Abstract
Objective
Ageing is accompanied by deterioration of multiple bodily functions and inflammation, which collectively contribute to frailty. We and others have shown that frailty co-varies with alterations in the gut microbiota in a manner accelerated by consumption of a restricted diversity diet. The Mediterranean diet (MedDiet) is associated with health. In the NU-AGE project, we investigated if a 1-year MedDiet intervention could alter the gut microbiota and reduce frailty.
Design
We profiled the gut microbiota in 612 non-frail or pre-frail subjects across five European countries (UK, France, Netherlands, Italy and Poland) before and after the administration of a 12-month long MedDiet intervention tailored to elderly subjects (NU-AGE diet).
Results
Adherence to the diet was associated with specific microbiome alterations. Taxa enriched by adherence to the diet were positively associated with several markers of lower frailty and improved cognitive function, and negatively associated with inflammatory markers including C-reactive protein and interleukin-17. Analysis of the inferred microbial metabolite profiles indicated that the diet-modulated microbiome change was associated with an increase in short/branch chained fatty acid production and lower production of secondary bile acids, p-cresols, ethanol and carbon dioxide. Microbiome ecosystem network analysis showed that the bacterial taxa that responded positively to the MedDiet intervention occupy keystone interaction positions, whereas frailty-associated taxa are peripheral in the networks.
Conclusion
Collectively, our findings support the feasibility of improving the habitual diet to modulate the gut microbiota which in turn has the potential to promote healthier ageing.
Keywords: diet, enteric bacterial microflora, ageing, inflammation, intestinal bacteria
Significance of this study.
What is already known about this subject?
Ageing is associated with deterioration of multiple bodily functions and inflammation, leading to the onset of frailty.
The onset of frailty is associated with changes in the gut microbiota that are linked with a restricted diversity diet.
The Mediterranean dietary regime is positively associated with health.
What are the new findings?
Adherence to the Mediterranean diet led to increased abundance of specific taxa that were positively associated with several markers of lower frailty and improved cognitive function, and negatively associated with inflammatory markers including C-reactive protein and interleukin-17.
These associations were independent of host factors such as age and body mass index.
Inferred microbial metabolite profiling indicated that the diet-modulated microbiome change was associated with an increase in short/branch chained fatty acid production and lower production of secondary bile acids, p-cresols, ethanol and carbon dioxide.
Microbiome ecosystem network analysis showed that the bacterial taxa enriched due to the MedDiet intervention occupy keystone interaction positions, whereas frailty-associated taxa are peripheral in the networks.
Significance of this study.
How might it impact on clinical practice in the foreseeable future?
Our findings support the feasibility of changing the habitual diet to modulate the gut microbiota which in turn has the potential to promote healthier ageing.
Our findings also provide a short list of candidate taxa that can be investigated further as live biotherapeutic agents for direct administration to older subjects to reduce the onset of frailty.
Introduction
Frailty that accompanies ageing involves failure of multiple physiological systems1 and a persistent activation of the innate immune inflammatory response.2 Frailty may include the development of chronic low-grade inflammation (ie, inflamm-ageing),3 loss of cognitive function,4 sarcopenia5 and the development of chronic diseases like diabetes and atherosclerosis.3 Modification of dietary patterns such as adopting a Mediterranean diet (MedDiet) has been suggested as a major therapeutic strategy to address frailty.6 The MedDiet regimen is characterised by increased consumption of vegetables, legumes, fruits, nuts, olive oil and fish and low consumption of red meat and dairy products and saturated fats.7 Adherence to a MedDiet is associated with reduced mortality, increased antioxidant activity, reduced incidences of several diseases, as well as reduced inflammation.8 Several studies have shown that increased adherence to the MedDiet is linked to reduced frailty.9 Beyond the negative association with disease, higher level adherence to the MedDiet has been associated with beneficial changes in gut microbiome composition, with reduction in proteobacterial abundance accompanied by increased levels of short chain fatty acid production.10 11 On a global basis, the majority of elderly people do not consume a MedDiet and, in fact, a major challenge in elderly healthcare is the consumption of a restricted diet which is associated with a low-diversity gut microbiome, especially in subjects in long-term residential care.12 13 In previous work we have used fine detail bioinformatic (bi-clustering) analysis to identify specific microbial taxa that are lost in incremental stages in the transition from high-diversity microbiome healthy subjects to low-diversity frail subjects.14 In a recently completed 6-month dietary intervention in which elderly individuals were supplemented with up to 20 g daily of five prebiotics, multiple gut microbial taxa responded to the prebiotic supplementation,15 but there was no change in overall microbiota alpha diversity and trends towards a reduction of inflammatory markers did not reach overall statistical significance. We thus reasoned that a more dramatic dietary intervention was required.
The NU-AGE dietary intervention project aimed to study the effect of the administration of a customised MedDiet for 12 months in a large cohort of more than 1200 elderly individuals aged 65–79 years,16 distributed across five different countries (Poland, Netherlands, UK, France and Italy). Baseline and post-intervention results from this study have reported gender- and country-specific differences for measured metabolite levels as well as body composition data. A significant association was observed between increased adherence to the NU-AGE MedDiet and enhanced global cognitive ability and episodic memory.17 Moreover, higher adherence has been shown to reduce the rate of bone loss in individuals with osteoporosis18 and to improve innate immune function,19 blood pressure and arterial stiffness.20 In the current study we have profiled the effect of the NU-AGE MedDiet on the gut microbial community of a subset of participants from the NU-AGE trial comprising 612 individuals (289 controls (145 men and 144 women) and 323 with the NU-AGE MedDiet (141 men and 182 women)). A variety of beneficial outcomes were correlated to microbiome alterations.
Methods
Study participants and dietary intervention
The NU-AGE study is a 1-year, randomised, multicentre, single-blind, controlled trial (registered with clinicaltrials.gov, NCT01754012). Details on the recruitment of participants and the dietary intervention and the collection of metadata corresponding to anthropometry, frailty and cognitive response have been previously described.17 21 Online supplementary table 1 provides descriptive statistics of participants by country for whom paired microbiome profiles at baseline and the final time point were available.
gutjnl-2019-319654supp001.pdf (39.8KB, pdf)
Measurement of inflammatory and adiposity related hormones
Methods for the measurements of inflammatory markers have been previously described.22 Online supplementary text 1 briefly summarises the techniques used for this purpose.
gutjnl-2019-319654supp002.pdf (48.2KB, pdf)
DNA extraction and 16S rRNA gene sequencing
Microbial DNA was extracted from stool samples using the repeated bead beating method as previously described,23 with some modifications.24 The detailed protocol adopted for the DNA and 16S rRNA gene sequencing is described in online supplementary text 2.
