Abstract
Plasmepsins are a group of diverse aspartic proteases in the malaria parasite Plasmodium. Their functions are strikingly multifaceted, ranging from hemoglobin degradation to secretory organelle protein processing for egress, invasion, and effector export. Some, particularly the digestive vacuole plasmepsins, have been extensively characterized, whereas others, such as the transmission-stage plasmepsins, are minimally understood. Some (e.g. plasmepsin V) have exquisite cleavage sequence specificity; others are fairly promiscuous. Some have canonical pepsin-like aspartic protease features, whereas others have unusual attributes, including the nepenthesin loop of plasmepsin V and a histidine in place of a catalytic aspartate in plasmepsin III. We have learned much about the functioning of these enzymes, but more remains to be discovered about their cellular roles and even their mechanisms of action. Their importance in many key aspects of parasite biology makes them intriguing targets for antimalarial chemotherapy. Further consideration of their characteristics suggests that some are more viable drug targets than others. Indeed, inhibitors of invasion and egress offer hope for a desperately needed new drug to combat this nefarious organism.
Keywords: aspartic protease, parasitology, plasmodium, protease, protozoan, hemoglobin, malaria, maturase, digestive vacuole, transmission, antimalarial chemotherapy
Introduction
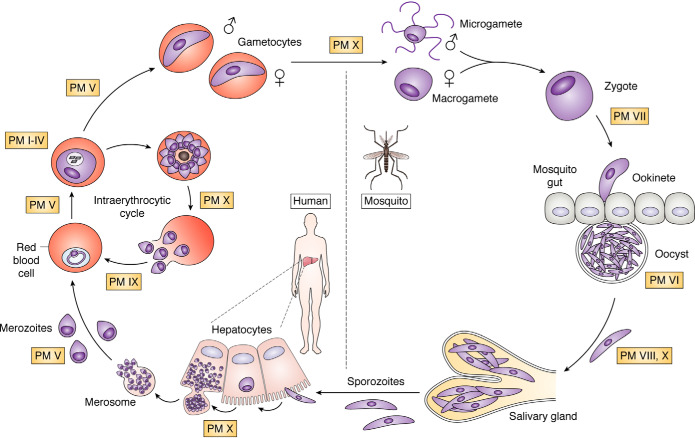
Malaria is caused by protozoan organisms of the genus Plasmodium. The disease is transmitted by infected female Anopheles mosquitos, which inject salivary gland sporozoites into the skin during bloodfeeding. These sporozoites make their way to the liver, replicate, and differentiate into infective merozoites. The merozoites egress into the bloodstream, where they invade red blood cells (RBCs) and set up a continuous intraerythrocytic cycle that amplifies their population, often to overwhelming numbers. Some differentiate into sexual-stage parasites, to be taken up by the next mosquito and develop in the mosquito midgut, ultimately migrating to the salivary glands for spread to a new victim (Fig. 1).
Figure 1.
Life cycle of the malaria parasite. Sporozoites from the salivary glands of an infected mosquito (bottom) make their way to the liver, infect hepatocytes, replicate to thousands of infective merozoites, and bud off as merosomes that rupture into the bloodstream. The merozoites invade RBCs, replicate, and multiply in the intraerythrocytic cycle. Some differentiate into male and female gametocytes that are taken up into the mosquito, where they develop. In the mosquito midgut, parasites egress from the RBCs as gametes, mate to form zygotes, and differentiate into ookinetes that traverse the midgut and become oocysts. They replicate and differentiate into sporozoites that migrate to the salivary glands, where they are ready for transmission to the next human upon mosquito bite. Points in the life cycle at which plasmepsins are thought to function are labeled.
Aspartic proteases called plasmepsins (Plasmodium pepsins, abbreviated PM) play important roles in each stage of Plasmodium development. Interest in the plasmepsins began when the digestive vacuole plasmepsins (I, II, III, and IV) were found to be important for intraerythrocytic hemoglobin degradation (1–5). There followed a major effort to make small-molecule inhibitors to these enzymes, especially PM II, the easiest to express and the first to have a crystal structure (6, 7). A poor correlation between ability of a compound to kill parasites and potency against isolated enzyme (8) suggested that digestive vacuole plasmepsin inhibition was not the mode of parasite killing for these molecules. This ultimately led to the realization that there must be other targets, likely other aspartic proteases, whose inhibition is responsible for the antiplasmodial properties. The search for these targets has uncovered myriad functions for these enzymes. Plasmepsins are involved in bulk protein degradation, secretory protein maturation, egress, invasion, endothelial adherence, and perhaps other processes. A number have been the subject of serious efforts as targets for drug development.
Plasmepsins (Fig. 2) belong to an ancient family of aspartic proteases—the A1 or pepsin-like family—that is widespread throughout eukaryotes. Among the 10 plasmepsins, the most closely related are the digestive vacuolar plasmepsins, PM I–IV. These proteases are spread across just 16 kilobases of chromosome 14 and share 50–70% amino acid identity. Outside of P. falciparum and related primate-infecting species, these proteases are represented by a single plasmepsin, called PM IV in Plasmodium and ASP1 in the related apicomplexan Toxoplasma gondii (9). PM V is the most diverged plasmepsin, sharing 19–23% amino acid identity with the other plasmepsins. Its structure is bolstered by seven disulfide bonds (compared with two in PM I–IV), bringing it into a separate aspartic protease subfamily from the other plasmepsins—subfamily A1B, with type member Nep1 of the pitcher plant Nepenthesia (10). Other apicomplexans also have a single PM V ortholog (ASP5 in Toxoplasma gondii), except for cryptosporidia, which have three (9). Regarding transmission-stage plasmepsins, PM VI and VIII form a clade, sharing 36% amino acid identity with each other. Each has a little-studied ortholog in T. gondii (ASP2 and ASP4 respectively). PM VII has distant homology to PM VI and VIII (31% identity); its uncharacterized Toxoplasma ortholog is ASP6. PM IX and PM X share 37% amino acid identity. Although the two are distinct across Plasmodium and exist on different chromosomes, they are represented by a single T. gondii aspartic protease, ASP3.
Figure 2.
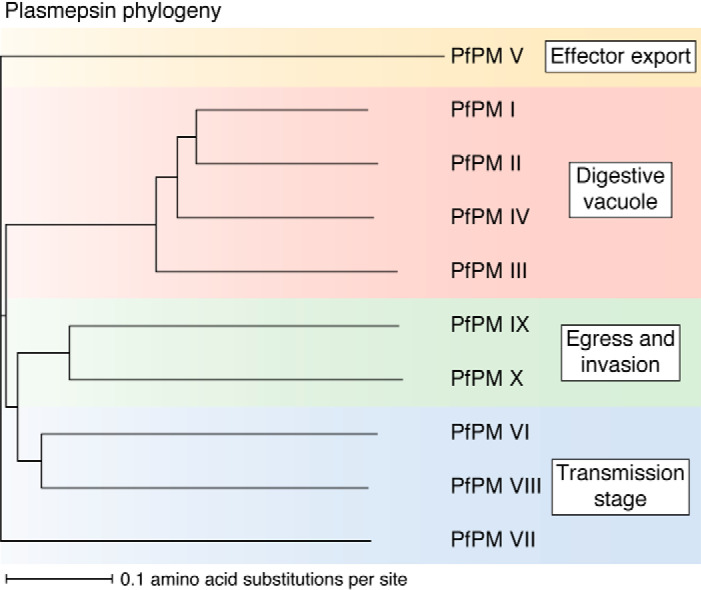
Plasmepsin phylogeny. Sequences for PMs I–X were obtained from PlasmoDB (release 46), aligned using MUSCLE (“Multiple Sequence Comparison by Log-Expectation”, EMBL) (189), and visualized using iTOL (Interactive Tree of Life) (190).
A note on nomenclature: In the literature, plasmepsins are denoted with Roman numerals or Arabic numerals, with or without a space before the number, and plasmepsin III is known as histo-aspartic protease or HAP or PM III (or PMIII or PM3 or PM 3). We suggest going back to a convention initiated in early publications of having Roman numerals after a space. We further suggest that HAP be referred to as PM III for consistency with the other plasmepsins and because its His32 has not been shown to be catalytic. Also, HAP is the name for a gamete fusion protein. Using PM III allows the digestive vacuole plasmepsins in aggregate to be called PM I–IV without ambiguity. An argument for the space before the Roman numeral is that PM V is often referred to in discussions of sending proteins out to the parasitophorous vacuolar membrane or PVM, and PMV gets confusing in this context. These issues are ultimately for those concerned with nomenclature to weigh in on, but in this review, we will use our preferred convention.
There are also nonplasmepsin aspartic proteases in the Plasmodium genome (see “Other aspartic proteases”). The microgametocyte surface protein (MiGS) has homology to PM V (21% amino acid identity and shared transmembrane domain; 17–20% to other plasmepsins). Additional aspartic proteases in the Plasmodium genome—signal peptide peptidase (SPP) and DNA damage-inducible 1 (Ddi1)—are quite distinct from the plasmepsins. SPP is a presenilin-type aspartic protease (family A22). Ddi1 (family A28) is the most diverged from the other aspartic proteases. It possesses a single catalytic aspartate and likely homodimerizes to complete the active site, an unusual architecture among eukaryotic aspartic proteases (although common to retroviral aspartic proteases).
Here, we review the state of knowledge on the plasmepsins, noting outstanding questions in the field. This review will focus on P. falciparum, the deadliest human malaria parasite, for which extensive biological studies have been performed; reference to other species will be made as relevant. Study of the diverse plasmepsins has taught us much about the biology of the malaria parasite and a little about the protein chemistry of aspartic proteases and is pointing the way to exciting new inhibitors that are being developed as antimalarial chemotherapeutic agents.
Digestive vacuole plasmepsins: Plasmepsins I–IV
Intraerythrocytic malaria parasites digest hemoglobin at a prodigious rate (11). This catabolic process provides nutrients for the parasite and has been proposed to maintain osmotic balance in the host cell (11–13). Hemoglobin is ingested by Plasmodium through an endocytic structure called the cytostome, which spans the plasma membrane and the vacuolar membrane surrounding the parasite (Fig. 3A) (14, 15). Hemoglobin-containing vesicles pinch off and fuse with the digestive vacuole, an acidic organelle containing 10 proteases that function in a semi-ordered pathway to degrade the hemoglobin to small peptides and amino acids (16–18) (Fig. 3B). Some of these peptides are thought to be exported out of the digestive vacuole for terminal degradation in the cytoplasm (17, 19, 20).
Figure 3.
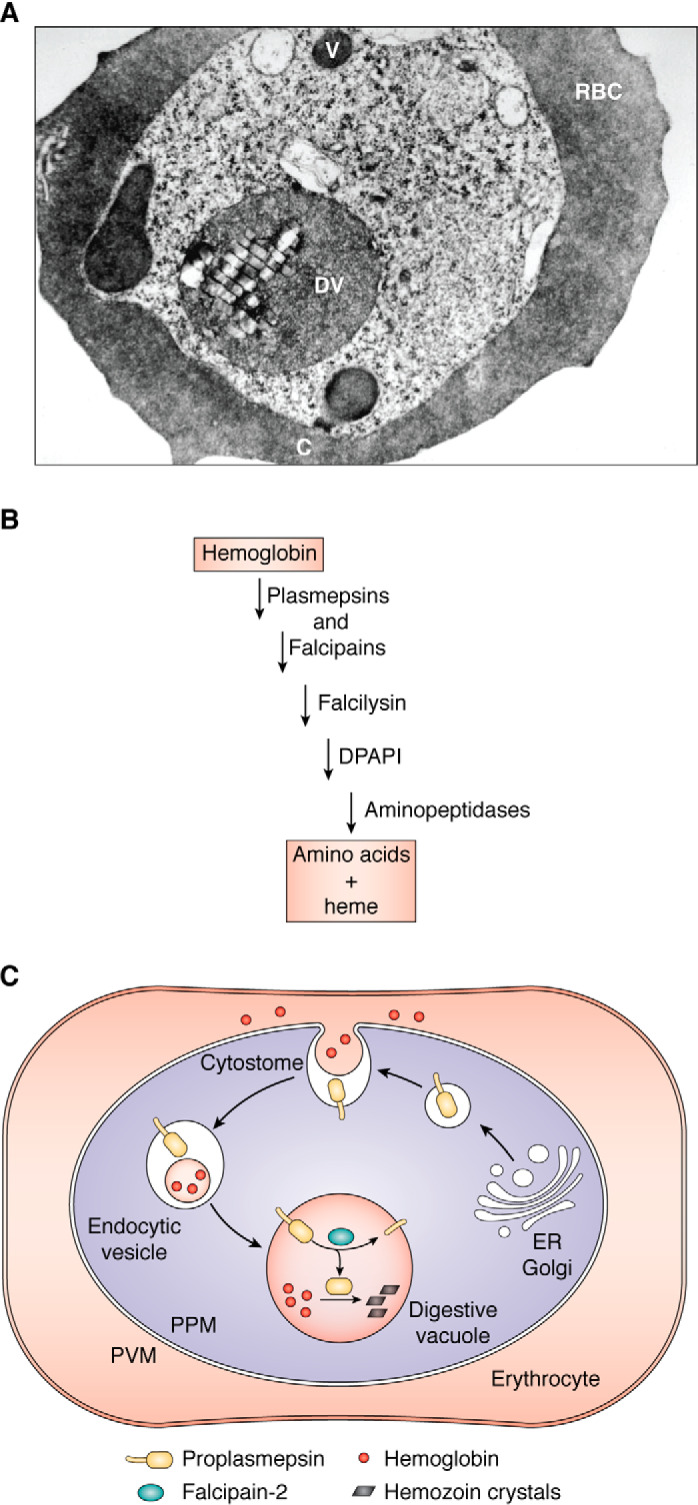
Hemoglobin ingestion and digestion. A, electron micrograph of a P. falciparum-infected erythrocyte. C, cytostome; V, vesicle; DV, digestive vacuole. Adapted from Ref. 16. This research was originally published in Proceedings of the National Academy of Sciences of the United States of America. Goldberg, D. E., Slater, A. F. G., Cerami, A., and Henderson, G. B. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:2931–2935. © United States National Academy of Sciences. B, semi-ordered pathway of hemoglobin degradation. Plasmepsins and falcipains are involved in the initial steps of catabolism and are partially redundant. Falcilysin recognizes oligopeptides. DPAP1 cleaves two residues off of the N terminus of hemoglobin fragments. Aminopeptidases finish the digestion, resulting in free amino acids. Heme is liberated during the initial steps of proteolysis; most is sequestered in the digestive vacuole as hemozoin. C, plasmepsin targeting pathway follows the hemoglobin internalization route. Plasmepsins are made in the ER and traverse the secretory system as type II integral membrane protein precursors (ball and stick). They traffic to the cytostome, a hemoglobin ingestion apparatus that spans the two membranes at the parasite surface. They are internalized with their substrate cargo (hemoglobin) and are delivered to the digestive vacuole surface, where they are cleaved by falcipain-2. PPM, parasite plasma membrane. Black bars, hemozoin crystals that accumulate after heme release.
