Summary
Reactive oxygen species—superoxide, hydrogen peroxide, and hydroxyl radicals—have long been suspected of constraining bacterial growth in important microbial habitats, and indeed of shaping microbial communities. Over recent decades, studies of paradigmatic organisms such as E. coli, Salmonella typhimurium, Bacillus subtilis, and Saccharomyces cerevisiae have pinpointed the biomolecules that oxidants can damage and the strategies by which microbes minimize their injuries. What is lacking is a good sense of the circumstances under which oxidative stress actually occurs. In this MiniReview several potential natural sources of oxidative stress are considered: endogenous ROS formation, chemical oxidation of reduced species at oxic-anoxic interfaces, H2O2 production by lactic acid bacteria, the oxidative burst of phagocytes, and the redox-cycling of secreted small molecules. While all of these phenomena can be reproduced and verified in the lab, the actual quantification of stress in natural habitats remains lacking—and, therefore, we have a fundamental hole in our understanding of the role that oxidative stress actually plays in the biosphere.
Keywords: hydrogen peroxide, superoxide, lactic acid bacteria, obligate anaerobiosis, SoxRS, OxyR
Introduction
Enzymes that scavenge superoxide (O2−) and hydrogen peroxide (H2O2) were discovered by serendipity. Their existence, in virtually all organisms, implies that these reactive oxygen species (ROS) comprise threats to fitness. In this Minireview I pose two key questions: What are the natural sources of these oxidants? In what circumstances do their levels rise high enough to impinge upon cell fate? I cannot provide definitive answers, but I summarize ideas that have arisen in the literature. The main point is that this issue is a key unknown in our understanding of oxidative stress.
To tolerate oxygen bacteria must cope with endogenous ROS formation
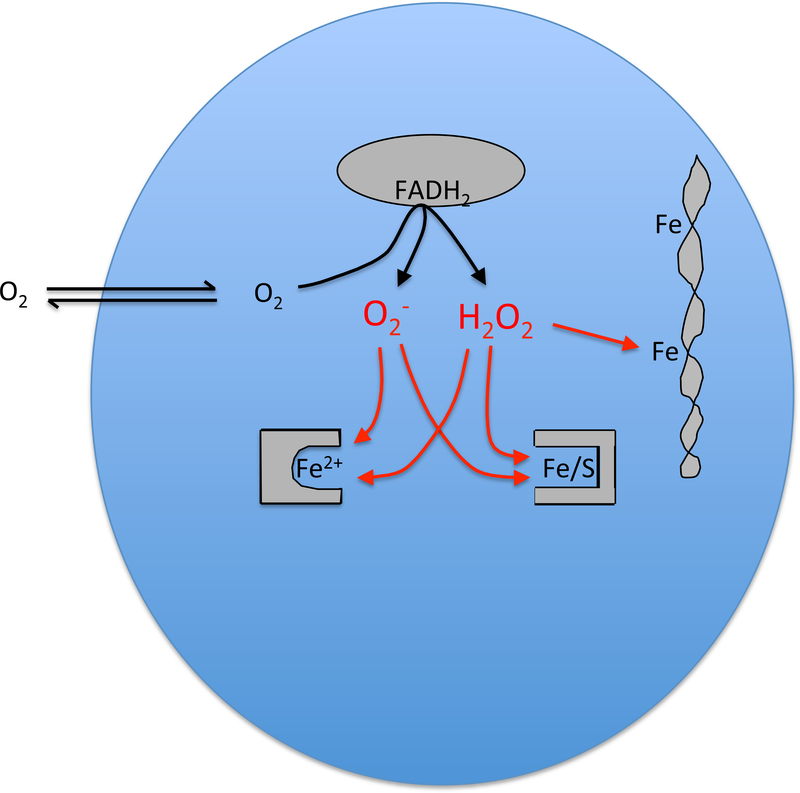
Mutants of E. coli that lack either superoxide dismutase or catalase and peroxidase exhibit distinctive growth defects, which led investigators to pinpoint the specific injuries that O2− and H2O2 can produce (Carlioz and Touati, 1986; Seaver and Imlay, 2001a) (Fig. 1). These species inappropriately oxidize and disable [4Fe-4S] dehydratases of the aconitase class (Kuo et al., 1987; Gardner and Fridovich, 1991; Flint et al., 1993; Jang and Imlay, 2007) as well as mononuclear Fe(II) enzymes such as ribulose-5-phosphase 3-epimerase (Sobota and Imlay, 2011; Gu and Imlay, 2013; Sobota et al., 2014). Both enzyme families use solvent-exposed iron atoms to directly bind metabolites; therefore, their metal centers are accessible to direct oxidation by either H2O2 or O2−. The oxidized metals dissociate from the enzymes, rendering them inactive and disabling the pathways to which they belong. Enzymes of these two families are requisite for core cellular processes: the TCA cycle, the pentose-phosphate pathway, the biosynthesis of branched-chain and aromatic amino acids, and others.
Figure 1. Endogenous oxidative stress.
The adventitious transfer of electrons from redox enzymes to oxygen generates a mixture of O2− and H2O2. These species oxidize the solvent-exposed iron centers of mononuclear Fe2+ enzymes and [4Fe-4S] dehydratases, provoking iron dissociation and activity loss. The H2O2 also reacts with the pool of loose iron, most notably leading to DNA damage from hydroxyl-radical production.
The existence of mutant phenotypes implied that these oxidants are formed continuously in the aerobic cell. Indeed, measurements of H2O2 efflux from catalase/peroxidase mutants have fixed the H2O2 production rate inside air-saturated cells at 10–15 μM/sec; O2− rates are estimated to be 5–10 μM/sec (Seaver and Imlay, 2001b). In wild-type (scavenger-proficient) cells, the balance between production and scavenging sets the steady-state level of H2O2 at an estimated 20–50 nM, while O2− is predicted to be about 0.2 nM (Imlay, 2013). The reaction rates of H2O2 and O2− with the metal centers of dehydratases and mononuclear enzymes (ca. 104 and 106 s−1 (Imlay, 2013)) therefore predict that the enzymes will be damaged every 30 min or so. The damaging events are reversible, however: clusters are reassembled with half-times of ca. 5 min (Gardner and Fridovich, 1992), and remetallation of mononuclear enzymes may be faster yet. This means that at any moment in wild-type cells, the bulk of these enzyme populations are in their active form, with a small minority being inactive because of a recent damaging event. On balance, their net activities are sufficiently high that their pathways are functional.
