Significance
The 3C protease has long been the focus of enterovirus antiviral development due to its conservation and critical role in the virus life cycle. Nevertheless, although antienterovirus drugs have a potentially enormous global market, the nucleophilic active site of 3C has frustrated drug discovery efforts. Here, we identified two disparate sites on 3C and unraveled the unconventional inhibitory mechanism of two antibodies that target these sites. We show that the regulatory role of 3C in enterovirus replication is another drug-targetable function besides its protease activity. These findings together with the antiviral effects of these antibodies suggest that these two sites on 3C may represent new chemotherapeutic intervention opportunities against enterovirus infections.
Keywords: pan-enterovirus antiviral target, X-ray crystallography, HDX-MS, allosteric inhibitor, genome replication regulation
Abstract
The existence of multiple serotypes renders vaccine development challenging for most viruses in the Enterovirus genus. An alternative and potentially more viable strategy for control of these viruses is to develop broad-spectrum antivirals by targeting highly conserved proteins that are indispensable for the virus life cycle, such as the 3C protease. Previously, two single-chain antibody fragments, YDF and GGVV, were reported to effectively inhibit human rhinovirus 14 proliferation. Here, we found that both single-chain antibody fragments target sites on the 3C protease that are distinct from its known drug site (peptidase active site) and possess different mechanisms of inhibition. YDF does not block the active site but instead noncompetitively inhibits 3C peptidase activity through an allosteric effect that is rarely seen for antibody protease inhibitors. Meanwhile, GGVV antagonizes the less-explored regulatory function of 3C in genome replication. The interaction between 3C and the viral genome 5′ noncoding region has been reported to be important for enterovirus genome replication. Here, the interface between human rhinovirus 14 3C and its 5′ noncoding region was probed by hydrogen–deuterium exchange coupled mass spectrometry and found to partially overlap with the interface between GGVV and 3C. Consistently, prebinding of GGVV completely abolishes interaction between human rhinovirus 14 3C and its 5′ noncoding region. The epitopes of YDF and GGVV, therefore, represent two additional sites of therapeutic vulnerability in rhinovirus. Importantly, the GGVV epitope appears to be conserved across many enteroviruses, suggesting that it is a promising target for pan-enterovirus inhibitor screening and design.
Enterovirus is the prototype and by far the largest genus within Picornaviridae (1, 2). Many important human pathogens belong to this genus, such as rhinovirus, which alone accounts for more than 50% of the common cold infections that impose a huge health and economic burden on society every year (3–5). Besides the common cold, rhinovirus infection can also lead to asthma and life-threatening chronic obstructive pulmonary disease exacerbation in young children and some adults (3, 5, 6). Moreover, coxsackievirus and poliovirus, which can lead to hand, foot, and mouth disease and paralyzing polio, respectively, are also close neighbors of rhinovirus in the Enterovirus genus (1). To date, vaccines are only available for enterovirus A71 (EV71) and poliovirus (7). For the other viruses in this genus, vaccine development is compounded by multiple serotypes (2). A more rational strategy might be to develop antivirals whose targets are conserved across different serotypes and are indispensable for the virus life cycle. Furthermore, if a conserved target site could be identified across different enteroviruses, a broad-acting antiviral against most, if not all, enteroviruses might even be feasible.
Indeed, extensive efforts have been made toward developing antivirals targeting different stages along the enterovirus life cycle, with virus attachment and entry, polyprotein processing, genomic RNA synthesis, and assembly of progeny virions being the foci of investigation (8). Following virus attachment, the genomic RNA of enterovirus is released into the cytoplasm of host cells and translated into a single polyprotein (8). This polyprotein then undergoes viral protease-mediated self-cleavage, first by 2Apro(2A) and then by 3Cpro(3C), to generate functional structural and nonstructural viral proteins for subsequent virus genome synthesis and encapsidation (8, 9). Besides viral proteins, the substrates of 2A and 3C also encompass many host proteins, such as MAP4 (microtubule-associated protein 4), eIF4G (eukaryotic translation initiation factor 4G), PABP (poly-A-binding protein), RIG-I (retinoic acid-inducible gene I), MAVS (mitochondrial antiviral-signaling protein) and TRIF (TIR-domain-containing adapter-inducing interferon-β) (10, 11). Through cleavage of these host factors, 2A and 3C help the virus hijack the host translation and transcription machinery and inhibit host antiviral responses (9). Hence, 2A and 3C are essential for the enterovirus life cycle, thereby making them suitable candidates for development of antivirals.
However, 3C carries out most of the viral and related host protein cleavage and is more conserved across different serotypes than 2A (10, 12). As such, 3C has been the primary focus for antiviral screening for several decades. The 3C is a cysteine protease with a catalytic triad composed of Cys, His, and Glu/Asp (13, 14). To date, most if not all inhibitors of 3C target its catalytic center and act as substrate analogs (7). Among them, two of the most potent inhibitors are rupintrivir (AG7088) and AG7404, which is a rupintrivir derivative with better oral bioavailability (15, 16). Both inhibitors are Michael acceptor-based peptidomimetics that irreversibly inhibit the 3C catalytic activity by forming a covalent bond with the catalytic Cys (15, 16). Rupintrivir is broadly effective against 48 human rhinovirus (HRV) serotypes and other related picornaviruses in vitro (17) and in the initial phase II trials of experimental rhinovirus infection (18); however, its clinical evaluation was discontinued after phase II/III trials of natural rhinovirus infection (https://www.clinicaltrialsarena.com/projects/ag7088/). Active-site protease inhibitors frequently bear electrophilic warheads that further manifest unfavorable pharmacokinetic properties and off-target promiscuity, whereas the binding sites of allosteric inhibitors are usually more attractive for drug-like compounds (19). Therefore, allosteric inhibitors are now actively pursued in the field as alternative approaches.
