CPX parameters using baseline measurements.
Multi-omic associations of peak VO2.
We applied linear regression models to find significant associations with peak VO2 at baseline and at each time point in recovery. To account for potential confounders, we adjusted for age, sex and race/ethnicity (Kaminsky et al., 2015) as well as for body mass index, fat mass or percent fat estimated with the NHANES formulas. The overlapping associations presented in Figure S6c and Table S4.1 were used for downstream analysis. We found that a large proportion of omic measures associated significantly with exercise capacity with 51.9% of complex lipids and 30.3% of metabolites on average across all time points (FDR < 0.05, Figure 5a). In addition, we calculated associations with peak VO2 scaled to estimated lean body mass (LBM) to further identify correlations independent of muscle mass. As expected, fewer associations were significant, but a number of key biological processes, in particular lipid metabolism, persisted as described below (Figure S6d). Table S4.2 shows associations based on both BW and LBM scaling.
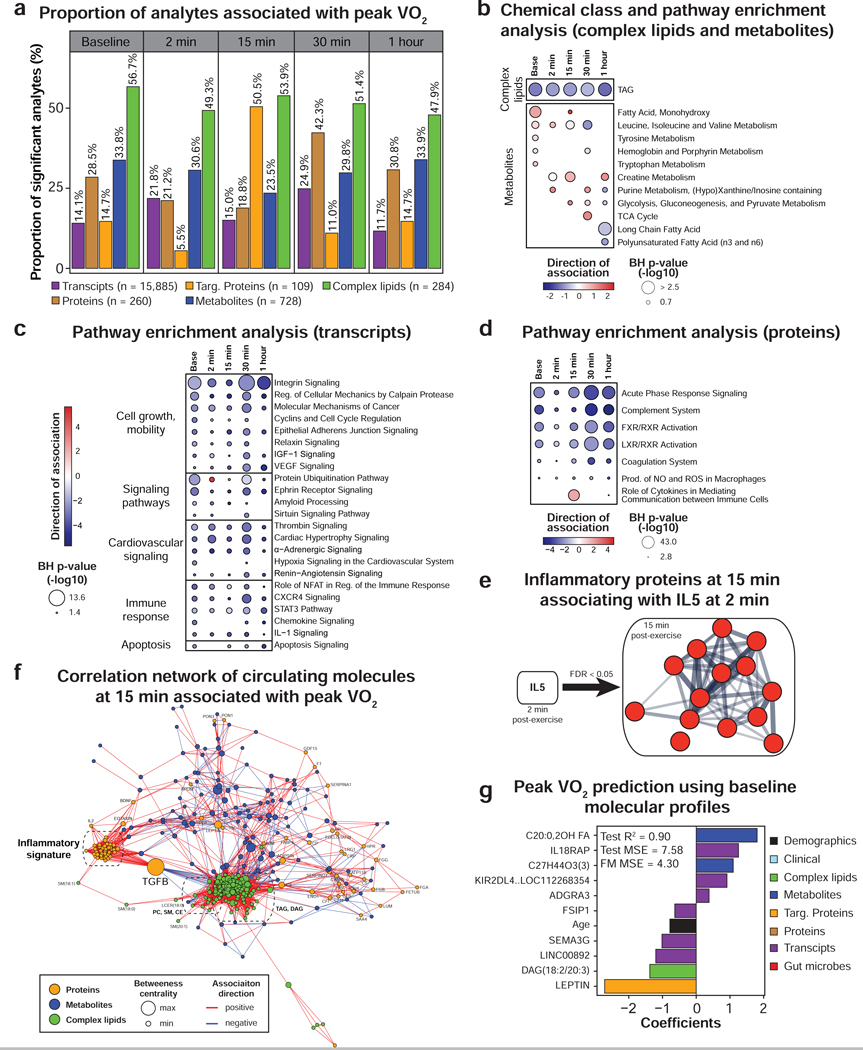
Figure 5. Multi-omic analysis of peak VO2.
(a) Proportion of analytes associated with peak VO2 (scaled by body weight) as determined by linear regression analysis. Only the molecules significant in three regression models adjusting for BMI or fat mass or percent fat were presented. Pathway/chemical class enrichment analysis of metabolites and complex lipids (b) as well as pathway analysis using PBMC gene expression (c) and circulating proteins (d). Pathway direction is the median beta coefficient of significant molecules in the pathway (blue: negative association, red: positive association). The dot size represents pathway significance. (e) Functional association network using the proteins from the “inflammatory fitness signature” at 15 min in recovery significantly associated with IL-5 at 2 min post-exercise (spearman correlation, FDR < 0.05). This analysis was performed using the web tool STRING. Line thickness indicates the strength of data support. Proteins are colored in red to signify a positive association with IL-5. (f) Pairwise spearman correlation networks of multi-omic measures significantly associated with peak VO2 at 15 min post-exercise. Nodes were color-coded by molecule type, their size represent the betweenness centrality and the edges were color-coded by association direction. (g) Molecules selected in the multi-omic peak VO2 prediction model and associated coefficients. MSE: mean square error, FM: full model. See also Figure S6 and Table S4.
Baseline associations.
Leptin was the strongest correlate of peak VO2 both at baseline and in recovery (negative association, FDR < 1.0E-05) (Table S4.1). Triglycerides and BCAA, known markers of poor metabolic health (obesity and type 2 diabetes) (Guasch-Ferre et al., 2016) were also associated with lower peak VO2 (Figure 5b and Table S4.3). In contrast, the transporter of thyroxine and retinol transthyretin (TTR), a biomarker of lean body mass (Ingenbleek and Bernstein, 2015), hydroxy-fatty acids, corticosterone, hippuric acid and bile pigments (i.e. biliverdin and bilirubin) were positively associated with peak VO2. Hydroxy-fatty acids and corticosterone are known to increase with exercise training (Droste et al., 2003; Nieman et al., 2013) and hippuric acid is a marker of gut microbiome diversity (Pallister et al., 2017). Bile pigments have potent antioxidant properties which may explain their association with fitness (Wegiel and Otterbein, 2012). Our proteomic analysis also revealed enrichment in the FXR/RXR and LXR/RXR activation pathways that are known to be activated by bile acids and regulate glucose and lipid metabolism (Claudel et al., 2005).
PBMC gene expression enrichment analysis revealed numerous pathways negatively associated with aerobic fitness mainly involving cell growth and mobility, immune response, signaling pathways, cardiovascular signaling, apoptosis and metabolism (Figure 5c and Table S4.4). Even though most pathways were enriched at baseline and at each time point in recovery, many pathways were the most significant 30 min post-exercise.
Of note many of the associations were stronger when scaled to BW than LBM. Novel findings include the negative associations of peak VO2 with the calpain and integrin pathways which are relevant in sarcopenia and skeletal muscle health (Bowen et al., 2015; Graham et al., 2015). Gene expression analysis also highlighted the inter-relationship between metabolic and immune health. In fact, many inflammatory pathways were negatively associated with peak VO2 such as ‘role of NFAT in regulation of the immune response’, ‘STAT3 pathway’ and ‘chemokine signaling’ (Hotamisligil, 2006). These findings were further supported by the enrichment of ‘acute phase response signaling’ as well as complement and coagulation systems in the proteomic profiles (Figure 5d and Table S4.5).
Associations in recovery.
Early in recovery (2–15 min), the strongest associations were with molecules involved in energy metabolism and included positive associations with glucose, malate, citrate, lactate, hypoxanthine and xanthine as well as negative associations with tryptophan, cystine, ornithine and allantoin (Figure 5b and Table S4.1). Later in recovery (30–60 min), positive associations were noted for cortisol and negative associations with long-chain and polyunsaturated free fatty acids and medium-chain acylcarnitines highlighting the importance of energy homeostasis. A pro-inflammatory signature at 15 minutes also emerged as positively associated with peak VO2. This was supported by a pathway enrichment of the ‘role of cytokines in mediating communication between immune cells’ (FDR = 7.9E-34, Figure 5d) and the association of 16 of 21 interleukins, members of the TNF superfamily (TNF-β, FASL, CD40L) and interferons (IFN-α and IFN-γ) with exercise capacity (Table S4.1). Similarly, many regulatory or growth factors positively (n = 9) and negatively (i.e. GDF-15) associated with peak VO2. These data indicate that a higher level of inflammatory and growth/protective factors at 15 min in recovery is a critical biological process of aerobic fitness. This signature was largely independent of muscle mass (Table S4.2).
We examined which molecules might be candidates for driving this “fitness inflammatory signature”. Among all circulating molecules at 2 min, IL-5 was the molecule that correlated with the highest number of proteins from the signature at 15 min (n = 14, FDR < 0.05, Figure 5e). These proteins were centered on IL-1β that is an important mediator of the inflammatory response. IL-5 has been shown to be responsible for the release of IL-1β by airway smooth muscle cells in the context of asthma (Hakonarson et al., 1999). Hence, IL-5 may regulate the “inflammatory signature” observed 15 min post-exercise. In addition, the analysis of molecules significantly associated with peak VO2 at 15 min (FDR < 0.05) revealed a central topology for transforming growth factor β (TGF-β), a master regulator of the immune system (Li and Flavell, 2008), at the interface between lipid metabolism (negative correlation with 18 individual TAG species) and inflammation (positive correlation with 35 immune proteins) (Figure 5f). Hence, TGF-β may also be involved in regulating the “fitness inflammatory signature”.
Multi-omic prediction of CPX parameters.
In addition to the multi-omic data generated in this study, we included in the predictive models clinical laboratory (Table S4.6) and gut microbiome data (Table S4.7) generated within 4 months of the exercise date (54.2 and 112.9 days on average, respectively). These measurements are relatively stable within this time range (Zhou et al., 2019). By identifying highly predictive molecules using a Bayesian network algorithm and ridge regression modeling, we built predictive models for peak VO2, VE/VCO2 and RER that had cross-validated R2 of 0.90, 0.75 and 0.81, respectively (Figure 5g and Tables S4.8–4.10). Adding omic data to the peak VO2 model significantly improved its performance when compared to age and BMI alone (R2 of 0.45) which are strong factors influencing fitness (Ribisl et al., 2007). Leptin was confirmed as a critical marker and other measures such as interleukin 18 receptor accessory protein (IL18RAP) emerged from this analysis. The model generated to predict VE/VCO2 from transcriptomic data alone was superior than any other single ome models (R2 = 0.87; Table S4.9) and RER could be moderately predicted with only lipidomic and transcriptomic data (R2 > 0.65) (Table S4.10). As expected, glucose level was important to predict RER and this analysis revealed potential markers including the proportion of Butyricimonas genus in the gut and plasma eicosapentaenoic acid (EPA). Interestingly, microbial strains were selected in these models highlighting important roles of the gut microbiome as was recently demonstrated with the performance-enhancing microbe genus Veillonella (Scheiman et al., 2019).
Differential response to exercise in insulin resistant participants.
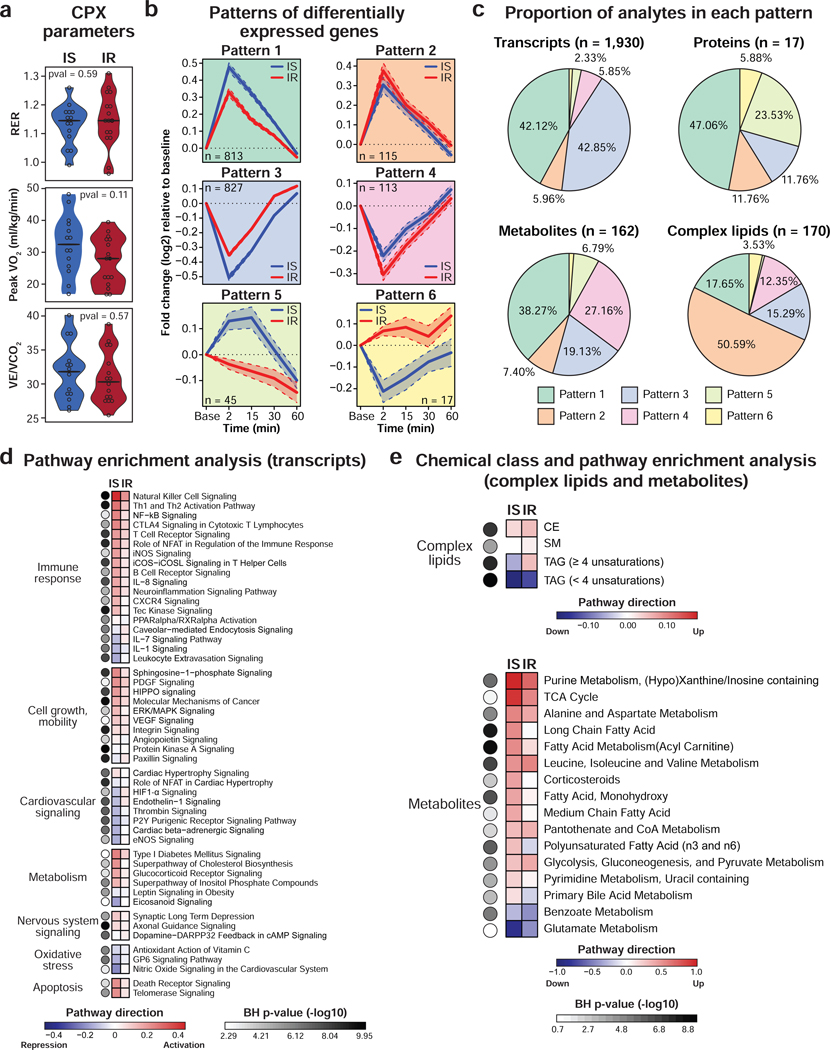
Participants presented a wide range of peripheral insulin resistance with 14 participants categorized insulin sensitive (IS) and 16 insulin resistant (IR, SSPG ≥ 150 mg/dl). Six other participants were excluded either because of diabetes mellitus status or absence of SSPG profile. We investigated the differential response to exercise in IR relative to IS participants. Individuals from both groups reached comparable RER at peak exercise (P = 0.59) (Figure 6a). Peak VO2 and VE/VCO2 were not significantly different despite a trend towards lower exercise capacity in IR participants (P = 0.11).
