Abstract
Emerging links between gut microbiota and diseases of aging point to possible shared immune, metabolic, and cellular damage mechanisms, operating long before diseases manifest. We conducted 16S rRNA sequencing of fecal samples collected from a subsample (n = 668) of Add Health Wave V, a nationally representative longitudinal study of adults aged 32–42. An overlapping subsample (n = 345) included whole-blood RNA-seq. We examined associations between fecal taxonomic abundances and dried blood spot-based markers of lipid and glucose homeostasis and C-reactive protein (measured in Wave IV), as well as gene expression markers of inflammation, cellular damage, immune cell composition, and transcriptomic age (measured in Wave V), using Bayesian hierarchical models adjusted for potential confounders. We additionally estimated a co-abundance network between inflammation-related genes and bacterial taxa using penalized Gaussian graphical models. Strong and consistent microbiota associations emerged for HbA1c, glucose, C-reactive protein, and principal components of genes upregulated in inflammation, DNA repair, and reactive oxygen species, with Streptococcus infantis, Pseudomonas spp., and Peptoniphilus as major players for each. This pattern was largely echoed (though attenuated) for immunological cell composition gene sets, and only Serratia varied meaningfully by transcriptomic age. Network co-abundance indicated relationships between Prevotella sp., Bacteroides sp., and Ruminococcus sp. and gut immune/metabolic regulatory activity, and Ruminococcus sp, Dialister, and Butyrivibrio crossotus with balance between Th1 and Th2 inflammation. In conclusion, many common associations between microbiota and major physiologic aging mechanisms are evident in early-mid adulthood and suggest avenues for early detection and prevention of accelerated aging.
Keywords: Gastrointestinal microbiota, Cellular senescence, Lipid metabolism, Glucose metabolism, Bayesian analysis
Commensal bacteria and other microorganisms in the human gut represent a co-evolved community, hypothesized to have a central role in the development and maintenance of many human physiological systems across the life course, including the metabolic, neuroendocrine, and immune systems (1). Relatedly, accumulating research has linked between-person variation in microbiota composition to conditions of aging, including cardiometabolic diseases (2), dementia (3), frailty (4), and cancers (5). Interconnected aging-related and inflammatory mechanisms proposed to underlie these conditions include accumulation of aged immune cells, metabolic dysregulation, and oxidative stress-mediated macromolecular damage (6).
Multiple intestinal bacterial metabolites, including toll-like receptor agonists, short-chain fatty acids (SCFAs), and secondary bile acids, have direct effects both on immune/inflammatory processes and metabolic functions like insulin sensitivity, as well as immune-mediated metabolic effects (7). Moreover, murine experiments have illustrated that commensals are necessary for regulating intestinal inflammation (8–10). Specifically, bacteria in the murine gut aid in regulating inflammatory responses by decreasing the level of responsiveness of pattern-recognition receptors (PPRs) to lipopolysaccharide (LPS) (8), a membrane component of Gram-negative bacteria which otherwise induces inflammation and insulin resistance (9). Murine gut commensals also participate in immune regulation by promoting differentiation of regulatory T-cells (Treg) and production of IL-10 (10), a major anti-inflammatory cytokine. Conversely, age-related chronic inflammation appears causally dependent on gut bacteria, as the raised circulating IL-6 seen in aged mice (similarly to humans) is absent in germ-free mice (11), whereas transfer of cecal contents from aged conventional mice to germ-free mice results in elevation of specific Th subsets consistent with inflammaging (12). Importantly, there is also evidence of bidirectionality in relationships between immune and metabolic health and the murine gut microbiota—for example, both genetically determined obesity (13) and immunodeficiency (14) can alter gut commensal composition.
In humans, there is preliminary evidence to support the role of gut bacteria in immune and metabolic function. A small randomized study showed that antibiotic exposure can alter response to influenza vaccine—causing decreased antibody production and raised systemic inflammation—a response echoing immunosenescence (15). Another randomized study found that a probiotic blend of Lactobacillus, Streptococcus, and Bifidobacterium spp. can decrease inflammatory markers in overweight adults (16). Moreover, an accumulating body of human research has linked gut microbiota composition to metabolic disorders, including type 2 diabetes, obesity, and associated systemic inflammation (17).
Thus far, most human research on how the microbiota affect physiologic aging and age-related disease has been conducted in older adults (18,19). However, biological aging likely begins early in the life course, with immune and metabolic signals of aging emerging as early as the 30s (20). Detailing relationships between gut microbiota and markers of immune and metabolic aging processes in younger populations is, therefore, an important step towards understanding how age-related changes in disease status and microbiota interact over the life course, ultimately influencing health status in older age. This study examines early adult associations between the gut microbiome and hallmarks of aging and immunity, including biomarkers of inflammation, immunosenescence, oxidative stress, and cardiometabolic health.
Methods
Study Design, Participants, Informed Consent
This analysis draws on data from the National Longitudinal Study of Adolescent to Adult Health (Add Health), a nationally representative longitudinal cohort of more than 20,000 individuals who were in grades 7–12 in 1994–1995, from 80 communities and 134 middle and high schools throughout the United States, with oversampling of highly educated African Americans and multiple Latinx and Asian-American ethnic subgroups. Add Health has followed participants through adolescence into adulthood in five waves; further design details of Add Health are published (21). In each wave, in-person interviews collected extensive information about demographics, the social environment, and health, with immune/inflammatory, cardiometabolic, and gene expression biomarker data added in Waves IV and V. This study draws on data from Waves IV and V, when participants were ages 24–32 and 32–42, respectively.
Wave V embedded an ancillary study to collect tongue and stool microbiome specimens on a subsample of participants. Wave V data collection was divided into four sampling groups (Samples 1, 2A, 2B, and 3); the eligible pool for the microbiome subsample was nationally representative and included those who accepted the microbiome pack from either the biomarker home visit in Sample 1 or the in-person interview in Sample 2B (n = 2,341). In sum, n = 763 (response rate, 32.5%) returned their mail-in self-collection kits, and n = 706 (92.5%) provided sufficient fecal material for sequencing. Both Add Health Wave V and the microbiome ancillary study were approved by the Institutional Review Board of the University of North Carolina-Chapel Hill, and all participants provided written or digital informed consent.