The importance of 3C in the enterovirus life cycle extends beyond its proteolytic functions. The 3C from different enteroviruses has been found to specifically interact with the cloverleaf structure in the 5′ noncoding region (NCR) of corresponding virus (20–24), thereby assembling a functional ribonucleoprotein complex. Disruption of the interaction between the 5′ NCR and viral 3C has been reported to abolish RNA replication and render polio and EV71 viruses nonviable (20, 22, 24–26). Nevertheless, compared with the proteolytic function of 3C, the nonproteolytic function of 3C in genomic RNA replication is much less explored. As a consequence, the determination of the actual interface between 3C and enterovirus genomic RNA remains elusive and has not been examined as an antiviral target, to the best of our knowledge.
Previously, two of us reported that two scFvs (single-chain antibody fragments), named YDF and GGVV, manifested outstanding inhibitory effects against human rhinovirus 14 (HRV14) proliferation in HeLa cell models (27). Although 3C was identified to be the common target of the two scFvs, the mechanism by which they inhibit HRV14 infection likely differs, as YDF inhibited the endopeptidase activity of 3C while GGVV did not (27). This finding suggests that GGVV may hinder HRV14 proliferation through a novel mechanism, possibly by antagonizing the aforementioned nonproteolytic function of 3C.
In this work, we determined cocrystal structures of HRV14 3C in complex with these two scFvs. As expected, the two scFvs bind to different epitopes on HRV14 3C. Interestingly, their structural epitopes do not overlap with the active site. Subsequent enzymatic kinetics and molecular dynamic simulation of 3C in the absence and presence of YDF revealed that YDF inhibits the protease activity of 3C through a noncompetitive, allosteric effect that is rarely seen for antiproteolytic antibody inhibitors. Meanwhile, we utilized hydrogen–deuterium exchange coupled mass spectrometry (HDX-MS) to interrogate the interaction between HRV14 3C and its 5′ NCR, and the revealed binding interface partially aligns with the GGVV binding site on HRV14 3C. Moreover, isothermal titration calorimetry (ITC) experiments show that binding of GGVV to 3C completely abolishes interaction between 3C and HRV14 5′ NCR, further suggesting that binding of GGVV to HRV14 3C may prevent the assembly of the aforementioned ribonucleoprotein complex and thus impede genomic RNA replication. Importantly, residues on 3C that mediate its interaction with GGVV are conserved across not only different rhinovirus serotypes but also several other enteroviruses, rendering the GGVV epitope on 3C as a highly promising target for pan-enterovirus antiviral screening and design. Taken together, this work identifies two alternative sites of therapeutic vulnerability on an important antiviral target and provides an important framework for further rational inhibitor development against HRV and other enteroviruses.
Results
Biochemical Characteristics of Antibodies YDF and GGVV.
We previously reported that two antibodies in an scFv format, YDF and GGVV, showed profound inhibitory effect against HRV14 proliferation in HeLa cells (27). These two antibodies (GGVV: IGHV1-69, IGKV1-27 and YDF: IGHV3-30-3, IGLV3-1) share relatively high sequence similarity (74%) in their framework regions with the major diversity in their complementarity-determining regions (CDRs) (SI Appendix, Fig. S1). The target of YDF and GGVV was identified to be HRV14 3C protease, with YDF inhibiting the endopeptidase activity of HRV14 3C, while GGVV does not (27).
Both YDF and GGVV bind HRV14 3C at a molar ratio of 1:1 with high affinity in vitro (SI Appendix, Fig. S2A) and form stable 1:1 complexes with HRV14 3C in solution (SI Appendix, Fig. S2C). The values of the dissociation constant (Kd) for YDF and GGVV are 82.3 ± 8.2 nM and 9.1 ± 1.1 nM, respectively, indicating that GGVV binds HRV14 3C more strongly than YDF (SI Appendix, Fig. S2A). Consistent with our previous report (25), YDF and GGVV manifested completely different inhibitory properties in enzymatic assays (SI Appendix, Fig. S2D). The endopeptidase activity of HRV14 3C decreased ∼65% when incubated with YDF at a 1:1 molar ratio, and a fivefold excess of YDF almost completely eliminated the enzymatic activity of HRV14 3C. To the contrary, GGVV did not impact protease activity. Hence, from these functional experiments, YDF and GGVV likely target different regions on HRV14 3C and inhibit HRV14 proliferation through different mechanisms.
Structures of HRV14 3C in Complex with YDF and GGVV.
To understand the detailed HRV14 inhibition mechanism of these two antibodies, we determined their cocrystal structures in complex with the 3C protease (Fig. 1 A and B). The HRV14 3C–YDF complex (3C-YDF) was crystallized in space group P21212, with two molecules per asymmetric unit. The final structure was refined to a resolution of 1.85 Å, with Rwork of 17.5% and Rfree of 20.8% (Table 1). The structure of HRV14 3C with GGVV (3C-GGVV) was determined in space group C2 with four molecules per asymmetric unit to a resolution of 2.40 Å, with Rwork of 18.6% and Rfree of 22.3% (Table 1). The core structure of HRV14 3C comprises four α-helices and 14 β-strands. YDF and GGVV both have the typical antibody fold. Structural superimposition shows that the overall fold of HRV14 3C protease in both complexes is similar, except for two flexible loops, residues 64 to 69 and 124 to 134, that are disordered in the 3C-GGVV structure.
Fig. 1.
YDF and GGVV bind to different epitopes on HRV14 3C protease. (A) Crystal structure of HRV14 3C in complex with YDF scFv, with HRV14 3C colored in light gray and VH and VL of YDF scFv colored in cyan and blue, respectively. Both the heavy-chain and light-chain CDRs on YDF participate in interaction with HRV14 3C. (B) Structure of HRV14 3C in complex with GGVV scFv, with HRV14 3C colored in dark gray and the VH and VL of YDF scFv colored in green and lime, respectively. Only the heavy chain of GGVV is involved in binding HRV14 3C. (C) Superimposition of the two complexes, color-coded as above. (D) YDF and GGVV bind to different surfaces on HRV14 3C, both of which are distal to the active site. The contact regions of YDF VH and VL on HRV14 3C are highlighted in cyan and blue, respectively, and the epitope of GGVV on HRV14 3C is highlighted in green. The three critical residues in the active site of HRV14 3C are shown as a red surface.
Table 1.
