Abstract
Background
MYC is an oncogenic driver of development and progression in triple-negative breast cancer (TNBC). Ornithine decarboxylase, the rate-limiting enzyme in polyamine metabolism, is a transcriptional target of MYC. We therefore hypothesized that a plasma polyamine signature may be predictive of TNBC development and progression.
Methods
Using liquid chromatography mass spectrometry, polyamine levels were determined in plasma samples from newly diagnosed patients with TNBC (n = 87) and cancer-free controls (n = 115). Findings were validated in plasma samples from an independent prospective cohort of 54 TNBC, 55 estrogen receptor negative (ER−) and progesterone receptor negative (PR−) and HER2 positive (HER2+), and 73 ER+ case patients, and 30 cancer-free control subjects. Gene expression data and clinical data for 921 and 2359 breast cancer tumors were obtained from The Cancer Genome Atlas repository and the Oncomine database, respectively. Relationships between plasma diacetylspermine (DAS) and tumor spermine synthase (SMS) mRNA expression with metastasis-free survival and overall survival were determined using Cox proportional hazard models; Fisher exact tests were used to assess risk of distant metastasis in relation to tumor SMS mRNA expression.
Results
An increase in plasma DAS, a catabolic product of spermine mediated through SMS, was observed in the TNBC subtype of breast cancer. Plasma levels of DAS in TNBC associated with increased risk of metastasis (plasma DAS value ≥ 1.16, hazard ratio = 3.06, 95% confidence interval [CI] = 1.15 to 8.13, two-sided P = .03). SMS mRNA expression in TNBC tumor tissue was also found to be predictive of poor overall survival (top 25th percentile hazard ratio = 2.06, 95% CI = 1.04 to 4.08, one-sided P = .04) and increased risk of distant metastasis in TNBC (comparison of lowest SMS quartile [reference] to highest SMS quartile relative risk = 1.90, 95% CI = 0.97 to 4.06, one-sided Fisher exact test P=.03).
Conclusions
Metabolomic profiling identified plasma DAS as a predictive marker for TNBC progression and metastasis.
MYC is an oncogenic driver of development and progression in triple-negative breast cancer (TNBC) (1–3). A downstream target of MYC is ornithine decarboxylase (ODC), a rate-limiting enzyme in polyamine metabolism (4,5). Polyamines have been implicated to play functional roles in promoting neoplastic transformation and growth (6,7). Therapeutic intervention strategies are currently being explored that target polyamine metabolism (8). With our evolved understanding of polyamine metabolism (8,9), there remains a need to uncover differences and similarities in polyamine metabolism between and within cancer type, leading to the discovery of polyamine metabolism-related cancer markers (8,10–13). Prior studies have demonstrated high levels of acetylated polyamines in breast cancer tumors; elevated levels of acetylated polyamines were associated with concurrent increases in spermidine and spermine N1-acetyltransferase (SAT1) activity and reduced activity of polyamine oxidase (14).
Herein, we tested whether a plasma polyamine signature would exhibit predictive value in TNBC. We further explored the expression and regulation of polyamine metabolizing enzymes in relation to diacetylspermine (DAS) synthesis and release and the association between polyamine metabolizing enzymes and TNBC immunophenotype.
Methods
Detailed methods are provided in the Supplementary Methods (available online).
Human Cohorts
All human plasma samples were obtained with written informed consent, and the studies were conducted under protocols approved and supervised by the University of Texas MD Anderson Institutional Review Board (LAB03-0479 [MDACC Cohort #1]; LAB90-049 and 2005–0388 [MDACC Cohort #2]). MDACC Cohort #1 consisted of a case-control cohort comprised of 87 TNBC breast cancer case patients and 115 cancer-free (minimum of 3-year follow-up) control subjects (Supplementary Table 1, available online). For MDACC Cohort #2, prospective plasma samples were collected from 197 women with newly diagnosed (0 to approximately 0.8 years) breast cancer (Supplementary Table 2, available online). Blood samples were drawn after diagnostic biopsy and prior to neoadjuvant chemotherapy, or prior to definitive surgery in patients who did not receive chemotherapy in the neoadjuvant setting. Control plasma samples (n = 30) were drawn from patients enrolled in a biomarker discovery trial to aid low-dose computed tomography-based screening for lung cancer. Plasma samples were selected from patients without lung nodules.
Transgenic Mouse Models
All animal experiments were performed under protocols approved and supervised by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee. Detailed information regarding the generation and maintenance of transgenic FVB/N-Tg(MMTV-PyVT)634Mul/J (PyMT) mice and bitransgenic MMTV-rt TA/Teto-NeuNT (Erbb2+) mice is provided in the Supplementary Materials (available online) (15,16). Metabolomics profiling was conducted on individual plasma samples from 10 PyMT tumor-bearing mice and 10 littermate controls as well as individual plasma samples from 15 bitransgenic MMTV-rt TA/Teto-NeuNT (Erbb2+) tumor-bearing mice and 20 littermate controls, respectively.
Metabolomics Analysis
Detailed information is provided in the Supplemental Material (available online).
Untargeted Metabolomic Analysis
Metabolomic profiling was conducted on a Waters Acquity ultra performance liquid chromatography (UPLC) system (Waters Corp. Milford, MA) with 2-D column regeneration (I class and H class) coupled to a Xevo G2-XS quadrupole time-of-flight mass spectrometer (Waters Corp. Milford, MA) as previously described (10,17).
Gene Expression Data and Networks
Gene expression data for breast cancer cell lines was obtained from Cancer Cell Line Encyclopedia (www.broadinstitute.org/ccle). Gene expression data and clinical data were downloaded from The Cancer Genome Atlas (TCGA) repository (https://tcga-data.nci.nih.gov/tcga/). Networks were visualized using cytoscape (18). Gene expression for the Curtis dataset (19), Hatzis dataset (20), and van de Vijver dataset (21) were from the Oncomine database (22).