Inside cells H2O2 also reacts with the pool of loose iron to generate hydroxyl radicals (Keyer and Imlay, 1996). These powerful oxidants can damage most biomolecules, including DNA. The contribution of oxidants to DNA damage—and to mutagenesis—has been proposed to be significant: mutation rates are lower, and the viability of repair mutants is greater, when E. coli is cultured in the absence of oxygen (Morimyo, 1982; Boling et al., 1984; Imlay and Linn, 1986; Sakai et al., 2006). However, this conclusion may be premature. Lab media routinely contain micromolar H2O2 that is formed by chemical and photochemical processes, and under many experimental conditions the flow of this H2O2 into the cell greatly outstrips the pace of H2O2 formation within it (Li and Imlay, 2018). Therefore exogenous H2O2 can be the predominant source of the intracellular H2O2 and DNA oxidation. The upshot is that DNA-oxidation phenotypes in lab media can be artifactual. The actual impact of endogenous oxidants is currently unclear and in principle may be quite small.
When defensive systems are not enough: endogenous ROS contributes to the phenomenon of obligate anaerobiosis
E. coli—and presumably all oxygen-tolerant bacteria—have acquired a mixture of scavenging and repair systems that adequately sustain the activities of oxidant-sensitive enzymes in the face of endogenous ROS production. At one time textbooks asserted that obligate anaerobes cannot tolerate oxygen because they lack superoxide dismutases and catalases. This idea is incorrect. Many obligate anaerobes actually have these scavenging enzymes, and the others rely upon superoxide reductases and peroxidases (Sheng et al., 2014). It is not for lack of evolution that anaerobes cannot thrive around oxygen.
However, it may still be true that ROS contribute to oxygen poisoning in these bacteria. The carbohydrate fermenting Bacteroides thetaiotaomicorn is a classic oxygen-sensitive bacterium: when fully aerated, growth stops, and it resumes only when anoxia is restored. Analysis showed that both branches of central metabolism are poisoned, due to the inactivation of the metalloenzymes fumarase and pyruvate:ferredoxin oxidoreductase (Pan and Imlay, 2001). The inactivation of fumarase, an aconitase-class [4Fe-4S] dehydratase, is striking because the same enzyme remains active inside air-tolerant bacteria (Flint et al., 1993; Liochev and Fridovich, 1993). The key difference? Aerated B. thetaiotaomicron generates intracellular ROS at 15 times the rate of aerated E. coli (Mishra and Imlay, 2013; Lu and Imlay, 2017; Lu et al., 2018). The reason is not clear. Biochemical studies have shown that redox enzymes can accidentally transfer electrons to oxygen instead of to their proper substrates, and it is supposed that the collective autoxidations of such enzymes are responsible for the O2− and H2O2 that are formed endogenously in aerated cells (Massey et al., 1969; Messner and Imlay, 1999; Imlay, 2013). The rate of ROS formation in aerated Bacteroides might therefore be especially high because it possesses redox enzymes that are especially reactive with molecular oxygen. The low-potential redox chains (Eo’ ~ −.4 V) in such anaerobes are plausible sources of these ROS, since they are thermodynamically competent to drive electrons onto oxygen (Eo’ = −0.16 V). To resolve this issue completely, it will be necessary to pinpoint the specific enzymes that are the predominant sources of internal ROS—a goal that has not yet been achieved in any organism.
It is important to note that ROS are not exclusively responsible for the oxic problems of obligate anaerobes: molecular oxygen itself can directly poison key enzymes that are exclusive to anaerobes. Oxygen forms adducts to glycyl-radical enzymes, leading to polypeptide cleavage (Wagner et al., 1992), and it seems likely that molecular oxygen itself can over-oxidize the low-potential metal centers of enzymes such as pyruvate:ferredoxin oxidoreductase, hydrogenase, and nitrogenase (Vita et al., 2008; Stiebritz and Reiher, 2012; Schlesier et al., 2015). These enzymes are rapidly damaged by the introduction of oxygen in vitro and in vivo.
Oxidative stress from the environment
In 1982 Demple and Halbrook reported that when E. coli was pre-exposed to 50 μM H2O2 it could survive a subsequent exposure to 5 mM H2O2, whereas naïve cells could not (Demple and Halbrook, 1983). The phenomenon was subsequently linked to the induction of a regulon under the control of an H2O2-activated transcription factor, named OxyR (Christman et al., 1985; Aslund et al., 1999; Choi et al., 2001). Hydrogen peroxide rapidly oxidizes its sensory cysteine residue, and in this form OxyR triggers synthesis of defensive proteins. Catalase and NADH peroxidase (AhpCF) are induced 10-fold, thereby helping to drive down the intracellular level of H2O2 (Zheng et al., 2001). Dps is a mini-ferritin that suppresses damage to DNA by sequestering the cellular pool of loose iron (Altuvia et al., 1994; Ilari et al., 2002; Park et al., 2005). SufABCDES comprises a secondary iron-sulfur-cluster assembly system that counteracts damage to the dehydratase clusters (Lee et al., 2004; Jang and Imlay, 2010). MntH imports manganese(II), which supplants iron(II) in mononuclear enzymes, providing nearly as much activity without the tendency toward oxidation by peroxide (Kehres et al., 2002; Anjem and Imlay, 2012). YaaA and Fur help suppress loose-iron levels, and HemF and HemH sustain heme synthesis to enable catalase induction (Varghese et al., 2007; Liu et al., 2011b; Mancini and Imlay, 2015). The roles of other members of the regulon are not yet completely clear. Thus the induced activities are well-matched to the types of injuries that H2O2 creates. Dosimetric studies indicate that OxyR is activated when cytoplasmic H2O2 reaches about 0.1–0.2 μM—appropriately close to the concentrations (0.3–0.5 μM) that begin to inactivate enzymes and disrupt growth (Seaver and Imlay, 2001b; Zheng et al., 2001; Sobota and Imlay, 2011).