Data collection and structure refinement statistics
| 3C-YDF | 3C-GGVV | |
| Data collection statistics | ||
| Beamline | SSRF, BL18U1 | Spring-8, BL41XU |
| Wavelength, Å | 0.9776 | 1.0000 |
| Space group | P21212 | C2 |
| Cell dimensions | ||
| a, b, c, Å | 107.3, 120.9, 71.5 | 193.8, 133.3, 78.8 |
| α, β, γ, ° | 90, 90, 90 | 90, 93.6, 90 |
| Resolution range, Å* | 50.0–1.85 (1.89–1.85) | 50.0–2.40 (2.46–2.40) |
| No. of observed reflections | 989,979 | 489,683 |
| No. of unique reflections | 80,144 | 77,385 |
| Completeness, %* | 99.6 (95.4) | 100.0 (100.0) |
| Rmeas, %*,† | 18.2 (52.8) | 23.9 (90.1) |
| Rpim, %*,‡ | 5.1 (18.4) | 9.3 (37.6) |
| <I/σ(I)>* | 20.0 (2.8) | 11.1 (2.3) |
| CC1/2*,§ | 0.983 (0.851) | 0.980 (0.854) |
| Redundancy* | 12.4 (7.8) | 6.3 (5.3) |
| Wilson B, Å2 | 20 | 27 |
| Refinement statistics | ||
| Reflections used in refinement* | 80,053 (2300) | 77,222 (1967) |
| Reflections used for Rfree* | 3973 (129) | 3842 (110) |
| Resolution range, Å* | 49.1–1.85 (1.87–1.85) | 48.4–2.40 (2.43–2.40) |
| Rwork, %*,¶, | 17.5 (19.5) | 18.6 (25.2) |
| Rfree, %*,¶ | 20.8 (22.4) | 22.3 (31.1) |
| Atoms | 6,813 | 12,416 |
| Protein | 6,281 | 11,966 |
| Solvent | 532 | 450 |
| Average B-values, Å2 | 23 | 32 |
| Protein | 23 | 32 |
| Solvent | 32 | 30 |
| Rmsd bond length, Å | 0.007 | 0.01 |
| Rmsd bond angle, ° | 0.91 | 1.08 |
| Ramachandran favored, % | 97.7 | 96.8 |
| Ramachandran allowed, % | 2.3 | 3.2 |
Statistics for the highest-resolution shell are shown in parentheses.
Rwork ; Rfree is defined as Rwork calculated from 5% of the reflections that were excluded from refinement.
As expected, the epitopes of YDF and GGVV on HRV14 3C are indeed distinct from each other (Fig. 1 C and D). Surprisingly, however, not only the epitope of GGVV but also that of YDF is nonoverlapping with the active site (Fig. 1D), with the GGVV binding site being more distally located on the opposite side of 3C from the active site. These structural results are consistent with our finding that GGVV does not affect the peptidase activity of HRV14 3C yet are harder to reconcile at first glance with the peptidase inhibition activity of YDF on HRV14 3C (SI Appendix, Fig. S2D).
YDF Is an Allosteric Inhibitor of HRV14 3C.
YDF uses its CDRH3 and three light-chain CDRs to make intimate interactions with residues 107 to 114 and loop 138 to 145 of HRV14 3C, burying 879 Å2 solvent-accessible surface area on HRV 14 3C (Fig. 2A). CDRH3 residues 98 to 100A of YDF form an antiparallel β-sheet with residues 110 to 114 on HRV14 3C (Fig. 2 B, Top). As such, extensive inter–β-strand main-chain H-bonding dominates in this region (Fig. 2 B, Top, black dashed lines). Consistent with the structural results, Ala mutations of single CDRH3 residues only lead to a moderate decrease in binding affinity in YDF (SI Appendix, Table S1). In addition to main-chain H-bonding, a side-chain salt bridge is formed between E1143C and K97CDRH3, and hydrophobic interactions among Y100CDRH3, F100BCDRH3, and I1123C also contribute to the interface (Fig. 2 B, Top). The importance of hydrophobic interaction was corroborated by the YDF triple mutant (Y100A/D100AA/F100BACDRH3), which lost 98% of the original binding (SI Appendix, Table S1).
Fig. 2.
YDF allosterically inhibits the endopeptidase activity of HRV14 3C. (A) The protein–protein interface between HRV14 3C and YDF. HRV14 3C is shown as gray ribbons, with loop 107 to 114 and loop 138 to 145 highlighted in purple and pink, respectively. CDRH3 and the three light-chain CDRs of YDF that are involved in binding HRV14 3C are highlighted in cyan and blue, respectively. (B) Close-up views of the interface between CDRH3 (YDF) and HRV14 3C (Top) and the interface between CDRLs (YDF) and HRV14 3C (Bottom). Key residues that are involved in hydrophilic or hydrophobic interactions on protein–protein interfaces are shown as stick models and color-coded as in A. Polar and electrostatic interactions are indicated with black dashed lines. (Bottom) Electrostatic surface of HRV14 3C is shown to illustrate the hydrophobic cage that is formed by Y139, A140, K142, and F108 on HRV14 3C. Hydrophobic surfaces are in whitish gray, basic in blue, and acidic in red. The CDRs residues are labeled following Kabat numbering. (C) Protease activity (micromoles per second) of HRV14 3C was plotted against the concentration of substrate (micromolar) in the absence or presence of different molar ratios of YDF. The presence of YDF reduces Vmax proportionally without affecting the apparent Km of HRV14 3C, indicating that YDF is a noncompetitive inhibitor of HRV14 3C. Values are means ± SD (n = 3 assays).
