Summary
Synthetic biology has promised and delivered on an impressive array of applications based on genetically modified microorganisms. While novel biotechnology undoubtedly offers benefits, like all new technology, precautions should be considered during implementation to reduce the risk of both known and unknown adverse effects. To achieve containment of transgenic microorganisms, confidence to a near scientific certainty that they cannot transfer their transgenic genes to other organisms, and that they cannot survive to propagate in unintended environments, is a priority. Here we present an in-depth summary of biological containment systems for micro-organisms published to date, including the production of a genetic firewall through genome recoding and physical containment of microbes using auxotrophies, regulation of essential genes and expression of toxic genes. The level of containment required to consider a transgenic organism suitable for deployment is discussed, as well as standards of practice for developing new containment systems.
Introduction
1.1. History of containment
Microorganisms have been used inadvertently in the processes behind food production for thousands of years, with various bacterial and fungal species playing key roles in brewing beer and the fermenting of wine, the baking of bread, and the process of turning milk to yoghurt. However, microbiology as a scientific discipline did not solidify until the 17th century, with the exploration of microorganisms as the possible vector for human and animal diseases. It was with these observations that the motivation to control and contain bacterial growth became apparent. Medical advances attributed to sterile and abiotic techniques played an integral part in the control of pathogens. In the 20th century, the discovery and use of antibiotics lead to a revolution in bacterial containment by the application of small molecules.
Today we use microbes for an astonishing range of functions. In addition to the aforementioned production of ethanol, CO2 and lactic acid for alcoholic drinks, bread and yoghurt respectively, both wild-type and engineered microbial species are used to produce a variety of industrially relevant products. Examples include; enzymes such as amylase (Sundarram & Murthy, 2014; Yoneda, 1980) and proteases (Razzaq et al., 2019); polysaccharides for use in food, cosmetics, and pharmaceutical products such as xanthan gum (Santos et al., 2000) and dextran (Sarwat et al., 2008); nutrients in the form of amino acids, nucleotides, vitamins and organic acids (Adrio & Demain, 2010); and pharmaceuticals themselves including insulin (Baeshen et al., 2014), chemotherapeutics (Łukasiewicz & Fol, 2018) and antibiotics (Clardy et al., 2009).
In addition to their use as biological factories, the advent of synthetic biology has allowed microbes to be manipulated by humans to facilitate desirable processes, such as the bio-remediation of polluted environments and for use as living therapeutics. Microbial bio-remediation generally involves the breakdown of an undesirable substrate, rather than concentrating on the production of a functional product. Applications include sewage treatment (Dhall et al., 2012), removal of pesticides (Paridah et al., 2016) and aromatic compounds (McClure & Venables, 1986) from soil, and clean-up of oil spills (Mapelli et al., 2017). Although microbes have been consciously used as therapeutics for over a century (Coley, 1893), the last decade has seen a plethora of possible applications presented. A non-exhaustive list includes diagnostic tools (Kotula et al., 2014; Riglar et al., 2017) and treatments for the human digestive system (Braat et al., 2006), a treatment for oral mucositis (Caluwaerts et al., 2010), HIV prevention (Lagenaur et al., 2011) and cancer immunotherapy (Zheng et al., 2017a). Various reviews have covered the novel applications that synthetic biology allows for bioremediation (de Lorenzo, 2009; Pieper & Reineke, 2000; Sayler & Ripp, 2000) and for living therapeutics (Riglar & Silver, 2018).
Despite the broad acceptance that there has been no known instance of a transgenic microbe being conferred a fitness advantage in the wild, frequent release of transgenic microbes into real world environments presents the possibility of such an individual manifesting. The responsibility of international governing bodies to understand and mitigate this potential problem has been established and ratified in legal treaties such as the Cartagena Protocol and the Rio Declaration on Environment and Development. Transgenic microbes capable of proliferating outside of a lab would lead to their uncontrolled propagation and persistence in an environment that may affect biological processes through interaction with indigenous populations, therefore reducing biodiversity and disrupting food webs. Increases in the risk of transfer of transgenic DNA through horizontal gene transfer could, in principle, lead to novel pathogens. (NIH FAQs, 2019; Risks SCoEaNIH, 2015).
Although the full risks associated with the spread of transgenic microbes are not known, it is this very uncertainty that makes it important to implement control and regulation over transgenic microbial growth. Proving absolutely that a particular transgenic microbe capable of proliferation in the wild poses no environmental risk upon release is inherently an effort in futility, as biological processes contain too many unknown interactions for a perfectly predictive model to be formed. In cases where such uncertainty is prevalent, the precautionary principle can be applied to help form guidelines for the implementation of such a novel technology (A. Stirling, 2007), with critics agreeing it can have a positive effect in such a situation (Peterson, 2007). For example, if a strain is developed capable of treating a Salmonella infection (Riglar et al., 2017), it is prudent to introduce biological safeguards that would reduce the possibility of such a microbe escaping its intended environment (F. Stirling et al., 2017) even if we cannot fully predict what, if any, negative consequences may arise from such an escape. It is our view that transgenic microbes offer a range of solutions to environmental and health related issues, and in the coming decades they should and probably will be implemented. To facilitate this, exercising rigorous precaution will both increase public trust (A. Stirling et al., 2018) and, most importantly, reduce the possibility of negative consequences.
For these reasons, the field of biocontainment has been of interest for decades. The Asilomar conference (Berg et al., 1975) laid out guidelines for the introduction of auxotrophies to prevent escape of transgenic microbes. With the advent of synthetic biology, it has been judged necessary to repeatedly revisit the topic, with a number of reviews appearing in the intervening years (Chari & Church, 2017; Diwo & Budisa, 2019; Lee et al., 2018; Moe-Behrens et al., 2013; Molin et al., 1993; Schmidt & De Lorenzo, 2012; Torres et al., 2016; Wright et al., 2013). The applications of transgenic microbes covered so far can broadly be divided into two categories; applications where the entire intended purpose can be carried out in a controlled setting, and those where the intended purpose is designed to take place in an uncontrolled environment. Microbes engineered for production predominantly fall into the former category, whereas the majority of the second category are those microbes engineered to facilitate a process. However, both categories face the same requirements, that i) that transgenic material is not spread to other species and ii) transgenic microbes are only viable in their intended environment. Containment systems to date predominantly address one or both of these two issues.
A key means to quantify the effectiveness of a containment system used throughout this review is to measure the ‘survival ratio’ it imparts on a population. The survival ratio is the term used to define the ratio of colony forming units (cfu) in non-permissible conditions to the cfu in permissible conditions. For a survival ratio of 10−4, a population of 10,000 bacteria would yield only 1 survivor on transition to non-permissible conditions. Although not all systems can be quantified in such a manner - such as the essentializer in (F. Stirling et al., 2017) - it is a useful tool for comparing different approaches and when assaying a systems effectiveness. In the United States, the National Institutes of Health has recommended a guideline providing that the “escape of the recombinant or synthetic nucleic acid molecule either via survival of the organisms or via transmission of the recombinant or synthetic nucleic acid molecule to other organisms should be less than 1 in 108 under specified conditions” (NIH FAQs, 2019, Apendix 1–1-B). A survival ratio of 10−8 would currently meet these requirements. For many applications, it is likely that this threshold is too high, which is discussed further in section 1.5.
