Abstract
Ultra-small microorganisms are ubiquitous in Earth’s environments. Ultramicrobacteria, which are defined as having a cell volume of <0.1 μm3, are often numerically dominant in aqueous environments. Cultivated representatives among these bacteria, such as members of the marine SAR11 clade (e.g., “Candidatus Pelagibacter ubique”) and freshwater Actinobacteria and Betaproteobacteria, possess highly streamlined, small genomes and unique ecophysiological traits. Many ultramicrobacteria may pass through a 0.2-μm-pore-sized filter, which is commonly used for filter sterilization in various fields and processes. Cultivation efforts focusing on filterable small microorganisms revealed that filtered fractions contained not only ultramicrocells (i.e., miniaturized cells because of external factors) and ultramicrobacteria, but also slender filamentous bacteria sometimes with pleomorphic cells, including a special reference to members of Oligoflexia, the eighth class of the phylum Proteobacteria. Furthermore, the advent of culture-independent “omics” approaches to filterable microorganisms yielded the existence of candidate phyla radiation (CPR) bacteria (also referred to as “Ca. Patescibacteria”) and ultra-small members of DPANN (an acronym of the names of the first phyla included in this superphyla) archaea. Notably, certain groups in CPR and DPANN are predicted to have minimal or few biosynthetic capacities, as reflected by their extremely small genome sizes, or possess no known function. Therefore, filtered fractions contain a greater variety and complexity of microorganisms than previously expected. This review summarizes the broad diversity of overlooked filterable agents remaining in “sterile” (<0.2-μm filtered) environmental samples.
Keywords: filterable microorganisms, ultramicrocells, ultramicrobacteria, candidate phyla radiation, minimal cell
How small may actual organisms be? This question has long fascinated scientists in various fields. Prokaryotic microorganisms (Archaea and Bacteria) constitute the smallest life forms. Bacterial cells range in volume from ultramicrobacteria (UMB; <0.1 μm3; Duda et al., 2012) to the typical bacterium Escherichia coli (1.6 μm3; Moore, 1999) and the giant bacterium Epulopiscium fishelsoni (3.0×106 μm3; Schulz and Jørgensen, 2001; note that the cells of Thiomargarita namibiensis are larger [2.2×108 μm3], but are occupied by a liquid vacuole, that is, they do not have large cytoplasmic bodies; Schulz et al., 1999). Thus, bacteria exhibit cell-size plasticity by varying cell volume by more than seven orders of magnitude in different species. UMB may pass through membrane filters down to 0.2-μm-pore-size, which is commonly used for filter sterilization in research laboratories as well as in medical, food, and industrial processes (Levy and Jornitz, 2006). In fact, efforts to culture microorganisms remaining in the 0.2-μm filtrate (hereafter called filterable microorganisms) of environmental samples have yielded diverse UMB members. The several isolates were affiliated with unique lineages, such as cosmopolitan freshwater Actinobacteria and Betaproteobacteria (Hahn, 2003; Hahn et al., 2003) as well as the candidate phylum termite group 1 (TG1) described as Elusimicrobia (Geissinger et al., 2009). The existence of UMB has expanded our knowledge of microbial life at the lower size limit.
In the last five years, filterable microorganisms have been attracting increasing interest with the discovery of other ultra-small members: the candidate phyla radiation (CPR) bacteria, also referred to as “Candidatus Patescibacteria” (hereafter described as CPR/Patescibacteria; Rinke et al., 2013; Brown et al., 2015), and some members of DPANN (an acronym of the names of the first phyla included in this superphyla, “Ca. Diapherotrites”, “Ca. Parvarchaeota”, “Ca. Aenigmarchaeota”, Nanoarchaeota, and “Ca. Nanohalorchaeota”; Rinke et al., 2013; Dombrowski et al., 2019). Several CPR members have an extremely small cell volume (approximately 0.01 μm3) that was unveiled by cryo-transmission electron microscopy imaging (Luef et al., 2015). Moreover, the emergence of these ultra-small prokaryotes has re-opened debate on the tree of life (Hug et al., 2016; Parks et al., 2018; Zhu et al., 2019). These members are ubiquitous in the environment and recent studies have provided insights into their contribution to the material cycle (e.g., carbon and nitrogen cycles; Danczak et al., 2017; Lannes et al., 2019). This review focuses on the phylogenetic diversity and complexity of filterable microorganisms in natural systems, with specific references to UMB and pleomorphic bacteria. Other reviews presented aspects of ultra-small microorganisms including CPR/Patescibacteria and DPANN members (e.g., terminology, biogeography, genomic diversity, and metabolic variety; Duda et al., 2012; Castelle et al., 2018; Ghuneim et al., 2018; Dombrowski et al., 2019). In this review, archaea with a cell volume of <0.1 μm3 are specifically referred to as ultramicroarchaea (UMA) to distinguish them from UMB.
Filterable microorganisms
To date, many studies have reported the presence of filterable microorganisms in various environments (mainly aqueous environments) including seawater (Haller et al., 2000; Elsaied et al., 2001; Lannes et al., 2019; Obayashi and Suzuki, 2019), lake water (Hahn, 2003; Hahn et al., 2003; Watanabe et al., 2009; Fedotova et al., 2012; Maejima et al., 2018; Vigneron et al., 2019), terrestrial aquifers (Miyoshi et al., 2005; Luef et al., 2015), glacier ice and the ice cover of lakes (Miteva and Brenchley, 2005; Kuhn et al., 2014), deep-sea hydrothermal fluids (Naganuma et al., 2007; Nakai et al., 2011), and soil and sand (Nakai et al., 2013). However, the use of membrane filters with a small pore size (approximately 0.2 μm) was traditionally recommended for the retention of bacteria in the field of marine microbial ecology in the 1960s (e.g., Anderson and Heffernan, 1965) and is still widely practiced today in various fields. The existence of very small microorganisms has been well recognized since the 1980s. The term “ultramicrobacteria” was first used by Torrella and Morita (1981) to describe very small coccoid cell forms of <0.3 μm in diameter from seawater. MacDonell and Hood (1982) subsequently isolated and characterized viable filterable microorganisms potentially belonging to the genera Vibrio, Aeromonas, Pseudomonas, and Alcaligenes from estuarine waters. They concluded that these filterable microorganisms represented a state of dormancy for adaptation to low nutrient conditions and were not completely novel bacteria. Other studies also reported that external factors reduced cell sizes, such as Staphylococcus aureus and Pseudomonas syringae (~50% reduction in size as described in Table 1; Watson et al., 1998; Monier and Lindow, 2003). Therefore, the cells of miniaturized microorganisms need to be distinguished from true UMB and are described in this review as “ultramicrocells”, which has the synonyms dwarf cells and midget cells, according to Duda et al. (2012). Schut et al. (1997) and Duda et al. (2012) subsequently defined a cell volume index of <0.1 μm3 as being characteristic of true UMB.
Table 1.