gutjnl-2019-319654supp003.pdf (56.4KB, pdf)
Bioinformatic and biostatistical analysis
Online supplementary text 3 provides a complete description of the methodology used for the bioinformatics and the multivariate statistical analysis of the amplicon sequence data. This includes preprocessing of sequenced reads,25 identification of and removal of chimaeras,26 27 taxonomic classification of Operational Taxonomic Units (OTUs),28 29 machine learning-based identification of microbiome taxa associated with the dietary intervention30 (described in online supplementary figure 1), identification of taxonomic modules using the iterative Binary Bi-clustering of Gene-sets (iBBiG) approach,31 association analysis of dietary adherence and diet-associated taxonomic markers with the various components of diet as well as with the markers of frailty and inflammation, computation of MedDiet-associated microbiome indices (described pictorially in online supplementary figure 2) and the association analysis of these indices with dietary components, inflammation and frailty, obtaining inferred metabolite profiles based on per-sample species abundances and previously curated mappings of experimentally validated species-to-metabolite links32 33 and generation and visualisation of co-occurrence networks and computation of centrality measures (see online supplementary text 3).30 34
gutjnl-2019-319654supp005.pdf (71.1KB, pdf)
gutjnl-2019-319654supp004.pdf (1.6MB, pdf)
Results
Diet and microbiome profiles co-vary and differ between countries at baseline
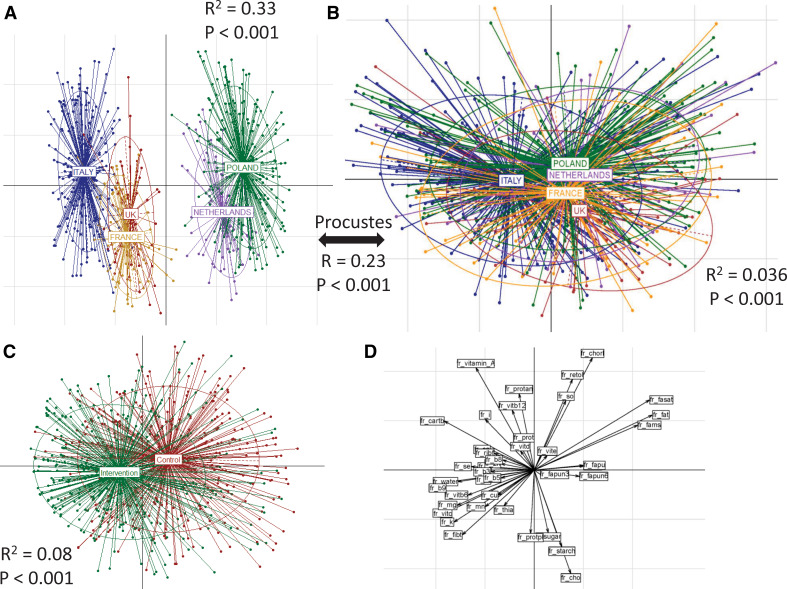
Overall, there were 612 individuals (across the control and intervention cohorts) for whom paired microbiome data were collected at both the baseline and 1 year (referred to as ‘final’) time points. While the age ranges of the individuals in the control and intervention cohorts across countries were similar, there was a marginally higher representation of women in the intervention cohort (Fisher's test p<0.12; online supplementary table 1). Principal coordinate analysis (PCoA) indicated significant dietary differences at baseline between the countries showing three distinct subgroups (figure 1A) (PERMANOVA p<0.001: R2=0.33): the first containing Italian subjects; the second containing UK and French subjects; and the third containing those from the Netherlands and Poland. This specific pattern of clustering was also observed at the level of PCoA (based on Spearman distances) using the 16S rDNA OTU profiles at baseline (figure 1B) (significant: PERMANOVA p<0.001; although with considerable overlaps: R2=0.036). While the Italian subjects had a distinct microbiome composition, those from UK/France and Poland/Netherlands were more like each other. Procrustes analysis of the food consumption and the OTU abundance profiles confirmed a significant association between diet and microbiome composition (figure 1A, B; online supplementary figure 3; Procrustes RV coefficient 0.23; p<0.001). Specific microbiome components drove country-specific separations at baseline (Mann–Whitney test FDR-corrected p<0.15; online supplementary figure 4A–B). As expected, the dietary variations within the intervention group were significantly different from the control group (envfit p<0.006) (figure 1C). These changes in the intervention group were primarily driven by an increase in the intake of fibres, vitamins (C, B6, B9, thiamine) and minerals (Cu, K, Fe, Mn, Mg), while changes in the controls were associated with an increase in fat intake (saturated fats and mono-unsaturated fatty acids) relative to the MedDiet intervention group (figure 1D).
Figure 1.
Baseline habitual diet and microbiota composition separate and co-vary by country, and the dietary intervention altered macronutrient profiles. Principal component analysis (PCoA) plots of (A) baseline dietary profiles and (B) baseline 16S microbiome profiles across the five different countries. For both, the PERMANOVA p values showing the significance of the association with the countries are also indicated. For the association between the dietary frequencies, the microbiome profiles, R2 and the significance values obtained using the Procrustes analysis are also shown. The results indicate that there are country-specific patterns in dietary habits which are also reflected in the microbiome profiles. (C) PCoA plots showing the distinct variations in the dietary patterns in the intervention and control cohorts. The PERMANOVA p values of these differences are also indicated. This reflects the effect of the dietary intervention to detect the specific dietary components driving these effects. Associations were computed between the intake frequencies of the components and the two PCoA axes (PCoA1 and PCoA2). These associations are plotted in (D). While the intervention group is primarily driven by an increase in consumption of fibres, vitamins (C, B6, B9, thiamine) and minerals (Cu, K, Fe, Mn, Mg), the changes in controls are associated with an increase in fats consumption.
Increasing adherence to the NU-AGE MedDiet influences specific components of the gut microbiome previously associated with health
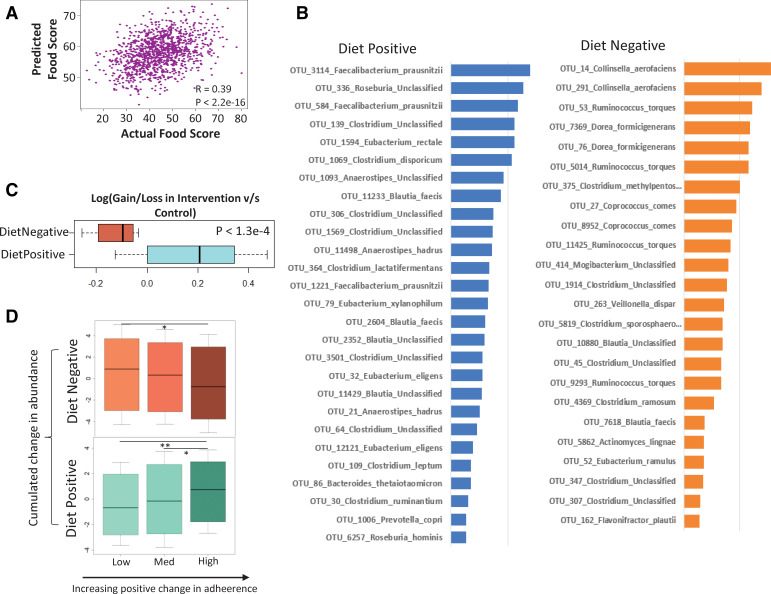
There were no significant changes in the global gut microbiota diversity in the subjects from individual countries in the intervention and control groups (online supplementary figure 5). However, we observed that, across the study, increasing adherence to the diet was associated with an attenuated loss of microbiome diversity (table 1). For a finer detailed microbiota–diet association analysis, we used adherence scores to the MedDiet, previously calculated based on the NU-AGE Food Based Dietary Guidelines (FBDG).35 These are recommendations that act as the basis for facilitating or measuring adherence to healthy eating initiatives or dietary interventions for improving public health. The NU-AGE FBDG covered 15 dietary goals including a vitamin D supplement that has been described in detail by Berendsen et al.35 We created Random Forest (RF) models to predict dietary adherence from microbiome profiles at both the baseline and final (1-year) time points. For both models, the correlations observed between the predicted food score (using the Random Forest model) and the actual food score were significant (baseline: R=0.27; p<1.2e-11; final: R=0.30; p<2.2e-14) (online supplementary figure 6A, B), indicating that there was a clear association between the microbiome and adherence to the MedDiet. For either time point, an optimal set of 75 OTUs provided the highest predictive performance (a total number of 129 OTUs combining both; see online supplementary figure 6C, D) to identify the microbiome response to the MedDiet. We refer to these as 'diet-responsive' OTUs/taxa/markers throughout this study. Overall, using this optimal set of OTUs, the correlation between the predicted and the observed adherence score was 0.39 (p<2.2 e-16) (figure 2A). The list of the top predictive OTUs along with their taxonomic classification (obtained using SPINGO28) is provided in online supplementary table 2.
Table 1.