OxyR turns out to be wide-spread among bacteria. Interestingly, still other bacteria use PerR, an iron-binding transcription factor of the Fur family (Lee and Helmann, 2006b). This transcription factor is activated when H2O2 reacts with its Fe(II) cofactor, creating a hydroxyl radical that irreversibly oxidizes an adjacent histidine residue. PerR represses many of the same genes that OxyR controls in E. coli; when H2O2 oxidizes PerR, its DNA-binding activity is lost, and the genes that it controls are expressed. The rate constant with which H2O2 oxidizes PerR is similar to that of OxyR, suggesting that these disparate bacteria have evolved to respond to similar amounts of H2O2 (Lee and Helmann, 2006a). This arrangement would make sense if OxyR and PerR serve to defend similar classes of enzymes against oxidation.
Aeration alone does not activate these transcription factors, because the basal levels of scavenging enzymes inside cells are sufficient to keep endogenous H2O2 well below the OxyR and PerR trigger points. Instead, it seems likely that they respond to larger influxes of H2O2 from the local environment. The cytoplasmic membrane is semipermeable to H2O2, and external concentrations > 2 μM are sufficient to drive internal concentrations up to the 0.2 μM dose that activates the responses (Li and Imlay, 2018). The key question is: When do these bacteria encounter these doses of H2O2?
Photochemically generated H2O2 in surface waters
Somewhat surprisingly, H2O2 is readily detected in both fresh and oceanic waters (Lesser, 2006; Mesle et al., 2017). The levels can rise to the low-micromolar range—enough to plausibly threaten bacteria and induce their stress responses. The diurnal cycling of H2O2 levels tipped off investigators that ultraviolet photochemistry lie at the root of the phenomenon (Wilson et al., 2000b; Wilson et al., 2000a). Dissolved organic compounds in these waters include chromophores that, when excited by solar radiation, transfer electrons from local reductants to molecular oxygen. The events recapitulate the flavin-dependent production of O2− that is used to visualize superoxide dismutase in activity gels (Beauchamp and Fridovich, 1971)—and also the steady accrual of micromolar H2O2 that occurs in complex growth media under room lights (Li and Imlay, 2018). Workers are exploring the involvement of ROS in solar disinfection protocols. It remains unclear whether the ROS generated in natural waters comprises enough of a stress to add structure to the microbial community.
H2O2 at oxic-anoxic interfaces
One of the genes regulated by E. coli OxyR is a periplasmic cytochrome c peroxidase (Ccp) (Partridge et al., 2007). The enzyme receives electrons from the respiratory chain and transfers them directly to H2O2 in the periplasm. Apparently the purpose of Ccp is not to clear H2O2, since H2O2 equilibration across the outer membrane is so fast that the enzyme cannot lower the periplasmic level below the environmental level. Instead, Ccp allows the cell to employ H2O2 as a terminal oxidant to support respiration (Khademian and Imlay, 2017). This function would be beneficial only when molecular oxygen, a superior acceptor, is unavailable—and accordingly Ccp is synthesized only when molecular oxygen is absent (Partridge et al., 2007).
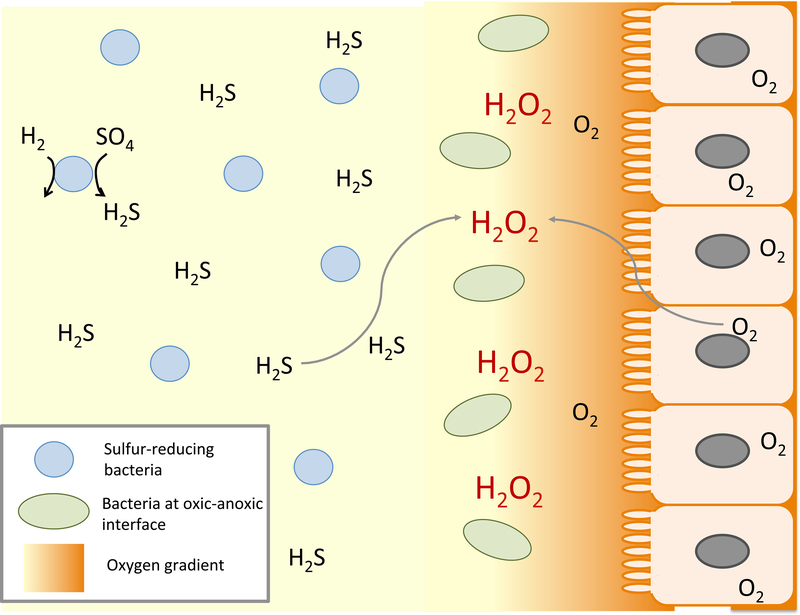
The peculiar aspect of this arrangement is its implication that H2O2 is present in some hypoxic environments. E. coli inhabits the margins of the large intestine, where it is ensconced in the mucoid periphery of the lumen (Fig. 2). Some oxygen diffuses into the intestine from the surrounding epithelial cells, but the levels drop by orders of magnitude just a few microns towards the interior (Espey, 2013). Deeper in the lumen oxygen is essential absent, and sulfate-reducing bacteria generate copious amounts of hydrogen sulfide (Macfarlane et al., 1992; Pitcher et al., 2000). One possibility is that as this sulfide diffuses towards the microoxic margins, it reacts chemically with the oxygen, producing H2O2. A similar situation might occur at oxic-anoxic interfaces in soil and at hydrothermal vents (Ogino et al., 2018), where low- and high-potential fluids run into one another. The apparent KM of Ccp is 5 μM, which is a good match to the extracellular dose that activates OxyR (Khademian and Imlay, 2017; Li and Imlay, 2018); this agreement lends further weight to the idea that low-micromolar concentrations of H2O2 are biologically relevant.
