Abstract
Structural characterization of misfolded protein aggregates is essential to understanding molecular mechanism of protein aggregation associated with various protein misfolding disorders. Here, we report structural analyses of ex vivo transthyretin aggregates extracted from human cardiac tissue. Comparative structural analyses of in vitro and ex vivo transthyretin aggregates using various biophysical techniques revealed that cardiac transthyretin amyloid has similar structural features to those of in vitro transthyretin amyloid. Our solid-state NMR studies showed that in vitro amyloid contains extensive native-like β-sheet structures, while other loop regions including helical structures are disrupted in the amyloid state. These results suggest that transthyretin undergoes a common misfolding and aggregation transition to native-like aggregation-prone monomers that self-assemble into amyloid precipitates in vitro and in vivo.
Graphical Abstract
Transthyretin (TTR) is a 55 kDa homo-tetrameric protein consisting of four 127-residue β-barrel subunits.1, 2 The primary role of TTR is to transport thyroid hormones and retinol binding protein in plasma and cerebrospinal fluid. TTR misfolding and aggregation is associated with numerous amyloidoses featuring cardiomyopathy and polyneuropathy.3, 4, 5, 6 For example, aggregation of wild type TTR (TTRwt) is implicated in senile systemic amyloidosis (SSA) that affects nearly 25 % of the population over the age of 80.7 In addition, more than 100 single point mutations have been identified and most of these TTR variants (TTRv) can undergo misfolding and aggregation, leading to onset of ATTR amyloidosis.
Previous extensive biochemical studies have revealed a detailed molecular mechanism of TTR misfolding and aggregation in vitro.8, 9, 10, 11, 12, 13, 14, 15 However, it remains unknown whether TTR undergoes the same misfolding and aggregation transition in vivo. In order to gain insights into the misfolding and aggregation in vivo, it is essential to examine TTR aggregates formed under physiological conditions (human tissue) and compare structural features of ex vivo and in vitro TTR amyloids. In this study, we carried out comparative structural analyses of in vitro as well as ex vivo TTR amyloid extracted from human cardiac tissues by using various biophysical techniques including FT-IR, transmission electron microscopy (TEM), and solid-state NMR.
The ex vivo TTRwt amyloid fibrils were extracted from cardiac tissue of an SSA patient.16 The cardiac extracts were examined with thioflavin T (ThT) fluorescence and polyacrylamide gel electrophoresis (PAGE). The enhanced ThT fluorescence confirmed the amyloid property of the cardiac extracts (Figure S1). SDS-PAGE analysis also showed that TTR is the major component of the amyloid consistent with previous structural analyses of the cardiac tissue deposited fibrils (Figure S2).16
Our previous mechanistic studies of TTR misfolding in vitro revealed that native-like aggregation-prone TTR monomers form dimers, which self-associate to form small spherical oligomers in vitro,12 as shown in Figure 1a and Figure S3. TTR aggregates extracted from human cardiac tissue was examined by TEM (Figure 1b and Figure S4) for comparative structural analysis. Small spherical oligomers, indistinguishable from in vitro oligomers, were also observed in ex vivo cardiac TTR amyloid.
Figure 1.
