Abstract
The principal function of archaeal and bacterial CRISPR-Cas systems is antivirus adaptive immunity. However, recent genome analyses identified a variety of derived CRISPR-Cas variants at least some of which appear to perform different functions. Here, we describe a unique repertoire of CRISPR-Cas-related systems that we discovered by searching archaeal metagenome-assemble genomes of the Asgard superphylum. Several of these variants contain extremely diverged homologs of Cas1, the integrase involved in CRISPR adaptation as well as casposon transposition. Strikingly, the diversity of Cas1 in Asgard archaea alone is greater than that detected so far among the rest of archaea and bacteria. The Asgard CRISPR-Cas derivatives also encode distinct forms of Cas4, Cas5, and Cas7 proteins, and/or additional nucleases. Some of these systems are predicted to perform defense functions, but possibly not programmable ones, whereas others are likely to represent previously unknown mobile genetic elements.
Introduction
CRISPR-Cas is an adaptive immunity system that protects bacteria and archaea from viruses and other invasive genetic elements.1–4 The cas genes generally can be partitioned into three sometimes overlapping modules that are responsible for consecutive stages of the immune response: (1) adaptation (i.e., incorporation of new spacers into CRISPR arrays), (2) processing of pre-CRISPR RNAs (pre-crRNAs) that generates mature crRNAs, and (3) interference when the crRNAs are employed as guides to recognize and cleave the cognate target DNA or RNA molecules.5,6 In addition, many CRISPR-Cas systems include various accessory genes that regulate different functions of the basic CRISPR machinery.6
In addition to full-fledged, interference-competent CRISPR-Cas systems—a variety of derived, defective variants that lack some of the essential components, often the active moiety of the interference module and also the adaptation module as well as the CRISPR array itself—have been identified by comparative genomics analysis.7,8 These derived CRISPR-Cas variants are generally thought to perform functions distinct from adaptive immunity and in some cases apparently not involved in defense at all. A striking example of such exaptation of derived CRISPR-Cas systems are the interference-defective subtype I-F and subtype V-K variants encoded in different groups of Tn7-like transposons that are involved in RNA-guided transposition.9–12 The functions of other derived variants remain poorly understood. Type IV CRISPR-Cas systems that are carried by numerous plasmids seem to be the most common of such defective variants that seem to have evolved via partial degradation of type III systems, including the loss of the interference module.6,13 The prevalence of spacers targeting plasmid DNA suggests that type IV systems are involved in inter-plasmid competition.14 Indeed, it has been demonstrated that a type IV-A CRISPR-Cas system from Pseudomonas aeruginosa mediates RNA-guided interference against a plasmid in vivo, although the mechanism of this interference remains obscure.15 Another notable case is the Halobacterial Repeat-Associated Mysterious Proteins (HRAMP) systems that are widespread in Halobacteria and consist of extremely diverged variants of Cas5 and Cas7 (two families of the RAMP superfamily) along with additional nucleases and uncharacterized conserved proteins.16 The HRAMP systems are not associated with CRISPR arrays and lack adaptation modules. Given the presence of the Cas5 and Cas7 proteins that form crRNA-binding complexes in Class1 CRISPR-Cas systems, it has been proposed that HRAMPs are involved in RNA-dependent, although probably not programmable, defense functions.16
Apart from the derived CRISPR-Cas variants, homologs of some individual cas genes, in particular cas1, cas2 and cas4, have been identified in the non-CRISPR context. The most prominent case is that of casposons, a distinct family of transposons that employ a Cas1 homolog as the integrase.17–21 In addition, some cas1 homologs are “solo” genes, without any conserved genomic context and therefore without any hint of the function.18 Some of the casposons also encode a Cas4-like protein, although the role of this nuclease in the transposon life cycle (if any) remains unknown. Furthermore, Cas4 homologs are encoded in various contexts, suggesting involvement in repair processes or other defense functions.22
Here, we report a surprising diversity of previously unknown CRISPR-Cas derivatives and highly diverged Cas1 homologs encoded in the genomes of many Asgard archaea. The Asgard archaea are a recently discovered and rapidly expanding, thanks to metagenomic sequencing, archaeal superphylum.23–27 They are best known for their apparent evolutionary relationships with eukaryotes, which are thought to share common ancestry with one of the Asgard lineages. Otherwise, however, the biology of this major group of archaea is poorly understood, in large part because of their recalcitrance to growth in culture.28
We sought to characterize the antivirus defense mechanisms in Asgard archaea and, in particular, the CRISPR-Cas systems. The presence of a unique repertoire of CRISPR-Cas derivatives and extremely divergent Cas1 variants might reflect some specific aspects of the Asgard mobilome that remain to be investigated.
