Abstract
Nucleic acids have been utilized to construct an expansive collection of nanoarchitectures varying in design, physicochemical properties, cellular processing and biomedical applications. However, the broader therapeutic adaptation of nucleic acid nanoassemblies in general, and RNA-based nanoparticles in particular, have faced several challenges in moving towards (pre)clinical settings. For one, the large-batch synthesis of nucleic acids is still under development, with multi-stranded and chemically modified assemblies requiring greater production capacity while maintaining consistent medical-grade outputs. Furthermore, the unknown immunostimulation by these nanomaterials poses additional challenges, necessary to be overcome for optimizing future development of clinically approved RNA nanoparticles.
Keywords: : immune stimulation, nanoparticles, NANPs, RNA nanoparticles, RNA nanotechnology, self-assembly, TNAs
RNA, an original carrier and regulator of the flow of genetic information from nucleic to amino acids, is now realized as a programmable, self-assembling and biodegradable polymer capable of encoding functional frameworks and nanodevices [1,2]. The inherent programmability of RNA (and DNA) into rationally designed multi-stranded nanoscaffolds results from canonical and noncanonical base pairings that allow for the rational design of nucleic acid nanoparticles (NANPs) of various topologies, compositions and functions [3–8]. Each NANP can be enabled with a variety of therapeutic, biosensing and targeting capabilities and as RNA and DNA oligonucleotides naturally hybridize with one another, the hybrid RNA/DNA NANPs diversify the functional range of resulting assemblies [9–12].
Through the tunable and biocompatible nature of NANPs, these nanoformulations show promise in their potential for clinical adaptation. Two RNAi-based nanoformulations, Onpattro® (patisiran) and Givlaari™ (givosiran), were recently approved by the US FDA, with 17 other RNAi therapies entering clinical trials [13] and many more in development [14,15]. Besides specific RNAi inducers, there are several other classes of clinically approved therapeutic nucleic acids [13], including DNA-based therapeutics (six therapies approved), antisense oligonucleotides (with five therapies approved), mRNAs (with five therapies in trials), aptamers (with one approved and seven in trials), ribozymes (four in trials) and gene editing therapeutics (three in trials). These therapies have just begun to emerge as clinical contenders after facing multiple challenges related to their potent immune activation, stability and large-scale production. Multifunctional therapeutic NANPs would consequently benefit from optimizing these aspects early in the design phase of production, so that the advanced preclinical studies do not encounter these challenges when the synthesis and physicochemical characterization of NANPs have been finalized. Addressing the numerous challenges faced in the production of RNA nanostructures will ultimately lead to a successive pipeline for the clinical translation of this innovative technology.
Scaling up NANP production
The exploration of multifunctional NANPs shows promising results with potential applications in therapy or biotechnology [16]. However, the translation of NANP nanotechnologies to clinic or industry is currently challenged with several hurdles, not least of which is the cost-efficient industrial-scale synthesis of pyrogen-free NANPs. The most current types of NANPs are typically assembled from more than one strand that can be synthesized by various methods, loosely categorized into synthetic/chemical, enzymatic (produced by in vitro transcription [IVT]), and recombinant (produced in cells) approaches (Table 1) [17]. Fundamental differences in the biological roles between DNA and RNA reflect their biogenesis, stability and metabolic turnover. While DNA, a guardian of genetic code, evolved to be fairly stable and efficiently replicated, RNA plays diverse roles with significantly shorter half-lives and, with the exception of some viruses, RNAs are not amenable to cellular replication. These biochemical discrepancies, therefore, generally determine the overall complexity of biotechniques suitable for DNA and RNA production.
Table 1. . Overview of NANP production methods.
| Synthesis | Characteristics | Multistrand NANP assembly† | Adaptability for NANP production |
|---|---|---|---|
| Chemical | - Phosphoramidite chemistry with solid-phase supports | No | The synthesis of pure short RNA or DNA oligonucleotides ready to use for NANP assembly is especially useful. This is an easy and versatile way to attach various chemical modifications (fluorophores, linkers, etc.) or produce DNA oligonucleotides for further enzymatic synthesis or cloning for biological applications |
| Enzymatic | - PCR - PCT - IVT |
NA NA Yes |
- PCR is predominantly used for amplification of DNA templates/parts for IVT or cloning, respectively. - PCT has not been yet evaluated for NANP production, but is potentially promising. - IVT is the current golden standard for production of individual RNA strands for NANP assemblies |
| Biological | - Phagemid amplification of ssDNA - Extracellular RNA production - Intracellular RNA production - Vesicular/viral RNA production |
Yes Yes Yes Yes |
- Amplification of various ssDNAs in phagemid particles can be done, as one strand is very cost effective with the currently largest yield. No similar variant is available for RNA. - Intracellular overexpression of DNA/RNA strands from recombinant vectors may provide a means of production of RNA in the cell, allowing for the transport of RNA to media or its export within a vesicle or virus. Co-transcription of multiple strands can lead to in vivo assembly of NANPs |
Possibility of multistrand NANP assembly during one pot production.