Fecal Microbiome Sample Collection and Processing
Stool specimens were collected via mail-in self-collection kit. Respondents were asked to dab fecal material on the “first wipe” toilet paper with dual cotton swabs (BBL CultureSwab EZ Sterile Kits BD, New Jersey) simultaneously before placing them in the tube containing desiccant and sealing with the supplied stopper. Specimen tubes were shipped directly to CU Boulder (Krauter lab) with transit times of <4 days and immediately stored frozen at −80°C. We used PCR to amplify the V4 region of the 16S rDNA using universal primers. Amplicons were sequenced using the Illumina MiSeq Personal Sequencer and amplicon sequence variants were determined using the Deblur pipeline and clustered into operational taxonomic units (OTUs) at 97% similarity, with closed-reference taxonomic assignment using GreenGenes. Further details of the microbial DNA processing, storage, sequencing, and data management are given in the Supplementary eMethods.
Measurement of Biomarkers and Medications
As part of Wave IV, dried blood spots were assayed for glucose (mg/dl), low-density lipoprotein (LDL), high-density lipoprotein (HDL), and high-sensitivity c-reactive protein (CRP), and HbA1c (%). Self-reported antidiabetic, antihyperlipidemic, and anti-inflammatory (COX-2 inhibitors, NSAIDS, inhaled corticosteroids, corticotropins/glucocorticoids, antirheumatic/antipsoriatics, or immunosuppressives) medications taken in the past 4 weeks were recorded at the same visit. Further details of the dried blood spot collection and assays are given in the Supplementary eMethods.
Measurement of Gene Expression
A separate random subsample of 5,000 participants in Wave V was selected for whole-blood RNA-seq transcriptome analysis (currently, n = 1,132 samples have been fully processed.) Details about the venous blood sampling, RNA isolation, and sequencing are given in the Supplementary eMethods. We created gene expression variables based on nine Molecular Signature Database gene sets the Hallmark sets for inflammation, reactive oxygen species (ROS), and DNA repair (22); as well as immunological signatures for naive versus effector CD8+; CD4+ versus CD8+; Th1 versus naive CD4+; Treg versus naive CD4+; naive versus KLRG1 high CD8+; and CD57+ versus CD57- NK (23). To construct these variables, we used the first principal component of the centered log-ratio transformed expression values of all genes in each expression set; a similar approach has been used previously (24). Additionally, we computed a measure of transcriptomic age, including 1,497 genes developed previously based on a large meta-analysis (25). This measure is a model-based prediction of the expected age of a person given their gene expression, which we computed using the linear regression coefficients provided in the supplementary data of the article by Peters et al. (25). The final variable used in analyses was the difference between their transcriptomic age and chronological age (hereafter, Δage).
Confounders
All models adjust for age, sex, and educational attainment (coded categorically as high school degree or less, some college or vocational/technical school, bachelor’s degree, or master’s/doctoral/professional degree); models for HDL and LDL additionally adjust for lipid medications in Wave IV; models for glucose and HbA1c additionally adjust for glucose medications in Wave IV; models for log-CRP additionally adjust for lipid and inflammation medications in Wave IV; models for all gene expression markers additionally adjust for medications for asthma, lipids, glucose, and psychiatric conditions in Wave V.
Statistical Analysis
All statistical analyses were conducted in R v. 3.5.1 (26), and code used to perform all analyses is available in a GitHub repository (27). This analysis focused on estimating associations between taxonomic abundances and biomarkers of immune and cardiometabolic aging, which were HbA1c, glucose, HDL, LDL, log-CRP, and the selected gene set principal component vectors, described above. Each biomarker was treated in a separate model. We implemented a Bayesian hierarchical log-linear modeling approach, implemented in the R packages “DESeq2” v 1.22.2 (28) and “ashr” v 2.2–32 (29). The model for the ith individual and jth taxon can be written as:
The response variable yij is the observed count of bacterial taxon j for person i, regressed on , the biomarker of interest, and potential confounders, Because we standardized all biomarkers to a variance of one, the biomarker regressor coefficient represents the log2 ratio change of a given taxon’s relative abundance that would be expected for a standard deviation change in biomarker value (hereafter, log2-fold change or logFC). Because the number of reads varies between samples, we use an offset in the model to accomplish normalization. As offset, instead of DESeq2’s default, we use a measure called the geometric mean of pairwise ratios (30), a function of the number of reads that accounts for the compositional and zero-inflated nature of microbiome sequencing data. Estimation of the dispersion parameter is improved by sharing information across samples; that is, each estimate is shrunken towards , a smooth function of the mean normalized count of taxon j. Details of the estimation procedure are given in references (28) and (29).
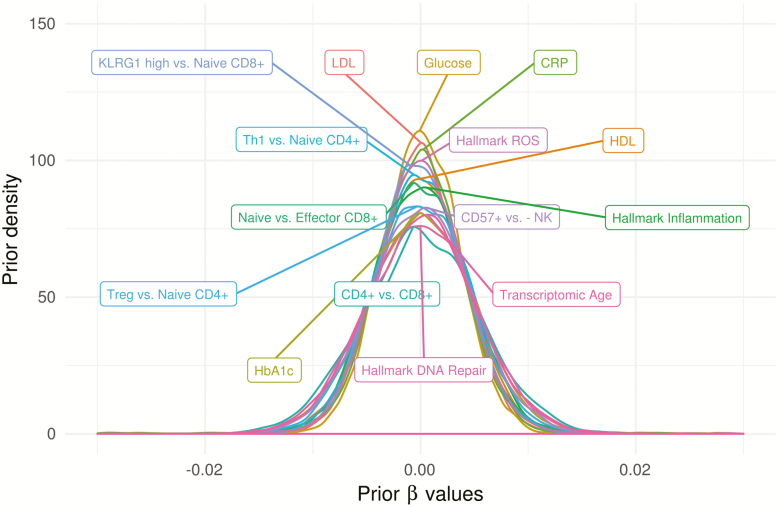
This is a Bayesian hierarchical model in the sense that a prior probability distribution, estimated from the data, is placed on each biomarker’s coefficient. Specifically, the prior on the vector is the best-fitting zero-mean Gaussian mixture estimated from the maximum likelihood estimates, (29). Importantly, this prior accomplishes two goals: (a) it encodes our belief that few taxa are likely to have strong associations with biomarkers, and (b) it strongly regularizes noisy estimates to prevent undue confidence in discoveries. Figure 1 shows samples from priors for for each biomarker, illustrating that these are strongly regularizing priors: the range of coefficients estimated using maximum likelihood is much wider (approximately −5 to 5) across taxonomic levels and biomarkers, with priors contributing on average 7.5 times as much information as the data.