Methods
The sequences of 41 metagenome-assembled genomes (MAGs) of Asgard archaea with high coverage were obtained from various environments, including marine sediments and seawater (Supplementary Table S1). The only currently available complete genome of an Asgard archaeon, the anaerobic archaeon MK-D1,28 was obtained from GenBank (GCF_008000775.1). The protein sequences encoded in the Asgard genomic contigs were assigned to the 2015 version of arCOGs29 using PSI-BLAST,30 with the arCOG alignments used as the position-specific scoring matrix sources, as previously described.31
Iterative profile searches using PSI-BLAST,30 with the cutoff e-value of 0.0001 were employed to search for distantly similar sequences in either the non-redundant (NR) database or the protein sequence database of 41 Asgard genomes. To detect distant sequence similarity, a CD-search32 with a cutoff e-value of 0.01 and low complexity filtering turned off and a HHpred search with default parameters33 were run against PDB, Pfam, and CDD profile databases. Protein secondary structure was predicted using Jpred 4.34
The set of 2,512 Cas1 domain sequences35 was augmented with 69 sequences from Asgard archaea and 20 sequences from subtype V-F36 and their homologs in NR. All protein sequences were clustered using MMSEQS237 (–min-seq-id 0.9) and aligned using MUSCLE.38 Alignments were iteratively compared to each other using HHSEARCH and aligned using HHALIGN.39 An approximate ML tree of Cas1 was reconstructed using FastTree with WAG evolutionary model and gamma-distributed site rates.40 A similarity dendrogram for TnpB and type V effectors was built, as previously described, using the same alignments of TnpB protein sequences35 combined with two alignments of Asgard-specific TnpB families.
The CRISPR-Cas loci were identified and annotated, as previously described, using custom profiles derived from multiple alignments of Cas proteins to search for cas genes.6 CRISPR arrays were detected using the minCED tool (https://github.com/ctSkennerton/minced)41 with default parameters. The search for protospacers homologous to spacers from Asgard CRISPR arrays was performed using BLASTN with 90% sequence identity, otherwise as previously described.42
Results
By using the previously developed approaches for the identification of CRISPR-Cas systems and CRISPR arrays,6 we detected only class 1 systems in four of the six major Asgard lineages (Odin, Thor, Loki, Hel) in at least 12 of the 41 analyzed MAGs (Supplementary Tables S1 and S2).27 These systems belong to subtypes I-E and D or subtypes III-A, B, and D. Most of these systems were found to be partial because of the small contig size (i.e., truncated at the end of a contig). These are “garden variety” CRISPR-Cas systems, closely resembling those found in other archaea and bacteria. A search for protospacers homologous to the spacers from these CRISPR-Cas systems yielded no hits to any known virus genomes but several hits to putative proviruses integrated in Asgard genomes (Supplementary Table S3), supporting the role of these systems in antivirus immunity.