1.2. Preventing the spread of transgenes
Every transgenic organism constitutes an example of one or more units of genetic material existing in an organism or in an arrangement that does not exist in nature. This enables interactions and transfers of genetic material that have not previously been possible. The various strata of life display both unique and interconnected mechanisms for spreading genetic material, and the intentional transfer of such material by humans is not only possible, but performed on a routine basis. The potential negative consequences of this can often be categorised as low risk, like the expression of the green fluorescent protein (GFP) from the jellyfish Aequorea victoria. The use of GFP is considered harmless because no adverse consequences have been observed from its use in transgenic strains (Lee et al., 2018; Moe-Behrens et al., 2013), and hypothetical problems that have been conceived of should transgenic strains expressing it be released are mild or uncertain. However this by no means indicates that if GFP were spread throughout an environment it did not originate from, that it is impossible for it to have a negative impact. Since it would be essentially impossible to prove the innocuousness of the spread of a transgene in all conceivable scenarios, a more feasible and assured approach is to control its inability to spread in the first place with a reasonable level of confidence.
Early attempts at preventing horizontal transfer of genetic material between microbes relied upon toxin-antitoxin systems expressed on plasmids and genomes (Diaz et al., 1994; Torres et al., 2000; Diaz et al., 2003) or host repression of a plasmid borne toxin (Knudsen & Karlstrom, 1991) to prevent the spread of transgenic plasmids to other microbes. A similar approach has been proposed that uses origins of replication that are split between a plasmid and the host genome to prevent plasmid spreading (Wright et al., 2013). These two techniques were combined (Wright et al., 2015), achieving a survival ratio of <10−3 for each method individually but the study did not report the survival ratio from the combined strain. These systems are summarized in Table 1. It is of note that the stability (see section 1.4) of these systems, ie. the capacity of the system to maintain its function when passaged in permissible conditions, is not reported for any system.
Table 1.
Containment systems designed to prevent the spread of transgenic plasmids.
| Year | Lead Author | Species | Conditional Origin | toxin/antitoxin system | Survival ratio | Stability |
|---|---|---|---|---|---|---|
| 1991 | SM. Knudsen | E. coli | RelF | 10^−5 | not tested | |
| 1994 | E. Diaz | E. coli | colicin E3 | 10^−4 | not tested | |
| 2000 | B.Torres | E. coli | EcoRI | 10^−4 | not tested | |
| 2003 | B.Torres | E. coli | EcoRI, colicin E3 | 10^−8 | not tested | |
| 2014 | O. Wright | E. coli | ColE2 | Kid or ζ | <10−3 | not tested |
To achieve a ‘hard lock’ (where the probability of the transfer and expression of transgenic material can be deemed low enough as to be negligible) it is necessary to address the ‘central dogma’ of biology and unpick some of its core truths. Currently, all natural organisms are based around the same genetic code. The four deoxyribose nucleotides are transcribed into the four ribose nucleotides, which are in turn translated using the ubiquitous triplet codon code into the twenty standard amino acids that make up all proteins. Several studies have sought to alter this dynamic in one way or another. By engineering a tRNA synthetase, it has been shown that a quadruplet codon scheme can be implemented in order to incorporate non-natural amino acids (Chatterjee et al., 2014; Hankore et al., 2019). In addition Escherichia coli has been engineered to survive on an artificial nucleotide (Marlière et al., 2011) or to incorporate non-natural amino acid pairs (Malyshev et al., 2014).
For containment applications, the most effective modification to the fundamental tenets of life upheld in the central dogma is to recode a genome so completely that it no longer requires the tRNA machinery for a specific codon or set of codons. This was first accomplished in bacteria with E. coli by removing all instances of the TAG stop codon and its corresponding tRNA (Lajoie et al., 2013), and again more recently removing TAG along with the serine encoding codons TCG and TCA (Fredens et al., 2019). Alternative methods have been published for the complete recoding of an organism such as an integrase based approach to achieve a 57 codon E. coli genome (Ostrov et al., 2016), a technique termed SIRCAS (Step-wise Integration of Rolling Circle Amplified Segments) used to remove two codons from the genome of Salmonella typhimurium (Lau et al., 2017) and an extensive international effort in Saccharomyces cerevisiae that successfully removed all instances of the TAG stop codon (Mitchell et al., 2017; Richardson et al., 2017). The different recoding approaches are summarised in Table 2 and in previous reviews (J. Kuo et al., 2018).
Table 2.
Recoding Approaches Attempted to Date
| Year | Lead Author | Species | % Recoded | Recoding Method | Codons Removed | Amino Acid |
|---|---|---|---|---|---|---|
| 2011 | F. Isaacs | E. coli | 100 | MAGE/CAGE | TAG | Stop |
| 2016 | N. Ostrov | E. coli | 100a | integrase-based segments approach | AGA, AGG, AGC, AGT, TTA, TTG, TAG | Arg |
| Ser | ||||||
| Leu | ||||||
| Stop | ||||||
| 2017 | Y.H. Lau | S. typhimurium | 4.5 | SIRCAS | TTA, TTG | Leu |
| 2017 | S. Richardsonb | S. cerevisiae | 100 | SWaP-In | TAG | Stop |
| 2019 | J. Fredens | E. coli | 100 | REXER | TCG, TCA, TAG | Ser |
| Stop |
This was achieved over 87 different strains.
The recoding of S. cerevisiae was a large collaborative effort that resulted in 7 publications in a special issue of Science (March 10, 2017), of which S. Richardson is one lead author.
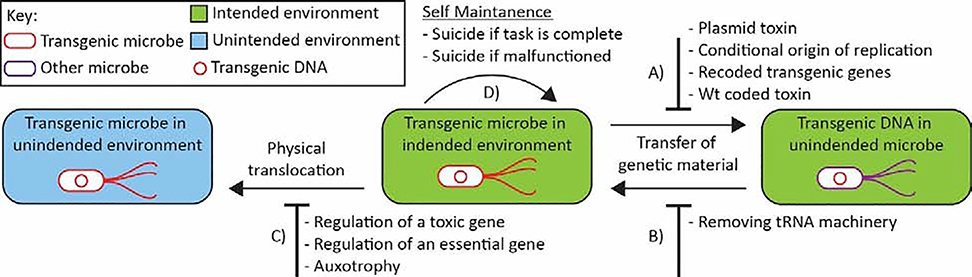
Freeing up a codon, and subsequently its associated tRNA, allows for four novel containment possibilities; 1) A recoded organism could have transgenic genes that are only translated in a functional manner by the host by inserting a recoded stop codon throughout the open reading frame, thus initiating early termination of translation in any organism not recoded in the same manner (Figure 1A). It should be noted this approach cannot prevent the transfer of transgenic DNA and subsequent mutations that remove recoded codons would allow expression. 2) A second approach could be to incorporate a broad range highly lethal toxin, such as an endonuclease, alongside any transgenic circuit. Such a toxin could be prevented from expression in the transgenic strain by incorporating canonical codon usage. Upon transfer to a wild-type strain, toxin expression would cause cell death. As transformation and transduction of genomic material rely upon homologous recombination, transgenic circuits would reliably be transferred in single units (Figure 1A). This approach prevents the transfer of the transgenic material in the first place, but has the drawback of a single mutation disabling the toxin in the transgenic strain would prevent containment system functioning. 3) A recoded organism without the full contingent of tRNA’s and their accompanying release factors is unable to translate most novel genetic material it acquires, blocking a potential avenue of evolution. (Figure 1B). 4) Novel tRNA synthetases that incorporate a non-natural amino acid can be used alongside an unassigned codon to engineer an auxotrophy, forming an effective containment system (Mandell et al., 2015; Rovner et al., 2015), explored further in section 1.3 (Figure 1C).
