Abstract
Extracellular vesicles (EVs) are phospholipid bilayer membrane-enclosed structures containing RNAs, proteins, lipids, metabolites, and other molecules, secreted by various cells into physiological fluids. EV-mediated transfer of biomolecules is a critical component of a variety of physiological and pathological processes. Potential applications of EVs in novel diagnostic and therapeutic strategies have brought increasing attention. However, EV research remains highly challenging due to the inherently complex biogenesis of EVs and their vast heterogeneity in size, composition, and origin. There is a need for the establishment of standardized methods that address EV heterogeneity and sources of pre-analytical and analytical variability in EV studies. Here, we review technologies developed for EV isolation and characterization and discuss paths toward standardization in EV research.
Keywords: extracellular vesicles, exosomes, standardization, characterization, isolation, molecular profiling
Highlights
Despite the substantial recent progress made in extracellular vesicle (EV) research, our understanding of the functional and mechanistic biology of EVs and their relevance to specific pathophysiological states remains limited.
Detailed characterization of the molecular composition of EVs and EV subpopulations remains a challenge.
Alternative, similar, or identical experimental approaches may often lead to substantially different EV profiling results in different laboratories.
Standard protocols for specimen procurement, collection, preprocessing, EV isolation, analytical characterization, and data analysis/interpretation need to be developed for specialized applications and analytical workflows, optimized, documented, cross-evaluated by several laboratories, and disseminated to further accelerate progress toward further understanding of EV biology and development of novel EV-based diagnostic and therapeutic approaches.
Biogenesis, Biological Significance, and Applications of EVs
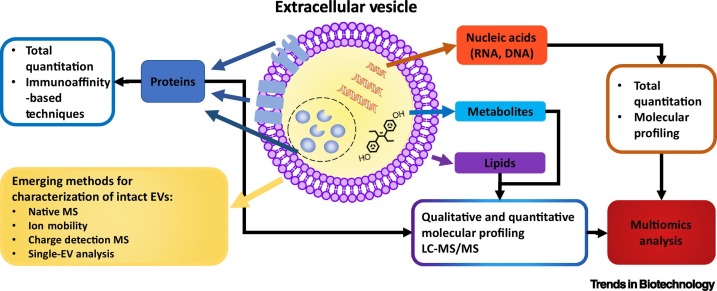
EVs are phospholipid bilayer membrane-enclosed biological entities present in a wide range of physiological fluids (Figure 1 , Key Figure). Secreted by various types of cells, EVs carry important biomolecules originating from their parent cells. There are several biogenesis pathways involved in the generation of EVs, according to which EVs can be subdivided into several types, including exosomes and microvesicles (MVs) [1., 2., 3.]. Exosomes (~30–150 nm) are generated through inward budding of endosomal membranes during their maturation into multivesicular endosomes (MVEs), while MVs (~100–1000 nm) are formed by outward budding of the plasma membrane [1]. However, they still share several common features, including: clustering of lipids and proteins onto endosomal/plasma membranes [4,5]; sequestration of cytosolic nucleic acids, proteins, and other biomolecules via various sorting machineries followed by budding and fission of small vesicles [4,5]; and intercellular trafficking of cargoes that involve docking, fusion, and uptake [4,5]. Vesicles formed during apoptosis are larger than 1000 nm and are termed apoptotic bodies [6]. The recently coined term ‘exomeres’ describes the smallest (<50 nm) non-membrane-bound nanoparticles and larger macromolecular complexes that can also be included under the umbrella term ‘EVs’ (Figure 2 ) [7,8].
Figure 1.
Key Figure. Structure, Biomolecular Cargo, and Characterization of Extracellular Vesicles (EVs).
Abbreviations: LC, liquid chromatography; MS, mass spectrometry.
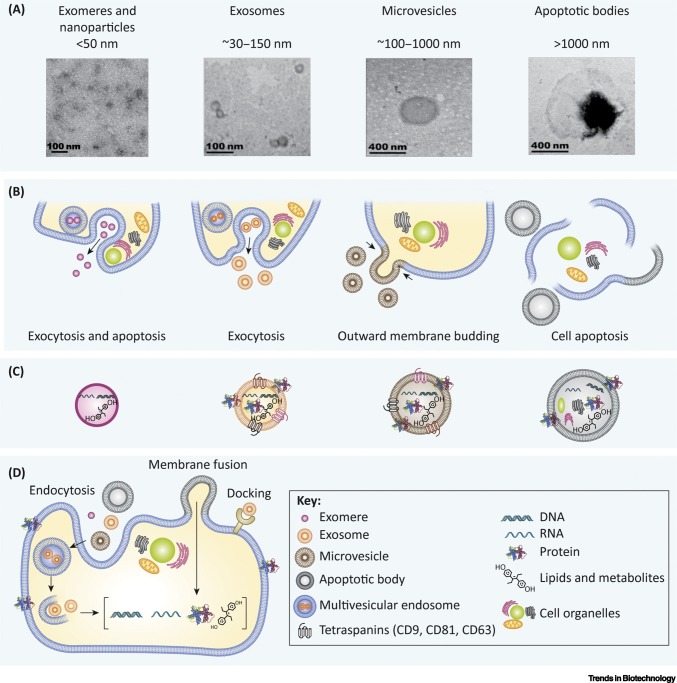
Figure 2.
Overview of Extracellular Vesicle (EV) Biogenesis, Secretion, and Uptake.
(A) Transmission electron microscopy images of EV subtypes (exomeres, exosomes, microvesicles, and apoptotic bodies) and their approximate sizes [1,6,214,215]. (B) EV biogenesis pathways. Exosomes are formed through inward budding of the cell membrane and the formation of multivesicular endosomes, which capture exosomes then fuse with the cell membrane and release exosomes through exocytosis [1]. Microvesicles are formed through outward budding of the cell membrane and apoptotic bodies are formed during cell apoptosis and death [1,6]. (C) EV subtype cargo. Each subtype of EVs contains a different cargo. Exosomes and microvesicles contain membrane proteins and tetraspanins, while apoptotic bodies also carry fragments of cell organelles from apoptosis [12,13]. (D) EV uptake occurs through the internalization of the EV into the cell by either docking or fusion of the membranes. Endosomes can also be created, and then release their EV content into the cell [4,5]. Reprinted, with permission, from referenced sources.
The sorting machinery is diverse in terms of the intracellular complexes involved and can be broadly classified into two main pathways, endosomal sorting complex required for transport (ESCRT) dependent and ESCRT independent. The ESCRT-dependent machinery acts in a stepwise manner where a series of ESCRT subcomplexes (e.g., TSG101, CHMP proteins) are involved [9,10]. The ESCRT-independent pathways involve the ceramide-dependent pathway, which generates membrane subdomains and tetraspanins such as CD63, CD81, and CD9 to form clusters on the membranes and induce inward budding of vesicles and the formation of EVs [11]. In addition, cytosolic proteins, such as heat shock 70 kDa protein (HSP70), are sequestered into exosomes derived from most cell types [12]. Nucleic acids (DNA, mRNA, and miRNA) are another important type of cargo molecules carried by EVs [13]. The sorting of miRNA is sequence dependent, leading to differential sorting of specific miRNAs into EVs [14., 15., 16.]. Overall, there are multiple pathways of EV formation, which leads to the release of a heterogeneous population that may vary widely in size, composition, and function. So far, it remains difficult to assign a particular pathway based on the subpopulation of isolated vesicles. Hence, there is a clear need for a better understanding of the factors that differentiate the biogenesis, sorting, and release of EVs.
The International Society for EVs (ISEV) recommends the use of ‘EV’ as an umbrella term for these types of vesicles, due to the difficulty in assigning an EV to a particular biogenesis pathway, unless the EV is caught in the course of release by live-imaging techniques. The ISEV also suggests classifying EVs by referring to their physical characteristics like size and density, their differing biochemical composition, and the amount of surface charge, among other characteristics [6].
Once EVs are released into the extracellular space, they undergo internalization by recipient cells (endocytosis, phagocytosis, or micropinocytosis), followed by transfer of the EVs’ genetic materials and proteins, which further interact with cellular signaling pathways of the target cells [9,17., 18., 19.]. EV cargo is used to deliver messages between immune cells [dendritic cells (DCs), B cells, T cells], leading to either immunosuppressive or immune-activating effects on the immune response [20]. The cells of the central nervous system (CNS) use EVs as a major route of signal transfer to other neuronal cells. More importantly, EVs have been found to have roles in the spreading of neurodegenerative diseases such as Parkinson’s and Alzheimer’s diseases between anatomically connected regions in the CNS [21,22]. In the cardiovascular system, cardiac fibroblast EVs are enriched with miR-21, a crucial paracrine signaling mediator of cardiac hypertrophy [23]. Furthermore, tumor-derived EVs have been reported to have a role in the hallmarks of cancer, such as tumor growth, invasiveness, evading apoptosis, immune cell modulation, resistance, and metastasis [24,25].
Bearing a variety of biological cargoes, EVs are deemed to reflect the physiological state of parent cells [26]. The components of EVs have been studied in the probing of pathophysiological states of the host as potential biomarkers for the diagnosis and monitoring of diseases [27]. For example, miRNA profiling of epithelially derived EVs from blood samples of colorectal cancer (CRC) patients showed elevated levels of 13 EpCAM+-EV miRNAs compared with healthy individuals [28]. In addition, EV miRNA profiling of stage I non-small cell lung cancer (NSCLC) patients was able to discriminate between various stages of cancer [29]. With respect to proteins, CD147 was found to be enriched in EVs secreted by CRC cell lines as well as in the serum of CRC patients [30]. The level of EVs positive for both CD9 and CD147 was significantly greater in cancer patients than in healthy donors [30]. Although these studies highlight the diagnostic potential of EVs, the challenges in EV isolation and characterization (discussed in ‘EV Isolation Methods’ and ‘EV Characterization: Challenges and Needs’) directly impact the specificity and sensitivity of EV biomarkers and their ability to translate into a robust diagnostic tool.
EVs have been exploited to transport a variety of therapeutic molecules, such as siRNA, miRNA, and small molecules [31]. Various methods, such as transfection, incubation, sonication, freeze–thaw cycles, and electroporation, have been developed for the loading of these molecules [32]. The choice of loading method depends on the type of cargo molecule; for example, for loading of small noncoding RNA (siRNA and miRNA), transfection of parent cells before isolation and electroporation of EVs post-isolation are currently the preferred methods. Small-molecule-based anticancer drugs such as doxorubicin and paclitaxel have been incorporated into EVs using incubation and have demonstrated enhanced antitumor activity in preclinical studies [33]. One of the unique advantages that engineered EVs offer compared with other delivery vectors is the ability to incorporate ligands, which can specifically target cancer cells and evade immune responses. The Kalluri laboratory has shown that CD47+ EVs loaded with siRNA against KRAS target only KRAS-mutated tumors and display enhanced therapeutic efficacy compared with liposomes carrying similar amounts of the agent [34]. Colleagues from the Amiji laboratory have used a novel approach of modulating the EV miRNA content derived from cancer cells to reprogram macrophages from M2 (protumor) to M1 (antitumor) [35]. In addition, EV immune signaling has been utilized to produce an antitumor effect [31]. A recent study investigated the potential synergistic effects of DC-derived EVs with or without a programmed cell death-1 (PD-1) antibody for the treatment of hepatocellular carcinoma with sorafenib [36]. Based on the potential of EVs as drug delivery vehicles and immunotherapies, several companies, such as Capricor Therapeutics, Codiak Biosciences, Evox Therapeutics, and Puretech Health, have started product development efforts to translate EV therapeutics to the clinic [37]. A search for the keywords ‘exosomes’ or ‘extracellular vesicles’ on the ClinicalTrials.gov website showed a number of clinical trials, which are recruiting patients for various types of disease including several types of cancer, neurodegenerative diseases, anxiety, diabetes, and COVID-19 [38]. Due to the surface expression of antigens from parent cells, DC-derived exosomes have been used for vaccine delivery and proved safe in multiple Phase I trials in different types of cancer [39]. Some promising results have been shown in Phase II clinical studies using these EVs loaded with tumor antigen as a vaccine against NSCLC in combination with cyclophosphamide [39]. Despite these interesting advances in therapeutic applications of EVs, there are several challenges in translating EV therapeutics to the clinic. One of the major challenges is the heterogeneity in content and composition inherent in the production of EVs by parent cells through various complex biogenesis pathways. This heterogeneity can lead to intrabatch and interbatch variabilities in the large-scale production of EVs. For the successful translation of EVs to the clinic, the identification of critical quality attributes (CQAs) (e.g., size, purity, molecular composition) that impact the potency and stability of the product is essential.
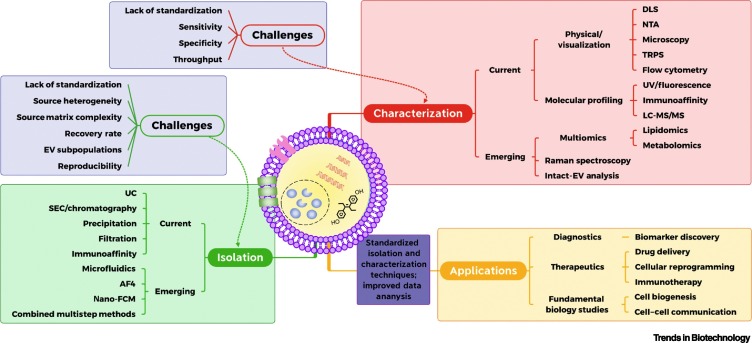
In this review, we provide a comprehensive overview of conventional and emerging approaches for the isolation and characterization of EVs, as well as the main challenges that these technologies encounter. Also, we highlight possible paths toward the standardization of the discussed approaches and identified potential benefits and disadvantages of these techniques with respect to dissemination and standardization. This critical evaluation of published reports and the provided assessment of the advantages and deficiencies of the described approaches will help the reader to learn about the current state of EV research and to identify the most efficient paths toward selecting, implementing, and standardizing specific technologies of interest in future studies in relevant research fields.