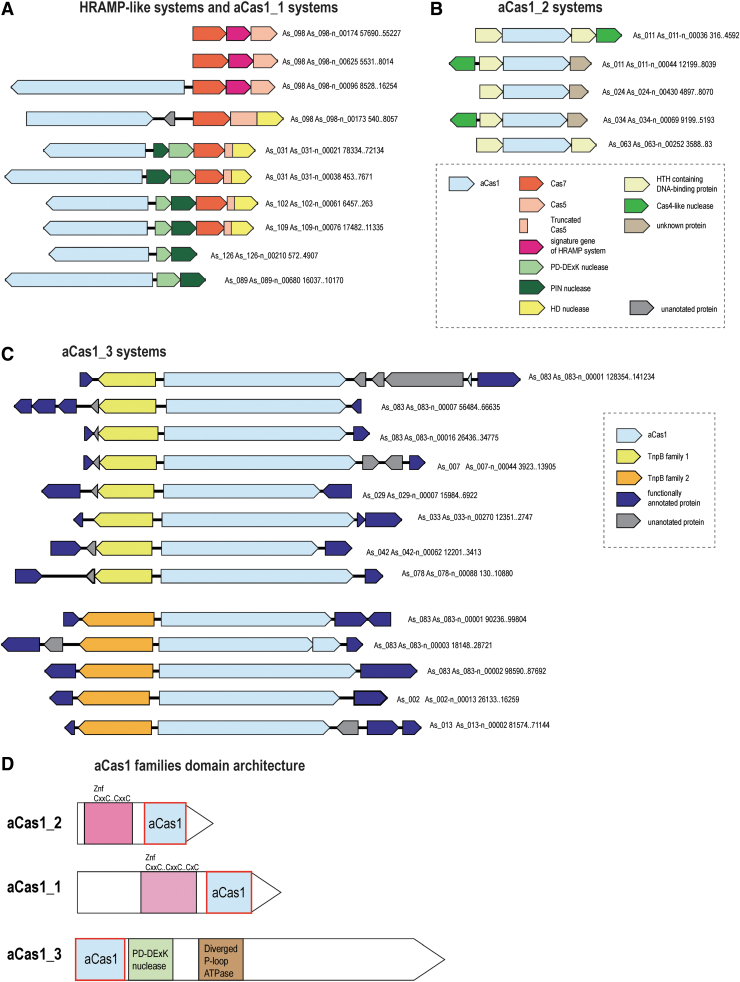
In addition to the typical Class 1 CRISPR-Cas systems, we identified unusual novel variants that have no counterparts outside the Asgard archaea. Notably, these unique CRISPR-Cas-related entities are more abundant in Asgard than typical CRISPR-Cas systems, with the latter comprising only about 30% of the CRISPR-related loci (see Supplementary Table S1). One group of such apparent derived CRISPR-Cas systems, present in several Asgard MAGs, is distantly related to the HRAMP systems and has been mentioned in the latest publication on CRISPR-Cas classification.6 The HRAMP systems contain three core genes, two of which—cas7 and cas5—are shared with class 1 CRISPR-Cas systems, where they are subunits of the crRNA-binding complex, and one gene remains uncharacterized; they are often also associated with additional nucleases.16 In the analyzed set of Asgard MAGs, there are at least three loci with the same organization as a typical HRAMP, but the majority of the related systems from Asgard have a unique organization (Fig. 1A and Supplementary Table S2). Most of these loci lack the uncharacterized core gene present of HRAMP and instead encode a HD nuclease domain fused to Cas5. Furthermore, this Cas5 domain is truncated and contains only the C-terminal region including a glycine-rich loop, the hallmark of the RAMP superfamily.43 By analogy to HRAMP, we provisionally denote these systems ARAMPs (Asgard RAMPs). The ARAMP loci also typically encode two nucleases: PD-DExK, predicted to cleave DNA, and PIN, a predicted RNase. Additionally, and unlike HRAMPs, several ARAMPs include a gene encoding a large protein that contains a domain distantly related to Cas1 (hereafter aCas1_1), the integrase involved in the insertion of new spacers into CRISPR arrays, as well as the transposition of the casposons.44 This protein often also contains a Zn finger and, in some cases, a DNA-binding helix-turn-helix (HTH) domain (Supplementary Fig. S1). Like HRAMP, none of the ARAMPs are adjacent to a CRISPR array. The ARAMPs or aCas1_1s are found in several MAGs of the Gerd, Heimdall, Hel, and Thor lineages of Asgard archaea, in most of which other known CRISPR-Cas systems were not identified (Fig. 1A and Supplementary Table S1 and S2). Although the function and mechanism of ARAMP are currently enigmatic, based on the presence of nucleases and remnants of a Class 1 effector complex, it appears highly likely that ARAMP is an RNA-dependent defense system, although perhaps not a programmable one.
FIG. 1.
Genomic organization of Asgard-specific derivatives of CRISPR-Cas systems and Cas1-containing modules. For each locus, the metagenome-assembled genome contig identifier and the respective nucleotide coordinates are indicated. The genes of a representative locus are shown by arrows. The arrows indicate the transcription direction of the respective gene. Genes are shown roughly to scale. Homologous genes are colored according to the respective insets. Genes are shown to the same scale in (A)–(C). (A) ARAMP: degraded Class1 CRISPR-Cas systems. (B) aCas1_2-containing modules. (C) aCas1_3-TnpB modules. (D) Domain architectures of aCas1 families. Domains are shown as rectangles drawn roughly to scale. The Cas1 homologous domains are highlighted by red borders.
