Abstract
The larvae of the wax moth Galleria mellonella and human oral keratinocytes were used to investigate the protective activity of the candidate oral probiotics Lactobacillus rhamnosus GG (LHR), Lactobacillus reuteri (LR), and Streptococcus salivarius K-12 (SS) against the periodontal pathogens Fusobacterium nucleatum (FN), Porphyromonas gingivalis (PG), and Aggregatibacter actinomycetemcomitans (AA). Probiotics were delivered to the larvae (i) concomitantly with the pathogen in the same larval pro-leg; (ii) concomitantly with the pathogen in different pro-legs, and (iii) before inoculation with the pathogen in different pro-legs. Probiotics were delivered as viable cells, cell lysates or cell supernatants to the oral keratinocytes concomitantly with the pathogen. The periodontal pathogens killed at least 50% of larvae within 24 h although PG and FN were significantly more virulent than AA in the order FN > PG > AA and were also significantly lethal to mammalian cells. The candidate probiotics, however, were not lethal to the larvae or human oral keratinocytes at doses up to 107 cells/larvae. Wax worm survival rates increased up to 60% for some probiotic/pathogen combinations compared with control larvae inoculated with pathogens only. SS was the most effective probiotic against FN challenge and LHR the least, in simultaneous administration and pre-treatment, SS and LR were generally the most protective against all pathogens (up to 60% survival). For P. gingivalis, LR > LHR > SS, and for A. actinomycetemcomitans SS > LHR and LR. Administering the candidate probiotics to human oral keratinocytes significantly decreased the toxic effects of the periodontal pathogens. In summary, the periodontal pathogens were variably lethal to G. mellonella and human oral keratinocytes and the candidate probiotics had measurable protective effects, which were greatest when administrated simultaneously with the periodontal pathogens, suggesting protective effects based on bacterial interaction, and providing a basis for mechanistic studies.
Keywords: periodontal pathogens, Galleria mellonella, probiotics, infection model, keratinocytes
Introduction
Periodontitis is a complex infectious disease associated with inflammation and the loss of periodontal attachment and bone support. It has several etiological and contributing factors such as the accumulation of biofilm and calculus (Wolf et al., 2005; Slots, 2017) and the presence of certain bacteria that have been identified as periodontal pathogens. There are several concepts of periodontal pathogenesis including the specific plaque hypothesis, which emphasizes the importance of specific bacteria (Nisha et al., 2017); the concept of keystone pathogens (Hajishengallis and Lamont, 2012) where a given bacterium exerts effects that are disproportionate to its abundance, and the polymicrobial synergy and dysbiosis model (Lamont and Hajishengallis, 2015; Nisha et al., 2017) all of which are significantly related to oral bacteria. Gram-negative anaerobic bacteria, in particular, have been implicated in the etiology of periodontitis. Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, and Porphyromonas gingivalis, as well as other bacterial species including Tannerella forsythia and Treponema denticola (Socransky et al., 1998) are considered to be the important contributors to periodontitis in humans (Slots et al., 1986; Dzink et al., 1988; Duncan et al., 1993; Sandros et al., 1994).
A. actinomycetemcomitans is reported to damage host tissue via the production of a leukotoxin (Johansson, 2011), and a cytolethal distending toxin (DiRienzo, 2014). F. nucleatum directly influences host responses and can also increase the infectivity of other pathogens via the induction of expression of the antimicrobial peptide β-defensin and pro-inflammatory cytokines in the oral epithelium (Krisanaprakornkit et al., 2000; Bhattacharyya et al., 2016; Ahn et al., 2017). P. gingivalis expresses two types of gingipains (Imamura, 2003), which are reportedly implicated in the progression of periodontal disease and have been strongly associated with the induction of inflammation and destruction of the host periodontium (Miyachi et al., 2007). Porphyromonas gingivalis has been linked to the perturbation of periodontal microbial homeostasis. Hence, this bacterium has been proposed as a keystone periodontal pathogen (Hajishengallis and Lamont, 2012).
The potential of putatively beneficial bacteria (probiotics) to prevent or treat periodontitis and other oral diseases has been investigated (Vivekananda et al., 2010; Ince et al., 2015; Bohora and Kokate, 2017; Brignardello-Petersen, 2017; Kobayashi et al., 2017). Previous investigations have demonstrated the ability of certain species of Lactobacillus to inhibit the growth of P. gingivalis and A. actinomycetemcomitans (Sookkhee et al., 2001; Kõll-Klais et al., 2005). Krasse et al. (2006) reported decreased gum bleeding and reduced gingivitis following the administration of a Lactobacillus reuteri-based candidate probiotic suggesting mechanisms including the production of bacteriocins such as reuterin, competition with oral pathogens, and anti-inflammatory activity (Krasse et al., 2006). Another study proposed that Lactobacillus rhamnosus GG could suppress bone loss in a mouse model of induced periodontitis (Gatej et al., 2018). The administration of S. salivarius K12 has been associated with reduced alveolar bone loss and resorption in a murine periodontitis model (Zhu et al., 2019).
