Significance
We demonstrate the ability of two-dimensional transition metal dichalcogenides (TMDs) such as molybdenum disulfide (MoS2) to modulate and direct cellular functions of human stem cells through photothermal modulation. The global gene expression profile of stem cells reveals significant influence on integrins, cellular migration, and wound healing by photothermal modulation of MoS2. The photostimulation of MoS2 may provide new approaches to regulate cellular migration and related functions.
Keywords: two-dimensional (2D) nanomaterials, whole-transcriptome sequencing, cell–nanoparticles interactions, human mesenchymal stem cells, cell adhesion
Abstract
Two-dimensional (2D) molybdenum disulfide (MoS2) nanomaterials are an emerging class of biomaterials that are photoresponsive at near-infrared wavelengths (NIR). Here, we demonstrate the ability of 2D MoS2 to modulate cellular functions of human stem cells through photothermal mechanisms. The interaction of MoS2 and NIR stimulation of MoS2 with human stem cells is investigated using whole-transcriptome sequencing (RNA-seq). Global gene expression profile of stem cells reveals significant influence of MoS2 and NIR stimulation of MoS2 on integrins, cellular migration, and wound healing. The combination of MoS2 and NIR light may provide new approaches to regulate and direct these cellular functions for the purposes of regenerative medicine as well as cancer therapy.
Two-dimensional (2D) nanomaterials are an emerging class of materials with anisotropic layered structure that have shown distinct physical and chemical properties (1–4). Recently, 2D transition metal dichalcogenides (TMDs) such as molybdenum disulfide (MoS2) have been explored for biomedical applications due to their photothermal characteristics (5–9). Specifically, the generation of ultrathin sheets via exfoliation of MoS2 enables the formation of direct bandgap nanosheets, which can be utilized for light-based therapies (8, 10–13). In addition, the absence of dangling bonds from terminal sulfur atoms of MoS2 nanomaterials render high physiological stability (6, 10). The photoresponsive ability of 2D MoS2 along with its chemical stability in biological microenvironments can be exploited for the photomodulation of cellular functions. Although some studies have investigated cellular compatibility of 2D MoS2 (14–17), no studies have explored their photomodulation ability.
Understanding cellular responses following treatment with 2D MoS2, with and without photostimulation, will assist in exploring their biomedical applications. Although molecular biology techniques such as PCR and microarrays can be used to evaluate the effect of this treatment on cells, they are limited in throughput. To overcome this limitation, “omics” techniques can provide global high-throughput readouts of various cellular and molecular processes and can help discern affected cell–nanomaterials interactions. Specifically, whole-transcriptome sequencing (RNA-seq) of treated cells can provide an unbiased and global perspective of stimulated cellular and molecular activity as well as pivotal signaling pathways triggered by exposure to the nanomaterial. RNA-seq can be used for determination and quantification of all expressed transcripts in cells by overcoming the limitations and biases of conventional PCR and microarrays analysis.
Here, we report interactions of exfoliated MoS2 with human mesenchymal stem cells (hMSCs) by uncovering molecular targets and affected signaling pathways. We first examine the interplay of 2D MoS2 with biomolecules under physiological conditions. Subsequently, we utilize RNA-seq to characterize the in-depth and unbiased effect of MoS2 on the gene expression program of hMSCs. RNA-seq reveals the steady-state expression of the transcriptome after the MoS2 treatment, as well as the photothermal effect of MoS2 at near-infrared wavelengths (NIR). This information assists in identifying the role of MoS2 and NIR exposure in influencing key signaling pathways.
Biophysical Characterization of MoS2 Nanosheets
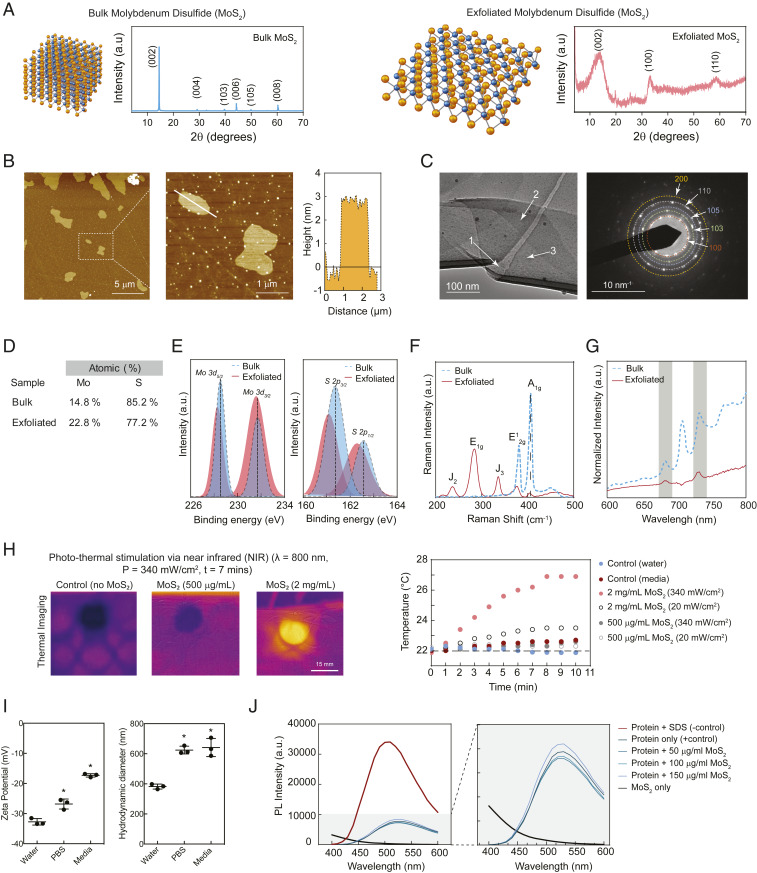
Bulk MoS2 is composed of atomically thin S–Mo–S sheets stacked by short-range van der Waals forces. Bulk MoS2 was chemically exfoliated following previously reported protocols (3, 18, 19). X-ray diffraction (XRD) of bulk MoS2 showed characteristic peaks at (002), (004), (103), (006), (105), and (008), while peaks broadened postexfoliation at (002), (100), and (110) as indexed by Joint Committee on Powder Diffraction File card number 37-1492 (Fig. 1A). The peaks of exfoliated MoS2 indicated a hexagonal structure. Using Bragg’s equation [2d sin(θ) = nλ], the interlayer spacing [(002) Miller indices spacing] of exfoliated MoS2 is 6.328 Å, which is larger than the interlayer spacing of bulk MoS2 (6.124 Å). The full width at half maximum of peak (002) was 0.089 rad for bulk MoS2, which increased to 2.493 rad for exfoliated MoS2. The crystallite sizes for bulk and exfoliated MoS2 were calculated using Scherrer equation as ∼120 nm and 3.5 nm, respectively.