Figure 1: Schematic of biological containment mechanisms.
A) Methods for addressing transfer and expression of genetic material in unintended hosts B) methods of preventing DNA from other bacteria being expressed in a transgenic strain C) methods for preventing the physical translocation of bacteria from their intended environment to an unintended environment, and D) methods for removing bacteria from their intended environment if they are no longer required. Physical forms of containment such as barriers and waste management are not included.
A major obstacle to the widespread adoption of genome recoding is the ability to devise recoding schemes for a diverse range of organisms. Published recoding efforts to date have concentrated on E. coli and S. cerevisiae, and to a lesser extent S. typhimurum, some of the most comprehensively studied model organisms. Recoding a genome requires an extensive knowledge of an organism’s essential and overlapping genes, as well as established protocols for engineering and growth. Although the cost and methods of DNA synthesis currently makes recoding on a routine basis difficult, falling costs will make this more viable in the future (James Kuo et al., 2018).
It is important to note that a recoded organism represents a divergence from all previously known forms of life, and while this is not inherently dangerous, additional caution should be employed when considering their biocontainment.
1.3. Containment of transgenic microbes to their intended environment
Applications of transgenic living microbes are undertaken in controlled environments. Here, a controlled environment is defined as one where physical barriers, effective waste management, and enforced regulation can easily be assured, including both laboratory research and industrial production. There are many published containment systems that have been designed to prevent transgenic microbes from escaping their intended environment. These generally fall into one or more of the following categories: allowing for the expression of an essential gene, regulation of a toxic gene, and supplementation of an auxotrophy (Figure 1C). In addition, regulation of a toxic gene has been shown to modulate the self-maintenance of a transgenic strain (Figure 1D). Table 3 compiles a reasonably exhaustive list of 35 containment systems, that take advantage of essential and toxic gene regulation, engineered auxotrophy or a combination of several mechanisms. It includes their mode(s) of containment, the signal(s) they respond to, their survival ratio, the evolutionary stability of the system when grown in permissible conditions and whether they are designed for use in a controlled or uncontrolled environment. Each row of the table represents a single containment system in a single strain. Papers that published multiple systems have generally been represented with whichever system had the lowest survival ratio, unless the modes of regulation are so disparate as to be considered separately of interest. Unless otherwise stated, the following systems are solely applicable to the processes described in Figure 1C. Although containment systems have been published for non-microbial purposes (Deans et al. 2007), they are not reported on in this review.
Table 3.
Containment systems designed to prevent escape of transgenic microbes.
| Year | Lead Author | Species | Auxotrophy | Essential gene | Toxic gene | Responds to | Survival ratio | Stability | Used in |
|---|---|---|---|---|---|---|---|---|---|
| 1987 | S. Molin | E. coli | hok | tryptophan | 10−4 | not tested | con | ||
| 1988 | A.K. Bej | E. coli | hok | IPTG | <10−6 | not tested | con | ||
| 1991 | A. Contreras | E. coli | gef | benzoates | 10−6 | not tested | un | ||
| 1991 | S.M. Knudsen | E. coli | relF | IPTG | 10−8 | not tested | con | ||
| 1992 | A.K. Bej | P. putida | gef | IPTG | 10−5 | not tested | con | ||
| 1993 | G. Recorbet | E. coli | sacB | sucrose | 10−3 | not tested | con | ||
| 1994 | I. Ahrenholtz | E. coli | nucA | temp. (cI857) | 10−5 | not tested | un | ||
| 1995 | SM. Knudsen | E. coli | relE | IPTG | 10−7 | not tested | con | ||
| 1996 | MT. Munthali | P. putida | colE3 | 3-methyl benzoate | N/A | not tested | un | ||
| 1997 | P. Szafranski | P. putida | streptavidin | 3-methyl benzoate | 10−7 | not tested | un | ||
| 1998 | L. Molina | P. putida | gef | 3-methyl benzoate | 10−8 | not tested | un | ||
| 2000 | P. Kristoffersen | S. cerevisiae | relE | galactose | N/A | not tested | con | ||
| 2001 | MC. Ronchel | P. putida | gef | 3-methyl benzoate | <10−9 | not tested | un | ||
| 2003 | L. Steidler | L. lactis | thymidine | thymidine | 10−7 | not tested | un | ||
| 2005 | A. Balan | S. cerevisiae | nucA | glucose | 10−5 | not tested | con | ||
| 2008 | W. Kong | S. typhimurium | asdA, murA | arabinose | 10−4 | not tested | con | ||
| 2009 | Q. Li | E. coli | nucA | arabinose | N/A | not tested | con | ||
| 2010 | JM. Callura | E. coli | ccdB/λ lysis/lexA3 | aTc, arabinose, IPTG | 10−3 | not tested | con | ||
| 2015 | CTY. Chan | E. coli | murC | ecoRI | aTc, IPTG | <10−5 | Unstable after four days | con | |
| 2015 | CTY. Chan | E. coli | murC | ecoR1 | IPTG, gal, cellobiose | <10−8 | Unstable after four days, | con | |
| 2015 | RR. Gallagher | E. coli | Biotin | ribA, glmS | ecoRI | aTc, IPTG | <10−12 | stable for >110 gen. | con |
| 2015 | Y. Cai | S. cerevisiae | HHTS, HHFS | galactose, estradiol | <10−10 | not tested | con | ||
| 2015 | DJ. Mandel | E. coli | bipA | bipA | <10−11 | not tested | con | ||
| 2015 | AJ. Rovner | E. coli | pAcF α | pAcF α, arabinose | <10−11 | not tested | con | ||
| 2015 | G. Lopez | E. coli | benzothazole | benzothiazole | 10−7 | not tested | con | ||
| 2016 | DI. piraner | E. coli | ccdB | temp. | 10−5 | not tested | un | ||
| 2016 | S. Huang | E. coli | blaM, cat1 | cell density | 10−5 | not tested | con | ||
| 2016 | R. Hirota | E. coli | Phosphite | phosphite | <10−12 | not tested | con | ||
| 2017 | N. Agmon | S. cerevisiae | SEC17 | estradiol | 10−8 | not tested | con | ||
| 2017 | F. Stirling | E. coli | ccdB | temp. | 10−5 | Stable for >140 gen. | un | ||
| 2017 | F. Stirling | E. coli | ccdB | cI and/or Cro | N/A | Stable for >140 gen. | un | ||
| 2018 | RL. Clark | S. sp. PCC7002 | CO2 | CO2 | 10−9 | not tested | con | ||
| 2019 | F. Stirling | E. coli | doc | pH | 10−6 | Stable for >100 gen. | un | ||
| 2019 | F. Stirling | E. coli | ccdB/doc | temp., pH | <10−11 | Stable for >100 gen. | un | ||
| 2019 | F. Stirling | E. coli | doc | pH2 | 10−4 | not tested | un |
Strains: Escherichia coli, Pseudomonas putida, Saccharomyces cerevisiae, Lactococcus lactis, Salmonella typhimurium, Synechococcus sp. PCC7002. Each row represents a single containment system in a single strain. For “Used in” column, con/un refers to controlled or uncontrolled environment.
