Abstract
We report a sensitive and versatile biosensing approach, LUCID (luminescence compact in-vitro diagnostics), for quantitative molecular and cellular analyses. LUCID uses upconversion nanoparticles (UCNPs) as luminescent reporters in mutually exclusive photo-excitation and read-out sequences implemented on a smartphone. The strategy improves imaging signal-to-noise ratios, eliminating interference from excitation sources and minimizing autofluorescence, and thus enables filter-less imaging. Here we developed a miniaturized detection system and optimized UCNPs for the system and biological applications. Nanoparticle luminescence lifetime was extended by controlling particles’ structure and composition. When tested with a range of biological targets, LUCID achieved high detect sensitivity (0.5 pM for protein and 0.1 pM for nucleic acids), differentiated bacterial samples, and allowed profiling of cells. In proof-of-concept clinical use, LUICD demonstrated effective screening of cancer cells in cervical brushing specimens, identifying patients at high risk for malignancy. These results suggest that LUCID could serve as a broadly applicable and inexpensive diagnostic platform.
Keywords: diagnostic, point-of-care, nanoparticles, luminescence, global health
Infections and cancer are leading causes of death in low and middle income countries (LMICs), with annual death tolls reaching over 7 million (infection, 4.7 million; cancer 2.4 million).1 Access to treatment of preventable and curable diseases has significantly improved in many LMIC settings, spurred by continuing global initiatives.2, 3 Reaping the full benefit of these advances mandates the access to affordable diagnostic tests to enable prompt initiation of treatment or expedited patient triaging. Limited infrastructure and geographical barriers, however, demands stringent performance metric on LMIC diagnostics.4, 5 Assays should yield actionable results preferably at the point-of-care (POC). Moreover, sensing systems should be easy to operate and maintain, and also be compact for use in mobile clinics. We further reason that ideal POC sensors should be platform technologies equipped to deal with diverse targets in the field settings. Different sensor types have been advanced for POC operations, yet typically limited to specific analytic targets. For example, low-cost membrane-based sensors allow for low-resolution, qualitative measurement of soluble molecules (e.g., serum proteins, nucleic acids).6–8 For cellular detection, miniaturized imaging devices have been developed,9–12 although their operation often rely on intensive computation,13 complex optics (e.g., fluorescence, illumination control),11, 14 or external servers.15
We set out to devise an end-user friendly, generalizable strategy for quantitative molecular tests. We specifically explored upconversion nanoparticles (UCNPs) in an imaging-based assay, both for soluble targets, bacterial and cellular detection. UCNPs have a distinct optical feature – they emit luminescent light even after photo-excitation is turned off,16–18 allowing for temporal decoupling between optical excitation and detection. Such imaging would be free from optical crosstalk and detector-saturation caused by high-intensity illumination. This in turn simplifies a system setup by enabling filter-less imaging. Based on this principle, here we report on a compact luminescence platform, termed LUCID (luminescence compact in-vitro diagnostics), for versatile POC molecular testing. We optimized both nanoparticles and their detection system. Specifically, we lengthened UCNPs’ luminescence lifetime by controlling particle composition and built a portable, filter-less imaging device around a smartphone. Applying the first prototype, we demonstrate highly sensitive detection across broad ranges of biological targets. LUCID detected soluble proteins (~0.5 pM) and nucleic acids (~0.1 pM) at high sensitivity, identified different bacterial species, and screened individual cancer cells. As for potential clinical applications, we used LUCID in the context of cervical cancer screening. Specimens obtained during colposcopy were subjected to LUCID for cellular detection, which reliably identified patients at high clinical risk for malignancy.
RESULTS AND DISCUSSION
LUCID strategy and system development
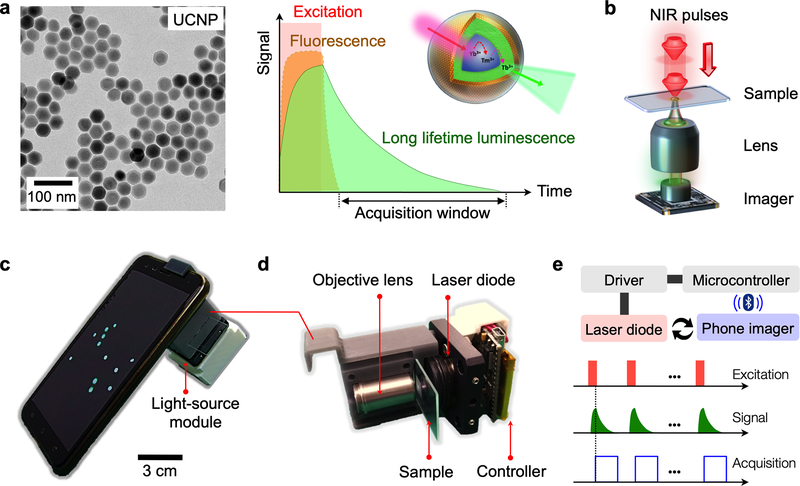
Photo-excited UCNPs emit both fluorescent and luminescent lights; fluorescence decays (<100 nsec) soon after excitation is removed, whereas luminescence typically persists (Fig. 1a). LUCID is designed to detect this long-lived luminescence only, which brings the following advantages: i) imaging becomes free of artifacts associated with fluorescence detection (e.g., no bleed-through of excitation light, autofluorescence, photobleaching); ii) repeated imaging can selectively amplify luminescence signal without increasing the background; and iii) samples are directly imaged without need for optical filter sets (Fig. 1b).
Figure 1. Luminescence upconversion in-vitro diagnostics (LUCID).
(a) The assay used upconversion nanoparticles (UCNPs; left) as reporters. Optical excitation and detection was temporally separated, as the particles can emit luminescent light after pulse excitation (right). (b) The LUICD scheme simplified the detection optical setup. An imager can directly image UCNP-labeled samples without using filter sets. (c) A mobile LUCID prototype was assembled by attaching an excitation module to a smartphone camera. (d) The excitation module consisted of a laser diode (emission, 980 nm), its control electronics, and beam shaping optics. (e) A microcontroller provided the triggering cues for particle excitation and image acquisition, ensuring that only luminescence signal was recorded. Multiple frames could be averaged to improve overall image quality. For the prototype shown in (c), the controller communicated with the phone via bluetooth to access its camera function.
These merits enabled us to devise a compact, mobile LUCID system (Fig. 1c and Fig. S1). Figure 1c shows a prototype built with a smartphone. We used a phone camera as a detector and interfaced it with an illumination source (Fig. 1d). A snap-on module, housing a laser diode, optical lenses, and a microcontroller, was assembled and mounted over the phone camera. The microcontroller was programmed to synchronize operations between the laser diode and the phone camera, ensuring image acquisitions when the diode was turned off (Fig. 1e). Specifically, the detection cycle started with NIR illumination (~5 msec). Excitation light was then turned off; the detector acquired luminescence signal after a short time-delay (1 msec). The integration time was set to ~3 × the luminescence lifetime (τ), capturing >90% of the luminescence light.
