Abstract
Over many years, extensive efforts have focused on the development and improvement of diagnostic and therapeutic strategies to reduce stroke-associated neurovascular damage, such as blood-brain-barrier dysfunction, brain edema, parenchymal inflammation, and neural cell death. However, the only clinically-applied pharmacological therapy to date for the treatment of acute ischemic stroke is thrombolysis. Due to the short therapeutic window of current thrombolytic therapy and the activation of various pathophysiological signaling cascades triggered after ischemic stroke, the development of new therapies is urgently required. Non-coding RNAs (ncRNAs) are defined as untranslated regulatory RNA molecules. Although ncRNAs with biological roles have been known for almost 60 years, they have within the past decade emerged as key mediators of posttranscriptional gene expression/function in pathological aspects of ischemic stroke. With properties of relative stability, specificity and reproducibility, ncRNAs are considered to be promising as bio-markers and better candidates than proteins and genes for early recognition of the onset of disease. In this update, we summarized the current knowledge for three groups of ncRNAs in stroke, focusing on the role of long non-coding RNAs and circular RNAs as biomarkers for stroke and as targets for regulating large sets of genes in related pathways after ischemic stroke.
Non-coding RNAs in Stroke Study
Stroke is a life-threatening disease and a major cause of long-term disability in both developing and developed countries. Ischemic stroke results in severe reduction of cerebral blood flow with a subsequent lack of oxygen and nutrients in affected brain tissue that could ultimately lead to neuronal cell death and brain infarction. The development of effective therapies in the clinical setting is limited by several reasons, including the rapid development of brain injury following ischemia, complex interplays among signaling pathways, and the treatment window for specific targets [1]. In humans, DNA sequences responsible for non-protein coding regions comprise at least 98% of the total genome. Non-coding RNAs can be roughly divided into two groups: small ncRNAs (< 200 bp, including rRNA, microRNA, snRNA, snoRNA, siRNA and piRNA) and long ncRNAs (lncRNAs) with > 200 bp in length [2]. Noncoding transcripts were once regarded as evolutionary junk or transcriptional noise. However, increasing evidence indicates that ncRNAs are important regulators of gene expression [3]. Among them, microRNAs (miRNAs) have been the most extensively studied for their functions in developmental and tissue-specific expression patterns, and also human diseases [2].
MicroRNAs and Stroke
MiRNAs are ubiquitous, endogenous, non-coding single-stranded RNA transcripts that are normally 19–25 nucleotides in length and act as post-transcriptional regulators of gene expression by binding to the 3’ untranslated regions of target messenger RNAs (mRNAs). In mammals, more than 50% of mRNAs are predicted to be the subject of miRNA-mediated control, and alterations in miRNA expression profiles have been observed in numerous pathological processes. It is now evident that miRNAs regulate expression of at least one-third of the human genome and play a critical role in a variety of normal biological processes, including cell differentiation, apoptosis, development and metabolism [4]. This gene regulatory function of miRNAs makes them intriguing candidates for disease biomarkers, since changes in their expression patterns are detected even before phenotypic projection of disease onset. The first demonstration that miRNAs could be useful as diagnostic biomarkers came from the 2002 publication by Calin et al. [5], who made the connection between the frequently deleted 13q14 locus, and the downregulation of the miR-15a/16 cluster encoded within this region in chronic lymphocytic leukemia patients [5]. Specific miRNA expression has also been shown in both brain tissue and blood following ischemic stroke [6]. Besides, circulating miRNA expression varies significantly in stroke patients as well as different stroke subtypes [7]. Consequently, there is much current interest in miRNAs both as novel biomarkers and as potential targets for therapeutic intervention of stroke. Recently, several research articles were published that focused on the role of miRNAs as biomarkers for stroke and as targets for regulating large sets of genes in related pathways of the stroke cascade. Current miRNA-based therapeutic applications in stroke, the underlying mechanisms of miRNA-based therapeutics, and pharmacological agents that protect against stroke injuries by targeting specific miRNAs are well summarized in recent review articles and a book chapter by Khoshnam et al. [8], Mirzaei et al. [7], Chandran et al. [9]. This update will focus more on the functional significance and mechanisms of two less studied groups of ncRNAs in cerebrovascular pathophysiology after stroke.
