Abstract
Efficient microbial synthesis of chemicals requires the coordinated supply of precursors and cofactors to maintain cell growth and product formation. Substrates with different entry points into the metabolic network have different energetic and redox statuses. Generally, substrate cofeeding could bypass the lengthy and highly regulated native metabolism and facilitates high carbon conversion rate. Aiming to efficiently synthesize the high-value rose-smell 2-phenylethanol (2-PE) in Y. lipolytica, we analyzed the stoichiometric constraints of the Ehrlich pathway and identified that the selectivity of the Ehrlich pathway and the availability of 2-oxoglutarate are the rate-limiting factors. Stepwise refactoring of the Ehrlich pathway led us to identify the optimal catalytic modules consisting of l-phenylalanine permease, ketoacid aminotransferase, phenylpyruvate decarboxylase, phenylacetaldehyde reductase, and alcohol dehydrogenase. On the other hand, mitochondrial compartmentalization of 2-oxoglutarate inherently creates a bottleneck for efficient assimilation of l-phenylalanine, which limits 2-PE production. To improve 2-oxoglutarate (aKG) trafficking across the mitochondria membrane, we constructed a cytosolic aKG source pathway by coupling a bacterial aconitase with a native isocitrate dehydrogenase (ylIDP2). Additionally, we also engineered dicarboxylic acid transporters to further improve the 2-oxoglutarate availability. Furthermore, by blocking the precursor-competing pathways and mitigating fatty acid synthesis, the engineered strain produced 2669.54 mg/L of 2-PE in shake flasks, a 4.16-fold increase over the starting strain. The carbon conversion yield reaches 0.702 g/g from l-phenylalanine, 95.0% of the theoretical maximal. The reported work expands our ability to harness the Ehrlich pathway for production of high-value aromatics in oleaginous yeast species.
Keywords: stoichiometric model, Yarrowia lipolytica, cofactor engineering, 2-phenylethanol, pathway selectivity, Ehrlich pathway
Metabolic engineering is the enabling technology to rewire cellular endogenous metabolism for the optimal production of native metabolites, or endow cells with the capacity for synthesizing nonnative chemicals.1 To date, there are numerous studies of engineering microbes for production of commodity chemicals and natural products, including the production of cannabinoids,2 artemisinin,3 taxol precursors,4,5 branched-chain alcohols,6,7 ornithine,8 arginine,9 advanced biofuels,10−13 and so on, from different host organisms. The primary goal of metabolic engineers is to achieve high titer, yield, and productivity (TYP) with improved cost-efficiency and process economics.14,15 In the past decades, a number of prominent metabolic engineering strategies and methodologies have been adopted to balance or redirect metabolic flux, such as modular pathway engineering,10 dynamic pathway regulation,16−19 cofactor engineering,20,21 and scaffold-guided spatial colocalization of metabolic enzymes,22,23et al. The spatiotemporal or ratiometric control of enzyme expression have been proven as efficient strategies to relieve metabolic burden and improve the TYP index.
However, the complex and lengthy synthetic routes from the starting carbons, such as glucose, commonly generate low yield and suboptimal productivity of desired products, which could be attributed to that complex biosynthesis requires the coordinated supply of precursors, ATPs, and reducing equivalents (NADPH/NADH) to maintain both cell metabolism and product formation.24,25 Substrates with different entry points into the metabolic network have different energetic and redox statuses,24,26 which leads to pathway-dependent product yields that critically affect cost-efficiency. Thus, providing microbes with multiple carbon sources can potentially mitigate these metabolic constraints, since it adds an additional degree of freedom to cellular systems that can be leveraged for product formation (namely, cosubstrate feeding).26 One recent example is to use the acetate-driven acetyl-CoA metabolic shortcut to bypass the lengthy and highly regulated glycolytic pathway for efficient polyketides27 and lipids production.24 Such substrate doping strategy may present a metabolic advantage over a single substrate and lead to metabolic optimality beyond what can be achieved in single substrate fermentations. In other words, single substrate strategies may not be suitable for particular metabolic engineering applications. Instead, metabolite doping or cosubstrate feeding can overcome intrinsic pathway limitations and achieve high carbon conversion efficiency and cost-efficiency.
2-Phenylethanol (2-PE) is a high-value compound widely used in the food, fragrance, and flavor industries. High-quality 2-PE is primarily extracted from the volatile oil components of rose flower, with a marketed price ranging from $150/kg to $200/kg, which suffers from economics and scalability issues for sustainable production.28 Chemically synthesized 2-PE, despite its low-cost and high conversion rate, is largely rejected by consumers and IFF society (International Flavors & Fragrances) due to safety and health issues.29 As a result, heterologous production of 2-PE from microbial host is considered as an economically viable alternative to plant extraction. Herein, Yarrowia lipolytica was chosen as the host strain because of its strong acetyl-CoA flux and high TCA metabolic activity and the ease of genetic toolbox,30 as demonstrated by its superior performance for production of advanced biofuels and oleochemicals.31 In addition, Y. lipolytica is also a “Generally Regarded As Safe” (GRAS) organism in the food and nutraceutical industry.32
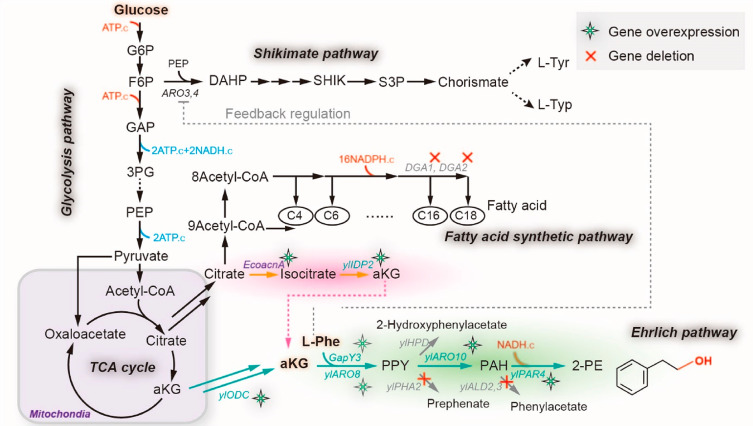
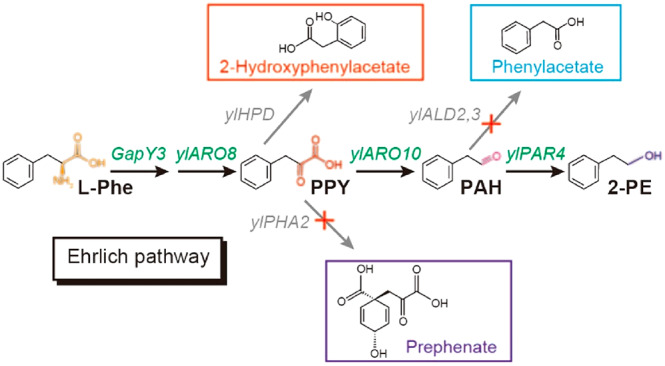
To demonstrate the utility of substrate cofeeding, we harnessed the Ehrlich pathway of Yarrowia lipolytica to optimize the biosynthesis of 2-phenylethanol (2-PE), with glucose and l-phenylalanine as the cosubstrates. The bioconversion process involves deamination, decarboxylation, and aldehyde reduction (Figure 1), with several aromatic byproducts branched out from this pathway (Figure 1). In particular, the deamination is an α-ketoglutarate (aKG)-coupled amino transfer reaction and aldehyde reduction is a NADH-dependent reduction reaction (Figure 1). Both the aKG and NADH, as cofactors, are derived from glucose. To improve the precursor flux and maximize 2-PE production, we have applied a stoichiometric model to determine the pathway yield and predict engineering targets (Supplementary Note). To improve the overall pathway specificity, we characterized and reconfigured all the biochemical reactions in Ehrlich pathway by stepwise pathway refactoring more than 20 enzymes, and further blocked the competing pathways to eliminate any byproducts. On the other hand, cytosolic 2-oxoglutarate (aKG) is the cofactor to complete the amine transfer reaction. But aKG is derived from the oxidative decarboxylation of isocitrate that is primarily compartmentalized in mitochondria, creating a bottleneck for efficient assimilation of l-Phe, which limits 2-PE production. To improve metabolite trafficking across the mitochondria membrane, we constructed an artificial aKG source pathway in the cytosol, by coupling a cytosolic aconitase with the overflowed citrate from mitochondria. In addition, we further improved the aKG supply and 2-PE production by mitigating fatty acid synthesis. As a result, the engineered strain (po1fk7P) produced 2669.54 mg/L of 2-PE with 0.702 g/g conversion yield from l-Phe, representing about 4.16-fold and 2.07-fold increase over the starting strain (641.59 mg/L of 2-PE production with 0.339 g/gl-phenylalanine), respectively. Taken together, refactoring critical enzymes and increasing metabolite trafficking as well as mitigating competitive pathways are effective strategies to improve the efficiency of overall pathway yield.