Development of nano-reagents
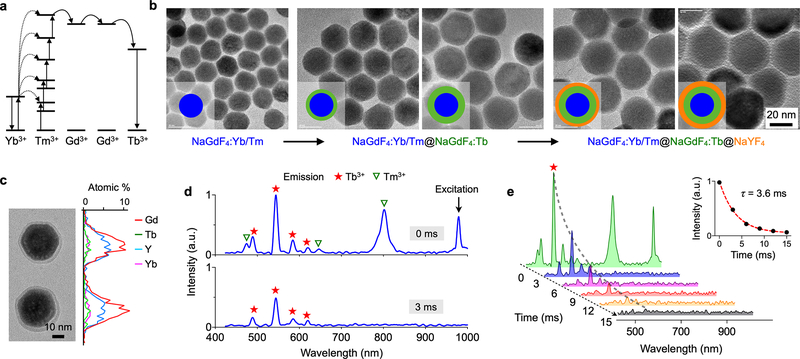
We next optimized UCNPs for LUCID applications, focusing on extending luminescence lifetime. Conventional UCNPs tend to have short luminescence lifetimes (~microseconds).19, 20 Further extending luminescence lifetime is desirable; this can relax requirements for high-speed electronics and reduce electronic noise through longer signal integration. With a typical semiconductor imager, we estimated that the required luminescence lifetime should be in the range of milliseconds (see Supplement Notes for details). As a luminescence source, we selected Tb3+ which is known to have long luminescence lifetime (several milliseconds).16, 21 The material, however, required deep UV (254 nm) excitation, which would increase device costs (i.e., UV light source) and raise safety concerns. To provide UV-level excitation from NIR, we embedded Tb3+ into a UCNP substrate. Specifically, we used a Yb3+/Tm3+ co-doped NaGdF4 as a core material; this particle can absorb 980 nm NIR light and emit deep UV photons through the upconversion process (Fig. 2a). A Tb3+ doped shell was then grown around the core as an emitting layer.
Figure 2. UCNPs optimized for LUCID.
(a) Energy diagram. Yb3+ functions as a sensitizer to absorb NIR photons. The energy is accumulated in Tm3+ and then transferred to Tb3+ through Gd3+. The particle finally emits photons with Tb3+ as an activator. (b) Synthetic route. We first synthesized a core particle (NaGdF4:Yb/Tm) and then grew a shell of emission layer (NaGdF4:Tb). The particle was further passivated with a protection layer (NaYF4). (c) Energy dispersive X-ray spectroscopy (EDS) line scan confirmed the incorporation of key elements (i.e., Gd, Tb, Y, Yb). (d) The emission spectra of the prepared UCNPs were measured. With the excitation light on, the particle showed emission peaks both from Tm3+ and Tb3+ (top). With the excitation off, the short-lived Tm3+ luminescence disappeared, but the signal from Tb3+ persisted (bottom). (e) Time-lapse detection revealed persistent luminescence from Tb3+. The intensity was fitted to a single-phase exponential decay function (inset). The decay time was 3.6 ± 0.4 msec.
Figure 2b shows the synthetic route. We first formed NaGdF4:Yb/Tm core particles (diameter, 20 nm) via a thermal decomposition method (see Methods and Experimental Section).22, 23 Next, we encased the core with a shell of NaGdF4:Tb. Through a dual-growth step, we thickened the shell (thickness, 5.8 nm) to enhance overall emission intensity. We finally passivated the particle with an inert NaYF4 shell to protect Tb3+ from vibrational quenching by solvents. The particle had the overall diameter of 37 nm, and showed high crystallinity (Fig. S2a) from epitaxial growth. Elemental mapping confirmed the incorporation of Tb3+ into the UCNP matrix (Fig. 2c and Fig. S2). Synthesized UCNPs were made water-soluble through ligand exchange.24, 25 We incubated oleic-acid capped UCNPs with excess amounts of poly(acrylic acid) (PAA). The carboxylic group on PAA was used for bioconjugation (Methods and Experimental Section).
We analyzed the optical properties of the prepared UCNPs. Aqueous suspensions of UCNPs were pulse-excited at 980 nm and resulting emission spectra were measured. During excitation, we observed emission peaks from both Tm3+ (core) and Tb3+ (shell). The core-emission (Tm3+) had two major emission peaks from internal transitions (475 nm and 800 nm). These peaks decayed fast (< 1 msec); the reported lifetimes were ~100 μsec (475 nm) and ~340 μsec (800 nm).26. In contrast, Tb3+ emission peaks persisted much longer after excitation was turned off (Fig. 2d). We determined Tb3+ luminescence lifetime via time-lapse measurements (Fig. 2e). The emission at 546 nm, which is characteristic of Tb3+, showed an exponential decay (Fig. 2e, inset) with the lifetime τ = 3.6 ± 0.4 msec. This value was about 8-fold higher than luminescence lifetime of conventional UCNPs (NaYF4:Yb/Tm; Fig. S3).
Platform characterization
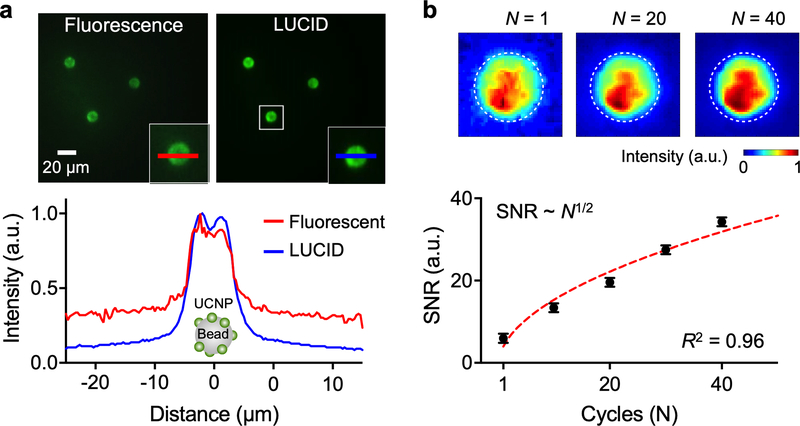
We first tested LUCID using UCNP-coated microbeads (diameter, 10 μm). We compared bead images obtained through conventional fluorescence detection and LUCID (Fig. 3a). Fluorescence images had lower signal-to-noise ratio (SNR = 2.6), likely due to the bleed-through from incident light. In contrast, LUCID was nearly background free, achieving higher SNR (= 7.3) even from single image acquisition (15 msec integration time). We further improved LUCID SNR through repeated time-gated imaging. For a given repetition number of N, the signal level would be linearly proportional to N, whereas system noise (mainly from dark currents of an imager) increases as N1/2. Overall SNR is thus expected to scale as N1/2. Repeated measurements on UCNP-coated microbeads confirmed such reasoning (Fig. 3b) with overall SNR increasing (~N1/2) with repeated excitation and integration. Note that this type of measurement was possible due to UCNP’s high photostability (i.e., no photobleaching).
Figure 3. Characterization of LUCID signal.
(a) Polystyrene microbeads (diameter 10 μm) were embedded with UCNPs and imaged in a conventional fluorescent and a LUCID detection modes (top). LUCID had lower background than fluorescent imaging, free from autofluorescence and scattering of incident light. The graph (bottom) shows intensity line scans of the same bead under the two detection modes. The signal level was normalized to its maximum. (b) The overall signal-to-noise ratio (SNR) of LUCID improved by repeating the luminescence imaging sequence (Fig. 1e). The SNR scaled as N1/2, where N is the cycle number. Each data point is from 5 beads, and data is displayed as mean ± s.d. The inset (top) shows pseudo-colored images of an UCNP-embedded microbead (dotted circle).