Fig. 1.
Physicochemical characterization of exfoliate 2D MoS2 nanosheets. (A) XRD of bulk and exfoliated MoS2. (B) AFM measures micrometer-sized nanosheets with nanometer thickness, confirming 2D shape. (C) TEM images of ultrathin MoS2 sheets displayed overlapping layers; each individual layer is numbered. Further, electron diffraction corroborated the formation of the 1T phase following lithium intercalation. (D) Atomic composition of bulk and exfoliated MoS2 determined via elemental analysis. (E) Crystallographic transformations from bulk to exfoliated MoS2 were monitored with XPS analysis with shifts from 2H to 1T phases observed. (F) Raman spectroscopy and (G) photoluminescent measurements likewise indicated structural modifications had occurred, seen through changes in vibrational energy bands and luminescent intensity, respectively. (H) Response to NIR light was determined using an IR camera and change in temperature over time. (I) Zeta potential and hydrodynamic size of exfoliated MoS2 in water, PBS, and media. (J) ANS assay shows protein structures are intact in the presence of exfoliated MoS2.
The morphology of MoS2 nanosheets was characterized by atomic force microscopy (AFM) and transmission electron microscopy (TEM). AFM measured layered sheets that were 2 to 7 µm in diameter and ∼3 nm thick (Fig. 1B). The theoretical calculation suggested the S–Mo–S atomic lattice thickness to be about 6.5 Å (8), which indicated the selected nanoflake in the magnified AFM image contained three to five sheets of monolayer MoS2. TEM images indicated smooth topography and crystalline structure of MoS2 (Fig. 1C). The selected area electron diffraction (SAED) pattern of monolayer MoS2 confirmed the formation of 1T phase after exfoliation. Characteristic SAED peaks agreed with XRD data indicating a hexagonal structure.
The elemental composition of bulk (14.8% Mo and 85.2% S) and exfoliated MoS2 (22.8% Mo and 77.8% S) was determined by X-ray photoelectron spectra (XPS) (Fig. 1D). The binding energies (BE) for molybdenum and sulfur core-level electrons revealed the crystallographic phases associated with bulk MoS2 transformed to a few layers (Fig. 1E). The BE for Mo 3d doublets (3d5/2 and 3d3/2) were at 228.5 and 231.7 eV for bulk MoS2, which, after exfoliation, shifted to 228.2 and 231.5 eV, respectively, indicating the 2H to 1T phase transformation. Similarly, the BE for S 2p doublets (2p3/2 and 2p1/2) were 161.4 and 162.6 eV, which correlate with the 2H phase and shifted to 161.1 and 162.3 eV, respectively, following exfoliation indicating 1T phase.
Raman spectroscopy confirmed the vibrational modes associated with the MoS2 lattices for both bulk and exfoliated sheets (Fig. 1F). As expected, the two characteristic modes, E12g corresponding to in-plane vibrations of sulfur atoms with respect to molybdenum and A1g corresponding to out-of-plane vibration of sulfur atoms, had energies of 380.8 and 406.5 cm−1, respectively, in bulk form. Upon exfoliation, the E12g shifted to 376.5 cm−1 and the A1g band shifted to 403.9 cm−1. The relative magnitude of out-of-plane (A1g) vibration was larger than in-plane vibration (E12g) in bulk. After layer exfoliation, the relative magnitude of in-plane vibration (E12g) was larger than out-of-plane vibration (A1g), corroborating more free sulfur atoms. Furthermore, due to the phase change from 2H to 1T, additional vibrational bands such as E1g, J2, and J3 were also observed for the exfoliated MoS2 sample (20).
Furthermore, photoluminescent intensity decreased for exfoliated MoS2 samples due to Li intercalation, resulting in a phase transition from semiconducting 2H to metallic 1T phase (Fig. 1G). This corroborates the observation of phase transition from 2H to 1T in the Raman spectra. The NIR responsiveness was evaluated to ensure photothermal effects could be utilized for cell stimulation. A solution of MoS2 (500 µg/mL and 2 mg/mL) was subjected to NIR light, and the solution temperature was monitored using a thermocouple and IR imaging. After 10 min of exposure to NIR light, the temperature of MoS2 solution (2 mg/mL) was increased by ∼5.5 °C and ∼1.5 °C for the high- (340 mW/cm2) and low- (20 mW/cm2) power NIR light, respectively (Fig. 1H). For low concentration of MoS2 (500 µL/mL) and water, no significant change in temperature was observed. This demonstrated that photothermal effects could be expected within a local cellular environment following NIR exposure to exfoliated MoS2 nanosheets.
Physicochemical Stability of MoS2 Nanosheets
The aqueous stability of exfoliated MoS2 nanosheets was evaluated in deionized water (diH2O), phosphate-buffered saline (PBS), and cell culture media containing fetal bovine serum. MoS2 nanosheets were stable in diH2O and PBS and showed strong negative zeta-potential measurements of −32.7 ± 1.1 mV and −29.2 ± 1.7 mV for diH2O and PBS, respectively (Fig. 1I). After the addition of protein, however, the surface of shear, or the interface of bound charges and diffuse layer, is shifted further from the surface, effectively reducing the zeta potential. This phenomenon was observed as the zeta potential significantly dropped to −17.3 ± 0.8 mV after subjecting MoS2 nanosheets to cell culture media. This indicated that while biomolecules and multivalent ions were strongly interacting with the nanomaterial surface, the repulsive forces between MoS2 nanosheets are reduced via charge screening, leading to eventual flocculation at longer timescales.
To improve nanosheet dispersions within these aqueous media environments, bath or probe sonication can be utilized. These mechanical methods to reduce interactions between sheets may also reduce the effective sheet size. To investigate the effects of type and duration of sonication, dynamic light scattering (DLS) was utilized on aqueous dispersions of MoS2. Samples lacking sonication displayed relatively large nanosheet sizes with an average hydrodynamic diameter of 6.9 ± 0.6 µm. Following sonication (2 min), there was a significant reduction in size to 2.6 ± 0.8 µm (bath sonication) and 0.6 ± 0.03 µm (probe sonication). Extending sonication (10 min) further reduced the hydrodynamic size to 1.0 ± 0.3 µm (bath sonication) and 0.38 ± 0.02 µm (probe sonication). Significant reduction in physical size within 10 min of sonication has important ramifications for sample preparation prior to cellular exposure. Specifically, even a brief sonication to improve dispersions of the nanosheets may result in physical changes to the MoS2. Smaller MoS2 nanosheet sizes will affect binding dynamics to proteins in the aqueous environment and subsequently to the cell membrane. Furthermore, reduction in hydrodynamic size as a result of sonication facilitates improved uptake of the nanosheets. Hence, we selected probe sonication for 10 min for MoS2 (∼400-nm-long and 3- to 4-nm-thick nanosheet) for all future studies.