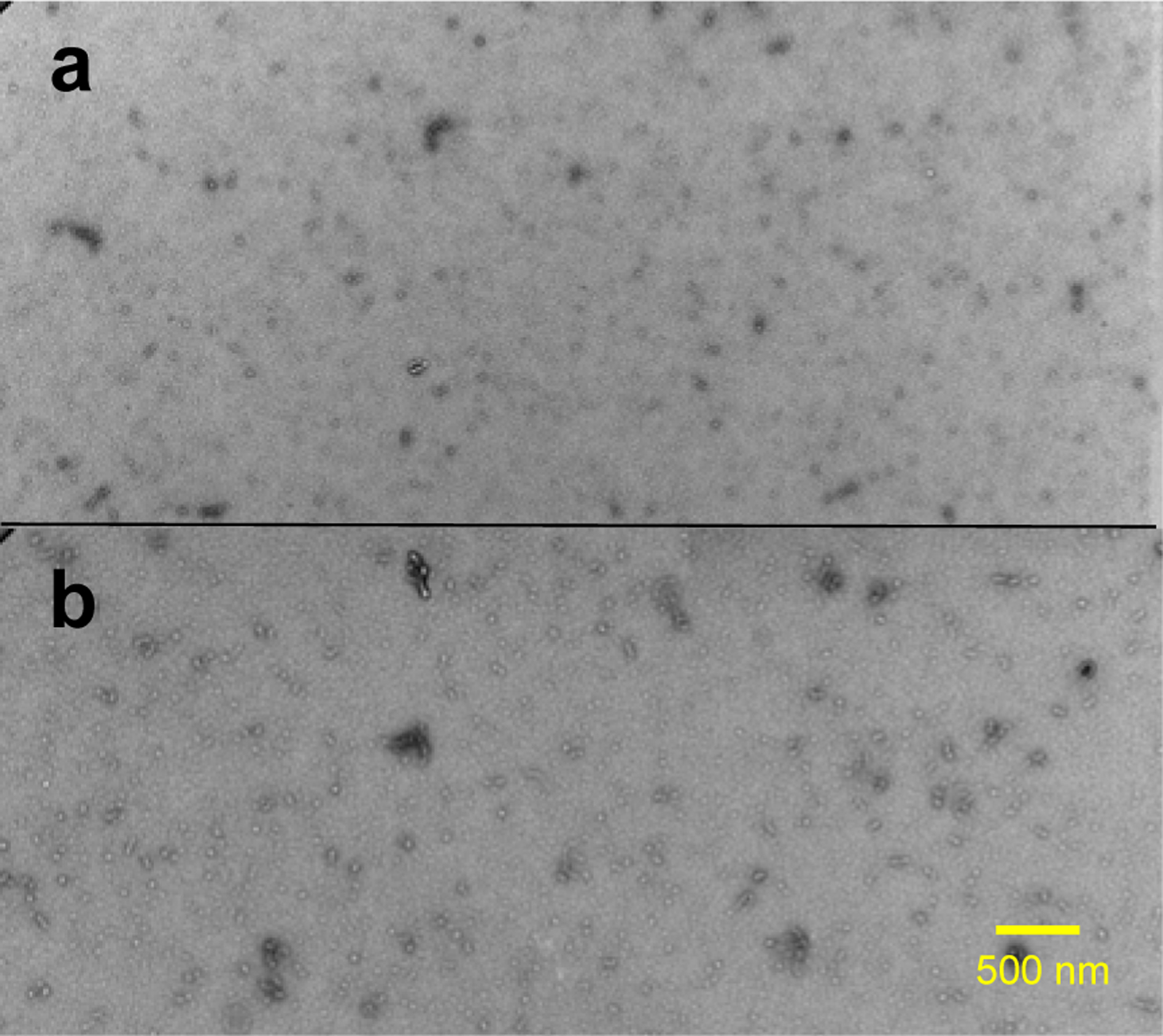
TEM images of in vitro (a) and ex vivo (b) TTR amyloid. The in vitro TTR amyloid was obtained by incubating recombinantly-generated monomeric TTR (mTTR; F87M/L110M) at a protein concentration of 0.5 mg/ml in PBS buffer (pH 7.4) at 37 °C for two days. The ex vivo TTR amyloid was extracted from human cardiac tissue (TTRwt).
Structural features of the in vitro and ex vivo TTR amyloids were further investigated using FT-IR (Figure 2 and Figure S5). Previous FT-IR studies showed that absorption peaks arising from the stretching vibrations of main-chain C=O groups (amide I) and from the N-H bending vibrations (amide II) in the peptide backbone are strongly sensitive to secondary structures and hydrogen bonding patterns in the amide backbone. Thus, FT-IR has been used to investigate structural features of amyloids formed by various aggregation-prone proteins.17, 18, 19 In our FT-IR studies of the in vitro and ex vivo TTR amyloids, the two amyloid states exhibited almost identical absorption peaks in the IR spectra, particularly the amide I (1600 – 1700 cm−1) and II (1500 – 1600 cm−1) bands (Figure 2), suggesting that the two amyloids exhibit similar structural characteristics. Protease K digestion analyses of the in vitro and ex vivo aggregates are also consistent with the TEM and FT-IR results (Figure S6).
Figure 2.