LUCID molecular assay formats
We next adopted LUCID to detect different types of biological targets. We developed assays for three different types of targets: i) soluble protein, ii) bacterial RNA and iii) whole cells. For small molecular targets (e.g., soluble protein, nucleic acid), we used microbeads (diameter, 10 μm) as a solid substrate to capture molecules and subsequently label them with UCNPs (Fig. S4a). This approach effectively enhances detectable luminescence signal by locally concentrating UCNPs. The assay also benefits from 3-dimensional diffusion of beads and target analytes, which shortens the traveling distance of targets before they are captured by affinity ligands. For cellular detection, we used cells as a substrate and directly labeled them with antibody-coated UCNPs.
We acquired multiple time-gated frames (N = 20) to produce a final image. For each frame, the laser diode was on for 5 msec; after 1 msec delay, signal was acquired for 10 msec. The total light illumination was short (~0.1 sec) with no apparent thermal effect on samples. After the image acquisition, computing routines automatically located UCNP-labeled objects, either beads or cells, according to their light intensity, and calculated total intensity per object. As an analytical measure, we used the mean intensity value from ≥25 beads or 50 cells (see Methods and Experimental Section). A low magnification lens (20×) was sufficient for imaging while extending the system’s field-of-view (350 × 350 μm2).
For small molecule detection, we used thrombin and bacterial DNA as model targets. We conjugated microbeads with capture ligands: aptamers for thrombin (Table S1) and oligonucleotides complementary to bacterial DNA. UCNPs were also modified with target-specific affinity ligands (see Methods and Experimental Section). To assess assay statistics, we generated calibration curves (signal vs. analyte concentration) using standard samples of known analyte concentrations (Figs. S4b and S4c). Negative controls were prepared by incubating beads with UCNPs but without targets. LUCID achieved high sensitivity with a limit-of-detection (LOD) of 0.5 pM for thrombin and 0.1 pM for DNA. The dynamic range spanned about three orders of magnitude.
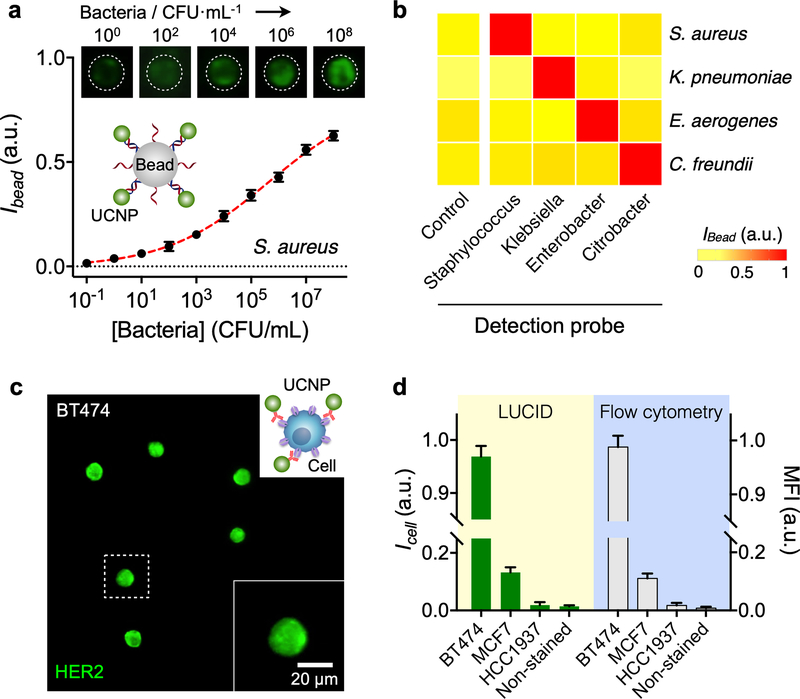
We extended the assay to pathogen analyses. A set of oligonucleotide pairs were prepared to detect different bacterial species. The oligonucleotides were complementary to the hypervariable region of 16S rRNA across different bacterial species (Table S2).27 RNA was extracted from bacteria samples and PCR-amplified to produce single-stranded DNA. Figure 4a shows a representative example, the detection of S. aureus. Titration experiments showed that the sensitivity was close to single colony forming units (CFUs). We next applied the assay to identify different bacterial species (Fig. 4b). LUCID showed high detection selectivity, superior to conventional SBYR green-based PCR (Fig. S5). This could be attributed to the sandwich hybridization employed in the UCNP assay; both capture and detection probes should bind to their target to emit luminescence signal.
Figure 4. Versatile LUCID assays.
(a) Bacteria detection. DNA targets from S. aureus were detected by LUCID. The detection limit reached down to single colony forming units (CFUs). Data are displayed as mean ± s.d. from triplicate measurements. The signal level was normalized to the maximum pixel intensity of the camera. (b) We further designed probe sets for four different bacteria genera (Staphylococcus, Klebsiella, Enterobacter, Citrobacter). LUCID displayed excellent signal contrast and achieved high specificity. For each sample, intensities from 25 beads were averaged. (c) Cell profiling. A panel of breast cancer cells (BT474, MCF7, HCC1937) were labeled with UCNPs conjugated with anti-HER2 antibodies, and imaged by LUCID. An image of BT474 cells is shown here; images of other cell lines are in Fig. S6. (d) LUCID results enabled quantitive cell profiling. The intensity (ICell) of UCNP-labeled cells was reflective of HER2 expression and correlated (R2 = 0.99) with the mean fluorescent intensity (MFI) from flow cytometry. Intensities from 50 (LUCID) and 104 (flow cytometry) cells were averaged, and the data are displayed as mean ± s.d.
We finally demonstrated cell profiling with LUCID. Human breast cancer cell lines (BT474, MCF7, HCC1937) were labeled with immuno-UCNPs recognizing the human epidermal growth factor receptor 2 (HER2)/neu. Differential luminescence signal was observed per cell line (Fig. 4c and Fig. S6), reflective of varying HER2 levels. The average luminescence signal (from 50 cells) correlated linearly with mean fluorescence intensity (MFI) by flow cytometry (R2 = 0.99; Fig. 4d).