The degradative process may start in transport vesicles and commences early after erythrocyte invasion (14). Heme is liberated by degradation and most is sequestered as a crystalline lattice of β-hematin dimers called hemozoin (21). Some antimalarial drugs like chloroquine appear to function by blocking heme sequestration, leading to toxic heme build-up (22). A network of proteins implicated in the hemoglobin endocytosis process has recently been defined and includes the artemisinin-resistance protein Kelch13 (23). This finding has yielded a cogent mechanistic explanation for artemisinin resistance. Mutations in Kelch13 and other proteins cause decreased hemoglobin ingestion and digestion, yielding less artemisinin-activating heme (23, 24).
Discovery
Involvement of aspartic proteases in hemoglobin digestion was suspected nearly 50 years ago from the characterization of crude parasite extracts (25, 26). The older literature contains numerous reports of hemoglobin-degrading acidic protease activities in various Plasmodium species, and some were shown to be blocked by the canonical aspartic protease inhibitor pepstatin A (25–34). Their cellular roles remained unclear until studies on isolated digestive vacuoles showed the ability to degrade hemoglobin (16, 35), and protease inhibitor profiling of hemoglobinase activity from highly purified digestive vacuoles revealed a central role for aspartic proteases (16). Soon thereafter, two aspartic proteases (now called PM I and II) were purified from large-scale digestive vacuole preparations and shown to cleave hemoglobin (1, 2, 36). After release of the P. falciparum genome, two more plasmepsins were shown to be present in the digestive vacuole, PM III and PM IV (4, 5, 37).
Function
There is extensive functional redundancy among the digestive vacuole plasmepsins and between these enzymes and the cysteine proteases called falcipains. Knockouts of individual P. falciparum plasmepsins have minor effects on parasite growth in culture (38–41), whereas knockout of all four leads to a substantial growth defect, although the knockout line is still viable (42). Knockout of the single Plasmodium berghei digestive vacuole plasmepsin yields a similar phenotype in the mouse model (43, 44). Cysteine protease inhibitors are more potent in the P. falciparum knockouts (38, 42), and knockouts of falcipain-2 are more sensitive to aspartic protease inhibitors (45), highlighting the redundancy between the two digestive vacuole protease families. Plasmepsins and falcipains are synergistic in biochemical assays of hemoglobin degradation (16, 36, 46), and cysteine/aspartic protease inhibitor synergism is observed in culture (47, 48) as well as in a rodent malaria model (48). A final level of redundancy is that digestive vacuole plasmepsins are activated by falcipains, but if the falcipains are impaired, plasmepsins can autoactivate (49) (see below). Further proteolysis of globin fragments by the metalloprotease falcilysin (50, 51), dipeptidyl aminopeptidase 1 (DPAP1) (52), aminopeptidase P (53), and aminopeptidase M1 (54) finishes the degradation process (Fig. 3). The digestive vacuole plasmepsins have been proposed to be in a complex with some of the downstream proteases as well as with a heme detoxification protein (HDP) in the digestive vacuole (55).
Curiously, recent reports have associated amplification of PM II and PM III with resistance to the antimalarial drug piperaquine in several lineages of Southeast Asian field isolates (56–58) Removal of these extra copies of PM II/PM III resensitized parasites to piperaquine, suggesting that this amplification contributes to the resistance phenotype (58). Additionally, deletion of PM II or PM III from a laboratory P. falciparum strain sensitized parasites to piperaquine in vitro (59). However, increasing PM II/PM III levels is not sufficient to impart piperaquine resistance in laboratory strains, hinting at a more complex picture, where interplay between PM II/PM III levels and other constituents of a strain's genetic background contributes to phenotypic resistance (60). Consistent with this, polymorphisms in the digestive vacuolar membrane transporter PfCRT are sufficient to impart piperaquine resistance in vitro, and parasite lines with these PfCRT mutations naturally lose PM II/PM III amplification while retaining piperaquine resistance in culture (61).
Structure and mechanism
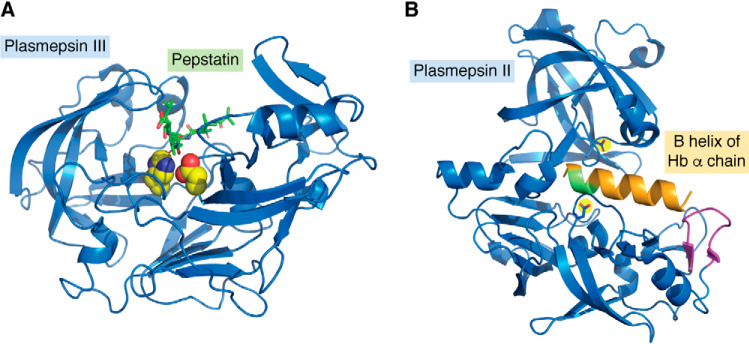
The structures of all four P. falciparum digestive vacuole plasmepsins and the singleton PM IV from several other species have been elucidated (7, 62–68). Each has the typical bilobal pepsin-family aspartic protease fold. These plasmepsins are capable of forming dimers in crystalline form and in solution (69, 70), although it is the monomer that has been shown to be active. The PM III dimer has a loop from one of the subunits that intrudes into the second subunit active site, where it coordinates a zinc ion via Asp215 and His32 (67). The canonical Asp215–Thr218 hydrogen bond is disrupted by the zinc coordination.
PM I, II, and IV appear to function as typical aspartic proteases, with their two aspartates participating in acid-base catalytic activation of a water molecule through a tetrahedral transition state (71, 72), but in contrast, the mechanism of PM III (Fig. 4A) is not fully understood (66). In PM III, Asp215 appears to function as a catalytic base, activating the water to form the hydrolytic nucleophile (67). It is not clear whether His32 is involved in catalysis. It could be the catalytic acid for resolution of the tetrahedral intermediate typical of aspartic protease action, in analogy to the standard aspartate in this position. The positioning of the statine hydroxyl between Asp215 and His32 in the transition-state inhibitor pepstatin-bound structure is supportive of this concept (67). A H32A mutant of recombinant PM III retained activity, however (73). It has been suggested that PM III could be a serine protease, using Ser35 as the nucleophile (74), but a S35A mutant retained activity (73), and the pepstatin active site interactions in the structure look more consistent with an aspartic protease mechanism (67). Concerning the possibility that it could be a zinc metalloprotease, native enzyme was not inhibited by EDTA (5), and only the dimeric apoenzyme structure had active site zinc (67), whereas it is the monomer that is active (70). A proposal was put forth that Asp215 acts as an acid as well as a base, but this model was based on a profound stabilization (several orders of magnitude) by His32 (75). Perhaps a different residue could be the stabilizer; Lys78 has been suggested (73). Weighing on this debate is the fact that the properties of native enzyme and the recombinant enzyme used for mutagenesis studies are very different. PM III was originally isolated from digestive vacuoles, had a sharp pH optimum of 5.5, and was well-inhibited by pepstatin (5). Three different isolation procedures gave similar results, and there was no detectable contamination by other plasmepsins (although trace contamination by a plasmepsin or another protease cannot be completely excluded). Recombinant protein expressed in Escherichia coli had similar properties (76). Later, recombinant protein was again expressed in E. coli for analysis of site-directed mutants mentioned above but had a broad pH optimum from 5 to 8.5, was poorly inhibited by pepstatin, and was potently inhibited by phenylmethylsulfonyl fluoride (73). A H215A mutant had no activity, but the S35A and H32A mutants were similar to WT. It is hard to reconcile these later results with the earlier data, unless there was a contaminating activity present in some of the samples, perhaps from the enterokinase used to generate mature recombinant enzyme. The existing data do not yet form a coherent picture of the mechanism of action of this unusual aspartic protease ortholog.
Figure 4.
A, crystal structure of PM III complexed with pepstatin. Shown is a ribbon structure (blue) with Asp215 and His32 highlighted in yellow with red and blue heteroatoms. Pepstatin is in green with red oxygens. The figure was constructed from PDB entry 3FNT. B, crystal structure of PM II (blue) with the B helix of the hemoglobin α chain modeled in orange. The α33–34 cleavage site is green; the helix-interacting loop is magenta; from PDB entry 1PSE. Created using PyMOL Molecular Graphics System, Version 2.3.
Specificity
An extensive series of studies has assessed specificity of cleavage of hemoglobin or synthetic peptides using native and recombinant digestive vacuole plasmepsins. The four P. falciparum enzymes and the single enzyme from the three other human parasite species (Plasmodium ovale, Plasmodium vivax, and Plasmodium malariae) as well as enzyme from rodent species (P. berghei and Plasmodium chabaudi) have been studied (4–6, 77–90). All four P. falciparum enzymes are capable of degrading hemoglobin and function at the acidic pH of the digestive vacuole (5). Other degradative proteases such as cathepsin E are not able to degrade native hemoglobin, so this is a special property of the digestive vacuole plasmepsins (91). There are minor differences between species, but activity on synthetic peptide libraries shows a general preference for large hydrophobic residues proximal to the cleavage site on the P (N-terminal) and P′ (C-terminal) sides, especially at the P1 and P3 positions, with somewhat broader tolerance at P1′ and P3′ and wide latitude at P2 and P2′. On native hemoglobin, time course studies using native PM I and PM II revealed an initial cleavage of hemoglobin (α2β2 tetramer) on the α chain between Phe33 and Leu34 (1, 36). Proteolysis proceeded with cleavages at a number of sites on both chains. Specificity of cleavage correlated well with the data just mentioned for synthetic peptides. Experiments using peptides corresponding to the scissile α-helix (helix B) revealed enhanced cleavage by PM II when the helix was extended N-terminally and could then interact with a loop on the PM II molecule (91). A model was put forth wherein the plasmepsin recognizes the beginning of the B helix on the α chain of hemoglobin and perches, waiting for the helix to breathe. The B helix is the weakest link on the protein because it has two helix-disrupting glycines. The protease is in position to cleave the 33–34 peptide bond as soon as the helix opens, helping unravel the tightly wound hemoglobin molecule so that further proteolysis can proceed (Fig. 4B).
Biosynthesis
Biosynthesis of digestive vacuole plasmepsins follows a circuitous route. The enzymes are made as transmembrane zymogens that traverse the secretory system, reach the parasite surface, are internalized via the cytostome along with their substrate hemoglobin, get delivered to the digestive vacuole membrane, and are then cleaved, releasing the mature enzyme into the vacuolar lumen (92–94) (Fig. 3C).
Maturation is carried out by the digestive vacuole cysteine proteases falcipain-2 and -3 (49). If those enzymes are inhibited, plasmepsin autoprocessing takes place, albeit at a slower rate and at a site 1 amino acid upstream from the normal cleavage site. Structures of P. falciparum PM II, PM III, and PM IV proenzymes as well as those of P. vivax PM IV have been solved (63, 95–97). The proenzyme of each is inactive because the propiece pushes apart the N- and C-terminal lobes of the protease, keeping the catalytic residues too far apart for activity. This differs from mammalian aspartic protease zymogens, for which the propiece binds to the substrate cleft to maintain inactivity until pH-dependent autoactivation can take place.
Key questions
What is the mechanism of PM III? Is His32 involved in catalysis? Is PM III even an active enzyme?
Systems biology of hemoglobin degradation: How do cleavages made by each plasmepsin and by other proteases synergize? Why maintain four plasmepsins in P. falciparum? Why do some parasites amplify the digestive vacuole plasmepsin gene region even further?
When is heme released during hemoglobin proteolysis, and how is this orchestrated with hemozoin formation?
Effector protein export: Plasmepsin V
After invasion, the parasite exports hundreds of proteins into the host RBC. These exported effectors enact a dramatic program of host modification, reconstituting a complex trafficking system in the RBC cytosol and altering the host cell's rigidity, nutrient permeability, and endothelial binding properties (98, 99). A significant portion of this export program appears dedicated to trafficking families of variable membrane adhesins to the RBC surface, creating adhesin-rich protrusions called “knobs” that mediate binding to the vascular endothelium (Fig. 5) (100). In P. falciparum, around 400 predicted gene products are annotated as likely to be exported (101, 102). Nearly 200 belong to three families of membrane adhesins (∼120 rifins, 50 PfEMP1s, and 30 stevors) (101, 103). Many of the remaining predicted proteins have no clear function and no homology to genes outside Plasmodium. We are still just beginning to understand the mechanisms by which Plasmodium manipulates its host through its exported proteins.
Figure 5.
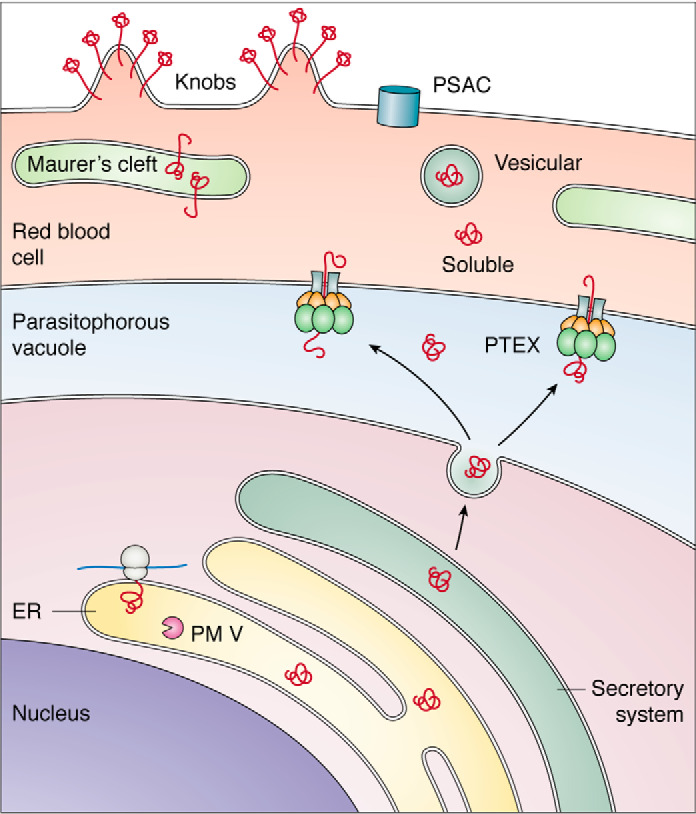
Biosynthesis and trafficking of exported proteins. Proteins are synthesized in the ER and traverse the secretory system (green) to the parasitophorous vacuole (blue), where they are recognized by the PTEX translocon and exported into the RBC (pink). In the RBC, effectors can be soluble, can reside in the Golgi-like Maurer's clefts that are established by the parasite in the host cell, can be vesicular, or can go to the RBC surface, forming nutrient acquisition channels (PSAC) or clustering variant surface cytoadhesins in knobs at the RBC surface.