These genes are essential in the presence of the antibiotics carbenicillin and chloramphenicol.
This containment system only expresses the toxin upon two, non-consecutive exposures to low pH.
Containment systems for a controlled environment
Of the 35 systems, 22 respond to one or more small molecules intended to be provided by human intervention, and are therefore designed to contain the transgenic strains to a controlled environment. Twelve systems consist of a basic design concentrating on a single molecule regulating a single toxic gene (Balan & Schenberg, 2005; Bej et al., 1992; Bej et al., 1988; Knudsen & Karlstrom, 1991; S. Knudsen et al., 1995; Kristoffersen et al., 2000; Li & Wu, 2009; Molin et al., 1987; Recorbet et al., 1993) or one or more essential genes (Agmon et al., 2017; Cai et al., 2015; Kong et al., 2008).
The last decade has seen a progression towards more complex containment systems. Three examples from the Collins lab (Callura et al., 2010; Chan et al., 2015) explore the capabilities of synthetic circuits to respond to multiple inputs controlling the expression of both toxic and essential genes, and are excellent examples of modular and customisable containment systems. Another example comes from the Isaacs lab (Gallagher et al., 2015), who combined an auxotrophy, suppression of two essential genes and the expression of a toxin to construct a containment system with a survival ratio below their detection limit of 10−12.
A unique containment system based on cell density (Huang et al., 2016) depends on collective expression of an antibiotic resistance gene whose expression is stimulated by the quorum sensing factor N-acyl homoserine lactone (AHL). At high cell densities, enough antibiotic resistance is expressed at a population level and released (upon cell lysis, expressed intracellularly, or excreted) to allow individual cells to survive. However if individual cells split off from the “microbial swarmbot”, insufficient resistance proteins would be expressed to allow survival. Although this system currently requires the continued application of antibiotics to provide the toxic function, it is suggested that future iterations could modulate the expression of toxic or essential genes, removing the requirement for supplementation and facilitating its use in an uncontrolled environment.
The final 5 systems designed to be used in a controlled environment are all auxotrophies, categorized as such because they require the uptake of the small molecule they are responding to for some aspect of their metabolism that cannot otherwise be self-synthesised. To construct a system that responded to atmospheric CO2 (Clark et al., 2018) the CO2 concentrating mechanism of the cyanobacteria Synechococcus sp. PCC7002 was removed. This only allowed survival when the transgenic strain was grown in an environment with at least 5% ambient CO2, with atmospheric levels of CO2 resulting in a survival ratio of 10−9. Another study engineered an E. coli phosphite auxotroph by removing all phosphate production pathways except via phosphite uptake, achieving a survival ratio below their detection limit of 10−12 (Hirota et al., 2017). A system for making Synthetic auxotroph’s based on a Ligand-Dependent Essential genes (SLiDE) was developed (Lopez & Anderson, 2015). SLiDE was used to develop a strain were the function of three essential genes was dependent upon the presence of the ligand benzothiazole. Finally, two synthetic auxotrophs were designed using the E. coli recoded strain with all instances of the TAG stop codon removed, mentioned in section 1.2 (Lajoie et al., 2013). Novel tRNA machinery was introduced to incorporate non-natural amino acids into the primary protein sequence of several essential genes (Mandell et al., 2015; Rovner et al., 2015). These last two methods both showed extremely robust containment, below their detection limit of 10−11.
Containment systems for an uncontrolled environment
The other 13 of the 35 systems in Table 1 were designed for application outside of a controlled environment. A thymidine auxotrophy containment system for Lactococcus lactis engineered to act as a therapeutic for Crohn’s disease when applied to the human digestive system showed a survival ratio of 10−7 both in vitro and when applied to a porcine model (Steidler et al., 2003). Although unable to propagate outside of a controlled environment, the functional application for this strain of expressing human interleukin-10 did not require DNA replication, allowing its application in an uncontrolled environment.
Five systems were designed for application in microbes engineered to facilitate the bioremediation of soil by the degradation of benzoates (Contreras et al., 1991; Molina et al., 1998; Munthali et al., 1996; Ronchel & Ramos, 2001; Szafranski et al., 1997). While the degradation target, benzoates, are present, toxin expression is repressed allowing survival. Because of this they respond to the presence of small molecule regulators, but will initiate population suicide upon the completion of their task or translocation from their intended location without the need for human intervention (Figure 1C and 1D).
A system was developed that was designed to terminate a bacterial population upon a loss of function mutation, based on the bacteriophage lambda cI/cro regulatory system (Stirling et al., 2017). In the presence of either cI or Cro, the toxin is repressed. However, in the absence of either, repression is relieved and the toxin is expressed. This system allows for the maintenance of functionality of a specific transgenic strain, checking to see if the transgenic element is present and terminating the strain if it is not (Figure 1D).
Three systems that responded exclusively to temperature have been published, each with a different mode of control. The first controls expression of the Serratia marcescens nuclease nucA with the temperature sensitive mutant of cI, cI857 (Ahrenholtz et al., 1994). At 42 °C, cI857 is unable to function, and the toxin is expressed resulting in a survival ratio of 10−5. The second controls expression of the DNA gyrase inhibitor ccdB using a mutant tlpA repressor that is activated below 36 °C. Using this system they were able to show the containment of a bacterial population to the mammalian gut with a survival ratio of 10−5 (Piraner et al., 2016). The third uses the regulatory region from cold shock protein A to control expression of the toxin ccdB, also achieving containment to the mouse gut with a survival ratio of around 10−5 (Stirling et al., 2017).
Finally, three systems that respond to pH have been reported (F. Stirling et al., 2020). The first controls the expression of the toxin Doc with the pH sensitive promoter Pasr, achieving a survival ratio of 10−6 when exposed to pH 5 conditions. The second combines the pH sensitive expression of Doc with the temperature sensitive expression of CcdB to achieve a survival ratio of below 10−11 when grown at 22 °C and at pH 5. The third and final containment system uses an excisionase based system to only express doc upon two, non-consecutive exposures to low pH, achieving a survival ratio of 10−4.
1.4. Evolutionary stability
Containment systems are only effective if they maintain functionality throughout the entire period of their intended use. Microbes exist across all domains of life, but are unified in their almost universal short generational lifespan and capacity to mutate and evolve at a rapid rate. This presents a problem for technology designed to contain and control the growth of engineered microbial species. To coin a term from the fictional Dr Ian Malcolm, “Life, uh, finds a way”. Any containment system that is designed to prevent microbial growth inherently has the potential to inflict a fitness defect, and therefore an evolutionary pressure to remove this fitness defect. For different forms of containment, this can occur in different manners.