Meanwhile, the light-chain CDRs of YDF contact residues 107 to 109 and loop 138 to 145 of HRV14 3C. Specifically, Y32CDRL1 and W91CDRL3 from YDF insert into a hydrophobic cluster formed by Y1393C, A1403C, K1423C (aliphatic part), and F1083C on HRV14 3C (Fig. 2 B, Bottom). When Y32CDRL1 and W91CDRL3 were mutated to Ala, binding to HRV14 3C was abrogated, highlighting the importance of these two aromatic residues in the interaction (SI Appendix, Table S1). Moreover, multiple hydrophilic or electrostatic interactions from side chain to side chain (Y1393C to Q50CDRL2, K1423C to D51CDRL2, N1073C to D92CDRL3) and side chain to main chain (Y32CDRL1 to A1403C, K53CDRL2 to D1383C, S93CDRL3 to T1093C) were also observed (Fig. 2 B, Bottom, black dashed lines), where Ala mutations in YDF generally resulted in a 15 to 75% affinity decrease, except for D51CDRL2(SI Appendix, Table S1). When D51CDRL2 was mutated to Ala, 3C binding was completely lost (SI Appendix, Table S1), probably due to the critical role of D51CDRL2 in maintaining the correct positioning of the K1423C side chain, which in turn is essential for complete assembly of the aforementioned hydrophobic cage in the heart of the interaction in this region (Fig. 2 B, Bottom).
Although YDF can inhibit the peptidase activity of HRV14 3C (SI Appendix, Fig. S2D), it does not form direct interactions with the three critical active site residues, that is, H40, E71, and C146 (Figs. 1D and 2B), suggesting that YDF might function as an allosteric inhibitor of HRV14 3C. We thus performed a detailed enzymatic kinetics study on HRV14 3C in the absence and presence of different concentrations of YDF (Fig. 2C). In the absence of YDF, the Michaelis constant (Km) and maximum velocity (Vmax) of HRV14 3C are 93.4 ± 8.0 μM and 0.48 ± 0.02 μM s−1, respectively. In the presence of YDF, the apparent Km of HRV14 3C only shows slight changes in all tested conditions, while the apparent Vmax of HRV14 3C decreased proportionally to the different concentrations of YDF (Fig. 2C and SI Appendix, Table S3). Notably, in the presence of 0.6 μM YDF (1.2-fold that of HRV14 3C), the measured Km of 3C is 85.4 ± 10.8 μM, while the measured Vmax decreased to 0.11 ± 0.01 μM s−1, indicating that HRV14 3C only retained about ∼25% of its original activity. Consistent with the kinetic results, the substrate cleft of HRV14 3C in the presence of YDF could accommodate the substrate analog (SI Appendix, Fig. S3 A and B). Moreover, occupation of the substrate cleft by rupintrivir did not affect the binding of YDF to HRV14 3C (SI Appendix, Fig. S2B). Hence, addition of YDF has no impact on substrate binding while reducing the 3C enzyme activity, further suggesting that YDF functions as an allosteric, noncompetitive inhibitor of HRV14 3C.
We also superimposed the structure of 3C-YDF onto the NMR solution structures of apo HRV14 3C and HRV14 3C in complex with a peptide mimetic inhibitor (14). The overall conformation of 3C is similar in all three structures with root-mean-square deviations (RMSDs) in the range of 0.9 to 1.0 Å for 178 Cα atoms (SI Appendix, Fig. S3C), indicating that no large conformational changes in 3C were induced by YDF binding. To better understand the allosteric inhibition of YDF, we performed molecular simulations of HRV14 3C in the absence and presence of YDF (SI Appendix, Fig. S4 A–C). No obvious root-mean-square fluctuation (RMSF) increases were observed in most regions of 3C upon YDF binding (SI Appendix, Fig. S4B), whereas residues 63 to 67, 93 to 96, 106 to 109, and 141 to 146 contributed the highest 10% RMSF decrease (SI Appendix, Fig. S4C, highlighted in dark brown). Notably, the RMSF decrease of residues 106 to 109 and 141 to 146 are consistent with their localization at the interface between YDF and HRV14 3C (Fig. 2). Previously, atomic displacement of residues 136 to 147 was found to decrease considerably upon inactivation of 3C (14). In this regard, the observed “rigidification” of residues 141 to 146 (SI Appendix, Fig. S4 B and C) indicates that this region was likely locked into an “rigidified inactive” state by YDF binding, which may account for the “activity loss” of HRV14 3C in the presence of YDF. How the RMSF of residues 63 to 67 and 93 to 96 are affected by binding of YDF is not clear as they are distant to either the YDF interface or the substrate cleft, but long-range perturbation between noncontact regions on 3C has been observed before by NMR experiments (14, 28).
GGVV Hinders Nonproteolytic Functions of HRV14 3C.
Compared to YDF, the contact region of GGVV on HRV14 3C is even more distant from the active site, mainly involving residues from helix α1, residues 84 to 89 around 310 helix η2, and residues 153 to 154 on the last β-turn of HRV14 3C (Fig. 3A). GGVV only uses residues from CDRH2 and CDRH3 for binding (Fig. 3A), burying 675 Å2 solvent-accessible surface area on HRV14 3C.
Fig. 3.
GGVV binds HRV14 3C only with two heavy-chain CDRs. (A) The protein–protein interfaces between HRV14 3C protease and GGVV. The three critical residues are shown as red stick models to illustrate the position of active site. HRV14 3C is shown as gray ribbons, while helix α1, 310 helix η2, and the β-turn between the β10 and β11 strands at the antibody–protein interface are highlighted in orange. CDRH3 and CDRH2 of GGVV that are involved in binding HRV14 3C are highlighted in green. (B and C) Close-up views of the interfaces between CDRH2 (GGVV) and HRV14 3C (B) and between CDRH3 (GGVV) and HRV14 3C (C). HRV14 3C is shown as an electrostatic surface, with helix α1, 310 helix η2, and the β-turn between the β10 and β11 strands as orange ribbons. Key residues that are involved in interactions at the antibody–protein interface are shown as stick models, labeled and color-coded as in A. Polar and electrostatic interactions are indicated with black dashed lines. The CDR residues of GGVV are labeled following Kabat numbering.