Figure 6. Differential response to acute exercise in insulin resistant participants.
(a) Violin plots showing CPX parameters in insulin sensitive (IS) and resistant (IR) participants as defined by the modified insulin suppression test (IR: steady-state plasma glucose (SSPG) ≥ 150 mg/dl). A two-sided Student t-test (normal distribution) or a Wilcoxon rank sum test was used for differential analysis. (b) Patterns of differentially expressed genes in IS and IR participants. The solid line represents the mean and the dashed line represents the 95% confidence interval. (c) Pie charts depicting the proportion of significant transcripts (FDR < 0.05), proteins (FDR < 0.20), metabolites (FDR < 0.10) and complex lipids (FDR < 0.20) in each of the six patterns as defined in (b). Pathway analysis using PBMC gene expression (d) and pathway/chemical class enrichment analysis of metabolites and complex lipids (e). Pathway direction is the median of max/min fold change relative to baseline of significant molecules in the pathway (blue: downregulated, red: upregulated). The color of the dots represents pathway significance. See also Figure S7 and Table S5.
Using linear mixed models adjusted for personal baseline, age, sex, race/ethnicity and BMI, we found 2,279 differential analytes across all omic datasets and categorized them based on their longitudinal trajectories into 6 distinct time-series patterns (Table S5.1). Patterns generated from PBMC transcripts are presented in Figure 6b, whereas results from proteins, metabolites and complex lipids are shown in Figure S7a,b. Patterns 1 and 2 contained upregulated analytes with a higher amplitude in IS and IR participants, respectively. Analytes in patterns 3 and 4 were downregulated and patterns 5 and 6 presented opposite trajectories. As an example, 1,930 PBMC genes were differentially expressed in IR participants (FDR < 0.05) with most genes belonging to patterns 1 and 3 (85% in total) (Figure 5c) suggesting a dampened response of immune cells. This observation was verified at the pathway level with a stronger response in IS subjects (Figure 6d and Table S5.2). Differential pathways belonged to various biological processes including inflammatory, cell growth and mobility, cardiovascular and metabolism. Differential inflammatory response in IR participants was evident with a milder activation of ‘natural killer cell signaling’ and ‘Th1 and Th2 activation pathway’. Similarly, activation of apoptosis (‘telomerase signaling’) and cell growth and mobility pathways (‘PDGF signaling’) were stronger in IS. These findings are consistent with other studies describing a diminished PBMC response to weight gain (Piening et al., 2018), viral infection and immunization (Zhou et al., 2019) in IR subjects reflecting higher basal inflammatory profile. Circulating proteins reinforced gene expression results with a smaller amount of MPO and NGAL immediately after exercise in IR individuals (Figure S7c). The inflammatory protein TNF-α had a similar amplitude of response in both groups but persisted for a longer time in IR and ultimately returned to baseline levels 1-hour post-exercise in comparison to 15 min for IS subjects (Figure S7d). The same pattern was observed for IL-6 suggesting abnormal cytokine re-absorption or neutralization in IR participants. More dramatically, pentraxin 3, a marker of acute inflammatory response, increased immediately post-exercise in IS whereas its level remained low in IR across the whole study (Figure S7c). The higher levels of this inflammatory factor at baseline in IR participants may explain this difference. In addition, some cardiovascular pathways involved in vasoconstriction (i.e. ‘endothelin-1 signaling’), response to hypoxia (i.e. ‘HIF-1α signaling’) and nitric oxide synthesis (i.e. ‘eNOS signaling’) were impaired in IR with a mild upregulation relative to a repression in IS.
Although most genes responded in the same direction, some responded in opposite directions (n = 45 and 17 in pattern 5 and 6, respectively). Pathway enrichment analysis revealed that ‘protein ubiquitination pathway’ (FDR = 3.9E-06) was upregulated in IS and downregulated in IR. Protein ubiquitination is a key mechanism involved in protein turnover regulation in skeletal muscle following exercise (Cunha et al., 2012) suggesting that IR individuals have altered proteasome activity in PBMCs.
Similar analysis using circulating metabolites revealed that many major biological pathways impacted by exercise were altered in IR participants (Figure 6e and Table S5.3). For instance, lipid (increase of acylcarnitines and medium-chain free fatty acids), carbohydrate (increase of glucose) and amino acid metabolism (decrease of those used for energy production) responded more strongly in IS resulting in a stronger accumulation of malate (TCA cycle), lactate, hypoxanthine and xanthine (ATP turnover) and alanine (muscle ammonia detoxification) (Figure S7e and Table S5.1). Consistent with these observations, metabolic hormones including gastric inhibitory polypeptide (GIP), leptin and ghrelin decreased more strongly in IS following exercise (Figure S7c). In contrast, insulin was among the few molecules that responded more strongly in IR participants. This might be expected since a higher amount of insulin is necessary for peripheral tissues to absorb circulating glucose in insulin resistant individuals. In addition, insulin secretion was delayed reaching a maximum 15 min post-exercise in IR vs. 2 min in IS. In contrast, cortisol response had a similar amplitude in IS and IR but returned to baseline 1 hour post-exercise in IR whereas it remained high in IS individuals. In addition, we observed an impaired energy homeostasis with insulin resistance. Long- and polyunsaturated free fatty acids are oxidized during exercise to produce energy and are resynthesized in recovery. The level of these molecules increased by 15 min post-exercise and remained high until 30 min in IS subjects whereas their level stayed low in IR participants suggesting an abnormal synthesis or utilization of these free fatty acids.
In contrast with transcripts, proteins and metabolites, most complex lipids (51%) accumulated more strongly in IR individuals following exercise (pattern 2, i.e. CE and SM) (Figure 6c,e and Table S5.3). Unsaturated TAG (4–12 unsaturations) accumulated immediately post-exercise in IR but not in IS participants, whereas saturated TAG (0–3 unsaturations) decreased more in IS suggesting a more efficient hydrolysis.
Multi-omic outlier analysis highlights personal abnormalities.
We examined the number of outlier molecules (FDR < 0.05) at baseline (absolute levels) and in response to exercise (fold change) across multi- omic datasets. Four participants presented outlier molecular profiles not initially suspected by their clinical features (Figure S8a,b). Participant ZLTUJTN was borderline anemic with increased red blood cell volume distribution (Table S4.6) and presented abnormal transcript profiles at baseline and in response to exercise involving metabolic (e.g. ‘iron homeostasis’, ‘heme biosynthesis’), immune (e.g. ‘interferon signaling’, ‘activation of IRF by cytosolic pattern recognition receptors’) and hypoxia pathways (e.g. ‘hypoxia signaling in cardiovascular system’, ‘iNOS signaling’) (Table S6). Follow-up analysis revealed alpha thalassemia carrier status. Participant ZVM4N7A had borderline elevated platelet counts and had evidence of impaired ‘NF-κB signaling’ (FDR = 6.3E-04) following exercise, consistent with a recently described interaction between NF-κB and platelet functions (Kojok et al., 2019). Finally, participant ZL63I8R had an outlier protein profile consisting of immune proteins, growth factors and proteins involved in hemostasis, blood clot and lipid metabolism (Figure S8c). No further clinical interpretation was made due to lack of clinical follow-up on this participant. Altogether, these results show that individuals can present large differences in their molecular composition and that deep individual molecular profiles may be useful to detect subclinical conditions.
DISCUSSION
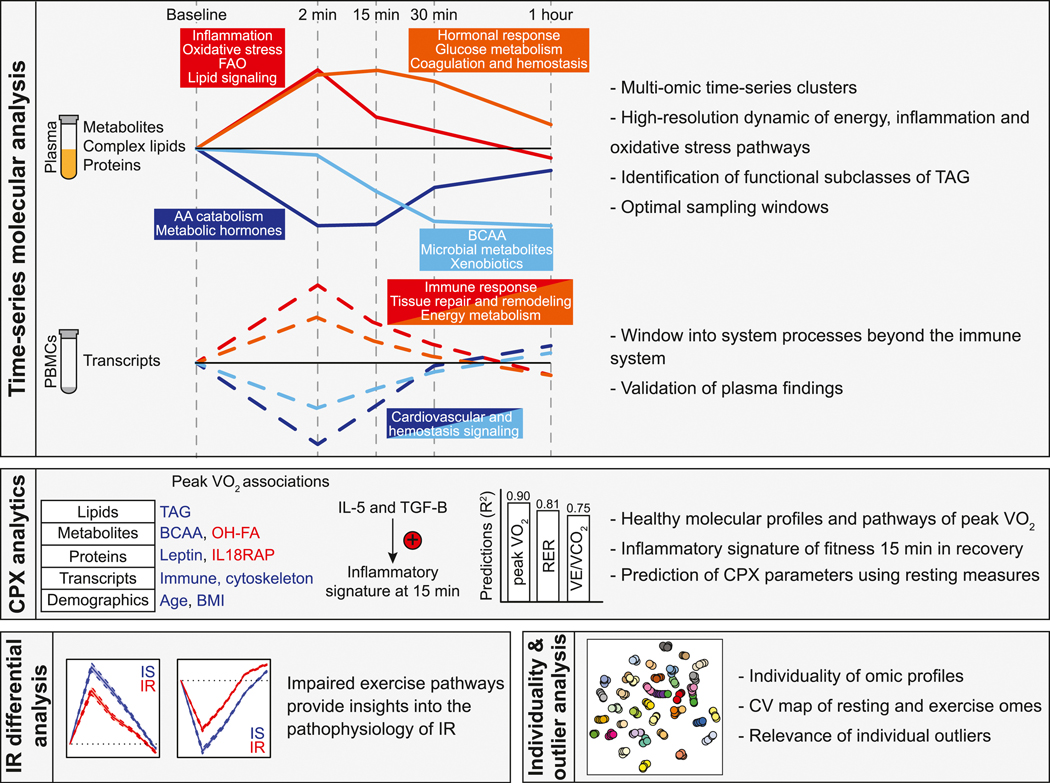
Deep longitudinal molecular profiling coupled with dense sampling revealed a detailed molecular choreography of acute exercise (Figure 7). These biological processes include early (i.e. energy metabolism, oxidative stress and immune response) and late events (i.e. energy homeostasis, tissue repair and remodeling). The high-resolution timing of bioenergetic molecules (i.e. acylcarnitines, medium- and long-chain fatty acids, amino acids and TAG) revealed novel insights into energy utilization and production pathways. Systematic regression analyses led to the discovery of a ‘fitness inflammatory signature’ 15 min post-exercise that was centered on IL-1β and potentially regulated by IL-5 and TGF-β. We also showed that a small number of resting blood-based analytes can potentially predict exercise testing parameters, including peak VO2 (a proxy for fitness) and to a lesser extent ventilatory efficiency. We have also found that the exercise response is influenced by insulin resistant status which modulates several key biological processes including inflammation and cardiovascular response pathways. Finally, molecular individuality at baseline and in response to exercise illustrated the clinical relevance of outlier molecules at an individual level.
Figure 7. Summary of the main discoveries.
Discoveries were classified in four categories: time-series molecular analysis, cardiopulmonary exercise (CPX) analytics, insulin resistance (IR) differential analysis, individuality and outlier analysis.
Longitudinal multi-omic profiling revealed thousands of molecules affected by an acute bout of exercise. These changes were not evident in the absence of exercise. Our time-series clustering and network analysis revealed complex interplays between various compartments (i.e. skeletal muscle, adipose tissue, immune cells, liver and cardiovascular system) and identified potential regulators of important biological processes involved in exercise. In particular, crosstalks between metabolism, oxidative stress and immunity were observed at multiple instances.
