Summary
Depression is a common disorder that affects women at twice the rate of men. Here we report that long non-coding RNAs (lncRNAs), a recently discovered class of regulatory transcripts, represent about one-third of the differentially expressed genes in the brains of depressed humans, and display complex region- and sex-specific patterns of regulation. We identified the primate-specific, neuronal-enriched gene, LINC00473, as downregulated in prefrontal cortex (PFC) of depressed females but not males. Using viral-mediated gene transfer to express LINC00473 in adult mouse PFC neurons, we mirrored the human sex-specific phenotype by inducing stress resilience solely in female mice. This sex-specific phenotype was accompanied by changes in synaptic function and gene expression selectively in female mice and, along with studies of human neuron-like cells in culture, implicates LINC00473 as a CREB effector. Together, our studies identify LINC00473 as a female-specific driver of stress resilience that is aberrant in female depression.
eTOC Blurb
Issler et al. demonstrate that long non-coding RNAs are robustly regulated in the brains of postmortem depressed humans in a brain-site and sex-specific manner. LINC00473 is highlighted as key regulator of mood in females only, where it acts in prefrontal cortex by regulating gene expression, neurophysiology and behavior.
Introduction
Depression is a devastating disorder and the leading cause of disability worldwide (WHO, 2017). Although many patients benefit from antidepressant medications, less than half show complete remission (Rush, 2007). The etiology and pathophysiology of depression remain incompletely understood. Along with genetic factors that represent ~35% of the risk for depression, environmental factors - predominantly stress exposure - are strongly associated with depression risk (Akil et al., 2018), (Otte et al., 2016). A more comprehensive appreciation of the molecular basis of depression will promote the development of novel and more effective pharmacotherapies.
Women are twice as likely to develop depression as men (Seedat et al., 2009), (Ferrari et al., 2013), yet the molecular mechanisms contributing to this sex difference remain poorly understood. Among women, depression tends to be more severe (Ferrari et al., 2013) and is characterized by partly different subsets of symptoms such as increased likelihood for co-morbid anxiety (Martin et al., 2013). Women can also respond differently to pharmacological interventions (Sramek et al., 2016). Sex differences in depression risk have been attributed to enhanced stress responses in women (Bangasser and Valentino, 2014) and to alterations in circulating ovarian hormones (Bale and Epperson, 2015), (Solomon and Herman, 2009). However, the brain mechanisms responsible for mediating the actions of these and other factors are not fully known (Issler and Nestler, 2018). Only recently have the molecular and cellular mechanisms underlying these differences begun to be systematically addressed (Hodes et al., 2015), (Labonte et al., 2017), (Seney et al., 2018), focusing on the role of transcription, particularly of protein-coding genes (PCGs).
Long non-coding RNAs (lncRNAs) are a recently discovered class of regulatory RNAs that play pivotal roles in epigenetic regulation (Ulitsky and Bartel, 2013). They are defined as containing more than 200 nucleotides and generally have similar structural properties as transcripts of PCGs (Rinn and Chang, 2012). LncRNAs play regulatory roles as decoys, scaffolds, or guides by interacting with DNA, RNAs (including microRNAs), or proteins (Quinn and Chang, 2016). LncRNAs are classified into several biotypes according to their genomic proximity to PCGs, for example, ones with no overlap with a PCG are named long intergenic noncoding RNAs (lincRNAs), or others that are coded from the opposite strand of a PCG are antisense RNAs (St Laurent et al., 2015). Notably, in the human genome there are more lncRNAs than PCGs (Zhao et al., 2016). Comparative genomic studies indicate that a third of lncRNAs have arisen within the primate lineage and that ~40% of lncRNAs are brain specific (Ransohoff et al., 2018), (Derrien et al., 2012). These observations suggest a key role for lncRNAs in the evolution of higher brain functions (Barry, 2014), although research focusing on the role of lncRNAs in mood and depression is in its infancy.
To evaluate the contribution of lncRNAs to depression, we bioinformatically analyzed lncRNAs within our recently published dataset of sex-specific transcriptional signatures in human depression (Labonte et al., 2017). We noted robust regulation of lncRNAs in depression in a brain-region and sex-specific manner. Our analysis identified LINC00473 as a key sex-specific target that is downregulated in prefrontal cortex (PFC) of depressed women only. Using parallel approaches in mouse models and human-derived neural-like cells in culture, we found that LINC00473, a primate-specific lncRNA, promotes behavioral resilience and alters neuroendocrine, neurophysiological, and transcriptional responses to stress specifically in females. This study opens a new avenue of research on the involvement of lncRNAs generally, and LINC00473 specifically, in sex-specific regulation of mood disorders.
Results
Brain-region and sex-specific regulation of lncRNAs in depression
To explore the potential role of lncRNAs in depression, we utilized a whole-genome RNA sequencing (RNA-seq) dataset recently generated by our laboratory from human postmortem brain samples from ~50 depressed and control human subjects of both sexes (Supplementary Table S1) (Labonte et al., 2017). In this dataset, the transcriptome of the following six brain regions was profiled: the ventromedial PFC (vmPFC), dorsolateral PFC (dlPFC), orbitofrontal cortex (OFC), nucleus accumbens (NAc), anterior insula (aINS), and ventral subiculum (vSUB) (Figure 1A). Our published analysis focused on PCGs (Labonte et al., 2017); here we turned our attention to lncRNAs.
Figure 1. Brain-region and sex-specific regulation of lncRNAs in depression.
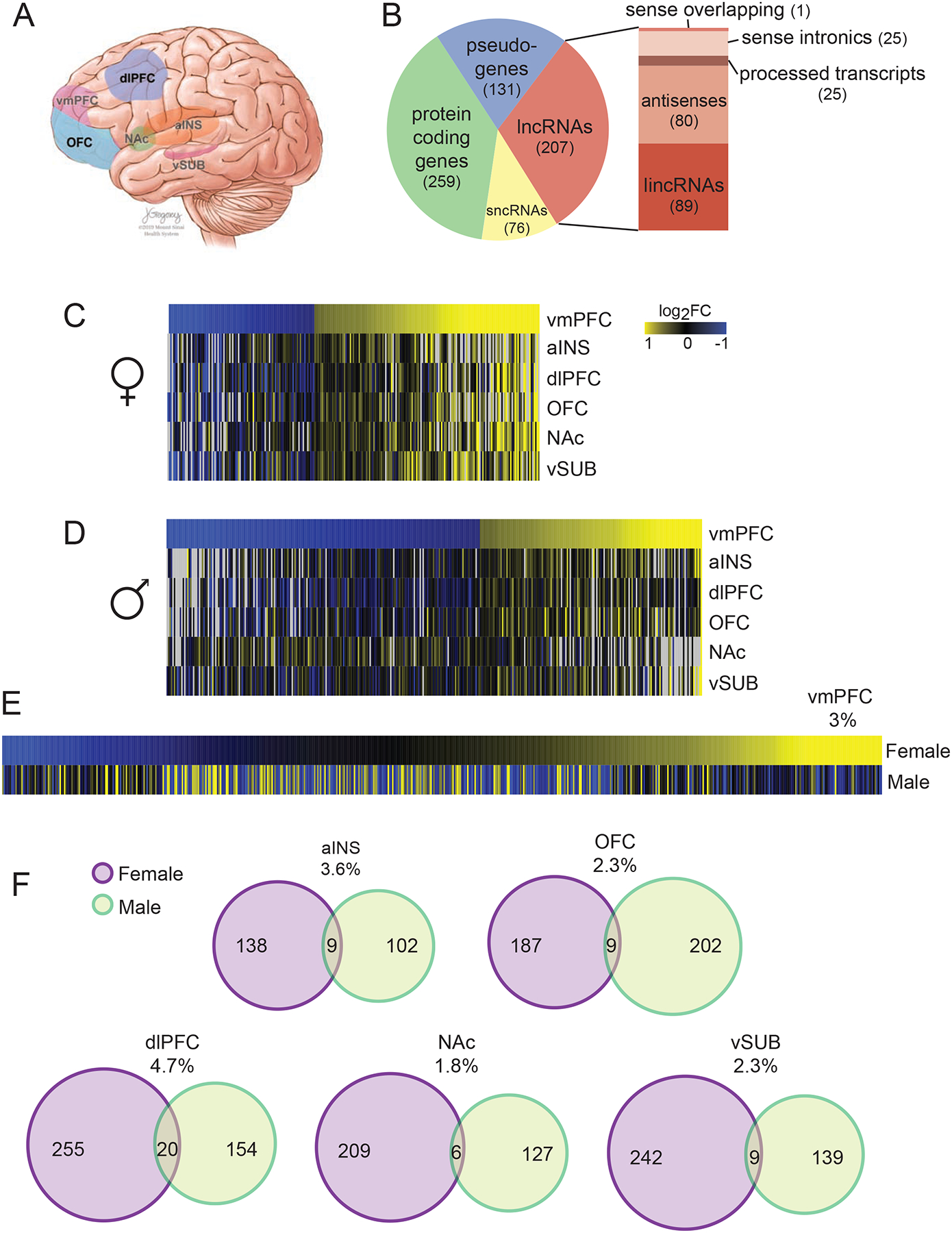
(A) Schematic illustration of the postmortem human brain regions used in the study from depressed and control subjects from both sexes. vmPFC, ventromedial prefrontal cortex; dlPFC, dorsolateral PFC; OFC, orbitofrontal cortex; NAc, nucleus accumbens; aINS, anterior insula; and vSUB, ventral subiculum. (B) Pie chart demonstrating the proportion of RNA biotypes that are differentially expressed in depression across brain regions and sexes. LncRNAs, long noncoding RNAs; snRNAs, short noncoding RNAs; and lincRNAs, long intergenic noncoding RNAs. Differential expression criteria: fold change >30% and P<0.05. Mean number of differently expressed lncRNAs is noted. (C–D) Heatmaps illustrating low similarly in the pattern of differentially expressed lncRNAs between brain regions in depressed subjects compared to controls in females (C) and males (D). Yellow, upregulation; blue, downregulation; black, no regulation; and gray, no expression. (E–F) Low overlap in lncRNAs regulated by depression between females and males in vmPFC (E) and other brain regions (F). % overlap and number of differentially expressed lncRNAs are noted. N=9–13 per group. RNA-seq data from Labonté et al. (2017). See also Figure S1.
We first examined the biotypes of the genes regulated across the brain regions analyzed and found that lncRNAs represent about 30% of the differentially expressed genes in human depression in both men and women (Figure 1B). The abnormally expressed lncRNAs were primarily of the antisense and lincRNA biotypes. We next compared the differentially expressed lncRNAs among the six brain regions and noted highly brain-region specific differential expression patterns in both sexes (Figure 1C,D). Subsets of lncRNAs are similarly regulated in multiple brain regions, yet others are oppositely regulated, not regulated, or even not expressed in other regions. We compared the regulation of lncRNAs in depression between the sexes and noted an extremely small overlap in identity and directionality of the regulated lncRNAs between the sexes (Figure 1E,F). Notably, in most of the brain regions, females had a larger number of lncRNAs regulated in depression, and there was only ~3% overlap between the lncRNAs regulated in depression in men vs. women. We repeated this analysis on an independent dataset of RNA-seq of rostral anterior cingulate cortex (rACC), which disregarded the sex of the subjects in their analyses (Zhou et al., 2018). In line with findings from our dataset, we identified numerous lncRNAs among the differentially expressed genes in rACC, with a very small (0.5%) overlap of the regulated lncRNAs between the two sexes (Figure S1A,B). These findings support our hypothesis that lncRNAs are highly regulated in depression pathophysiology and in a dramatic sex-specific fashion.
To further explore this hypothesis and to identify key depression-related lncRNAs, we performed a correlation analysis of the expression levels of lncRNAs and those of PCGs between the sexes in our human dataset. LncRNA and PCG transcripts that exhibit coordinated expression might be considered “guilty by association” and potentially take part in common biological processes (Rinn and Chang, 2012). We found that the number of correlated lncRNA-PCG pairs is higher in female compared to male brain (Wilcoxon test Z=−2.20558, P=0.0274), suggesting stronger lncRNA-PCG interactions in females. To identify key lncRNA targets, we filtered the list of lncRNAs that have any correlated PCG according to four parameters: 1) abundance (at least 1 read on average across all samples), 2) differential expression in depression (fold change >30% and P<0.05), 3) enrichment in neurons (Zhang et al., 2016), and 4) expression in neural progenitor cells (NPCs) (Hoffman et al., 2017) (Figure 2A). This pinpointed LINC00473, which is significantly correlated with 35 PCGs in the female brain, is robustly expressed, is downregulated in several brain regions of depressed females, and is expressed in NPCs. Several of the PCGs that associate with LINC00473 have previously been implicated in depression, including activity-regulated cytoskeleton-associated protein (ARC) (Li et al., 2015), nuclear receptor subfamily 4 group A member 1 (NR4A1) (Orsetti et al., 2008), early growth response 1 (EGR1) (Covington et al., 2010)), EGR2 (Orsetti et al., 2008), and dual specificity phosphatase 6 (DUSP6) (Labonte et al., 2017). Furthermore, LINC00473 is downregulated in depression across brain regions among females, but not males (Figure 2B). To validate the RNA-seq results we performed qPCR on vmPFC samples and confirmed its downregulation in depression in females, with a weak trend only seen in males (Two- way ANOVA, F group(1,42)=7.661, P=0.008, post hoc Fisher LSD, q(42)=2.154, P=0.037) (Figure 2C). Furthermore, our analysis of an independent RNA-seq dataset (Zhou et al., 2018) confirmed LINC00473 downregulation in rACC of depressed females but not males (t=−2.629, P=0.008) (Figure S2A). Finally, we confirmed this sex-specific regulation using RNA fluorescence in situ hybridization (FISH) in the rACC in a third independent cohort (Figures 2D and S2B) (Supplementary Table S2), where LINC00473 expression was downregulated only in depressed females compared to control subjects (Two-way ANOVA, F group(1,35)=5.453, P=0.0254, post hoc Fisher LSD, q(35)=2.389, P=0.0224).