Figure 1.
Central carbon metabolism and Ehrlich pathway for synthesizing 2-PE in Y. lipolytica. l-Phe, l-phenylalanine; PPY, phenylpyruvate; PAH, phenylacetaldehyde; GapY3, l-phenylalanine specific permease; ylARO8, l-phenylalanine transaminases; ylHPD, 4-hydroxyphenylpyruvate dioxygenase; ylPHA2, prephenate dehydratase; ylARO10, phenylpyruvate decarboxylase; ylPAR4, phenylacetaldehyde reductases; ALD2,3, aldehyde dehydrogenase; G6P, glucose-6-phosphate; F6P, fructose 6-phosphate; GAP, glyceraldehyde-3-phosphate; 3PG, 3-phospho-glycerate; PEP, phosphoenolpyruvate; aKG, 2-oxoglutarate; DAHP, 3-deoxy-arabino-heptulonate-7-phosphate; SHIK, shikimate ; S3P, shikimate-3-phosphate; l-Tyr, l-tyrosine; l-Typ, L-tryptophan; ARO3,4, 3-deoxy-7-phosphoheptulonate synthase; ylODC, 2-oxodicarboxylate carrier; ylIDP2, cytosolic isocitrate dehydrogenase; EcoacnA, aconitate hydratases; DGA1,2, diacylglycerol acyltransferase;
Results and Discussion
Characterization of Ehrlich Pathway for 2-PE Synthesis in Y. lipolytica
Microbially produced 2-PE is mainly obtained by two routes, namely, the de novo pathway from glucose and bioconversion from l-phenylalanine by the Ehrlich pathway. Owing to the hard-wired, tightly complex feedback regulation33 and lengthy reaction steps (>20 steps) of the de novo pathway, bioconversion by the Ehrlich pathway is considered as the preferred biological route to synthesize 2-PE.28 In the Ehrlich pathway (Figure 1), l-phenylalanine is converted to 2-PE through four enzyme-dependent processes: (i) extracellular l-phenylalanine is internalized by amino permeases; (ii) l-phenylalanine is transaminated to phenylpyruvate by amino transferase with 2-oxoglutarate (aKG) as the amine-receptor; (iii) phenylpyruvate is further decarboxylated to phenylacetaldehyde by phenylpyruvate decarboxylases; (iv) and finally, phenylacetaldehyde is reduced to 2-PE by alcohol dehydrogenases or phenylacetaldehyde reductase with NADH as cofactor.
In Y. lipolytica, transaminases and the phenylpyruvate decarboxylase are ylARO8 (encoded by gene YALI0E20977g) or ylARO9 (encoded by gene YALI0C05258g) and ylARO10 (encoded by gene YALI0D06930g), respectively. However, the specific l-phenylalanine permease and phenylacetaldehyde alcohol dehydrogenases have not been characterized. To determine the ability of Y. lipolytica in producing 2-PE, Y. lipolytica po1g (leu–) harboring an empty plasmid pYXLP′ (strain po1g pYXLP′) was used as control to incubate with different concentrations of l-phenylalanine (0, 2, 4, 6, 8, and 10 g/L) in shake flasks. Our results (Figure 2a) indicate that the highest titer of 2-PE reached 720.43 mg/L with 0.342 g/gl-phenylalanine yield by adding 10 g/L l-phenylalanine, accompanying with 197.59 mg/L of byproduct phenylacetate. Considering economic efficiency (incomplete conversion of l-Phe), 4 g/L of l-phe was able to support 641.59 mg/L (Figure 2b) of 2-PE production with 0.339 g/gl-phenylalanine yield (Figure 2c). Thus, in the following work, 4 g/L of l-phe was determined for bioconversion.
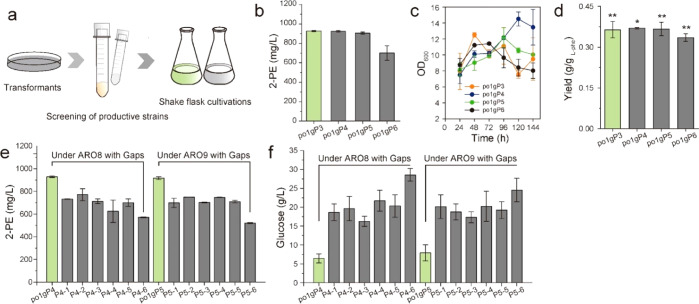
Figure 2.
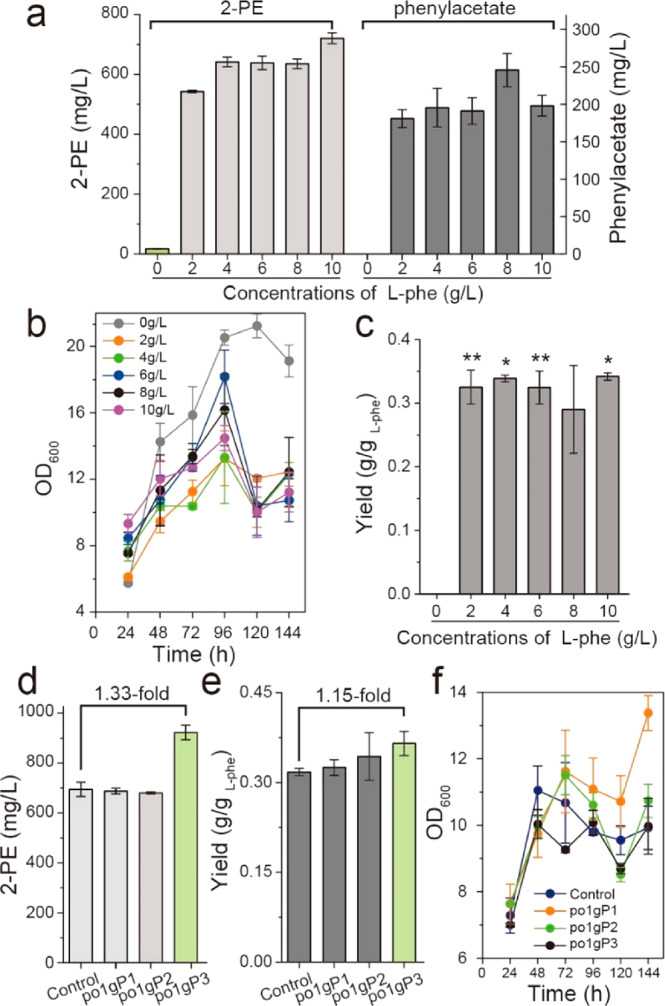
Initial assessment of using Y. lipolytica to produce 2-PE from l-phenylalanine via the Ehrlich pathway. (a–c) 2-PE and phenylacetate titer, time profiles of cell growth, and 2-PE yield of adding different concentrations l-phenylalanine (0, 2, 4, 6, 8, and 10 g/L). (d–f) 2-PE titer, yield, and cell growth of respective overexpressing genes ylARO8, ylARO9, and ylARO10. All experiments were performed in triplicate and error bars show standard deviation (SD). The * indicates p < 0.01, and ** indicates p < 0.05 (two-tailed test).
