Abstract
Streptococcus pyogenes (group A streptococcus; GAS) causes 600 million cases of pharyngitis annually worldwide. There is no licensed human GAS vaccine despite a century of research. Although the human oropharynx is the primary site of GAS infection, the pathogenic genes and molecular processes used to colonize, cause disease, and persist in the upper respiratory tract are poorly understood. Using dense transposon mutant libraries made with serotype M1 and M28 GAS strains and transposon-directed insertion sequencing, we performed genome-wide screens in the nonhuman primate (NHP) oropharynx. We identified many potentially novel GAS fitness genes, including a common set of 115 genes that contribute to fitness in both genetically distinct GAS strains during experimental NHP pharyngitis. Targeted deletion of 4 identified fitness genes/operons confirmed that our newly identified targets are critical for GAS virulence during experimental pharyngitis. Our screens discovered many surface-exposed or secreted proteins — substrates for vaccine research — that potentially contribute to GAS pharyngitis, including lipoprotein HitA. Pooled human immune globulin reacted with purified HitA, suggesting that humans produce antibodies against this lipoprotein. Our findings provide new information about GAS fitness in the upper respiratory tract that may assist in translational research, including developing novel vaccines.
Keywords: Infectious disease
Keywords: Bacterial infections
In vivo selection identifies Streptococcus pyogenes genes that promote pharyngitis.
Introduction
Streptococcus pyogenes (group A streptococcus; GAS) is a significant human pathogen with extensive health and economic impact worldwide (1). Globally, GAS causes over 700 million infections each year, including 600 million cases of pharyngitis (1). Despite a century of research, no licensed human vaccine is available (2–22). The human oropharynx is the primary site for GAS infection, and GAS is the leading cause of bacterial pharyngitis. However, current understanding of GAS pathogenesis in the human oropharynx is limited.
High-throughput genome-wide screens using techniques such as signature-tagged mutagenesis (23) and transposon insertion site sequencing technologies (TIS-Seq, Tnseq, and transposon-directed insertion sequencing [TraDIS]) (24–27) are powerful tools to study the genetic basis of bacterial virulence. The results of these types of genetic studies can assist translational research efforts, including development of vaccines and therapeutics. Genome-wide transposon mutagenesis screens have been successfully used to identify novel GAS genes contributing to fitness in human blood and saliva ex vivo, in mouse subcutaneous infection, and in nonhuman primate (NHP) necrotizing myositis and female urogenital tract infection (28–33). However, a genome-wide investigation of the GAS factors contributing to primate pharyngitis has not been conducted using transposon mutagenesis strategies.
Analysis of GAS pathogenesis in the oropharynx requires an experimental infection model that faithfully recapitulates human pharyngitis. Mouse infection models have been used to study GAS pathogenesis and have yielded important information (34–38). However, the NHP pharyngitis model is the gold standard experimental animal infection model for many reasons. First, GAS pharyngitis in cynomolgus macaques mirrors oropharyngeal disease in humans, and as a consequence we have successfully used this model for GAS molecular pathogenesis investigations for more than 20 years (39–47). Pharyngitis in cynomolgus macaques, like humans, follows a course of initial colonization and proliferation, acute symptomatic disease, and chronic asymptomatic carriage and recovery (41). GAS-infected cynomolgus macaques have fever, decreased appetite, and an inflammatory response characterized by elevated levels of segmented neutrophils, C-reactive protein, and cytokines (41). Second, certain GAS virulence factors are specific for human and NHP host molecules and have little or no activity against mouse homologs (48, 49). Third, the oropharynx of NHPs is anatomically and physiologically similar to the human oropharynx. For example, like humans, NHPs have tonsils, whereas mice do not. Based on a clinically validated scoring method used in human children, GAS-infected monkeys develop tonsillitis and pharyngitis with erythema, edema, and exudate (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137686DS1) (50). Fourth, the NHP oropharynx also permits the recovery of a sufficient number of bacterial cells from each animal to generate statistically valid results. Fifth, the NHP model permits interactions with intact host innate and acquired immune functions to be assessed. Taken together, cynomolgus macaques provide the most relevant animal experimental model for studying GAS-oropharyngeal interactions and bacterial fitness.
Here we report what is likely the first use of the NHP pharyngitis infection model coupled with transposon mutant screens to identify GAS genes that contribute to bacterial fitness in the primate oropharynx. Using saturated transposon mutant libraries that were generated in 2 genetically divergent GAS strains that represent common causes of human pharyngitis and invasive infections, we identified potentially novel genes required for GAS fitness in pharyngitis. Our findings have extensive implications for translational research and vaccine development.
Results
Genome-wide screens identify GAS genes that contribute to fitness in an NHP pharyngitis model.
We screened the M1 and M28 GAS transposon mutant libraries in the upper respiratory tract (URT) of our NHP model to discover genes that contribute to GAS fitness during pharyngitis. For each screen, 6 cynomolgus macaques were infected in the URT with each transposon mutant library. As expected based on previous studies (40–47), all animals developed signs and symptoms mimicking human pharyngitis. Throat swabs were obtained from each animal at 24 hours postinfection. GAS cells recovered from the throat swabs collected constituted the output mutant pools. TraDIS was used to analyze the change in mutant composition of the output bacteria pools relative to the input pools. Genes with a significantly decreased mutant frequency in the output mutant pools relative to the input mutant pools were regarded as potentially required for growth and persistence in the URT.
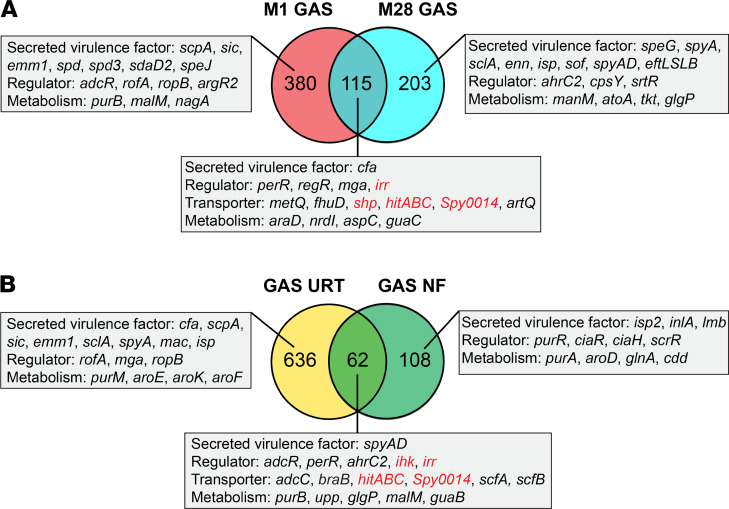
TraDIS identified 495 M1 GAS genes and 318 M28 GAS genes that contribute to GAS fitness in the primate oropharynx (Figure 1 and Supplemental Tables 1 and 2). In aggregate, 698 fitness genes were identified. These identified fitness genes encode many known GAS virulence factors, such as C5a peptidase (ScpA) (51, 52), streptococcal inhibitor of complement (Sic) (53, 54), antiphagocytic M protein (55), secreted DNAses (Spd, Spd3, and SdaD2) (45, 56, 57), surface-exposed adhesins and invasins (SclA, Enn, serum opacity factor [Sof], Streptococcus pyogenes adhesion and division [SpyAD], and EftLSLB) (33, 58–63), and secreted toxins (SpeJ, SpeG, and SpyA) (49, 64) (Figure 1). A common set of 115 genes was found to be important for GAS fitness for both M1 and M28 organisms (Figure 1 and Supplemental Table 3). These include genes encoding two-component regulator Irr-Ihk (65), stand-alone transcriptional regulator Mga (66), CAMP factor (67, 68), amino acid permease (M5005_Spy0014) (32), streptococcal heme-associated protein (Shp) (69), and a putative ABC transporter (HitA, HitB, and HitC) (31) (Figure 1A). Recently, using the same M1 and M28 GAS transposon insertion mutant libraries and TraDIS, we identified 170 GAS genes that are required during NF and myositis in NHPs (32). Comparing the GAS fitness genes required in the URT and during NF showed that 62 genes are important in both environments (Figure 1B). These include genes encoding SpyAD (62), zinc transporter regulator AdcR (70), peroxide regulator PerR (71), and the ScfAB permease that has been shown to contribute to GAS soft tissue infection (31). Of note, Irr-Ihk two-component regulators, permease Spy0014, and ABC transporter HitABC were identified as important for both URT infection and NF (Figure 1B).
Figure 1. TraDIS analysis of GAS genes contributing to primate URT infection.
(A) Comparison of M1 and M28 GAS genes contributing to fitness in the NHP URT. (B) Comparison of GAS genes required in the NHP URT and those important for NHP necrotizing fasciitis (NF). Representative fitness genes assigned to each category are shown. Genes selected for further validation and investigation are highlighted in red.
