Abstract
The ongoing COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in more than 7,000,000 infections and 400,000 deaths worldwide to date. Antibody development efforts mainly revolve around the extensively glycosylated SARSCoV-2 spike (S) protein, which mediates the host cell entry by binding to the angiotensin-converting enzyme 2 (ACE2). In the context of vaccine design, similar to many other viruses, the SARS-CoV-2 spike utilizes a glycan shield to thwart the host immune response. Here, we built a full-length model of glycosylated SARS-CoV-2 S protein, both in the open and closed states, augmenting the available structural and biological data. Multiple microsecond-long, all-atom molecular dynamics simulations were used to provide an atomistic perspective on the glycan shield and the protein structure, stability, and dynamics. End-to-end accessibility analyses outline a complete overview of the vulnerabilities of the glycan shield of SARS-CoV-2 S protein, which can be harnessed for vaccine development. In addition, a dynamic analysis of the main antibody epitopes is provided. Finally, beyond shielding, a possible structural role of N-glycans at N165 and N234 is hypothesized to modulate and stabilize the conformational dynamics of the spike’s receptor binding domain, which is responsible for ACE2 recognition. Overall, this work presents hitherto unseen functional and structural insights into the SARS-CoV-2 S protein and its glycan coat, which may be exploited by therapeutic efforts targeting this essential molecular machine.
Introduction
COVID-19 is an infectious respiratory disease that started in Wuhan, China, near the end of 2019 and has now spread worldwide as a global pandemic.1 This is not the first time that a coronavirus (CoV) has posed a threat to human health. SARS-CoV-2 (the virus that causes COVID-19) is in the same family of viruses, Coronaviridae, as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) related coronaviruses, which have resulted in previous epidemics.2–4 Owing to the lack of immunity, COVID-19 has already caused a catastrophic loss of human life worldwide5 as well as significant economic damage.6
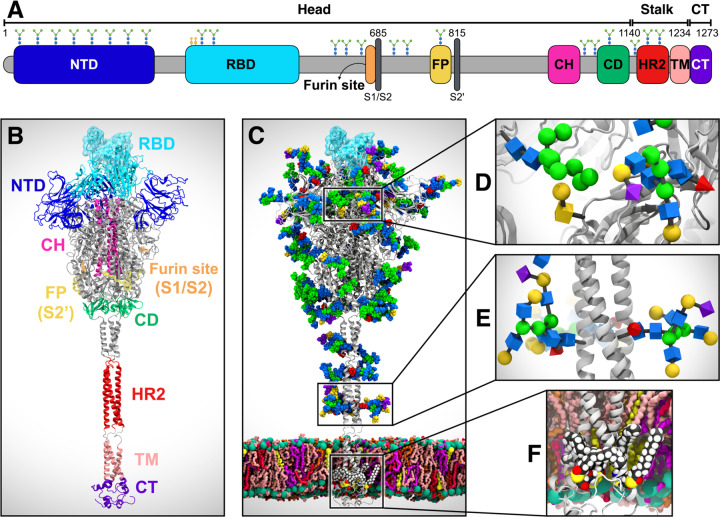
Coronaviruses, including SARS-CoV-2, are lipid-enveloped positive-sense RNA viruses. Together with the host-derived membrane, a set of structural proteins provide an organizational scaffold that wraps and contains the positive-sense viral RNA. Among them, the most critical is the spike, or S, protein, which is conserved to varying degrees across the Coronaviridae family and plays a key role in the virus’ initial attachment to and fusion with the host cell. The S protein is a class I fusion protein, synthesized as a single 1273 amino acid polypeptide chain, which associates as a trimer. Each trimer can be divided into three main topological domains, namely the head, stalk, and cytoplasmic tail (CT), where we can identify the S1 and S2 subunits (Figure 1A). One particularly interesting feature of the SARS-CoV-2 S protein is its adoption of a novel furin cleavage site (S1/S2), likely cleaved by the TMPRSS protease,7 between S1 and S2; this site is believed to prime and activate the spike for infection.8,9 A second proteolytic cleavage at S2ʹ releases the fusion peptide (FP), which penetrates and primes the host cell membrane for fusion.10 A number of recently published structural studies provide an atomic or a near-atomic understanding of the head portion of SARS-CoV-2 spike, which comprises multiple domains (Figure 1A).11,12 The S1 subunit contains an N-terminal domain (NTD) and the receptor binding domain (RBD), where the receptor binding motif (RBM) is responsible for the interaction with the angiotensin-converting enzyme 2 (ACE2) receptor to gain entry into the host.13 The S2 subunit has been aptly described as “a metastable spring-loaded fusion machine”14 because it plays a key role in integrating the viral and host cell membranes. It contains the FP, the central helix (CH), and the connecting domain (CD). Additional domains within the S2 subunit that are not resolved via cryo-EM or X-ray experiments include the heptad repeat 2 (HR2) and the transmembrane (TM) domains forming the stalk, and the CT (Figure 1A).
Figure 1. System overview.
(A) A sequence of the full-length spike (S) protein contains the N-terminal domain (NTD, 16–291), receptor binding domain (RBD, 330–530), furin cleavage site (S1/S2), fusion peptide (FP, 788–806), central helix (CH, 987–1034), connecting domain (CD, 1080–1135), heptad repeat 2 (HR2, 1163–1210) domain, transmembrane domain (TD, 1214–1234), and cytoplasmic tail (CT, 1235–1273). Representative icons for N-glycans (blue and green) and O-glycan (yellow) are also depicted according to their position in the sequence. (B) Assembly of the head, stalk, and CT domains into a full-length model of the S protein. (C) Fully glycosylated and palmitoylated model of the Open system. (D-F) Magnified view of the N-/O-glycans rendered using the Symbol Nomenclature for Glycans (SNFG) (D, E) and S-palmitoylation of the cytoplasmic tail (F).
Another key structural feature of the S protein that eludes detailed experimental structural characterization is its extensive glycosylation, shown in Figure 1C. Protein glycosylation plays a crucial role in viral pathogenesis,15–17 as demonstrated by the characteristically thick N-glycan coating of the viral fusion proteins.18–21 In the HIV-1 envelope spike (Env), for example, the protein-accessible surface area is almost entirely covered in N-glycans.20,22 These are so densely packed that they account for more than half of the protein’s molecular weight.23 The biological roles of the N-glycans expressed on the surface of viral envelope glycoproteins are very diverse16 and are all inextricably linked to their nature. Viral entry through membrane fusion is initiated by envelope glycoproteins through molecular recognition events involving cell surface receptors, which are often mediated by specific N-glycan epitopes.16,24,25 Moreover, a highly dense coating of non-immunogenic or weakly immunogenic complex carbohydrates on otherwise dangerously exposed viral proteins constitutes a perfect camouflage (or shield) to evade the immune system.16,18,19,26,27 To this end, the HIV-1 Env glycan shield, which is largely structured by oligomannose (Man5–9) N-glycans,18,20,26,28 has been shown to be quite effective in allowing the virus to thwart the immune system;15,16 it has also been found to be responsible for the virus’ interactions with DC-SIGN C-type lectins.29 Contrary to HIV-1 Env, the betacoronaviruses SARS and MERS S proteins are not shielded as effectively.15 Furthermore, both SARS-CoV and SARSCoV-2 spikes present a rather different glycosylation pattern from that of HIV-1 Env, with a large presence of complex N-glycans relative to oligomannose type.11,15,30,31 More specifically, the SARS-CoV-2 trimer has 22 predicted N-glycosylation sites per protomer,11,12 of which at least 17 have been found to be occupied,11,31 and 2 predicted O-glycosylation sites (see Figures 1C–1E).31
In this work, we present multiple microsecond-long, all-atom, explicitly solvated molecular dynamics (MD) simulations of the full-length SARS-CoV-2 S glycoprotein embedded in the viral membrane, with a complete glycosylation profile consistent with the currently available glycomic data.11,31 The MD simulations discussed here augment and extend the available structural and biological data to provide an atomically detailed perspective on the full-length SARS-CoV-2 S protein structure, stability, and dynamics. Analyses of the glycan shield indicate different accessibility for the RBM according to the respective RBD conformation (“up” or “down”), highlighting how glycans have direct implications for the recognition of and binding to ACE2 and ultimately for the invasion of the host cell. Unique insights into the shielding of our full-length model of the S protein disclose the lack of vulnerabilities of the stalk, particularly for large molecules. On the contrary, the head region appears to be a more viable target for antibody development, as extensively shown by epitope-specific accessibility analyses. Finally, in addition to acting as a shield, our simulations reveal that the peculiar glycosylation of the SARS-CoV-2 S protein may also play an essential functional role. More specifically, we identified that the N-glycans at N234 and N165 modulate the conformational dynamics of the S protein and stabilize the RBD in the “up” conformation. Notably, these observations converge on a larger set of simulations performed with different force field combinations and system set-ups. Overall, this work provides an atomic-level glimpse into the SARS-CoV-2 S protein, highlighting the importance of glycans in the evasion of the host cell humoral response and in the modulation of the RBD structural dynamics. These insights might be harnessed in the therapeutics efforts aimed at fighting the pandemic threat.
Results & Discussion
All-Atom MD Simulations of the Full-Length Model of the SARS-CoV-2 S Protein in Open and Closed State
In this work, we built a complete, full-length model of the glycosylated SARS-CoV-2 S protein in both closed and open states, hereafter referred to as “Closed” and “Open,” respectively. Closed is based on cryo-EM structure 6VXX, where all RBDs are in a “down” conformation.12 Open is based on cryo-EM structure 6VSB, where the RBD within chain A (RBD-A) is in an “up” conformation.32 A detailed view of chains A, B, and C and the RBD “up/down” conformations as in Open/Closed is shown in Figure S1 of the Supporting Information (SI). These models were built in three steps based on three main topological domains, as described in the Materials and Methods: the “head,” comprising S1/S2 subunits until residue 1140; the “stalk,” comprising HR2 and TM domains (residues 1141–1234); and the CT (residues 1235–1273) (Figure 1A).32 The modeled constructs were fully glycosylated at N-/O-glycosylation sites11,31 and further refined with cysteine palmitoylation within the CT (Figures 1C–1E).33,34 The full-length structures were embedded into an equilibrated all-atom membrane bilayer with a composition mimicking the endoplasmic reticulum Golgi intermediate compartment (ERGIC), where the virus buds; subsequently, explicit water molecules and ions were added, affording two ~1.7 million atoms final systems (Figure 1B).