IVT: In vitro transcription; NANP: Nucleic acid nanoparticle; PCR: Polymerase chain reaction; PCT: Polymerase chain transcription; ssDNA: Single-stranded DNA.
Chemical synthesis
Currently, the golden approach for the chemical synthesis of nucleic acids and their chemical analogues is the automated method taking advantage of phosphoramidite chemistry with solid-phase supports. Nucleoside phosphoramidites enable the 3′ to 5′ sequential extension of nucleic acid chains (opposite to enzymatic synthesis) via a cyclic reaction and currently support synthesis of DNA oligos of up to 200 bases long. Synthesis of RNA, however, requires additional protection for the ribose 2′-OH group maintained throughout the whole process and the loss of the protecting groups leads to cleavage of the RNA phosphodiester chain and the formation of a 5′-2′ linkage, a problem that increases with oligonucleotide length [18]. Despite the existence of various protecting groups, chemical synthesis of RNAs results in sequences well below 100 bases in length. While longer RNAs can be ligated from shorter RNA strands with the aid of DNA splints complementary to both ligated ends [19,20], the additional enzymatic step and purification lower the final yields of RNA products. All these issues make the straightforward chemical synthesis of oligos expensive and time consuming for mass production of NANPs. As for cost, commercial synthesis for the lowest (0.025–0.05 μmole) scale of production, with basic desalting for purification for a 45 nt-long DNA oligonucleotide costing between €10 and 20, while the price for the synthesis of an RNA oligonucleotide of the same amount and quality would skyrocket to at least €300.
Enzymatic in vitro production
As exemplified by polymerase chain reaction (PCR), an enzymatic approach has the potential for fast and exponential amplification of target DNA sequences. The PCR amplification of a DNA template is mediated by thermostable DNA polymerases through thermal cycling reactions. Analogical RNA amplification via PCR is currently impossible due to the lack of reliable heat-resistant RNA polymerases (RNAP). Potentially, DNA polymerases can be engineered to use ribonucleoside triphosphate (rNTPs) as a substrate to synthesize RNA while retaining their thermostability [21] but the chemical instability of RNAs at elevated temperatures will remain an issue.
Instead, the standard enzymatic synthesis of RNA is IVT mediated by RNAPs isolated from T7, T3 or SP6 bacteriophages. In comparison with cellular RNAPs, the advantage of bacteriophage-derived RNAPs is their single polypeptide chain composition, allowing for their simple recombinant production. T7 RNAP transcribes RNA complementary to a user-designed DNA template [22]. Despite the potential to yield mg/ml quantities within a few hours, IVT often results in 3′-end heterogeneity of the RNA strand, which imposes a potential risk for incomplete NANP assembly and off-target effects. Recently, the combination of two improvements, C2′-methoxy-modified DNA templates in the last two positions at the 5′-end and the addition of dimethyl sulfoxide, resulted in increased 3′-end homogeneity of transcripts [23–25]. Another approach promoting homogeneity of 5′- and 3′- ends of RNAs is through cleavage of the phosphodiester backbone at a specific site by cis-acting self-cleaving ribozymes fused to both ends of the transcript [26]. Overall, IVT is a very promising approach since it allows for the one-pot cotranscriptional assembly of individual NANPs made of RNA and chemically modified analogs [27–30].
Biological-intracellular overexpression
While the enzymatic production of NANPs (or individual oligos in their composition) offers synthesis under defined reaction conditions, large-scale NANP preparations would require a high concentration of processing enzymes, making the operation too costly. In comparison with IVT, intracellular synthesis of recombinant nucleic acids represents a price-effective approach. Recombinant expression of nucleic acids in microbial cells in a manner similar to recombinant proteins is a viable strategy but requires several considerations. For one, DNA expressed in cells is in a duplex form and the subsequent production of ssDNA strands would require additional intracellular or postisolation treatment which may be ineffective for mass production.