The use of alternative animal models for studying pathogenic microorganisms has increased in recent years (Swanson and Hammer, 2000; Casadevall, 2005; Chamilos et al., 2011) for several reasons, including throughput, cost, and ethics (Ball et al., 1995). In addition to the fact that microbial virulence mechanisms may be common between different hosts (Rahme et al., 1995), larvae are simple to work with, inherently replicable, and have a short life cycle in comparison to higher animals (Tsai et al., 2016). The presence of an innate immune system in invertebrates (Boman and Hultmark, 1987; Tsai et al., 2016) is an additional advantage over the use of alternatives such as cell culture.
Galleria mellonella, the caterpillar (larva) of the greater wax moth (Lepidoptera: pyralidae) is widely used as a non-mammalian animal model system to study host-pathogen interactions using a variety of microorganisms including bacteria (Morton et al., 1983; Miyata et al., 2003; Fedhila et al., 2006; Aperis et al., 2007; Seed and Dennis, 2008) and fungi (Mylonakis et al., 2005). Use of the G. mellonella model to study the protective effects of candidate probiotics has received some research attention where for example, it has been applied in the evaluation of candidate probiotics to impair Pseudomonas aeruginosa biofilm formation (Berrios et al., 2018), to reduce virulence in Candida albicans (Vilela et al., 2015; de Oliveira et al., 2017; Rossoni et al., 2017, 2018), and to protect against gastrointestinal pathogens such as Listeria monocytogenes and Escherichia coli (Scalfaro et al., 2017). Significant correlations between observed virulence in G. mellonella and mammalian models have been previously reported (Jander et al., 2000; Mylonakis et al., 2005), and G. mellonella host immune defense mechanisms have been proposed to broadly resemble those of humans (Nathan, 2014; Kohler, 2015). Host responses of the larvae can be assessed through both the cellular response mediated by phagocytic cells and the humoral immune response pathway mediated by antimicrobial peptides (AMPs) (Kavanagh and Reeves, 2004; Scalfaro et al., 2017).
The current study aimed to study the interactions between candidate probiotics (L. rhamnosus GG, L. reuteri, and S. salivarius K12) and periodontopathogens (A. actinomycetemcomitans, F. nucleatum, and P. gingivalis) in oral keratinocytes and a lower animal infection model (G. mellonella).
Materials and Methods
Bacterial Strains and Culture Preparations
Candidate probiotics comprised Lactobacillus rhamnosus Goldin and Gorbach (GG) (ATCC 53103), Lactobacillus reuteri ATCC 55730, and Streptococcus salivarius K-12. The periodontal pathogens were Fusobacterium nucleatum ATCC 10953, Porphyromonas gingivalis ATCC 33277, and Aggregatibacter actinomycetemcomitans ATCC 33384. Aggregatibacter actinomycetemcomitans was grown in Tryptic Soya Agar and broth supplemented with 0.6% yeast extract (TSA and TSB) and incubated in a 5% CO2 atmosphere. All other bacteria were grown using Wilkins-Chalgren broth or agar (Oxoid, Basingstoke, United Kingdom) at 37°C and incubated in an anaerobic cabinet (atmosphere, 10:10:80, H2, CO2, N2). All bacteria were incubated at 37°C. For both in vitro and in vivo experiments, 10 ml volumes of each bacterium were prepared by growing cells overnight either anaerobically or in a 5% CO2 environment. A 100-fold dilution of overnight culture was made in sterile broth, which was incubated until the desired growth phase was reached according to constructed growth curves. Cells were harvested by centrifugation at 3220 × g for 15 min and the pelleted cells were washed and re-suspended in sterile phosphate buffer saline (PBS) (0.01 M PBS; NaCl 8 g/L, KCL 0.2 g/L, Na2HPO4 1.42 g/L, and KH2PO4 0.24 g/L, pH 7.4). These steps were repeated twice, and the cell suspension adjusted to a final optical density at 600 nm of 0.1. Each larva received aliquots of 5 μl of this bacterial suspension injected directly to the hemocoel. For experiments using lysates and cell-free extracts, 10 ml of 108 Colony Forming Units (CFU)/ml of the appropriate strain was centrifuged. The supernatant was reserved to use as a cell-free extract. The pellet was washed, concentrated in 1 ml of Wilkins Chalgren broth, and lysed using a bead beater (FastPrep FP120; Thermo Electron Corporation). Samples were filter sterilized to remove any whole bacteria remaining.
Galleria Infection Model
G. mellonella were obtained from Live Foods Direct, Sheffield, United Kingdom. The larvae were in the last instar stage (shedding of the exoskeleton), and were selected based on their weight (275–300 mg), the presence of a fresh cream color, and no gray markings. All larvae were used within 3 days from shipment.
Well-Diffusion Test
The test organisms were grown to the stationary phase (108 CFU/ml). Cultures of pathogenic bacteria were diluted 1:100 in Wilkins Chalgren agar. After careful mixing, 20 ml agar plates were poured and left to set. Once set, cup cuts were aseptically made within the agar (8 mm wells) and filled with 100 μl of the probiotic cell culture (∼1.5 × 108 CFU/ml) of each tested probiotic organism. The plates were incubated at 37°C anaerobically for 48 h and the diameter of the zone of inhibition produced measured using calipers.
