Abstract
The ongoing outbreak of a new coronavirus (2019‐nCoV, or severe acute respiratory syndrome coronavirus 2 [SARS‐CoV‐2]) has caused an epidemic of the acute respiratory syndrome known as coronavirus disease (COVID‐19) in humans. SARS‐CoV‐2 rapidly spread to multiple regions of China and multiple other countries, posing a serious threat to public health. The spike (S) proteins of SARS‐CoV‐1 and SARS‐CoV‐2 may use the same host cellular receptor, angiotensin‐converting enzyme 2 (ACE2), for entering host cells. The affinity between ACE2 and the SARS‐CoV‐2 S protein is much higher than that of ACE2 binding to the SARS‐CoV S protein, explaining why SARS‐CoV‐2 seems to be more readily transmitted from human to human. Here, we report that ACE2 can be significantly upregulated after infection of various viruses, including SARS‐CoV‐1 and SARS‐CoV‐2, or by the stimulation with inflammatory cytokines such as interferons. We propose that SARS‐CoV‐2 may positively induce its cellular entry receptor, ACE2, to accelerate its replication and spread; high inflammatory cytokine levels increase ACE2 expression and act as high‐risk factors for developing COVID‐19, and the infection of other viruses may increase the risk of SARS‐CoV‐2 infection. Therefore, drugs targeting ACE2 may be developed for the future emerging infectious diseases caused by this cluster of coronaviruses.
Keywords: 2019‐nCoV, ACE2, COVID‐19, IFN, ISG, SARS‐CoV‐2
Highlights
Virus infection and inflammatory cytokines can stimulate angiotensin‐converting enzyme 2 (ACE2) expression. ACE2 is upregulated by the activation of RNA‐sensing pathways. ACE2 is a novel interferon‐stimulated gene (ISG). The increase in ACE2 induced by various viruses and inflammatory cytokines may facilitate severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection and spreading.
1. INTRODUCTION
The outbreak of a novel coronavirus (2019‐nCoV or severe acute respiratory syndrome coronavirus 2 [SARS‐CoV‐2]) was first reported in Wuhan, Hubei, China. 1 According to the World Health Organization (WHO), on 24 February 2020, about 77 269 viral infected patients and at least 2596 deaths were confirmed globally. The SARS‐CoV‐2 genome shows high similarity to some bat coronaviruses as well as severe acute respiratory syndrome coronavirus 1 (SARS‐CoV‐1) and Middle East respiratory syndrome coronavirus (MERS‐CoV), which have caused two large‐scale pandemics in the last two decades. 2 These viruses all belong to the betacoronavirus genus, a type of positive‐stranded RNA viruses. 2 , 3 Similar to SARS‐CoV‐1 and MERS‐CoV, SARS‐CoV‐2 can spread from human to human and causes fever, severe respiratory illness, and a series of unidentified pneumonia diseases. 4 , 5 , 6 , 7 Compared with SARS‐CoV‐1 and MERS, SARS‐CoV‐2 seems to be more readily transmitted from human to human, having spreading to multiple countries and regions and leading to a WHO declaration of a Public Health Emergency of International Concern. 4 , 8 , 9
Certain coronaviruses utilize spike (S) proteins to select and enter target cells. Both the SARS‐CoV‐1 S protein and the SARS‐CoV‐2 S protein engage angiotensin‐converting enzyme 2 (ACE2) as their entry receptor. 2 , 8 , 10 ACE2 is recognized as the cellular entry receptor of SARS‐CoV‐1 and SARS‐CoV‐2, which plays an indispensable role in the infection of both viruses. 2 , 10 ACE2 is a type I membrane protein expressed in the lungs, heart, kidney, and intestines. 11 Enhanced expression of ACE2 accelerates SARS‐CoV‐1 infection and spreading, while the silencing of ACE2 blocks its entry into cells. 10 Although it is believed that lungs are the target organ of SARS‐CoV‐2, many nonrespiratory symptoms have also been reported, suggesting the involvement of other organs during the disease. 12 Organs and tissues with high ACE2 abundance, such as the lungs, kidneys, and small intestine, are the infection targets of SARS‐CoV‐2. 11 These organs and tissues commonly exhibit high ACE2 expression levels, such as the kidney and small intestine. SARS‐CoV‐1 infects multiple cell types in several different organs; immune cells and the pulmonary epithelium are among the main sites of injury. 13 Acute kidney injury is a predictor of high mortality in SARS patients. 14 In the lungs, human ACE2 primarily occurs in type II and type I alveolar epithelial cells. ACE2 is mainly expressed in AT2 cells and is also detected in AT1 cells, airway epithelial cells, fibroblasts, endothelial cells, and macrophages. 12 Taken together, these findings reveal that the expression level of ACE2 is extremely important for the successful infection of SARS‐CoV‐1 and SARS‐CoV‐2.