Multiple replicas of all-atom MD simulations of the Open (6x) and Closed (3x) systems were run on the NSF Frontera computing system at the Texas Advanced Computing Center (TACC) with NAMD 2.14,35 achieving benchmarks of ~60 ns/day on 256 nodes for cumulative extensive sampling of ~4.2 and ~1.7 μs, respectively (Table S1, Movie S1). Note that, since RBD-A within Open is captured in a metastable state,32 Open was simulated for a longer time than Closed.
Topological domain-specific root-mean-square-deviation (RMSD) relative to the starting structures was used to monitor the simulations and structural stability (Figures S2–S3). Overall, the two systems showed structural convergence of the extracellular topological domains (head and stalk) within 400 ns. No significant difference was observed in the RMSD values of the stalk between the systems (Figure S3). Besides displaying a replica-specific convergence of RMSD values, the triple-stranded coiled-coil structural motif of the stalk persists throughout the simulations. Notably, as a result of secondary structure prediction (see Material and Methods), the stalk model includes two loops that break the alpha-helix within each strand (Figure 1B). Along with linked N-glycans, these loops confer critical dynamic properties to the stalk, allowing it to accommodate the head’s wiggling motions while remaining stably anchored to the membrane. The RMSD values of the CT domain show extremely large deviations and a mixed range of dynamic behaviors across replicas (Figure S3). While the first section of CT remains anchored to the inner-leaflet of the lipid bilayer through the palmitoylated cysteines, the rest is highly flexible and solvent exposed.
The root-mean-square-fluctuation (RMSF) values of the glycans were measured to determine their average flexibility around their mean position across the entire trajectory (Figure S4). These values indicate a diverse range of mobility for specific sites, which is mostly dependent on solvent exposure, branching, and sequence. Complex-type N-glycans located on the NTD exhibit larger RMSF values than oligomannose and other N-glycans populating the head, including the ones in the RBD at N331 and N343. Interestingly, the O-glycans linked in the immediate vicinity of the RBD, namely at T323 and S325, show the lowest RMSF values. This is mostly attributable to their small size; however, their position may be critical for modulating RBD conformational changes, although we note that both positions are not consistently occupied. Finally, the large tetrantennary N-glycans in the stalk region at N1173 and N1194, modeled consistently with experimental data,11 show the most extensive fluctuations, suggesting their good shielding potential.
The stability of the membrane bilayer was also monitored by tracking the variation in a series of structural properties over time, including area per lipid, membrane thickness, order parameters, and phospholipid tilt angles. Equilibrium area per lipid and P–P membrane thickness plots are shown in Figure S5. These results are in agreement with known POPC and previously reported binary POPC + X trends, where X is any other lipid added to the mix.36–42
Glycan Shielding of the Full-Length SARS-CoV-2 S Protein
In the context of vaccine design, it is critical to consider all the strategies developed by viruses to evade the host immune response. Within this framework, many viruses use a glycan shield to mask the immunogenic epitopes targeted by neutralizing antibodies.11,15 SARS-CoV-2 S protein glycosylation is extensive and capable of thwarting the host humoral recognition.11,15,43 In addition to the spike head and RBD,44,45 another possible attractive target for antibodies is the stalk, as also shown in influenza virus studies.46 Whereas 19 N-glycans essentially camouflage the spike head region, only 3 N-glycosylation sites (N1158, N1174, and N1194) are present on the stalk. Exploiting the extensive sampling derived from all-atom simulations of our full-length model of SARS-CoV-2 S glycoprotein, we provide an unprecedented end-to-end overview of the spike’s glycan shield (Figure 2). In Figure 2A, each blue bush-like structure represents an ensemble of equally interspersed conformations of a single glycan sampled along the trajectory for a total of 300 superimposed poses. Considering the different timescales of the dynamics of these N-glycans (nanoseconds), relative to the antibody/spike binding process (microseconds to milliseconds), this representation provides a realistic view of the shielding that molecules targeting the spike might encounter upon binding.
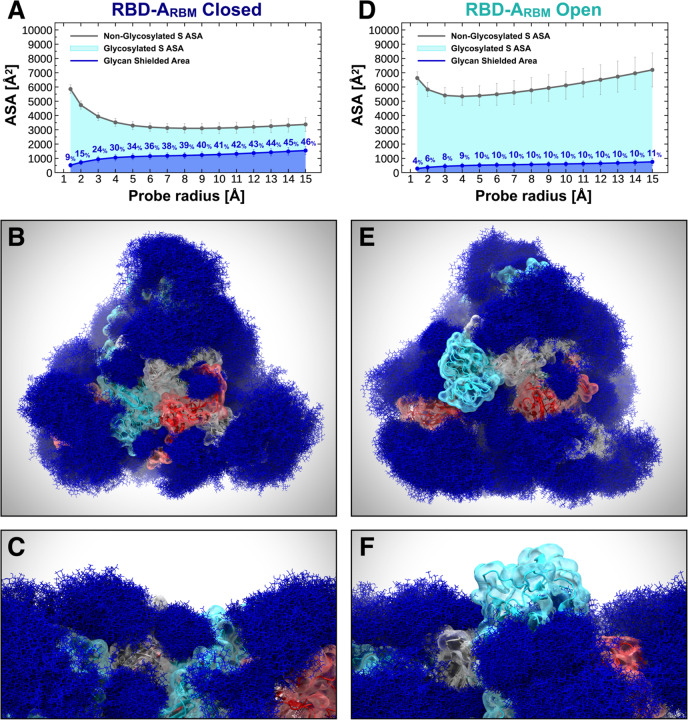
Figure 2. Glycan shield of the SARS-CoV-2 S protein.
(A) Molecular representation of Open. Glycans at several frames (namely, 300 frames, one every 30 ns from one replica) are represented with blue lines, and the RBD within chain A is highlighted with a cyan transparent surface. (B-C) Accessible surface area of the head (B) and stalk (C) and the area shielded by glycans at multiple probe radii from 1.4 (water molecule) to 15 A. The values have been averaged across replicas and are reported with standard deviation. The area shielded by the glycans is presented in blue (rounded % values are reported), whereas the gray line represents the accessible area of the protein in the absence of glycans. Highlighted in cyan is the area that remains accessible in the presence of glycans, which is also graphically depicted on the structure in the panels located above the plots.
To quantify this shielding effect, we calculated the S protein’s accessible surface area (ASA) that is covered by glycans on both the head and stalk regions. We also screened a wide range of probe radii, from 1.4 to 15 Å, to measure the effectiveness of the shield with respect to molecules of different sizes. A value of 1.4 Å is commonly set to approximate the radius of a water molecule, whereas larger radii can be used to describe larger moieties, ranging from small molecules at 2–5 Å to larger peptide- and antibody-sized molecules at 10–15 Å.43,47
Our results indicate that the head is overall less shielded by the glycans than the stalk at all probe radii (Figures 2B and 2C). Interestingly, the stalk is almost completely inaccessible to large molecules such as antibodies, with glycan coverage equal to 90% of the protein accessible area for 15-Å-radius probe. On the contrary, the head represents an easier target as its glycan camouflaging is insufficient (62%) to cover its larger surface area. When smaller probes are screened (1.4–3 Å radius), the stalk and the head result to be similarly shielded at an average of 26% and 20%, respectively, suggesting that small molecules can equally penetrate either area. ASA average and standard deviation values for the head and stalk are provided in Tables S2 and S3, respectively. Overall, taking glycosylation into account, the stalk appears as a potentially more difficult therapeutic target than the head, despite being a highly conserved domain among betacoronaviruses. Its smaller surface is well-protected by large sialylated and fucosylated tetrantennary glycans, which are found to be almost 100% effective in shielding large molecules. Interestingly, glycans at N1174 and N1198 have been found to always be tetrantennary, 100% fucosylated (N1174 and N1198), and 100% sialylated (N1198).11,15 However, small drugs might still interfere with the fusion process by binding to the HR2 domain of the stalk. In contrast, the head shows a higher vulnerability, which can be harnessed for vaccine development.
Glycan Shield of the Receptor Binding Domain
As discussed in the previous section, the glycan shield plays a critical role in hiding the S protein surface from molecular recognition. However, to effectively function, the spike needs to recognize and bind to ACE2 receptors as the primary host cell infection route. For this reason, the RBM must become fully exposed and accessible.48 In this scenario, the glycan shield works in concert with a large conformational change that allows the RBD to emerge above the N-glycan coverage. Here, we quantify the ASA of the RBM within RBD-A, corresponding to the RBD/ACE2-interacting region (residues 400–508), at various probe radii in both the Open and Closed systems (Figures 3A and 3D, full data in Tables S4–S6.). As expected, the ASA plots show a significant difference between the “down” (Closed) and “up” (Open) RBD conformations, with the RBM area covered by glycans being remarkably larger in the former. When RBD-A is in the “up” conformation, its RBM shows an average (across all radii) of onlŷ9% surface area covered by glycans, compared with ~35% in the Closed system (Figures 3A and 3D). This difference is further amplified when considering a larger probe radius of 15 Å, with a maximum of 11% and 46% for Open and Closed, respectively. Interestingly, for smaller probes (1.4–3 Å) the shielding becomes weak in both systems, with an average of 6% and 16% for Open and Closed, respectively.
Figure 3. Glycan shield of the RBD ACE2-interacting region.
The accessible surface area of the RBM-A and the area shielded by neighboring glycans in the Closed (A) and Open (D) systems are plotted at multiple probe radii from 1.4 (water molecule) to 15 A. The values have been averaged across replicas and are reported with standard deviation. In blue is the area of the RBM-A covered by the glycans (rounded % values are reported), whereas the gray line is the accessible area in the absence of glycans. Highlighted in cyan is the RBM-A area that remains accessible in the presence of glycans, which is also graphically depicted on the structure in the panels located below the plots. (B-F) Molecular representation of Closed and Open systems from top (B and E) and side (C and F) views. Glycans (blue lines) are represented at several frames equally interspersed along the trajectories (300 frames along 0.55 ns for Closed and 1.0 μs for Open), while RBD-A is presented with cyan cartoons and transparent surface. Chains B and C are shown in red and gray cartoons, respectively, and as transparent surface.