High adherence to a MedDiet attenuates the loss of diversity of the gut microbiome
| Low adherence | Medium adherence | High adherence | ||||||||||
| Estimate | Standard error | Z value | P value | Estimate | Standard error | Z value | P value | Estimate | Standard error | Z value | P value | |
| Intercept | 387.11 | 113.53 | 3.41 | 0.00065*** | 283.04 | 117.05 | 2.42 | 0.016* | 412.97 | 97.48 | 4.24 | 2.3e-5** |
| Time point | −9.51 | 4.85 | −1.96 | 0.049* | −9.34 | 4.99 | −1.87 | 0.061(.) | −3.84 | 4.93 | −0.78 | 0.44 |
| Gender | -2 | 12.34 | −0.16 | 0.87 | 16.97 | 12.62 | 1.34 | 0.179 | −7.40 | 10.93 | −0.68 | 0.5 |
| Age | −0.19 | 1.57 | −0.12 | 0.90 | 1.21 | 1.64 | 0.74 | 0.46 | −0.54 | 1.37 | −0.39 | 0.69 |
A significant decline in diversity was observed across the time points in the low adherence group (as indicated in the estimate value).
Data tabulated are from regression analysis of the change in gut microbial diversity across the time points (baseline vs final), taking age and gender as the confounders in the three adherence change groups.
The decline attenuated from being marginally significant in the medium adherence group to non-significant in the high adherence group. Please refer to the Methods section for the definition of ‘low’, ‘medium’ and ‘high’ adherence groups of individuals.
The notations used for the p-values of significance are **P < 0.01; *P < 0.05 and; ***P < 0.10
Figure 2.
Identification of diet responsive taxa by machine learning. (A) Correlation between the actual and predicted diet scores obtained using the random Forest approach. (B) Ranked feature importance scores of the top marker Operational Taxonomic Units (OTUs) responding positively and negatively to diet, along with their taxonomic affiliations (see Methods section for the selection of the top markers significantly associated with the food score). Top markers having a significant positive or negative association with diet scores were tagged as ‘DietPositive’ and ‘DietNegative’, respectively. The two groups show distinct taxonomic classifications. While DietPositive markers have an over-representation of species like Faecalibacterium prausnitzii, Eubacterium and Roseburia, DietNegative markers are characterised by the presence of Ruminococcus torques, Collinsella aerofaciens, Coprococcus comes, Dorea formicigenerans, Clostridium ramosum. The associations of the different groups with the adherence scores are also reflected in the changes across the time points between the intervention and control cohorts (as shown in C). (C) Boxplot showing the log-fold change in the gain/loss ratios of the various taxa (ie, the number of individuals in which a given OTU is increased divided by the number of individuals in which it is decreased across the time points) in the intervention cohorts compared with non-intervention in the two groups. While the DietPositive OTUs had a relatively positive increase in the intervention cohort (compared with the non-intervention group), changes in the DietNegative indicated a significant decrease with the intervention. (D) Boxplots showing the variation in the across time point changes in the DietPositive and the DietNegative OTUs in groups of individuals obtained after dividing them into three tertile groups (low, medium and high) based on increasing positive changes in adherence to the NU-AGE diet. The p values of the significance of the association are indicated as ****p<0.0001, ***p<0.001, **p<0.01 and *p<0.05.
gutjnl-2019-319654supp006.pdf (62.9KB, pdf)
A total of 44 top predictive OTUs had a positive association with adherence scores (enriched with increasing adherence to diet) and 45 had a negative association (depleted with MedDiet adherence) (see Methods section). We refer to these as ‘DietPositive’ and ‘DietNegative’ OTUs, respectively. A subset of these OTUs (with defined taxonomic classifications) along with their absolute feature importance scores is shown in figure 2B. The sets of taxa comprising these two groups are distinct. The DietPositive OTUs were mainly assigned to Faecalibacterium prausnitzii, along with Roseburia (R. hominis), Eubacterium (E. rectale, E. eligens, E. xylanophilum), Bacteroides thetaiotaomicron, Prevotella copri and Anaerostipes hadrus. A majority of these taxa have previously reported positive health associations including production of short chain fatty acids (SCFAs) and anti-inflammatory properties as well as negative associations with diseases like type 2 diabetes and colorectal cancer.36–38 F. prausnitzii had also been negatively associated with frailty onset in the elderly.39 In contrast, the DietNegative OTUs mainly belonged to Ruminococcus torques, Collinsella aerofaciens, Coprococcus comes, Dorea formicigenerans, Clostridium ramosum, Veillonella dispar, Flavonifractor plautii and Actinomyces lingnae. Increase in the abundances of R. torques, C. aerofaciens, C. ramosum and V. dispar have been associated with type 2 diabetes and colorectal cancer, atherosclerosis, cirrhosis and inflammatory bowel disease.38 40–43 These findings collectively suggest that adherence to the MedDiet has the potential to modulate the microbiome in a direction positively associated with health.
Notably, in spite of country-specific microbiome composition differences at baseline (figure 1B; online supplementary figure 4A, B) and different dietary adherences (online supplementary figure 7A) (as also reported by previous studies on this cohort),35 44 the diet-responsive taxa identified across the entire cohort were largely shared across the different nationalities—that is, their association with diet was not specific for any country (see online supplementary text 4; online supplementary figure 4A, B; online supplementary figure 7). Their associations with MedDiet adherence were further validated by their pattern of abundance variation in both the intervention and control cohorts, as well as in individuals ranked by increasing adherence to the diet (see online supplementary text 5; online supplementary figure 8, figure 2C, D).
gutjnl-2019-319654supp007.pdf (27.5KB, pdf)
gutjnl-2019-319654supp008.pdf (41.3KB, pdf)
Next we investigated the co-occurring modules within the gut microbiomes. These modules are analogous to ‘guilds’ within the microbiomes that have similar or associated functional properties. We used iBBiG to identify modules in the gut microbiome,31 an approach we previously used to identify granular differences in the microbiome as a function of healthy ageing in the ELDERMET cohort14 (see online supplementary text 3; online supplementary figure 9; online supplementary tables 3 and 4). iBBiG identified six overlapping taxonomic modules (named A to F) within the NU-AGE dataset. Notably, we identified a specific module C which was significantly over-abundant in individuals with increased frailty and also increased in representation in the set of DietNegative OTUs (see online supplementary text 6; online supplementary figure 9C, D). This indicates that module C is similar to the long-stay-like modules we identified in ELDERMET individuals.14 However, the specific enrichment of module C in the set of OTUs depleted with MedDiet adherence indicated the likelihood that the MedDiet successfully modulated the gut microbiome in a manner negatively associated with frailty.
gutjnl-2019-319654supp009.pdf (29.5KB, pdf)
gutjnl-2019-319654supp010.pdf (23.2KB, pdf)
gutjnl-2019-319654supp011.pdf (42.3KB, pdf)
Adherence to the NU-AGE MedDiet intervention modulated the microbiome in a manner negatively associated with frailty and inflammation
A major objective of the NU-AGE dietary intervention was reduction of frailty and inflamm-ageing. The study subjects were categorised into Non-Frail (or apparently healthy), Pre-Frail and Frail groups based on Fried scores.21 While the DietNegative taxa showed a stepwise significant decrease with the three frailty groupings (ie, Frail>Pre-Frail>Non-Frail), DietPositive taxa showed a significantly higher abundance in healthy (Non-Frail) individuals compared with Frail individuals (see online supplementary figure 10A). The DietPositive taxa showed a significantly positive change in individuals with reduced frailty (see online supplementary text 7; online supplementary figure 10B). During the intervention period, within the control cohort there was a marginally significant increase in the proportion of individuals with increased frailty (compared with the intervention group) (Fisher's test p<0.06; online supplementary figure 10C). However, we could not observe a direct association between dietary adherence scores and frailty (online supplementary figure 10D). We hypothesised that the effect of dietary adherence on frailty could be indirect, whereby increasing adherence to a Mediterranean diet could modulate the microbiome (potentially with some non-responders), and that this microbiome response could have a direct association with an attenuation of, or reduced risk of, frailty and improvements in other measures of well-being.