LUCID application with clinical specimens
To explore LUCID’s potential for clinical application, we identified effective yet feasible cervical cancer screening as an unmet need in point-of-care settings. Cervical cancer incurs high incidence and mortality rates in many resource-limited countries28 which often lack robust early screening infrastructures. We reasoned that the portable LUCID system could effectively triage suspicious or high-risk cases. This decentralized strategy could potentially offset pathology bottlenecks and reduce repeated patient visits to central clinics, often complicated by geographical and/or socioeconomic constraint.29
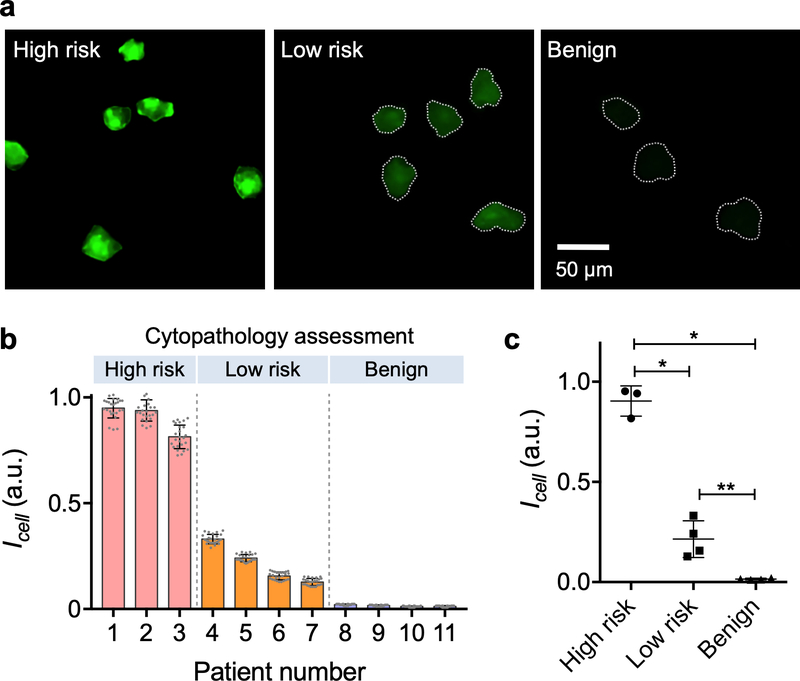
We obtained excess cervical specimens through brushings or biopsies during colposcopic evaluation. Eleven patients with previously abnormal Pap smear results were recruited. We labeled cellular samples with a cocktail of UCNPs specific to EpCAM, CD44 or Trop2; these markers were chosen for their elevated expression in cervical cancer as described in literature and Human Protein Atlas.30–32 LUCID resolved individual cells (Fig. 5a) and was able to quantify their luminescence intensities. Patient specimens were also independently assessed via conventional cytopathology, which classified them into three groups: ‘high risk,’ ‘low risk’ or ‘benign’ (Fig. 5b, top row). On average, we observed higher luminescence signal with increased clinical risk (Fig. 5b). Notably, the mean intensity per cell (Icell) could serve as a classification metric (Fig. 5c), significantly different among risk groups (Sidak’s multiple comparison test).
Figure 5. LUCID screening of cervical cancer.
(a) Human cervical brushing samples were labeled with a cocktail of UCNPs specific to EpCAM, CD44, and Trop-2. Representative LUCID images from high-risk (cervical intraepithelial neoplasia/CIN 2, 3), low-risk (CIN 1) and benign patient samples are shown. (b) LUCID profiling results from 11 patient samples are displayed in a waterfall plot. For each sample, intensities from 25 labeled cells were measured; risk levels were independently assessed by cytopathology. Increased risk levels were found associated with higher LUCID signals. (c) The average cellular intensity was significantly different among high risk, low risk and benign groups (Sidak’s multiple comparisons test): *p < 0.0001; **p = 0.0094.
CONCLUSIONS
Deploying low-cost mobile diagnostics can mitigate healthcare disparities in LMIC settings where pathology bottlenecks, limited resources, and logistic barriers delay disease diagnoses and often lead to over/under-treatment.33 Such tests would also benefit underserved populations in developed countries; in the US, for example, a quarter of the population live in rural areas, but only 10% of physicians practice there.34 The developed LUCID platform can help tackle diagnostic challenges in such regions: i) sensing is robust against autofluorescence and optical scattering; ii) filter-free, simple optics renders the assay system cost-effective and easy to maintain, and iii) the platform can perform onsite detection of diverse molecular targets. The underlying technical innovation sought to engineer nanoparticles with long-lived luminescence and integrate them into compact, time-gated imaging devices. Our pilot studies confirmed that LUCID’s analytical results are comparable or superior to costlier laboratory tests demanding greater skill sets (i.e., qPCR, flow cytometry, cytopathology).
We used upconversion nanoparticles (UCNPs) that have emerged as a new class of nanomaterials for bio-sensing.35, 36 Doped with lanthanide ions, UCNPs absorb multiple near-infrared (NIR) photons, and emit visible or ultraviolet (UV) light. The parity forbidden nature of 4f-4f transitions of lanthanide ions delays the spontaneous photon emission, opening a temporal window to excitation-free imaging. Luminescence is also highly stable without photobleaching or blinking.37, 38 These favorable optical properties have motivated diverse applications of UCNPs for in vivo imaging.17, 20, 39–42 One drawback of UCNPs is their low quantum yields (QYs; generally <1%).43 We expect our Tb-doped UCNPs have a similar QY, although the exact value was yet to be determined. UCNP QYs increase with higher excitation power,44 but at the risk of photothermal damage on samples. In LUCID, we compensated for UCNPs’ low QY through repeated time-gated imaging (Fig. 1e), exploiting particles’ high photostability. Indeed, signal quality progressively improved with acquisition numbers (Fig. 3b). Furthermore, by syncing the light source to the camera, the total acquisition time was <2 sec.
The current system can be further refined, particularly through particle engineering. First, we envision synthesizing a panel of UCNPs for multiplexed molecular detection. The luminescence color can be tuned by varying the composition of lattice atoms and dopant ions.16, 45–47 UCNPs also have a narrower width of emission peak (10–20 nm) than quantum dots or organic dyes.48, 49 Combined, these features would allow LUCID to resolve different particles types through simultaneous multicolor detection. Second, we could render UCNPs excitable at longer wavelengths (>1100 nm) wherein the quantum efficiency of semiconductor imagers becomes negligible (due to indirect bandgap of silicon). For example, the excitation wavelength of 1530 nm has been demonstrated when UCNPs were doped with Er and Tm.50 Under this condition, LUCID could directly detect particles even with continuous illumination, which will make the system even simpler and easier to use. Third, one could explore the use of phosphorescent particles that exhibit long-lasting (minutes to hours) afterglow. The same time-gated imaging can detect these particles but with longer integration time than that of UCNPs; this would help further enhancing SNR. One technical challenge is establishing new fabrication methods, different from physical grinding of crystals,51 to synthesize mono-disperse, bioconjugatable phosphorescent particles.52 These advances in nanoengineering will propel LUICD as an affordable and versatile diagnostic tool to improve and decentralize health care delivery in resource-limited settings.
METHODS AND EXPERIMENTAL SECTION
LUCID system setup
The excitation light source consisted of a 980 nm laser diode (LDM-0980–002w-35, Roithner LaserTechnik), an aspheric lens (focal length ƒ = 11 mm, numerical aperture NA = 0.25; C22MB, Thorlabs), and an objective lens (20×, Olympus). Two types of signal detector, a CMOS imager (2592 × 1944 pixels; DFM 72BUC02-ML; The Image Source) or a smartphone (Galaxy S5, Samsung) with a CMOS imager (1334 × 750 pixels), were used. A microcontroller unit (Arduino MEGA 2560) was programmed to control the light source and the camera. The laser diode was turned on for 5 msec to excite UCNPs, and turned off. Following 1 msec delay, the camera was on for 10 msec to acquire a luminescence image, and the data were transferred to a computer (~70 msec). The accusation time for a single cycle was thus <90 msec. We repeated the cycle for 20 times. The total illumination time per image was thus ~0.1 sec, which led to the incident light energy ~20 mJ per sample. Considering the typical sample geometry (350 × 350 × 10 μm3) and the absorption coefficient of water (α = 5 m−1 at λ = 980 nm), the energy absorbed by water would be on the order of 10−6 J ( = 10 μm × 5 m−1× 20 mJ), and the resulting increase in the sample temperature is on the order of 0.1 °C. The light-source was powered by battery (9V D-cell) or through USB connection.
