Abstract
Genetically encodable fluorescent biosensors provide spatiotemporal information on their target analytes in a label-free manner, which has enabled the study of cell biology and signaling in living cells. Over the past three decades, fueled by the development of a wide palette of fluorescent proteins, protein-based fluorescent biosensors against a broad array of targets have been developed. Recently, with the development of fluorogenic RNA aptamer-dye pairs that function in live cells, RNA-based fluorescent (RBF) biosensors have emerged as a complementary class of biosensors. Here we review the current state-of-the-art for fluorogenic RNA aptamers and RBF biosensors for imaging small molecules and RNAs, and highlight some emerging opportunities.
Keywords: Fluorogenic RNA aptamer, RBF biosensors, molecular imaging, RNA imaging
Graphical Abstract
Introduction
To study cell signaling and other dynamic and spatiotemporally controlled processes in cells requires some method of molecular tracking. Whereas metabolic incorporation strategies and tagging strategies provide different means to label a molecule, a biosensor that binds to an endogenous molecule provides a means for label-free detection. Furthermore, genetically encodable biosensors that yield fluorescent or bioluminescent signals are amenable for live cell imaging experiments. In addition, the reversible nature of the binding event allows biosensors to capture dynamic and transitory processes, e.g. rise and fall of levels.
With the advent of natural and engineered fluorescent and bioluminescent proteins, protein-based biosensors utilizing fluorescence turn-on, fluorescence resonance energy transfer (FRET), bioluminescence resonance energy transfer (BRET), and other mechanisms have been developed that exploit protein domains that undergo conformational change in response to target analytes. Protein-based biosensors have been applied for real-time imaging of small molecules, redox factors, ions, pH, voltage, proteins, and protein modifications in live cells [1]. However, one general limitation for genetically encodable biosensors is the availability of suitable binding domains. Not only do the target binding domains have to retain stability, affinity, and specificity in the context of the constructed biosensor, but importantly they have to undergo ligand-dependent conformational change in order to alter signal. This latter property is a key feature that often is difficult to engineer.
An alternative class of biomolecules that is capable of molecular recognition and genetically encodable is RNA. For example, riboswitches are natural well-structured RNA folds that undergo conformational change in response to specific analytes, including small molecules, redox factors, and ions [2], and natural and engineered RNA regulatory elements undergo structure switching in response to specific RNAs [3]. Thus, with the recent advent of fluorogenic RNA aptamer-dye pairs that are amenable for application in live cells, these two RNA classes have become a rich and novel source of binding domains for genetically encoded RNA-based fluorescent (RBF) biosensors. This review focuses on recent and exciting progress on RBF biosensor development by first giving an overview of fluorogenic RNA aptamer-dye pairs, presenting notable design approaches and applications for RBF biosensors for small molecules, ions, and RNAs, and concluding with some potential opportunities and challenges for this field.
Fluorogenic RNA aptamer-dye pairs for live cell imaging applications
To our knowledge, the first RBF biosensors were developed based on the malachite green fluorescent dye and its aptamer [4]. Detection of small molecules [5] and nucleic acids [6] was shown in vitro, but application of this system to live cell imaging was limited by the cytotoxicity and nonspecific binding of malachite green. In order to be useful for live cell imaging, a fluorogenic dye compound should be highly soluble in water, low in molecular weight, cell permeable, and not toxic to cells. Another critical property is the dye should specifically bind to its RNA aptamer and not to other cellular components, especially other high abundant nucleic acids.
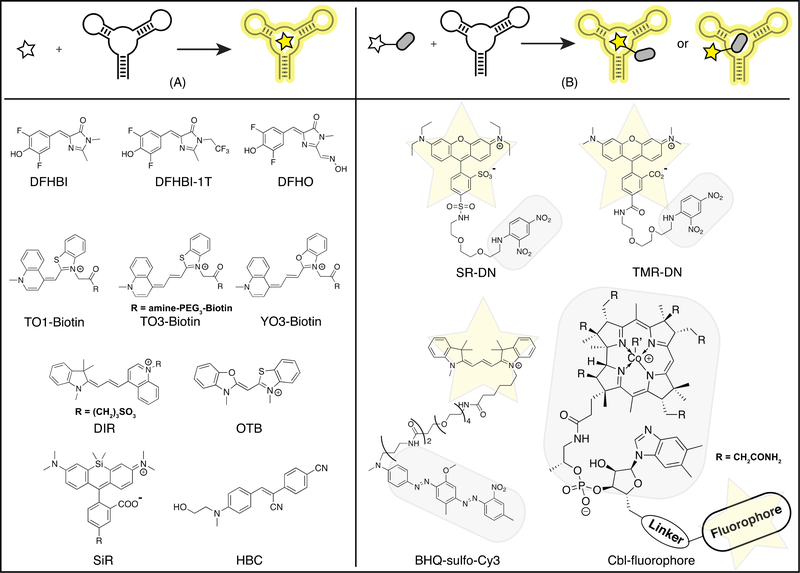
Current fluorogenic RNA aptamer-dye pairs can be divided into two design categories, single dyes (Figure 1A) and fluorophore-quencher conjugates (Figure 1B). Single dye systems employ fluorescent compounds that exhibit low fluorescence quantum yield in their free states in water, but become highly fluorescent when bound by the RNA aptamer. Fluorophore-quencher conjugate systems use fluorescent dyes that exhibit high fluorescence quantum yield in their free states in aqueous solution, but exhibit low fluorescence when conjugated to a quencher. Binding of the RNA aptamer to either the fluorophore or quencher moiety restores high fluorescence by interfering with the quenching mechanism.
Figure 1.