Note that the RBD region that do not directly interact with ACE2 remains shielded by the glycans in both “up” and “down” conformations (Figure S6). This region is equally protected regardless of the RBD conformation mostly owing to the presence of N-glycans bound to the RBD itself at N331 and N343 and to the N-glycans at N165 and N234 (Figure S7). Ultimately, this analysis shows that the RBM is always accessible when RBD is “up”, whereas it is very well-camouflaged when “down” (Figures 6B–6C, 6E, and 6F). This suggests that the glycan shield of this critical domain is effectively paired with its “down-to-up” conformational change, allowing the RBM to transiently emerge from the glycan shield and bind to ACE2 receptors. Furthermore, while antibody targeting the RBD might be ineffective when the RBD is “down”, small molecules could more easily circumvent the glycan coverage. This is in agreement with structural data reporting the “up” conformation as a requirement for C-terminal domain-1 (CTD1) neutralization by host antibodies.49
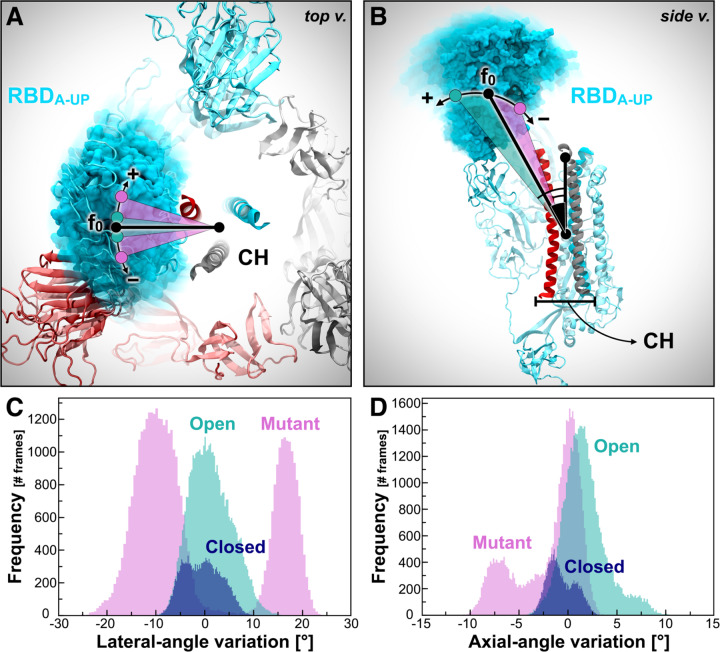
Figure 6. RBD-A lateral and axial angle fluctuations.
(A, B) RBD-A lateral angle (A) and axial angle (B), where chains A, B, and C of the spike are represented as ribbons colored in cyan, red, and gray, respectively. Positive and negative variations with respect to the initial frame (0) are indicated with the “+” and “−” symbols, respectively. (C, D) Distributions of RBD-A lateral angle (C) and axial angle (D) fluctuations with respect to the initial frame in Closed (blue), Open (teal), and Mutant (magenta).
Overview of Neutralizing Antibody Epitope Accessibility
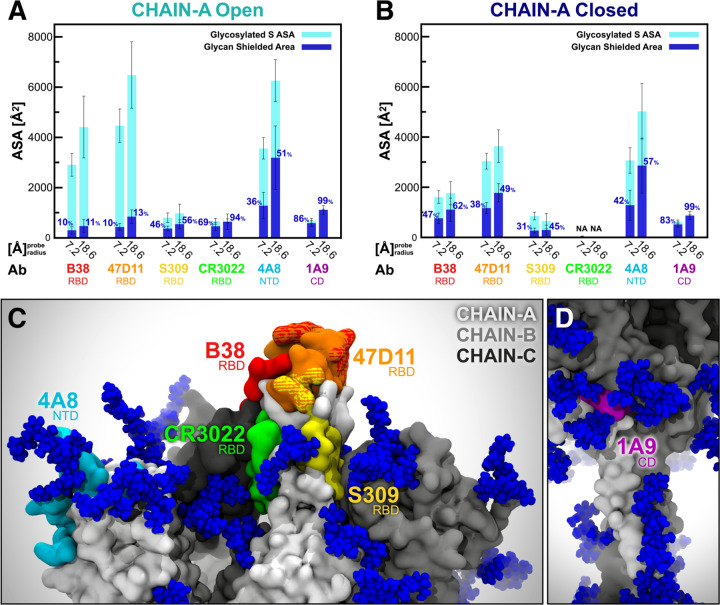
Several SARS-CoV-2 antibodies targeting the S protein have been identified (Table S7).50–62 The majority of these antibodies recognize epitopes on the RBD, while only a few have been shown to address the antigenic regions within the NTD and CD (Figures 4C, 4D, and S8). Among the RBD antibodies, B38 and 47D11 interact with the RBM at the RBD/ACE2 interface,58,59 whereas S309 and CR3022 target the side/bottom part of the RBD.60–62 In addition, 4A8 and 1A9 have been found to engage with the NTD and CD, respectively.51,57 To quantify the effects of glycan shielding on these epitopes, we calculated each epitope’s ASA at two probe radii, 7.2 and 18.6 Å, which approximate the size of antibody hypervariable loop and variable fragments domains, respectively (Figures 4A and 4B, full data provided in Table S8).43 In Open (RBD “up”), epitopes on the RBD/ACE-2 interface (B38 and 47D11) show large ASA that is minimally shielded by glycans (10%/11% and 10%/13%, respectively) (Figure 4A). Antibodies in this region exploit the vulnerability of the S protein when RBD is in the “up” conformation. Conversely, in Closed, the shielding of the epitopes remarkably increases to 47%/62% (B38) and 38%/49% (47D11) (Figure 4B). When the RBD is in the “down” conformation, the RBM is buried by the other two neighboring RBDs, which already reduce its overall accessibility by ~40%. These values are in agreement with the RBM ASA trends shown in Figure 3.
Figure 4. Accessibility of neutralizing antibody epitopes.
The accessible surface area of antibody epitopes and the area shielded by neighboring glycans are plotted at probe radii 7.2 and 18.6 A for chain A in Open (A) and Closed (B) systems. The area of the protein covered by the glycans is depicted in blue (rounded % values are reported), whereas highlighted in cyan is the epitope area that remains accessible in the presence of glycans. The values have been averaged across replicas and are reported with standard deviation. (C) Side view of the top region of S protein in the Open system, where the antibody epitopes are highlighted following the color scheme used in panels A and B. (D) Magnified view of 1A9 epitope (highlighted in purple) located within CD.
The S309 epitope, located on the side of the RBD and near the N-glycan at N343, shows an interesting behavior. When including glycan N343 as a shielding factor, the epitope is covered up to 45%/56% of its total area. However, this glycan has been shown to be incorporated into the recognized epitope, which would considerably increase the antigenic region targeted by S309.60 Interestingly, no substantial differences in shielding are observed between Open and Closed because this epitope is mostly located on the RBD side, which remains exposed even in the “down” conformation. Considering the bottom part of the RBD, the epitope recognized by the CR3022 antibody is found to be almost completely shielded in Open (69%/94%) and not accessible at all in Closed. This is in agreement with structural data showing that the cryptic epitope engaged by CR3022 is only available when the RBD is both “up” and rotated.61,62 Remarkably, this epitope partially overlaps with VHH72, an antibody found to neutralize SARS-CoV-2 S pseudotyped viruses.52 Finally, the 4A8 and 1A9 epitopes located within the NTD and CD, respectively, are not affected by the conformational changes of the RBD.51,57 Whereas the epitope recognized by 4A8 is about 36%/51% shielded by glycans, the one targeted by 1A9 is almost completely covered at 86%/99%. These results probe further questions on 4A8 and 1A9 binding mode in the presence of glycans.
Role of N-Glycans at N234 and N165 in the Structural Mechanism of the S Protein
SARS-CoV and SARS-CoV-2 S glycoproteins share 76% sequence identity,12 and 18 out of 22 N-glycosylation sites found on SARS-CoV-2 are conserved from SARS-CoV. Based on the analysis of our simulations of both the full-length model of SARS-CoV-2 S protein and the S protein head alone in the Open conformation, the latter discussed in Section 3 of SI, we identified two N-glycans at N165 and N234 in the NTD, which, given their strategic position and structure, we hypothesized may play a role in the structural stability of the S protein in its active Open conformation. Therefore, an additional third system, “Mutant,” was generated from Open by mutating the N-glycans at N165 and N235 into alanine. Subsequently it was also simulated through all-atom MD for six replicas totaling ~4.2 μs (Table S1). Our simulations show that in Closed, the N-glycan at N234, modeled as Man9 in agreement with glycoanalytic data,11 is pointing away from the core of the S protein and is directed toward the solvent (Figure 5A); in contrast, in Open, where the RBD of chain A is “up,” the N234 Man9 is directed inward (Figure 5B) and is inserted into the large volume left vacant by the opening of the RBD. Because the glycans were built by structural alignment on the linked GlcNAc (or chitobiose) residues, wherever available, the relative position of the existing GlcNAc or chitobiose may affect the orientation of the entire N-glycan. Indeed, the N-glycan core is quite rigid.63 Additional simulations of the open SARS-CoV-2 S head started from an alternative open cryo-EM structure, namely 6VYB,64 present the N234 GlcNAc in a slightly different orientation. These simulations show that the N234 Man9 progressively inserts itself to reach the core of the trimer through interactions with the surface of the closed RBD domain located across from it (details in Section 3 of SI and Movie S2). The highly conserved N-glycan at N165 also inserts itself between NTD-B and RBD-A, either making extensive interactions with RBD-A or occupying the volume left vacant following its opening (Figure 5C).
Figure 5. N234A and N165A mutations show increased instability of RBD-A in the “up” state.
(A-B) Top view of the S protein as in the Closed (A) and Open (B) systems. Protein is represented with cartoons, colored in cyan, red, and gray for chains A, B and C, respectively. O-mannose N-glycans at position N234 are depicted with VdW spheres, where GlcNAc is colored in blue and Man in green. In Closed (A), all the N-glycans at N234 are tangential to a hypothetical circle going through N234. In Open (B), the N-glycan at N234 of chain B moves inward, filling in the vacancy under RBD-A in the “up” conformation. (C) Side view of the S protein (surface representation) in Open, where the RBD of chain A (RBD-A, cyan) is stabilized by N-glycans at N165 and N234 in the “up” conformation. Same color scheme as panels A and B is applied. (D) PCA plot showing PC1 vs. PC2 of RBD-A (residues 330–530) in Closed, Open, and Mutant in blue, teal, and magenta, respectively. A large variance is observed for Mutant.