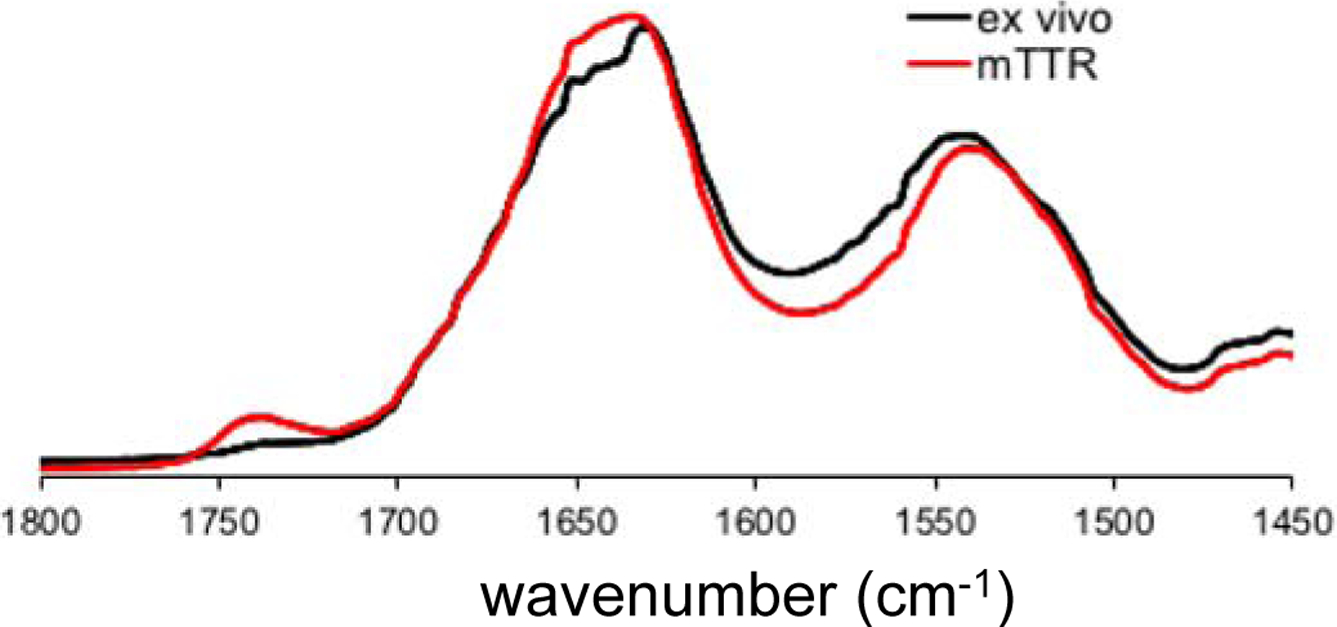
Amide I and II bands of the FT-IR spectra for in vitro mTTR (red) and ex vivo (black) TTR amyloid.
Our combined structural studies indicate that in vitro mTTR amyloid formed at pH 7.4 has similar structural properties to those of ex vivo cardiac TTR amyloid. However, previous mechanistic studies including our structural studies have utilized mildly acidic conditions (pH 4 – 5) to induce amyloid formation of TTRwt.12, 15, 20, 21 Thus, structural characteristics of the mTTR and TTRwt amyloids formed at pH 7.4 and 4.4, respectively, were compared by using solid-state NMR (Figure S7). The 13C chemical shift of backbone (Cα) and aliphatic sidechains are highly sensitive to local environments such as secondary structures and ϕ/ψ dihedral angles. Thus, the two-dimensional (2D) 13C-13C correlation NMR experiment is an ideal tool for comparative structural studies of biological molecules. In the overlaid 2D 13C-13C correlation NMR spectra, NMR cross-peaks between the backbone (50 – 65 ppm) and sidechain carbons (10 – 70 ppm), and between sidechain carbons (10 – 50 ppm) overlap well. The nearly identical 2D spectra for the two TTR amyloids clearly indicate that the TTR amyloid states have similar molecular conformations. These solid-state NMR results suggest that TTR has similar misfolding and aggregation pathways at both pH 7.4 and 4.4 and the mechanistic studies of TTR misfolding and aggregation at pH 4.4 may also provide valuable insight into in vivo TTR misfolding and aggregation.