As previously noted, the hydrodynamic size of MoS2 was influenced by the presence of ions (PBS and media) and protein (media). The adsorption of protein on the MoS2 nanosheet will influence cellular adhesion and internalization due to formation of protein corona on the nanosheet. To evaluate these changes in protein conformation, an 8-anilinonapthalene-1-sulfonic acid (ANS) assay (21) was utilized (Fig. 1J). ANS binds selective cationic amino acid residues (such as arginine and lysine) via formation of an ion pair between the positively charged residues and the sulphonyl groups of the ANS. The formation of these ion pairs results in increased fluorescence emission, and thus changes in conformation can be detected by measuring the relative fluorescence intensity. When a model protein with positive charge at buffered pH, lysozyme (isoelectric point –11.35) is subjected to sodium dodecyl sulfate (SDS), a known disruptor of protein structure, a significant change in protein conformation was observed, as indicated by an increase in fluorescence signal. However, subjecting protein to MoS2 nanosheets resulted in a minimal change in fluorescence signal and therefore protein conformation. Furthermore, following an increase in the concentration of MoS2 nanosheet from 50 to 150 µg/mL, no significant impact on fluorescent signal was observed, indicating maintenance of native protein structure over a range of relevant concentrations. The sequestering proteins with intact native structure would enable clustering of receptors on the cell membrane and stimulate downstream intracellular pathways (22, 23).
Cellular Interaction of MoS2 Nanosheets
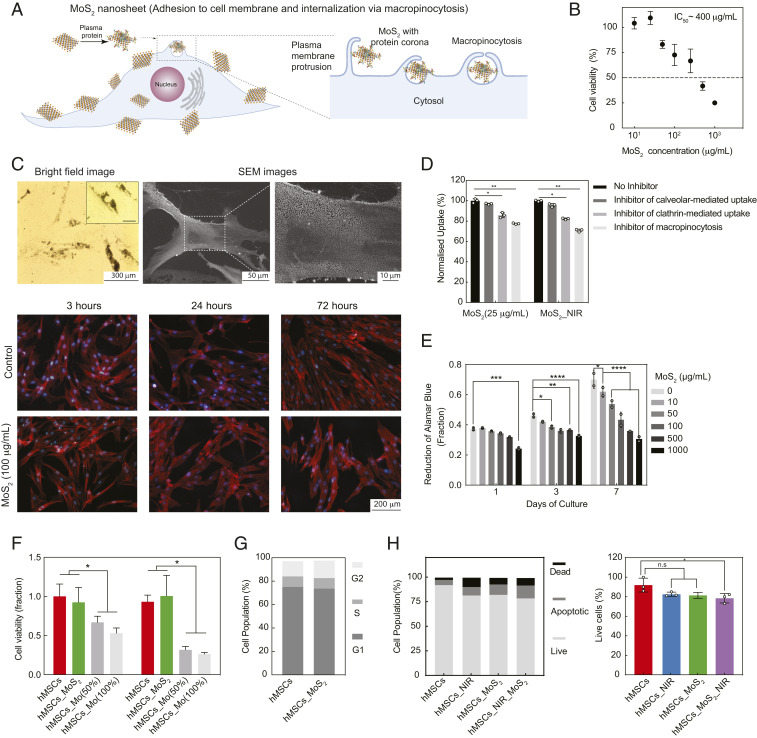
As soon as MoS2 is subjected to physiological fluids, protein corona formed on its surface due to high surface area and charge characteristics. We used gel electrophoresis to observed formation of protein corona. Most of serum protein was adsorbed on MoS2 and no significant difference in protein adsorption was observed due to NIR treatment of MoS2. The colloidal stability of MoS2 in physiological conditions will enable significant interactions of MoS2 nanosheets with the cell membrane. It is expected that due to 2D characteristics, MoS2 can become internalized by cells (Fig. 2A). We investigated cellular compatibility of MoS2 by determining the half inhibitory concentration (IC50) using metabolic activity assay. MoS2 nanosheets showed high cytocompatibility at lower concentrations (<100 µg/mL) after 7 d of culture (Fig. 2B). The IC50 of MoS2 was observed to be ∼400 µg/mL.
Fig. 2.
Cellular interactions of 2D MoS2 with hMSCs. (A) A protein corona forms when 2D MoS2 is suspended in media and adheres to the cell surface and is subsequently internalized by cells. (B) IC50 of MoS2 determined using metabolic assays. (C) Cellular internalization of MoS2 determined using optical, electron, and fluorescence microscopy. (D) MoS2 adhere to cell membrane and internalize via micropinocytosis and clathrin-mediated endocytosis. (E) Effect of MoS2 on metabolic activity over 7 d. (F) Effect of Mo and MoS2 nanosheet at equivalent molarities on cell viability (day 3 and day 7 on the left and right, respectively). (G) Effect of MoS2 on cell cycle. (H) Cell viability when treated with NIR, MoS2, and MoS2_NIR. *P < 0.05; not significant (n.s.) > 0.05.
Bright-field imaging indicated substantial binding of MoS2 nanosheets (100 µg/mL) to cell surfaces (Fig. 2C). Electron microscopy was used to evaluate the extent of membrane coating by these nanomaterials following introduction into the media. Interestingly, scanning electron microscopy (SEM) images of the cell body depicted homogeneous coverage of submicron-sized material across the membrane. The uniform coverage of MoS2 on the cell surface is expected to improve the likelihood of photothermal activation of cells. However, a slight decrease in cell spreading was observed due to MoS2 exposure (100 µg/mL) over a period of 72 h (SI Appendix, Fig. S1).
To determine the uptake mechanism of MoS2 nanosheet, an inhibition assay was performed using endocytosis pathway-specific inhibitors (chlorpromazine hydrochloride to inhibit clathrin-mediated, nystatin to inhibit caveolar-mediated, and wortmannin to inhibit macropinocytosis) (Fig. 2D). The results indicated that the primary mechanisms of nanosheets uptake were clathrin (16 ± 3% inhibition, P < 0.05, one-way ANOVA) and macropinocytosis (26 ± 4% inhibition, P < 0.01, one-way ANOVA). No significant difference was observed using caveolar inhibitor as 4 ± 2% inhibition was observed. It is also important to note that adhered MoS2 nanosheets may increase side scattering, the metric utilized to monitor uptake, which may inflate uptake values and result in relatively modest inhibition using pathway-specific inhibitors. Long-term studies indicate that hMSCs readily proliferated over a course of 7 d in the presence of various concentration of MoS2, indicating cytocompatibility even at higher concentrations (>IC50) (Fig. 2E).