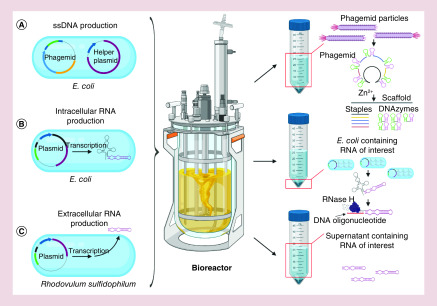
This challenge is essential to overcome for the production of DNA origami, which has been shown to conditionally interact in cell cultures as well as in animal models [31]. DNA origami is assembled from a long ssDNA shaped by short staple oligos. Recently, it has been shown that long and short components can be encoded on one phagemid. A phagemid is a combination of bacteriophage and plasmid serving as a cloning vector with the ability to be packaged into a filamentous bacteriophage. This approach offers a very high rate of ssDNA amplification. The proposed strategy exploits the bacteriophage to carry ssDNA-containing staple sequences interleaved with two Zn2+-dependent DNA-cleaving DNA enzymes and a scaffold sequence as a backbone (Figure 1A). The released phagemid particles are separated from the bacterial cells and ssDNA is isolated. The staple sequences and scaffold strand are excised from the construct after induction by ZnCl2. Subsequently, the blend of staples and scaffolds undergoes self-assembly in large volume. Finally, assembled DNA nanoparticles are precipitated. From 2 l of reaction volume, authors gained approximately 160 mg of NANPs including residual chemicals. The total cost of synthesis was calculated to be approximately €23/mg (2 l) with an estimation of €0.18/mg for 800 l, which would be 1000-times less than the comparative amount synthesized by solid-phase chemistry [32]. Additional works have developed even more customizable phagemids [33,34].
Figure 1. . Three strategies for large-scale preparation of nucleic acids composing nucleic acid nanoparticles.
(A) Production of all ssDNA components for DNA origami structures. Currently the most effective method for the manufacturing of single-stranded nucleic acids relies on the production of engineered bacteriophages. Individual origami components (staples and scaffold) are excised from phagemid after induction of DNAzymes by Zn2+. (B) Intracellular transcription of user-designed RNAs. To prevent degradation, the RNA of interest is expressed in fusion with protecting scaffold. The RNA is isolated from total cellular RNA and subsequently the scaffold is enzymatically removed. Alternatively, RNA can be transcribed in fusion with inducible glmS ribozyme (not shown). The ribozyme is designed to display the boxB RNA, the high-affinity tag to the N-peptide derived from bacteriophage λ. The λN peptide is fused to glutathione S-transferase in Glutathione–Sepharose. Subsequently, the captured chimeric ribozyme is activated by addition of glucosamine-6-phosphate and RNA of interest is cleaved off and eluted [35]. (C) The microbes that export RNA to its environment with undetectable levels of RNases can be used for the production of recombinant RNAs without the support of RNA scaffolds.
Intracellularly, RNA is mostly transcribed as single-stranded, suggesting that expression of heterologous RNA is straightforward: a vector encoding the RNA construct is transferred into host cells where it is maintained and passed to daughter cells while transcribing chosen RNAs. At some point, cells are pelleted, lysed and RNAs of interest are pulled out using affinity tags [36]. The major drawbacks of recombinant RNA synthesis in organisms are quick degradation by intracellular RNases and heterogeneity of the transcripts. Fusion of the RNA of interest to an RNA scaffold such as transfer RNA or 5S ribosomal RNA is a standard strategy aiming to protect RNA constructs against bacterial endogenous ribonucleases. If necessary, upon purification, the scaffold structures can be cleaved away with the help of RNase H and DNA oligonucleotides complementary to the scaffold sequences flanking the RNA of interest, which may be a limiting step for large-scale production [37]. Interestingly, in mammalian cells, RNAs fused to transfer RNA are unstable and require a different scaffold [38].
The heterogeneity of RNA strands can be solved by incorporating ribozymes into the transcripts. Recently, a new class of so-called twister ribozymes was discovered [39]. Their self-cleavage under physiological conditions is significantly faster than standard ribozymes, which prevents degradation before the creation of homogenous ends. Based on twister ribozymes, a new system named Tornado (Twister-Optimized RNA for Durable Overexpression) was recently introduced. The Tornado system is characterized by the rapid self-cleavage of Twister ribozymes located on both sides of the RNA of interest. The exposed sequences close to the 5′ and 3′ ends form stems with several loose bases on both termini that are subsequently ligated by the endogenous RNA ligase RtcB. The final structure is a highly stable circularized RNA with the potential to accommodate several functionalities such as RNA-based fluorescent structures, metabolite RNA sensors or RNA aptamers [40].