To determine the structural consequences of removing these two glycans, we compared the conformational landscape of the RBD in Open, Mutant, and Closed systems using principal component analysis (PCA). Scatter plot projections of RBD-A dynamics onto the first two eigenvectors (PC1 vs. PC2), i.e. the two motions with the largest variance in the trajectories (63% and 18%, respectively), clearly indicate that the RBD-A in Mutant explores a larger conformational space than in Open, which in turn is considerably larger than in Closed (Figure 5D). Although the difference between Open and Closed is immediately explained by the RBD-A state (“up/down”), the PCA landscape strongly suggests a stabilizing role of the N-glycans at N165 and N234, supporting the RBD in its “up” conformation. Analysis of the RMSD values along the trajectory shows that even a single point N234A mutation contributes to destabilize the RBD “up” conformation (Figure S2). This is further confirmed by additional independent simulations of the spike head alone, as described in Section 3 of SI.
Notably, RBD-B is in the “down” conformation in all three systems (Figure S1), and its dynamics are affected by the vacant volume left by RBD-A in the “up” conformation. Indeed, RBD-B shows an increased degree of mobility in Open with respect to Mutant along PC1 (Figure S9). Finally, there are no differences between the three systems for RBD-C, where the sampled PCA space remains unaffected by the presence or absence of the N-glycans at N165 and N234 or by the RBD-A in the “up” conformation (Figure S9).
To obtain further insights into the dynamic behavior of the RBD and examine the differences in the explored conformational spaces revealed by PCA, we monitored the fluctuations of the two most dominant motions determined by PCA: PC1 and PC2. PC1 reports an in-plane motion of the RBD along a hypothetical circle centered on the central helices of the spike (Figure 6A), which we call the “lateral angle.” PC2 identifies an RBD tilting motion either away from or toward the central helices of the spike (Figure 6B), which we call the “axial angle.” Full details are outlined in the Material and Methods section.
In agreement with the PCA plot in Figure 5C, the RBD-A lateral angle displays a well-defined single distribution centered on zero in Open, whereas it shows a bimodal population in Mutant (Figure 6C). This analysis shows that the presence of the N-glycans at N165 and N234 in NTD-B are crucial to stabilize RBD-A in its “up” conformation, preventing it from undergoing disorganized motions. Interestingly, the axial-angle analysis results display a similar behavior of RBD-A in Open and Mutant, showing a long tail distribution in both systems (Figure 6D). However, Mutant exhibits a significant negative trend, whereas Open shows a positive trend. This suggests that the N-glycans at N165 and N234 may not only have a critical stabilizing role but also contribute to or facilitate/support the opening of RBD (see Figure 6D).
Finally, to better characterize the stabilizing role of the N-glycans at N165 and N234, we examined their interaction with the S protein through hydrogen bond analysis. N234 Man9 deeply extends into the large pocket created by the opening of the RBD-A (Figure 7C), makes stable hydrogen bonds with Asp 198 in chain B, and interacts as deep as Glu 988 on CH of chain B. This same, stable interaction was also observed in the independent simulation study of the N234-Man9 S head, as shown in Section 3 of SI. Both of these hydrogen bonds are stable for more than 50% of the 4.2 μs trajectory of the Open system, with the majority of hydrogen bonds formed with CH of chain B (see Figure 7A). The N-glycan at N165 is more exposed to the solvent relative to the Man9 glycan at N234; nevertheless, it is seen to frequently engage in interactions with the “up” conformation of RBD-A (~80% frequency) with a variable hydrogen bonding pattern across simulation replicas (Figures 7B and 7D). The per-replica hydrogen bond networks observed for the N-glycans at N234 and N165 are shown in Figure S10.
Figure 7. Hydrogen bond interactions of N-glycans at N234 and N165.
The main hydrogen bond interactions of N-glycans at N234 (A) and N165 (B) within the Open system are shown as occupancy across all replicas (% frames). (C) A snapshot capturing Man9 glycan (licorice representation) at N234 establishing multiple hydrogen bonds with S protein residues (licorice representation) belonging to RBD-A (cyan surface), NTD-B (red surface), CH-B (red cartoons), and CH-C (gray cartoons). GlcNAc and Man carbons are colored in blue and green, respectively. (D) Molecular representation of Man5 glycan at N165 interacting with RBD-A. Multiple (1000) equally interspersed configurations (selected across all replicas) of the glycan at N165 are simultaneously shown. The glycan is represented as colored licorices (GlcNAc in blue, Man in green), whereas RBD-A and NTD-B are represented as cyan and red surfaces, respectively.
Altogether, the heterogeneous conformational dynamics revealed by PCA and by angle and hydrogen bond analyses show that the structural stability of the RBD depends on the presence or absence of N-glycans at N165 and N234. Furthermore, Cα atoms’ RMSD values indicate that even a single point N234A mutation may be sufficient to destabilize the RBD “up” conformation (Figure S2 and Section 3 of SI). Notably, the RBD has only two linker loops connecting it with the receptor-binding C-terminal domain 2 (CTD2); thus, the domain is primed to undergo large conformational rearrangements, such as the ACE2-induced hinge motion.65 Our analysis suggests that, similarly to the MERS spike protein,66 the SARS-CoV-2 spike RBD “up” state is probably metastable within the conformational “up” and “down” ensemble and that it needs to be supported by the N-glycans at N234 and N165. Indeed, our results show that glycan-deleting mutations at these positions cause the RBD to experience a higher level of conformational fluctuations when “up,” with motion trending toward the “down” state. In addition, in the absence of N-glycans at N234 and N165, the RBD might rotate along its main axis or undergo a lateral tilt motion, exposing cryptic epitopes.45
Conclusions
Our work presents multiple microsecond-long, all-atom, explicitly solvated MD simulations of the full-length model of the glycosylated SARS-CoV-2 S protein embedded in a viral membrane. We show how the time-averaged glycan shield covers a vast amount of the S protein surface area and how it changes depending on the conformational state of the protein between open and closed states; moreover, we present an analysis of the glycan shield within the context of the known SARS-CoV-2 neutralizing antibodies. Interestingly, our simulated models suggest a role – beyond shielding – for glycans at N234 and N165. Simulations of the mutant forms of the S protein corroborate their potential role in modulating the conformational landscape of the S trimer and allow us to define the principal components of motion of the RBD as a large-scale axial and lateral tilt. Overall, our work sheds new light on the full structure of this critical target and points to opportunities and challenges for small molecules and vaccine design. We make available through the NSF MolSSI and BioExcel COVID-19 Molecular Structure and Therapeutics site (https://covid.molssi.org/) our structures, models, input files, and trajectories to enable other groups to use and explore this dynamic system in atomic detail.67
Methods
Model systems.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) glycoprotein is a large, glycosylated homotrimer, where each of its three identical monomers (residues 16–1273) can be divided into three main topological domains: the “head,” comprising S1 and S2 subunits until residue 1140; the “stalk,” composed of heptad repeat 2 (HR2) and transmembrane (TM) domains (residues 1141–1234); and the cytoplasmic tail (CT) (residues 1235–1273) (Figure 1A).32 Experimental structures of SARS-CoV-2 S have been resolved in two main conformational states, open and closed, that were used in this study to build two complete, fully glycosylated models, referred to in this manuscript as “Open” and “Closed”, respectively. The Closed system is based on a cryo-EM structure of the S protein solved at 2.80-Å average resolution (PDB ID: 6VXX),12 where all receptor binding domains (RBDs) are in the “down” conformation. The Open system is instead built upon a cryo-EM structure of the S protein solved at 3.46-Å average resolution (PDB ID: 6VSB),32 where only one RBD (chain A) is in the “up” conformation. A third system, called “Mutant”, was also generated from the Open system upon mutation of N-glycans at N165 and N234 into alanine within all the three monomers. Although the cryo-EM structures of the S protein already provide critical information about its structure, they are usually incomplete and/or have been modified to increase protein stability.32,66 For example, the introduction of two consecutive prolines in the central helix and/or of an engineered C-terminal foldon trimerization domain68 has been adopted as a common strategy to stabilize the S protein for cryo-EM.32,66 In addition, highly flexible protein regions (loops) and glycans beyond the first three sugars often remain unresolved owing to resolution limits. Therefore, several modeling steps were required to produce a full-length model of the wild type protein as described below.
Missing loops modeling.
The employed cryo-EM structures of the S protein reveal several missing gaps corresponding to flexible loops ranging from 3 to 38 residues. To generate a complete construct, missing gaps were modeled as disordered loops using Modeller9.19.69 Keeping the cryo-EM coordinates fixed, 50 models were independently generated for each monomer, from which the top models were selected and reassembled to recreate the full trimeric head. The alignment between the cryo-EM structure and the FASTA sequence of SARS-CoV-2 spike (QHD43416.1)70 used by Modeller was generated using Clustal Omega.71 The top models were further visually inspected to discard those in which loops were entangled in a knot or clashed with the rest of the structure. Finally, the stabilizing proline mutations from the cryo-EM structures were mutated back to wild type, and modeling artifacts were detected and corrected prior to simulations.
HR2 and TM domain (stalk) modeling.
Both cryo-EM structures employed to build our models were stabilized using an engineered C-terminal foldon trimerization domain.68 Therefore, the stalk region of the S protein from residues 1147–1234, including the HR2 and TM domains, had to be constructed. Using the Jpred4 server,72 the secondary structure of the stalk sequence was predicted as three helical segments connected by two unstructured loops (Figure S11). Given the amphipathic nature of the helical segments, the three chains were assembled into an alpha-helical coiled-coil trimeric bundle using Modeller9.19.69 A coiled-coil crystal structure, where the smaller and more hydrophobic residues are positioned inside the bundle and the polar residues are solvent-exposed, served as a template (PDB ID: 2WPQ).73 Each alpha-helix was broken into three segments separated by two loops according to Jpred4 secondary structure predictions.
CT modeling.