Validating the contribution of 4 loci to GAS fitness in the NHP oropharynx.
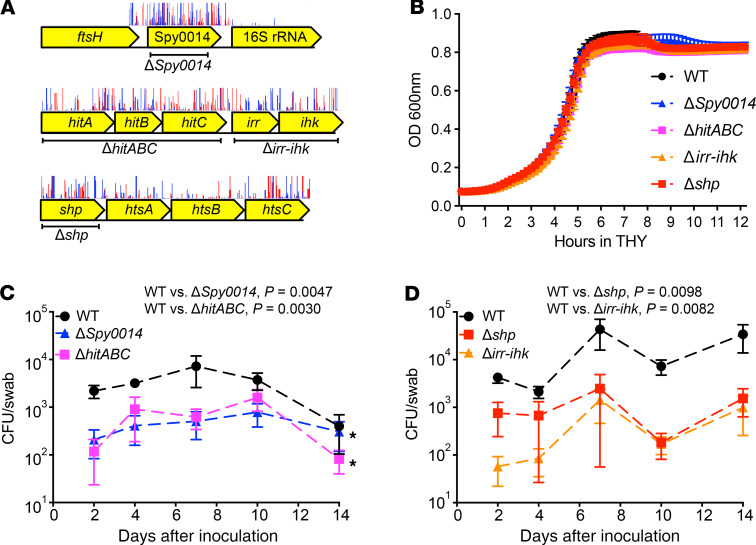
To confirm the TraDIS screen results in vivo, we validated the role of 4 gene loci identified by our screen as promoting GAS survival in the NHP oropharynx — Spy0014 (putative amino acid permease), hitABC (putative ABC efflux pump), irr-ihk (two-component system), and shp (streptococcal heme-associated protein) (Figure 2A). Targeted gene deletion was used to generate isogenic mutant strains ΔSpy0014, ΔhitABC, Δirr-ihk, and Δshp. None of the isogenic gene deletion mutant strains had a significant growth defect in rich culture medium (THY) (Figure 2B). We infected 4 NHPs with parental WT M1 strain MGAS2221 and each isogenic gene deletion mutant strain (ΔSpy0014, ΔhitABC, Δirr-ihk, and Δshp) in the URT. Compared with the WT parental strain, each mutant strain was significantly less able to survive and persist in the URT (Figure 2, C and D). Relative to the WT parental strain, significantly fewer CFU of each isogenic mutant strain were recovered from the NHP oropharynx (Figure 2, C and D).
Figure 2. Validation of the contribution of Spy0014, hitABC, irr-ihk, and shp to GAS fitness in the primate URT.
(A) Schematic showing the gene neighborhood of the studied loci, with the deleted regions of each isogenic mutant strain noted. Also shown are transposon insertion sites of each gene determined by TraDIS. Red vertical spikes are forward reads; blue vertical spikes are reverse reads. Read orientations indicate the direction of the transposon insertion. (B) Growth of the isogenic gene deletion mutant strains in rich medium THY. (C and D) Significantly decreased ability of the isogenic mutant strains to colonize the NHP URT, compared with WT parental strain MGAS2221. P values for CFU recovery were determined across the 14-day experiment by 2-way ANOVA with correction for multiple comparisons by the method of Benjamini, Krieger, and Yekutiel. Data are presented as the mean ± standard error of the mean of 4 experiments.
Growth of isogenic gene deletion mutant strains in human saliva, serum, and whole blood ex vivo.
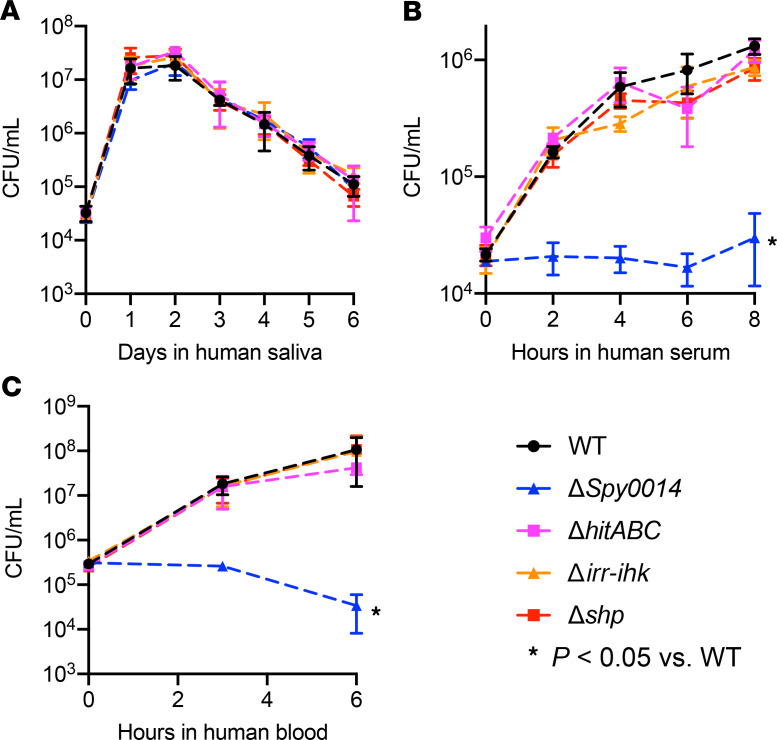
To better understand the role of the validated genes in pathogen-host interactions, we compared the growth of the WT parental and isogenic gene deletion mutant strains ΔSpy0014, ΔhitABC, Δirr-ihk, and Δshp in human saliva. This body fluid is ubiquitous in the human oropharynx and is the first substance GAS encounters in the URT. Compared with the WT parental strain, none of the isogenic mutant strains had a significant growth defect in human saliva (Figure 3A).
Figure 3. Growth of GAS strains in human body fluids.
Growth of the WT parental strain MGAS2221 and the isogenic gene deletion mutant strains in human saliva (A), human serum (B), and human whole blood (C). *P < 0.05 determined by 1-way ANOVA. Data are presented as the mean ± standard deviation of the mean of 4 experiments.
To next test the hypothesis that these genes may contribute to GAS fitness in the URT by interacting with innate host immune functions, we examined the growth of the WT and isogenic mutant strains in human serum and human whole blood ex vivo. We found that the isogenic mutant strain ΔSpy0014 had a significant growth defect in both human serum and human whole blood (Figure 3, B and C).
Spy0014 encodes a unique amino acid permease that affects GAS virulence and SpeB protease activity.
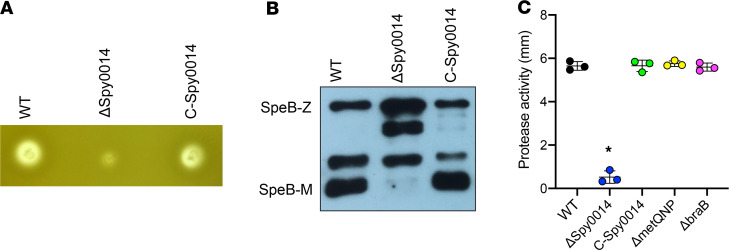
Spy0014 encodes an inferred amino acid permease, and our previous studies suggest it participates in histidine acquisition (32). Our results here suggested this permease is crucial for GAS fitness in the NHP URT (Figure 2). To study the virulence role of Spy0014, we examined the production of major GAS virulence factors made by the isogenic ΔSpy0014 deletion mutant strain. Compared with the WT parental strain MGAS2221, the ΔSpy0014 mutant strain had no significant difference in the production of virulence factors NADase and streptolysin O (data not shown). However, this mutant strain had significantly decreased SpeB protease activity (Figure 4). Western immunoblotting showed that the SpeB protein was made, but the maturation of the zymogen to the mature secreted protease was decreased in the ΔSpy0014 mutant strain. Complementing this mutant strain with the Spy0014 gene (with its native promoter) in trans restored WT protease activity and SpeB maturation (Figure 4).
Figure 4. Spy0014 contributes to maturation of GAS cysteine protease SpeB.
(A) SpeB protease activity of the isogenic Spy0014 deletion mutant strain and the complemented mutant strain (C-Spy0014). (B) Western immunoblot showing the extent of SpeB maturation in the WT parental strain, the Spy0014 deletion strain, and the complemented mutant strain. SpeB-Z, immature zymogen. SpeB-M, mature protease. (C) Contribution of amino acid transporters to GAS SpeB protease activity. *P < 0.05 determined by 1-way ANOVA. Data are presented as the mean ± standard deviation of the mean of 3 experiments.