Myeloperoxidase (MPO) - a marker of oxidative stress secreted by neutrophils - was among the most increased protein in the acute recovery phase of exercise (cluster 1) where it presented a strong central connectivity bridging pro-inflammatory (i.e. NGAL, IL-7), growth/protective factors (i.e. IL-1ra) as well as metabolic factors (i.e. acylcarnitines). Experimental studies document an increase in skeletal muscle MPO in both trained and untrained rats, a response that is attenuated by training (Morozov et al., 2006). The increase in MPO activity is not limited to skeletal muscle but also involves the heart and the liver (Belcastro et al., 1996). In more intense exercise such as marathons, the increase in MPO activity also coincides with an increase in circulating cardiac biomarkers of injury, wall stress and inflammation (Melanson et al., 2006). In our study, the degree of MPO elevation was not significantly related to the respiratory exchange ratio or maximal oxygen consumption, suggesting a non-linear relationship between oxidative stress and exercise intensity. Other key molecules bridging inflammation and metabolism included tumor necrosis factor alpha (TNF-α) secreted by adipose tissue and vascular cell adhesion molecule-1 (VCAM-1). To a lesser extent, growth differentiating factor-15 (GDF-15), which is a strong marker of survival in cardiometabolic disease (Wallentin et al., 2013), was also increased in the early phase of exercise recovery. In addition, we reported a delayed increase of a myriad of steroid hormones and corticosteroids post-exercise (cluster 2) that are essential to energy homeostasis. Among others, the rise of cortisol was concomitant with circulating level of glucose and several factors facilitating glucose cellular absorption such as insulin and fatty acid binding proteins 3 and 4 (FABP3 and FABP4) as well as downstream factors reflecting energy production (i.e. TCA cycle constituents and markers of ATP turnover). FABP3 has been proposed as an early marker of myocardial ischemia (Tanaka et al., 1991) and the observation of its increase post-exercise invites further investigation on its potential value during exercise testing.
Our study also adds granularity on the potential role of species of acylcarnitines, free fatty acids, complex lipids and amino acids. Depending on their number of carbons and unsaturations, free fatty acids were divided among different time clusters with longer chain fatty acids decreasing as they are metabolized. In contrast, medium-chain free fatty acid and by extension medium-chain acylcarnitines accumulated reflecting partial FAO in skeletal muscle. Ketone bodies were found to increase at later time points post-exercise (15–60 min) presumably originating from fatty acid oxidation in the liver (Laffel, 1999). Phase 2 clinical trials in heart failure suggest a potential beneficial role of ketone bodies as energetic substrate (Nielsen et al., 2019) representing an active area of investigation. We have also found that the trajectory of some complex lipids was dependent on their carbon and unsaturation levels. Even though most triglycerides (TAG) decrease in recovery and are presumably hydrolyzed for energy production (Ranallo and Rhodes, 1998), a subset containing long- chain polyunsaturated fatty acids increased immediately post-exercise (cluster 1). This finding may reflect pro- or anti-inflammatory signaling since these TAG contain arachidonic acid, eicosapentaenoic acid and docosahexaenoic acid (Calder, 2013). We also detected other complex lipids following the same trajectory including cholesterol esters and sphingomyelins. Interestingly, these processes were primarily observed in insulin resistant participants indicating a dysregulation in complex lipid metabolism at the class and fatty acid composition levels. Amino acids also presented different temporal profiles depending on their type, i.e. essential versus non-essential amino acids, and whether they are catabolized for energy generation (i.e. branched chain amino acids) or produced due to heightened cellular metabolism (i.e. alanine and glutamine). We also observed an increase of neuroactive metabolites acetylcholine and kynurenic acid (KYNA) in response to exercise which links physical activity to mental health. KYNA is a product of tryptophan metabolism, is produced in exercise-stimulated skeletal muscle and has been shown to deliver antidepressant activity (Agudelo et al., 2014).
Transcriptomic analysis of PBMCs revealed changes beyond the immune and oxidative stress pathways providing a window to systemic biological processes. In particular, dynamic transcripts informed on tissue repair and remodeling, cardiovascular health, vascular and epithelial growth factors, metabolism and apoptosis. Tissue repair and remodeling was evident via the upregulation of the insulin growth factor-1 (IGF-1) signaling pathway that activates by phosphorylation serine/threonine kinases (p70S6K) which in turn increases protein synthesis and repairs the damaged muscle (Schiaffino and Mammucari, 2011). Cardiovascular processes included endothelin-1, thrombin as well as nitric oxide signaling. It is also interesting to note a decrease in the mTOR signaling pathways following exercise; attenuation of the liver mTORC1 pathway is often linked with increased lifespan (Sengupta et al., 2010).
While most analytes presented a rather homogenous response to exercise across participants, certain molecules had a high inter-individual variability such as metabolic hormone ghrelin as well as interleukins (IL- 13 and IL-23) and nerve growth factor (NGF). Such molecules potentially carry valuable clinical information. Systematic outlier analysis highlighted participants with abnormal molecular profiles relative to the cohort that were associated with potential clinical conditions and informed on dysregulated biological processes associated with alpha thalassemia carrier status and anemia. Hence, personalized molecular exercise profiling may prove to be valuable for disease detection and understanding how exercise impacts disease-related pathways.
Baseline metabolic health status of participants was an important determinant of exercise capacity. As an example, high baseline leptin, BCAAs, TAGs and low abundance of the microbial metabolite hippuric acid -reflecting poor metabolic health - were associated with lower peak VO2. These findings extend studies of low (LCR) and high capacity runners (HCR) rats describing skeletal muscle energy plasticity as a critical aspect of oxygen consumption (Koch et al., 2011; Overmyer et al., 2015). Several pathways previously associated with mitochondrial function, cell adhesion and extracellular matrix were enriched in skeletal muscles and differed between HCR and LCR lines (Ren et al., 2016). Interestingly, two of the most significant pathways in association with peak VO2 in our study (i.e. calpain and integrin pathways) are involved in muscle regulation. The calpain pathway has been associated with sarcopenia, an important mechanism of frailty in the aging population (Bowen et al., 2015). The integrin pathway mediates the control of insulin-like growth factor receptor (IGF1R) signaling and in turn regulates the muscular response to exercise (Legate et al., 2009). Our study also revealed a complex role of post-exercise inflammatory response in aerobic fitness with a higher level of inflammatory and growth/regulatory factors at 15 min in recovery that strongly associated with higher peak VO2. Our data suggest that this ‘fitness inflammatory signature’ was centered on IL-1β and may be regulated by IL-5 and/or transforming growth factor β (TGF-β), a novel potential trigger of exercise-induced inflammation. Finally, our analysis of omic associations with oxygen consumption highlights the importance of indexing as the strength of associations varies depending on indexing to total body weight or lean body mass.
Participants in our study were selected to span a wide range of insulin resistance which allowed investigation of differential response to exercise providing insights into the pathophysiology of metabolic conditions. Significant differences were noted in all the main biological processes impacted by exercise including inflammatory, oxidative stress, vascular, hypertrophic and cell growth pathways. In addition to a reduced inflammatory response in insulin resistant participants, often related to a higher baseline activation (Piening et al., 2018; Zhou et al., 2019), we observed a reduced efficiency to oxidize free fatty acids, produce energy and restore energy homeostasis. We also detected differential response in glutamate metabolism which is implicated in coronary heart disease (Qi et al., 2013). Furthermore, cardiovascular signaling showed marked differences (often in opposite direction) and included, endothelin-1, a vasoconstrictor and therapeutic target in vascular biology (Marasciulo et al., 2006), thrombin, a critical enzyme in the coagulation pathway (Yanagisawa, 1994) and cardiac beta-adrenergic signaling found to be reduced in heart failure syndrome or autonomic disorders (Lymperopoulos et al., 2013).
This work should be assessed in the context of its limitations. Our cohort was relatively small (n = 36) and generally of older age and consisted of a diverse group of participants (BMI, insulin resistance status). However, to our knowledge our deep phenotyping combined with a personalized exercise test is one of the most comprehensive molecular studies ever performed. Importantly, most of the data is open access. Other large initiatives underway, of note, Molecular Transducers of Physical Activity Consortium (NCT03960827) will scale the concepts presented here with muscle and fat biopsy.
In conclusion, our study provides an in-depth and integrated multi-omic profiling of the response to exercise. The translation potential of the study resides in the discovery of promising resting biomarker signature of aerobic fitness as well as in demonstrating the value of exercise molecular testing in identifying key differences in the mechanisms of insulin resistance. Ongoing studies will help develop standardization for exercise omic testing as well as refine reference ranges for clinical or mechanistic studies.
STAR METHODS
LEAD CONTACT AND MATERIAL AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Snyder (mpsnyder@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Participant Recruitment and IRB Consent.
Study participants were enrolled as “healthy volunteers” in the framework of the NIH integrated Human Microbiome Project 2 (iHMP) (Zhou et al., 2019). Inclusion and exclusion criteria are described in detail elsewhere (Schussler-Fiorenza Rose et al., 2019). Among the iHMP cohort, 36 subjects provided informed written consent to participate in the exercise study under a research study protocol approved by the Stanford University Institutional Review Board (IRB 23602) and 33 individuals agreed to make all of their data open access. Participants were screened for contraindications for exercise testing and comorbidities with a basic health questionnaire and didn’t have an active infection, history of cancer or autoimmune disease. No participant had evidence of stress-induced wall motion abnormality during stress testing, severe clinical heart failure or symptomatic atherosclerosis. Detailed demographic information can be found in Table S1.1. Thirty out of 36 participants underwent the modified insulin suppression test to determine steady-state plasma glucose (SSPG) levels as described (Schussler-Fiorenza Rose et al., 2019) and classify the participants as insulin sensitive (n = 14, SSPG < 150 mg/dl) or insulin resistant (n = 16, SSPG ≥ 150 mg/dl). The remaining six individuals didn’t perform the test because of medical contraindications. The cohort was composed of normoglycemic (n = 16), prediabetic (n = 16) and diabetic (n = 4) individuals as determined by fasting plasma glucose (FPG) and hemoglobin A1C (HbA1C) levels measured within 2 months of the exercise date (prediabetic range: 100 mg/dl ≤ FPG < 126 mg/dl or 5.7% ≤ HbA1C < 6.5%; diabetic range FPG ≥ 126 mg/dl or HbA1C ≥ 6.5%).
METHOD DETAILS
Study Design.
Overnight-fasted participants (10–12 hours) arrived at Stanford Clinical Translational Research Unit (CTRU) at 7:00 am in the morning. Resting vital signs including heart rate, blood pressure, oxygen saturation, height and weight as well as blood glucose were recorded. Blood was collected from participants at baseline (7:15 am) and echocardiography as well as vascular ultrasound were performed at rest (7:45 am). Afterwards, the study subjects underwent symptom-limited cardiopulmonary exercise (CPX) testing (8:00 am) and received a stress echocardiography. Additional blood samples were collected longitudinally post-exercise.
Transthoracic Echocardiography.
Participants underwent transthoracic echocardiography using commercially available echocardiographic systems (iE33; Philips Medical Imaging, Eindhoven, the Netherlands). Post-stress images were acquired immediately post-exercise, as per international consensus guidelines and all participants had satisfactory imaging. Digitized echocardiographic studies were analyzed on Xcelera workstations in accordance with published guidelines of the American Society of Echocardiography (ASE) (Lang et al., 2015). Left ventricular diameters were indexed on height, while mass and volumes were indexed on body surface area. Left ventricular ejection fraction (LVEF) was calculated by modified biplane Simpson’s rule of apical imaging (Wilson et al., 1998). Left ventricular global longitudinal strain (LV GLS) was calculated from apical imaging on manual tracings of the mid wall with the formula for Lagrangian Strain % = 100 x (Lt - L0)/L0), as previously described (Smith, 2016). With tissue doppler imaging (TDI), we used peak myocardial early diastolic velocity at the lateral mitral annulus and the assessment of trans mitral to TDI early diastolic velocity ratio (E/e’) (Lee et al., 2010; McClelland et al., 2015). Left atrial volume was calculated by the biplane disk summation technique and indexed to body surface area as described by the ASE (Lang et al., 2015).
Vascular Ultrasound.
Screening for subclinical atherosclerosis was performed using vascular ultrasound of the carotid and femoral arteries with a 9.0 MHz Philips linear array probe and iE33 xMATRIX echocardiography system (Philips, Andover, MA, USA). Vascular stiffness was assessed using central pulse wave velocity (PWV). PWV was calculated with the formula distance (m)/transit time (s) by assessing the flow of the carotid and femoral arteries separately and normalizing with electrocardiogram (Calabia et al., 2011). No participant had severe atherosclerotic plaque exceeding 30% in diameter stenosis or abdominal aortic aneurysms.
Symptom-limited Cardiopulmonary Exercise Testing.
Symptom-limited cardiopulmonary exercise (CPX) testing was performed according to individualized ramp-treadmill protocols (Myers and Froelicher, 1993). Participants were encouraged to exercise to maximal exercise capacity with a target duration of 8–12 minutes following the ramp protocol tuned to individuals cardiovascular fitness as determined by a questionnaire. All participants ceased exercise due to dyspnea and/or fatigue and none experienced chest pain or terminated the study due to arrhythmia. Ventilatory efficiency (VE), oxygen consumption (VO2), volume of carbon dioxide production (VCO2) and other CPX variables were acquired breath-by-breath and averaged over 10 second intervals (Omnia CPET, CosMed USA, Concord, CA, USA). A respiratory exchange ratio (RER; VCO2/VO2) > 1.05, heart rate (HR) > 85% of predicted maximum and Rating of Perceived Exertion (RPE 6–20; Borg Perception, Hasselby, Sweden) were determined to indicate peak effort. Peak oxygen uptake VO2 was calculated as the highest VO2 levels. VE and VCO2 responses throughout exercise were used to calculate the VE/VCO2 slope via least squares linear regression (y = mx + b, m = slope) (Arena et al., 2003).
Blood Collection and Sample Preparation.