Refactoring the Upstream Ehrlich Pathway to Improve 2-PE Yield
As suggested by the mathematical model (Supplementary Note S1), improving the catalytic efficiency of the Ehrlich pathway and increasing aKG supplementation are the key determinants to improve 2-PE yield from l-phenylalanine. To refactor the Ehrlich pathway, we adopted a stepwise pathway engineering strategy. First, the native ylARO8, ylARO9, and ylARO10 genes in Ehrlich pathway were overexpressed in Y. lipolytica po1g under the control of strong constitutive TEF-intron promoter. Shaking flask cultivation (Figure 2d, and Figure 2e,f for cell growth and 2-PE yield) indicates that overexpression of ylARO8 and ylARO9 (strain po1gP1 and po1gP2) has little effect on the 2-PE production. However, overexpression of ylARO10 (strain po1gP3) increased 2-PE titer to 922.86 mg/L, about 33% higher than the control strain (Figure 2d), suggesting that the reaction catalyzed by phenylpyruvate decarboxylase ylARO10 is a limiting-step in Ehrlich pathway. Thus, the ineffectiveness of overexpressing downstream genes (ylARO8 or ylARO9) is likely due to the limited performance of the upstream ylARO10. Specifically, ylARO8 and ylARO9 are both transaminases, which catalyze the transamination of l-phenylalanine with different amine-receptors:
l-Phenylalanine + 2-Oxoglutarate → Phenylpyruvate + l-Glutamate (catalyzed by YlARO8)
l-Phenylalanine + Pyruvate → Phenylpyruvate + l-Alanine (catalyzed by YlARO9)
Apparently, different amine-receptors (that is 2-oxoglutarate and pyruvate) will lead to distinct outcomes (that is l-glutamate and l-alanine) that may affect 2-PE production. Therefore, we next constructed strains carrying pYLXP′-ylARO10-ylARO8 and pYLXP′-ylARO10-ylARO9 to identify the optimal transaminases, and obtained po1gP4 and po1gP5. However, screening of these two strains showed no significant differences in 2-PE production (Figure 3b, and Figure 3c,d for cell growth and 2-PE yield). We also overexpressed a transcriptional activator (encoded by YALI0C18645) to up-regulate the expression of ylARO8 or ylARO9.29 However, cultivation of this strain (po1gP6) showed deteriorated 2-PE production (Figure 3b). With this, we speculated that cellular uptake of l-phe might be the bottleneck limiting 2-PE titer.
Figure 3.
Characterization of upstream module of the Ehrlich pathway: l-phenylalanine specific permeases and l-phe: 2-oxoglutarate transaminase. (a) Flow-chart of shake flask cultivations of engineered strains. (b–d) 2-PE titers, cell growth, and 2-PE yield of respective overexpressing genes ylARO10 and ylARO8, ylARO10 and ylARO9, and ylARO10 and C18645. (e,f) 2-PE titers and 96h residual glucose of screening l-phenylalanine specific permeases with CSM fermentation medium in shake cultivations. All experiments were performed in triplicate and error bars show standard deviation (SD). The * indicates p < 0.01, and ** indicates p < 0.05 (two-tailed test).
To facilitate the cellular uptake for l-phe, we assessed a number of l-phe permease, including BAP and Gap1 from S. cerevisiae,34 PheP from E. coli, and three Y. lipolytica native Gap1 homologues, encoded by YALI0C17237g (named as GapY1), YALI0B16522g (GapY2), and YALI0B19800g (GapY3), respectively. Beyond our expectation, all the tested l-phe permeases showed decreased 2-PE production compared with control po1gP4 and po1gP5 when coupling with expression of ylARO8-ylARO10 or ylARO9-ylARO10 (Figure 3e). Noticeably, abundant residual glucose was detected at 96 h (Figure 3f), suggesting the central carbon metabolism of these strains was strongly inhibited. This result is consistent with the finding that high level of endogenous l-phe induces a strong down regulation of cellular metabolism in S. cerevisiae.35 We reasoned that a similar effect might exist in Y. lipolytica. Thus, we dropped out all other amino acids and only used YNB-glucose with 4 g/L l-phe as the nitrogen source. As a result, the highest 2-PE titer reached 1632.73 mg/L with 0.547 g/gl-phenylalanine yield in strain po1gP4–6 (Figure 4a,b) after 144 h cultivation, indicating GapY3 is a potent l-phe permease. Nevertheless, differences of 2-PE titers in strains po1gP4–6 and po1gP5–6 (1607.46 mg/L with 0.533 g/gl-phenylalanine) were insignificant, which may be attributed to the poor substrate specificity of transaminases ylARO8 and ylARO9.
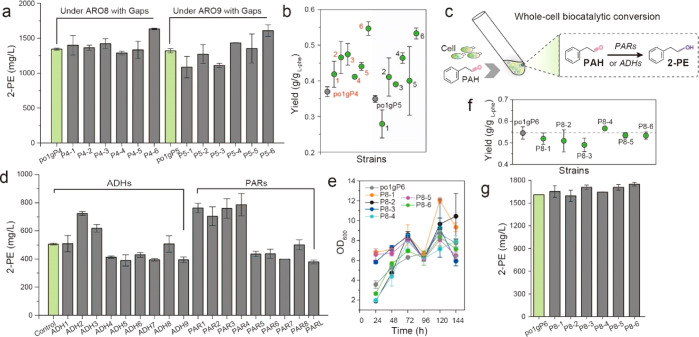
Figure 4.
Characterization of the downstream module of the Ehrlich pathway: phenylacetaldehyde reductases and alcohol dehydrogenases. (a,b) 2-PE titers and yield by rescreening l-phenylalanine specific permeases. Red 1, strain P4–1; Red 2, strain P4–2; Red 3, strain P4–3; Red 4, strain P4–4; Red 5, strain P4–5; Red 6, strain P4–6; Black 1, strain P5–1; Black 2, strain P5–2; Black 3, strain P5–3; Black 4, strain P5–4; Black 5, strain P5–5; Black 6, strain P5–6. (c) Manipulations of whole-cell biocatalytic conversion of phenylacetaldehyde. PAH, phenylacetaldehyde. (d–g) 2-PE titers, cell growth and yield of screening and identifying the optimized phenylacetaldehyde reductases or alcohol dehydrogenases. P8–1, strain po1gP8–1; P8–2, strain po1gP8–2; P8–3, strain po1gP8–3; P8–4, strain po1gP8–4; P8–5, strain po1gP8–5; P8–6, strain po1gP8–6. All experiments were performed in triplicate and error bars show standard deviation (SD).
Refactoring the Downstream Ehrlich Pathway to Improve 2-PE Yield
To refactor the optimal alcohol dehydrogenase (ADH) or phenylacetaldehyde reductase (PAR), we screened the entire genome of Y. lipolytica with putative rose phenylacetaldehyde reductase PARL as the template, and identified eight PARs with high similarity (>70%), encoded by YALI0D08844g (PAR1), YALI0F09097g (PAR2), YALI0F24937g (PAR3), YALI0D07062g (PAR4), YALI0D12386g (PAR5), YALI0C20251g (PAR6), YALI0D11616g (PAR7), and YALI0D08778g (PAR8), respectively. We also codon-optimized and synthesized the rose-derived PARL as a positive control. The resulting 9 strains, with overexpression of PAR1, PAR2, PAR3, PAR4, PAR5, PAR6, PAR7, PAR8, and PARL, were incubated with 1 g/L phenylacetaldehyde (Figure 4c), leading to 2-PE titers of 762.71, 703.62, 759.37, 786.58, 435.61, 437.33, 399.11, 501.10, and 379.28 mg/L (Figure 4d), respectively. The highest 2-PE titer was produced by strain PAR4 (YALI0D07062g). Interestingly, the rose-derived PARL performed worst in our test.
