Abstract
Depletion of CD4+ T cells during HIV-1 infection is mostly mediated by inflammatory cells via indirect but not clearly defined mechanisms. In this report, we used single-cell RNA-Seq (scRNA-Seq) technology to study HIV-induced transcriptomic change in innate immune cells in lymphoid organs. We performed scRNA-Seq on hCD45+hCD3–hCD19– human leukocytes isolated from spleens of humanized NOD/Rag2–/–γc–/– (NRG) mice transplanted with human CD34+ hematopoietic stem progenitor cells (NRG-hu HSC mice). We identified major populations of innate immune cells, including plasmacytoid dendritic cells (pDCs), myeloid dendritic cells (mDCs), macrophages, NK cells, and innate lymphoid cells (ILCs). HIV-1 infection significantly upregulated genes involved in type I IFN inflammatory pathways in each of the innate immune subsets. Interestingly, we found that TRAIL was upregulated in the innate immune populations, including pDCs, mDCs, macrophages, NK cells, and ILCs. We further demonstrated that blockade of the TRAIL signaling pathway in NRG-hu HSC mice prevented HIV-1–induced CD4+ T cell depletion in vivo. In summary, we characterized HIV-induced transcriptomic changes of innate immune cells in the spleen at single-cell levels, identified the TRAIL+ innate immune cells, and defined an important role of the TRAIL signaling pathway in HIV-1–induced CD4+ T cell depletion in vivo.
Keywords: AIDS/HIV
Keywords: Innate immunity, T cells
Investigation of HIV-induced transcriptomic changes of innate immune cells in the spleen at single-cell level identified TRAIL+ innate cells in CD4+ T cell depletion.
Introduction
HIV-1 infection leads to depletion and dysregulation of CD4+ T cells in patients (1). It has been reported that aberrant activation and dysregulation of innate immune responses contribute to systemic immune activation and CD4+ T cell depletion. In particular, HIV-1 infection leads to persistent activation of plasmacytoid dendritic cells (pDCs) and sustained IFN-I production in vivo that contributes to immune activation and CD4+ T cell depletion as well as T cell exhaustion (2–9). HIV-1 infection–induced monocyte activation is associated with both AIDS- and non-AIDS-defining diseases (10). Chronic HIV-1 infection also alters the population distribution and functional capacity of NK cells (11). In addition, we and others have reported that HIV-1 infection causes depletion and functional impairment of innate lymphoid cells (ILCs), which may contribute to systemic immune activation and CD4+ T cell depletion (12–14). These studies suggest that a dysregulated innate immune response plays an important role in HIV-1 pathogenesis. Further understanding of how HIV infection leads to alterations of innate immune cells is critical for the development of new therapeutic interventions of HIV-associated diseases.
Mice reconstituted with a functional human immune system provide a valuable platform to study HIV-1 infection, pathogenesis, and therapy in vivo (15–18). We and others have reported that major populations of human innate immune cells, including pDCs, myeloid dendritic cells (mDCs), monocytes/macrophages, and ILCs, are developed in NOD/SCID γc−/− (NSG) and NOD/Rag2–/–γc–/– (NRG) mice transplanted with human CD34+ hematopoietic stem progenitor cells (NSG- and NRG-hu HSC) or human HSCs and thymus tissue (hu-BLT and hu-Thy/HSC) (5, 19–22). Importantly, the innate immunity of humanized mice is functionally similar to that of human or nonhuman primates in vitro and in vivo (21, 22). Moreover, we have demonstrated recently that depletion of human pDCs or blocking the IFN-I signaling pathway rescues T cell number and function during HIV-1 infection in vivo, even in the presence of higher levels of viral replication (3, 8). These studies support the use of humanized mice to study human innate immune responses to HIV-1 infection in vivo and investigate their contribution to HIV-1 pathogenesis.
Advances in single-cell RNA-Seq (scRNA-Seq) have enabled novel comprehensive analysis of the immune system in an unbiased way at the single-cell level (23, 24). In the present report, we used scRNA-Seq to identify and reconstruct the transcriptomic change in innate immune cells from HIV-1–infected NRG-hu HSC mice at single-cell levels in lymphoid organs. Our findings reveal that HIV-1 infection significantly changed transcriptomic profiles of each innate immune subset. Moreover, we discovered that the TRAIL was significantly upregulated in subsets of the innate immune populations and demonstrated that TRAIL signaling functionally contributes to CD4+ T cell depletion in vivo.
Results
scRNA-Seq analysis of human CD45+CD3–CD19– leukocytes from the spleens of NRG-hu HSC mice.
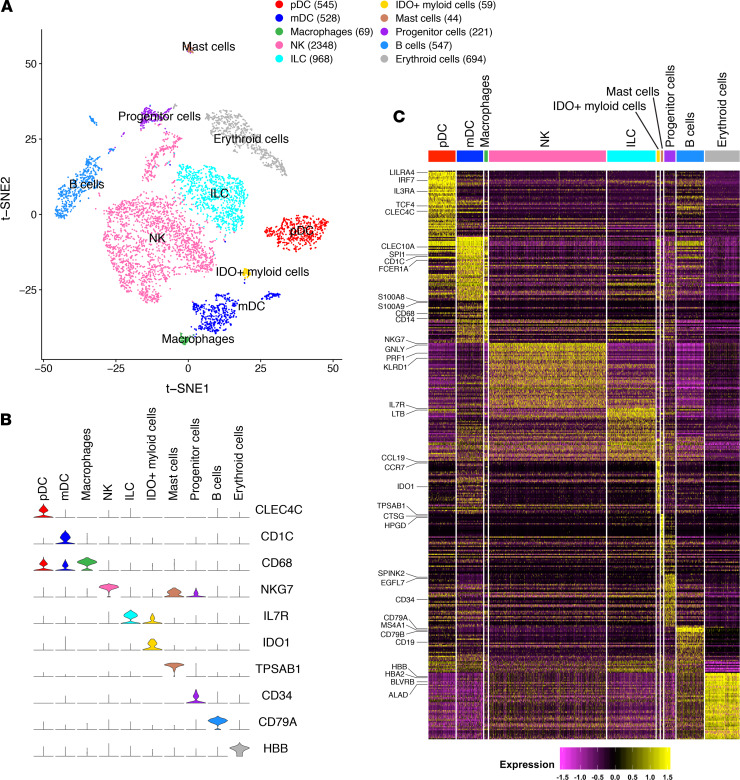
To characterize the human innate immune cells and their transcriptomic profiles developed in humanized mice in a comprehensive and unbiased way, we performed scRNA-Seq on human CD45+CD3–CD19– cells isolated from spleens of NRG-hu HSC mice. We obtained single-cell transcriptomes of 6023 human innate immune cells from 2 mice and performed clustering analyses to examine cellular characteristics and heterogeneity. We analyzed the expression difference of genes between each single cluster and all other cells to identify cluster marker genes. We used t-distributed stochastic neighbor embedding (t-SNE) visualization of cells to reveal 10 major clusters (Figure 1A), based on marker gene expression for pDCs (e.g., enrichment of CLEC4C, TCF4) (25), mDCs (CD1C, SPI1) (26), macrophages (CD68, CD14) (27), NK cells (NKG7, PRF1) (28), ILCs (IL7R) (29), IDO+ myeloid cells (IDO1), mast cells (TPSAB1, CTSG) (30), CD34+ progenitor cells (CD34), B cells (CD79A, CD19, due to incomplete depletion by the CD19 antibody), and erythroid cells (HBB, HBA2) (31), which may not have been completely lysed by the red blood cell lysis procedure (Figure 1, B and C; and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.135344DS1). The results indicate that major human innate immune subsets were developed in NRG-hu HSC mice, consistent with their responses to pathogen-associated molecular patterns, which are similar to those of human leukocytes (21).