The CT of the S protein (residues 1235–1273) was modeled using the i-TASSER software.74–76 i-TASSER generated five models with confidence scores of −1.25, −2.65, −3.15, −4.33, and −1.33 (C-score range [−5, 2]). Out of these, model 3 was selected because it revealed a helical domain between residues 1238 and 1245, where two cysteines were shown to be palmitoylated in another betacoronavirus, MHV-A59.33,34 The corresponding cysteines in SARSCoV-2 were C1240 and C1241. The remaining sequence of CT was predicted to be intrinsically disordered. Both cysteines (C1240 and C1241) were palmitoylated using lipid-tail functionality available within Glycan Reader in CHARMM-GUI.77,78
Glycosylation.
SARS-CoV-2 S protein features 22 N-glycan sequons (N-X-S/T) per monomer, which have been found to be heterogeneously populated in different glycoanalytic studies.11,31,79 Interestingly, two O-glycans have also been characterized at positions T323 and S325.31 Our modeled constructs have been fully N-/O-glycosylated using the Glycan Reader & Modeler tool80 integrated into Glycan Reader77 in CHARMM-GUI.78 An asymmetric (i.e., not specular across monomers) site-specific glycoprofile has been derived according to glycoanalytic data reported by Watanable et al.11 for N-glycans and by Shajahan et al.31 for O-glycans. Detailed per-chain descriptions of the site-specific glycoprofile of the S protein systems simulated in this work are shown in Tables S9–S11 of the SI. In the Mutant system, N165A and N234A mutations were introduced to remove the respective N-glycans. This was performed with PSFGEN during system setup. In summary, 70 glycans (22 × 3monomers N-glycans and 2chainA + 1chainB + 1chainC O-glycans) have been added in the Open and Closed systems, whereas 64 have been added in the Mutant system. Our modeled N-glycans account for oligo-mannose from Man5GlcNAc2 to Man9GlcNAc2, complex and hybrid types, displaying one to four antennas. Additional modifications, such as fucosylation and sialylation, have also been site-specifically considered, as reported in Watanabe et al.11 We remark that the oligosaccharides (GlcNAc-/GlcNAc2-/ManGlcNAc2-) originally solved in the cryo-EM structures have been generally retained or used as a basic scaffold, when possible, to build the full glycans. However, owing to steric clashes arising at particularly buried sites (for example, N122), a manipulation of glycan dihedrals and/or asparagine side chain has been necessary to fit in all the glycans.
Membrane modeling.
The lipid composition of the membrane patch was selected based on the lipoprofiles81,82 of the endoplasmic reticulum and trans-Golgi network, organelles in which the coronavirus membranes are known to be constructed.83,84 A symmetric 220 Å × 220 Å lipid bilayer patch was generated using CHARMM-GUI’s input generator.78 The lipids were packed to an area per lipid of 70 Å2 with the following ratio of phospholipids and cholesterol: POPC (47%), POPE (20%), CHL (15%), POPI (11%), and POPS (7%). The IUPAC names corresponding to these abbreviations are given in Table S12. The area per lipid value was selected based on the suggested CHARMM-GUI areas; the equilibrium values for this system were calculated and can be found in Figure S5.
System preparation.
Upon functionalization (i.e., glycosylation and palmitoylation) of the glycoproteins through the Glycan Reader module available within CHARMM-GUI, further modifications to the structures were necessary and formatting issues were manually solved. Differently from SARS-CoV, where the S1/S2 site is cleaved prior to fusion (i.e., at the host cell surface), in SARS-CoV-2 the cleavage occurs when the virus is assembled.12 Therefore, all the three constructs were modeled in their cleaved form, i.e. with the furin site cleaved between residues R685 and S686. However, for convenience, S1 and S2 subunits within each protomer have been assigned to the same chain (referred to as A/B/C), following the scheme used in 6VSB,32 where RBD of chain A is in the “up” conformation. Glycans were attributed segnames from G1 to G70 (G1–G64 for Mutant), as reported in Tables S9–S11 of the SI. Protonation states were assessed using PROPKA385 in the presence and absence of glycans, without registering any critical differences. The generated models were parametrized using PSFGEN and CHARMM36 all-atom additive force fields for protein, lipids, and glycans.86 Parameters for palmitoylated cysteine were taken from Jang et al.,87 whereas ions were treated using Beglov and Roux force fields.88 The systems were fully solvated with explicit water molecules described using the TIP3P model.89 The total number of atoms is 1,693,017 for the Open system (size: 23.2 nm × 23.2 nm × 36.7 nm), 1,658,797 for the Closed system (size: 23.2 nm × 23.2 nm × 35.9 nm), and 1,693,069 for the Mutant system (size: 23.2 nm × 23.2 nm × 36.7 nm).
Molecular dynamics (MD) simulations.
All-atom MD simulations were conducted on the Frontera computing system at the Texas Advanced Computing Center (TACC) using NAMD 2.14.35 The systems were initially relaxed through a series of minimization, melting (for the membrane), and equilibration cycles. During the first cycle, the protein, glycans, lipid heads (P atom for POPC, POPI, POPE, and POPS and O3 atom for CHL), solvent, and ions were kept fixed and the systems were subjected to an initial minimization of 10000 steps using the conjugate gradient energy approach. Subsequently, to allow the lipids tails to equilibrate, the temperature was incrementally changed from 10 to 310 K for 0.5 ns at 1 fs/step (NVT ensemble). The following simulation cycle was run at 2 fs/step, 1.01325 bar, and 310 K (NPT ensemble). Next, the systems were simulated with only the protein and glycans harmonically restrained at 5 kcal/mol to allow the full environment to relax in 2500 minimization steps and 0.5-ns simulations. Finally, all the restraints were released, and the systems were equilibrated for additional 0.5 ns. From this point, the production run was started, and frames were saved every 100 ps. Production MD simulations were run in triplicates for ~1 μs for Open and Mutant and ~0.6 μs for Closed (Table S1). To further explore the conformational space of the RBD in the “up” conformation, additional adaptive sampling simulations were run for Open and Mutant, which were also performed in triplicates for ~0.4 μs. Whereas velocities were randomly reinitialized, the initial coordinates were selected after principal component analysis (PCA) of RBD-A (Figure 2D). In detail, the minimum (replica 4), mean (replica 5), and maximum (replica 6) along PC1 were identified and the corresponding frames used a starting point for adaptive sampling simulations.
All simulations were performed using periodic boundary conditions and particle-mesh Ewald90 electrostatics for long-range electrostatic interactions with maximum grid spacing of 2 Å and evaluation every 3 time steps. Non-bonded van der Waals interactions and short-range electrostatic interactions were calculated with a cutoff of 12 Å and a switching distance of 10 Å. The SHAKE algorithm91 was employed to fix the length of all hydrogen-containing bonds, enabling the use of 2-fs integration time steps. All simulations were performed under the NPT ensemble using a Langevin thermostat92 (310 K) and a Nosé-Hoover Langevin barostat93,94 (1.01325 bar) to achieve temperature and pressure control, respectively.
Accessible surface area (ASA).
ASA was calculated using the measure sasa command implemented in VMD,95 which is based on the Shrake and Rupley algorithm,96 in combination with in-house Tcl scripts. Three separate ASA analyses were conducted by taking into account the S protein head (residues 16–1140), stalk (residues 1141–1234), and receptor binding motif (RBM) of the RBD (residues 400–508), respectively. The area covered by glycans (i.e., the glycan shield) was obtained after the subtraction of the ASA of the considered domain in the absence of glycans with the ASA in the presence of glycans. This value was calculated along the trajectory with a stride of 150, 20, and 20 frames between each assessment for head, stalk, and RBM-A, respectively. For each system (Open and Closed), the values were averaged across all the respective replicas and standard deviation was computed. Apart from the standard 1.4-Å probe, this analysis was repeated for 14 different (1-Å-interspersed) values of probe radius (from 2 to 15 Å). Note that additional ASA analyses on the whole RBD-A (residues 330–530) and on its noninteracting region (residues 330–399 and 509–530) were also analogously performed (see SI). Similarly, epitope-specific ASA analyses were conducted on the chain A of Open and Closed systems using only 7.2 and 18.6 Å as probe radii. ASA evaluations were conducted with a stride of 20 frames. The residues considered for each epitope are listed in Table S7.
PCA.
PCA was performed using the sklearn.decomposition.PCA function in the Scikit-learn library using python3.6.9.97 First, all simulations were aligned with mdtraj98 onto the same initial coordinates using Cα atoms of chain-A central scaffold (residues 747–783 and 920–1032). Next, simulation coordinates of RBD-A (residues 330–530) from all systems (Open, Mutant, and Closed) and replicas were concatenated and used to fit the transformation function. Subsequently, the fitted transformation function was applied to reduce the dimensionality of each system simulation RBD-A Cα coordinates. Note that it is important that all systems are transformed into the same PC space to evaluate the simulation variance across systems.
Angles calculation.
The lateral angle and axial angle were calculated using in-house Tcl scripts along with VMD.95 The axial angle is defined by three points corresponding to (i) the center of mass (COM) of RBD β-sheets (residues 394–403, 507–517, and 432–437), (ii - vertex) the COM of the central helices (residues 987–1032), and (iii) the COM of the top section of the central helices (residues 987–993). The lateral angle is described by three points corresponding to the (i) COM of RBD β-sheets at frame 0, (ii - vertex) COM of the top section of the central helices, and (iii) COM of RBD β-sheets at frame n. Both angles were evaluated at each frame along the trajectories as a variation (positive or negative) with respect to their initial value. The trajectories were aligned by the S protein central scaffold (residues 747–783 and 920–1032) including the central helices using the coordinates at frame 0 as a reference. Importantly, whereas the axial angle was calculated in a three-dimensional space defined by xyz coordinates, the lateral angle was assessed by considering the projection of the COMs onto a two-dimensional space defined only by xy coordinates. In this way, the lateral angle only accounts for lateral tilt/shift of the RBD, discarding any other motion along z.
Hydrogen bonds calculation.
Hydrogen bonds were calculated using the measure hbonds command implemented in VMD95 in combination with in-house Tcl scripts. Hydrogen bonds criteria were set as 3.5 Å for distance between heavy atoms and as 45° for angle between Acc-Don-Hyd. All frames across all replicas were considered for this analysis. Occupancy (%) was determined by counting the number of frames in which a specific hydrogen bond was formed with respect to the total number of frames.
Root-mean-square-deviation (RMSD).