Figure 2. Neuronal LINC00473 is downregulated in depressed females.
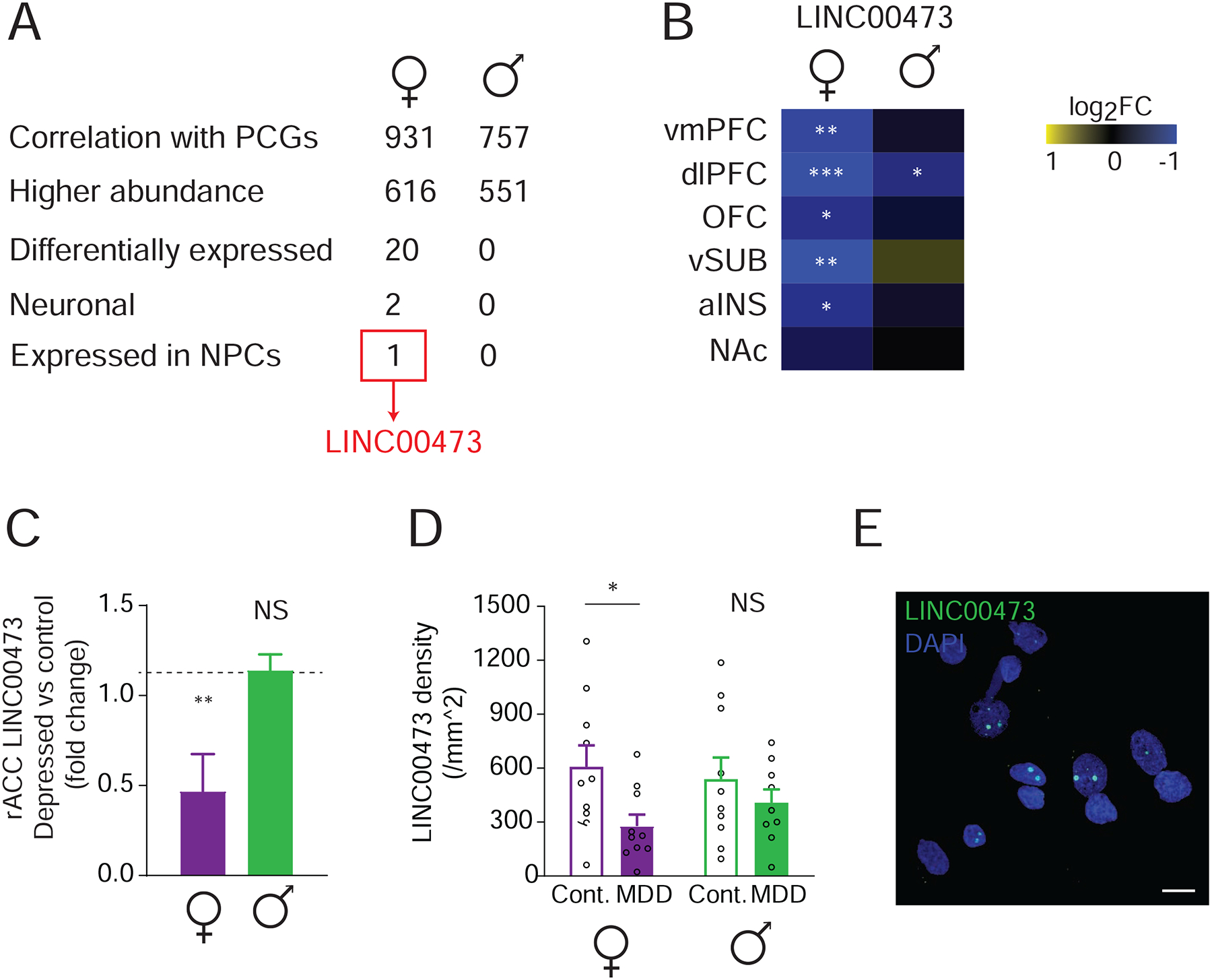
(A) Filtering of lncRNA hits from correlation analysis by abundance, regulation in depression, and cell-type specificity leads to LINC00473. (B) LINC00473 is downregulated in depressed females, but not males, across several brain regions. Stars indicate significance in MDD vs. control, *P<0.05, **P<0.01, ***P<0.001. (C) LINC00473 is downregulated in the rACC of depressed females compared to controls, an effect not seen in males. N=5–7 for females, N=19 for males. Data re-analyzed from (Zhou et al., 2018). (D) Quantification of LINC00473 mRNA expression by RNA FISH in rostral anterior cingulate cortex (rACC) from an independent cohort shows downregulation of LINC00473 in depressed females only. N=10 per group. (E) Representative LINC00473 FISH indicates nuclear enrichment characterized by two main puncta per nucleus. Scale bar=10 μM. *P<0.05. Bar graphs show mean ± SEM. See also Figure S2.
LINC00473 is expressed in primates only, with no apparent homolog expressed in rodents (Kent et al., 2002). It is found on chromosome 6 in humans, and expressed in the human brain and several other organs (GTEx, 2013) (Figure S2C). In brain, LINC00473 is expressed primarily in neurons at moderate levels (Zhang et al., 2016) (Figure S2D), predominantly in glutamatergic pyramidal neurons (p= 3.1072E-08) (Kozlenkov et al., 2016) (Figure S2E), and enriched within the cell nucleus (Figure 2E,F) (Mas-Ponte et al., 2017). There are no baseline sex differences in expression levels of LINC00473 in published datasets (GTEx, 2013) (Figure S2C), in the brains of control subjects in our RNA-seq dataset, or in the FISH results. Interestingly, studies show that LINC00473 is robustly induced by cAMP and CREB signaling (Liang et al., 2016), (Chen et al., 2016) and by neuronal activity (Pruunsild et al., 2017), (Ataman et al., 2016) in cultured cells derived from humans. Otherwise, LINC00473 has not been studied in neuropsychiatric syndromes or in the context of sex differences.
LINC00473 expression in mouse PFC neurons promotes behavioral, physiological, and transcriptional changes selectively in female mice
To test a causal role for LINC00473 in depression, we turned to chronic stress models in mice, with the notion that, even though LINC00473 is not normally expressed in rodents, its expression might nevertheless induce homologous functions and thereby regulate depression-related behavioral responses. Notably, this approach of using mice for studying mechanisms of action of primate-specific lncRNAs was fruitful in liver for cardiometabolic traits (Ruan et al., 2020). We generated neurotropic Herpes Simplex Virus (HSV) vectors expressing LINC00473 and eGFP (LINC00473) or eGFP only (GFP) as a control, and stereotactically delivered the HSVs bilaterally into medial PFC (mPFC) of adult male or female mice. This manipulation induced expression of LINC00473 to a similar extent in both sexes (Figure S3A–C). Since LINC00473 is downregulated in human female depression, we hypothesized that expressing it in female mice would promote stress resilience. To test this hypothesis, we used chronic social defeat stress (CSDS), which has been validated extensively in male mice (Krishnan et al., 2007), and more recently in female mice (Takahashi et al., 2017). We utilized an accelerated version of the protocol to match the short (4 days) time line of HSV-mediated transgene expression (Figure 3A). As predicted, socially defeated male mice (Two-way ANOVA F stress(1,40)=4.470, P=0.041) and female GFP mice (Two-way ANOVA F virus (1,31)=4.470, P=0.0088, post hoc Fisher LSD CSDS:GFP vs. Control:GFP t31=2.273, P=0.030) showed increased social avoidance as indicated by decreased social interaction (Figure 3B–D). We found that LINC00473 rescued social interaction to control levels only among female mice (post hoc Fisher LSD CSDS:LINC00473 vs. CSDS:GFP t31=3.169, P=0.0033) (Figure 3C), supporting a female-specific pro-resilient effect of LINC00473. We collected blood from the control and CSDS mice and probed serum corticosterone (CORT) levels (Figure S3D). The behavioral resilience observed in LINC00473-stressed females was associated with reduced levels of circulating CORT (Two-way ANOVA F stress(1,29)=8.380, P=0.007, post hoc Fisher LSD Control:GFP vs. CSDS:LINC00473 t29=2.405, P=0.022; Control:LINC00473 vs. CSDS:LINC00473 t29=3.13, P<0.004) (Figure S3E), while no differences were observed in male mice between the groups (Figure S3F).
Figure 3. LINC00473 expression in adult mouse mPFC neurons promotes stress resilience in females only.
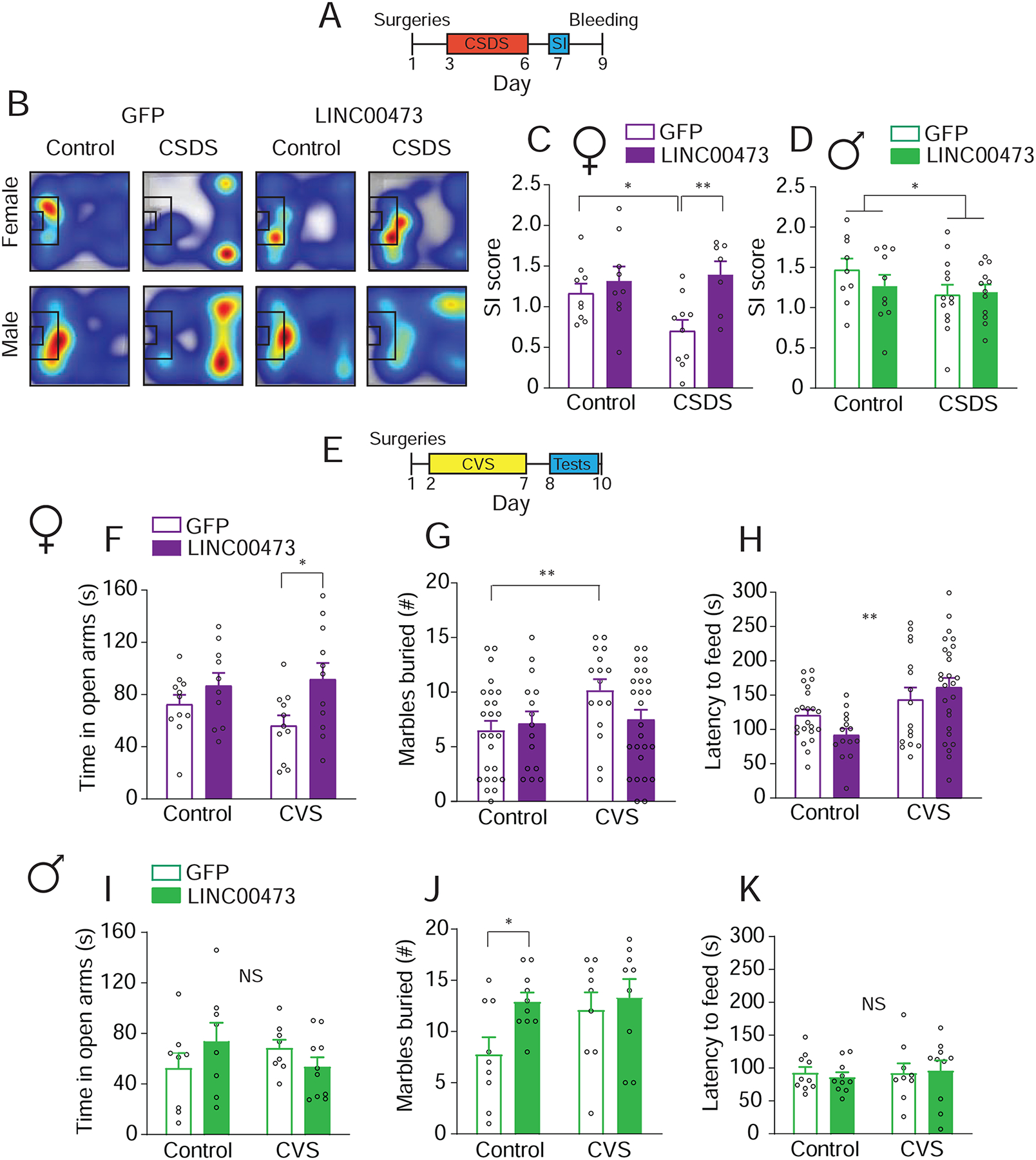
(A) Schematic illustration of experimental timeline of LINC00473 expression followed by accelerated chronic social defeat stress (CSDS) and testing. SI, social interaction. (B) Representative heatmaps of tracks of mice in the SI test. Warmer colors indicate longer duration of time spent in the region. The black rectangle is the interaction zone. (C) Female mice expressing LINC00473 have similar SI scores to non-stressed mice, while GFP controls exhibit decreased SI after CSDS. N=7–10 per group. (D) Male mice exposed to CSDS have decreased SI compared to control, regardless of viral treatment. N=9–13 per group. (E) Schematic illustration of experimental time line of LINC00473 expression followed by chronic variable stress (CVS) and behavioral testing. (F–G) Female mice expressing LINC00473 and exposed to CVS spent similar amounts of time as non-stressed mice in the open arms of the elevated plus maze (N=10–11 per group) (F) and buried similar numbers of marbles (N=15–35 per group) (G), unlike the stress GFP mice. (H) LINC00473 control mice have the shortest latency to feed in the novelty suppressed feeding test compared to other groups. N=15–26 per group. (I–K) No differences were seen between the groups of male mice in the elevated plus maze (I) and novelty suppressed feeding test (K). (J) Male mice expressing LINC00473 bury more marbles than GFP mice at baseline. (J). N=8–10 per group. * P<0.05, **P<0.01. Bar graphs show mean ± SEM. See also Figure S3.