Immune Cell Signature Analyses
Detailed information is provided in the Supplementary Methods (available online). Specific immune cell infiltration was computationally inferred using RNA-sequencing data based on gene sets overexpressed in 1 of 24 immune cell types according to Bindea et al. (23).
Statistical Analyses
Detailed information is provided in Supplementary Methods (available online). Cox proportional hazard models were performed using R statistical software (https://www.r-project.org/). To test for the proportionality of hazard assumption of a Cox regression, we used the method of Patricia and Grambsch (24). Log-rank statistic-based methods as described by Contal and O’Quigley (25) were used to the determine optimal cutoff point for plasma DAS to distinguish TNBC subjects who later developed distant metastasis from those who did not. Kaplan-Meier survival analyses were performed using R Version 1.1.442. Univariate analyses were conducted using the Kruskal-Wallis test for comparisons with more than two groups; group-specific differences were determined using Dunn’s multiple comparison test. For two-class comparisons, statistical significance was determined using the Wilcoxon rank sum test. Receiver operating characteristic curves were generated using R. The 95% confidence intervals (CIs) presented for individual performance of each biomarker were based on the bootstrap procedure in which we resampled with replacement separately for the controls and the diseased 1000 bootstrap samples. Spearman correlation analyses were performed to assess relationships between continuous variables. All statistical tests were two-sided unless specified otherwise. χ2 tests for trend were used to assess the goodness of the fit between spermine synthase (SMS) mRNA expression quartiles and risk of metastasis in the van de Vijver dataset (21). The Fischer exact test was one-sided because we hypothesize that elevated SMS mRNA expression is associated with worse outcome as opposed to variation in SMS mRNA expression being associated with worse outcome.
Results
Profiling of Polyamine Levels in Plasma of Breast Cancer Patients
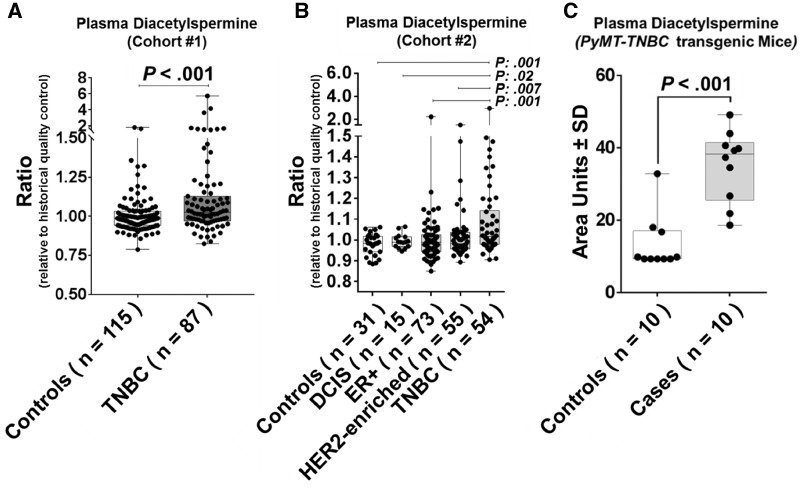
Metabolic profiling was conducted using plasma samples collected from newly diagnosed patients with TNBC (n = 87) and cancer-free control subjects (n = 115) (Cohort #1) (Supplementary Table 1, available online). Multiple polyamine metabolites were identified and quantified (Supplementary Table 3, available online). Plasma DAS, a terminal product of polyamine metabolism, was statistically significantly elevated in TNBC cases compared with cancer-free controls (TNBC cases: mean relative abundance [SD] = 1.20 [0.68]; cancer-free controls: mean relative abundance [SD] = 1.01 [0.13]; two-tailed Wilcoxon rank sum test P < .001) (Figure 1A). Classification performance of DAS yielded receiver operating characteristic area under the curve of 0.64 (95% CI = 0.57 to 0.72) with 15.0% sensitivity at 95.0% specificity when comparing TNBC against cancer-free controls (Supplementary Figure 1A, available online). Next, we compared plasma levels of DAS in an independent prospective cohort of newly diagnosed breast cancer patients that consisted of 54 TNBC, 55 estrogen receptor negative (ER−) and progesterone receptor negative (PR−) and HER2 positive (HER2+), and 73 ER+ cases as well as 15 cases diagnosed with ductal carcinoma in situ and 30 cancer-free controls (Cohort #2) (Supplementary Table 2, available online). Cases were followed prospectively from time of diagnosis for cancer-related outcomes including recurrence, metastasis, and survival. Plasma DAS was statistically significantly elevated in TNBC cases compared with cancer-free controls (TNBC cases: mean relative abundance [SD] = 1.11 [0.30]; cancer-free controls: mean relative abundance [SD] = 0.98 [0.05]; two-tailed Wilcoxon rank-sum test P = .001) or to subjects with ductal carcinoma in situ (mean relative abundance [SD] = 0.99 [0.03]; two-tailed Wilcoxon rank-sum test P = .02) or with non-TNBC breast cancer (ER+/PR+: mean relative abundance [SD] = 1.01 [0.16]; HER2-enriched: mean relative abundance [SD] = 1.02 [0.12]; two-tailed Wilcoxon rank-sum test P = .001 and .007 for TNBC vs ER+/PR+ and TNBC vs HER2-enriched, respectively) (Figure 1B). Classification performance of DAS yielded an receiver operating characteristic area under the curve of 0.70 (95% CI = 0.59 to 0.81) with 37.0% sensitivity at 95.0% specificity when comparing TNBC vs cancer-free controls (Supplementary Figure 1B, available online). We additionally analyzed plasma levels of DAS in MMTV-PYMT –TNBC transgenic mice (n = 10) and doxycycline-induced bitransgenic MMTV-rt TA/Teto-NeuNT (Erbb2+) mice (n = 15) as well as their respective littermate controls (n = 10 and 20, respectively) (15,16). The plasma level of DAS was statistically significantly higher in PYMT-TNBC (mean area units [SD] = 35.19 [9.81] vs 13.42 [7.60]; two-tailed Wilcoxon rank-sum test P < .001), but not bitransgenic MMTV-rt TA/Teto-NeuNT (Erbb2+) mice, as compared with littermate controls in concordance with plasma DAS findings from human subjects with TNBC (Figure 1C;Supplementary Figure 1C, available online).