We also identified distant homologs of aCas1-1 (hereafter aCas1_2) that are not linked to a Cas7–Cas5 RNA-binding complex but instead are strongly associated with a Cas4-like DNase and an HTH-domain-containing DNA-binding protein. aCas1_2 is shorter than aCas1_1 and has a distinct Zn finger domain (Supplementary Fig. S1). These variants are also found in the MAGs of Heimdall, Hel, and Gerd lineages (Fig. 1B and Supplementary Table S2). Given the absence of RNA-binding proteins, this could be a defense system that functions at the DNA level.
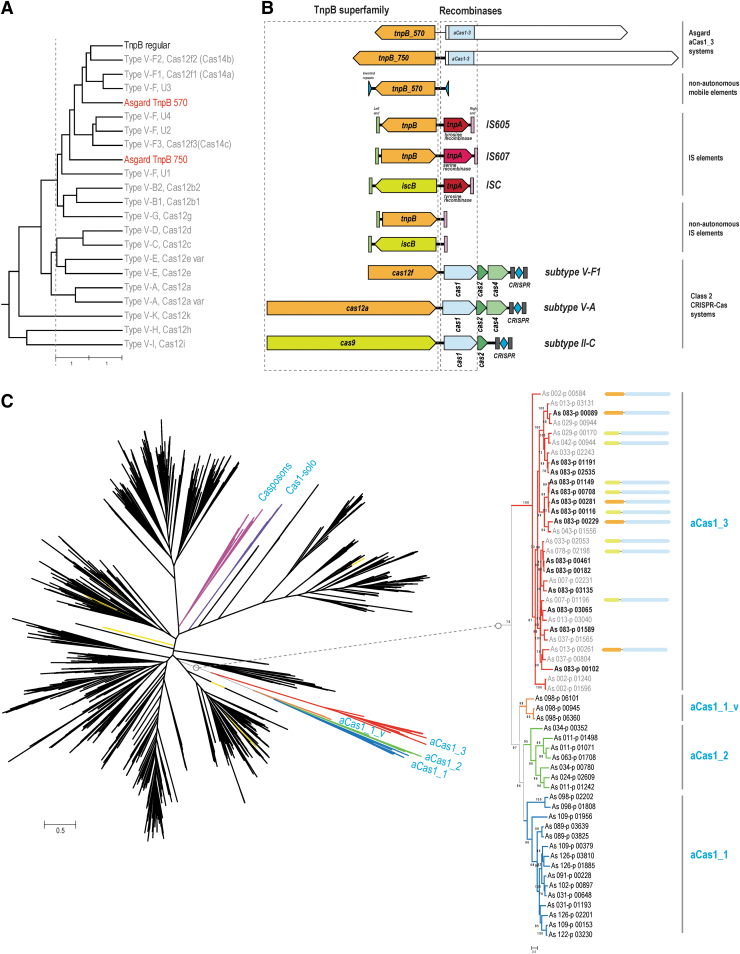
Another unusual group of CRISPR-Cas-related elements was identified, mostly in Thorarchaeota but also in several Heimdall MAGs in the course of examination of “dark matter islands,” genomic loci enriched in unannotated genes.45 This variant is centered around a large protein (aCas1_3) that contains three identifiable but highly diverged domains, namely the catalytic domain of Cas1, a PD-DExK nuclease and a P-loop ATPase that is often inactivated, as indicated by the substitution of the aspartate critical for catalysis in the Walker B site46 (Fig. 1C and D, Supplementary Fig. S1, and Supplementary Table S2). These three domains are located in the N-terminal region of the large protein, whereas the remaining portion (about 800 aa) contains no identifiable domains and shows no detectable sequence similarity to any proteins in the current databases. Nevertheless, this region is predicted to adopt an alpha/beta secondary structure, suggesting that it consists of globular domains (Supplementary Fig. S1). This gene is strongly linked to genes encoding TnpB-like family proteins of two distinct subfamilies, both with intact catalytic RuvC-like motifs, indicating that these are active nucleases (Supplementary Fig. S1). The TnpB-like proteins of both subfamilies are larger than typical transposon-encoded TnpB (∼570 and ∼750 aa, respectively; hereafter, TnpB-570 and TnpB-750 families), which is comparable to the size of the smallest type V effectors (Fig. 2A).47 The aCas1_3-TnpB modules are not accompanied by CRIPSR repeats, suggesting that these might not be functional CRISPR-Cas systems. Several TnpB-like proteins of both families are encoded by stand-alone genes. For stand-alone TnpB-570 family genes, we identified traces of recent transposition events in AS_002 MAG and detected flanking inverted terminal repeats, suggesting that these genes represent non-autonomous transposable elements (Supplementary Fig. S1). No tyrosine recombinase or serine recombinase genes were identified in this MAG, which makes aCas1_3 the best candidate for the role of the recombinase responsible for the in trans transposition of these elements. The functionality of aCas1_3-TnpB modules remains unclear, given that they share some features with both transposable (IS) elements and type V CRISPR-Cas systems (Fig. 2B). Considering the presence of several copies of this module (albeit not identical) in some of the Thorarchaeota MAGs (up to three of each variety in As_083 genome; Supplementary Table S2) and the clear evidence of transposition of the stand-alone TnpBs-570, the more plausible hypothesis appears to be that this is a novel IS-like mobile genetic element (MGE).