EV Isolation: Challenges and Needs
Sources of EVs: EV Heterogeneity and Complexity
EVs have been isolated from various physiological fluids. Blood is one of the most abundant sources of EVs, with an estimated concentration of 5–15 ×108 particles/ml [40., 41., 42.]. One of the challenging aspects of the isolation of EVs from blood is that it contains lipoproteins [<35 nm, density 1.06–1.20 g/cm3 very-low-, low-, and high-density lipoprotein (VLDL, LDL, HDL)] and chylomicrons [75–1100 nm, density <0.930 g/cm3; a.k.a. ultralow-density lipoproteins (ULDLs)], which overlap in size and density with EVs [43] and cannot be completely removed by conventional isolation methods [e.g., ultracentrifugation (UC)] [44,45]. Other factors that impact the amount, purity and, heterogeneity of EVs from blood include sample collection, handling, storage conditions, stability, anticoagulants, volume of blood collection, time of blood collection, and the age, sex, disease state, and fed/fast status of the animal/patient [46,47]. In cases of advanced-stage ovarian cancer, hemorrhagic malignant ascites will appear [48] containing abundant EVs secreted by various cell types along with cells, cytosolic components, and extracellular matrix (ECM) fragments [27,49]. These additional components not only contribute to the chemical composition of EV subpopulations but also alter the physical properties, such as the viscosity of the blood, increasing the difficulty of isolating high-purity EV populations. Further heterogeneity arises from the biogenesis of the various vesicles released from the cells.
Scalability and Throughput
Most of the traditional laboratory-scale methods used for EV isolation employ multiple steps of isolation, which poses challenges for scaling up the processing to large volumes, and are low throughput. Recently, tangential flow filtration (TFF) coupled with chromatography-based methods have been developed for the large-scale and current Good Manufacturing Practice (cGMP)-based production of EVs [50]. This additional separation step enabled the more efficient removal of protein contaminants and resulted in a yield of EVs similar to that with UC-based protocols. Similarly, size-exclusion-based EV isolation protocols can be readily scaled up (see ‘EV Isolation Methods’).
Recommendations for Standardization of Sample Collection, Handling, and Storage
Given the complexity and heterogeneity of biological fluids, it becomes essential to minimize pre-isolation and pre-analytical variables through the standardization of sample collection, storage, and handling. Some of the recommendations and measures to minimize artifacts for downstream isolation and analysis are listed here.
Sample Collection, Sample Matching, Sample Size, and Data Collection
For the isolation of EVs from cultured cells, it is recommended to use either serum-free medium, if possible, or EV-free serum as a growth supplement in cell culture medium [51]. For clinical blood sample collection, it is important to use a needle of a gauge that minimizes shear forces responsible for platelet activation and the release of platelet- and red blood cell-derived EVs [52] and to discard the first bolus of the collected blood, where the cell debris caused by the venipuncture can contaminate the EV isolate [46]. For each specimen that is collected, parent cells are also recommended to be collected whenever possible, enabling molecular matching, profiling, differential analysis of cellular and EV components, and the determination of specific molecular signatures associated with changes in the EV source, such as the stage of the disease. Fed/fasting status and the time of sample collection (morning/night) affect EV levels in samples, but more studies are needed to determine the optimal parameters before a recommendation can be made [46]. When designing experiments for the discovery of biomarkers and profiling of EV components, it becomes essential to have a sample size that gives a statistical power of at least 80% [53]. Finally, information regarding the age, sex, race, disease state, and treatment state of the patient/animal should be collected to understand the impact of each of these parameters on the profiling of the EV content [54].
Sample Handling and Processing
Most published studies reporting the isolation of EVs from blood suggest using plasma for isolation, as serum contains platelet-derived EVs, which are mostly released during clotting [47]. Furthermore, the choice of anticoagulant (heparin, citrate, EDTA) can affect the results. A number of studies suggest the use of EDTA as an anticoagulant for downstream EV-RNA analysis, as it prevents the formation of EV-cell aggregates, inhibits platelet-derived release, and because heparin inhibits PCR reactions [55]. Initial centrifugation to separate components such as cells [red blood cells (RBCs), white blood cells (WBCs), platelets] from EVs should be performed at speeds that minimize the release of cellular components. Also, precautions must be taken to minimize processing time to avoid sample degradation due to temperature and enzymatic activities such as RNase and protease hydrolysis.
Sample Stability and Storage
A number of studies have been performed on various biofluids, such as urine, blood, and bronchoalveolar lavage (BAL) to evaluate the impact of various storage temperatures (4°C, −20°C, and −80°C) and freeze–thaw cycles (one to ten) on the size, composition, and functionality of EV isolates, which have been reviewed previously [56]. Overall, current evidence suggests that −80°C is the best-suited temperature to preserve EV contents for downstream molecular profiling. However, freeze–thaw cycles should be minimized, as EV aggregation and lysis may occur, resulting in overestimation of EV size, underestimation of EV count, and loss of cargo during isolation procedures. Furthermore, when EVs intended for drug delivery applications go through freeze–thaw cycles, loss of cargo leads to loss of potency [57].
EV Isolation Methods
High-efficiency isolation of EVs or EV subpopulations and separation from contaminating proteins and other possible matrix contaminants is needed to ensure accurate inferences of the biological activity and function of EVs or a specific EV subpopulation of interest. Here we describe current and commonly used methods of EV isolation and their advantages and disadvantages (summarized in Table 1 ).
Table 1.
Overview of the Described EV Isolation Methods and Their Main Advantages and Disadvantages
| Method | Principle | Advantage | Disadvantage | Scalabilitya | Costa | Refs |
|---|---|---|---|---|---|---|
| UC | Isolation by differential centrifugation | Low protein contamination | Low throughput, isolates similarly sized particles, potential damage to EVs | + | $$$$ | [60., 61., 62., 63.] |
| Density gradient | Separates EVs by density after initial isolation by UC or alternative techniques | Increased purity | Low throughput, lower yield | ++ | $$$ | [58,59] |
| SEC | Separates by hydrodynamic volume | Reduced contamination with high-abundance proteins, gentle | Low resolution and dilution of EV isolates | ++++ | $$ | [44,69,70] |
| Filtration | Uses membranes with specific pore sizes | Simple, time efficient, and relatively gentle | Low sample recovery, extrusion effects, possible irreproducibility | ++++ | $ | [72., 73., 74., 75.] |
| Immunoaffinity-based isolation strategies | Capture EVs using antibodies | Increased purification efficiency, target specific population | Costly, nonspecific binding | ++ | $$$$ | [76., 77., 78.] |
| Commercial reagents | Precipitate EVs using polymers | High yield, simple workflows | High protein contamination, various degrees of compatibility with profiling techniques | ++ | $$$ | [87., 88., 89.] |
| Microfluidics | Based on physical, mechanical, and/or surface chemistry properties | Low sample volumes, low cost, low consumption, high throughput, high size selectivity | Prone to clogging, possible irreproducibility | ++ | $$$ | [91., 92., 93., 94., 95., 96.] |
| AF4 | Laminar flow | Gentle, isolation of EV subpopulations | Low resolution, possible irreproducibility | ++ | $$$$ | [7,8,98,99] |
| Nano-FCM | Flow-cytometry based | High fidelity sorting | Swarm detection, simultaneous detection of multiple EVs, inadequate size assessment | ++ | $$$$ | [100,101] |
Potential for scalability and cost rankings shown in arbitrary units using a range of 0–4 units.
UC
The current ‘gold-standard’ technique is UC, which separates and concentrates EVs from other specimen constituents according to their density [51,58]. EVs typically have a density of 1.13–1.19 g/ml [59]. This process begins with the pelleting out of any cells and cell debris and then transfer of the supernatant to the ultracentrifuge and centrifuging twice at 100 000 g or higher speeds, first to pellet EVs and then, after rinsing in phosphate buffered saline (PBS), to pellet EVs with reduced protein contamination [51,60]. This method, however, may also isolate MVs and microparticles of various size and composition, including viruses, lipoprotein particles, and protein complexes [61,62]. UC is also low throughput, requires expensive specialized equipment, and can damage EV membranes or cause aggregation during processing [52,63]. The purity of UC-based EV isolates and EV populations can be further increased using an additional density gradient step [58,59]. Sucrose gradients and commercial OptiPrep density gradients are commonly used in this method [64,65]. A drawback of this method is that additional purification might be needed to separate EV subpopulations from each other, from other microparticles with similar densities, and from the density gradient matrix [66].
Size-Exclusion Chromatography
Size-exclusion chromatography (SEC) separates the components of a sample based on the hydrodynamic volume. SEC is typically performed with a Sepharose CL-2B or similar stationary-phase column where fractions are eluted with PBS [40,67]. SEC is simple, robust, and scalable, does not require expensive equipment, and can be used with mild elution conditions, allowing various applications [67,68]. In addition, SEC may be an efficient EV isolation method for downstream proteomic analysis, in that high-abundance protein reduction is comparable with or better than UC [40,69,70]. However, it is limited by the low resolution and dilution of EV isolates. There is ample evidence that shows the presence of contaminants in SEC vesicle isolates, including lipoprotein particles, viral particles, free proteins, and protein complexes from biological matrices [44]. Nevertheless, both the yield and the EV:protein ratio are typically significantly higher for SEC-based isolation than for the UC-based method [69]. However, the same study showed that UC isolates provided higher yields of smaller EVs and contained less apolipoprotein (APO) particles than SEC isolates. Additionally, SEC stationary phases may exhibit nonspecific interactions with analytes (i.e., ion-exchange interactions), which can result in changes in separation selectivity [71].
Filtration
Filtration uses matrices defined by molecular mass and size exclusion range, usually with cellulose filters [72]. After the initial filtration, additional ultrafiltration and wash steps are typically included to remove contaminants smaller than a specific size and concentrate the vesicle sample [73]. Centrifugation-based filters (Centricon) have been shown to recover three times as many particles as pressure-driven membranes (BioMax). Polyethersulfone nanomembranes have also been used successfully [74]. An advanced version – TFF – is less prone to clogging than conventional filtration due to the lower chance of cake formation. Compared with UC, filtration offers several advantages, including mild pressure, time efficiency, effective purification, and scalability. Additionally, isolation is greatly dependent on the quality of filter membranes and the uniformity of the membrane pore size distribution. Moreover, filtration may alter the structural integrity of EVs due to extrusion effects and lead to EV losses on the filter membrane. Therefore, the applicability of this approach to physiological fluids with high complexity and high dynamic range may be restrained by a low recovery rate and insufficient efficiency of separation from high-abundance contaminants [72,75].
Immunoaffinity-Based Isolation Strategies
Immunoaffinity-based isolation strategies use highly specific antibody–antigen interactions to target specific populations of EVs, reducing contaminant EVs and microparticles. EVs have been isolated using antibodies in many studies [76., 77., 78.]. In one method, biotinylated antibodies are captured on streptavidin-coated magnetic beads to isolate specific EV subpopulations [79,80]. Another method uses paper-based immunoaffinity devices for EV isolation by conjugating antibodies to chromatography paper, followed by scanning electron microscopy (SEM) or ELISA [81]. Antibodies for known markers of specific diseases, as well as heparin, have been used to purify EVs and assess their diagnostic potential [82., 83., 84., 85.]. Immunoaffinity isolation reduces the isolation time and increases the purification specificity, but is costly and often plagued by nonspecific binding, competitive inhibition, and cross-reactivity of antibodies. In addition, antibodies have a short lifetime, and the specific release of EVs from stationary phases could be problematic [86]. Several immunoaffinity-based microfluidics approaches, which can isolate and characterize EVs in an integrated platform, are discussed in ‘Characterization of Proteins in EVs’, ‘Immunoaffinity-Based Techniques’.
Commercial Reagents
Recently, several commercial kits have been introduced to isolate EVs with yields comparable with UC-based techniques without the need for any specialized equipment [87]. Polymer precipitation-based commercial EV isolation kits are available, including ExoQuick™ Exosome Precipitation Solution (Systems Bioscience), miRCURYTM (Exiqon), and the Total Exosome Isolation Reagent (TEIR) (Invitrogen) [73,87., 88., 89.]. Nonprecipitation kits like PureExo and MagCapture are also commercially available [87,89,90]. Another kit is Exo-FlowTM, which uses antibody-labeled magnetic beads to target surface proteins. Overall, such kits also usually result in a high level of protein contamination, require long isolation protocols, and can be expensive. In addition, the proprietary formulations of such kits may interfere with downstream experiments. A comparison study of ExoquickTM, miRCURYTM, TEIR, and UC showed similar EV size distributions in isolates [87].
Emerging Techniques
As EV research is a fairly new field, novel EV isolation techniques are constantly being developed. Here, we discuss several approaches that have recently emerged.
Microfluidics
Microfluidic EV isolation techniques include immunoaffinity capture [91,92] and capture based on the physical or mechanical characteristics of EVs (size, density, compressibility, viscoelasticity, etc.) [93,94]. Physical property-based methods are either pressure or electrophoretically driven, with the latter less prone to clogging of pores or channels [95]. ExoChip captures EVs with a polydimethylsiloxane surface functionalized with anti-CD63 antibodies, followed by fluorescent carbocyanine dye staining for rapid quantification of EVs [96]. Another microfluidic filtration device utilizes porous polymer monoliths as filter membranes tuned to different geometries and pore sizes [94,95]. Viscoelastic flow and acoustic isolation systems are also among the novel nondestructive label-free microfluidic techniques that can be potentially useful for EV isolation and size sorting [93]. These methods are advantageous because of the low sample volumes, low cost, low consumption, high-throughput, and high precision.
Asymmetric Flow Field-Flow Fractionation (AF4)
AF4 is a gentle separation technique that does not require a stationary phase that may alter, retain, and degrade EV samples. The separation is accomplished in a thin film (submillimeter) of a laminar flow confined in a narrow chamber with a membrane at the bottom, where a force field is applied perpendicular to the laminar flow [8,97]. Unlike chromatography-based separation methods, AF4 has a programmable cross-flow intensity, which can be optimized during separation to increase the efficiency [98]. AF4 has become attractive for the fractionation of EV subpopulations [8,99]. Recent studies suggest that AF4, in combination with sensitive molecular assays, can serve as an efficient isolation technique for specific EV and particle subpopulations based on their size, thereby addressing the complexity of EV heterogeneity in physiological fluids [99]. Online detectors, including UV, multi-angle light scattering (MALS), and dynamic light scattering (DLS), were effectively used with AF4 whereby Zhang and coworkers fractionated small EVs into distinct subclasses – small exosomes (Exo-S, 60–80 nm), large exosomes (Exo-L, 90–120 nm), and exomeres (~35 nm) – from various cell types [7,8].