Similarly, we also investigated the performances of alcohol dehydrogenases (ADHs) in Y. lipolytica. Nine Y. lipolytica ADHs annotated by Genbank (https://www.ncbi.nlm.nih.gov/genbank/) and GRYC (http://gryc.inra.fr/) were tested, including ADH1 (YALI0D25630g), ADH2 (YALI0E17787g), ADH3 (YALI0A16379g), ADH4 (YALI0A15147g), ADH5 (YALI0E07766g), ADH6 (YALI0E15818g), ADH7 (YALI0D02167g), ADH8 (YALI1C17782g), and ADH9 (SFA1). The resulting strains were tested and only suboptimal level of 2-PE were detected in these strains compared to the strain harboring the optimal PARs (Figure 4d), and the best strain (ADH2) produced 723.74 mg/L 2-PE, which is about 10% lower than strain PAR4.
After analyzing the downstream PAR and ADH modules, we chose six efficient candidates for further investigation, including ADH2, ADH3, PAR1, PAR2, PAR3, and PAR4. These PARs and ADHs were combined with the optimal upstream module ylARO10-ylARO8-GapY3, leading to strains po1gP7–1, po1gP7–2, po1gP7–3, po1gP7–4, po1gP7–5, and po1gP7–6. When tested in shake flask with YNB-glucose media and 4 g/L l-phenylalanine, these strains produced 2-PE at 1126.62, 902.12, 1109.89, 1166.23, 960.96, and 1378.98 mg/L, respectively (Supplementary Figure S3), representing no improvements in 2-PE titer compared to control po1gP4–6 (ylARO10-ylARO8-GapY3, 1610.76 mg/L). We speculated that simultaneous overexpression of four genes (>16 000 bp) may lead to genetic instability (loss of plasmid or unequal distribution/propagation of plasmid) in Y. lipolytica. To test this hypothesis, we linearized these plasmids and integrated the long DNA cassettes at the pBR docking site of po1g chromosome, leading to strains po1gP8–1, po1gP8–2, po1gP8–3, po1gP8–4, po1gP8–5, and po1gP8–6, respectively. Shake flask screening indicates that po1gP8–6 produced 1750.46 mg/L of 2-PE with 0.534 g/g yield (Figure 4f,g, and Figure 4e for cell growth), the highest titer obtained, indicating PAR4 is the optimal downstream module that reduces phenylacetaldehyde to 2-PE. So far, the four steps of the Ehrlich pathway (permeation, transamination, decarboxylation, and reduction) have been fully refactored, and the refactored pathway represents 2.4-fold increase in 2-PE titer over the starting strain (∼710 mg/L).
Blocking Competing Pathways to Improve 2-PE Titer
To further improve 2-PE yield, we attempted to delete the competing pathways. For the convenience of the genetic manipulations, we first knocked out gene ku70 in po1f (named po1fk) to inhibit nonhomologous end joining (NHEJ) and improve the frequency of homologous recombination.
As mentioned above, the major byproduct is phenylacetate, which is the oxidized product of phenylacetaldehyde catalyzed by aldehyde dehydrogenases (ALD2 and ALD3). Thus, we sequentially knocked out both ALD2 (YALI0D07942g) and ALD3 (YALI0F04444g) in po1fk, obtaining strain po1fk1. Then the optimal upstream and downstream Ehrlich module ylARO10-ylARO8-GapY3-PAR3 was integrated at the genome pBR docking site of po1fk1, obtaining strain po1fk1P. Although shake flask cultivation of po1fk1P showed no improvement in 2-PE titer, the byproduct phenylacetate was significantly decreased to 124.49 mg/L (Figure 5b, 532.48 mg/L phenylacetate in po1gP8–6).
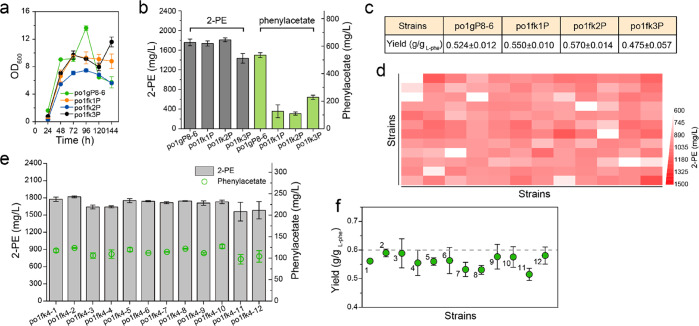
Figure 5.
Blocking competitive pathways and integration of the Ehrlich pathway in 26s rDNA sites to improve 2-PE production. (a–c) Cell growth, 2-PE titers, phenylacetate titers, and 2-PE yield of blocking competitive pathways. (d) Screening of the productive strains based on 24-well deep plate cultivations. (e,f) 2-PE titer and yield by further validating productive strains with integration of the Ehrlich pathway in flask cultivations. 1, strain po1fk4–1; 2, strain po1fk4–2; 3, strain po1fk4–3; 4, strain po1fk4–4; 5, strain po1fk4–5; 6, strain po1fk4–6; 7, strain po1fk4–7; 8, strain po1fk4–8; 9, strain po1fk4–9; 10, strain po1fk4–10; 11, strain po1fk4–11; 12, strain po1fk4–12. All experiments were performed in triplicate and error bars show standard deviation (SD).
In addition, prephenate dehydratase PHA2 catalyzes the anaplerotic reaction from phenylpyruvate to prephenate (Figure 1). Deletion of gene PHA2 in strain po1fk1 generated strain po1fk2. Chromosomally integrated (integrated at pBR docking site) Ehrlich module (ylARO10-ylARO8-GapY3-PAR3) in po1fk2 (strain po1fk2P) leads to an improved 2-PE titer of 1809.65 mg/L with the yield at 0.570 g/gl-phenylalanine compared to po1fk1P (Figure 5b,c, 1735.78 mg/L with yield at 0.550 g/gl-phenylalanine), but a significant decrease in biomass was observed in po1fk2P (Figure 5a). On the other hand, 4-hydroxyphenylpyruvate dioxygenase (encoded by gene ylHPD) could convert phenylpyruvate to 2-hydroxyphenylacetate. Therefore, we further knocked out ylHPD in strain po1fk2, obtaining strain po1fk3. However, the chromosomally integrated (integrated at pBR docking site) Ehrlich module in po1fk3 (strain po1fk3P) leads to decreased 2-PE production and increased phenylacetate titer, which were 1432.43 and 226.17 mg/L (Figure 5b), respectively. This result suggests that ylHPD cannot be knocked out in this scenario. It is not clear why the deletion of gene ylHPD has a negative result, possibly due to the incorrect annotation of the reaction directionality of ylHPD.
We have refactored the Ehrlich pathway and blocked the byproducts synthesis, and integrated the optimal gene module ylARO10-ylARO8-GapY3-PAR3 to the pBR docking site of the triple knockout strain (po1fk2, ALD2, ALD3, and PHA2 were knocked out). In previous work, our group has established a 26s rDNA/Cre-loxP-based iterative gene integration and marker-curation method.36 With more than 200 copies of ribosome DNAs in Y. lipolytica, this technique allows genes of interests to be randomly integrated into the genome with multiple copies. Thus, we next sought to apply this method and further optimize the Ehrlich pathway, including genes ylARO10, ylARO8, GapY3, and PAR4. To obtain a productive strain, we screened 144 genome-integrated transformants by 24-well deep plate cultivation in glucose-YNB medium with 4 g/L l-phenylalanine (Figure 5d). About 45% of the genome-integrated produced 2-PE at 900–1000 mg/L, less than 5% of the screened strain produced 2-PE over 1200 mg/L. Then the top-performed strains were further validated in shake flasks (Figure 5e,f). Strain po1fk4–2 produced the highest 2-PE of 1817.46 mg/L with the yield at 0.590 g/gl-phenylalanine.