FIG. 2.
Phylogenomic analysis of Asgard aCas1 and TnpB. (A) Unweighted pair group method with arithmetic mean similarity dendrogram for the TnpB superfamily. The dendrogram was generated from the distance matrix constructed using pairwise HHalign scores between the respective TnpB family alignments. (B) Comparisons of aCas1_3 modules with IS elements and class 2 CRISPR-Cas systems. The organization of the genes in the genomic loci encoding representative systems of each type are shown. The genes are denoted by arrows drawn roughly to scale. Inverted terminal repeats are shown as blue triangles, and left and right ends of the IS elements are shown as green and light red rectangles, respectively, and are not to scale. (C) Phylogenetic tree of the Cas1 superfamily. The alignment of 2599 representative Cas1 sequences was obtained using the previously described procedure (see Methods). The approximate ML tree was reconstructed using FastTree, with the WAG substitution model and gamma-distributed site rates. Asgard aCas1_1 (and a minor variant aCas1_1_v), aCas1_2 and aCas1_3 clades as well as casposases and Cas1-solo that are not associated with CRISPR-Cas systems are colored coded. Small yellow branches scattered throughout the tree denote Asgard Cas1 homologs associated with typical CRISPR-Cas systems. The subtree in the right shows the expanded aCas1 clade. Protein identifiers for each leaf are indicated. The gene arrangements in the respective genomic loci are mapped to the aCas1_3 branch. Genes are shown as in Figure 1C.
As discussed above, Asgard archaea encode three distinct groups of Cas1 homologs that are more diverged from both the CRISPR-associated Cas1 and the casposases than any previously identified proteins within the Cas1 superfamily. We constructed a phylogenetic tree of the entire Cas1 superfamily, including the Asgard homologs (Fig. 2C). The tree contains a well-supported clade that includes all three aCas1 families; this clade splits into two branches, one of which consists of aCas1-3 and the other that includes aCas1-1 and aCas1-2 along with a minor, distinct variant (aCas1_1_v). One of the genes encoding aCas1_1_v (As_098-p_00945), like most of aCas1 genes, is located next to an aRAMP locus. Mapping of the genomic neighborhoods of aCas1_3 to the tree showed that aCas1_3 did not co-evolve with the two TnpB families. It appears that aCas1 genes freely combined with genes of the two TnpB families, suggesting interchangeable functionalities (Fig. 2C).