Figure 1.
Plasma levels of diacetylspermine (DAS) in human cohorts and mouse model of breast cancer. A) Box and whisker plots depicting the relative plasma levels of DAS in MDACC Cohort #1 consisting of 87 TNBC cases and 115 cancer-free controls. B) Box and whisker plots depicting the relative plasma levels of DAS in MDACC Cohort #2 consisting of cancer-free women (n = 30) and women diagnosed with DCIS (n = 15), ER+ (n = 73), HER2-enriched (n = 55), or triple-negative (n = 54) breast cancer. Statistical significance was determined by Kruskal Wallis test; pairwise comparisons were determined by two-sided Wilcoxon rank-sum test. P values to the right of the respective lines correspond to pairwise comparisons between the respective groups. C) Box and whisker plots depicting the relative abundance of DAS in plasma of PyMT-TNBC transgenic mice (n = 10) and littermate controls (n = 10), respectively. Missing values, due to analyte abundances being below the limit of detection, were imputed as the minimum value observed among all samples analyzed. Statistical significance was determined by two-sided Wilcoxon rank-sum test. DCIS = ductal carcinoma in situ; ER = estrogen receptor; HQC = historical quality control; TNBC = triple-negative breast cancer.
Plasma DAS and Risk of Future Distant Metastasis in TNBC Cases
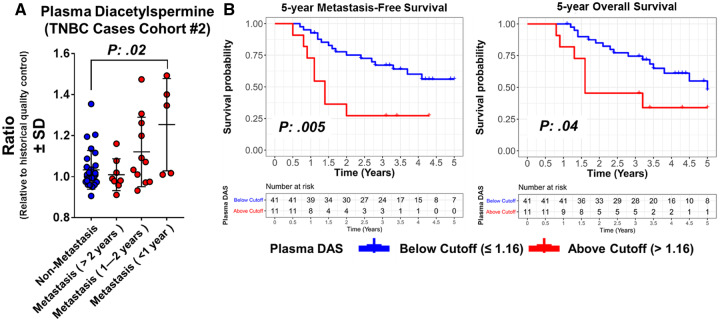
Next, we interrogated whether plasma DAS may predict tumor progression and metastasis in our prospective breast cancer cohort (Cohort #2). Plasma DAS was statistically significantly elevated in women who presented with limited stage disease TNBC and who subsequently developed recurrence and metastasis within 1 year (mean relative abundance [SD] = 1.25 [0.23] vs 1.03 [0.09]; two-tailed Wilcoxon rank sum test P = .02) (Figure 2A). Using Cox proportional hazard models, we assessed the association between plasma DAS levels with 5-year metastasis-free survival and 5-year overall survival in TNBC cases. As a continuous variable, plasma DAS was statistically significantly associated with worse 5-year distant metastasis-free survival (hazard ratio [HR] = 31.1, 95% CI = 2.32 to 417.20; two-sided P = .009) and worse 5-year overall survival (HR = 21.5, 95% CI = 1.16 to 399.30; two-sided P = .04), independent of menopausal status or stage (Table 1). Next, using log-rank test statistics from the Cox model (25), we calculated an optimal cutoff point for plasma DAS to yield the greatest difference between TNBC cases that went on to develop future distant metastasis from those that did not. This resulted in a cutoff value of 1.16. In multivariable analyses, adjusted for menopausal status and stage, TNBC cases with a plasma DAS value greater than 1.16 exhibited statistically significantly worse 5-year metastasis-free survival as compared with those with a plasma DAS cutoff value of no more than 1.16 (HR = 3.06, 95% CI = 1.15 to 8.13; two-sided P = .03) (Table 1). Notably, nonproportionality hazard model tests yielded statistically nonsignificant P values. Representative Kaplan-Meier survival curves illustrating 5-year metastasis-free survival and 5-year overall survival for TNBC cases with plasma DAS above or below the cutoff point are shown in Figure 2B.
Figure 2.
Association between plasma levels of DAS and development of future distant metastasis. A) Relative plasma levels (SD) of DAS in TNBC cases that did or did not go on to develop future distant metastasis. Data are derived from MDACC Cohort #2. Statistical significance was determined by two-sided Wilcoxon rank-sum test. B) Kaplan-Meier survival curve indicating 5-year metastasis-free survival and 5-year overall survival in TNBC cases from MDACC Cohort #2 stratified by plasma DAS levels ≤1.16 or >1.16. Optimal cutoff values for plasma DAS were derived using two-sided log-rank test statistics-based methods as described by Contal and O’Quigley (25). DAS = diacetylspermine; HQC = historical quality control; TNBC = triple-negative breast cancer.
Table 1.