Figure 1. scRNA-Seq analysis of human CD45+CD3–CD19– immune cells in spleens of NRG-hu HSC mice.
(A) t-SNE analysis of hCD45+hCD3–hCD19– splenocyte clusters from NRG-hu HSC mice based on 6023 single-cell transcriptomes. Cell counts for plasmacytoid dendritic cells (pDC), myeloid dendritic cells (mDC), macrophages, NK cells (NK), innate lymphoid cells (ILC), IDO+ myeloid cells, mast cells, CD34+ progenitor cells, B cells, and erythroid cells are indicated in parentheses. (B) Violin plots showing expression of representative marker genes for each cell type cluster. (C) Heatmap of marker gene expression in individual cells of each cell type cluster.
HIV-1 infection significantly alters the transcriptomic profile of human innate immune cells from spleens of NRG-hu HSC mice.
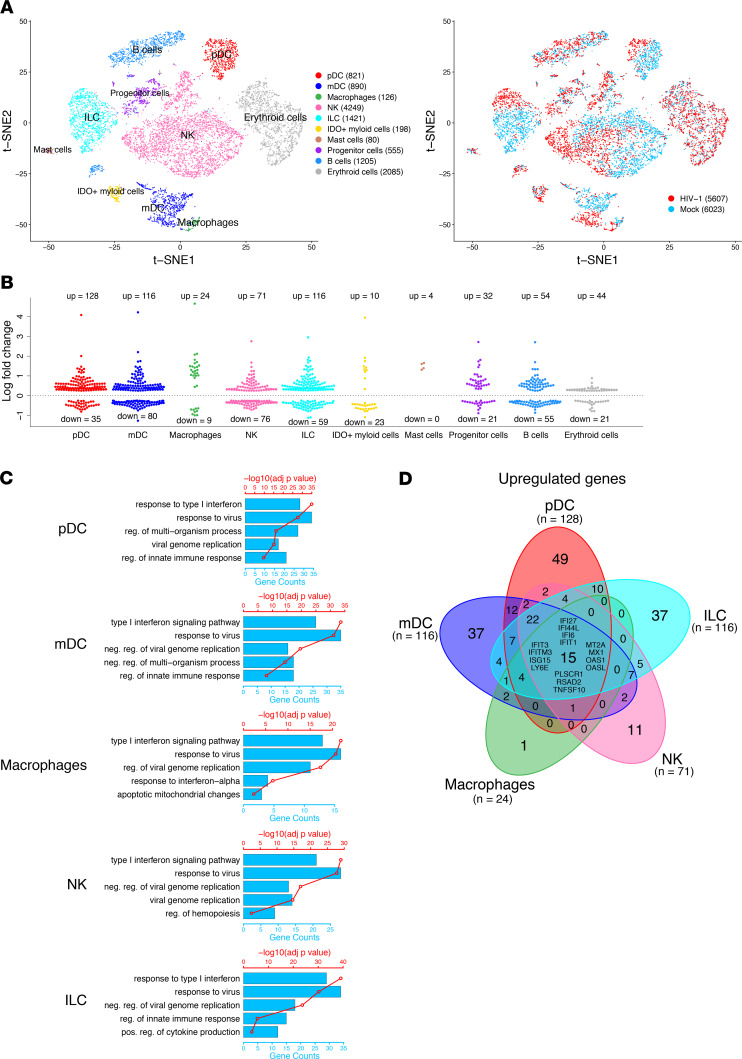
We then determined how HIV-1 infection changed the innate immune cells by detecting their transcriptome at the single-cell level. We performed scRNA-Seq with human CD45+CD3–CD19– cells isolated from spleens of NRG-hu HSC mice 3 weeks after HIV-1 infection. We obtained a total of 5607 human single-cell transcriptomes from 2 HIV-1–infected mice and then compared them with the 6023 cells from the 2 mock control mice. By t-SNE analysis of cells from the mock and HIV-1–infected mice, we detected all 10 clusters of human innate immune cells derived from both mock and HIV-1–infected mice (Figure 2A). HIV-1 infection changed the transcriptome of human innate immune cells, including pDC (128 upregulated genes and 35 downregulated genes), mDC (116 upregulated genes and 80 downregulated genes), macrophage (24 upregulated genes and 9 downregulated genes), NK cell (71 upregulated genes and 76 downregulated genes), and ILC populations (116 upregulated genes and 59 downregulated genes) at the single-cell level, as indicated by the segregation and differential gene expression of HIV-1–infected cells and mock control cells (Figure 2A, right; Figure 2B; and Supplemental Table 2). By Gene Ontology (GO) enrichment profile analysis, we showed that most of the upregulated genes in the innate immune subsets are related to the type I IFN signaling pathway, response to virus, and negative regulation of viral replication (Figure 2C). By Venn diagram analysis of genes differentially modulated by HIV-1 infection, we showed that there were 15 genes that were upregulated in all innate immune cell subsets, and all 15 genes are IFN-stimulated genes (Figure 2D). These results indicate that HIV-1 infection triggered IFN-I signaling in each innate immune cell subset, consistent with our previous reports that IFN-I plays a major role in HIV-1 pathogenesis (3, 4, 12, 14). Interestingly, the pathways downregulated by HIV-1 infection in each of the subsets were related to cellular metabolic processes, suggesting that HIV-1 infection alters the metabolism of human innate immune cells in vivo (Supplemental Figure 1).
Figure 2. HIV-1–induced alterations of human innate immune cells by scRNA-Seq analysis of human cells in spleens of NRG-hu HSC mice.