Discussion
The unexpected findings presented here show that Asgard archaea encompass a unique repertoire of highly diverged, derived CRISPR-Cas variants as well as putative novel MGE that share domains with CRISPR-Cas. These observations support and expand the previously noted trends in the evolution of CRISPR-Cas systems, namely their apparent recruitment for functions different from adaptive immunity as well as the evolutionary entanglement with MGE.10,48,49
Of special interest is the tight association between aCas1_3 and TnpB-like nucleases. TnpB appears to be the evolutionary ancestor of Cas12 proteins, the effectors of type V CRISPR-Cas systems.47,49 The Cas12 proteins of the multiple subtypes of type V seem to have evolved from different TnpB subfamilies on multiple independent occasions.47 The likely evolutionary intermediates are large TnpB-like proteins that acquired various additional still poorly characterized domains on the evolutionary route to mature Cas12 effectors. These putative evolutionary intermediates have been shown to function as CRISPR effectors that, however, prefer single-stranded DNA or RNA substrates in contrast to the typical effector proteins, such as Cas12a or Cas12b, that cleave dsDNA.36,50 Strikingly, in the case of aCas1_3 in Asgard archaea, we observed what appears to be yet another independent association between TnpB and a Cas protein homolog, in this case a large protein containing a diverged Cas1 domain. In previously analyzed bacterial and archaeal genomes, the TnpB proteins are encoded either on their own by non-autonomous transposons or together with RayT-like tyrosine or serine superfamily transposases, TnpA, in less abundant autonomous transposons51 (Fig. 2B). Unlike most DDE superfamily transposases (so denoted after their triad of catalytic amino acids), these two families of transposases do not require terminal inverted sequences.51 In Asgard genomes, we identified inverted terminal repeats flanking non-autonomous TnpB-570 transposons, indicating that transposition of these elements is unlikely to involve TnpA that indeed was not detected in most of Asgard MAGs. By contrast, the Cas1-like transposases of the casposons (casposase) does require inverted terminal repeats.21 Thus, the tight link between aCas1_3 and TnpB_570 strongly suggests that aCas1-3 functions as the transposase of these novel MGE that might also act in trans.
Perhaps the most remarkable aspect of these findings is the major expansion of the diversity of Cas1 in terms of both sequence and domain architecture of proteins containing the Cas1 domain (Figs. 1D and 2). Strikingly, the Asgard MAGs alone encompass a greater diversity of Cas1-containing proteins than the rest of bacteria and archaea taken together. The apparent monophyly of all aCas1 families implies a major expansion of the Cas1 superfamily in Asgard archaea. The lack of association of these Cas1 homologs with other cas genes (with the exception of some ARAMP modules) suggests that these proteins function as transposases (recombinases) in Asgard-specific MGE. The unique complex domain architectures of Asgard Cas1 homologs imply that these recombinases employ novel molecular mechanisms. Finally, it is perhaps worth noting that notwithstanding the apparent evolutionary affinity of the Asgard archae with eukaryotes, there is nothing eukaryote-like in Asgard-specific CRISPR-Cas-related elements: these are wonders of the archaeal world. Experimental characterization of the derived CRISPR-Cas variants and unique Cas1 homologs of Asgard archaea should illuminate both the functional plasticity of CRISPR-Cas and Asgard biology.
Supplementary Material
Author Disclosure Statement
No competing financial interests exist.
Funding Information
K.S.M., Y.I.W., S.A.S., and E.V.K. are supported by the Intramural Research Program of the National Institutes of Health of the USA (National Library of Medicine). M.L. and Y.L. are supported by National Natural Science Foundation of China (Grant No. 91851105, 31970105 and 31700430).
Supplementary Material
References
- 1. Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol 2017;2:17092 DOI: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 2. Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 2016;353:aad5147 DOI: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 3. Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc Lond B Biol Sci 2016;371: DOI: 10.1098/rstb.2015.0496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hille F, Richter H, Wong SP, et al. The biology of CRISPR-Cas: backward and forward. Cell 2018;172:1239–1259 [DOI] [PubMed] [Google Scholar]
- 5. Makarova KS, Wolf YI, Koonin EV. The basic building blocks and evolution of CRISPR-cas systems. Biochem Soc Trans 2013;41:1392–1400. DOI: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 2020;18:67–83. DOI: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol 2014;12:317–326. DOI: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 8. Faure G, Makarova KS, Koonin EV. CRISPR-Cas: complex functional networks and multiple roles beyond adaptive immunity. J Mol Biol 2019;431:3–20. DOI: 10.1016/j.jmb.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 9. Peters JE, Makarova KS, Shmakov S, et al. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc Natl Acad Sci U S A 2017;114:E7358–E7366. DOI: 10.1073/pnas.1709035114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faure G, Shmakov SA, Yan WX, et al. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 2019;17:513–525. DOI: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strecker J, Ladha A, Gardner Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019;365:48–53. DOI: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klompe SE, Vo PLH, Halpin-Healy TS, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 2019;571:219–225. DOI: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 13. Özcan A, Pausch P, Linden A, et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat Microbiol 2018;4:89–96. DOI: 10.1038/s41564-018-0274-8. [DOI] [PubMed] [Google Scholar]