Fluorogenic RNA aptamers-dye pairs. Above, schematic representations of two types of fluorogenic RNA aptamers based on their paired dyes: single dyes (A) and fluorophore-quencher conjugates (B). Yellow star, fluorophore; Gray rounded rectangle, quencher. Below, chemical structures of representative dyes. DFHBI, 3,5-difluoro-4-hydroxybenzylidene imidazolinone [8]; DFHBI-1T, DFHBI with a 1,1,1-trifluoroethyl substituent [9]; DFHO, 3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-oxime [15]; TO, thiazole orange [18]; YO, oxazole yellow [20]; DIR, dimethylindole red [7]; OTB, oxazole thiazole blue [7]; SiR, silicon rhodamine [21]; HBC, (4-((2-hydroxyethyl)(methyl)amino)-benzylidene)-cyanophenyl-acetonitrile [22]; SR, Sulforhodamine B; DN, dinitroaniline; TMR, 5-carboxytetramethylrhodamine; MN, mononitroaniline [25,26]; BHQ, black hole quencher; Cbl, cobalamin [23].
Most single dye systems have been developed and used as fusion tags for intracellular imaging of RNAs (Figure 1A). A notable exception is the DIR2s aptamer, an in vitro selected aptamer that binds and activates either dimethylinodole red (DIR) or oxazole thiazole blue (OTB) dyes, giving two well-resolved emission colors [7]. This system was fused to an aptamer that binds the cell-surface EGF receptor protein and enabled live cell imaging of the surface protein, presenting an alternative application when the dye is not cell-permeable. The key development goals for current fluorogenic RNA systems are to improve signal-to-background for reducing the number of copies of tag needed and imaging lower abundance transcripts, to enable multi-color imaging, and to enable advanced methods such as single-molecule and super-resolution fluorescence microscopy. These goals also are desirable for biosensing applications.
The first single dye system successfully developed for live cell imaging to our knowledge was the Spinach aptamer with 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), an analogue of the GFP chromophore [8]. This system has been improved with the development of DFHBI analogues with different spectral properties [9] and with next-generation variants of Spinach (e.g. Spinach2 [10], Broccoli [11], iSpinach [12], baby Spinach [13]) with increased brightness, intracellular stability, and folding fidelity, lower salt dependence, and smaller tag size. The same group recently developed an aptamer-dye pair with yellow fluorescence and slower photobleaching, the Corn aptamer with 3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-oxime (DFHO), an analogue of the DsRed chromophore [14,15]. Spinach and Corn systems have been employed in both RNA imaging and biosensor applications [8,15–17•].
Another well-developed single dye system is the Mango aptamer with the thiazole orange (TO1)-biotin conjugate [18]. Several Mango aptamer variants (e.g. Mango-II to Mango-IV) with improved intracellular brightness have been engineered, and this system is distinguished by very high affinities to the dye-biotin conjugate (Kd ~ 1–11 nM) [19•]. Low concentrations of the dye-biotin conjugate can be used in live cell imaging experiments, which reduces background fluorescence from nonspecific nucleic acid interactions. Other biotin conjugates to related dyes, red-shifted thiazole orange (TO3) and oxazole yellow (YO3), also were found to bind the Mango aptamer [18,20•]. The former enables far-red fluorescent RNA imaging and the latter was applied to develop a FRET biosensor incorporating both Spinach and Mango aptamers.
Other promising single dye systems recently have been advanced for RNA tagging and are briefly summarized because they have not yet been applied to RBF biosensors. The silicon rhodamine aptamer (SiRA) binds to silicon rhodamines (SiR) and activates fluorescence through potentially stabilizing the fluorescent zwitterionic state over the nonfluorescent spirolactone [21•]. This system exhibited exceptional brightness in the far-red to near-infrared and enabled super-resolution imaging of mRNA in live Escherichia coli. The Pepper aptamer binds with high affinity to a novel family of fluorogenic dyes, (4-((2-hydroxyethyl)(methyl)amino)-benzylidene)-cyanophenyl-acetonitrile (HBC), that yield a broad range of fluorescence from cyan to red [22••]. This impressively bright system was applied to live cell imaging of RNA polymerase II transcripts and is compatible with advanced microscopy techniques such as two-photon imaging and super-resolution imaging.
Since fluorescence activation of single dyes can be difficult to predict or to engineer, fluorophore-quencher conjugates present a promising alternative (Figure 1B). One general benefit of this strategy is that existing bright, fluorescent dyes can be used, such as fluoresceins, rhodamines, cyanines, and Atto dyes. In cases where the RNA aptamer binds to the quencher moiety, there is an added advantage that the fluorophore can be easily swapped. It has been shown that fluorescence unquenching and overall signal brightness in these systems can be improved by altering the spectral overlap between fluorophore and quencher, the linker length, and the fluorophore group [23•–26]. One potential issue for these systems is the necessarily larger size of fluorophore-quencher conjugates, which affects cell permeability and may require cellular delivery.
The SRB-2 aptamer binds several structurally related fluorophores, including Sulforhodamine B (SR) and 5-carboxytetramethylrhodamine (TMR). Binding to the RNA was shown to disrupt quenching by the dinitroaniline (DN) or mononitroaniline (MN) group, resulting in fluorescence turn-on [25,26]. The black hole quencher (BHQ) aptamer and DN-binding aptamer bind to the respective quenchers, permitting potentially any fluorophore to be conjugated, although with variable turn-on efficiency. Each of these aptamers have been applied in dual-color imaging and to make different RBF biosensors [27•, 28, 29•, 30•].
Whereas development of all fluorogenic systems described above involved in vitro selection of the aptamer, the Riboglow system was engineered from a natural bacterial cobalamin (Cbl) riboswitch aptamer that binds to Cbl as the quencher [23•]. Cbl is shown to be an effective quencher of a wide variety of fluorophores, including the extremely bright Atto and cyanine class dyes. The fluorophore-quencher compounds are delivered via bead loading for live-cell imaging of mRNA translocation.