Nano-Flow Cytometry (Nano-FCM)
A high-resolution flow cytometry (FCM)-based method was developed to sort EVs by the systematic analysis of background reference noise. Using this method, a pool of carboxyfluorescein succinimidyl ester and cell-trace violet-stained EVs derived from immune and tumor cell lines were sorted with fidelities of 78% and 99%, respectively [100]. The drawbacks of EV FCM include coincidences of two or more EVs being detected simultaneously, or swarm detection, causing overestimation of the signal and inappropriate measurements, particularly when dealing with submicron-size particles [101]. There is also a possibility of the introduction of artifacts caused by contamination with lipoprotein particles as well as the use of inadequate size standards [102].
Combined Techniques
To increase the specificity or purity of EV isolation, multiple methods can be combined. Examples of UC followed by a density gradient step, SEC or AF4, have been reported [64,103]. Another combination method is ultrafiltration followed by liquid chromatography (LC), which has been shown to isolate significantly more EVs than UC and preserves their biophysical properties [104,105].
EV Characterization: Challenges and Needs
LDLs have diameters similar to those of exosomes, and HDLs fall in the density range of all EVs. It is highly possible that EV isolates will contain lipoproteins when traditional protocols are used. The elimination of contaminating proteins would improve characterization results by reducing possible contaminants. However, what makes the situation more challenging is that EV recovery and protein yields can be low and are further decreased by losses at subsequent steps of isolation and sample processing [106]. Also, while there is a handful of frequently used protein markers, finding either a universal panel of markers for all possible EV types or unique panels for specific EV populations remains a significant challenge in the field. Designing simple, effective, and cost-efficient means to assess the purity of EV isolates will enable biological and clinical applications and will facilitate much-needed standardization in the EV field.
Methods for EV Characterization
Physicochemical Characterization Methods
These methods are summarized in Table 2 and Figure 3 .
Table 2.
Advantages and Disadvantages of the Described EV Characterization Methods
| Method | Principle | Advantage | Disadvantage | Potential for quantitationa | Costa | Refs |
|---|---|---|---|---|---|---|
| Fluorescence/confocal microscopy | Fluorescence/light radiation | Nondestructive analysis, EV uptake/degradation can be monitored, semiquantitative | Lengthy procedure, dye aggregates result in overestimation, reporters need to be specific | ++ | $$$ | [108., 109., 110.,119., 120., 121., 122., 123., 124., 125., 126.] |
| TEM and cryo-TEM | Electron radiation | Direct imaging of EVs, nondestructive | High computational cost, challenging sample preparation, low throughput, not quantitative, reproducibility might be an issue | + | $$$$ | [113., 114., 115.,135] |
| AFM | Hooke’s law | High resolution, provides details of EV morphology | Low throughput, specialized equipment | + | $$$ | [108., 109., 110.,117,118] |
| DLS | Brownian motion | Simple and fast | Nonideal for heterogeneous and polydisperse samples, low resolution | ++ | $$ | [113., 114., 115.,119., 120., 121., 122., 123., 124., 125., 126.] |
| NTA | Light scattering/Brownian motion; dark microscopy and fluorescence | Size and concentration measured simultaneously | Biased toward smaller particles | +++ | $$$ | [117,118,135] |
| FCM and nano-FCM | Fluorescence/light scattering, Coulter principle | No sample preparation necessary, fast, EV specific, reproducible, quantitative, low sample volume | Size standards do not correlate correctly, restrictions on lower size limits of detection | +++ | $$$$ | [128,133] |
| RPS | Coulter principle | High throughput, measures concentration, size, and charge simultaneously, low sample volume | Biased toward larger particles, nonstandardized settings, cannot determine particle type, requires frequent calibration; reproducibility and robustness can be an issue | +++ | $$ | [135,141] |
| RS | Light scattering | Reports chemical composition (for simple sample systems), no sample preparation, small sample volume | Challenging data interpretation, medium throughput, characterization of composition is challenging for complex samples | + | $$ | [128,133,135] |
| FLOWER | Immunoaffinity interactions and resonance frequency shifts | Requires further evaluation | Requires further evaluation | ++ | $$ | [135,141,143] |
| SP-IRI | Immunoaffinity interactions and interference of light | Requires further evaluation | Requires further evaluation | ++ | $$ | [135,144,145] |
Potential for quantitation and cost rankings shown in arbitrary units using a range of 0–4 units.
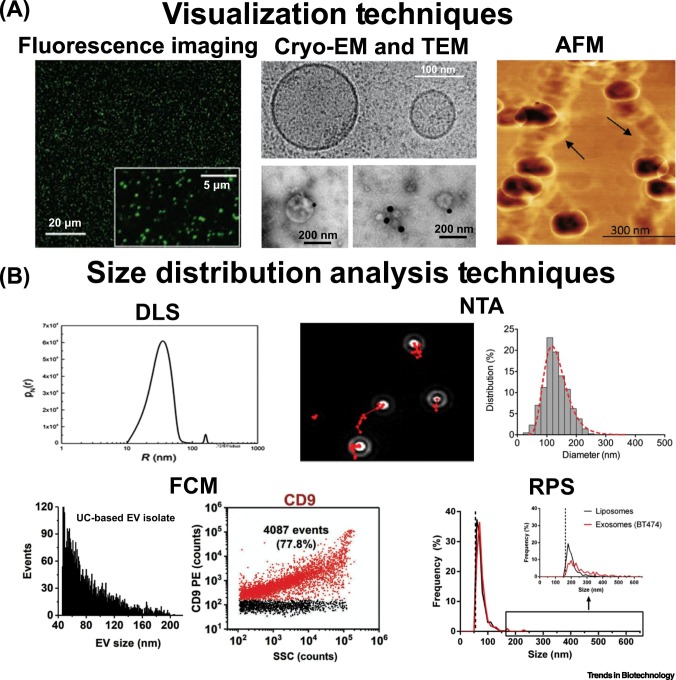
Figure 3.
Overview of Extracellular Vesicle (EV) Characterization Techniques.
(A) Visualization techniques that allow the observation of EVs and recording of images, including fluorescence imaging [216], cryoelectron microscopy (cryo-EM) that compares a microvesicle (>100 nm) with an exosome (~100 nm) [216] along with transmission electron microscopy (TEM) with EVs labeled with CD9-biotin/streptavidin-gold nanoparticles [217], and atomic force microscopy (AFM) [118]. (B) Size distribution analysis techniques that measure the size of sample particles, including dynamic light scattering (DLS) [218], nanoparticle tracking analysis (NTA) [180], flow cytometry (FCM) [134], and resistive pulse sensing (RPS) [219]. Reprinted, with permission, from referenced sources.
Microscopy and Imaging
Using confocal microscopy, the release, uptake, and exchange of EVs can be monitored by fluorescently labeled cell membranes with ~200-nm resolution [107., 108., 109.]. Another method involves EVs labeled with a reporter that are injected into a living specimen and imaged by various methods including MRI, single-photon emission computed tomography/positron emission tomography (SPECT/PET), or fluorescence-mediated tomography (FMT) [110,111]. These imaging methods can be used to determine which tissues’ and organs’ EVs are taken up, and the degradation of EVs by the host can be monitored [110].
The advantages of using these methods to monitor EVs are as follows: it is possible to determine where they end up in the host organism; EVs can be observed as they are created and absorbed by cells; and images can be acquired during animal studies without using invasive sampling techniques [110]. However, there are some disadvantages. A common dye used to label EVs (PKH67) can outlast EVs or aggregate, forming micelles, which can result in false positives, causing the total EV population to be overestimated [108,110]. Other challenges include the need for highly specific reporters and labels that are stable over time [110] and obtaining reliable EV concentrations by fluorescence.
Transmission electron microscopy (TEM) is an imaging method used to identify particles in a sample. EVs are fixed on a sample grid and stained with reagents such as uranyl acetate or osmium tetroxide before analysis [112]. Electron radiation increases the resolution to subnanometer resolution. Both 2D and 3D images can be created using various computer programs [113]. TEM and cryo-TEM (cryogenic conditions) can also be used to observe the formation of EVs through blebbing and the shedding of EVs from cells, allowing comparison of EV formation between different cell types or conditions [114].
Advantages of cryo-TEM include the ability to directly observe samples, reduce damage up to sixfold [113], minimize changes in morphology during sample preparation because dehydration is not required [113], and eliminate the need for staining procedures. However, the drawbacks of using TEM/cryo-TEM include a high demand for computational power to convert images to 3D [113], challenging sample preparation and handling methods, low signal-to-noise ratio due to ice artifacts (cryo-TEM) [115], high cost, and low throughput. It is also difficult to tell whether an EV is entering or exiting the cell (endocytosis vs exocytosis). Some larger EVs might be excluded from the cryo-TEM sample due to preparation techniques. Also, TEM is not reliably quantitative [114]. Therefore, other techniques should be used if a concentration is necessary. Recently, software (TEM ExosomeAnalyzer) was created that automates the analysis of TEM images. However, it does not currently work with cryo-TEM images [116]. Such software tools would reduce the variability that comes with subjective human judgment.
Besides TEM, atomic force microscopy (AFM) is also a high-resolution technique for EV imaging. In one study, AFM was used to characterize platelet-derived EVs, which were captured on a CD41 antibody-functionalized mica surface [117]. The results showed reproducible measurements of the numbers and size distribution of CD41-positive EVs; the higher resolution allowed AFM to detect EVs as small as several nanometers, which is far more sensitive than regular FCM [117]. AFM is able to provide substructural details in addition to the surface topology of EVs. In one study, as the imaging force changed from <1 nN to ~2 nN at a phase-modulated tapping mode, round-shaped EVs showed a gradual deformation where a distinctive phase appeared in the centered depression. This indicates heterogeneous density and components present in EVs, which agrees with previous findings in varied EV cargoes [118]. As with TEM, limited throughput and specialized equipment requirements are the drawbacks of AFM.
DLS
DLS, also known as photon correlation spectroscopy or quasi-elastic light scattering, is a technique to determine the size distribution of particles suspended in a fluid medium. DLS primarily measures the velocity of the Brownian motion of macromolecular structures in solution due to collisions with solvent molecules [119,120]. Size measurement by DLS offers a simple and time-saving technique that provides information about the mean size and the dispersity of the particle size distribution [a.k.a. the polydispersity index (PDI)] for particles ranging from 1 nm to 6 μm [121]. DLS has been used in several studies involving the characterization of EVs isolated from biofluids such as blood and BAL [122,123]. DLS is ideal for the size measurement of monodisperse sample populations and monitoring aggregation because the scattering intensity is proportional to d 6 where d is the diameter of the particle. In the case of biological fluids, the sample is polydisperse due to the presence of differently sized vesicles, particles, and biomolecules, which range from a few nanometers to several thousand nanometers [124,125]. It becomes essential to remove large particles (>1 μm), such as cell debris and dead cells, as well as aggregated proteins and protein complexes. Low resolution is an important limitation of DLS, where the peak resolution is best only when the sizes of the particles differ by at least a factor of three (e.g., 50 nm and 150 nm) and results in broader polydispersity ranges and larger PDI values when particles have a close particle size distribution [126].
Nanoparticle Tracking Analysis (NTA)
NTA is a dark-field microscopy technique that can effectively measure particle size distribution and concentration in a liquid suspension based on the Brownian motion of the particles tracked using laser light scattering (reviewed in [127]). Particles scatter the light on laser illumination and their Brownian motion is live imaged by an optical microscope equipped with a video camera. By tracking the mean squared displacement of a single particle, the software can determine its theoretical hydrodynamic diameter using the Stokes–Einstein equation. Compared with DLS, where light scattering and particle size distribution are biased toward large-sized particles, NTA provides better resolution for heterogeneous mixtures of particles that vary in size, by measuring both the light-scattering intensity and the size of individual particles. Additionally, NTA allows the detection of EVs labeled with stable fluorophores. The refractive index (RI) of the analyte (e.g., high RI of colloidal gold vs low RI of cell-derived vesicles) determines the size range that can be detected by NTA. The smallest detectable EV size is typically around 50 nm, but the signal-to-noise ratio and the amount of scattered light should also be factored in to improve the accuracy of measurements. The upper limit is approximately 1 μm, due to the Brownian motion becoming too limited to track accurately. The accuracy of the quantitative analysis of EVs is also a challenge.
FCM
FCM can be used for biomarker discovery or to size EVs by measuring either light scattering or the fluorescence of the sample particles. Samples can be labeled with fluorescent tags and/or antibodies (anti-CD63, anti-CD81, or anti-CD9) to track EVs [128,129]. The sample is passed through multiple lasers of differing wavelength [130], scattering light. The amount and direction of the forward scatter (FSC) or fluorescence, along with the duration, indicates the size of the EVs measured [129], while the side scatter (SSC) indicates the internal complexity. SSC is more sensitive than FSC, which makes it more appropriate for the analysis of low-concentration samples [131]. It has also been reported that using a lower wavelength (405 nm) increases the SSC sensitivity and resolution for all EV sizes tested, with a more significant effect on smaller EVs [132]. To improve EV detection, lipophilic fluorophores or EV-specific fluorescently labeled antibodies can be incorporated into the EV membrane [130,133]. Qualitative and quantitative analysis of specific target proteins can help to indicate what subpopulations of EVs are most prevalent, which may be important for the diagnosis of various diseases [129].
The advantages of FCM include the applicability to unprocessed biological samples, fast analysis time, reproducibility, and quantitative output [133]. However, FCM cannot detect particles below 200 nm on most flow cytometers [133]. To lower the limit of detection, nano-FCM was developed. The same principles apply; however, there are changes to the angles of the light scatter collected, which allow detection down to 40 nm [46,100,130,133,134]. The swarm effect, as described previously, can also affect measurements [128]. Unfortunately, light-scattering-bead standards do not correctly correlate with EV size due to differences in the RIs. Because EVs have a lower RI than polymer size standards, the measured size of an EV could be about double the size of the corresponding size standard [100,133,135,136]. Silica beads have a RI that is more similar to EVs than polystyrene beads, thus they may serve as better standards [135]. Standard alternatives to silica and polystyrene beads were also introduced for EV FCM analysis. For example, liposomes or hollow organosilica beads scatter light similarly to EVs [137,138]. Also, differences in the hardware from different manufacturers can cause variabilities, which should be taken into account when comparing data from different flow cytometers [131].