Human Oral Keratinocyte Cell Culture
Human oral keratinocytes (HOKs, Sciencell Research Laboratories, United States) were used to assess the effect of probiotics on their viability. HOKs were maintained in oral keratinocyte medium (OKM, Sciencell Research Laboratories, United States) supplemented with oral keratinocyte growth supplement (OKGS) and 100 U/ml of both penicillin and streptomycin (OKM, Sciencell Research Laboratories, United States). The medium was substituted twice weekly and cells were incubated in a humid atmosphere with 5% CO2 at 37°C. Cells were cultured in T-25 or T-75 vented culture flasks and 24 well plates (Corning, Sigma, United States). The cells were plated, at a density of 10 × 104 cells per cm2, in 1 ml of the appropriate medium either in 12 or 24 well plates according to the experiment and used after 24 h incubation at 37°C at ∼90–100% confluence. Cells were exposed to 108 CFU/ml of each probiotic cell suspension for 24 h. Viability was determined using the trypan blue exclusion assay (Prince et al., 2012). Uninfected cells were included as a control. Probiotic lysates and cell-free extracts (100 μl) were added simultaneously with the periodontopathogen to the human oral keratinocytes.
Galleria mellonella Pathogenicity and Protection Assays
A modified version of the assay described by Ramarao et al. (2012) was performed. Larvae of G. mellonella were incubated for 30 min at room temperature before injection. Overnight cultures of each microorganism were centrifuged (3220 × g, 15 min) and suspended in PBS. This was repeated twice. Cultures were adjusted to an OD600 nm of 0.1. For intrahemocoelic injection, bacterial suspensions were prepared with final concentrations in the range of 104 CFU/ml to 108 CFU/ml. Volumes of 5 μl of each strain, cell-free extract or lysate were delivered directly to the hemocoel through an injection in the rear left pro-leg using a 26-gauge needle Hamilton microsyringe (Sigma, United Kingdom). Sterile PBS (5 μl) was injected into the “trauma” control group and additionally, a “no treatment” control group was added. The right pro-leg was used as the injection site. Different sites were used for pathogenic and probiotic strains to reduce the risk of injection site infection. Infected larvae were incubated in a petri dish in groups of 10 at 37°C in the dark for the duration of the experiment (5–7 days).
Determination of Larval Mortality
Larval mortality was determined daily over a week. Larvae that had turned black and that were not moving in response to a gentle shaking of the dish or touching with a pipette tip were considered dead. Dead larvae were removed from the petri dish and the death was recorded. The experimental endpoint was designated by either the death of all the larvae in the tested groups or the conversion of larvae into pupae. Pupae were identified via a color change to white (Jorjao et al., 2018). Five Petri dishes containing 10 larvae each were assigned to each experiment and control groups (50 larvae total for each sample). Dead G. mellonella were placed into sterile Universal bottles and homogenized in 10 ml of sterile PBS. This suspension was then serially diluted, and spot plated onto Wilkins Chalgren agar to calculate bacterial load per individual larva. The experiments were terminated once two of the control individuals had died or pupated.
Statistical Analyses
Galleria mellonella data were plotted as survival curves using the Kaplan–Meier estimator in Microsoft Excel 2010. The survival values were considered significantly different if the p-value was < 0.05. For cell culture work paired student’s T-tests done using Microsoft Excel 2010. Results were considered significant if p ≤ 0.05.
Results
In vitro Antibacterial Activity of Selected Probiotics
In vitro testing of antibacterial activity using an agar well-diffusion assay indicated that all three tested pathogens were inhibited by the three investigated probiotics to varying degrees. A. actinomycetemcomitans showed the greatest sensitivity to all probiotics with significantly larger zones of inhibition produced than for other pathogens. Streptococcus salivarius K-12 was the most effective against A. actinomycetemcomitans (Table 1). For both F. nucleatum and P. gingivalis the effect of all probiotics was comparable with P. gingivalis exhibiting slightly more sensitivity (Table 1).
TABLE 1.
Inhibition of periodontal pathogens by suspensions of candidate probiotics.
| Periodontal pathogen |
Candidate probiotic |
||
| L. rhamnosus | L. reuteri | S. salivarius K-12 | |
| P. gingivalis | 19 ± 1 | 15 ± 2 | 20 ± 2 |
| F. nucleatum | 11 ± 2 | 15 ± 1 | 20 ± 0 |
| A. actinomycetemcomitans | 33 ± 2 | 24 ± 0 | 41 ± 2 |
Determined using agar well-diffusion assays. Values are zones of inhibition (mm) and represent mean ± SEM from three separate experiments.
The Susceptibility of Human Oral Keratinocytes and G. mellonella to Periodontal Pathogens
Data in Figures 1–3 show when control HOKs were incubated for 24 h, ∼97% ± 0.2 of the cells remained viable; whereas the percentage of cells that remained viable following 24 h inoculation with the periodontopathogens was significantly (p > 0.01) lower. P. gingivalis decreased the viability of treated cell monolayers to 40% (Figure 1), A. actinomycetemcomitans decreased the viability of cell monolayers to 51% (Figure 2), and F. nucleatum decreased the viability of treated cell monolayers to 34% (Figure 3).
FIGURE 1.