The interface between HRV14 3C and CDRH2 of GGVV is dominated by hydrophobic interactions. On HRV14 3C, residues F63C, L103C, and the aliphatic part of K133C from helix α1, and R843C, I863C, and F893C around 310 helix η2 together form a deep hydrophobic groove, into which F54CDRH2 from GGVV snugly inserts its aromatic side chain (Fig. 3B). In fact, the hydrophobic packing contributed by F54 at the tip of hydrophobic CDRH2 of VH1-69 antibodies is a key contributor to the interaction. Its importance is demonstrated upon mutation to Ala, where interaction between GGVV and HRV14 3C is almost completely abrogated (SI Appendix, Table S2). V53CDRH2 is also involved in this hydrophobic packing interaction (Fig. 3B and SI Appendix, Table S2). On the rim of the hydrophobic groove, R123C makes side-chain to side-chain interactions with T56CDRH2 and side-chain to main-chain interactions with F54CDRH2 (Fig. 3B), while R843C salt bridges with D55CDRH2, all of which further strengthen the interaction between GGVV and HRV14 3C. Besides CDRH2, CDRH3 also contributes to the interface through hydrophobic packing of V100 with P23C, T1533C, and F63C from HRV14 3C, and hydrophilic interactions of its backbone atoms with E53C and G1543C (Fig. 3C and SI Appendix, Table S2). We also performed a simulation of HRV14 3C in the absence and presence of GGVV (SI Appendix, Fig. S4 D–F), which indicated that binding of GGVV has no effect on the substrate cleft. Hence, consistent with GGVV’s having no impact on the peptidase activity of HRV14 3C (SI Appendix, Fig. S2D), the cocrystal structure of 3C-GGVV further suggests that GGVV hinders HRV14 by antagonizing nonproteolytic functions of 3C.
GGVV Blocks Interaction between HRV14 3C and HRV14 Genomic RNA.
Interactions between 3C and the 5′ NCR of the corresponding genomic RNA have been reported for different enteroviruses, including HRV, poliovirus, coxsackievirus B3 (CVB3), and EV71, and appears to play a regulatory role in their genomic RNA replication (20–24). The 5′ NCR of HRV14 genomic RNA forms a cloverleaf structure of which a 72-nucleotide (nt) RNA truncate has been reported to be sufficient for binding 3C (29). We therefore in vitro-translated the 72-nt 5′ NCR region of HRV14 and utilized HDX-MS to map the binding between HRV14 3C and the 72-nt RNA. From the HDX-MS results, peptides 9 to 16 amino acids (aa), 76 to 84 aa, 85 to 93 aa, 86 to 93 aa, 151 to 157 aa, and 151 to 160 aa manifested markedly decreased hydrogen-to-deuterium exchange in the presence of 72-nt RNA, suggesting that residues from these peptide fragments are located at the interface between HRV14 3C and genomic RNA (Fig. 4 B–D and SI Appendix, Fig. S6). Based on the HDX-MS results, the viral RNA binding regions on 3C were mapped onto the structure of HRV14 3C (Fig. 4A, blue regions). Previous mutagenesis assays conducted with poliovirus, HRV14, and EV71 suggested that residues 12 to 13 aa, 82 to 89 aa, and 153 to 156 aa are pivotal for the interaction between 3C and genomic RNA of the corresponding virus (24–26, 29, 30). Notably, the 3C–RNA interface revealed here by HDX-MS is highly consistent with these mutagenesis data (Fig. 4 B–D and SI Appendix, Fig. S6). However, while some of these studies suggested that residues 6 and 31 to 32 are also involved in genomic RNA binding (26, 30), our HDX-MS results indicate that this does not seem to be the case for HRV14 3C (Fig. 4A, light gray regions and SI Appendix, Fig. S7). Although suggested by mutagenesis studies (26), whether the last α-helix is located at the HRV14 3C-viral RNA interface is not known due to poor peptide coverage in this region (Fig. 4A, dark gray regions and SI Appendix, Fig. S5).
Fig. 4.
GGVV blocks HRV14 3C binding to HRV genomic RNA. (A) The structure of HRV14 3C is shown as a surface representation, onto which the HDX difference heat map for HRV14 3C in complex with 72-nt RNA compared to the apo form is mapped. For heat-map color coding, red indicates increased HDX and blue indicates decreased HDX in 3C-RNA relative to apo 3C, light gray indicates no significant changes, and dark gray represents regions that were not detected in the HDX experiment. CDRH2 and CDRH3 of GGVV are shown as green stick models to illustrate the paratopes of GGVV. (B–D) Deuterium uptake plots and mass spectra of indicated peptides from HRV14 3C in the absence and presence of 72-nt RNA. Note that the HDX in peptides 9 to 16, 85 to 93, and 151 to 160 decreased considerably in the presence of 72-nt RNA, suggesting that residues in these peptides are located at the interface between HRV14 3C and 72-nt RNA. (Left) The deuterium uptake data are plotted as percent deuterium uptake verse time on a logarithmic scale. (Right) Mass spectra of indicated peptides at different labeling time points, with the mass spectra of undeuterated samples shown as controls. (E) ITC results of RNA titrated into 3C protease, 3C-YDF complex, and 3C-GGVV complex clearly demonstrate that the interaction between HRV14 3C and the 5′ NCR of HRV genomic RNA is completely abrogated in the presence of GGVV.
According to the HDX-MS results and the 3C-GGVV crystal structure, the interface between HRV14 viral RNA and 3C overlaps with that of GGVV and 3C. We therefore hypothesized that GGVV binding may inhibit the interaction between HRV14 3C and its genomic RNA. We interrogated the binding by ITC between HRV14 3C and the 72-nt RNA in the presence or absence of GGVV. The ITC results demonstrate that the 72-nt RNA binds HRV14 3C at 1:1 molar ratio with a Kd of 1.28 ± 0.12 μM (Fig. 4 E, Left). While titrating HRV14 3C in the presence of YDF yields a similar stoichiometry and a similar Kd of 1.38 ± 0.12 μM (Fig. 4 E, Middle), no binding was observed between 72-nt RNA and 3C in the presence of GGVV (Fig. 4 E, Right). Hence, interaction between HRV14 3C and viral RNA was specifically inhibited by prebinding of GGVV. Considering that interactions between 3C and viral RNA are pivotal for the viability of corresponding enterovirus (20, 22, 24–26), these findings indicate that GGVV can interrupt the life cycle of HRV14 by blocking interaction of 3C and 5′ NCR of its genomic RNA.