Intravenous blood from the upper forearm was drawn from overnight-fasted participants at baseline (before exercise) as well as 2 min, 15 min, 30 min, and 1h post- exercise. A subset of the participants (n = 15) also donated a blood sample fasted the next morning. Samples at baseline and 1-hour time points were collected in the CTRU while samples collected 2 min, 15 min and 30 min post-exercise were collected in the exercise laboratory. Additional samples were collected and analyzed at later time points (i.e. 2, 4 and 6 hours) but the effect of exercise at these time points was difficult to determine due to the confounding impact of food consumption after the 1- and 4-hour time points. For time points 2 min, 15 min and 30 min, the intravenous cannula was flushed with 5 ml of normal saline after each blood draw to prevent obstruction and contamination. Specimens were immediately placed on ice after collection to avoid sample deterioration and processed together immediately after collection of the last sample (9:00 am). Blood was collected in a purple top tube vacutainer (BD), layered onto Ficoll media and spun at 2,000 rpm for 25 min at 24°C. The top layer EDTA-plasma was pipetted off, aliquoted and immediately frozen at −80°C. The peripheral blood mononuclear cells (PBMC) layer was collected, counted via cell counter and aliquots of PBMCs were further pelleted and flash-frozen. Multi-level molecular profiling was performed on all blood samples including gene expression from PBMCs (transcriptomics), proteins (targeted and untargeted proteomics), metabolites (untargeted metabolomics), and complex lipids (semi-targeted lipidomics) from plasma. Transcriptomics, metabolomics and targeted proteomics were performed on fresh EDTA-plasma aliquots while untargeted proteomics and lipidomics were performed on EDTA-plasma that went through one freeze-thaw cycle. In order to assess natural molecular deviation in absence of exercise, a subset of the cohort (n = 14) participated in a control experiment in the absence of exercise and donated blood samples following the exact same protocol as described above. Intravenous blood samples were collected in purple top tube vacutainer (BD) at the same time of the day and at the same intervals. EDTA-plasma and PBMC cell pellets were then processed and aliquoted. The 2 min time point was not collected due to the close proximity and presumably similarity with the baseline time point.
RNA sequencing from Peripheral Blood Mononuclear Cells (PBMCs).
RNA extraction and library preparation.
The transcriptome was evaluated by RNA sequencing (RNA-seq) from bulk PBMCs. PBMCs were thawed on ice, and subsequently lysed and processed to DNA, RNA and protein fractions using silica-membrane spin columns from the AllPrep DNA/RNA/Protein kit (cat# 80004, Qiagen, Chatsworth, CA, USA). PBMCs were processed in a randomized order. A Bravo NGS Workstation (Agilent, Santa Clara, CA, USA) was used to perform automated preparation of strand-specific RNA-seq libraries using the TruSeq Stranded Total RNA with Ribo-Zero Gold kit (cat# RS-122–2301, Illumina, San Diego, CA, USA). According to manufacturer’s protocol, total RNA was depleted of mitochondrial and cytoplasmic ribosomal RNA followed by fragmentation and random priming to synthesize cDNA fragments. Barcoded sequencing adapters were ligated to cDNA inserts and enriched using PCR to create the final cDNA libraries. Qualitative and quantitative assessment of libraries was performed using a Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA). Quantified, barcoded libraries were normalized and mixed at equimolar concentrations into a multiplexed sequencing library.
RNA sequencing and data processing.
Pooled libraries were sequenced on a HiSeq 4000 sequencer (Illumina, San Diego, CA, USA) to a depth of 30 million reads per sample using a paired-end 100 base pair run configuration. Four samples were sequenced in each pool to correct for potential batch effect and longitudinal samples from the same participants were mixed in the same pool. Sequencing data were demultiplexed and converted into fastq files using Illumina’s bcl2fastq conversion software (v2.20). Quality and adapter trimming along with filtering of rRNA reads was performed using BBDuk (v37.22). The decontaminated reads were mapped to personal genomes using STAR aligner (v2.5.1b) by modifying the GRCh38 reference genome at variant sites called for each participant through exome sequencing. Gene quantification was performed using the tool htseq-count from the Python package HTSeq (v0.9.1). The GENCODE v28 annotation was used to define genomic features where each gene is considered as the union of all its exons. After normalization of read counts to the sequencing depth in each sample, genes with an average expression below 10 were discarded. Missing values were imputed using the k-nearest neighbors’ method (‘impute.knn’ function) in the R package ‘impute’ (v1.52.0). Two datasets were generated, one containing read counts normalized to the sequencing depth in each sample (original), and another that was further processed by applying the variance- stabilizing transformation (VST) in R package ‘DESeq2’ (v3.9).
Untargeted Proteomics from Plasma by Sequential Window Acquisition of all Theoretical (SWATH)-MS.
Sample preparation and data acquisition.
Plasma samples were thawed on ice, prepared and analyzed in a randomized order. Tryptic peptides were generated from 8 μg of undepleted plasma proteins and separated on a NanoLC 425 System (Sciex, Redwood City, CA, USA). 5 μl/min flow was used with trap-elute setting using a ChromXP C18 trap column 0.5 × 10 mm, 5 μm, 120 Å (cat# 5028898, Sciex, Redwood City, CA, USA). Tryptic peptides were eluted from a ChromXP C18 column 0.3 × 150 mm, 3 μm, 120 Å (cat# 5022436, Sciex, Redwood City, CA, USA) using a 43-minute gradient from 4–32% B with 1-hour total run. Mobile phase solvents consisted of 92.9% water, 2% acetonitrile, 5% dimethyl sulfoxide, 0.1% formic acid (A) and 92.9% acetonitrile, 2% water, 5% dimethyl sulfoxide, 0.1% formic acid (B). MS analysis was performed using SWATH acquisition on a TripleTOF 6600 System equipped with a DuoSpray Source and 25 μm I.D. electrode (Sciex, Redwood City, CA, USA). Variable Q1 window SWATH Acquisition methods (100 windows) were built in high sensitivity MS/MS mode with Analyst TF Software (v1.7). A quality control (QC) consisting of an equimolar pool of all the samples in the study was injected at the beginning and end of each batch. Samples were run in two batches and QC data were used to control for batch effect. Longitudinal samples from the same participants were run in the same batch.
Data processing.
Peak groups from individual runs were statistically scored with pyProphet tool (v2.0.1) and all runs were aligned using TRIC strategy (Rost et al., 2016). A final data matrix was produced with 1% FDR at peptide level and 10% FDR at protein level. Protein abundances were computed as the sum of the three most abundant peptides (top3 method). Batch effect was corrected by applying median-normalization and proteins detected in less than 2/3 of the samples were discarded. Missing values were imputed by drawing from a random distribution of low values in the corresponding sample (Tyanova et al., 2016). Untargeted protein levels were reported as spectral counts.
Targeted Proteomics from Plasma by Immunoassays.
Plasma samples were thawed on ice, prepared and analyzed in a randomized order. Levels of circulating cytokines and growth factors were measured in plasma using a 63-plex Luminex antibody-conjugated bead capture assay (eBiosciences/Affymetrix). Metabolic hormones were measured using MILLIPLEX MAP Human Metabolic Hormone Magnetic Bead Panel - Metabolism Multiplex Assay (HMHEMAG-34K, Millipore, Burlington, MA, USA). Cardiovascular risk markers were measured using MILLIPLEX MAP Human Cardiovascular Disease (CVD) Magnetic Bead Panel (1 to 4) - Cardiovascular Disease Multiplex Assay (HCVD1MAG-67K, HCVD2MAG-67K, HCVD3MAG-67K, HCVD4MAG-67K, Millipore, Burlington, MA, USA). Experiments were performed by the Stanford Human Immune Monitoring Center (HIMC) according to the manufacturer’s recommendations and read using a Luminex 200 instrument with a lower bound of 20 beads per sample per analyte. Custom assay control beads by Radix Biosolutions were added to all wells for Human 63-plex assay. Longitudinal samples from the same participant were analyzed on the same plate. Inter-plate variability was corrected using the median of inter- plate ratios for four representative samples analyzed in each plate. Raw mean fluorescence intensity (MFI) values were used for the analysis. Missing values (bead count below 20) were imputed using the k-nearest neighbors’ method (‘impute.knn’ function) in the R package ‘impute’ (v1.52.0). Targeted protein levels were reported as MFI.
Untargeted Metabolomics from Plasma by Liquid Chromatography (LC)-MS.
Sample preparation and data acquisition.
Plasma samples were thawed on ice, prepared and analyzed randomly as previously described (Contrepois et al., 2015). Briefly, metabolites were extracted using 1:1:1 acetone:acetonitrile:methanol, evaporated to dryness under nitrogen and reconstituted in 1:1 methanol:water before analysis. Metabolic extracts were analyzed four times using HILIC and RPLC separation in both positive and negative ionization modes. Data were acquired on a Q Exactive plus mass spectrometer for HILIC and a Q Exactive mass spectrometer for RPLC (Thermo Scientific, San Jose, CA, USA). Both instruments were equipped with a HESI-II probe and operated in full MS scan mode. MS/MS data were acquired on quality control samples (QC) consisting of an equimolar mixture of all samples in the study. HILIC experiments were performed using a ZIC-HILIC column 2.1 × 100 mm, 3.5 μm, 200Å (cat# 1504470001, Millipore, Burlington, MA, USA) and mobile phase solvents consisting of 10 mM ammonium acetate in 50/50 acetonitrile/water (A) and 10 mM ammonium acetate in 95/5 acetonitrile/water (B). RPLC experiments were performed using a Zorbax SBaq column 2.1 × 50 mm, 1.7 μm, 100Å (cat# 827700–914, Agilent Technologies, Santa Clara, CA, USA) and mobile phase solvents consisting of 0.06% acetic acid in water (A) and 0.06% acetic acid in methanol (B). Data quality was ensured by (i) injecting 6 and 12 pool samples to equilibrate the LC-MS system prior to run the sequence for RPLC and HILIC, respectively, (ii) injecting a pool sample every 10 injections to control for signal deviation with time, and (iii) checking mass accuracy, retention time and peak shape of internal standards in each sample.
Data processing.
Data from each mode were independently analyzed using Progenesis QI software (v2.3, Nonlinear Dynamics). Metabolic features from blanks and that didn’t show sufficient linearity upon dilution in QC samples (r < 0.6) were discarded. Only metabolic features present in >2/3 of the samples were kept for further analysis. Inter- and intra-batch variation was corrected using the LOESS (locally estimated scatterplot smoothing Local Regression) normalization method on QC injected repetitively along the batches (span = 0.75). Data were acquired in three and two batches for HILIC and RPLC modes, respectively. Missing values were imputed by drawing from a random distribution of low values in the corresponding sample (Tyanova et al., 2016). Data from each mode were merged and metabolites were formally identified by matching fragmentation spectra and retention time to analytical-grade standards when possible or matching experimental MS/MS to fragmentation spectra in publicly available databases. We used the Metabolomics Standards Initiative (MSI) level of confidence to grade metabolite annotation confidence (level 1 - level 4) (Table S1.3). Level 1 represents formal identifications where the biological signal matches accurate mass, retention time and fragmentation spectra of an authentic standard run on the same platform. For level 2 identification, the biological signal matches accurate mass and fragmentation spectra available in METLIN database. Acylcarnitines, free fatty acids and complex lipids don’t necessarily all have MS/MS data in public databases but were annotated based on expected signature fragments. Level 3 represents putative identifications that are the most likely name based on previous knowledge of blood composition. Level 4 consists in unknown metabolites. After careful annotation of the metabolite dataset, a total of 728 metabolites were measured and categorized in classes and pathways based on the KEGG database where possible. Some metabolites elute in multiple peaks and are indicated with a number in parenthesis following the metabolite name ordered by elution time. Metabolite abundances were reported as spectral counts. 664/728 metabolites (91.2%) could be detected in the metabolomics dataset from the control experiment.
Semi-targeted Lipidomics from Plasma using the Lipidyzer Platform.
Sample preparation.
Plasma samples were thawed on ice, prepared and analyzed in a randomized order. Plasma lipids were extracted using a biphasic separation protocol with ice-cold methanol, methyl tert-butyl ether (MTBE) and water (Contrepois et al., 2018). Briefly, 300 μl of methanol spiked-in with internal standards (cat# 5040156, Sciex, Redwood City, CA, USA) was added to 40 μl of plasma and vortexed for 20 s. Lipids were solubilized by adding 1,000 μl of MTBE and incubated under agitation for 30 min at 4°C. After addition of 250 μl of ice-cold water, the samples were vortexed for 1 min and centrifuged at 14,000 g for 5 min at 20°C. The upper phase containing the lipids was then collected, dried down under nitrogen, reconstituted with 200 μl of methanol and stored at −20°C. The day of the experiment, lipids were dried down under nitrogen and reconstituted with 300 μl of 10 mM ammonium acetate in 9:1 methanol:toluene.
Data acquisition and processing.