Previous structural studies showed that TTR amyloid formed at pH 4.4 had extensive native-like β-sheet structures.20, 22, 23, 24, 25, 26, 27 In this study, more detailed solid-state NMR experiments were carried out to clearly identify the native-like β-structured regions and other regions that undergo misfolding transition. The 2D 13C-13C correlation NMR spectra were acquired and compared for the native (red) and amyloid (black) states of TTR (Figure 3). The NMR spectra for the two states overlap well, suggesting that the amyloid state contains extensive native-like structures. It is also notable that numerous cross-peaks from native TTR disappear in the amyloid state, indicative of structural changes during misfolding and aggregation.
Figure 3.
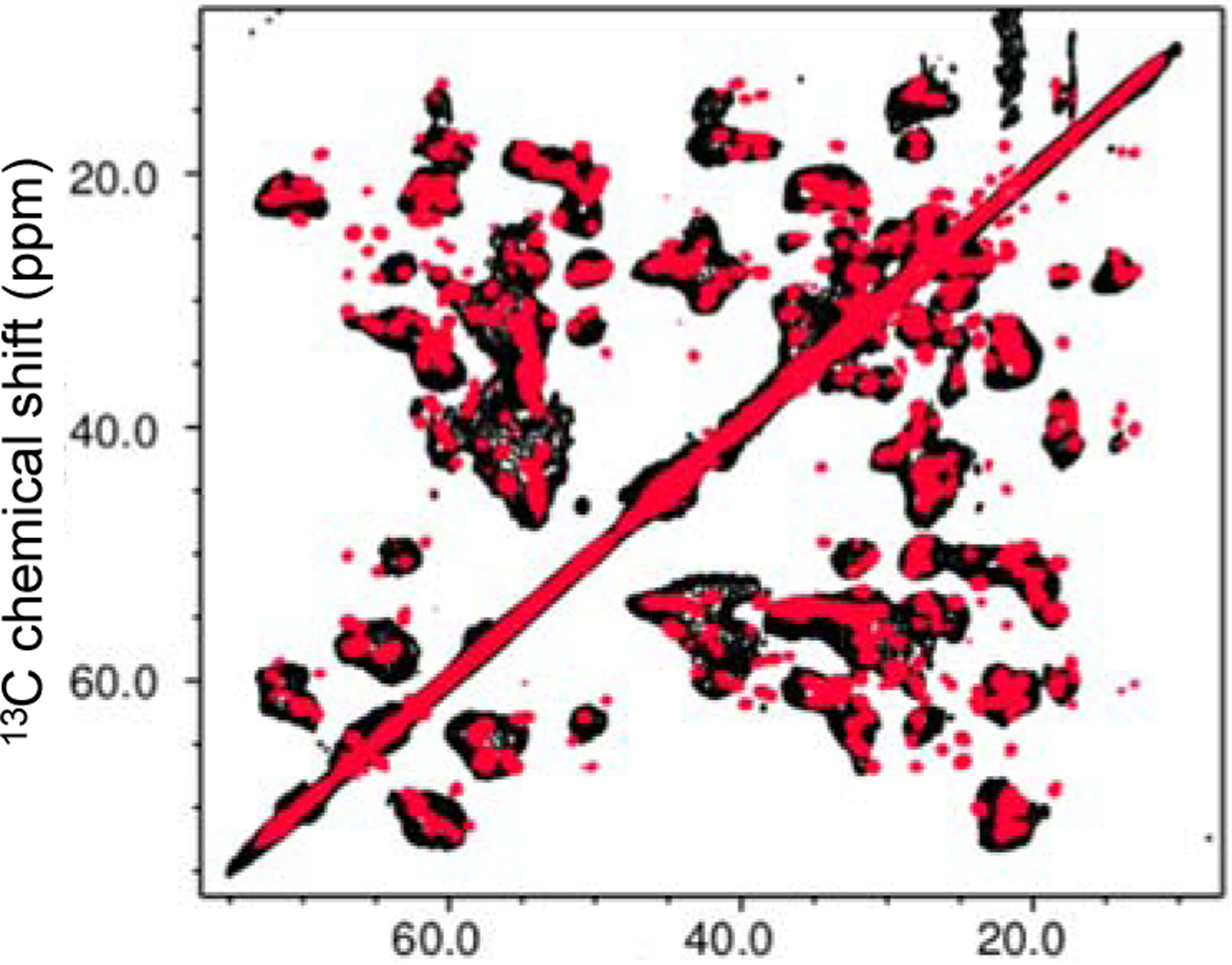
Overlaid two-dimensional 13C-13C correlation solid-state NMR spectra for the native (red) and amyloid (black) states of TTRwt formed at pH 4.4 obtained at a proton frequency of 800 MHz. A dipolar-assist rotational resonance (DARR) with 20 ms mixing time was used for the mixing scheme.