We next sought to validate our findings in a second chronic stress model, chronic variable stress (CVS), in which female mice are more sensitive than males (Hodes et al., 2015; Labonté et al., 2017) (Figure 3E). Female mice expressing LINC00473 in mPFC neurons displayed no baseline differences in the elevated plus maze (Figure 3F) or marble burying test (Figure 3G) compared to GFP-expressing control mice. However, after CVS, LINC00473-expressing female mice spent more time in the open arms (Two-way ANOVA F virus(1,39)=6.875, P=0.0124; post hoc Fisher LSD CVS:GFP vs. CVS:LINC00473 t39=2.675, P=0.011) and buried fewer marbles compared to the stressed GFP group (Two-way ANOVA F interaction(1,78)=4.573, P=0.0356; post hoc Fisher LSD CVS:GFP vs. CVS:LINC00473 t78=2.300, P=0.0241). Both of these behavioral measures suggest reduced anxiety-related behavior. In the novelty suppressed feeding test, which is more stressful at baseline than the elevated plus maze or marble burying tests due to the use of food deprivation (Two-way ANOVA F stress(1,74)=11.96, P=0.0009), we noted differences at baseline where LINC00473-expressing female mice displayed the shortest latency to feed (survival analysis, Mantel-Cox Test, Chi square3=14.47 p=0.0023) (Figure 3H), indicative of reduced anxiety- and depression-like behavior. A similar pro-resilient effect was seen in the tail suspension test in female mice (Figure S3G). When we performed the same experiments in male mice, we noted no effects of LINC00473 expression in the elevated plus maze (Figure 3I), novelty suppressed feeding (Figure 3K), or tail suspension tests (Figure S3H), however, we observed increased marble burying indicative of increased anxiety-related behavior (Two-way ANOVA F virus(1,33)=4.285, P=0.0463; post hoc Fisher LSD Control:GFP vs. CVS:LINC00473 t33=2.394, P=0.0225) (Figure 3J). Interestingly, we noted that CVS led to a decrease in body weigh only in males expressing LINC00473 (Two-way ANOVA F virus(1,34)=9.219, P=0.0046; post hoc Fisher LSD CVS:GFP vs. CVS:LINC00473 t34=2.331, P=0.0258) (Figure S3M). In neither sex were there effects of LINC00473 expression on sucrose preference or forced swim test measures (Figure S3K,L,N,&O). Combined, expression of LINC00473 in mPFC had a pro-resilient role in female, but not male, mice. These results are in line with our human findings, where lower levels of LINC00473 are associated with female, but not male, depression.
To explore one possible mechanism by which expression of the activity-induced LINC00473 (Pruunsild et al., 2017), (Ataman et al., 2016) in mPFC promotes stress resilience in female mice specifically, we probed its effects on synaptic function. LINC00473 was expressed in mPFC neurons in vivo by use of our HSV vectors, after which brain slices were prepared and intracellular electrophysiological recordings were obtained from mPFC pyramidal neurons expressing LINC00473 plus GFP or GFP alone in both sexes. We found that LINC00473 expression decreased both the frequency (One-way ANOVA F(2,18)=4.709, P=0.0227; post hoc Fisher LSD Non-infected vs. LINC00473 t18=2.843, P=0.0108; GFP vs. LINC00473 t18=2.636, P=0.0168) (Figure 4A,B) and amplitude (One-way ANOVA F(2,19)=5.022, P=0.0177; post hoc Fisher LSD GFP vs. LINC00473 t19=3.166, P=0.0051) of spontaneous excitatory postsynaptic currents (sEPSCs) in females (Figure 4C) compared to GFP-infected or non-infected neurons. By contrast, sEPSCs in males were not different between the LINC00473 and GFP groups (Figure 4D–F). There was no effect of LINC00473 expression on membrane excitability in either sex (Figure S4). These results suggest that LINC00473 affects the pre- and postsynaptic properties of mPFC pyramidal neurons selectively in female mice, which parallels its sex-specific effect in controlling depression- and anxiety-like behavior.
Figure 4. LINC00473 expression modulates sEPSCs in mPFC pyramidal neurons of female but not male mice.
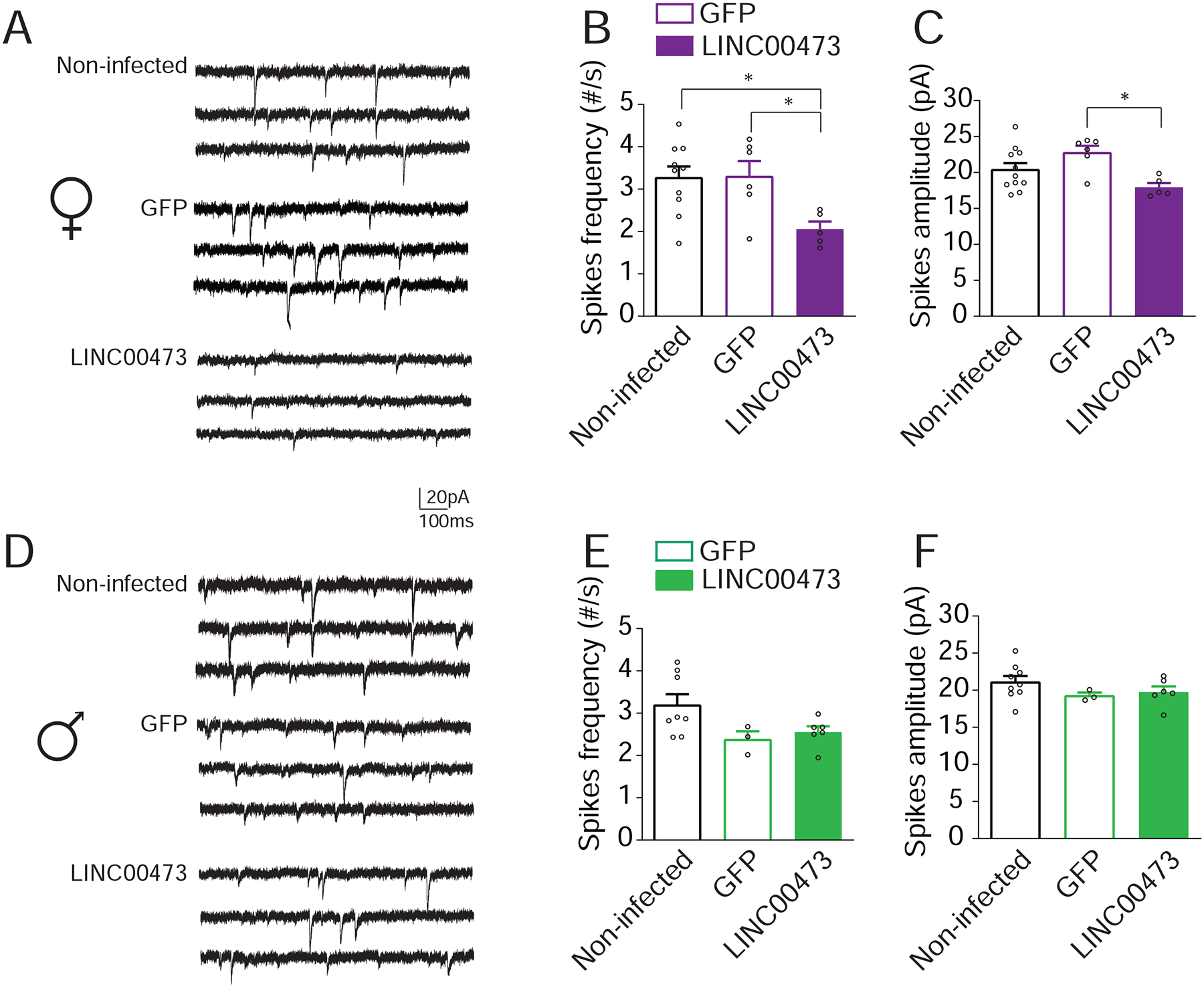
(A, D) Representative membrane potential of pyramidal neurons from either female (A) or male (D) mPFC in slices obtained from non-infected (top panel), GFP (middle panel), or LINC00473 expressing (lower panel) mice. Lower spiking frequency (B) and amplitude (C) of spontaneous excitatory postsynaptic currents (sEPSCs) in female mouse neurons expressing LINC00473 compared to controls. (N=5–10 per group). No difference in the frequency (D) or amplitude (E) of sEPSCs was seen in male mice. N=3–8 mice per group. * P<0.05. Bar graphs show mean ± SEM. See also Figure S4.
Nuclear lncRNAs can regulate gene expression through diverse mechanisms (Quinn and Chang, 2016), therefore, we tested the effect of LINC00473 expression in mPFC neurons using RNA-seq. We profiled RNAs in mPFC of female mice expressing LINC00473 or GFP only in this brain region, either at baseline or after CVS (Figure 5A). We found that CVS induces altered expression of ~5x more genes in the GFP group compared to the LINC00473 group (Figure 5B, Supplementary Table S4). These data reveal that the transcriptional effects of CVS in mPFC are dramatically blunted by LINC00473 expression (Figure 5C), which parallels the dampened effects of CVS seen under these same conditions at the behavioral level.
Figure 5. LINC00473 expression in adult mPFC neurons blunts gene expression changes induced by CVS.
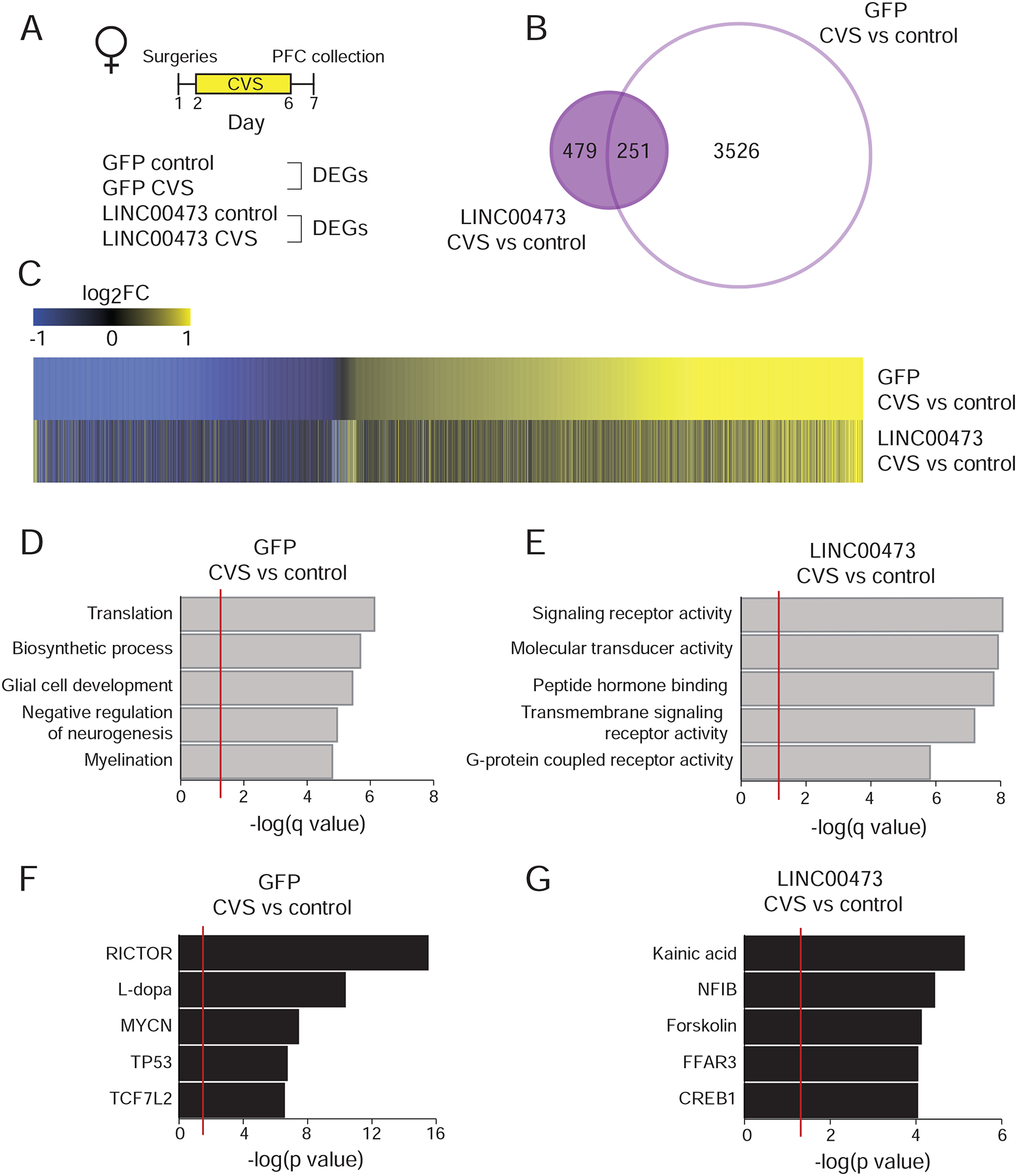
(A) Schematic representation of experimental design and groups used for analyzing differentially expressed genes (DEGs) comparing CVS to control groups for each viral treatment, LINC00473 vs. GFP. (B) The number of DEGs induced by CVS in GFP controls is ~5x higher than in the LINC00473 group. (C) This blunting is also apparent on heatmaps, which also identify a subset of genes that are regulated uniquely in the LINC00473 group. (D-E) GO terms enriched for genes regulated by CVS vs. control focusing on genes uniquely regulated in the GFP (D) or LINC00473 (E) group. Number of DEGs in each term is indicated in brackets. (F-G) Predicted upstream regulators of genes uniquely regulated in GFP (F) or LINC00473 (G) groups by CVS compared to control. N=3–5 mice per group. DEG cutoff is fold change >20% and P<0.05.