As the cellular receptor of SARS‐CoV‐1 and SARS‐CoV‐2, ACE2 plays an essential role during the infection of these viruses. 2 , 10 The ACE2 level is the decisive factor in SARS‐CoV‐2 infection. 2 These observations drove us to explore whether ACE2 is a virus responsive gene. We analyzed the datasets of virus‐infected or inflammatory cytokine‐stimulated cells or tissues. The results clearly showed that ACE2 can be upregulated by infection by various viruses or stimulation by inflammatory cytokines. However, whether the upregulation of ACE2 by the virus is directly caused by virus infection‐induced inflammatory cytokinesis is currently unknown. Here, we analyzed the microarray datasets of virus‐infected or inflammatory stimulated animals and cells, performed several experimental studies, and finally clearly showed that virus infection including infection by MERS‐CoV, SARS‐CoV‐1 and SARS‐CoV‐2, or interferon (IFN) stimulation can upregulate ACE2 expression, which suggests that SARS‐CoV‐2 may augment its infection and spread via the upregulation of ACE2.
2. MATERIALS AND METHODS
2.1. Cell culture and transfection
HEK293 and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% heat‐inactivated fetal bovine serum (FBS; Gibco). Poly (I:C) (Sigma P1530) was transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific). Plasmids were transfected into HEK293 and HEK293T cells by polyethylenimine (Polysciences, Inc, Germany).
2.2. Plasmids
pcDNA6B‐RIG‐IN‐FLAG, pcDNA6B‐MAVS‐FALG, pcDNA6B‐TBK1‐FLAG, and pcDNA6B‐TRIF‐FLAG were constructed using standard molecular cloning methods as described previously. 15 , 16 The 1582‐bp promoter region of human ACE2 was cloned into pGL3‐Basic (Promega) to construct a luciferase reporter (pACE2‐luc) using the following primers: ACE2 promoter‐F (5′‐3′), GGGGTACCTGCGCTCAGAGAGAACGCTT; ACE2 promoter‐R (5′‐3′), CCCTCGAGGCCACTGTGCCCAGCCAT.
2.3. Recombinant proteins
Recombinant proteins of human IFN‐α (P5637), human IFN‐β (P5660), human IFN‐γ (P5664), human TNF‐α (P5318), human IL‐6 (P5138), human IL‐1α (P5102), and human IL‐1β (P5106) were purchased from Beyotime (China).
2.4. Reverse‐transcription quantitative polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Invitrogen). First‐strand complementary DNA (cDNA) was synthesized with a Transcriptor First Strand cDNA Synthesis Kit (Takara, Dalian, China). Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) assays and analysis were performed using a Roche LightCycler 96 system. SYBR Green‐based RT‐qPCR was used according to the manuals of the SYBR Premix Ex Taq kit (Takara). The primers used for RT‐qPCR were as follows: GAPDH‐F (5′‐3′), GGAGCGAGATCCCTCCAAAAT; GAPDH‐R (5′‐3′), GGCTGTTGTCATACTTCTCATGG; ACE2‐F (5′‐3′), GGAGTTGTGATGGGAGTGAT; ACE2‐R (5′‐3′), GATGGAGGCATAAGGATTTT; IFN‐β‐F (5′‐3′), TTGCTCTCCTGTTGTGCTTC; IFN‐β‐R (5′‐3′), AAGCCTCCCATTCAATTGCC; ISG56‐F (5′‐3′), CTAAGCAAAACCCTGCAGAAC; ISG56‐R (5′‐3′), TCAGGCATTTCATCGTCATC.
2.5. Virus infection
Vesicular stomatitis virus (VSV)‐enhanced green fluorescent protein (eGFP) infection was described in our previous publications. 15 , 16 Briefly, target cells were washed twice with DMEM at 37°C, after which the virus, diluted in serum‐free medium, was added to the cells. Virus‐medium complexes were incubated with the cells in normal culture conditions. After 1 to 2 hours, virus‐medium complexes were discarded and replaced with a culture medium containing 10% FBS.
2.6. Luciferase reporter assays
To determine whether the innate immune signaling pathway affects the promoter activities of ACE2, 2 × 105 HEK293T cells cultured in 24‐well plates were transfected with the pACE2‐luc luciferase reporter plasmid and the expression vector plasmids pcDNA6B‐RIG‐IN‐FLAG, pcDNA6B‐MAVS‐FALG, pcDNA6B‐TBK1‐FLAG, or pcDNA6B‐TRIF‐FLAG as described in our previous studies. 15 , 16 The pRL‐TK Renilla luciferase reporter was cotransfected to serve as an internal control. The cells were harvested and lysed at 36 hours after transfection for the assessment of luciferase activities using the Dual‐Luciferase Reporter Assay System (Promega). Briefly, cell lysates were loaded into a 96‐well plate for the measurement of luciferase activity in a Centro XS3 LB 960 microplate luminometer (Berthold Technologies, Germany). Relative luciferase activity was calculated by normalizing firefly luciferase activity to that of Renilla luciferase.