An overview of ultra-small and filterable microorganisms in the environment
| Taxa | Phylum (and class for Proteobacteria) |
Isolation source | Cell shape | Cell size (length×width and/or volume) | Genome size (Mbp) | Physiological and ecological trait(s) or its potential | Reference |
|---|---|---|---|---|---|---|---|
| Ultramicrocells | |||||||
| Staphylococcus aureus 8325-4 | Firmicutes | derivative of S. aureus NCTC8325 (patient’s strain) | cocci | cell size reduction from 0.69±0.08 to 0.41±0.08 μm |
n.d. | host cell invasion, starvation-associated cell size reduction | Watson et al. (1998) |
| Pseudomonas syringae pv. syringae B728a |
Proteobacteria
(γ-proteobacteria) |
snap bean leaflet | rods | cell length reduction from ~2.5 to ~1.2 μm | 6.09 | host cell invasion, leaf environment-induced cell size reduction | Monier and Lindow (2003); Feil et al. (2005) |
| Obligate ultramicrobacteria and related candidates | |||||||
| “Candidatus Pelagibacter ubique” HTCC1062 |
Proteobacteria
(α-proteobacteria) |
coastal sea | curved rods | 0.01 μm3 | 1.31 | glycine auxotrophy, rhodopsin-based photometabolism, utilization of one-carbon compounds | Rappé et al. (2002); Tripp (2013); Giovannoni (2017) |
| “Candidatus Fonsibacter ubiquis” LSUCC0530 |
Proteobacteria
(α-proteobacteria) |
coastal lagoon | curved rods | 1.0×0.1 μm | 1.16 | glycine auxotrophy, rhodopsin-based photometabolism, tetrahydrafolate metabolism** | Henson et al. (2018) |
| Sphingopyxis alaskensis RB2256 |
Proteobacteria
(α-proteobacteria) |
fjord estuary | short rods | 0.05–0.09 μm3 | 3.35 | utilization of various amino acids, resistance to heat shock, H2O2, and ethanol |
Eguchi et al. (1996); Schut et al. (1997) |
| Aurantimicrobium minutum KNCT | Actinobacteria | freshwater river | curved rods | 0.7–0.8×0.3 μm; 0.04–0.05 μm3 |
1.62 | rhodopsin-based photometabolism** | Nakai et al. (2015, 2016b) |
| Rhodoluna lacicola MWH-Ta8T | Actinobacteria | freshwater lake | curved rods | 0.85×0.30 μm; 0.053 μm3 |
1.43 | rhodopsin-based photometabolism |
Hahn et al. (2014); Keffer et al. (2015) |
| Rhodoluna limnophila 27D-LEPIT | Actinobacteria | freshwater pond | short rods | 0.49×0.28 μm | 1.40 | nitrate uptake and nitrite excretion system** | Pitt et al. (2019) |
| “Candidatus Planktophila rubra” IMCC25003 | Actinobacteria | freshwater lake | curved rods | 0.041 μm3 | 1.35 | catalase-dependent growth | Kim et al. (2019) |
| “Candidatus Planktophila aquatilis” IMCC26103 | Actinobacteria | freshwater lake | curved rods | 0.061 μm3 | 1.46 | catalase-dependent growth | Kim et al. (2019) |
| Polynucleobacter necessarius subsp. asymbioticus QLW-P1DMWA-1T |
Proteobacteria
(β-proteobacteria) |
freshwater pond | straight rods | 0.7–1.2×0.4–0.5 μm | 2.16 | utilization of low-molecular-weight substrates |
Hahn et al. (2012); Meincke et al. (2012) |
| Opitutus sp. VeCb1 | Verrucomicrobia | rice paddy soil | ellipsoids | 0.49×0.33 μm; 0.030 μm3 |
n.d. | utilization of sugars and sugar polymers, strict fermentative metabolism, oxygen tolerance |
Janssen et al. (1997); Chin et al. (2001) |
| Facultative ultramicrobacteria | |||||||
| Endomicrobium proavitum Rsa215 | Elusimicrobia | gut homogenate of Reticulitermes santonensis | cocci, rods showing budding cell division | 0.3–0.5 μm (for cocci); 0.5–3.5×0.15–0.30 μm (for rods) | 1.59 | nitrogen fixation | Zheng and Brune (2015); Zheng et al. (2016) |
| Chryseobacterium solincola NF4 | Bacteroidetes | lake sediment | cocci, rods showing budding cell division or cell septation | 0.004–0.04 μm3 (for cocci); 0.1–0.3 μm3 (for rods) | ~1.7 | ectoparasite of Bacillus subtilis |
Suzina et al. (2011); Duda et al. (2012) |
| Slender filamentous bacteria | |||||||
| Hylemonella gracilis CB |
Proteobacteria
(β-proteobacteria) |
freshwater | spirals | 0.12 μm3 (smallest width=0.2 μm) | n.d. | n.d. | Wang et al. (2007, 2008) |
| Oligoflexus tunisiensis Shr3T | Proteobacteria (Oligoflexia)* | desert sand | pleomorphic (rods, filaments, spirals, and spherical [or curled] cells) | various lengths×0.4–0.8 μm (for filaments) | 7.57 | multidrug resistance, incomplete denitrification** | Nakai et al. (2014, 2016a) |
| Silvanigrella aquatica MWH-Nonnen-W8redT | Proteobacteria (Oligoflexia)* | freshwater lake | pleomorphic (rods, filaments, and spirals) | 3.6×0.6 μm (for rods) | 3.51 | antimicrobial peptides, plasmid-encoded type IV secretion systems** | Hahn et al. (2017) |
| Silvanigrella paludirubra SP-Ram-0.45-NSY-1T | Proteobacteria (Oligoflexia)* | freshwater pond | pleomorphic (rods and filaments) | various lengths | 3.94 | utilization of limited substrates | Pitt et al. (2020) |
| Fluviispira multicolorata 33A1-SZDPT | Proteobacteria (Oligoflexia)* | freshwater creek | pleomorphic (rods and filaments) | various lengths | 3.39 | violacein-like production | Pitt et al. (2020) |
| CPR/Patescibacteria bacteria | |||||||
| WWE3-OP11-OD1 bacteria | candidate division WWE3, “Candidatus Microgenomates” (OP11), “Candidatus Parcubacteria” (OD1) | deep aquifer | cocci or oval-shaped | 0.009±0.002 μm3 | 0.69–1.05 | potential interaction with other bacterial cells via pili-like structures | Luef et al. (2015) |
| “Candidatus Sonnebornia yantaiensis” | “Candidatus Parcubacteria” (OD1) | ciliated protist Paramecium bursaria | straight rods | 1.6–1.9×0.5–0.6 μm | n.d. | endoplasmic symbiont of the ciliate P. bursaria | Gong et al. (2014) |
| TM7x bacterium | “Candidatus Saccharibacteria” (TM7) | human oral cavity | cocci | 0.2–0.3 μm | 0.71 | ectosymbiont of Actinomyces odontolyticus | He et al. (2015) |
| DPANN archaea | |||||||
| Nanoarchaeum equitans | Nanoarchaeota | submarine hot vent | cocci | 0.4 μm | ~0.5 | ectosymbiont of Ignicoccus hospitalis | Huber et al. (2002) |
| “Candidatus Nanopusillus acidilobi” | Nanoarchaeota | hot spring | cocci | 0.1–0.3 μm | 0.61 | ectosymbiont of Acidilobus species | Wurch et al. (2016) |
| “Candidatus Nanoclepta minutus” Ncl-1 | Nanoarchaeota | hot spring | flagellated cocci | ~0.2 μm | 0.58 | ectosymbiont of Zestosphaera tikiterensis | John et al. (2019) |
| “Candidatus Nanosalina” sp. J07AB43 | “Candidatus Nanohaloarchaeota” | hypersaline lake | cocci-like | 0.6 μm | 1.23 | possible free-living lifestyle | Narasingarao et al. (2012) |
| “Candidatus Nanosalinarum” sp. J07AB56 | “Candidatus Nanohaloarchaeota” | hypersaline lake | cocci-like | 0.6 μm | 1.22 | possible free-living lifestyle | Narasingarao et al. (2012) |
| ARMAN-2, -4, and -5 | “Candidatus Micrarchaeota” | acid mine drainage | cocci | ~0.5 μm | ~1.0 | potential interaction with Thermoplasmatales cells via pili-like structures | Baker et al. (2010) |
| “Candidatus Mancarchaeum acidiphilum” Mia14 | “Candidatus Micrarchaeota” | acid mine drainage | n.d. | n.d. | 0.95 | ectoparasite of Cuniculiplasma divulgatum | Golyshina et al. (2017) |
n.d.: no data.
* The proteobacterial class Oligoflexia is classified in the candidate phylum “Bdellovibrionota” in the Genome Taxonomy Database (GTDB).