We explored the genes regulated by CVS in the GFP and LINC00473 groups using gene ontology (GO) analysis (Eden et al., 2009). In the GFP groups, the top GO terms are linked to stress response, including changes in glia and neurogenesis (Figure 5D), while the GO terms associated with CVS in the context of LINC00473 expression were very different and included signaling receptor activity and peptide hormones binding (Figure 5E). Ingenuity Pathway Analysis (IPA) (QIAGEN) was used to search for upstream regulators of the genes altered by CVS in the GFP and LINC00473 groups. The top predicted upstream regulators of differential gene expression following CVS in the GFP group included factors previously implicated in stress regulation (Figure 5F) such as Rictor (Carson et al., 2013), (Kaska et al., 2017), while the top upstream regulators in the LINC00473 group included forskolin and CREB1 (Figure 5G). The cAMP–CREB pathway has an established role in depression (Carlezon et al., 2005), including within the mPFC specifically (Lorsch et al., 2019), and it has been demonstrated that LINC00473 is induced by this pathway in cultured cells as well (Liang et al., 2016), (Chen et al., 2016).
Together, these findings demonstrate that expressing the primate-specific gene LINC00473 in mPFC neurons in mice can affect behavioral responses of the mice to stress, the neurophysiological properties of mPFC pyramidal neurons, and gene expression profiles within this brain region. Of note, the RNA-seq data show that LINC00473 expression perturbs a conserved signaling pathway (i.e., cAMP–CREB) as seen in human cells.
LINC00473 expression alters gene transcription preferentially in female human neural-like cells
We next examined the actions of LINC00473 in cells that express it endogenously and therefore turned to NPCs derived from male or female healthy human subjects (Hoffman et al., 2017). We utilized antisense oligonucleotides (ASOs), an approach shown previously to knockdown (KD) nuclear levels of specific lncRNAs (Lennox and Behlke, 2016), in our case to KD LINC00473 as a way to mimic its downregulation in female depression (Figure 6A). We screened multiple ASOs to identify ones that potently KD LINC00473 expression in both female (one sample t- test t2=14.19, P=0.0049) (Figure 6B) and male (t2=19.88, P=0.0025) cell lines (Figure 6C) compared to control ASOs, and then subjected the samples to RNA-seq. We found that 5x more genes are significantly regulated by LINC00473 KD in female-derived than in male-derived NPCs (Figure 6D, Supplementary Table S5). There is also very little overlap between the transcriptional changes caused by LINC00473 KD in female vs male NPCs (Figure 6D). These data provide further evidence for greater regulatory effects of LINC00473 in females than males.
Figure 6. LINC00473 knockdown in human female NPCs has a stronger effect on gene expression than in male NPCs.
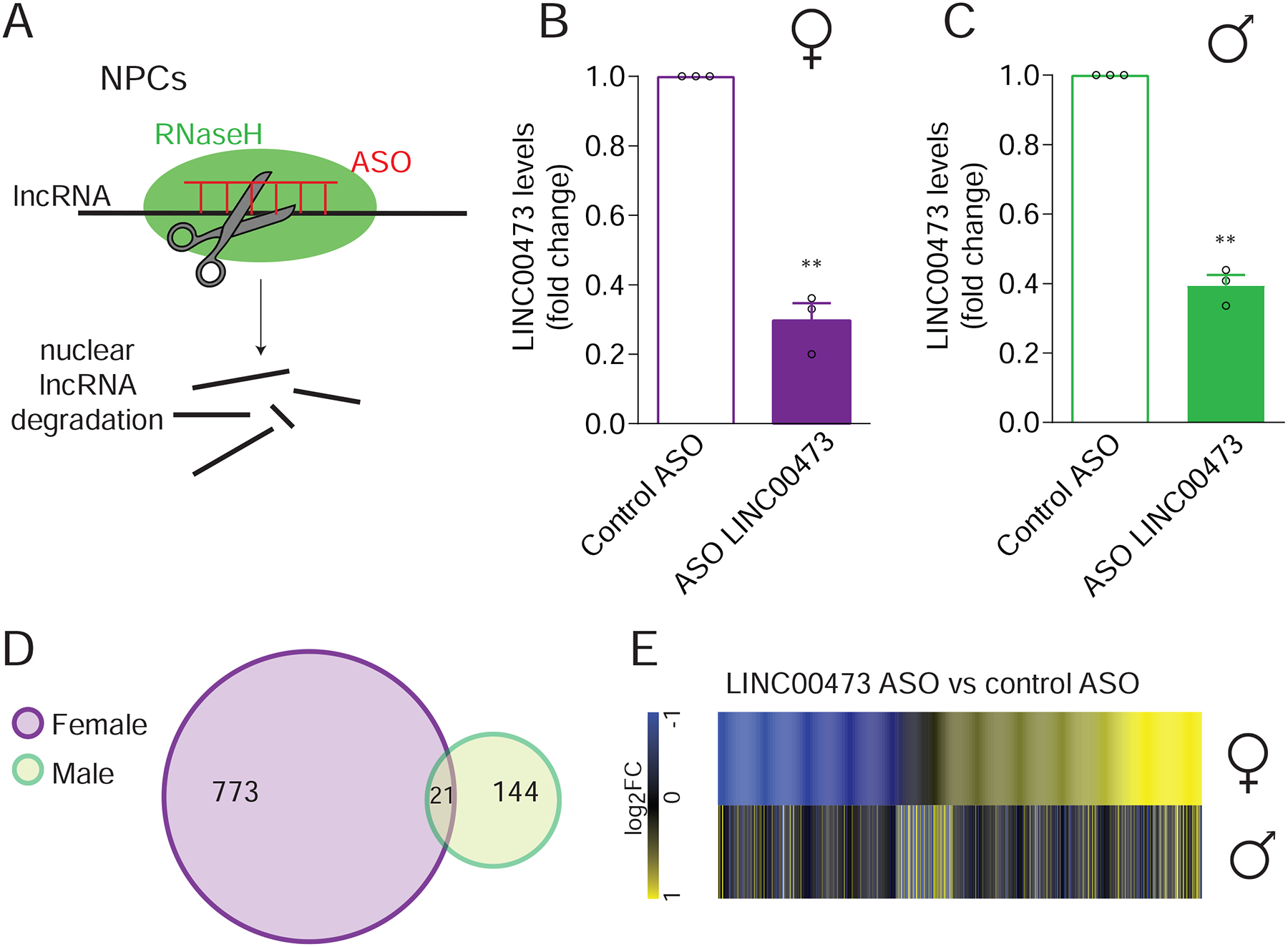
(A) Schematic representation of lncRNA knockdown (KD) by antisense oligonucleotides (ASOs). (B–C) Validation of LINC00473 KD in female- (B) and male- (C) derived neural progenitor cells (NPCs). (D–E) Five times more genes are regulated by LINC00473 KD in female NPCs than in male (D) and there is a small overlap in the regulated genes between the sexes (E). DEG cutoff is fold change >30% and P<0.05. N=3 lines per group. *P<0.05; ** P<0.01. Bar graphs show mean ± SEM.
Finally, we sought to identify direct partners for LINC00473 in human cells. Since LINC00473 is predominately nuclear (Figures 2E and S2F) and regulates gene expression (Figures 4 and 5), we hypothesized that LINC00473 might interact directly with DNA. We therefore performed chromatin isolation by RNA purification (ChIRP) followed by DNA-sequencing (DNA-seq). We used a human neuroblastoma female-derived SH-SY5Y cell line which, unlike human NPCs, endogenously expresses LINC00473 at sufficient levels for this assay. We tested the effects of inducing endogenous LINC00473 expression by stimulating cAMP-CREB signaling with the adenylyl cyclase activator, forskolin (One-way ANOVA F(4,14)=34.75, P<0.0001) (Figure S5A). We precipitated substantial amounts of LINC00473 both in control (DMSO) and forskolin-treated cells (Figure 7A) in a specific manner (Figure 7B,C). We then sequenced DNA precipitated using probes for LINC00473 or LacZ as control, and input DNA for background. In the DMSO condition, we identified binding sites for LINC00473 predominately in intergenic areas (Figure 7D, Supplementary Table S6). In contrast, in forskolin-treated samples there was enhanced binding in gene bodies and promoters (Figure 7E, Supplementary Table S6). Notably, among the genes to which LINC00473 binds in the forskolin condition, there is enrichment for genes that have been implicated in brain function in general, and in depression- or anxiety-related behavior specifically (Figure 7F). Such LINC00473-precipitated and depression-linked genes include those encoding CREB binding protein (CREB-BP) which is a transcriptional co-activator of CREB and other transcription factors (Malki et al., 2012), (Datson et al., 2012), centrosomal protein 350 (CEP350) (Dunn et al., 2016) which has a role in microtubule binding, microtubule associated protein 2 (MAP2) which regulates neurogenesis during development and controls dendritic function in adulthood (Cereseto et al., 2006), (Pei et al., 1998), and ERB-B2 receptor tyrosine kinase 4 (ERBB4) which is a member of the EGFR subfamily of receptor tyrosine kinases (Bi et al., 2015), (Chung et al., 2018), (Dang et al., 2016).
Figure 7. Induction of endogenous LINC00473 expression recruits its binding to depression-related genomic loci.
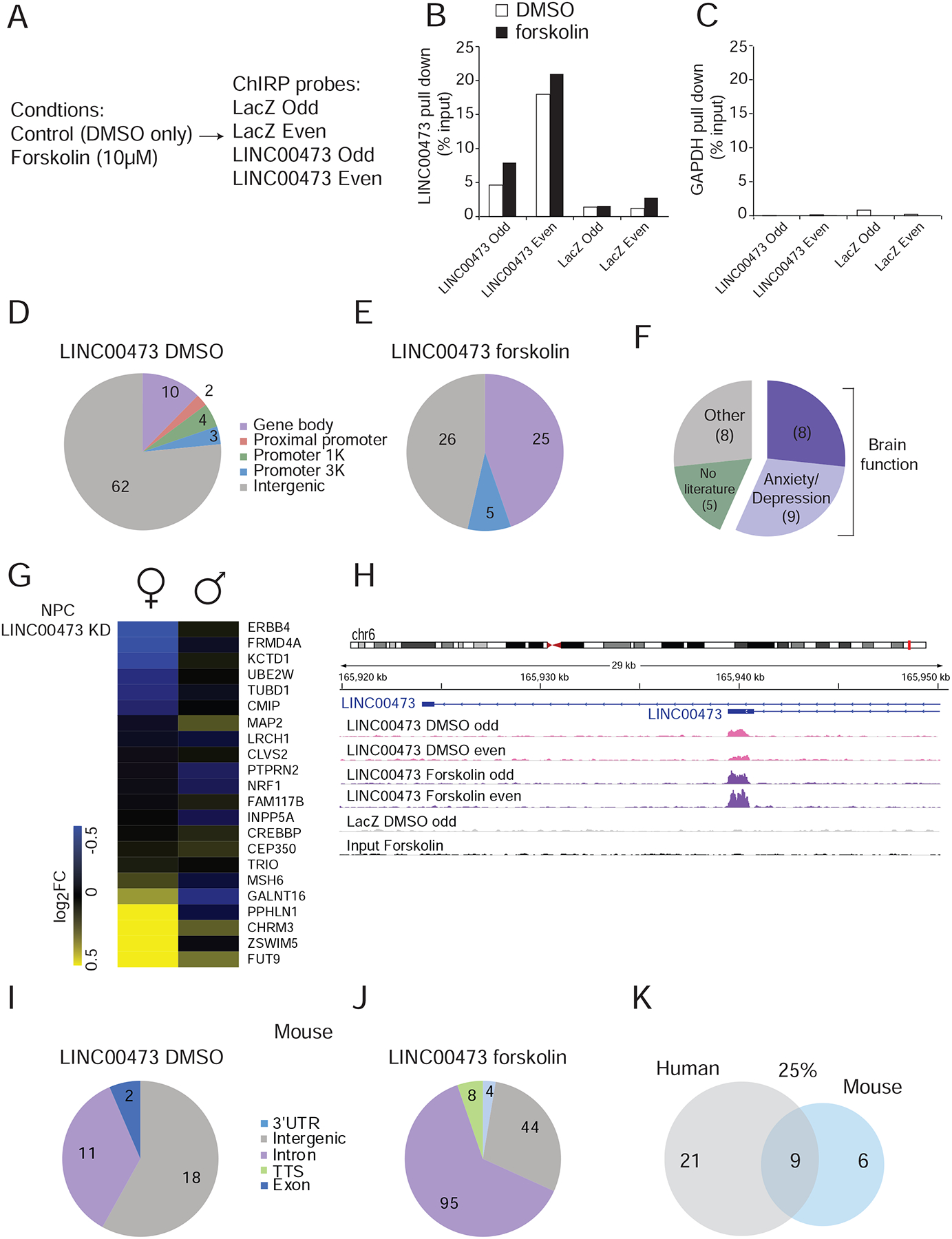
(A) Experimental conditions in LINC00473 ChIRP performed in SH-SY5Y cells. Odd and even represent replicates of tiling antisense biotinylated oligonucleotides for LINC00473. (B–C) Validation of efficacy (B) and specificity (C) of LINC00473 pull down. (D–E) LINC00473 binds mainly in intergenic regions under control DMSO conditions (D), with increased LINC00473 binding to gene bodies and promoters following treatment with forskolin which induces LINC00473 (E). (F) Enrichment for genes with a functional role in brain and implicated in depression that are bound by LINC00473 in the forskolin condition. (G) Stronger regulation of LINC00473-forskolin ChIRP hits in female NPCs with LINC00473 KD than in male. (H) Binding of LINC00473 to its own genomic locus both under DMSO and forskolin conditions. (I,J) Number of corresponding sequences in the mouse genome that are identical to sequences in the human genome that bind LINC00473 in the DMSO (I) or foskolin (J) conditions. (K) 25% of the genomic sites bound by LINC00473 in human cells treated with forskolin have matching DNA sequences in the mouse genome. See also Figure S5.