Figure 1.
Adaptive prior distributions for β for each phenotype, for models estimated at the genus level, illustrating zero-centering and strong regularization such that very low prior probability is placed on absolute values greater than 0.01 for a standard deviation change in continuous variables. Although only genus priors are shown, priors at other taxonomic levels are nearly identical.
We estimated these models for all taxa present in the dataset after collapsing separately at the species, genus, family, and phylum levels, and after filtering out low-prevalence taxa; in sum, we performed 3,645 statistical tests. We did not perform multiplicity correction. Instead, we rely on the s-value, the posterior probability that the sign of a coefficient is incorrect, as a continuous descriptive measure corresponding to relative confidence in a discovery. This value is estimated directly from the posterior distribution and represents the proportion of the probability mass lying on the same side of zero as the posterior mean. Because of the strong shrinkage, this value does not require multiplicity correction and is more conservative than the Benjamini–Hochberg false discovery rate in our data (not shown). In some cases, we use cutoff thresholds of the s-value and/or logFC, which are selected arbitrarily to select a manageable number of findings for discussion and/or highlighting in figures. As such, these cutoffs should be understood as heuristic rather than confirmatory.
Bacteria / Blood Gene Expression Network
To estimate a co-occurrence network among bacterial taxa levels and inflammation-related gene expression levels, we used Gaussian graphical modeling (GGM), implemented in the R package “huge” v 1.3.0 (31). GGMs have been used extensively to infer interaction networks in multi-omics data (32) and in microbiome data alone (33), but to the best of our knowledge, this is the first study to apply GGMs to combined 16S and RNA-Seq data. GGMs estimate pairwise covariances between nodes (genes and taxa), conditional on all other nodes, using regularization to shrink noisy estimates towards zero. To be as granular as possible, we considered only raw OTUs, not higher levels of taxonomic classification. Given the small sample size for this analysis (n = 345), we selected a limited set of genes broadly related to inflammation pathways by searching Gene Ontology for acute or chronic inflammation, response to antigenic stimuli or wounding, leukocyte activation & migration, neuroinflammation, and mediators of cytokine, histamine, serotonin, nitric oxide, and respiratory burst pathways (34). The search revealed 138 unique genes, of which 122 were expressed in any of our samples; these 122 were used to estimate the network (see Supplementary Table S1, (27) for the full list). Some covariate values were missing for between 3% and 12% of participants, so we first performed single imputation using fully conditional specification for age, sex, all medication variables, educational attainment, and all dried blood spot values used in the main analysis, performed using the R package “mice” v. 3.1.0 (35). To further reduce the risk of chance discoveries, we randomly split the data into a discovery set of n = 177 (used for initial estimation) and a replication set of n = 168.
Before splitting, we transformed the data according to the centered log-ratio (log of the count, divided by the geometric mean of counts in a sample), performed separately for OTUs and genes. We then filtered out OTUs and genes present in fewer than 10 individuals in the full data set. We then applied the nonparanormal transformation (a nonparametric transformation which relaxes the multivariate Gaussian assumption in GGMs (36)) to the entire data matrix. We adjusted for potential confounders by taking residuals from a linear regression model for each nonparanormal, centered log ratio-transformed OTU and gene regressed on age, sex, educational attainment, medications for lipids, glucose, and inflammation in Wave IV, and medications for lipids, glucose, asthma, and mental health in Wave V. These residuals were the ultimate input into the model.
Model selection in GGMs requires selection of a penalty parameter denoted , which controls the level of sparsity in the resulting network. We performed model selection in the discovery set as follows: to select the GGM penalty , we first selected the values for which the degree distribution fit to a scale-free topology had R2 greater than 0.6 (using the R package “WCGNA” v. 1.68 (37)). This initial step restricts the set of biologically plausible networks. Then we fit GGMs to a grid of values in this range using the Meinshausen-Buhlmann method and selected the model based on the STARS criterion, which maximizes the edge stability under 100 bootstrap resamples. After model selection on the discovery set, we fit a GGM to the replication data using the same value of . We considered the final network to be only the replicated edges.
Results
Descriptive Statistics
The complete microbiome subsample of Add Health Wave V includes fecal samples from n = 706 participants, and n = 668 after removing 38 samples with less than 1,000 reads. Of these, n = 345 also overlapped with the peripheral blood gene expression subsample. Because some participants were missing covariate data, final analytic sample sizes range from n = 565–632 for dried blood spot and n = 342 for gene expression measures. Although Add Health is nationally representative, the microbiome subsample has a higher proportion female (65% vs. 50%), non-Hispanic White (69% vs. 53%), and college graduates (46 vs. 31%) compared to the total sample. Restricting attention to the microbiome subsample, a substantial number (30%) had HbA1c > 5.6%, indicating higher than average diabetes risk (mean = 5.6, SD = 0.8), and a large percentage (44%) had elevated (>3 mg/L) CRP (geometric mean = 2.1, SD = 4.2). Because Δage (25) is scaled to the age distribution in the sample, the mean of Δage was 0; 95% of the sample fell in the range (-3.8, +5.7).
Correlations between biomarkers and the general marker of Shannon diversity (adjusted for confounders) were small (all with absolute value < 0.05 or with large standard errors), and we did not observe large shifts in compositionally robust distances for any of the aging/disease biomarkers (R2 < 0.35%, distance-based redundancy analysis on Aitchison distances, also adjusted for confounders).