In order to identify the residues that undergo structural changes, three-dimensional solid-state NMR experiments (NCACX, NCOCX, and CANCO)28, 29 were performed for the sequential resonance assignment of the native TTR (Figure S8). The chemical shift changes in the 2D DARR spectra were mapped into the crystal structure of native TTR (Figure 4). The residues with cross-peaks that disappeared in the amyloid spectrum are colored in green, and the overlapped regions are colored in yellow. The cross-peaks in the boundary in Figure 3 and Figure S9 (* in green) were indicated as red, and the loops in purple could not be assigned presumably due to high flexibility of the loop regions in the native state.
Figure 4.
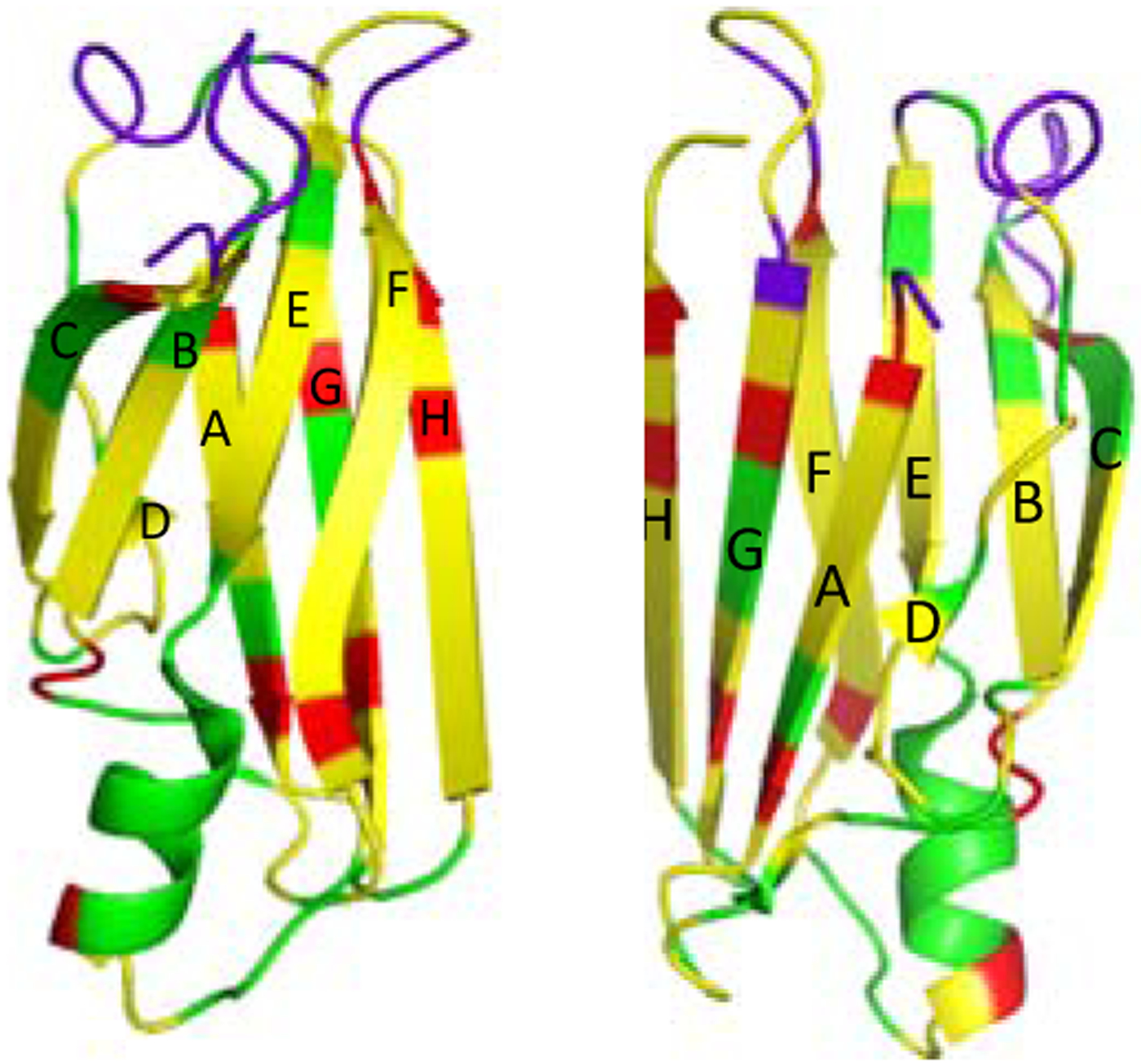
Crystal structure of the monomeric form of native TTR with colors for the residues that are absent (green). The cross-peaks in boundary (Figure S8) were indicated in red. The crystal structure was drawn in two different views.