In vivo-transcribed RNA NANPs that are assembled from more than one strand can perform various functions [41,42]. Current works have studied NANPs designed to be functional in situ and it is unknown if individual strands of multistrand assemblies can be effectively isolated in a stoichiometric ratio. Therefore, the composition of NANPs from several strands is a significant disadvantage for their cellular production. While isolated NANPs adopt their final structures after thermal annealing, this process obviously cannot be realized in vivo. However, it may be favorable to have RNA NANPs that readily fold also in vivo into the designed shape. To comply with this, it is necessary to design nanostructures that exhibit high thermodynamic stability with favorable folding kinetics. The potential solutions for these problems were experimentally proven by modeling various single-component RNA NANPS that folded in a sequential and hierarchical pathway to designed nanostructures. As a first, during their transcription in Escherichia coli, arising RNA strands form stable hairpins where a prevailing number of RNA bases are engaged in forming duplex structures (Figure 1B). The final designed nanostructure is then promoted by unpaired RNA residues that form long-range tertiary interactions. The amount of in vivo-transcribed NANPs was estimated to be up to 11% from total RNA, implying the potential for large-scale production [43].
In comparison with the intracellular accumulation of RNAs, the extracellular release of RNA to cell culture is an alternative approach with the potential for continuous industrial manufacturing systems without the necessity to collect producing cells. Isolation of RNA from the cell culture medium would not require exhausting time and laborious RNA extraction methods. The inevitable condition for extracellular production of RNAs is the absence of RNases in the growth medium. This crucial prerequisite together with cells' natural ability to secrete RNAs in the medium was observed in the marine bacterium Rhodovulum sulfidophilum (Figure 1C). RNase-free medium additionally allows for expression of linear RNA structures without any camouflaging scaffold or circularization. However, despite optimization of the promoter sequence and growth conditions, the yields for RNAs in trials have been reported below 200 μg/l [44–46]. Nevertheless, future advances in synthetic biology could successfully realize feasible methodology of large-scale production of extracellular RNA.
The comparison of cost–effectiveness between enzymatic and biological production of NANPs or their constituent strands is highly complicated with many variables. Considering the following simplest scenario, by using, for example, AmpliScribe™ T7-Flash™ Transcription Kit or HiScribe™ T7 IVT Kit (both 50 rxns for €250–300), the cost for synthesis of a 400 nt RNA oligonucleotide would be up to €6 with a yield of approximately 160 μg of RNA per one reaction (based on the advertised graphs). On the other side, based on the comparison of band densities, Li and colleagues determined that one of their NANPs (400 nts long) accounted for 11% of the total RNA isolated from E. coli [43]. With approximation that one E. coli contains 0.1 pg of total RNA and there are approximately 8.108.ml-1. Optical density (OD-1) cells, we can extrapolate that 2 l of cultures would yield approximately 160 mg of total RNA with 11% of NANP share, making approximately 16 mg. To synthesize the same concentration by IVT, we would need 100 reactions (two kits for €500–600). In contrast to that, for 2 l of bacterial culture, we need 40 g of lysogeny broth (LB) medium (1 kg/€130) and 0.2 g ampicillin (5 g/€60) together for €8.
Safety considerations in immune stimulation
In addition to scaling up NANP production, a foremost consideration in their optimization toward clinical use is safety regarding formulations of NANPs. The production of NANPs requires care to ensure that the resulting assemblies are not contaminated in a way that can become harmful to either the samples or the subject, which is of critical importance to their ability to perform therapeutic roles. Bacterial endotoxins are notorious contaminants of nanoformulations and contributors to toxic immune responses, emphasizing the need for screening prior to application [47]. However, while straightforward measures can be taken to minimize the chances of contamination in small-scale preparations, the potential for endotoxin exposure in large-scale NANP production is considerably greater, especially if bacterial systems themselves or recombinant enzymes are the means of NANP preparation [48].