LncRNAs and Stroke
LncRNAs are loosely classified and the largest class of longer (200 nt) ncRNAs. They are diverse and numerous; to date, over 50,000 human lncRNAs have been identified [10]. This number keeps increasing with advances in more sensitive RNA sequencing, and improved epigenomic and computational prediction techniques. Recently, a number of studies focused on lncRNAs for their crucial roles in molecular function, including RNA processing, transcriptional and post-transcriptional modulation of gene expression, nuclear organization and serving as precursors to small RNAs [11]. The complex secondary and higher order structures of lncRNAs make them quite versatile for protein and target recognition. It is now evident that lncRNAs are involved in many crucial biological processes, such as cell survival, proliferation, differentiation, chromatin remodeling, and organogenesis [11].
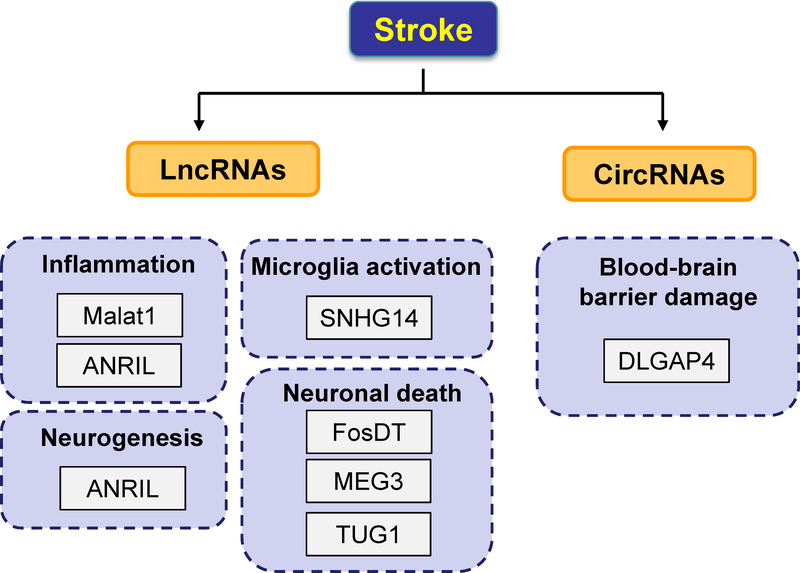
Accumulating evidence has shown that lncRNAs play a critical role in the pathogenesis of ischemic stroke. LncRNA profiles have been reported in rodent focal cerebral ischemia models [12, 13] as well as in cultured mouse brain microvascular endothelial cells (BMECs) following an in vitro mimic of ischemic stroke conditions [14]. Altered lncRNA levels were also found in blood samples of rodent stroke models [15] and stroke patients [15, 16] and can serve as potential biomarkers. In the study by Dykstra-Aliello et al. [16], lncRNA expression was assessed in 266 whole blood RNA samples drawn from ischemic stroke patients and healthy controls and matched with vascular risk factor controls. A total of 299 lncRNAs were found differentially expressed between control and stroke males, whereas 97 lncRNAs were found differentially expressed between control and stroke females [16]. In addition, there were some differentially expressed lncRNAs mapped close to genomic locations of previously identified putative stroke-risk genes [16]. We and others are among the first to identify the functional significance of individual lncRNAs in ischemic stroke. A recent study by Mehta et al. showed that lncRNA FosDT promotes ischemic brain injury by interacting with REST-associated chromatin modifying proteins [17], suggesting lncRNAs can be therapeutically targeted to minimize post-stroke brain damage. Notably, we have defined that lncRNA Malat1 was involved in the protection of cerebral microvessels after ischemic insult [14]. Malat1 levels increased in cultured brain microvascular endothelial cells (BMECs) following oxygen-glucose deprivation (OGD) and in cerebral microvessels in mice subjected to transient middle cerebral artery occlusion (MCAO) [18]. Furthermore, silencing of Malat1 by locked nucleic acid (LNA)-GapmeR (single-stranded oligonucleotides designed to specifically silencing Malat1) significantly increased OGD-induced endothelial damage through upregulating the levels of pro-apoptotic factor Bim and proinflammatory cytokines MCP-1, IL-6 and E-selectin [18]. Other lncRNAs including SNHG14 [19], TUG1 [20], MEG3 [21], ANRIL [22, 23] were also reported to mediate ischemia-induced neuronal death, neurogenesis and neurological recovery (summarized in Figure 1).