Synthesis of NaGdF4:Yb/Tm core UCNPs
We synthesized UCNPs according to the previously reported methods with minor modifications.23 0.5 mmol of gadolinium(III) acetate hydrate (99.9%, Aldrich), 0.49 mmol of ytterbium(III) acetate tetrahydrate (99.9%, Aldrich), and 0.01 mmol of thulium(III) acetate hydrate (99.9%, Aldrich) were mixed with 10 mL of oleic acid (technical grade, 90%, Aldrich) and 15 mL of 1-octadecene (technical grade, 90%, Aldrich) in a 100 mL three-neck round bottom flask. The reaction mixture was heated at 150 °C with stirring for 40 min to remove residual water and oxygen, and then cooled to room temperature. 10 mL of methanol solution containing 2.5 mmol of NaOH (98%, Aldrich) and 3.3 mmol of NH4F (98%, Aldrich) was added to the reaction vessel. The reaction mixture was stirred for 30 min at 50 °C, and then heated to 100 °C under vacuum with stirring for 30 min to remove methanol. The temperature was then elevated to 280 °C at the ramping rate of 10 °C/min, and then kept constant for 90 min under Ar. The resulting solution was then cooled to room temperature. The UCNPs were precipitated by adding ethanol and retrieved by centrifugation. The purification procedure was repeated two more times to remove excess surfactant and solvent. The purified UCNPs were dispersible in organic solvents such as hexane and chloroform.
Synthesis of NaGdF4:Yb/Tm@NaGdF4:Tb core-shell UCNPs
0.85 mmol of gadolinium(III) acetate hydrate and 0.15 mmol of terbium(III) acetate hydrate (99.9%, Aldrich) were mixed with 10 mL of oleic acid and 15 mL of 1-octadecene in a 100 mL three-neck round bottom flask. The reaction mixture was heated at 150 °C with stirring for 40 min to remove residual water and oxygen, and then cooled to room temperature. A solution of core UCNPs in hexane was injected to the reaction vessel. 10 mL of methanol solution containing 2.5 mmol of NaOH and 3.3 mmol of NH4F was added into the reaction vessel. The reaction mixture was stirred for 30 min at 50 °C, and then heated to 100 °C under vacuum with stirring for 30 min to remove methanol. Then the reaction mixture was heated to 280 °C at a ramping rate of 10 °C/min, and then kept at that temperature for 90 min under Ar. The resulting solution was then cooled to room temperature. The prepared UCNPs were collected and washed as described above. The shell coating was repeated one more time to form thick Tb-doped shell.
Synthesis of NaGdF4:Yb/Tm@NaGdF4:Tb@NaYF4 core-multi shell UCNPs
1 mmol of yttrium(III) acetate hydrate (99.9%, Aldrich) was mixed with 7.5 mL of oleic acid and 17.5 mL of 1-octadecene in a 100 mL three-neck round bottom flask. We applied the same reaction condition as for Tb-doped shell coating, but used NaGdF4:Yb/Tm@NaGdF4:Tb UCNPs as seed particles.
Characterization of UCNPs
The shape, structure, and composition of synthesized nanoparticles were analyzed using a transmission electron microscope (TEM; JEOL 2100, JOEL USA). High resolution TEM (HRTEM) images and energy dispersive X-ray spectroscopy (EDS) were obtained using a JEOL 2100F equipped with an EDS detector (X-MAX 80T, Oxford). Optical properties of particles were measured by a fluorescent plate reader (TECAN Safire 2).
Ligand exchange with poly(acrylic acid)
The surface modification of UCNPs was performed using poly(acrylic acid) (PAA, Mw 1800, Aldrich).25 2 mL of UCNPs in chloroform (15 mg/mL) was slowly added to the 2 mL of PAA in ethanol (150 mg/mL) with overnight stirring. The solution was centrifuged at 20,000 ×g for 10 min. After washing with ethanol and deionized water for several times, the resulting UCNPs were redispersed in deionized water under sonication.
UCNP-embedded beads
22 μL of polystyrene beads (Polybead® Carboxylate Microspheres, 10 μm, Polysciences) was added into a mixture of butanol (9.5 mL) and chloroform (0.5 mL). After 30 min incubation with stirring, 20 μL of UCNPs in THF (10 mg/mL) was added to the bead solution. After 60 min incubation, the beads were washed using ethanol several times (3,000 ×g for 5 min), and dispersed in deionized water.
Capture probe conjugation on the beads
10 mg of streptavidin coated polystyrene beads (SuperAvidin™ Coated Microspheres, 9.94 μm, Bangs Laboratories) were washed 3 times (3,000 ×g for 3 min) using Tris buffer (20 mM pH 7.5 Tris, 1 mM EDTA, 1 M NaCl, 0.0005% Triton X-100) and dispersed in 1 mL of Tris buffer. The beads were incubated with 40 μL of biotinylated capture probes (40 nmol, Integrated DNA Technologies) for 60 min at room temperature. After incubation, the beads were washed 4 times using Tris buffer and dispersed in 1 mL of Tris buffer. The amount of oligonucleotide probes per bead was quantified using Qubit ssDNA assay kit and Qubit 2.0 fluorometer (Thermo Fisher Scientific). The average number of capture ligand was ~106 per bead.
Conjugation of oligonucleotide probes on UCNPs
50 μL of detection probes (50 nmol, Integrated DNA Technologies) and 0.1 mL of EDC (1 mg, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride, Thermo Fisher Scientific) were added to 0.5 mL of PAA coated UCNPs (~30 mg), and they were incubated for 60 min at room temperature. 0.1 mL of EDC (1 mg) was added to the reaction mixture, and incubated overnight. After incubation, the nanoparticles were centrifuged at 20,000 ×g for 10 min. The precipitated UCNPs were re-dispersed in Tris-Tween 20 (125 mL of 1 M pH 8 Tris, 0.1 g of Tween 20, 375 mL of deionized water) under sonication. The purification process was repeated 4 times, and UCNPs were dispersed in pH 8 Tris-HCl (20 mM) under sonication. The amount of oligonucleotide probes per nanoparticle (10~20 per particle) was quantified using Qubit ssDNA assay kit and Qubit 2.0 fluorometer (Thermo Fisher Scientific).
Conjugation of antibodies on UCNPs
The following antibodies were purchased: anti-HER2 (clone 24D2, Biolegend); anti-EpCAM (clone MAB9601, R&D Systems); anti-CD44 (clone IM7, Biolegend); Trop 2 (clone 162–46.2, Abcam). 20 μL of antibody (1 mg/mL) and 0.1 mL of EDC (2 mg/mL) were added to 0.5 mL of PAA coated UCNPs (~20 mg), and they were incubated for 2 h at room temperature. After incubation, the nanoparticles were centrifuged at 20,000 ×g for 10 min. The precipitated UCNPs were redispersed in 1 mL of 1× PBS.