Earlier studies have mentioned that MoS2 degrades in physiological conditions particularly in the presence of peroxidases (24). The effect of Molybdenum (Mo) was evaluated on cell viability by directly exposing 50 and 100% molar concentration of Mo to cells (Fig. 2F). A significant reduction in cell viability was observed due to Mo exposure, while a molar equivalent of MoS2 did not impact cell viability, indicating that Mo is not released from MoS2 in physiological conditions. Other indicators of cellular viability such as cell cycle showed no change as result of MoS2 exposure (Fig. 2G). These results were used to determine the concentration of MoS2 nanosheets (25 to 100 µg/mL) for subsequent photothermal activation.
To further establish the cytocompatibility of MoS2, cellular apoptosis after exposure to MoS2 was measured using Annexin V assay. The cells were treated with MoS2 nanosheets (25 µg/mL) for a period of 24 h with and without NIR exposure. The data indicated that there was no significant difference in live cell populations between control and treated cells, irrespective of NIR treatment (Fig. 2H). The increase in NIR intensity (340 mW/cm2) can be used for photothermal therapy, which will be useful for cancer treatment. Specifically, cells in the selected region can be remotely ablated by focusing NIR light (SI Appendix, Fig. S2). However, here we have used low-intensity NIR (20 mW/cm2) exposure for photothermal modulation.
Global Transcriptomic Profile of Stem Cells Treated with MoS2 Nanosheets
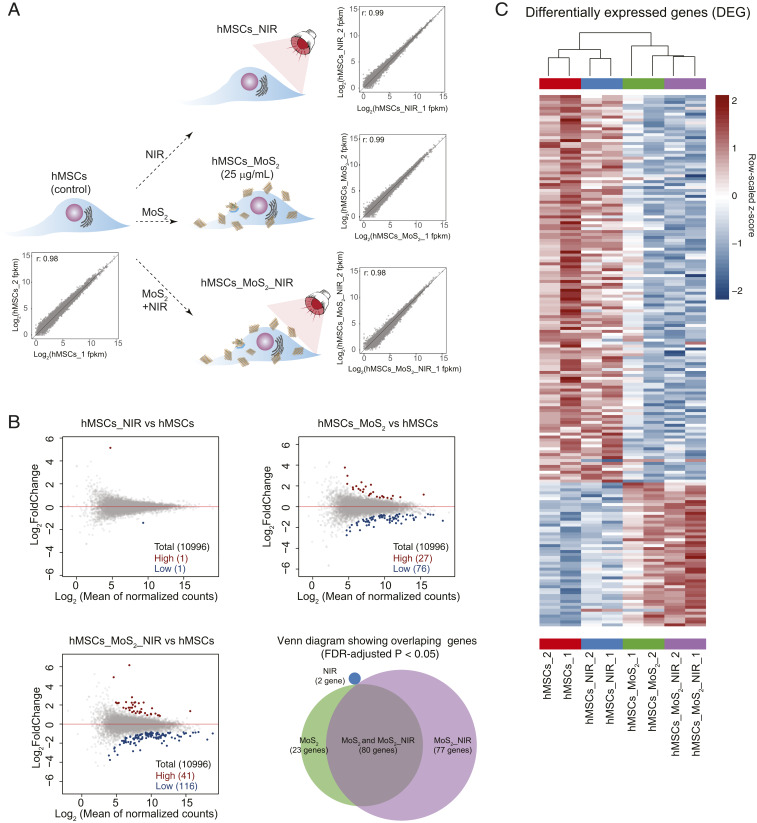
The changes in the steady-state expression levels of cellular transcriptome after treatment with external agents such as nanoparticles and radiation can be determined using RNA-seq (25). To investigate the effect of NIR (20 mW/cm2), MoS2 (25 µg/mL), and NIR stimulation of MoS2 (MoS2_NIR), hMSCs were cultured under four different conditions including untreated hMSCs as control (Fig. 3A). A lower concentration of MoS2 (25 µg/mL) was selected to avoid molecular perturbation due to cellular stress. After 7 d of treatment, the expressed messenger RNA (mRNA) of treated cells were collected and sequenced (Materials and Methods and SI Appendix, Materials and Methods). This early-stage time point was utilized to uncover initial molecular changes, as well as to capture a snapshot of cell–nanomaterial interactions as MoS2 remained on the cell surface up until 7 d.
Fig. 3.
Understanding the effect of NIR, MoS2, and MoS2_NIR on hMSCs using RNA-seq. (A) Pearson correlation coefficient of expressed genes between replicates of different conditions (r = 0.98 for hMSC controls, r = 0.99 for hMSCs_MoS2, r = 0.99 for hMSCs_NIR, and r = 0.98 for hMSCs_MoS2_NIR). (B) MA plot highlighting differentially regulated genes (FDR-adjusted P < 0.05) of the treatment groups against the untreated hMSCs and extent of expression change (gray: all of the expressed genes, red: up-regulated genes, blue: down-regulated genes). Venn diagram indicating differential genes overlap between pairwise comparison of treatment groups. (C) Heat map of row normalized FPKM z-scores of differentially expressed genes across all pairwise comparisons against the untreated hMSCs (red: up-regulated, blue: down-regulated).
Using RNA-seq aligner (26), the sequenced reads were aligned to the reference genome (hg38). The gene expression levels for every gene in every sample was normalized to obtain fragments per kilobase of transcript per million reads (FPKM). The correlation of expressed gene levels (FPKM) for replicate samples subjected to different conditions indicated a high degree of consistency (r = 0.98 for hMSC controls, r = 0.99 for hMSCs_MoS2, r = 0.99 for hMSCs_NIR, and r = 0.98 for hMSCs_MoS2_NIR). The read count for gene expression was modeled by generalized linear models (GLMs) (27), revealing significant gene expression changes between different treatment conditions compared to untreated hMSCs (Fig. 3B and Dataset S1). NIR exposure to hMSCs (hMSCs_NIR) displayed minimal effect on the gene expression program, as only two differentially expressed genes (DEGs, false discovery rate [FDR]-adjusted P < 0.05) were observed. Meanwhile application of MoS2 and MoS2_NIR resulted in more robust changes in the gene expression program of hMSCs. Specifically, hMSCs treated with MoS2 and MoS2_NIR showed significant (FDR-adjusted P < 0.05) change in their gene expression profile, 103 and 157 genes, respectively. Interestingly, our previous work investigating 2D nanomaterial treatment of hMSCs had demonstrated a higher number of DEGs (28), indicating that the chemical and physical characteristics of MoS2 affects a more selective group of cellular pathways. The DEGs in hMSCs_MoS2 and hMSCs_MoS2_NIR showed a decent overlap (80 genes).