RMSD of protein Cα atoms was computed using the measure rmsd command implemented in VMD95 in combination with in-house Tcl scripts. Different alignments were done before RMSD calculations using the initial coordinates of Cα atoms as a reference. In particular, for RBD-A RMSDs, Cα atoms of the S protein central scaffold (residues 747–783 and 920–1032) were used as a reference for alignment, whereas for the head, stalk, and CT RMSDs, the trajectories were aligned onto the Cα atoms of the residues of the respective regions.
Root-mean-square-fluctuations (RMSF).
RMSF was calculated using in-house python scripts along with mdtraj.98 RMSF was computed for each glycan for all chains across all replicas in Closed, Open, and Mutant. The trajectories were aligned onto the initial coordinates using the Cα atoms of the entire protein as a reference.
Supplementary Material
Acknowledgements
This work was supported by NIH GM132826, NSF RAPID MCB-2032054, an award from the RCSA Research Corp., a UC San Diego Moore’s Cancer Center 2020 SARS-COV-2 seed grant, and the Irish Research Council. LC is funded by a Visible Molecular Cell Consortium fellowship. We are grateful for the efforts of the Texas Advanced Computing Center (TACC) Frontera team and for the compute time made available through a Director’s Discretionary Allocation (made possible by the National Science Foundation award OAC-1818253), and also to the Irish Centre for High-End Computing (ICHEC) for computational resources. We thank Prof. Michael Feig and Dr. Lim Heo (Michigan State University), Prof. Syma Khalid (University of Southampton), Prof. Carlos Simmerling and his research group (SUNY Stony Brook), Prof. Ben Neuman (Texas A&M University), Prof. Greg Voth and Dr. Viviana Monje-Galvan (University of Chicago), Prof. Adrian Mulholland and his research group (University of Bristol), Prof. Julien Michel (University of Edinburgh) and his research group, Prof. Jean-Philip Piquemal (Sorbonne University), and Dr. Reda Rawi (NIH Vaccine Research Center) for system structure checks and helpful discussions.
References
- (1).Chan J. F. W.; Yuan S.; Kok K. H.; To K. K. W.; Chu H.; Yang J.; Xing F.; Liu J.; Yip C. C. Y.; Poon R. W. S.; Tsoi H. W.; Lo S. K. F.; Chan K. H.; Poon V. K. M.; Chan W. M.; Ip J. D.; Cai J. P.; Cheng V. C. C.; Chen H.; Hui C. K. M.; Yuen K. Y. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet 2020, 395 (10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chan J. F.-W.; Kok K.-H.; Zhu Z.; Chu H.; To K. K.-W.; Yuan S.; Yuen K.-Y. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. Emerg. Microbes Infect. 2020, 9 (1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Cheng V. C. C.; Lau S. K. P.; Woo P. C. Y.; Yuen K. Y. Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin. Microbiol. Rev. 2007, 20 (4), 660 LP – 694. 10.1128/CMR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cui J.; Li F.; Shi Z. L. Origin and Evolution of Pathogenic Coronaviruses Nature Reviews Microbiology. Nature Publishing Group; March 1, 2019, pp 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Remembering the Nearly 100,000 Lives Lost to Coronavirus in America - The New York Times https://www.nytimes.com/interactive/2020/05/24/us/us-coronavirus-deaths-100000.html (accessed May 25, 2020).
- (6).McKee M.; Stuckler D. If the World Fails to Protect the Economy, COVID-19 Will Damage Health Not Just Now but Also in the Future Nature Medicine. Nature Research; April 9, 2020, pp 640–642. 10.1038/s41591-020-0863-y [DOI] [PubMed] [Google Scholar]
- (7).Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181 (2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Davidson A. D.; Williamson M. K.; Lewis S.; Shoemark D.; Carroll M. W.; Heesom K.; Zambon M.; Ellis J.; Lewis P. A.; Hiscox J. A.; Matthews D. A. Characterisation of the Transcriptome and Proteome of SARS-CoV-2 Using Direct RNA Sequencing and Tandem Mass Spectrometry Reveals Evidence for a Cell Passage Induced in-Frame Deletion in the Spike Glycoprotein That Removes the Furin-like Cleavage Site. bioRxiv 2020, 2020.03.22.002204. 10.1101/2020.03.22.002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Belouzard S.; Chu V. C.; Whittaker G. R. Activation of the SARS Coronavirus Spike Protein via Sequential Proteolytic Cleavage at Two Distinct Sites. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (14), 5871–5876. 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Apellániz B.; Huarte N.; Largo E.; Nieva J. L. The Three Lives of Viral Fusion Peptides Chemistry and Physics of Lipids. Elsevier Ireland Ltd; 2014, pp 40–55. 10.1016/j.chemphyslip.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science 2020. 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 1–12. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang Q.; Zhang Y.; Wu L.; Niu S.; Song C.; Zhang Z.; Lu G.; Qiao C.; Hu Y.; Yuen K. Y.; Wang Q.; Zhou H.; Yan J.; Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 1–11. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Alejandra Tortorici M.; Walls A. C.; Lang Y.; Wang C.; Li Z.; Koerhuis D.; Boons G. J.; Bosch B. J.; Rey F. A.; de Groot R. J.; Veesler D. Structural Basis for Human Coronavirus Attachment to Sialic Acid Receptors. Nat. Struct. Mol. Biol. 2019, 26 (6), 481–489. 10.1038/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Watanabe Y.; Berndsen Z. T.; Raghwani J.; Seabright G. E.; Allen J. D.; McLellan J. S.; Wilson I. A.; Bowden T. A.; Ward A. B.; Crispin M. Vulnerabilities in Coronavirus Glycan Shields despite Extensive Glycosylation. bioRxiv 2020, 2020.02.20.957472. 10.1101/2020.02.20.957472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Watanabe Y.; Bowden T. A.; Wilson I. A.; Crispin M. Exploitation of Glycosylation in Enveloped Virus Pathobiology Biochimica et Biophysica Acta - General Subjects. Elsevier B.V. October 1, 2019, pp 1480–1497. 10.1016/j.bbagen.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Raman R.; Tharakaraman K.; Sasisekharan V.; Sasisekharan R. Glycan–Protein Interactions in Viral Pathogenesis Current Opinion in Structural Biology. Elsevier Ltd; October 1, 2016, pp 153–162. 10.1016/j.sbi.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Doores K. J.; Bonomelli C.; Harvey D. J.; Vasiljevic S.; Dwek R. A.; Burton D. R.; Crispin M.; Scanlan C. N. Envelope Glycans of Immunodeficiency Virions Are Almost Entirely Oligomannose Antigens. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (31), 13800–13805. 10.1073/pnas.1006498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Crispin M.; Ward A. B.; Wilson I. A. Structure and Immune Recognition of the HIV Glycan Shield. Annu. Rev. Biophys. 2018, 47 (1), 499–523. 10.1146/annurev-biophys-060414-034156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Stewart-Jones G. B. E.; Soto C.; Lemmin T.; Chuang G. Y.; Druz A.; Kong R.; Thomas P. V.; Wagh K.; Zhou T.; Behrens A. J.; Bylund T.; Choi C. W.; Davison J. R.; Georgiev I. S.; Joyce M. G.; Kwon Y. Do; Pancera M.; Taft J.; Yang Y.; Zhang B.; Shivatare S. S.; Shivatare V. S.; Lee C. C. D.; Wu C. Y.; Bewley C. A.; Burton D. R.; Koff W. C.; Connors M.; Crispin M.; Baxa U.; Korber B. T.; Wong C. H.; Mascola J. R.; Kwong P. D. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 2016, 165 (4), 813–826. 10.1016/j.cell.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yang M.; Huang J.; Simon R.; Wang L. X.; MacKerell A. D. Conformational Heterogeneity of the HIV Envelope Glycan Shield. Sci. Rep. 2017, 7 (1), 1–15. 10.1038/s41598-017-04532-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wei X.; Decker J. M.; Wang S.; Hui H.; Kappes J. C.; Wu X.; Salazar-Gonzalez J. F.; Salazar M. G.; Kilby J. M.; Saag M. S.; Komarova N. L.; Nowak M. A.; Hahn B. H.; Kwong P. D.; Shaw G. M. Antibody Neutralization and Escape by HIV-1. Nature 2003, 422 (6929), 307–312. 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- (23).Go E. P.; Herschhorn A.; Gu C.; Castillo-Menendez L.; Zhang S.; Mao Y.; Chen H.; Ding H.; Wakefield J. K.; Hua D.; Liao H.-X.; Kappes J. C.; Sodroski J.; Desaire H. Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble Gp140. J. Virol. 2015, 89 (16), 8245–8257. 10.1128/jvi.00628-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Cunningham A. L.; Harman A. N.; Donaghy H. DC-SIGN “AIDS” HIV Immune Evasion and Infection. Nat. Immunol. 2007, 8 (6), 556–558. 10.1038/ni0607-556 [DOI] [PubMed] [Google Scholar]
- (25).Goncalves A.-R.; Moraz M.-L.; Pasquato A.; Helenius A.; Lozach P.-Y.; Kunz S. Role of DC-SIGN in Lassa Virus Entry into Human Dendritic Cells. J. Virol. 2013, 87 (21), 11504–11515. 10.1128/jvi.01893-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bonomelli C.; Doores K. J.; Dunlop D. C.; Thaney V.; Dwek R. A.; Burton D. R.; Crispin M.; Scanlan C. N. The Glycan Shield of HIV Is Predominantly Oligomannose Independently of Production System or Viral Clade. PLoS One 2011, 6 (8). 10.1371/journal.pone.0023521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Altman M. O.; Angel M.; Košík I.; Trovão N. S.; Zost S. J.; Gibbs J. S.; Casalino L.; Amaro R. E.; Hensley S. E.; Nelson M. I.; Yewdell J. W. Human Influenza a Virus Hemagglutinin Glycan Evolution Follows a Temporal Pattern to a Glycan Limit. MBio 2019. 10.1128/mBio.00204-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Behrens A. J.; Crispin M. Structural Principles Controlling HIV Envelope Glycosylation Current Opinion in Structural Biology. Elsevier Ltd; June 1, 2017, pp 125–133. 10.1016/j.sbi.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Geijtenbeek T. B. H.; Kwon D. S.; Torensma R.; Van Vliet S. J.; Van Duijnhoven G. C. F.; Middel J.; Cornelissen I. L. M. H. A.; Nottet H. S. L. M.; KewalRamani V. N.; Littman D. R.; Figdor C. G.; Van Kooyk Y. DC-SIGN, a Dendritic Cell-Specific HIV-1-Binding Protein That Enhances Trans-Infection of T Cells. Cell 2000, 100 (5), 587–597. 10.1016/S0092-8674(00)80694-7 [DOI] [PubMed] [Google Scholar]