- 14. Pinilla-Redondo R, Mayo-Muñoz D, Russel J, et al. Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res 2020;48:2000–2012. DOI: 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowley VM, Catching A, Taylor HN, et al. A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo. CRISPR J 2019;2:434–440. DOI: 10.1089/crispr.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Makarova KS, Karamycheva S, Shah SA, et al. Predicted highly derived class 1 CRISPR-Cas system in Haloarchaea containing diverged Cas5 and Cas7 homologs but no CRISPR array. FEMS Microbiol Lett 2019;366: DOI: 10.1093/femsle/fnz079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krupovic M, Koonin EV. Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems. Curr Opin Microbiol 2016; 31: 25–33. DOI: 10.1016/j.mib.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krupovic M, Makarova KS, Forterre P, et al. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biology 2014;12:36 DOI: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beguin P, Charpin N, Koonin EV, et al. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res 2016;44:10367–10376. DOI: 10.1093/nar/gkw821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hickman AB, Dyda F. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res 2015;43:10576–10587. DOI: 10.1093/nar/gkv1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hickman AB, Kailasan S, Genzor P, et al. Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas. Elife 2020;9 DOI: 10.7554/eLife.50004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hudaiberdiev S, Shmakov S, Wolf YI, et al. Phylogenomics of Cas4 family nucleases. BMC Evol Biol 2017;17:232 DOI: 10.1186/s12862-017-1081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spang A, Saw JH, Jørgensen SL, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015;521:173–179. DOI: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017;541:353–358. DOI: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 25. Williams TA, Cox CJ, Foster PG, et al. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol 2020;4:138–147. DOI: 10.1038/s41559-019-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MacLeod F, Kindler GS, Wong HL, et al. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol 2019;5:48–61. DOI: 10.3934/microbiol.2019.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai M, Liu Y, Yin X, et al. Diverse Asgard archaea including the novle phylum Gerdarchaeota participate in organic matter degradation. Sci China Life Sci 2020 Mar 16 [Epub ahead of print]; DOI: 10.1007/sl1427-020-1679-1 [DOI] [PubMed] [Google Scholar]
- 28. Imachi H, Nobu MK, Nakahara N, et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 2020;577:519–525. DOI: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Makarova KS, Wolf YI, Koonin EV. Archaeal Clusters of Orthologous Genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 2015;5:818–840. DOI: 10.3390/life5010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makarova KS, Wolf YI, Koonin EV. Towards functional characterization of archaeal genomic dark matter. Biochem Soc Trans 2019;47:389–398. DOI: 10.1042/BST20180560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marchler-Bauer A, Derbyshire MH, Gonzales NR, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res 2015;43:D222–226. DOI: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005;21:951–960. DOI: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 34. Drozdetskiy A, Cole C, Procter J, et al. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 2015;43:W389–394. DOI: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 2018;1:325–336. DOI: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harrington LB, Burnstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018;362:839–842. DOI: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Steinegger M, Soding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 2017;35:1026–1028. DOI: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 38. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meier A, Soding J. Context similarity scoring improves protein sequence alignments in the midnight zone. Bioinformatics 2015;31:674–681. DOI: 10.1093/bioinformatics/btu697. [DOI] [PubMed] [Google Scholar]
- 40. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490 DOI: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bland C, Ramsey TL, Sabree F, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 2007;8:209 DOI: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shmakov SA, Sitnik V, Makarova KS, et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. MBio 2017;8 DOI: 10.1128/mBio.01397-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 2011;6:38 DOI: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krupovic M, Beguin P, Koonin EV. Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr Opin Microbiol 2017;38:36–43. DOI: S1369-5274(16)30171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Makarova KS, Wolf YI, Forterre P, et al. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles 2014;18:877–893. DOI: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walker JE, Saraste M, Runswick MJ, et al. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1982;1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 2017;15:169–182. DOI :10.1038/nrmicro.2016.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol Evol 2017;9:2812–2825. DOI: 10.1093/gbe/evx192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans R Soc Lond B Biol Sci 2019;374:20180087 DOI: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yan WX, Hunnewell P, Alfonse LE, et al. Functionally diverse type V CRISPR-Cas systems. Science 2018;363:88–91. DOI: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 2014;38:865–891. DOI: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.