Discussion
The HRV-encoded protease 3C is central to its life cycle, as well as that of many other viruses in the same genus. Besides cleaving the single viral polyprotein into functional structural and nonstructural viral proteins for virus progeny assembly (8, 9), 3C also cleaves multiple host proteins to subvert host translation, transcription, and antiviral systems (10, 11). At the same time, 3C takes on another important nonproteolytic function where, together with 3Dpol and cellular cofactor PCBP2, it interacts with the cloverleaf structure in the 5′ NCR of the corresponding virus and thus plays a regulatory role in genomic RNA replication (20–24). For its indispensable roles and high sequence conservation in these viruses, 3C has long been a prime target for development of enterovirus antiviral therapeutics.
Here, we discovered that two specific inhibitory antibodies, YDF and GGVV, recognize different antigenic determinants in 3C and each antagonizes different physiological functions of 3C. YDF serves as an allosteric noncompetitive inhibitor of HRV14 3C and decreases its peptidase activity. In contrast, GGVV blocks the interaction between HRV14 3C and 5′ NCR of HRV genomic RNA, thereby interfering with genome replication of HRV14.
Protease inhibitors constitute an important class of antiviral agents and have had revolutionarily success in the treatment of HIV/AIDS and hepatitis C infection (31). As such, many potential antienterovirus agents screened are targeted toward the peptidase activity of 3C (7). Unlike active-site protease inhibitors that frequently bear electrophilic warheads and manifest unfavorable pharmacokinetic properties, the binding sites of allosteric inhibitors are usually more attractive for drug-like compounds (19). In this regard, the YDF epitope reported here might serve as an alternative target site for inhibiting the protease activity of 3C. From an historical perspective, protease inhibitors usually are synthetic small molecules or peptidomimetics that function as substrate analogs to block the active sites of the corresponding proteases (31). Allosteric small-molecule inhibitors have also been described, albeit less frequently (19, 32). Besides small molecules and peptidomimetics, antiproteolytic antibodies are also considered important modulators of therapeutically relevant proteases, especially when small-molecule inhibitors fail to deliver desired high specificity (33). To date, several antiproteolytic antibodies against serine proteases have been reported. Most of these antibodies act in a competitive manner, blocking the substrate binding either through direct steric clash (34–36) or allosterically altering the substrate cleft (37–39). Unlike serine proteases, we are not aware of any reports on antiproteolytic antibodies against cysteine proteases to date, probably due to the oxidation state of the catalytic cysteine being hard to maintain during library screening. YDF, identified from intracellular screening, then, appears to be a previously undisclosed allosteric and noncompetitive antibody inhibitor against a cysteine protease. Recently, another method of selecting protease inhibitory antibodies in the reducing periplasmic of Escherichia coli was reported, which may facilitate anticysteine protease antibody screening in the future (40).
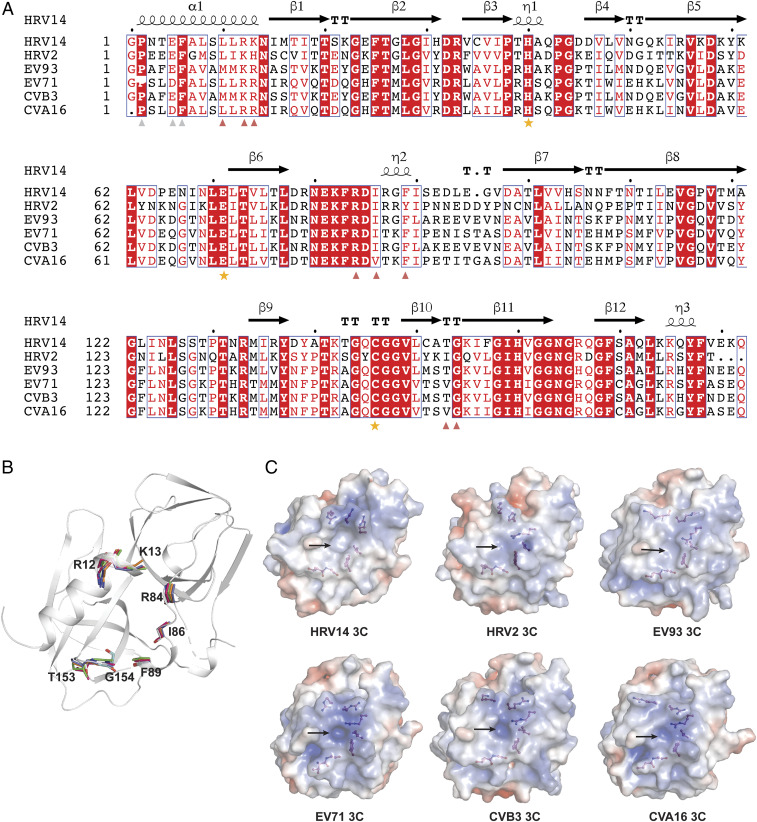
Besides unfavorable pharmacokinetic properties, rupintrivir’s failure in phase II/III trials may relate to its need to be administered within 24 h of infection, as a protease active-site blocker (https://www.clinicaltrialsarena.com/projects/ag7088/). On the other hand, virus genomic RNA replication impacts the entire lifetime of the infection and thus is another valuable target for antiviral development. Disruption of the interaction between viral 5′ NCR and 3C has been found to cause viral infectivity loss in poliovirus and EV71 virus (20, 22, 24–26). To this end, the RNA binding site appears to be an alternative, highly promising drug target site on 3C. However, compared to the proteolytic function of 3C, investigation of its regulatory role in genomic RNA replication is still in infancy where detailed structural and mechanistic studies are not available yet. As such, the actual interface between 3C and enterovirus genomic RNA remains obscure, as in vitro RNA binding-based studies lead to uncertainties at several regions (26, 30). Here, we used HDX-MS to map the RNA binding interface on the crystal structure of HRV14 (Fig. 4). Our results confirmed the importance of the “RK” (corresponding to residues 12 to 13), “KFRDI,” and “VGK” motifs in 5′ NCR binding, while potentially eliminating any role for residues 1 to 8 and residues 31 to 32 in direct interaction with the 5′ NCR. Moreover, our finding that the antiviral effect of GGVV (27) was conferred by abolishing the interaction between 3C and 5′ NCR (Fig. 4) serves as a critical proof of concept that the RNA-binding site is indeed another promising drug target on 3C. We therefore anticipate that our findings here will aid in future development of antiviral compounds targeting the replication regulation activity of 3C. Meanwhile, targeting the interface between 3C and viral genomic RNA for inhibitor design has yet another advantage. The residues on HRV14 3C that are involved in both genomic RNA and GGVV binding are conserved among HRV14, HRV2, CVA16, CVB3, EV71, and enterovirus 93 (EV93) (Fig. 5) and are also important for the interactions between 3C and genomic RNA of HRV14, poliovirus, and EV71 (21, 24, 26, 30). Thus, the interface between 3C and viral genomic RNA may even have the potential to serve as a pan-enterovirus target site for broad-spectrum inhibitor development.