- (30).Watanabe Y.; Allen J. D.; Wrapp D.; McLellan J. S.; Crispin M. Site-Specific Analysis of the SARS-CoV-2 Glycan Shield. bioRxiv 2020. 10.1101/2020.03.26.010322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shajahan A.; Supekar N. T.; Gleinich A. S.; Azadi P. Deducing the N- and OGlycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. bioRxiv 2020, 2020.04.01.020966. 10.1101/2020.04.01.020966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wrapp D.; Wang N.; Corbett K. S.; Goldsmith J. A.; Hsieh C.-L.; Abiona O.; Graham B. S.; McLellan J. S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 1263 (March), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Shulla A.; Gallagher T. Role of Spike Protein Endodomains in Regulating Coronavirus Entry. J. Biol. Chem. 2009, 284 (47), 32725–32734. 10.1074/jbc.M109.043547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Thorp E. B.; Boscarino J. A.; Logan H. L.; Goletz J. T.; Gallagher T. M. Palmitoylations on Murine Coronavirus Spike Proteins Are Essential for Virion Assembly and Infectivity. J. Virol. 2006, 80 (3), 1280–1289. 10.1128/jvi.80.3.1280-1289.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable Molecular Dynamics with NAMD. Journal of Computational Chemistry. 2005. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Ghysels A.; Krämer A.; Venable R. M.; Teague W. E.; Lyman E.; Gawrisch K.; Pastor R. W. Permeability of Membranes in the Liquid Ordered and Liquid Disordered Phases. Nat. Commun. 2019, 10 (1), 1–12. 10.1038/s41467-019-13432-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Shahane G.; Ding W.; Palaiokostas M.; Orsi M. Physical Properties of Model Biological Lipid Bilayers: Insights from All-Atom Molecular Dynamics Simulations. J. Mol. Model. 2019, 25 (3), 1–13. 10.1007/s00894-019-3964-0 [DOI] [PubMed] [Google Scholar]
- (38).Saito H.; Morishita T.; Mizukami T.; Nishiyama K.-I.; Kawaguchi K.; Nagao H. Molecular Dynamics Study of Binary POPC Bilayers: Molecular Condensing Effects on Membrane Structure and Dynamics. J. Phys 2018, 12022 10.1088/1742-6596/1136/1/012022 [DOI] [Google Scholar]
- (39).Venable R. M.; Brown F. L. H.; Pastor R. W. Mechanical Properties of Lipid Bilayers from Molecular Dynamics Simulation Chemistry and Physics of Lipids. Elsevier Ireland Ltd; November 1, 2015, pp 60–74. 10.1016/j.chemphyslip.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Janosi L.; Gorfe A. A. Simulating POPC and POPC/POPG Bilayers: Conserved Packing and Altered Surface Reactivity. J. Chem. Theory Comput. 2010, 6 (10), 3267–3273. 10.1021/ct100381g [DOI] [PubMed] [Google Scholar]
- (41).Berkowitz M. L. Detailed Molecular Dynamics Simulations of Model Biological Membranes Containing Cholesterol Biochimica et Biophysica Acta - Biomembranes. Elsevier; January 1, 2009, pp 86–96. 10.1016/j.bbamem.2008.09.009 [DOI] [PubMed] [Google Scholar]
- (42).Róg T.; Pasenkiewicz-Gierula M. Cholesterol Effects on a Mixed-Chain Phosphatidylcholine Bilayer: A Molecular Dynamics Simulation Study. Biochimie 2006, 88 (5), 449–460. 10.1016/j.biochi.2005.10.005 [DOI] [PubMed] [Google Scholar]
- (43).Grant O. C.; Montgomery D.; Ito K.; Woods R. J. Analysis of the SARS-CoV-2 Spike Protein Glycan Shield: Implications for Immune Recognition. 10.1101/2020.04.07.030445 [DOI] [PMC free article] [PubMed]
- (44).Jiang S.; Hillyer C.; Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends in Immunology. 2020. 10.1016/j.it.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Yuan M.; Wu N. C.; Zhu X.; Lee C.-C. D.; So R. T. Y.; Lv H.; Mok C. K. P.; Wilson I. A. A Highly Conserved Cryptic Epitope in the Receptor-Binding Domains of SARSCoV-2 and SARS-CoV. Science (80-. ). 2020. 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ng S.; Nachbagauer R.; Balmaseda A.; Stadlbauer D.; Ojeda S.; Patel M.; Rajabhathor A.; Lopez R.; Guglia A. F.; Sanchez N.; Amanat F.; Gresh L.; Kuan G.; Krammer F.; Gordon A. Novel Correlates of Protection against Pandemic H1N1 Influenza A Virus Infection. Nat. Med. 2019. 10.1038/s41591-019-0463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Urbanowicz R. A.; Wang R.; Schiel J. E.; Keck Z.; Kerzic M. C.; Lau P.; Rangarajan S.; Garagusi K. J.; Tan L.; Guest J. D.; Ball J. K.; Pierce B. G.; Mariuzza R. A.; Foung S. K. H.; Fuerst T. R. Antigenicity and Immunogenicity of Differentially Glycosylated Hepatitis C Virus E2 Envelope Proteins Expressed in Mammalian and Insect Cells. J. Virol. 2019, 93 (7). 10.1128/jvi.01403-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science (80-. ). 2020, 367 (6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gui M.; Song W.; Zhou H.; Xu J.; Chen S.; Xiang Y.; Wang X. Cryo-Electron Microscopy Structures of the SARS-CoV Spike Glycoprotein Reveal a Prerequisite Conformational State for Receptor Binding. Cell Res. 2017. 10.1038/cr.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Brouwer P. J. M.; Caniels T. G.; K. van der Straten; Snitselaar J. L.; Aldon Y.; Bangaru S.; Torres J. L.; Okba N. M. A.; Claireaux M.; Kerster G.; Bentlage A. E. H.; Haaren M. M. van; Guerra D.; Burger J. A.; Schermer E. E.; Verheul K. D.; Velde N. van der; Kooi A. van der; Schooten J. van; Breemen M. J. van; Bijl T. P. L.; Sliepen K.; Aartse A.; Derking R.; Bontjer I.; Kootstra N. A.; Wiersinga W. J.; Vidarsson G.; Haagmans B. L.; Ward A. B.; Bree G. J. de; Sanders R. W.; Gils M. J. van. Potent Neutralizing Antibodies from COVID-19 Patients Define Multiple Targets of Vulnerability. bioRxiv 2020, 2020.05.12.088716. 10.1101/2020.05.12.088716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chi X.; Yan R.; Zhang J.; Zhang G.; Zhang Y.; Hao M.; Zhang Z.; Fan P.; Dong Y.; Yang Y.; Chen Z.; Guo Y.; Zhang J.; Li Y.; Song X.; Chen Y.; Xia L.; Fu L.; Hou L.; Xu J.; Yu C.; Li J.; Zhou Q.; Chen W. A Potent Neutralizing Human Antibody Reveals the N-Terminal Domain of the Spike Protein of SARS-CoV-2 as a Site of Vulnerability. bioRxiv 2020, 2020.05.08.083964. 10.1101/2020.05.08.083964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wrapp D.; De Vlieger D.; Corbett K. S.; Schepens B.; Saelens X.; Mclellan J. S. Article Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell 2020, 181, 1004–1015.e15. 10.1016/j.cell.2020.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zost S. J.; Gilchuk P.; Case J. B.; Binshtein E.; Chen R. E.; Reidy J. X.; Trivette A.; Nargi R. S.; Sutton R. E.; Suryadevara N.; Williamson L. E.; Chen E. C.; Jones T.; Day S.; Myers L.; Hassan A. O.; Kafai N. M.; Winkler E. S.; Fox J. M.; Steinhardt J. J.; Ren K.; Loo Y.-M.; Kallewaard N. L.; Martinez D. R.; Schäfer A.; Gralinski L. E.; Baric R. S.; Thackray L. B.; Diamond M. S.; Carnahan R. H.; Crowe J. E. Potently Neutralizing Human Antibodies That Block SARS-CoV-2 Receptor Binding and Protect Animals. bioRxiv 2020, 2020.05.22.111005. 10.1101/2020.05.22.111005 [DOI] [Google Scholar]
- (54).Ejemel M.; Li Q.; Hou S.; Schiller Z. A.; Wallace A. L.; Amcheslavsky A.; Yilmaz N. K.; Toomey J. R.; Schneider R.; Close B. J.; Chen D.-Y.; Conway H. L.; Saeed M.; Cavacini L. A.; Klempner M. S.; Schiffer C. A.; Wang Y. IgA MAb Blocks SARS-CoV-2 Spike-ACE2 Interaction Providing Mucosal Immunity. bioRxiv 2020, 2020.05.15.096719. 10.1101/2020.05.15.096719 [DOI] [Google Scholar]