2.7. Patients and clinical specimens
Twenty‐one nasopharyngeal swab specimens were collected from 14 patients with coronavirus disease (COVID‐19) who were confirmed to be SARS‐CoV‐2 RNA‐positive and SARS‐CoV‐2‐specific immunoglobulin G (IgG)‐positive in Jinan Infectious Disease Hospital (Shandong, China) as described in our previous studies. 17 Eight nasopharyngeal swab specimens were collected as controls from eight healthy individuals who were confirmed to be SARS‐CoV‐2 RNA‐negative. Total RNA was extracted from nasopharyngeal swab specimens as described in our previous studies. 17 This study was approved by the ethics commissions of Jinan Infectious Disease Hospital (Shandong, China).
2.8. Statistical analysis
Three independent experiments were performed in all studies. Two‐tailed unpaired Student t test conducted with GraphPad Prism 7.0 was used to perform statistical analysis. P < .05 was considered statistically significant in all experiments.
3. RESULTS
3.1. Virus infection induces ACE2 upregulation
We retrieved datasets submitted to the microarray database of the National Center for Biotechnology Information and analyzed the ACE2 expression levels during virus infection. In the lungs of C57Bl/6 mice infected with 102 PFU SARS‐CoV‐1, the ACE2 messenger RNA (mRNA) level was significantly increased compared with the mock group (Figure 1A). 18 , 19 In primary human airway epithelial cells, MERS‐CoV infection increased ACE mRNA levels at 24, 36, and 48 hours after infection (Figure 1B). These results revealed that ACE could be upregulated by infection with SARS‐CoV‐1 and MERS‐CoV. Next, we evaluated ACE expression after infection with other RNA viruses. Microarray data showed that the ACE2 mRNA level was strongly induced in human airway epithelial cells from four donors infected with rhinovirus (Figure 2). 20 In wild‐type H1N1 influenza‐infected human bronchial epithelial cells, ACE2 expression was dramatically increased to approximately five‐fold higher than the level in the control group (Figure 3). 21 These lines of evidence indicate that ACE2 is a virus‐responsive gene and that its expression can be stimulated by various viruses. Overall, ACE2 expression can be upregulated by virus infection.
Figure 1.
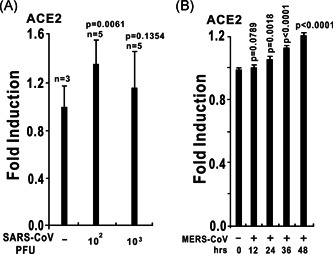
The expression of angiotensin‐converting enzyme 2 (ACE2) is upregulated by coronaviruses. A, ACE2 expression is upregulated in the lungs of severe acute respiratory syndrome coronavirus (SARS‐CoV) MA15‐infected C57Bl/6 mice. At 24 hours after the intranasal instillation of 102 or 103 PFU of SARS‐CoV MA15 in 50 µL of phosphate buffered saline (PBS) or mock infection with PBS alone, lungs were harvested, and total RNA was isolated and subjected to microarray analysis. ACE2 expression data were retrieved from the Gene Expression Omnibus (GEO) microarray database (National Center for Biotechnology Information [NCBI] accession: PRJNA149057; GEO: GSE33266). B, ACE2 expression is upregulated in primary human airway epithelial cells in response to wild‐type Middle East respiratory syndrome coronavirus (MERS‐CoV; icMERS‐CoV EMC2012). Human airway epithelial cells were infected with a multiplicity of infection of 5 PFU per cell. Infected samples were collected at the time points as indicated above. ACE2 expression data were retrieved from the GEO microarray database (NCBI accession: PRJNA391962; GEO: GSE100504)
Figure 2.
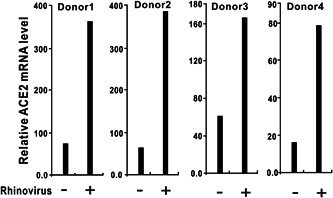
ACE2 expression is upregulated in human airway epithelial cells in response to rhinovirus infection. Primary epithelial cells from four donors were exposed to rhinovirus or medium alone (used as a control). Twenty‐four hours after infection, gene expression was assessed using Affymetrix U133 plus 2.0 human GeneChips. ACE2 expression data were retrieved from the GEO microarray database (NCBI accession: PRJNA137767; GEO: GSE27973). ACE2, angiotensin‐converting enzyme 2; GEO, Gene Expression Omnibus; NCBI, National Center for Biotechnology Information
Figure 3.