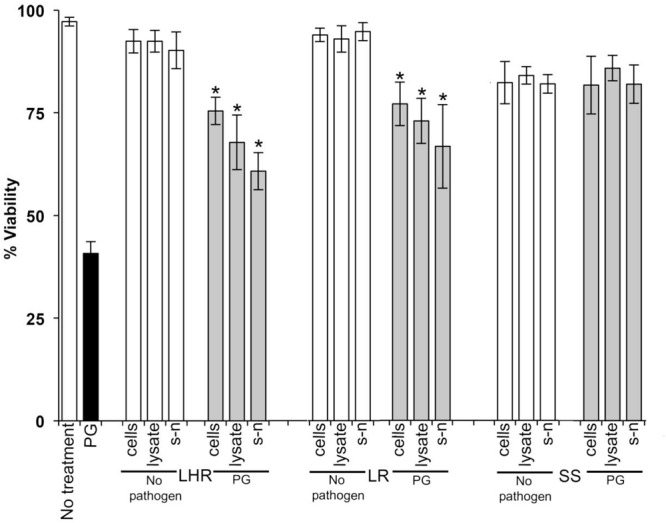
The protective effects of candidate probiotics for human oral keratinocytes (HOKs) against P. gingivalis. All probiotic cells, cell lysates, and culture supernatants protected HOKs against P. gingivalis. HOKs were inoculated with either live L. rhamnosus GG (LHR), L. reuteri (LR), or S. salivarius K-12 (SS) cells, lysates and supernatants (s-n) (white bars) or simultaneously in combination with P. gingivalis (PG) (gray bars). All experiments were performed for a minimum of three biological replicates, with three technical replicates each time. All data are shown as mean values plus/minus standard deviations. Results expressed as the mean ± SEM, *p < 0.05.
FIGURE 3.

The protective effects of candidate probiotics for human oral keratinocytes against F. nucleatum. All probiotic cells, cell lysates, and culture supernatants protect HOKs against F. nucleatum. HOKs were inoculated with either live L. rhamnosus GG (LHR), L. reuteri (LR), or S. salivarius K−12 (SS) lysates and supernatants (s-n) (white bars) or simultaneously in combination with F. nucleatum (FN) (gray bars). All experiments were performed for a minimum of three biological replicates, with three technical replicates each time. All data are shown as mean values plus/minus standard deviations. Results expressed as the mean ± SEM, *p < 0.05.
FIGURE 2.

The protective effects of candidate probiotics for human oral keratinocytes against A. actinomycetemcomitans. All probiotic cells, cell lysates, and culture supernatants protect HOKs against A. actinomycetemcomitans. HOKs were inoculated with either live L. rhamnosus GG (LHR), L. reuteri (LR), or S. salivarius K-12 (SS) cells, lysates and supernatants (s-n) (white bars) or simultaneously in combination with A. actinomycetemcomitans (AA) (gray bars). All experiments were performed for a minimum of three biological replicates, with three technical replicates each time. All data are shown as mean values plus/minus standard deviations. Results expressed as the mean ± SEM, *p < 0.05.
In the G. mellonella model (Figure 4), no mortality was observed in either control (non-treated control or PBS control). All three pathogens caused the death of at least 50% of larvae by the experimental endpoint. However, P. gingivalis and F. nucleatum caused significantly higher mortality (p < 0.05) than A. actinomycetemcomitans in the order F. nucleatum > P. gingivalis > A. actinomycetemcomitans (Figure 4). Bacterial lysates and broth culture filtrates of pathogens reduced the viability of larvae, but the mortality rate was less than with live bacteria. Lysates were more lethal than cell-free extracts. These data show that HOKs and G. mellonella are susceptible to infection with selected periodontal pathogens. By contrast, none of the candidate probiotics (L. rhamnosus GG, L. reuteri, and S. salivarius K-12) induced significant mortality in HOKs or G. mellonella (Figures 1–4).
FIGURE 4.
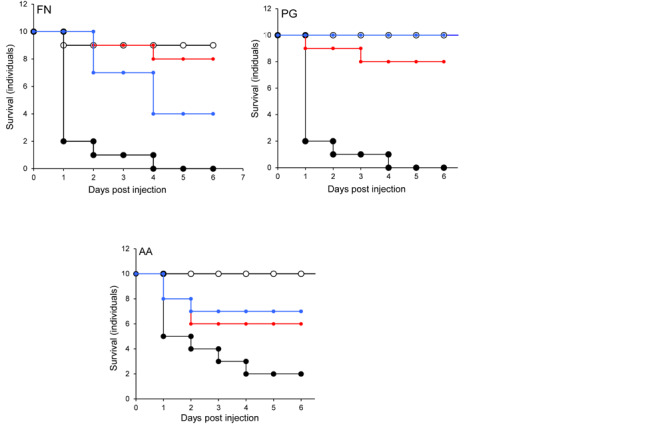
The lethal effect of periodontal pathogens in an invertebrate model. Kaplan-Meier plots of survival of G. mellonella larvae after challenge with periodontal pathogens. F. nucleatum (FN), Porphyromonas gingivalis (PG), or A. actinomycetemcomitans (AA). Viable bacterial suspension (black symbols), cell-free culture supernatant (blue symbols), bacterial lysate (red symbols), and PBS (white symbols). All experiments were done twice on 2 consecutive weeks, with different batches of larvae and with three replica plates where each plate contains 10 larvae. All three periodontal pathogens caused at least 50% larvae morality at the endpoint of the experiment. The mortality effects of F. nucleatum and P. gingivalis were significantly greater than A. actinomycetemcomitans (p < 0.05) in the order F. nucleatum > P. gingivalis > A. actinomycetemcomitans. Bacterial lysates and cell-free culture supernatants contributed to the reduction of larvae viability, but the lethal effects were lower than for viable bacteria.