Differential Abundance
After removing samples with <1,000 reads, samples had a mean ± SD 11,543 ± 13,216 reads, with an average of 173 unique taxa detected per sample. After filtering taxa to keep only those with a count greater than 3 in n = 25 or more samples, we detected a total of 11 phyla, 59 families, 95 genera, and 72 species. We applied this same filter before calculating all the results that follow.
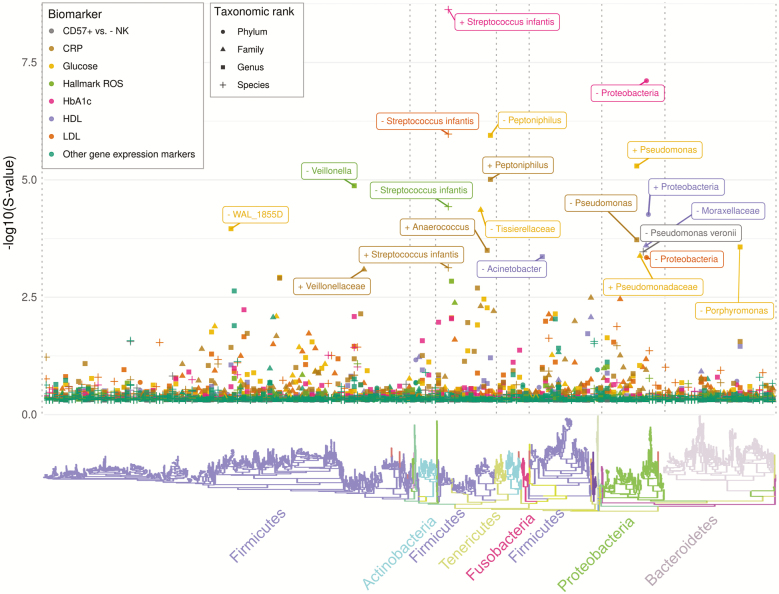
Figure 2 shows a Manhattan plot of the negative log10-transformed s-value (posterior probability of sign error), for reads collapsed at the species, genus, family, and phylum level, arranged according to the phylogenetic tree to illustrate the phylogenetic distribution of estimates. Focusing on the s-value (vs. the log2-fold change estimate) highlights the associations for which we have the highest level of confidence in the sign (direction of association)—this does not suggest these are large associations, only that they are estimated with precision. Overall, we have high confidence in the sign for many taxa—for example, there are 47 taxon-biomarker associations for which we are more than 99% certain of the sign (ie, direction of association), given our modeling assumptions hold. The majority of confidently estimated associations are for taxa that belong to the phylum Firmicutes (followed by Proteobacteria) and the majority are at the genus level (eg, 55% of those with s-values less than 5% are genus-level associations, but the same pattern holds regardless of the threshold). Most of the highly confident associations are for HbA1c, glucose, LDL, HDL, and CRP (all measured in Wave IV), although a few are for the ROS and CD57+ versus - NK gene sets (measured in Wave V). Notably, confidence in the associations with most Wave 5 immunological gene expression signatures is much lower (Th1 vs. naive CD4+, Treg vs. naive CD4+, and CD4+ vs. CD8+ each had all s-values greater than 23%), which may suggest these are noisy measures in this dataset, or may reflect the sample size available for gene expression.
Figure 2.
Phylogenetic distribution of posterior sign confidence for log-linear model coefficients for all phenotypes and at all taxonomic levels. A large –log10 S-value (y-axis) indicates low posterior probability of incorrectly declaring an association to be positive or negative. A non-random distribution across the phylogenetic tree is evident, with most high-confidence estimates in Firmicutes and Proteobacteria. To maximize figure readability, we labeled taxa with –log10(S-value) greater than 3 (less than 1 in 1,000 chance of sign error). The direction of association is indicated by + or − in the label.
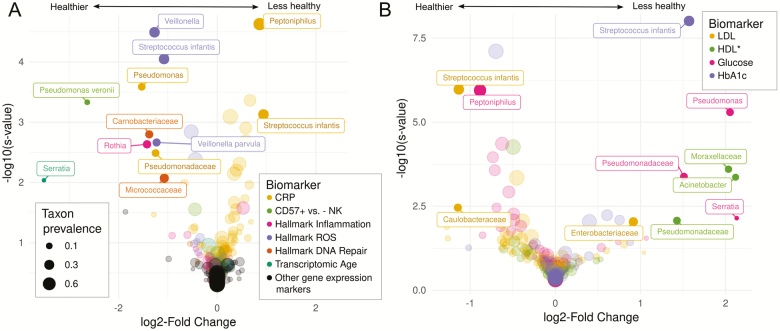
Figure 3 shows volcano plots, where strength of association (log2-fold change [logFC]) is elucidated. For figure readability, attention is restricted to associations with relatively large magnitude and precision; defined as those with an estimated logFC with an absolute value greater than 0.8 and s-value less than 0.01. That is, these are taxa for which we are 99% sure of the direction (given assumptions), and where a standard deviation change in the blood biomarker is estimated to be associated with at least 20.8 = 1.75 times greater or lesser relative abundance of the taxon.
Figure 3.
Volcano plots, showing beta estimates (posterior mode of log2-fold change) on the x-axis and posterior sign confidence (−log10(s-value)) on the y axis, for (A) immune and gene expression markers, and (B) metabolic markers. We required an absolute value of beta greater than 1 and a sign error probability less than 0.01 in order to be labeled. These cutoffs are selected to maximize figure readability. *HDL has been inverse coded so that higher values indicate lower HDL, to ease interpretation of taxa as related to health versus disease.
An overall pattern in Figure 3 is that all strong associations between gut bacteria and DNA repair- and ROS-related gene expression signatures in Wave V are negative, suggesting many gut bacteria may be specifically associated with reduced cellular aging. In contrast, there are many gut bacteria positively and negatively associated with Wave IV HbA1c, LDL, glucose, and CRP, suggesting some harmful and some beneficial associations of certain common gut commensals with metabolic health. Another pattern evident in Figure 3 is that nearly every clade that demonstrates a strong association with more than one biomarker points to potential contradictory effects. For example, Pseudomonas and Pseudomonadaceae correlate with healthier CRP and CD57+ profiles, but worse lipid and glucose measures. A similar pattern is visible for S. infantis and Peptoniphilus.