Lipid extracts were analyzed using the Lipidyzer platform that comprises a 5500 QTRAP System equipped with a SelexION differential mobility spectrometry (DMS) interface (Sciex, Redwood City, CA, USA) and a high flow LC-30AD solvent delivery unit (Shimazdu, Kyoto, Japan). A full description of the method is available elsewhere (Contrepois et al., 2018). Briefly, lipid molecular species were identified and quantified using multiple reaction monitoring (MRM) and positive/negative ionization switching. Two acquisition methods were employed covering 10 lipid classes; method 1 had SelexION voltages turned on while method 2 had SelexION voltages turned off. Lipidyzer data were reported by the Lipidomics Workflow Manager (LWM) software which calculates concentrations for each detected lipid as average intensity of the analyte MRM/average intensity of the most structurally similar internal standard (IS) MRM multiplied by its concentration. Lipid abundances were reported as concentrations in nmol/g. Lipids detected in less than 2/3 of the samples were discarded and missing values were imputed by drawing from a random distribution of low values in the corresponding sample (Tyanova et al., 2016). Data quality was ensured by i) tuning the DMS compensation voltages using a set of lipid standards (cat# 5040141, Sciex, Redwood City, CA, USA) after each cleaning, more than 24 hours of idling or 3 days of consecutive use, ii ) performing a quick system suitability test (QSST) (cat# 50407, Sciex, Redwood City, CA, USA) before each batch to ensure acceptable limit of detection for each lipid class, and iii) triplicate injection of lipids extracted from a reference plasma sample (cat# 4386703, Sciex, Redwood City, CA, USA) at the beginning of the batch. The data were acquired in eight batches. Two datasets were generated, one containing all individual lipid species (original) and a second one containing concatenated TAG information and LPC, LPE, FFA classes were discarded (redundant with metabolomic dataset). In the latter, the median was calculated from individual TAG with the same total number of carbons and unsaturations (TAG_med).
16S Microbiome Sequencing from Stool.
Stool samples were collected within 4 months of the day of exercise (112.9 days on average) as part of the NIH integrated Human Microbiome Project 2 study. DNA was first extracted from stool in line with the Human Microbiome Project’s (HMP) Core Sampling Protocol A (hmpdacc.org) and then sequenced on a MiSeq system using 2 × 300 bp paired-end sequencing (Illumina, San Diego, CA, USA). Raw sequences were processed using Illumina’s software, assigned to Operational Taxonomic Units (OTU) by Usearch against GreenGenes database (May 2013 version) and final taxonomic assignment was performed using RDP-classifier (Zhou et al., 2019).
Clinical Laboratory Tests.
Clinical laboratory tests were performed within 2 months of the day of exercise (54.2 days on average) as part of the NIH integrated Human Microbiome Project 2 study (Zhou et al., 2019).
QUANTIFICATION AND STATISTICAL ANALYSIS
When not specified otherwise, the transcriptomic dataset used was transformed with variance-stabilization (VST), the lipid dataset contained individual TAG species (original), and all missing values were imputed.
Fuzzy c-mean clustering.
Fuzzy c-mean clustering was performed using the R package ‘Mfuzz’ (v2.20.0) after log2-transformation and Z-score scaling of the data. We calculated the minimum centroid distance for a range of cluster numbers and the optimal number was chosen using the ‘elbow’ method.
t-distributed stochastic neighbor embedding (tSNE) dimensionality reduction.
tSNE scatterplots were generated after log2-transformation and Z-score scaling of the data using the R package ‘Rtsne’ (v0.15) with the following parameters: perplexity = 5, theta = 0.05.
Inter-individual variability.
Coefficients of variation (CV) were calculated for each analyte on non-imputed datasets to avoid any potential bias. CV = standard deviation/mean*100. The transcriptomic dataset used was normalized to sequencing depth (original). Inter-individual variability in response to exercise was determined as the median CV for each analyte across all time points post-exercise relative to baseline. Technical variability for each assay was determined by calculating the CV across all quality control samples in the study. The boxplots show the first (lower edge of the box), median (middle line) and third (upper edge of the box) quartiles. The upper whisker is the third quartile + 1.5 x (interquartile range) and the lower whisker is the first quartile - 1.5 x (interquartile range).
Linear models to find analytes that change in response to exercise.
Linear models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were computed in R using the lm base function. Linear models were applied on log2-transformed data and the lipid dataset used was the concatenated TAG version (TAG_med). One-way ANOVA testing (two-sided) was then used to calculate significance at each time point relative to baseline. P values were corrected for multiple hypothesis using the Benjamini-Hochberg method and analytes with FDR below 0.05 were considered significant.
Pathway dynamic analysis.
Differential expression analysis between each two consecutive time points was performed using the same models as described above. Between each two consecutive time points, significant transcripts (FDR < 0.05) were used to define the largest connected component of the Protein-Protein Interaction network (PPI, STRING database (Szklarczyk et al., 2015)). In these networks, each node represents a protein and an edge between two nodes represents an experimentally validated protein-protein interaction. Subsequently, we identified the highest-scoring functional module from the PPI subgraph using BioNet R package (v1.46.0) (Beisser et al., 2010). BioNet identifies functional modules by accounting for network topology as well as the node’s significance P value. The genes with the highest-scoring module were used for pathway enrichment analysis as described below.
Pathway/chemical class enrichment analysis.
We used the Ingenuity pathway analysis (IPA, Qiagen) platform to search for enriched pathways using differentially expressed genes in PBMCs and circulating proteins. All the detected transcripts or proteins were used as background. Significance of pathways was determined by the hypergeometric test (one-sided) in IPA. For metabolites and complex lipids, enrichment was calculated using the Kolmogorov–Smirnov method (one-sided, Figure 2) (Barupal and Fiehn, 2017) or using the Fisher exact t-test (one-sided, Figure 5) in R with pathway/chemical class annotation of all detected metabolites/lipids. P values were corrected for multiple hypothesis using the Benjamini-Hochberg method and pathways with FDR below 0.05 (transcripts, proteins) and 0.20 (metabolites, complex lipids) were considered significant. Pathway directions were calculated as follows: 1) median of fold change values relative to baseline for significant molecules in the pathway (Figures 2 and 4), 2) median of beta coefficients for significant molecules in the pathway (Figure 5), 3) median of max (if up) or min (if down) fold change values relative to baseline for significant molecules in the pathway (Figure 6). Beta coefficients were provided by regression models described below.
Linear regressions to find associations of key changes induced by exercise with demographics and physiological measures.
Linear regressions were conducted for selected molecules against demographics and physiological measures as detailed in Table S2.4 using the lm base function in R. Lean body mass, fat mass and percent fat were calculated using the equations derived from the National Health and Nutrition Examination Survey (NHANES) (Lee et al., 2017). Associations with a P value below 0.001 were considered significant. The lipid dataset used was TAG_med.
Correlation network analysis.
Pairwise Spearman’s rank correlations were calculated using the R package ‘Hmisc’ (v4.1–1) and weighted, undirected networks were plotted with ‘igraph’ (v1.2.1). Correlations with Bonferroni adjusted P values below 0.05 (Figure 3) and 0.01 (Figure 5) were included and displayed via the fruchterman-reingold method. Only the main networks were plotted. Nodes were color-coded by assay and their size represent the maximum (clusters 1 and 2) or minimum (clusters 3 and 4) median fold change in response to exercise (Figure 3) and the betweenness centrality (Figure 5) as calculated by the betweenness function in ‘igraph’.
Differential allele-specific expression (ASE) with exercise.
Read mapping bias was removed by following the WASP pipeline. The GATK tool ASEReadCounter was used to count allele specific reads at exonic heterozygous sites. For each individual, only SNPs supported by at least 20 reads in each time point and bi- allelic (0.1 ≤ allelic ratio ≤ 0.9) in at least one time-point were considered. Only SNP shared by at least 10 heterozygous individuals were considered. We tested for differences in ASE with time using the beta-binomial generalized linear mixed models implemented in EAGLE (v2.0) (Knowles et al., 2017). In short, for each exonic SNP, we model the alternative allele count for heterozygous individual i, at time t using a Beta-Binomial distribution, i.e. yit ~ BB(nit, σ(φi βt + ui), c), where nit is the total number of reads for i in t, σ() is the logistic function, φi represents the phase between the causal cis-SNP and the exonic SNP and is treated as a latent variable taking values in {−1,+1}. We learn a prior π = P(φi = +1) across all individuals and marginalize (sum) over the possible values of π. βt is the effect of time on the reference allele proportion. ui ~ N(0,v) is a per individual, per exonic SNP random effect. We use variational Bayes EM to approximately integrate over all ui while optimizing with respect to the other parameters. Last, c is the concentration parameter which we learn per exonic SNP using maximum a posteriori probability estimation with a Gamma (1.0001, 10E−4) prior. We use a likelihood ratio test to test the global hypothesis of no differences in ASE between baseline and any time point. We corrected for multiple testing across SNPs using the BH procedure (FDR < 0.05).
Linear regressions to find analytes associated with peak VO2.
Peak VO2 was scaled to body weight. Three linear regressions were conducted at each time point using the lm base function in R and were adjusted for age, sex, ethnicity/race, batch information as well as body mass index or fat mass or percent fat. An additional linear regression adjusted for age, sex, ethnicity/race, batch information was performed on peak VO2 scaled by lean body mass. Fat mass, percent fat and lean body mass were calculated using the equations from NHANES (Lee et al., 2017). P values were corrected using BH method and analytes with FDR below 0.05 were considered significant. The lipid dataset used was TAG_med.
CPX parameter prediction models.
Prediction models were trained using baseline multi-omic measurements (pre-exercise). Clinical laboratory results and gut microbiome data were from the iHMP dataset at the closest healthy visit to the exercise day. No clinical and gut microbiome data were available for participant ZJBOZ2X, hence this participant was excluded from the analysis. Prediction modeling was performed by first selecting predictive analytes using a Bayesian network algorithm and then building the model using ridge regression.
Feature selection.
All data (demographics, clinical, complex lipids, metabolites, targeted and untargeted proteins, transcripts) except gut microbiome percent data were log2-transformed. All features were standardized to zero mean with unit variance. Output CPX parameters were not transformed or scaled. Identification of predictive features was performed using the Max-Min Parents and Child algorithm (MMPC) in the R package ‘MXM’ (v1.4.1). Feature selection was performed using leave-one-out cross-validation, where 34 training sets were constructed and each training set excludes the data from a different participant. The MMPC algorithm was run on each training set and predictive features were selected if they were used in ≥ 20% of the models generated from the training sets.
Ridge regression modeling.
We performed leave-one-out cross-validation to maximize available training data. For each training set, we optimize the hyperparameter by performing a grid search and selecting the model that minimizes test error. The predicted output value is the value from the cross-validation iteration in which that output data point and its associated features are excluded from the training set. We use these predicted values to calculate mean square error (MSE) and R2. The value of the hyperparameter used was the average of the hyperparameters which minimized test error during cross-validation.
Linear mixed models to find analytes responding differently to exercise in insulin resistant participants.
Linear mixed models were conducted at each time point using the ‘lme4’ package (v1.1–21) in R and were adjusted for personal baseline, age, sex, BMI, race/ethnicity and batch information. ‘lmerTest’ package in R (v3.1–0) was used to compute P values at each time point post-exercise. P values were corrected using BH method. The lipid dataset used was TAG_med.
Multi-omic outlier analysis.
Outlier analysis was performed on non-imputed datasets to avoid any potential bias. Multi-omic data were log2-transformed and all features were standardized to zero mean with unit variance. Absolute levels and delta values relative to baseline were used for the ‘baseline’ and ‘in response to exercise’ analyses, respectively. P values were calculated assuming a normal distribution and were corrected for multiple hypothesis using the Benjamini-Hochberg procedure. Analytes with FDR below 0.05 at baseline were considered outliers. For outliers in response to exercise, two significant outliers across the four time points (2, 15, 30 min and 1-hour time points) were required.
DATA AND CODE AVAILABILITY
Raw and processed omic data (transcriptome, targeted and untargeted proteome, metabolome, lipidome) are hosted on our portal at http://hmp2-data.stanford.edu/index.phpunder the substudy Exercise. Microbiome and clinical laboratory data are provided in Tables S4.6 and S4.7, respectively.
Supplementary Material
Figure S1. Quality controls for data generation and exercise protocol, molecular changes in response to exercise and molecular individuality. (a) Principal component analysis across omic assays. The study samples were intermixed suggesting limited batch effect and the quality control samples (QCs) clustered together indicating good technical reproducibility. Each dot represents a sample colored by batch information. Black and grey dots are QCs. For metabolomics and untargeted proteomics, QCs consist in an equimolar mix of all the samples in the study. For lipidomics, QCs are lipid extracts from a reference plasma sample. QC1–4 in targeted proteomic experiments represent individual samples in the study analyzed in each plate. Transcriptomic experiment didn’t contain QC. (b) 2D visualization of all multi-omic analytes using t-distributed stochastic neighbor embedding (tSNE) algorithm. Each dot represents a single sample colored by participants. (c) Bar graph representing the proportion of analytes across molecule types significantly changing in response to exercise (FDR < 0.05). (d) Principal component analysis across omic assays. Each dot represents a sample colored by participants. Related to Figure 1.
Figure S2. Inter-individual variability. (a) Technical variability for complex lipids, metabolites and proteins using quality control samples as described in Figure S1a. We calculated that 95.2% of metabolites, 91.7% of complex lipids and 65.8% of proteins had a coefficient of variation (CV) below 20% across QCs. (b) Inter- individual variability at all time points using absolute levels across molecule types. Inter-individual variability by lipid class (c) and metabolic class (d). (e) Inter-individual variability at all time points using analyte levels relative to baseline. Related to Figure 1 and Table S3.