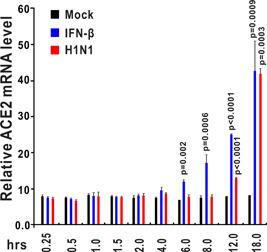
ACE2 expression is upregulated in human bronchial epithelial cells (HBECs) after interferon‐β (IFN‐β) stimulation and H1N1 influenza infection. HBECs were treated with IFN‐β (1000 U/mL) or wild‐type H1N1 influenza (A/PR/8/34). Control samples were incubated with fresh medium under the same conditions. HBECs were harvested at the indicated time points (0.25, 0.5, 1, 1.5, 2, 4, 6, 8, 12, and 18 hours after treatment). Total RNA was extracted and analyzed using an Affymetrix High Throughput Array. ACE2 expression data were retrieved from GEO microarray database (NCBI accession: PRJNA121751; GEO: GSE19392). ACE2, angiotensin‐converting enzyme 2; GEO, Gene Expression Omnibus; NCBI, National Center for Biotechnology Information
3.2. Interferons stimulate ACE2 expression
We hypothesized that ACE2 induction by viruses was caused by virus infection‐induced inflammatory cytokines. After the analysis of the microarray database, we observed that ACE2 expression could be stimulated by type I IFNs including IFN‐α and IFN‐β, and type III IFNs, such as IFN‐γ (Figures 4 and 5). In human bronchial epithelial cells stimulated by IFN‐β (1000 U/mL), ACE2 was significantly upregulated at 6, 8, 12, and 18 hours after treatment, especially at 18 hours (Figure 3), and ACE2 was increased more than four‐fold compared with the mock group at 18 hours after IFN‐β stimulation. 21 ACE2 could also be significantly induced by IFN‐γ stimulation in human primary keratinocytes (Figure 4). 22 Taken together, these findings indicate that ACE2 mRNA is upregulated in response to IFN stimulation.
Figure 4.
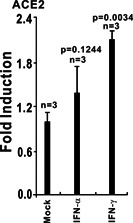
ACE2 expression is upregulated in human primary keratinocytes exposed to IFN‐α and IFN‐γ. Primary keratinocytes from three donors were either untreated (control) or exposed to the IFN‐α or IFN‐γ cytokines. Twenty‐four hours after treatment, total RNA was extracted for subsequent microarray analysis. ACE2 expression data were retrieved from the GEO microarray database (NCBI accession: PRJNA153023; GEO: GSE36287). ACE2, angiotensin‐converting enzyme 2; GEO, Gene Expression Omnibus; IFN, interferon; NCBI, National Center for Biotechnology Information
Figure 5.
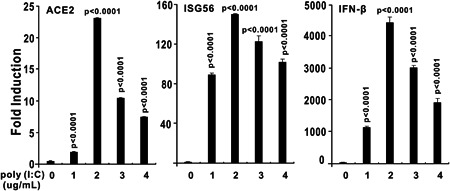
Transfection of poly (I:C) induces ACE2 expression. HEK293 cells were cultured in six‐well plates and transfected with poly (I:C) as indicated above using Lipofectamine 2000. HEK293 cells were collected for RNA isolation and reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) analysis at 9 hours after transfection. The results in each panel are representative of three independent experiments. The graphs show the means ± SD (n = 3). The results in each panel are representative of three independent experiments. ACE2, angiotensin‐converting enzyme 2
3.3. Transfection of poly (I:C) induces ACE2 expression
Poly (I:C) is a mimic of viral RNA that stimulates the activation of innate immune signaling pathways. When delivered into HEK293 cells, poly (I:C) significantly induced the upregulation of both ACE2 and IFN‐stimulated gene 56 (ISG56, also known as IFIT1) according to RT‐qPCR assessment (Figure 5). Poly (I:C) mimics viral RNA and activates immune signaling pathways of RIG‐I/MDA‐5‐MAVS, which lead to IFN and inflammatory cytokine production, so we explored whether RNA virus infection can induce ACE2 expression. As an RNA virus, VSV‐eGFP can slightly induce ACE2 mRNA upregulation in HEK293 cells (Figure 6).
Figure 6.