Here, we report associations as logFC estimate (posterior mode) and a 90% highest posterior density interval in brackets. Several short-chain fatty acid-producing clades in Firmicutes show substantial associations with several biomarkers. For example, Streptococcus infantis is a prevalent (35%), low abundance (<1% average) acetate producer that is associated with higher HbA1c (1.6 [1.2, 2.0]) and CRP (0.9 [0.6, 1.3]), but lower LDL (−1.1 [−1.5, −0.8]) and ROS (−1.1 [−1.4, −0.7]). Peptoniphilus is also a common (54%) low abundance (<1% average) butyrate-producing Firmicutes member associated with lower glucose (−0.9 [−1.2, −0.6]) and higher CRP (0.9 [0.6, 1.1]). Additionally, several Proteobacteria clades appear. Pseudomonas, its family Pseudomonadaceae, or the species P. veronii have associations with healthier profiles of CRP (Pseudomonas: −1.5 [−2.2, −0.9]; Pseudomonadaceae: −1.2 [−2.0, −0.6]) and CD57+ versus - NK (P. veronii: −2.6 [−3.5, −1.8]), but worse profiles of naive versus KLRG1 high CD8+ (P. veronii: −1.9 [−3.0, −0.9]), hallmark inflammation gene expression (P. veronii: 1.6 [0.6, 2.6]), glucose (Pseudomonas: 2.1 [1.4, 2.7]), and HDL (Pseudomonadaceae: −1.4 [−2.2, −0.8], Pseudomonas: −1.2 [−1.8, 0.0]). Serratia and its family Enterobacteriaceae appear associated with higher LDL (Serratia: 1.0 [0.0, 2.4]; Enterobacteriaceae: 0.9 [0.4, 1.4]) and glucose (Serratia: 2.1 [0.7, 3.5]), but lower Δage (−3.5 [−5.0, −2.2]). Pseudomonas and Serratia are both relatively rare (<14%) members of Proteobacteria, which is indeed the only confidently estimated phylum-level association (not labeled in Figure 3)—this phylum correlates with healthier profiles of HbA1c (−0.7 [−0.9, −0.5]), LDL (−0.5 [−0.7, −0.4]) and HDL (0.5 [0.3, 0.7]), and is also weakly related to lower Δage (−0.2 [−0.3, 0.0]). Notably absent from this list are well-studied butyrate-producing anaerobes in Clostridium clusters IV and XIVa making up significant portions of individuals’ microbiomes, such as Faecalibacterium, Ruminococcus, Roseburia, Blautia, and Coprocuccus, all of which have logFC estimates close to zero and narrow posterior intervals for all biomarkers (Supplementary Table S2, (27)). Also notably, many immunological gene expression markers (CD4+ vs. CD8+, Th1 vs. naive CD4+, and Treg vs. naive CD4) did not demonstrate associations with sufficient confidence or size to be highlighted in either figure, although their microbiome signatures frequently correlated strongly with other markers in expected directions (Supplementary eFigure 1). All taxon-biomarker associations are listed in Supplementary Table S2, (27).
Bacteria / Gene Expression Network
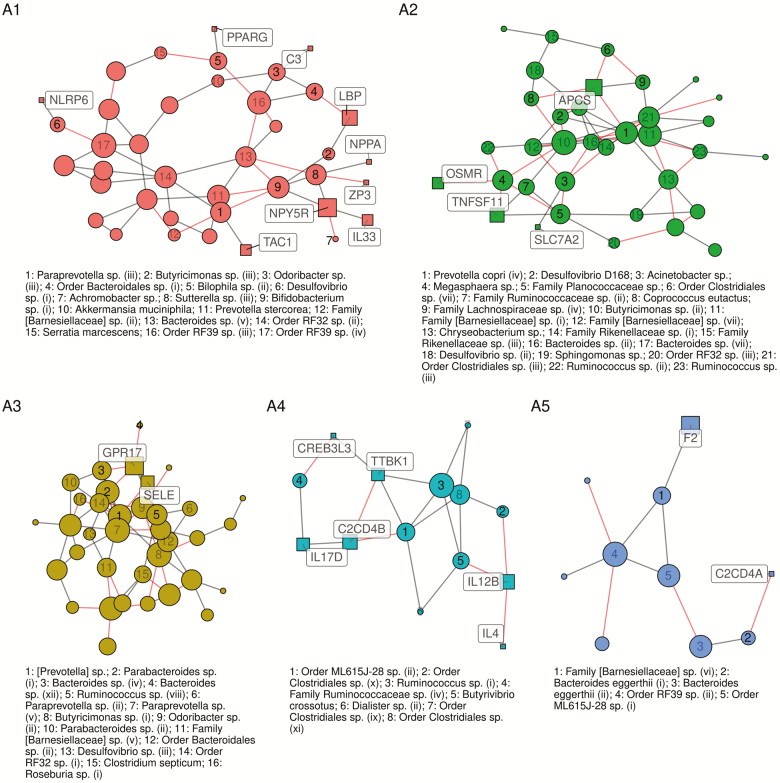
The network fit to the discovery data (n = 177) revealed a sparse (edge density = 2.2%, average degree = 7.8) network with 1364 total edges, 149 of which were between OTUs and genes. The final network (discovery ∩ replication) had 575 edges and 53 OTU-gene edges, for an overall replication rate of 42% and an OTU-gene edge replication rate of 36%, with greater sparsity as a result (edge density = 1.7%, average degree=4.4), and strong modularity (68%, Louvain’s community detection). Of 10 clusters identified with more than two nodes, five clusters (A1–A5) contained both OTUs and genes, four (B1–B4) contained only OTUs, and two (C1–C2) contained only genes. Note that these clusters are not truly separate; clusters A1–A5 each share between 1 and 13 edges with one another (A1 and A2 are especially well connected), suggesting many biological processes in common. Because primary interest is in gene-bacteria interconnectedness, we focus attention on clusters A1–A5 containing both OTUs and genes, which are illustrated in Figure 4 (the entire network is shown in Supplementary eFigure 2, and all members are listed in Supplementary eTables 3 and 4, (27)).