gutjnl-2019-319654supp012.pdf (39.7KB, pdf)
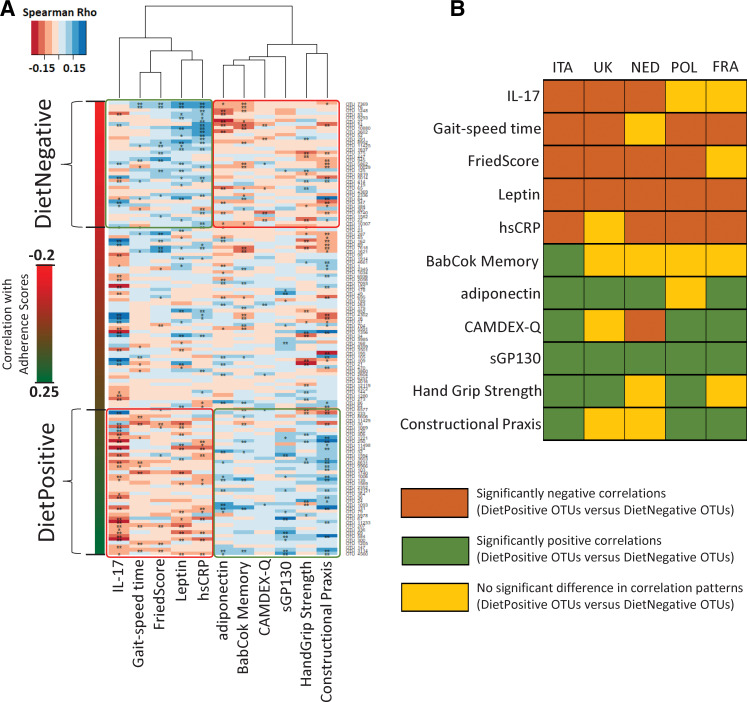
To investigate this, we computed the associations between the diet-responsive OTU markers and specific indices of frailty, cognitive function and inflammation across the entire study cohort (see online supplementary table 5 for list of metadata tested). The objective was to test if diet-responsive taxa showed significantly different trends of association with these indices (see online supplementary figure 11). Overall, although the absolute values of the associations were relatively weak, we observed significant differences in the association patterns of DietPositive and DietNegative OTUs for five different cytokines/biomarkers (namely, pro-inflammatory high-sensitivity C reactive protein (hsCRP) and interleukin 17 (IL-17), anti-inflammatory sGP130 as well as adiponectin and leptin); three frailty-associated measures (Fried Score, Hand Grip Strength and Gait Speed Time); BabCock Memory Score and Constructional Praxis (both associated with cognitive function). The most notable observation, however, was the pattern of these associations. The DietPositive OTU markers had consistent negative associations (significantly lower than the DietNegative markers) with the inflammatory markers hsCRP and IL-17 levels as well as with Fried Scores and Gait Speed Time (both measures associated with increased frailty) (figure 3A). In contrast, their associations were consistently positive with measures of improved cognitive function (eg, Constructional Praxis, BabCock Memory Score), reduced frailty (Hand Grip Strength) and two of the cytokines (adiponectin and sGP130) (a trend exactly opposite to that for DietNegative OTU markers). While the role of adiponectin as an anti-inflammatory marker is well documented,45 sGP130 is a negative regulator of the pro-inflammatory trans IL-6 signalling pathway.46 Notably, in spite of the country-specific variations in dietary intake, microbiome scores and adherence scores, each of these associations (with the exception of BabCock Memory Scores) could be replicated (both in terms of direction as well as the significance of the associations) in at least three of the countries (six of the 10 associations replicated in four of the five countries) (figure 3B). These results clearly indicate that adherence to the NU-AGE MedDiet is associated with modulation of the microbiome in a manner that is relatively consistent (across the countries) and is in turn associated with reduced frailty, improved cognitive function and reduced inflammation.
Figure 3.
Consistent association of diet responsive taxa with different measures of frailty, cognitive function and inflammation. (A) Heatmap showing the variation of the association patterns (obtained using Spearman rhos) of the adherence associated marker Operational Taxonomic Units (OTUs) (arranged from top to bottom in increasing order of their correlations with the adherence scores) with the selected measures of frailty, cognitive function and the pro/anti-inflammatory cytokine levels. For each cell, colours indicate the Spearman rho values (as shown). **Significant association with FDR-corrected p value <0.15. *Marginal association with nominal p value <0.05. The DietPositive and DietNegative OTUs are also demarcated. Specific differences could be observed between the association pattern of the different measures and the DietPositive and DietNegative OTUs. For certain measures such as high-sensitivity C reactive protein (hsCRP) levels, interleukin 17 (IL-17) levels and gait speed time, DietPositive OTUs were observed to have significantly more negative correlations as compared to DietNegative OTUs. For the other measures associated with reduced frailty and improved cognitive function, as well as adiponectin and sGP130 levels, an exact opposite trend was observed. (B) Heat plot showing the replication of these trends individually within each of the country-specific cohorts. Brown indicates those cases where the correlations of the DietPositive OTUs were significantly more negative than the DietNegative group, green indicates those cases with the opposite trend and yellow indicates those cases of no significant change.
gutjnl-2019-319654supp013.pdf (49.9KB, pdf)
Microbiome response, accompanied by specific beneficial changes in the gut metabolic profiles, is the key intermediate between dietary adherence and health
Based on the preceding findings, it seemed likely that a microbiome associated with dietary adherence was more important for improved health status than merely adherence to the diet itself. Testing this hypothesis required the computation of measurable ‘microbiome scores/indices’ (analogous to the dietary adherence scores) that would take into account the variations associated with individual marker OTUs. Switching to the NU-AGE MedDiet is characterised by changes in the consumption pattern of specific dietary components—namely, an increase in the consumption of fibres (vegetables, fruits), carbohydrates (wholegrains), plant proteins (legumes), polyunsaturated fatty acids (fish) and vitamins such as vitamin C (fruits) and a concomitant decrease in the consumption of fats, alcohol, sodium and sugar (sweets).35 We first validated the diet-responsive OTUs (identified based on their association with the overall NU-AGE FBDG scores) by checking their associations with the consumption patterns with the different food components (partial Spearman correlations taking into account age, body mass index, gender, country and polypharmacy as confounders). We observed that OTU markers with an increasing positive association with FBDG adherence scores showed increasing positive correlations with fibre, vitamin C, vitamin D, plant proteins and carbohydrates and increasing negative associations with the components alcohol, fats and sugar whose consumption was decreased during the MedDiet change35 (see online supplementary figure 12). Thus, the above results indicate that the associations of the marker OTUs were not only with the overall FBDG scores, but also with individual dietary components whose modulations were associated with the NU-AGE MedDiet intervention (even after taking into account all host-associated confounding factors like age, body mass index, gender, country and polypharmacy). This validated the association of the dietary markers with the dietary intervention. Further, each of the diet-responsive OTUs had a specific degree of correlation with the dietary adherence scores and specific trends of association with the dietary components (see online supplementary figure 12). Based on the overall correlations of the diet-associated marker OTUs with the adherence scores as well as the abundances marker OTUs in a given sample, we calculated a sample-specific diet-modulated microbiome index (see Methods section; online supplementary figure 2). As expected, the microbiome index was positively correlated with the overall adherence scores and also captured the overall association patterns of the individual marker OTUs (ie, positive associations with fibre, carbohydrate, plant proteins, vitamin C, polyunsaturated fatty acids and negative associations with fats, alcohol and sugar), indicating its validity as a proxy for the taxonomic markers associated with consumption of the MedDiet (see online supplementary figures 12,13; online supplementary text 8).