A Nonnative 2-Oxoglutarate (aKG) Shuttle to Improve Metabolite Trafficking
As suggested by the mathematical model (Supplementary Note S1), we next sought to improve the precursor (2-oxoglutarate, aKG) flux. Noteworthily, Y. lipolytica is known for the strong TCA metabolic activity.37 Indeed, we detected 4.84 g/L of citrate at 72 h of cultivation in strain po1fk4–2 (Figure 6b). Thus, rewiring the overflowed citrate flux to cytosolic aKG is a feasible strategy to increase precursor (aKG) supply and improve 2-PE production. Nevertheless, Y. lipolytica lacks the cytosolic aconitate hydratase and isocitrate dehydrogenase. To construct the cytosolic pathway from citrate to aKG (Figure 6a), we introduced two aconitate hydratases, including CitB from Bacillus subtilis and AcnA from E. coli, and a cytosolic isocitrate dehydrogenase ScIDP2 from S. cerevisiae.38 Additionally, by searching the genome of Y. lipolytica with ScIDP2 as template, we retrieved a putative cytosolic isocitrate dehydrogenase ylIDP2 (encoded by gene YALI0F04095g). Subsequently, four nonnative aKG source pathways, including citB-scIDP2, citB-ylIDP2, acnA-scIDP2, and acnA-ylIDP2, were introduced into po1fk4–2 by plasmid pYLXP′. The resulting strains po1fk4P1, po1fk4P2, po1fk4P3, and po1fk4P4 produced 1733.45, 2146.45, 1737.58, and 1842.85 mg/L of 2-PE with yield of 0.574, 0.606, 0.597, and 0.591 g/gl-phenylalanine in shake flasks (Figure 6c,d,e), respectively. It indicates the coexpression of citB and ylIDP2 could effectively convert cytosolic citrate to aKG. In addition, we also overexpressed the mitochondrial 2-oxodicarboxylate carrier ylODC1 (encoded by gene YALI0D02629g) to export 2-oxoglutarate (aKG) out of the mitochondrial matrix to the cytosol (strain po1fk4P5, po1fk4–2 harboring plasmid pYLXP′-citB-ylIDP2-ylODC). As expected, the 2-PE titer and yield in po1fk4P5 were further increased, reaching 2269.08 mg/L and 0.632 g/gl-phenylalanine (Figure 7a,b,c), respectively.
Figure 6.
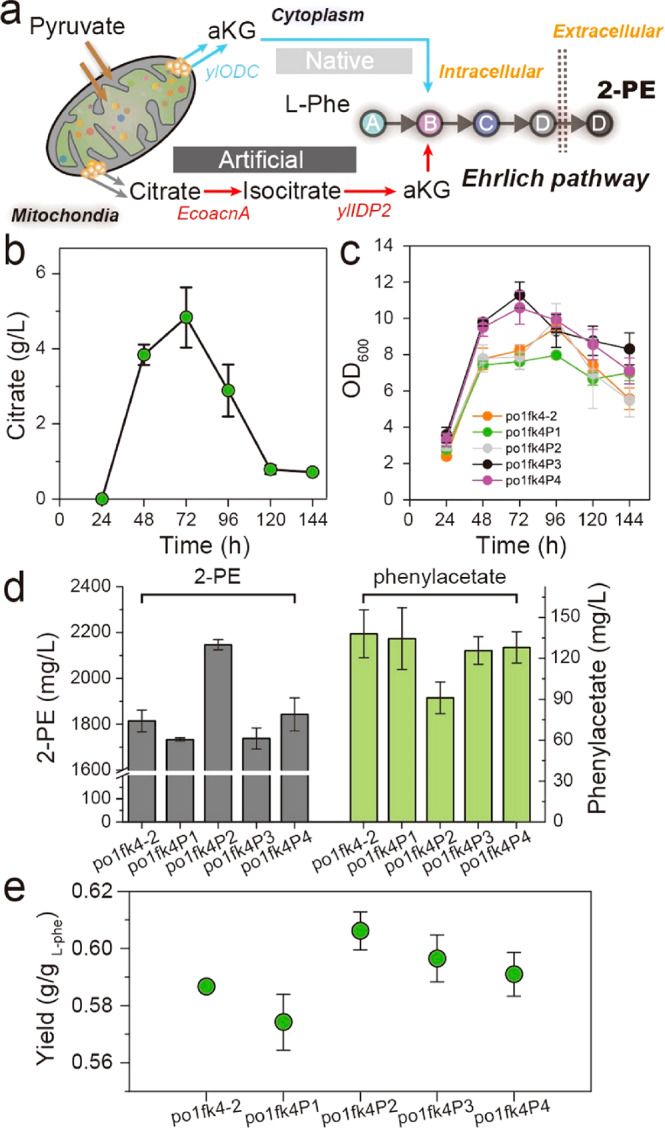
Overcoming cellular compartmentalization of 2-oxoglutarate (aKG) to increase aKG availability. (a) Construction of cytosolic source for 2-oxoglutarate synthesis by coupling bacterial aconitase with the overflowed citrate from Krebs cycle. (b) Citrate titers of strain po1fk4–2. (c–e) 2-PE titers, cell growth and yield of strains po1fk4P1, po1fk4P2, po1fk4P3, and po1fk4P4. All experiments were performed in triplicate and error bars show standard deviation (SD).
Figure 7.
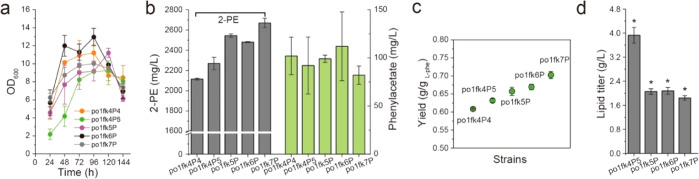
Improving 2-oxoglutarate trafficking across the mitochondria membrane by overexpressing the mitochondrial 2-oxodicarboxylate carrier ylODC1 and improving 2-PE titer by blocking fatty acid synthesis. (a–d) 2-PE titers, cell growth, yield, and lipid titers of strains po1fk4P5, po1fk5P, po1fk6P, and po1fk7P. All experiments were performed in triplicate and error bars show standard deviation (SD). The * indicates p < 0.01 (two-tailed test).
On the other hand, Y. lipolytica is known to accumulate up to 30–60% dry cell weight as lipid.10,39 As shown in Figure 1b, production of 1 mol of fatty acids (stearic acids C18:0) will consume 16 mol of NADPH and 9 mol of citrate (acetyl-CoA). As a result, the synthesis of lipid competes with aconitase (aconitate hydratases) and PARs for cytosolic citrate and NADPH. Specially, diacylglycerol acyltransferases have been identified as the rate-limiting steps to lipid synthesis via the Kennedy pathway in Y. lipolytica.40,41 Thus, we pursued further to knock out the diacylglycerol acyltransferases, encoded by ylDGA1 (YALI0E32769g) and ylDGA2 (YALI0D07986g), to mitigate fatty acid synthesis. Shake flask cultivation of the resulting strain po1fk5P (po1fk4–2 ΔylDGA1::loxP harboring pYLXP′-citB-ylIDP2-ylODC), po1fk6P (po1fk4–2 ΔylDGA2::loxP harboring pYLXP′-citB-ylIDP2-ylODC), and po1fk7P (po1fk4–2 ΔylDGA2 ΔylDGA1::loxP harboring pYLXP′-citB-ylIDP2-ylODC) all led to significant improvements in 2-PE titers and yield (Figure 7a,b,c). Specifically, strain po1fk7P produced 2669.54 mg/L of 2-PE with yield at 0.702 g/gl-phenylalanine (Figure 7a,b,c), which was 1.11-fold and 1.18-fold higher than strain po1fk4P5. Furthermore, to validate the impacts of ylDGA1 and ylDGA2 deletion on lipid production, we measured the lipid titer in strains po1fk4P5, po1fk5P, po1fk6P, and po1fk7P (Figure 7d), reaching 3.93, 2.06, 2.08, and 1.84 g/L, respectively. These results confirmed that fatty acid synthesis was indeed mitigated by deleting genes ylDGA1 and ylDGA2. In conclusion, by constructing a cytosolic aKG pathway and expression of the dicarboxylic acid transporter (ylODC) as well as mitigating fatty acids synthesis, we efficiently improved the aKG flux and 2-PE tier and yield.