Resistive Pulse Sensing (RPS)
RPS is a high-throughput method that measures the size, concentration, and charge of particles in solution based on the Coulter effect [135]. Changes in the applied electric current are measured when particles pass through a submicron-sized pore in a membrane with electrolyte solutions on both sides. The size of the particle is determined by the ratio of the change in current to the background current. The length of the disturbance is related to the volume of the particle and the blockade rate is related to the concentration [139]. The zeta potential can be measured by relating the blockade event signals to both the voltage and the pressure [140]. In instruments from Spectradyne LLC, samples pass through a solid pore or aperture, while Izon Science’s instruments have a tunable, stretchable pore that can be optimized for specific experiments [135].
RPS is an attractive method for EV characterization due to its ability to measure the size (down to 40 nm), concentration, and zeta potential at the same time [135]. The technique can also be tuned to a particular size range and requires about 40 μl of the sample. However, similar to NTA, RPS cannot determine the particle type or chemical makeup of a sample, making it difficult to determine whether the sample contains EVs, protein aggregates, or other nonmembranous particles (e.g., lipoproteins, debris) [135,139,141]. Another issue is that RPS and NTA demonstrate results that are inconsistent in the detection of larger (>150 nm) and smaller (<150 nm) EVs: RPS may be biased toward more efficient detection of larger particles, while NTA may be biased toward smaller particles [135]. The challenge is to determine which approach is more accurate, or whether the two methods could be used in tandem to obtain a more accurate characterization of EVs in the sample. Polystyrene or silica beads, and potentially other standards, are used to calibrate the RPS instrument, which provides grounds for initial standardization [135,138]. Beads with different surface chemistries or liposomes are used to calibrate the zeta potential [140]. The technique requires frequent calibration. In many studies, the instrument was calibrated before every sample to ensure correct measurements of EV samples [135,139., 140., 141.].
Emerging Methods
Raman spectroscopy (RS) is based on inelastic light scattering in which the photon energy is transferred either to or from the molecules of EVs, resulting in a wavelength shifted from that of the incident light. The amount of energy transferred is proportional to the shift in the wavelengths of the scattered photons, which is dependent on the molecular arrangement of the sample particle. Measurement of the inelastically scattered photons results in a spectral fingerprint, similar in appearance to an IR spectrum [135]. RS has previously been used on EVs from mammalian cell lines, bacteria, and human samples [135].
The method is often able to provide some information about the chemical composition of a sample or at least the major constituents of the sample in one measurement with minimal sample preparation and with only a small sample volume (<50 μl) required [135]. Although RS appears to allow EV identification, it is challenging practically to obtain informative spectra due to the chemical complexity of EVs. The throughput of RS measurements is rather moderate, which makes it challenging to use in clinical settings [135]. RS can be used to make relatively quick measurements of EVs without the need for targeted protein biomarkers [142]. RS has been used in industry for small molecules; however, a study by Gualerzi and colleagues suggested that RS can also be used for EV analysis [142]. RS was demonstrated to have the potential to identify the origin of a specific vesicle (e.g., bone marrow, adipose tissue, dermal fibroblasts). The method developed in this study shows that EV characterization can be performed in bulk, on an industrial scale, before they are used for either in vivo or in vitro clinical applications. This approach can be applied to increase the number of EV-based products offered by the biotechnology and biopharma industries [142].
Frequency-locked optical whispering evanescent resonance (FLOWER) uses an antibody-coated silica microtoroid whose resonant frequency is probed by a laser. The sample flows over the microtoroid optical resonators and EVs bind to the antibodies. Each time an analyte binds, the resonant frequency shifts and a count of EV concentration is acquired [135]. The change in amplitude corresponds to the diameter of the particle that became bound or unbound from the microtoroid [143]. This method still requires further evaluation, but it has the potential to identify EVs.
Single-particle interferometric reflectance imaging (SP-IRI), sold as the ExoView system, is based on the interference of particles (labeled with fluorescent antibodies) immobilized on the sensor surface with light that reflects off the surface. The interference can be correlated with size [144]. This technique has mostly been applied to viruses but is beginning to be applied to EVs as well [145].
Biochemical and Molecular Characterization Methods
These methods are summarized in Table 3 and Figure 4 .
Table 3.
Overview of Biochemical EV Characterization Techniques and Their Main Advantages and Disadvantages
| Technique | Analyte type | Principle | Advantage | Disadvantage | Sample volume | Limit of detection | Potential for quantitationa | Costa | Refs |
|---|---|---|---|---|---|---|---|---|---|
| UV-Vis spectrophotometry | RNA, protein, or intact EVs | Absorbance of UV or visible light | Simple, cheap | Limited sensitivity, compromised quantitative accuracy, and interference from other molecular species in complex matrices | <2 μl | 2 ng/μl | ++ | $ | [149] |
| Electrophoresis | RNA and protein | Electrophoretic mobility, hydrodynamic volume | Low detection limit | Low quantitative accuracy, limited resolution |
~10 μl | 50 pg/μl | +++ | $ | [149] |
| RiboGreen assay | RNA | Fluorescence | Very low detection limit, excellent linearity, high throughput | Interference from interaction with other nucleic acids | 20–100 μl | 1–200 ng | ++++ | $$ | [149] |
| qRT-PCR | RNA | Transcription and amplification using primers and PCR | Low sample volume, low detection limit, quantitative, high throughput | Limited to analysis of known target RNA sequences | Low μl level | pg to fg | ++++ | $$ | [88,150,151] |
| NGS/RNA-seq | RNA | PCR, transcription to cDNA, and sequencing with fluorescent nucleobases | RNA sequence can be determined, can detect low-abundance transcripts and differentiate isoforms | Low throughput, expensive, possible biases from isolation method, library preparation, data processing | low μl level | <100 ng | +++ | $$$$ | [75,149,152., 153., 154.] |
| Microarrays | RNA | Hybridization of DNA probes to target sequences | Simultaneous measurement of thousands of transcripts | Expensive, specialized equipment, low specificity, lack of control over analyzed transcripts | 1 nl | pM | ++++ | $$$$ | [155., 156., 157., 158.] |
| NanoString | RNA | Hybridization of RNA to capture probe and fluorescent reporter probe | High accuracy since the method does not involve RT and amplification | Limited by applying only to known target sequences | ~30 μl | 100 ng | ++++ | $$$$ | [159., 160., 161.] |
| BCA, Bradford (Coomassie), and fluorometric assays | Protein | Colorimetric and reagent binding | Simple, reliable, high throughput | Protein composition can affect results, contamination reduces accuracy | Low μl level | ~20 μg/ml | +++ | $$ | [203] |
| Western blot, ELISA, microfluidic devices | Protein | Immunoaffinity/antibody interactions | Capable of detecting EV-specific proteins | Lack of specificity, cross-reactivity, unpredictable quality, short shelf life, expensive | Low μL level | Low ng level | +++ | $$$ | [30,76,82,169., 170., 171., 172., 173.] |
| SEA | EV | Immunoaffinity and fluorescence | Capable of analyzing one EV at a time, high signal-to-noise ratio | Low throughput | Low μl level | 1 EV | +++ | $$$ | [174,175] |
| μNMR | EV | Immunoaffinity, MNP labeling, and NMR | Quantitative, little interference | Requires specialized equipment | Low μL level | Low EV counts | ++ | $$$$ | [176] |
| SPR | EV | Surface-electron oscillation under light | Quantitative, high sensitivity, does not damage analytes, fast, low sample volume | Signal interference in complex nonhomogeneous samples | Low μL level | 670 aM | ++ | $$$ | [177., 178., 179., 180.] |
| iMEX detection | Protein | Magnetic bead capture and oxidation | Fast, low sample volume, portable, high sensitivity | Requires specialized equipment | 10 μl | ~104 EV counts | +++ | $$$ | [181,182] |
| MS-based molecular profiling | Proteins, metabolites, and lipids | Determines mass over charged values for analytes or their fragments | Capable of determining chemical structure, PTMs, glycan, quantitation, high sensitivity | Complicated sample preparation, biases from choice of sample processing and data processing workflows | <5 μl | pM to fM | ++++ | $$$$ | [8,185,186,191,192] |
Potential for quantitative analysis and cost rankings shown in arbitrary units using a range of 0–4 units.
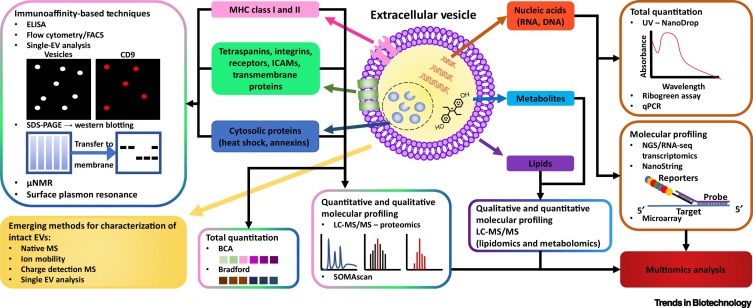
Figure 4.
Overview of Biochemical Techniques Used for Extracellular Vesicle (EV) Characterization.
Abbreviations: BCA, bicinchoninic acid; FACS, fluorescence-activated cell sorting; ICAM, intercellular adhesion molecule; LC, liquid chromatography; MS, mass spectrometry; NGS, next-generation sequencing;
μNMR, microfluidic NMR.
Characterization of RNA in EVs
RNA represents one of the most important biomolecules associated with EV research. Early studies analyzing EV-associated nucleic acid cargo, particularly small RNAs, miRNAs, and mRNAs, have revealed the horizontal transfer of RNA in cell–cell communication [13]. Since then, a variety of EV-encapsulated miRNAs have been shown to serve as highly specific biomarker candidates for various pathological conditions [146]. miRNAs are the most extensively studied EV-associated RNA species; however, a variety of other RNA species, such as fragments of ribosomal RNA, Y-RNA, and tRNA, have been reported, and they can be important for functional assays [75,147,148]. Overall, a number of studies that used deep sequencing to profile EV-RNA from various biological fluids and cell types indicate that a major proportion of EV-RNA comprises small RNA (<200 bp), with heterogeneous subpopulations [148]. In this section, we provide a brief overview of the analytical methods most widely used for the quantitation and comprehensive profiling of EV-RNA (Figure 4).
In the UV-Vis method, spectrophotometers, including low-volume detection cell instruments (e.g., NanoDrop), can be effectively used for the quantification of nucleic acids and proteins by measuring the UV-light absorbance. However, the lower limit of detection of a low-volume detection cell instrument like NanoDrop is around 2 ng/μl, which may pose a challenge when the yield or concentration of RNA and protein from EVs is low. Since EV subpopulations and content are very heterogeneous, microvolume UV-spectrophotometric measurements may not be accurate unless a specific type of RNA is being quantified with the use of an appropriate extinction coefficient [149].
Fluorescence-based methods include electrophoresis-based techniques, the RiboGreen assay, and quantitative reverse transcription (qRT)-PCR-based assays.
In electrophoresis-based techniques, a microfluidic chip is used for the electrophoretic analysis of nucleic acids such as DNA and RNA. The EV sample is lysed and combined with a fluorescent dye that binds to the nucleic acids. Separation of various lengths/sizes of nucleic acids is achieved based on their respective electrophoretic mobilities. The concentration is estimated based on the measured fluorescence intensity of the peaks in the electropherogram. This technique has a lower detection limit of 50 pg/μl (Agilent) and is suitable for low-yield EV-RNA isolations. As this technique gives the size distribution profile of RNA, the relative amounts of various RNAs can be estimated. One caveat of chip-based techniques is that the quality of RNA is commonly estimated using rRNA standards, while full-length rRNA may be absent in EV subpopulations [149].
The RiboGreen assay employs a sensitive fluorescent dye that binds to the phosphate backbone of RNA and produces a concentration-dependent fluorescent signal using excitation and emission wavelengths. These assays have a very low detection limit of 1–200 ng and have excellent linearity, which enables accurate quantitation. This assay highlighted by the ISEV allows concentration measurements of EV-RNA in a high-throughput manner using a fluorescent plate reader [149].
qRT-PCR-based assays are used when the levels of a particular sequence of RNA or DNA need to be quantified. This technique measures the increase in fluorescent signal probes that bind to the nucleic acid as a particular transcript is amplified using reverse transcription (RT) of RNA to cDNA, followed by amplification of the cDNA using sequence-specific primers and PCR. Advantages include a low sample volume (microliter level) requirement, a high sensitivity/low detection limit (picogram to femtogram level), the ability to quantify relatively and absolutely, and high-throughput (96/384-well plates) quantification of multiple genes in a single sample. Several previous studies relied on qRT-PCR to conduct comparative profiling of miRNAs in EVs isolated by UC, filtration, and precipitation [88,150]. Chevillet and coworkers aimed to quantify the stoichiometric relationship between the number of EVs and the number of miRNAs per EV. qPCR-based miRNA quantification from EV-RNA samples isolated from five different sources revealed that, on average, there was far less than one molecule of a given miRNA (even the most abundant miRNA species) per EV [151]. However, this technique, unlike the previous two, does not measure the total amount of RNA and is useful only for the detection and quantification of known and specific sequences of RNA.
Next-generation sequencing (NGS/RNA-seq) is one of the most advanced and powerful techniques for the comprehensive profiling of nucleic acids. Several previous studies reported the feasibility and applications of sequencing small RNAs in EVs [152,153]. The method involves three steps, which are: (i) library preparation, using PCR amplification, transcription of cDNA from RNA, ligation of DNA adapters, and hybridization onto the sequencer, which is coated with complementary oligo sequences and amplified by bridge amplification; (ii) sequencing by synthesis with fluorescent nucleobases; and (iii) data processing, which involves target sequence coverage and alignment with the reference sequence. The strength of RNA-seq lies in its ability to identify and comprehensively profile RNA subtypes in various EV subpopulations. A recent study performed to characterize the extracellular RNA (exRNA) released from human glioma stem cells using NGS found that the RNA profiles of MVs, exosomes, and ribonucleoproteins (RNPs) differ substantially [75]. Additionally, the amount of miRNA species in exosomes was reported to be higher than in MVs or RNPs, which supports the hypothesis of the loading of specific miRNA sequences into exosomes. Both RNA-seq and microarrays performed similarly in clinical endpoint prediction, but RNA-seq was more efficient in detecting low-abundance transcripts, distinguishing biologically critical isoforms, and enabling the identification of genetic variants [154]. RNA-seq by NGS is prone to biases that arise from a number of factors, which include the selection of methods, kits, and vendor platforms for RNA isolation, the preparation of libraries, ligation, and sequencing (e.g., HiSeq or MiSeq, SOliD, Ion Torrent), as well as bioinformatics pipelines and parameters in data processing in normalization methods [149]. Additionally, sequencing methods are time consuming and expensive.