For an auxotroph based containment system, an engineered strain would require the capacity to produce the metabolite that it is auxotrophic for, or negate the necessity for it in the first place, which can be achieved in four ways. 1) By taking on genes or operons from other strains that confer this capacity, through horizontal gene transfer or sexual reproduction, most likely from related species. 2) Evolution of pathways that are already intrinsic to the engineered strain that become capable of producing the required metabolite. 3) Evolution or modifications to the composition of the other organisms in the surrounding environment that increase production and/or excretion of the metabolites in question, allowing an environment that was initially non-permissible to become viable. 4) In the case of the systems mentioned that rely upon essential genes requiring ligand cofactors or nnAA for function/ translation (Lopez & Anderson, 2015; Mandell et al., 2015; Rovner et al., 2015), SNPs and small mutations to the genes in question can result in removing those requirements.
Escaping from a system of containment that uses the regulation of an essential gene can come about through three potential mechanisms. 1) Mutation of the regulation of the gene in question, making it no longer dependent upon whatever factor was designed to control expression. 2) Evolution of other pathways within the cell to fulfil the role the essential gene confers, negating the essential genes necessity. 3) Taking on the same or similar genes from other organisms that are regulated in an alternative manner.
Population control based upon the expression of a toxic gene has four possible modes of escape. 1) Mutation of the ORF of the toxin, negating its function 2) Mutation to the regulation of the toxin, either in its promoter or any transcriptional factors or in the regulation of the antitoxin (if present, see below) 3) Mutation in the target of the toxin. 4) Uptake of a resistance gene through horizontal gene transfer or sexual reproduction. Option 1, and in many cases option 2, represents a loss of function mutation, meaning the range of possible mutations that achieve the effect of escape is vastly wider than that required to confer a gain of function, which all other modes of escape mentioned above are some form of. Another factor that contributes to the instability of a containment system based on regulation of a toxic gene is the inherent leakiness of most regulatory systems. Promoters can best be described as up or down regulated rather than off or on, as even when repressed or not induced, low levels of expression can be observed. To counteract the fitness defect imparted from leaked expression of a toxic gene in permissible conditions, an antitoxin should be included in the design of the system. Most natural toxins evolved to function alongside their cognate antitoxin, and this dynamic can be exploited by expressing antitoxin at low levels to negate the effect of a leaked toxin (Gallagher et al., 2015; F. Stirling et al., 2017, 2020).
Of the 35 containment systems reported in Table 3, only 7 provide data of their long-term stability when passaged in permissible conditions. The multi-layered containment system (Gallagher et al., 2015), consisting of a biotin auxotrophy as well as the arabinose mediated regulation of the two essential genes ribA and glmS and the toxin ecoRI nuclease, was shown to be stable after passaging in permissible conditions for at least 110 generations. This system includes the ecoRI methyltransferase, the antitoxin to ecoRI nuclease. The Deadman and Passcode containment systems (Chan et al., 2015) also regulate the expression of ecoRI, as well as the essential gene murC, this time without the presence of ecoRI methyltransferase. Neither maintained their respective level of containment after a four-day period of growth, with an increase in survival ratio of 4–5 orders of magnitude. Passcode was additionally passaged for four days in E. coli MDS42pdu ΔrecA (Csörgo et al., 2012), a strain lacking recombinogenic and mobile genomic elements. This reduced the Passcode escapee rate by 3–5 logs over the four-day period. This practice is appropriate for certain purposes, although the fitness defect observed in E.coli MDS42pdu ΔrecA will prevent its widespread application.
The individual and combined essentializer, cryodeath and acidTRP containment systems (F. Stirling et al., 2017, 2020) all use a toxin antitoxin based approach, resulting in stable growth for over 100 generations. In addition, the construction of these systems explored a method for intelligent design of libraries with promoter variance to achieve the desired balance of expression between toxin and antitoxin.
1.5. Containment standard practices
Currently publications on containment systems are disparate in their reporting techniques. Not all reports display a quantifiable survival ratio by comparing cfu at permissible conditions to cfu at non-permissible conditions, and instead rely on metrics such as a growth curve to show growth disparities. Although it is easy to infer an effect from a growth curve, the fact that OD measurements do not differentiate well between living and dead cells means that a true survival ratio is hard to calculate. We recommend that wherever possible the survival ratios for containment systems should always be calculated by plating dilutions of culture in both permissible and non-permissible conditions and comparing cfu.
In most cases, individual studies report unique limits to the sensitivity of their survival assays. This limit predominantly comes from the total cfu that are given the possibility to grow in non-permissible conditions. Using the traditional techniques of spreading culture on agar, the upper limit of plating a bacterial culture is reached at about 1011-1012 cfu (Gallagher et al., 2015; Hirota et al., 2017; Mandell et al., 2015; Rovner et al., 2015; F. Stirling et al., 2020). Beyond this, the concentration of cells is so high as to cause clumping that prevents accurate assays. Increasing the area of agar plates used circumvents this, but quickly becomes prohibitive in the area of plates required. We recommend that when reporting the survival ratio for a containment system, it should become standard practice to reach this limit of detection. For some containment mechanisms, lower survival ratios could feasibly be detected using technology such as a morbidostat (Toprak et al., 2013). A morbidostat continually monitors the growth rate of a bacterial culture, automatically adjusting the concentration of a small molecule (often an antibiotic but theoretically any small molecule regulator) in response to culture density. By incrementally increasing/decreasing the concentration of a containment system regulator, an effective population of greater than 1016 can be exposed to a non-permissible condition. Although this capacity is currently not easily accessible for all labs, widespread adoption of this technique may become necessary as the field develops. It is difficult to claim a transgenic strain is effectively contained without improvements to the detection limit reached by plating techniques, unless such an environmental release involved a population several orders of magnitude less than the established survival ratio of the containment systems involved.
The majority of studies did not report data on the stability of their respective containment systems. For all containment systems, no matter the application, evolutionary stability is an essential quality, and can easily be assayed by passaging in permissible conditions while calculating the number of generations that pass. A comparison of survival ratios before and after this growth period allows for a simple display of evolutionary stability. We recommend that determining the stability of a strain over 100 generations should become standard practice when reporting a new containment system. This is sufficient to allow at least one adaptive sweep to pass through the population, allowing beneficial mutations to take over (Maddamsetti et al., 2015, Novick & Szilard, 1950). Additional experiments comparing the growth rates of engineered strains with parent strains, either individually or in competitive co-culture growth assays, would also be informative. Further theory on the capacity of asexual strains to maintain a circuit that confers only a very slight fitness disadvantage can be found in (Stirling et al., 2017).
Although NIH guidelines currently recommend a survival ratio of no greater than 10−8 for the containment of transgenic microbes, it is readily apparent that this standard is too high for many applications of transgenic microbes. A containment system should only be considered functional if its survival ratio is such that over the time period a transgenic microbe is to be deployed, the probability of an escape event occurring is negligible. For a transgenic population of microbes, if Y cells are expected to physically translocate away from their intended environment over the lifespan of the application, the survival ratio of a containment system must be orders of magnitude less than 10−Y. Any application of transgenic microbes where greater than 108 cells are expected to be deployed will require a survival ratio lower than the current guidelines for effective containment. Future recommendations could reflect the number of cells expected to survive, relating to both the population released and the escape rate of the containment systems involved.