The expanded palette of fluorogenic RNA-dye pairs means that novel RBF biosensors can be made with different designs (e.g. FRET-based), tailored spectral properties, and multiplexed applications in mind. Some orthogonality has been demonstrated in live-cell imaging experiments for Spinach-Mango [20•], Spinach-DNB [30•], DNB-SRB-2 [25] and DNB-BHQ [29•] aptamer pairs. With the general biosensor design strategies discussed in the following sections, we expect that most of the novel RNA-dye pairs can be engineered into functional biosensors for molecular and RNA imaging.
RNA-based fluorescent biosensors for molecular imaging
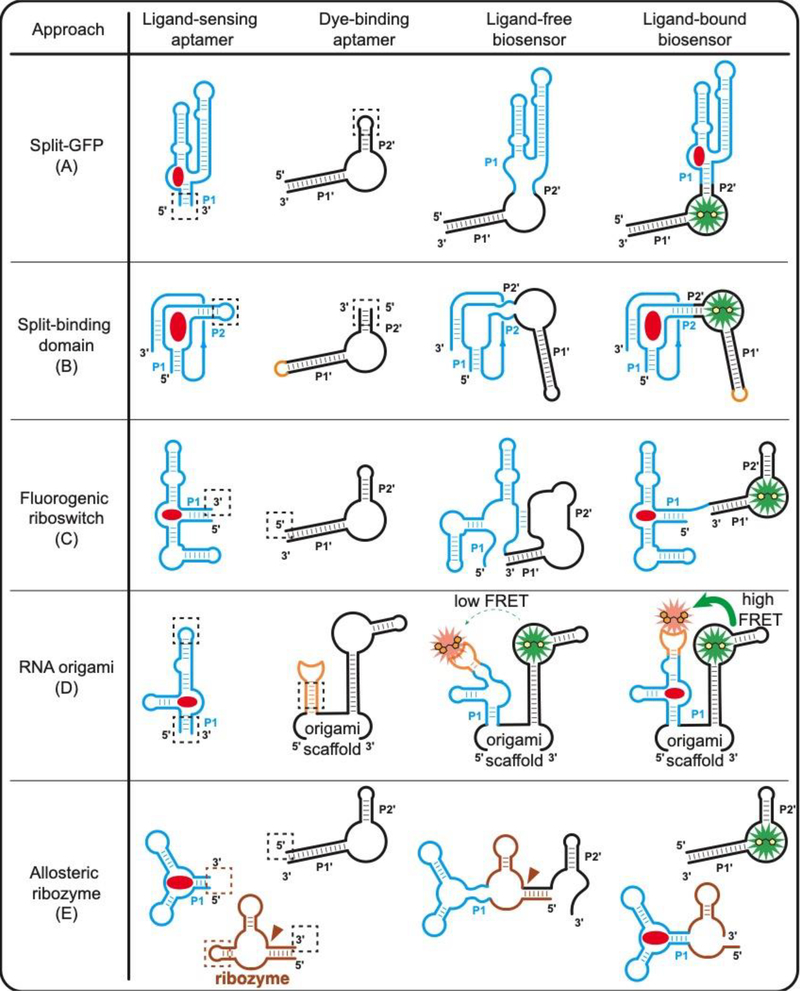
To date, almost all published examples utilize the Spinach-DFHBI family of dye-binding aptamers, which were the first developed and employ commercially available dyes. The first RBF biosensors were designed in analogous fashion to the (1) split-GFP approach, and this remains the most common and generalizable strategy (Figure 2A) [16,31]. The biosensor comprises a contiguous RNA sequence with the dye-binding aptamer “split” by the ligand binding domain, which acts through a transducer stem to reconstitute the dye-binding domain in the presence of target analyte. However, unlike split-GFP systems, which often remain reconstituted, it was shown that depletion of the analyte leads to loss of fluorescence, which enables dynamic monitoring of the small molecule target [16,31]. This type of biosensor also has been shown to accommodate circular permutations of the ligand-binding domain, which permits alternative stems to be used as the transducer [32].
Figure 2.
Design strategies of RNA-based fluorescent biosensors for molecular sensing. Functional RBF biosensors can be generated either by (A) splitting the dye-binding aptamer [16] or (B) splitting the ligand-sensing aptamer [33•]. (C) Fluorogenic riboswitches can be generated by replacing the regulatory expression platform in natural riboswitches with a dye-binding aptamer [34]. (D) Ligand-sensing aptamer can be inserted into an established RNA origami scaffold for ligand-dependent FRET signal change [20•]. (E) Allosteric ribozymes can be fused to a dye-binding aptamer for ligand-dependent release of the fluorogenic aptamer [36]. Red circle, target ligand; dashed square, fusion section between the ligand-sensing domain and the signal reporter domain. Brown triangle, self-cleavage site of ribozyme.
Alternative strategies for designing RBF biosensors exploit the unique ease of designing RNA secondary structures and expand the ligand-binding domain structures that can be adapted into RBFs (Figure 2B–E): (2) Split-binding domain approach: the biosensor comprises a contiguous RNA sequence with the ligand binding domain “split” by a dye-binding aptamer, which inverts the classic strategy. In general, this approach enables biosensors to be developed using binding domains where the 5’ and 3’ ends are far apart, and requires using a circularly permuted dye-binding aptamer. The SAH biosensor was developed using this method with cpSpinach2 [33•]. (3) Fluorogenic riboswitch approach: the structure-switching mechanism of riboswitches can be exploited by replacing the gene regulatory expression platform in the natural riboswitch with a dye-binding aptamer. The TPP biosensor was developed using detailed mechanistic information about the riboswitch [34]. (4) RNA origami approach: The first FRET-based RBF biosensor (apta-FRET) was developed by integrating a SAM-III riboswitch aptamer domain to control the orientation of Spinach and Mango aptamers positioned on each arm of a single-stranded RNA origami called 2H-AE. Upon binding to SAM, the Mango-SAM-III arm is proposed to switch to a rigid conformation, leading to higher FRET efficiency between Spinach-DFHBI-1T and Mango-YO3-biotin [20•]. (5) Allosteric ribozyme approach: allosteric ribozymes are developed by fusing ligand-sensing aptamers to a ribozyme and have been applied to control expression of fluorescent proteins [35]. A recent report applied this principle to develop a cyclic di-GMP biosensor that controls expression of fluorogenic RNA aptamers as alternate reporters [36].