** Putative physiological traits are inferred from their genomic and plasmid annotation.
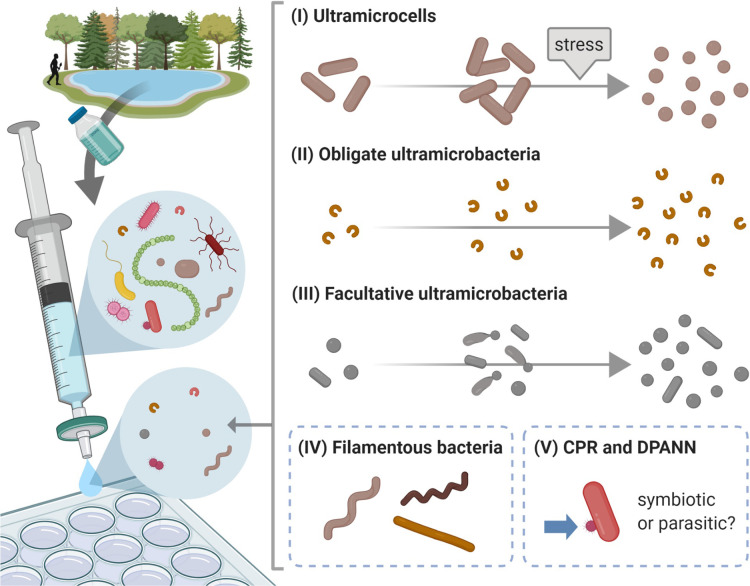
Based on previous studies, filterable microorganisms have been classified into five groups (Fig. 1): (I) ultramicrocells that are miniaturized microorganisms because of external factors (e.g., environmental stress) as described above; (II) obligate UMB that maintain small cell volumes (<0.1 μm3) regardless of their growth conditions; (III) facultative UMB that contain a small proportion of larger cells with a cell volume >0.1 μm3 (note that the definitions of the terms “obligate” and “facultative” UMB follow those of Duda et al. [2012]); (IV) slender filamentous bacteria; and (V) ultra-small members among CPR/Patescibacteria bacteria and DPANN archaea. In contrast to UMB strains, the cell shapes and morphological characteristics of members in group V are largely unknown under different environmental or culture conditions because all of the members of CPR and DPANN are uncultivated, with a few exceptions of members belonging to the phyla “Ca. Saccharibacteria” (former TM7) and Nanoarchaeota (e.g., Huber et al., 2002; He et al., 2015). Incidentally, the groups presented in this review do not include filterable cell-wall-less mycoplasmas as well as “nanobacteria” or “nannobacteria” as microfossils, which are often referred to in geological literature (Folk, 1999), or as calcium carbonate nanoparticles in the human body, as reported in medical literature (Martel and Young, 2008). Representative cases of groups II to V are described below and Table 1 shows a summarized list.
Fig. 1.
Diagram showing filterable microorganisms in the environment. (I) ultramicrocells; (II) obligate ultramicrobacteria; (III) facultative ultramicrobacteria; (IV) slender filamentous bacteria; (V) ultra-small members of CPR bacteria (also referred to as “Candidatus Patescibacteria”) and DPANN archaea indicated by the arrow in this Figure. See details in the text. This figure was created with BioRender (https://biorender.com/).
Obligate UMB
Obligate UMB are often reported from aqueous environments. One of the most prominent representatives is “Candidatus Pelagibacter ubique” HTCC1062, which is a SAR11 clade bacterium that is ubiquitous in marine environments. Previous studies found that SAR11 members consistently dominated ribosomal RNA gene clone libraries derived from seawater DNA and estimated their global population size as 2.4×1028 cells—approximately 25% of all prokaryotic cells—in oceans (Giovannoni et al., 1990; Morris et al., 2002). Despite their ubiquitous and abundant presence, it was not possible to isolate them. However, the first cultivated strain HTCC1062 was established in 2002 using a high-throughput dilution-to-extinction culturing (HTC) technique (Rappé et al., 2002). This HTC technique involves cultivation with serial dilutions of natural seawater samples into very low nutrient media (Connon and Giovannoni, 2002). The cell volume (approximately 0.01 μm3) of “Ca. P. ubique” was reported as one of the smallest free-living cells known. Subsequent studies characterized the SAR11 clade with the small, streamlined genomes (<1.5 Mbp) described below, an unusual mode of glycine auxotrophy, a light-dependent proton pump known as proteorhodopsin, and the ability to utilize various one-carbon compounds (reviewed in Tripp, 2013; Giovannoni, 2017). The SAR11 clade is highly divergent with multiple ecotypes and has freshwater members known as LD12 classified in SAR11 subclade IIIb (Grote et al., 2012). An LD12 cultivated representative, “Ca. Fonsibacter ubiquis” strain LSUCC0530, was subsequently established (Henson et al., 2018), and its genomic characteristics promoted the hypothesis that gene losses for osmolyte uptake were related to the evolutionary transition, or metabolic tuning, of freshwater SAR11 (LD12) from a salt to freshwater habitat.
Another marine ultramicrobacterium, Sphingopyxis alaskensis (formerly known as Sphingomonas alaskensis) RB2256 was intensively investigated before the study of the SAR11 clade (e.g., Eguchi et al., 1996; Schut et al., 1997). This strain was also characterized as an obligate UMB (Duda et al., 2012). When the cultivation of this strain transitioned from low-carbon to highly-enriched media, the cell volume of S. alaskensis remained at <0.1 μm3 in most media; however, larger elongated cells, not UMB cells, were observed in trypticase soy agar medium (Vancanneyt et al., 2001). Furthermore, this strain possesses a larger genome of 3.3 Mb (DDBJ/ENA/GenBank accession no. CP000356) than other UMB (Table 1).
Other prominent representatives of obligate UMB are freshwater actinobacterial strains. Typically, actinobacteria are among the numerically dominant groups in freshwater and their cells are found in smaller size fractions (Glöckner et al., 2000; Sekar et al., 2003). Hahn et al. (2003) first isolated nine filterable UMB of the class Actinobacteria from freshwater habitats and newly described a novel phylogenetic cluster (Luna cluster). This isolation was achieved by the “filtration-acclimatization” method of filter separation combined with an acclimatization procedure, which is a stepwise transition from low substrate conditions to artificial culture conditions. The important features of Luna cluster strains are their wide distribution in freshwater systems (Hahn and Pöckl, 2005) and their small cell sizes are stable and maintained in nutrient-rich media (Hahn et al., 2003). Our group also isolated an ultamicrosize actinobacterium related to Luna strains from river water in Japan and named it Aurantimicrobium minutum KNCT (Fig. 2; Nakai et al., 2015). This strain showed high 16S rRNA gene sequence similarity (>99%) to strains isolated from freshwater systems in other places in Japan as well as in Austria, Australia, China, Nicaragua, and Uganda (accession nos. AB278121, AB599783, AJ507461, AJ507467, AJ565412, AJ565413, and AJ630367), suggesting its cosmopolitan distribution in freshwater.
Fig. 2.
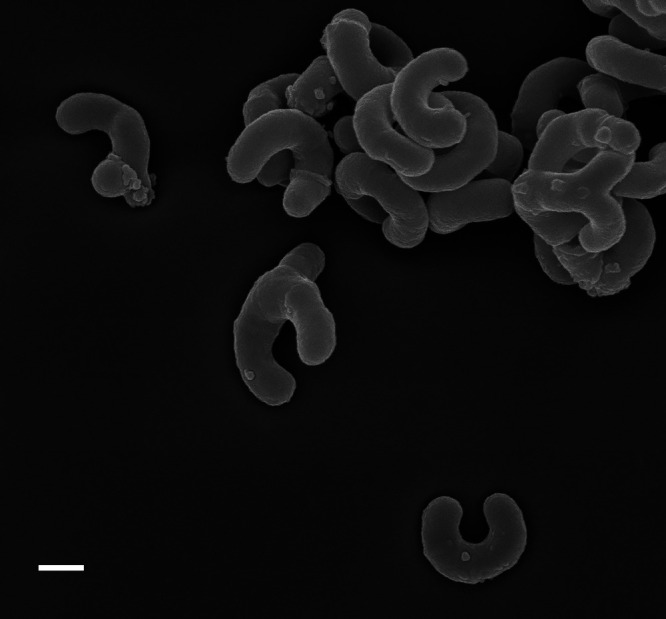
Scanning electron micrograph of c-shaped cells of Aurantimicrobium minutum KNCT. Cells were cultured in organic NSY (nutrient broth, soytone, and yeast extract; Hahn et al., 2004) medium for two weeks. Scale bar: 200 nm. This micrograph is an unpublished figure from the author; other micrographs of this species are shown in Nakai et al. (2013, 2015).
The other freshwater bacterium belonging to the Luna cluster, Rhodoluna lacicola MWH-Ta8T, was also described as an obligate UMB (Hahn et al., 2014); an additional three Rhodoluna strains smaller than R. lacicola were subsequently reported (Pitt et al., 2019). From an eco-physiological point of view, the genomes of freshwater actinobacteria possess rhodopsin photosystems (Neuenschwander et al., 2018), while R. lacicola has an unconventional proton-pumping rhodopsin that requires external supplementation with the cofactor retinal (Keffer et al., 2015). The underlying cause is considered to be an inability to biosynthesize the cofactor (Neuenschwander et al., 2018), suggesting that R. lacicola obtains retinal from the surrounding environment. One potential source in freshwater appears to be retinoids produced and released by cyanobacteria (Ruch et al., 2005; Wu et al., 2013).