Figure 2. Plausible sources of oxidative stress at the oxic-anoxic interface near the intestinal epithelium.
Oxygen influx from the epithelium collides with sulfide generated by luminal bacteria, potentially generating H2O2 through direct reaction. Lactic acid bacteria near the epithelium are also likely to excrete H2O2 as a direct metabolic product, threatening by-stander bacteria.
Alternative possibilities are that intestinal H2O2 is generated by lactic acid bacteria, which can also thrive at such interfaces, or that H2O2 is released by phagocytic cells along the epithelial layer of the intestine. These sources are considered in more detail below. The most striking take-home message is that the intuitive idea, that H2O2 is most abundant in the most oxic environments, may be wrong.
Lactic acid bacteria spew H2O2 into their environment
Lactic acid bacteria (LAB) are primarily carbohydrate fermenters who have made metabolic compromises that enable them to thrive in low-iron habitats. They lack full respiratory chains, but many can nevertheless exploit oxygen as a direct electron acceptor by using soluble lactate and pyruvate oxidases (Spellerberg et al., 1996; Seki et al., 2004). When LAB do so, they produce additional ATP through acetate kinase (Pericone et al., 2003). The product of these oxidases is H2O2. In laboratory cultures of LAB, the H2O2 concentration in the cells and the medium can rise to the millimolar range (Tong et al., 2007; Liu et al., 2011a). The oxidases are expresses in human plaque samples (Zhu et al., 2014), for example, raising the prospect that the excreted H2O2 might shape the oral microbial community.
How do LAB avoid poisoning themselves? They lack [4Fe-4S] dehydratases; aconitase, for example, is absent by virtue of the lack of a TCA cycle. LAB famously accumulate millimolar concentrations of intracellular manganese (Archibald, 1986; Daly et al., 2004), and it is likely that they metallate their mononuclear enzymes with Mn rather than Fe. Thus they constitutively employ the same defensive tactic that E. coli activates only under H2O2 duress (Anjem et al., 2009; Anjem and Imlay, 2012). A Dps homolog keeps free-iron levels low enough that DNA damage is suppressed (Xu et al., 2014). Collectively these devices avoid damage to iron-sulfur dehydratases, mononuclear enzymes, and DNA, so that lactic acid bacteria are largely immune to the H2O2 that they generate.
It has long been suspected that the H2O2 that LAB excrete may suppress the growth of competing bacteria. The effect has been documented in mixed lab cultures (Pericone et al., 2000; Tong et al., 2007; Bao et al., 2017). However, these experiments represent richly fed bacteria in closed environments; in natural habitats H2O2 may be convected away and or degraded by scavenging-proficient cells. Much less H2O2 may accumulate (Margolis, 2009). The accurate measurement of H2O2 in natural LAB environments will be fundamental to understanding its impact upon flora.
A final interesting contrast between LAB and E. coli can be considered. E. coli has only one high-flux enzyme that generates H2O2 stoichiometrically: a monoamine oxidase that catalyzes the first step in phenylethylamine catabolism (Kumar and Imlay, 2013). The key distinction is that, unlike the oxidases in LAB, the E. coli oxidase is localized in the periplasm. Consequently the H2O2 that it produces rapidly leaves the cell through the outer membrane porins and thereby avoids damaging the cytoplasmic iron enzymes.
ROS as an antimicrobial weapon wielded by phagocytes
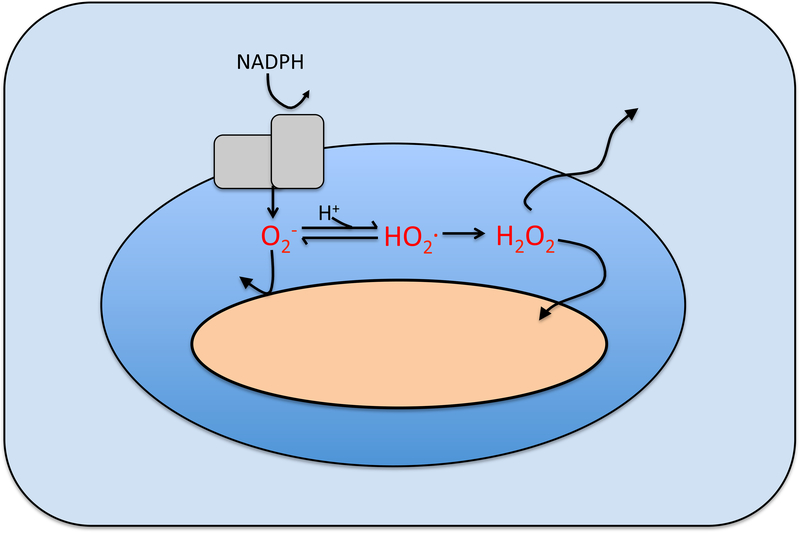
Mammalian phagocytes and plants respond to bacteria by engulfing them and then dousing them with superoxide that the generate with a dedicated NADPH oxidase (Bedard et al., 2007; Kawahara et al., 2007) (Fig. 3). Its role in suppressing microbial growth is clear, as humans and mice that lack the enzyme are vulnerable to infection (Shiloh et al., 1999; Thomas, 2017). However, it has been surprisingly hard to figure out exactly how phagocytic ROS production suppresses microbial growth (Slauch, 2011).
Figure 3. Oxidant formation in phagosomes.
The O2− produced by the host NADPH oxidase cannot penetrate the cytoplasmic membranes of captive bacteria; either it, or its more-reactive protonated form HO2., are believed to injure the extracytoplasmic surface of bacteria. Dismutation produces H2O2 that can penetrate membranes. Tentative calculations suggest that O2−, HO2., and H2O2 levels may be in the ranges of 10–50 μM, 0.1–4 μM, and 1–4 μM, respectively, depending upon phagosomal pH (Imlay, 2009). Modeling of fluxes in neutrophils predicts similar levels (Winterbourn et al., 2006).