Acknowledgements
We thank Kevin Esvelt, John Glass, Tom Ellis, James Collins and George Church for giving extensive feedback during the writing process. This work was supported by Defense Advanced Research Projects Agency grant HR0011-15-C-0094 and funds from the Wyss Institute for Biologically Inspired Engineering. F.S. acknowledges funding from NIH training grant [5T32GM007598].
Footnotes
Declaration of Interest
P.A.S is a cofounder of 64-x, a company focused on genome recoding.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1.6 References
- Adrio JL, & Demain AL (2010). Recombinant organisms for production of industrial products. Bioengineered Bugs, 1(2), 116–131. 10.4161/bbug.1.2.10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N, Tang Z, Yang K, Sutter B, Ikushima S, Cai Y, … Boeke JD (2017). Low escape-rate genome safeguards with minimal molecular perturbation of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America, 114(8), E1470–E1479. 10.1073/pnas.1621250114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrenholtz I, Lorenz MG, & Wackernagel W (1994). A conditional suicide system in Escherichia coli based on the intracellular degradation of DNA. Applied and Environmental Microbiology, 60(10), 3746–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeshen NA, Baeshen MN, Sheikh A, Bora RS, Ahmed MMM, Ramadan HAI, … Redwan EM (2014). Cell factories for insulin production. Microbial Cell Factories, 13(1), 1–9. 10.1186/s12934-014-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan A, & Schenberg ACG (2005). A conditional suicide system for Saccharomyces cerevisiae relying on the intracellular production of the Serratia marcescens nuclease. Yeast, 22(3), 203–212. 10.1002/yea.1203 [DOI] [PubMed] [Google Scholar]
- Bej AK, Molin S, Perlin M, & Atlas RM (1992). Maintenance and killing efficiency of conditional lethal constructs in Pseudomonas putida. Journal of Industrial Microbiology, 10(2), 79–85. 10.1007/BF01583839 [DOI] [PubMed] [Google Scholar]
- Bej AK, Perlin MH, & Atlas RM (1988). Model suicide vector for containment of genetically engineered microorganisms. Applied and Environmental Microbiology, 54(10), 2472–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg P, Baltimore D, Brenner S, Roblin RO, & Singer MF (1975). Summary statement of the asilomar conference on recombinant DNA molecules. Proceedings of the National Academy of Sciences of the United States of America, 72(6), 1981–1984. 10.1073/pnas.72.6.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon JP, … Steidler L (2006). A Phase I Trial With Transgenic Bacteria Expressing Interleukin-10 in Crohn’s Disease. Clinical Gastroenterology and Hepatology, 4(6), 754–759. 10.1016/j.cgh.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Cai Y, Agmon N, Choi WJ, Ubide A, Stracquadanio G, Caravelli K, … Boeke JD (2015). Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes. Proceedings of the National Academy of Sciences of the United States of America, 112(6), 1803–1808. 10.1073/pnas.1424704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, & Collins JJ (2010). Physiology Using Synthetic Riboregulators. Proceedings of the National Academy of Sciences, 107(36), 15898–15903. 10.1073/pnas.1009747107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caluwaerts S, Vandenbroucke K, Steidler L, Neirynck S, Vanhoenacker P, Corveleyn S, … Rottiers P (2010). AG013, a mouth rinse formulation of Lactococcus lactis secreting human Trefoil Factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncology, 46(7), 564–570. 10.1016/j.oraloncology.2010.04.008 [DOI] [PubMed] [Google Scholar]
- Chan CTY, Lee JW, Cameron DE, Bashor CJ, & Collins JJ (2015). “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nature Chemical Biology, 12(2), 82–86. 10.1038/nchembio.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, & Church GM (2017). Beyond editing to writing large genomes. Nature Reviews Genetics, 18(12), 749–760. 10.1038/nrg.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Lajoie MJ, Xiao H, Church GM, & Schultz PG (2014). A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem, 15(12), 1782–1786. 10.1002/cbic.201402104 [DOI] [PubMed] [Google Scholar]
- Clardy J, Fischbach MA, & Currie CR (2009). The natural history of antibiotics. Current Biology, 19(11), 1–8. 10.1016/j.cub.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RL, Gordon GC, Bennett NR, Lyu H, Root TW, & Pfleger BF (2018). High-CO2 Requirement as a Mechanism for the Containment of Genetically Modified Cyanobacteria . ACS Synthetic Biology, 7(2), 384–391. 10.1021/acssynbio.7b00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley WB (1893). The treatment of malignant tumors by repeated inoculations of erysipelas: With a report of ten original cases. Clinical Orthopaedics and Related Research. [PubMed] [Google Scholar]
- Contreras A, Molin S, & Ramos JL (1991). Conditional-suicide containment system for bacteria which mineralize aromatics. Applied and Environmental Microbiology, 57(5), 1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csörgo B, Fehér T, Tímár E, Blattner FR, & Pósfai G (2012). Low-mutation-rate, reduced-genome Escherichia coli: An improved host for faithful maintenance of engineered genetic constructs. Microbial Cell Factories, 11, 1–13. 10.1186/1475-2859-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo V (2009). Recombinant bacteria for environmental release: What went wrong and what we have learnt from it. Clinical Microbiology and Infection, 15(SUPPL. 1), 63–65. 10.1111/j.1469-0691.2008.02683.x [DOI] [PubMed] [Google Scholar]
- Deans TL, Cantor CR, & Collins JJ (2007). A Tunable Genetic Switch Based on RNAi and Repressor Proteins for Regulating Gene Expression in Mammalian Cells. Cell, 130(2), 363–372. 10.1016/j.cell.2007.05.045 [DOI] [PubMed] [Google Scholar]
- Dhall P, Kumar R, & Kumar A (2012). Biodegradation of sewage wastewater using autochthonous bacteria. TheScientificWorldJournal, 2012. 10.1100/2012/861903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E, Munthali M, de Lorenzo V, & Timmis KN (1994). Universal barrier to lateral spread of specific genes among microorganisms. Molecular Microbiology, 13(5), 855–861. 10.1111/j.1365-2958.1994.tb00477.x [DOI] [PubMed] [Google Scholar]