gutjnl-2019-319654supp014.pdf (13.3KB, pdf)
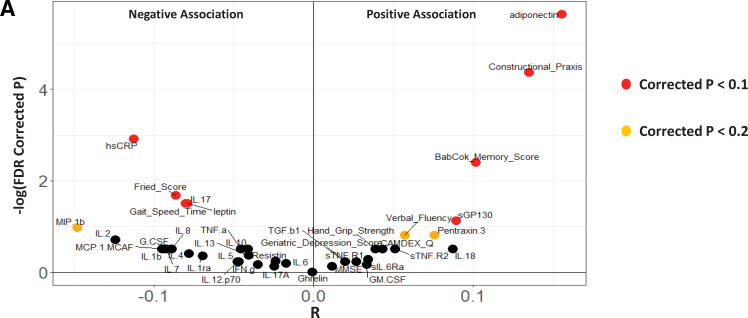
We then checked the association of this index with the different measures of frailty, cognitive function and inflammation (across the entire cohort), considering the adherence scores as a confounder. Ten of the 11 associations with measures of improved cognition, reduced frailty and inflammation could be reproduced. We also observed additional negative associations with the inflammation-related cytokines interleukin (IL)-2 and macrophage inflammatory protein (MIP)-1b, and positive associations with verbal fluency (figure 4). These results show that the diet-modulated microbiome components are associated with frailty, inflammation and cognitive function independent of the adherence scores (ie, these are not indirect consequences of associations with dietary adherence). We had previously shown that these associations were stable across the different countries (figure 3B). We next checked the effect of confounders (such as age, body mass index, gender, disease pathophysiologies and medication intake) on the extent of diet–taxon associations. Individuals with multiple diseases, specifically those with diabetes, heart attack and inflammatory disorders, were observed to have significantly lower microbiome scores compared with non-diseased controls (lower but marginally significant for cancer) (see online supplementary figure 14A–E; online supplementary text 9; online supplementary table 6). However, the pattern of association of the microbiome index with seven of the 10 inflammatory markers and frailty indices (identified in figure 4) largely remained invariant, even after taking into account all confounders including age, body mass index, gender, polypharmacy and different disease pathophysiologies (see online supplementary text 9; online supplementary figure 15).
Figure 4.
MedDiet microbiome index correlates with reduced frailty, improved cognitive function and reduced inflammation, independent of the adherence scores. Violin plot showing the association (partial Spearman correlations) of the different measures of frailty, cognitive function and inflammatory marker levels with the MedDiet-modulated microbiome index after taking into account the adherence scores as a confounder. The x axis shows the Spearman rho values and the y axis indicates the −log (base 10) of the p values. Most negatively associated measures are expected to be at the extreme left of the plot, the most positively associated measures are expected to be at the extreme right of the plot. Points are coloured based on the significance of the obtained associations (red indicates associations with FDR-corrected p<0.1, orange indicates associations with FDR-corrected p<0.2). The MedDiet microbiome index is observed to be associated with several measures associated with reduced frailty, reduced inflammation and improved cognitive function and this association is independent of the adherence scores.
gutjnl-2019-319654supp015.pdf (27.6KB, pdf)
gutjnl-2019-319654supp017.pdf (39.8KB, pdf)
Even with respect to the across time point changes, while change in dietary adherence scores were significantly associated with change in the microbiome index, it was the change in the microbiome index that was positively associated with improvement in cognitive function, physical well-being and negatively associated with inflammatory markers like hsCRP (see online supplementary text 10; online supplementary figure 16). Positive changes in microbiome indices were also associated with positive changes in the levels of the anti-inflammatory cytokine IL-10 and negative changes in the ratio of hsCRP to anti-inflammatory cytokine levels, further indicating the negative association of the diet-associated microbiome index with inflammatory cytokine levels (see online supplementary figure 16C and online supplementary text 10).
gutjnl-2019-319654supp016.pdf (32.4KB, pdf)
The positive influence of the diet-modulated microbiome change on health status is likely to be driven by specific microbial metabolites. Given that faecal metabolomic data were unavailable for the individuals, we predicted the functional metabolic profiles of the gut microbiome using the corresponding 16S species composition profiles (see Methods section). Correlating the across time point changes in the abundances of these predicted metabolic profiles with the microbiome index change identified dramatic differences across the microbiome response landscape (see online supplementary figure 17). A positive microbiome change was associated with an increase in the microbial consumption of fibre-associated non-starch polysaccharides (probably indicative of Mediterranean diet change). In contrast, a negative change was associated with an increase in microbial simple sugar consumption. A negative microbiome response was also accompanied by a predicted increase in the microbial consumption of tauro- and glyco- derivatives of bile acids (such as taurocholate or glycochenodeoxycholate) to secondary bile acids (lithocholate, deoxycholate) through cholate and chenodeoxycholate (see online supplementary figure 18A). Bile acid dysregulation is associated with different disease conditions,47 specifically the increase in production of lithocholic and deoxycholic acid has been associated with colorectal cancer.48 In contrast, increased production of both branched chain fatty acids (BCFAs) and SCFAs are associated with a positive microbiome response. A positive association of SCFAs with host health is well recognised.49 Previous studies measuring the metabolomic changes associated with intake of a MedDiet have also observed a similar increase in SCFA levels,10 50 as well as an exactly similar link wherein MedDiet-like dietary modulations (increased fibre intake and decreased fat intake) were observed to be positively associated with faecal SCFA levels and negatively associated with faecal secondary bile acids.51 52 Furthermore, in the current study we had data for the measured plasma levels of cholic acid (CA), glycochenodeoxycholic acid (GDCDA) and chenodeoxycholic acid (CDCA) for a subset of individuals belonging to the Italian and Polish cohorts. For GCDCA and CDCA, correlating the plasma levels of these bile acids with the abundances of the diet-associated markers revealed trends that the DietPositive OTUs had significantly more positive associations with GCDCA levels and more negative associations with CDCA levels compared with DietNegative OTUs (online supplementary figure 18B). By grouping this subset of individuals into three terciles based on their GDCDA/CDCA ratios, we observed that individuals with an increasing GCDCA/CDCA ratio were associated with a significantly positive change in their diet-associated microbiome index (online supplementary figure 18C). These results confirm the predicted metabolite profiles wherein individuals with increasing diet-associated microbiome indices were predicted to have decreased microbial conversion of GDCDA to CDCA (and thereafter to lithocholic acid (LCA) and deoxycholic acid (DCA)), thereby resulting in higher GDCDA/CDCA levels. Thus, some of the key global changes (in bile acid and SCFA levels) we detect and that we predicted to be linked with diet-associated microbiome response have been reported in the literature across multiple studies as well as the plasma level analysis. The only conflicting trend was with CA levels which were observed to show the pattern opposite to that expected. However, it could be because the measurements were on serum samples (in contrast to faecal levels) and CA/CDCA are produced by both the liver and the microbiota (see online supplementary figure 18C).
A negative microbiome response was also associated with other detrimental metabolites like p-cresol, ethanol and carbon dioxide, whose relative overproduction is associated with onset of colorectal cancer, insulin resistance, non-alcoholic fatty liver disease, cytotoxicity and small intestinal bacterial overgrowth.53–57 Notably, at baseline the diet-associated microbiome index was observed to be negatively associated with multiple diseases including hypertension, diabetes and cancer (online supplementary figure 14). Thus, although inferred rather than measured, the data indicate that metabolic change associated with a positive microbiome response beneficially impacts host health.