Current RBF biosensors have been developed that respond to cofactors (e.g. SAM, TPP, ATP, etc.) [16,34], metabolites (e.g. SAH, ADP, adenine, guanine, etc.) [16,34], signaling molecules (e.g. cyclic dinucleotides, neurotransmitters) [31,36–37,38•,39–41•], drugs (e.g. tetracycline) [30•], and ions (e.g. Ag+) [42]. The majority of these biosensors use natural riboswitch binding domains, because of their extraordinary affinity and specificity, robust folding in the intracellular environment, and structure-switching properties. In vitro selected aptamers could be utilized, as exemplified by the biosensors for adenosine, ADP, and tetracycline [16,30•]. However, most in vitro selected aptamers may not be suitable as binding domains for the same reason that antibodies are not used in protein-based biosensors: they are selected for binding, not conformational change.
To expand the scope of ligands while retaining the advantageous properties of natural riboswitches, novel ligand sensing domains have been engineered via either structure-based design [31,33•] or structure-based selection of riboswitch scaffolds [41•]. The former strategy was employed to develop an RBF biosensor capable of detecting an innate immune signal in mammalian cell lysates. The latter strategy was employed to develop RBF biosensors for 5-hydroxy-L-tryptophan (5HTP) and 3,4-dihydroxy-Lphenylalanine (L-DOPA), the precursors of neurotransmitter serotonin and dopamine.
RBF biosensors have enabled the detection of many small molecules for which no other biosensor tool exists to target, and the ease of producing them through in vitro transcription empowers both in vitro and in vivo applications. In vitro applications that have been demonstrated include: (1) Discovery and characterization of riboswitch-ligand interactions: RBF biosensors provide a convenient way to screen for ligands that bind to the riboswitch and to determine relative binding affinities. We discovered the 3’,3’-cGAMP riboswitch using biosensors [37]. We also have characterized riboswitch mutants [43] and shown that 42 out of 53 (79%) bioinformatically predicted riboswitch sequences function in the same biosensor context [39]. (2) High-throughput enzyme activity assays: RBF biosensors serve as robust reagents for screening enzyme activity, exhibiting better sensitivity and selectivity than antibody-based assays in some cases. We showed that the SAH biosensor can be used to identify methyltransferase inhibitors [33•] and the 2’,3’-cGAMP biosensor can be used to identify modulators of the human cGAS enzyme [40].
In vivo applications that have been demonstrate include: (1) Discovery of novel enzyme activities: RBF biosensors provide a way to implement cell-based screens for enzyme activity. This approach has been particularly effective to study overexpressed signaling enzymes, including membrane-bound ones. For example, we discovered enzyme classes that produce and degrade the cyclic dinucleotide signal 3’,3’-cGAMP using biosensors [44,45•]. (2) Validation of enzyme agonists or antagonists: We also have shown that RBF biosensors can detect the modulation of endogenous enzyme activities in live cells. We have visualized chemical inhibition of quorum signal production and activation of cyclic di-GMP signal production [33,46••]. (3) Biosensing under anaerobic conditions: RBF biosensors bind synthetic dyes so do not require chromophore maturation like the majority of fluorescent protein-based biosensors. We have shown that the cyclic di-GMP biosensor works in cells grown under anaerobic conditions [39]. (4) Evaluation of the efficacy of therapeutic agents: Silver is a widely used disinfection reagent and an RBF biosensor for silver ion has been applied to measure the cellular flux and silver ion release of silver nanoparticles [42].
RNA-based fluorescent biosensors for RNA imaging
While fluorogenic aptamer-dye pairs are highly useful as fusion tags for tracking mRNAs in live cells, there are shorter RNA species whose function, processing, and translocation may be affected by tagging approaches. For example, microRNAs (miRNAs) are 21–24 nucleotides in size and play key roles in regulating development, epigenetics and immune response in both plants and metazoans [47]. Thus, for imaging native RNAs in a label-free manner, especially small RNAs such as miRNAs, there is a demand for trans-acting sensors.
RBF biosensors that target specific RNAs present a similar strategy to classic molecular beacon and RNA-FISH (fluorescence in situ hybridization) methods. However, a major difference is that RBF biosensors are genetically encodable, with an exogenously added dye or dye-quencher. This is advantageous for live cell imaging experiments, as cellular delivery of small molecule fluorophores is easier than for dye-conjugated RNA probes. CRISPR protein-based sensors also have been developed that permit imaging of native RNAs [48,49]. These systems are analogous to the MS2-fusion tagging strategy, except with guide RNA specified targeting of the CRISPR complex. In order to visualize cytoplasmic mRNA transcripts, background fluorescence must be reduced by confining unbound fluorescent proteins in the nucleus or keeping fluorescent protein expression low via negative feedback.