Species-Dependent Protective Effects of Candidate Probiotics in G. mellonella and Human Oral Cell Lines Challenged With Periodontal Pathogens
When S. salivarius K-12 was injected with either F. nucleatum or A. actinomycetemcomitans there was a higher larval viability 24 h post-injection (p ≤ 0.01) than when G. mellonella was injected with the pathogens alone (Figure 5). However, S. salivarius K-12 did not protect larvae from the effects of P. gingivalis (Figure 5). L. reuteri afforded some protection against P. gingivalis but had a limited effect against F. nucleatum and A. actinomycetemcomitans. L. rhamnosus GG had some protective effect on G. mellonella viability when injected in a mixture with F. nucleatum and P. gingivalis but increased the mortality of larvae when injected in a mixture with A. actinomycetemcomitans (Figure 5). Probiotic protection in G. mellonella according to bacterial species was in the following order: for F. nucleatum SS > LR > LHR, for P. gingivalis LR > LHR > SS, and for A. actinomycetemcomitans, SS > LHR = LR. Administering probiotic strains, their supernatants and lysates simultaneously with F. nucleatum (Figure 3) or P. gingivalis (Figure 1) to human oral keratinocytes significantly (p < 0.05) increased the viability compared to when infected with the pathogen alone. Protection against A. actinomycetemcomitans inoculation in HOKs (Figure 2) was variable and was only statistically significant (p < 0.05) when lysates or supernatants were administered (p < 0.05).
FIGURE 5.
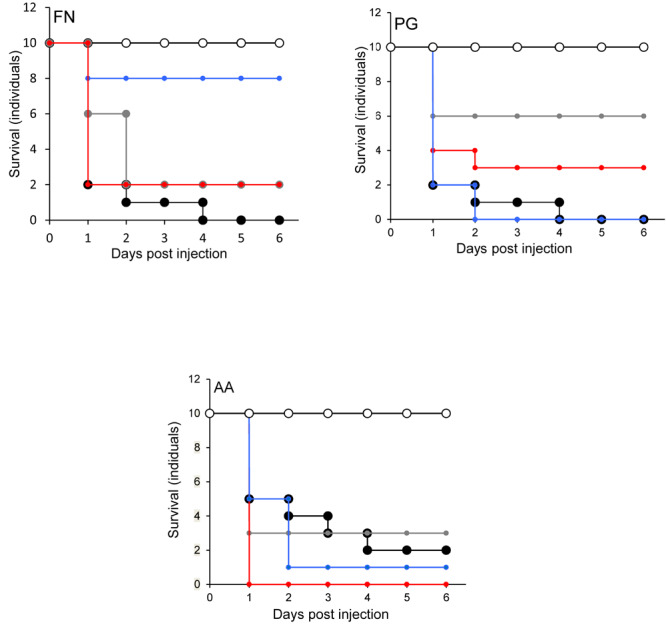
Protection of larvae conferred by candidate probiotics following simultaneous administration of periodontal pathogens into the same larval proleg. Pathogen only (black symbols) and PBS (white symbols). Pathogen administered simultaneously with L. rhamnosus (gray symbols), L. reuteri (red symbols), S. salivarius (blue symbols). All experiments were done twice on 2 consecutive weeks, with different batches of larvae and with three replica plates where each plate contains 10 larvae. S. salivarius K-12 had a protective effect (p ≤ 0.01) when administered simultaneously with F. nucleatum but not for A. actinomycetemcomitans or P. gingivalis. L. reuteri conferred protection against P. gingivalis, limited protection against F. nucleatum, and was not protective against A. actimomycetemcomitans. L. rhamnosus GG had some protective effect on larvae when was injected with F. nucleatum and P. gingivalis but reduced the larval viability when injected combined with A. actinomycetemcomitans.
Effect of Injecting of Pathogen and Probiotic Simultaneously in Different Pro-legs on G. mellonella Mortality
The effects of injecting pathogens and probiotics into separate prolegs were evaluated. This was to exclude any effects due to an inhibitory interaction between microorganisms. The viability of larvae injected with F. nucleatum was increased when injected simultaneously with all probiotics. The most effective was S. salivarius K-12 (Figure 6). For P. gingivalis, L. rhamnosus GG reduced the lethal effect of G. mellonella (Figure 6), while with the other two probiotics the viability was lower than the larvae injected with P. gingivalis alone. L. reuteri decreased conferred protection against A. actinomycetemcomitans, as did L. rhamnosus GG and S. salivarius K-12 to a lesser extent (Figure 6). Probiotic protection according to bacterial species occurred in the following order: SS > LR > LHR for F. nucleatum, LR > LHR > SS for P. gingivalis, and LR > LHR = SS for A. actinomycetemcomitans.
FIGURE 6.