DietPositive OTUs are keystone species in the gut microbial community
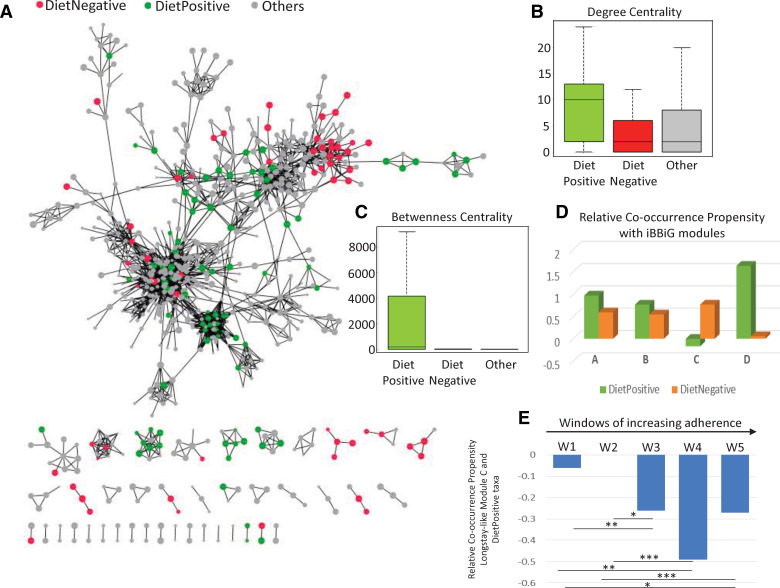
Finally, we evaluated the role of the diet-responsive taxa in the overall microbiome community structure, represented by networks defined by the Reboot Approach (see Methods section).30 A co-occurrence network provides a representation of nodes and edges (interconnecting lines) between these nodes, wherein the nodes represent the taxa (in this case, the OTUs) and the edges between the nodes represent a significant co-occurrence relationship between them (across a provided set of observations or samples). The placement of the taxa within a co-occurrence network indicates the relative importance of the taxa in the stability of the community. We first obtained the co-occurrence network for all the samples across time points for both cohorts. The major component of the co-occurrence network is a conglomeration of clusters of taxa, with other taxa acting as interlinking hubs. However, the positioning of the majority of DietPositive and DietNegative taxa was strikingly different. The DietPositive taxa were either located centrally at the hubs of the network or as linking nodes within the major subnodes (figure 5A). This shows the centrality of these taxa in the gut community structure, a phenomenon termed 'keystone species'.58 In contrast, the majority of the DietNegative taxa were placed at the periphery of the network. We probed this observation by computing two centrality measures for each taxon in the network: 'degree centrality', which is the number of nodes connected to a given node, and 'betweenness centrality', which is the number of paths connecting any two nodes that pass through a given node. DietPositive taxa had a significantly higher degree of betweenness centrality compared with the DietNegative taxa or the non-associated markers (figure 5B, C). We regenerated the network within each of the different countries as well as across overlapping windows of samples of increasing dietary adherence (see online supplementary figure 19; online supplementary figure 20; online supplementary figure 21A). Despite major differences in the overall structure of the individual networks, the placement patterns of the taxa as well as their relative importance within the gut microbial networks were invariant irrespective of the country. The DietPositive taxa had significantly higher centrality measures irrespective of the nationality and the dietary adherence of the individuals. As expected, there were also distinct patterns of interactions for the DietPositive and DietNegative groups of taxa, specifically with respect to the iBBiG identified frailty-associated module C, which had negative co-occurrence propensities with the DietPositive group (figure 5D). Interestingly, the strength of the co-occurrence propensities became significantly more negative with increasing adherence to the diet (figure 5E). This was not observed for any of the other taxonomic modules (see online supplementary figure 21B).
Figure 5.
Bacterial taxa that respond positively to Mediterranean diet intervention occupy keystone interaction nodes for peripheral frailty-associated taxa in microbiome networks. (A) Representation of the Operational Taxonomic Unit (OTU) co-occurrence network obtained for all the samples across the time points and cohorts with the DietPositive, DietNegative and non-correlated OTUs shown in green, red and grey colours, respectively. The network shows two distinct characteristics of the DietPositive and DietNegative markers (or OTUs). While the DietNegative markers (barring a few exceptions) are observed to occur as the peripheral nodes in the network, the DietPositive markers mostly act as either the centrally connected hub nodes or as interconnecting nodes between the hubs, indicating their centrality to the microbiome. This is also reflected in the comparison of the degree and betweenness centrality measures shown as boxplots in (B) and (C), respectively. (D) Relative co-occurrence propensity (calculated as the logged ratio of the number of positive edges to the number of negative edges) between the DietPositive and DietNegative OTUs with those belonging to the different iterative Binary Bi-clustering of Gene-sets (iBBiG) modules. It was observed that, specifically for the frailty-associated longstay-like module C, while the DietNegative markers showed a positive co-occurrence, the DietPositive markers showed a negative association, further indicating that taxa that respond positively to the diet negatively associate with those that are associated with frailty. (E) The negative association was further investigated by building networks for the five overlapping windows of samples W1–W5 (see Methods section), with increasing adherence to the diet. Relative co-occurrence propensity between the DietPositive and the module C across networks obtained for the overlapping windows of samples with increasing adherence to the diet. With increasing adherence to the diet, the relative co-occurrence propensity between the DietPositive OTUs and those belonging to the module C becomes increasingly negative. The p values of the significance of association are indicated as ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05.
Discussion
The current results provide a systemic view of the effect of consuming the NU-AGE MedDiet on the microbiome and subsequently on biomarkers of health in the elderly. A significant challenge for the current study was the high level of microbiome variability across individuals in five countries, resulting in a low signal-to-noise ratio which translated to weaker taxonomic signals for association with metadata. Analysis using traditional methodologies are useful and provide statistical rigour, even if the assumption of independent variables is not a true reflection of the community structure in the microbiota. However, due to the multitude and disparate nature of the microbiota structure configurations across individuals, combined with the relatively small effect of diet over a year of life in an established gut microbiota community as well as other aspects such as the subjective nature of the dietary measurements that is expected for community-dwelling individuals and the assumption that dietary measurements accurately measure the actual dietary change, the traditional statistical methodologies are unable to identify the taxa associated with the statistically significantly lower loss of diversity associated with adherence to the MedDiet. To establish the diet-responsive taxa and generate a diet-associated microbiome index, we applied a novel leave-one-out-cross-validation machine-learning methodology to predict the adherence score for each individual with good accuracy and used these predictive models to probe and identify the specific taxonomic signals that best predict increased adherence to the MedDiet.
We observed that increased adherence to the MedDiet modulates specific components of the gut microbiota that were associated with a reduction in risk of frailty, improved cognitive function and reduced inflammatory status. For reasons described above, these associations for some of the diet-modulated microbiome markers could only be observed at relatively weaker thresholds (rho <−0.09 and rho >0.07, FDR-corrected p values <0.2). This allowed for the visualisation and re-examination of the most predictive OTUs. However, the striking observation was the consistency of associations of the diet-modulated microbiome markers with biological markers of ageing (independent of nationality). The formulation and calculation of a single sample-specific microbiome index clarified these associations even further. For a single sample, this index provided a quantitative summary of the abundance patterns of the diet-responsive markers (the higher the value, the higher the abundance of DietPositive taxa and vice versa), thereby addressing the sample-specific variability associated with the individual markers. We showed that they were not only associated with dietary compliance but were consistently associated with frailty and inflammatory markers, thereby confirming their importance for health maintenance independent of other anthropometric confounders like age, body mass index and gender. In fact, the apparent lack of a direct link of the adherence score with frailty further hints that the response of an individual to the diet could be mediated by the change in the microbiome. Besides these associations, the keystone nature of the DietPositive markers within the gut microbiome remains remarkably stable across multiple nationalities. These keystone properties of the DietPositive markers add support to the so-called Anna Karenina principle59 of microbiomes which posits that microbiomes of healthy individuals are similar and the unhealthy individuals are each aberrant in their own way. By protecting the ‘core’ of the gut microbial community, adherence to the diet could facilitate the retention of a stable community state in the microbiome, providing resilience and protecting from changes to alternative states that are found in unhealthy subjects.