The comparative solid-state NMR structural analyses of the native and amyloid states of TTR indicate that the loop regions (AB, CD, EF and GH loops) and EF helix undergo structural changes or become disordered during amyloid formation. The AB and GH loops are located in the tetrameric interfacial regions.30 Thus, the dissociation of the tetramers into monomers may lead to the changes in local environments of the loop regions, resulting in the chemical shift changes. In addition, sidechain interactions between the strands C and B, and DE loop appear disrupted in the amyloid state (Figure S10a). It is also interesting to note that the chemical shifts of the residues in A108 in the strand G are affected during misfolding and aggregation (Figure 4 and Figure S10b). The residue A108 is involved in the formation of the binding pocket in native tetrameric TTR. The dissociation of the native tetramer into aggregation-prone monomers is the initial step in the aggregation process, and thus the disruption of the binding pocket may result in the chemical shift changes of the residues in strand G.
On the other hand, a majority of cross-peaks overlap well in the overlaid spectra. Most of the cross-peaks come from the residues in the two β-sheets (CBEF and DAGH), suggesting that the overall native-like β-sheet structures appear to remain unchanged in the amyloid state (Figure 4). These results are consistent with our previous structural studies using selective 13CO/13Cα labeling schemes that revealed the two β-sheet structures are maintained in the amyloid state of TTR.20
Understanding the molecular mechanism of protein misfolding and aggregation is essential to developing therapeutic approaches. Previous mechanistic studies of TTR misfolding and aggregation have provided detailed insights into the molecular basis for misfolding and aggregation process in vitro. Under the amyloidogenic condition of mildly acidic pH of 4 – 5, tetrameric native TTR is dissociated into aggregation-prone monomers that self-assemble into small oligomers and subsequently amyloid precipitates.11, 21 Comparative structural analyses of ex vivo and in vitro TTR amyloids are essential to investigating whether a similar TTR misfolding and aggregation process takes place in vivo. Previous studies showed that TTR forms a heterogeneous mixture of amyloids including thick and long filaments as well as less-ordered aggregates in vivo depending on the age of onset and mutations.31, 32 Very recently, a cryo-EM structure of TTR fibrils extracted from ATTRV30M cardiac tissues was reported.33 The structural studies revealed that the TTRV30M adopts distinctive non-native conformations in the ex vivo filaments, suggesting that the TTR variant protein becomes unfolded during amyloid formation. The filamentous aggregates were, however, not observed in the cardiac extracts from a patient with ATTRwt amyloidosis used in this study. Moreover, the ex vivo cardiac TTRwt amyloid resembles in vitro TTR amyloid with extensive native-like β-structures. These observations suggest that TTR misfolding and aggregation may take place through multiple pathways in vivo depending on the disease phenotypes and mutations.
In summary, we report solid-state NMR studies that suggest in vitro TTR amyloid formed at pH 4.4 contains extensive native-like β-sheet structures, while most of the native loop structures are changed in the amyloid state, consistent with previous structural studies20, 22, 23, 34. In addition, the in vitro TTR amyloid has similar structural characteristics to those of ex vivo cardiac TTR amyloid, suggesting that the TTR misfolding and aggregation observed at mildly acidic conditions may occur in vivo as well. Structural characterization of ex vivo TTR variant amyloids associated with distinct disease phenotypes will be of great importance to understand tissue-selective deposition of TTR amyloid, which is currently underway in our laboratory.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grant (NS097490, KHL). We thank Dr. Pender for assistance in the FT-IR experiments. Solid-state NMR spectra were acquired at the National High Magnetic Field Laboratory (Tallahassee, FL), which is supported by NSF DMR-1644779 and the State of Florida.