To study whether other GAS amino acid transporters also affect SpeB activity, we examined the protease activity made by the branched-chain amino acid transporter mutant ΔbraB and the methionine transporter mutant ΔmetQNP (32). In contrast to the results observed for the ΔSpy0014 mutant strain, these 2 amino acid transporters had no significant decrease in SpeB protease activity (Figure 4). Collectively, these results supported that Spy0014 is a unique amino acid permease that contributes to the virulence and SpeB protease activity of GAS.
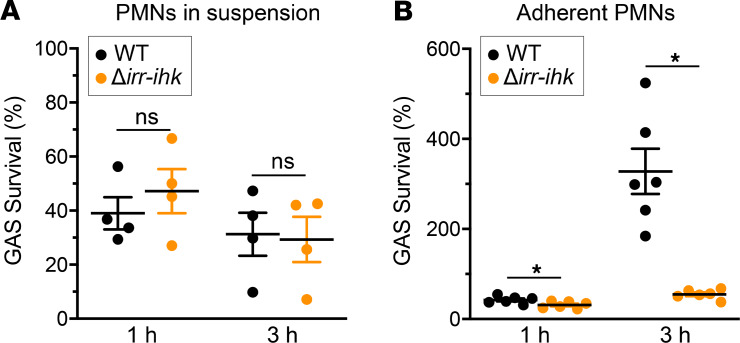
Irr-Ihk two-component system protects GAS against killing by adherent polymorphonuclear leukocytes.
Previous studies suggested the response regulator Irr contributes to evasion of GAS from host innate immunity (65, 72). We examined the effect of deleting both irr and ihk on GAS susceptibility to killing by human polymorphonuclear leukocytes (PMNs). There was no significant difference in survival between the WT parental strain and the Δirr-ihk deletion mutant strain during interaction with PMNs in suspension (Figure 5A), a finding consistent with our data for GAS survival in human blood (above). In striking contrast, the Δirr-ihk deletion mutant strain was significantly less able to survive when phagocytosis was synchronized and bacteria were exposed to adherent PMNs, compared with the WT strain (Figure 5B).
Figure 5. Irr-Ihk two-component system protects against killing by adherent human PMNs ex vivo.
Killing of WT and irr-ihk–knockout GAS strains by human PMNs in suspension (A) or adherent cells (B). *P ≤ 0.02 using a paired 2-tailed t test (Prism 7 for Windows, GraphPad Software, Inc); ns, not significant. Data are presented as the mean ± standard error of the mean of 4 (A) or 6 (B) separate experiments as indicated.
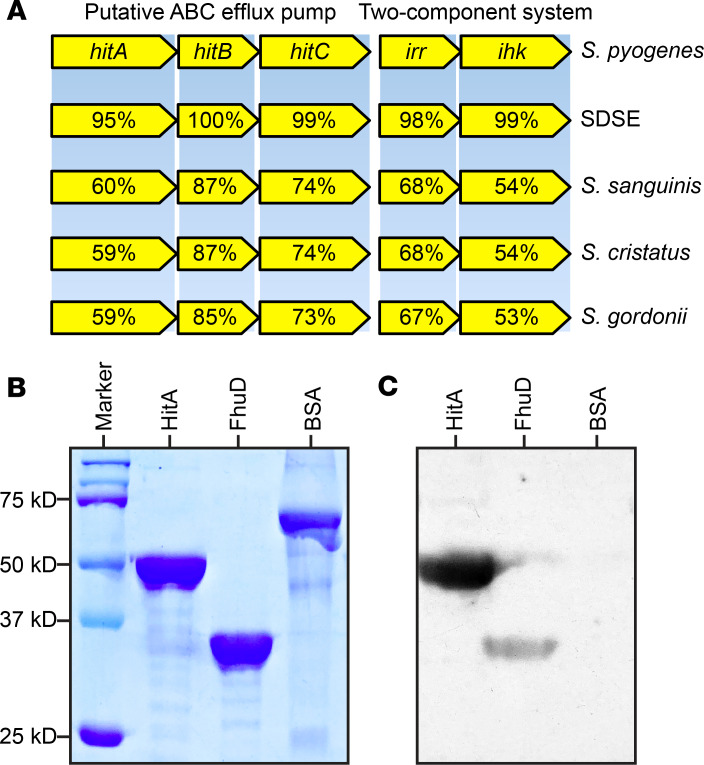
Restricted distribution of the hitABC transporter genes in streptococcal species and translational implication of HitA.
The ABC transporter genes hitABC contributed significantly to GAS pharyngitis in NHPs (Figure 2). Nucleotide BLAST (http://www.ncbi.nlm.nih.gov/blast/) results using publicly available genome sequences found that close homologs of these ABC transporter genes are present in only 4 additional streptococcal species, namely Streptococcus dysgalactiae subspecies equisimilis (SDSE), Streptococcus sanguinis, Streptococcus cristatus, and Streptococcus gordonii (Figure 6A). Interestingly, each of these organisms also colonizes the oropharynx of humans (73–75). Further, as with GAS, homologs of hitABC in these 4 streptococcal species are located adjacent to a two-component system locus with homology to irr and ihk (Figure 6A). Previous studies suggest that substrate-binding lipoprotein encoded by hitA (HitA) is a surface-displayed protein (76). As the first step toward understanding whether HitA may be a viable vaccine target, we overexpressed and purified the recombinant proteins HitA and FhuD, a known GAS surface-displayed protein (77) (Figure 6B). Western immunoblot analysis showed that pooled human immune globulin reacts with HitA and FhuD, but not with the control protein, BSA (Figure 6C). These results suggest that humans produce specific antibodies against HitA. Further investigations are needed to address whether antibodies against HitA elicit protection against GAS infections in the primate URT.
Figure 6. Distribution of hitABC transporter genes in streptococcal species and immunoreactivity of recombinant HitA.
(A) Distribution of the hitABC transporter genes and the adjacent irr-ihk two-component regulatory genes in other streptococcal species (Streptococcus dysgalactiae subspecies equisimilis [SDSE] strain GGS124, Streptococcus sanguinis strain SK36, Streptococcus cristatus strain NCTC12479, and Streptococcus gordonii strain NCTC7868). Percentages refer to amino acid identities compared with proteins made by serotype M1 GAS reference strain MGAS2221. (B) SDS-PAGE gel showing the purified recombinant GAS lipoproteins HitA and FhuD. BSA was added as a control. (C) Immunoreactivity of purified HitA and FhuD with pooled human immune globulin.
Discussion
In this study, we performed the first genome-wide transposon mutagenesis screens designed to discover GAS genes that contribute to pharyngitis in primates. Our screens identified many genes previously not known to contribute to GAS fitness in the primate URT and thereby help fill a critical knowledge gap in our understanding of this disease that affects an estimated 600 million people annually (1).
Our TraDIS screens identified many proven and putative secreted virulence factors that are potentially important for GAS URT infection (Figure 1A). Some of the secreted proteins have already been considered vaccine candidates (78). These include M protein (emm1) (79, 80), streptococcal C5a peptidase (scpA), serum opacity factor (sof) (81), and SpyAD (spyAD). Importantly, we recently reported that SpyAD is also crucial for GAS virulence during necrotizing myositis and female genital tract infections in NHPs (32, 33). The critical roles in multiple infection models suggest SpyAD could be an attractive vaccine target (62, 82). Further investigations are needed to test whether immunizing NHPs with proteins we identified in this study could evoke protective immunity against GAS pharyngitis.
Some transposon mutagenesis screens using animal models harvest a whole organ or whole lesion as a strategy to recover large numbers of mutants in the output pool (31, 32, 83–87). As an alternative strategy, which does not require the animal to be sacrificed, we collected and pooled 10 swabs from the posterior oropharynx of each animal. We used an identical 10-swab strategy in our recent successful cynomolgus macaque female genital tract TraDIS study (33). The high number of recovered CFU indicates that the output mutant pools are likely representative of the in vivo condition. That said, we acknowledge it is of course possible that not every mutant will be represented in the output pool. Regardless, validating the genome-wide screen results using isogenic gene deletion mutant strains is useful for individual genes that are of particular interest.
To validate our screen results, we examined the virulence role of 4 loci using isogenic gene deletion mutant strains. We showed that compared with the WT parental strain, isogenic mutant strains (ΔSpy0014, ΔhitABC, Δirr-ihk, and Δshp) were significantly less able to initiate infection and grow to high numbers of CFU during the 14 days of infection, thereby validating the TraDIS screen result (Figure 2, C and D). Consistent with our previous observations, the growth of WT GAS strain reaches a peak on day 7 in the NHP URT (47). Interestingly, for some mutant strains (Δirr-ihk and Δshp), the change of CFU is somewhat consistent with the WT strain (Figure 2D). Although the CFU are lower than the WT, the growth of these mutant strains also peaks on day 7 (Figure 2D). A previous longitudinal analysis of GAS pharyngitis using the URT model and 20 NHPs identified 3 phases of infection: colonization phase, acute phase, and asymptomatic phase (41). The acute phase starts after day 5 and key infection indicators, such as C-reactive protein, pharyngitis values, and GAS CFU, peak during this period (41). It is possible that mutant strains such as Δirr-ihk and Δshp could still cause acute phase infection, but they are significantly less fit than the WT strain in the URT and therefore cannot grow to high CFU. The molecular basis of GAS acute phase infection warrants further investigations.