The oxidase directly generates superoxide. Because superoxide is anionic (O2−) at neutral pH, it cannot cross bacterial membranes (Lynch and Fridovich, 1978; Korshunov and Imlay, 2002). Phagosomal acidification can partially protonate the superoxide to neutral HO2., which might in principle penetrate into the cytoplasm, but genetic studies generally support the view that superoxide acts upon an as-yet unidentified target either on the cell surface or in the periplasm (Craig and Slauch, 2009). Metalloenzymes of the aconitase or Rpe classes are not present in those regions, raising the possibility that HO2.—a much more reactive oxidant than O2−—might attack a novel target. This idea has proven difficult to pursue, since it is hard to recreate the predicted micromolar levels of HO2. (Imlay, 2009) in experimental systems.
Superoxide spontaneously dismutates to H2O2, and one might wonder whether H2O2 formed in the phagosome exerts toxic effects upon bacteria. However, modeling suggests that H2O2 efflux from the macrophage phagosome is so rapid that, despite the impressive pace of its formation, the steady-state concentration of H2O22 H will rise no higher than a few micromolar (Imlay, 2009). A similar result has been predicted for neutrophil phagosomes (Winterbourn et al., 2006). As was discussed, bacteria can cope with this level of H2O2, as long as they can activate their OxyR response (Li and Imlay, 2018).
This analysis considers bacteria trapped inside isolated macrophages; the extruded H2O2 is treated as disappearing from the system. However, it is conceivable that H2O2 might accumulate on the macroscopic level in inflamed tissue, potentially rising to higher concentrations. An abscess is a plausible example. Actual measurements of H2O2 will be needed to resolve this point. The surprising bottom line is that we still do not know the mechanism by which phagocytic O2− and/or H2O2 suppress microbial growth, nor whether their presence affects by-stander bacteria.
Redox-cycling antibiotics impose intracellular O2− stress
In their initial investigations into the function of SOD, Hassan and Fridovich used redox-cycling compounds to elevate the rate of O2− production inside E. coli (Hassan and Fridovich, 1979). The synthetic viologen paraquat is still commonly used for this purpose: it penetrates into bacteria, oxidizes their redox enzymes, and transfers the electrons to oxygen. The resultant O2− flux is enough to disable ROS-sensitive enzymes and block growth even of wild-type cells (Kuo et al., 1987).
We now realize that such experiments recapitulate natural phenomena. Although paraquat is a synthetic compound, both plants and bacteria synthesize and secrete phenazines and quinones that do the same thing (Turner and Messenger, 1986; Paiva et al., 2003; Inbaraj and Chignell, 2004). Their significance is reflected in the fact that enteric bacteria manifest a response regulon, governed by SoxRS (Greenberg et al., 1990; Tsaneva and Weiss, 1990), that has evolved to defend the cell against such compounds. Although this response was initially believed to be triggered by O2− itself, more recent data indicate that the system is activated when the redox-cycling compounds directly oxidize the [2Fe-2S] cluster of the SoxR regulator (Gu and Imlay, 2011). The activated protein then induces a regulon (Pomposiello et al., 2001) that includes drug-efflux pumps, proteins that diminish cell permeability, and defensive enzymes including SOD (Aiba et al., 1987; Ma et al., 1995; Lee et al., 2009). Interestingly, the system appears to have been laterally inherited and modified from SoxR regulons in the bacteria that produce these compounds, where SoxR controls their secretion and does not involve defensive enzymes (Dietrich et al., 2008).
Thus the new view is that such compounds are natural sources of ROS stress. Their benefit to the producing organisms, however, is a matter of ongoing discussion. On the one hand, they can suppress the growth of competitors. A persuasive example is that walnut trees provide bare ground for their nuts by poisoning undergrowth with the juglone in their fallen leaves. However, workers have proposed alternative functions. The secreted compounds can rescue iron-poor bacteria by reducing and solubilizing iron in mineral deposits (Wang et al., 2011). They can also carry electrons from bacteria embedded deep in anoxic biofilms to outer, oxic regions, thereby enabling respiration at a distance (Glasser et al., 2017). In these views, the toxicity that redox-cycling compounds impose upon by-stander bacteria such as E. coli is largely an accident—albeit one against which the bacterium must defend itself.
Do heat, metals, solvents, and clinical antibiotics create oxidative stress?
In contemporary toxicology it seems that oxidative stress is easy to suspect but hard to prove. Microbiologists have proposed that ROS underlie the toxic effects of many stresses that have no obvious connection to oxidants—from toxin/antitoxin systems (Kolodkin-Gal et al., 2008)and nanoparticle surfaces (Applerot et al., 2012), to high salinity (Mishra et al., 2009) and hydrostatic pressure (Aertsen et al., 2005). The list is so long as to invite caution (Imlay, 2015). The problem is two-fold: at the experimental level, ROS themselves are evanescent and difficult to detect directly; at the conceptual level, it might seem plausible that any stress that physically impairs redox enzymes could cause them to inadvertently transfer electrons to oxygen. How can ROS suspicions be tested?
A popular approach is the use of redox-sensitive dyes. By 2007 skeptical reviewers were able to cite > 2000 papers in which such dyes had been used to diagnose oxidative stress inside living cells (Wardman, 2007). The number is surely far higher now. Various dyes have been shown to be oxidized by hydroxyl radicals—or O2−, or H2O2—in vitro, and the hope is that their oxidation in vivo can be regarded as evidence of the same oxidant (Kalyanaraman et al., 2012). However, problems have arisen. When coupled to cell-sorting, these analyses have sometimes been confounded by changes in bacterial morphology (Renggli et al., 2013; Paulander et al., 2014). Most of the experiments do not carefully quantify dye loading in stressed cells versus their unstressed controls; since stress may alter membrane packing, or influx and efflux energetics, he amount of dye inside sample cells may differ from that in control cells. It is worth considering that a ratiometric sensor may be more appropriate (Bilan et al., 2013; Lin et al., 2013). And rarely have control experiments rigorously established that the dyes are not vulnerable to the oxidation by high-valent metal centers, or countervailing reduction by cellular thiols (Imlay, 2015). These concerns are not intended to refute claims but to invite care when considering the evidence.