Fig. 5.
Key residues on 3C involved in both GGVV and genomic RNA interactions are conserved across a panel of enteroviruses. (A) Sequence alignment of HRV14 3C with its orthologs from HRV2, EV93, EV71, CVB3, and CVA16. The secondary structure of HRV14 3C is shown above the sequence and the three critical active-site residues are indicated with orange stars. Residues on HRV14 3C involved in both GGVV and 72-nt RNA binding are marked by red triangles, and residues involved in GGVV binding but not 72-nt RNA binding are marked by light gray triangles. (B) The structure of HRV14 3C (gray) is shown as a cartoon representation onto which its orthologs from HRV2 (PDB ID code 5FX5), EV93 (PDB ID code 3Q3X), EV71 (PDB ID code 3SJK), CVB3 (PDB ID code 2ZTZ), and CVA16 (PDB ID code 3SJ8) are superimposed. The backbone of the residues that are marked by red triangle in A are shown as stick models to highlight their structural conservation in space across HRV14 (gray), HRV2 (cyan), EV93 (orange), EV71 (green), CVB3 (blue), and CVA16 (hot pink). (C) The 3C proteases from HRV14, HRV2, EV93, EV71, CVB3, and CVA16 were rendered as an electrostatic surface representation (−5 to +5 kT/e), where hydrophobic surfaces are in whitish gray, basic in blue, and acidic in red. The residues marked by red triangle in A are shown as stick-and-ball models. Note that these residues are located around a similar pocket (marked with an arrow) on 3C in all six structures.
The common cold is a frequent and seasonally recurring disease that causes great economic burden on society every year in terms of healthcare expenses and disease-related productivity losses (41). Although over-the-counter symptomatic treatments for the common cold are available, there is no cure so far for this disease. Hence, a successful common-cold treatment would greatly impact the global market (41). However, development hurdles are also high as effectivity, high specificity (minimal off-target side effects), and commercial viability all have to be met. Although the canonical targeting site on 3C has frustrated drug discovery efforts, the two unconventional sites and corresponding virus inhibition mechanisms uncovered here may represent new therapeutic opportunities on this seemingly ideal drug target 3C. Future translation of these two sites may be developed through several different routes. On the one hand, engineering antibody-based peptidomimetics is one conceivable way given the recent success in design of peptidic ligands against the stem of influenza hemagglutinin (42). On the other, high-throughput fragment-based approaches for small molecule discovery could be considered, such as the tethering (43) and the DNA-encoded library (DEL) methods (44). The tethering method, considering its site-specific potential and ability to identify weak binders, seems quite suitable for identifying inhibitors that bind to the relatively flat allosteric site (YDF epitope) on 3C. Meanwhile, the concave shape of the GGVV epitope (especially the CDRH2 binding site) may render it attractive for small molecules wherein the DEL approach seems appropriate and may enable the sampling of larger chemical space.
Materials and Methods
HRV14 3C protease and antibody YDF were expressed in E. coli BL21 (DE3) cells and antibody GGVV was expressed in High Five cells (BTI-TN-5B1-4). All proteins were purified via affinity chromatography and size-exclusion chromatography. The 1 to 72 nt of the 5′ NCR region of HRV14 genomic RNA was transcribed in vitro using the HiScribe T7 High Yield RNA Synthesis kit (New England Biolabs). The enzymatic activity of HRV 3C protease was measured using the HRV 3C Protease Activity Assay kit (Abcam). ITC assays were performed by titrating 3C or 3C-Rupintrivir with antibody YDF or GGVV and titrating 3C, 3C-YDF, or 3C-GGVV with 72-nt RNA. The crystal structures of 3C-YDF and 3C-GGVV were resolved by molecular replacement using the HRV 3C protease structure (Protein Data Bank [PDB] ID code 2IN2) and an scFv structure (PDB ID code 2GHW) as the search models. HDX-MS experiments were performed on 3C and 3C-RNA using method similar to that previously described ref. 45. Molecular dynamics simulations were all performed using Desmond from Schrodinger Suite 2019-1 (https://www.schrodinger.com/). Detailed descriptions of materials and methods can be found in SI Appendix.
Data and Materials Availability.
The atomic coordinates and structure factors of 3C-YDF and 3C-GGVV protein complexes have been deposited in the PDB with accession nos. 6KYZ and 6KZ0, respectively.
Supplementary Material
Acknowledgments
We thank Prof. Dennis Wolan at The Scripps Research Institute for insightful discussions and suggestions, the staffs of beamline BL18U1 at the National Facility for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility, beamline BL41XU at Super Photon Ring-8 GeV (Spring-8) for assisting with X-ray data collection, the National Center for Protein Science Shanghai for technical support of ITC experiments, and the Bioinformatics Platform and the Analytical Chemistry Platform at Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University for technical and instrument support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.K. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors of 3C-YDF and 3C-GGVV protein complexes have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID codes 6KYZ and 6KZ0, respectively).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918844117/-/DCSupplemental.