A key finding of our study is that the putative amino acid permease Spy0014 contributes to GAS infection in the NHP URT. Our previous study suggests that Spy0014 participates in the acquisition of histidine (32), consistent with the idea that acquisition of amino acids from the host is crucial for GAS fitness in the URT. However, Spy0014 is not required for optimal GAS growth in human saliva ex vivo, the first environment GAS encounters during URT infection (Figure 3A) (30). In contrast, this gene is crucial for GAS growth in human serum and whole blood, which are more nutrient rich than saliva and contain bactericidal components, such as complement proteins and white blood cells (88–91). These results suggest nutrient acquisition may not be the only role of Spy0014. Importantly, we also found that Spy0014 is required for proper maturation of SpeB cysteine protease, a key virulence factor of GAS that is known to inhibit complement activation and neutrophil opsonophagocytosis (47, 48). We think it likely that the contribution of Spy0014 to GAS fitness and pathogenesis during pharyngitis is multifaceted. We also note that speB was not identified by TraDIS screens as required for GAS fitness during URT infection. This result is expected because the nature of the TraDIS screen is mixed infection with a large population with many distinct mutant strains, most of which will have an intact and functional speB gene. In this setting, mutant strains can be “trans-complemented” by secreted products made by the WT-like strains.
PMNs are a key component of innate immunity against GAS infections (65). Compared with the WT parental strain, the isogenic mutant strain lacking the Irr-Ihk two-component regulatory system is significantly attenuated in resisting the killing by adherent PMNs that are adherent to 96-well plates precoated with human serum. However, relative to the WT strain, this mutant is not attenuated in ability to resist killing by PMNs in suspension. In line with this observation, we found that Irr-Ihk is not required for GAS growth in human whole blood (Figure 3B). These observations may shed light on the virulence role of the Irr-Ihk two-component system. It is well documented that PMNs that migrate from the blood and attach to surfaces are phenotypically very different from PMNs in circulation (92–97). For example, PMNs that are adherent to biological surfaces are primed for enhanced function in response to subsequent stimuli and thus have significantly increased phagocytosis (96, 97). It is possible that the Irr-Ihk two-component system contributes to GAS virulence during pharyngitis by resisting the killing of activated PMNs adherent to the retropharyngeal or other epithelial cell surfaces. PMNs adherent to the serum-coated plates may better recapitulate the physiological state of upper respiratory PMNs during GAS pharyngitis.
We discovered that a poorly characterized ABC transporter, HitABC, contributes significantly to GAS fitness during experimental NHP pharyngitis (Figure 2). The 3 genes encoding this ABC transporter (hitABC) are located immediately upstream of irr and ihk that encode a two-component regulatory system (TCS) (Figure 4). The Irr-Ihk TCS is known to contribute to virulence by protecting GAS from neutrophil-mediated killing, although the exact mechanism is not known (49, 50). Importantly, our data show that Irr-Ihk also contributes to GAS fitness during pharyngitis (Figure 2). It is unclear whether the HitABC transporter and the Irr-Ihk two-component system are functionally related. Interestingly, transcriptomic analysis suggests polycistronic expression of hitABC and irr-ihk (51). Further, we showed previously that hitABC and irr-ihk are both upregulated when GAS is exposed to human whole blood ex vivo and during NHP necrotizing myositis in vivo (52). The polycistronic expression and upregulation of these genes in whole blood and during necrotizing myositis suggest that HitABC and Irr-Ihk might be functionally related. In line with this idea, we reported that hitABC and irr-ihk are both crucial for GAS virulence in an NHP model of necrotizing myositis (32, 98). Because the Irr-Ihk TCS contributes to GAS’s ability to resist being killed by neutrophils and neutrophil-derived antimicrobial peptides (72), further functional studies are needed to determine whether the HitABC transporter serves as an efflux pump for neutrophil-derived antimicrobial peptides.
Although the exact function of the HitABC transporter in GAS is unknown, we found that close homologs of this ABC transporter are present only in SDSE, Streptococcus sanguinis, Streptococcus cristatus, and Streptococcus gordonii. Each of these species colonizes the human oropharynx (43–45). Interestingly, in these species, homologs of hitABC are also located contiguous with genes encoding a two-component system with homology to ihk and irr, suggesting a functional connection between this ABC transporter and the Irr-Ihk system. It would be useful to determine whether homologs of hitABC also contribute to oropharyngeal colonization of SDSE, an emerging human pathogen with a disease spectrum similar to that of GAS (53). The hitA gene encodes the inferred substrate-binding lipoprotein of the HitABC transporter. HitA is a proven GAS surface-displayed protein (76) that may be useful as a vaccine or therapeutic target. Consistent with this idea, Western immunoblot analysis of purified recombinant proteins (Figure 4) showed pooled human immune globulin is reactive with HitA and FhuD, indicating that antibodies arise during streptococcal infection or colonization.
Using an isogenic mutant strain and the NHP model of infection, we showed the significant contribution of the shp gene to GAS URT infection (Figure 2D). Shp, which is an essential component of GAS heme uptake machinery, is encoded by shp (99). Host heme is the key iron source for bacterial pathogens, and iron acquisition is essential for the growth and virulence of GAS (100–104). Thus, it is not unexpected that shp is required for GAS URT infection. However, we note that the shp deletion strain is not significantly impaired for growth in human saliva, human whole blood, or serum ex vivo. It is well established that during infection, the innate immune system reduces the free iron in the host by increasing the level of ferric-binding proteins in the serum and releasing apo-lactoferrin into the infection site (105–109). Therefore, heme acquisition is crucial for many bacterial pathogens during infection. However, because the human saliva, whole blood, and serum we used for ex vivo growth assays was collected from healthy donors with no infection, the free iron in these body fluids might be adequate for the growth of GAS. The critical role of Shp-mediated heme acquisition might be bypassed in these settings. Shp is located on the bacterial cell surface and is accessible to specific antibodies raised against the purified recombinant Shp (69). Therefore, Shp was considered a viable vaccine target. In this regard, immunization with Shp protein was reported to protect mice from GAS skin and systemic infections (103). Since shp is required for the fitness of GAS in the primate URT, it would be of interest to study whether immunization with Shp could evoke protection against GAS URT infection.
To summarize, we screened transposon mutant libraries made in serotype M1 and M28 GAS strains for genes contributing to fitness in the URT tract of NHPs, the gold standard model of pharyngitis. These 2 M protein serotypes of GAS are commonly among the top 5 M types responsible for human pharyngitis in many countries surveyed (1). NHP infection with each of the 4 isogenic mutant strains confirmed the crucial contribution of the GAS genes identified by the TraDIS screens. Our findings provide extensive new information about the genes that contribute to GAS fitness in the URT of primates. This research also complements work conducted with other transposon mutant screens, including those designed to identify GAS genes contributing to growth in vitro, human saliva and blood ex vivo, and animal (mouse and NHP) models of soft tissue infection, muscle infection, and genital tract infection (29–33). The discoveries presented herein may ultimately lead to better prevention and treatment of GAS pharyngitis.
Methods
Bacterial strains.
Strain MGAS2221 is genetically representative of a virulent serotype M1 GAS clone that arose in the 1980s and spread rapidly worldwide to cause pandemic pharyngitis and invasive infections (42). Strain MGAS27961 is genetically representative of a serotype M28 clone that is prevalent in the United States and elsewhere (110). Each of the 2 strains has WT alleles of all major transcriptional regulators known to affect virulence, such as covR and covS, ropB, mga, and rocA. These 2 GAS strains were used as parental strains for generating transposon insertion mutant libraries and subsequent isogenic mutant strains.
Transposon insertion mutant libraries and culture conditions.
The mutant libraries generated in serotype M1 strain MGAS2221 and M28 strain MGAS27961 using transposon plasmid pGh9:ISS1 were recently described (30, 111). The strains were grown in Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) with 0.5 μg/mL erythromycin.
Preparation of transposon mutant library frozen stocks for NHP infection.
The transposon mutant library stock culture (100 μL, serotype M1 or M28 GAS library) was inoculated into 500 mL THY supplemented with 0.5 μg/mL erythromycin and cultured at 37°C for 8 hours. Bacteria in the expanded transposon library were pelleted by centrifugation, washed 3 times with PBS, and suspended in 20 mL PBS supplemented with 20% glycerol. The suspended mutant library was aliquoted into cryogenic tubes (1 mL/tube) and stored at –80°C until use in NHP infections.