Besides contamination of the NANP formulation by bacterial components, the second main consideration in the safety of therapeutic NANPs is their direct interaction with the host’s immune system. Though advantageous due to their biodegradability, there are also multiple preexisting pathways in mammalian cells which have evolved to specifically recognize foreign nucleic acids as a defense mechanism against invading pathogens. Pattern recognition receptors are employed by cells for policing motifs which are known to indicate the presence of pathogens in the cellular environment, such as unmethylated CpG DNAs and viral dsRNAs, to name a few [49]. Due to NANPs’ composition, the immune stimulation brought about by their introduction has been shown to employ these same routes of recognition [50]. Endosomal (i.e., Toll-like receptors 3, 7, 8 and 9) and cytoplasmic receptors (i.e., RIG-I-like, MDA5, cGAS, IFI16, etc.) play key roles in the pattern recognition of exogeneous NANPs. However, the interactions each NANP has with these key immune receptors have in turn been shown to largely depend on the physicochemical properties of the NANP, such as composition (DNA and/or RNA), topology (varying between globular, planar and fibrous structures) and functionalization with therapeutic nucleic acids [4,5,50,51]. Thus, different routes of sensing and innate immune response activation are triggered based on the design of the NANP introduced. These new trends in the structure–activity relationship of NANPs, explored conventionally and with the aid of machine learning approaches [5], have not only elucidated a hurdle which has kept previous formulations at bay from advancing into the clinical setting, but are now expected to serve as favorable mechanisms for designing NANPs equipped to modulate immune responses [52,53].
Another descriptor of how NANPs are interpreted by the immune system comes with the vehicle used for their delivery. As a result of their negatively charged phosphate backbone, nucleic acids or NANPs by themselves are unable to traffic across the cell membrane and therefore, carrier-free NANPs are completely immunoquiescent [51]. While NANPs are not able to interact with any nucleic acid-specific intracellular immune receptors without entering the cell, they are also not able to be processed for intracellular therapeutic function without doing so and therefore can present endless opportunities for extracellular action and imaging. For intracellular function, NANPs require a carrier [54–57], which could include a secondary delivery agent or could be a targeting functionality on the NANP itself, such as an aptamer or small molecule [2,58–61]. In addition to a vast library of nanoscale platforms functioning as nucleic acid delivery agents, viral vectors have also been shown to be efficient carriers, though they impose many additional safety concerns [13]. These variable modes of delivery, which have different routes of trafficking, are likely to result in variable immune pathways despite similarity in NANPs, which adds an additional level of customization to potential immune modulation. Carriers are beneficial because they protect the NANP from serum RNases and DNases as without the carrier, the NANP could be rapidly degraded in serum. The carrier is also a useful means of modulating the physicochemical properties of the NANP, as it is able to be made with specific charges and sizes to compensate for the overwhelming negative charge and direct NANPs’ biodistribution [58]. A variety of biologically based carriers which are lipid-based or extracellular vesicles are a promising avenue for maintaining biocompatibility and offering patient specificity while achieving high payloads. Inorganic carriers, mesoporous silica nanoparticles and dendrimers have also been used for siRNA and NANP delivery (Figure 2) [4,60].
Figure 2. . Introduction of chemical modifications.
Chemical modifications to the backbone, sugar and base as well as fine-tuning the dimensionality and composition of nucleic acid nanoparticles in combination with choice of a particular delivery carrier (i.e., extracellular vesicles, mesoporous silica nanoparticles, inorganic nanoparticles, dendrimers and lipid-based nanoparticles) is a modular approach that allows for transport into the cell for recognition by desired immune receptors and corresponding downstream signaling of an immune response.
EV: Extracellular vesicle; MSN: Mesoporous silica nanoparticles.
Control over the biodistribution and targeting of NANPs is especially beneficial for their applications in vivo, where selective tissue targeting allows for more potent therapies without off-target effects. Many examples of antisense oligonucleotides are currently in clinical trials which exhibit high tissue specificity based on their chemical modifications [62].
Long-term stability
Though nucleic acids are primarily repositories of genetic information, the instability of RNA as a reactive but transient multifunctional molecule is also translated into the instability of RNA-composed NANPs, affecting their long-term activity or storage. The presence of the 2′-OH group leads to RNA’s fragility manifested by spontaneous cleavage of the phosphodiester backbone, mostly in single-stranded flexible regions, through a self-cyclization reaction that is catalyzed by basic solutions and the presence of divalent cations. The catalysis by divalent cations is particularly relevant, as a large number of tertiary structural motifs of RNA tend to be stabilized by these cations, especially Mg2+. Additionally, when intended for clinical applications, a major impediment is that NANPs encounter nucleases within the patient’s bloodstream, leading to a low overall retention time [30].