Discovery
Initial evidence of aspartic proteases acting on processes other than hemoglobin degradation came nearly 2 decades ago, when antibodies raised against newly annotated aspartic proteases labeled parasites outside the digestive vacuole (104). PM V was characterized as a constitutively expressed endoplasmic reticulum (ER)-resident integral membrane protein (105). In 2004, the Haldar and Cowman groups independently discovered that many proteins destined for export contain a pentameric amino acid motif ∼15–30 amino acids downstream from the signal sequence, termed the Plasmodium export element (PEXEL) with the consensus sequence RXLX(E/Q/D) (106, 107). This element was shown to be necessary and sufficient to export fluorescent reporters. In 2008, Chang et al. (108) reported that exported proteins are cleaved in the ER, after the conserved Leu in the PEXEL sequence. The advent of tools for reverse genetics in P. falciparum enabled the assignment of the PEXEL-processing function to PM V (109, 110). Development of peptidomimetic inhibitors of PM V has driven further discovery, validating PM V as essential for parasite survival in RBCs during the asexual cycle and gametocytogenesis (111–113). In addition to uncovering novel parasite biology, these inhibitors have enabled the determination of a high-resolution PM V structure, opening the door to biochemical and pharmacological investigation of this essential enzyme (114). A number of tools have now been turned toward the study of PM V, including inducible Di-Cre excision (115) and post-transcriptional depletion (111, 116, 117), as well as expression, purification, and activity assessment of PM V expressed from E. coli (110, 118–121), from insect cells (114), and from parasite culture (109, 110). This has enabled rapid progress in our knowledge of this enzyme and its development as a potential antimalarial target.
Function
PEXEL-containing proteins are translated into the ER, immediately processed by PM V after the PEXEL Leu, and then acetylated at the new N terminus by an unknown N-acetyltransferase (108–110, 122, 123). Preventing PEXEL processing by mutating PEXEL or depleting/inhibiting PM V blocks protein export, indicating that PM V serves as a gatekeeper for the export pathway (106, 107, 111, 117). Processed PEXEL proteins are secreted into the parasitophorous vacuole and then exported across the vacuolar membrane into the host cell via the Plasmodium translocon for exported proteins (PTEX) (124–127) (Fig. 5). A number of proteins lack PEXEL yet are still exported, termed PEXEL-negative exported proteins (PNEPs). Their N termini are believed to confer export competency similarly to that of the mature PEXEL N terminus (128). Most known PNEPs are involved in trafficking adhesins to the RBC surface or are adhesins themselves.
PM V is not the only gatekeeper to protein export; several mutated PEXEL reporters are correctly cleaved by PM V but still retained in the parasitophorous vacuole (101, 123). Two non-mutually exclusive models have been put forward to explain this. First, export-destined proteins may contain trafficking information in addition to PEXEL that targets them for export. This information would have to be C-terminal to PEXEL, as the N-terminal sequence is removed by PM V. Exported reporter studies have greatly limited the possible locations of such information. The N terminus of PEXEL proteins, containing the PEXEL motif followed by just 11 amino acids, supports export of a GFP fusion (106, 107). Following PEXEL cleavage, the only detectable sequence conservation is the P2′ residue (largely restricted to Gln, Glu, or Asp). Residues beyond P2′ are important for export, as an inserted PEXEL was not sufficient to re-target the normally vacuole-resident SERA5 to the RBC, but if the subsequent 18 amino acids from the PEXEL protein PfEMP3 were added, export was restored (101). However, the properties of these sequences that support export have remained elusive; they have no obvious conserved sequence, structure, or biochemical properties; and Boddey et al. (101) reported the surprising finding that even replacing the post-P2′ amino acids with all Ala supports protein export, leaving us to wonder what signaling information this sequence could contain. A variant of this model would be that secreted proteins bind HSP101 and get exported by default, unless they are too tightly folded (129) or contain an N-terminal sequence that prevents chaperone recognition.
The second model posits that following PEXEL cleavage, PM V hands off export-destined cargo to an ER chaperone that ushers it through the export pathway (109). In this model, PM V is required not just for cleavage, but also for some downstream steps. Tests of this model were attempted with constructs that would create a post-PEXEL N terminus independent of PM V using either an engineered signal peptidase or a fused viral protease; these gave discordant results, with the signal peptidase-generated N terminus unable to support export, whereas the viral protease-generated N terminus did support export (110, 130).
Another avenue for elucidating the post-PM V path is to determine its interacting partners, a task that was carefully undertaken by Danushka et al. (131). They found that PM V interacts with a noncatalytic component of the signal peptidase complex SPC25 and, if cross-linked, with the ER translocon Sec61, its accessory proteins for post-translational import Sec62/63, as well as signal peptidase. They posited that secretory proteins follow one of two distinct paths: either they are cleaved by the canonical signal peptidase complex, or they are recognized and cleaved by a distinct signal-processing complex consisting of PM V and SPC25 (131) (Fig. 6). Perhaps there are chaperones that recognize this complex selectively, although their identity is not apparent at this time. A variant of the chaperone hand-off model is that PM V could be in a subregion of the ER with a direct route to the PTEX translocon at the PVM (110, 132). Subregions of the PVM have recently been described, with each region hosting distinct secretory proteins (133, 134). However, there is no information yet about whether these PVM subregions are fed by distinct regions of the ER.
Figure 6.
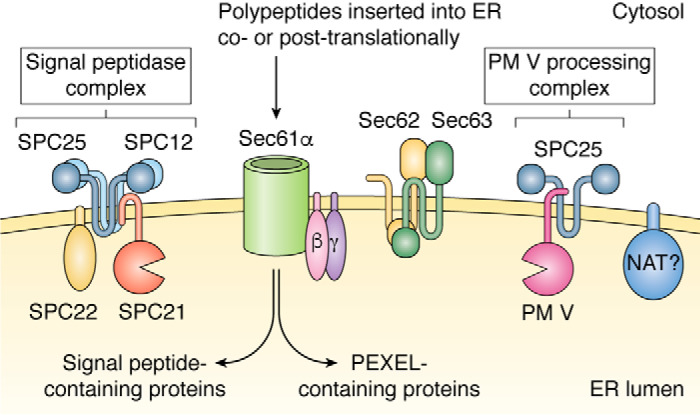
Model for signal processing in the ER. Secretory proteins are cleaved immediately after translation either by the canonical signal peptidase (SPC21) complex (left) or a noncanonical PM V–processing complex (right). NAT, putative N-acetyltransferase.
There is some evidence that PM V additionally functions as a secretory maturase for dense granule proteins. Dense granules are small secretory vesicles that discharge shortly after parasite invasion into the parasitophorous vacuole surrounding the parasite. In addition to the canonical PEXEL, PM V cleaves the “relaxed PEXEL” (RXLXXE) of the dense granule protein RESA (101, 117). Following PEXEL cleavage, RESA is diverted to the dense granules of the forming daughter merozoites. When these merozoites re-invade new RBCs, RESA is secreted into the vacuolar lumen and rapidly exported via PTEX into the host cell (124, 125). Inhibition of PM V shortly before egress is rapidly deleterious in newly invaded ring-stage parasites (117). RESA itself is nonessential, and whether PM V cleaves additional dense granule proteins is not yet clear. In T. gondii, the PM V ortholog ASP5 cleaves dense granule proteins at a PEXEL-like motif (135–137). When ASP5 processing is blocked, substrates still traffic to the dense granules but are unable to fulfill their roles in modifying the parasitophorous vacuole or host cell (136). If PM V does act as a dense granule protein maturase in both Toxoplasma and Plasmodium, perhaps this is its ancestral function, a concept that would be especially interesting given recent findings on the translocon, PTEX. This complex is made up of three core components: an AAA+-ATPase, Hsp101; a channel-forming protein, Exp2; and an adaptor protein, PTEX150 (126). Hsp101 is a ubiquitous chaperone that has been co-opted by Plasmodium to unfold and thread proteins through the translocon. Exp2, in Plasmodium and Toxoplasma, forms a nutrient pore to get essential substances such as glucose and amino acids from the host cell (138, 139). This protein has also been deployed by Plasmodium for a second function—protein export. To dock Hsp101 to the Exp2 pore for protein export, the organism has come up with a novel adapter, PTEX150 (127). Thus, we suggest that Plasmodium has cobbled together a protein export system from mostly preexisting components, and further, it has recruited another preexisting protein, PM V, to prepare proteins for export. It is not at all clear why most exported proteins need to be made as preproteins and processed by PM V co-translationally unless the cleavage motif (PEXEL) is an important component of the export recognition system.
Specificity
Unlike the digestive vacuole proteases, PM V has a highly specific recognition sequence, cleaving only after the conserved Leu of the consensus PEXEL sequence RXLX(E/Q/D) (108, 122, 123). Mutation of the highly conserved PEXEL residue P1 or P3 ablates cleavage (with the notable exception of P1 Ile, which can inconsistently support some PM V cleavage) (101, 106, 107, 140). PM V does not cleave the PNEPs SBP1, PfEMP1, and REX2 (101) or a fluorogenic globin-derived peptide cleaved by PM I–IV (105). Functional PEXELs are found 15–30 amino acids downstream of the signal sequence (141); however, whether this location is required for PM V cleavage has not been reported.
Structure
The structure of the P. vivax PM V bound to a peptidomimetic inhibitor has been determined and reveals that PM V largely resembles the digestive vacuole plasmepsins, with a few notable differences (114) (Fig. 7). Like PM I-IV, PM V is produced with a poorly conserved N-terminal extension speculated to be a self-inhibiting prodomain. However, processing of PM V has not been detected (105), and the full-length recombinant protein has been reported to be active (118, 119). As in other plasmepsins, the PM V active site is covered by a flap that may serve to regulate substrate access. The PM V flap is several amino acids longer than the analogous region of the digestive vacuole plasmepsins, a difference that could perhaps underlie PM V's unique substrate specificity. Along the flap (but not predicted to contact the substrate) is an unpaired Cys, which has been implicated in PM V's sensitivity to Hg2+ (118, 120, 121). Connected directly to the flap is an insert made up of two cysteine pairs folded into a cloverleaf-like structure called a “nepenthesin insert,” which it shares with Nep1 from the pitcher plant Nepenthesia. The nepenthesin insert is present in PM V across the genus, but absent from other plasmepsins. Its location adjoined to the flap over the active site tempts us to speculate that the nepenthesin insert may respond to some environmental condition or binding partner and regulate access to the active site. Alternatively, the nepenthesin insert could be a set distance from the active site and be involved in measuring distance from the N-end of potential substrates (142). No clear hint comes from its ortholog Nep1, a broad-substrate digestive protease present in the low pH of the pitcher plant lumen (143). Another unusual feature is a helix-turn-helix motif, again not present in other plasmepsins. Whereas this is often a nucleic acid-interacting motif, it is not clear what purpose such a motif might have in the ER lumen, where all but the nonessential C-terminal tail of PM V is found (109, 144). Projecting away from the active site of the enzyme is a poorly structured insert whose length and sequence varies among different Plasmodium species and isolates from different regions (145, 146); mutagenesis studies on recombinant enzyme have not yet revealed a function for this structure (146). Further work has the potential to both uncover novel parasite biology and also unearth secrets of biochemistry broadly applicable to other enzymes outside of Plasmodium.
Figure 7.
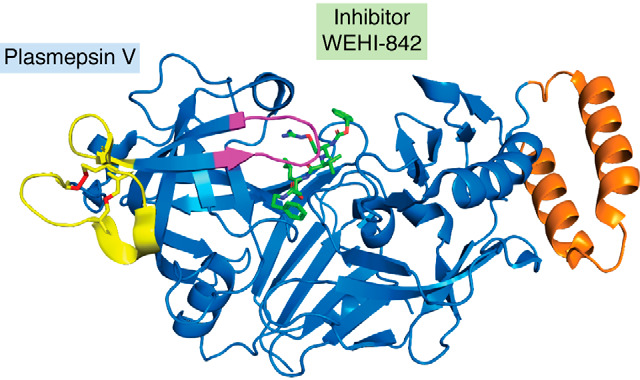
Crystal structure of PM V with the inhibitor WEHI-842 bound (green). Highlighted are the flap over the active site (magenta), nepenthesin loop (yellow, with Cys-Cys bonds in red), and helix-turn-helix (orange); from PDB entry 4ZL4.
Key questions
What do signature features of PM V do (helix-turn-helix motif, nepenthesin loop, unpaired cysteine in the flap)?
What is the basis of exported protein targeting, and how is PM V involved? Is N-acetylation a required part of this, and is it coordinated with PM V action?
What are the essential PM V substrates required for parasitophorous vacuole establishment?
Transmission-stage plasmepsins: Plasmepsins VI, VII, and VIII
PM VI, VII, and VIII are expressed only in transmission-stage parasites and have roles in midgut sporozoite development and function (104). They have been studied mostly in the rodent malaria parasite P. berghei, where each has been disrupted with no effect in the RBC cycle. PM VI knockout parasites produce oocysts that fail to develop into sporozoites, resulting in a transmission block (147). PM VII can be detected in the cytoplasm of P. falciparum zygotes and ookinetes, but its function is not known (148). The protein is not present in P. falciparum gametocytes, suggesting that it is produced after fertilization. Knockout has no effect on any stage of the P. berghei life cycle (149). PM VIII knockout parasites form oocysts in the mosquito midgut (150). However, there is a drastic decrease in the number of salivary gland and hemolymph sporozoites when PM VIII is absent, due to a defect in egress from oocysts. The few sporozoites produced have a defect in gliding motility. Consequently, transmission of these parasites to the host is blocked. The egress phenotype mirrors that seen with PM X in intraerythrocytic parasites (see below), but mechanistic details for PM VIII function remain to be elucidated. More work is needed to understand the function of these important enzymes.
Key questions
What are the specific roles and substrates of these proteases?
Do the inhibitors being developed against other plasmepsins also target the transmission-stage plasmepsins?
What is the role of other plasmepsins, such as PM X, in the transmission stages?
Egress/invasion plasmepsins: Plasmepsins IX and X
An obvious consequence of the malaria parasite's intraerythrocytic lifestyle is that it needs to get into the host cell and, when it is finished replicating and dividing, needs to get back out (Fig. 8A). Invasion is a multistep process (151, 152) involving initial low-affinity recognition of the red blood cell by the parasite, reorientation of the invasive merozoite, and discharge of secretory organelles. This allows high-affinity binding to erythrocyte surface proteins and to parasite proteins that the organism has secreted onto the host cell membrane. This is followed by erythrocyte cytoskeletal reorganization and signaling cascades, tight junction formation, actin-mediated entry, and finally resealing once inside. The whole invasion process takes less than a minute. Egress is also a complex phenomenon (153–155). The parasite must porate and disrupt the surrounding parasitophorous vacuolar membrane, disassemble the host cell cytoskeleton, porate, and finally rupture the erythrocyte membrane, which it does explosively, releasing new infective merozoites into the circulation. The egress process takes about 10 min. Proteases play key roles in egress and invasion; in invasion, proteolytic processing is essential for invasion protein function; in egress, there is a proteolytic cascade, triggered by the cGMP-dependent discharge of proteases into the parasitophorous vacuole (154). Recent evidence suggests that aspartic proteases play important roles in both entry and exit from the erythrocyte.
Figure 8.