Other common tests of whether a phenomenon is driven by oxidants include the use of “antioxidants”—usually thiols or ascorbate—in suppressing it, or measurements of TBARS as evidence of lipid peroxidation. Thiols may be effective scavengers of extracellular H2O2, but they are not much more reactive with intracellular hydroxyl radicals than is everything else inside the cell, and they barely react with superoxide at all (Winterbourn and Metodiewa, 1999). Tests for TBARS (thiobarbituric-acid reactive species) are easy to perform but are not specific to peroxidized lipids; the latter issue is especially pertinent to bacteria, as they have no polyunsaturated fatty acids, which are requisite according to standard models of lipid peroxidation (Bielski et al., 1983).
So what are the most reliable markers of oxidative stress? Induction of the OxyR regulon is a good one, as it trades upon the cell’s own detection system for H2O2. Damage to [4Fe-4S] dehydratases and mononuclear enzymes are known effects. And to approach the problem head-on, the rate of H2O2 production can be directly determined with catalase/peroxidase mutants, if they are available. In the view of the author, these techniques provide the approaches that are currently the most specific and the least prone to artifacts.
Concluding remarks
Over the past forty years, much has been learned about the mechanisms by which O2− and H2O2 can poison bacteria and about the tactics by which bacteria strive to defend themselves. The point of this MiniReview is that we know much less about the natural circumstances in which oxidative stress occurs. In real environments H2O2 stress is likely to be low-micromolar in dose but protracted in time, and laboratory experiments should seek to replicate this regime to obtain the most pertinent outcomes. Meanwhile, we will need to determine H2O2 levels in natural samples if we are to know whether oxidative stress is a real occurrence there. And multiple, direct assays are needed to resolve uncertainties as to whether non-oxidative stresses trigger ROS formation.
Originality-significance statement.
The mechanisms by which oxidants can damage cells are increasingly understood, but the natural circumstances under which they do so remain poorly defined. This manuscript reviews what is known about the level of threat imposed by oxidants that are generated by endogenous, chemical, and biological-warfare processes. The point is to emphasize a key gap in our understanding.
References
- Aertsen A, Spiegeleer PD, Vanoirbeek K, Lavilla M, and Michiels CW (2005) Induction of oxidative stress by high hydrostatic pressure in Escherichia coli. Appl Environ Microbiol 71: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H, Matsuyama S, Mizuno T, and Mizushima S (1987) Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J Bacteriol 169: 3007–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Almiron M, Huisman G, Kolter R, and Storz G (1994) The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13: 265–272. [DOI] [PubMed] [Google Scholar]
- Anjem A, and Imlay JA (2012) Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem 287: 15544–15556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjem A, Varghese S, and Imlay JA (2009) Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol 72: 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applerot G, Lellouche J, Lipovsky A, Nitzan Y, Lubart R, Gedanken A, and Banin E (2012) Understanding the antibacterial mechanism of CuO nanoparticles: revelaing the route of induced oxidative stress. Small 8: 3326–3337. [DOI] [PubMed] [Google Scholar]
- Archibald F (1986) Manganese: its acquisition by and function in the lactic acid bacteria. Crit Rev Microbiol 13: 63–109. [DOI] [PubMed] [Google Scholar]
- Aslund F, Zheng M, Beckwith J, and Storz G (1999) Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA 96: 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Yang J, Soet J.J.d., Liu H, Gao X, Loveren C.v., and Deng D (2017) Factors influencing the competition between Streptococcus oligofermentans and Streptococcus mutans in dual-species biofilms. Caries Res 51: 507–514. [DOI] [PubMed] [Google Scholar]
- Beauchamp C, and Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287. [DOI] [PubMed] [Google Scholar]
- Bedard K, Lardy B, and Krause K-H (2007) NOX family NADPH oxidases: Not just in mammals. Biochemie 89: 1107–1112. [DOI] [PubMed] [Google Scholar]
- Bielski BHJ, Arudi RL, and Sutherland MW (1983) A study of the reactivity of HO2/O2- with unsaturated fatty acids. J Biol Chem 258: 4759–4761. [PubMed] [Google Scholar]
- Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, Gadella TWJ et al. (2013) HyPer-3: A genetically encoded H2O2 probe with improved performance for ratiometric andfluorescence lifetime imaging. ACS Chem Biol 8: 535–542. [DOI] [PubMed] [Google Scholar]
- Boling M, Adler H, and Masker W (1984) Restoration of viability to an Escherichia coli mutant deficient in the 5’ to 3’ exonuclease of DNA polymerase I. J Bacteriol 160: 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, and Touati D (1986) Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J 5: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, and Ryu S (2001) Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113. [DOI] [PubMed] [Google Scholar]
- Christman MF, Morgan RW, Jacobson FS, and Ames BN (1985) Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41: 753–762. [DOI] [PubMed] [Google Scholar]
- Craig M, and Slauch J (2009) Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4: e4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Gaidamakova EK, Matrosova VY, Valilenko A, Zhai M, Venkateswaran A et al. (2004) Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306: 1025–1028. [DOI] [PubMed] [Google Scholar]
- Demple B, and Halbrook J (1983) Inducible repair of oxidative DNA damage in Escherichia coli. Nature 304: 466. [DOI] [PubMed] [Google Scholar]
- Dietrich LE, Teal TK, Price-Whelan A, and Newman DK (2008) Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321: 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espey MG (2013) Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Rad Biol Med 55: 130–140. [DOI] [PubMed] [Google Scholar]
- Flint DH, Tuminello JF, and Emptage MH (1993) The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem 268: 22369–22376. [PubMed] [Google Scholar]
- Gardner PR, and Fridovich I (1991) Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem 266: 1478–1483. [PubMed] [Google Scholar]
- Gardner PR, and Fridovich I (1992) Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J Biol Chem 267: 8757–8763. [PubMed] [Google Scholar]
- Glasser NR, Saunders SH, and Newman DK (2017) The colorful world of extracellular electron shuttles. Annu Rev Microbiol 71: 731–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Monach P, Chou JH, Josephy PD, and Demple B (1990) Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA 87: 6181–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, and Imlay JA (2011) The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol 79: 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, and Imlay JA (2013) Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol 89: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan HM, and Fridovich I (1979) Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys 196: 385–395. [DOI] [PubMed] [Google Scholar]