NRG-hu HSC mice were infected with HIV-1 and terminated at 3 weeks after infection. Splenocytes were enriched for hCD45+hCD3–hCD19– cells for single-cell transcriptomics assay. (A) t-SNE analysis of hCD45+hCD3–hCD19– innate immune clusters from spleens of 2 mock and 2 HIV-1–infected NRG-hu HSC mice reconstituted with CD34+ cells from the same donor. Different colors indicate distinct cell types on the left. Blue dots indicate cells from mock mice, while red dots indicate cells from HIV-1–infected mice on the right. (B) Distribution of upregulated and downregulated genes in HIV-1–infected NRG-hu HSC mice compared with mock control mice for each cell type. (C) GO term analysis of pathways upregulated by HIV-1 in pDCs, mDCs, macrophages, NK cells, and ILCs. The red points connected by a red line indicate the –log10 Benjamini-Hochberg adjusted P values for enrichment of the upregulated genes for the specified biological process with the scale on the top axis. The blue bars indicate the number of upregulated genes found in the specified biological process, with the scale on the bottom axis. (D) Venn diagram showing overlap of genes upregulated by HIV-1 for each of the 5 innate immune subsets. The differentially modulated genes common to all 5 innate immune subsets are listed in the center region of each Venn diagram.
HIV-1 infection induces TRAIL expression on all major innate immune subpopulations.
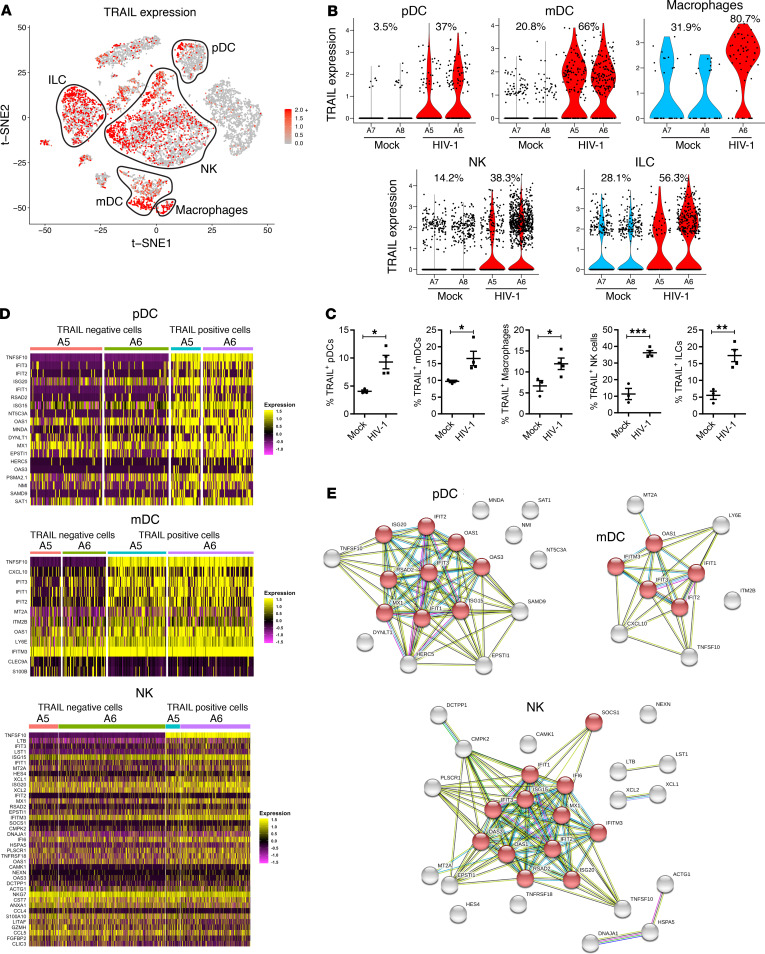
The hallmark of AIDS pathogenesis is a progressive depletion of CD4+ T cells. However, the mechanism by which HIV-1 infection leads to CD4+ T cell depletion in vivo is still not clearly defined. We thus determined whether and how HIV-1 infection–induced changes in innate immune cells contribute to CD4+ T cell depletion. Interestingly, we found that the TNF superfamily member 10 (TNFSF10) gene, which encodes TRAIL, was significantly induced by HIV-1 in pDCs, mDCs, macrophages, and NK cells as well as ILC cells (Figure 3, A and B, and Supplemental Table 2). TRAIL was among the top 25 differentially expressed genes in each of the major innate immune subsets (Supplemental Figure 2). The percentage of TRAIL+ pDCs increased from 3.5% in mock mice to 37% in HIV-1–infected mice (Figure 3B). HIV-1 infection also led to a 2- to 3-fold increase of TRAIL expression in mDCs (from 21% to 66%), macrophages (from 32% to 81%), NK cells (from 14% to 38%), and ILCs (from 28% to 56%) (Figure 3B). We did not observe induction of other cell death ligands, such as FAS ligand (FASLG) and TNF-α in innate immune cells by scRNA-Seq (Supplemental Figure 3). Using flow cytometry, we confirmed the upregulation of TRAIL protein expression in pDCs, mDCs, macrophages, NK cells, and ILCs after HIV-1 infection (Figure 3C and Supplemental Figure 4). No significant induction of soluble TRAIL was detected in the plasma of NRG-hu HSC mice by HIV-1 infection (Supplemental Figure 5).
Figure 3. HIV-1 infection induces TRAIL expression in a subset of all innate immune cells.
(A) TRAIL (TNFSF10) expression in human innate immune cells from mock and HIV-1–infected mice by t-SNE visualization. (B) Violin plots of normalized unique molecular identifier (UMI) showing distribution of TRAIL (TNFSF10) gene expression in different innate immune cells from mock or HIV-1–infected mice. A5, A6, A7, and A8 are the mouse identification numbers; the percentages shown on the violin plots correspond to percentage of TRAIL+ cells. No macrophage cluster in mouse A5 due to the limited input cell number and low percentage of macrophages in this sample. (C) TRAIL protein expression detected by flow cytometry on each of the innate immune cell subsets from spleens of mock and HIV-1–infected mice 5 weeks after HIV-1 infection. Mock, n = 3; HIV-1, n = 4. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, by 2-tailed Student’s t test. (D) Heatmaps showing differentially expressed genes between TRAIL+ and TRAIL– cells in HIV-1–infected mice for pDCs, mDCs, and NK cells. (E) STRING association networks of upregulated genes in TRAIL+ cells in HIV-1–infected mice for pDC, mDC, and NK cell types. Red nodes indicate proteins found in the IFN-α/β (IFN-I) signaling pathway.