Microarrays are a well-established technique frequently employed to study global profiles of hundreds of genes in biomedical samples and enables differential analysis of RNA/DNA samples. This technique is based on the principle of hybridization of DNA probes to the complementary target gene sequences in the samples [155]. The probes are deposited onto a chip in the array format using various methods, such as ink jetting and microspotting [156]. In a recent study performed on the EV-RNA produced by mast cells using the data from a microarray combined with NGS, four different clusters from two distinct exRNA signatures [high-density (HD) and low-density (LD) exRNA] were shown. Pearson correlation and principal component analysis suggest that the different structures that are present in the HD and LD fractions correspond to fundamentally different RNA cargo [157]. High throughput and the simultaneous measurement of thousands of mRNA transcripts for gene expression or genomic DNA fragments to enable copy number variation analysis are among the major advantages of microarrays. However, a secondary confirmation step, such as qRT-PCR, is generally employed to verify some of the key genes depending on the purpose and scope of the experiment [158]. The disadvantages of microarrays include the high cost, access to specialized equipment, and numerous probe designs based on sequences of low specificity, as well as the dependence of the pools of transcripts selected for the analysis on the sets of probes designed by the manufacturers of microarray platforms [157].
NanoString is a gene expression profiling method that has been employed to study the biogenesis of EV-miRNA and its mechanism and to identify biomarkers from biofluids in diseases such as glioblastoma multiforme (GBM) and bladder cancer [159,160]. The technique is based on the hybridization of RNA to a capture probe complementary to the target gene sequence. The probe has a biotin tag on the 3′ end to immobilize it to streptavidin beads. The target gene is also complementary to the fluorescently labeled reporter probe with a unique molecular barcode on the 5′ end. After the hybridization step, the unbound probes are washed off and the complementary pairs are immobilized onto a glass slide and imaged. The NanoString assay does not count the intensity of the barcode but counts the number of barcodes. One fundamental sample preparation step that differentiates NanoString from NGS and microarray techniques is that the assay does not involve RT and amplification steps, which eliminates errors introduced by these steps. Additionally, this technique is less time consuming than NGS. NanoString has been employed to profile the modification of tumor miRNA content by transfection of parent cells using hyaluronic acid nanoparticles loaded with wt-p53 and miR-125b [161]. NanoString-based miRNA panels have been employed to study the distinct cell phenotypes. Comprehensive analysis of cellular and EV-miRNA showed that the miRNA signature separated GBM stem cells (GSCs) and GSC-derived EVs and further miRNA profiles for both the cells and EVs, which were aligned with previously determined gene expression analysis of the cells [160]. However, unlike NGS, NanoString employs a panel of genes for which sequences are known in the samples, which can limit the profiling and differential analysis for unknown RNA sequences.
In summary, each RNA profiling technique mentioned above has its own unique advantages and disadvantages and can be used as a complementary technique in combination with others to provide the information needed for more comprehensive EV characterization. The suitability of the various RNA detection methods for the quantification of EV-RNA has been summarized in an ISEV proposition paper [149]. Some of the challenges associated with the characterization of EV-RNA are listed below.
One challenge is the quantity of initial sample available. Inferring credible information from various RNA characterization techniques relies heavily on the accurate quantitation of RNA, which in turn depends on the quality and integrity of the RNA. Compared with RNA isolation yields from cellular RNA, EVs have been shown to have several-fold lower yields, especially when dealing with in vivo or patient samples.
Second, a number of studies show that the choice of EV isolation method affects both the quantity and the quality of the EV-RNA characterization [162,163].
Third, a comprehensive study of small RNA sequences performed on various biofluids, using various EV isolations and RNA isolations, suggests that variables such as laboratory-to-laboratory variability and the choice of RNA isolation method are the biggest sources of variability in target RNA profiles such as miRNA, and that different mechanisms might underlie the loading of various RNA biotypes into exRNA carrier subclasses (EVs, RNPs, and HDLs). Therefore, the optimum exRNA isolation method should be selected according to the targeted RNA biotype [164,165].
Fourth, one of the challenges in profiling RNA from a complex mixture such as blood is being able to detect specific RNA patterns, as multiple classes of EVs carrying individual RNA signatures can be secreted from one cell type. Further, the distribution of RNA types varies by biofluid and donor/cell line [165,166].
Some experimental guidelines and practices have been suggested in various publications to standardize RNA characterization. First, assessing the EV-RNA quality is one of the most important standardization steps [149]. Second, another important step is assessing the type of carrier associated with the miRNA (EV-associated vs lipoprotein-associated miRNA). Third, an optimal method for EV-RNA isolation should be identified, depending on the target RNA population. The method should have a higher proportion of highly expressed miRNAs and better reproducibility than the other methods of RNA isolation. The RNA-to-protein ratio in exosomes is significantly higher than that in MVs, according to a study that evaluated RNA in various EV subtypes [75,167]. Therefore, having a robust EV isolation method that isolates the target RNA and EV populations with high reproducibility is essential. Fourth, the demonstration of protection from nucleases after proteinase treatment is an effective way to ensure that a given nucleic acid species is enclosed inside an EV rather than adhered to its surface or co-isolated. Fifth, for functional testing, cellular transfection of miRNA-loaded EVs is recommended to be performed to evaluate the transfer of the miRNA and the downstream functional effects of the miRNA, such as translation inhibition effects and transcription-level reprogramming profiles. Finally, technical and biological replicates should be included in the experimental workflow wherever possible.
Characterization of Proteins in EVs
Proteins are an important class of molecules that are transported by EVs and are integral constituents of EV structures (Figure 4). Analysis of EV protein composition is crucial to understanding the mechanisms of their biogenesis and their functions. For the purpose of quality control (QC) in EV protein characterization, the current leaders in the field recommend demonstration of the presence of at least one protein from the following classes: transmembrane or glycosylphosphatidylinositol (GPI)-anchored proteins associated with plasma membranes and/or endosomes [e.g., tetraspanins (CD63, CD81, CD82)], and cytosolic proteins recovered in EVs [e.g., ESCRT-I/II/III and the accessory protein ALIX, the heat shock proteins HSC70 (HSPA8) and HSP84 (HSP90AB1)]. Furthermore, the absence of co-isolating APOs and high-abundance proteins in a specific system (e.g., albumin for plasma) in EV isolates should be demonstrated [6].
Protein detection and quantification of the total protein amount in EV isolates: Total protein measurement assays include colorimetric, reagent-based bicinchoninic acid (BCA), and Bradford (Coomassie dye) assays or fluorescent reagent-based fluorometric assays. The main advantage of these assays is the ability to measure the EV proteins in a simple, reliable, and high-throughput manner using a plate reader. However, since these assays are based on the labeling of specific amino acid residues, the composition of EV proteins can affect the quantitation of the samples. Furthermore, when EV isolates are contaminated with high-abundance matrix proteins (e.g., albumin), overestimation of the total protein concentration can occur, reducing the accuracy [6]. This is due to the assumption that the assay’s output (absorbance per unit concentration) is uniform across all proteins in the sample. The ratios of high-abundance proteins to low-abundance proteins will lead to such over- or underestimation [168].
Immunoaffinity-based techniques: Western blotting and ELISA are conventional antibody-based techniques representing targeted methods, which can be used for the detection of EV-specific proteins such as tetraspanins, MHC, and the generic EV markers mentioned above. In the case of western blotting, sample preparation involves lysis of EV samples using a detergent, followed by the denaturation and separation of various proteins based on their size using gel electrophoresis (SDS-PAGE). In both immunoaffinity-based techniques, the specificity is achieved by employing antibodies that bind to the epitopes present on the target protein with high affinity. Western blotting has significant processing time (>10 h) but can provide information about the size and abundance of proteins. By contrast, ELISA is a higher-throughput method due to requiring less processing time (~4 h) as well as the use of 96-well plates [76]. In addition, both of these techniques are hampered by lack of specificity/cross-reactivity, unpredictable quality, short shelf life, and the high cost of antibodies.
A variety of microfluidic approaches have also been developed [82,169., 170., 171., 172., 173.]. Most employ immunomagnetic beads to capture EVs through binding specific EV surface proteins. Some microfluidic chips are distinct in that they combine the on-chip separation of exosomes with a multiplexed assay of various protein markers [169,170]. The ExoSearch chip technique showed several advantages over other methods for EV characterization. First, flexible scalability can be provided by applying a continuous sample flow in the microliter to milliliter volume range. In addition, specific subtypes of EVs could potentially be isolated when using different antibodies immobilized on the beads. Moreover, a multiplexed immunoassay against one sample can greatly reduce the analysis time.
In addition to immunomagnetic configuration, photosensitizer immunobeads can be employed for EV protein characterization. Yoshioka and colleagues introduced donor and acceptor beads to a luminescent proximity assay for the detection of EV proteins, termed ExoScreen [30]. Streptavidin allows the donor beads to trap EVs through specific biotinylated antibodies. The acceptor beads are conjugated with a second antibody that will recognize certain epitopes on the EV surface and will be excited by a singlet oxygen and emit amplified fluorescent light.
Single-EV analysis (SEA): Currently, the majority of EV characterization analyses are performed using highly heterogeneous EV isolates. The main problem with bulk measurements is that they only provide global properties and ignore the heterogeneities in the studied specimen. Therefore, SEA may provide additional information. Lee and colleagues recently introduced a novel SEA method that employed immunofluorescent microscopy [174]. Biotinylated EVs were first caught by the NeutrAvidin coated on the inner surface of a microfluidic chamber, immunostained with fluorescent probes conjugated to antibodies of interest, and imaged. Due to the fixation of EVs, a much higher signal-to-noise ratio from each detection was achieved. Moreover, H2O2 could be used to quench the fluorophores after imaging to allow the second round of staining–imaging. Thus, multiplexed assays of single EVs were achieved. Unlike traditional global EV detection methods, the results of this SEA approach showed that EV subpopulations contain vastly different markers. For example, the most widely used EV markers, the tetraspanins CD9, CD63, and CD81, were present in only 4.8%, 54%, and 26% of EV fractions, respectively [174]. CD9 and CD81 were also suggested to be mutually exclusively expressed [175]. Thus, SEA constitutes a very strong candidate for EV compositional and functional heterogeneity studies and other subtle biological explorations in the future.
Microfluidic NMR (μNMR): Proton NMR (1H NMR) represents another promising detection mechanism for EVs. Shao and coworkers designed an on-chip μNMR detection system that integrated immunocapture with EV profiling [176]. The microfluidic platform preconcentrated EVs before μNMR detection, contributing to an increased detection sensitivity, which was critical in overcoming the limitations in sensitivity due to the small size of EVs when using μNMR alone. To be detected by NMR, magnetic nanoparticles (MNPs) were used to label EVs through specific protein markers on the surface, such as CD63. Through magnetic labeling, EVs became superparamagnetic when placed into the NMR microcoil, resulting in accelerated decay of the 1H NMR signal, the rate of which was proportional to the amount of MNPs present in the system. Therefore, specifically targeted EVs can be quantified. Using this technology, comparative protein marker analyses between GBM-derived EVs and those from host cells were performed. This μNMR sensing approach held a distinctive advantage in that it lessened the cumbersome work of EV purification, because most naturally occurring entities in biological systems are not ferromagnetic and thus cause little interference.
Surface plasmon resonance (SPR): SPR sensing has also been used for EV detection. SPR is an optical technique that relies on a large number of free electrons on the metallic sensing surface, where the free electrons collectively oscillate under the action of the incident light field (i.e., the effect called surface plasma). The change of the RI of the adjacent medium on the metal film would change the plasmon resonance frequency, which causes a shift of the extinction spectrum, resulting in the detection of the target substance [177,178]. SPR-based EV protein analysis was improved by switching the conventional reflection configuration to a transmission mode using a set of periodic nanohole arrays, termed nanoplasmonic exosome (nPLEX) technology [179]. Various capturing/probe antibodies were immobilized on the sensing surface, which allowed the continuous isolation, probing, and quantification of EVs in a one-stop manner. The spectral intensity of optical transmittance resulted in a redshift on specific binding of EVs to the sensing surface, which was proportional to the mass density of the captured EVs on the surface and allowed quantitative profiling. Furthermore, the authors integrated the sensing array with parallel microfluidic channels to enable the independent analysis of up to 12 subtypes of exosomes. Compared with western blotting and ELISA, the nPLEX assay possesses much higher sensitivity, by 104 and 102 times, respectively. The lower limit of EV detection by nPLEX is ~3000 counts (670 aM) [180]. Due to the nature of SPR, nPLEX does not cause any damage to the analytes nor require any labels. Also, the total analysis time could be less than 30 min, and only a few microliters of a sample are required [179].
Electrochemical detection: Electrochemical sensing provides another promising mechanism for protein detection in EVs, where high sensitivity levels can be attained by amplifying redox reporter signals. Recently, integrated magnetic–electrochemical exosome (iMEX) detection orthogonally brings immunomagnetic isolation/enrichment and electrochemical detection together into a single platform to yield fast and simplified analyses of EVs, which consumes only a few microliters of the sample [181]. Magnetic beads are first functionalized with EV antibodies for capturing. Then, another horseradish peroxide (HRP)-conjugated antibody and 3,3′,5,5′-tetramethylbenzidine (TMB), a substrate generating electrical current when oxidized by HRP, are applied to detect specific EV markers. iMEX has also been used to detect T cell-derived exosome surface CD3 in urine to monitor cellular rejection in kidney transplantation patients [182]. The magnetic enrichment procedure, along with the enzymatic signal amplification, provides high sensitivity. The lower limit of detection is ~104 vesicles and the dynamic range spans over four orders of magnitude. Furthermore, the device is a portable unit containing eight standalone microfluidic channels, which can offer multiplexed, on-the-spot clinical EV marker detection. Typically, using iMEX, an entire multiple-marker-detecting assay can be finished in under 1 h and consumes only 10 μl of the sample.