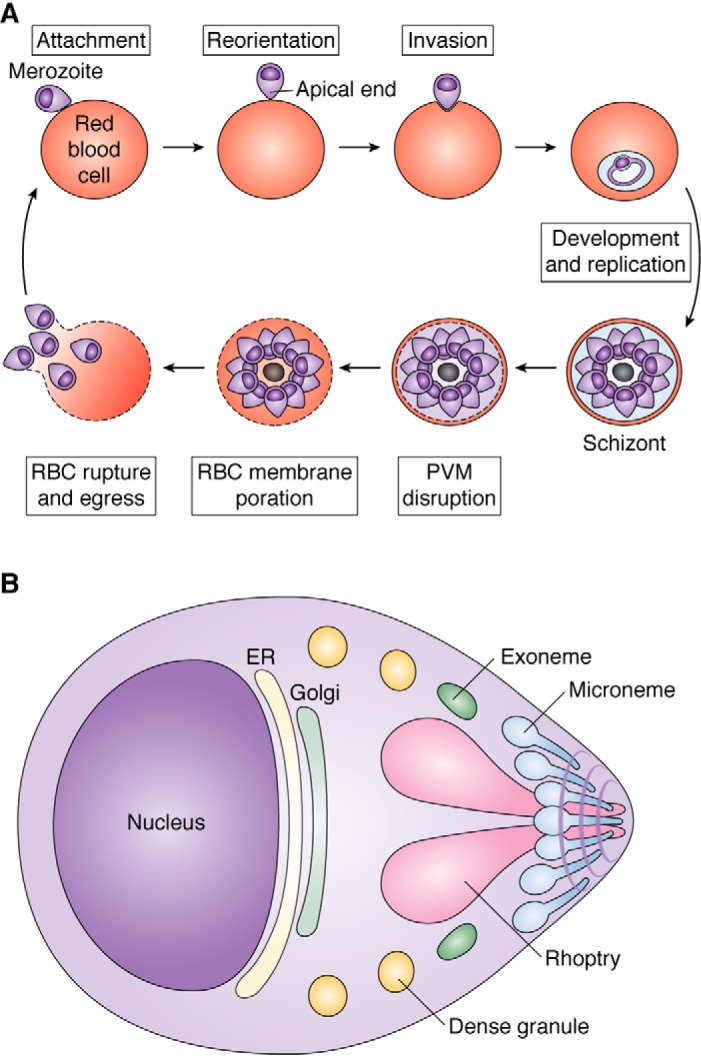
Plasmodium invasion and egress. A, an infective merozoite (top left) attaches to an RBC, reorients so that its apical end is in contact with the host cell, and invades. It develops, replicates, and divides, forming a schizont (bottom right). When ready to egress, it disrupts the PVM and then the RBC membrane, and finally merozoites rupture out of the RBC explosively (bottom left). B, schematic of a merozoite, showing secretory organelles whose function is influenced by plasmepsins. Exonemes discharge to initiate egress. Then micronemes discharge to secrete RBC adhesion ligands onto the cell surface. Next, rhoptries discharge to prepare the parasite and host cell for invasion and to condition the PVM. Finally, dense granules discharge to effect maturation of the parasitophorous vacuole.
PM IX is expressed in late-stage schizonts and is localized to rhoptries (Fig. 8B), secretory organelles that contain adhesins for erythrocyte invasion and proteins for setting up the PVM upon invasion (156). This organelle is highly segregated, with the invasion ligands at the apical end (neck) and other proteins at the basal end (bulb). Upon depletion/disruption of P. falciparum PM IX, invasion of red blood cells is severely compromised (156, 157). Depletion/disruption of PM IX also blocks cleavage of the rhoptry-associated protein 1 (RAP1) and the apical sushi protein (ASP). Peptides corresponding to the cleavage sites of both proteins are cleaved by recombinant PM IX. The activity was observed at pH 6.4 (157). RON3, a rhoptry protein inserted into the PVM during or just after invasion, is also thought to be a PM IX substrate from inhibitor studies (158). Rhoptry morphogenesis and attachment defects were observed in the PM IX-depleted strain (156). These defects presumably explain the invasion phenotype observed. The substrates of PM IX responsible for the phenotype are still not known, because RAP1 is not essential (159) and ASP is predicted to be nonessential by a piggy bac transposon mutagenesis screen (160). Maturation of PM IX appears to be autocatalytic (158).
PM X is involved in both egress and invasion (156–158). In asexual P. falciparum parasites, PM X is localized to small oval-shaped vesicles called exonemes (156). The other protein that has been localized to this organelle is the serine protease subtilisin-like protease 1 (SUB1). PM X processes SUB1 to activate it. Knockdown or chemical inhibition of PM X results in the accumulation of SUB1 precursor (156–158). Mature SUB1 is required for the degradation of both the parasitophorous vacuole and red cell membranes to allow dissemination of merozoites from a mother schizont. To initiate the egress cascade, SUB1 activates cysteine proteases called SERAs and merozoite surface proteins called MSPs (154). Full block of PM X traps parasites within the PVM, whereas partial block allows egress from this membrane but prevents escape from the RBC membrane. Presumably, a higher level of activated SUB1 is required for its effects on the erythrocyte. PM X also processes the surface adhesin sheddase SUB2 as well as erythrocyte binding-like (EBL) and Rh family members that are all involved in invasion (158). In sexual stages of P. falciparum, PM X is expressed in gametes, zygotes, and ookinetes and in P. berghei, it is implicated by inhibitor studies in egress from gametocytes and mosquito midgut invasion (148, 157). PM X processes the midgut invasion protein cell-traversal protein for ookinetes and sporozoites (CelTOS) (157). Inhibitors also prevent progression from the liver to erythrocytes, although different compounds either block merosome formation (hence egress of schizonts in hepatocytes (157)) or have an effect on merozoite competence (158). It is worth noting that compounds used in these studies inhibit both PM IX and PM X. PM IX is not expressed in liver or sexual stages, however, suggesting that action may be specific to PM X in these portions of the life cycle, although some effects through action on another aspartic protease may be suggested by the fact that different small molecules have been reported to have somewhat different effects on mosquito and liver-stage parasites.
Recombinant PM X is active at pH 5.5 and can cleave a variety of synthetic peptide substrates, including ones based on SUB1, EBA-175, EBA-181, and Rh2 (156–158). It has a strong preference for cleavage between two hydrophobic residues, not dissimilar from the digestive vacuole plasmepsins, and prefers a polar P2 residue and Glu, Gln, or Asp for P2′ (158). PM X can cleave PM IX substrates, suggesting overlapping specificity (158). PM X is autocatalytic, but in parasites not all of the PM X gets processed; they retain some as proenzyme (66 kDa) or as active 51- and 44-kDa versions (156–158). An ER calcium-binding protein called PfERC is required for PM X maturation in an unknown fashion (161).
In Toxoplasma gondii, one aspartic protease called ASP3 is closely related to both PM IX and PM X (9). It is localized in an endosomal compartment (post-Golgi). This protein processes microneme and rhoptry proteins and is implicated in egress and invasion and hence is thought of as a PM IX and PM X chimera (162). Mutations in the conserved Phe344 confer resistance to the small-molecule inhibitor 49c in cultured Toxoplasma and for isolated ASP3, PM IX, or PM X (163). The cleavage sites for ASP3 substrates are similar to the peptide cleavage sites for the PM IX and PM X that were tested, adding to the concept of their relatedness and similar roles in the two organisms. PM V, IX, and X all share a function as secretory organelle protein maturases (Fig. 8B).
Key questions
What are the key substrates of PM IX required for rhoptry morphogenesis?
How does PM X cleave and activate the SUB1 precursor that is bound to its propiece?
Where does PM X interact with and process some microneme and rhoptry proteins? It has only been found in exonemes thus far.
Other aspartic proteases
Four other putative aspartic proteases are encoded in the Plasmodium genome but were not appreciated when the plasmepsins were named (164). MiGS is most homologous to PM V but is expressed in male gametocytes and microgametes (165, 166). It appears to play a role in formation of the secretory osmiophilic bodies of male gametocytes. After secretion onto the surface, it also plays a role in exflagellation of male microgametes that go on to fertilize the female gametes. Like the other plasmepsins, it is predicted to have a classical pepsin-like fold; however, one of the catalytic sites has clearly undergone substantial mutation, with the conserved DTGS mutated to LTNS. Whether MiGS folds in an unexpected way to bring another catalytic residue into the active site, acts as a homodimer, or acts as a pseudoprotease is unclear.
Ddi1 (DNA damage-inducible protease 1) is another aspartic protease with a Plasmodium ortholog. In other eukaryotes, it is important for the cell cycle through ubiquitin/proteasome interactions (167). It has only one catalytic aspartate and homodimerizes to form the active enzyme. In malaria parasites, all that is known is that its gene is refractory to knockout in P. berghei and P. falciparum (160, 168, 169).
SPP is an intramembrane aspartic protease that processes signal peptides after release from secretory proteins in the ER. Early reports based on antibody and inhibitor studies suggested that the Plasmodium ortholog of SPP might be a micronemal protein involved in invasion (170, 171). Further reports, however, instead localized this protein to the ER and demonstrated roles in signal peptide proteolysis and ER stress responses (172–174). More detailed cellular analysis of SPP-selective inhibitor treatment showed inhibition of intraerythrocytic parasite development rather than invasion (172). Heterologous expression studies have confirmed the ability to cleave intramembrane signal peptides (173, 175, 176). Potent SPP inhibitors that kill cultured intraerythrocytic parasites (171–173, 177) and also block liver-stage development (173, 178) have been described. SPP appears to be an attractive drug target.
Finally, a gene present on chromosome 14 (PF3D7_1465600) next to PM VIII and annotated as an aspartic protease (164) has very distant homology to PM VIII, but the predicted protein is missing both catalytic motifs and is unlikely to be an active protease. It is expressed most highly in male gametocytes and may also be expressed in asexual parasites and other life-cycle stages (179). The gene is predicted to be essential in P. falciparum but not in P. berghei from genome-scale mutational analyses (160, 168). Its function is completely unknown.
Key questions
How does MiGS work in exflagellation? Is it an active enzyme? What are its substrates?
What is the role of Ddi1 in the parasite? What are associated proteins and substrates?
Can selective SPP inhibitors be developed? What is the role of SPP in the ER stress response?
Plasmepsins as drug targets
Thousands of inhibitors have been designed against plasmepsins. Additionally, clinically used HIV protease inhibitors such as lopinavir have been investigated in detail, although their clinical antimalarial efficacy is likely to be marginal. The inhibitor literature has been reviewed elsewhere (8, 180, 181) and is beyond the scope of this article. Here, we will instead focus on implications of the biology of plasmepsins for drug development.
Digestive vacuole plasmepsins play an important role in hemoglobin degradation. Knockout lines are markedly impaired in growth both in culture (P. falciparum) (42) and in vivo (P. berghei- rodent model) (43, 44). In fact, the attenuated P. berghei are cleared and can protect from subsequent challenge with heterologous parasites. That being said, knockout parasites are still viable, and an anti-digestive vacuole plasmepsin drug would need a very long half-life to suppress growth over multiple cycles for parasite clearance. It is unlikely that a digestive vacuole plasmepsin-specific agent will come to fruition, although a compound that blocked digestive vacuole plasmepsins in addition to an essential plasmepsin (CWHM-117 is an example (156)) would likely get an efficacy boost from its action on hemoglobin degradation.
PM V is another interesting enzyme to consider as a target. The process of protein export is essential (124, 125, 138), as presumably is dense granule secretion that helps form the parasitophorous vacuole after invasion, in which PM V likely plays a role (117). Blocking PM V could potentially hit the parasite at two different points in its developmental cycle, which is an advantage for treating an infection that can be asynchronous. Tempering enthusiasm for this target is the observation that PM V must be knocked down nearly completely to have any effect on parasite viability. The implication is that a drug would have to have exceptional potency to be effective. Current recommended EC50 values for parasite killing by candidate compounds are in the low nanomolar range. A compound would likely have to have a mid-picomolar ki for PM V to achieve efficacy—not impossible, but a very high bar.
Plasmepsin VI, VII, and VIII are potential drug targets for insect stages of the parasite. A drug against an enzyme in these stages would not cure a patient but could prevent transmission of the disease. Dosing patients to get a curative concentration in the mosquito is a potential hurdle. Not enough is known about these enzymes to have a clear idea how to move forward. PM VII appears to be dispensable and therefore would be low on the list.
PM IX is a candidate drug target. It is essential for blood-stage parasites (156, 157) and is inhibited by the small-molecule inhibitors 49c and WM382 (157, 158), although both compounds are also potent against PM X. PM X is also essential for blood-stage parasites and can be potently blocked by the dual-inhibitory compounds mentioned above, as well as by aminohydantoins (156–158). Each of these chemotypes is active in vivo using rodent malaria models, and 49c and WM382 also have been shown to interrupt transmission to mosquitos as well as egress from hepatocytes. The extra-erythrocytic phenotypes are likely due to a block of PM X, although an important role for the inhibition of other plasmepsins such as PM VI–VIII cannot be excluded. Dual inhibition of PM IX and X or of PM X and an insect-stage PM would be a bonus for an antimalarial because of heightened difficulty in selecting resistance or ability to treat more than one stage (157, 158). However, keeping a drug development program on-target for two different enzymes while avoiding inhibition of host orthologs such as cathepsin D is a challenging task (182, 183). Inhibition of PM X may be enough to hit multiple life cycle stages by itself. Drug development for PM X is proceeding at a rapid pace (158) (https://www.mmv.org/research-development/mmv-supported-projects).
Key questions
HIV protease inhibitors like lopinavir kill malaria parasites in culture. What is their killing target? The digestive vacuole plasmepsins, Ddi1, SPP, and hexose transporter PfHT1 have all been proposed (169, 184–188).
Do PM X inhibitors work in mosquito stages by blocking PM X, and/or do they target other enzymes, such as MiGS or PM VI–VIII?
Will a drug against a target that is active during just a few hours of the life cycle show sustained efficacy?
Is hemoglobin digestion a viable target?
Conclusions
Plasmepsins participate throughout the complex biology of the malaria parasite (Table 1). The digestive vacuole plasmepsins are the most extensively studied, and we have a reasonable picture of their biosynthesis and roles in hemoglobin degradation. We do not yet understand the mechanism of PM III, with its histidine instead of a canonical aspartate. Plasmepsin V is the next best-studied plasmepsin. Its role in exported effector protein processing is fairly clear, but its function in dense granule protein processing is not well-established. The protein has a number of unusual domains whose functions are not clear at this time. Plasmepsins VI–VIII as well as the PM V paralog MiGS play roles in the insect stages; PM X may as well. Studies of these protease activities in the mosquito are just beginning. PM IX and PM X have defined roles in erythrocytic invasion and egress, but their enzymology is not yet defined. Many of these enzymes afford opportunities for antimalarial drug discovery, and we can look forward to exciting new drug candidates in the not too distant future.
Table 1.