Conclusions
Efficient microbial synthesis of chemicals necessitates systematic debottlenecking, refactoring and optimization of the native or nonnative metabolic pathways. With 2-PE as a tested molecule, we analyzed the stoichiometric constraints of the Ehrlich pathway toward efficient synthesis of the target molecules in Y. lipolytica. The proposed metabolic model reveals potential engineering targets for high yield production of 2-PE with cosubstrate feeding of glucose and l-phenylalanine. Specifically, guided by the model, we identified that the catalytic efficiency of the Ehrlich pathway and the precursor 2-oxoglutarate (aKG) supplement might be the rate-limiting steps. Stepwise refactoring the Ehrlich pathway and overcoming the cellular compartmentalization of precursor (aKG) significantly improved 2-PE production. By blocking the precursor-competing pathways and mitigating fatty acids synthesis, the engineered strain produced 2669.54 mg/L of 2-PE with yield at 0.702 g/g in shake flasks, which were 4.16 and 2.07-fold of the starting strain, respectively. The strategies reported in this study should be adding value and guide us to engineer other complex metabolic pathways for various biomanufacturing applications.
Further investigations on relieving 2-PE toxicity and promoting cell fitness are viable solutions to improve 2-PE production. Major considerations should include 2-PE tolerance/toxicity, medium composition, and optimal fermentation processes. In general, mutagenesis coupled with high throughput screening is an efficient and convenient way to evolve tolerance phenotype.42 In addition, small amounts of organic nitrogen sources, such as peptone and yeast extract, benefit cell growth without inhibiting the Ehrlich pathway.43 Moreover, it was found that both Ca2+ and Mg2+ salts protect the cell and facilitate 2-PE production by increasing membrane stability and integrity.44 Besides, by adding organic solvents as overlays during fermentation, 2-PE titer could be increased to 12.6 g/L in the concentrated organic layer.45 These strategies will enable us to build a sustainable 2-PE platform at low cost and high efficiency.
Materials and Methods
Strains, Plasmid, Primers, and Chemicals
All strains of engineered Y. lipolytica, including the genotypes of manipulations, recombinant plasmids, and primers, have been listed in Supplementary Table S1 and S2. Chemicals used in this study include phenylacetaldehyde, l-phenylalanine, phenylacetate, 2-phenylethanol, 2-oxoglutarate, and citrate, which were all purchased from Sigma-Aldrich.
Shake Flask and 24-Wells Deep-Plate Cultivations
All engineered strains were underwent optimal screening before shake flask cultivations (see Yeast Transformation and Screening of High-Producing Strains, Figure 3a). For shake flask cultivations, seed culture was carried out in the shaking tube with 2 mL seed culture medium at 30 °C and 250 r.p.m. for 48 h. Then, 0.8 mL of seed culture was inoculated into the 250 mL flask containing 35 mL of fermentation medium and grown under the conditions of 30 °C and 250 r.p.m. for 144 h. One milliliter of cell suspension was sampled every 24 h for OD600, glucose, 2-PE, l-phenylalanine, and penylacetate measurements. For performing 24-well deep-plate cultivation, engineered Y. lipolytica strains were first cultured in 96-wells pales with 150 μL seed culture medium at 30 °C and 1000 r.p.m. for 36 h. Sequentially, 40 μL of seed culture was inoculated to the 24-well deep plate with 2 mL fermentation medium at 30 °C and 1000 r.p.m. for 96 h, and finally, 1 mL of cell suspension was sampled for 2-PE, l-phenylalanine, and penylacetate measurements.
The seed culture medium used in this study was the yeast complete synthetic media regular media CSM containing the following: glucose 20.0 g/L, yeast nitrogen base (without ammonium sulfate) 1.7 g/L, ammonium sulfate 5.0 g/L, and CSM-Leu or CSM-Ura 0.74 g/L. Two types of fermentation medium were used in our work, including nitrogen-limited media CSM and YNB. The nitrogen-limited media CSM contained the following: glucose 40.0 g/L, yeast nitrogen base (without ammonium sulfate) 1.7 g/L, ammonium sulfate 1.1 g/L, CSM-Leu 0.74 g/L, and appropriate l-phenylalanine. The nitrogen-limited media YNB contained the following: glucose 40.0 g/L, yeast nitrogen base (without ammonium sulfate) 1.7 g/L, leucine or uracil 0.2 g/L, and appropriate l-phenylalanine.
Whole-Cell Bioconversion of Phenylacetaldehyde
To prepare the whole-cell biocatalyst, cells were harvested during the exponential growth phase (48 h) from the shake flask cultivation. Then, cells were washed twice with 100 mM phosphate buffer (pH 7.0), and resuspended to an OD600 of 4 in the same buffer. Next, whole-cell biocatalytic conversion of phenylacetaldehyde was performed in 20-ml glass tube containing 1 mL of cell suspension (OD600 = 4) and 1 mL phenylacetaldehyde–water solution (2 g/L phenylacetaldehyde) at 30 °C and 250 r.p.m. for 4 h. One hundred microliters of cell suspension was sampled every 1 h for 2-PE and penylacetate measurements.
Yeast Transformation and Screening of High-Producing Strains
The standard protocols of Y. lipolytica transformation by the lithium acetate method were described as previously reported.36,46 In brief, 1 mL of cells was harvested during the exponential growth phase (16–24 h) from 2 mL YPD medium (yeast extract 10 g/L, peptone 20 g/L, and glucose 20 g/L) in the 14 mL shake tube, and washed twice with 100 mM phosphate buffer (pH 7.0). Then, cells were resuspended in 105 μL transformation solution, containing 90 μL 50% PEG4000, 5 μL lithium acetate (2 M), 5 μL boiled single stand DNA (salmon sperm, denatured), and 5 μL DNA products (including 200–500 ng of plasmids, lined plasmids or DNA fragments), and incubated at 39 °C for 1 h, then spread on selected plates. It should be noted that the transformation mixtures needed to be vortexed for 15 s every 15 min during the process of 39 °C incubation. The selected markers, including leucine and uracil, were used in this study. All engineering strains after genetic manipulations were performed optimized screening by the shaking tube cultivations, and the optimal strain was used to perform shaking flask (these data have been shown in Supporting Information).
Expression Vectors Construction and Pathway Assembly
The YaliBrick plasmid pYLXP′ was used as the expression vector in this study.47,48 Plasmid constructions were performed by using preciously described methods.36,49 In brief, recombinant plasmids of pYLXP′-XX (a single gene expression) were obtained by Gibson Assembly method50 using linearized pYLXP′ (digested by SnaBI and KpnI) and the appropriate PCR-amplified DNA fragment. Multigenes assembly was achieved by restriction enzyme digestion subcloning based on the application of isocaudamers AvrII and NheI.47,51 All genes were respectively expressed by the TEF promoter with intron sequence and XPR terminator. The modified DNA fragments and plasmids were sequenced by Quintarabio. The endonucleases used in this research were purchased from Thermo Fisher Scientific or NEB.
Gene Knockout and 26s rDNA Genomic Integration of Ehrlich Pathway
A marker-free gene knockout method based on Cre-lox recombination system was used as previously reported.52 For performing gene knockout, the upstream and downstream sequences (both 1000 bp) flanking the deletion targets were PCR-amplified. These two fragments, the loxP-URA-loxP cassette (digested from plasmid pYLXP′-loxp-URA by AvrII and salI), and the residual plasmid backbone of pYLXP′-loxp-URA were joined by the Gibson Assembly method, obtaining the gene knockout plasmids pYLXP′-loxP-URA-XX. The obtained plasmids were sequenced by Quintarabio. Next, the gene knockout cassettes were PCR-amplified from construction plasmids pYLXP′-loxp-URA-XX, and further transformed into Y. lipolytica. The positive transformants were determined by colony PCR. Next, plasmid pYLXP′-Cre was introduced into the positive transformants and promoted the recombination of loxP sites, which recycle the selected marker. Finally, the intracellular plasmid pYLXP′-Cre was evicted by incubation at 30 °C for 48 h.