Some of the challenges associated with the characterization of EV proteins include the following. Currently, there are no specific universal sets of proteins that can be used for detection and characterization of EVs, in general, and EV subpopulations. All EVs do not originate from the same biological source; therefore, EV subpopulations may have different protein surface markers associated with them, as well as carry different protein cargo [6]. Additionally, several studies have identified specific EV subpopulation protein markers, demonstrating that EVs can be classified based on their protein content. However, these studies used various isolation methods and different cellular sources. Due to the differences in the experimental designs and approaches, the specific sets of proteins that are definitively associated with each subpopulation of EVs remain to be determined [6]. Finally, detected protein abundance levels cannot be directly correlated with the EV count. EVs carry a wide range of proteins at highly variable abundances, depending on their source and biogenesis pathways [183].
Some experimental guidelines and practices that have been suggested include the following. First, it has been suggested that APOA1/2, APOB, and albumin (human or bovine) should be used for negative markers for serum-derived EVs as well as EVs isolated from cells cultured in the presence of bovine serum. Since such high-abundance proteins in the matrix can be co-isolated with EVs, it is recommended to report the level of these markers that need to be depleted to reach the desired purity of EV isolates [6]. Second, to identify specific proteins for each EV subpopulation it is recommended to use the same biological origin and the same isolation method to ensure the reproducibility of the analysis for a fair comparison [6,184]. Third, when culturing cells for use in EV studies, it is recommended to culture cells with fetal bovine serum (FBS) that has been EV depleted prior to addition to the cell culture medium. EV-depleted FBS is commercially available; however, the vendors often do not disclose the depletion protocol. Also, the storage and freeze–thaw conditions of EV-depleted FBS and FBS-based medium can lead to protein aggregation. Therefore, it is recommended to include a depletion step in-house [6].
Proteomics, Metabolomics, Lipidomics, and Multiomics Techniques for EV Characterization
Proteomics profiling holds great significance in EV research, and mass spectrometry (MS)-based proteomics techniques are currently the most widely used. In the study focused on fractionation of EV subclasses by AF4, Zhang and coworkers used nano-LC–electrospray ionization (ESI) MS/MS to profile signature proteins for each class. The functional analysis of these proteins revealed that exomeres were significantly enriched in proteins involved in metabolic processes, including protein synthesis and carbohydrate metabolism. The Exo-S fraction was enriched with proteins related to membrane vesicle biogenesis and transport, receptor signaling, and protein secretion, while the Exo-L EV fraction was enriched with proteins related to multiorganism organelle organization, the mitotic spindle, and interleukin (IL)-2/Stat 5 and G protein signaling [8]. Sample processing for MS-based EV proteomic analyses involves a few steps that typically require several hours, but the technology allows high-throughput, comparative, qualitative, and quantitative studies for hundreds and thousands of EV-derived analytes (Figure 4).
Beyond proteomic identification and quantification, profiling of post-translational modifications (PTMs) might provide a more in-depth insight into the biogenesis, cargo sorting, biomarker finding, and uptake mechanisms of EVs. Both lectin microarrays and MS have been used for EV glycosylation analysis, with lectin microarray assays being nondestructive and more unbiased toward the characterization of specific classes of glycans. While MS-based glycomic techniques currently target only N-glycans, the MS-based approaches are able to provide information to decipher glycan structures, including carbohydrate-based linkages and composition [185,186]. Using matrix-assisted laser desorption/ionization-time of flight/time of flight (MALDI-TOF/TOF) in an ovarian carcinoma cell-derived EV glycomics study, both high-mannose and complex-type N-linked glycans with various numbers of mannose residues or N-acetylglucosamine (GlcNAc) antennae, bisecting GlcNAc, and fucose residues on the chitobiose core were identified [187]. LC-MS/MS is a highly efficient tool for phosphoproteomics investigations. Neutral loss scanning mode was used to pinpoint 19 phosphorylation sites in 14 human urine-derived EV proteins [188]. Parallel reaction monitoring (PRM)-based label-free quantitative MS has also been used to identify phosphoproteins in blood-derived EVs as candidate biomarkers for breast cancer [189].
SOMAscan is an affinity-based proteomics analysis technique where slow-off-rate modified aptamers are used for multiplexed, highly sensitive, and specific protein detection in human blood and other biomatrices, which has been instrumental in finding novel protein biomarker candidates [190]. This platform was applied to biomarker identification in prostate cancer cell-derived EVs and found over 300 candidate proteins [191]. SOMAscan was also used for relative abundance analysis of 1128 proteins from cerebrospinal fluid (CSF)-derived EVs to find potential biomarkers for CNS inflammatory diseases [192]. One drawback of SOMAscan is that the results are largely dependent on data-processing algorithms and normalization, which might lead to severe inconsistencies in findings from different laboratories. Also, the method relies on the readout of the binding assay, which greatly depends on the specificity of binding, which can be compromised in highly complex matrices like biological fluids. Moreover, the method is limited to quantitative characterization of the binding between a specific aptamer and an assumed target analyte without the ability to confirm the identity of the bound species by structural characterization.
Along with protein and lipid EV components, EV metabolites are also expected to be of high functional importance, and metabolomics profiling can probe deeper into EVs’ structural features. Although a new field, EV metabolomics is rapidly growing in demand, due to the fact that metabolites can be transferred as nutrients and signals between cells via EVs [193]. Metabolites can be extracted from EVs by organic solvents, separated based on their various properties (mass, charge, polarity, hydrophobicity), and then detected using MS- or NMR-based techniques [193,194]. One study showed that there are active metabolic processes in the EV by comparing EVs from GBM cells with their parent cells [194]. However, there are currently few metabolomic biomarkers. Explorations of metabolomic changes in EVs and the cells and tissues of their origin can potentially be used to expand the toolbox for clinical diagnostics [194]. Recent advantages in metabolomics include the improved sensitivity that the instrumentation offers, although these methods can be costly and not readily accessible.
Studying the EV cargo using MS-based techniques has led to a better understanding of EVs and their biogenesis and structural features. There have been several deficiencies in proteomic and MS-based molecular profiling studies of EVs for a number of years. The main challenges include the accurate quantification of proteins, characterization of PTMs, and the identification of multiomics workflows to characterize EVs and to better understand the biology of EVs [195]. MS-based quantitation techniques are promising, and once appropriate standards and target proteins/peptides have been determined, quantitation should be able to be performed accurately. Currently, data-independent and targeted proteomics approaches have been applied to solve the quantification-related issues. A study performed by Chen and coworkers used Tandem Mass Tags (TMT) reagents to chemically label samples and reported 36 upregulated proteins and 22 downregulated proteins in exosomes released by CRC tumor cells into plasma compared with exosomes in healthy-volunteer plasma. The analysis showed that the upregulated proteins are involved in the protumorigenic microenvironment for metastasis [196]. In a metabolomic study, Puhka and coworkers used targeted based LC-MS/MS to profile the metabolites from urine EVs in prostate cancer patients compared with urine from healthy patients [197]. This study provided a foundation for enriched metabolites in EVs and new normalization techniques that have not been implemented before. Due to the low abundance in biological samples, EV proteomics and metabolomics force the need for novel techniques during sample preparation, data acquisition, and data analysis.
In combination, proteomics, metabolomics, and lipidomics profiling (a.k.a. multiomics) can provide more comprehensive information about EV molecular composition and give greater insight into EV biology and classification. MS-based techniques can be used to identify different types of small molecules in EVs, including various classes of lipids, organic acids, amino acids, sugars, and nucleotides. These small molecules can provide important information about distinct biomarkers carried in EVs [198]. Using differential and gradient UC, lipoproteins can be fractionated based on their respective densities, with HDLs and EV pellets falling to the bottom of the sample tube and lower-density lipoproteins migrating toward the top [45]. These fractions contain various classes of lipids, including phospholipids, glycerophospholipids, sphingolipids, cholesteryl esters, and glycerolipid species, along with other proteins. Analysis of these different fractions showed the correlation between proteins and lipids among EV compositions [199]. It has been shown that lipids play a critical role in signal transduction in cellular growth and facilitate intercellular signaling [200]. An emerging method for EV analysis is coupling ion-mobility spectrometry (IMS), which separates ions based on their mobility in a carrier gas and an applied electric field [i.e., the collision cross-section (CCS) and the charge of the ion] before MS analysis. This method can be used to analyze lipids, metabolites, and glycans and has previously been used to study viruses [198,201,202]. Combining lipidomics, metabolomics, and proteomics with transcriptomics and EV morphology and size distribution profiling for EV populations will result in the most comprehensive knowledge about specific structural features of EVs of different origins and types (Figure 4).
Concluding Remarks
A major advance in the field of EV research during the past decade was largely enabled using already existing analytical techniques and purposefully developed novel technologies for EV isolation and characterization. While this progress resulted in an increased depth of knowledge about the structural features, functions, and biogenesis of EVs, our understanding of EVs’ functional and mechanistic biology, as well as their relevance to specific pathophysiological states, remains limited. Also, the detailed characterization of the molecular composition of each type of EV and of EV subpopulations remains a challenge mainly due to the deficiencies in current techniques for the isolation of high-purity homogeneous EVs and their specific subtypes from complex matrices such as physiological fluids. Alternative, as well as similar or identical experimental approaches used in different laboratories, often lead to substantially different EV profiling results. There is a significant need for improved performance and reproducibility of techniques for EV isolation and comprehensive characterization. Designing and conducting consortium-type studies across multiple laboratories will accelerate progress toward standardizing experimental approaches and procedures for reporting data (see Outstanding Questions). Improved standardized techniques will enable a comprehensive characterization of EV types and further advance our understanding of EV biology to develop novel EV-based diagnostic and therapeutic approaches (Figure 5 ).
Outstanding Questions.
How can straightforward, robust, and reproducible isolation and detailed characterization of EVs and EV subpopulations from complex matrices such as physiological fluids across multiple laboratories be enabled?
How can EV isolation approaches that would enable effective and robust isolation from limited specimens be miniaturized for potential fast and sensitive diagnostic applications?
What are the biological and technical challenges currently impeding the clinical translation of EVs?
What specific CQAs can help in assessing the reproducibility of the final product and the batch–batch variability in the large-scale manufacture of EV-based therapeutic products?
How can a detailed catalog of molecular features (nucleic acids, proteins/proteoforms, lipids, and metabolites) be developed to characterize EV populations of various properties, origins, and functions?
What are the mechanisms to stimulate and support multi-laboratory consortium-style efforts for the development and evaluation of novel analytical technologies for EV isolation and characterization?
What EV structural features can serve as the strongest predictors and high-specificity/high-selectivity markers of specific pathological changes?
Alt-text: Outstanding Questions
Figure 5.
Summary of Technologies, Challenges, and Applications in Extracellular Vesicle (EV) Research.
Abbreviations: AF4, asymmetric flow field-flow fractionation; DLS, dynamic light scattering; LC, liquid chromatography; MS, mass spectrometry; nano-FCM, nano-flow cytometry; NTA, nanoparticle tracking analysis; TRPS, tunable resistive pulse sensing;
UC, ultracentrifugation.
The following areas of technology advancement will be critically important for moving EV research forward more efficiently. In studies related to EVs from physiological fluids, sample procurement, collection, and preprocessing prior to EV isolation are essential areas that critically require major efforts devoted to further development and standardization. A set of standard protocols for specimen collection needs to be optimized, documented in a very detailed way, evaluated by several laboratories, and specialized for several main workflows of downstream analyses (e.g., proteomics, transcriptomics, TEM). Such protocols should be very specific and include requirements and recommendations to minimize preanalytical variables that can be possibly dependent on fasting status, hydration status, the time of day for specimen collection, the type of physiological fluid, and the type of the specimen’s fraction (e.g., plasma, serum), with step-by-step instructions for specimen collection, processing, and storage (e.g., the choice of an anticoagulant or other additives, sample cleanup procedures, duration and speed of centrifugation, storage temperature and suggested duration).
For EV isolation, the priorities include developing protocols for reliable and reproducible isolation of both total EVs and specific EV subpopulations based on morphological (i.e., size and shape), biophysical (e.g., surface charge, membrane stiffness), biochemical composition (e.g., membranous, nonmembranous, presence/absence of specific surface and possibly internal markers), biological (cells/tissues of origin), and functional (i.e., specific biological activity) properties. Additionally, developing reproducible and straightforward EV isolation techniques that can be used in a clinical setting is a major priority. Finally, miniaturization of EV isolation approaches that would enable effective and robust isolation from either limited samples (e.g., finger-prick blood specimens, archived specimens from longitudinal studies, neonatal, children’s, animal, scarce microneedle aspirates, other small biological specimens) is also of high priority to enable potential fast and sensitive diagnostic applications. New modalities for separation might need to be developed for the efficient EV isolation required by the above-outlined areas of applications. Robust isolation protocols will also improve batch-to-batch reproducibility and potentially may address some of the heterogeneity concerns in EV samples.
For application of EVs as drug delivery vehicles, large-scale-manufacturing EV isolation techniques such as TFF and SEC are utilized to produce clinical-grade EVs. In such cases, the development of cGMP protocols that include a thorough characterization of a number of process parameters, such as parent/source cell type, bioreactor conditions, and chromatography conditions, is essential to understand the CQAs and reproducibility of the final product. When such CQAs are identified, an acceptable range for variables such as defined size, purity, surface marker profile, content loading, and identity needs to be implemented for release testing and to understand the batch–batch variability and consistency of the EV manufacturing process [204].
In the area of biochemical characterization of EVs, enabling the following capabilities appears especially attractive: detailed molecular cataloging of EV populations of various properties, origins, and functions; sensitive quantitative characterization of PTMs of EV proteins (e.g., glycosylation, acetylation, phosphorylation); informative multiomics characterization of the same EV isolates from the same specimen; high-sensitivity deep molecular profiling of EV isolates from limited samples (finger-prick blood specimens, neonatal specimens, tear fluid; see the above-listed examples); and highly specific and sensitive detection and quantitation of target EV molecules of interest [205] in EV isolates, nonprocessed physiological fluids (e.g., whole blood, CSF), minimally processed biological fluids (e.g., plasma, serum), and even archived samples derived from physiological fluids (e.g., dried blood spots, small aliquots of longitudinally collected cohort studies) that can enable the detection and quantitation of rare EVs in the bulk of other, irrelevant EV populations at high sensitivity and specificity.