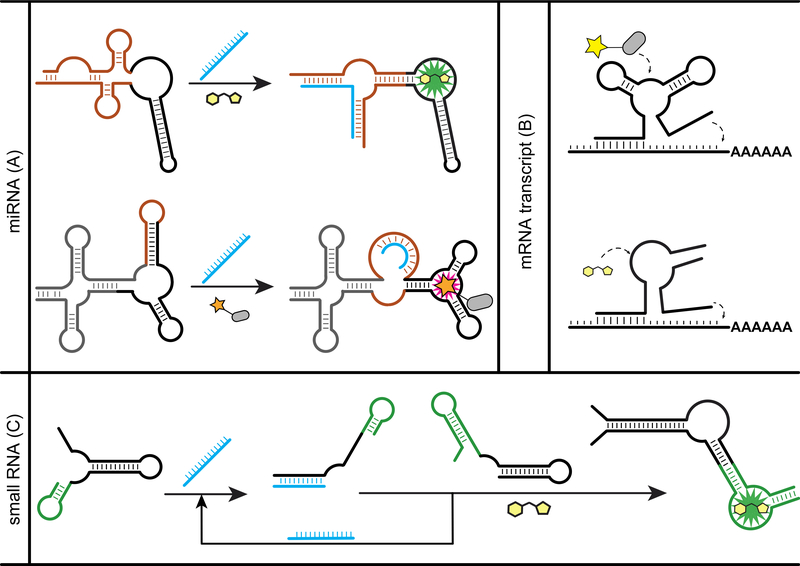
Again, one strategy to design RBF biosensors for imaging RNAs is analogous to the split-binding domain approach (Figure 3A). Rather than a ligand-binding aptamer, the sensing domain in this case is a complementary sequence to the target RNA of interest. The critical aspect for this design strategy is that there must be a structural change when the RNA-sensing domain binds its target to reconstitute the dye-binding domain. Secondary structure prediction is required but remains particularly challenging for structure-switching RNAs [50•].
Figure 3.
Examples of RBF biosensors for RNA sensing. (A) Split-binding domain strategy was applied on Spinach and SRB-2 aptamer to generate biosensors for miRNA [52•,27•]. (B) Destabilizing the fluorogenic aptamer by shortening a stem or splitting led to functional biosensors for mRNA [28,53]. (C) For sensitive RNA detection, the catalytic hairpin assembly was used to amplify the signal triggered by target RNA [55].
An early example of a miRNA biosensor was called Pandan and based on stem-loop insertion into a circularly permuted Spinach aptamer [51]. Although Pandan functions in vitro, the in vivo performance was not reported. The FASTmiR sensors employed a similar design and were developed to target human miR122 and Arabidopsis thaliana miR171 [52•]. The miR122 sensor was multiplexed and applied to report miR122 expression level in mammalian cells. The miR171 sensor was applied as an in vitro assay for directly quantifying miR171 levels in A. thaliana total leaf and flower RNA extract. To achieve ratiometric imaging, a miRNA biosensor based on the SRB-2 aptamer was co-expressed with GFP encoded on the same transfected plasmid [27•]. Levels of miR-21 could be reliably quantified in different mammalian cell lines.
A simpler design was developed for imaging mRNAs (Figure 3A). The fluorogenic aptamer first is destabilized to achieve low background, so that target RNA binding to two complementary regions flanking the domain is required to reconstitute the dye-binding domain. A family of RNA targeting aptamers (RT-aptamers) was developed by flanking the BHQ aptamer with two targeting sequences of 9 nt each, and were applied to image mRNAs such as β-actin, ARFIP2, CTTN and CYFIP2 [28]. Using a similar design but based on the Spinach aptamer, biosensors for imaging mRNAs in E. coli have been developed against targets such as mreB, dnaJ, dnaK and rpoH [53].
More recently, a splitting sensor strategy was implemented and shown to lower fluorescence background and thus increase sensitivity of detection. The biosensor was designed by flanking the Broccoli aptamer with two targeting sequences and then splitting the biosensor into separate RNA halves. Thus, target mRNA binding is required to reconstitute the dye-binding domain. mRNAs such as β-actin, CFL1 and GAPDH were visualized in live HeLa and HuMSC cells [54•]. Alternatively, a hairpin assembly circuit for detection of target RNA was developed that catalyzes the reconstitution of split Broccoli (Figure 3C), and was applied to detect changes in endogenous small RNA SgrS in live E. coli [55].
Conclusions
The emerging field of RNA-based fluorescent biosensors has seen rapid progress and creative contributions by many research groups in the development of fluorogenic aptamers and biosensor designs. Going forward, we consider that the greatest opportunity in the next stage of the field lies in establishing and disseminating these tools to address important questions in cell biology. Towards this goal of visualizing spatiotemporal dynamics of molecular targets, the field will have to tackle general issues such as increasing signal-to-noise, accounting for expression variability, and improving biosensor kinetics. Approaches to date include optimizing intracellular RNA stability through circularization [56••] and ratiometric systems employing two dye compounds [20•], but we look forward to many more advancements to come.
We consider that one of the grand challenges for the field is to enable other researchers to design their own biosensors. A practical aspect is that while RNA aptamer or biosensor constructs are readily shared by request and often made publicly available through Addgene, one limitation to non-chemists is the availability of synthetic dyes and dye-quencher compounds. RNA aptamer paired with degron-tagged fluorescent proteins could provide an “all encoded” solution [57•], but the potential of this system for biosensor engineering has yet to be explored. Another important aspect for enabling growth of the field is to explain the general design strategies, which we have presented in this review. In our opinion, it also is important to consider whether increasing the complexity of a given biosensor design is warranted and would improve utility for cell biology studies. Finally, we anticipate that there will be advances in the computational design of RNA targeting biosensors that will reduce experimental trial-and-error, and that the scope of binding domains will be greatly expanded through natural discovery and exciting advances to the in vitro / in vivo / in silico evolution of riboswitches [41,50,58,59••].
Acknowledgements
Work on RBF biosensors in the Hammond lab is supported by the National Institutes of Health (grant number GM124589 to M.C.H.) and Office of Naval Research (grant number N00014-19-1-2043 to M.C.H.).
Footnotes
Declaration of interests
The authors have filed a patent application for a fluorescent biosensor for methyltransferase assays.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yichi Su, Department of Neurobiology, Stanford University.
Ming C. Hammond, Department of Chemistry and Henry Eyring Center for Cell & Genome Sciences, University of Utah.