Thrombin detection
Capture beads were dispersed in Tris-HCl buffer (50 mM pH 7.4 Tris-HCl, 1 mM MgCl2, 5 mM KCl, 0.1% BSA). 40 μL of capture beads were incubated with 10 μL of thrombin solution (Human alpha-Thrombin, Haematologic Technologies) for 30 min at room temperature. After incubation, the beads were purified 4 times using 5 μm centrifugal filter, and they were dispersed in 40 μL of Tris-HCl buffer. The 40 μL of beads were incubated with 60 μL of detection UCNPs for 30 min at room temperature. Excess UCNPs were removed via centrifugation (1 min, 60 ×g) using 5 μm centrifugal filter. Collected beads were dispersed in 50 μL of Tris-HCl buffer.
DNA detection
40 μL of capture beads were diluted with 50 μL of Tris buffer. The beads were incubated with 10 μL of synthetic DNA specific for S. aureus (Integrated DNA Technologies) for 30 min at room temperature. After incubation, the beads were purified 4 times using 0.45 μm centrifugal filter (94 ×g for 3 min), and were dispersed in 40 μL of Tris buffer. The beads were incubated with 60 μL of detection UCNPs for 30 min at room temperature. Excess UCNPs were removed via centrifugation (1 min, 60 ×g) using 5 μm centrifugal filter. Beads were then dispersed in 100 μL of Tris-buffered saline (25 mM Tris, 150 mM NaCl).
Bacterial detection
Four different bacteria species were purchased from the American Type Culture Collection and grown to mid-log phase in suitable media: Staphylococcus aureus (#25923) in Staphylococcus broth (BD Biosciences); Klebsiella pneumoniae (#43816) in trypticase soy broth; Enterobacter aerogenes (#13048), Citrobacter freundii (#6879) in nutrient broth (BD Biosciences). For RNA extraction, bacteria were centrifuged at 6,000 ×g for 10 min and pellets were treated with the pre-heated Trizol (Life Technologies). The resuspended cells were transferred to 2 mL Safe-Lock tubes (Eppendorf) containing sterilized disruptor beads (0.1 mm, Scientific Industries), and lysed using a vortex mixer. After centrifugation, the supernatant was transferred to a new tube and mixed with an equal volume of ethanol. The mixture was processed by Direct-zol RNA kits (Zymo Research), following manufacturer’s protocol. With the extracted RNA samples, the representative single-stranded cDNA was synthesized using random priming with Promega’s Reverse Transcription System per manufacturer’s protocol. The asymmetric PCR was then carried out in a total volume of 25 μL containing 2.5 μL of cDNA, 0.8 μM of excess primer and 0.08 μM of limiting primer (Supplementary Table 2), 1× PCR reaction buffer (20 mM Tris-HCl, 20 mM KCl, 5 mM (NH4)2SO4, and 2 mM MgCl2), 0.2 mM of each dNTP, and 2 U Maxima Hot Start Taq DNA polymerase (Thermo Scientific). The reaction tubes were heated to 94 °C for 2 min, followed by 35 or 40 cycles of 5 s at 94 °C, 15 s at 56 °C, and 15 s at 72 °C, and then a final 10 min extension step at 72 °C.
SYBR green-based quantitative PCR (qPCR)
The qPCR was carried out on 7500 Fast Real-time PCR system (Life Technologies) in a total reaction volume of 20 μL containing 2.5 μL of cDNA derived from in-vitro cultured bacteria (~106 CFU/mL), 0.5 μM of each primer (Supplementary Table 2), 1× SYBR Select Master mix (Life Technologies). Thermal cycling was then carried out with the following conditions: Initiation (95 ºC, 2 min); 40 cycles of denaturation (95 ºC, 5 sec); annealing (56 ºC, 15 sec); extension (72 ºC, 30 sec). The threshold cycle (Ct) values were obtained using the 7500 Fast software. No-template control (NTC) remained undetected, not crossing the established threshold for 40 cycles, which was arbitrarily given a Ct value of 41. The ΔCt was calculated by subtracting the Ct value of the specimen from the Ct value of NTC.
Cell preparation for LUCID
A panel of human breast cancer cell lines were purchased (ATCC) and grown in the vendor-recommended media: BT474 (Hybri-Care Medium 46-X with 0.15% NaHCO3, ATCC); MCF7 (Eagle’s Minimum Essential Medium 30–2003 with 0.1% human recombinant insulin, ATCC); HCC1937 (RPMI-1640, Corning). All media were supplemented with 10% heat inactivated fetal bovine serum (Gibco) and 1% penicillin-streptomycin (5,000 U/mL, Gibco). Cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. At about 80% confluence, cells were washed, trypsinized, and resuspended to a concentration of ~106 cells/mL in PBS containing 1% BSA. Cells were incubated with 200 μg/mL HER2 conjugated UCNP for 20 min at room temperature, washed via centrifugation (350 ×g, 5 min), and resuspended in PBS. About 10 μL of cell solution (~105 cells/mL) were placed on a plastic slide for LUCID imaging.
Flow cytometry
About 105 cells were used for flow cytometry. Cells were suspended in 1% BSA PBS and incubated with 1 μg/mL anti-human HER2 (BioLegend, 324401) antibody for 30 min at room temperature. Cells were then triple washed with 1% BSA PBS, incubated with 1 μg/mL FITC anti-mouse IgG antibody (BioLegend, 406001), and triple washed for flow cytometry measurements. Control samples were prepared and labeled similarly using 1 μg/mL mouse IgG1, κ isotype antibody (BioLegend, 401401) and 1 μg/mL FITC anti-mouse IgG antibody. Fluorescent signal was measured by CytoFLEX (Beckman Coulter). The mean fluorescence intensity of 10,000 cells was determined using FlowJo software.
Clinical samples
The clinical study was approved by the Partners Healthcare Institutional Review Board. We obtained informed consents from adult women referred to the MGH Colposcopy Clinic for cervical biopsy or loop electrosurgical excision procedure (LEEP). One clinical provider (M.A.) performed all cervical procedures and provided excess or otherwise discarded ectocervical or endocervical brushing or biopsy specimens. Study specimens were collected conventionally before loop excision to avoid potential tissue alteration. Cells were rinsed in sterile PBS and concentrated via centrifugation (300 × g, 5 min). Pelleted cells were treated with collagenase 1 and 2 (5 – 10 mg/mL; Sigma Aldrich) at 37 °C, gently agitated and dispersed. Treated cells were washed in PBS and targeted with a cocktail of three UCNP probes, each specific to EpCAM, CD44, and Trop-2. After 20 min incubation at room temperature, samples were washed via centrifugation (350 ×g, 5 min). Cell pellets were resuspended in PBS (100 μL). We used about 10 μL of samples for LUCID. All experiments were conducted blind to the clinical interpretation of risk.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in parts by U.S. NIH Grants R01CA229777 (H.L.), U01CA233360 (H.L., C.M.C), MGH Scholar Fund (H.L.), DoD-W81XWH1910199 (H.L.), DoD-W81XWH1910194 (H.L.), and MGH Fund for Medical Discovery Fellowship (H.-Y.L.); Taiwan MOST Grants 107-2112-M-008-001 (C.-H.H.) and NSC104-2917-I-564-086 (H.-Y.L.); NRF Grant 2017R1C1B1010703 (Y.I.P.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.