Figure 4.
Estimated conditional correlation network between OTUs and a curated set of inflammation-related genes expressed in whole blood, estimated using a penalized Gaussian graphical model and adjusting for age, sex, and medications by residualizing. Colors correspond to clusters flagged by Levain’s community detection algorithm. Cluster identifiers indicated in large text are also referenced in Supplementary eTables 3 and 4, (27). Size of nodes corresponds to degree—larger nodes have more connections than smaller nodes. Square nodes are genes, circular nodes are bacterial taxa.
Cluster A1 highlights several important regulators of intestinal metabolism and type 1 inflammatory response. These include NLRP6 and PPARG, both of which are involved in regulating gut homeostasis, and which directly correlate with Desulfovibrio sp. and Bilophila sp., respectively. NPPA and NPY5R both suppress inflammatory cytokines and are involved in cardiovascular regulation; these appear positively related to Sutterella sp. and Bifidobacterium sp. This cluster also includes two inducers of the acute phase response, C3 and LBP, the latter of which is the pro-inflammatory receptor of the gram-negative surface glycoprotein lipopolysaccharide, which connects directly to Butyricimonas sp.
Similarly, cluster A3 includes GPR17, a negative regulator of Th2 response, and SELE, a neutrophil adhesion mediator activated by IL-1 and TNF-a. These two genes are positively correlated, as expected, and also negatively correlated with a large group of bacterial taxa, predominantly members of Bacteroides, Parabacteroides, Prevotella, and Ruminococcus. Relatedly, cluster A4 highlights several Clostridiales sp., Ruminococcaceae sp., Ruminococcus sp, Dialister, and Butyrivibrio crossotus, and a member of Order ML615J-28. These taxa potentially represent a cluster of major immune activity—the group correlates either directly or indirectly with downregulation of IL12B and upregulation of IL-4, major upstream inducers of Th1 and Th2 cell differentiation, respectively. Cluster A4 and A5 contain the orthologous C2CD4A and C2CD4B, which are induced by IL-1b and likely involved in type 2 diabetes; both clusters also contain members of Order ML615J-28, suggesting its common involvement in this pathway.
In contrast, cluster A2 includes OSMR, SLC7A2, and TNFSF11, all of which are upregulated in the type 2 inflammation associated with IgE-mediated hypersensitivities. OSMR and TNFSF11, in particular, have been implicated in inflammatory bowel disease (38). Prevotella copri appears to be a central player in this group, with Megasphaera sp., Desulfovibrio sp, and Ruminococcaceae sp. closely involved. The large number of negative covariance estimates in cluster A2 suggests that several of these taxa are in competition and may be involved in downregulating IgE-mediated pro-inflammatory activity. In short, clusters A3–A5 highlight potential microbial mechanisms of balance between type 1 and type 2 inflammation response, whereas cluster A2 highlights taxa potentially relevant to allergic and atopic activation.
Discussion
To the best of our knowledge, this study is the first to explore relations between biomarkers of physiologic aging and gut microbiota composition in a population-based sample of young or middle-aged adults. We found that multiple taxa show strong and consistent associations with multiple biomarkers of interconnected aging mechanisms, suggesting that microbiota are linked to aging earlier in the life course. Alpha- or beta-diversity-based shifts in composition according to biomarker levels were not evident, which suggests that shifts in the gut microbiota influencing (or influenced by) physiologic aging at this life stage are on the level of specific taxa rather than broad ecosystem-wide changes. Generally, the taxa showing the strongest relationships with metabolic, inflammation, and macromolecular damage markers (S. infantis, Pseudomonadaceae, Veillonella, Serratia) were not core members classically associated with central ecosystem services (such as Clostridium clusters VI and XVIa), but rarer groups that have been less studied in the context of intestinal ecosystems. In contrast, bacteria highlighted in our estimated bacteria-gene expression network do include members of these core groups, many of which have previously shown markedly lower abundance in older versus younger adults, including Prevotella, Bacteroides, Ruminococcaceae, Lachnospiraceae (39), as well as Bifidobacterium, which has been associated with extreme longevity (40).
One contribution of our study is simultaneous examination of biomarkers related to interconnected aging mechanisms of metabolism, macromolecular damage, inflammation, and immunosenescence, as well as an overall transcriptomic marker of aging. The general correspondence between the most strongly associated taxa for nearly all these systems, and between gene expression and more conventional markers, allows us to highlight taxa potentially involved in cellular aging processes, and thus age-related disease, with relative confidence. Specifically, Veillonella and S. infantis are each associated with markers of inflammation, macromolecular damage, and metabolism; Peptoniphilus with the latter two; and Serratia with Δage and metabolic markers. Of note, some of these associations contradict one another: for example, S. infantis appears protective for LDL and hallmark inflammation gene expression, but potentially harmful for CRP and HbA1c; many taxa follow a similar pattern. Similarly, our finding of the phylum Proteobacteria associated with salutary parameters of LDL, HDL, and HbA1c is inconsistent with the hypothesis that this pathobiont-enriched clade upregulates inflammaging (12,18), possibly reflecting functional diversity within this phylum or different effects at varying life stages. Additionally, network results at the level of specific genes and specific OTUs, intended to be as granular as possible, highlight strikingly different taxa than the analysis collapsed at clade and pathway levels. This may again reflect functional diversity of such core clades as Ruminococcus, Prevotella, and Lachnospiraceae, which, when collapsed, may attenuate the respective associations of their specific, unclassified OTUs. This may highlight a limitation of 16S taxonomic assignment, which can be addressed by more granular methods such as shotgun sequencing.