To overcome the barrier of chemical instability and increase the availability of NANPs, chemical modifications to the bases, phosphate groups and 2′-OH have been explored and new frameworks including peptide nucleic acids, polycarbamate nucleic acids and locked nucleic acids have been demonstrated. Though biological functions such as processing through RNA interference can still be retained, these modifications can greatly alter the folding profile of nucleic acids, which contributes to changes in the assembly of NANPs [16]. Furthermore, chemical modifications are a distinctive challenge in the efforts to upscale production. To enzymatically produce chemically modified RNAs, the yield from a mutant polymerase is generally much lower than its naturally occurring counterpart and the purification of each strand prior to its incorporation into a NANP assembly takes a toll on the total yield of NANPs. To overcome this, the cotranscriptional assembly of NANPs is especially useful. 2′-F-modified RNA rings and cubes have been shown to assemble in a one-pot reaction which offers a greater and more cost-effective yield [30]. There is still much work to be done in incorporating the known library of chemical modifications into NANP assemblies to overcome the issue of stability and potentially alter immune recognition. Additionally, besides chemical stability, the thermodynamic stability of NANPs should be considered in the design process so that smaller concentrations of treatments may retain their integrity when introduced in vivo. The 2′-F modification of 3WJ motif-containing NANPs have demonstrated that the combination of this modification with the incorporated motif enhanced resistance to degradation [63]. Modifications improving biological stability should also positively impact the long-term shelf-life of NANPs, increasing their robustness and affordability for downstream clinical translation.
Future perspective
The last two decades of positive experimental perspectives in the development of NANP technology have also revealed several complications toward their clinical progression. One of these hurdles is the requirement of mass NANP production to provide larger scale volumes of high batch-to-batch consistency. While this is a major challenge for nucleic acid synthesis in general, there is the equally necessary demand for large-scale synthesis of chemically modified nucleic acids as well as multistrand NANP assemblies. As seen in large-scale production of other bioactive compounds, biological systems are the most promising for the manufacturing of large quantities of NANPs. Besides the current approaches relying on the harvesting of producing bacteria, other engineered cellular systems may be developed including mammalian. In addition, produced NANPs have the potential to be exported to media after being enclosed in vesicles or virus-like particles. Such an approach would allow for the continual collection of particles containing NANPs as well as protection against nucleases. More advanced systems can produce targetable vesicle–NANP complexes. Developments to NANP designs themselves could also introduce programmable means for production and isolation. Dynamic RNA structures are intrinsically passive without an induced signal required for activation to promote their functions [64,65]. These functional elements are already used in NANP production, for example, ribozymes in self-scission during rolling-circle transcription or dissection of a single strand into several [27,32]. However, we may expect the employment of more complex combinatorial systems, including logic gates, in NANP synthesis. It is very probable for future directions of NANP management that DNA/RNA processing with intracellular enzymes other than DNA or RNA polymerases will continue to emerge, as has recently been demonstrated [40]. Furthermore, even upon the successful delivery of nanoparticle formulations into the cytoplasm of target cells, intracellular interactions of NANPs with the cellular environment, including cell surveillance systems, remains the final barrier. Recent work in the field has started to elucidate relationships between NANP design and immune responses. We believe that the culmination of our mutual work will lead to downstream NANP formulations with intentional immunomodulatory capabilities.