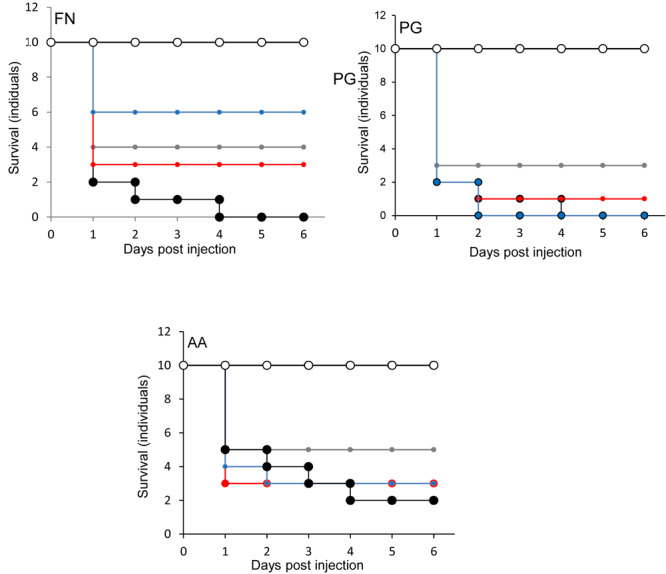
Protection of larvae conferred by candidate probiotics following simultaneous administration of periodontal pathogens into the different larval prolegs. Pathogen only (black symbols) and PBS (white symbols). Pathogen administered simultaneously with L. rhamnosus (gray symbols), L. reuteri (red symbols), or S. salivarius (blue symbols). All experiments were done twice on 2 consecutive weeks, with different batches of larvae and with three replica plates where each plate contains 10 larvae. The greatest protection against F. nucleatum was conferred by S. salivarius K-12. For P. gingivalis and A. actinomycetemcomitans. L. rhamnosus GG was the most protective bacterium.
Effect of Probiotic Pre-treatment on Survival of G. mellonella Inoculated With Periodontal Pathogens
None of the larvae were killed when F. nucleatum was injected into larvae that were pre-injected with L. rhamnosus GG or S. salivarius K-12 (Figure 7). Furthermore, there was a highly significant decrease (p < 0.001) in the mortality of larvae pre-treated with L. reuteri (Figure 7). The effects of injecting P. gingivalis on larvae that were pre-treated for 24 h with probiotics was total protection for larvae pre-treated with L. reuteri whereas the least protection was conferred by L. rhamnosus GG (Figure 7). For A. actinomycetemcomitans the best protection was conferred by S. salivarius K-12 followed by L. reuteri and L. rhamnosus GG was the least effective (Figure 7) in terms of the increase of viability of larvae compared to larvae treated by pathogen only (p < 0.01). Probiotic protection according to bacterial species was in the following order: LHR > SS > LR for F. nucleatum, LR > SS > LHR for P. gingivalis, and SS > LR > LHR for A. actinomycetemcomitans.
FIGURE 7.
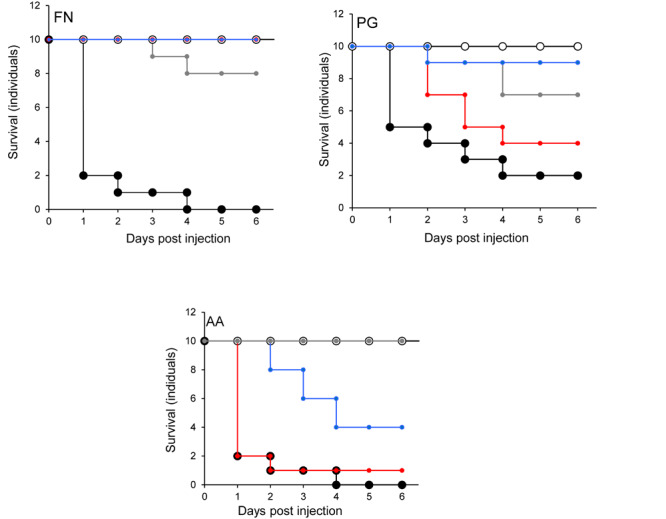
Protection of larvae conferred by prior inoculation with candidate probiotics and administration of periodontal pathogens 24 h later, into the different larval prolegs. Pathogen only (black symbols) and PBS (white symbols). L. rhamnosus (gray symbols), L. reuteri (red symbols), or S. salivarius (blue symbols). All experiments were done twice on 2 consecutive weeks, with different batches of larvae and with three replica plates where each plate contains 10 larvae. Prior-injection of larvae with L. rhamnosus GG or S. salivarius K-12 was highly protective against F. nucleatum. L. reuteri had the most effective protection against the effects of P. gingivalis, while S. salivarius afforded the greatest protective effect against A. actinomycetamcomitans.