We were unsurprised to find many taxonomic associations with conventional markers of lipid and glucose homeostasis, as a growing body of literature suggests there are causal links there (17,41). However, immune and inflammation pathways have been consistently observed in animal models but less examined in humans, particularly in younger adults. Strong associations with CRP and inflammation gene expression that echo those with metabolic markers support the hypothesis that inflammation is a central mechanism of the effect of microbiota on metabolism (6). Moreover, a few taxa (primarily Pseudomonas veronii, whose role in the human gut is largely uncharacterized, and Succinivibrio spp., producers of pro-inflammatory succinate) appear associated with genes upregulated in lymphoid cell markers of immunosenescence, CD57+ NK, and KLRG1, markers which are known to be associated with CMV activation (42). Of note, among markers of T-cell senescence examined, these two markers appeared most strongly associated with the bacterial taxa assessed here. These findings are salient because CD57+NK and KLRG1 are key markers of immunosenescence and associated with terminal differentiation, suggesting that certain microbiota may drive cellular mechanisms of aging even in middle adulthood. As animal research has shown effects of commensal metabolites on T cell populations (43), future studies directly measuring immune cell populations are warranted to continue to examine this pathway in humans.
Strengths and Limitations
Although this is an observational study, our underlying scientific question is about causal effects of gut bacteria on physiological aging. While never perfect in this regard, observational studies can contribute information to causal questions, when certain assumptions are met (44). One assumption here, causal consistency (a.k.a. well-defined intervention), is challenged in 16S taxonomy because most taxa are only confidently classified at the genus level, while bacteria are often extremely functionally diverse even below the species level. This assumption can be relaxed by the addition of shotgun metagenomics, metatranscriptomics, and/or metabolomics, which come closer to relevant functional specificity. Moreover, a major issue that has been largely ignored in microbiome research (45) has been the compositional nature of sequencing data, for which common statistical procedures produce associations which, at best, correspond to effects that are likely not of interest (46). Our work improves on this by using robust denominators developed under Aitchison’s framework of compositional data analysis (30). To help meet the assumption of conditional exchangeability (ie, no unmeasured confounding), we adjusted for a number of possible confounders, but many likely remain. For example, we did not have good measures for diet, which likely influences bacterial levels as well as immune and metabolic parameters through pathways other than bacteria. We may have partially addressed this by adjusting for educational attainment, which is strongly correlated with diet in the United States. Moreover, identifying causal effects of specific bacteria (eg, for use as potential probiotics) is unrealistic in observational data because gut bacteria exhibit strong pairwise correlations whose causal structure is typically unknown, and thus many confounders (ie, other bacteria) cannot be controlled.
Another threat to conditional exchangeability is lack of clear temporal ordering in our data: metabolic panel markers were measured roughly 10 years before the microbiome, and gene expression was measured nearly concurrently; therefore, we cannot ascertain the direction of association for these measures. Despite this, we have some confidence that our data capture effects of the microbiota for two reasons. (a) The powerful action of commensals in training the immune system in the first several years of life is thought to lead to a durable and homeostatic community that largely persists throughout adulthood (47), except for environmental shocks such as major dietary changes (48)—indeed, longitudinal evidence suggests that gut microbial taxonomic composition is relatively stable within persons in adulthood (49). (b) While findings from microbiome studies have been difficult to compare because of differences in normalization and analytic approaches, our findings are consistent with previous results indicating that butyrate and acetate producers appear to play major roles in metabolism and inflammation (19,41). Despite this, there are known feedback loops between gut bacteria and metabolic and immune processes (13,14); thus, our measures at best capture both (a) effects of gut bacteria, and (b) alteration of the community in response to human bodily processes, including immunity.
Lastly, several of our measurements have notable limitations. Although our Gene Ontology search terms likely captured a large portion of inflammation-related genes, given our limited sample size, we were unable to capture the complete construct of immunity in our network analysis, resulting in some simplifications and selectivity of immune markers we hypothesized to be most closely related to aging. Additionally, glucose, LDL, and HDL are reported in Add Health as deciles, and transcriptomic age is standardized to the sample age distribution; therefore, these measurements contain only relative (rather than absolute) information about any given individual. Dried blood spot-based measures are highly reliable relative to whole-blood analysis for HbA1c and CRP, although perhaps less so for lipid homeostasis (50). Finally, high-throughput 16S rRNA sequencing and RNA-seq are measurements known to contain error, which may be compounded by large variation in library size typical of these data (see Supplementary eFigure 3); we make the critical assumption that this error is random and is thus less likely to produce systematic bias in our effect estimates.
Conclusion
Our study adds to the evidence that gut bacteria may influence physiologic mechanisms underlying a wide range of age-related diseases and biological phenotypes. Of note, we present some of the first data to characterize these associations far earlier in the life course than the majority of age-related disease onset and age-driven health declines. Our results, therefore, suggest that strategies aimed at repairing or improving gut microbiome community dynamics at or before this life stage may be useful avenues to explore for prevention of premature aging, whether through, for example, probiotic supplementation, targeted diet changes, or vaccines. Moreover, our results point to a few diverse groups of taxa potentially warranting further investigation for the physiologic aging potential of the microbiome in early to mid-adulthood. Replication of these findings by future studies with richer confounder control, larger sample sizes, longitudinal follow-up, and direct immunological measurements would build further support that gut microbiota could aid in the early detection and prevention of accelerated aging and age-related disease.
Funding
Research reported in this article was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health grant numbers P01HD031921, R01HD060726, R01HD087061; National Institute on Aging grant number R01AG042794-01A1, and the National Institute on Minority Health and Health Disparities grant number R01MD013349. We are grateful to the Carolina Population Center for training support (grant number T32HD091058) and for general support (grant number P2C HD050924).
Supplementary Material
Acknowledgments
The authors would like to acknowledge Grace Noppert, PhD, for her critical feedback and expertise in developing this manuscript. This research uses data from Add Health, a program project directed by Kathleen Mullan Harris and designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill, and funded by grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations.
Conflict of Interest
None reported.