Freshwater actinobacteria, including UMB strains, were previously shown to be phylogenetically diverse and subsequent studies yielded nine lineages (acI, acTH1, acSTL, Luna1, acIII, Luna3, acTH2, acIV, and acV; Newton et al., 2011). Among these lineages, acI containing multiple tribes is considered to be the most successful and ubiquitous group in the environment (Zwart et al., 2002; Warnecke et al., 2004; Kang et al., 2017), although pure cultures had not been established despite various cultivation trials. However, Kim et al. (2019) recently reported the first two pure acI cultures with very small sizes (volume, 0.04–0.06 μm3; Table 1), which are assumed to be obligate UMB. A key factor for their growth was the supplementation of a “helper” catalase, an enzyme that degrades hydrogen peroxide (H2O2), to the culture medium. Previous studies showed that H2O2 generated in medium affected the culture efficiency of microorganisms sensitive to oxidative stress (Kawasaki and Kamagata, 2017) and that the growth of the cyanobacterium Prochlorococcus was promoted by the presence of H2O2-scavenging microbes (Morris et al., 2011). These findings demonstrated that a catalase-supplemented cultivation strategy may facilitate the successful isolation of previously uncultured freshwater UMB.
Freshwater habitats also harbor another obligate UMB belonging to the genus Polynucleobacter in the class Betaproteobacteria. Similar to some actinobacteria described earlier, UMB members of this genus also showed a cosmopolitan distribution in freshwater systems (Hahn, 2003). The relative abundance of the subspecies named PnecC was high, ranging between <1% and 67% (average 14.5%) of total bacterial numbers, in more than 130 lakes studied in Central Europe, as assessed by fluorescent in situ hybridization (Jezberová et al., 2010). Culture experiments and genomic characterization suggested that PnecC bacteria in nature can utilize low-molecular-weight products derived from photooxidation and/or the direct enzymatic cleavage of high-molecular-weight substrates, such as humic substances (Watanabe et al., 2009; Hahn et al., 2012). Certain PnecC strains sharing ≥99% similarity in 16S rRNA gene sequences differed in their ecophysiological and genomic features (e.g., the presence/absence of iron transporter genes), suggesting cryptic diversity among the abundant lineage not covered by 16S rRNA gene-based typing (Hahn et al., 2016).
The obligate UMB inhabiting sea and freshwaters described above were characterized by minute cell sizes, but also small genome sizes (<2 Mbp) with a low genomic guanine-cytosine (GC) content: this genome “streamlining” is considered to reflect an adaptation to nutrient-limited conditions (e.g., SAR11 members; 1.16–1.46 Mb; Giovannoni et al., 2005; Grote et al., 2012; Henson et al., 2018) (Table 1). This phenomenon of a reduced genome size with gene loss also indicates metabolic dependencies on co-existing microorganisms in nature, as described by the “Black Queen Hypothesis” (Morris et al., 2012). As another example, the reconstructed genomes of ultra-small and uncultivated marine actinobacteria (“Candidatus Actinomarinidae”) were very small (<1 Mb) and had a very low GC content of 33% (Ghai et al., 2013). In addition, known obligate UMB of different lineages, such as “Ca. P. ubique” (Alphaproteobacteria), Polynucleobacter strains (Betaproteobacteria), and A. minutum and R. lacicola (Actinobacteria), showed similar “c-shaped” (curved-rod) cells (Table 1; A. minutum for Fig. 2; Hahn, 2003). This unique shape may be advantageous for the efficient acquisition of substances because of their increased surface-to-volume ratio of cells or grazing resistance against bacteriovorus protists for planktonic life in waters.
In contrast to aquatic environments, limited information is currently available on UMB, including the obligate type, from soil habitats. Janssen et al. (1997) previously reported anaerobic obligate UMB with very small ellipsoid to nearly spherical shapes (e.g., Opitutus sp. VeCb1 with a cell volume of 0.030 μm3) belonging to the Verrucomicrobiales lineage from rice paddy soil using dilution culture techniques. Nakai et al. (2013) isolated and cultivated filterable strains from soil and sand suspensions; however, obligate UMB were not found among these strains. High-throughput sequencing of the 16S rRNA gene revealed that the smaller size fractions in soils were more likely to harbor rare or poorly characterized bacterial and archaeal taxa, such as Acidobacteria, Gemmatimonadetes, Elusimicrobia, Verrucomicrobia, and Crenarchaeota (Portillo et al., 2013). However, further studies are needed to clarify whether the members detected in the small fractions contain UMB.
Facultative UMB
Facultative UMB that contain a small proportion of larger cells with a cell volume >0.1 μm3 have not yet been characterized in detail (Table 1) because morphological changes throughout the growth cycle have only been examined in a limited number of UMB. Endomicrobium proavitum Rsa215 (now deposited as DSM29378T=JCM32103T) belonging to the phylum Elusimicrobia appears to be a well-studied example of facultative UMB. The phylum Elusimicrobia (former termite group 1 candidate phylum) was initially established with the cultivated ultramicrobacterium of Elusimicrobium minutum strain Pei191T from the 0.2 μm-filtered filtrate—originally prepared as a growth promoting supplement for gut bacteria—of the gut homogenates of a scarab beetle larva (Geissinger et al., 2009; Herlemann et al., 2009). E. proavitum Rsa215 was isolated from the filtrate of the gut homogenate and was identified as a free-living bacterium of a novel class-level lineage in Elusimicrobia (Zheng et al., 2016). E. proavitum has an unusual cell cycle that involves different cell forms, i.e., cocci, rods, and budding-like cells, during the cell cycle. Under laboratory cultivation conditions, before growth commences, the cell population is comprised of a large population of UMB coccoid cells with a few rod-shaped cells (~3.5 μm in length); small cocci are formed from a bud-like swelling at one pole of the rod-shaped cells during growth. Although its morphological variation in the host gut currently remains unclear, cell characteristics as observed in the laboratory result in the classification of facultative UMB. Another important trait for E. proavitum is the ability to fix nitrogen gas with a group IV nitrogenase, which was considered to harbor functions other than nitrogen fixation (Dos Santos et al., 2012).
Slender filamentous bacteria
In addition to ultramicrocells and UMB, slender filamentous bacteria have frequently been found in 0.2 μm-filtered fractions of environmental samples. Slender spirillum-shaped Hylemonella gracilis was isolated from filtrates of freshwater samples (e.g., Hahn et al., 2004; Nakai et al., 2013) and passes through membrane filters with small pore sizes of not only 0.22–0.45 μm, but also 0.1 μm (Wang et al., 2007). The smallest widths of H. gracilis cells are approximately 0.2 μm and close to filter pore sizes, which may allow its slender cells to “squeeze” through these pores. Regarding the quality control and assessment of filter sterilization, Wang et al. (2008) proposed that filterable slender bacteria, such as H. gracilis with small cell widths, may be used for the microbiological validation of membrane filters instead of Brevundimonas diminuta, which is the current standard strain tested.
During a screening of UMB, our group isolated a slender filamentous bacterium from the filtrate of a suspension of desert sands collected in Tunisia, and described Oligoflexus tunisiensis Shr3T, which represents the eighth novel class named Oligoflexia within the phylum Proteobacteria (Nakai et al., 2014; 2016a). The cell shape of this species is mainly slender, filamentous, and of variable lengths, but shows a pleomorphism with other shapes, such as a spiral, spherical (or curled), or curved rod morphology (Fig. 3; Nakai and Naganuma, 2015). This polymorphic flexibility of cells with small widths down to 0.4 μm appears to be related to their ability to pass through membrane filters; however, it has not yet been clarified whether each morphological shape is associated with a resting state or other states. Regarding filamentous formation, this shape may be related to resistance to protozoan grazing, as reported in previous studies (e.g., Jürgens et al., 1999; Suzuki et al., 2017a). The environmental sequences closely related (>97%) to the 16S rRNA gene sequence of O. tunisiensis were recovered from paddy soil, cyanobacterial bloom in lake water, bioreactors, and human skin using culture-independent approaches; however, their detection frequency was low, with at most ~0.6% (Nakai and Naganuma, 2015). Thus, O. tunisiensis and its relatives appear to be rare species, and their ecological roles are currently unclear; one possible role for O. tunisiensis may be incomplete denitrification to nitrous oxide, as inferred from its genome sequence (Nakai et al., 2016a).
Fig. 3.