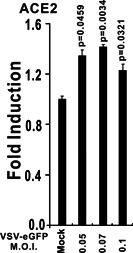
Vesicular stomatitis virus (VSV)‐enhanced green fluorescent protein (eGFP) infection induces ACE2 expression. HEK293 cells were cultured in six‐well plates and infected with VSV‐eGFP as indicated above. After 6 hours, HEK293 cells were collected for RNA isolation and RT‐qPCR analysis. The graphs show the means ± SD (n = 3). The results in each panel are representative of three independent experiments. ACE2, angiotensin‐converting enzyme 2; RT‐qPCR, reverse‐transcription quantitative polymerase chain reaction
3.4. Activation of RNA‐sensing pathway induces ACE2 production
Toll‐like receptor 3 (TLR3), which predominantly localizes to endosomes, is a sensor of viral RNA, and RIG‐I and MDA‐5 are cytoplasmic RNA sensors which may serve as the immune sensor of SARS‐CoV‐2. Because both poly (I:C) and RNA viruses induce ACE2 expression, we next evaluated whether the RNA‐sensing pathway affects ACE expression. Activation of the RIG‐I/MDA‐5‐MAVS signaling pathway by the overexpression of MAVS can strongly induce the expression of ACE2 in HEK293T cells. TRIF, an adaptor protein of the TLR3 pathway, also induced the expression of ACE2 in HEK293T cells (Figure 7). Overexpression of TBK1, which is downstream of both TLR3 and RIG‐I/MDA‐5 signaling pathways can strongly upregulate ACE2 mRNA level (Figure 7). Taken together, the results indicated that the expression of human ACE2 can be induced by innate immune pathways involving TLR3‐TRIF and RIG‐I/MDA‐5‐MAVS signaling cascades.
Figure 7.
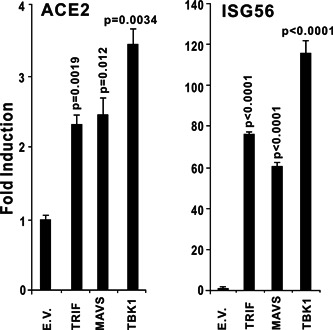
Overexpression of TRIF, MAVS, or TBK1 induces ACE2 expression. TRIF and MAVS are the essential adaptors of Toll‐like receptor 3 (TLR3) and RIG‐I/MDA‐5, respectively. TBK1 is an crucial activator of TLR3 and RIG‐I/MDA‐5 pathways downstream of TRIF and MAVS. HEK293 cells were cultured in 24‐well plates. After 24 hours, the plasmids indicated above were transfected into HEK293T cells. Thirty‐six hours after transfection, total RNA was isolated for subsequent RT‐qPCR analysis. The graphs show the means ± SD (n = 3). The results in each panel are representative of three independent experiments. ACE2, angiotensin‐converting enzyme 2; RT‐qPCR, reverse‐transcription quantitative polymerase chain reaction
3.5. The promoter of ACE2 was activated by RNA‐sensing pathways
Using the ChIP‐seq data from ENCODE from the UCSC Genome Browser (https://genome.ucsc.edu/ENCODE/) and the Signaling Pathways Project (https://www.signalingpathways.org/ominer/query.jsf), we found that the putative promoter region (from −2000 to +500 bp of the transcription start site of ACE2) contains several immune‐ and cytokine‐responsive transcription factor binding sites, including sites for c‐Jun, signal transducer and activator of transcription 1 (STAT1), STAT3, STAT5, interferon regulator factor (IRF1), IRF2, and IRF8.
We cloned the putative promoter region (from the transcription start site of human ACE2 −1281 to +300 bp) into the luciferase reporter vector pGL3‐Basic to construct the pACE2‐luc reporter and investigated the regulatory properties of ACE2 (Figure 8A). Dual‐luciferase reporter assays showed that the ACE2 promoter could be strongly activated by RIG‐IN, MAVS, TRIF, and TBK1 (Figure 8B).
Figure 8.