We next determined whether TRAIL-expressing innate immune cells had a unique transcriptome compared with TRAIL– cells in spleens of HIV-1–infected mice. Differential gene expression analysis of TRAIL+ and TRAIL– cells revealed that TRAIL+ cells expressed higher levels of IFN-stimulated genes, such as IFIT3, ISG15, and other IFN-stimulated genes, among pDCs, mDCs, and NK cells (Figure 3D and Supplemental Table 3). STRING network analysis (32) of the upregulated genes in TRAIL+ cells revealed that IFN-I signaling pathways were preferentially activated in TRAIL-expressing pDCs, mDCs, and NK cells (Figure 3E), while the gene transcription between TRAIL+ and TRAIL– macrophages and TRAIL+ and TRAIL– ILCs was not significantly changed (Supplemental Table 3).
Blockade of TRAIL/DR5 signaling prevents CD4+ T cell depletion induced by HIV-1 infection in vivo.
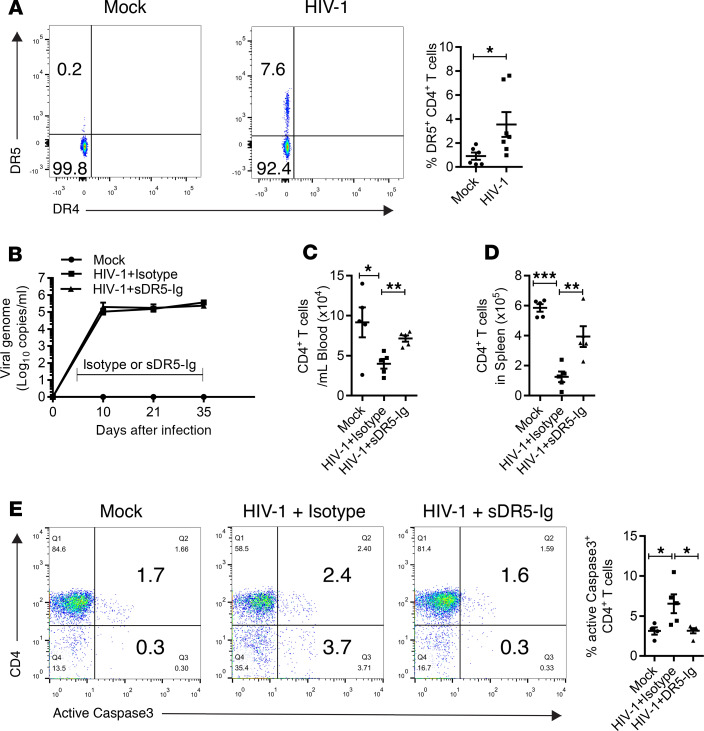
In a previous study, we showed that HIV-1 infection induced CD4+ T cell apoptosis in vivo, as indicated by active caspase-3 expression (2). TRAIL is a proapoptotic ligand with an immune effector function to promote the eradication of infected or malignant cells (33). The data above suggest that HIV-induced TRAIL expression on innate immune cells may contribute to CD4+ T cell depletion in lymphoid organs. We thus checked whether the death receptors of TRAIL were induced on CD4+ T cells after HIV-1 infection in vivo. We found that HIV-1 infection upregulated the expression of TRAIL receptor death receptor 5 (DR5) but not death receptor 4 (DR4) on CD4+ T cells from spleens of humanized mice (Figure 4A). To block the TRAIL/DR5 signaling axis, we used a soluble form of DR5 fused with human IgG-Fc (sDR5-Ig) that could prevent TRAIL-induced cell death (34–36). We treated HIV-1–infected NRG-hu HSC mice with sDR5-Ig twice a week starting from day 5 through day 35 after infection and analyzed whether the treatment affected HIV-1 replication and prevented CD4+ T cell depletion in vivo. Although sDR5-Ig treatment did not affect HIV-1 replication in vivo (Figure 4B), it efficiently prevented HIV-induced depletion of CD4+ T cells and CD8+ T cells from both peripheral blood and spleens (Figure 4, C and D; and Supplemental Figure 6A). Furthermore, we detected elevated expression of active caspase-3, a surrogate marker of apoptosis, in CD4+ T cells after HIV-1 infection. We found that sDR5-Ig significantly reduced HIV-induced caspase-3 activation in CD4+ T cells from the spleen, indicating that blocking the TRAIL/DR5 signaling pathway prevented CD4+ T cell apoptosis (Figure 4E). However, the sDR5-Ig treatment did not rescue the function of T cells, which was impaired by HIV-1 infection (Supplemental Figure 6B). In summary, we identified at the single-cell level that TRAIL was upregulated in multiple human innate immune subsets and demonstrated that activation of the TRAIL signaling pathway contributed to CD4+ T cell depletion during HIV-1 infection in vivo.
Figure 4. Blocking TRAIL signaling in vivo prevents HIV-1–induced CD4+ T cell depletion.
(A) Representative FACS plots and summarized data show expression of DR5 on CD4+ T (CD3+CD8–) cells from mock and HIV-1–infected mice 3–5 weeks after HIV-1 infection. Combined data from 2 independent experiments. Mock, n = 5; HIV-1, n = 7. Data represent mean ± SEM. *P < 0.05, by 2-tailed Student’s t test. (B–E) NRG-hu HSC mice infected with HIV-1 were treated with sDR5-Ig or control Ig twice a week (200 μg/mouse, intraperitoneal injection) from 5 to 35 days after infection. (B) Plasma HIV-1 RNA levels are shown at indicated time points after HIV-1 infection. (C and D) Numbers of CD4+ T cells (CD3+CD8–) in peripheral blood (C) and spleens (D) at termination (day 35) are summarized. (E) Representative FACS plots and summarized data showing the percentage of splenic CD4+ T cells (CD3+CD8–) expressing active caspase-3. Mock, n = 5; HIV-1+control Ig, n = 5; HIV-1+DR5-Ig, n = 5. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, by 1-way ANOVA and Bonferroni’s post hoc test.
Discussion
The mechanisms by which HIV infection depletes CD4+ T cells and damages the immune system are not clearly defined. They involve multiple innate immune cells as well as adaptive immune cells. Advances in scRNA-Seq technology have allowed for comprehensive analysis of the immune system in an unbiased way at the single-cell level. In the present study, we investigated how HIV-1 infection changed the transcriptome of human innate immune cells in lymphoid organs in NRG-hu HSC mice at the single-cell level by scRNA-Seq. We report that HIV-1 infection significantly changed the transcriptome of each of the innate immune cell subsets. Most interestingly, we found that the TRAIL was significantly upregulated in the innate immune populations and blockade of the TRAIL signaling pathway prevented HIV-1–induced CD4+ T cell depletion in vivo.