Association between plasma DAS and 5-year metastasis-free survival in TNBC cases*
| Variable | Univariate |
Multivariable† |
||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P‡ | HR (95% CI) | P‡ | |||
| 5-year metastasis-free survival | ||||||
| Menopausal status | ||||||
| Pre | Referent | Referent | ||||
| Post | 0.88 (0.39 to 1.98) | .75 | 1.4 (0.57 to 3.46) | .47 | ||
| Stage | ||||||
| II | Referent | Referent | ||||
| III | 1.95 (0.77 to 4.95) | .16 | 1.3 (0.48 to 3.54) | .60 | ||
| Plasma DAS | 27.21§ (3.01 to 246.10) | .003 | 31.1 (2.32 to 417.20) | .009 | ||
| 5-year overall survival | ||||||
| Menopausal status | ||||||
| Pre | Referent | Referent | ||||
| Post | 0.99 (0.43 to 2.27) | .98 | 1.51 (0.59 to 3.88) | .39 | ||
| Stage | ||||||
| II | Referent | Referent | ||||
| III | 1.65 (0.67 to 4.06) | .28 | 1.16 (0.44 to 3.08) | .77 | ||
| Plasma DAS | 14.8 (1.32 to 167.00) | .03 | 21.5 (1.16 to 399.30) | .04 | ||
| 5-year metastasis-free survival | ||||||
| Menopausal status | ||||||
| Pre | Referent | Referent | ||||
| Post | 0.88 (0.39 to 1.98) | .75 | 1.18 (0.49 to 2.81) | .71 | ||
| Stage | ||||||
| II | Referent | Referent | ||||
| III | 1.95 (0.77 to 4.95) | .16 | 1.45 (0.54 to 3.92) | .46 | ||
| Plasma DAS | ||||||
| Below cutoff (≤1.16) | Referent | Referent | ||||
| Above cutoff (>1.16) | 3.29 (1.39 to 7.79) | .007 | 3.06 (1.15 to 8.13) | .03 | ||
| 5-year overall survival | ||||||
| Menopausal status | ||||||
| Pre | Referent | Referent | ||||
| Post | 0.99 (0.434 to 2.27) | .98 | 1.28 (0.528 to 3.11) | .585 | ||
| Stage | ||||||
| II | Referent | Referent | ||||
| III | 1.65 (0.666 to 4.06) | .28 | 1.28 (0.484 to 3.36) | .623 | ||
| Plasma DAS | ||||||
| Below cutoff (≤1.16) | Referent | Referent | ||||
| Above cutoff (>1.16) | 2.58 (1.05 to 6.31) | .038 | 2.57 (0.924 to 7.17) | .071 | ||
Cox proportional hazard models using plasma DAS as a continuous variable or using a plasma DAS cutoff value of 1.16. Optimal cutoff values for plasma DAS were derived using log-rank statistic-based methods as described by Contal and O’Quigley (25). CI = confidence interval; DAS = diacetylspermine; HR = hazard ratio; TNBC = triple-negative breast cancer.
Menopausal status and staging were included as covariates in multivariable Cox proportional hazard models.
Two-sided P value.
Per unit log2 increase.
Polyamine Metabolism in Different Molecular Subtypes of Breast Cancer
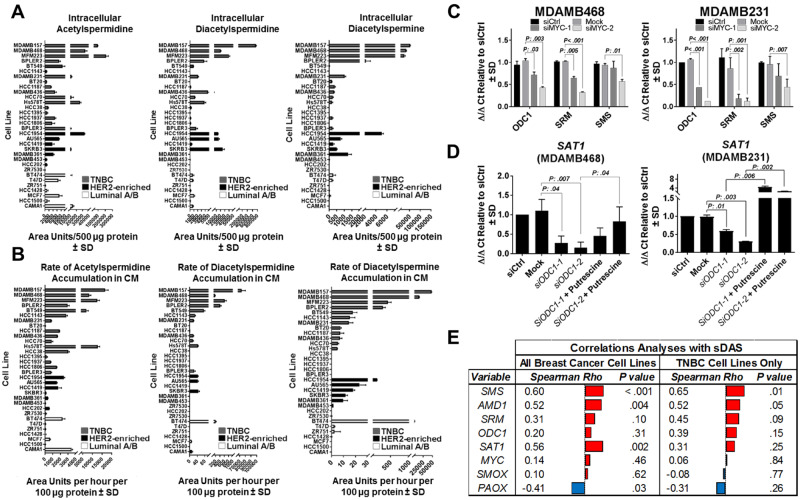
We performed metabolomics profiling on whole cell lysates and a time-course analysis (baseline, 1, 2, 4, and 6 hours post conditioning; see Supplementary Methods, available online) of conditioned media of 32 breast cancer cell lines. TNBC cell lines exhibited substantially higher levels of intracellular acetylated polyamines, including DAS, and elevated rates (area units per hour per 100 µg of protein) of their accumulation in conditioned media compared with other subtypes (Figure 3, A–B). Intracellular pools of spermidine were statistically significantly higher in TNBC compared with non-TNBC cell lines (two-sided Wilcoxon rank-sum test P = .03); no difference in intracellular spermine was observed between TNBC and non-TNBC cell lines (Supplementary Figure 2A, available online).
Figure 3.
Regulation of polyamine metabolism in breast cancer cell lines. A–B) Relative intracellular levels (area units per 500 µg of protein) of DAS (A) and rates (area units per hour per 100 µg of protein) of DAS accumulation in temporally collected (baseline, 1, 2, 4, and 6 hrs) conditioned media (B) of 32 breast cancer cell lines. Each cell line was analyzed in biological triplicate. C) Relative fold change in mRNA expression (2-Δ/Δ CT) of ODC1, SRM, and SMS following transient knockdown of MYC in TNBC cell lines MDAMB468 and MDAMB231. D) Relative fold change in mRNA expression (2-Δ/Δ CT) of SAT1 following transient knockdown of ODC1 and rescue by putrescine in TNBC cell lines MDAMB468 and MDAMB231. E) Spearman correlations depicting association between rates by which DAS accumulates in conditioned media (sDAS) and gene expression of polyamine-metabolizing enzymes. AMD1 = adenosylmethionine decarboxylase 1; MYC = Myc proto-oncogene protein; DAS = diacetylspermine; ER = estrogen receptor; ODC1 = ornithine decarboxylase 1; PAOX = polyamine oxidase; PR = progesterone receptor; SAT1 = spermidine/spermine N1-acetyltransferase 1; SMOX = spermine oxidase; SMS = spermine synthase; SRM = spermidine synthase; TNBC = triple-negative breast cancer.