Figure 1. Contributory roles of lncRNAs and circRNAs in the pathogenesis of stroke.
Dysregulated lncRNAs and circRNAs contribute to the pathophysiology of ischemic stroke by mediating common pathophysiological mechanisms including inflammation, blood-brain barrier damage, microglial activation, and neuronal death as well as neuroprotective mechanisms such as neurogenesis.
Circular RNAs and Stroke
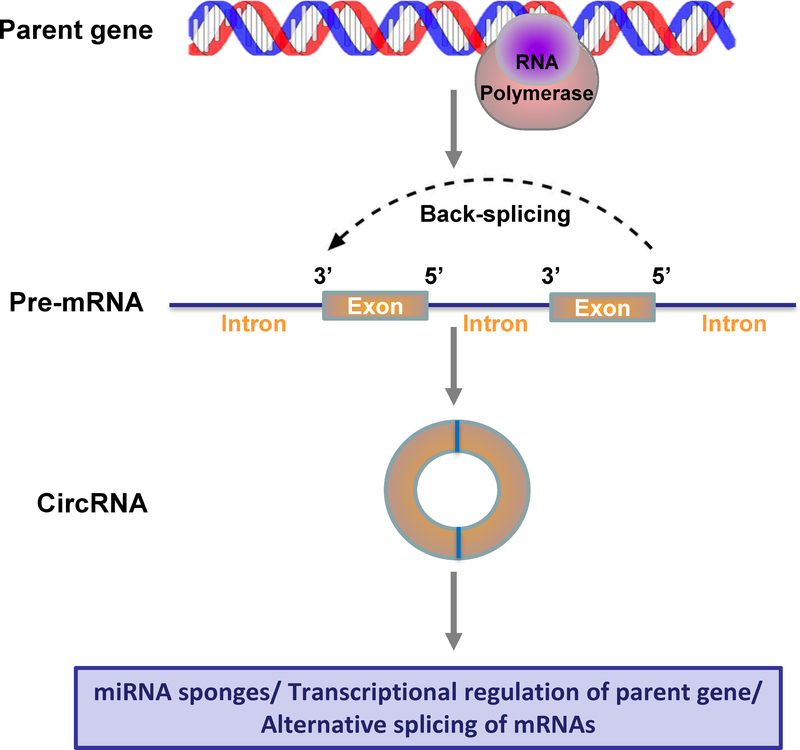
Another subgroup of ncRNAs form circular structures when the 5’ and 3’ ends of a single precursor messenger RNA molecule are joined together by covalent bonds (Figure 2). They were therefore named circular RNAs (circRNAs). Due to the absence of the defined 5’ caps and 3’ poly-A tails, circRNAs are resistant to Ribonuclease R (RNase R) treatment (which degrades essentially all linear forms of RNAs), thus making circRNAs extremely stable with a half-life exceeding 48 hours [24]. Unlike lncRNAs, circRNAs are evolutionary conserved in mammalian and human cells [24]. These features confer numerous physiological functions to circRNAs, including binding to miRNAs as competing endogenous RNAs, transcriptional regulation of parent genes, promoting rolling circle translation, and alternative splicing of mRNAs [25]. CircRNAs have also been reported to play important roles in pathophysiological processes, including heat senescence, Alzheimer’s disease, cardiac hypertrophy and failure [26]. However, little is known about their function in brain injury and repair after stroke.
Figure 2. Circular RNA biogenesis and functional mechanisms.
CircRNAs are produced from precursor mRNA (pre-mRNA) by back-splicing circularization in higher eukaryotes. Different from the canonical pre-mRNA splicing process that is catalyzed by the spliceosomal machinery to remove introns and join exons leading to the formation of linear RNA transcript with 5’−3’ polarity, most circRNAs are formed during back-splicing in which a downstream 5’ donor site of an exon is joined to an upstream 3’ acceptor site by covalent bonds. The ligated 5’ and 3’ ends of circRNAs make them resistant to exoribonuclease degradation, and therefore are more stable than linear mRNAs.