Figure S3. Natural molecular variation in absence of exercise and inter-day molecular variation. (a) Proportion of analytes changing in absence of exercise among the analytes that are changing with exercise. Linear models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed using a subset of 14 individuals that participated in both the exercise and the control studies. We found 30/108 proteins, 224/664 metabolites and 561/710 complex lipids changing significantly in response to exercise (FDR < 0.10) among which 3 proteins, 12 metabolites and 0 complex lipid also changed significantly without exercise over the 1-hour time frame. Longitudinal trajectories of targeted proteins (b) and metabolites (c) that changed without exercise (FDR < 0.10). The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). Of note, 1 targeted protein (vWF) was not measured in the control set because it was no longer commercialized as part of the CVD kit. 664/728 metabolites (91.2%) could be detected in the control set. (d) Proportion of analytes changing the next morning among the analytes that are changing with exercise in a subset of the cohort (n = 15). Linear models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed. We found 3,637/15,855 transcripts, 8/109 proteins, 159/728 metabolites and 157/710 complex lipids changing significantly in response to exercise (FDR < 0.05) among which 15 metabolites and 95 complex lipids (94/95 were TAG) were different the next morning. (e) Inter-day variability relative to the variability induced by exercise. For each analyte that was different the next morning (n = 110), we calculated the ratio of the maximum/minimum fold change with exercise over the fold change the next morning. The distribution of ratios shows that the variability induced by exercise is much larger than the inter-day variability (> 2-fold difference). TAG were included in this analysis. Related to Figure 1.
Figure S4. Cluster compositions and longitudinal trajectories of selected metabolic pathways/classes in response to exercise. (a) Cluster compositions in metabolite and complex lipid classes as well as protein functions. Longitudinal trajectories of significant metabolites: (b) branched-chain ketoacids, (c) caffeine metabolism, (d) bile acids, (e) urea cycle, (f) cofactors and vitamins, (g) choline metabolism, (h) lysine metabolism, (i) histidine metabolism, (j) tryptophan metabolism and (k) gamma-glutamyl dipeptides. The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). Related to Figures 2–3 and Table S2.
Figure S5. Pathway dynamics between each two consecutive time points and population-level differential allele-specific expression (ASE) by time. Pathway/chemical class enrichment analysis of circulating plasma metabolites (a) and pathway analysis using significant PBMC transcripts(b). Pathway direction is the median log2 fold change relative to the previous time point of significant molecules in each pathway (blue: downregulated, red: upregulated). The dot size represents pathway significance. (c) QQ plot of likelihood-ratio test (LRT) P values for differential ASE with time analysis. (d) Distribution of allelic ratios across time for all SNPs with significant differential ASE by time effects. Related to Figures 2,4 and Table S2.
Figure S6. Peak VO2 in the cohort relative to a reference population (FRIEND registry) and molecular associations with peak VO2 (a) Peak VO2 values categorized by age and sex. Blue and green dots represent the mean values from the FRIEND registry and error bars represent one standard deviation. (b) Distribution of peak VO2 normalized to reference values from the FRIEND registry. (c) Proportion of analytes associated with peak VO2 (scaled by BW) as determined by linear regression analyses adjusted for BMI, fat mass or percent fat (FDR < 0.05). (d) Proportion of analytes associated with peak VO2 scaled by BW and LBM as determined by linear regression analyses (FDR < 0.05). Related to Figure 5 and Tables S4.
Figure S7. Differential response to acute exercise in insulin resistant participants. Patterns of differential metabolites (FDR < 0.10) (a) and complex lipids (FDR < 0.20) (b) in IR participants. Linear mixed models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed. The solid line represents the mean and the dashed line represents the 95% confidence interval. (c) Heatmap of significant proteins as determined by the linear mixed model (FDR < 0.20) representing the median log2 fold change relative to baseline in the cohort. Proteins were grouped in patterns. Differential response to exercise for (d) TNF-α and IL-6 as well as key metabolites (e). Related to Figure 6 and Table S5.
Figure S8. Multi-omic outlier analysis. (a) Number of outliers across omes per participant at baseline and in response to exercise (FDR < 0.05). Participants with significantly different number of outliers in comparison to the population (FDR < 0.05) are represented with a solid bar. (b) Heatmap of clinical laboratory results as well as key rest and stress parameters representing the BH-corrected P values for each measure in each participant. Outlier measures (FDR < 0.05) are outlined in red. Participant ZLTUJTN was mildly anemic with borderline high RBC (5.92 M/μl, normal range: 4.40–5.90), borderline low hemoglobin (13.4 g/dl, normal range: 13.5–17.7), high RDW (20.5%, normal range: 11.5–14.5%) and low MCV (72.8fl/red cell, normal range: 82.0– 98.0), MCH (22.6 pg/cell, normal range: 27.0 to 34.0) and MCHC (31.0 g/dl, normal range: 32.0 – 36.0). Participant ZVM4N7A had a borderline high mean platelet count over 7 laboratory visits ± 5 months relative to the exercise date (405 K/μl, normal range: 150–400). (c) Functional association network of outlier proteins (FDR < 0.05) in response to exercise in individual ZL63I8R. This analysis was performed using the web tool STRING. Edges correspond to known, predicted or other interactions. Proteins in blue are downregulated and proteins in red are upregulated.Related to Table S6.
Table S1. Cohort characteristics and listing of reported analytes. (1) Participants baseline demographics, echocardiographic and cardiopulmonary exercise characteristics. (2) Listing of targeted proteins (immune, cardiovascular and metabolic). (3) Listing of all plasma and cellular analytes detected in the study (Related to Figure 1).
Table S2. Molecular and pathway changes in response to acute exercise. (1) Multi-omic changes in response to acute exercise and cluster information (Related to Figures 1–4 and S1, S4). (2) Lipid class and metabolic pathways changed in response to exercise (Related to Figure 2). (3) Metabolomic pathway dynamics (Related to Figure S5). (4) Association of key changes induced by exercise with various parameters. (5) PBMC RNA pathways changed in response to exercise (Related to Figure 4). (6) Transcriptomic pathway dynamics (Related to Figure S5).
Table S3. Molecular individuality. (1) Inter-individual variability using absolute levels. (2) Pathway enrichment analysis using variable genes (CV > 100%). (3) Inter-individual variability using fold change to exercise (Related to Figures 1 and S2).
Table S4. Molecular and pathway associations with peak VO2. (1) Multi-omic associations with peak VO2 normalized by body weight and adjusted for body composition. (2) Molecular associations with peak VO2 scaled by body weight and lean body mass. (3) Lipid classes and metabolic pathways associated with peak VO2. (4) RNA pathways associated with peak VO2. (5) Protein pathways associated with peak VO2. (6) Clinical laboratory results (closest to exercise date). (7) Gut microbiome data (closest to exercise date). (8) Peak VO2, (9) VE/VCO2 slope and (10) RER prediction models (Related to Figures 5 and S6).
Table S5. Differential response to exercise in insulin resistant participants. (1) Molecules responding differentially to acute exercise in IR participants and associated patterns. (2) Differential PBMC pathways in IR participants. (3) Differential lipid classes and metabolic pathways in IR participants (Related to Figures 6 and S7).
Table S6. Pathway enrichment analysis using outlier transcripts (Related to Figure S8).
Highlights:
-
-
Time-series analysis reveals an orchestrated molecular choreography of exercise
-
-
Multi-level omic associations identify key biological processes of peak VO2
-
-
Prediction models highlight resting blood biomarkers of fitness
-
-
Exercise omics provides insights into the pathophysiology of insulin resistance
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Health (NIH) Human Microbiome Project (HMP) 1U54DE02378901, S10OD020141 as well as SDRC grant P30DK116074. We would like to acknowledge the support from the Deanship of Scientific Research (DSR), King Abdulaziz University (KAU), and Jeddah the Ministry of Education for Saudi Arabia. K.M. received support from an Australian Government Research Training Program (RTP) Scholarship. We thank Yael Rosenberg-Hasson and the Human Immune Monitoring Center (HIMC) at Stanford for generating targeted proteomic data. We also would like to thank the iPOP participants who generously gave their time and biological samples.
Footnotes
Declaration of Interests
M.P.S. is a cofounder and on the advisory board of Personalis, SensOmics, January, Filtricine, Qbio, Protos and Mirive. M.P.S. is on the advisory board of Genapsys and Tailai. M.P.S is an inventor on provisional patent number 62/897,908 ‘Surrogate of VO2 MAX Test’. K.C. and F.H. are also listed as inventors. E.A.A is a cofounder of Personalis, Deepcell, SVEXA and on the advisory board of Apple, SequenceBio and Foresite Labs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agudelo LZ, Femenia T, Orhan F, Porsmyr-Palmertz M, Goiny M, Martinez-Redondo V, Correia JC, Izadi M, Bhat M, Schuppe-Koistinen I, et al. (2014). Skeletal muscle PGC-1alpha1 modulates kynurenine metabolism and mediates resilience to stress-induced depression. Cell 159, 33–45. [DOI] [PubMed] [Google Scholar]
- Albouaini K, Egred M, Alahmar A, and Wright DJ (2007). Cardiopulmonary exercise testing and its application. Heart 93, 1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena R, Myers J, Aslam SS, Varughese EB, and Peberdy MA (2003). Technical considerations related to the minute ventilation/carbon dioxide output slope in patients with heart failure. Chest 124, 720–727. [DOI] [PubMed] [Google Scholar]
- Arena R, Myers J, Aslam SS, Varughese EB, and Peberdy MA (2004). Peak VO2 and VE/VCO2 slope in patients with heart failure: a prognostic comparison. Am Heart J 147, 354–360. [DOI] [PubMed] [Google Scholar]
- Arena R, and Sietsema KE (2011). Cardiopulmonary exercise testing in the clinical evaluation of patients with heart and lung disease. Circulation 123, 668–680. [DOI] [PubMed] [Google Scholar]
- Barupal DK, and Fiehn O. (2017). Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci Rep 7, 14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisser D, Klau GW, Dandekar T, Muller T, and Dittrich MT (2010). BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics 26, 1129–1130. [DOI] [PubMed] [Google Scholar]
- Belcastro AN, Arthur GD, Albisser TA, and Raj DA (1996). Heart, liver, and skeletal muscle myeloperoxidase activity during exercise. J Appl Physiol (1985) 80, 1331–1335. [DOI] [PubMed] [Google Scholar]
- Bird SR, and Hawley JA (2016). Update on the effects of physical activity on insulin sensitivity in humans. BMJ Open Sport Exerc Med 2, e000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen TS, Schuler G, and Adams V. (2015). Skeletal muscle wasting in cachexia and sarcopenia: molecular pathophysiology and impact of exercise training. J Cachexia Sarcopenia Muscle 6, 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevetti G, De Caterina M, Martone VD, Ungaro B, Corrado F, Silvestro A, de Cristofaro T, and Scopacasa F. (2001). Exercise increases soluble adhesion molecules ICAM-1 and VCAM-1 in patients with intermittent claudication. Clin Hemorheol Microcirc 24, 193–199. [PubMed] [Google Scholar]
- Calabia J, Torguet P, Garcia M, Garcia I, Martin N, Guasch B, Faur D, and Valles M. (2011). Doppler ultrasound in the measurement of pulse wave velocity: agreement with the Complior method. Cardiovasc Ultrasound 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder PC (2013). Omega-3 polyunsaturated fatty acids and inflammatory processes: nutrition or pharmacology? Br J Clin Pharmacol 75, 645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson LA, Tighe SW, Kenefick RW, Dragon J, Westcott NW, and Leclair RJ (2011). Changes in transcriptional output of human peripheral blood mononuclear cells following resistance exercise. Eur J Appl Physiol 111, 2919–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xun K, Chen L, and Wang Y. (2009). TNF-alpha, a potent lipid metabolism regulator. Cell Biochem Funct 27, 407–416. [DOI] [PubMed] [Google Scholar]
- Claudel T, Staels B, and Kuipers F. (2005). The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol 25, 2020–2030. [DOI] [PubMed] [Google Scholar]
- Connolly PH, Caiozzo VJ, Zaldivar F, Nemet D, Larson J, Hung SP, Heck JD, Hatfield GW, and Cooper DM (2004). Effects of exercise on gene expression in human peripheral blood mononuclear cells. J Appl Physiol (1985) 97, 1461–1469. [DOI] [PubMed] [Google Scholar]
- Contrepois K, Jiang L, and Snyder M. (2015). Optimized Analytical Procedures for the Untargeted Metabolomic Profiling of Human Urine and Plasma by Combining Hydrophilic Interaction (HILIC) and Reverse- Phase Liquid Chromatography (RPLC)-Mass Spectrometry. Mol Cell Proteomics 14, 1684–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois K, Mahmoudi S, Ubhi BK, Papsdorf K, Hornburg D, Brunet A, and Snyder M. (2018). Cross-Platform Comparison of Untargeted and Targeted Lipidomics Approaches on Aging Mouse Plasma. Sci Rep 8, 17747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha TF, Moreira JB, Paixao NA, Campos JC, Monteiro AW, Bacurau AV, Bueno CR Jr., Ferreira JC, and Brum PC (2012). Aerobic exercise training upregulates skeletal muscle calpain and ubiquitin-proteasome systems in healthy mice. J Appl Physiol (1985) 112, 1839–1846. [DOI] [PubMed] [Google Scholar]