Plasmodium aspartic proteases: Temporal and spatial location and function
IE, infected erythrocyte; ERAD, endoplasmic reticulum-associated protein degradation.
| Plasmepsin | Stage where functional | Location | Function |
|---|---|---|---|
| I–IV (IV only for non-falciparum) | Intraerythrocytic (IE) | Digestive vacuole | Hemoglobin degradation |
| V | IE, liver | ER | Effector protein export, dense granule protein maturation |
| VI | Oocyst | ? | ? |
| VII | Zygote, Ookinete | ? | ? |
| VIII | Oocyst | ? | Sporozoite motility and egress |
| IX | IE (late) | Rhoptries | Invasion |
| X | IE (late) | Exonemes | Egress/invasion |
| DdiI | IE | Nucleus | Cell cycle? |
| MiGS | Male gametocytes and gametes | Osmiophilic bodies, cell surface | Exflagellation |
| SPP | IE, liver | ER | ERAD |
EuPathDB Accession numbers
Plasmepsins: I (PF3D7_1407900), II (PF3D7_1408000), III (PF3D7_1408100), IV (PF3D7_1407800, PBANKA_1034400), V (PF3D7_1323500), VI (PF3D7_0311700), VII (PF3D7_1033800), VIII (PF3D7_1465700), IX (PF3D7_1430200), X (PF3D7_0808200); other proteases: MiGS (PF3D7_1234400), DDI1 (PF3D7_1409300), SPP (PF3D7_1457000), falcipain 2 (PF3D7_1115700), falcipain 3 (PF3D7_1115400), DPAP1 (PF3D7_1116700), aminopeptidase P (PF3D7_1454400), aminopeptidase M1 (PF3D7_1311800), SUB1 (PF3D7_0507500), SUB2 (PF3D7_1136900), TgAsp1 (TGME49_201840), TgAsp2 (TGME49_262940), TgAsp3 (TGME49_246550), TgAsp4 (TGME49_209620), TgAsp5 (TGME49_242720), TgAsp6 (TGME49_272510); other proteins: HDP (PF3D7_1446800), RAP1 (PF3D7_1410400), ASP (PF3D7_0405900), AMA1 (PF3D7_1133400).
Data availability
All data are contained within the manuscript.
Acknowledgments
We thank Dr. Prasenjit Bhaumik (Indian Institute of Technology, Mumbai) for helpful comments and Dr. Robert Potter (Washington University) for help with phylogenetic analysis. We acknowledge PlasmoDB as a source for analysis of genes discussed herein.
Author contributions—D. E. G. conceptualization; D. E. G. resources; D. E. G. supervision; D. E. G. funding acquisition; A. S. N., A. J. P., E. S. I., and D. E. G. validation; A. S. N., A. J. P., E. S. I., and D. E. G. writing-original draft; A. S. N., A. J. P., E. S. I., and D. E. G. writing-review and editing.
Funding and additional information—This work was supported by National Institutes of Health Grants AI138447 and AI112508. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- RBC
- red blood cell
- PM
- plasma membrane
- MiGS
- microgametocyte surface protein
- SPP
- signal peptide peptidase
- PEXEL
- Plasmodium export element
- PTEX
- Plasmodium translocon for exported proteins
- PNEP
- PEXEL-negative exported protein
- ER
- endoplasmic reticulum
- RAP1
- rhoptry-associated protein 1
- ASP
- apical sushi protein
- DPAP1
- dipeptidyl aminopeptidase 1
- PDB
- Protein Data Bank.
References
- 1. Goldberg D. E., Slater A. F. G., Beavis R., Chait B., Cerami A., and Henderson G. B. (1991) Hemoglobin degradation in the human malaria pathogen Plasmodium falciparum: a catabolic pathway initiated by a specific aspartic protease. J. Exp. Med. 173, 961–969 10.1084/jem.173.4.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Francis S. E., Gluzman I. Y., Oksman A., Knickerbocker A., Mueller R., Bryant M. L., Sherman D. R., Russell D. G., and Goldberg D. E. (1994) Molecular characterization and inhibition of a Plasmodium falciparum aspartic hemoglobinase. EMBO J. 13, 306–317 10.1002/j.1460-2075.1994.tb06263.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dame J. B., Reddy G. R., Yowell C. A., Dunn B. M., Kay J., and Berry C. (1994) Sequence, expression and modeled structure of an aspartic proteinase from the human malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 64, 177–190 10.1016/0166-6851(94)90024-8 [DOI] [PubMed] [Google Scholar]
- 4. Dame J. B., Yowell C. A., Omara-Opyene L., Carlton J. M., Cooper R. A., and Li T. (2003) Plasmepsin 4, the food vacuole aspartic proteinase found in all Plasmodium spp. infecting man. Mol. Biochem. Parasitol. 130, 1–12 10.1016/S0166-6851(03)00137-3 [DOI] [PubMed] [Google Scholar]
- 5. Banerjee R., Liu J., Beatty W., Pelosof L., Klemba M., and Goldberg D. E. (2002) Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci. U.S.A. 99, 990–995 10.1073/pnas.022630099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hill J., Tyas L., Phylip L. H., Kay J., Dunn B. M., and Berry C. (1994) High level expression and characterisation of Plasmepsin II, an aspartic proteinase from Plasmodium falciparum. FEBS Lett. 352, 155–158 10.1016/0014-5793(94)00940-6 [DOI] [PubMed] [Google Scholar]
- 7. Silva A. M., Lee A. Y., Gulnik S. V., Maier P., Collins J., Bhat T. N., Collins P. J., Cachau R. E., Luker K. E., Gluzman I. Y., Francis S. E., Oksman A., Goldberg D. E., and Erickson J. W. (1996) Structure and inhibition of plasmepsin II, a hemoglobin-degrading enzyme from Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 93, 10034–10039 10.1073/pnas.93.19.10034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyers M. J., and Goldberg D. E. (2012) Recent advances in plasmepsin medicinal chemistry and implications for future antimalarial drug discovery efforts. Curr. Top. Med. Chem. 12, 445–455 10.2174/156802612799362959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shea M., Jäkle U., Liu Q., Berry C., Joiner K. A., and Soldati-Favre D. (2007) A family of aspartic proteases and a novel, dynamic and cell-cycle-dependent protease localization in the secretory pathway of Toxoplasma gondii. Traffic 8, 1018–1034 10.1111/j.1600-0854.2007.00589.x [DOI] [PubMed] [Google Scholar]
- 10. Rawlings N. D., Waller M., Barrett A. J., and Bateman A. (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 42, D503–D509 10.1093/nar/gkt953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Francis S. E., Sullivan D. J. Jr., and Goldberg D. E. (1997) Hemoglobin metabolism in the malaria parasite Plasmodium falciparum. Annu. Rev. Microbiol. 51, 97–123 10.1146/annurev.micro.51.1.97 [DOI] [PubMed] [Google Scholar]
- 12. Lew V. L., Tiffert T., and Ginsburg H. (2003) Excess hemoglobin digestion and the osmotic stability of Plasmodium falciparum-infected red blood cells. Blood 101, 4189–4194 10.1182/blood-2002-08-2654 [DOI] [PubMed] [Google Scholar]
- 13. Sherman I. W., and Tanigoshi L. (1970) Incorporation of 14C-amino-acids by malaria (Plasmodium lophurae) IV. In vivo utilization of host cell haemoglobin. Int. J. Biochem. 1, 635–637 10.1016/0020-711X(70)90033-9 [DOI] [Google Scholar]
- 14. Abu Bakar N., Klonis N., Hanssen E., Chan C., and Tilley L. (2010) Digestive-vacuole genesis and endocytic processes in the early intraerythrocytic stages of Plasmodium falciparum. J. Cell Sci. 123, 441–450 10.1242/jcs.061499 [DOI] [PubMed] [Google Scholar]
- 15. Aikawa M., Hepler P. K., Huff C. G., and Sprinz H. (1966) The feeding mechanism of avian malarial parasites. J. Cell Biol. 28, 355–373 10.1083/jcb.28.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goldberg D. E., Slater A. F. G., Cerami A., and Henderson G. B. (1990) Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc. Natl. Acad. Sci. U.S.A. 87, 2931–2935 10.1073/pnas.87.8.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kolakovich K. A., Gluzman I. Y., Duffin K. L., and Goldberg D. E. (1997) Generation of hemoglobin peptides in the acidic digestive vacuole of Plasmodium falciparum implicates peptide transport in amino acid production. Mol. Biochem. Parasitol. 87, 123–135 10.1016/S0166-6851(97)00062-5 [DOI] [PubMed] [Google Scholar]
- 18. Rudzinska M. A., Trager W., and Bray R. S. (1965) Pinocytotic uptake and the digestion of hemoglobin in malaria parasites. J. Protozool. 12, 563–576 10.1111/j.1550-7408.1965.tb03256.x [DOI] [PubMed] [Google Scholar]
- 19. Lewis I. A., Wacker M., Olszewski K. L., Cobbold S. A., Baska K. S., Tan A., Ferdig M. T., and Llinás M. (2014) Metabolic QTL analysis links chloroquine resistance in Plasmodium falciparum to impaired hemoglobin catabolism. PLoS Genet. 10, e1004085 10.1371/journal.pgen.1004085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee A. H., Dhingra S. K., Lewis I. A., Singh M. K., Siriwardana A., Dalal S., Rubiano K., Klein M. S., Baska K. S., Krishna S., Klemba M., Roepe P. D., Llinás M., Garcia C. R. S., and Fidock D. A. (2018) Evidence for regulation of hemoglobin metabolism and intracellular ionic flux by the Plasmodium falciparum chloroquine resistance transporter. Sci. Rep. 8, 13578 10.1038/s41598-018-31715-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sigala P. A., and Goldberg D. E. (2014) The peculiarities and paradoxes of plasmodium heme metabolism. Annu. Rev. Microbiol. 68, 259–278 10.1146/annurev-micro-091313-103537 [DOI] [PubMed] [Google Scholar]
- 22. Sullivan D. J. (2017) Quinolines block every step of malaria heme crystal growth. Proc. Natl. Acad. Sci. U.S.A. 114, 7483–7485 10.1073/pnas.1708153114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birnbaum J., Scharf S., Schmidt S., Jonscher E., Hoeijmakers W. A. M., Flemming S., Toenhake C. G., Schmitt M., Sabitzki R., Bergmann B., Fröhlke U., Mesén-Ramírez P., Blancke Soares A., Herrmann H., Bártfai R., and Spielmann T. (2020) A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 367, 51–59 10.1126/science.aax4735 [DOI] [PubMed] [Google Scholar]
- 24. Yang T., Yeoh L. M., Tutor M. V., Dixon M. W., McMillan P. J., Xie S. C., Bridgford J. L., Gillett D. L., Duffy M. F., Ralph S. A., McConville M. J., Tilley L., and Cobbold S. A. (2019) Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 29, 2917–2928.e5 10.1016/j.celrep.2019.10.095 [DOI] [PubMed] [Google Scholar]
- 25. Levy M. R., Siddiqui W. A., and Chou S. C. (1974) Acid protease activity in Plasmodium falciparum and P. knowlesi and ghosts of their respective host red cells. Nature 247, 546–549 10.1038/247546a0 [DOI] [PubMed] [Google Scholar]
- 26. Levy M. R., and Chou S. C. (1973) Activity and some properties of an acid proteinase from normal and Plasmodium berghei-infected red cells. J. Parasitol. 59, 1064–1070 10.2307/3278644 [DOI] [PubMed] [Google Scholar]
- 27. Sherman I. W., and Tanigoshi L. (1981) The proteases of Plasmodium: a cathepsin D-like enzyme from Plasmodium lophurae. in The Biochemistry of Parasites, pp. 137–149, Elsevier, Amsterdam [Google Scholar]
- 28. Gyang F. N., Poole B., and Trager W. (1982) Peptidases from Plasmodium falciparum cultured in vitro. Mol. Biochem. Parasitol. 5, 263–273 10.1016/0166-6851(82)90034-2 [DOI] [PubMed] [Google Scholar]
- 29. Aissi E., Charet P., Bouquelet S., and Biguet J. (1983) Endoprotease in Plasmodium yoelii nigeriensis. Comp. Biochem. Physiol. B 74, 559–566 10.1016/0305-0491(83)90229-8 [DOI] [PubMed] [Google Scholar]
- 30. Sato K., Fukabori Y., and Suzuki M. (1987) Plasmodium berghei: a study of globinolytic enzyme in erythrocytic parasite. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 264, 487–495 10.1016/s0176-6724(87)80072-x [DOI] [PubMed] [Google Scholar]
- 31. Hempelmann E., and Wilson R. J. M. (1980) Endopeptidases from Plasmodium knowlesi. Parasitology 80, 323–330 10.1017/S0031182000000780 [DOI] [PubMed] [Google Scholar]
- 32. Bailly E., Savel J., Mahouy G., and Jaureguiberry G. (1991) Plasmodium falciparum: isolation and characterization of a 55-kDa protease with a cathepsin D-like activity from P. falciparum. Exp. Parasitol. 72, 278–284 10.1016/0014-4894(91)90147-O [DOI] [PubMed] [Google Scholar]
- 33. Vander Jagt D. L., Hunsaker L. A., and Campos N. M. (1986) Characterization of a hemoglobin-degrading, low molecular weight protease from Plasmodium falciparum. Mol. Biochem. Parasitol. 18, 389–400 10.1016/0166-6851(86)90095-2 [DOI] [PubMed] [Google Scholar]
- 34. vander Jagt D. L., Hunsaker L. A., Campos N. M., and Scaletti J. V. (1992) Localization and characterization of hemoglobin-degrading aspartic proteinases from the malarial parasite Plasmodium falciparum. Biochim. Biophys. Acta 1122, 256–264 10.1016/0167-4838(92)90401-X [DOI] [PubMed] [Google Scholar]
- 35. Choi I., and Mego J. L. (1987) Intravacuolar proteolysis in Plasmodium falciparum digestive vacuoles is similar to intralysosomal proteolysis in mammalian cells. Biochim. Biophys. Acta 926, 170–176 10.1016/0304-4165(87)90234-0 [DOI] [PubMed] [Google Scholar]
- 36. Gluzman I. Y., Francis S. E., Oksman A., Smith C. E., Duffin K. L., and Goldberg D. E. (1994) Order and specificity of the Plasmodium falciparum hemoglobin degradation pathway. J. Clin. Invest. 93, 1602–1608 10.1172/JCI117140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coombs G. H., Goldberg D. E., Klemba M., Berry C., Kay J., and Mottram J. C. (2001) Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 17, 532–537 10.1016/S1471-4922(01)02037-2 [DOI] [PubMed] [Google Scholar]
- 38. Liu J., Gluzman I. Y., Drew M. E., and Goldberg D. E. (2005) The role of Plasmodium falciparum food vacuole plasmepsins. J. Biol. Chem. 280, 1432–1437 10.1074/jbc.M409740200 [DOI] [PubMed] [Google Scholar]
- 39. Omara-Opyene A. L., Moura P. A., Sulsona C. R., Bonilla J. A., Yowell C. A., Fujioka H., Fidock D. A., and Dame J. B. (2004) Genetic disruption of the Plasmodium falciparum digestive vacuole plasmepsins demonstrates their functional redundancy. J. Biol. Chem. 279, 54088–54096 10.1074/jbc.M409605200 [DOI] [PubMed] [Google Scholar]
- 40. Bonilla J. A., Moura P. A., Bonilla T. D., Yowell C. A., Fidock D. A., and Dame J. B. (2007) Effects on growth, hemoglobin metabolism and paralogous gene expression resulting from disruption of genes encoding the digestive vacuole plasmepsins of Plasmodium falciparum. Int. J. Parasitol. 37, 317–327 10.1016/j.ijpara.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 41. Liu J., Istvan E. S., Gluzman I. Y., Gross J., and Goldberg D. E. (2006) Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc. Natl. Acad. Sci. U.S.A. 103, 8840–8845 10.1073/pnas.0601876103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonilla J. A., Bonilla T. D., Yowell C. A., Fujioka H., and Dame J. B. (2007) Critical roles for the digestive vacuole plasmepsins of Plasmodium falciparum in vacuolar function. Mol. Microbiol. 65, 64–75 10.1111/j.1365-2958.2007.05768.x [DOI] [PubMed] [Google Scholar]