REFERENCES
- (1).Global Health Estimates 2016: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2016.; World Health Organization: Geneva, 2018. [Google Scholar]
- (2).Robertson J; Barr R; Shulman LN; Forte GB; Magrini N Essential Medicines for Cancer: Who Recommendations and National Priorities. Bull World Health Organ 2016, 94, 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Klein EY; Van Boeckel TP; Martinez EM; Pant S; Gandra S; Levin SA; Goossens H; Laxminarayan R Global Increase and Geographic Convergence in Antibiotic Consumption Between 2000 and 2015. Proc Natl Acad Sci U S A 2018, 115, E3463–E3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Drain PK; Hyle EP; Noubary F; Freedberg KA; Wilson D; Bishai WR; Rodriguez W; Bassett IV Diagnostic Point-of-Care Tests in Resource-Limited Settings. Lancet Infect Dis 2014, 14, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Urdea M; Penny LA; Olmsted SS; Giovanni MY; Kaspar P; Shepherd A; Wilson P; Dahl CA; Buchsbaum S; Moeller G; Hay Burgess DC Requirements for High Impact Diagnostics in the Developing World. Nature 2006, 444 Suppl 1, 73–79. [DOI] [PubMed] [Google Scholar]
- (6).Li J; Macdonald J Multiplexed Lateral Flow Biosensors: Technological Advances for Radically Improving Point-of-Care Diagnoses. Biosens Bioelectron 2016, 83, 177–192. [DOI] [PubMed] [Google Scholar]
- (7).Pollock NR; Rolland JP; Kumar S; Beattie PD; Jain S; Noubary F; Wong VL; Pohlmann RA; Ryan US; Whitesides GM A Paper-Based Multiplexed Transaminase Test for Low-Cost, Point-of-Care Liver Function Testing. Sci Transl Med 2012, 4, 152ra129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Singh AT; Lantigua D; Meka A; Taing S; Pandher M; Camci-Unal G Paper-Based Sensors: Emerging Themes and Applications. Sensors (Basel) 2018, 18, 2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).D’Ambrosio MV; Bakalar M; Bennuru S; Reber C; Skandarajah A; Nilsson L; Switz N; Kamgno J; Pion S; Boussinesq M; Nutman TB; Fletcher DA Point-of-Care Quantification of Blood-Borne Filarial Parasites With a Mobile Phone Microscope. Sci Transl Med 2015, 7, 286re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Breslauer DN; Maamari RN; Switz NA; Lam WA; Fletcher DA Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS One 2009, 4, e6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhu H; Mavandadi S; Coskun AF; Yaglidere O; Ozcan A Optofluidic Fluorescent Imaging Cytometry on a Cell Phone. Anal Chem 2011, 83, 6641–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Im H; Castro CM; Shao H; Liong M; Song J; Pathania D; Fexon L; Min C; Avila-Wallace M; Zurkiya O; Rho J; Magaoay B; Tambouret RH; Pivovarov M; Weissleder R; Lee H Digital Diffraction Analysis Enables Low-Cost Molecular Diagnostics on a Smartphone. Proc Natl Acad Sci U S A 2015, 112, 5613–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Coskun AF; Ozcan A Computational Imaging, Sensing and Diagnostics for Global Health Applications. Curr Opin Biotechnol 2014, 25, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lee SA; Erath J; Zheng G; Ou X; Willems P; Eichinger D; Rodriguez A; Yang C Imaging and Identification of Waterborne Parasites Using a Chip-Scale Microscope. PLoS One 2014, 9, e89712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Boppart SA; Richards-Kortum R Point-of-Care and Point-of-Procedure Optical Imaging Technologies for Primary Care and Global Health. Sci Transl Med 2014, 6, 253rv2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang F; Deng R; Wang J; Wang Q; Han Y; Zhu H; Chen X; Liu X Tuning Upconversion Through Energy Migration in Core-Shell Nanoparticles. Nat Mater 2011, 10, 968–973. [DOI] [PubMed] [Google Scholar]
- (17).Lu Y; Lu J; Zhao J; Cusido J; Raymo FM; Yuan J; Yang S; Leif RC; Huo Y; Piper JA; Paul Robinson J; Goldys EM; Jin D On-the-Fly Decoding Luminescence Lifetimes in the Microsecond Region for Lanthanide-Encoded Suspension Arrays. Nat Commun 2014, 5, 3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wen S; Zhou J; Zheng K; Bednarkiewicz A; Liu X; Jin D Advances in Highly Doped Upconversion Nanoparticles. Nat Commun 2018, 9, 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ding M; Chen D; Ma D; Liu P; Song K; Lu H; Ji Z Tuning the Upconversion Luminescence Lifetimes of Kyb2F7 :Ho3+ Nanocrystals for Optical Multiplexing. ChemPhysChem 2015, 16, 3784–3789. [DOI] [PubMed] [Google Scholar]
- (20).Lu Y; Zhao J; Zhang R; Liu Y; Liu D; Goldys EM; Yang X; Xi P; Sunna A; Lu J Tunable Lifetime Multiplexing Using Luminescent Nanocrystals. Nat Photonics 2014, 8, 32–36. [Google Scholar]
- (21).Zheng W; Zhou S; Chen Z; Hu P; Liu Y; Tu D; Zhu H; Li R; Huang M; Chen X Sub-10 nm Lanthanide-Doped CaF2 Nanoprobes for Time-Resolved Luminescent Biodetection. Angew Chem Int Ed Engl 2013, 52, 6671–6676. [DOI] [PubMed] [Google Scholar]
- (22).Su Q; Han S; Xie X; Zhu H; Chen H; Chen CK; Liu RS; Chen X; Wang F; Liu X The Effect of Surface Coating on Energy Migration-Mediated Upconversion. J Am Chem Soc 2012, 134, 20849–20857. [DOI] [PubMed] [Google Scholar]
- (23).Wang F; Deng R; Liu X Preparation of Core-Shell NaGdF4 Nanoparticles Doped With Luminescent Lanthanide Ions to Be Used as Upconversion-Based Probes. Nat Protoc 2014, 9, 1634–1644. [DOI] [PubMed] [Google Scholar]
- (24).Yi G-S; Chow G-M Water-Soluble NaYF4: Yb, Er (Tm)/NaYF4/polymer Core/Shell/Shell Nanoparticles with Significant Enhancement of Upconversion Fluorescence. Chem Mater 2007, 19, 341–343. [Google Scholar]
- (25).You M; Zhong J; Hong Y; Duan Z; Lin M; Xu F Inkjet Printing of Upconversion Nanoparticles for Anti-Counterfeit Applications. Nanoscale 2015, 7, 4423–4431. [DOI] [PubMed] [Google Scholar]
- (26).Wang Y; Deng R; Xie X; Huang L; Liu X Nonlinear Spectral and Lifetime Management in Upconversion Nanoparticles by Controlling Energy Distribution. Nanoscale 2016, 8, 6666–6673. [DOI] [PubMed] [Google Scholar]
- (27).Chung HJ; Castro CM; Im H; Lee H; Weissleder R A Magneto-DNA Nanoparticle System for Rapid Detection and Phenotyping of Bacteria. Nat Nanotechnol 2013, 8, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schiffman M; Solomon D Clinical Practice. Cervical-Cancer Screening with Human Papillomavirus and Cytologic Cotesting. N Engl J Med 2013, 369, 2324–2331. [DOI] [PubMed] [Google Scholar]
- (29).Adesina A; Chumba D; Nelson AM; Orem J; Roberts DJ; Wabinga H; Wilson M; Rebbeck TR Improvement of Pathology in Sub-Saharan Africa. Lancet Oncol 2013, 14, e152–7. [DOI] [PubMed] [Google Scholar]
- (30).Dall P; Heider KH; Hekele A; von Minckwitz G; Kaufmann M; Ponta H; Herrlich P Surface Protein Expression and Messenger RNA-Splicing Analysis of CD44 in Uterine Cervical Cancer and Normal Cervical Epithelium. Cancer Res 1994, 54, 3337–3341. [PubMed] [Google Scholar]
- (31).Varughese J; Cocco E; Bellone S; Ratner E; Silasi DA; Azodi M; Schwartz PE; Rutherford TJ; Buza N; Pecorelli S; Santin AD Cervical Carcinomas Overexpress Human Trophoblast Cell-Surface Marker (Trop-2) and Are Highly Sensitive to Immunotherapy with HRS7, a Humanized Monoclonal Anti-Trop-2 Antibody. Am J Obstet Gynecol 2011, 205, 567.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Went PT; Lugli A; Meier S; Bundi M; Mirlacher M; Sauter G; Dirnhofer S Frequent EpCAM Protein Expression in Human Carcinomas. Hum Pathol 2004, 35, 122–128. [DOI] [PubMed] [Google Scholar]
- (33).Sayed S; Cherniak W; Lawler M; Tan SY; El Sadr W; Wolf N; Silkensen S; Brand N; Looi LM; Pai SA; Wilson ML; Milner D; Flanigan J; Fleming KA Improving Pathology and Laboratory Medicine in Low-Income and Middle-Income Countries: Roadmap to Solutions. Lancet 2018, 391, 1939–1952. [DOI] [PubMed] [Google Scholar]
- (34).The Distribution of the U.S. Primary Care Workforce. Https://www.ahrq.gov/research/findings/factsheets/primary/pcwork3/index.html. Accessed August 18th, 2019.