Regulation of Diacetylspermine Biosynthesis
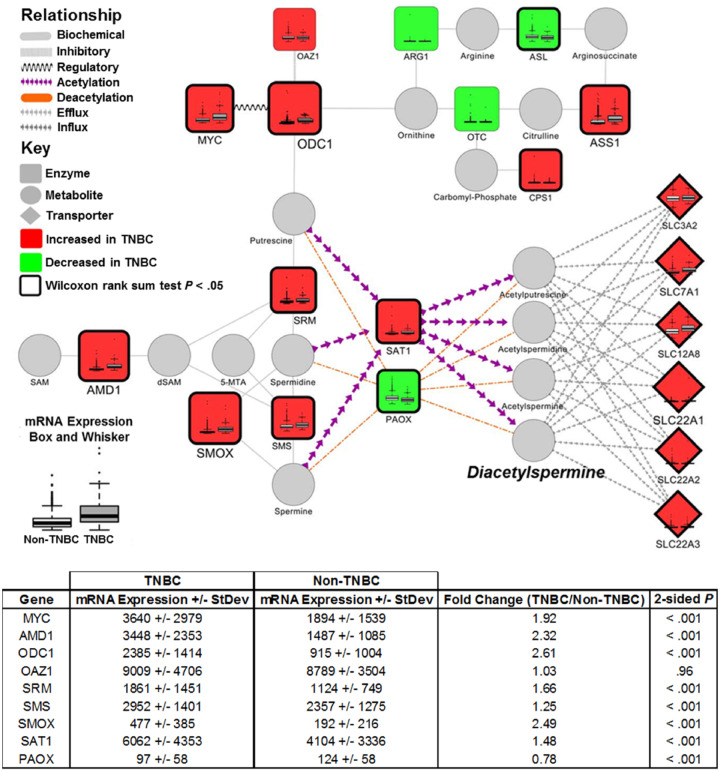
To explore the extent to which elevated polyamine flux and secretion of DAS are indeed mediated through oncogenic MYC, we first evaluated mRNA expression of polyamine metabolizing enzymes (PMEs) using gene expression profiles obtained from TCGA. As compared with non-TNBCs, TNBC tumors exhibited statistically significant elevations (two-sided Wilcoxon rank-sum test P < .001) in mRNA expression of genes central to the biosynthesis and catabolism of polyamines (Figure 4); only polyamine oxidase, which catalyzes the oxidation of acetylated polyamines, was statistically significantly (two-sided Wilcoxon rank-sum test P < .001) reduced (Figure 4). Expectantly, gene expression of MYC was statistically significantly elevated (mean mRNA expression [SD] = 3640 [2979] vs 1894 [1539]; two-sided Wilcoxon rank-sum test P < .001) in TNBC compared with non-TNBCs (Figure 4) (1). Next, we performed knockdown of MYC in TNBC cell lines MDAMB468 and MDAMB231 and confirmed reduced mRNA expression of ODC1, SRM, and SMS (Figure 3C), consistent with previous findings by others (26). Notably, reduced mRNA expressions of SRM and SMS were not due to depletion of polyamines, because knockdown of ODC1 did not impact mRNA expression of either SRM or SMS (Supplementary Figure 2B, available online). Knockdown of MYC in hormone-receptor positive (HR+) and HER2-enriched breast cancer cell lines MCF-7 and AU565 also resulted in reduced mRNA expression of ODC1 and, to a lesser extent, SRM (Supplementary Figure 2C, available online), respectively. No statistically significant difference was observed in SMS mRNA expression following knockdown of MYC in MCF7 (HR+) and AU565 (HER2-enriched) breast cancer cell lines (Supplementary Figure 2C, available online).
Figure 4.
Gene expression of polyamine-related enzymes in breast cancer tumors stratified into TNBC and non-TNBC. Biochemical network depicting mRNA expression of enzymes involved in the biosynthesis and catabolism of polyamines between TNBC and non-TNBC breast cancers. Data derived from The Cancer Genome Atlas. Node size and color indicate magnitude and directionality (red: increased; green: decreased) of mRNA expression for the respective encoding gene between TNBC and non-TNBC breast cancers. Thickened black border indicates statistical significance (Wilcoxon rank-sum test P < .05). Box and whisker plots illustrate differences in distribution of mRNA expression for the respective encoding gene between TNBC and non-TNBC breast cancers. Table beneath indicates mean mRNA expression (SD), fold change, and P value (two-sided Wilcoxon rank-sum test) for respective genes central to polyamine metabolism in comparison of TNBC and non-TNBC tumors. AMD1 = adenosylmethionine decarboxylase 1; MYC = Myc proto-oncogene protein; OAZ1 = ornithine decarboxylase antizyme 1; ODC1 = ornithine decarboxylase 1; PAOX = polyamine oxidase; SAT1 = spermidine/spermine N1-acetyltransferase 1; SMS = spermine synthase; SMOX = spermine oxidase; SRM = spermidine synthase; TNBC = triple-negative breast cancer.
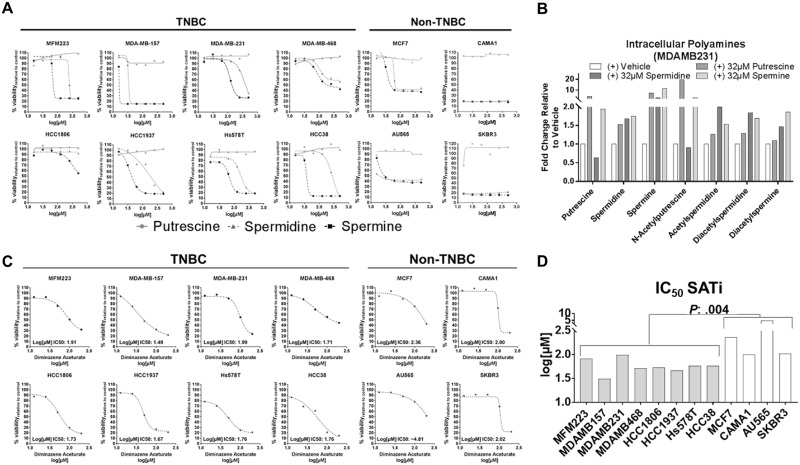
Maintenance of intracellular polyamine pools is regulated by a highly dynamic network that orchestrates both its biosynthesis and catabolism. Knockdown of ODC1 in both TNBC cell lines (MDAMB468 and MDAMB231) and non-TNBC cell lines (MFC7 and AU565) resulted in reduced mRNA expression of SAT1, an effect that was reversed through the addition of extracellular putrescine, the direct metabolic product of ODC1 (Figure 3D;Supplementary Figure 2D, available online). Treatment of MDAMB231 cells with exogenous putrescine, spermidine, or spermine resulted in the increased biochemical elongation of both putrescine and spermidine to spermine, as well as increased conversion of these polyamines to their respective acetylated derivatives (Figure 5B). Collectively, our results implicate that polyamine catabolism is regulated by intracellular polyamine pools and that high levels of intracellular DAS and its subsequent extracellular secretion serves to maintain intracellular polyamine pools, including spermine (27,28). To this effect, treatment of eight TNBC cell lines and four non-TNBC cell lines (MCF7 + CAMA1 [HR+] and AU565 + SKBR3 [HER2-enriched]) with exogenous spermidine or spermine, but not putrescine, induced dose-dependent reductions in cell viability (Figure 5A). Treatment of the same breast cancer cell lines (eight TNBC and four non-TNBC) with diminazene aceturate, an SAT1 inhibitor (29), reduced cell viability (Figure 5C). Non-TNBC cell lines were less sensitive to diminazene aceturate as compared with TNBC cell lines (Figure 5D). Our findings provide supportive evidence that oncogenic MYC drives increased expression of PMEs (1), which results in an increase in cancer polyamine flux and extracellular secretion.