- 43. Spaccapelo R., Janse C. J., Caterbi S., Franke-Fayard B., Bonilla J. A., Syphard L. M., Di Cristina M., Dottorini T., Savarino A., Cassone A., Bistoni F., Waters A. P., Dame J. B., and Crisanti A. (2010) Plasmepsin 4-deficient Plasmodium berghei are virulence attenuated and induce protective immunity against experimental malaria. Am. J. Pathol. 176, 205–217 10.2353/ajpath.2010.090504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin J. W., Spaccapelo R., Schwarzer E., Sajid M., Annoura T., Deroost K., Ravelli R. B. G., Aime E., Capuccini B., Mommaas-Kienhuis A. M., O'Toole T., Prins F., Franke-Fayard B. M. D., Ramesar J., Chevalley-Maurel S., et al. (2015) Replication of Plasmodium in reticulocytes can occur without hemozoin formation, resulting in chloroquine resistance. J. Exp. Med. 212, 893–903 10.1084/jem.20141731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sijwali P. S., and Rosenthal P. J. (2004) Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 101, 4384–4389 10.1073/pnas.0307720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moon S. U., Kang J. M., Kim T. S., Kong Y., Sohn W. M., and Na B. K. (2011) Plasmodium vivax: collaborative roles for plasmepsin 4 and vivapains in hemoglobin hydrolysis. Exp. Parasitol. 128, 127–132 10.1016/j.exppara.2011.02.015 [DOI] [PubMed] [Google Scholar]
- 47. Bailly E., Jambou R., Savel J., and Jaureguiberry G. (1992) Plasmodium falciparum: differential sensitivity in vitro to E-64 (cysteine protease inhibitor) and pepstatin A (aspartyl protease inhibitor). J. Protozool. 39, 593–599 10.1111/j.1550-7408.1992.tb04856.x [DOI] [PubMed] [Google Scholar]
- 48. Semenov A., Olson J. E., and Rosenthal P. J. (1998) Antimalarial synergy of cysteine and aspartic protease inhibitors. Antimicrob. Agents Chemother. 42, 2254–2258 10.1128/AAC.42.9.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drew M. E., Banerjee R., Uffman E. W., Gilbertson S., Rosenthal P. J., and Goldberg D. E. (2008) Plasmodium food vacuole plasmepsins are activated by falcipains. J. Biol. Chem. 283, 12870–12876 10.1074/jbc.M708949200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eggleson K. K., Duffin K. L., and Goldberg D. E. (1999) Identification and characterization of falcilysin, a metallopeptidase involved in hemoglobin catabolism within the malaria parasite Plasmodium falciparum. J. Biol. Chem. 274, 32411–32417 10.1074/jbc.274.45.32411 [DOI] [PubMed] [Google Scholar]
- 51. Murata C. E., and Goldberg D. E. (2003) Plasmodium falciparum falcilysin: a metalloprotease with dual specificity. J. Biol. Chem. 278, 38022–38028 10.1074/jbc.M306842200 [DOI] [PubMed] [Google Scholar]
- 52. Klemba M., Gluzman I., and Goldberg D. E. (2004) A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J. Biol. Chem. 279, 43000–43007 10.1074/jbc.M408123200 [DOI] [PubMed] [Google Scholar]
- 53. Ragheb D., Bompiani K., Dalal S., and Klemba M. (2009) Evidence for catalytic roles for Plasmodium falciparum aminopeptidase P in the food vacuole and cytosol. J. Biol. Chem. 284, 24806–24815 10.1074/jbc.M109.018424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ragheb D., Dalal S., Bompiani K. M., Ray W. K., and Klemba M. (2011) Distribution and biochemical properties of an M1-family aminopeptidase in Plasmodium falciparum indicate a role in vacuolar hemoglobin catabolism. J. Biol. Chem. 286, 27255–27265 10.1074/jbc.M111.225318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chugh M., Sundararaman V., Kumar S., Reddy V. S., Siddiqui W. A., Stuart K. D., and Malhotra P. (2013) Protein complex directs hemoglobin-to-hemozoin formation in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 110, 5392–5397 10.1073/pnas.1218412110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Amato R., Lim P., Miotto O., Amaratunga C., Dek D., Pearson R. D., Almagro-Garcia J., Neal A. T., Sreng S., Suon S., Drury E., Jyothi D., Stalker J., Kwiatkowski D. P., and Fairhurst R. M. (2017) Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect. Dis. 17, 164–173 10.1016/S1473-3099(16)30409-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Witkowski B., Duru V., Khim N., Ross L. S., Saintpierre B., Beghain J., Chy S., Kim S., Ke S., Kloeung N., Eam R., Khean C., Ken M., Loch K., Bouillon A., et al. (2017) A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect. Dis. 17, 174–183 10.1016/S1473-3099(16)30415-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bopp S., Magistrado P., Wong W., Schaffner S. F., Mukherjee A., Lim P., Dhorda M., Amaratunga C., Woodrow C. J., Ashley E. A., White N. J., Dondorp A. M., Fairhurst R. M., Ariey F., Menard D., Wirth D. F., and Volkman S. K. (2018) Plasmepsin II–III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat. Commun. 9, 1769 10.1038/s41467-018-04104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mukherjee A., Gagnon D., Wirth D. F., and Richard D. (2018) Inactivation of plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob. Agents Chemother. 62, e02309–17 10.1128/AAC.02309-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loesbanluechai D., Kotanan N., de Cozar C., Kochakarn T., Ansbro M. R., Chotivanich K., White N. J., Wilairat P., Lee M. C. S., Gamo F. J., Sanz L. M., Chookajorn T., and Kümpornsin K. (2019) Overexpression of plasmepsin II and plasmepsin III does not directly cause reduction in Plasmodium falciparum sensitivity to artesunate, chloroquine and piperaquine. Int. J. Parasitol. Drugs Drug Resist. 9, 16–22 10.1016/j.ijpddr.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ross L. S., Dhingra S. K., Mok S., Yeo T., Wicht K. J., Kümpornsin K., Takala-Harrison S., Witkowski B., Fairhurst R. M., Ariey F., Menard D., and Fidock D. A. (2018) Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 9, 3314 10.1038/s41467-018-05652-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Asojo O. A., Gulnik S. V., Afonina E., Yu B., Ellman J. A., Haque T. S., and Silva A. M. (2003) Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J. Mol. Biol. 327, 173–181 10.1016/S0022-2836(03)00036-6 [DOI] [PubMed] [Google Scholar]
- 63. Bernstein N. K., Cherney M. M., Yowell C. A., Dame J. B., and James M. N. G. (2003) Structural insights into the activation of P. vivax plasmepsin. J. Mol. Biol. 329, 505–524 10.1016/S0022-2836(03)00444-3 [DOI] [PubMed] [Google Scholar]
- 64. Clemente J. C., Govindasamy L., Madabushi A., Fisher S. Z., Moose R. E., Yowell C. A., Hidaka K., Kimura T., Hayashi Y., Kiso Y., Agbandje-McKenna M., Dame J. B., Dunn B. M., and McKenna R. (2006) Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor. Acta Crystallogr. D Biol. Crystallogr. 62, 246–252 10.1107/S0907444905041260 [DOI] [PubMed] [Google Scholar]
- 65. Madabushi A., Chakraborty S., Fisher S. Z., Clemente J. C., Yowell C., Agbandje-McKenna M., Dame J. B., Dunn B. M., and McKenna R. (2005) Crystallization and preliminary X-ray analysis of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 228–231 10.1107/S1744309105001405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bhaumik P., Gustchina A., and Wlodawer A. (2012) Structural studies of vacuolar plasmepsins. Biochim. Biophys. Acta 1824, 207–223 10.1016/j.bbapap.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bhaumik P., Xiao H., Parr C. L., Kiso Y., Gustchina A., Yada R. Y., and Wlodawer A. (2009) Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum. J. Mol. Biol. 388, 520–540 10.1016/j.jmb.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bhaumik P., Horimoto Y., Xiao H., Miura T., Hidaka K., Kiso Y., Wlodawer A., Yada R. Y., and Gustchina A. (2011) Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum. J. Struct. Biol. 175, 73–84 10.1016/j.jsb.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu J., Istvan E. S., and Goldberg D. E. (2006) Hemoglobin-degrading plasmepsin II is active as a monomer. J. Biol. Chem. 281, 38682–38688 10.1074/jbc.M608535200 [DOI] [PubMed] [Google Scholar]
- 70. Xiao H., Briere L. A. K., Dunn S. D., and Yada R. Y. (2010) Characterization of the monomer-dimer equilibrium of recombinant histo-aspartic protease from Plasmodium falciparum. Mol. Biochem. Parasitol. 173, 17–24 10.1016/j.molbiopara.2010.04.008 [DOI] [PubMed] [Google Scholar]
- 71. Friedman R., and Caflisch A. (2007) The protonation state of the catalytic aspartates in plasmepsin II. FEBS Lett. 581, 4120–4124 10.1016/j.febslet.2007.07.033 [DOI] [PubMed] [Google Scholar]
- 72. Xie D., Gulnik S., Collins L., Gustchina E., Suvorov L., and Erickson J. W. (1997) Dissection of the pH dependence of inhibitor binding energies for an aspartic protease: direct measurement of the protonation states of the catalytic aspartic acid residues. Biochemistry 36, 16166–16172 10.1021/bi971550l [DOI] [PubMed] [Google Scholar]
- 73. Parr C. L., Tanaka T., Xiao H., and Yada R. Y. (2008) The catalytic significance of the proposed active site residues in Plasmodium falciparum histoaspartic protease. FEBS J. 275, 1698–1707 10.1111/j.1742-4658.2008.06325.x [DOI] [PubMed] [Google Scholar]
- 74. Andreeva N., Bogdanovich P., Kashparov I., Popov M., and Stengach M. (2004) Is histoaspartic protease a serine protease with a pepsin-like fold? Proteins Struct. Funct. Genet. 55, 705–710 10.1002/prot.20078 [DOI] [PubMed] [Google Scholar]
- 75. Bjelic S., and Åqvist J. (2004) Computational prediction of structure, substrate binding mode, mechanism, and rate for a malaria protease with a novel type of active site. Biochemistry 43, 14521–14528 10.1021/bi048252q [DOI] [PubMed] [Google Scholar]
- 76. Xiao H., Sinkovits A. F., Bryksa B. C., Ogawa M., and Yada R. Y. (2006) Recombinant expression and partial characterization of an active soluble histo-aspartic protease from Plasmodium falciparum. Protein Expr. Purif. 49, 88–94 10.1016/j.pep.2006.02.022 [DOI] [PubMed] [Google Scholar]
- 77. Luker K. E., Francis S. E., Gluzman I. Y., and Goldberg D. E. (1996) Kinetic analysis of plasmepsins i and II, aspartic proteases of the plasmodium falciparum digestive vacuole. Mol. Biochem. Parasitol. 79, 71–78 10.1016/0166-6851(96)02651-5 [DOI] [PubMed] [Google Scholar]
- 78. Moon R. P., Tyas L., Certa U., Rupp K., Bur D., Jacquet C., Matile H., Loetscher H., Grueninger-Leitch F., Kay J., Dunn B. M., Berry C., and Ridley R. G. (1997) Expression and characterisation of plasmepsin I from Plasmodium falciparum. Eur. J. Biochem. 244, 552–560 10.1111/j.1432-1033.1997.00552.x [DOI] [PubMed] [Google Scholar]
- 79. Westling J., Yowell C. A., Majer P., Erickson J. W., Dame J. B., and Dunn B. M. (1997) Plasmodium falciparum, P. vivax, and P. malariae: a comparison of the active site properties of plasmepsins cloned and expressed from three different species of the malaria parasite. Exp. Parasitol. 87, 185–193 10.1006/expr.1997.4225 [DOI] [PubMed] [Google Scholar]
- 80. Tyas L., Gluzman I., Moon R. P. R. P., Rupp K., Westling J., Ridley R. G. R. G., Kay J., Goldberg D. E. D. E., and Berry C. (1999) Naturally-occurring and recombinant forms of the aspartic proteinases plasmepsins I and II from the human malaria parasite Plasmodium falciparum. FEBS Lett. 454, 210–214 10.1016/S0014-5793(99)00805-4 [DOI] [PubMed] [Google Scholar]
- 81. Westling J., Cipullo P., Hung S.-H., Saft H., Dame J. B., and Dunn B. M. (1999) Active site specificity of plasmepsin II. Protein Sci. 8, 2001–2009 10.1110/ps.8.10.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Humphreys M. J., Moon R. P., Klinder A., Fowler S. D., Rupp K., Bur D., Ridley R. G., and Berry C. (1999) The aspartic proteinase from the rodent parasite Plasmodium berghei as a potential model for plasmepsins from the human malaria parasite, Plasmodium falciparum. FEBS Lett. 463, 43–48 10.1016/s0014-5793(99)01597-5 [DOI] [PubMed] [Google Scholar]
- 83. Wyatt D. M., and Berry C. (2002) Activity and inhibition of plasmepsin IV, a new aspartic proteinase from the malaria parasite, Plasmodium falciparum. FEBS Lett. 513, 159–162 10.1016/S0014-5793(02)02241-X [DOI] [PubMed] [Google Scholar]
- 84. Siripurkpong P., Yuvaniyama J., Wilairat P., and Goldberg D. E. (2002) Active site contribution to specificity of the aspartic proteases plasmepsins I and II. J. Biol. Chem. 277, 41009–41013 10.1074/jbc.M204852200 [DOI] [PubMed] [Google Scholar]
- 85. Li T., Yowell C. A., Beyer B. B., Hung S. H., Westling J., Lam M. T., Dunn B. M., and Dame J. B. (2004) Recombinant expression and enzymatic subsite characterization of plasmepsin 4 from the four Plasmodium species infecting man. Mol. Biochem. Parasitol. 135, 101–109 10.1016/j.molbiopara.2004.01.010 [DOI] [PubMed] [Google Scholar]
- 86. Beyer B. B., Johnson J. V., Chung A. Y., Li T., Madabushi A., Agbandje-McKenna M., McKenna R., Dame J. B., and Dunn B. M. (2005) Active-site specificity of digestive aspartic peptidases from the four species of Plasmodium that infect humans using chromogenic combinatorial peptide libraries. Biochemistry 44, 1768–1779 10.1021/bi047886u [DOI] [PubMed] [Google Scholar]
- 87. Martins T. M., Domingos A., Berry C., and Wyatt D. M. (2006) The activity and inhibition of the food vacuole plasmepsin from the rodent malaria parasite Plasmodium chabaudi. Acta Trop. 97, 212–218 10.1016/j.actatropica.2005.11.001 [DOI] [PubMed] [Google Scholar]
- 88. Xiao H., Tanaka T., Ogawa M., and Yada R. Y. (2007) Expression and enzymatic characterization of the soluble recombinant plasmepsin I from Plasmodium falciparum. Protein Eng. Des. Sel. 20, 625–633 10.1093/protein/gzm066 [DOI] [PubMed] [Google Scholar]
- 89. Liu P., Marzahn M. R., Robbins A. H., Gutiérrez-de-Terán H., Rodríguez D., McClung S. H., Stevens S. M. Jr., Yowell C. A., Dame J. B., McKenna R., and Dunn B. M. (2009) Recombinant plasmepsin 1 from the human malaria parasite Plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis. Biochemistry 48, 4086–4099 10.1021/bi802059r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu P., Robbins A. H., Marzahn M. R., McClung S. H., Yowell C. A., Stevens S. M. Jr., Dame J. B., and Dunn B. M. (2015) Enzymatic characterization of recombinant food vacuole plasmepsin 4 from the rodent malaria parasite plasmodium berghei. PLoS ONE 10, e0141758 10.1371/journal.pone.0141758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Istvan E. S., and Goldberg D. E. (2005) Distal substrate interactions enhance plasmepsin activity. J. Biol. Chem. 280, 6890–6896 10.1074/jbc.M412086200 [DOI] [PubMed] [Google Scholar]