The hierarchical clustering of DEGs highlights the concordance of effect of MoS2 and MoS2_NIR treatments compared to untreated control (hMSCs) and hMSCs_NIR (Fig. 3C) across samples. Overall, our results indicated that the effect of MoS2 on hMSCs is relatively moderate compared to previously reported 2D nanomaterials (28). We suspect that the relatively large size and chemical stability of MoS2 nanomaterials limited the cell–nanomaterials interaction. However, our study suggests that NIR-stimulated MoS2 nanomaterials may be used to regulate gene expression on demand.
Photothermal Modulation of Stem Cell Gene Expression
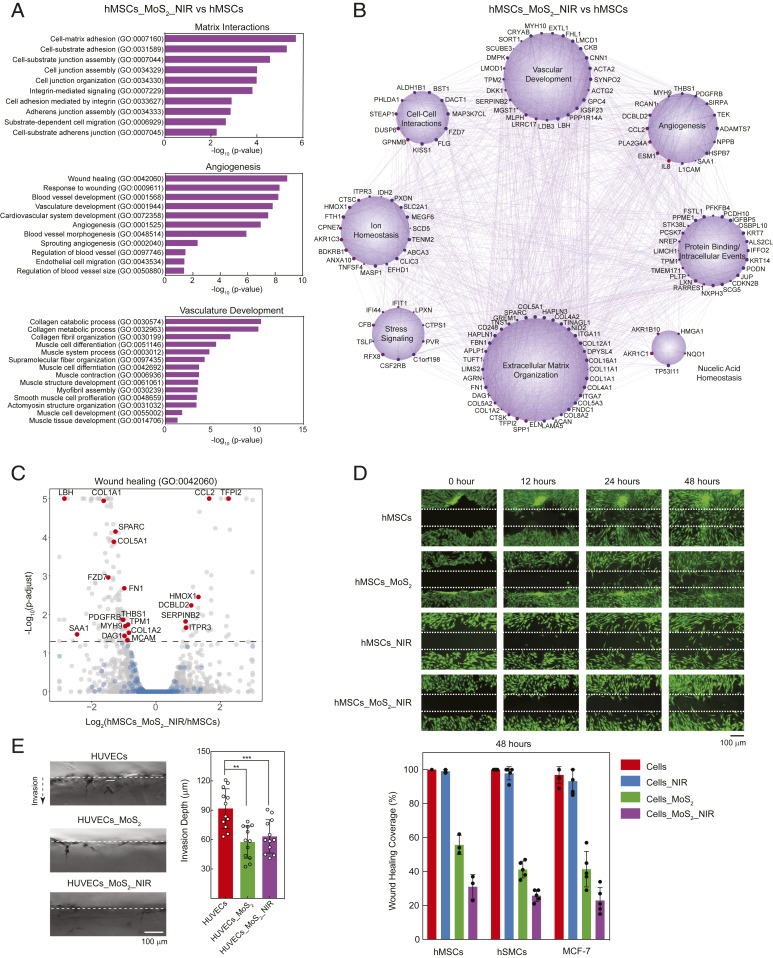
In order to determine the pathways significantly impacted by MoS2 and MoS2_NIR exposure, we utilized a variety of gene set enrichment techniques including GOStats (29) and Cytoscape (30, 31). Gene ontology (GO) enrichment analysis identifies cellular functions, processes, and components that show enrichment for DEGs affected by nanomaterials and NIR treatment. While MoS2 treatment alone showed enrichment for 106 GO terms (P < 0.05), MoS2_NIR treatment resulted in 218 GO terms (P < 0.05). Focusing largely on GO terms with high significance (P value < 0.05), we encountered key terms specific to extracellular matrix organization (GO:0030198) as well as angiogenesis (GO:0001525) and wound healing (GO:0042060) (Fig. 4A). REVIGO analysis (32), a data management technique to reduce redundancies among terms from clustered genes, refined enrichments within the MoS2/NIR treatment group to macromolecule metabolism, extracellular matrix (ECM) organization, response to stimulus, wound healing, and regulation of motility and proliferation (SI Appendix, Fig. S3). These reduced terms correlated well with our GO terms of high significance and demonstrated a clarified progression of cellular events following nanomaterial treatment and NIR stimulation.
Fig. 4.
Functional effect of NIR, MoS2, and MoS2_NIR on cells. (A) GO terms obtained by gene enrichment analysis of DEGs (FDR-adjusted P < 0.05) due to MoS2_NIR treatment. (B) GeneMANIA analysis produced a network of coexpression interactions for differentially expressed genes within the hMSCs_MoS2_NIR population (red: up-regulated, blue: down-regulated; size increases with significance). (C) A volcano plot for the wound healing (GO:0042060), gray: all of the expressed genes, blue: genes associated with the GO term with no significant change in expression, red: genes associated with the GO term that show significantly differential expression due to treatment. (D) Scratch assay used to determine the effect of NIR, MoS2, and MoS2_NIR on migration, cell proliferation, and wound healing of hMSCs, hSMCs, and MCF-7. The dotted lines represent scratch generated using pipette tip. The cell migration across the wound area was determined by imaging the sample after 12, 24, and 48 h. (E) Three-dimensional invasion assay demonstrating ability of HUVECs treated with MoS2 and MoS2_NIR to invade collagen gel.
We likewise created a gene network from coexpression data to examine clusters of these differentially expressed genes (FDR-adjusted P < 0.05) using Cytoscape (30, 31). These clusters grouped genes based on known pathways and signaling function, thereby generating a visualization of genetic interdependencies among multiple cellular processes (Fig. 4B). Out of the pool of genes identified from differential analysis, eight clear clusters of cell function and behavior emerged pertaining to angiogenesis, vascular development, cell–cell interactions, ion homeostasis, stress signaling, ECM organization, nucleic acid homeostasis, and protein binding/intracellular signaling. Specifically, for MoS2-treated hMSCs, there were clear dependencies between genes expressed within angiogenic/vascular development clusters with that of the extracellular matrix.