We next overlapped genes that bind LINC00473 in the forskolin condition in the ChIRP experiment with genes whose expression levels are altered by LINC00473 KD in the NPC experiment described in Figure 6. We found a subset of forskolin-LINC00473-ChIRP hits that are also regulated in female NPCs upon LINC00473 KD, with significantly fewer of these genes affected in male-derived NPCs (Figure 7G). Using HOMER motif analysis, we identified DNA motifs that are common to LINC00473 genomic binding sites in either the DMSO (Figure S5B) or forskolin (Figure S5C) condition. The motifs identified under either condition do not correspond to known transcription factor binding motifs. Moreover, the only overlap between DMSO and forskolin conditions with respect to LINC00473 binding is its own genomic loci (Figure 7H), indicating a potential auto-regulatory role for this lincRNA. Finally, we identified the corresponding sites in the mouse genome that match the coordinates to which LINC00473 binds in human cells (LiftOver). This analysis revealed that many of the sites identified by ChIRP in human cells have parallel sites in mouse (Figure 7I & J). In fact, 25% of LINC00473 binding sites within PCGs in the forskolin condition are identical in the mouse (Figure 7K). This analysis reveals that the mouse genome contains a significant fraction of homologous sites to which LINC00473 binds in the human genome, and provides the basis for the functional effects we observe at the behavioral, electrophysiological, and molecular levels upon LINC00473 expression in mouse PFC neurons.
Discussion
In this study, we show the dramatic regulation of lncRNAs in human depression with a complex pattern seen across brain regions and sexes. We identified a target lncRNA with a sex-specific role in depression, LINC00473, which is downregulated in mPFC and several other forebrain regions in depressed women only. Viral-mediated expression of LINC00473 in mouse mPFC neurons increased behavioral resilience in two chronic stress paradigms in females only. This behavioral resilience was associated with blunted CORT levels, decreased sEPSC frequency and amplitude of mPFC pyramidal neurons, and dramatically blunted effects of chronic stress on gene expression. We found that the genes regulated by LINC00473 include components of cAMP–CREB signaling, a pathway known to regulate LINC00473 in cultured cells (Liang et al., 2016), (Chen et al., 2016). When we induced endogenous LINC00473 expression in human-derived neuroblastoma cells with forskolin, we found that LINC00473 binds predominantly to genes involved in brain function, including several implicated previously in depression and anxiety. Together, this study provides a fundamentally new view of the molecular adaptations in brain that contribute to depression in a sex-specific manner and may lead to the identification of novel targets for sex-specific treatments of this syndrome.
Our overall hypothesis, that lncRNAs participate in depression pathogenesis, is supported by two recent GWASs: numerous lncRNA genomic loci are associated with depression (Wray et al., 2018), (Howard et al., 2019), (Zeng et al., 2017) and single nucleotide polymorphisms within lncRNA genes were individually linked with depression risk (Delacretaz et al., 2015), (Ye et al., 2017). Descriptive studies profiling lncRNAs in humans have found differences in rACC (Zhou et al., 2018) and blood (Liu et al., 2014), (Cui et al., 2017), (Seki et al., 2019) of depressed patients. Similar profiles have been observed in rodent hippocampus after stress exposure (Wang et al., 2019), (Li et al., 2017), (Roy et al., 2018). However, none of these prior studies accounted for sex of the subjects or provided a mechanistic link between lncRNA expression and sex-specific risk for depression. Notably, LINC00473 was not highlighted in any of these studies. This is unsurprising given the sex-specific role of LINC00473 found here. Given that the study of lncRNAs in depression is a burgeoning field, we provide both an important resource for further exploration of additional lncRNA targets and a frame work of the variety of experimental approaches that can be utilized for probing specific lncRNA candidates and their sex-specific role in depression.
Our analysis reveals that lncRNAs represent a large proportion of all of the regulated transcripts in human depression in a brain region- and sex-specific manner. This regional selectivity is consistent with the highly tissue-specific expression patterns that characterize lncRNAs in general (Cabili et al., 2011), (Derrien et al., 2012). Sex-specific regulation of PCGs in the brain in depression was noted by us (Labonte et al., 2017) and others (Seney et al., 2018), but these studies did not examine lncRNAs. In support of our present findings, Seney et al. (2018) reported an analysis of our dataset that suggested that the top transcripts opposingly regulated by depression between the sexes were enriched for lncRNAs. Some of the effects we report may be due to baseline sex differences in lncRNA expression levels, however, that is not the case for LINC00473 whose expression in brain is equivalent between females and males at baseline. Little is known about regulation of lncRNAs by sex at the genome-wide level, for example, there are reports of sex-specific expression patterns of lncRNAs in mouse liver (Melia and Waxman, 2019) and brain (Reinius et al., 2010), however, this has not yet been demonstrated in human brain. One of the most studied lncRNAs, Xist, is highly sexually dimorphic, female-specific, and plays a pivotal role in chromosome X inactivation (Sahakyan et al., 2018). Our findings thus support the importance of further investigations of the sex-specific regulation of lncRNAs in brain. Future studies should include determining the potential effects of psychotropic drugs such as antidepressant medications and alcohol, as well as the effects of depression severity and other clinical variables on lncRNA regulation.
It has been suggested that LINC00473 is an oncogene as it is upregulated in several types of cancers (Bai et al., 2019), (Chen et al., 2016), (Dinh et al., 2017), (Zhu et al., 2018), (Zhang and Song, 2018), (Wang et al., 2018), (Chen et al., 2018a), (Shi et al., 2017), (Han et al., 2018), (Zhang et al., 2017). Interestingly, studies investigating the regulatory mechanisms of LINC00473 in cancer have identified several targets that have also been implicated in stress responses. Some of these genes encode microRNAs (miRNAs), a subset of which have competitive interactions with lncRNAs (Quinn and Chang, 2016). For example, miR-34 which is upregulated by stress in amygdala (Haramati et al., 2011) reduces the stability of LINC00473 (Shi et al., 2017), suggesting a potential mechanism for the reduced levels observed in our dataset. Altering levels of LINC00473 may have downstream effects as well. miR-15, which is inhibited by LINC00473 in cell culture (Wang et al., 2018), is upregulated by stress in amygdala (Volk et al., 2016), and others have shown that LINC00473 activates β-catenin signaling in cell culture (Han et al., 2018), which has been linked to stress resilience in brain (Dias et al., 2014). LINC00473 is robustly upregulated by cAMP in cell culture (Liang et al., 2016) and its promoter harbors CRE binding sites that facilitate this induction by CREB binding (Liang et al., 2016), (Chen et al., 2016). CREB signaling is strongly associated with depression (Carlezon et al., 2005) and with stress resilience in mPFC specifically (Lorsch et al., 2019).
We report that endogenous LINC00473 is enriched within the nucleus of rACC neurons (Figure 2E). This finding matches reports in the LncATLAS database (Mas-Ponte et al., 2017) and other studies performed in different tissues (Chen et al., 2016), (Chen et al., 2018b), (Wu et al., 2018), (Chujo et al., 2017). Nuclear localization suggests the involvement lncRNAs in nuclear processes, such as scaffolding or regulation of transcription (Quinn and Chang, 2016). Indeed, we show here that altering LINC00473 expression levels, either in mouse mPFC neurons or in human NPCs, has a robust effect on gene expression, with much stronger effects seen in female-derived cells. Interestingly, similar to our FISH results (Figure 2E), other groups noted as well that LINC00473 is localized within distinct nuclear structures in two main puncta per nucleus (Chen et al., 2016), (Chujo et al., 2017). This localization pattern might be explained by our ChIRP results, indicating binding of LINC00473 to its own genomic loci, suggesting a potential auto-regulatory feedback loop of LINC00473.
Using viral-mediated gene delivery we found that expression of LINC00473 in mPFC neurons, despite it being a primate-specific lncRNA, promotes stress resilience, an effect seen in female mice only despite equivalent expression levels achieved in females and males. Such pro-resilient behavioral effects were documented in two chronic stress models, CSDS and CVS, both widely used in the field. The various behavioral outcomes measured in the present study suggest that LINC00473 reduces both depression- and anxiety-like behaviors induced by stress exposure. This is interesting because human females tend to have a greater degree of comorbidity of depression with anxiety than males (Martin et al., 2013). We used 6 days of CVS to fit within the short timeline of HSV expression. This relatively short stress exposure is sufficient to induced certain stress effects, as we report for marble burying and novelty suppressed feeding, but was not strong enough to affect forced swim or sucrose preference behavior. We also found a baseline sex difference in circulating CORT levels, with higher levels in females (Figure S3D–F), that matches reports in the literature (Kalil et al., 2013). LINC00473 expression in mPFC neurons affected circulating CORT levels in female mice, an effect not seen in male mice. Together, the mouse models reveal a female-specific effect of LINC00473 expression in mPFC consistent with LINC00473 downregulation in mPFC and several other brain regions of depressed females only.
We found that LINC00473 decreases sEPSC amplitude and frequency of mPFC pyramidal neurons in female mice only. These changes are interpreted as reflecting alterations in both pre- and postsynaptic domains. These findings are in line with our earlier studies of DUSP6, the knockdown of which in mPFC neurons promotes behavioral resilience and decreases sEPSP frequency in female mice only (Labonte et al., 2017). LINC00473 is induced in human-induced neurons in culture upon electrical activation (Pruunsild et al., 2017), (Ataman et al., 2016), suggesting a homeostatic mechanism by which neural activity induces LINC00473 which then feeds back to dampen neural activity. In addition, our ChIRP study showed that LINC00473 binds to genes that encode proteins with a role in synaptic function, such as MAP2 (Sanchez et al., 2000) and trio rho guanine nucleotide exchange factor (TRIO) (Miller et al., 2013), among several others, thus providing putative mechanisms by which LINC00473 controls synaptic function.
The RNA-seq experiment in female mouse mPFC showed that the behavioral stress resilience seen in mice expressing LINC00473 is associated with dramatically blunted induction of gene expression changes by chronic stress. The genes affected by LINC00473 include a subset which are predicted to be regulated by the CREB pathway. As mentioned above, there is ample evidence, including our results (Figure S5A), showing that LINC00473 is upregulated by CREB signaling. In addition, it is very well established that CREB signaling is altered in depression in a brain-site specific manner (Carlezon et al., 2005). We recently reported that downregulation of CREB in mPFC promotes stress susceptibility in both sexes (Lorsch et al., 2019). This is consistent with our finding of decreased LINC00473 expression in depressed women, and the pro-resilient effect of expressing LINC00473 in female mice. Our ChIRP study found that, upon forskolin treatment and consequent LINC00473 induction, LINC00473 binds the CREBBP genomic locus, suggesting that LINC00473 potentially affects CREB action. Moreover, comparative genomic analysis suggests that the LINC00473 sequence originated from partial duplication of the phosphodiesterase 10A (PDE10A) genomic locus. PDE10A is also associated with CREB signaling as it is an enzyme that degrades cAMP (Soderling et al., 1999). Moreover there is evidence for sex-specific roles of PDE10A function (Porcu et al., 2013) (Hsu et al., 2011), with primate-specific splice variants of the gene associating with bipolar disorder (MacMullen et al., 2016). Collectively, these findings propose that LINC00473 arose in primates to mediate additional effects of CREB on its gene targets, and to possibly control mood particularly in females.
It will be important to fully understand the molecular basis by which LINC00473 affects gene expression and downstream consequences (synaptic activity and behavior) in females only. Further studies are required to determine whether these sex-specific actions reflect distinct chromatin architecture at gene targets for LINC00473 in females vs. males or the sex-specific expression of other molecules (RNAs or proteins) that control LINC00473 action. Regardless of the underlying mechanisms, LINC00473 joins several PCGs (e.g., DUSP6, EMX1, ESR1) which also have been shown to control depression-related behavioral outcomes in a sex-selective fashion (Labonté et al., 2017; (Lorsch et al., 2018).
In summary, studying the mechanisms of action of primate-specific genes in psychiatric disorders is challenging. To address this, we used a combination of approaches, including expressing the primate-specific LINC00473 in adult mouse mPFC neurons, knocking down the gene in human NPCs, and controlling expression of the endogenous gene in cultured cells derived from humans. The cell culture studies made it possible to examine the effects of manipulating endogenous LINC00473 within the relevant genomic context, while the animal models allowed studying the gene in vivo in stress models with direct relevance to depression. While both approaches offered insight into the mechanisms underlying the pro-resilient and female-specific effects of LINC00473, it is likely that the molecular basis of LINC00473 action also involves additional primate-specific effects. Our results provide a roadmap for others to investigate how primate-specific lncRNAs influence complex brain disorders and provide a repository of sex-specific lncRNAs for future studies. Taken together, this work suggests that the complex primate brain uses lncRNAs to facilitate regulation of higher brain function including mood, with malfunction of these processes contributing to pathological conditions such as depression and anxiety in a sex-specific manner.