NHP pharyngitis infection model used for TraDIS analysis.
Cynomolgus macaques (2.8 to 6.5 years of age, 2.9 to 3.5 kg of body weight) were used for the study. The animals used for the initial fitness screen and for mutant validation were GAS culture negative, and they had negligible anti–streptolysin O titers, suggesting they had no recent history of GAS exposure. The animals used in this study were not used in earlier studies. For the input mutant library pool of each M protein serotype strain, 6 NHPs were infected in the oropharynx with 1010 CFU. To infect the posterior oropharynx, animals were sedated with ketamine and held in a semisupine position (approximately 45° angle relative to the examination table) with the head tilted back to align the nasopharynx, posterior oropharynx, and URT. The 1-mL GAS inoculum was slowly instilled from a needleless syringe by dripping into the bilateral nares. The fluid was inhaled into the respiratory tract by the animal’s natural breathing movements. The veterinary team confirmed GAS uptake by direct observation and palpation of swallowing during the instillation. At 24 hours postinoculation, signs and symptoms of GAS disease were confirmed by measuring vital signs and observing the posterior oropharynx (Supplemental Figure 1). Output mutant pools were collected by throat swabs at 24 hours postinfection. Ten throat swabs per animal were collected. Briefly, for each swab, 2 circumferential sweeps around the posterior oropharynx and tonsils were done. All 10 swabs from each animal were immersed in the same 40 mL of THY containing 0.5 μg/mL erythromycin (used to inhibit growth of non-GAS bacteria). To recover the output mutant pools, swabs in THY were cultured at 37°C for 8 hours. GAS cells were pelleted by centrifugation, washed with PBS, and suspended in 2 mL of PBS with 20% glycerol. Output mutant pools were stored at –80°C for subsequent TraDIS analysis. All animal studies were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute (protocol AUP-1217-0058).
DNA preparation and massively parallel sequencing.
The mutant library genomic DNA preparation and DNA sequencing were performed according to procedures described previously for TraDIS analysis (30, 111). The PCR-amplified libraries were sequenced with a NextSeq550 instrument (Illumina) using a single-end 75-cycle protocol.
Processing of TraDIS sequencing reads and data analysis.
Processing of TraDIS sequencing reads and data analysis were performed according to previously described procedures (30). Briefly, the multiplexed raw Illumina reads obtained from the input and output mutant pools were parsed with FASTX Barcode Splitter (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html). The resulting demultiplexed sequencing reads were analyzed with the TraDIS toolkit (27). tradis_comparison.R was used to compare the reads mapped per gene between the input pools and the output pools. GAS genes with significantly altered mutant frequency (log2 fold change greater than 1 or less than –1, and P value less than 0.05) in the output mutant pools were interpreted as contributing to GAS fitness.
Isogenic mutant strain construction.
Isogenic deletion mutant strains were constructed from WT parental strain MGAS2221, the organism used for construction of the serotype M1 transposon mutant library. Methods used to construct and confirm isogenic mutant strains ΔSpy0014, ΔhitABC, and Δirr-ihk were described in our previous studies (32). Complementation of Spy0014 in trans was performed by introducing the Spy0014 gene and its native promoter into strain ΔSpy0014. Briefly, primers Spy0014-fwd and Spy0014-rev (Supplemental Table 4) were used to amplify a DNA fragment containing Spy0014 and its promoter. The amplicon was cloned into plasmid pDC123 and transformed into mutant strain ΔSpy0014 to generate the complemented strain C-Spy0014. Δshp was constructed by allelic exchange using procedures previously described (47). Briefly, primer sets shp-1/2 and shp3/4 (Supplemental Table 4) were used to amplify 2 DNA fragments flanking the shp gene. The 2 amplicons were cloned into suicide vector pBBL740 and then transformed into parental strain MGAS2221. Allelic exchange was accomplished by passaging the plasmid integrant onto nonselective agar. The genome of all mutant strains was sequenced to rule out introduction of spurious mutations.
Infection of NHPs in the URT with isogenic mutant strains.
To confirm the fitness role of candidate genes in the NHP oropharynx, and thereby validate the TraDIS screen data, the virulence of the WT parental strain MGAS2221 and the 4 isogenic deletion mutant strains was assessed in the NHP pharyngitis infection model. Animals randomly assigned to different strain treatment groups were inoculated in the URT with 108 CFU of 1 strain (i.e., WT or isogenic mutant). Each strain was tested in 4 NHPs.
Growth of GAS strains in human saliva, whole blood, and serum.
GAS saliva growth assay was performed using pooled saliva from 5 healthy donors as previously described (30, 112). The growth and persistence of GAS strains were monitored for 6 days. Growth of GAS in human blood was performed using a previously described method (29), with whole blood collected from 4 healthy donors. To assess GAS growth and persistence in human serum, 104 CFU/mL of each GAS strain was inoculated into pooled human serum (MilliporeSigma) and incubated at 37°C for 8 hours. Aliquots of each tube were taken at indicated time points (Figure 3B) and were diluted and plated on THY agar plates. GAS colonies were enumerated the following day.
SpeB protease activity assay.
SpeB protease activity assay was performed by stabbing GAS strains into trypticase soy agar–milk agar plates (113). SpeB protease activity was analyzed by measuring the casein hydrolysis zone following an overnight incubation at 37°C.
Western immunoblot analysis of SpeB.
Western immunoblot assessment of SpeB production and maturation was performed as previously described (114). Briefly, GAS strains were grown to stationary phase and pelleted by centrifugation. The cell-free culture supernatant was collected and used for Western immunoblot. Equal volumes of the samples were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with polyclonal anti-SpeB rabbit antibodies (from M. Kumaraswami, Houston Methodist Research Institute and Houston Methodist Hospital, Houston, Texas, USA; ref. 115).
Expression and purification of recombinant proteins HitA (Spy1728) and FhuD (Spy0323).
Recombinant GAS lipoproteins HitA and FhuD were expressed in Escherichia coli (E. coli) without the secretion signal sequences. Briefly, primer sets hitA-1/2 and fhuD-1/2 were used to amplify the HitA and FhuD genes using genomic DNA from strain MGAS2221. The amplicons were cloned between the NdeI and XhoI restriction sites of plasmid vector pET28a (Novagen) and transformed into E. coli strain BL21. Overexpression of the recombinant proteins was accomplished by inducing BL21 cells with 0.5 mM isopropyl-β-d-thiogalactopyranoside grown at 20°C for 16 hours. To purify the recombinant proteins, E. coli cells were lysed with a French press, and the soluble fraction of the lysate was loaded onto a 5-mL HisTrap FF column (GE Healthcare) and eluted with a 20 to 500 mM imidazole gradient in the lysis buffer. Fractions containing the recombinant proteins were pooled, concentrated with an Amicon concentrator unit (MilliporeSigma), and further purified by gel filtration chromatography. The purity of each protein was more than 90% based on SDS-PAGE analysis (Figure 4).
Western immunoblot analysis of recombinant proteins.
Western immunoblot analysis was performed as previously described (78). Briefly, purified recombinant proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. After blocking, the membrane was probed with pooled human immune globulin (0.2 μg/mL, Grifols) followed by goat anti–human IgG horseradish peroxidase–conjugated secondary antibody (1721033, Bio-Rad, diluted at 1:4000).
GAS-PMN assays.
Human PMNs were isolated from heparinized venous blood using a published method (116). GAS from a frozen stock was cultured overnight in THY broth and then subcultured to the early exponential phase of growth (OD600 ~0.2) for use. For assays performed with PMNs in suspension, GAS (106 CFU) was combined with 106 PMNs in RPMI 1640 buffered with 10 mM HEPES with 10% human serum in a 1.5-mL tube (600 μL final volume). PMNs were omitted from tubes for time-matched control assays. Assay tubes were rotated gently for 1 or 3 hours, at which time PMNs were lysed in H2O (pH 11), and aliquots of each tube were diluted and plated on THY agar plates. GAS colonies were enumerated the following day, and percentage of survival was determined with the equation: GAS+PMN / GAS–PMN × 100.
Alternatively, assays were performed with adherent PMNs using a published method to synchronize phagocytosis (65). Briefly, serum opsonized GAS (106 CFU, preopsonized with 50% autologous human serum for 20 minutes at 37°C) was combined with 106 PMNs (200 μL final volume) in a 96-well flat-bottom culture plate that had been precoated with 20% human serum for 1 hour. Samples were centrifuged gently at 450 g for 5 minutes at 4°C to synchronize phagocytosis. Plates were transferred to 37°C for 1 or 3 hours, and phagocytosis was terminated with addition of cold saponin (0.1% final solution). Samples were diluted in H2O (pH 11) and aliquots plated on THY agar plates. Colonies were enumerated the following day, and percentage of survival was determined as described above.