- Droste SK, Gesing A, Ulbricht S, Muller MB, Linthorst AC, and Reul JM (2003). Effects of long-term voluntary exercise on the mouse hypothalamic-pituitary-adrenocortical axis. Endocrinology 144, 3012–3023. [DOI] [PubMed] [Google Scholar]
- Fischer CP (2006). Interleukin-6 in acute exercise and training: what is the biological relevance? Exerc Immunol Rev 12, 6–33. [PubMed] [Google Scholar]
- Force ERST, Palange P, Ward SA, Carlsen KH, Casaburi R, Gallagher CG, Gosselink R, O’Donnell DE, Puente-Maestu L, Schols AM, et al. (2007). Recommendations on the use of exercise testing in clinical practice. Eur Respir J 29, 185–209. [DOI] [PubMed] [Google Scholar]
- Gjevestad GO, Holven KB, and Ulven SM (2015). Effects of Exercise on Gene Expression of Inflammatory Markers in Human Peripheral Blood Cells: A Systematic Review. Curr Cardiovasc Risk Rep 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbidi S, and Laher I. (2014). Exercise induced adipokine changes and the metabolic syndrome. J Diabetes Res 2014, 726861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin ML, Harris JE, Hernandez A, and Gladden LB (2007). Blood lactate measurements and analysis during exercise: a guide for clinicians. J Diabetes Sci Technol 1, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham ZA, Gallagher PM, and Cardozo CP (2015). Focal adhesion kinase and its role in skeletal muscle. J Muscle Res Cell Motil 36, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J, and Hu FB (2016). Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 39, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Baglaenko Y, Arora J, Hannes S, Luo Y, Amariuta T, Teslovich N, Rao DA, Ermann J, Jonsson H, et al. (2019). Allele-specific expression changes dynamically during T cell activation in HLA and other autoimmune loci. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakonarson H, Maskeri N, Carter C, Chuang S, and Grunstein MM (1999). Autocrine interaction between IL-5 and IL-1beta mediates altered responsiveness of atopic asthmatic sensitized airway smooth muscle. J Clin Invest 104, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson J. (1991). Effect of exercise on amino acid concentrations in skeletal muscle and plasma. J Exp Biol 160, 149–165. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS (2006). Inflammation and metabolic disorders. Nature 444, 860–867. [DOI] [PubMed] [Google Scholar]
- Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, and Sauer K. (2007). Positive regulation of Itk PH domain function by soluble IP4. Science 316, 886–889. [DOI] [PubMed] [Google Scholar]
- Ingenbleek Y, and Bernstein LH (2015). Plasma Transthyretin as a Biomarker of Lean Body Mass and Catabolic States. Adv Nutr 6, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilma B, Eichler HG, Stohlawetz P, Dirnberger E, Kapiotis S, Wagner OF, Schutz W, and Krejcy K. (1997). Effects of exercise on circulating vascular adhesion molecules in healthy men. Immunobiology 197, 505–512. [DOI] [PubMed] [Google Scholar]
- Kaminsky LA, Arena R, and Myers J. (2015). Reference Standards for Cardiorespiratory Fitness Measured With Cardiopulmonary Exercise Testing: Data From the Fitness Registry and the Importance of Exercise National Database. Mayo Clin Proc 90, 1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NA, Burley VJ, and Blundell JE (1994). Exercise-induced suppression of appetite: effects on food intake and implications for energy balance. Eur J Clin Nutr 48, 715–724. [PubMed] [Google Scholar]
- Kinugawa T, Kato M, Ogino K, Osaki S, Tomikura Y, Igawa O, Hisatome I, and Shigemasa C. (2003). Interleukin-6 and tumor necrosis factor-alpha levels increase in response to maximal exercise in patients with chronic heart failure. Int J Cardiol 87, 83–90. [DOI] [PubMed] [Google Scholar]
- Kjaer M. (1998). Hepatic glucose production during exercise. Adv Exp Med Biol 441, 117–127. [PubMed] [Google Scholar]
- Klok MD, Jakobsdottir S, and Drent ML (2007). The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes Rev 8, 21–34. [DOI] [PubMed] [Google Scholar]
- Knowles DA, Davis JR, Edgington H, Raj A, Fave MJ, Zhu X, Potash JB, Weissman MM, Shi J, Levinson DF, et al. (2017). Allele-specific expression reveals interactions between genetic variation and environment. Nat Methods 14, 699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch LG, Kemi OJ, Qi N, Leng SX, Bijma P, Gilligan LJ, Wilkinson JE, Wisloff H, Hoydal MA, Rolim N, et al. (2011). Intrinsic aerobic capacity sets a divide for aging and longevity. Circ Res 109, 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojok K, El-Kadiry AE, and Merhi Y. (2019). Role of NF-kappaB in Platelet Function. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusudo T, Kontani Y, Kataoka N, Ando F, Shimokata H, and Yamashita H. (2011). Fatty acid-binding protein 3 stimulates glucose uptake by facilitating AS160 phosphorylation in mouse muscle cells. Genes Cells 16, 681–691. [DOI] [PubMed] [Google Scholar]
- Ladenvall P, Persson CU, Mandalenakis Z, Wilhelmsen L, Grimby G, Svardsudd K, and Hansson PO (2016). Low aerobic capacity in middle-aged men associated with increased mortality rates during 45 years of follow-up. Eur J Prev Cardiol 23, 1557–1564. [DOI] [PubMed] [Google Scholar]
- Laffel L. (1999). Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab Res Rev 15, 412–426. [DOI] [PubMed] [Google Scholar]
- Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, et al. (2015). Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Eur Heart J Cardiovasc Imaging 16, 233–270. [DOI] [PubMed] [Google Scholar]
- Lee DH, Keum N, Hu FB, Orav EJ, Rimm EB, Sun Q, Willett WC, and Giovannucci EL (2017). Development and validation of anthropometric prediction equations for lean body mass, fat mass and percent fat in adults using the National Health and Nutrition Examination Survey (NHANES) 1999–2006. Br J Nutr 118, 858–866. [DOI] [PubMed] [Google Scholar]
- Lee KK, Cipriano LE, Owens DK, Go AS, and Hlatky MA (2010). Cost-effectiveness of using high- sensitivity C-reactive protein to identify intermediate- and low-cardiovascular-risk individuals for statin therapy. Circulation 122, 1478–1487. [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, and Fassler R. (2009). Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev 23, 397–418. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Zhao X, Weigert C, Simon P, Fehrenbach E, Fritsche J, Machann J, Schick F, Wang J, Hoene M, et al. (2010). Medium chain acylcarnitines dominate the metabolite pattern in humans under moderate intensity exercise and support lipid oxidation. PLoS One 5, e11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis GD, Farrell L, Wood MJ, Martinovic M, Arany Z, Rowe GC, Souza A, Cheng S, McCabe EL, Yang E, et al. (2010). Metabolic signatures of exercise in human plasma. Sci Transl Med 2, 33ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, and Flavell RA (2008). Contextual regulation of inflammation: a duet by transforming growth factor- beta and interleukin-10. Immunity 28, 468–476. [DOI] [PubMed] [Google Scholar]
- Lymperopoulos A, Rengo G, and Koch WJ (2013). Adrenergic nervous system in heart failure: pathophysiology and therapy. Circ Res 113, 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, and Spiegel S. (2014). Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasciulo FL, Montagnani M, and Potenza MA (2006). Endothelin-1: the yin and yang on vascular function. Curr Med Chem 13, 1655–1665. [DOI] [PubMed] [Google Scholar]
- McClelland RL, Jorgensen NW, Budoff M, Blaha MJ, Post WS, Kronmal RA, Bild DE, Shea S, Liu K, Watson KE, et al. (2015). 10-Year Coronary Heart Disease Risk Prediction Using Coronary Artery Calcium and Traditional Risk Factors: Derivation in the MESA (Multi-Ethnic Study of Atherosclerosis) With Validation in the HNR (Heinz Nixdorf Recall) Study and the DHS (Dallas Heart Study). J Am Coll Cardiol 66, 1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melanson SE, Green SM, Wood MJ, Neilan TG, and Lewandrowski EL (2006). Elevation of myeloperoxidase in conjunction with cardiac-specific markers after marathon running. Am J Clin Pathol 126, 888–893. [DOI] [PubMed] [Google Scholar]
- Millard AL, Valli PV, Stussi G, Mueller NJ, Yung GP, and Seebach JD (2013). Brief Exercise Increases Peripheral Blood NK Cell Counts without Immediate Functional Changes, but Impairs their Responses to ex vivo Stimulation. Front Immunol 4, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov VI, Tsyplenkov PV, Golberg ND, and Kalinski MI (2006). The effects of high-intensity exercise on skeletal muscle neutrophil myeloperoxidase in untrained and trained rats. Eur J Appl Physiol 97, 716–722. [DOI] [PubMed] [Google Scholar]
- Myers J, and Froelicher VF (1993). Exercise testing. Procedures and implementation. Cardiol Clin 11, 199–213. [PubMed] [Google Scholar]
- Negro M, Giardina S, Marzani B, and Marzatico F. (2008). Branched-chain amino acid supplementation does not enhance athletic performance but affects muscle recovery and the immune system. J Sports Med Phys Fitness 48, 347–351. [PubMed] [Google Scholar]
- Nielsen R, Moller N, Gormsen LC, Tolbod LP, Hansson NH, Sorensen J, Harms HJ, Frokiaer J, Eiskjaer H, Jespersen NR, et al. (2019). Cardiovascular Effects of Treatment With the Ketone Body 3- Hydroxybutyrate in Chronic Heart Failure Patients. Circulation 139, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman DC, Shanely RA, Gillitt ND, Pappan KL, and Lila MA (2013). Serum metabolic signatures induced by a three-day intensified exercise period persist after 14 h of recovery in runners. J Proteome Res 12, 4577–4584. [DOI] [PubMed] [Google Scholar]
- Odobasic D, Kitching AR, and Holdsworth SR (2016). Neutrophil-Mediated Regulation of Innate and Adaptive Immunity: The Role of Myeloperoxidase. J Immunol Res 2016, 2349817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GP, Hurtado-Oliveros J, Chung HY, Knoll K, Neumann T, Muller HJ, Herbsleb M, Kohl M, Busch M, Sossdorf M, et al. (2015). Plasma Neutrophil Gelatinase-Associated Lipocalin Is Primarily Related to Inflammation during Sepsis: A Translational Approach. PLoS One 10, e0124429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Evans CR, Qi NR, Minogue CE, Carson JJ, Chermside-Scabbo CJ, Koch LG, Britton SL, Pagliarini DJ, Coon JJ, et al. (2015). Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab 21, 468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallister T, Jackson MA, Martin TC, Zierer J, Jennings A, Mohney RP, MacGregor A, Steves CJ, Cassidy A, Spector TD, et al. (2017). Hippurate as a metabolomic marker of gut microbiome diversity: Modulation by diet and relationship to metabolic syndrome. Sci Rep 7, 13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippou A, Maridaki M, Theos A, and Koutsilieris M. (2012). Cytokines in muscle damage. Adv Clin Chem 58, 49–87. [DOI] [PubMed] [Google Scholar]
- Piening BD, Zhou W, Contrepois K, Rost H, Gu Urban GJ, Mishra T, Hanson BM, Bautista EJ, Leopold S, Yeh CY, et al. (2018). Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell Syst 6, 157–170 e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Qi Q, Prudente S, Mendonca C, Andreozzi F, di Pietro N, Sturma M, Novelli V, Mannino GC, Formoso G, et al. (2013). Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA 310, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom-Aizik S, Zaldivar F Jr., Leu SY, and Cooper DM (2009). Brief bout of exercise alters gene expression in peripheral blood mononuclear cells of early- and late-pubertal males. Pediatr Res 65, 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo RF, and Rhodes EC (1998). Lipid metabolism during exercise. Sports Med 26, 29–42. [DOI] [PubMed] [Google Scholar]
- Reihmane D, Jurka A, and Tretjakovs P. (2012). The relationship between maximal exercise-induced increases in serum IL-6, MPO and MMP-9 concentrations. Scand J Immunol 76, 188–192. [DOI] [PubMed] [Google Scholar]
- Ren YY, Koch LG, Britton SL, Qi NR, Treutelaar MK, Burant CF, and Li JZ (2016). Selection-, age-, and exercise-dependence of skeletal muscle gene expression patterns in a rat model of metabolic fitness. Physiol Genomics 48, 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribisl PM, Lang W, Jaramillo SA, Jakicic JM, Stewart KJ, Bahnson J, Bright R, Curtis JF, Crow RS, Soberman JE, et al. (2007). Exercise capacity and cardiovascular/metabolic characteristics of overweight and obese individuals with type 2 diabetes: the Look AHEAD clinical trial. Diabetes Care 30, 2679–2684. [DOI] [PubMed] [Google Scholar]
- Rost HL, Liu Y, D’Agostino G, Zanella M, Navarro P, Rosenberger G, Collins BC, Gillet L, Testa G, Malmstrom L, et al. (2016). TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat Methods 13, 777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarullo FM, Fazio G, Brusca I, Fasullo S, Paterna S, Licata P, Novo G, Novo S, and Di Pasquale P. (2010). Cardiopulmonary Exercise Testing in Patients with Chronic Heart Failure: Prognostic Comparison from Peak VO2 and VE/VCO2 Slope. Open Cardiovasc Med J 4, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiman J, Luber JM, Chavkin TA, MacDonald T, Tung A, Pham LD, Wibowo MC, Wurth RC, Punthambaker S, Tierney BT, et al. (2019). Meta-omics analysis of elite athletes identifies a performance- enhancing microbe that functions via lactate metabolism. Nat Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, and Mammucari C. (2011). Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussler-Fiorenza Rose SM, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, Dagan- Rosenfeld O, Ganz AB, Dunn J, Hornburg D, et al. (2019). A longitudinal big data approach for precision health. Nat Med 25, 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting- induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Sethi JK, and Hotamisligil GS (1999). The role of TNF alpha in adipocyte metabolism. Semin Cell Dev Biol 10, 19–29. [DOI] [PubMed] [Google Scholar]