To evaluate the activation of stress related pathways such mitogen-activated protein kinase cascade due to NIR, MoS2, and MoS2_NIR treatment, we evaluated production of phospho-MEK1/2. MEK1/2 are dual-specificity protein kinases that mediate the phosphorylation of tyrosine and then threonine in ERK1 or ERK2. As no significant production of pMEK1/2 was observed, treatment with NIR, MoS2, and MoS2_NIR did not result in activation of cellular stress-related signaling pathways (SI Appendix, Fig. S4). We also evaluated the differentiation ability of hMSCs after NIR, MoS2, and MoS2_NIR treatment; however, no significant change in expression of early osteogenic marker such as alkaline phosphate was observed (SI Appendix, Fig. S5). Overall, these results indicate that NIR, MoS2, and MoS2_NIR treatment do not cause any cellular stress.
MoS2 and NIR Exposure Suppresses Cell Migration by Modulating Cellular Adhesion
By evaluating the genes comprising highly significant GO terms, we may identify genes responsible for observable changes in cell behavior. Among the cluster of genes related to wound healing (GO:0042060), we noted the majority of significantly DEGs between control cells and those receiving MoS2_NIR were down-regulated (14 out of 20) (Fig. 4C). Among these, the limb bud and heart development gene (LBH) was down-regulated after cells were introduced to MoS2 and MoS2_NIR (log2fold: −1.799 and −2.846) (SI Appendix, Fig. S6). LBH plays a prominent role in cardiogenesis as a transcriptional activator. In addition, LBH interacts with a multitude of genes implicated in a variety of processes, including vascular development and transcriptional events inside the nucleus.
Due to this significant shift in signaling and impact on cell machinery, we investigated the ability of cells to migrate in a simulated wound-healing experiment. Compared to control populations or those receiving NIR exposure alone, hMSCs exposed to MoS2 and MoS2_NIR migrated into a simulated wound at significantly reduced rates (Fig. 4D and SI Appendix, Fig. S7). We further validated this effect on two different cells, human smooth muscle cells (hSMCs) and Michigan Cancer Foundation-7 (MCF-7) breast cancer cells, with both types displaying significantly decreased migration following MoS2 and MoS2_NIR treatment. This indicated that the reduction in cell migration due to MoS2 and MoS2_NIR was not cell-type-specific and utilized universal cell machinery that is affected by MoS2. To further strengthen this hypothesis, we performed three-dimensional invasion assay to determine the effect of MoS2 nanosheets and NIR stimulation on human umbilical vein endothelial cells (HUVECs) migration within a collagen matrix (33). HUVECs treated with MoS2 and MoS2_NIR showed significant decrease in invasion ability as determined by the invasion depth as compared to control (Fig. 4E).
All these experimental results validated our observations from RNA-seq data. Specifically, GO term enrichment analysis indicated highly related terms, including regulation of cell motility (GO:2000145) and negative regulation of locomotion (GO:0040013). Within these GO terms for the MoS2_NIR group, genes like PODN (log2fold: −1.526), GREM1 (−1.232), CCL2 (1.651), and HMOX1 (1.320) were all significantly differentially expressed. Each of these markers plays a distinct role in ECM binding or cytoskeletal remodeling during cell locomotion. These genes, among others, may drive the changes in cell motility following MoS2_NIR treatment. For example, GREM1 is known to stimulate cellular motility and induce potent angiogenic response in vivo (34, 35). Due to its role in breast cancer motility, invasion, and angiogenesis, GREM1 is considered an attractive molecular target in cancer therapeutics. Suppression of GREM1 expression due to MoS2_NIR treatment emphasizes its therapeutic potential in cancer. In addition, MoS2_NIR treatment upregulated HMOX1, which has shown to inhibit migration and cell invasion in breast cancer. It is important to note that spatial rearrangement of cells is necessary for angiogenesis. Based on these results, we therefore believe that MoS2 and MoS2_NIR treatment may limit the angiogenic potential of cells involved in vascular development or wound-healing responses by modulating cellular motility.
MoS2 and Concurrent NIR Exposure Modulate Integrin Signaling
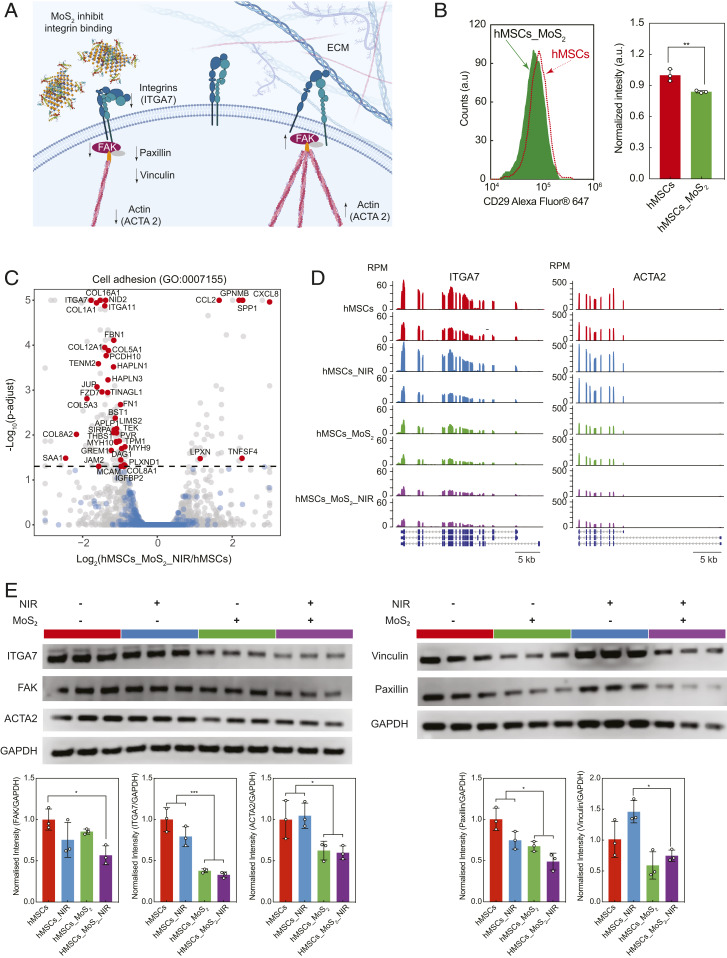
The reduction in cell motility due to MoS2 and MoS2_NIR treatment might be affected by integrin signaling (Fig. 5A). Integrin belongs to a glycoprotein family consisting of a noncovalently linked α- and β-subunits. They form heterodimers upon interactions with ECM components and play critical roles in signal transduction and functions such as cellular migration, differentiation, proliferation, and apoptosis. One of the potential approaches to investigate the interaction between MoS2 nanosheets and integrin molecules is to quantify the amount of integrin beta-1 (CD29) using flow cytometry. Beta-1 integrins recognize the sequence R–G–D in a wide array of ligands and can form heterodimers with integrin alpha-7. Integrin alpha-7/beta-1 is known to regulate cell adhesion, matrix deposition, cell motility, and angiogenesis. The flow cytometry data showed a significant decrease in unbound integrin beta-1 levels in cells treated with MoS2 nanosheets (Fig. 5B and SI Appendix, Fig. S8), thus indicating that nanosheets inhibit integrin function. Although the flow experiments support our primary hypothesis that integrin signaling is involved, antibody blocking and knock-down studies are appropriate methods to further confirm this hypothesis.