Advances in sequencing technologies such as NGS have helped in understanding the heterogeneity of the EV transcriptome. However, there remains a need for standardization of the complex workflow involved in the analysis of EV samples so the process is robust and repeatable across various laboratories. The information from these advanced techniques should be able to inform the choice of analytical methods depending on the type of RNA species for routine QC purposes. While the high-performance transcriptomics approaches in EV characterization are becoming well matured, the proteomics, lipidomics, and metabolomics approaches have yet to catch up in their performance, reproducibility, and ruggedness. The following novel analytical technologies appear especially promising to enable further progress in EV research: SEA [174,175]; native MS of intact macromolecular structures of high-molecular-mass macromolecular complexes, including intact EVs [206., 207., 208., 209.]; and charge detection MS of high-molecular-mass macromolecular complexes that can be made applicable to the characterization of individual EVs [210., 211., 212.]. Moreover, the direct, unambiguous EV characterization that combines analysis of intact EVs (e.g., imaging, mass and/or charge, ζ-potential, size measurements) with molecular profiling of the same EVs or at least EVs from the same sample (e.g., charge detection MS, native MS analysis, immunoaffinity-based SEA) will be instrumental in the future development of the EV research and knowledge base (Figure 4).
Additionally, further advances in data interpretation and multiparametric data analysis that can include multiple levels of information about the patient, the patient’s own and family health history and personal metadata, the treatment history, sample collection, processing, and analysis conditions and outcomes [213] are also of high importance for the further progress of the EV field of research and applications.
To conclude, advances in various aspects of EV research such as isolation, analytical characterization, and standardization of methods would greatly enhance the mechanistic understanding of EV biology and cargo components. These insights can help in further translating the application of EVs for efficient diagnostic purposes and therapeutic strategies.
Acknowledgments
This work was supported by the National Institutes of Health under award numbers 1R01GM120272 (A.R.I.) and 5R01CA218500 (A.R.I.). We acknowledge Thermo Fisher Scientific for their support through a technology alliance and SCIEX for their collaborative support.
References
- 1.Harding C. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 1984;35:256–263. [PubMed] [Google Scholar]
- 2.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Niel G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.Cai H. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;1162:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 6.Thery C. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H., Lyden D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019;14:1027–1053. doi: 10.1038/s41596-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018;20:332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathieu M. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 10.Hurley J.H. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trajkovic K. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 12.Geminard C. Degradation of AP2 during reticulocyte maturation enhances binding of hsc70 and Alix to a common site on TFR for sorting into exosomes. Traffic. 2004;5:181–193. doi: 10.1111/j.1600-0854.2004.0167.x. [DOI] [PubMed] [Google Scholar]
- 13.Valadi H. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Villarroya-Beltri C. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbings D.J. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 16.Shurtleff M.J. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5 doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raposo G. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng D. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 19.Escrevente C. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zitvogel L. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat. Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 21.Rajendran L. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.J. Cell-to-cell transmission of non-prion protein aggregates. Nat. Rev. Neurol. 2010;6:702–706. doi: 10.1038/nrneurol.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang C. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milane L. Exosome mediated communication within the tumor microenvironment. J. Control. Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 25.Ruivo C.F. the biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77:6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 26.Stremersch S. Therapeutic and diagnostic applications of extracellular vesicles. J. Control. Release. 2016;244:167183. doi: 10.1016/j.jconrel.2016.07.054. [DOI] [PubMed] [Google Scholar]
- 27.Sharma S. Tumor-derived exosomes in ovarian cancer – liquid biopsies for early detection and real-time monitoring of cancer progression. Oncotarget. 2017;8:104687–104703. doi: 10.18632/oncotarget.22191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostenfeld M.S. miRNA profiling of circulating EpCAM+ extracellular vesicles: promising biomarkers of colorectal cancer. J. Extracell. Vesicles. 2016;5:31488. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin X. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017;23:5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 30.Yoshioka Y. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiklander O.P.B. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vader P. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;106:148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Pascucci L. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: a new approach for drug delivery. J. Control. Release. 2014;192:262–270. doi: 10.1016/j.jconrel.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 34.Kamerkar S. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su M.J. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci. Rep. 2016;6:30110. doi: 10.1038/srep30110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi S. Dendritic cells pulsed with exosomes in combination with PD-1 antibody increase the efficacy of sorafenib in hepatocellular carcinoma model. Transl. Oncol. 2018;11:250–258. doi: 10.1016/j.tranon.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zipkin M. Exosome redux. Nat. Biotechnol. 2019;37:1395–1400. doi: 10.1038/s41587-019-0326-5. [DOI] [PubMed] [Google Scholar]
- 38.Gudbergsson J.M. Systematic review of targeted extracellular vesicles for drug delivery – considerations on methodological and biological heterogeneity. J. Control. Release. 2019;306:108–120. doi: 10.1016/j.jconrel.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Markov O. Immunotherapy based on dendritic cell-targeted/-derived extracellular vesicles – a novel strategy for enhancement of the anti-tumor immune response. Front. Pharmacol. 2019;10:1152. doi: 10.3389/fphar.2019.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baranyai T. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Revenfeld A.L. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin. Ther. 2014;36:830–846. doi: 10.1016/j.clinthera.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Wu M. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10584–10589. doi: 10.1073/pnas.1709210114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karimi N. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. 2018;75:2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sodar B.W. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016;6:24316. doi: 10.1038/srep24316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuana Y. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danielson K.M. Diurnal variations of circulating extracellular vesicles measured by nano flow cytometry. PLoS One. 2016;11 doi: 10.1371/journal.pone.0144678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witwer K.W. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kipps E. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat. Rev. Cancer. 2013;13:273–282. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed N., Stenvers K.L. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front. Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson D.C. Scalable, cGMP-compatible purification of extracellular vesicles carrying bioactive human heterodimeric IL-15/lactadherin complexes. J. Extracell. Vesicles. 2018;7:1442088. doi: 10.1080/20013078.2018.1442088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thery C. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;3:22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 52.Coumans F.A.W. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 53.Skates S.J. Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. J. Proteome Res. 2013;12:5383–5394. doi: 10.1021/pr400132j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q. Biopreserv Biobank; 2018. Obtaining High-Quality Blood Specimens for Downstream Applications: A Review of Current Knowledge and Best Practices. [DOI] [PubMed] [Google Scholar]
- 55.Yokota M. Effects of heparin on polymerase chain reaction for blood white cells. J. Clin. Lab. Anal. 1999;13:133–140. doi: 10.1002/(SICI)1098-2825(1999)13:3<133::AID-JCLA8>3.0.CO;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeyaram A., Jay S.M. Preservation and storage stability of extracellular vesicles for therapeutic applications. AAPS J. 2017;20:1. doi: 10.1208/s12248-017-0160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kusuma G.D. To protect and to preserve: novel preservation strategies for extracellular vesicles. Front. Pharmacol. 2018;9:1199. doi: 10.3389/fphar.2018.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lasser C. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012;59 doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nocera A.L. Exosomes mediate interepithelial transfer of functional P-glycoprotein in chronic rhinosinusitis with nasal polyps. Laryngoscope. 2017;127:E295–E300. doi: 10.1002/lary.26614. [DOI] [PubMed] [Google Scholar]
- 60.Fraser K. Characterization of single microvesicles in plasma from glioblastoma patients. Neuro-Oncology. 2019;21:606–615. doi: 10.1093/neuonc/noy187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liga A. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15:2388–2394. doi: 10.1039/c5lc00240k. [DOI] [PubMed] [Google Scholar]
- 62.Mueller S.K. Exosome function in aerodigestive mucosa. Nanomedicine. 2018;14:269–277. doi: 10.1016/j.nano.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 63.Linares R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles. 2015;4:29509. doi: 10.3402/jev.v4.29509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tauro B.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Nocera A.L. Exosome swarms eliminate airway pathogens and provide passive epithelial immunoprotection through nitric oxide. J. Allergy Clin. Immunol. 2018;143:1525–1535.e1. doi: 10.1016/j.jaci.2018.08.046. [DOI] [PubMed] [Google Scholar]
- 66.Greening D.W. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 67.Boing A.N. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blans K. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles. 2017;6:1294340. doi: 10.1080/20013078.2017.1294340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takov K. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J. Extracell. Vesicles. 2019;8:1560809. doi: 10.1080/20013078.2018.1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lozano-Ramos I. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles. 2015;4:27369. doi: 10.3402/jev.v4.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tengattini S. Chromatographic Approaches for purification and analytical characterization of extracellular vesicles: recent advancements. Chromatographia. 2019;82:415–424. [Google Scholar]
- 72.Taylor D.D., Shah S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods. 2015;87:3–10. doi: 10.1016/j.ymeth.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 73.Lobb R.J. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheruvanky A. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am. J. Physiol. Renal Physiol. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei Z. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 2017;8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ueda K. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014;4:6232. doi: 10.1038/srep06232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clayton A. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 2001;247:163–174. doi: 10.1016/s0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 78.du Cheyron D. Urinary measurement of Na+/H+ exchanger isoform 3 (NHE3) protein as new marker of tubule injury in critically ill patients with ARF. Am. J. Kidney Dis. 2003;42:497–506. doi: 10.1016/s0272-6386(03)00744-3. [DOI] [PubMed] [Google Scholar]
- 79.Sharma P. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles. 2018;7:1435138. doi: 10.1080/20013078.2018.1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mathivanan S. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen C. Paper-based devices for isolation and characterization of extracellular vesicles. J. Vis. Exp. 2015;98 doi: 10.3791/52722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fang S. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One. 2017;12 doi: 10.1371/journal.pone.0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nilsson J. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br. J. Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skog J. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balaj L. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015;5:10266. doi: 10.1038/srep10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Konoshenko M.Y. Isolation of extracellular vesicles: general methodologies and latest trends. Biomed. Res. Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Helwa I. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rekker K. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 89.Schageman J. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed. Res. Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patel G.K. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019;9:5335. doi: 10.1038/s41598-019-41800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen C. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shao H. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo S.C. Microfluidics-based on-a-chip systems for isolating and analysing extracellular vesicles. J. Extracell. Vesicles. 2018;7:1508271. doi: 10.1080/20013078.2018.1508271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rho J. Magnetic nanosensor for detection and profiling of erythrocyte-derived microvesicles. ACS Nano. 2013;7:11227–11233. doi: 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davies R.T. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 96.Kanwar S.S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891–1900. doi: 10.1039/c4lc00136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petersen K.E. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal. Bioanal. Chem. 2014;406:7855–7866. doi: 10.1007/s00216-014-8040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fraunhofer W., Winter G. The use of asymmetrical flow field-flow fractionation in pharmaceutics and biopharmaceutics. Eur. J. Pharm. Biopharm. 2004;58:369–383. doi: 10.1016/j.ejpb.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 99.Sitar S. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 2015;87:9225–9233. doi: 10.1021/acs.analchem.5b01636. [DOI] [PubMed] [Google Scholar]
- 100.Morales-Kastresana A. High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. J. Extracell. Vesicles. 2019;8:1597603. doi: 10.1080/20013078.2019.1597603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kormelink T.G. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytometry A. 2016;89:135–147. doi: 10.1002/cyto.a.22644. [DOI] [PubMed] [Google Scholar]
- 102.Pospichalova V. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vaswani K. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod. Biol. 2017;17:341–348. doi: 10.1016/j.repbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 104.Nordin J.Z. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine. 2015;11:879–883. doi: 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 105.Muller L. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods. 2014;411:55–65. doi: 10.1016/j.jim.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kreimer S. Mass-spectrometry-based molecular characterization of extracellular vesicles: lipidomics and proteomics. J. Proteome Res. 2015;14:2367–2384. doi: 10.1021/pr501279t. [DOI] [PubMed] [Google Scholar]
- 107.Cox G., Sheppard C.J.R. Practical limits of resolution in confocal and non-linear microscopy. Microsc. Res. Tech. 2004;63:18–22. doi: 10.1002/jemt.10423. [DOI] [PubMed] [Google Scholar]
- 108.Lai C.P. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015;6:7029. doi: 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zomer A. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lai C.P. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483–494. doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Niers J. Single reporter for targeted multimodal in vivo imaging. J. Am. Chem. Soc. 2012;134:5149–5156. doi: 10.1021/ja209868g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rikkert L.G. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles. 2019;8:1555419. doi: 10.1080/20013078.2018.1555419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Milne J.L.S. Cryo-electron microscopy: a primer for the non-microscopist. FEBS J. 2012;280:28–45. doi: 10.1111/febs.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koifman N.A. A direct-imaging cryo-EM study of shedding extracellular vesicles from leukemic monocytes. J. Struct. Biol. 2017;198:177–185. doi: 10.1016/j.jsb.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 115.Cizmar P., Yuana Y. Detection and characterization of extracellular vesicles by transmission and cryo-transmission electron microscopy. Methods Mol. Biol. 2017;1660:221–232. doi: 10.1007/978-1-4939-7253-1_18. [DOI] [PubMed] [Google Scholar]