- Diwo C, & Budisa N (2019). Alternative biochemistries for alien life: Basic concepts and requirements for the design of a Robust biocontainment system in genetic isolation. Genes, 10(1). 10.3390/genes10010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredens J, Wang K, de la Torre D, Funke LFH, Robertson WE, Christova Y, … Chin JW (2019). Total synthesis of Escherichia coli with a recoded genome. Nature. 10.1038/s41586-019-1192-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RR, Patel JR, Interiano AL, Rovner AJ, & Isaacs FJ (2015). Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Research, 43(3), 1945–1954. 10.1093/nar/gku1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankore ED, Zhang L, Chen Y, Liu K, Niu W, & Guo J (2019). Genetic Incorporation of Noncanonical Amino Acids Using Two Mutually Orthogonal Quadruplet Codons [Research-article]. ACS Synthetic Biology, 8(5), 1168–1174. 10.1021/acssynbio.9b00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota R, Abe K, Katsuura ZI, Noguchi R, Moribe S, Motomura K, … Kuroda A (2017). A novel biocontainment strategy makes bacterial growth and survival dependent on phosphite. Scientific Reports, 7(March), 1–10. 10.1038/srep44748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee AJ, Tsoi R, Wu F, Zhang Y, & Leong KW (2016). Coupling spatial segregation with synthetic circuits to control bacterial survival. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen SM, & Karlstrom OH (1991). Development of efficient suicide mechanisms for biological containment of bacteria. Applied and Environmental Microbiology, 57(1), 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S, Saadbye P, Hansen LH, Collier A, Jacobsen BL, Schlundt J, & Karlstrom OH (1995). Development and testing of improved suicide functions for biological containment of bacteria. Applied and Environmental Microbiology, 61(3), 985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Wanda S, Zhang X, Bollen W, Tinge SA, Roland KL, & Curtiss R (2008). Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. 105(27), 9361–9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotula JW, Kerns SJ, Shaket LA, Siraj L, Collins JJ, Way JC, & Silver PA (2014). Programmable bacteria detect and record an environmental signal in the mammalian gut. Proceedings of the National Academy of Sciences of the United States of America, 111(13), 4838–4843. 10.1073/pnas.1321321111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen P, Jensen GB, Gerdes K, & Piškur J (2000). Bacterial toxin-antitoxin gene system as containment control in yeast cells. Applied and Environmental Microbiology, 66(12), 5524–5526. 10.1128/AEM.66.12.5524-55262000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J, Stirling F, Lau YH, Shulgina Y, Way JC, & Silver PA (2018). Synthetic genome recoding: new genetic codes for new features. Current Genetics, 64(2). 10.1007/s00294-017-0754-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo James, Stirling F, Lau YH, Shulgina Y, Way JC., & Silver PA (2018). Synthetic genome recoding: new genetic codes for new features. Current Genetics. 10.1007/s00294-017-0754-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni H-R, Haimovich AD, Kuznetsov G, … Isaacs FJ (2013). Genomically Recoded Organisms Expand Biological Functions. Science, 342(6156), 357–360. 10.1126/science.1241459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YH, Stirling F, Kuo J, Karrenbelt MAP, Chan YA, Riesselman A, … Silver PA (2017). Large-scale recoding of a bacterial genome by iterative recombineering of synthetic DNA. Nucleic Acids Research. 10.1093/nar/gkx415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur Laurel A, Sanders-Beer Brigitte E, Brichacek Beda, Ranajit Pal X., & Yang Liu Liu, Yu Rosa, Venzon David, Lee Peter P, and H. H D. (2011). Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunol, 4(6), 648–657. 10.1038/nature22814.Trans-kingdom [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Chan CTY, Slomovic S, & Collins JJ (2018). Next-generation biocontainment systems for engineered organisms. Nature Chemical Biology, 14(6), 530–537. 10.1038/s41589-018-0056-x [DOI] [PubMed] [Google Scholar]
- Li Q, & Wu YJ (2009). A fluorescent, genetically engineered microorganism that degrades organophosphates and commits suicide when required. Applied Microbiology and Biotechnology, 82(4), 749–756. 10.1007/s00253-009-1857-3 [DOI] [PubMed] [Google Scholar]
- Lopez G, & Anderson JC (2015). Synthetic Auxotrophs with Ligand-Dependent Essential Genes for a BL21(DE3) Biosafety Strain . ACS Synthetic Biology, 4(12), 1279–1286. 10.1021/acssynbio.5b00085 [DOI] [PubMed] [Google Scholar]
- Łukasiewicz K, & Fol M (2018). Microorganisms in the Treatment of Cancer: Advantages and Limitations. Journal of Immunology Research, 2018. 10.1155/2018/2397808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddamsetti R, Lenski RE, & Barrick JE (2015). Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli. 200(June), 619–631. 10.1534/genetics.115.176677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, … Romesberg FE (2014). A semi-synthetic organism with an expanded genetic alphabet. Nature, 509(7500), 385–388. 10.1038/nature13314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, … Church GM (2015). Biocontainment of genetically modified organisms by synthetic protein design. Nature. 10.1038/nature14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli F, Scoma A, Michoud G, Aulenta F, Boon N, Borin S, … Daffonchio D (2017). Biotechnologies for Marine Oil Spill Cleanup: Indissoluble Ties with Microorganisms. Trends in Biotechnology, 35(9), 860–870. 10.1016/j.tibtech.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Marlière P, Patrouix J, Döring V, Herdewijn P, Tricot S, Cruveiller S, … Mutzel R (2011). Chemical evolution of a bacterium’s genome. Angewandte Chemie - International Edition, 50(31), 7109–7114. 10.1002/anie.201100535 [DOI] [PubMed] [Google Scholar]
- McClure NC, & Venables WA (1986). Adaptation of pseudomonas putida mt-2 to growth on aromatic amines. Journal of General Microbiology. [DOI] [PubMed] [Google Scholar]
- Mitchell LA, Wang A, Stracquadanio G, Kuang Z, Wang X, Yang K, … Boeke JD (2017). Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science, 355(6329). 10.1126/science.aaf4831 [DOI] [PubMed] [Google Scholar]
- Moe-Behrens GHG, Davis R, & Haynes KA (2013). Preparing synthetic biology for the world. Frontiers in Microbiology, 4(JAN), 1–10. 10.3389/fmicb.2013.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S, Boe L, Jensen LB, Kristensen CS, Givskov M, Ramos JL, & Bej AK (1993). Suicidal genetic elements and their use in biological containment of bacteria. Annu Rev Microbiol, 47, 139–166. 10.1146/annurev.mi.47.100193.001035 [DOI] [PubMed] [Google Scholar]
- Molin S, Klemm P, Poulsen L, Biehl H, Gerdes K, & Andersson P (1987). Conditional suicide system for containment of bacteria and plasmids. Nature Biotechnology, 5(December), 1315–1318. [Google Scholar]
- Molina L, Ramos C, Ronchel MC, Molin S, & Ramos JL (1998). Construction of an efficient biologically contained Pseudomonas putida strain and its survival in outdoor assays. Applied and Environmental Microbiology, 64(6), 2072–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munthali MT, Timmis KN, & Diaz E (1996). Use of colicin E3 for biological containment of microorganisms. Applied and Environmental Microbiology, 62(5), 1805–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH FAQs. (2019). FAQs NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules. NIH Guidelines, 2(April), 1–7. [Google Scholar]
- NOVICK A, & SZILARD L (1950). Experiments with the Chemostat on spontaneous mutations of bacteria. Proceedings of the National Academy of Sciences of the United States of America, 36(12), 708–719. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1063276&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, … Church GM (2016). Design, synthesis, and testing toward a 57-codon genome. Science, 353(6301), 819–822. 10.1126/science.aaf3639 [DOI] [PubMed] [Google Scholar]