- (55).Tai W.; Zhang X.; He Y.; Jiang S.; Du L. Identification of SARS-CoV RBD-Targeting Monoclonal Antibodies with Cross-Reactive or Neutralizing Activity against SARS-CoV-2. Antiviral Res. 2020, 179, 104820 10.1016/j.antiviral.2020.104820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wu Y.; Li C.; Xia S.; Tian X.; Kong Y.; Wang Z.; Gu C.; Zhang R.; Tu C.; Xie Y.; Yang Z.; Lu L.; Jiang S.; Ying T. Identification of Human Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe 2020. 10.1016/j.chom.2020.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zheng Z.; Monteil V. M.; Maurer-Stroh S.; Yew C. W.; Leong C.; Mohd-Ismail N. K.; Arularasu S. C.; Chow V. T. K.; Lin R. T. P.; Mirazimi A.; Hong W.; Tan Y.-J. Monoclonal Antibodies for the S2 Subunit of Spike of SARS-CoV Cross-React with the Newly-Emerged SARS-CoV-2. bioRxiv 2020, 2020.03.06.980037. 10.1101/2020.03.06.980037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Wu Y.; Wang F.; Shen C.; Peng W.; Li D.; Zhao C.; Li Z.; Li S.; Bi Y.; Yang Y.; Gong Y.; Xiao H.; Fan Z.; Tan S.; Wu G.; Tan W.; Lu X.; Fan C.; Wang Q.; Liu Y.; Zhang C.; Qi J.; Gao G. F.; Gao F.; Liu L. A Noncompeting Pair of Human Neutralizing Antibodies Block COVID-19 Virus Binding to Its Receptor ACE2. Science (80-. ). 2020, eabc2241. 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Wang C.; Li W.; Drabek D.; Okba N. M. A.; van Haperen R.; Osterhaus A. D. M. E.; van Kuppeveld F. J. M.; Haagmans B. L.; Grosveld F.; Bosch B. J. A Human Monoclonal Antibody Blocking SARS-CoV-2 Infection. Nat. Commun. 2020, 11 (1), 1–6. 10.1038/s41467-020-16256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Pinto D.; Park Y.; Beltramello M.; Walls A. C.; Tortorici M. A.; Bianchi S.; Jaconi S.; Culap K.; Zatta F.; Marco A. De; Peter A.; Guarino B.; Spreafico R.; Cameroni E.; Case J. B.; Chen R. E.; Havenar-daughton C.; Snell G.; Telenti A.; Virgin H. W.; Lanzavecchia A.; Diamond M. S.; Fink K.; Veesler D.; Corti D. Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody. Nature 2020, 1–10. 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- (61).Yuan M.; Wu N. C.; Zhu X.; Lee C.-C. D.; So R. T. Y.; Lv H.; Mok C. K. P.; Wilson I. A. A Highly Conserved Cryptic Epitope in the Receptor-Binding Domains of SARSCoV-2 and SARS-CoV. bioRxiv 2020, 2020.03.13.991570. 10.1101/2020.03.13.991570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Huo J.; Zhao Y.; Ren J.; Zhou D.; Duyvesteyn H. M.; Ginn H. M.; Carrique L.; Malinauskas T.; Ruza R. R.; Shah P. N.; Tan T. K.; Rijal P.; Coombes N.; Bewley K.; Radecke J.; Paterson N. G.; Supasa P.; Mongkolsapaya J.; Screaton G. R.; Carroll M.; Townsend A.; Fry E. E.; Owens R. J.; Stuart D. I. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. bioRxiv 2020, 2020.05.05.079202. 10.1101/2020.05.05.079202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Harbison A. M.; Brosnan L. P.; Fenlon K.; Fadda E. Sequence-to-Structure Dependence of Isolated IgG Fc Complex Biantennary N-Glycans: A Molecular Dynamics Study. Glycobiology 2019, 29 (1), 94–103. 10.1093/glycob/cwy097 [DOI] [PubMed] [Google Scholar]
- (64).Walls A. C.; Park Y. J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181 (2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Song W.; Gui M.; Wang X.; Xiang Y. Cryo-EM Structure of the SARS Coronavirus Spike Glycoprotein in Complex with Its Host Cell Receptor ACE2. PLOS Pathog. 2018, 14 (8), e1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Pallesen J.; Wang N.; Corbett K. S.; Wrapp D.; Kirchdoerfer R. N.; Turner H. L.; Cottrell C. A.; Becker M. M.; Wang L.; Shi W.; Kong W. P.; Andres E. L.; Kettenbach A. N.; Denison M. R.; Chappell J. D.; Graham B. S.; Ward A. B.; McLellan J. S. Immunogenicity and Structures of a Rationally Designed Prefusion MERS-CoV Spike Antigen. Proc. Natl. Acad. Sci. U. S. A. 2017. 10.1073/pnas.1707304114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Amaro R. E.; Mulholland A. J. A Community Letter Regarding Sharing Biomolecular Simulation Data for COVID-19. J. Chem. Inf. Model. 2020, 0–6. 10.1021/acs.jcim.0c00319 [DOI] [PubMed] [Google Scholar]
- (68).Miroshnikov K. A.; Marusich E. I.; Cerritelli M. E.; Cheng N.; Hyde C. C.; Steven A. C.; Mesyanzhinov V. V. Engineering Trimeric Fibrous Proteins Based on Bacteriophage T4 Adhesins; 1998; Vol. 11. [DOI] [PubMed] [Google Scholar]
- (69).Šali A.; Blundell T. L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234 (3), 779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- (70).Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; Yuan M. L.; Zhang Y. L.; Dai F. H.; Liu Y.; Wang Q. M.; Zheng J. J.; Xu L.; Holmes E. C.; Zhang Y. Z. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579 (7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7 (1), 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Drozdetskiy A.; Cole C.; Procter J.; Barton G. J. JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 2015, 43, 389–394. 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Hartmann M. D.; Ridderbusch O.; Zeth K.; Albrecht R.; Testa O.; Woolfson D. N.; Sauer G.; Dunin-Horkawicz S.; Lupas A. N.; Alvarez B. H. A Coiled-Coil Motif That Sequesters Ions to the Hydrophobic Core. Proc. Natl. Acad. Sci. U. S. A. 2009. 10.1073/pnas.0907256106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Zhang Y. I-TASSER Server for Protein 3D Structure Prediction. BMC Bioinformatics 2008, 9 (1), 1–8. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Roy A.; Kucukural A.; Zhang Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5 (4), 725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Yang J.; Roy A.; Zhang Y. Protein-Ligand Binding Site Recognition Using Complementary Binding-Specific Substructure Comparison and Sequence Profile Alignment. Struct. Bioinforma. 2013, 29 (20), 2588–2595. 10.1093/bioinformatics/btt447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Jo S.; Song K. C.; Desaire H.; MacKerell A. D.; Im W. Glycan Reader: Automated Sugar Identification and Simulation Preparation for Carbohydrates and Glycoproteins. J. Comput. Chem. 2011. 10.1002/jcc.21886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Jo S.; Kim T.; Iyer V. G.; Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 2967–2970. 10.1002/jcc [DOI] [PubMed] [Google Scholar]
- (79).Zhang Y.; Zhao W.; Mao Y.; Wang S.; Zhong Y.; Su T.; Gong M.; Lu X.; Cheng J.; Yang H. Site-Specific N-Glycosylation Characterization of Recombinant SARS-CoV-2 Spike Proteins Using High-Resolution Mass Spectrometry. bioRxiv 2020. 10.1101/2020.03.28.013276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Park S. J.; Lee J.; Qi Y.; Kern N. R.; Lee H. S.; Jo S.; Joung I.; Joo K.; Lee J.; Im W. CHARMM-GUI Glycan Modeler for Modeling and Simulation of Carbohydrates and Glycoconjugates. Glycobiology 2019. 10.1093/glycob/cwz003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Van Meer G.; Voelker D. R.; Feigenson G. W. Membrane Lipids: Where They Are and How They Behave. Nature Reviews Molecular Cell Biology. February 2008, pp 112–124. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Casares D.; Escribá P. V.; Rosselló C. A. Membrane Lipid Composition: Effect on Membrane and Organelle Structure, Function and Compartmentalization and Therapeutic Avenues. Int. J. Mol. Sci. 2019, 20 (9). 10.3390/ijms20092167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Krijnse-Locker J.; Ericsson M.; Rottier P. J. M.; Grittiths G. Characterization of the Budding Compartment of Mouse Hepatitis Virus: Evidence That Transport from the RER to the Golgi Complex Requires Only One Vesicular Transport Step. [DOI] [PMC free article] [PubMed]
- (84).Klumperman J.; Krijnse Locker J.; Meijer A.; Horzinek M. C.; Geuze And, H. J.; Rottier2 P. J. M. Coronavirus M Proteins Accumulate in the Golgi Complex beyond the Site of Virion Budding; 1994. [DOI] [PMC free article] [PubMed]
- (85).Olsson M. H. M.; SØndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical p K a Predictions. J. Chem. Theory Comput. 2011. 10.1021/ct100578z [DOI] [PubMed] [Google Scholar]
- (86).Huang J.; Mackerell A. D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013. 10.1002/jcc.23354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Jang H.; Muratcioglu S.; Gursoy A.; Keskin O.; Nussinov R. Membrane-Associated Ras Dimers Are Isoform-Specific: K-Ras Dimers Differ from H-Ras Dimers. Biochem. J. 2016. 10.1042/BCJ20160031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Beglov D.; Roux B. Finite Representation of an Infinite Bulk System: Solvent Boundary Potential for Computer Simulations. J. Chem. Phys. 1994. 10.1063/1.466711 [DOI] [Google Scholar]
- (89).Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983. 10.1063/1.445869 [DOI] [Google Scholar]
- (90).Darden T.; York D.; Pedersen L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98 (12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- (91).Ryckaert J. P.; Ciccotti G.; Berendsen H. J. C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23 (3), 327–341. 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- (92).Turq P.; Lantelme F.; Friedman H. L.; Lantelme F. Brownian Dynamics with Hydrodynamic Interactions. J. Chem. Phys. 1977, 66, 3045 10.1063/1.434317 [DOI] [Google Scholar]
- (93).Martyna G. J.; Tobias D. J.; Klein M. L. Constant Pressure Molecular Dynamics Algorithms. J. Chem. Phys. 1994, 101 (5), 4177–4189. 10.1063/1.467468 [DOI] [Google Scholar]
- (94).Feller S. E.; Zhang Y.; Pastor R. W.; Brooks B. R. Constant Pressure Molecular Dynamics Simulation: The Langevin Piston Method. J. Chem. Phys. 1995, 103 (11), 4613–4621. 10.1063/1.470648 [DOI] [Google Scholar]
- (95).Humphrey W.; Dalke A.; Schulten K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- (96).Shrake A.; Rupley J. A. Environment and Exposure to Solvent of Protein Atoms. Lysozyme and Insulin. J. Mol. Biol. 1973. 10.1016/0022-2836(73)90011-9 [DOI] [PubMed] [Google Scholar]
- (97).Pedregosa F.; Michel V.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Vanderplas J.; Cournapeau D.; Pedregosa F.; Varoquaux G.; Gramfort A.; Thirion B.; Grisel O.; Dubourg V.; Passos A.; Brucher M.; Perrot M.; Duchesnay É. Scikit-Learn: Machine Learning in Python; 2011; Vol. 12. [Google Scholar]
- (98).McGibbon R. T.; Beauchamp K. A.; Harrigan M. P.; Klein C.; Swails J. M.; Hernández C. X.; Schwantes C. R.; Wang L. P.; Lane T. J.; Pande V. S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015, 109 (8), 1528–1532. 10.1016/j.bpj.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.