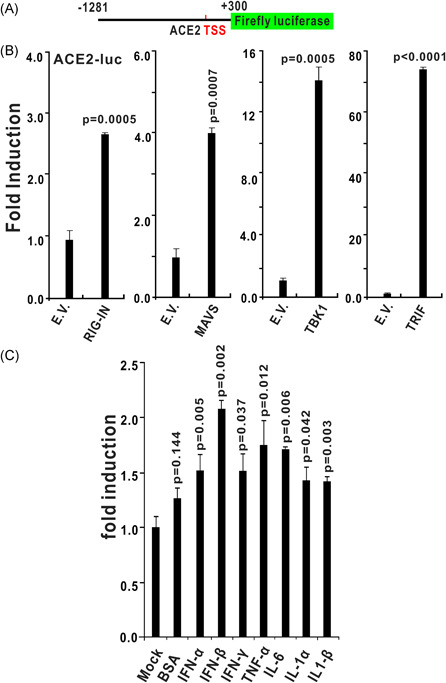
The promoter of ACE2 is activated by RNA‐sensing pathways and inflammatory cytokine stimulation. A, Schematic diagram of the human ACE2 luciferase reporter. Nucleotides −1281 to +300 (with transcript start site set as 1) of the human ACE2 promoter region were cloned into the pGL3‐Basic luciferase reporter to construct ACE‐luc. B, Induction patterns of the human ACE2 promoter by RIG‐IN (a truncated active form of RIG‐I), MAVS, TRIF, and TBK1. HEK293T cells were transfected with plasmids containing the empty vector (EV), RIG‐IN, MAVS, TRIF, or TBK1 together with ACE‐luc. C, The promoter activities of ACE2 was induced by recombinant protein of IFN‐α (10 ng/mL), IFN‐β (10 ng/mL), IFN‐γ (10 ng/mL), TNF‐α (10 ng/mL), IL‐6 (10 ng/mL), IL‐1α (10 ng/mL), or IL‐1β (10 ng/mL). The graphs show the means ± SD (n = 3). The results in each panel are representative of three independent experiments. ACE2, angiotensin‐converting enzyme 2; IFN, interferon
3.6. The promoter activities of ACE2 was induced by inflammatory cytokines
One of the clinical features of patients with COVID‐19 is the upregulation of inflammatory cytokines. Next, we treated HEK293 cells receiving pACE2‐luc reporter plasmids with recombinant protein of IFN‐α, IFN‐β, IFN‐γ, TNF‐α, IL‐6, IL‐1α, or IL‐1β; 12 hours later, dual‐luciferase reporter assays were performed. The results indicated that all above inflammatory cytokines can significantly induce the promoter activities of ACE2. The induction folds of ACE2 promoter by these cytokines is not high, which is consistent with two recent studies showing that ACE2 is slightly upregulated by some inflammatory cytokines. 23 , 24
Combined with the above results showing that ACE2 can be induced by the stimulation of IFN‐β and IFN‐γ (Figures 3 and 4), we proposed that ACE2 is a novel IFN‐stimulated gene (ISG) that downstream of RNA‐sensing pathways of TLR3 and RIG‐I/MDA‐5 and the upregulation of ACE2 by virus infection and cytokine stimulation would accelerate SARS‐CoV‐2 infection and COVID‐19 development.
3.7. ACE2 is upregulated in SARS‐CoV‐2‐infected patients
To investigate whether ACE2 is upregulated in SARS‐CoV‐2‐infected individuals, the nasopharyngeal swab specimens containing epithelial cells were collected from patients with COVID‐19. Total RNA was extracted and reverse‐transcribed into cDNA. All nasopharyngeal swab specimens collected from patients with COVID‐19 were confirmed to be SARS‐CoV‐2 RNA‐positive as described in our previous studies 17 ; and all nasopharyngeal swab specimens collected from healthy individuals were confirmed to be SARS‐CoV‐2 RNA‐negative. We detected the expression of ACE2 in these cDNA samples using RT‐qPCR. The results showed that the expression levels of ACE2 are approximately 3.6‐fold higher in nasopharyngeal swab specimens from patients with COVID‐19 than those from healthy individuals (Figure 9). These results indicated that ACE2 is upregulated in SARS‐CoV‐2‐infected patients with COVID‐19.
Figure 9.
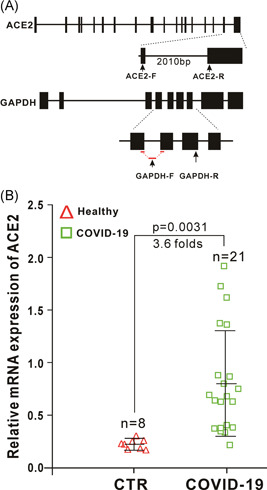
ACE2 is upregulated in severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)‐infected patients. A, Schematic diagrams of the RT‐qPCR primer positions of human ACE2 and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH). The RT‐qPCR primers of ACE2 located in two different exons with a large intron between them to make sure that the primers can only amplify a fragment of ACE2 from complementary DNA (cDNA) only but not genomic DNA. The forward primer of GAPDH (GAPDH‐F) span an exon‐exon boundary as indicated, which only amplify a fragment of GAPDH from cDNA only but not genomic DNA. B, Expression of ACE2 in nasopharyngeal swab specimens from healthy (control, CTR) and SARS‐CoV‐2‐infected individuals. Nasopharyngeal swab specimens collected from patients with COVID‐19 were confirmed to be SARS‐CoV‐2 RNA‐positive. Expression of ACE2 in theses samples was detected by RT‐qPCR. ACE2, angiotensin‐converting enzyme 2; RT‐qPCR, reverse‐transcription quantitative polymerase chain reaction
4. DISCUSSION
SARS‐CoV‐2 infection exhibits an alarming transmission speed, and the corresponding pathological phenomenon is still poorly understood. SARS‐CoV‐2 utilizes cell membrane receptors for entering cells and causes infectious diseases. The expression level of ACE2 is crucial for the infection of SARS‐CoV‐1 and SARS‐CoV‐2. 2 , 10 It is of great interest to investigate the expression patterns of ACE2 during virus infection and immune activation. After the analysis of microarray database, we found that the expression levels of ACE2 were upregulated by the infection of various viruses, including SARS‐CoV‐1, MERS‐CoV, rhinovirus, and influenza virus (Figures 1, 2, 3). Additionally, ACE2 can be strongly induced by IFN‐β in human bronchial epithelial cells and slightly upregulated by IFN‐α and IFN‐γ in human primary keratinocytes (Figures 3 and 4). In some cell types, such as CD4 T cells, the expression levels of ACE2 are downregulated by IFN stimulation, suggesting diverse regulatory mechanisms are involved in the regulation of ACE2 expression in distinct cell types by immune responses. 23 In Vero E6 cells, SARS‐CoV‐1 infection may cause ACE2 downregulation, which engages in the release of soluble ACE2 into the cellular supernatants upon binding with SARS‐CoV‐1 S protein. This downregulation of ACE2 by SARS‐CoV‐1 S protein occurs in protein levels, 25 which is different from the upregulation of ACE2 in transcription levels by IFNs and virus infections.