- Paridah M., Moradbak A, Mohamed A., Owolabi F. abdulwahab taiwo, Asniza M., & Abdul Khalid SH. (2016). Pesticides - Recent Trends in Pesticide Residue Assay. Intech, i(tourism), 13 [Google Scholar]
- Peterson M (2007). The precautionary principle should not be used as a basis for decision-making. Talking Point on the precautionary principle. EMBO Reports, 8(4), 305–308. 10.1038/sj.embor.7400947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper DH, & Reineke W (2000). Engineering bacteria for bioremediation. Current Opinion in Biotechnology, 11, 262–270. [DOI] [PubMed] [Google Scholar]
- Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A, & Shapiro MG (2016). Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat Chem Biol, advance on(November), 1–8. 10.1038/nchembio.2233 [DOI] [PubMed] [Google Scholar]
- Razzaq A, Shamsi S, Ali A, Ali Q, Sajjad M, Malik A, & Ashraf M (2019). Microbial proteases applications. Frontiers in Bioengineering and Biotechnology, 7(JUN), 1–20. 10.3389/fbioe.2019.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recorbet G, Robert C, Givaudan A, Kudla B, Normand P, & Faurie G (1993). Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Applied and Environmental Microbiology, 59(5), 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SM, Mitchell LA, Stracquadanio G, Yang K, Dymond JS, DiCarlo JE, … Bader JS (2017). Design of a synthetic yeast genome. Science, 355(6329), 1040–1044. [DOI] [PubMed] [Google Scholar]
- Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, … Silver PA (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nature Biotechnology, 35(7), nbt.3879. 10.1038/nbt.3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar DT, & Silver PA (2018). Engineering bacteria for diagnostic and therapeutic applications. Nature Reviews Microbiology, 16(4), 214–225. 10.1038/nrmicro.2017.172 [DOI] [PubMed] [Google Scholar]
- Risks SCoEaNIH. (2015). Opinion on Synthetic Biology II. Risk Assessment Methodologies and Safety Aspects. European Commission. [Google Scholar]
- Ronchel MC, & Ramos JL (2001). Dual System to Reinforce Biological Containment of Recombinant Bacteria Designed for Rhizoremediation. Applied and Environmental Microbiology, 67(6), 2649–2656. 10.1128/AEM.67.6.2649-2656.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, … Isaacs FJ (2015). Recoded organisms engineered to depend on synthetic amino acids. Nature, 518(7537), 89–93. 10.1038/nature14095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos VE, Casas JA, & Go E (2000). Xanthan gum: production, recovery, and properties. 18 10.1016/S0734-9750(00)00050-1 [DOI] [PubMed] [Google Scholar]
- Sarwat F, Qader SAU, Aman A, & Ahmed N (2008). Production & characterization of a unique dextran from an indigenous Leuconostoc mesenteroides CMG713. International Journal of Biological Sciences, 4(6), 379–386. 10.7150/ijbs.4.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G, Biotechnology, S. R.-C. O. in, & 2000, U (2000). Field applications of genetically engineered microorganisms forbioremediation processes. Elsevier, 11, 286–289. Retrieved from https://www.sciencedirect.com/science/article/pii/S0958166900000975 [DOI] [PubMed] [Google Scholar]
- Schmidt M, & De Lorenzo V (2000). Synthetic bugs on the loose: Containment options for deeply engineered (micro)organisms. New Scientist, 165(2221), 18 10.1016/j.copbio.2016.01.006 [DOI] [PubMed] [Google Scholar]
- Schmidt M, & De Lorenzo V (2012). Synthetic constructs in/for the environment: Managing the interplay between natural and engineered Biology. FEBS Letters, Vol. 586, pp. 2199–2206. 10.1016/j.febslet.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B, … Remaut E (2003). Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol, 21(7), 785–789. 10.1038/nbt840 [DOI] [PubMed] [Google Scholar]
- Stirling A (2007). Risk, precaution and science: towards a more constructive policy debate. Talking point on the precautionary principle. EMBO Reports, 8(4), 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling A, Hayes KR, & Delborne J (2018). Towards inclusive social appraisal: risk, participation and democracy in governance of synthetic biology. 12(Suppl 8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling F, Bitzan L, O’Keefe S, Redfield E, Oliver JWK, Way J, & Silver PA (2017). Rational Design of Evolutionarily Stable Microbial Kill Switches. Molecular Cell, 68(4), 686–697.e3. 10.1016/j.molcel.2017.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling F, Naydich A, Bramante J, Barocio R, Certo M, Wellington H, … Silver P (2020). Synthetic cassettes for pH-mediated sensing, counting and containment. Cell Reports, 30(9), 740902 10.1101/740902 [DOI] [PubMed] [Google Scholar]
- Sundarram A, & Murthy TPK (2014). α -Amylase Production and Applications : A Review. Journal of Applied & Environmental Microbiology, 2(4), 166–175. 10.12691/jaem-2-4-10 [DOI] [Google Scholar]
- Szafranski P, Mello CM, Sano T, Smith CL, Kaplan DL, & Cantor CR (1997). A new approach for containment of microorganisms: Dual control of streptavidin expression by antisense RNA and the T7 transcription system. Proceedings of the National Academy of Sciences of the United States of America, 94(4), 1059–1063. 10.1073/pnas.94.4.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toprak E, Veres A, Yildiz S, Pedraza JM, Chait R, Paulsson J, & Kishony R (2013). Building a morbidostat: An automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nature Protocols, 8(3), 555–567. 10.1038/nprot.2013.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres B, Jaenecke S, Timmis KN, García JL, & Díaz E (2000). A gene containment strategy based on a restriction-modification system. Environmental Microbiology, 2(5), 555–563. 10.1046/j.1462-2920.2000.00138.x [DOI] [PubMed] [Google Scholar]
- Torres B, Jaenecke S, Timmis KN, García JL, & Díaz E (2003). A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology, 149(12), 3595–3601. 10.1099/mic.0.26618-0 [DOI] [PubMed] [Google Scholar]
- Torres L, Krüger A, Csibra E, Gianni E, & Pinheiro VB (2016). Synthetic biology approaches to biological containment: Pre-emptively tackling potential risks. Essays in Biochemistry, 60(4), 393–410. 10.1042/EBC20160013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright O, Delmans M, Stan GB, & Ellis T (2015). GeneGuard: A modular plasmid system designed for biosafety. ACS Synthetic Biology, 4(3), 307–316. 10.1021/sb500234s [DOI] [PubMed] [Google Scholar]
- Wright O, Stan GB, & Ellis T (2013). Building-in biosafety for synthetic biology. Microbiology (United Kingdom), 159(PART7), 1221–1235. 10.1099/mic.0.066308-0 [DOI] [PubMed] [Google Scholar]
- Yoneda Y (1980). Increased production of extracellular enzymes by the synergistic effect of genes introduced into Bacillus subtilis by stepwise transformation. Applied and Environmental Microbiology, 39(1), 274–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JH, Nguyen VH, Jiang SN, Park SH, Tan W, Hong SH, … Min JJ (2017). Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Science Translational Medicine. 10.1126/scitranslmed.aak9537 [DOI] [PubMed] [Google Scholar]