Discussion
We have utilized a waxworm larval pathogenicity model, and human oral keratinocytes to study interactions between candidate probiotics and periodontal pathogens. A. actinomycetemcomitans, F. nucleatum, and P. gingivalis were variably lethal to the larvae and were significantly lethal in oral keratinocytes. The candidate probiotics were not significantly pathogenic in all infection models investigated. Inoculation of larvae with the candidate probiotica before pathogen challenge gave measurable but partial protection against the periodontopathogens. Protection in mammalian culture was conferred by bacterial cells, their lysates, and cell-free extracts. There are two main mechanisms by which this protection could be conferred. The first involves direct inhibition of the periodontal pathogens by the candidate probiotics. In this respect, we observed an antagonistic effect of the probiotics against the periodontopathogens in in vitro tests, where A. actinomycetemcomitans was inhibited to the greatest extent. Previous studies have demonstrated the susceptibility of this bacterium to probiosis both in vitro (Jaffar et al., 2016, 2018) and in vivo (Morales et al., 2018). However, protection of the waxworm could also be conferred by host-dependent mechanisms. The lethality of the pathogens in the G. mellonella model followed the order F. nucleatum > P. gingivalis > A. actinomycetemcomitans. F. nucleatum, and P. gingivalis demonstrated significantly greater pathogenicity than A. actinomycetemcomitans. This finding is broadly in agreement with the Socranksy complexes (Socransky et al., 1998) where P. gingivalis and F. nucleatum form part of the red and orange complexes respectively, which have been strongly associated with periodontal disease. The toxic effects of spent culture fluid and lysates were also investigated. Both were shown to affect the viability of the larvae but to a lesser extent than with inoculation with live bacteria. This could involve extra-cellular virulence factors in addition to those that require a viable cell to be present. For example, the production of sialidase in P. gingivalis (Frey et al., 2019), the induction of inflammation by F. nucleatum (Han, 2015), and the production of toxins in A. actinomycetemcomitans (Belibasakis et al., 2019).
All of the candidate probiotic strains tested conferred a degree of protection to infection. However, the extent of this protection was dependent on the species of probiotic and the pathogen under test. This is in agreement with other studies demonstrating strain and species-dependent effects of probiotics when targeted toward periodontal pathogens (Teughels et al., 2013; Montero et al., 2017). The direct protection observed could be due to competition for adhesion, acid production, production of bacteriocins, and biosurfactants (Spurbeck and Arvidson, 2010; Kohler et al., 2012; Orsi et al., 2014; Sabia et al., 2014).
The effects of simultaneously administering the probiotic strain and the pathogen to G. mellonella but in a different pro-legs were investigated to differentiate between direct competitive effects (as seen in Table 1) and immunomodulation. Data in Figure 6 suggest that there was a protective effect comparable to that observed when the probiotics were administered to the same site. This supports the hypothesis that the probiotics protect the larvae from infection via a mechanism distinct from direct competition, such as immunomodulation (Toshimitsu et al., 2017). Data in Figure 7 show the effects of administering the probiotics 24 h before pathogen inoculation. Competitive inhibition of pathogens by probiotic strains has previously been indicated as important in terms of observing a significant and relevant probiotic effect (Zhu et al., 2010; Munoz-Quezada et al., 2013). Figure 7 shows that for F. nucleatum injected 24 h after Lactobacillus rhamnosus administration, F. nucleatum injected after S. salivarius K12 and P. gingivalis injected after L. reuteri administration larval mortality was comparable to that observed in the PBS control group. This could be due to immunomodulation in G. mellonella that subsequently inhibited infection with the periodontopathogens. This observation is in agreement with a previous study that demonstrated that the experimental co-infection of G. mellonella with L. acidophilus and C. albicans reduced the number of yeast cells in the larval hemolymph and increased the survival of larvae (Ribeiro et al., 2017).
The G. mellonella larvae represent a cost-effective simple in vivo model as a preliminary investigative tool for screening the potential protective probiotic effects against periodontal and potentially other pathogens. Selected probiotic candidates showed varied probiotic activity against F. nucleatum, P. gingivalis and A. actinomycetemcomitans in both the larval model an in human oral keratinocytes, warranting further investigation of the mechanisms of interaction and applicability to human health.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
RM performed the laboratory-based analyses, contributed to data analysis, and co-wrote the manuscript. TC co-wrote and contributed to data analysis. CO’N, RL, and AM designed the study, supervised the project, performed the data analysis, and co-wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a scholarship from the Faculty of Pharmacy, The University of Tripoli through the Ministry of Higher Education of Libya to RM.