HIV-1 infection in humanized mice provides a valuable platform to study HIV-1 disease or AIDS pathogenesis in vivo. We and others have reported that major human innate immune populations, including pDCs, mDCs, and monocytes as well as ILCs, are functionally developed in both NRG-hu HSC and NRG-hu Thy/HSC mice (5, 20–22). In the present study with scRNA-Seq, we identified major subsets of human innate immune cells in the spleens of NRG-hu HSC mice. These results further support the use of NRG-hu HSC mice to study human innate immunity in vivo.
How HIV-1 infection changes the function of human immune cells in lymphoid organs in vivo at the single-cell level is not clearly documented yet. We report here, using scRNA-Seq, that HIV-1 infection significantly changed the transcriptome of each of the human innate immune cells in lymphoid organs. GO analysis showed that the IFN signaling pathway was significantly upregulated in each of the innate immune subsets during HIV-1 infection. We have reported previously that persistent pDC activation and IFN-I signaling lead to T cell depletion and dysfunction in vivo (2, 3, 8). These innate immune cells altered by HIV-1 infection may indirectly contribute to CD4+ T cell depletion and HIV-1 pathogenesis.
Previous studies suggest that most of the dying CD4+ T cells in lymphoid organs of HIV-infected hosts are bystander CD4+ T cells that were not directly infected (37). Various mechanisms have been proposed to account for the death of these bystander cells, including apoptosis induced by various host factors (e.g., FASLG, TNF-α, and TRAIL) (7, 34, 37–39). Treatment of human dendritic cells and CD4+ T cells with HIV-1 in vitro induces TRAIL expression (38, 39), and TRAIL-expressing cells are reported to induce CD4+ T cell apoptosis in vitro (7) and in HIV-1–infected hu-PBL-NOD/SCID mice (40). Previous studies in SIV-infected nonhuman primates suggest that TNF-α signaling may lead to CD4+ T cell depletion in lymphoid tissues (41) and that blockade of IFN-I signaling during the acute phase of SIV infection leads to higher levels of virus replication and accelerated CD4+ T cell depletion (42). However, we and others have reported that blocking IFN-I signaling during chronic HIV-1 infection in humanized mice rescues CD4+ T cells and enhances anti–HIV-1 immunity (2–4) and blockade of IFN-I signaling during chronic SIV infection suppresses specific inflammatory pathways (43). Recent studies suggest that cell death by pyroptosis may also drive CD4+ T cell depletion during HIV-1 infection in vitro (44). However, the role of pyroptosis in CD4+ T cell depletion during HIV-1 infection in vivo is not clear. We report here, using scRNA-Seq technology, that HIV-1 infection significantly upregulated TRAIL in a subset of the innate immune populations, including pDCs, mDCs, macrophages, NK cells, and ILC cells. Importantly, we demonstrate that blockade of the TRAIL/DR5 signaling pathway in vivo by a soluble DR5 fusion protein prevented HIV-1–induced CD4+ T cell depletion in NRG-hu HSC mice. It is possible that the DR5-Ig fusion protein may also trigger FcR-dependent elimination of TRAIL-expressing cells in vivo. Interestingly, the rescued T cells were still impaired in function. These results suggest that the TRAIL signaling pathway plays a critical role in HIV-1–induced CD4+ T cell apoptosis in vivo. It is reported that TRAIL is an IFN-I–stimulated gene (38, 45). We observed that TRAIL-expressing innate immune cells in HIV-1–infected mice also preferentially expressed IFN-I signature genes. It will be interesting to identify which subsets of TRAIL+ innate cells play a major role in inducing CD4+ T cell death in vivo in future studies. It has been reported that plasma levels of soluble TRAIL are increased in patients infected with HIV (39). Furthermore, an alternative spliced form of TRAIL named TRAIL-short can be induced by HIV-1 infection and is reported to protect against CD4+ T cell death during acute HIV-1 infection (46, 47). In the present study, we did not observe any significant induction of soluble TRAIL in the plasma of NRG-hu HSC mice at 3–5 weeks after HIV-1 infection. Our results suggest that surface TRAIL on innate immune cells in the spleen may predominantly contribute to CD4+ T cell depletion in HIV-1–infected humanized mice. It is possible that soluble TRAIL may only be produced during long persistent HIV infection. It will be interesting to further explore the mechanisms in humanized mice in future studies. In addition, the CD4+ T cell number from HIV-1–infected mice treated with sDR5-Ig, although increased in comparison to isotype control treatment group, was still lower than that in mock mice, which suggests that mechanisms other than the TRAIL pathway may also contribute to CD4+ T cell depletion in vivo (41).
It is important to point out, however, that the human immunity developed in humanized mice is not fully functional, as found in immunocompetent hosts (18, 48). The human T cells developed in NRG-hu HSC mice are produced in the mouse thymus and presumed to be educated predominately in the context of mouse MHC (49–52). Thus, it is not suitable to study the effect of TRAIL blockade on the number and function of human MHC-restricted anti-HIV T cells. In addition, B cells and gut-associated lymphoid tissues are not fully developed in humanized mice, which has limited the general application of humanized mice.
In summary, we characterized human innate immune cell subsets in lymphoid organs by scRNA-Seq during HIV-1 infection in vivo. We identified the pathogenic TRAIL+ innate immune cells that contributed to HIV-1 immunopathogenesis in vivo and functionally proved that the TRAIL/DR5 signaling pathway contributed to CD4+ T cell depletion by HIV-1 in vivo. Our study has not only established the foundation to study human immunopathogenic mechanisms by scRNA-Seq in humanized mice, but also identified a pathogenic pathway of HIV-1 immunopathogenesis in vivo.
Methods
Generation of NRG-hu HSC mice.
NRG mice were obtained from The Jackson Laboratory. Humanized NRG-hu HSC mice were generated by intrahepatic injection of 2 × 105 CD34+ hematopoietic progenitor cells derived from human fetal liver and transplanted into newborn (2–5 days old) NRG mice as previously reported (8). Engraftment of human immune cells in peripheral blood was detected by flow cytometry 10–12 weeks after transplantation. Mice with human reconstitution levels higher than 20% (human CD45+ cells) were used. All mice were housed and bred in a specific pathogen–free environment.
HIV-1 infection of humanized mice.
The HIV-1 JR-CSF Infectious Molecular Clone (pYK-JRCSF) (catalog 2708) was obtained through the NIH AIDS Reagent Program. The virus was generated by transfection of 293T cells (ATCC) with the pYK-JRCSF plasmid. Mice were anesthetized and infected with 10 ng p24/mouse HIV-1 (JR-CSF) (generated in 293T cells as stated above) through retro-orbital injection.