Fig. 5.
Role of integrin expression on cell adhesion due to NIR, MoS2, and MoS2_NIR treatment. (A) Schematic showing the potential interaction between MoS2 nanosheets and integrin molecules at the cell surface. (B) Quantification of unbound integrin beta-1 (CD29) in cells treated with and without MoS2 nanosheets. (C) A volcano plot for the cell adhesion (GO:0007155), gray: all of the expressed genes, blue: genes associated with the GO term with no significant change in expression, red: genes associated with the GO term that show significant difference in expression due to treatment. (D) Gene track showing normalized mRNA expression of integrin alpha-7 (ITGA7) and smooth muscle alpha-2 actin (ACTA2) for hMSCs, hMSCs_NIR, hMSCs_ MoS2, and hMSCs MoS2_NIR. (E) Protein expression level of ITGA7, FAK, vinculin, paxillin, and smooth muscle actin (ACTA2) determined using Western blot for hMSCs, hMSCs_NIR, hMSCs_ MoS2, and hMSCs MoS2_NIR. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is internal control. *P < 0.05, ***P < 0.001.
To identify a potential mechanism by which MoS2 nanosheets regulate cellular motility, we looked at another important GO term that showed a high number of differentially expressed genes: cell adhesion (GO:0007155) (Fig. 5C). A large number of genes in this GO term were down-regulated (23 of 28) due to MoS2_NIR treatment compared to control. Some of the prominent genes in this GO term are related to extracellular matrix proteins (COL11A1, COL4A1, and FN1), cell–ECM adhesion receptors (ITGA11, ITGA7, IGFBP2, IGFBP5, NID2, and HAPLN3), and cytoskeletal rearrangement (ACTA2, ACTG2, MYH9, and MYH10) (SI Appendix, Fig. S9). In particular, the down-regulation of integrin subunits (ITGA7, log2fold: −1.772 and ITGA11, log2fold: −1.405) in conjugation with the cytoskeletal protein (ACTA2, log2fold: −1.506) indicates that MoS2 nanosheets may be interfering with surface receptor-mediated focal adhesion formation (Fig. 5D).
To further validate this hypothesis, we quantified the protein synthesis of integrin alpha-7 (ITGA7), focal adhesion kinase (FAK), smooth muscle actin (ACTA2), vinculin, and paxillin (Fig. 5E). FAK, vinculin, and paxillin are known to play key roles in integrin-dependent cellular adhesion, speeding, and motility (36, 37). The results indicate the ITGA7 synthesis was significantly decreased in cells treated with MoS2 and MoS2_NIR. Interestingly, no significant difference was observed due to photothermal modulation on ITGA7 production. The trend in ITGA7 expression in different conditions is similar to results observed in wound-healing experiments, indicating the influence of MoS2 and MoS2_NIR on cellular migration. The expression of other proteins involved in focal adhesion such as FAK, vinculin, paxillin, and ACTA2 showed significant decrease in expression in MoS2_NIR compared to control (hMSCs). To further visualize the effect of MoS2 nanosheets on focal adhesion and subsequent cellular spreading, the cellular area was measured after treating with MoS2 nanosheets (SI Appendix, Fig. S1). We noted a gradual decrease in cellular spreading with an increase in MoS2 concentration, indicating reduced integrin-mediated cell adhesion.
Conclusions
Chemically exfoliated MoS2 is an emerging biomaterial with unique photoresponsive ability. We investigated the biological interactions of MoS2 nanosheets with proteins and cells. The high surface area of MoS2 nanosheets facilitates protein adsorption and cellular adhesion/internalization. Once localized to the cell surface, the MoS2 nanosheets can be exploited with NIR light to generate thermal stimulation. The effect of MoS2 nanosheets and subsequent stimulation with NIR light were investigated via RNA-seq to provide a global and unbiased snapshot of gene expression of the cells. No significant effect of NIR exposure alone was observed on gene expression of hMSCs. However, treatment with MoS2 nanosheets and subsequent stimulation with NIR light influenced ∼103 and 157 genes in hMSCs, respectively. Most of these genes were related to cellular migration and wound healing. Further validation using a wound-healing assay, integrin signaling, and formation of focal adhesion kinase confirmed the role of MoS2 and photothermal modulation on cellular functions. Overall, the combination of MoS2 and NIR light may provide a novel tool to modulate cellular activity for potential applications in regenerative medicine and cancer therapeutics.
Materials and Methods
See SI Appendix for detailed materials and methods. Detailed methods for nanoparticle characterization, in vitro studies, and RNA-seq can be found in SI Appendix, Materials and Methods.
Synthesis and Characterization.
The bulk MoS2 was chemically exfoliated following previously reported protocols to obtain MoS2 nanosheets (3, 18, 19). The structure of MoS2 nanosheets were evaluated using XRD (Bruker D8 Advanced), AFM (Bruker Dimension Icon Nanoscope), TEM (JEOL JEM-2010), XPS (Omicron XPS system with Argus detector), Raman spectroscopy (LabRam HR confocal Raman microscope; Horiba Inc.), DLS (Zetasizer Nano ZS, Malvern Instrument), and zeta potential. Cellular interactions of MoS2 nanosheet was determined using hMSCs (obtained from Tissue Culture Core, Institute of Regenerative Medicine, Texas A&M University Health Science Center). The following in vitro studies were performed: metabolic activity (Alamar Blue assay), cell cycle (propidium iodide), apoptosis assay (Annexin V), and cytosketal staining (actin-phalloidin and nucleus-DAPI staining).
RNA-Seq and Analysis.