Annotated Bibliography
- 1.Greenwald EC, Mehta S, Zhang J: Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem Rev 2018, 118:11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hallberg ZF, Su Y, Kitto RZ, Hammond MC: Engineering and In Vivo Applications of Riboswitches. Annu Rev Biochem 2017, 86:515–539. [DOI] [PubMed] [Google Scholar]
- 3.Villa JK, Su Y, Contreras LM, Hammond MC: Synthetic Biology of Small RNAs and Riboswitches. Microbiol Spectr 2018, 6, doi: 10.1128/microbiolspec.RWR-0007-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babendure JR, Adams SR, Tsien RY: Aptamers Switch on Fluorescence of Triphenylmethane Dyes. J Am Chem Soc 2003, 125:14716–14717. [DOI] [PubMed] [Google Scholar]
- 5.Stojanovic MN, Kolpashchikov DM: Modular aptameric sensors. J Am Chem Soc 2004, 126:9266–9270. [DOI] [PubMed] [Google Scholar]
- 6.Kolpashchikov DM: Binary malachite green aptamer for fluorescent detection of nucleic acids. J Am Chem Soc 2005, 127:12442–12443. [DOI] [PubMed] [Google Scholar]
- 7.Tan X, Constantin TP, Sloane KL, Waggoner AS, Bruchez MP, Armitage BA: Fluoromodules Consisting of a Promiscuous RNA Aptamer and Red or Blue Fluorogenic Cyanine Dyes: Selection, Characterization, and Bioimaging. J Am Chem Soc 2017, 139:9001–9009. [DOI] [PubMed] [Google Scholar]
- 8.Paige JS, Wu KY, Jaffrey SR: RNA Mimics of Green Fluorescent Protein. Science 2011, 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song W, Strack RL, Svensen N, Jaffrey SR: Plug-and-play fluorophores extend the spectral properties of spinach. J Am Chem Soc 2014, 136:1198–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strack RL, Disney MD, Jaffrey SR: A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods 2013, 10:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filonov GS, Moon JD, Svensen N, Jaffrey SR: Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc 2014, 136:16299–16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Autour A, Westhof E, Ryckelynck M: iSpinach: a fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res 2016, 44:2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, Ferré-D’Amaré AR: Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol 2014, 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner KD, Sjekloa L, Song W, Filonov GS, Jaffrey SR, Ferré-D’Amaré AR: A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat Chem Biol 2017, 13:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song W, Filonov GS, Kim H, Hirsch M, Li X, Moon JD, Jaffrey SR: Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat Chem Biol 2017, 13:1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR: Fluorescence imaging of cellular metabolites with RNA. Science 2012, 335:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H, Jaffrey SR: A Fluorogenic RNA-Based Sensor Activated by Metabolite-Induced RNA Dimerization. Cell Chem Biol 2019, doi: 10.1016/j.chembiol.2019.09.013.• The authors show a novel variant of the split GFP approach for biosensing by induced dimerization of the Corn aptamer, which binds the dye DFHO at the dimer interface. A circularized Corn biosensor with reduced photobleaching and improved cellular stability was applied to monitor dynamics of S-adenosyl-L-methionine (SAM) metabolism in live mammalian cells.
- 18.Dolgosheina EV, Jeng SCY, Panchapakesan SSS, Cojocaru R, Chen PSK, Wilson PD, Hawkins N, Wiggins PA, Unrau PJ: RNA Mango aptamer-fluorophore: A bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol 2014, 9:2412–2420. [DOI] [PubMed] [Google Scholar]
- 19.Autour A, Jeng SCY, Cawte AD, Abdolahzadeh A, Galli A, Panchapakesan SSS, Rueda D, Ryckelynck M, Unrau PJ: Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat Commun 2018, 9:658.• A cleverly designed microfluidics-based selection strategy was employed by the authors to develop second-generation Mango II-IV aptamers that bind the red-shifted TO1-biotin dye with higher affinity and brighter fluorescence than EGFP. These short RNA tags were incorporated into small RNAs in a single copy for live mammalian cell imaging.
- 20.Jepsen MDE, Sparvath SM, Nielsen TB, Langvad AH, Grossi G, Gothelf KV, Andersen ES: Development of a genetically encodable FRET system using fluorescent RNA aptamers. Nat Commun 2018, 9:18.• Apta-FRET was the first proof-of-concept demonstration of RNA-based FRET biosensor design. Using Spinach-DFHBI-1T and Mango-YO3-biotin pairs to improve spectral overlap in an RNA origami scaffold, the authors engineered ratiometric sensors for RNAs and small molecules, and showed FRET signal in live E. coli.
- 21.Wirth R, Gao P, Nienhaus GU, Sunbul M, Jäschke A: SiRA: A Silicon Rhodamine-Binding Aptamer for Live-Cell Super-Resolution RNA Imaging. J Am Chem Soc 2019, 141:7562–7571.• The silicon rhodamine-binding RNA aptamer was developed with fluorescence turn-on driven by a proposed equilibrium shift to favor the bright zwitterionic state. The near-IR fluorogenic RNA system showed remarkable photostability and the authors applied a 5x SiRA tag to co-visualize GFP mRNA and protein in live E. coli via STED super-resolution microsocopy.
- 22.Chen X, Zhang D, Su N, Bao B, Xie X, Zuo F, Yang L, Wang H, Jiang L, Lin Q, et al. : Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol 2019, doi: 10.1038/s41587-019-0249-1.•• The Pepper aptamer was developed to bind with high affinity to the novel dye scaffold HBC, which showed exceptional fluorescence turn-on and permit facile spectral tuning of the dye from cyan to red. The authors applied 4x or 8x Pepper-HBC tags to co-visualize BFP mRNA and protein in mammalian cells using confocal microscopy, two-photon microscopy, and flow cytometry, and tagged sgRNAs with Pepper to visualize genomic loci via CRISPR display.