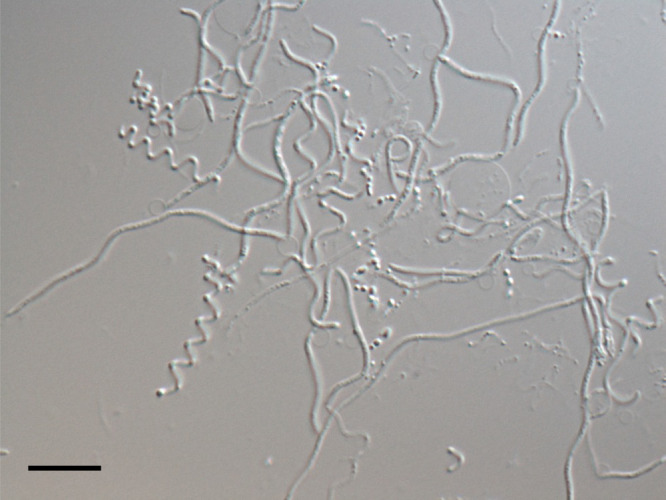
Micrograph of pleomorphic cells of Oligoflexus tunisiensis Shr3T. Cells were cultured in R2A medium for more than two weeks. This micrograph is slightly modified from the figure originally published in Nakai and Naganuma (2015). Scale bar: 10 μm.
Despite the potential rarity of its occurrence, the size filtration method led to the isolation of an additional slender filamentous strain, Silvanigrella aquatica MWH-Nonnen-W8redT, with a pleomorphic morphology in the class (Hahn et al., 2017). Hahn et al. (2017) reclassified the order Bdellovibrionales, including Bdellovibrio spp. known as small “bacteria-eating” bacteria (reviewed in Sockett, 2009), from the class Deltaproteobacteria to the class Oligoflexia based on in-depth phylogenetic analyses. Incidentally, 0.45-μm filtrates of environmental samples are frequently used for the enrichment culture of Bdellovibrio predatory bacteria. In the Genome Taxonomy Database (GTDB) based on genome phylogeny (https://gtdb.ecogenomic.org/; Parks et al., 2018), the class Oligoflexia belongs to the candidate phylum “Bdellovibrionota”, named after the genus Bdellovibrio, and not the phylum Proteobacteria; its taxonomic assignment will be discussed in future studies. Oligoflexia very recently gained two more species, Fluviispira multicolorata 33A1-SZDPT and Silvanigrella paludirubra SP-Ram-0.45-NSY-1T, from freshwater habitats (Pitt et al., 2020). Silvanigrella spp. are phylogenetically closely aligned with “Candidatus Spirobacillus cienkowskii” (Pitt et al., 2020), which is an uncultured pathogen of water fleas (Daphnia spp.) described morphologically almost 130 years ago (Metchnikoff, 1889). Since Silvanigrella spp. are isolated from the filtrates of micropore filtration, size fractionation may be an effective method for isolating the uncultivated pathogen as well as additionally overlooked agents in Oligoflexia. A detailed comparison within members of this class will also be important for pursuing the evolutionary acquisition and divergence of predatory and pathogenic behaviors.
Diverse ultra-small members and their potentials
Metagenomic investigations on microbial communities have generated genomes for an astounding diversity of bacteria and archaea; CPR/Patescibacteria inhabiting groundwater has attracted increasing attention in recent years. Traditionally, certain types of groundwater bacteria were known to pass through a micropore filter (e.g., Shirey and Bissonnette, 1991). Additionally, Miyoshi et al. (2005) phylogenetically characterized filterable microorganisms captured by 0.1-μm-pore-sized filters from deep aquifers of the Tono uranium mine, Japan and then discovered candidate divisions OD1 and OP11 (now recognized as candidate phyla “Ca. Parcubacteria” and “Ca. Microgenomates”, respectively) enriched by approximately 44% in 16S rRNA gene clones from the filtered fraction. The specific occurrence of “Ca. Parcubacteria” (OD1) in the 0.2-μm filtrate was also detected in deep-sea hydrothermal fluid (Naganuma et al., 2007). It was previously unclear whether members of these candidate divisions were UMB. In subsequent studies using cryo-imaging, ultra-small cells (approximately 0.009±0.002 μm3) were reported in the filtrate of an aquifer water near Colorado, USA, which were enriched with the candidate divisions WWE3, OD1, and OP11, all recently belonging to CPR/Patescibacteria (Luef et al., 2015).
Metagenomics was then used to reconstruct the genomes of filterable members in the aquifer system, representing >35 candidate phyla named CPR (Brown et al., 2015). This highly diversified group of uncultivated bacteria may subdivide the domain Bacteria (Hug et al., 2016); however, this scenario remains controversial (e.g., Parks et al., 2018; Zhu et al., 2019). Importantly, measurements of replication rates (Brown et al., 2016; Suzuki et al., 2017b) and cryo-transmission electron microscopy images showing a dividing cell (Luef et al., 2015) indicated that the extremely small cells of CPR/Patescibacteria are metabolically active and not simply ultramicrocells during starvation. Moreover, CPR/Patescibacteria genomes have been recovered from other environments, such as highly alkaline groundwater (Suzuki et al., 2017b; Sato et al., 2019), lakes (Vigneron et al., 2019), soil (Starr et al., 2018), and marine sediment (Orsi et al., 2018) as well as the human microbiome (He et al., 2015) and dolphin mouse (Dudek et al., 2017), suggesting a wide distribution across environments. Besides describing ultra-small life forms with high phylogenetic novelty, genomic analyses of CPR/Patescibacteria members have provided information on their small genomes, fermentative metabolism, and other unusual features (e.g., self-splicing introns varying in length and proteins encoded within their 16S rRNA genes; Brown et al., 2015; Castelle et al., 2018). Divergent 16S rRNA gene sequences prevent many specific phyla (e.g., ~50% of “Ca. Microgenomates” [OP11] and 60% of candidate division WWE3) from being detected by typical PCR surveys with the universal bacterial primer set 515F and 806R (Brown et al., 2016). The small genome sizes observed (often <1 Mb) appear to be a reflection of a symbiotic lifestyle and/or high in situ selection pressure in a stable environment, rather than the genome streamlining of free-living obligate UMB, as described earlier, assuming streamlining characteristics (e.g., highly conserved core genomes with few pseudogenes; Giovannoni et al., 2014). Although the CPR/Patescibacteria genomes studied to date possess incomplete biosynthetic pathways for their cellular building blocks (e.g., nucleotides and fatty acids; Castelle et al., 2018), the possibility of their ability to de novo synthesize them by unknown pathways cannot be ruled out. Furthermore, their host-associated distribution was reported: “Candidatus Sonnebornia yantaiensis” of “Ca. Parcubacteria” (OD1) as an endoplasmic symbiont of the protist (Gong et al., 2014) and TM7x bacterium of “Ca. Saccharibacteria” (TM7) attached to Actinomyces odontolyticus (He et al., 2015), as shown in Table 1.
The features of small cell sizes and small genomes observed in CPR/Patescibacteria are shared by some members of the DPANN archaea, particularly Nanoarchaeota (Huber et al., 2002), “Ca. Nanohalorchaeota” (Narasingarao et al., 2012), and so-called ARMAN (archaeal Richmond Mine acidophilic nano-organisms; Baker et al., 2010). DPANN including these UMA has been expanded by the addition of novel phylum-level groups, and, at the time of writing, encompasses at least ten different lineages (reviewed in Dombrowski et al., 2019). In several cases, except for the members of “Ca. Nanohalorchaeota”, as with CPR/Patescibacteria, DPANN-affiliated UMA showed an ectosymbiotic localization: Nanoarchaeum equitans attached to Ignicoccus hospitalis (Huber et al., 2002), “Ca. Nanopusillus acidilobi” and its host Acidilobus species (Wurch et al., 2016), and “Ca. Mancarchaeum acidiphilum” Mia14 (ARMAN-2-related organism) and its host Cuniculiplasma divulgatum (Golyshina et al., 2017) (other data in Table 1). Additionally, DPANN organisms lack the ability to biosynthesize their building blocks (Castelle et al., 2018). Although it is still unclear whether these symbiotic or parasitic lifestyles represent a way of life for the CPR/Patescibacteria and DPANN groups, the cases described above indicate that several members of these groups appear to be important in organism-organism interactions.