The standard protocol of 26s rDNA genomic integration also has been described in previous reported.36 For 26s rDNA genomic integration of the Ehrlich pathway, the expression plasmid pYLXP′-ylARO10-ylARO8-GapY3-PAR4 was digested by AvrII and NotI, and the fragment containing the Ehrlich pathway was inserted into linearized pYLXP′-loxp-URA digested by Nhel and NotI, getting pYLXP′-loxp-URA-ylARO10-ylARO8-GapY3-PAR4. The homologous arms of 26s rDNA, including 26s rDNA 1s and 2s, were PCR-amplified by appropriate primers. And next, these two homologous-arm fragments, Ehrlich pathway with the loxP-URA-loxP cassette (digested from plasmid pYLXP′-loxp-URA-ylARO10-ylARO8-GapY3-PAR4 by AvrII and NotI), and the residual plasmid backbone of pYLXP′-loxp-URA-ylARO10-ylARO8-GapY3-PAR4 were joined by Gibson Assembly method, obtaining the 26s rDNA genomic integration plasmids prDNAloxP-ylARO10-ylARO8-GapY3-PAR4. Then, the 26s rDNA genomic integration cassette of the Ehrlich pathway was gotten by digesting prDNAloxP-ylARO10-ylARO8-GapY3-PAR4 with AvrII and NotI, and the integration cassette was further transformed into Y. lipolytica. In addition, the manipulation of recycling the selected marker was similar to gene knockout.
Quantification of Cell Density, 2-PE, Penylacetate, l-Phenylalanine, Glucose, Citrate, Fatty Acid, and Y2-PE Calculation
Cell densities were monitored by measuring the optical density at 600 nm (OD600). The concentrations of 2-PE, penylacetate, and l-phenylalanine were measured by high-performance liquid chromatography (HPLC) through Agilent HPLC 1220 equipped with a ZORBAX Eclipse Plus C18 column (4.6 × 100 mm, 3.5 μm, Agilent) and a VWD detector. The analysis was performed at 215 nm under 40 °C column temperature with a mobile phase comprising 50% (v/v) methanol in water at a flow rate of 0.5 mL/min. The concentrations of glucose and citrate were also measured by Agilent HPLC 1220 equipped with a Supelcogel Carbohydrate column (Sigma, USA) and a refractive index detector. H2SO4 (5 mM) was used as the mobile phase at a flow rate of 0.6 mL/min at 40 °C. The method of quantification of fatty acid was used as previously reported.39 Y2-PE is the 2-PE yield relative to the consumption of l-phenylalanine.
Acknowledgments
This work was supported by the Cellular & Biochemical Engineering Program of the National Science Foundation under grant no. 1805139 and the Bill & Melinda Gates Foundation under grant no. OPP1188443. YG would like to thank the School of Biotechnology in Jiangnan University for funding support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.9b00468.
Supplementary Table S1. Strains and plasmids; Supplementary Table S2. Primers and Oligos; Supplementary Table S3. Strain performance; Supplementary Note. Stoichiometric model to assess Ehrlich pathway efficiency; Supplementary Figure S1. Stoichiometric models reveal that 2-PE yield is driven by selectivity of the Ehrlich pathway and supply of 2-oxoglutarate (aKG); Supplementary Figure S2. 2-PE titers of screening l-phenylalanine specific permeases with the addition of final concentration of 4g/L l-phenylalanine into CSM fermentation medium (48 h) in shake cultivations; Supplementary Figure S3. 2-PE titers of strain strains po1gP7–1, po1gP7–2, po1gP7–3, po1gP7–4, po1gP7–5, and po1gP7–6 in shake cultivations (PDF)
Author Contributions
PX and YG conceived the topic. YG developed the models, performed genetic engineering and fermentation experiments with input from JM and YZ. YG and PX wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Nielsen J.; Keasling J. D. (2016) Engineering Cellular Metabolism. Cell 164, 1185–1197. 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Luo X.; Reiter M. A.; d’Espaux L.; Wong J.; Denby C. M.; Lechner A.; Zhang Y.; Grzybowski A. T.; Harth S.; Lin W.; Lee H.; Yu C.; Shin J.; Deng K.; Benites V. T.; Wang G.; Baidoo E. E. K.; Chen Y.; Dev I.; Petzold C. J.; Keasling J. D. (2019) Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126. 10.1038/s41586-019-0978-9. [DOI] [PubMed] [Google Scholar]
- Westfall P. J.; Pitera D. J.; Lenihan J. R.; Eng D.; Woolard F. X.; Regentin R.; Horning T.; Tsuruta H.; Melis D. J.; Owens A.; Fickes S.; Diola D.; Benjamin K. R.; Keasling J. D.; Leavell M. D.; McPhee D. J.; Renninger N. S.; Newman J. D.; Paddon C. J. (2012) Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. U. S. A. 109, E111–E118. 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajikumar P. K.; Xiao W. H.; Tyo K. E.; Wang Y.; Simeon F.; Leonard E.; et al. (2010) Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science 330, 70–74. 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Mutanda I.; Wang K.; Yang L.; Wang J.; Wang Y. (2019) Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 10, 4850. 10.1038/s41467-019-12879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S.; Hanai T.; Liao J. C. (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Sawaya M.; Eisenberg D.; Liao J. (2008) Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U. S. A. 105, 20653–20658. 10.1073/pnas.0807157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.; Zhou Y. J.; Krivoruchko A.; Huang M.; Liu L.; Khoomrung S.; Siewers V.; Jiang B.; Nielsen J. (2015) Modular pathway rewiring of Saccharomyces cerevisiae enables high-level production of L-ornithine. Nat. Commun. 6, 8224. 10.1038/ncomms9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. H.; Kim H. U.; Kim T. Y.; Park J. S.; Kim S. S.; Lee S. Y. (2018) Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nat. Commun. 5, 4618. 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- Xu P.; Gu Q.; Wang W. Y.; Wong L.; Bower A. G. W.; Collins C. H.; Koffas M. A. G. (2013) Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 4, 8. 10.1038/ncomms2425. [DOI] [PubMed] [Google Scholar]
- Xu P.; Qiao K.; Ahn W. S.; Stephanopoulos G. (2016) Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. U. S. A. 113, 10848–10853. 10.1073/pnas.1607295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P.; Qiao K.; Stephanopoulos G. (2017) Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnol. Bioeng. 114, 1521–1530. 10.1002/bit.26285. [DOI] [PubMed] [Google Scholar]
- Price J. V.; Chen L.; Whitaker W. B.; Papoutsakis E.; Chen W. (2016) Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc. Natl. Acad. Sci. U. S. A. 113, 12691–12696. 10.1073/pnas.1601797113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y.; Lv X.; Liu Y.; Li J.; Du G.; Chen J.; Rodrigo L.-A.; Liu L. (2019) Synthetic redesign of central carbon and redox metabolism for high yield production of N-acetylglucosamine in Bacillus subtilis. Metab. Eng. 51, 59–69. 10.1016/j.ymben.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Xu P.; Bhan N.; Koffas M. A. G. (2013) Engineering plant metabolism into microbes: from systems biology to synthetic biology. Curr. Opin. Biotechnol. 24, 291–299. 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Xu P. (2018) Production of chemicals using dynamic control of metabolic fluxes. Curr. Opin. Biotechnol. 53, 12–19. 10.1016/j.copbio.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Gupta A.; Reizman I. M. B.; Reisch C. R.; Prather K. L. J. (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279. 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong S. J.; Gupta A.; Prather K. L. J. (2018) Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. A115, 2964–2969. 10.1073/pnas.1716920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Bowen C. H.; Liu D.; Zhang F. (2016) Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat. Chem. Biol. 12, 339–344. 10.1038/nchembio.2046. [DOI] [PubMed] [Google Scholar]