Figure 5.
Viability curves in breast cancer cell lines following treatment with exogenous polyamines or the spermidine/spermine N(1)-acetyltransferase 1 inhibitor diminazene aceturate. A–B) Viability curves for eight TNBC and four non-TNBC cell lines following 48-hour treatment with vehicle, putrescine, spermidine, or spermine. Viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. B) Intracellular levels of polyamines in MDAMB231 TNBC cells following 6-hour treatment with 32 µM of putrescine, spermidine, spermine, or vehicle. C) Viability curves for eight TNBC and four non-TNBC cell lines following 48-hour treatment with the SAT1 inhibitor diminazene aceturate. Viability was assessed by MTS assay. D) Half maximal inhibitory concentration (IC50) values for SAT1 inhibitor in breast cancer cell lines. Statistical significance was determined by Wilcoxon rank-sum test P < .05 comparing IC50 values between TNBC (gray bars) and non-TNBC (white bars) cell lines. SAT1 = spermidine/spermine N1-acetyltransferase 1; TNBC = triple-negative breast cancer.
Association Between Tumor Spermine Synthase Gene Expression and Tumor Immune Cell Infiltrates and Prognosis in TNBC
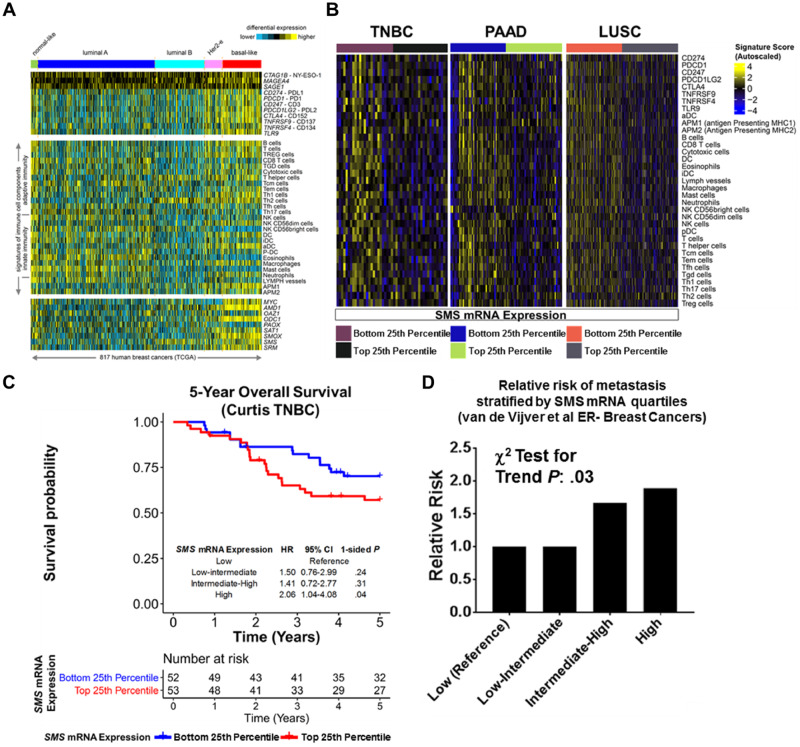
Reduced tumor immune cell infiltration in TNBC is associated with poor overall survival and reduced distant metastasis-free survival compared with immune-rich tumors (30–33). In our prospective breast cancer cohort, elevated plasma DAS was associated with increased risk of developing future metastasis (Figure 2, A–B). We therefore hypothesized that elevated plasma DAS may associate with a subtype of TNBC characterized with a distinct tumor immunophenotype and poor overall survival. To this end, we first screened gene expression profiles of polyamine and checkpoint-blockade–related genes as well as gene expression signatures reflective of immune cell infiltrates (23) from 817 human breast cancers in TCGA, the results of which revealed distinct immune signatures associated with the different breast cancer subtypes (Figure 6A).
Figure 6.
Association between mRNA expression of spermine synthase (SMS) and immune infiltrates, overall survival, and metastasis. A) Differential expression heat maps for checkpoint blockade-related genes as well as immune cell gene signatures across 817 TCGA breast cancers, with cases ordered by Pam50 molecular subtype, of molecular features highlighted in part A. Expression values and signatures are normalized to SD from the median of breast cancer cases. B) Heat maps depict TCGA gene expression profiles for checkpoint blockade-related genes as well as immune cell gene signatures in TNBC, PAAD, and LUSC tumors stratified by the bottom 25th percentile and top 25th percentile of SMS mRNA expression. Immune cell gene signatures are based off those of Bindea et al. (23). C) Kaplan-Meier survival curves for overall survival stratified by SMS mRNA expression (bottom 25th percentile vs top 25th percentile) in the Curtis et al. TNBC cohort (19). D) Association between relative risk of future metastasis and SMS mRNA expression in ER− tumors stratified into SMS gene expression quartiles. Gene expression was derived from Oncomine (22) for the van de Vijver et al. breast cohort (21). For the van de Vijver et al. dataset, the bottom 25th percentile was used as the reference point; statistical significance was determined by two-sided χ2 test for trend. LUSC = lung squamous cell carcinoma; PAAD = pancreatic adenocarcinoma; SMS = spermine synthase; TCGA = The Cancer Genome Atlas; TNBC = triple-negative breast cancer; ER = estrogen receptor.