Isolation of mCD45–hCD3–hCD19–hCD45+ splenocytes.
2 × 106 splenocytes from humanized mice 3 weeks after HIV-1 or mock infection were first incubated with biotin-labeled anti-mouse CD45 (0.5 μg/106 cells/50 μl) and anti-human CD3 (0.5 μg/106 cells/50 μl) and CD19 (0.5 μg/106 cells/50 μl) antibodies in staining buffer (1× PBS with 2% FBS and 2 mM EDTA) on ice for 20 minutes. The cells were then washed and resuspended in 90 μl staining buffer and incubated with 10 μl Streptavidin MicroBeads (Miltenyi Biotech) for 20 minutes for negative selection using LS columns by manual MACS (Miltenyi Biotech) per the manufacturer’s recommendations. The mCD45–hCD3–hCD19– population of splenocytes was then incubated with biotin-labeled anti-human CD45 antibody (0.5 μg/106 cells/100 μl) for 20 minutes, washed, and resuspended in staining buffer and incubated with 10 μl Streptavidin MicroBeads for 20 minutes, and then positive selected using MS columns by manual MACS (Miltenyi Biotech) per the manufacturer’s recommendations. Following selection, the cell number of each sample was manually counted and adjusted to 1 × 106 cells/mL in 1× PBS with 0.04% bovine serum albumin for subsequent scRNA-Seq via 10× Genomics platform.
Single-cell transcriptome library preparation and sequencing.
The single-cell transcriptome library was prepared per the manufacturer’s recommendations. Briefly, single cells were loaded to 10× Genomics Chromium chip (10× Genomics) to generate single-cell Gel Bead-in-emulsion (GEM). scRNA-Seq libraries were prepared using GemCode Single-Cell Gel Bead and Library Kit (10× Genomics). GEM reverse transcription was performed with the following conditions: 55°C for 2 hours, 85°C for 5 minutes; held at 4°C. After reverse transcription, GEMs were broken, and the single-strand cDNA was cleaned up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific) and the SPRIselect Reagent Kit (0.6× SPRI; Beckman Coulter). cDNA was amplified with the following condition: 98°C for 3 minutes; 14 cycles of 98°C for 15 seconds, 67°C for 20 seconds, and 72°C for 1 minute; 72°C for 1 minute; held at 4°C. cDNA product was cleaned up using the SPRIselect Reagent Kit. Indexed sequencing libraries were constructed with the reagents in the GemCode Single-Cell 3’ Library Kit (10× Genomics) (53). Sequencing were performed on Illumina NextSeq 500 with NextSeq 500/550 v2.5 kits.
Quality control and data analysis of scRNA-Seq.
Raw sequencing reads were processed and mapped to the hg19 human reference transcriptome using the Cell Ranger version 1.1.0 pipeline from 10× Genomics for individual scRNA-Seq data sets: 2 HIV-1–infected and 2 mock samples from NRG-hu HSC mice. The Seurat (version 3) R package (54, 55) was used to analyze the scRNA-Seq data.
First each sample data set was analyzed separately in Seurat. The processed Unique Molecular Identifier count matrix was loaded in Seurat. Genes with counts in fewer than 3 cells were excluded. We filtered low-quality cells by removing cells with unique gene counts over 2500 or less than 200, along with cells with > 5% mitochondrial counts. The data were log normalized using the Seurat NormalizeData function with the scale factor set to the default value of 10,000. Supplemental Table 4 lists the genes detected in the scRNA-Seq analysis. The 2000 variable genes were determined using the Seurat FindVariableFeatures function. The Seurat ScaleData function was used to scale the data and remove unwanted sources of heterogeneity by regressing on the number of molecules per cell and the percentage of mitochondrial gene content. Principle component analysis was conducted using the variable genes. The cells were clustered using the Seurat FindNeighbors function using the first 15 principle components, followed by the Seurat FindClusters function using a resolution of 0.8. t-SNE analysis was performed using the first 15 principle components to allow for the visualization of the clusters in a t-SNE plot. The average gene expression (calculated using the Seurat AverageExpression function) of known cell type–specific markers for each cluster was used to manually assign a cell type to each cluster.
Following this individual sample analysis, we merged different combinations of these samples for further analysis: (a) the 2 mock samples from NRG-hu HSC mice and (b) 2 HIV-1–infected and 2 mock samples from NRG-hu HSC mice. After loading the individual sample data sets into Seurat, the data sets were merged using Seurat’s merge function. The cell type assignment from the Seurat analysis of the individual samples was added to the merged data object for each cell. Normalization, scaling, and t-SNE analysis of the merged data object were performed as described above. Markers for each cell type were determined using the Seurat FindAllMarkers function using a Wilcoxon rank-sum test with default parameter values, except that min.pct was set to 0.25 and only.pos was set to TRUE. The Seurat FindMarkers function using a Wilcoxon rank-sum test was used to identify differentially expressed genes between HIV-1 and mock populations from a particular cell type as well as between TRAIL+ (TNFSF10) and TRAIL– HIV-1 cells from a particular cell type. Markers with an adjusted P value less than 0.05 were classified as significant.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (56) (GSE148796).
GO analysis.
GO enrichment analysis for the biological process class was performed on the sets of differentially expressed genes between HIV-1 and mock NRG-hu HSC mouse samples using the enrichGO function in the clusterProfiler (version 3.12.0) R package (57). Redundant enriched GO terms were removed using the simplify function in clusterProfiler. GO terms were considered to be significantly enriched if the Benjamini-Hochberg corrected P value was less than 0.05.
STRING analysis.
STRING protein-protein association networks were constructed using STRING version 11.0 (32).
Flow cytometry.
FITC-conjugated anti–human CD14 (63D3) and –IFN-γ (4S.B3), PE-conjugated anti–human TRAIL (RIK-2) and –IL-2 (MQ1-17H12), PerCP/Cy5.5-conjugated anti–human CD3 (RPA-T4), –CD4 (RPA-T4), –CD11c (3.9), –CD14 (63D3), –CD16 (3G8), –CD19 (HIB19), –CD20 (2H7), –CD56 (5.1H11), –CD123 (6H6), PE/Cy7-conjugated anti–human CD4 (RPA-T4), –CD8 (HIT8a), APC-conjugated anti–human DR5 [DJR2-4 (7-8)], –CD11c (3.9), –CD56 (5.1H11), and APC/Cy7-conjugated anti–human CD45 (HI30) were purchased from BioLegend. PE-conjugated anti-human active caspase-3 (C92-605) was purchased from BD Pharmingen. Pacific orange–conjugated anti-mouse CD45 (30-F11), PE/Texas red–conjugated anti-human CD3 (7D6), –CD19 (SJ25-C1), –CD56 (MEM-188), and the LIVE/DEAD Fixable Yellow Dead Cell Stain Kit were purchased from Invitrogen. For surface staining, single-cell suspensions prepared from spleens of humanized mice were stained with surface markers and analyzed on a CyAn ADP flow cytometer (Dako). For intracellular staining, cells were first stained with surface markers and then fixed and permeabilized with cytofix/cytoperm buffer (BD Biosciences), followed by intracellular staining. Data were analyzed using Summit4.3 software (Dako).