- 92. Francis S. E., Banerjee R., and Goldberg D. E. (1997) Biosynthesis and maturation of the malaria aspartic hemoglobinases plasmepsins I and II. J. Biol. Chem. 272, 14961–14968 10.1074/jbc.272.23.14961 [DOI] [PubMed] [Google Scholar]
- 93. Banerjee R., Francis S. E., and Goldberg D. E. (2003) Food vacuole plasmepsins are processed at a conserved site by an acidic convertase activity in Plasmodium falciparum. Mol. Biochem. Parasitol. 129, 157–165 10.1016/S0166-6851(03)00119-1 [DOI] [PubMed] [Google Scholar]
- 94. Klemba M., Beatty W., Gluzman I., and Goldberg D. E. (2004) Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J. Cell Biol. 164, 47–56 10.1083/jcb200307147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bernstein N. K., Cherney M. M., Loetscher H., Ridley R. G., and James M. N. G. (1999) Crystal structure of the novel aspartic proteinase zymogen proplasmepsin II from Plasmodium falciparum. Nat. Struct. Biol. 6, 32–37 10.1038/4905 [DOI] [PubMed] [Google Scholar]
- 96. Bhaumik P., Xiao H., Hidaka K., Gustchina A., Kiso Y., Yada R. Y., and Wlodawer A. (2011) Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum. Biochemistry 50, 8862–8879 10.1021/bi201118z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Recacha R., Jaudzems K., Akopjana I., Jirgensons A., and Tars K. (2016) Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins. Acta Crystallogr. F Struct. Biol. Commun. 72, 659–666 10.1107/S2053230X16011663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Spillman N. J., Beck J. R., and Goldberg D. E. (2015) Protein export into malaria parasite–infected erythrocytes: mechanisms and functional consequences. Annu. Rev. Biochem. 84, 813–841 10.1146/annurev-biochem-060614-034157 [DOI] [PubMed] [Google Scholar]
- 99. Matthews K. M., Pitman E. L., and de Koning-Ward T. F. (2019) Illuminating how malaria parasites export proteins into host erythrocytes. Cell. Microbiol. 21, e13009 10.1111/cmi.13009 [DOI] [PubMed] [Google Scholar]
- 100. Maier A. G., Rug M., O'Neill M. T., Brown M., Chakravorty S., Szestak T., Chesson J., Wu Y., Hughes K., Coppel R. L., Newbold C., Beeson J. G., Craig A., Crabb B. S., and Cowman A. F. (2008) Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 134, 48–61 10.1016/j.cell.2008.04.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Boddey J. A., Carvalho T. G., Hodder A. N., Sargeant T. J., Sleebs B. E., Marapana D., Lopaticki S., Nebl T., and Cowman A. F. (2013) Role of plasmepsin V in export of diverse protein families from the Plasmodium falciparum exportome. Traffic 14, 532–550 10.1111/tra.12053 [DOI] [PubMed] [Google Scholar]
- 102. Heiber A., Kruse F., Pick C., Grüring C., Flemming S., Oberli A., Schoeler H., Retzlaff S., Mesén-Ramírez P., Hiss J. A., Kadekoppala M., Hecht L., Holder A. A., Gilberger T.-W., and Spielmann T. (2013) Identification of new PNEPs indicates a substantial non-PEXEL exportome and underpins common features in Plasmodium falciparum protein export. PLoS Pathog. 9, e1003546 10.1371/journal.ppat.1003546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., and Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12 10.1186/gb-2006-7-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Banerjee R., Liu J., Beatty W., Pelosof L., Klemba M., Goldberg D. E. (2002) Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc. Natl. Acad. Sci. U.S.A. 99, 990–995 10.1073/pnas.022630099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Klemba M., and Goldberg D. E. (2005) Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Mol. Biochem. Parasitol. 143, 183–191 10.1016/j.molbiopara.2005.05.015 [DOI] [PubMed] [Google Scholar]
- 106. Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estraño C., and Haldar K. (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 10.1126/science.1102737 [DOI] [PubMed] [Google Scholar]
- 107. Marti M., Good R. T., Rug M., Knuepfer E., and Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 10.1126/science.1102452 [DOI] [PubMed] [Google Scholar]
- 108. Chang H. H., Falick A. M., Carlton P. M., Sedat J. W., DeRisi J. L., and Marletta M. A. (2008) N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115 10.1016/j.molbiopara.2008.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Russo I., Babbitt S., Muralidharan V., Butler T., Oksman A., and Goldberg D. E. (2010) Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 463, 632–636 10.1038/nature08726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Boddey J. A., Hodder A. N., Günther S., Gilson P. R., Patsiouras H., Kapp E. A., Pearce J. A., de Koning-Ward T. F., Simpson R. J., Crabb B. S., and Cowman A. F. (2010) An aspartyl protease directs malaria effector proteins to the host cell. Nature 463, 627–631 10.1038/nature08728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sleebs B. E., Lopaticki S., Marapana D. S., O'Neill M. T., Rajasekaran P., Gazdik M., Günther S., Whitehead L. W., Lowes K. N., Barfod L., Hviid L., Shaw P. J., Hodder A. N., Smith B. J., Cowman A. F., and Boddey J. A. (2014) Inhibition of Plasmepsin V activity demonstrates its essential role in protein export, PfEMP1 display, and survival of malaria parasites. PLos Biol. 12, e1001897 10.1371/journal.pbio.1001897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sleebs B. E., Gazdik M., O'Neill M. T., Rajasekaran P., Lopaticki S., Lackovic K., Lowes K., Smith B. J., Cowman A. F., and Boddey J. A. (2014) Transition state mimetics of the Plasmodium export element are potent inhibitors of Plasmepsin V from P. falciparum and P. vivax. J. Med. Chem. 57, 7644–7662 10.1021/jm500797g [DOI] [PubMed] [Google Scholar]
- 113. Jennison C., Lucantoni L., O'Neill M. T., McConville R., Erickson S. M., Cowman A. F., Sleebs B. E., Avery V. M., Boddey J. A., O'Neill M. T., McConville R., Erickson S. M., Cowman A. F., Sleebs B. E., Avery V. M., and Boddey J. A. (2019) Inhibition of plasmepsin V activity blocks Plasmodium falciparum gametocytogenesis and transmission to mosquitoes. Cell Rep. 29, 3796–3806.e4 10.1016/j.celrep.2019.11.073 [DOI] [PubMed] [Google Scholar]
- 114. Hodder A. N., Sleebs B. E., Czabotar P. E., Gazdik M., Xu Y., O'Neill M. T., Lopaticki S., Nebl T., Triglia T., Smith B. J., Lowes K., Boddey J. A., and Cowman A. F. (2015) Structural basis for plasmepsin V inhibition that blocks export of malaria proteins to human erythrocytes. Nat. Struct. Mol. Biol. 22, 590–596 10.1038/nsmb.3061 [DOI] [PubMed] [Google Scholar]
- 115. Boonyalai N., Collins C. R., Hackett F., Withers-Martinez C., and Blackman M. J. (2018) Essentiality of Plasmodium falciparum plasmepsin V. PLoS ONE 13, e0207621 10.1371/journal.pone.0207621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gambini L., Rizzi L., Pedretti A., Taglialatela-Scafati O., Carucci M., Pancotti A., Galli C., Read M., Giurisato E., Romeo S., and Russo I. (2015) Picomolar inhibition of plasmepsin V, an essential malaria protease, achieved exploiting the prime region. PLoS ONE 10, e0142509 10.1371/journal.pone.0142509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Polino A. J., Nasamu A. S., Niles J. C., and Goldberg D. E. (2020) Assessment of biological role and insight into druggability of the Plasmodium falciparum protease plasmepsin V. ACS Infect Dis. 6, 738–746 10.1021/acsinfecdis.9b00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xiao H., Bryksa B. C., Bhaumik P., Gustchina A., Kiso Y., Yao S. Q., Wlodawer A., and Yada R. Y. (2014) The zymogen of plasmepsin v from Plasmodium falciparum is enzymatically active. Mol. Biochem. Parasitol. 197, 56–63 10.1016/j.molbiopara.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Boonyalai N., Sittikul P., and Yuvaniyama J. (2015) Plasmodium falciparum plasmepsin V (PfPMV): insights into recombinant expression, substrate specificity and active site structure. Mol. Biochem. Parasitol. 201, 5–15 10.1016/j.molbiopara.2015.05.004 [DOI] [PubMed] [Google Scholar]
- 120. Loymunkong C., Sittikul P., Songtawee N., Wongpanya R., and Boonyalai N. (2019) Yield improvement and enzymatic dissection of Plasmodium falciparum plasmepsin V. Mol. Biochem. Parasitol. 231, 111188 10.1016/j.molbiopara.2019.111188 [DOI] [PubMed] [Google Scholar]
- 121. Sittikul P., Songtawee N., Kongkathip N., and Boonyalai N. (2018) In vitro and in silico studies of naphthoquinones and peptidomimetics toward Plasmodium falciparum plasmepsin V. Biochimie 152, 159–173 10.1016/j.biochi.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 122. Boddey J. A., Moritz R. L., Simpson R. J., and Cowman A. F. (2009) Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10, 285–299 10.1111/j.1600-0854.2008.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Osborne A. R., Speicher K. D., Tamez P. A., Bhattacharjee S., Speicher D. W., and Haldar K. (2010) The host targeting motif in exported Plasmodium proteins is cleaved in the parasite endoplasmic reticulum. Mol. Biochem. Parasitol. 171, 25–31 10.1016/j.molbiopara.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Elsworth B., Matthews K., Nie C. Q., Kalanon M., Charnaud S. C., Sanders P. R., Chisholm S. A., Counihan N. A., Shaw P. J., Pino P., Chan J.-A., Azevedo M. F., Rogerson S. J., Beeson J. G., Crabb B. S., et al. (2014) PTEX is an essential nexus for protein export in malaria parasites. Nature 511, 587–591 10.1038/nature13555 [DOI] [PubMed] [Google Scholar]
- 125. Beck J. R., Muralidharan V., Oksman A., and Goldberg D. E. (2014) PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature 511, 592–595 10.1038/nature13574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. de Koning-Ward T. F., Gilson P. R., Boddey J. A., Rug M., Smith B. J., Papenfuss A. T., Sanders P. R., Lundie R. J., Maier A. G., Cowman A. F., and Crabb B. S. (2009) A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 10.1038/nature08104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ho C.-M., Beck J. R., Lai M., Cui Y., Goldberg D. E., Egea P. F., and Zhou Z. H. (2018) Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75 10.1038/s41586-018-0469-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Grüring C., Heiber A., Kruse F., Flemming S., Franci G., Colombo S. F., Fasana E., Schoeler H., Borgese N., Stunnenberg H. G., Przyborski J. M., Gilberger T.-W., and Spielmann T. (2012) Uncovering common principles in protein export of malaria parasites. Cell Host Microbe 12, 717–729 10.1016/j.chom.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 129. Matthews K. M., Kalanon M., and de Koning-Ward T. F. (2019) Uncoupling the threading and unfoldase actions of Plasmodium HSP101 reveals differences in export between soluble and insoluble proteins. mBio 10, e01106–19 10.1128/mBio.01106-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tarr S. J., Cryar A., Thalassinos K., Haldar K., and Osborne A. R. (2013) The C-terminal portion of the cleaved HT motif is necessary and sufficient to mediate export of proteins from the malaria parasite into its host cell. Mol. Microbiol. 87, 835–850 10.1111/mmi.12133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Marapana D. S., Dagley L. F., Sandow J. J., Nebl T., Triglia T., Pasternak M., Dickerman B. K., Crabb B. S., Gilson P. R., Webb A. I., Boddey J. A., and Cowman A. F. (2018) Plasmepsin V cleaves malaria effector proteins in a distinct endoplasmic reticulum translocation interactome for export to the erythrocyte. Nat. Microbiol. 3, 1010–1022 10.1038/s41564-018-0219-2 [DOI] [PubMed] [Google Scholar]
- 132. Goldberg D. E., and Cowman A. F. (2010) Moving in and renovating: exporting proteins from Plasmodium into host erythrocytes. Nat. Rev. Microbiol. 8, 617–621 10.1038/nrmicro2420 [DOI] [PubMed] [Google Scholar]
- 133. Garten M., Beck J. R., Roth R., Tenkova-Heuser T., Heuser J., Bleck C. K. E., Goldberg D. E., and Zimmerberg J. (2019) Contacting domains that segregate lipid from solute transporters in malaria parasites. bioRxiv 10.1101/863993 [DOI] [PMC free article] [PubMed]
- 134. Nessel T., Beck J. M., Rayatpisheh S., Jami-Alahmadi Y., Wohlschlegel J. A., Goldberg D. E., and Beck J. R. (2020) EXP1 is required for organization of EXP2 in the intraerythrocytic malaria parasite vacuole. Cell. Microbiol. 22, e13168 10.1111/cmi.13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Curt-Varesano A., Braun L., Ranquet C., Hakimi M.-A. A., and Bougdour A. (2016) The aspartyl protease TgASP5 mediates the export of the Toxoplasma GRA16 and GRA24 effectors into host cells. Cell. Microbiol. 18, 151–167 10.1111/cmi.12498 [DOI] [PubMed] [Google Scholar]
- 136. Hammoudi P.-M., Jacot D., Mueller C., Di Cristina M., Dogga S. K., Marq J.-B., Romano J., Tosetti N., Dubrot J., Emre Y., Lunghi M., Coppens I., Yamamoto M., Sojka D., Pino P., and Soldati-Favre D. (2015) Fundamental roles of the Golgi-associated Toxoplasma aspartyl protease, ASP5, at the host-parasite interface. PLoS Pathog. 11, e1005211 10.1371/journal.ppat.1005211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Coffey M. J., Dagley L. F., Seizova S., Kapp E. A., Infusini G., Roos D. S., Boddey J. A., Webb A. I., and Tonkin C. J. (2018) Aspartyl protease 5 matures dense granule proteins that reside at the host-parasite interface in Toxoplasma gondii. mBio 9, e01796–18 10.1128/mBio.01796-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Garten M., Nasamu A. S., Niles J. C., Zimmerberg J., Goldberg D. E., and Beck J. R. (2018) EXP2 is a nutrient-permeable channel in the vacuolar membrane of Plasmodium and is essential for protein export via PTEX. Nat. Microbiol. 3, 1090–1098 10.1038/s41564-018-0222-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Gold D. A., Kaplan A. D., Lis A., Bett G. C. L., Rosowski E. E., Cirelli K. M., Bougdour A., Sidik S. M., Beck J. R., Lourido S., Egea P. F., Bradley P. J., Hakimi M.-A., Rasmusson R. L., and Saeij J. P. J. (2015) The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe 17, 642–652 10.1016/j.chom.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Schulze J., Kwiatkowski M., Borner J., Schlüter H., Bruchhaus I., Burmester T., Spielmann T., and Pick C. (2015) The Plasmodium falciparum exportome contains non-canonical PEXEL/HT proteins. Mol. Microbiol. 97, 301–314 10.1111/mmi.13024 [DOI] [PubMed] [Google Scholar]