References
- 1. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heianza Y, Ma W, Manson JE, Rexrode KM, Qi L. Gut microbiota metabolites and risk of major adverse cardiovascular disease events and death: a systematic review and meta-analysis of prospective studies. J Am Heart Assoc. 2017;6. doi: 10.1161/JAHA.116.004947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ticinesi A, Tana C, Nouvenne A, Prati B, Lauretani F, Meschi T. Gut microbiota, cognitive frailty and dementia in older individuals: a systematic review. Clin Interv Aging. 2018;13:1497–1511. doi: 10.2147/CIA.S139163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson MA, Jeffery IB, Beaumont M, et al. Erratum to: signatures of early frailty in the gut microbiota. Genome Med. 2016;8:21. doi: 10.1186/s13073-016-0275-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4 [DOI] [PubMed] [Google Scholar]
- 7. Zmora N, Bashiardes S, Levy M, Elinav E. The role of the immune system in metabolic health and disease. Cell Metab. 2017;25:506–521. doi: 10.1016/j.cmet.2017.02.006 [DOI] [PubMed] [Google Scholar]
- 8. Alenghat T, Osborne LC, Saenz SA, et al. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504:153–157. doi: 10.1038/nature12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31:817–844. doi: 10.1210/er.2009-0030 [DOI] [PubMed] [Google Scholar]
- 10. Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 11. Thevaranjan N, Puchta A, Schulz C, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455–466.e4. doi: 10.1016/j.chom.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fransen F, van Beek AA, Borghuis T, et al. Aged gut microbiota contributes to systemical inflammaging after transfer to germ-free mice. Front Immunol. 2017;8:1385. doi: 10.3389/fimmu.2017.01385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hagan T, Cortese M, Rouphael N, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rajkumar H, Mahmood N, Kumar M, Varikuti SR, Challa HR, Myakala SP. Effect of probiotic (VSL#3) and omega-3 on lipid profile, insulin sensitivity, inflammatory markers, and gut colonization in overweight adults: a randomized, controlled trial. Mediators Inflamm. 2014;2014:348959. doi: 10.1155/2014/348959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H. Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr Rev. 2018;39:133–153. doi: 10.1210/er.2017-00192 [DOI] [PubMed] [Google Scholar]
- 18. Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 20. Parker D, Sloane R, Pieper CF, et al. Age-Related adverse inflammatory and metabolic changes begin early in adulthood. J Gerontol A Biol Sci Med Sci. 2019;74:283–289. doi: 10.1093/gerona/gly121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris KM. Design Features of Add Health. Chapel Hill, NC: University of North Carolina at Chapel Hill; 2011. https://www.cpc.unc.edu/projects/addhealth/documentation/guides/DesignPaperWIIV.pdf. Accessed February 25, 2020. [Google Scholar]
- 22. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Godec J, Tan Y, Liberzon A, et al. Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity. 2016;44:194–206. doi: 10.1016/j.immuni.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burt JB, Demirtaş M, Eckner WJ, et al. Hierarchy of transcriptomic specialization across human cortex captured by structural neuroimaging topography. Nat Neurosci. 2018;21:1251–1259. doi: 10.1038/s41593-018-0195-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peters MJ, Joehanes R, Pilling LC, et al. ; NABEC/UKBEC Consortium The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. http://www.R-project.org/. Accessed February 25, 2020. [Google Scholar]
- 27. Renson A. eTables and R code for ‘Early signals of gut microbiome aging: immune and cardiometabolic phenotypes in young adulthood’. GitHub Repository. 2020. https://github.com/audreyrenson/addhealthmb.10.5281/zenodo.3804462. [Google Scholar]
- 28. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18:275–294. doi: 10.1093/biostatistics/kxw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L, Reeve J, Zhang L, Huang S, Wang X, Chen J. GMPR: A robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600. doi: 10.7717/peerj.4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao T, Liu H, Roeder K, Lafferty J, Wasserman L. The huge package for high-dimensional undirected graph estimation in R. J Mach Learn Res. 2012;13:1059–1062. [PMC free article] [PubMed] [Google Scholar]
- 32. Hawe JS, Theis FJ, Heinig M. Inferring interaction networks from multi-omics data. Front Genet. 2019;10:535. doi: 10.3389/fgene.2019.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012;4. doi: 10.1101/cshperspect.a006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buuren SV, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Soft. 2011;45(3):1–68. doi: 10.18637/jss.v045.i03 [DOI] [Google Scholar]
- 36. Liu H, Lafferty J, Wasserman L. The nonparanormal: semiparametric estimation of high dimensional undirected graphs. J Mach Learn Res. 2009;10:2295–2328. [PMC free article] [PubMed] [Google Scholar]
- 37. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. West NR, Hegazy AN, Owens BMJ, Bullers SJ, Linggi B, Buonocore S, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. doi: 10.1038/nm.4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biagi E, Franceschi C, Rampelli S, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016 [DOI] [PubMed] [Google Scholar]
- 40. Santoro A, Ostan R, Candela M, et al. Gut microbiota changes in the extreme decades of human life: a focus on centenarians. Cell Mol Life Sci. 2018;75:129–148. doi: 10.1007/s00018-017-2674-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Q, Lin SL, Kwok MK, Leung GM, Schooling CM. The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a Mendelian randomization study. Am J Epidemiol. 2018;187:1916–1922. doi: 10.1093/aje/kwy096 [DOI] [PubMed] [Google Scholar]
- 42. Xu W, Larbi A. Markers of T cell senescence in Humans. Int J Mol Sci. 2017;18:1742. doi: 10.3390/ijms18081742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hernan MA, Robins JM.. Causal Inference. Oxfordshire, UK: Taylor & Francis; 2019. [Google Scholar]
- 45. Renson A, Herd P, Dowd JB. Sick individuals and sick (microbial) populations: challenges in epidemiology and the microbiome. Annu Rev Public Health. 2020;41:63–80. doi: 10.1146/annurev-publhealth-040119-094423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arnold K, Berrie L, Tennant P, Gilthorpe M. A causal inference perspective on the analysis of compositional data. Int J Epidemiol. 2020. doi: 10.1093/ije/dyaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O’Toole PW, Claesson MJ. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–291. doi: 10.1016/j.idairyj.2009.11.010 [DOI] [Google Scholar]
- 48. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crimmins E, Kim JK, McCreath H, Faul J, Weir D, Seeman T. Validation of blood-based assays using dried blood spots for use in large population studies. Biodemography and social biology. 2014;60:38–48. 10.1080/19485565.2014.901885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.