RNA-seq was performed on hMSCs-treated with NIR (20 mW/cm2), MoS2 nanosheet, and both, using a Nova seq platform (Illumina Nova sEq. 6000) utilizing TruSeqRNA preparation and 75 paired-end read length. Sequenced reads were aligned to the human reference genome (hg38) using a RNA-seq aligner, Spliced Transcripts Alignment to a Reference (STAR) (26). Expression of a gene was determined by counting the number of uniquely mapped reads overlapping the coding exons normalized by gene length in FPKM. Genes expressed with >1 FPKM in at least half of the samples of any condition were only used for the analysis. The gene expression read counts were modeled as a negative binomial distribution in GLMs (27) to determine the DEGs. All of the analysis was performed using R. The GO enrichment analysis was done using GOStats Bioconductor package (29). For network formation (Cytoscape) (31) and GO term enrichment (GeneMANIA and ClueGO), only genes with FDR-adjusted P < 0.05 were selected. REVIGO (32) was performed to visualize GO clustering. Western analysis was performed using the iBind system (iBind; Invitrogen).
Statistical Analysis.
Determination of statistical significance between multiple groups was completed via ANOVA with the Tukey method. Significant P values were considered <0.05 unless otherwise noted. All analysis was completed in GraphPad Prism.
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE141456).
Supplementary Material
Acknowledgments
A.K.G. acknowledges financial support from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health Director’s New Innovator Award (DP2 EB026265). We thank Dr. Holly Gibbs for her assistance with experiments utilizing the NIR laser. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency. Some of the schematics are prepared using https://biorender.com/.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition:The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE141456).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914345117/-/DCSupplemental.
References
- 1.Novoselov K. S., Mishchenko A., Carvalho A., Castro Neto A. H., 2D materials and van der Waals heterostructures. Science 353, aac9439 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Chimene D., Alge D. L., Gaharwar A. K., Two-dimensional nanomaterials for biomedical applications: Emerging trends and future prospects. Adv. Mater. 27, 7261–7284 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Chhowalla M. et al., The chemistry of two-dimensional layered transition metal dichalcogenide nanosheets. Nat. Chem. 5, 263–275 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Gaharwar A. K. et al., 2D nanoclay for biomedical applications: Regenerative medicine, therapeutic delivery, and additive manufacturing. Adv. Mater. 31, e1900332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T. et al., Drug delivery with PEGylated MoS2 nano-sheets for combined photothermal and chemotherapy of cancer. Adv. Mater. 26, 3433–3440 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Chou S. S. et al., Chemically exfoliated MoS2 as near-infrared photothermal agents. Angew. Chem. Int. Ed. Engl. 52, 4160–4164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Late D. J. et al., Sensing behavior of atomically thin-layered MoS2 transistors. ACS Nano 7, 4879–4891 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Radisavljevic B., Radenovic A., Brivio J., Giacometti V., Kis A., Single-layer MoS2 transistors. Nat. Nanotechnol. 6, 147–150 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Jin K., Xie L. M., Tian Y., Liu D. M., Au-modified monolayer MoS2 sensor for DNA detection. J. Phys. Chem. C 120, 11204–11209 (2016). [Google Scholar]
- 10.Nguyen E. P. et al., Electronic tuning of 2D MoS2 through surface functionalization. Adv. Mater. 27, 6225–6229 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Frindt R. F., Single crystals of Mos2 several molecular layers thick. J. Appl. Phys. 37, 1928 (1966). [Google Scholar]
- 12.Joensen P., Frindt R. F., Morrison S. R., Single-layer Mos2. Mater. Res. Bull. 21, 457–461 (1986). [Google Scholar]
- 13.Schumacher A., Scandella L., Kruse N., Prins R., Single-layer Mos2 on mica–Studies by means of scanning force microscopy. Surf. Sci. 289, L595–L598 (1993). [Google Scholar]
- 14.Jaiswal M. K., Singh K. A., Lokhande G., Gaharwar A. K., Superhydrophobic states of 2D nanomaterials controlled by atomic defects can modulate cell adhesion. Chem. Commun. (Camb.) 55, 8772–8775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaiswal M. K. et al., Vacancy-driven gelation using defect-rich nanoassemblies of 2D transition metal dichalcogenides and polymeric binder for biomedical applications. Adv. Mater. 29, 1702037 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Teo W. Z., Chng E. L. K., Sofer Z., Pumera M., Cytotoxicity of exfoliated transition-metal dichalcogenides (MoS2, WS2, and WSe2 ) is lower than that of graphene and its analogues. Chemistry 20, 9627–9632 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Appel J. H. et al., Low cytotoxicity and genotoxicity of two-dimensional MoS2 and WS2. ACS Biomater. Sci. Eng. 2, 361–367 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Eda G. et al., Photoluminescence from chemically exfoliated MoS2. Nano Lett. 11, 5111–5116 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Yuwen L. et al., Rapid preparation of single-layer transition metal dichalcogenide nanosheets via ultrasonication enhanced lithium intercalation. Chem. Commun. (Camb.) 52, 529–532 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Cheng P., Sun K., Hu Y. H., Memristive behavior and ideal memristor of 1T phase MoS2 nanosheets. Nano Lett. 16, 572–576 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Gasymov O. K., Glasgow B. J., ANS fluorescence: Potential to augment the identification of the external binding sites of proteins. Biochim. Biophys. Acta 1774, 403–411 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischer C. C., Payne C. K., Secondary structure of corona proteins determines the cell surface receptors used by nanoparticles. J. Phys. Chem. B 118, 14017–14026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shang L., Nienhaus K., Nienhaus G. U., Engineered nanoparticles interacting with cells: Size matters. J. Nanobiotechnology 12, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurapati R. et al., Enzymatic biodegradability of pristine and functionalized transition metal dichalcogenide MoS2 nanosheets. Adv. Funct. Mater. 27, 1605176 (2017). [Google Scholar]
- 25.Nel A. E. et al., Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 8, 543–557 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrow J. K. et al., Widespread changes in transcriptome profile of human mesenchymal stem cells induced by two-dimensional nanosilicates. Proc. Natl. Acad. Sci. U.S.A. 115, E3905–E3913 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falcon S., Gentleman R., Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Shannon P. et al., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohl M., Wiese S., Warscheid B., Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howell D. W., Peak C. W., Bayless K. J., Gaharwar A. K., 2D nanosilicates loaded with proangiogenic factors stimulate endothelial sprouting. Adv. Biosyst. 2, 1800092 (2018). [Google Scholar]
- 34.Mitola S. et al., Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood 116, 3677–3680 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Karagiannis G. S., Berk A., Dimitromanolakis A., Diamandis E. P., Enrichment map profiling of the cancer invasion front suggests regulation of colorectal cancer progression by the bone morphogenetic protein antagonist, gremlin-1. Mol. Oncol. 7, 826–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C., Focal adhesion: A focal point in current cell biology and molecular medicine. Cell Adhes. Migr. 1, 13–18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X., Guan J.-L., Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 63, 610–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE141456).