We also observed that the promoter region of ACE2 contains several immune‐ and cytokine‐responsive transcription factor binding sites such as sites for c‐Jun, STAT1, STAT3, STAT5, IRF1, and IRF8 (Figure 8A). We evaluated whether ACE2 is upregulated by the relative innate immune signaling cascades. Both luciferase reporter assays and RT‐qPCR analysis demonstrated that the TLR3 and RIG‐I/MDA‐5 pathways can upregulate ACE2 expression (Figures 7 and 8). TLR3 recognizes viral dsRNA in the endosomes and RIG‐I/MDA‐5 senses cytosolic dsRNA, both of which would induce the production of IFNs and other inflammatory cytokines (Figure 10). 26 We further explored whether the inflammatory cytokines can stimulate ACE2 promoter activities using luciferase reporter assays. We found that IFN‐α, IFN‐β, IFN‐γ, TNF‐α, IL‐6, IL‐1α, or IL‐1β can significantly induce the promoter activities of ACE2 (Figure 8C). Thus, we propose that ACE2 may act as a novel ISG that is induced by IFNs and RNA‐sensing pathways (Figure 10). Importantly, we also observed that ACE2 is upregulated in SARS‐CoV‐2‐infected individuals. To our knowledge, this is the first report that clearly showed the expression levels of ACE2 is higher in patients with COVID‐19 than that in healthy individuals (Figure 9). After this manuscript was posted at bioRxiv on 27 February 2020 (https://www.biorxiv.org/content/10.1101/2020.02.24.963348v1), other groups independently found that ACE2 is an ISG in human airway epithelial cells and upregulated by viral infections. 23 , 24 Overall, the available results indicate that ACE2 can be upregulated by infection by various viruses and stimulation by IFNs, which indicates that ACE2 is a novel ISG that can be stimulated by viruses and IFNs (Figure 10).
Figure 10.
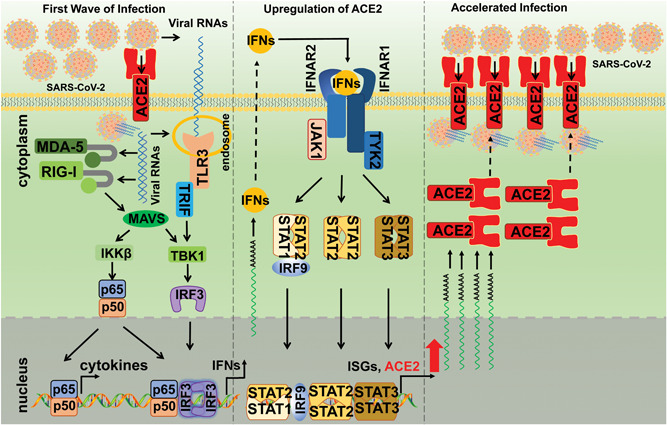
Proposed working model of ACE2 during SARS‐CoV‐2 infection. In the first wave of infection, SARS‐CoV‐2 enters host cells through ACE2 and releases its viral RNAs, which are subsequently sensed by TLR3 and RIG‐I/MDA‐5. The activation of TLR3 and RIG‐I/MDA‐5 leads to the activation of innate immune signaling pathways, resulting in the production of IFNs and other inflammatory cytokines. Then, ACE2 was upregulated by these cytokines through the Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway or other pathways, which subsequently mediated the enhanced entry of SARS‐CoV‐2 into host cells. ACE2, angiotensin‐converting enzyme 2; IFN, interferon; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; TLR3, Toll‐like receptor 3
We also showed that the viral RNA mimic poly (I:C) can induce the upregulation of ACE2, IFN‐β, and ISG56 (Figure 5); importantly, as an RNA virus, SARS‐CoV‐2 viral RNAs are expected to be sensed by TLR3 and RIG‐I/MDA‐5, which will then activate the innate immune signaling cascades and induce the production of IFNs and other inflammatory cytokines, ultimately, leading to the upregulation of ACE2, which may facilitate the entry of SARS‐CoV‐2 into host cells (Figure 10). Importantly, the induction of ACE2 by inflammatory cytokines also implies that the “cytokine storm” caused by SARS‐CoV‐2 not only damages host tissues but may also accelerate the spread of the virus (Figure 10). 5 , 6
Thus, this study provides the first evidence that SARS‐CoV‐1 and SARS‐CoV‐2 can upregulate the cellular receptor ACE2 to facilitate their infection and spread (Figures 9 and 10). Therefore, reducing ACE2 expression by using small interfering RNAs or other drugs may be a direction for drug development. Because this cluster of coronaviruses may use the ACE2 same cellular receptor for entry, the development of drugs that can reduce ACE2 expression may be useful for future emerging infectious diseases. Our study suggests that people infected by other viruses, such as influenza virus, or those with high inflammatory cytokine levels may be more susceptible to SARS‐CoV‐2. This study also indicated that targeting the expression of ACE2 is a direction for drug development and the control of SARS‐CoV‐2 infection.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