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
ThT fluorescence emission spectrum of ex vivo TTRwt amyloid. SDS-PAGE of cardiac extracts. TEM images of in vitro and ex vivo TTR amyloids. FT-IR spectra of in vitro mTTR and ex vivo TTR amyloids. Overlaid 2D DARR for mTTR and TTRwt amyloids. 3D strip plots for the sequential assignments. Crystal structure of monomeric form of native TTR showing side chain interactions. Overlaid 2D DARR for the native and amyloid states of TTRwt. (PDF)
The authors declare no competing financial interest.
Accession Codes
Transthyretin: UniProtKB entry P02766
REFERENCES
- 1.Buxbaum JN, and Reixach N (2009) Transthyretin: The servant of many masters. Cell Mol. Life Sci 66, 3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbert J, Wilcox JN, Pham KT, Fremeau RT, Zeviani M, Dwork A, Soprano DR, Makover A, Goodman DS, and Zimmerman EA (1986) Transthyretin: A choroid plexus-specific transport protein in human brain. the 1986 S. weir mitchell award. Neurology. 36, 900–911. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson DR, Pastore RD, Yaghoubian R, Kane I, Gallo G, Buck FS, and Buxbaum JN (1997) Variant-sequence transthyretin (isoleucine 122) in late-onset cardiac amyloidosis in black americans. N. Engl. J. Med 336, 466–473. [DOI] [PubMed] [Google Scholar]
- 4.Connors LH, Lim A, Prokaeva T, Roskens VA, and Costello CE (2003) Tabulation of human transthyretin (TTR) variants, 2003. Amyloid. 10, 160–184. [DOI] [PubMed] [Google Scholar]
- 5.Saraiva MJ (1995) Transthyretin mutations in health and disease. Hum. Mutat 5, 191–196. [DOI] [PubMed] [Google Scholar]
- 6.Kelly JW (1996) Alternative conformations of amyloidogenic proteins govern their behavior. Curr. Opin. Struct. Biol 6, 11–17. [DOI] [PubMed] [Google Scholar]
- 7.Cornwell GG, Murdoch WL, Kyle RA, Westermark P, and Pitkanen P (1983) Frequency and distribution of senile cardiovascular amyloid. A clinicopathologic correlation. Am. J. Med 75, 618–623. [DOI] [PubMed] [Google Scholar]
- 8.Hurshman AR, White JT, Powers ET, and Kelly JW (2004) Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 43, 7365–7381. [DOI] [PubMed] [Google Scholar]
- 9.Palaninathan SK, Mohamedmohaideen NN, Snee WC, Kelly JW, and Sacchettini JC (2008) Structural insight into pH-induced conformational changes within the native human transthyretin tetramer. J. Mol. Biol 382, 1157–67. [DOI] [PubMed] [Google Scholar]
- 10.Connelly S, Choi S, Johnson SM, Kelly JW, and Wilson IA (2010) Structure-based design of kinetic stabilizers that ameliorate the transthyretin amyloidoses. Curr. Opin. Struct. Biol 20, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai ZH, Colon W, and Kelly JW (1996) The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 35, 6470–6482. [DOI] [PubMed] [Google Scholar]
- 12.Dasari AKR, Hughes RM, Wi S, Hung I, Gan Z, Kelly JW, and Lim KH (2019) Transthyretin aggregation pathway toward the formation of distinct cytotoxic oligomers. Sci. Rep 9, 33–018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galant NJ, Westermark P, Higaki JN, and Chakrabartty A (2017) Transthyretin amyloidosis: An under-recognized neuropathy and cardiomyopathy. Clin. Sci. (Lond) 131, 395–409. [DOI] [PubMed] [Google Scholar]
- 14.Saelices L, Johnson LM, Liang WY, Sawaya MR, Cascio D, Ruchala P, Whitelegge J, Jiang L, Riek R, and Eisenberg DS (2015) Uncovering the mechanism of aggregation of human transthyretin. J. Biol. Chem 290, 28932–28943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Dyson HJ, and Wright PE (2018) Kinetic analysis of the multistep aggregation pathway of human transthyretin. Proc. Natl. Acad. Sci. U. S. A 115, E6201–E6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingsbury JS, Theberge R, Karbassi JA, Lim A, Costello CE, and Connors LH (2007) Detailed structural analysis of amyloidogenic wild-type transthyretin using a novel purification strategy and mass spectrometry. Anal. Chem 79, 1990–1998. [DOI] [PubMed] [Google Scholar]