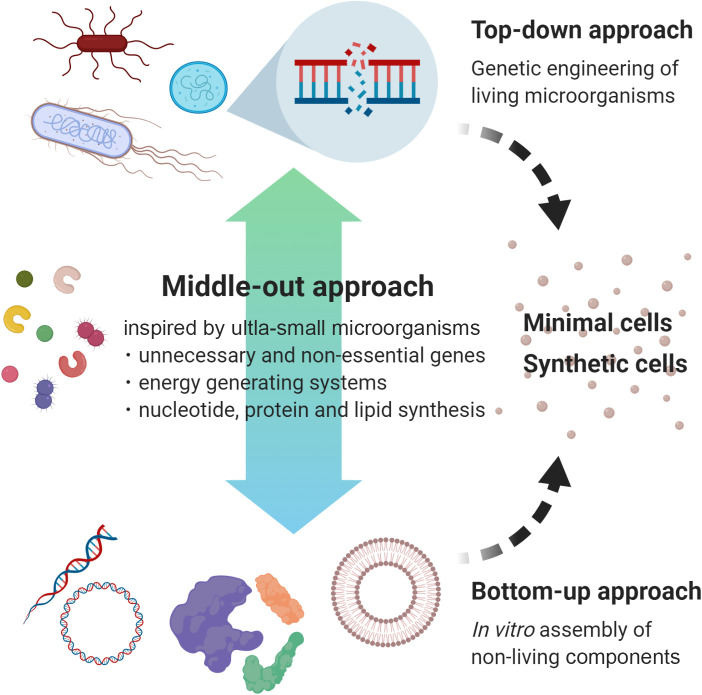
The characterization of ultra-small life forms may provide a new perspective for minimal cells and synthetic cells. In the field of synthetic biology, the top-down approach has been employed to reduce and simplify the genomes of microbial cells by genetic engineering, and then to identify essential genes for living systems; the bottom-up approach, which is the opposite of the top-down approach, has been used to examine what is sufficient for living systems by assembling non-living components, such as nucleic acids, proteins, and lipids (e.g., Matsuura et al., 2011; Xu et al., 2016). In this context, DeWall and Cheng (2011) pointed out that the small genomes of microorganisms in nature may be models for the identification of a minimal genome. Since the ultra-small members described here as well as free-living obligate UMB already harbor small and sometimes streamlined genome structures (<2 Mb) through the loss of unnecessary components, the “middle-out” approach, referring to the metabolic pathway of these members (Fig. 4), which effectively combines traditional top-down and bottom-up approaches, will be useful for the rational design of artificial cells.
Fig. 4.
A schematic diagram of the “middle-out” approach toward the development of minimal cells or synthetic cells. This approach, inspired by the unusual biology of ultra-small life forms, may provide a new perspective to traditional top-down or bottom-up approaches. This figure was created with BioRender (https://biorender.com/).
Conclusions
Numerous cultivation efforts have clearly shown that some previously uncultured members remain viable in small-size fractions. Some obligate UMB are ubiquitous and dominant in water systems and may play important roles in natural microbiome functions. In parallel, the advent of high-throughput sequencing technology has greatly expanded our knowledge of ultra-small microbial diversity. Future studies are required to shed light on small microorganisms hidden in various environmental samples (e.g., soils and sediments) other than aqueous environments, and on the ecophysiological traits and biogeochemical roles of these members, including CPR/Patescibacteria and DPANN. Further studies on “extreme” microorganisms at the lower size limit will undoubtedly lead to new conundrums about life on Earth.
Citation
Nakai, R. (2020) Size Matters: Ultra-small and Filterable Microorganisms in the Environment. Microbes Environ 35: ME20025.
https://doi.org/10.1264/jsme2.ME20025
Acknowledgement
I would like to thank Dr. K. Takai (JAMSTEC) and one anonymous reviewer for their helpful comments and suggestions on an earlier draft of this review.
References
- Anderson J.I.W., and Heffernan W.P. (1965) Isolation and characterization of filterable marine bacteria1. J Bacteriol 90: 1713–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.J., Comolli L.R., Dick G.J., Hauser L.J., Hyatt D., Dill B.D., et al. (2010) Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci U S A 107: 8806–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.T., Hug L.A., Thomas B.C., Sharon I., Castelle C.J., Singh A., et al. (2015) Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523: 208–211. [DOI] [PubMed] [Google Scholar]
- Brown C.T., Olm M.R., Thomas B.C., and Banfield J.F. (2016) Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34: 1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle C.J., Brown C.T., Anantharaman K., Probst A.J., Huang R.H., and Banfield J.F. (2018) Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16: 629–645. [DOI] [PubMed] [Google Scholar]
- Chin K.J., Liesack W., and Janssen P.H. (2001) Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int J Syst Evol Microbiol 51: 1965–1968. [DOI] [PubMed] [Google Scholar]
- Connon S.A., and Giovannoni S.J. (2002) High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68: 3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danczak R.E., Johnston M.D., Kenah C., Slattery M., Wrighton K.C., and Wilkins M.J. (2017) Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWall M.T., and Cheng D.W. (2011) The minimal genome—a metabolic and environmental comparison. Briefings Funct Genomics 10: 312–315. [DOI] [PubMed] [Google Scholar]
- Dombrowski N., Lee J.-H., Williams T.A., Offre P., and Spang A. (2019) Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol Lett 366: fnz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos P.C., Fang Z., Mason S.W., Setubal J.C., and Dixon R. (2012) Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda V.I., Suzina N.E., Polivtseva V.N., and Boronin A.M. (2012) Ultramicrobacteria: formation of the concept and contribution of ultramicrobacteria to biology. Microbiology 81: 379–390. [PubMed] [Google Scholar]
- Dudek N.K., Sun C.L., Burstein D., Kantor R.S., Aliaga Goltsman D.S., Bik E.M., et al. (2017) Novel microbial diversity and functional potential in the marine mammal oral microbiome. Curr Biol 27: 3752–3762.e6. [DOI] [PubMed] [Google Scholar]
- Eguchi M., Nishikawa T., Macdonald K., Cavicchioli R., Gottschal J.C., and Kjelleberg S. (1996) Responses to stress and nutrient availability by the marine ultramicrobacterium Sphingomonas sp. strain RB2256. Appl Environ Microbiol 62: 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaied H.E., Sato M., and Naganuma T. (2001) Viable Cytophaga-like bacterium in the 0.2 μm-filtrate seawater. Syst Appl Microbiol 24: 618–622. [DOI] [PubMed] [Google Scholar]
- Fedotova A.V., Belova S.E., Kulichevskaya I.S., and Dedysh S.N. (2012) Molecular identification of filterable bacteria and archaea in the water of acidic lakes of northern Russia. Microbiology 81: 281–287. [Google Scholar]
- Feil H., Feil W.S., Chain P., Larimer F., DiBartolo G., Copeland A., et al. (2005). Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci U S A 102: 11064–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk R.L. (1999) Nannobacteria and the precipitation of carbonate in unusual environments. Sediment Geol 126: 47–55. [Google Scholar]
- Geissinger O., Herlemann D.P.R., Mörschel E., Maier U.G., and Brune A. (2009) The ultramicrobacterium “Elusimicrobium minutum” gen. nov., sp. nov., the first cultivated representative of the termite group 1 phylum. Appl Environ Microbiol 75: 2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R., Mizuno C.M., Picazo A., Camacho A., and Rodriguez-Valera F. (2013) Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep 3: 2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuneim L.-A.J., Jones D.L., Golyshin P.N., and Golyshina O.V. (2018) Nano-sized and filterable bacteria and archaea: biodiversity and function. Front Microbiol 9: 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S.J., Britschgi T.B., Moyer C.L., and Field K.G. (1990) Genetic diversity in Sargasso Sea bacterioplankton. Nature 345: 60–63. [DOI] [PubMed] [Google Scholar]
- Giovannoni S.J., Tripp H.J., Givan S., Podar M., Vergin K.L., Baptista D., et al. (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242. [DOI] [PubMed] [Google Scholar]
- Giovannoni S.J., Cameron Thrash J., and Temperton B. (2014) Implications of streamlining theory for microbial ecology. ISME J 8: 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S.J. (2017) SAR11 bacteria: the most abundant plankton in the oceans. Ann Rev Mar Sci 9: 231–255. [DOI] [PubMed] [Google Scholar]
- Glöckner F.O., Zaichikov E., Belkova N., Denissova L., Pernthaler J., Pernthaler A., and Amann R. (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol 66: 5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golyshina O.V., Toshchakov S.V., Makarova K.S., Gavrilov S.N., Korzhenkov A.A., La Cono V., et al. (2017) ‘ARMAN’ archaea depend on association with euryarchaeal host in culture and in situ. Nat Commun 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Qing Y., Guo X., and Warren A. (2014) “Candidatus Sonnebornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea). Syst Appl Microbiol 37: 35–41. [DOI] [PubMed] [Google Scholar]
- Grote J., Thrash J.C., Huggett M.J., Landry Z.C., Carini P., Giovannoni S.J., and Rappé M.S. (2012) Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3: e00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W. (2003) Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl Environ Microbiol 69: 5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Lünsdorf H., Wu Q., Schauer M., Höfle M.G., Boenigk J., and Stadler P. (2003) Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol 69: 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Stadler P., Wu Q.L., and Pöckl M. (2004) The filtration–acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods 57: 379–390. [DOI] [PubMed] [Google Scholar]