- 141. Hiss J. A., Przyborski J. M., Schwarte F., Lingelbach K., and Schneider G. (2008) The Plasmodium export element revisited. PLoS ONE 3, e1560 10.1371/journal.pone.0001560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Goldberg D. E. (2015) Plasmepsin V shows its carnivorous side. Nat. Struct. Mol. Biol. 22, 647–648 10.1038/nsmb.3077 [DOI] [PubMed] [Google Scholar]
- 143. Athauda S. B. P., Matsumoto K., Rajapakshe S., Kuribayashi M., Kojima M., Kubomura-Yoshida N., Iwamatsu A., Shibata C., Inoue H., and Takahashi K. (2004) Enzymic and structural characterization of nepenthesin, a unique member of a novel subfamily of aspartic proteinases. Biochem. J. 381, 295–306 10.1042/BJ20031575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tarr S. J., and Osborne A. R. (2015) Experimental determination of the membrane topology of the Plasmodium protease plasmepsin V. PLoS ONE 10, e0121786 10.1371/journal.pone.0121786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Rawat M., Vijay S., Gupta Y., Tiwari P. K., and Sharma A. (2013) Imperfect duplicate insertions type of mutations in plasmepsin V modulates binding properties of PEXEL motifs of export proteins in Indian Plasmodium vivax. PLoS ONE 8, e60077 10.1371/journal.pone.0060077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Sappakhaw K., Takasila R., Sittikul P., Wattana-Amorn P., Assavalapsakul W., and Boonyalai N. (2015) Biochemical characterization of plasmepsin V from Plasmodium vivax Thailand isolates: substrate specificity and enzyme inhibition. Mol. Biochem. Parasitol. 204, 51–63 10.1016/j.molbiopara.2016.01.003 [DOI] [PubMed] [Google Scholar]
- 147. Ecker A., Bushell E. S. C., Tewari R., and Sinden R. E. (2008) Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 70, 209–220 10.1111/j.1365-2958.2008.06407.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Li F., Bounkeua V., Pettersen K., and Vinetz J. M. (2016) Plasmodium falciparum ookinete expression of plasmepsin VII and plasmepsin X. Malar. J. 15, 111 10.1186/s12936-016-1161-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mastan B. S., Kumari A., Gupta D., Mishra S., and Kumar K. A. (2014) Gene disruption reveals a dispensable role for Plasmepsin VII in the Plasmodium berghei life cycle. Mol. Biochem. Parasitol. 195, 10–13 10.1016/j.molbiopara.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 150. Mastan B. S., Narwal S. K., Dey S., Kumar K. A., and Mishra S. (2017) Plasmodium berghei plasmepsin VIII is essential for sporozoite gliding motility. Int. J. Parasitol. 47, 239–245 10.1016/j.ijpara.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 151. Gilson P. R., and Crabb B. S. (2009) Morphology and kinetics of the three distinct phases of red blood cell invasion by Plasmodium falciparum merozoites. Int. J. Parasitol. 39, 91–96 10.1016/j.ijpara.2008.09.007 [DOI] [PubMed] [Google Scholar]
- 152. Cowman A. F., Tonkin C. J., Tham W. H., and Duraisingh M. T. (2017) The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 22, 232–245 10.1016/j.chom.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hale V. L., Watermeyer J. M., Hackett F., Vizcay-Barrena G., van Ooij C., Thomas J. A., Spink M. C., Harkiolaki M., Duke E., Fleck R. A., Blackman M. J., and Saibil H. R. (2017) Parasitophorous vacuole poration precedes its rupture and rapid host erythrocyte cytoskeleton collapse in Plasmodium falciparum egress. Proc. Natl. Acad. Sci. U.S.A. 114, 3439–3444 10.1073/pnas.1619441114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Thomas J. A., Tan M. S. Y., Bisson C., Borg A., Umrekar T. R., Hackett F., Hale V. L., Vizcay-Barrena G., Fleck R. A., Snijders A. P., Saibil H. R., and Blackman M. J. (2018) A protease cascade regulates release of the human malaria parasite Plasmodium falciparum from host red blood cells. Nat. Microbiol. 3, 447–455 10.1038/s41564-018-0111-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Glushakova S., Beck J. R., Garten M., Busse B. L., Nasamu A. S., Tenkova-Heuser T., Heuser J., Goldberg D. E., and Zimmerberg J. (2018) Rounding precedes rupture and breakdown of vacuolar membranes minutes before malaria parasite egress from erythrocytes. Cell. Microbiol. 20, e12868 10.1111/cmi.12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Nasamu A. S., Glushakova S., Russo I., Vaupel B., Oksman A., Kim A. S., Fremont D. H., Tolia N., Beck J. R., Meyers M. J., Niles J. C., Zimmerberg J., and Goldberg D. E. (2017) Plasmepsins IX and X are essential and druggable mediators of malaria parasite egress and invasion. Science 358, 518–522 10.1126/science.aan1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Pino P., Caldelari R., Mukherjee B., Vahokoski J., Klages N., Maco B., Collins C. R., Blackman M. J., Kursula I., Heussler V., Brochet M., and Soldati-Favre D. (2017) A multistage antimalarial targets the plasmepsins IX and X essential for invasion and egress. Science 358, 522–528 10.1126/science.aaf8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Favuzza P., de Lera Ruiz M., Thompson J. K., Triglia T., Ngo A., Steel R. W. J., Vavrek M., Christensen J., Boyce C., Guo Z., Hu M., Khan T., Murgolo N., Zhao L., Penington J. S., et al. (2020) Dual plasmepsin-targeting antimalarial agents disrupt multiple stages of the malaria parasite life cycle. Cell Host Microbe 27, 642–658.e12 10.1016/j.chom.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Baldi D. L., Andrewsm K. T., Waller R. F., Roos D. S., Howard R. F., Crabb B. S., and Cowman A. F. (2000) RAP1 controls rhoptry targeting of RAP2 in the malaria parasite Plasmodium falciparum. EMBO J. 19, 2435–2443 10.1093/emboj/19.11.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., Udenze K., Bronner I. F., Casandra D., Mayho M., Brown J., Li S., Swanson J., Rayner J. C., Jiang R. H. Y., and Adams J. H. (2018) Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science 360, eaap7847 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Fierro M. A., Asady B., Brooks C. F., Cobb D. W., Villegas A., Moreno S. N. J., and Muralidharan V. (2020) An endoplasmic reticulum CREC family protein regulates the egress proteolytic cascade in malaria parasites. mBio 11, e03078–19 10.1128/mBio.03078-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Dogga S. K., Mukherjee B., Jacot D., Kockmann T., Molino L., Hammoudi P.-M., Hartkoorn R. C., Hehl A. B., and Soldati-Favre D. (2017) A druggable secretory protein maturase of Toxoplasma essential for invasion and egress. Elife 6, e27480 10.7554/eLife.27480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Mukherjee B., Tessaro F., Vahokoski J., Kursula I., Marq J. B., Scapozza L., and Soldati-Favre D. (2018) Modeling and resistant alleles explain the selectivity of antimalarial compound 49c towards apicomplexan aspartyl proteases. EMBO J. 37, e98047 10.15252/embj.201798047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Weissbach T., Golzmann A., Bennink S., Pradel G., and Julius Ngwa C. (2017) Transcript and protein expression analysis of proteases in the blood stages of Plasmodium falciparum. Exp. Parasitol. 180, 33–44 10.1016/j.exppara.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 165. Tachibana M., Ishino T., Takashima E., Tsuboi T., and Torii M. (2018) A male gametocyte osmiophilic body and microgamete surface protein of the rodent malaria parasite Plasmodium yoelii (PyMiGS) plays a critical role in male osmiophilic body formation and exflagellation. Cell. Microbiol. 20, e12821 10.1111/cmi.12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kehrer J., Frischknecht F., and Mair G. R. (2016) Proteomic analysis of the plasmodium berghei gametocyte egressome and vesicular bioid of osmiophilic body proteins identifies merozoite trap-like protein (MTRAP) as an essential factor for parasite transmission. Mol. Cell. Proteomics 15, 2852–2862 10.1074/mcp.M116.058263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gabriely G., Kama R., Gelin-Licht R., and Gerst J. E. (2008) Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol. Biol. Cell. 19, 3625–3637 10.1091/mbc.e07-05-0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Bushell E., Gomes A. R., Sanderson T., Anar B., Girling G., Herd C., Metcalf T., Modrzynska K., Schwach F., Martin R. E., Mather M. W., McFadden G. I., Parts L., Rutledge G. G., Vaidya A. B., et al. (2017) Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272.e8 10.1016/j.cell.2017.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Onchieku N. M., Mogire R., Ndung'u L., Mwitari P., Kimani F., Matoke-Muhia D., Kiboi D., and Magoma G. (2018) Deciphering the targets of retroviral protease inhibitors in Plasmodium berghei. PLoS ONE 13, e0201556 10.1371/journal.pone.0201556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Li X., Chen H., Oh S. S., and Chishti A. H. (2008) A presenilin-like protease associated with Plasmodium falciparum micronemes is involved in erythrocyte invasion. Mol. Biochem. Parasitol. 158, 22–31 10.1016/j.molbiopara.2007.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Li X., Chen H., Bahamontes-Rosa N., Kun J. F. J., Traore B., Crompton P. D., and Chishti A. H. (2009) Plasmodium falciparum signal peptide peptidase is a promising drug target against blood stage malaria. Biochem. Biophys. Res. Commun. 380, 454–459 10.1016/j.bbrc.2009.01.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Marapana D. S., Wilson D. W., Zuccala E. S., Dekiwadia C. D., Beeson J. G., Ralph S. A., and Baum J. (2012) Malaria parasite signal peptide peptidase is an ER-resident protease required for growth but not for invasion. Traffic 13, 1457–1465 10.1111/j.1600-0854.2012.01402.x [DOI] [PubMed] [Google Scholar]
- 173. Harbut M. B., Patel B. A., Yeung B. K. S., McNamara C. W., Bright A. T., Ballard J., Supek F., Golde T. E., Winzeler E. A., Diagana T. T., and Greenbaum D. C. (2012) Targeting the ERAD pathway via inhibition of signal peptide peptidase for antiparasitic therapeutic design. Proc. Natl. Acad. Sci. U.S.A. 109, 21486–21491 10.1073/pnas.1216016110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Baldwin M., Russo C., Li X., and Chishti A. H. (2014) Plasmodium falciparum signal peptide peptidase cleaves malaria heat shock protein 101 (HSP101). Implications for gametocytogenesis. Biochem. Biophys. Res. Commun. 450, 1427–1432 10.1016/j.bbrc.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Nyborg A. C., Ladd T. B., Jansen K., Kukar T., and Golde T. E. (2006) Intramembrane proteolytic cleavage by human signal peptide peptidase like 3 and malaria signal peptide peptidase. FASEB J. 20, 1671–1679 10.1096/fj.06-5762com [DOI] [PubMed] [Google Scholar]
- 176. Ran Y., Ladd G. Z., Ceballos-Diaz C., Jung J. I., Greenbaum D., Felsenstein K. M., and Golde T. E. (2015) Differential inhibition of signal peptide peptidase family members by established γ-secretase inhibitors. PLoS ONE 10, e0128619 10.1371/journal.pone.0128619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Hirano J., Okamoto T., Sugiyama Y., Suzuki T., Kusakabe S., Tokunaga M., Fukuhara T., Sasai M., Tougan T., Matsunaga Y., Yamashita K., Sakai Y., Yamamoto M., Horii T., Standley D. M., et al. (2017) Characterization of SPP inhibitors suppressing propagation of HCV and protozoa. Proc. Natl. Acad. Sci. U.S.A. 114, E10782–E10791 10.1073/pnas.1712484114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Parvanova I., Epiphanio S., Fauq A., Golde T. E., Prudêncio M., and Mota M. M. (2009) A small molecule inhibitor of signal peptide peptidase inhibits Plasmodium development in the liver and decreases malaria severity. PLoS ONE 4, e5078 10.1371/journal.pone.0005078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., et al. (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 10.1093/nar/gkn814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Wegscheid-Gerlach C., Gerber H.-D., and Diederich W. E. (2010) Proteases of Plasmodium falciparum as potential drug targets and inhibitors thereof. Curr. Top. Med. Chem. 10, 346–367 10.2174/156802610790725461 [DOI] [PubMed] [Google Scholar]
- 181. Cheuka P. M., Dziwornu G., Okombo J., and Chibale K. (2020) Plasmepsin inhibitors in antimalarial drug discovery: medicinal chemistry and target validation (2000 to present). J. Med. Chem. 63, 4445–4467 10.1021/acs.jmedchem.9b01622 [DOI] [PubMed] [Google Scholar]
- 182. Munsamy G., Ramharack P., and Soliman M. E. S. (2018) Egress and invasion machinery of malaria: an in-depth look into the structural and functional features of the flap dynamics of plasmepsin IX and X. RSC Adv. 8, 21829–21840 10.1039/c8ra04360d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Munsamy G., and Soliman M. E. S. (2019) Unveiling a new era in malaria therapeutics: a tailored molecular approach towards the design of plasmepsin IX inhibitors. Protein J. 38, 616–627 10.1007/s10930-019-09871-2 [DOI] [PubMed] [Google Scholar]
- 184. Parikh S., Gut J., Istvan E., Goldberg D. E., Havlir D. V., and Rosenthal P. J. (2005) Antimalarial activity of human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 49, 2983–2985 10.1128/AAC.49.7.2983-2985.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Skinner-Adams T. S., McCarthy J. S., Gardiner D. L., Hilton P. M., and Andrews K. T. (2004) Antiretrovirals as antimalarial agents. J. Infect. Dis. 190, 1998–2000 10.1086/425584 [DOI] [PubMed] [Google Scholar]
- 186. Schwake C., Baldwin M. R., Bachovchin W., Hegde S., Schiemer J., Okure C., Levin A. E., Vannier E., Hanada T., and Chishti A. H. (2019) HIV protease inhibitors block parasite signal peptide peptidases and prevent growth of Babesia microti parasites in erythrocytes. Biochem. Biophys. Res. Commun. 517, 125–131 10.1016/j.bbrc.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kraft T. E., Armstrong C., Heitmeier M. R., Odom A. R., and Hruz P. W. (2015) The glucose transporter PfHT1 is an antimalarial target of the HIV protease inhibitor lopinavir. Antimicrob. Agents Chemother. 59, 6203–6209 10.1128/AAC.00899-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kumar S., and Suguna K. (2018) Crystal structure of the retroviral protease-like domain of a protozoal DNA damage-inducible 1 protein. FEBS Open Bio. 8, 1379–1394 10.1002/2211-5463.12491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Letunic I., and Bork P. (2019) Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.