- 23.Braselmann E, Wierzba AJ, Polaski JT, Chromiński M, Holmes ZE, Hung ST, Batan D, Wheeler JR, Parker R, Jimenez R, et al. : A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells. Nat Chem Biol 2018, 14:964–971.• In the Riboglow system, cobalamin (Cbl) functions as an effective quencher in a variety of Cbl-dye conjugates, and the authors engineered natural Cbl riboswitch aptamers as the RNA tag. Using beads loading, the Riboglow system was applied in live-cell imaging of mRNA in stress granules and U1 snRNA in U-bodies. This is the first fluorogenic RNA system developed from natural riboswitches, as opposed to in vitro selection.
- 24.Sunbul M, Jäschke A: Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed 2013, 52:13401–13404. [DOI] [PubMed] [Google Scholar]
- 25.Arora A, Sunbul M, Jäschke A: Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res 2015, 43:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunbul M, Jäschke A: SRB-2: A promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res 2018, 46:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying ZM, Wu Z, Tu B, Tan W, Jiang JH: Genetically Encoded Fluorescent RNA Sensor for Ratiometric Imaging of MicroRNA in Living Tumor Cells. J Am Chem Soc 2017, 139:9779–9782.• An RBF biosensor was developed for live-cell imaging of native miRNA miR-21 using the SR-binding aptamer paired with the SR-DN dye-quencher conjugate in the split GFP approach. Different levels of miR-21 could be detected by biosensor fluorescence normalized to co-expressed GFP and results correlated with RT-PCR results.
- 28.Sato SI, Watanabe M, Katsuda Y, Murata A, Wang DO, Uesugi M: Live-cell imaging of endogenous mRNAs with a small molecule. Angew Chem Int Ed 2015, 54:1855–1858. [DOI] [PubMed] [Google Scholar]
- 29.Yatsuzuka K, Sato SI, Pe KB, Katsuda Y, Takashima I, Watanabe M, Uesugi M: Live-cell imaging of multiple endogenous mRNAs permits the direct observation of RNA granule dynamics. Chem Commun 2018, 54:7151–7154.• Another DNB quencher-binding aptamer was developed and adapted to an RBF biosensor for native mRNAs using the split-binding domain approach. Combining the DNB system with the BHQ system previously developed by the authors enabled dual-color live-cell imaging of β-actin and cortactin mRNA accumulation in stress granules.
- 30.Wu R, Karunanayake Mudiyanselage A, Shafiei F, Zhao B, Bagheri Y, Yu Q, McAuliffe K, Ren K, You M: Genetically Encoded Ratiometric RNA-based Sensors for Quantitative Imaging of Small Molecules in Living Cells. Angew Chem Int Ed 2019, doi: 10.1002/anie.201911799.• The split GFP strategy was applied to generate RBF biosensors using the DNB quencher-binding aptamer originally developed by Jäschke (ref. 25). Inserting the DNB biosensor and the Broccoli aptamer in the two arms of the F30 scaffold enabled ratiometric measurements that normalizes for biosensor expression and quantifies analyte concentrations in live E. coli.
- 31.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC: RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc 2013, 135:4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong J, Hsieh YF, Truong L, Jia G, Hammond MC: Designing fluorescent biosensors using circular permutations of riboswitches. Methods 2018, 143:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su Y, Hickey SF, Keyser SGL, Hammond MC: In Vitro and in Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl- l -homocysteine (SAH). J Am Chem Soc 2016, 138:7040–7047.• An RBF biosensor was developed for high-throughput in vitro methyltransferase assays and shown to have better affinity and selectivity for SAH than commercial antibodies. The authors employed the split binding domain design and circularly permuted Spinach2 for the first time, and visualized real-time chemical inhibition of quorum signal biosynthesis in vivo.
- 34.You M, Litke JL, Jaffrey SR: Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proc Natl Acad Sci U S A 2015, 112:E2756–E2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frommer J, Appel B, Müller S: Ribozymes that can be regulated by external stimuli. Curr Opin Biotechnol 2015, 31:35–41. [DOI] [PubMed] [Google Scholar]
- 36.You M, Litke JL, Wu R, Jaffrey SR: Detection of Low-Abundance Metabolites in Live Cells Using an RNA Integrator. Cell Chem Biol 2019, 26:471–481.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellenberger CA, Wilson SC, Hickey SF, Gonzalez TL, Su Y, Hallberg ZF, Brewer TF, Iavarone AT, Carlson HK, Hsieh YF, et al. : GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci U S A 2015, 112:5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kellenberger CA, Chen C, Whiteley AT, Portnoy DA, Hammond MC: RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP. J Am Chem Soc 2015, 137:6432–6435.• An RBF biosensor for cyclic di-AMP was developed that functions in both E. coli and Listeria monocytogenes, thus showcasing these biosensors are useful in both Gram negative and positive bacteria. These biosensors were applied to show that archaeal enzymes have diadenylate cyclase (DAC) activity, thus demonstrating all three domains of life have cyclic dinucleotide signaling.
- 39.Wang XC, Wilson SC, Hammond MC: Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Res 2016, 44:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose D, Su Y, Marcus A, Raulet DH, Hammond MC: An RNA-Based Fluorescent Biosensor for High-Throughput Analysis of the cGAS-cGAMP-STING Pathway. Cell Chem Biol 2016, 23:1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter EB, Polaski JT, Morck MM, Batey RT: Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat Chem Biol 2017, 13:295–301.• The authors applied in vitro selection to natural riboswitch aptamer scaffolds in order to generate novel aptamers that bind 5HTP and L-DOPA, the former of which was made into a 5HTP/serotonin biosensor with the malachite green aptamer. This study establishes the advantages of basing the initial RNA pool on natural RNA motifs.