Rybak-Wolf et al. analyzed circRNA expression in human and mouse brains and found that circRNAs are highly abundant in the brains compared to other tissues and were generally upregulated during neuronal differentiation and development [27]. This study suggested that circRNAs may participate in many aspects of brain function and pathologies of neurological disorders. In a later clinical study, Bazan et al. reported that in patients presenting with an acute carotid-related ischemic stroke event, the serum ratio of circulating circR-284 to miR-221 is elevated, indicating the potential for circRNAs to serve as diagnostic biomarkers for carotid-related cerebrovascular ischemia [28]. Raghu’s group profiled the expression of 14,236 circRNAs in the mouse brain after transient MCAO followed by 6, 12, and 24 hours reperfusion and found that 283 were altered more than 2-fold compared with the sham group [29]. They also performed a series of bioinformatics analysis and identified that some of the altered circRNAs contain microRNA binding sites. Further, the major biological and molecular functions controlled by these circRNAs, including metabolic processing, cell communication, biological regulation, and binding to proteins, ions, and nucleic acids were altered after MCAO [29]. For the first time, this study showed that circRNAs are sensitive to cerebral ischemia, and their altered function might promote the post-stroke pathophysiology. In agreement with Raghu’s study, Bai et al. reported that circDLGAP4 functions as an endogenous microRNA-143 sponge to inhibit miR-143 activity, resulting in increased expression of the miR-143 target HECTD1 [30]. They also found that the plasma levels of circDLGAP4 were significantly decreased in acute ischemic stroke patients and in a mouse stroke model [30]. These two recent studies both suggest the potential of circRNAs as targets for therapeutic interventions in stroke and as biomarkers for disease activity.
Challenges and Perspectives
Imaging technologies such as magnetic resonance imaging (MRI) remain the major diagnosis approach for patients suffering stroke. Unlike in myocardial infarction, specific plasma/serum markers that may be used to diagnose and/or assess the severity of ischemic brain injury have not yet been established. Although it has been reported that several protein markers are elevated in stroke patients, they are not specific for ischemic brain injury. Therefore, a quick and reliable ncRNA screening may be useful for the early diagnosis of stroke and also the prediction of stroke outcome for patients. Studies on the regulatory mechanisms of ncRNAs in experimental stroke models will lead to a better understanding of the roles of ncRNAs, which may provide a promising screening tool for a faster and more accurate prediction and diagnosis of different subtypes of stroke.
Along with intensive studies of ncRNAs, there are still challenges for using ncRNAs as biomarkers for the diagnosis and prognosis of stroke. First, current studies on identifying ncRNAs as biomarkers of ischemic stroke have been relatively small-scale to date. More independent and large cohort studies may help validate the results and drive reliable conclusions. Second, most miRNAs and lncRNAs are quickly degraded by the abundant RNases in the circulation/endocytic compartment of cells, and are therefore less reliable as biomarkers of diagnosis or prognosis in stroke patients. On the other hand, circRNAs have a relatively longer half-life compared with miRNAs and lncRNAs, which make them ideal candidates as biomarkers for diagnosis and prognosis of stroke. It should be noted that many microRNAs and lncRNAs are known to be differentially expressed in neurons, astrocytes, microglia, and oligodendrocytes in the central nerve system, as well as in brain vascular endothelial cells and pericytes. Although it is not currently known whether circRNAs are also expressed in a cell-specific manner, understanding the functions of ncRNAs in specific cell types under normal and pathophysiological conditions could improve specificity and efficacy of ncRNA-based therapeutic strategies. It may also help reduce unwanted side effects. Despite the challenges, ncRNA-based therapeutics have become promising strategies for stroke. Much remains to be studied to in order to better understand how ncRNAs exert their effects via hundreds of targets, develop novel approaches to identify candidate ncRNAs, design chemical formulations or modifications for better delivery of ncRNAs, and find methods to reduce unwanted side effects. Successful clinical trials and clinically-effective ncRNA-based drugs will certainly advance the stroke field.
Acknowledgements
This work was supported by the National Institute of Health Grants: NS094930, NS091175, NS086820 (K.J. Yin).
Footnotes
The authors declare no competing financial interests.