Footnotes
Financial & competing interests disclosure
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM120487 (to KA Afonin). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All figures were created with Biorender.com. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Jasinski D, Haque F, Binzel DW, Guo P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 11(2), 1142–1164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panigaj M, Johnson MB, Ke W. et al. Aptamers as modular components of therapeutic nucleic acid nanotechnology. ACS Nano 13(11), 12301–12321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ke W, Hong E, Saito RF. et al. RNA-DNA fibers and polygons with controlled immunorecognition activate RNAi, FRET and transcriptional regulation of NF-kappaB in human cells. Nucleic Acids Res. 47(3), 1350–1361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rackley L, Stewart JM, Salotti J. et al. RNA Fibers as optimized nanoscaffolds for siRNA coordination and reduced immunological recognition. Adv. Funct. Mater. 28(48), 1805959 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson MB, Halman JR, Satterwhite E. et al. Programmable nucleic acid based polygons with controlled neuroimmunomodulatory properties for predictive QSAR modeling. Small 13(42), 1701255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Afonin KA, Viard M, Koyfman AY. et al. Multifunctional RNA nanoparticles. Nano Lett. 14(10), 5662–5671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Lee T, Dziubla T. et al. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today 10(5), 631–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parlea L, Bindewald E, Sharan R. et al. Ring catalog: a resource for designing self-assembling RNA nanostructures. Methods 103, 128–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonin KA, Desai R, Viard M. et al. Co-transcriptional production of RNA–DNA hybrids for simultaneous release of multiple split functionalities. Nucleic Acids Res. 42(3), 2085–2097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afonin KA, Viard M, Martins AN. et al. Activation of different split functionalities upon re-association of RNA-DNA hybrids. Nature Nanotechnol. 8(4), 296–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roark B, Tan JA, Ivanina A. et al. Fluorescence blinking as an output signal for biosensing. ACS Sensors 1(11), 1295–1300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dao BN, Viard M, Martins AN, Kasprzak WK, Shapiro BA, Afonin KA. Triggering RNAi with multifunctional RNA nanoparticles and their delivery. DNA RNA Nanotechnol. 1(1), 27–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weng Y, Huang Q, Li C. et al. Improved nucleic acid therapy with advanced nanoscale biotechnology. Mol. Ther. Nucleic Acids 19, 581–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams D, Gonzalez-Duarte A, O'Riordan WD. et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379(1), 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Chan A, Liebow A, Yasuda M. et al. Preclinical development of a subcutaneous ALAS1 RNAi therapeutic for treatment of hepatic porphyrias using circulating RNA quantification. Mol. Ther. Nucleic Acids 4, e263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu Y, Pi F, Sharma A. et al. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 66, 74–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baronti L, Karlsson H, Marusic M, Petzold K. A guide to large-scale RNA sample preparation. Anal. Bioanal. Chem. 410(14), 3239–3252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caruthers MH. A brief review of DNA and RNA chemical synthesis. Biochem. Soc. Trans. 39(2), 575–580 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Kershaw CJ, O'Keefe RT. Splint ligation of RNA with T4 DNA ligase. Methods Mol. Biol. 941, 257–269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA 12(11), 2014–2019 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cozens C, Pinheiro VB, Vaisman A, Woodgate R, Holliger P. A short adaptive path from DNA to RNA polymerases. Proc. Natl Acad. Sci. USA 109(21), 8067–8072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckert B, Masquida B. Synthesis of RNA by in vitro transcription. Methods Mol. Biol. 703, 29–41 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5(9), 1268–1272 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Zhang Y. Dimethyl sulfoxide targets phage RNA polymerases to promote transcription. Biochem. Biophys. Res. Commun. 333(3), 664–670 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Helmling C, Keyhani S, Sochor F, Furtig B, Hengesbach M, Schwalbe H. Rapid NMR screening of RNA secondary structure and binding. J. Biomol. NMR 63(1), 67–76 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Avis JM, Conn GL, Walker SC. Cis-acting ribozymes for the production of RNA in vitro transcripts with defined 5′ and 3′ ends. Methods Mol. Biol. 941, 83–98 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Jasinski DL, Binzel DW, Guo P. One-pot production of RNA nanoparticles via automated processing and self-assembly. ACS Nano 13(4), 4603–4612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'hara J, Marashi D, Morton S. et al. Optimization of the split-spinach aptamer for monitoring nanoparticle assembly involving multiple contiguous RNAs. Nanomaterials 9(3), 378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kireeva ML, Afonin KA, Shapiro BA, Kashlev M. Cotranscriptional production of chemically modified RNA nanoparticles. Methods Mol. Biol. 1632, 91–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, Shapiro BA. Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett. 12(10), 5192–5195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335(6070), 831–834 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, Dietz H. Biotechnological mass production of DNA origami. Nature 552(7683), 84–87 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Nafisi PM, Aksel T, Douglas SM. Construction of a novel phagemid to produce custom DNA origami scaffolds. Synth. Biol. (Oxf.) 3(1), ysy015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engelhardt FaS, Praetorius F, Wachauf CH. et al. Custom-Size, functional and durable DNA origami with design-specific scaffolds. ACS Nano 13(5), 5015–5027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Tomasso G, Lampron P, Dagenais P, Omichinski JG, Legault P. The ARiBo tag: a reliable tool for affinity purification of RNAs under native conditions. Nucleic Acids Res. 39(3), e18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponchon L, Dardel F. Large scale expression and purification of recombinant RNA in Escherichia coli. Methods 54(2), 267–273 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F. A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat. Protoc. 4(6), 947–959 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Filonov GS, Kam CW, Song W, Jaffrey SR. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem. Biol. 22(5), 649–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 10(1), 56–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litke JL, Jaffrey SR. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37(6), 667–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333(6041), 470–474 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Alam KK, Tawiah KD, Lichte MF, Porciani D, Burke DH. A fluorescent split aptamer for visualizing RNA-RNA assembly in vivo. ACS Synth. Biol. 6(9), 1710–1721 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Zheng M, Wu S. et al. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat. Commun. 9(1), 2196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki H, Ando T, Umekage S, Tanaka T, Kikuchi Y. Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl. Environ. Microbiol. 76(3), 786–793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki H, Umekage S, Tanaka T, Kikuchi Y. Artificial RNA aptamer production by the marine bacterium Rhodovulum sulfidophilum: improvement of the aptamer yield using a mutated transcriptional promoter. J. Biosci. Bioeng. 112(5), 458–461 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Pereira P, Pedro AQ, Tomas J. et al. Advances in time course extracellular production of human pre-miR-29b from Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 100(8), 3723–3734 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Neun BW, Dobrovolskaia MA. Detection of endotoxin in nano-formulations using limulus amoebocyte lysate (LAL) assays. J. Vis. Exp. (143), (2019) (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 48.Dobrovolskaia MA. Nucleic acid nanoparticles at a crossroads of vaccines and immunotherapies. 24(24), 4620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marques JT, Williams BRG. Activation of the mammalian immune system by siRNAs. Nature Biotechnol. 23(11), 1399–1405 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Hong E, Halman JR, Shah A. et al. Toll-like receptor-mediated recognition of nucleic acid nanoparticles (NANPs) in human primary blood cells. Molecules 24(6), 1094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong E, Halman JR, Shah AB, Khisamutdinov EF, Dobrovolskaia MA, Afonin KA. Structure and composition define immunorecognition of nucleic acid nanoparticles. Nano Lett. 18(7), 4309–4321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandler M, Johnson MB, Panigaj M, Afonin KA. Innate immune responses triggered by nucleic acids inspire the design of immunomodulatory nucleic acid nanoparticles (NANPs). Curr. Opin. Biotechnol. 63, 8–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandler M, Afonin KA. Smart-responsive nucleic acid nanoparticles (NANPs) with the potential to modulate immune behavior. Nanomaterials (Basel) 9(4), 611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parlea L, Puri A, Kasprzak W. et al. Cellular delivery of RNA nanoparticles. ACS Comb. Sci. 18(9), 527–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim T, Afonin KA, Viard M. et al. In silico, in vitro and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol. Ther. Nucleic Acids 2, e80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta K, Mattingly SJ, Knipp RJ. et al. Oxime ether lipids containing hydroxylated head groups are more superior siRNA delivery agents than their nonhydroxylated counterparts. Nanomedicine (Lond.) 10(18), 2805–2818 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta K, Afonin KA, Viard M. et al. Bolaamphiphiles as carriers for siRNA delivery: from chemical syntheses to practical applications. J. Control. Rel. 213, 142–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Halman JR, Kim K-T, Gwak S-J. et al. A cationic amphiphilic co-polymer as a carrier of nucleic acid nanoparticles (NANPs) for controlled gene silencing, immunostimulation and biodistribution. Nanomedicine 23, 102094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobrovolskaia MA, Mcneil SE. Strategy for selecting nanotechnology carriers to overcome immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics. Expert Opin. Drug Deliv. 12(7), 1163–1175 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Juneja R, Lyles Z, Vadarevu H, Afonin KA, Vivero-Escoto JL. Multimodal polysilsesquioxane nanoparticles for combinatorial therapy and gene delivery in triple-negative breast cancer. ACS Appl. Mater. Interfaces 11(13), 12308–12320 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Cruz-Acuña M, Halman JR, Afonin KA, Dobson J, Rinaldi C. Magnetic nanoparticles loaded with functional RNA nanoparticles. Nanoscale 10(37), 17761–17770 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Seth PP, Tanowitz M, Bennett CF. Selective tissue targeting of synthetic nucleic acid drugs. J. Clin. Invest. 129(3), 915–925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 6(10), 658–667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bindewald E, Afonin KA, Viard M, Zakrevsky P, Kim T, Shapiro BA. Multistrand structure prediciton of nucleic acid assemblies and design of RNA switches. Nano Lett. 16(3), 1726–1735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zakrevsky P, Parlea L, Viard M, Bindewald E, Afonin KA, Shapiro BA. Preparation of conditional RNA switch. Methods Mol. Biol. 1632, 303–324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]