- 42.Yu Q, Shi J, Mudiyanselage APKKK, Wu R, Zhao B, Zhou M, You M: Genetically encoded RNA-based sensors for intracellular imaging of silver ions. Chem Commun 2019, 55:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren A, Wang XC, Kellenberger CA, Rajashankar KR, Jones RA, Hammond MC, Patel DJ: Structural basis for molecular discrimination by a 3’,3’-cGAMP sensing riboswitch. Cell Rep 2015, 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hallberg ZF, Wang XC, Wright TA, Nan B, Ad O, Yeo J, Hammond MC: Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3’, 3’-cGAMP). Proc Natl Acad Sci U S A 2016, 113:1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright TA, Jiang L, Park JJ, Anderson WA, Chen G, Hallberg ZF, Nan B, Hammond MC: Second messengers and divergent HD GYP phosphodiesterases regulate 3’,3’-cGAMP signaling. Mol Microbiol 2019, doi: 10.1111/mmi.14412• This study demonstrates a recent application of RBF biosensors for in vivo screening of enzyme activities. The authors developed a biosensor-based flow cytometry assay in order to discover the first cGAMP-specific phosphodiesterases.
- 46.Yeo J, Dippel AB, Wang XC, Hammond MC: In Vivo Biochemistry: Single-Cell Dynamics of Cyclic Di-GMP in Escherichia coli in Response to Zinc Overload. Biochemistry 2018, 57:108–116.•• This study features a second-generation RBF biosensor for cyclic di-GMP with superior brightness, affinity, and fast turn-on kinetics. The authors demonstrate real-time measurement of endogenous cyclic di-GMP signaling activity in response to zinc in live E. coli using both flow cytometry and fluorescent microscopy.
- 47.Gebert LFR, MacRae IJ: Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019, 20:21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. : RNA targeting with CRISPR-Cas13. Nature 2017, 550:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW: Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 2016, 165:488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu MJ, Andreasson JOL, Kladwang W, Greenleaf W, Das R: Automated Design of Diverse Stand-Alone Riboswitches. ACS Synth Biol 2019, 8:1838–1846.• RiboLogic was developed by the authors for in silico design of conditional MS2 coat protein-binding riboswitches that respond to small molecules and miRNAs, which could be made into fluorescent biosensors using MS2-GFP. This work showcases the potential of in silico design but also points out that the correlation to performance can be improved.
- 51.Aw SS, Tang MX, Teo YN, Cohen SM: A conformation-induced fluorescence method for microRNA detection. Nucleic Acids Res 2016, 44:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang K, Doyle F, Wurz ZE, Tenenbaum SA, Hammond RK, Caplan JL, Meyers BC: FASTmiR: an RNA-based sensor for in vitro quantification and live-cell localization of small RNAs. Nucleic Acids Res 2017, 45:e130.• FASTmiR biosensors based on the Spinach aptamer were developed for quantifying various miRNAs in total RNA extracts with sensitivity comparable to Northern blotting. The authors also applied FASTmiR to visualize expression of miR-122 in different mammalian cell lines.
- 53.Ong WQ, Citron YR, Sekine S, Huang B: Live Cell imaging of endogenous mRNA Using RNA-Based fluorescence “turn-on” probe. ACS Chem Biol 2017, 12:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Luo Y, Xie X, Hu X, Song H, Zhao Y, Shi J, Wang L, Glinsky G, Chen N, et al. : In Situ Spatial Complementation of Aptamer-Mediated Recognition Enables Live-Cell Imaging of Native RNA Transcripts in Real Time. Angew Chem Int Ed 2018, 57:972–976.• By splitting the Broccoli aptamer into two halves and flanking each fragment with an RNA recognition sequence, the authors developed an aptamer-initiated fluorescence complementation (AiFC) method. Either transfection of the split RNA probes or plasmids encoding the probes permit live-cell imaging of native mRNAs.
- 55.Karunanayake Mudiyanselage APKK, Yu Q, Leon-Duque MA, Zhao B, Wu R, You M: Genetically Encoded Catalytic Hairpin Assembly for Sensitive RNA Imaging in Live Cells. J Am Chem Soc 2018, 140:8739–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Litke JL, Jaffrey SR: Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol 2019, 37:667–675.•• This study describes Tornado, a highly efficient method based on Twister ribozyme and RNA ligase RtcB to circularize RNA constructs in live mammalian cells. The authors applied Tornado to generate circular fluorogenic aptamers that display enhanced cellular stability and thus greater fluorescence signal. This advance should be generally useful for RBF biosensors.
- 57.Wu J, Zaccara S, Khuperkar D, Kim H, Tanenbaum ME, Jaffrey SR: Live imaging of mRNA using RNA-stabilized fluorogenic proteins. Nat Methods 2019, 16:862–865.• The authors develop the first fully genetically encodable fluorogenic RNA-protein system. The Tat-degron peptide leads to proteasomal degradation of the tagged fluorescent protein, except binding to the TAR RNA sequence blocks degron recognition. This system is also called Pepper, because different colors are enabled through Tat-degron tagged fluorescent proteins.
- 58.Gotrik M, Sekhon G, Saurabh S, Nakamoto M, Eisenstein M, Soh HT: Direct Selection of Fluorescence-Enhancing RNA Aptamers. J Am Chem Soc 2018, 140:3583–3591. [DOI] [PubMed] [Google Scholar]
- 59.Boussebayle A, Torka D, Ollivaud S, Braun J, Bofill-Bosch C, Dombrowski M, Groher F, Hamacher K, Suess B: Next-level riboswitch development— implementation of Capture-SELEX facilitates identification of a new synthetic riboswitch. Nucleic Acids Res 2019, 47:4883–4895.•• The authors adapted the Capture-SELEX strategy to develop a novel conformationally switchable RNA aptamer that responds to paromomycin but not to its close structural analog, neomycin. Following in vivo screening, the paromycin riboswitch was shown to selectively repress fluorescent reporter expression in yeast.