- Ilari A, Ceci P, Ferrari D, Rossi G, and Chiancone E (2002) Iron incorporation into E. coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem 277: 37619–37623. [DOI] [PubMed] [Google Scholar]
- Imlay JA (2009) Oxidative Stress In EcoSal--Escherichia coli and Salmonella: Cellular and Molecular Biology http://wwwecosalorg. Bock A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nystrom T, Slauch JM, Squires CL, and Ussery D (eds). Washington, D.C.: ASM Press. [Google Scholar]
- Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA (2015) Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr Opin MIcrobiol 24: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, and Linn S (1986) Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol 166: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbaraj JJ, and Chignell CF (2004) Cytotoxic action of juglone and plumbagin: a mechanistic study using HaCaT keratinocytes. Chem Res Toxicol 17: 55–62. [DOI] [PubMed] [Google Scholar]
- Jang S, and Imlay JA (2007) Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem 282: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, and Imlay JA (2010) Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol 78: 1448–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B, Darley-Usmar V, Davies KJA, Dennery PA, Forman HJ, Grisham MB et al. (2012) Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Rad Biol Med 52: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Quinn MT, and Lambeth JD (2007) Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. MBC Evolutionary Biology 7: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Janakiraman A, Slauch JM, and Maguire ME (2002) Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184: 3151–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, and Imlay JA (1996) Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA 93: 13635–13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademian M, and Imlay JA (2017) Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci USA 114: E6922–E6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Sat B, Keshet A, and Engelberg-Kulka H (2008) The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol 6: e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov SS, and Imlay JA (2002) A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of phagocytosed bacteria. Mol Microbiol 43: 95–106. [DOI] [PubMed] [Google Scholar]
- Kumar SR, and Imlay JA (2013) How Escherichia coli tolerates profuse hydrogen peroxide formation by a catabolic pathway. J Bacteriol 195: 4569–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CF, Mashino T, and Fridovich I (1987) α,β-dihydroxyisovalerate dehydratase: a superoxide-sensitive enzyme. J Biol Chem 262: 4724–4727. [PubMed] [Google Scholar]
- Lee J-W, and Helmann JD (2006a) Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem 281: 23567–23578. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yeo WS, and Roe JH (2004) Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hdyrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol Microbiol 51: 1745–1755. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee KL, Yeo WS, Park SJ, and Roe JH (2009) SoxRS-mediated lipopolysaccharide modification enhances resistance against multiple drugs in Escherichia coli. J Bacteriol 191: 4441–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, and Helmann JD (2006b) The PerR transcription factor senses H2O2 by metal-catalyzed histidine oxidation. Nature 440: 363–367. [DOI] [PubMed] [Google Scholar]
- Lesser MP (2006) Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol 68: 253–278. [DOI] [PubMed] [Google Scholar]
- Li X, and Imlay JA (2018) Improved measurements of scant hydrogen peroxide enable experiments that define its threshold of toxicity for Escherichia coli. Free Rad Biol Med 120: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin VS, Dickinson BC, and Chang CJ (2013) Boronate-based fluorescent probes: imaging hydrogen peroxide in living systems. Meth Enzymol 526: 19–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev SI, and Fridovich I (1993) Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch Biochem Biophys 301: 379–384. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramsey MM, Chen X, Koley D, Whiteley M, and Bard AJ (2011a) Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci USA 108: 2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bauer SC, and Imlay JA (2011b) The YaaA protein of the Escherichia coli OxyR regulon lessens hydrogen peroxide toxicity by diminishing the amount of intracellular unincorporated iron. J Bacteriol 193: 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, and Imlay JA (2017) The fumarate reductase of Bacteroides thetaiotaomicron, unlike that of Escherichia coli, is configured so that it does not generate reactive oxygen species. mBio 8: e01873–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Sethu R, and Imlay JA (2018) Endogenous superoxide is a key effector of the oxygen sensitivity of a model obligate anaerobe. Proc Natl Acad Sci USA 115: e3266–e3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch R, and Fridovich I (1978) Permeation of the erythrocyte stroma by superoxide radical. J Biol Chem 253: 4697–4699. [PubMed] [Google Scholar]
- Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, and Hearst JE (1995) Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol 16: 45–55. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR, and Cummings JH (1992) Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72: 57–64. [DOI] [PubMed] [Google Scholar]
- Mancini S, and Imlay JA (2015) The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol 96: 744–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis E (2009) Hydrogen peroxide-mediated interference competition by Streptococcus pneumoniae has no significant effect on Staphylococcus aureus nasal colonization of neonatal rats. J Bacteriol 191: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V, Strickland S, Mayhew SG, Howell LG, Engel PC, Matthews RG et al. (1969) The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem Biophys Res Commun 36: 891–897. [DOI] [PubMed] [Google Scholar]
- Mesle MM, Beam JP, Jay ZJ, Bodle B, Bogenschutz E, and Inskeep WP (2017) Hydrogen peroxide cycling in high-temperature acidic geothermal springs and potential implications for oxidative stress response. Front Mar Sci 4: 130. [Google Scholar]
- Messner KR, and Imlay JA (1999) The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem 274: 10119–10128. [DOI] [PubMed] [Google Scholar]
- Mishra S, and Imlay JA (2013) An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol Microbiol 90: 1356–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra Y, Chaurasia N, and Rai LC (2009) AhpC (alkyl hydroperoxide reductase) from Anabaena sp. PCC 7120 protects Escherichia coli from multiple abiotic stresses. Biochem Biophys Res Commun 381: 606–611. [DOI] [PubMed] [Google Scholar]