- Hahn M.W., and Pöckl M. (2005) Ecotypes of planktonic actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl Environ Microbiol 71: 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Scheuerl T., Jezberová J., Koll U., Jezbera J., Šimek K., et al. (2012) The passive yet successful way of planktonic life: genomic and experimental analysis of the ecology of a free-living polynucleobacter population. PLoS One 7: e32772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Schmidt J., Taipale S.J., Doolittle W.F., and Koll U. (2014) Rhodoluna lacicola gen. nov., sp. nov., a planktonic freshwater bacterium with stream-lined genome. Int J Syst Evol Microbiol 64: 3254–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Jezberová J., Koll U., Saueressig-Beck T., and Schmidt J. (2016) Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. ISME J 10: 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Schmidt J., Koll U., Rohde M., Verbarg S., Pitt A., et al. (2017) Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales Int J Syst Evol Microbiol 67: 2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller C.M., Rölleke S., Vybiral D., Witte A., and Velimirov B. (2000) Investigation of 0.2 μm filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol Ecol 31: 153–161. [DOI] [PubMed] [Google Scholar]
- He X., McLean J.S., Edlund A., Yooseph S., Hall A.P., Liu S.-Y., et al. (2015) Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A 112: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson M.W., Lanclos V.C., Faircloth B.C., and Thrash J.C. (2018) Cultivation and genomics of the first freshwater SAR11 (LD12) isolate. ISME J 12: 1846–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann D.P.R., Geissinger O., Ikeda-Ohtsubo W., Kunin V., Sun H., Lapidus A., et al. (2009) Genomic analysis of “Elusimicrobium minutum,” the first cultivated representative of the phylum “Elusimicrobia” (formerly termite group 1). Appl Environ Microbiol 75: 2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., and Stetter K.O. (2002) A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417: 63–67. [DOI] [PubMed] [Google Scholar]
- Hug L.A., Baker B.J., Anantharaman K., Brown C.T., Probst A.J., Castelle C.J., et al. (2016) A new view of the tree of life. Nat Microbiol 1: 16048. [DOI] [PubMed] [Google Scholar]
- Janssen P.H., Schuhmann A., Mörschel E., and Rainey F.A. (1997) Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol 63: 1382–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezberová J., Jezbera J., Brandt U., Lindström E.S., Langenheder S., and Hahn M.W. (2010) Ubiquity of Polynucleobacter necessarius subsp. asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environ Microbiol 12: 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John E.S., Liu Y., Podar M., Stott M.B., Meneghin J., Chen Z., et al. (2019). A new symbiotic nanoarchaeote (Candidatus Nanoclepta minutus) and its host (Zestosphaera tikiterensis gen. nov., sp. nov.) from a New Zealand hot spring. Syst Appl Microbiol 42: 94–106. [DOI] [PubMed] [Google Scholar]
- Jürgens K., Pernthaler J., Schalla S., and Amann R. (1999) Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol 65: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I., Kim S., Islam M.R., and Cho J.C. (2017) The first complete genome sequences of the acI lineage, the most abundant freshwater Actinobacteria, obtained by whole-genome-amplification of dilution-to-extinction cultures. Sci Rep 7: 42252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., and Kamagata Y. (2017) Phosphate-catalyzed hydrogen peroxide formation from agar, gellan, and κ-carrageenan and recovery of microbial cultivability via catalase and pyruvate. Appl Environ Microbiol 83: e01366-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keffer J.L., Hahn M.W., and Maresca J.A. (2015) Characterization of an unconventional rhodopsin from the freshwater actinobacterium Rhodoluna lacicola. J Bacteriol 197: 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kang I., Seo J.-H., and Cho J.-C. (2019) Culturing the ubiquitous freshwater actinobacterial acI lineage by supplying a biochemical ‘helper’ catalase. ISME J 13: 2252–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E., Ichimura A.S., Peng V., Fritsen C.H., Trubl G., Doran P.T., and Murray A.E. (2014) Brine assemblages of ultrasmall microbial cells within the ice cover of Lake Vida, Antarctica. Appl Environ Microbiol 80: 3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannes R., Olsson-Francis K., Lopez P., and Bapteste E. (2019) Carbon fixation by marine ultrasmall prokaryotes. Genome Biol Evol 11: 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, R.V., and Jornitz, M.W. (2006) Types of filtration. In Sterile Filtration. Jornitz, M.W. (ed.) Berlin, Heidelberg: Springer, pp. 1–26. [Google Scholar]
- Luef B., Frischkorn K.R., Wrighton K.C., Holman H.-Y.N., Birarda G., Thomas B.C., et al. (2015) Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6: 6372. [DOI] [PubMed] [Google Scholar]
- MacDonell M.T., and Hood M.A. (1982) Isolation and characterization of ultramicrobacteria from a Gulf Coast estuary. Appl Environ Microbiol 43: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y., Kushimoto K., Muraguchi Y., Fukuda K., Miura T., Yamazoe A., et al. (2018) Proteobacteria and Bacteroidetes are major phyla of filterable bacteria passing through 0.22 μm pore size membrane filter, in Lake Sanaru, Hamamatsu, Japan. Biosci Biotechnol Biochem 82: 1260–1263. [DOI] [PubMed] [Google Scholar]
- Martel J., and Young J.D.-E. (2008) Purported nanobacteria in human blood as calcium carbonate nanoparticles. Proc Natl Acad Sci U S A 105: 5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, T., Ichihashi, N., Sunami, T., Kita, H., Suzuki, H., and Yomo, T. (2011) Evolvability and self-replication of genetic information in liposomes. In The Minimal Cell. Luisi, P., and Stano, P. (eds). Dordrecht: Springer, pp. 275–287. [Google Scholar]
- Meincke L., Copeland A., Lapidus A., Lucas S., Berry K.W., Del Rio T.G., et al. (2012) Complete genome sequence of Polynucleobacter necessarius subsp. asymbioticus type strain (QLW-P1DMWA-1T). Stand Genomic Sci 6: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchnikoff E. (1889) Contributions á l’etude du pleomorphisme des bacteriens. Ann Inst Pasteur 3: 61–68. [Google Scholar]
- Miteva V.I., and Brenchley J.E. (2005) Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl Environ Microbiol 71: 7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Iwatsuki T., and Naganuma T. (2005) Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol 71: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier J.-M., and Lindow S.E. (2003) Pseudomonas syringae responds to the environment on leaves by cell size reduction. Phytopathology 93: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Moore, P.B. (1999) A biophysical chemist’s thoughts on cell size. In Size Limits of Very Small Microorganisms: Proceedings of a Workshop. Washington, DC: National Academies Press, pp. 16–20. [PubMed] [Google Scholar]
- Morris J.J., Johnson Z.I., Szul M.J., Keller M., and Zinser E.R. (2011) Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6: e16805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.J., Lenski R.E., and Zinser E.R. (2012) The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R.M., Rappé M.S., Connon S.A., Vergin K.L., Siebold W.A., Carlson C.A., and Giovannoni S.J. (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810. [DOI] [PubMed] [Google Scholar]
- Naganuma T., Miyoshi T., and Kimura H. (2007) Phylotype diversity of deep-sea hydrothermal vent prokaryotes trapped by 0.2- and 0.1-μm-pore-size filters. Extremophiles 11: 637–646. [DOI] [PubMed] [Google Scholar]
- Nakai R., Abe T., Takeyama H., and Naganuma T. (2011) Metagenomic analysis of 0.2-μm-passable microorganisms in deep-sea hydrothermal fluid. Mar Biotechnol 13: 900–908. [DOI] [PubMed] [Google Scholar]
- Nakai R., Shibuya E., Justel A., Rico E., Quesada A., Kobayashi F., et al. (2013) Phylogeographic analysis of filterable bacteria with special reference to Rhizobiales strains that occur in cryospheric habitats. Antarct Sci 25: 219–228. [Google Scholar]
- Nakai R.*, Nishijima M.*, Tazato N., Handa Y., Karray F., Sayadi S., et al. (*equal contribution) (2014) Oligoflexus tunisiensis gen. nov., sp. nov., a Gram-negative, aerobic, filamentous bacterium of a novel proteobacterial lineage, and description of Oligoflexaceae fam. nov., Oligoflexales ord. nov. and Oligoflexia classis nov. Int J Syst Evol Microbiol 64: 3353–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R., Baba T., Niki H., Nishijima M., and Naganuma T. (2015) Aurantimicrobium minutum gen. nov., sp. nov., a novel ultramicrobacterium of the family Microbacteriaceae, isolated from river water. Int J Syst Evol Microbiol 65: 4072–4079. [DOI] [PubMed] [Google Scholar]