HIV-1 genomic RNA detection in plasma.
HIV-1 RNA was extracted from the plasma with the QIAamp Viral RNA Mini Kit (QIAGEN). Then, the RNA was reverse transcribed and quantitatively detected by real-time PCR using the TaqMan Fast Virus 1-Step PCR kit (Thermo Fisher Scientific). The primers used for detecting the HIV Gag gene were (5′-GGTGCGAGAGCGTCAGTATTAAG-3′ and 5′-AGCTCCCTGCTTGCCCATA-3′). The probe (FAM-AAAATTCGGTTAAGGCCAGGGGGAAAGAA-QSY7) used for detection was purchased from Applied Biosystems. The reactions were set up following manufacturer’s guidelines and run on the QuantStudio 6 Flex PCR system (Applied Biosystems).
sDR5-Ig preparation.
The sDR5-Ig was generated by fusing the extracellular domain of DR5 (amino acids 58–184) with the constant region of IgG1, and expressed by transient transfection of 293T cells as described previously (35, 58).
Detection of soluble TRAIL.
Soluble TRAIL levels in frozen plasma of mock and HIV-1–infected mice 3 or 5 weeks after infection were detected by the Human TRAIL/TNFSF10 Quantikine ELISA Kit (R&D Systems). The samples 3 weeks after infection corresponded to the cell isolation and scRNA-Seq samples.
Statistics.
Statistical analysis for scRNA-Seq data was described above. In Figure 3C and Figure 4, significance levels of data were determined using Prism5 (GraphPad Software). Experiments were analyzed by 2-tailed Student’s t test or by 1-way ANOVA and Bonferroni’s post hoc test according to the assumptions of the test, as indicated for each experiment. A P value less than 0.05 was considered significant.
Study approval.
All animal studies were approved by the University of North Carolina Institutional Animal Care and Use Committee (ID 17-071) and were conducted following NIH guidelines for housing and care of laboratory animals. Human fetal livers (gestational age of 16–20 weeks) were obtained from medically indicated or elective termination of pregnancy through a nonprofit intermediary working with outpatient clinics (Advanced Bioscience Resources). Written informed consent of the maternal donors was obtained in all cases, under regulations governing the clinic. The project was reviewed by the University of North Carolina’s Office of Human Research Ethics, which determined that the submission does not constitute human subjects research, as defined under federal regulations [45 CFR 46.102 (d or f) and 21 CFR 56.102(c)(e)(l)].
Author contributions
LC, HY, and LS conceived and designed the study. LC, HY, and PL performed experiments. JAW, ZH, and LC analyzed the scRNA-Seq data. GL generated the humanized mice. XNX generated and provided the sDR5-Ig protein. LC, JAW, and LS analyzed and interpreted the data and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Y. Wu for technical support, the UNC Lineberger Translational Genomics Lab for all sequencing work and assistance on bioinformatics analysis, and the UNC flow cytometry core for assistance with flow cytometry. This study was supported in part by NIH grants (DK119937 and AI138797) to LS.
Version 1. 05/14/2020
In-Press Preview
Version 2. 06/04/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(11):e135344.https://doi.org/10.1172/jci.insight.135344.
Contributor Information
Liang Cheng, Email: chengl84@email.unc.edu.
Haisheng Yu, Email: yuhaisheng_ibp@hotmail.com.
John A. Wrobel, Email: john_wrobel@med.unc.edu.
Guangming Li, Email: gmli@email.unc.edu.
Peng Liu, Email: liupz@med.unc.edu.
Zhiyuan Hu, Email: zhiyuan_hu@med.unc.edu.
Xiao-Ning Xu, Email: x.xu@imperial.ac.uk.
Lishan Su, Email: lsu@med.unc.edu.
References
- 1.Moir S, Chun TW, Fauci AS. Pathogenic mechanisms of HIV disease. Annu Rev Pathol. 2011;6:223–248. doi: 10.1146/annurev-pathol-011110-130254. [DOI] [PubMed] [Google Scholar]
- 2.Cheng L, et al. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest. 2017;127(1):269–279. doi: 10.1172/JCI90745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng L, et al. Type I interferons suppress viral replication but contribute to T cell depletion and dysfunction during chronic HIV-1 infection. JCI Insight. 2017;2(12):94366. doi: 10.1172/jci.insight.94366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhen A, et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest. 2017;127(1):260–268. doi: 10.1172/JCI89488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Jiang Q, Li G, Jeffrey J, Kovalev GI, Su L. Efficient infection, activation, and impairment of pDCs in the BM and peripheral lymphoid organs during early HIV-1 infection in humanized rag2−/−γ C−/− mice in vivo. Blood. 2011;117(23):6184–6192. doi: 10.1182/blood-2011-01-331173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrat FJ, Su L. A pathogenic role of plasmacytoid dendritic cells in autoimmunity and chronic viral infection. J Exp Med. 2019;216(9):1974–1985. doi: 10.1084/jem.20181359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stary G, et al. Plasmacytoid dendritic cells express TRAIL and induce CD4+ T-cell apoptosis in HIV-1 viremic patients. Blood. 2009;114(18):3854–3863. doi: 10.1182/blood-2009-04-217927. [DOI] [PubMed] [Google Scholar]
- 8.Li G, et al. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog. 2014;10(7):e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su L. Pathogenic Role of type I interferons in HIV-induced immune impairments in humanized mice. Curr HIV/AIDS Rep. 2019;16(3):224–229. doi: 10.1007/s11904-019-00444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS. 2014;28(15):2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scully E, Alter G. NK cells in HIV disease. Curr HIV/AIDS Rep. 2016;13(2):85–94. doi: 10.1007/s11904-016-0310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Plasmacytoid dendritic cells promote HIV-1-induced group 3 innate lymphoid cell depletion. J Clin Invest. 2015;125(9):3692–3703. doi: 10.1172/JCI82124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kløverpris HN, et al. Innate lymphoid cells are depleted irreversibly during acute HIV-1 infection in the absence of viral suppression. Immunity. 2016;44(2):391–405. doi: 10.1016/j.immuni.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, et al. Infection and depletion of CD4+ group-1 innate lymphoid cells by HIV-1 via type-I interferon pathway. PLoS Pathog. 2018;14(1):e1006819. doi: 10.1371/journal.ppat.1006819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsden MD, Zack JA. Humanized mouse models for human immunodeficiency virus infection. Annu Rev Virol. 2017;4(1):393–412. doi: 10.1146/annurev-virology-101416-041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masse-Ranson G, Mouquet H, Di Santo JP. Humanized mouse models to study pathophysiology and treatment of HIV infection. Curr Opin HIV AIDS. 2018;13(2):143–151. doi: 10.1097/COH.0000000000000440. [DOI] [PubMed] [Google Scholar]