Statistics.
Results of CFU recovery from NHPs are expressed as mean ± SEM, with statistically significant differences determined by 2-way ANOVA with correction for multiple comparisons by the method of Benjamini, Krieger, and Yekutiel. Results of GAS-PMN assays are presented as the mean ± standard error of the mean of 4 experiments. Statistically significant differences were determined using a 2-tailed Student’s t test. Results of SpeB protease activity are expressed as mean ± standard deviation, with statistically significant differences determined by 1-way ANOVA. A P value of less than 0.05 was considered statistically significant.
Study approval.
For studies with human PMNs, venous blood was obtained from healthy volunteers in accordance with a protocol (01-I-N055) approved by the Institutional Review Board for human subjects, NIH. All volunteers provided informed consent before participation in the study. Studies with human saliva, serum, and whole blood were performed in accordance with Houston Methodist Research Institute protocols PRO00003833 and PRO00004933. All animal studies were approved by the Institutional Animal Care and Use Committee at Houston Methodist Research Institute (protocol AUP-1217-0058).
Author contributions
LZ, RJO, SBB, LJ, ZS, TP, ARP, and FRD generated, analyzed, and interpreted data. ASW provided scholarly input on the TraDIS studies. MOS, SLK, and CCC provided expert technical assistance. JMM conceptualized and designed the study. All authors contributed to writing the manuscript. All authors reviewed and approved the final manuscript.
Supplementary Material
Acknowledgments
This study was supported by the Fondren Foundation, Houston Methodist Hospital and Research Institute, NIH grants AI139369 and AI146771 (to JMM) and AI32956 (to TP), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH (to FRD).
Version 1. 06/04/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(11):e137686.https://doi.org/10.1172/jci.insight.137686.
Contributor Information
Luchang Zhu, Email: lzhu2@houstonmethodist.org.
Matthew Ojeda Saavedra, Email: mojedasaavedra@houstonmethodist.org.
Samantha L. Kubiak, Email: sammiek21@gmail.com.
Andrew S. Waller, Email: andrew.waller@aht.org.uk.
Zhizeng Sun, Email: zhizeng.sun@bcm.edu.
Timothy Palzkill, Email: timothyp@bcm.edu.
Frank R. DeLeo, Email: fdeleo@niaid.nih.gov.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Dale JB. Group A streptococcal vaccines. Infect Dis Clin North Am. 1999;13(1):227–243. doi: 10.1016/s0891-5520(05)70052-0. [DOI] [PubMed] [Google Scholar]
- 3.Fox EN, Pachman LM, Wittner MK, Dorfman A. Primary immunization of infants and children with group A streptococcal M protein. Zentralbl Bakteriol Orig. 1970;214(3):352–363. [PubMed] [Google Scholar]
- 4.Massell BF, Michael JG, Amezcua J, Siner M. Secondary and apparent primary antibody responses after group A streptococcal vaccination of 21 children. Appl Microbiol. 1968;16(3):509–518. doi: 10.1128/AEM.16.3.509-518.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rantz LA, Randall E, Rantz HH. Immunization of human beings with group A hemolytic streptococci. Am J Med. 1949;6(4):424–432. doi: 10.1016/0002-9343(49)90210-7. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt WC. Type-specific antibody formation in man following injection of streptococcal M protein. J Infect Dis. 1960;106:250–255. doi: 10.1093/infdis/106.3.250. [DOI] [PubMed] [Google Scholar]
- 7.Watson RF, Rothbard S, Swift HF. Type-specific protection and immunity following intranasal inoculation of monkeys with group a hemolytic streptococci. J Exp Med. 1946;84(2):127–142. doi: 10.1084/jem.84.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beachey EH, Seyer JM, Dale JB. Protective immunogenicity and T lymphocyte specificity of a trivalent hybrid peptide containing NH2-terminal sequences of types 5, 6, and 24 M proteins synthesized in tandem. J Exp Med. 1987;166(3):647–656. doi: 10.1084/jem.166.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beachey EH, Seyer JM, Dale JB, Simpson WA, Kang AH. Type-specific protective immunity evoked by synthetic peptide of Streptococcus pyogenes M protein. Nature. 1981;292(5822):457–459. doi: 10.1038/292457a0. [DOI] [PubMed] [Google Scholar]
- 10.Bessen D, Fischetti VA. Passive acquired mucosal immunity to group A streptococci by secretory immunoglobulin A. J Exp Med. 1988;167(6):1945–1950. doi: 10.1084/jem.167.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bessen D, Fischetti VA. Influence of intranasal immunization with synthetic peptides corresponding to conserved epitopes of M protein on mucosal colonization by group A streptococci. Infect Immun. 1988;56(10):2666–2672. doi: 10.1128/IAI.56.10.2666-2672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox EN, Waldman RH, Wittner MK, Mauceri AA, Dorfman A. Protective study with a group A streptococcal M protein vaccine. Infectivity challenge of human volunteers. J Clin Invest. 1973;52(8):1885–1892. doi: 10.1172/JCI107372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poirier TP, Kehoe MA, Beachey EH. Protective immunity evoked by oral administration of attenuated aroA Salmonella typhimurium expressing cloned streptococcal M protein. J Exp Med. 1988;168(1):25–32. doi: 10.1084/jem.168.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polly SM, Waldman RH, High P, Wittner MK, Dorfman A. Protective studies with a group A streptococcal M protein vaccine. II. Challange of volenteers after local immunization in the upper respiratory tract. J Infect Dis. 1975;131(3):217–224. doi: 10.1093/infdis/131.3.217. [DOI] [PubMed] [Google Scholar]
- 15.Waldman RH, Lee JD, Polly SM, Dorfman A, Fox EN. Group A streptococcal M protein vaccine: protection following immunization via the respiratory tract. Dev Biol Stand. 1975;28:429–434. [PubMed] [Google Scholar]
- 16.Bessen D, Fischetti VA. Synthetic peptide vaccine against mucosal colonization by group A streptococci. I. Protection against a heterologous M serotype with shared C repeat region epitopes. J Immunol. 1990;145(4):1251–1256. [PubMed] [Google Scholar]
- 17.Dale JB. Multivalent group A streptococcal vaccine designed to optimize the immunogenicity of six tandem M protein fragments. Vaccine. 1999;17(2):193–200. doi: 10.1016/S0264-410X(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 18.Vashishtha A, Fischetti VA. Surface-exposed conserved region of the streptococcal M protein induces antibodies cross-reactive with denatured forms of myosin. J Immunol. 1993;150(10):4693–4701. [PubMed] [Google Scholar]
- 19.Kotloff KL, et al. Safety and immunogenicity of a recombinant multivalent group a streptococcal vaccine in healthy adults: phase 1 trial. JAMA. 2004;292(6):709–715. doi: 10.1001/jama.292.6.709. [DOI] [PubMed] [Google Scholar]
- 20.Martins TB, et al. Comprehensive analysis of antibody responses to streptococcal and tissue antigens in patients with acute rheumatic fever. Int Immunol. 2008;20(3):445–452. doi: 10.1093/intimm/dxn004. [DOI] [PubMed] [Google Scholar]
- 21.McNeil SA, et al. Safety and immunogenicity of 26-valent group a streptococcus vaccine in healthy adult volunteers. Clin Infect Dis. 2005;41(8):1114–1122. doi: 10.1086/444458. [DOI] [PubMed] [Google Scholar]
- 22.Steer AC, et al. Status of research and development of vaccines for Streptococcus pyogenes. Vaccine. 2016;34(26):2953–2958. doi: 10.1016/j.vaccine.2016.03.073. [DOI] [PubMed] [Google Scholar]
- 23.Shea JE, Santangelo JD, Feldman RG. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr Opin Microbiol. 2000;3(5):451–458. doi: 10.1016/S1369-5274(00)00120-X. [DOI] [PubMed] [Google Scholar]
- 24.Veeranagouda Y, Didier M. Transposon Insertion Site Sequencing (TIS-Seq): an efficient and high-throughput method for determining transposon insertion site(s) and their relative abundances in a PiggyBac transposon mutant pool by next-generation sequencing. Curr Protoc Mol Biol. 2017;120:21.35.1–21.35.11. doi: 10.1002/cpmb.47. [DOI] [PubMed] [Google Scholar]