- Nakai R., and Naganuma T. (2015) Oligoflexia, the newest class of the phylum Proteobacteria, consisting of only one cultured species and uncultured bacterial phylotypes from diverse habitats. J Phylogenet Evol Biol 3: 141. [Google Scholar]
- Nakai R., Fujisawa T., Nakamura Y., Baba T., Nishijima M., Karray F., et al. (2016. a) Genome sequence and overview of Oligoflexus tunisiensis Shr3T in the eighth class Oligoflexia of the phylum Proteobacteria. Stand Genomic Sci 11: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R., Fujisawa T., Nakamura Y., Nishide H., Uchiyama I., Baba T., et al. (2016. b) Complete genome sequence of Aurantimicrobium minutum type Strain KNCT, a planktonic ultramicrobacterium isolated from river water. Genome Announc 4: e00616-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P., Podell S., Ugalde J.A., Brochier-Armanet C., Emerson J.B., Brocks J.J., et al. (2012) De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander S.M., Ghai R., Pernthaler J., and Salcher M.M. (2018) Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J 12: 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R.J., Jones S.E., Eiler A., McMahon K.D., and Bertilsson S. (2011) A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi Y., and Suzuki S. (2019) High growth potential of transiently 0.2-μm-filterable bacteria with extracellular protease activity in coastal seawater. Plankton Benthos Res 14: 276–286. [Google Scholar]
- Orsi W.D., Richards T.A., and Francis W.R. (2018) Predicted microbial secretomes and their target substrates in marine sediment. Nat Microbiol 3: 32–37. [DOI] [PubMed] [Google Scholar]
- Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.-A., and Hugenholtz P. (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36: 996–1004. [DOI] [PubMed] [Google Scholar]
- Pitt A., Schmidt J., Koll U., and Hahn M.W. (2019) Rhodoluna limnophila sp. nov., a bacterium with 1.4 Mbp genome size isolated from freshwater habitats located in Salzburg, Austria. Int J Syst Evol Microbiol 69: 3946–3954. [DOI] [PubMed] [Google Scholar]
- Pitt A., Koll U., Schmidt J., and Hahn M.W. (2020) Fluviispira multicolorata gen. nov., sp. nov. and Silvanigrella paludirubra sp. nov., isolated from freshwater habitats. Int J Syst Evol Microbiol 70: 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo M.C., Leff J.W., Lauber C.L., and Fierer N. (2013) Cell size distributions of soil bacterial and archaeal taxa. Appl Environ Microbiol 79: 7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé M.S., Connon S.A., Vergin K.L., and Giovannoni S.J. (2002) Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633. [DOI] [PubMed] [Google Scholar]
- Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.-F., et al. (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Ruch S., Beyer P., Ernst H., and Al-Babili S. (2005) Retinal biosynthesis in Eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol Microbiol 55: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Sato T., Yoshiya K., and Maruyama S. (2019) History of the Hadean “Living Microfossil” OD1 and ultra-reducing environments. J Geogr (Tokyo, Japan) 128: 571–596. [Google Scholar]
- Schulz H.N., Brinkhoff T., Ferdelman T.G., Mariné M.H., Teske A., and Jørgensen B.B. (1999) Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284: 493. [DOI] [PubMed] [Google Scholar]
- Schulz H.N., and Jørgensen B.B. (2001) Big bacteria. Annu Rev Microbiol 55: 105–137. [DOI] [PubMed] [Google Scholar]
- Schut F., Gottschal J.C., and Prins R.A. (1997) Isolation and characterisation of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol Rev 20: 363–369. [Google Scholar]
- Sekar R., Pernthaler A., Pernthaler J., Warnecke F., Posch T., and Amann R. (2003) An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol 69: 2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirey J.J., and Bissonnette G.K. (1991) Detection and identification of groundwater bacteria capable of escaping entrapment on 0.45-micron-pore-size membrane filters. Appl Environ Microbiol 57: 2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett R.E. (2009) Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol 63: 523–539. [DOI] [PubMed] [Google Scholar]
- Starr E.P., Shi S., Blazewicz S.J., Probst A.J., Herman D.J., Firestone M.K., and Banfield J.F. (2018) Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 6: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzina N.E., Duda V.I., Esikova T.Z., Shorokhova A.P., Gafarov A.B., Oleinikov R.R., et al. (2011) Novel ultramicrobacteria, strains NF4 and NF5, of the genus Chryseobacterium: Facultative epibionts of Bacillus subtilis. Microbiology 80: 535. [PubMed] [Google Scholar]
- Suzuki K., Yamauchi Y., and Yoshida T. (2017. a) Interplay between microbial trait dynamics and population dynamics revealed by the combination of laboratory experiment and computational approaches. J Theor Biol 419: 201–210. [DOI] [PubMed] [Google Scholar]
- Suzuki S., Ishii S., Hoshino T., Rietze A., Tenney A., Morrill P.L., et al. (2017. b) Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at The Cedars. ISME J 11: 2584–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrella F., and Morita M.Y. (1981) Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl Environ Microbiol 41: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp H.J. (2013) The unique metabolism of SAR11 aquatic bacteria. J Microbiol 51: 147–153. [DOI] [PubMed] [Google Scholar]
- Vancanneyt M., Schut F., Snauwaert C., Goris J., Swings J., and Gottschal J.C. (2001) Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int J Syst Evol Microbiol 51: 73–79. [DOI] [PubMed] [Google Scholar]
- Vigneron A., Cruaud P., Langlois V., Lovejoy C., Culley A.I., and Vincent W.F. (2019) Ultra-small and abundant: Candidate phyla radiation bacteria are potential catalysts of carbon transformation in a thermokarst lake ecosystem. Limnol Oceanogr Lett 5:212–220. [Google Scholar]
- Wang Y., Hammes F., Boon N., and Egli T. (2007) Quantification of the filterability of freshwater bacteria through 0.45, 0.22, and 0.1 μm pore size filters and shape-dependent enrichment of filterable bacterial communities. Environ Sci Technol 41: 7080–7086. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hammes F., Düggelin M., and Egli T. (2008) Influence of size, shape, and flexibility on bacterial passage through micropore membrane filters. Environ Sci Technol 42: 6749–6754. [DOI] [PubMed] [Google Scholar]
- Warnecke F., Amann R., and Pernthaler J. (2004) Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6: 242–253. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Komatsu N., Ishii Y., and Negishi M. (2009) Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS Microbiol Ecol 67: 57–68. [DOI] [PubMed] [Google Scholar]
- Watson S.P., Clements M.O., and Foster S.J. (1998) Characterization of the starvation-survival response of Staphylococcus aureus. J Bacteriol 180: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Jiang J., and Hu J. (2013) Determination and occurrence of retinoids in a eutrophic lake (Taihu Lake, China): cyanobacteria blooms produce teratogenic retinal. Environ Sci Technol 47: 807–814. [DOI] [PubMed] [Google Scholar]
- Wurch L., Giannone R.J., Belisle B.S., Swift C., Utturkar S., Hettich R.L., et al. (2016) Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment. Nat Commun 7: 12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Hu S., and Chen X. (2016) Artificial cells: from basic science to applications. Mater Today (Oxford, U K) 19: 516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., and Brune A. (2015). Complete genome sequence of Endomicrobium proavitum, a free-living relative of the intracellular symbionts of termite gut flagellates (phylum Elusimicrobia). Genome Announc 3: e00679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Dietrich C., Radek R., and Brune A. (2016) Endomicrobium proavitum, the first isolate of Endomicrobia class. nov. (phylum Elusimicrobia)—an ultramicrobacterium with an unusual cell cycle that fixes nitrogen with a Group IV nitrogenase. Environ Microbiol 18: 191–204. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Mai U., Pfeiffer W., Janssen S., Asnicar F., Sanders J.G., et al. (2019) Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea. Nat Commun 10: 5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart G., Crump B.C., Kamst-van Agterveld M.P., Hagen F., and Han S.-K. (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28: 141–155. [Google Scholar]