References
- 1.van der Linden L., Wolthers K. C., van Kuppeveld F. J., Replication and inhibitors of enteroviruses and Parechoviruses. Viruses 7, 4529–4562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmenberg A. C., Gern J. E., Classification and evolution of human rhinoviruses. Methods Mol. Biol. 1221, 1–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tapparel C., Siegrist F., Petty T. J., Kaiser L., Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 14, 282–293 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Jacobs S. E., Lamson D. M., St George K., Walsh T. J., Human rhinoviruses. Clin. Microbiol. Rev. 26, 135–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner R. B., Rhinovirus: More than just a common cold virus. J. Infect. Dis. 195, 765–766 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Jamieson K. C., Warner S. M., Leigh R., Proud D., Rhinovirus in the pathogenesis and clinical course of asthma. Chest 148, 1508–1516 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Bauer L., Lyoo H., van der Schaar H. M., Strating J. R., van Kuppeveld F. J., Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr. Opin. Virol. 24, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baggen J., Thibaut H. J., Strating J. R. P. M., van Kuppeveld F. J. M., The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 16, 368–381 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jensen L. M., Walker E. J., Jans D. A., Ghildyal R., Proteases of human rhinovirus: Role in infection. Methods Mol. Biol. 1221, 129–141 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Laitinen O. H. et al., Enteroviral proteases: Structure, host interactions and pathogenicity. Rev. Med. Virol. 26, 251–267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lei X., Xiao X., Wang J., Innate immunity evasion by enteroviruses: Insights into virus-host interaction. Viruses 8, E22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanway G., Structure, function and evolution of picornaviruses. J. Gen. Virol. 71, 2483–2501 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Matthews D. A. et al., Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell 77, 761–771 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Bjorndahl T. C., Andrew L. C., Semenchenko V., Wishart D. S., NMR solution structures of the apo and peptide-inhibited human rhinovirus 3C protease (Serotype 14): Structural and dynamic comparison. Biochemistry 46, 12945–12958 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Matthews D. A. et al., Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. U.S.A. 96, 11000–11007 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragovich P. S. et al., Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 8. Pharmacological optimization of orally bioavailable 2-pyridone-containing peptidomimetics. J. Med. Chem. 46, 4572–4585 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Patick A. K. et al., In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 43, 2444–2450 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden F. G. et al., Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 47, 3907–3916 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauske P., Ottmann C., Meltzer M., Ehrmann M., Kaiser M., Allosteric regulation of proteases. ChemBioChem 9, 2920–2928 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Andino R., Rieckhof G. E., Baltimore D., A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63, 369–380 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Leong L. E., Walker P. A., Porter A. G., Human rhinovirus-14 protease 3C (3Cpro) binds specifically to the 5′-noncoding region of the viral RNA. Evidence that 3Cpro has different domains for the RNA binding and proteolytic activities. J. Biol. Chem. 268, 25735–25739 (1993). [PubMed] [Google Scholar]
- 22.Xiang W., Harris K. S., Alexander L., Wimmer E., Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69, 3658–3667 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zell R., Sidigi K., Bucci E., Stelzner A., Görlach M., Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8, 188–201 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih S. R. et al., Mutations at KFRDI and VGK domains of enterovirus 71 3C protease affect its RNA binding and proteolytic activities. J. Biomed. Sci. 11, 239–248 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Hämmerle T., Molla A., Wimmer E., Mutational analysis of the proposed FG loop of poliovirus proteinase 3C identifies amino acids that are necessary for 3CD cleavage and might be determinants of a function distinct from proteolytic activity. J. Virol. 66, 6028–6034 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andino R., Rieckhof G. E., Achacoso P. L., Baltimore D., Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12, 3587–3598 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie J. et al., Prevention of cell death by antibodies selected from intracellular combinatorial libraries. Chem. Biol. 21, 274–283 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Chan Y. M., Moustafa I. M., Arnold J. J., Cameron C. E., Boehr D. D., Long-range communication between different functional sites in the picornaviral 3C protein. Structure 24, 509–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker P. A., Leong L. E., Porter A. G., Sequence and structural determinants of the interaction between the 5′-noncoding region of picornavirus RNA and rhinovirus protease 3C. J. Biol. Chem. 270, 14510–14516 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Blair W. S. et al., Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA 4, 215–225 (1998). [PMC free article] [PubMed] [Google Scholar]
- 31.Agbowuro A. A., Huston W. M., Gamble A. B., Tyndall J. D. A., Proteases and protease inhibitors in infectious diseases. Med. Res. Rev. 38, 1295–1331 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Shen A., Allosteric regulation of protease activity by small molecules. Mol. Biosyst. 6, 1431–1443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesan R., Eigenbrot C., Kirchhofer D., Structural and mechanistic insight into how antibodies inhibit serine proteases. Biochem. J. 430, 179–189 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Farady C. J., Egea P. F., Schneider E. L., Darragh M. R., Craik C. S., Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition. J. Mol. Biol. 380, 351–360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider E. L. et al., A reverse binding motif that contributes to specific protease inhibition by antibodies. J. Mol. Biol. 415, 699–715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y. et al., Structural insight into distinct mechanisms of protease inhibition by antibodies. Proc. Natl. Acad. Sci. U.S.A. 104, 19784–19789 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganesan R. et al., Unraveling the allosteric mechanism of serine protease inhibition by an antibody. Structure 17, 1614–1624 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Kromann-Hansen T. et al., Allosteric inactivation of a trypsin-like serine protease by an antibody binding to the 37- and 70-loops. Biochemistry 52, 7114–7126 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Hinkofer L. C. et al., A monoclonal antibody (MCPR3-7) interfering with the activity of proteinase 3 by an allosteric mechanism. J. Biol. Chem. 288, 26635–26648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez T. et al., Functional selection of protease inhibitory antibodies. Proc. Natl. Acad. Sci. U.S.A. 116, 16314–16319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heikkinen T., Järvinen A., The common cold. Lancet 361, 51–59 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadam R. U. et al., Potent peptidic fusion inhibitors of influenza virus. Science 358, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erlanson D. A. et al., Site-directed ligand discovery. Proc. Natl. Acad. Sci. U.S.A. 97, 9367–9372 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner S., Lerner R. A., Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. U.S.A. 89, 5381–5383 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B., Stjepanovic G., Shen Q., Martin A., Hurley J. H., Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492–498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.