- 25.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19(12):2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barquist L, et al. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics. 2016;32(7):1109–1111. doi: 10.1093/bioinformatics/btw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kizy AE, Neely MN. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect Immun. 2009;77(5):1854–1865. doi: 10.1128/IAI.01306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Breton Y, et al. Genome-wide identification of genes required for fitness of group A Streptococcus in human blood. Infect Immun. 2013;81(3):862–875. doi: 10.1128/IAI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L, Charbonneau ARL, Waller AS, Olsen RJ, Beres SB, Musser JM. Novel genes required for the fitness of Streptococcus pyogenes in human saliva. mSphere. 2017;2(6):e00460-17. doi: 10.1128/mSphereDirect.00460-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Breton Y, et al. Genome-wide discovery of novel M1T1 group A streptococcal determinants important for fitness and virulence during soft-tissue infection. PLoS Pathog. 2017;13(8):e1006584. doi: 10.1371/journal.ppat.1006584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu L, et al. Gene fitness landscape of group A streptococcus during necrotizing myositis. J Clin Invest. 2019;129(2):887–901. doi: 10.1172/JCI124994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu L, et al. Genome-wide screens identify group A streptococcus surface proteins promoting female genital tract colonization and virulence. Am J Pathol. 2020;190(4):862–873. doi: 10.1016/j.ajpath.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam FM, Turner CE, Smith K, Wiles S, Sriskandan S. Inactivation of the CovR/S virulence regulator impairs infection in an improved murine model of Streptococcus pyogenes naso-pharyngeal infection. PLoS One. 2013;8(4):e61655. doi: 10.1371/journal.pone.0061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park HS, Costalonga M, Reinhardt RL, Dombek PE, Jenkins MK, Cleary PP. Primary induction of CD4 T cell responses in nasal associated lymphoid tissue during group A streptococcal infection. Eur J Immunol. 2004;34(10):2843–2853. doi: 10.1002/eji.200425242. [DOI] [PubMed] [Google Scholar]
- 36.Dileepan T, Linehan JL, Moon JJ, Pepper M, Jenkins MK, Cleary PP. Robust antigen specific th17 T cell response to group A Streptococcus is dependent on IL-6 and intranasal route of infection. PLoS Pathog. 2011;7(9):e1002252. doi: 10.1371/journal.ppat.1002252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park HS, Francis KP, Yu J, Cleary PP. Membranous cells in nasal-associated lymphoid tissue: a portal of entry for the respiratory mucosal pathogen group A streptococcus. J Immunol. 2003;171(5):2532–2537. doi: 10.4049/jimmunol.171.5.2532. [DOI] [PubMed] [Google Scholar]
- 38.Zeppa JJ, Kasper KJ, Mohorovic I, Mazzuca DM, Haeryfar SMM, McCormick JK. Nasopharyngeal infection by Streptococcus pyogenes requires superantigen-responsive Vβ-specific T cells. Proc Natl Acad Sci U S A. 2017;114(38):10226–10231. doi: 10.1073/pnas.1700858114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watson ME Jr, Neely MN, Caparon MG. Animal models of Streptococcus pyogenes infection. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, Oklahoma, USA: University of Oklahoma Health Sciences Center; 2016. Accessed May 18, 2020. https://www.ncbi.nlm.nih.gov/books/NBK333421/
- 40.Virtaneva K, et al. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71(4):2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 2005;102(25):9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasser W, et al. Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci U S A. 2014;111(17):E1768–E1776. doi: 10.1073/pnas.1403138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shea PR, et al. Interactome analysis of longitudinal pharyngeal infection of cynomolgus macaques by group A Streptococcus. Proc Natl Acad Sci U S A. 2010;107(10):4693–4698. doi: 10.1073/pnas.0906384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitkiewicz I, Nagiec MJ, Sumby P, Butler SD, Cywes-Bentley C, Musser JM. Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc Natl Acad Sci U S A. 2006;103(43):16009–16014. doi: 10.1073/pnas.0607669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumby P, et al. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A. 2005;102(5):1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sumby P, Tart AH, Musser JM. A non-human primate model of acute group a Streptococcus pharyngitis. Methods Mol Biol. 2008;431:255–267. doi: 10.1007/978-1-60327-032-8_20. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015;125(9):3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcum JA, Kline DL. Species specificity of streptokinase. Comp Biochem Physiol, B. 1983;75(3):389–394. doi: 10.1016/0305-0491(83)90345-0. [DOI] [PubMed] [Google Scholar]
- 49. Proft T, Fraser JD. Streptococcal superantigens: biological properties and potential role in disease. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, Oklahoma, USA: University of Oklahoma Health Sciences Center; 2016. Accessed May 18, 2020. https://www.ncbi.nlm.nih.gov/books/NBK333435/
- 50.Feinstein AR, Levitt M. The role of tonsils in predisposing to streptococcal infections and recurrences of rheumatic fever. N Engl J Med. 1970;282(6):285–291. doi: 10.1056/NEJM197002052820601. [DOI] [PubMed] [Google Scholar]
- 51.Ji Y, McLandsborough L, Kondagunta A, Cleary PP. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64(2):503–510. doi: 10.1128/IAI.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lynskey NN, et al. Multi-functional mechanisms of immune evasion by the streptococcal complement inhibitor C5a peptidase. PLoS Pathog. 2017;13(8):e1006493. doi: 10.1371/journal.ppat.1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frick IM, Shannon O, Neumann A, Karlsson C, Wikstrom M, Bjorck L. Streptococcal inhibitor of complement (SIC) modulates fibrinolysis and enhances bacterial survival within fibrin clots. J Biol Chem. 2018;293(35):13578–13591. doi: 10.1074/jbc.RA118.001988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernie-King BA, Seilly DJ, Willers C, Würzner R, Davies A, Lachmann PJ. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunology. 2001;103(3):390–398. doi: 10.1046/j.1365-2567.2001.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horstmann RD, Sievertsen HJ, Knobloch J, Fischetti VA. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci U S A. 1988;85(5):1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchanan JT, et al. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16(4):396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 57.Uchiyama S, Andreoni F, Schuepbach RA, Nizet V, Zinkernagel AS. DNase Sda1 allows invasive M1T1 Group A Streptococcus to prevent TLR9-dependent recognition. PLoS Pathog. 2012;8(6):e1002736. doi: 10.1371/journal.ppat.1002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasmussen M, Edén A, Björck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun. 2000;68(11):6370–6377. doi: 10.1128/IAI.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsatsaronis JA, et al. Streptococcal collagen-like protein A and general stress protein 24 are immunomodulating virulence factors of group A Streptococcus. FASEB J. 2013;27(7):2633–2643. doi: 10.1096/fj.12-226662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Podbielski A, Hawlitzky J, Pack TD, Flosdorff A, Boyle MD. A group A streptococcal Enn protein potentially resulting from intergenomic recombination exhibits atypical immunoglobulin-binding characteristics. Mol Microbiol. 1994;12(5):725–736. doi: 10.1111/j.1365-2958.1994.tb01060.x. [DOI] [PubMed] [Google Scholar]
- 61.Timmer AM, et al. Serum opacity factor promotes group A streptococcal epithelial cell invasion and virulence. Mol Microbiol. 2006;62(1):15–25. doi: 10.1111/j.1365-2958.2006.05337.x. [DOI] [PubMed] [Google Scholar]
- 62.Gallotta M, et al. SpyAD, a moonlighting protein of group A Streptococcus contributing to bacterial division and host cell adhesion. Infect Immun. 2014;82(7):2890–2901. doi: 10.1128/IAI.00064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramachandran V, et al. Two distinct genotypes of prtF2, encoding a fibronectin binding protein, and evolution of the gene family in Streptococcus pyogenes. J Bacteriol. 2004;186(22):7601–7609. doi: 10.1128/JB.186.22.7601-7609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoff JS, DeWald M, Moseley SL, Collins CM, Voyich JM. SpyA, a C3-like ADP-ribosyltransferase, contributes to virulence in a mouse subcutaneous model of Streptococcus pyogenes infection. Infect Immun. 2011;79(6):2404–2411. doi: 10.1128/IAI.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voyich JM, et al. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci U S A. 2003;100(4):1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Casal J, Caparon MG, Scott JR. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173(8):2617–2624. doi: 10.1128/JB.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gase K, Ferretti JJ, Primeaux C, McShan WM. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect Immun. 1999;67(9):4725–4731. doi: 10.1128/IAI.67.9.4725-4731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurosawa M, Oda M, Domon H, Saitoh I, Hayasaki H, Terao Y. Streptococcus pyogenes CAMP factor attenuates phagocytic activity of RAW 264.7 cells. Microbes Infect. 2016;18(2):118–127. doi: 10.1016/j.micinf.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 69.Lei B, et al. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect Immun. 2002;70(8):4494–4500. doi: 10.1128/IAI.70.8.4494-4500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015;43(1):418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brenot A, King KY, Caparon MG. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol. 2005;55(1):221–234. doi: 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 72.Voyich JM, et al. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J Immunol. 2004;173(2):1194–1201. doi: 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- 73.Brandt CM, Spellerberg B. Human infections due to Streptococcus dysgalactiae subspecies equisimilis. Clin Infect Dis. 2009;49(5):766–772. doi: 10.1086/605085. [DOI] [PubMed] [Google Scholar]