- 17.Victor Garcia J. Humanized mice for HIV and AIDS research. Curr Opin Virol. 2016;19:56–64. doi: 10.1016/j.coviro.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Su L. HIV-1 immunopathogenesis in humanized mouse models. Cell Mol Immunol. 2012;9(3):237–244. doi: 10.1038/cmi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng L, Ma J, Li G, Su L. Humanized mice engrafted with human HSC only or HSC and thymus support comparable HIV-1 replication, immunopathology, and responses to ART and immune therapy. Front Immunol. 2018;9:817. doi: 10.3389/fimmu.2018.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melkus MW, et al. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med. 2006;12(11):1316–1322. doi: 10.1038/nm1431. [DOI] [PubMed] [Google Scholar]
- 21.Cheng L, et al. Human innate responses and adjuvant activity of TLR ligands in vivo in mice reconstituted with a human immune system. Vaccine. 2017;35(45):6143–6153. doi: 10.1016/j.vaccine.2017.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meixlsperger S, et al. CD141+ dendritic cells produce prominent amounts of IFN-α after dsRNA recognition and can be targeted via DEC-205 in humanized mice. Blood. 2013;121(25):5034–5044. doi: 10.1182/blood-2012-12-473413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358(6359):58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 25.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 26.Breton G, Zheng S, Valieris R, Tojal da Silva I, Satija R, Nussenzweig MC. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J Exp Med. 2016;213(13):2861–2870. doi: 10.1084/jem.20161135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chistiakov DA, Killingsworth MC, Myasoedova VA, Orekhov AN, Bobryshev YV. CD68/macrosialin: not just a histochemical marker. Lab Invest. 2017;97(1):4–13. doi: 10.1038/labinvest.2016.116. [DOI] [PubMed] [Google Scholar]
- 28.Turman MA, Yabe T, McSherry C, Bach FH, Houchins JP. Characterization of a novel gene (NKG7) on human chromosome 19 that is expressed in natural killer cells and T cells. Hum Immunol. 1993;36(1):34–40. doi: 10.1016/0198-8859(93)90006-M. [DOI] [PubMed] [Google Scholar]
- 29.Björklund ÅK, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17(4):451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 30.Motakis E, et al. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood. 2014;123(17):e58–e67. doi: 10.1182/blood-2013-02-483792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabanova S, Kleinbongard P, Volkmer J, Andrée B, Kelm M, Jax TW. Gene expression analysis of human red blood cells. Int J Med Sci. 2009;6(4):156–159. doi: 10.7150/ijms.6.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummins N, Badley A. The TRAIL to viral pathogenesis: the good, the bad and the ugly. Curr Mol Med. 2009;9(4):495–505. doi: 10.2174/156652409788167078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandhi RT, Chen BK, Straus SE, Dale JK, Lenardo MJ, Baltimore D. HIV-1 directly kills CD4+ T cells by a Fas-independent mechanism. J Exp Med. 1998;187(7):1113–1122. doi: 10.1084/jem.187.7.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mongkolsapaya J, et al. Lymphocyte inhibitor of TRAIL (TNF-related apoptosis-inducing ligand): a new receptor protecting lymphocytes from the death ligand TRAIL. J Immunol. 1998;160(1):3–6. [PubMed] [Google Scholar]
- 36.Screaton GR, Mongkolsapaya J, Xu XN, Cowper AE, McMichael AJ, Bell JI. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr Biol. 1997;7(9):693–696. doi: 10.1016/S0960-9822(06)00297-1. [DOI] [PubMed] [Google Scholar]
- 37.Finkel TH, et al. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med. 1995;1(2):129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 38.Herbeuval JP, et al. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood. 2005;106(10):3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herbeuval JP, et al. TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected patients and its in vitro production by antigen-presenting cells. Blood. 2005;105(6):2458–2464. doi: 10.1182/blood-2004-08-3058. [DOI] [PubMed] [Google Scholar]
- 40.Miura Y, et al. Critical contribution of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) to apoptosis of human CD4+ T cells in HIV-1-infected hu-PBL-NOD-SCID mice. J Exp Med. 2001;193(5):651–660. doi: 10.1084/jem.193.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabb B, et al. Reduced inflammation and lymphoid tissue immunopathology in rhesus macaques receiving anti-tumor necrosis factor treatment during primary simian immunodeficiency virus infection. J Infect Dis. 2013;207(6):880–892. doi: 10.1093/infdis/jis643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nganou-Makamdop K, et al. Type I IFN signaling blockade by a PASylated antagonist during chronic SIV infection suppresses specific inflammatory pathways but does not alter T cell activation or virus replication. PLoS Pathog. 2018;14(8):e1007246. doi: 10.1371/journal.ppat.1007246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doitsh G, et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505(7484):509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato K, et al. Antiviral response by natural killer cells through TRAIL gene induction by IFN-alpha/beta. Eur J Immunol. 2001;31(11):3138–3146. doi: 10.1002/1521-4141(200111)31:11<3138::AID-IMMU3138>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 46.Natesampillai S, et al. TRAILshort Protects against CD4 T cell death during acute HIV infection. J Immunol. 2019;203(3):718–724. doi: 10.4049/jimmunol.1900271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnepple DJ, et al. Isolation of a TRAIL antagonist from the serum of HIV-infected patients. J Biol Chem. 2011;286(41):35742–35754. doi: 10.1074/jbc.M111.274639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL. Humanized mice for immune system investigation: progress, promise and challenges. Nat Rev Immunol. 2012;12(11):786–798. doi: 10.1038/nri3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Traggiai E, et al. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 2004;304(5667):104–107. doi: 10.1126/science.1093933. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa F, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106(5):1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shultz LD, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Kovalev GI, Su L. HIV-1 infection and pathogenesis in a novel humanized mouse model. Blood. 2007;109(7):2978–2981. doi: 10.1182/blood-2006-07-033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mongkolsapaya J, et al. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol. 1999;6(11):1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.