The positive impact of these microbial taxa on host health was further indicated by predictive metabolite profiling, where increasing adherence to the diet specifically selects for taxa that are enriched in the production of SCFAs/BCFAs, while selecting against those associated with bile acid dysregulation and production of proposed deleterious metabolites like acetone, p-cresol, ethanol and carbon dioxide. Although this is an in-silico prediction, an increase of SCFA production with MedDiet consumption (or with specific components of the MedDiet) has been previously shown.10 50–52 The study by Pagliai et al, which compared the microbiome and metabolome changes on MedDiet and vegetarian diets, reported a significant positive association of carbohydrate consumption (which is increased in the MedDiet) with faecal levels of SCFA butyrate and a significant negative association of lipid and fat intake (decreased in the MedDiet) with levels of the SCFAs propionate and acetate. Negative associations of the SCFAs were also observed with levels of the inflammatory cytokine IL-17.50 Although the study by Pagliai et al observed no significant differences in the levels of BCFAs on the MedDiet (in contrast to the in-silico predictive metabolite analysis performed in our current study), a negative association of the BCFA levels with fat intake was also observed (in line with our current findings). The links between the MedDiet-associated microbiome modulation, SCFA production and the carcinogenic secondary bile acid production are precisely in line with findings from two previous studies on African Americans and rural Africans.51 52 Interestingly, across the secondary bile acid production landscape, while the faecal metabolome results from the study by O’Keefe et al confirmed the predicted metabolite changes with respect to CA and the carcinogenic secondary bile acids DCA and LCA, the plasma metabolite levels (in the current study) confirm the predicted changes with respect to CDCA and GCDCA (although plasma metabolite levels are not expected to exactly reflect the faecal metabolome). Thus, results obtained from these studies largely complement each other and resonate with our current findings, and the predicted downregulation of the other potentially detrimental metabolite production provides an informed list of candidate compounds that can be further verified by targeted metabolomic profiling in future studies.
The interplay of diet, microbiome and host health is a complex phenomenon influenced by several factors. It is also probably a multistep process dictated by specific mechanistic rules. While the results of this study shed light on some of the rules of this three-way interplay, several factors such as age, body mass index, disease status and initial dietary patterns may play a key role in determining the extent of success of these interactions. Interestingly, the beneficial effects of MedDiet intervention mediated through the microbiome are not restricted to elderly subjects, as evidenced by the study by Meslier et al 60 (this volume; co-submitted to Gut for back-to-back publication) showing that a similar intervention in obese subjects resulted in multiple health-related shifts in the gut microbiome and metabolome independently of energy intake. Notwithstanding this theoretical and practical reinforcement, the strategy of promoting health in the elderly by maintaining a long-term MedDiet (or supplementation of specific ingredients) may be impractically expensive or logistically impossible in many countries where these ingredients are neither staple nor available year-round. In some older subjects with problems like dentition, saliva production, dysphagia or irritable bowel syndrome, adapting a MedDiet may not be a realistic option. Our definition here of MedDiet-responsive taxa that correlate with health, plus our recent identification of taxa associated with healthy ageing in a large metacohort of 2500 subjects,61 provides a short list of candidate taxa for development as live biotherapeutic agents for direct administration to older subjects to reduce onset of frailty.
Footnotes
TSG, SR, IBJ and AS contributed equally.
Contributors: TSG, IBJ, POT, CF and AS conceived and designed the analysis. MN, AS, MC, EG, AJ, MC, ST, EGZ, GDAH, CE, NM, CMB, EP-G, AM, JK, BP, BC, MN, MM-F and others collected and/or contributed data. TSG, IBJ and SR performed the analysis. TSG, IBJ and POT wrote the paper assisted by SR, AS, PB, EGZ and WMDeV. WMDeV, SF-T, AC, PB, CF, POT and others reviewed the analysis and the manuscript.
Funding: The authors are funded in part by Science Foundation Ireland (APC/SFI/12/RC/2273) in the form of a research centre, APC Microbiome Ireland. IBJ was supported by a Science Foundation Ireland grant (13/SIRG/2128). The NU-AGE project is supported by the European Union’s Seventh Framework Programme under grant agreement no. 266 486 (‘NU-AGE: New dietary strategies addressing the specific needs of the elderly population for healthy ageing in Europe’).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. The majority of results corresponding to the current study are included in the article or uploaded as supplementary information. Other data are available on request from the authors.
References
- 1. Clegg A, Young J, Iliffe S, et al. Frailty in elderly people. Lancet 2013;381:752–62. 10.1016/S0140-6736(12)62167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cevenini E, Monti D, Franceschi C. Inflamm-ageing. Curr Opin Clin Nutr Metab Care 2013;16:14–20. 10.1097/MCO.0b013e32835ada13 [DOI] [PubMed] [Google Scholar]
- 3. Franceschi C, Bonafè M, Valensin S, et al. Inflamm-aging. an evolutionary perspective on immunosenescence. Ann N Y Acad Sci 2000;908:244–54. 10.1111/j.1749-6632.2000.tb06651.x [DOI] [PubMed] [Google Scholar]
- 4. Sugimoto T, Sakurai T, Ono R, et al. Epidemiological and clinical significance of cognitive frailty: a mini review. Ageing Res Rev 2018;44:1–7. 10.1016/j.arr.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 5. Wilson D, Jackson T, Sapey E, et al. Frailty and sarcopenia: the potential role of an aged immune system. Ageing Res Rev 2017;36:1–10. 10.1016/j.arr.2017.01.006 [DOI] [PubMed] [Google Scholar]
- 6. An R, Wilms E, Masclee AAM, et al. Age-dependent changes in GI physiology and microbiota: time to reconsider? Gut 2018;67:2213–22. 10.1136/gutjnl-2017-315542 [DOI] [PubMed] [Google Scholar]
- 7. Trichopoulou A, Martínez-González MA, Tong TY, et al. Definitions and potential health benefits of the Mediterranean diet: views from experts around the world. BMC Med 2014;12:112 10.1186/1741-7015-12-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sofi F, Cesari F, Abbate R, et al. Adherence to Mediterranean diet and health status: meta-analysis. BMJ 2008;337:a1344 10.1136/bmj.a1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kojima G, Avgerinou C, Iliffe S, et al. Adherence to Mediterranean diet reduces incident frailty risk: systematic review and meta-analysis. J Am Geriatr Soc 2018;66:783–8. 10.1111/jgs.15251 [DOI] [PubMed] [Google Scholar]
- 10. De Filippis F, Pellegrini N, Vannini L, et al. High-Level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016;65:1812–21. 10.1136/gutjnl-2015-309957 [DOI] [PubMed] [Google Scholar]
- 11. Mitsou EK, Kakali A, Antonopoulou S, et al. Adherence to the Mediterranean diet is associated with the gut microbiota pattern and gastrointestinal characteristics in an adult population. Br J Nutr 2017;117:1645–55. 10.1017/S0007114517001593 [DOI] [PubMed] [Google Scholar]
- 12. Claesson MJ, Jeffery IB, Conde S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–84. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 13. O'Toole PW, Jeffery IB. Gut microbiota and aging. (1095-9203 (Electronic)). [DOI] [PubMed] [Google Scholar]
- 14. Jeffery IB, Lynch DB, O'Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J 2016;10:170–82. 10.1038/ismej.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tran TTT, Cousin FJ, Lynch DB, et al. Prebiotic supplementation in frail older people affects specific gut microbiota taxa but not global diversity. Microbiome 2019;7:39 10.1186/s40168-019-0654-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berendsen A, Santoro A, Pini E, et al. A parallel randomized trial on the effect of a healthful diet on inflammageing and its consequences in European elderly people: design of the NU-AGE dietary intervention study. Mech Ageing Dev 2013;134:523–30. 10.1016/j.mad.2013.10.002 [DOI] [PubMed] [Google Scholar]