STAR Methods
Resource availability
Lead Contact
Further information and requests should be directed to the lead contact, Eric J. Nestler (eric.nestler@mssm.edu).
Materials Availability
Plasmids and viruses generated in this study are available by contacting the lead contact.
Data Availability
The RNA-seq and ChIRP datasets generated during this study are available at GEO (GSE138677).
Experimental models and subjects details Mice
Experimental animals were eight-week-old C57BL/6J male or female mice (Jackson Laboratory). Mice were habituated to the animal facility for 1 week before any manipulations, and were maintained at 23–25°C with a 12 h light/dark cycle (lights on 7:00 A.M.) with ad libitum access to food and water. Experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Mount Sinai. Female estrous phase was monitored by microscopy of vaginal smears at the end of all experiments to minimize the confounding effects of the stress involved.
Human subjects
Human postmortem RNA-seq data from depressed and control subjects were collected, analyzed, and reported as part of a published study (Labonte et al., 2017). This earlier paper focused solely on protein-coding genes (PCGs). Briefly, brain tissue was obtained from the Douglas Bell Canada Brain Bank Québec. Subject information is listed in Supplementary Table S1. Males and females were group-matched for age, pH, and PMI. Inclusion criteria for both cases and controls were French-Canadian European origin and sudden death.
Florescent in situ hybridization (FISH) was performed on an independent cohort of male and female depressed cases and controls (N=10 per group per sex) (Supplementary Table S2). The samples were obtained by The University of Texas Southwestern Medical Center at Dallas
Neural progenitor cells (NPCs)
NPCs were derived from human iPSCs as described (Hoffman et al., 2017). Three lines derived from healthy control males (lines NSB690, NSB553 and NSB2607) and females (lines NSB 3182, NSB3188 and NSB 3121) were used. NPCs were maintained in NPC media (Dulbecco’s Modified Eagle Medium/Ham’s F12 Nutrient Mixture (ThermoFisher Scientific), 1x N2, 1x B27-RA (ThermoFisher Scientific) and 20 ng ml−1 FGF2, cultured at high density on Matrigel (BD Biosciences) and split approximately 1:4 once a week with Accutase (Millipore).
Method Details
Chronic social defeat stress (CSDS)
Experiments utilized published procedures for CSDS to induce depressive-like behaviors in male (Krishnan et al., 2007) and female (Takahashi et al., 2017) mice. For males, retired breeder CD1 male mice were screened for aggression. C57BL/6J mice were subjected to a 5-min defeat by a novel CD1 aggressor mouse and were then housed across a ventilated divider to allow for sensory contact. Control mice were housed in cages separated from other control mice by a ventilated divider and were rotated to a different cage daily. For the females, ERα-Cre mice were injected into the ventrolateral ventromedial hypothalamus with rAAV2/hSyn-DIO-hM3D(Gq)-mCherry (Krashes et al., 2011) to express an excitatory DREADD. Mice were screened for aggressive behavior toward females. Male aggressors were housed individually and females were paired and housed with another female from the same stress condition, all in standard mouse cages. Aggressors were injected with CNO (1 mg/kg, i.p.) 30 min before the encounter. A C57BL/6J female was introduced into the aggressor’s cage for 5 min and after the physical defeat, the female was returned to her home cage. For both sexes an accelerated social defeat paradigm (twice daily defeats for 5 d for 10 total defeats) was utilized to capture behavior within the timeframe of HSV expression (Krishnan et al., 2007). After the final defeat, both defeated and control mice were housed individually for behavioral testing.
Social interaction (SI)
Testing for social avoidance behavior was assessed with a novel CD1 mouse in a two-stage social interaction (SI) test under red lighting (Berton et al., 2006). In the first 2.5-min test, the experimental mouse was allowed to freely explore an arena (44×44 cm) containing a plexiglass and wire mesh enclosure (10×6 cm) centered against one wall of the arena. In the second 2.5 min test, the experimental mouse was immediately returned to the arena with a novel CD1 mouse enclosed in the plexiglass wire mesh cage. Time spent in the ‘interaction zone’ (14×26 cm) surrounding the CD1 mouse was measured by video tracking software (Ethovision, Noldus). An SI ratio was calculated as time spent exploring the interaction zone with the CD1 mouse present, divided by time spent exploring the interaction zone with CD1 mouse absent.
Chronic variable stress (CVS)
A shortened CVS protocol was used to induced depression- and anxiety-like behavior with a time line that matches the expression of HSVs (Labonte et al., 2017), (Hodes et al., 2015). The protocol consists of exposure to three different stressors repeated over a period of six days and alternated to prevent habituation. The mice were exposed to one of the following stressors for one hour daily: 100 random mild foot shocks at 0.45 mA, a tail suspension, or restraint stress within a ventilated 50 mL conical tube. Mice were group housed during the stress exposure and individually housed during the behavioral assessment. Unstressed mice were used as controls.
Novelty suppressed feeding
The novelty suppressed feeding test assesses primarily anxiety-like behaviors (Wang et al., 2008), (Lorsch et al., 2018). Mice were food deprived for 24 h before testing and were habituated to the experimental room for 1 h prior to testing. Mice were placed in the corner of a 50 × 50 × 20 cm box covered with bedding with a single food pellet placed in the center of the arena. The latency to eat was recorded during 5 min of testing. Mice were then transferred to their home cage and their latency to eat was recorded there as well.
Sucrose preference
Mice were individually housed and were given a choice between two 50-mL conical tubes with either water or 1% sucrose solution for 24 h (Pena et al., 2017). The bottles were weighed and sucrose preference was calculated by determining the percentage of sucrose consumption divided by total liquid intake.
Elevated plus maze
For the elevated plus maze, which assesses anxiety-like behaviors (Sun et al., 2015), mice were acclimated to the testing room for 1 h before testing. Animals were tested in a cross-shaped maze consisting of two 12 × 50 cm open arms and two closed arms with 40 cm high walls under red light for 5 min. Behavior was tracked using an automated system (Ethovision, Noldus).
Marble burying
The marble burying test for anxiety-like behaviors (Anderson et al., 2019) was conducted under red light conditions. After one hour of acclimation to the testing room, mice were put in a standard mouse cage filled with 15 cm of corn cob bedding topped with 20 glass marbles. After 15 min, the mice were removed and the number of marbles fully or partiality buried was counted.
Forced swim test
The forced swim test was performed following 1 h of habitation to the testing room. Mice were tested in a 4 L glass beaker, containing 3 L of water at 25±1°C for 6 min. The behavior was video tracked using an automated system (Ethovision, Noldus) (Pena et al., 2017).
Tail suspension test
The tail suspension test was performed following 1 h of habitation to the testing room. Mice were taped from there tail for a 10 min video recorded trail. Time spent immobile was manually scored by an observer blind to the mice group.
Corticosterone assay
Trunk blood was collected 5 h after lights on from both male and female mice exposed to CSDS or control conditions. The blood was allowed to clot for 2 h at room temperature, centrifuged at 2000xg for 20 min at 4°C, and serum was removed and stored at −80°C. Corticosterone was quantified by an enzyme-linked immunosorbent assay (Parameter Corticosterone Assay KGE009, R&D Systems). All samples were analyzed in duplicate and were within the standard curve.
Viral constructs
LINC00473 was prepared (Gene Script) and Gateway (Invitrogen) cloned into a p1005+ HSV vector. This vector expresses eGFP with a CMV promoter while the gene of interest is driven by the IE4/5 promoter. Empty p1005+ expressing eGFP only was used as the control. Viral vector expression was confirmed in vivo by epifluorescence microscopy and qPCR.
Stereotactic surgery
Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) and positioned in a small-animal stereotactic instrument (Kopf Instruments). The skull surface was exposed and 33-g syringe needles (Hamilton) were used to bilaterally infuse 0.5 μl of HSV vector at a rate of 0.1 μl/min. mPFC coordinates relative to the Bregma were: AP + 1.8 mm; ML +0.75 mm and DV −2.7 mm with 15° angle.
Electrophysiology
Mice were injected with HSVs expressing LINC00473 plus GFP or GFP alone two days before electrophysiological recordings. Virally-infected neurons were distinguished from non-infected neurons by their GFP signals under epifluorescence microscopy. Recordings were restricted to pyramidal neurons that were determined based on morphological criteria and firing patterns (Yang et al., 1996). Spontaneous EPSCs (sEPSCs) of pyramidal neurons were recorded for 3 min in the voltage-clamp mode, with a K-based internal solution (in mM: 130 K-methanesulfate, 10 KCl, 10 HEPES, 0.4 EGTA, 2.0 MgCl2, 3 MgATP, 0.5 Na3GTP, 7.5 phosphocreatine, pH 7.4; 285 mOsm). Frequency and amplitude of sEPSCs were analyzed with Minianalysis Program (Synaptosoft).
Antisense oligonucleotides (ASOs) for LINC00473
Five sequences of antisense LNA GapmeRs (Exiqon) targeting LINC00473 were ordered and screened for knockdown efficiency in SH-SY5Y cells compared to negative control A (Cat. #: 300610) and positive control targeting GAPDH (Cat. #: 300632–411) ASOs. See Supplementary Table S3 for list of ASOs. RNAiMAX and Opti-MEM (Thermo Fisher Scientific) were used for transfection according to manufacturer instructions. RT-qPCR was used for probing LINC00473 downregulation. The most effective probes were then validated in male and female NPCs.
ChIRP
Chromatin isolation by RNA purification (ChIRP) was performed to identify the DNA binding sites of LINC00473 (Chu et al., 2012). Antisense oligonucleotide probes for LINC00473 (n = 13) were designed using an online tool (LGC Biosearch Technologies; see Supplementary Table S3 for list of probes) and synthesized (IDT) to include 3′-biotinylated ends along with probes for LacZ as control. The probes were numerically labeled according to the sequential position on the RNA as “even” and “odd”. SH-SY5Y human female-derived neuroblastoma cells were grown to confluence and cross-linked with 1% glutaraldehyde 6 h after adding forskolin (10 μM) or DMSO as control. Next, the cells were lysed and sonicated for 120 min (Bioruptor). Shearing was validated using a DNA high sensitivity Bioanalyzer (Agelint). The chromatin obtained from 10 million cells per condition was hybridized with the biotinylated antisense probes overnight at 37°C. Streptavidin-conjugated magnetic beads were used to capture the biotinylated probes, followed by five stringent washes. From the ChIRP chromatin, RNA was purified from the beads using a miRNeasy kit (Qiagen). DNA was purified after RNase A, RNase H, and proteinase K treatment. RNA was subjected to RT and qPCR to detect LINC00473 enrichment; DNA was subjected to high-throughput sequencing.
Human postmortem FISH
Florescent in situ hybridyzation (FISH) was performed on an independent cohort of male and female depressed cases (N=10 per group) (Supplementary Table S2). The samples were obtained by The University of Texas Southwestern Medical Center at Dallas with consent from the next of kin. Collection criteria included <24 hour postmortem interval (PMI), and no direct head trauma or other medical illness. The brains were taken from the scull, put on ice, dissected, and flash frozen. Samples were stored at −80°C in plastic under vacuum seal. Diagnosis was performed based on hospital and medical records and interview with next of kin. This information was reviewed by four clinicians that came to a consensus diagnosis using DSM-IV criteria. Subjects were matched to controls for age, pH, RNA integrity number (RIN), and PMI.
FISH was performed using RNAscope in human rostral anterior cingulate cortex (rACC) layers II-III. Fixed-frozen tissue samples were cryostat sectioned (14 μm) and mounted on SuperFrost Plus slides. RNAscope Multiplex Fluorescent reagent kit and probes for LINC00473 and positive (HS-PPIB, Cat. #: 313901) and negative (DapB, Cat. #: 310043) controls (ACD) and fluorophore (TSA) were used according to manufacturer instructions. Briefly, the slides were subjected to pretreatment, including hydrogen peroxide, epitope retrieval in 100°C water bath and digestion enzyme. Next, the sections received LINC00473 probe in parallel to positive and negative controls and hybridized for 2 hours at 40°C. Amplification and detection was performed using TSA Fluorescein (1:750) prior to counterstain with Dapi. Confocal images were acquired using a Zeiss LSM800 confocal microscope. High-resolution and high-magnification tiling images were taken from the region of interest (ROI): cortical layers II-III. Quantification was performed with Fiji/ImageJ (NIH). The number of puncta was extracted and density was calculated by dividing the size of ROI.
RNA extraction
For in vivo samples, mice were cervically dislocated and the brains were rapidly removed. Bilateral 1 mm-thick 14 g microdissections of mPFC were taken from fresh slices and flash-frozen on dry ice. Total RNA was isolated with TriZol reagent (Invitrogen) and purified with RNeasy Micro Kits (Qiagen). For cell culture samples, a RNAeasy kit mini (Qiagen) was used for total RNA. All samples were tested for concentration and purified using a NanoDrop (Thermo Fisher). Samples used for RNA- seq were analyzed using a bioanalyzer RNA Nano chip (Agilent) for integrity.