- Shah SH, and Hunter WG (2017). Realizing the Potential of Metabolomics in Heart Failure: Signposts on the Path to Clinical Utility. JACC Heart Fail 5, 833–836. [DOI] [PubMed] [Google Scholar]
- Shinkai S, Shore S, Shek PN, and Shephard RJ (1992). Acute exercise and immune function. Relationship between lymphocyte activity and changes in subset counts. Int J Sports Med 13, 452–461. [DOI] [PubMed] [Google Scholar]
- Smith DA (2016). In adults without CVD, the MESA score, including coronary artery calcium, predicted 10-y risk for CHD events. Ann Intern Med 164, JC35. [DOI] [PubMed] [Google Scholar]
- Stakiw J, Bowman M, Hegadorn C, Pruss C, Notley C, Groot E, Lenting PJ, Rapson D, Lillicrap D, and James P. (2008). The effect of exercise on von Willebrand factor and ADAMTS-13 in individuals with type 1 and type 2B von Willebrand disease. J Thromb Haemost 6, 90–96. [DOI] [PubMed] [Google Scholar]
- Sutton JR, Toews CJ, Ward GR, and Fox IH (1980). Purine metabolism during strenuous muscular exercise in man. Metabolism 29, 254–260. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43, D447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak AG, Herder C, Rathmann W, Brunner EJ, and Kivimaki M. (2012). Prediabetes: a high-risk state for diabetes development. Lancet 379, 2279–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani AG, and Beinlich CJ (1991). Pantothenic acid in health and disease. Vitam Horm 46, 165–228. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Hirota Y, Sohmiya K, Nishimura S, and Kawamura K. (1991). Serum and urinary human heart fatty acid-binding protein in acute myocardial infarction. Clin Biochem 24, 195–201. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, and Cox J. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Ulven SM, Foss SS, Skjolsvik AM, Stadheim HK, Myhrstad MC, Raael E, Sandvik M, Narverud I, Andersen LF, Jensen J, et al. (2015). An acute bout of exercise modulate the inflammatory response in peripheral blood mononuclear cells in healthy young men. Arch Physiol Biochem 121, 41–49. [DOI] [PubMed] [Google Scholar]
- Vijayaraghava A, Doreswamy V, Narasipur OS, Kunnavil R, and Srinivasamurthy N. (2015). Effect of Yoga Practice on Levels of Inflammatory Markers After Moderate and Strenuous Exercise. J Clin Diagn Res 9, CC08–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallentin L, Zethelius B, Berglund L, Eggers KM, Lind L, Lindahl B, Wollert KC, and Siegbahn A. (2013). GDF-15 for prognostication of cardiovascular and cancer morbidity and mortality in men. PLoS One 8, e78797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton DE, Nicol CW, and Bredin SS (2006). Health benefits of physical activity: the evidence. CMAJ 174, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel B, and Otterbein LE (2012). Go green: the anti-inflammatory effects of biliverdin reductase. Front Pharmacol 3, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, and Kannel WB (1998). Prediction of coronary heart disease using risk factor categories. Circulation 97, 1837–1847. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M. (1994). The endothelin system. A new target for therapeutic intervention. Circulation 89, 1320–1322. [DOI] [PubMed] [Google Scholar]
- Zhang J, Light AR, Hoppel CL, Campbell C, Chandler CJ, Burnett DJ, Souza EC, Casazza GA, Hughen RW, Keim NL, et al. (2017). Acylcarnitines as markers of exercise-associated fuel partitioning, xenometabolism, and potential signals to muscle afferent neurons. Exp Physiol 102, 48–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Sailani MR, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T, et al. (2019). Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature 569, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quality controls for data generation and exercise protocol, molecular changes in response to exercise and molecular individuality. (a) Principal component analysis across omic assays. The study samples were intermixed suggesting limited batch effect and the quality control samples (QCs) clustered together indicating good technical reproducibility. Each dot represents a sample colored by batch information. Black and grey dots are QCs. For metabolomics and untargeted proteomics, QCs consist in an equimolar mix of all the samples in the study. For lipidomics, QCs are lipid extracts from a reference plasma sample. QC1–4 in targeted proteomic experiments represent individual samples in the study analyzed in each plate. Transcriptomic experiment didn’t contain QC. (b) 2D visualization of all multi-omic analytes using t-distributed stochastic neighbor embedding (tSNE) algorithm. Each dot represents a single sample colored by participants. (c) Bar graph representing the proportion of analytes across molecule types significantly changing in response to exercise (FDR < 0.05). (d) Principal component analysis across omic assays. Each dot represents a sample colored by participants. Related to Figure 1.
Figure S2. Inter-individual variability. (a) Technical variability for complex lipids, metabolites and proteins using quality control samples as described in Figure S1a. We calculated that 95.2% of metabolites, 91.7% of complex lipids and 65.8% of proteins had a coefficient of variation (CV) below 20% across QCs. (b) Inter- individual variability at all time points using absolute levels across molecule types. Inter-individual variability by lipid class (c) and metabolic class (d). (e) Inter-individual variability at all time points using analyte levels relative to baseline. Related to Figure 1 and Table S3.
Figure S3. Natural molecular variation in absence of exercise and inter-day molecular variation. (a) Proportion of analytes changing in absence of exercise among the analytes that are changing with exercise. Linear models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed using a subset of 14 individuals that participated in both the exercise and the control studies. We found 30/108 proteins, 224/664 metabolites and 561/710 complex lipids changing significantly in response to exercise (FDR < 0.10) among which 3 proteins, 12 metabolites and 0 complex lipid also changed significantly without exercise over the 1-hour time frame. Longitudinal trajectories of targeted proteins (b) and metabolites (c) that changed without exercise (FDR < 0.10). The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). Of note, 1 targeted protein (vWF) was not measured in the control set because it was no longer commercialized as part of the CVD kit. 664/728 metabolites (91.2%) could be detected in the control set. (d) Proportion of analytes changing the next morning among the analytes that are changing with exercise in a subset of the cohort (n = 15). Linear models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed. We found 3,637/15,855 transcripts, 8/109 proteins, 159/728 metabolites and 157/710 complex lipids changing significantly in response to exercise (FDR < 0.05) among which 15 metabolites and 95 complex lipids (94/95 were TAG) were different the next morning. (e) Inter-day variability relative to the variability induced by exercise. For each analyte that was different the next morning (n = 110), we calculated the ratio of the maximum/minimum fold change with exercise over the fold change the next morning. The distribution of ratios shows that the variability induced by exercise is much larger than the inter-day variability (> 2-fold difference). TAG were included in this analysis. Related to Figure 1.
Figure S4. Cluster compositions and longitudinal trajectories of selected metabolic pathways/classes in response to exercise. (a) Cluster compositions in metabolite and complex lipid classes as well as protein functions. Longitudinal trajectories of significant metabolites: (b) branched-chain ketoacids, (c) caffeine metabolism, (d) bile acids, (e) urea cycle, (f) cofactors and vitamins, (g) choline metabolism, (h) lysine metabolism, (i) histidine metabolism, (j) tryptophan metabolism and (k) gamma-glutamyl dipeptides. The dots represent the mean log2 fold change relative to baseline and the bars the standard error of the mean (SEM). Related to Figures 2–3 and Table S2.
Figure S5. Pathway dynamics between each two consecutive time points and population-level differential allele-specific expression (ASE) by time. Pathway/chemical class enrichment analysis of circulating plasma metabolites (a) and pathway analysis using significant PBMC transcripts(b). Pathway direction is the median log2 fold change relative to the previous time point of significant molecules in each pathway (blue: downregulated, red: upregulated). The dot size represents pathway significance. (c) QQ plot of likelihood-ratio test (LRT) P values for differential ASE with time analysis. (d) Distribution of allelic ratios across time for all SNPs with significant differential ASE by time effects. Related to Figures 2,4 and Table S2.
Figure S6. Peak VO2 in the cohort relative to a reference population (FRIEND registry) and molecular associations with peak VO2 (a) Peak VO2 values categorized by age and sex. Blue and green dots represent the mean values from the FRIEND registry and error bars represent one standard deviation. (b) Distribution of peak VO2 normalized to reference values from the FRIEND registry. (c) Proportion of analytes associated with peak VO2 (scaled by BW) as determined by linear regression analyses adjusted for BMI, fat mass or percent fat (FDR < 0.05). (d) Proportion of analytes associated with peak VO2 scaled by BW and LBM as determined by linear regression analyses (FDR < 0.05). Related to Figure 5 and Tables S4.
Figure S7. Differential response to acute exercise in insulin resistant participants. Patterns of differential metabolites (FDR < 0.10) (a) and complex lipids (FDR < 0.20) (b) in IR participants. Linear mixed models adjusted for personal baseline, age, sex, body mass index and race/ethnicity were employed. The solid line represents the mean and the dashed line represents the 95% confidence interval. (c) Heatmap of significant proteins as determined by the linear mixed model (FDR < 0.20) representing the median log2 fold change relative to baseline in the cohort. Proteins were grouped in patterns. Differential response to exercise for (d) TNF-α and IL-6 as well as key metabolites (e). Related to Figure 6 and Table S5.
Figure S8. Multi-omic outlier analysis. (a) Number of outliers across omes per participant at baseline and in response to exercise (FDR < 0.05). Participants with significantly different number of outliers in comparison to the population (FDR < 0.05) are represented with a solid bar. (b) Heatmap of clinical laboratory results as well as key rest and stress parameters representing the BH-corrected P values for each measure in each participant. Outlier measures (FDR < 0.05) are outlined in red. Participant ZLTUJTN was mildly anemic with borderline high RBC (5.92 M/μl, normal range: 4.40–5.90), borderline low hemoglobin (13.4 g/dl, normal range: 13.5–17.7), high RDW (20.5%, normal range: 11.5–14.5%) and low MCV (72.8fl/red cell, normal range: 82.0– 98.0), MCH (22.6 pg/cell, normal range: 27.0 to 34.0) and MCHC (31.0 g/dl, normal range: 32.0 – 36.0). Participant ZVM4N7A had a borderline high mean platelet count over 7 laboratory visits ± 5 months relative to the exercise date (405 K/μl, normal range: 150–400). (c) Functional association network of outlier proteins (FDR < 0.05) in response to exercise in individual ZL63I8R. This analysis was performed using the web tool STRING. Edges correspond to known, predicted or other interactions. Proteins in blue are downregulated and proteins in red are upregulated.Related to Table S6.
Table S1. Cohort characteristics and listing of reported analytes. (1) Participants baseline demographics, echocardiographic and cardiopulmonary exercise characteristics. (2) Listing of targeted proteins (immune, cardiovascular and metabolic). (3) Listing of all plasma and cellular analytes detected in the study (Related to Figure 1).
Table S2. Molecular and pathway changes in response to acute exercise. (1) Multi-omic changes in response to acute exercise and cluster information (Related to Figures 1–4 and S1, S4). (2) Lipid class and metabolic pathways changed in response to exercise (Related to Figure 2). (3) Metabolomic pathway dynamics (Related to Figure S5). (4) Association of key changes induced by exercise with various parameters. (5) PBMC RNA pathways changed in response to exercise (Related to Figure 4). (6) Transcriptomic pathway dynamics (Related to Figure S5).
Table S3. Molecular individuality. (1) Inter-individual variability using absolute levels. (2) Pathway enrichment analysis using variable genes (CV > 100%). (3) Inter-individual variability using fold change to exercise (Related to Figures 1 and S2).
Table S4. Molecular and pathway associations with peak VO2. (1) Multi-omic associations with peak VO2 normalized by body weight and adjusted for body composition. (2) Molecular associations with peak VO2 scaled by body weight and lean body mass. (3) Lipid classes and metabolic pathways associated with peak VO2. (4) RNA pathways associated with peak VO2. (5) Protein pathways associated with peak VO2. (6) Clinical laboratory results (closest to exercise date). (7) Gut microbiome data (closest to exercise date). (8) Peak VO2, (9) VE/VCO2 slope and (10) RER prediction models (Related to Figures 5 and S6).
Table S5. Differential response to exercise in insulin resistant participants. (1) Molecules responding differentially to acute exercise in IR participants and associated patterns. (2) Differential PBMC pathways in IR participants. (3) Differential lipid classes and metabolic pathways in IR participants (Related to Figures 6 and S7).
Table S6. Pathway enrichment analysis using outlier transcripts (Related to Figure S8).
Data Availability Statement
Raw and processed omic data (transcriptome, targeted and untargeted proteome, metabolome, lipidome) are hosted on our portal at http://hmp2-data.stanford.edu/index.phpunder the substudy Exercise. Microbiome and clinical laboratory data are provided in Tables S4.6 and S4.7, respectively.