- (35).Auzel F Upconversion and Anti-Stokes Processes with F and D Ions in Solids. Chem Rev 2004, 104, 139–173. [DOI] [PubMed] [Google Scholar]
- (36).Chen G; Qiu H; Prasad PN; Chen X Upconversion Nanoparticles: Design, Nanochemistry, and Applications in Theranostics. Chem Rev 2014, 114, 5161–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wu S; Han G; Milliron DJ; Aloni S; Altoe V; Talapin DV; Cohen BE; Schuck PJ Non-Blinking and Photostable Upconverted Luminescence from Single Lanthanide-Doped Nanocrystals. Proc Natl Acad Sci U S A 2009, 106, 10917–10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Park YI; Kim JH; Lee KT; Jeon K-S; Na HB; Yu JH; Kim HM; Lee N; Choi SH; Baik S-I Nonblinking and Nonbleaching Upconverting Nanoparticles as an Optical Imaging Nanoprobe and T1 Magnetic Resonance Imaging Contrast Agent. Adv Mater 2009, 21, 4467. [Google Scholar]
- (39).Sun Y; Zhu X; Peng J; Li F Core-Shell Lanthanide Upconversion Nanophosphors as Four-Modal Probes for Tumor Angiogenesis Imaging. ACS Nano 2013, 7, 11290–11300. [DOI] [PubMed] [Google Scholar]
- (40).Kwon OS; Song HS; Conde J; Kim HI; Artzi N; Kim JH Dual-Color Emissive Upconversion Nanocapsules for Differential Cancer Bioimaging in Vivo. ACS Nano 2016, 10, 1512–1521. [DOI] [PubMed] [Google Scholar]
- (41).Chen S; Weitemier AZ; Zeng X; He L; Wang X; Tao Y; Huang AJY; Hashimotodani Y; Kano M; Iwasaki H Near-Infrared Deep Brain Stimulation Via Upconversion Nanoparticle–Mediated Optogenetics. Science 2018, 359, 679–684. [DOI] [PubMed] [Google Scholar]
- (42).Zhao J; Chu H; Zhao Y; Lu Y; Li L A Nir Light Gated DNA Nanodevice for Spatiotemporally Controlled Imaging of MicroRNA in Cells and Animals. J Am Chem Soc 2019, 141, 7056–7062. [DOI] [PubMed] [Google Scholar]
- (43).Boyer J-C; Van Veggel FCJM Absolute Quantum Yield Measurements of Colloidal NaYF4: Er3+, Yb3+ Upconverting Nanoparticles. Nanoscale 2010, 2, 1417–1419. [DOI] [PubMed] [Google Scholar]
- (44).Wilhelm S Perspectives for Upconverting Nanoparticles. ACS Nano 2017, 11, 10644–10653. [DOI] [PubMed] [Google Scholar]
- (45).Wang F; Liu X Upconversion Multicolor Fine-Tuning: Visible to Near-Infrared Emission From Lanthanide-Doped NaYF4 Nanoparticles. J Am Chem Soc 2008, 130, 5642–5643. [DOI] [PubMed] [Google Scholar]
- (46).Liu Y; Tu D; Zhu H; Li R; Luo W; Chen X A Strategy to Achieve Efficient Dual-Mode Luminescence of Eu3+ in Lanthanides Doped Multifunctional NaGdF4 Nanocrystals. Adv Mater 2010, 22, 3266–3271. [DOI] [PubMed] [Google Scholar]
- (47).Wang F; Han Y; Lim CS; Lu Y; Wang J; Xu J; Chen H; Zhang C; Hong M; Liu X Simultaneous Phase and Size Control of Upconversion Nanocrystals through Lanthanide Doping. Nature 2010, 463, 1061–1065. [DOI] [PubMed] [Google Scholar]
- (48).Bruchez M; Moronne M; Gin P; Weiss S; Alivisatos AP Semiconductor Nanocrystals as Fluorescent Biological Labels. Science 1998, 281, 2013–2016. [DOI] [PubMed] [Google Scholar]
- (49).Wang F; Banerjee D; Liu Y; Chen X; Liu X Upconversion Nanoparticles in Biological Labeling, Imaging, and Therapy. Analyst 2010, 135, 1839–1854. [DOI] [PubMed] [Google Scholar]
- (50).Chen Q; Xie X; Huang B; Liang L; Han S; Yi Z; Wang Y; Li Y; Fan D; Huang L; Liu X Confining Excitation Energy in Er3+ -Sensitized Upconversion Nanocrystals Through Tm3+ -Mediated Transient Energy Trapping. Angew Chem Int Ed Engl 2017, 56, 7605–7609. [DOI] [PubMed] [Google Scholar]
- (51).Zhen X; Tao Y; An Z; Chen P; Xu C; Chen R; Huang W; Pu K Ultralong Phosphorescence of Water-Soluble Organic Nanoparticles for in Vivo Afterglow Imaging. Adv Mater 2017, 29, 1606665. [DOI] [PubMed] [Google Scholar]
- (52).Miao Q; Xie C; Zhen X; Lyu Y; Duan H; Liu X; Jokerst JV; Pu K Molecular Afterglow Imaging with Bright, Biodegradable Polymer Nanoparticles. Nat Biotechnol 2017, 35, 1102–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.