RT and qPCR
RNA amount was normalized across samples and cDNA was created using iScript (Bio-Rad). See Supplementary Table S3 for the sequences of primers used. Real-time PCR reactions were run in triplicate and SYBR-green was used in a QuantStudio 7 (ThermoFisher) qPCR machine. The 2-ΔΔCt method was used to calculate relative gene expression normalized to controls with Hprt1 as control gene. See Key Resources Table for primer sequences.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Clozapine N-oxide | Enzo Life Sciences | Cat. #: BML-NS105 |
| Lipofectamine RNAiMAX | Thermo Fisher | Cat. #: 13778150 |
| Critical Commercial Assays | ||
| RNeasy Micro Kits | Qiagen | Cat. #: 74004 |
| iScript | BioRad | Cat. #: 1708891 |
| Fast SYBR™ Green Master Mix | Applied Biosystems | Cat. #: 4334973 |
| ScriptSeq Complete Kit | Epicentre | Cat. #: BHMR1224 |
| Corticosterone Assay | R&D Systems | Cat. #: KGE009 |
| RNAscope Multiplex Fluorescent reagent kit version 2 | Advanced Cell Diagnostics | Cat. #: 323110 |
| Fluorescein plus evaluation kit | Perkin Elmer | Cat. #: 741E001KT |
| NEBNext Ultra DNA Library Prep kit | New England Biolabs | Cat. #: E7645S/L |
| Deposited Data | ||
| RNA-Seq Human depression postmortem | (Labonté et al., 2017) | N/A |
| RNA-Seq Human depression postmortem | (Zhou et al., 2018) | N/A |
| RNA-seq LINC00473 KD in NPCs | This Study | GSE138677 |
| RNA-seq LINC00473 expression in mouse | This Study | GSE138677 |
| ChIRP seq LINC00473 SHSY-5Y cells | This Study | GSE138677 |
| Experimental Models: Cell Lines | ||
| Neuron Progenitor Cells (NPCs) | (Hoffman et al., 2017) | N/A |
| SH-SY5Y | ATCC | ATCC CRL-2266 |
| Experimental Models: Organisms/Strains | ||
| C57Bl6/j | Jackson Laboratory | Cat. #: 000664 |
| Oligonucleotides sequences listed in Supplementary Table S3 | ||
| HS-LINC00473 probes (RNAScope) | Advanced Cell Diagnostics | Cat. #: 464821 |
| Recombinant DNA | ||
| p1005-LINC00473-eGFP | This study | N/A |
| p1005-eGFP | This study | N/A |
| rAAV2/hSyn-DIO-hM3D(Gq)-mCherry | (Krashes et al., 2011) | Addgene, Cat. #: 44361-AAV2 |
| Software and Algorithms | ||
| Ethovision XT | Noldus | https://www.noldus.com/ethovision-xt |
| Fiji/ImageJ | NIH | https://imagej.net/Fiji/Downloads |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| R | GNU package | https://www.r-project.org/ |
| QuantStudio 7 | Thermo Fisher | https://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-6-7-flex-software.html |
Sequencing library preparation
RNA libraries were prepared separately from female mouse mPFC infected with LINC00473 or GFP, and from male and female NPCs transfected with LINC00473 ASO or control ASO. The successful manipulation of LINC00473 was confirmed in the samples used for sequencing. Libraries were made using 1 μg of purified RNA using the ScriptSeq Complete Kit (Epicentre). Briefly, the cDNA was synthesized from ribosomal depleted RNA and then fragmented total RNA using random hexamers, followed by terminal tagging. The libraries were PCR amplified and then purified using AMPure XP beads (Beckman Coulter, Brea, CA). Barcode bases (6 bp) were introduced at one end of the adaptors during PCR amplification steps. Quality and concentration of libraries were confirmed on a Bioanalyzer (Agilent) and libraries were sequenced on an Illumina Hi-seq machine with 150-bp paired-end reads in Genewiz. Samples were multiplexed to produce >40 million reads per sample. Sample size was n=3–5 independent samples per group.
ChIRP-seq libraries were prepared from samples from DMSO- or forskolin-treated SH-SY5Y cells purified with two sets of probes (‘odd’ and ‘even’) for LINC00473 or LacZ and input samples. The libraries were made using the entire precipitation product with the NEBNext Ultra DNA Library Prep kit for Illumina (New England BioLabs), without size selection, as recommended for low-input samples. Barcodes were used to multiplex samples to achieve >60 million reads/sample. Library size and quality were assessed on a BioAnalyzer prior to sequencing on an Illumina Hi-seq machine with 150- bp paired-end reads at Genewiz.
RNA-seq analysis of human postmortem brain, mouse brain, and NPCs
Human postmortem RNA-seq data from our published study (Lebonte et al., 2017) include forty-eight subjects with tissue collected from six brain regions—orbitofrontal cortex (OFC; BA11), dorsolateral PFC (BA8/9; dlPFC), cingulate gyrus 25 (BA25; cg25; vmPFC), anterior insula (aINS), nucleus accumbens (NAc), and ventral subiculum (vSUB). Tissue was dissected by histopathologists using reference neuroanatomical maps at 4°C followed by flash-freezing in isopentane at −80°C. The study was approved by the research ethics boards of McGill University. Psychiatric history and socio-demographics were obtained for both cases and controls by “psychological autopsies” performed by clinicians with informants best acquainted with the deceased. Diagnoses were obtained using DSM-IV criteria by means of SCID-I interviews adapted for psychological autopsies. In this original RNA-seq analysis (Labonté et al., 2017)—in addition to phenotype (depressed vs. control), sex and brain region—RIN, age and alcohol had a significant effect on the overall variance in the dataset, whereas antidepressant use did not. All of these factors were included in the statistical models as covariates. By contrast, other drugs, cause of death, history of child abuse or other early life adversity and toxicology findings were not significantly associated with global variance of gene expression.
The correlation analysis between lncRNA and PCG expression data was converted to logCPM and normalized across brain regions for each sex separately, combining the depressed and control subjects to obtain sufficient statistical power. Permutation analysis of the same number of random genes iterated 10,000 times was used to calculate a threshold for significant Pearson correlation at R>0.58771 or R<−0.59516.
For RNA-seq differential expression analysis, raw reads were aligned to the mouse genome (mm10) for mouse PFC or to the human genome (hg38) for NPC data using HISAT2 (Kim et al., 2015). The average mapping rates for mouse and human studies were 84% and 89%, respectively. Quality control was performed using FastQC (Andrews, 2010) and principal component analysis was used to identify outliers. Normalization and differential expression analysis were performed using DESeq2 (Love et al., 2014). Raw data was deposited in GEO (GSE138677). Significance was set at uncorrected P<0.05 for broad pattern identification. A fold-change (FC) threshold was set at >1.3. See Supplementary Table S4 and S5 for genes differential expression lists. A similar approach was used for re-analyzing the published data of lncRNA regulation in depression (Zhou et al., 2018), but including sex as variable, which was not performed in the original publication. Heatmaps were generated using Morpheus and gene ontology analysis was performed using GOrilla (Eden et al., 2009). Upstream regulator predictions were made using Ingenuity Pathway Analysis (IPA, Qiagen). Comparison of RNA-seq data from mouse PFC and from human NPCs showed very little overlap between the two datasets, consistent with the knowledge that NPCs are highly immature cells with a very distinct transcriptome. Accordingly, very little overlap was seen between RNAs regulated by LINC00473 expression in mouse PFC vs. those regulated by LINC00473 knockdown in human NPCs, although we did identify a subset of RNAs regulated oppositely under these two conditions (not shown).
ChIRP-seq data analysis
ChIRP-seq data was checked for quality using FastQC. Raw sequencing reads were then aligned to the human hg19 genome using default settings of Bowtie where ~92% were successfully aligned. Raw data was deposited in GEO (GSE138677). The alignment data were visualized using the Integrative Genomics Viewer (IGV) programPeak-calling was performed using MACS with a FDR cutoff of 0.05. Bedtools was used to identify peaks common between data generated using the ‘odd’ and ‘even’ probes requiring at least 20% overlap. See Supplementary Table S6 for picks lists. HOMER (Heinz et al., 2010) was used to identify and predict DNA binding motifs. LiftOver tool was used to convert the human genome peaks to mouse genome following by annotation using HOMER.
Quantification and statistical analysis
All statistics were performed using Prizm 8. Outliers were defined as more than 2.5 standard deviations from the group mean and were removed from the analysis. Main effects and interactions were determined using one- or two-way ANOVA with Fisher LSD post hoc analysis. Student’s t test was used for comparisons of only two groups. All significance thresholds were set at P<0.05. Data in figures represent mean ± SEM, with sample size in figure legends and detailed statistic values listed in Results.
Supplementary Material
Highlights.
lncRNAs are robustly altered in depression in a sex- and brain-site specific manner
LINC00473 is downregulated in cortex of depressed female, but not in males
LINC00473 expression in mouse cortex promotes stress resilience in females only
LINC00473 regulates gene expression and physiology in a sex-specific manner
Acknowledgments
This work was funded by R01MH51399 and P50MH096890 (EJN), the Hope for Depression Research Foundation, and by a NARSAD Young Investigator Award (OI). We wish to thank Igor Ulitsky for his insight regarding LINC00473 evolution and Jill Gregory for her illustration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests
Additional resources
Supplemental spreadsheets
Supplementary Table Supplementary Table S4. List of genes differentially expressed in mouse mPFC by CVS in female mice expressing GFP or LINC00473. Related to Figure 5.
Supplementary Table S5. List of genes differentially expressed in human NPCs from both sexes by LINC00473 ASO vs control ASO. Related to Figure 6.
Supplementary Table S6. List of picks from LINC00473 ChIRP-seq in SH-SY5Y cells in DMSO (control) or forskolin treatments. Related to Figure 7.
References
- Akil H, Gordon J, Hen R, Javitch J, Mayberg H, McEwen B, Meaney MJ, and Nestler EJ (2018). Treatment resistant depression: A multi-scale, systems biology approach. Neurosci Biobehav Rev 84, 272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EM, Sun H, Guzman D, Taniguchi M, Cowan CW, Maze I, Nestler EJ, and Self DW (2019). Knockdown of the histone di-methyltransferase G9a in nucleus accumbens shell decreases cocaine self-administration, stress-induced reinstatement, and anxiety. Neuropsychopharmacology 44, 1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). FastQC: a quality control tool for high throughput sequence data.
- Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M, Yap EL, Malik AN, Mei K, Rubin AA, Spiegel I, et al. (2016). Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Zhao WY, Li WJ, Ying ZW, and Jiang DQ (2019). Long noncoding RNA LINC00473 indicates a poor prognosis of breast cancer and accelerates tumor carcinogenesis by competing endogenous sponging miR-497. Eur Rev Med Pharmacol Sci 23, 3410–3420. [DOI] [PubMed] [Google Scholar]
- Bale TL, and Epperson CN (2015). Sex differences and stress across the lifespan. Nat Neurosci 18, 1413–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangasser DA, and Valentino RJ (2014). Sex differences in stress-related psychiatric disorders: neurobiological perspectives. Front Neuroendocrinol 35, 303–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry G (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry 19, 410–416. [DOI] [PubMed] [Google Scholar]
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, et al. (2006). Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science 311, 864–868. [DOI] [PubMed] [Google Scholar]
- Bi LL, Sun XD, Zhang J, Lu YS, Chen YH, Wang J, Geng F, Liu F, Zhang M, Liu JH, et al. (2015). Amygdala NRG1-ErbB4 is critical for the modulation of anxiety-like behaviors. Neuropsychopharmacology 40, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA Jr., Duman RS, and Nestler EJ (2005). The many faces of CREB. Trends Neurosci 28, 436–445. [DOI] [PubMed] [Google Scholar]
- Carson RP, Fu C, Winzenburger P, and Ess KC (2013). Deletion of Rictor in neural progenitor cells reveals contributions of mTORC2 signaling to tuberous sclerosis complex. Hum Mol Genet 22, 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereseto M, Reines A, Ferrero A, Sifonios L, Rubio M, and Wikinski S (2006). Chronic treatment with high doses of corticosterone decreases cytoskeletal proteins in the rat hippocampus. Eur J Neurosci 24, 3354–3364. [DOI] [PubMed] [Google Scholar]
- Chen H, Yang F, Li X, Gong ZJ, and Wang LW (2018a). Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem Biophys Res Commun 499, 702–710. [DOI] [PubMed] [Google Scholar]
- Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al. (2016). cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest 126, 2267–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Lin S, Li JL, Ni W, Guo R, Lu J, Kaye FJ, and Wu L (2018b). CRTC1-MAML2 fusion-induced lncRNA LINC00473 expression maintains the growth and survival of human mucoepidermoid carcinoma cells. Oncogene 37, 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Quinn J, and Chang HY (2012). Chromatin isolation by RNA purification (ChIRP). J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Yamazaki T, Kawaguchi T, Kurosaka S, Takumi T, Nakagawa S, and Hirose T (2017). Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. Embo J 36, 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung DW, Chung Y, Bazmi HH, and Lewis DA (2018). Altered ErbB4 splicing and cortical parvalbumin interneuron dysfunction in schizophrenia and mood disorders. Neuropsychopharmacology 43, 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington HE 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, LaPlant Q, Mouzon E, Ghose S, Tamminga CA, et al. (2010). Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci 30, 16082–16090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, and Zhang L (2017). Long noncoding RNA expression in peripheral blood mononuclear cells and suicide risk in Chinese patients with major depressive disorder. Brain Behav 7, e00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang R, Cai H, Zhang L, Liang D, Lv C, Guo Y, Yang R, Zhu Y, and Jiang P (2016). Dysregulation of Neuregulin-1/ErbB signaling in the prefrontal cortex and hippocampus of rats exposed to chronic unpredictable mild stress. Physiol Behav 154, 145–150. [DOI] [PubMed] [Google Scholar]
- Datson NA, Speksnijder N, Mayer JL, Steenbergen PJ, Korobko O, Goeman J, de Kloet ER, Joels M, and Lucassen PJ (2012). The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus 22, 359–371. [DOI] [PubMed] [Google Scholar]
- Delacretaz A, Preisig M, Vandenberghe F, Saigi Morgui N, Quteineh L, Choong E, Gholam-Rezaee M, Kutalik Z, Magistretti P, Aubry JM, et al. (2015). Influence of MCHR2 and MCHR2-AS1 Genetic Polymorphisms on Body Mass Index in Psychiatric Patients and In Population-Based Subjects with Present or Past Atypical Depression. PLoS One 10, e0139155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonte B, Ribeiro E, et al. (2014). beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TA, Vitucci EC, Wauthier E, Graham RP, Pitman WA, Oikawa T, Chen M, Silva GO, Greene KG, Torbenson MS, et al. (2017). Comprehensive analysis of The Cancer Genome Atlas reveals a unique gene and non-coding RNA signature of fibrolamellar carcinoma. Sci Rep 7, 44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EC, Wiste A, Radmanesh F, Almli LM, Gogarten SM, Sofer T, Faul JD, Kardia SL, Smith JA, Weir DR, et al. (2016). Genome-Wide Association Study (Gwas) and Genome-Wide by Environment Interaction Study (Gweis) of Depressive Symptoms in African American and Hispanic/Latina Women. Depress Anxiety 33, 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, and Yakhini Z (2009). GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, Vos T, and Whiteford HA (2013). Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med 10, e1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx C (2013). The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han PB, Ji XJ, Zhang M, and Gao LY (2018). Upregulation of lncRNA LINC00473 promotes radioresistance of HNSCC cells through activating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci 22, 7305–7313. [DOI] [PubMed] [Google Scholar]
- Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, Hornstein E, and Chen A (2011). MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci 31, 14191–14203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, et al. (2015). Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J Neurosci 35, 16362–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM, Stahl EA, Rapoport J, Sklar P, and Brennand KJ (2017). Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat Commun 8, 2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, et al. (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YT, Liao G, Bi X, Oka T, Tamura S, and Baudry M (2011). The PDE10A inhibitor, papaverine, differentially activates ERK in male and female rat striatal slices. Neuropharmacology 61, 1275–1281. [DOI] [PubMed] [Google Scholar]
- Issler O, and Nestler EJ (2018). The molecular basis for sex differences in depression susceptibility. Current Opinion in Behavioral Sciences 23, 1–6. [Google Scholar]
- Kalil B, Leite CM, Carvalho-Lima M, and Anselmo-Franci JA (2013). Role of sex steroids in progesterone and corticosterone response to acute restraint stress in rats: sex differences. Stress 16, 452–460. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Koda S, Ye C, Rogan SC, Adams AC, Cusher DS, Maratos-Flier E, Roth BL and Lowell BB (2011). Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 4, 1424–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaska S, Brunk R, Bali V, Kechner M, and Mazei-Robison MS (2017). Deletion of Rictor in catecholaminergic neurons alters locomotor activity and ingestive behavior. Neuropharmacology 117, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlenkov A, Wang M, Roussos P, Rudchenko S, Barbu M, Bibikova M, Klotzle B, Dwork AJ, Zhang B, Hurd YL, et al. (2016). Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic Acids Res 44, 2593–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, et al. (2007). Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell 131, 391–404. [DOI] [PubMed] [Google Scholar]
- Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, Scarpa JR, Moy G, Loh YE, Cahill M, et al. (2017). Sex-specific transcriptional signatures in human depression. Nat Med 23, 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, and Behlke MA (2016). Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Cao F, Li S, Huang S, Li W, and Abumaria N (2017). Profiling and Co-expression Network Analysis of Learned Helplessness Regulated mRNAs and lncRNAs in the Mouse Hippocampus. Front Mol Neurosci 10, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pehrson AL, Waller JA, Dale E, Sanchez C, and Gulinello M (2015). A critical evaluation of the activity-regulated cytoskeleton-associated protein (Arc/Arg3.1)’s putative role in regulating dendritic plasticity, cognitive processes, and mood in animal models of depression. Front Neurosci 9, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Deng WB, Liu YF, Liang YX, Fan ZM, Gu XW, Liu JL, Sha AG, Diao HL, and Yang ZM (2016). Non-coding RNA LINC00473 mediates decidualization of human endometrial stromal cells in response to cAMP signaling. Sci Rep 6, 22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li X, Sun N, Xu Y, Meng Y, Yang C, Wang Y, and Zhang K (2014). Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS One 9, e93388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch ZS, Hamilton PJ, Ramakrishnan A, Parise EM, Salery M, Wright WJ, Lepack AE, Mews P, Issler O, McKenzie A, et al. (2019). Stress resilience is promoted by a Zfp189-driven transcriptional network in prefrontal cortex. Nat Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch ZS, Loh YE, Purushothaman I, Walker DM, Parise EM, Salery M, Cahill ME, Hodes GE, Pfau ML, Kronman H, et al. (2018). Estrogen receptor alpha drives pro-resilient transcription in mouse models of depression. Nat Commun 9, 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMullen CM, Vick K, Pacifico R, Fallahi-Sichani M, and Davis RL (2016). Novel, primate-specific PDE10A isoform highlights gene expression complexity in human striatum with implications on the molecular pathology of bipolar disorder. Transl Psychiatry 6, e742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki K, Lourdusamy A, Binder E, Paya-Cano J, Sluyter F, Craig I, Keers R, McGuffin P, Uher R, and Schalkwyk LC (2012). Antidepressant-dependent mRNA changes in mouse associated with hippocampal neurogenesis in a mouse model of depression. Pharmacogenet Genomics 22, 765–776. [DOI] [PubMed] [Google Scholar]
- Martin LA, Neighbors HW, and Griffith DM (2013). The experience of symptoms of depression in men vs women: analysis of the National Comorbidity Survey Replication. JAMA Psychiatry 70, 1100–1106. [DOI] [PubMed] [Google Scholar]
- Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, and Johnson R (2017). LncATLAS database for subcellular localization of long noncoding RNAs. Rna 23, 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T, and Waxman DJ (2019). Sex-Biased lncRNAs Inversely Correlate With Sex-Opposite Gene Coexpression Networks in Diversity Outbred Mouse Liver. Endocrinology 160, 989–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Yan Y, Eipper BA, and Mains RE (2013). Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist 19, 255–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsetti M, Di Brisco F, Canonico PL, Genazzani AA, and Ghi P (2008). Gene regulation in the frontal cortex of rats exposed to the chronic mild stress paradigm, an animal model of human depression. Eur J Neurosci 27, 2156–2164. [DOI] [PubMed] [Google Scholar]
- Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, Mohr DC, and Schatzberg AF (2016). Major depressive disorder. Nat Rev Dis Primers 2, 16065. [DOI] [PubMed] [Google Scholar]
- Pei Q, Burnet PJ, and Zetterstrom TS (1998). Changes in mRNA abundance of microtubule-associated proteins in the rat brain following electroconvulsive shock. Neuroreport 9, 391–394. [DOI] [PubMed] [Google Scholar]
- Pena CJ, Kronman HG, Walker DM, Cates HM, Bagot RC, Purushothaman I, Issler O, Loh YE, Leong T, Kiraly DD, et al. (2017). Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356, 1185–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR, Bos SD, Deelen J, den Heijer M, Freathy RM, et al. (2013). A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 9, e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Bengtson CP, and Bading H (2017). Networks of Cultured iPSC-Derived Neurons Reveal the Human Synaptic Activity-Regulated Adaptive Gene Program. Cell Rep 18, 122–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, and Chang HY (2016). Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17, 47–62. [DOI] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, and Khavari PA (2018). The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinius B, Shi C, Hengshuo L, Sandhu KS, Radomska KJ, Rosen GD, Lu L, Kullander K, Williams RW, and Jazin E (2010). Female-biased expression of long non-coding RNAs in domains that escape X-inactivation in mouse. BMC Genomics 11, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, and Chang HY (2012). Genome regulation by long noncoding RNAs. Annu Rev Biochem 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Wang Q, and Dwivedi Y (2018). Long Noncoding RNA-Associated Transcriptomic Changes in Resiliency or Susceptibility to Depression and Response to Antidepressant Treatment. Int J Neuropsychopharmacol 21, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan X, Li P, Chen Y, Shi Y, Pirooznia M, Seifuddin F, Suemizu H, Ohnishi Y, Yoneda N, Nishiwaki M, et al. (2020). In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun 11, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush AJ (2007). STAR*D: what have we learned? Am J Psychiatry 164, 201–204. [DOI] [PubMed] [Google Scholar]
- Sahakyan A, Yang Y, and Plath K (2018). The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol 28, 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C, Diaz-Nido J, and Avila J (2000). Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog Neurobiol 61, 133–168. [DOI] [PubMed] [Google Scholar]
- Seedat S, Scott KM, Angermeyer MC, Berglund P, Bromet EJ, Brugha TS, Demyttenaere K, de Girolamo G, Haro JM, Jin R, et al. (2009). Cross-national associations between gender and mental disorders in the World Health Organization World Mental Health Surveys. Arch Gen Psychiatry 66, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Yamagata H, Uchida S, Chen C, Kobayashi A, Kobayashi M, Harada K, Matsuo K, Watanabe Y, and Nakagawa S (2019). Altered expression of long noncoding RNAs in patients with major depressive disorder. J Psychiatr Res 117, 92–99. [DOI] [PubMed] [Google Scholar]
- Seney ML, Huo Z, Cahill K, French L, Puralewski R, Zhang J, Logan RW, Tseng G, Lewis DA, and Sibille E (2018). Opposite Molecular Signatures of Depression in Men and Women. Biol Psychiatry 84, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Yang Y, Yu J, Meng F, Zhang T, and Gao Y (2017). The long noncoding RNA LINC00473, a target of microRNA 34a, promotes tumorigenesis by inhibiting ILF2 degradation in cervical cancer. Am J Cancer Res 7, 2157–2168. [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Bayuga SJ, and Beavo JA (1999). Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc Natl Acad Sci U S A 96, 7071–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MB, and Herman JP (2009). Sex differences in psychopathology: of gonads, adrenals and mental illness. Physiology & behavior 97, 250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sramek JJ, Murphy MF, and Cutler NR (2016). Sex differences in the psychopharmacological treatment of depression. Dialogues Clin Neurosci 18, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, and Kapranov P (2015). The Landscape of long noncoding RNA classification. Trends Genet 31, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, Koo JW, Korb E, Bagot RC, Ahn FH, et al. (2015). ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med 21, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Chung JR, Zhang S, Zhang H, Grossman Y, Aleyasin H, Flanigan ME, Pfau ML, Menard C, Dumitriu D, et al. (2017). Establishment of a repeated social defeat stress model in female mice. Sci Rep 7, 12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, and Bartel DP (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk N, Pape JC, Engel M, Zannas AS, Cattane N, Cattaneo A, Binder EB, and Chen A (2016). Amygdalar MicroRNA-15a Is Essential for Coping with Chronic Stress. Cell Rep 17, 1882–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, David DJ, Monckton JE, Battaglia F, and Hen R (2008). Chronic fluoxetine stimulates maturation and synaptic plasticity of adult-born hippocampal granule cells. J Neurosci 28, 1374–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang X, Sheng L, Qiu C, and Luo R (2018). LINC00473 promotes the Taxol resistance via miR-15a in colorectal cancer. Biosci Rep 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Roy B, and Dwivedi Y (2019). Co-expression network modeling identifies key long non-coding RNA and mRNA modules in altering molecular phenotype to develop stress-induced depression in rats. Transl Psychiatry 9, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2017). Depression and Other Common Mental Disorders: Global Health Estimates.
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Xu Y, Zou Y, Zuo Q, Huang S, Wang S, Lu X, He X, Wang J, Wang T, et al. (2018). Long Noncoding RNA 00473 Is Involved in Preeclampsia by LSD1 Binding-Regulated TFPI2 Transcription in Trophoblast Cells. Mol Ther Nucleic Acids 12, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CR, Seamans JK, and Gorelova N (1996). Electrophysiological and morphological properties of layers V-VI principal pyramidal cells in rat prefrontal cortex in vitro. J Neurosci 16, 1904–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Rao S, Du T, Hu H, Liu Z, Shen Y, and Xu Q (2017). Intergenic variants may predispose to major depression disorder through regulation of long non-coding RNA expression. Gene 601, 21–26. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Navarro P, Shirali M, Howard DM, Adams MJ, Hall LS, Clarke TK, Thomson PA, Smith BH, Murray A, et al. (2017). Genome-wide Regional Heritability Mapping Identifies a Locus Within the TOX2 Gene Associated With Major Depressive Disorder. Biol Psychiatry 82, 312–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang Y, Li X, Xia X, Li N, He R, He H, Han C, and Zhao W (2017). ZBTB7A Enhances Osteosarcoma Chemoresistance by Transcriptionally Repressing lncRNALINC00473-IL24 Activity. Neoplasia 19, 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, and Song Y (2018). LINC00473 predicts poor prognosis and regulates cell migration and invasion in gastric cancer. Biomed Pharmacother 107, 1–6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, et al. (2016). NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44, D203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lutz PE, Wang YC, Ragoussis J, and Turecki G (2018). Global long non-coding RNA expression in the rostral anterior cingulate cortex of depressed suicides. Transl Psychiatry 8, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W, and Liu G (2018). LINC00473 antagonizes the tumour suppressor miR-195 to mediate the pathogenesis of Wilms tumour via IKKalpha. Cell Prolif 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIRP datasets generated during this study are available at GEO (GSE138677).