- 17.Zandomeneghi G, Krebs MR, McCammon MG, and Fandrich M (2004) FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 13, 3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran SD, and Zanni MT (2014) How to get insight into amyloid structure and formation from infrared spectroscopy. J. Phys. Chem. Lett 5, 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarroukh R, Goormaghtigh E, Ruysschaert JM, and Raussens V (2013) ATR-FTIR: A “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta 1828, 2328–2338. [DOI] [PubMed] [Google Scholar]
- 20.Lim KH, Dasari AK, Hung I, Gan Z, Kelly JW, Wright PE, and Wemmer DE (2016) Solid-state NMR studies reveal native-like beta-sheet structures in transthyretin amyloid. Biochemistry. 55, 5272–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lashuel HA, Lai ZH, and Kelly JW (1998) Characterization of the transthyretin acid denaturation pathways by analytical ultracentrifugation: Implications for wild-type, V30M, and L55P amyloid fibril formation. Biochemistry. 37, 17851–17864. [DOI] [PubMed] [Google Scholar]
- 22.Lim KH, Dasari AKR, Ma R, Hung I, Gan Z, Kelly JW, and Fitzgerald MC (2017) Pathogenic mutations induce partial structural changes in the native beta-sheet structure of transthyretin and accelerate aggregation. Biochemistry. 56, 4808–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim KH, Dasari AK, Hung I, Gan Z, Kelly JW, and Wemmer DE (2016) Structural changes associated with transthyretin misfolding and amyloid formation revealed by solution and solid-state NMR. Biochemistry. 55, 1941–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serag AA, Altenbach C, Gingery M, Hubbell WL, and Yeates TO (2002) Arrangement of subunits and ordering of beta-strands in an amyloid sheet. Nat. Struct. Biol 9, 734–739. [DOI] [PubMed] [Google Scholar]
- 25.Laidman J, Forse GJ, and Yeates TO (2006) Conformational change and assembly through edge beta strands in transthyretin and other amyloid proteins. Acc. Chem. Res 39, 576–583. [DOI] [PubMed] [Google Scholar]
- 26.Jahn TR, Parker MJ, Homans SW, and Radford SE (2006) Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol 13, 195–201. [DOI] [PubMed] [Google Scholar]
- 27.Chiti F, and Dobson CM (2009) Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol 5, 15–22. [DOI] [PubMed] [Google Scholar]
- 28.BALDUS M, PETKOVA AT, HERZFELD J, and GRIFFIN RG (1998) Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol. Phys 95, 1197–1207. [Google Scholar]
- 29.Sperling LJ, Berthold DA, Sasser TL, Jeisy-Scott V, and Rienstra CM (2010) Assignment strategies for large proteins by magic-angle spinning NMR: The 21-kDa disulfide-bond-forming enzyme DsbA. J. Mol. Biol 399, 268–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blake CC, Geisow MJ, Oatley SJ, Rerat B, and Rerat C (1978) Structure of prealbumin: Secondary, tertiary and quaternary interactions determined by fourier refinement at 1.8 A. J. Mol. Biol 121, 339–356. [DOI] [PubMed] [Google Scholar]
- 31.Adams D, Koike H, Slama M, and Coelho T (2019) Hereditary transthyretin amyloidosis: A model of medical progress for a fatal disease. Nat. Rev. Neurol 15, 387–404. [DOI] [PubMed] [Google Scholar]
- 32.Ihse E, Rapezzi C, Merlini G, Benson MD, Ando Y, Suhr OB, Ikeda S, Lavatelli F, Obici L, Quarta CC, Leone O, Jono H, Ueda M, Lorenzini M, Liepnieks J, Ohshima T, Tasaki M, Yamashita T, and Westermark P (2013) Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid. 20, 142–150. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt M, Wiese S, Adak V, Engler J, Agarwal S, Fritz G, Westermark P, Zacharias M, and Fandrich M (2019) Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat. Commun 10, 5008–019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasari AKR, Hung I, Gan Z, and Lim KH (2019) Two distinct aggregation pathways in transthyretin misfolding and amyloid formation. Biochim. Biophys. Acta Proteins Proteom 1867, 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