PHW conceptualized the study. MWZ and JZ performed the experiments. CY and JD contributed to the analysis of microarray data. XMJ and LW collected the clinical specimens and tested SARS‐CoV‐2 RNA and SARS‐CoV‐2‐specific IgG. PHW wrote the manuscript. All of the authors contributed in revising the manuscript and approved the final version for publication.
ACKNOWLEDGMENT
This study was supported by grants from COVID‐19 Emergency Tackling Research Project of Shandong University (grant no. 2020XGB03 to PHW).
Zhuang M‐W, Cheng Y, Zhang J, et al. Increasing host cellular receptor—angiotensin‐converting enzyme 2 expression by coronavirus may facilitate 2019‐nCoV (or SARS‐CoV‐2) infection. J Med Virol. 2020;92:2693–2701. 10.1002/jmv.26139
Meng‐Wei Zhuang, Yun Cheng, and Jing Zhang contributed equally to this study.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382(13):1199‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li W, Moore MJ, Vasilieva N, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019‐nCov infection: a bioinformatics analysis based on single‐cell transcriptomes. bioRxiv. 2020. [Google Scholar]
- 12. Wei Lin, Zhang Yan, Joshua D, et al. Single‐cell analysis of ACE2 expression in human kidneys and bladders reveals a potential route of 2019‐nCoV infection. bioRxiv. 2020. [Google Scholar]
- 13. Gu J, Gong E, Zhang B, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu KH, Tsang WK, Tang CS, et al. Acute renal impairment in coronavirus‐associated severe acute respiratory syndrome. Kidney Int. 2005;67(2):698‐705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang PH, Fung SY, Gao WW, et al. A novel transcript isoform of STING that sequesters cGAMP and dominantly inhibits innate nucleic acid sensing. Nucleic Acids Res. 2018;46(8):4054‐4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang PH, Ye ZW, Deng JJ, et al. Inhibition of AIM2 inflammasome activation by a novel transcript isoform of IFI16. EMBO Rep. 2018;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang B, Wang L, Kong X, et al. Long‐term coexistence of SARS‐CoV‐2 with antibody response in COVID‐19 patients. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gralinski LE, Sheahan TP, Morrison TE, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Totura AL, Whitmore A, Agnihothram S, et al. Toll‐like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6(3):e00638‐00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Proud D, Hudy MH, Wiehler S, et al. Cigarette smoke modulates expression of human rhinovirus‐induced airway epithelial host defense genes. PLoS One. 2012;7(7):e40762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shapira SD, Gat‐Viks I, Shum BO, et al. A physical and regulatory map of host‐influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139(7):1255‐1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swindell WR, Xing X, Stuart PE, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One. 2012;7(3):e34594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016‐1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith JC, Sausville EL, Girish V, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS‐CoV‐2 receptor ACE2 in the respiratory tract. Dev Cell. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glowacka I, Bertram S, Herzog P, et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol. 2010;84(2):1198‐1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang PH, He JG. Nucleic acid sensing in invertebrate antiviral immunity. Int Rev Cell Mol Biol. 2019;345:287‐360. [DOI] [PubMed] [Google Scholar]