- Wang M.; Chen B.; Fang Y.; Tan T. (2017) Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 35, 1032–1039. 10.1016/j.biotechadv.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Wu G.; Yan Q.; Jones J. A.; Tang Y. J.; Fong S. S.; Koffas M. A. G. (2016) Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol. 34, 652–664. 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Dueber J. E.; Wu G. C.; Malmirchegini G. R.; Moon T. S.; Petzold C. J.; Ullal A. V.; Prather K. L. J.; Keasling J. D. (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759. 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Delebecque C. J.; Lindner A. B.; Silver P. A.; Aldaye F. A. (2011) Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science 333, 470–474. 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Park J. O.; Liu N.; Holinski K. M.; Emerson D. F.; Qiao K.; Woolston B. M.; Xu J.; Lazar Z.; Islam M. A.; Vidoudez C.; Girguis P. R.; Stephanopoulos G. (2019) Synergistic substrate cofeeding stimulates reductive metabolism. Nat. Metab. 1, 643–651. 10.1038/s42255-019-0077-0. [DOI] [PubMed] [Google Scholar]
- Lv Y.; Qian S.; Du G.; Chen J.; Zhou J.; Xu P. (2019) Coupling feedback genetic circuits with growth phenotype for dynamic population control and intelligent bioproduction. Metab. Eng. 54, 109–116. 10.1016/j.ymben.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Liu N.; Santala S.; Stephanopoulos G. (2020) Mixed carbon substrates: a necessary nuisance or a missed opportunity?. Curr. Opin. Biotechnol. 62, 15–21. 10.1016/j.copbio.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Liu H.; Marsafari M.; Wang F.; Deng L.; Xu P. (2019) Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab. Eng. 56, 60–68. 10.1016/j.ymben.2019.08.017. [DOI] [PubMed] [Google Scholar]
- Liu C.; Zhang K.; Cao W.; Zhang G.; Chen G.; Yang H.; Wang Q.; Liu H.; Xian M.; Zhang H. (2018) Genome mining of 2-phenylethanol biosynthetic genes from Enterobacter sp. CGMCC 5087 and heterologous overproduction in Escherichia coli. Biotechnol. Biofuels 11, 305. 10.1186/s13068-018-1297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.; Yan W.; Zhang W.; Dong W.; Ma J.; Ochsenreither K.; Jiang M.; Xin F. (2019) Current status and perspectives of 2-phenylethanol production through biological processes. Crit. Rev. Biotechnol. 39, 235–248. 10.1080/07388551.2018.1530634. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Edwards H.; Xu P. (2020) CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab. Eng. Commun. 10, e00112 10.1016/j.mec.2019.e00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Mawgoud A. M.; Markham K. A.; Palmer C. M.; Liu N.; Stephanopoulos G.; Alper H. S. (2018) Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 50, 192–208. 10.1016/j.ymben.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Groenewald M.; Boekhout T.; Neuvéglise C.; Gaillardin C.; van Dijck P. W. M.; Wyss M. (2014) Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 40, 187–206. 10.3109/1040841X.2013.770386. [DOI] [PubMed] [Google Scholar]
- Martinez-Avila O.; Sanchez A.; Font X.; Barrena R. (2018) Bioprocesses for 2-phenylethanol and 2-phenylethyl acetate production: current state and perspectives. Appl. Microbiol. Biotechnol. 102, 9991–10004. 10.1007/s00253-018-9384-8. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Jiang M.; Guo X.; Liu Z.; He X. (2018) Reconstruction of metabolic module with improved promoter strength increases the productivity of 2-phenylethanol in Saccharomyces cerevisiae. Microb. Cell Fact. 17, 60. 10.1186/s12934-018-0907-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S. J., van’t Klooster J. S., Bianchi F., and Poolman B. (2017) Growth inhibition by amino acids in Saccharomyces cerevisiae. bioRxiv, 2017, 222224. 10.1101/222224, accessed 2017/12/01. [DOI] [PMC free article] [PubMed]
- Lv Y.; Edwards H.; Zhou J.; Xu P. (2019) Combining 26s rDNA and the Cre-loxP System for Iterative Gene Integration and Efficient Marker Curation in Yarrowia lipolytica. ACS Synth. Biol. 8, 568–576. 10.1021/acssynbio.8b00535. [DOI] [PubMed] [Google Scholar]
- Markham K. A.; Alper H. S. (2018) Synthetic Biology Expands the Industrial Potential of Yarrowia lipolytica. Trends Biotechnol. 36, 1085–1095. 10.1016/j.tibtech.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Bojunga N.; Entian K. D. (1999) Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262, 869–875. 10.1007/s004380051152. [DOI] [PubMed] [Google Scholar]
- Xu P.; Li L. Y.; Zhang F. M.; Stephanopoulos G.; Koffas M. (2014) Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. U. S. A. A111, 11299–11304. 10.1073/pnas.1406401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A.; Haddouche R.; Kabran P.; Dulermo T.; Chardot T.; Nicaud J. M. (2012) Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 93, 1523–37. 10.1007/s00253-011-3506-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajdos P.; Ledesma-Amaro R.; Nicaud J. M.; Certik M.; Rossignol T. (2016) Overexpression of diacylglycerol acyltransferase in Yarrowia lipolytica affects lipid body size, number and distribution. FEMS Yeast Res. 16, fow062. 10.1093/femsyr/fow062. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang H.; Lu X.; Zong H.; Zhuge B. (2019) Advances in 2-phenylethanol production from engineered microorganisms. Biotechnol. Adv. 37, 403–409. 10.1016/j.biotechadv.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Cui Z.; Yang X.; Shen Q.; Wang K.; Zhu T. (2011) Optimisation of biotransformation conditions for production of 2-phenylethanol by a Saccharomyces cerevisiae CWY132 mutant. Nat. Prod. Res. 25, 754–759. 10.1080/14786419.2010.529441. [DOI] [PubMed] [Google Scholar]
- Ciesarova Z.; Smogrovicova D.; Domeny Z. (1996) Enhancement of yeast ethanol tolerance by calcium and magnesium. Folia Microbiol. 41, 485–8. 10.1007/BF02814663. [DOI] [PubMed] [Google Scholar]
- Stark D.; Munch T.; Sonnleitner B.; Marison I. W.; von Stockar U. (2002) Extractive bioconversion of 2-phenylethanol from L-phenylalanine by Saccharomyces cerevisiae. Biotechnol. Prog. 18, 514–23. 10.1021/bp020006n. [DOI] [PubMed] [Google Scholar]
- Lv Y.; Marsafari M.; Koffas M.; Zhou J.; Xu P. (2019) Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 8, 2514–2523. 10.1021/acssynbio.9b00193. [DOI] [PubMed] [Google Scholar]
- Wong L.; Engel J.; Jin E.; Holdridge B.; Xu P. (2017) YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab. Eng. Commun. 5, 68–77. 10.1016/j.meteno.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L., Holdridge B., Engel J., and Xu P. (2019) Genetic Tools for Streamlined and Accelerated Pathway Engineering in Yarrowia lipolytica. In Microbial Metabolic Engineering: Methods and Protocols (Santos C. N. S., and Ajikumar P. K., Eds.) pp 155–177, Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- Marsafari M.; Xu P. (2020) Debottlenecking mevalonate pathway for antimalarial drug precursor amorphadiene biosynthesis in Yarrowia lipolytica. Metab. Eng. Commun. 10, e00121 10.1016/j.mec.2019.e00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Young L.; Chuang R.-Y.; Venter J. C.; Hutchison C. A. III; Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Xu P.; Vansiri A.; Bhan N.; Koffas M. (2012) ePathBrick: A Synthetic Biology Platform for Engineering Metabolic Pathways in E. coli. ACS Synth. Biol. 1, 256–266. 10.1021/sb300016b. [DOI] [PubMed] [Google Scholar]
- Fickers P.; Le Dall M. T.; Gaillardin C.; Thonart P.; Nicaud J. M. (2003) New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 55, 727–737. 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.