- 74.Frandsen EV, Pedrazzoli V, Kilian M. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol. 1991;6(3):129–133. doi: 10.1111/j.1399-302X.1991.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 75.Handley P, Coykendall A, Beighton D, Hardie JM, Whiley RA. Streptococcus crista sp. nov., a viridans streptococcus with tufted fibrils, isolated from the human oral cavity and throat. Int J Syst Bacteriol. 1991;41(4):543–547. doi: 10.1099/00207713-41-4-543. [DOI] [PubMed] [Google Scholar]
- 76.Rodríguez-Ortega MJ, et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol. 2006;24(2):191–197. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- 77.Lei B, Liu M, Chesney GL, Musser JM. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis. 2004;189(1):79–89. doi: 10.1086/380491. [DOI] [PubMed] [Google Scholar]
- 78. Dale JB, et al. Current approaches to group A streptococcal vaccine development. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. Oklahoma City, Oklahoma, USA: University of Oklahoma Health Sciences Center; 2016. Accessed May 18, 2020. https://www.ncbi.nlm.nih.gov/books/NBK333413/
- 79.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci. Vaccine. 2011;29(46):8175–8178. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dale JB, et al. Potential coverage of a multivalent M protein-based group A streptococcal vaccine. Vaccine. 2013;31(12):1576–1581. doi: 10.1016/j.vaccine.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Courtney HS, Hasty DL, Dale JB. Serum opacity factor (SOF) of Streptococcus pyogenes evokes antibodies that opsonize homologous and heterologous SOF-positive serotypes of group A streptococci. Infect Immun. 2003;71(9):5097–5103. doi: 10.1128/IAI.71.9.5097-5103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bensi G, et al. Multi high-throughput approach for highly selective identification of vaccine candidates: the Group A Streptococcus case. Mol Cell Proteomics. 2012;11(6):M111.015693. doi: 10.1074/mcp.M111.015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wong SM, Bernui M, Shen H, Akerley BJ. Genome-wide fitness profiling reveals adaptations required by Haemophilus in coinfection with influenza A virus in the murine lung. Proc Natl Acad Sci U S A. 2013;110(38):15413–15418. doi: 10.1073/pnas.1311217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCarthy AJ, Stabler RA, Taylor PW. Genome-wide identification by transposon insertion sequencing of Escherichia coli K1 genes essential for in vitro growth, gastrointestinal colonizing capacity, and survival in serum. J Bacteriol. 2018;200(7):e00698-17. doi: 10.1128/JB.00698-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang N, Ozer EA, Mandel MJ, Hauser AR. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio. 2014;5(3):e01163–e01114. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Armbruster CE, et al. Genome-wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog. 2017;13(6):e1006434. doi: 10.1371/journal.ppat.1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levy O. Antimicrobial proteins and peptides of blood: templates for novel antimicrobial agents. Blood. 2000;96(8):2664–2672. [PubMed] [Google Scholar]
- 89.Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011;343(1):227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Battistone GC, Burnett GW. The free amino acid composition of human saliva. Arch Oral Biol. 1961;3:161–170. doi: 10.1016/0003-9969(61)90133-9. [DOI] [PubMed] [Google Scholar]
- 91.McMenamy RH, Lund CC, Oncley JL. Unbound amino acid concentrations in human blood plasmas. J Clin Invest. 1957;36(12):1672–1679. doi: 10.1172/JCI103568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5(14):1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 93.González-Amaro R, Sánchez-Madrid F. Cell adhesion molecules: selectins and integrins. Crit Rev Immunol. 1999;19(5-6):389–429. [PubMed] [Google Scholar]
- 94.Nathan CF. Neutrophil activation on biological surfaces. Massive secretion of hydrogen peroxide in response to products of macrophages and lymphocytes. J Clin Invest. 1987;80(6):1550–1560. doi: 10.1172/JCI113241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hofman P, et al. Increased Escherichia coli phagocytosis in neutrophils that have transmigrated across a cultured intestinal epithelium. Infect Immun. 2000;68(2):449–455. doi: 10.1128/IAI.68.2.449-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simons MP, Nauseef WM, Apicella MA. Interactions of Neisseria gonorrhoeae with adherent polymorphonuclear leukocytes. Infect Immun. 2005;73(4):1971–1977. doi: 10.1128/IAI.73.4.1971-1977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu T, Porter AR, Kennedy AD, Kobayashi SD, DeLeo FR. Phagocytosis and killing of Staphylococcus aureus by human neutrophils. J Innate Immun. 2014;6(5):639–649. doi: 10.1159/000360478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kachroo P, et al. New pathogenesis mechanisms and translational leads identified by multidimensional analysis of necrotizing myositis in primates. mBio. 2020;11(1):e03363-19. doi: 10.1128/mBio.03363-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu M, Lei B. Heme transfer from streptococcal cell surface protein Shp to HtsA of transporter HtsABC. Infect Immun. 2005;73(8):5086–5092. doi: 10.1128/IAI.73.8.5086-5092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carver PL. The battle for iron between humans and microbes. Curr Med Chem. 2018;25(1):85–96. doi: 10.2174/0929867324666170720110049. [DOI] [PubMed] [Google Scholar]
- 101.Cook LCC, Chatterjee N, Li Y, Andrade J, Federle MJ, Eichenbaum Z. Transcriptomic analysis of Streptococcus pyogenes colonizing the vaginal mucosa identifies hupY, an MtsR-regulated adhesin involved in heme utilization. mBio. 2019;10(3):e00848-19. doi: 10.1128/mBio.00848-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song Y, et al. The heme transporter HtsABC of group A Streptococcus contributes to virulence and innate immune evasion in murine skin infections. Front Microbiol. 2018;9:1105. doi: 10.3389/fmicb.2018.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X, Lu C, Zhang F, Song Y, Cai M, Zhu H. Streptococcal heme binding protein (Shp) promotes virulence and contributes to the pathogenesis of group A Streptococcus infection. Pathog Dis. 2017;75(7):ftx085. doi: 10.1093/femspd/ftx085. [DOI] [PubMed] [Google Scholar]
- 104.Bates CS, Montañez GE, Woods CR, Vincent RM, Eichenbaum Z. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect Immun. 2003;71(3):1042–1055. doi: 10.1128/IAI.71.3.1042-1055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parrow NL, Fleming RE, Minnick MF. Sequestration and scavenging of iron in infection. Infect Immun. 2013;81(10):3503–3514. doi: 10.1128/IAI.00602-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sword CP. Mechanisms of pathogenesis in Listeria monocytogenes infection. I. Influence of iron. J Bacteriol. 1966;92(3):536–542. doi: 10.1128/JB.92.3.536-542.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quenee LE, et al. Hereditary hemochromatosis restores the virulence of plague vaccine strains. J Infect Dis. 2012;206(7):1050–1058. doi: 10.1093/infdis/jis433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arezes J, et al. Hepcidin-induced hypoferremia is a critical host defense mechanism against the siderophilic bacterium Vibrio vulnificus. Cell Host Microbe. 2015;17(1):47–57. doi: 10.1016/j.chom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller HK, Schwiesow L, Au-Yeung W, Auerbuch V. Hereditary hemochromatosis predisposes mice to Yersinia pseudotuberculosis infection even in the absence of the type III secretion system. Front Cell Infect Microbiol. 2016;6:69. doi: 10.3389/fcimb.2016.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lin F, et al. Cell wall composition and candidate biosynthesis gene expression during rice development. Plant Cell Physiol. 2016;57(10):2058–2075. doi: 10.1093/pcp/pcw125. [DOI] [PubMed] [Google Scholar]
- 111.Charbonneau ARL, et al. Defining the ABC of gene essentiality in streptococci. BMC Genomics. 2017;18(1):426. doi: 10.1186/s12864-017-3794-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shelburne SA, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci U S A. 2005;102(44):16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Olsen RJ, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci U S A. 2010;107(2):888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Do H, et al. Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway. Nat Commun. 2019;10(1):2586. doi: 10.1038/s41467-019-10556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Makthal N, Gavagan M, Do H, Olsen RJ, Musser JM, Kumaraswami M. Structural and functional analysis of RopB: a major virulence regulator in Streptococcus pyogenes. Mol Microbiol. 2016;(99):1119–1133. doi: 10.1111/mmi.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci U S A. 2002;99(10):6901–6906. doi: 10.1073/pnas.092148299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.