- 17. Marseglia A, Xu W, Fratiglioni L, et al. Effect of the NU-AGE diet on cognitive functioning in older adults: a randomized controlled trial. Front Physiol 2018;9:349 10.3389/fphys.2018.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jennings A, Cashman KD, Gillings R, et al. A Mediterranean-like dietary pattern with vitamin D3 (10 µg/d) supplements reduced the rate of bone loss in older Europeans with osteoporosis at baseline: results of a 1-y randomized controlled trial. Am J Clin Nutr 2018;108:633–40. 10.1093/ajcn/nqy122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maijo M, Ivory K, Clements SJ, et al. One-year consumption of a Mediterranean-like dietary pattern with vitamin D3 supplements induced small scale but extensive changes of immune cell phenotype, co-receptor expression and innate immune responses in healthy elderly subjects: results from the United Kingdom arm of the NU-AGE trial. Front Physiol 2018;9:997 10.3389/fphys.2018.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jennings A, Berendsen AM, de Groot LCPGM, et al. Mediterranean-style diet improves systolic blood pressure and arterial stiffness in older adults. Hypertension 2019;73:578–86. 10.1161/HYPERTENSIONAHA.118.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santoro A, Pini E, Scurti M, et al. Combating inflammaging through a Mediterranean whole diet approach: the NU-AGE project's conceptual framework and design. Mech Ageing Dev 2014;136-137:3–13. 10.1016/j.mad.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 22. Santoro A, Guidarelli G, Ostan R, et al. Gender-specific association of body composition with inflammatory and adipose-related markers in healthy elderly Europeans from the NU-AGE study. Eur Radiol 2019;29:4968–79. 10.1007/s00330-018-5973-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004;36:808–12. 10.2144/04365ST04 [DOI] [PubMed] [Google Scholar]
- 24. Costea PI, Zeller G, Sunagawa S, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 2017;35:1069–76. 10.1038/nbt.3960 [DOI] [PubMed] [Google Scholar]
- 25. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 27. Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allard G, Ryan FJ, Jeffery IB, et al. SPINGO: a rapid species-classifier for microbial amplicon sequences. BMC Bioinformatics 2015;16:324 10.1186/s12859-015-0747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 2012;8:e1002606 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gusenleitner D, Howe EA, Bentink S, et al. iBBiG: iterative binary bi-clustering of gene sets. Bioinformatics 2012;28:2484–92. 10.1093/bioinformatics/bts438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noronha A, Modamio J, Jarosz Y, et al. The virtual metabolic human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res 2019;47(D1):D614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sung J, Kim S, Cabatbat JJT, et al. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun 2017;8:15393 10.1038/ncomms15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berendsen AAM, van de Rest O, Feskens EJM, et al. Changes in dietary intake and adherence to the NU-AGE diet following a one-year dietary intervention among European older adults: results of the NU-AGE randomized trial. Nutrients 2018;10:E1905 10.3390/nu10121905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–83. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 37. Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 38. Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017;66:70–8. 10.1136/gutjnl-2015-309800 [DOI] [PubMed] [Google Scholar]
- 39. Jackson MA, Jackson M, Jeffery IB, et al. Signatures of early frailty in the gut microbiota. Genome Med 2016;8:8 10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gomez-Arango LF, Barrett HL, Wilkinson SA, et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 2018;9:189–201. 10.1080/19490976.2017.1406584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kovatcheva-Datchary P, Shoaie S, Lee S, et al. Simplified intestinal microbiota to study microbe-diet-host interactions in a mouse model. Cell Rep 2019;26:3772–83. 10.1016/j.celrep.2019.02.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59–64. 10.1038/nature13568 [DOI] [PubMed] [Google Scholar]
- 43. Karlsson FH, Fåk F, Nookaew I, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012;3:1245 10.1038/ncomms2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giampieri E, Ostan R, Guidarelli G, et al. A novel approach to improve the estimation of a diet adherence considering seasonality and short term variability - the NU-AGE Mediterranean diet experience. Front Physiol 2019;10:149 10.3389/fphys.2019.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ouchi N, Walsh K. Adiponectin as an anti-inflammatory factor. Clin Chim Acta 2007;380:24–30. 10.1016/j.cca.2007.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest 2011;121:3375–83. 10.1172/JCI57158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chávez-Talavera O, Tailleux A, Lefebvre P, et al. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 2017;152:1679–94. 10.1053/j.gastro.2017.01.055 [DOI] [PubMed] [Google Scholar]
- 48. Tsuei J, Chau T, Mills D, et al. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med 2014;239:1489–504. 10.1177/1535370214538743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sanna S, van Zuydam NR, Mahajan A, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 2019;51:600–5. 10.1038/s41588-019-0350-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pagliai G, Russo E, Niccolai E, et al. Influence of a 3-month low-calorie Mediterranean diet compared to the vegetarian diet on human gut microbiota and SCFA: the CARDIVEG study. Eur J Nutr 2019;11 10.1007/s00394-019-02050-0 [DOI] [PubMed] [Google Scholar]
- 51. Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr 2013;98:111–20. 10.3945/ajcn.112.056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Keefe SJD, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun 2015;6:6342 10.1038/ncomms7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bone E, Tamm A, Hill M. The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr 1976;29:1448–54. 10.1093/ajcn/29.12.1448 [DOI] [PubMed] [Google Scholar]
- 54. Elshaghabee FMF, Bockelmann W, Meske D, et al. Ethanol production by selected intestinal microorganisms and lactic acid bacteria growing under different nutritional conditions. Front Microbiol 2016;7:47 10.3389/fmicb.2016.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Baskaran S, Rajan DP, Balasubramanian KA. Formation of methylglyoxal by bacteria isolated from human faeces. J Med Microbiol 1989;28:211–5. 10.1099/00222615-28-3-211 [DOI] [PubMed] [Google Scholar]
- 56. Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab 2014;20:753–60. 10.1016/j.cmet.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 57. Ghoshal UC, Shukla R, Ghoshal U. Small intestinal bacterial overgrowth and irritable bowel syndrome: a bridge between functional organic dichotomy. Gut Liver 2017;11:196–208. 10.5009/gnl16126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med 2019;216:20–40. 10.1084/jem.20180448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2017;2:17121 10.1038/nmicrobiol.2017.121 [DOI] [PubMed] [Google Scholar]
- 60. Ercolini D, De Filippis F, Roager HM Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020;69:1258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ghosh D, Jeffery O' Toole. Adjusting for age improvesidentification of gut microbiome alterations in multiple diseases. eLife In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319654supp001.pdf (39.8KB, pdf)
gutjnl-2019-319654supp002.pdf (48.2KB, pdf)
gutjnl-2019-319654supp003.pdf (56.4KB, pdf)
gutjnl-2019-319654supp005.pdf (71.1KB, pdf)
gutjnl-2019-319654supp004.pdf (1.6MB, pdf)
gutjnl-2019-319654supp006.pdf (62.9KB, pdf)
gutjnl-2019-319654supp007.pdf (27.5KB, pdf)
gutjnl-2019-319654supp008.pdf (41.3KB, pdf)
gutjnl-2019-319654supp009.pdf (29.5KB, pdf)
gutjnl-2019-319654supp010.pdf (23.2KB, pdf)
gutjnl-2019-319654supp011.pdf (42.3KB, pdf)
gutjnl-2019-319654supp012.pdf (39.7KB, pdf)
gutjnl-2019-319654supp013.pdf (49.9KB, pdf)
gutjnl-2019-319654supp014.pdf (13.3KB, pdf)
gutjnl-2019-319654supp015.pdf (27.6KB, pdf)
gutjnl-2019-319654supp017.pdf (39.8KB, pdf)
gutjnl-2019-319654supp016.pdf (32.4KB, pdf)