References
- 1.Yin KJ, et al. , miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis, 2010. 38(1): p. 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin KJ, Hamblin M, and Chen YE, Non-coding RNAs in cerebral endothelial pathophysiology: emerging roles in stroke. Neurochem Int, 2014. 77: p. 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cech TR and Steitz JA, The noncoding RNA revolution-trashing old rules to forge new ones. Cell, 2014. 157(1): p. 77–94. [DOI] [PubMed] [Google Scholar]
- 4.Wienholds E and Plasterk RH, MicroRNA function in animal development. FEBS Lett, 2005. 579(26): p. 5911–22. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, et al. , Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A, 2002. 99(24): p. 15524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeyaseelan K, Lim KY, and Armugam A, MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke, 2008. 39(3): p. 959–66. [DOI] [PubMed] [Google Scholar]
- 7.Mirzaei H, et al. , MicroRNA: Relevance to stroke diagnosis, prognosis, and therapy. J Cell Physiol, 2018. 233(2): p. 856–865. [DOI] [PubMed] [Google Scholar]
- 8.Khoshnam SE, Winlow W, and Farzaneh M, The Interplay of MicroRNAs in the Inflammatory Mechanisms Following Ischemic Stroke. J Neuropathol Exp Neurol, 2017. 76(7): p. 548–561. [DOI] [PubMed] [Google Scholar]
- 9.Chandran R, Mehta SL, and Vemuganti R, Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int, 2017. 111: p. 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer MK, et al. , The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 2015. 47(3): p. 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Hamblin MH, and Yin KJ, The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol, 2017. 14(12): p. 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattarai S, et al. , Discovery of novel stroke-responsive lncRNAs in the mouse cortex using genome-wide RNA-seq. Neurobiol Dis, 2017. 108: p. 204–212. [DOI] [PubMed] [Google Scholar]
- 13.Dharap A, Nakka VP, and Vemuganti R, Effect of focal ischemia on long noncoding RNAs. Stroke, 2012. 43(10): p. 2800–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, et al. , Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol, 2016. 277: p. 162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. , Long Noncoding RNA H19 Promotes Neuroinflammation in Ischemic Stroke by Driving Histone Deacetylase 1-Dependent M1 Microglial Polarization. Stroke, 2017. 48(8): p. 2211–2221. [DOI] [PubMed] [Google Scholar]
- 16.Dykstra-Aiello C, et al. , Altered Expression of Long Noncoding RNAs in Blood After Ischemic Stroke and Proximity to Putative Stroke Risk Loci. Stroke, 2016. 47(12): p. 2896–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta SL, Kim T, and Vemuganti R, Long Noncoding RNA FosDT Promotes Ischemic Brain Injury by Interacting with REST-Associated Chromatin-Modifying Proteins. J Neurosci, 2015. 35(50): p. 16443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. , Long Non-coding RNA Malat1 Regulates Cerebrovascular Pathologies in Ischemic Stroke. J Neurosci, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi X, et al. , Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145–5p/PLA2G4A in cerebral infarction. Neuroscience, 2017. 348: p. 98–106. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, et al. , LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem Biophys Res Commun, 2017. 485(1): p. 167–173. [DOI] [PubMed] [Google Scholar]
- 21.Yan H, et al. , Long noncoding RNA MEG3 activation of p53 mediates ischemic neuronal death in stroke. Neuroscience, 2016. 337: p. 191–199. [DOI] [PubMed] [Google Scholar]
- 22.Bai Y, et al. , Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke, 2014. 45(2): p. 383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. , Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke, 2012. 43(1): p. 14–21. [DOI] [PubMed] [Google Scholar]
- 24.Jeck WR and Sharpless NE, Detecting and characterizing circular RNAs. Nat Biotechnol, 2014. 32(5): p. 453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LL, The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol, 2016. 17(4): p. 205–11. [DOI] [PubMed] [Google Scholar]
- 26.Floris G, et al. , Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol, 2017. 54(7): p. 5156–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rybak-Wolf A, et al. , Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell, 2015. 58(5): p. 870–85. [DOI] [PubMed] [Google Scholar]
- 28.Bazan HA, et al. , Carotid Plaque Rupture Is Accompanied by an Increase in the Ratio of Serum circR-284 to miR-221 Levels. Circ Cardiovasc Genet, 2017. 10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta SL, Pandi G, and Vemuganti R, Circular RNA Expression Profiles Alter Significantly in Mouse Brain After Transient Focal Ischemia. Stroke, 2017. 48(9): p. 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, et al. , Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J Neurosci, 2018. 38(1): p. 32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]