- Morimyo M (1982) Anaerobic incubation enhances the colony formation of a polA recB strain of Escherichia coli K-12. J Bacteriol 152: 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Maegawa S, Shigeno S, Fujikura K, and Toyohara H (2018) Highly sensitive avoidance plays a key role in sensory adaptation to deep-sea hydrothermal vent environments. PLoS ONE 13: e0189902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paiva SR, Figueiredo MR, Aragão TV, and Kaplan MAC (2003) Antimicrobial activity in vitro of plumbagin isolated from Plumbago species. Mem Inst Oswaldo Cruz 98: 959–961. [DOI] [PubMed] [Google Scholar]
- Pan N, and Imlay JA (2001) How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron? Mol Microbiol 39: 1562–1571. [DOI] [PubMed] [Google Scholar]
- Park S, You X, and Imlay JA (2005) Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc Natl Acad Sci USA 102: 9317–9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JD, Poole RK, and Green J (2007) The Escherichia coli yhjA gene, encoding a predicted cytochrome c peroxidase, is regulated by FNR and OxyR. Microbiology 153: 1499–1507. [DOI] [PubMed] [Google Scholar]
- Paulander W, Wang Y, Folkesson A, Charbon G, Lobner-Olesen A, and Ingmer H (2014) Bactericidal antibiotics increase hydroxyphenyl fluorescein signal by altering cell morphology. PLoS One 9: e92231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone CD, Overweg K, Hermans PWM, and Weiser JN (2000) Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus penumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68: 3990–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericone CD, Park S, Imlay JA, and Weiser JN (2003) Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J Bacteriol 185: 6815–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher MCL, Beatty ER, and Cummings JH (2000) The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomposiello PJ, Bennik MH, and Demple B (2001) Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183: 3890–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renggli S, Keck W, Jenal U, and Ritz D (2013) Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J Bacteriol 195: 4067–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Nakanishi M, Yoshiyama K, and Maki H (2006) Impact of reactive oxygen species on spontaneous mutagenesis in Escherichi coli. Genes to Cells 11: 767–778. [DOI] [PubMed] [Google Scholar]
- Schlesier J, Rohde M, Gerhardt S, and Einsle O (2015) A conformational switch triggers nitrogenase protection from oxygen damage by Shethna protein II (FeSII). J Am Chem Soc 138: 239–247. [DOI] [PubMed] [Google Scholar]
- Seaver LC, and Imlay JA (2001a) Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183: 7173–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, and Imlay JA (2001b) Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol 183: 7182–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Iida K, Saito M, Nakayama H, and Yoshida S (2004) Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J Bacteriol 186: 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller A-F, Teixeira M, and Valentine JS (2014) Superoxide dismutases and superoxide reductases. Chem Rev 114: 3854–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh MU, MacMicking JD, Nicholson S, Brause JE, Potter S, Marino M et al. (1999) Phenotype of mice and macrophages defiient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10: 29–38. [DOI] [PubMed] [Google Scholar]
- Slauch JM (2011) How does the oxidative burst of macrophages kill bacteria? Mol Microbiol 80: 580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota JM, and Imlay JA (2011) Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci USA 108: 5402–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota JM, Gu M, and Imlay JA (2014) Intracellular hydrogen peroxide and superoxide poison 3-deoxy-D-arabinoheptulosonate 7-phosphate synthase, the first committed enzyme in the aromatic biosynthetic pathway of Escherichia coli. J Bacteriol 196: 1980–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, and Masure HR (1996) Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19: 803–813. [DOI] [PubMed] [Google Scholar]
- Stiebritz MT, and Reiher M (2012) Hydrogenases and oxygen. Chem Sci 3: 1739–1751. [Google Scholar]
- Thomas DC (2017) The phagocyte respiratory burst: Historical perspectives and recent advances. Immunology Letters 192: 88–96. [DOI] [PubMed] [Google Scholar]
- Tong H, Chen W, Merritt J, Qi F, Shi W, and Dong X (2007) Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol 63: 872–880. [DOI] [PubMed] [Google Scholar]
- Tsaneva IR, and Weiss B (1990) soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol 172: 4197–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM, and Messenger AJ (1986) Occurrence, biochemistry, and physiology of phenazine pigment production. Adv Microb Physiol 27: 211–275. [DOI] [PubMed] [Google Scholar]
- Varghese S, Wu A, Park S, Imlay KRC, and Imlay JA (2007) Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol 64: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita N, Hatchikian EC, Nouailler M, Dolla A, and Pieulle L (2008) Disulfide bond-dependent mechanism of protection against oxidative stress in pyruvate-ferredoxin oxioreductase of anaerobic Desulfovibrio bacteria. Biochemistry 47: 957–964. [DOI] [PubMed] [Google Scholar]
- Wagner AF, Frey M, Neugebauer FA, Schafer W, and Knappe J (1992) The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci U S A 89: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, and Newman DK (2011) Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193: 3606–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardman P (2007) Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: Progress, pitfalls, and prospects. Free Rad Biol Med 43: 995–1022. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Hinman NW, and Sheridan RP (2000a) Hydrogen peroxide formation and decay in iron-rich geothermal waters: the relative roles of abiotic and biotic mechanisms. Photochem Photobiol 71: 691–699. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Hinman NW, Cooper WJ, and Brown CF (2000b) Hydrogen peroxide cycling in surface geothermal waters of Yellowstone National Park. Environ Sci Technol 34: 2655–2662. [Google Scholar]
- Winterbourn CC, and Metodiewa D (1999) Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Rad Biol Med 27: 322–328. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB, Livesey JH, and Kettle AJ (2006) Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome. Implications for microbial killing. J Biol Chem 281: 39860–39869. [DOI] [PubMed] [Google Scholar]
- Xu Y, Itzek A, and Kreth J (2014) Comparison of genes requires for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. MIcrobiology 160: 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, and Storz G (2001) DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Xu Y, Ferretti JJ, and Kreth J (2014) Probing oral microbial functionality--expression of spxB in plaque samples. PLoS ONE 9: e86685. [DOI] [PMC free article] [PubMed] [Google Scholar]