The rate of DAS secretion into conditioned media (DASCM) among TNBC cell lines as well as expression of PME genes indicated that SMS mRNA expression was the strongest positive predictor for DASCM (Figure 3E). We therefore hypothesized that elevated SMS mRNA expression would associate with reduced immune cell infiltrates in TNBC. The choice of SMS in particular is that SAT1, in contrast to SMS, does not exclusively catalyze acetylate spermine but can also catalyze acetylation of putrescine and spermidine (Figure 4). SMS mRNA was statistically significantly inversely associated with mRNA signatures (23) reflective of antigen-presenting MHC class II immune cells and T cells, a finding that was conserved in several TNBC datasets in contrast to luminal A or B breast tumors (Figure 6B; Supplementary Figure 3, A–C, Supplementary Table 4, available online). Moreover, the inverse association between SMS gene expression and gene-based signatures of antigen-presenting MHC class II cells and infiltrating T cells was found to be conserved in pancreatic adenocarcinoma and lung squamous cell carcinoma providing rationale to consider it as a broadly relevant feature reflective of the tumor-immuno microenvironment (Figure 6B;Supplementary Table 2, available online). The SMS mRNA analysis in a breast cancer cohort enriched in TNBC cases (19) yielded statistically significantly worse 5-year survival in the highest quartile compared with the lowest SMS quartile (HR = 2.06, 95% CI = 1.04 to 4.08; P = .04) (Figure 6C). SMS mRNA level in ER− breast cancers in another cohort was also positively associated with metastasis (χ2 test for Ptrend= .03; comparison of lowest SMS quartile [reference] to highest SMS quartile relative risk = 1.90, 95% CI = 0.97 to 4.0; one-sided Fisher exact test P = .03) in (21) (Figure 6D). Thus, both DAS plasma levels and SMS gene expression in tumors were predictive of future metastasis and poor overall survival.
Discussion
The TNBC subtype of breast cancer exhibits an aggressive phenotype with poor outcome. Identification of TNBC at an early stage and further stratifying metastatic potential among subjects would provide substantial benefit (34,35). At present, no clinically accepted biomarkers exist that can reliably predict the development and progression of TNBC. The findings reported herein directly address this unmet clinical need and provide a potential breakthrough that would enable classification of TNBC patients most at risk for developing metastasis. Importantly, the predictive marker we have identified is testable in blood.
MYC is an established oncogenic driver in TNBC development and progression. MYC is a reported upstream transcriptional regulator of ODC1, AMD1, and SRM (5,6,26); ODC1 and AMD1 serve as critical rate-limiting enzymes in polyamine biosynthesis (5,6,26,36). Using metabolic profiling of plasma samples from two independent human breast cancer cohorts, mouse breast cancer models, and breast cancer cell lines, we identified and validated elevated levels of DAS as being a prominent feature of TNBC, consistent with elevated levels of MYC expression. We further demonstrated the potential prognostic value for plasma DAS for TNBC progression. To the best of our knowledge, this is the first study to demonstrate that elevated plasma DAS associates with a distinct molecular subtype of breast cancer and is also associated with increased risk of future metastasis in the context of TNBC.
We additionally provide evidence that an elevation of plasma DAS in women diagnosed with TNBC may associate with distinct subtypes of TNBC tumors that are characterized by low immune infiltrate. Defining the mechanism underlying this relationship and the extent to which plasma DAS generally reflects immune-poor tumors will require extensive additional analysis of matched plasma and tissue samples with sufficient sample number to achieve statistically relevant validation. Nevertheless, in the current study, we emphasize that there is good concordance between SMS mRNA expression and rates of DAS secretion in vitro (as demonstrated for TNBC cell lines) and that high SMS mRNA expression is consistently and reproducibly associated with reduced immune-related gene signatures, poor overall survival, and metastasis. Our immune-related gene signature findings are based on in silico inference; previous studies have shown good concordance between these in silico infiltrate predictions and the bona fide presence of immune infiltrate as assessed by immunohistochemistry (23).
A limitation of the current study is the lack of external validation for the prognostic value of plasma DAS for predicting future distant metastasis. Additionally, as evident by our observations, not every subject exhibits elevated plasma levels of DAS. This is consistent with our observations that the DAS secretion rate is variable among TNBC cell lines, suggesting additional mechanistic complexity that requires further exploration.
In conclusion, our findings suggest the utility of DAS as a predictive marker for TNBC development and progression. We further demonstrate an inverse association between tumor SMS and tumor immune cell infiltration in the context of TNBC. Further studies are warranted.
Funding
This work was supported by a faculty fellowship from the University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment (JFF) and the National Institutes of Health CA125123 (CJC).
Notes
The authors declare no conflicts of interest.
Author contributions: JFF: analyzed all metabolomics data, data analysis and generation of figures and tables, conception and construction of study, manuscript writing; JV: general guidance throughout study, provided key insight, manuscript editing; AF: performed in vitro experiments; ST: general guidance throughout study, manuscript editing; JBD: metabolomics analyses; EM: metabolomics analyses; PW: metabolomics analyses; CY: metabolomics analyses, bioinformatics assistance; MC: general guidance throughout study, manuscript editing; CJC: provided key data with respect to immune signatures; KD: assistance with statistical analyses; JPL: assistance with statistical analyses; EI: assistance with statistical analyses; CP: general guidance throughout study, assistance with statistical analyses; HK: general guidance throughout study, manuscript editing; MD: provided plasma samples for mouse breast cancer models, manuscript editing; BA: provided plasma samples for human cohorts, manuscript editing; SH: critical review of manuscript, guidance and mentorship throughout study, manuscript writing and editing.