- 116.Kotrbová A. TEM ExosomeAnalyzer: a computer-assisted software tool for quantitative evaluation of extracellular vesicles in transmission electron microscopy images. J. Extracell. Vesicles. 2019;8:1560808. doi: 10.1080/20013078.2018.1560808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuana Y. Atomic force microscopy: a novel approach to the detection of nanosized blood microparticles. J. Thromb. Haemost. 2010;8:315–323. doi: 10.1111/j.1538-7836.2009.03654.x. [DOI] [PubMed] [Google Scholar]
- 118.Sharma S. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano. 2010;4:1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stetefeld J. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cummins H.Z., Pike E.R. Plenum Press; 1974. Photon Correlation and Light Beating Spectroscopy. [Google Scholar]
- 121.Maguire C.M. Characterisation of particles in solution – a perspective on light scattering and comparative technologies. Sci. Technol. Adv. Mater. 2018;19:732–745. doi: 10.1080/14686996.2018.1517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gercel-Taylor C. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012;428:44–53. doi: 10.1016/j.ab.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Kesimer M., Gupta R. Physical characterization and profiling of airway epithelial derived exosomes using light scattering. Methods. 2015;87:59–63. doi: 10.1016/j.ymeth.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Labrie A. Characterization of platelet concentrates using dynamic light scattering. Transfus. Med. Hemother. 2013;40:93–100. doi: 10.1159/000350362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lawrie A.S. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009;96:206–212. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 126.Kaszuba M. Resolving concentrated particle size mixtures using dynamic light scattering. Part. Part. Syst. Charact. 2007;24:159–162. [Google Scholar]
- 127.Dragovic R.A. Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kormelink T.G. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytometry A. 2015;89:135–147. doi: 10.1002/cyto.a.22644. [DOI] [PubMed] [Google Scholar]
- 129.Tian Y. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 130.Choi D. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano. 2019;13:10499–10511. doi: 10.1021/acsnano.9b04480. [DOI] [PubMed] [Google Scholar]
- 131.Poncelet P. Tips and tricks for flow cytometry-based analysis and counting of microparticles. Transfus. Apher. Sci. 2015;53:110–126. doi: 10.1016/j.transci.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 132.McVey M.J. Improved resolution in extracellular vesicle populations using 405 instead of 488 nm side scatter. J. Extracell. Vesicles. 2018;7:1454776. doi: 10.1080/20013078.2018.1454776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arraud N. Fluorescence triggering: a general strategy for enumerating and phenotyping extracellular vesicles by flow cytometry. Cytometry A. 2015;89:184–195. doi: 10.1002/cyto.a.22669. [DOI] [PubMed] [Google Scholar]
- 134.Tian Y. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles. 2019;9:1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buzás E.I. Single particle analysis: methods for detection of platelet extracellular vesicles in suspension (excluding flow cytometry) Platelets. 2017;28:249–255. doi: 10.1080/09537104.2016.1260704. [DOI] [PubMed] [Google Scholar]
- 136.Gorgens A. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J. Extracell. Vesicles. 2019;8:1587567. doi: 10.1080/20013078.2019.1587567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Simonsen J.B. Unique calibrators derived from fluorescence-activated nanoparticle sorting for flow cytometric size estimation of artificial vesicles: possibilities and limitations. Cytometry A. 2019;95:917–924. doi: 10.1002/cyto.a.23797. [DOI] [PubMed] [Google Scholar]
- 138.Varga Z. Hollow organosilica beads as reference particles for optical detection of extracellular vesicles. J. Thromb. Haemost. 2018;16:1646–1655. doi: 10.1111/jth.14193. [DOI] [PubMed] [Google Scholar]
- 139.Vogel R. A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing. J. Extracell. Vesicles. 2016;5:31242. doi: 10.3402/jev.v5.31242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vogel R. High-resolution single particle zeta potential characterisation of biological nanoparticles using tunable resistive pulse sensing. Sci. Rep. 2017;7:17479. doi: 10.1038/s41598-017-14981-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Akers J.C. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in clinical cerebrospinal fluids (CSF) PLoS One. 2016;11 doi: 10.1371/journal.pone.0149866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gualerzi A. Raman spectroscopy uncovers biochemical tissue-related features of extracellular vesicles from mesenchymal stromal cells. Sci. Rep. 2017;7:9820. doi: 10.1038/s41598-017-10448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ozgur E. Ultrasensitive detection of human chorionic gonadotropin using frequency locked microtoroid optical resonators. Anal. Chem. 2019;91:11872–11878. doi: 10.1021/acs.analchem.9b02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Daaboul G.G. Digital detection of exosomes by interferometric imaging. Sci. Rep. 2016;6:37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bachurski D. Extracellular vesicle measurements with nanoparticle tracking analysis – an accuracy and repeatability comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles. 2019;8:1596016. doi: 10.1080/20013078.2019.1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kinoshita T. MicroRNAs in extracellular vesicles: potential cancer biomarkers. J. Hum. Genet. 2017;62:67–74. doi: 10.1038/jhg.2016.87. [DOI] [PubMed] [Google Scholar]
- 147.Nolte-’t Hoen E.N. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Turchinovich A. Transcriptome of extracellular vesicles: state-of-the-art. Front. Immunol. 2019;10:202. doi: 10.3389/fimmu.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mateescu B. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Andreu Z. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J. Extracell. Vesicles. 2016;5:31655. doi: 10.3402/jev.v5.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chevillet J.R. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Amorim M.G. A total transcriptome profiling method for plasma-derived extracellular vesicles: applications for liquid biopsies. Sci. Rep. 2017;7:14395. doi: 10.1038/s41598-017-14264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huang X. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao S. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Murphy D. Gene expression studies using microarrays: principles, problems, prospects. Adv. Physiol. Educ. 2002;26:256–270. doi: 10.1152/advan.00043.2002. [DOI] [PubMed] [Google Scholar]
- 156.Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr. Protoc. Mol. Biol. 2013;22:22.1. doi: 10.1002/0471142727.mb2201s101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jaksik R. Microarray experiments and factors which affect their reliability. Biol. Direct. 2015;10:46. doi: 10.1186/s13062-015-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jaluria P. A perspective on microarrays: current applications, pitfalls, and potential uses. Microb. Cell Factories. 2007;6:4. doi: 10.1186/1475-2859-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Armstrong D.A. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol. Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Godlewski J. MicroRNA signatures and molecular subtypes of glioblastoma: the role of extracellular transfer. Stem Cell Rep. 2017;8:1497–1505. doi: 10.1016/j.stemcr.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Trivedi M. Modification of tumor cell exosome content by transfection with wt-p53 and microRNA-125b expressing plasmid DNA and its effect on macrophage polarization. Oncogenesis. 2016;5:e250. doi: 10.1038/oncsis.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Buschmann D. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles. 2018;7:1481321. doi: 10.1080/20013078.2018.1481321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Van Deun J. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Srinivasan S. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462.e16. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sork H. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 2018;8:10813. doi: 10.1038/s41598-018-28485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lässer C. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017;14:58–72. doi: 10.1080/15476286.2016.1249092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lunavat T.R. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells – evidence of unique microRNA cargos. RNA Biol. 2015;12:810–823. doi: 10.1080/15476286.2015.1056975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brady P.N., Macnaughtan M.A. Evaluation of colorimetric assays for analyzing reductively methylated proteins: biases and mechanistic insights. Anal. Biochem. 2015;491:43–51. doi: 10.1016/j.ab.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He M. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773–3780. doi: 10.1039/c4lc00662c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao Z. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489–496. doi: 10.1039/c5lc01117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cai S. Immuno-modified superparamagnetic nanoparticles via host–guest interactions for high-purity capture and mild release of exosomes. Nanoscale. 2018;10:14280–14289. doi: 10.1039/c8nr02871k. [DOI] [PubMed] [Google Scholar]
- 172.Xu H.Y. Magnetic-based microfluidic device for on-chip isolation and detection of tumor-derived exosomes. Anal. Chem. 2018;90:13451–13458. doi: 10.1021/acs.analchem.8b03272. [DOI] [PubMed] [Google Scholar]
- 173.Reategui E. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018;9:175. doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee K. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12:494–503. doi: 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gyuris A. Physical and molecular landscapes of mouse glioma extracellular vesicles define heterogeneity. Cell Rep. 2019;27:3972–3987.e6. doi: 10.1016/j.celrep.2019.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shao H.L. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zeng S.W. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem. Soc. Rev. 2014;43:3426–3452. doi: 10.1039/c3cs60479a. [DOI] [PubMed] [Google Scholar]
- 178.Park J. Analyses of intravesicular exosomal proteins using a nano-plasmonic system. ACS Photonics. 2018;5:487–494. doi: 10.1021/acsphotonics.7b00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Im H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shao H. New technologies for analysis of extracellular vesicles. Chem. Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jeong S. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano. 2016;10:1802–1809. doi: 10.1021/acsnano.5b07584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Assaker J. Integrated magnetic–electrochemical exosome (iMEX) technology to detect rejection in kidney transplant recipients. Am. J. Transplant. 2017;17:750. [Google Scholar]
- 183.Hartjes T.A. Extracellular vesicle quantification and characterization: common methods and emerging approaches. Bioengineering. 2019;6:7. doi: 10.3390/bioengineering6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ludwig N. Challenges in exosome isolation and analysis in health and disease. Int. J. Mol. Sci. 2019;20:4684. doi: 10.3390/ijms20194684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Williams C. Glycosylation of extracellular vesicles: current knowledge, tools and clinical perspectives. J. Extracell. Vesicles. 2018;7:1442985. doi: 10.1080/20013078.2018.1442985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Alley W.R., Novotny M.V. Structural glycomic analyses at high sensitivity: a decade of progress. Annu Rev Anal Chem (Palo Alto, Calif) 2013;6:237–265. doi: 10.1146/annurev-anchem-062012-092609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Escrevente C. Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gonzales P.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen I.H. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Candia J. Assessment of variability in the SOMAscan assay. Sci. Rep. 2017;7:14248. doi: 10.1038/s41598-017-14755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Webber J. Proteomics analysis of cancer exosomes using a novel modified aptamer-based array (SOMAscanTM) platform. Mol. Cell. Proteomics. 2014;13:1050–1064. doi: 10.1074/mcp.M113.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Welton J.L. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J. Extracell. Vesicles. 2017;6:1369805. doi: 10.1080/20013078.2017.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Williams C. Metabolomics applied to the study of extracellular vesicles. Metabolites. 2019;9:276. doi: 10.3390/metabo9110276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cuperlovic-Culf M. Analysis and simulation of glioblastoma cell lines-derived extracellular vesicles metabolome. Metabolites. 2020;10:88. doi: 10.3390/metabo10030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rosa-Fernandes L. A perspective on extracellular vesicles proteomics. Front. Chem. 2017;5:102. doi: 10.3389/fchem.2017.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen Y.Y. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int. J. Cancer. 2017;140:900–913. doi: 10.1002/ijc.30496. [DOI] [PubMed] [Google Scholar]
- 197.Puhka M. Metabolomic profiling of extracellular vesicles and alternative normalization methods reveal enriched metabolites and strategies to study prostate cancer-related changes. Theranostics. 2017;7:3824–3841. doi: 10.7150/thno.19890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zebrowska A. Metabolome of exosomes: focus on vesicles released by cancer cells and present in human body fluids. Int. J. Mol. Sci. 2019;20:3461. doi: 10.3390/ijms20143461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen S. Lipidomic characterization of extracellular vesicles in human serum. J. Circ. Biomark. 2019;8 doi: 10.1177/1849454419879848. 1849454419879848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nishida-Aoki N. Lipidomic analysis of cells and extracellular vesicles from high- and low-metastatic triple-negative breast cancer. Metabolites. 2020;10:67. doi: 10.3390/metabo10020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pease L.F., III Physical analysis of virus particles using electrospray differential mobility analysis. Trends Biotechnol. 2012;30:216–224. doi: 10.1016/j.tibtech.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 202.Pagel K., Harvey D.J. Ion mobility-mass spectrometry of complex carbohydrates: collision cross sections of sodiated N-linked glycans. Anal. Chem. 2013;85:5138–5145. doi: 10.1021/ac400403d. [DOI] [PubMed] [Google Scholar]
- 203.Vergauwen G. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci. Rep. 2017;7:2704. doi: 10.1038/s41598-017-02599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Rohde E. Manufacturing and characterization of extracellular vesicles from umbilical cord-derived mesenchymal stromal cells for clinical testing. Cytotherapy. 2019;21:581–592. doi: 10.1016/j.jcyt.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 205.Oliveira G.P., Jr. Detection of extracellular vesicle RNA using molecular beacons. iScience. 2020;23:100782. doi: 10.1016/j.isci.2019.100782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Synowsky S.A. Probing genuine strong interactions and post-translational modifications in the heterogeneous yeast exosome protein complex. Mol. Cell. Proteomics. 2006;5:1581–1592. doi: 10.1074/mcp.M600043-MCP200. [DOI] [PubMed] [Google Scholar]
- 207.Heck A.J. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 208.Ivanov A.R. ASMS 2015, 63rd ASMS Conference on Mass Spectrometry and Allied Topics. Springer; 2015. Analysis of proteins, protein complexes and proteomes under non-denaturing conditions using sheathless capillary electrophoresis coupled with native mass spectrometry. [Google Scholar]
- 209.Chorev D.S. Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry. Science. 2018;362:829–834. doi: 10.1126/science.aau0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brown B.A. Charge detection mass spectrometry measurements of exosomes and other extracellular particles enriched from bovine milk. Anal. Chem. 2020;92:3285–3292. doi: 10.1021/acs.analchem.9b05173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kafader J.O. Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat. Methods. 2020;17:391–394. doi: 10.1038/s41592-020-0764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Worner T.P. Resolving heterogeneous macromolecular assemblies by Orbitrap-based single-particle charge detection mass spectrometry. Nat. Methods. 2020;17:395–398. doi: 10.1038/s41592-020-0770-7. [DOI] [PubMed] [Google Scholar]
- 213.Welsh J.A. FCMPASS software aids extracellular vesicle light scatter standardization. Cytometry A. 2019 doi: 10.1002/cyto.a.23782. Published online June 28, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhang Q. Transfer of functional cargo in exomeres. Cell Rep. 2019;27:940–954.e6. doi: 10.1016/j.celrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Comelli L. Characterization of secreted vesicles from vascular smooth muscle cells. Mol. BioSyst. 2014;10:1146–1152. doi: 10.1039/c3mb70544g. [DOI] [PubMed] [Google Scholar]
- 216.Corso G. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule–single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J. Extracell. Vesicles. 2019;8:1663043. doi: 10.1080/20013078.2019.1663043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Thane K.E. Improved methods for fluorescent labeling and detection of single extracellular vesicles using nanoparticle tracking analysis. Sci. Rep. 2019;9:12295. doi: 10.1038/s41598-019-48181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Palmieri V. Dynamic light scattering for the characterization and counting of extracellular vesicles: a powerful noninvasive tool. J. Nanopart. Res. 2014;16:2583. [Google Scholar]
- 219.Lane R.E. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci. Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]