The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication.
We acknowledge Corinthia Emery for her contributions to the in vitro work and Angelica Gutierrez Barrera for assistance in obtaining clinical information. We additionally acknowledge the Robert Duncan Foundation.
Supplementary Material
References
- 1. Xu J, Chen Y, Olopade OI.. MYC and breast cancer. Genes Cancer. 2010;1(6):629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fallah Y, Brundage J, Allegakoen P, et al. MYC-driven pathways in breast cancer subtypes. Biomolecules. 2017;7(4):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gatza ML, Lucas JE, Barry WT, et al. A pathway-based classification of human breast cancer. Proc Natl Acad Sci USA. 2010;107(15):6994–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bello-Fernandez C, Packham G, Cleveland JL.. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA. 1993;90(16):7804–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachmann AS, Geerts D.. Polyamine synthesis as a target of MYC oncogenes. J Biol Chem. 2018;293(48):18757–18769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Funakoshi-Tago M, Sumi K, Kasahara T, et al. Critical roles of Myc-ODC axis in the cellular transformation induced by myeloproliferative neoplasm-associated JAK2 V617F mutant. PLoS One. 2013;8(1):e52844.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casero RA Jr, Marton LJ.. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6(5):373–390. [DOI] [PubMed] [Google Scholar]
- 8. Casero RA Jr, Murray Stewart T, Pegg AE.. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 2018;18(11):681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murray-Stewart TR, Woster PM, Casero RA Jr.. Targeting polyamine metabolism for cancer therapy and prevention. Biochem J. 2016;473(19):2937–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fahrmann JF, Bantis LE, Capello M, et al. A plasma-derived protein-metabolite multiplexed panel for early-stage pancreatic cancer. J Natl Cancer Inst. 2018;111(4):372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wikoff WR, Hanash S, DeFelice B, et al. Diacetylspermine is a novel prediagnostic serum biomarker for non-small-cell lung cancer and has additive performance with pro-surfactant protein B. J Clin Oncol. 2015;33(33):3880–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fahrmann JF, Grapov DD, Wanichthanarak K, et al. Integrated metabolomics and proteomics highlight altered nicotinamide- and polyamine pathways in lung adenocarcinoma. Carcinogenesis. 2017;38(3):271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller-Fleming L, Olin-Sandoval V, Campbell K, et al. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol. 2015;427(21):3389–3406. [DOI] [PubMed] [Google Scholar]
- 14. Cervelli M, Bellavia G, Fratini E, et al. Spermine oxidase (SMO) activity in breast tumor tissues and biochemical analysis of the anticancer spermine analogues BENSpm and CPENSpm. BMC Cancer. 2010;10(1):555.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pitteri SJ, Faca VM, Kelly-Spratt KS, et al. Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. J Proteome Res. 2008;7(4):1481–1489. [DOI] [PubMed] [Google Scholar]
- 16. Pitteri SJ, Kelly-Spratt KS, Gurley KE, et al. Tumor microenvironment-derived proteins dominate the plasma proteome response during breast cancer induction and progression. Cancer Res. 2011;71(15):5090–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang T, Fahrmann JF, Lee H, et al. JAK/STAT3-regulated fatty acid beta-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 2018;27(1):136–150.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. [DOI] [PubMed] [Google Scholar]
- 22. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. [DOI] [PubMed] [Google Scholar]
- 24. Patricia M, Grambsch T.. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81(3):12. [Google Scholar]
- 25. Contal C, O'Quigley J.. An application of changepoint methods in studying the effect of age on survival in breast cancer. Comput Stat Data Anal. 1999;30(3):253–270. [Google Scholar]
- 26. Hogarty MD, Norris MD, Davis K, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008;68(23):9735–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Butcher NJ, Broadhurst GM, Minchin RF.. Polyamine-dependent regulation of spermidine-spermine N1-acetyltransferase mRNA translation. J Biol Chem. 2007;282(39):28530–28539. [DOI] [PubMed] [Google Scholar]
- 28. Fogel-Petrovic M, Vujcic S, Brown PJ, et al. Effects of polyamines, polyamine analogs, and inhibitors of protein synthesis on spermidine-spermine N1-acetyltransferase gene expression. Biochemistry. 1996;35(45):14436–14444. [DOI] [PubMed] [Google Scholar]
- 29. Libby PR, Porter CW.. Inhibition of enzymes of polyamine back-conversion by pentamidine and berenil. Biochem Pharmacol. 1992;44(4):830–832. [DOI] [PubMed] [Google Scholar]
- 30. Karn T, Jiang T, Hatzis C, et al. Association between genomic metrics and immune infiltration in triple-negative breast cancer. JAMA Oncol. 2017;3(12):1707–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Teijido P, Cabal ML, Fernandez IP, et al. Tumor-infiltrating lymphocytes in triple negative breast cancer: the future of immune targeting. Clin Med Insights Oncol. 2016;10(suppl 1):31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loi S, Adams S, Schmid P, et al. LBA13 relationship between tumor infiltrating lymphocyte (TIL) levels and response to pembrolizumab (pembro) in metastatic triple-negative breast cancer (mTNBC): results from KEYNOTE-086. Ann Oncol. 2017;28(suppl 5):v605–v649. [Google Scholar]
- 33. Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9(5):R65.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hudis CA, Gianni L.. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16(suppl 1):1–11. [DOI] [PubMed] [Google Scholar]
- 35. Tseng LM, Hsu NC, Chen SC, et al. Distant metastasis in triple-negative breast cancer. Neoplasma. 2013;60(03):290–294. [DOI] [PubMed] [Google Scholar]
- 36. Zabala-Letona A, Arruabarrena-Aristorena A, Martin-Martin N, et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature. 2017;547(7661):109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.