References
- Ahn S. H., Chun S., Park C., Lee J. H., Lee S. W., Lee T. H. (2017). Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection. PLoS One 12:e0188755. 10.1371/journal.pone.0188755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aperis G., Fuchs B. B., Anderson C. A., Warner J. E., Calderwood S. B., Mylonakis E. (2007). Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microb. Infect. 9 729–734. 10.1016/j.micinf.2007.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball M., Goldberg A., Fentem J., Broadhead C., Burch R., Festing M. (1995). The three rs: the way forward, the report and recommendation of ECVAM (The european center for the validation of alternative methods). Altern. Lab. Anim. 23 836–866. [Google Scholar]
- Belibasakis G. N., Maula T., Bao K., Lindholm M., Bostanci N., Oscarsson J., et al. (2019). Virulence and pathogenicity properties of Aggregatibacter actinomycetemcomitans. Pathogens 8 222. 10.3390/pathogens8040222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrios P., Fuentes J. A., Salas D., Carreno A., Aldea P., Fernandez F., et al. (2018). Inhibitory effect of biofilm-forming Lactobacillus kunkeei strains against virulent Pseudomonas aeruginosa in vitro and in honeycomb moth (Galleria mellonella) infection model. Benef. Microb. 9 257–268. 10.3920/BM2017.0048 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S., Ghosh S. K., Shokeen B., Eapan B., Lux R., Kiselar J., et al. (2016). FAD-I, a Fusobacterium nucleatum cell wall-associated diacylated lipoprotein that mediates human beta defensin 2 induction through toll-like receptor-1/2 (TLR-1/2) and TLR-2/6. Infect. Immun. 84 1446–1456. 10.1128/IAI.01311-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohora A., Kokate S. (2017). Evaluation of the role of probiotics in endodontic treatment: a preliminary study. J. Int. Soc. Prev. Commun. Dent. 7 46–51. 10.4103/2231-0762.200710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman H. G., Hultmark D. (1987). Cell-free immunity in insects. Annu. Rev. Microbiol. 41 103–126. [DOI] [PubMed] [Google Scholar]
- Brignardello-Petersen R. (2017). Probiotics as adjuvant to scaling and root planning seem to improve periodontal parameters after 3 months of treatment. J. Am. Dent. Assoc. 148:e10. 10.1016/j.adaj.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Casadevall A. (2005). Host as the variable: model hosts approach the immunological asymptote. Infect. Immun. 73 3829–3832. 10.1128/IAI.73.7.3829-3832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G., Samonis G., Kontoyiannis P. (2011). Drosophila melanogaster as a model host for the study of microbial pathogenicity and the discovery of novel antimicrobial compounds. Curr. Pharm. Design 17 1246–1253. 10.2174/138161211795703744 [DOI] [PubMed] [Google Scholar]
- de Oliveira F. E., Rossoni R. D., De Barros P. P., Begnini B. E., Junqueira J. C., Jorge A. O. C., et al. (2017). Immunomodulatory effects and anti-Candida activity of lactobacilli in macrophages and in invertebrate model of Galleria mellonella. Microb. Pathog. 110 603–611. 10.1016/j.micpath.2017.08.006 [DOI] [PubMed] [Google Scholar]
- DiRienzo J. M. (2014). Breaking the gingival epithelial barrier: role of the Aggregatibacter actinomycetemcomitans cytolethal distending toxin in oral infectious disease. Cells 3 476–499. 10.3390/cells3020476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M., Nakao S., Skobe Z., Xie H. (1993). Interactions of Porphyromonas gingivalis with epithelial cells. Infect. Immun. 61 2260–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzink J., Socransky S., Haffajee A. (1988). The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15 316–323. 10.1111/j.1600-051x.1988.tb01590.x [DOI] [PubMed] [Google Scholar]
- Fedhila S., Daou N., Lereclus D., Nielsen-Leroux C. (2006). Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62 339–355. 10.1111/j.1365-2958.2006.05362.x [DOI] [PubMed] [Google Scholar]
- Frey A. M., Satur M. J., Phansopa C., Honma K., Urbanowicz P. A., Spencer D. I. R., et al. (2019). Characterization of Porphyromonas gingivalis sialidase and disruption of its role in host-pathogen interactions. Microbiology 165 1181–1197. 10.1099/mic.0.000851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatej S. M., Marino V., Bright R., Fitzsimmons T. R., Gully N., Zilm P., et al. (2018). Probiotic Lactobacillus rhamnosus GG prevents alveolar bone loss in a mouse model of experimental periodontitis. J. Clin. Periodontol. 45 204–212. 10.1111/jcpe.12838 [DOI] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27 409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. W. (2015). Fusobacterium nucleatum: a commensal-turned pathogen. Curr. Opin. Microbiol. 23 141–147. 10.1016/j.mib.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T. (2003). The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74 111–118. 10.1902/jop.2003.74.1.111 [DOI] [PubMed] [Google Scholar]
- Ince G., Gursoy H., Ipci S. D., Cakar G., Emekli-Alturfan E., Yilmaz S. (2015). Clinical and biochemical evaluation of lozenges containing Lactobacillus reuteri as an adjunct to non-surgical periodontal therapy in chronic periodontitis. J. Periodontol. 86 746–754. 10.1902/jop.2015.140612 [DOI] [PubMed] [Google Scholar]
- Jaffar N., Ishikawa Y., Mizuno K., Okinaga T., Maeda T. (2016). Mature biofilm degradation by potential probiotics: Aggregatibacter actinomycetemcomitans versus Lactobacillus spp. PLoS One 11:e0159466. 10.1371/journal.pone.0159466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffar N., Okinaga T., Nishihara T., Maeda T. (2018). Enhanced phagocytosis of Aggregatibacter actinomycetemcomitans cells by macrophages activated by a probiotic Lactobacillus strain. J. Dairy Sci. 101 5789–5798. 10.3168/jds.2017-14355 [DOI] [PubMed] [Google Scholar]
- Jander G., Rahme L. G., Ausubel F. M. (2000). Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 182 3843–3845. 10.1128/jb.182.13.3843-3845.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. (2011). Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins 3 242–259. 10.3390/toxins3030242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjao A. L., Oliveira L. D., Scorzoni L., Figueiredo-Godoi L. M. A., Cristina A. P. M., Jorge A. O. C., et al. (2018). From moths to caterpillars: ideal conditions for Galleria mellonella rearing for in vivo microbiological studies. Virulence 9 383–389. 10.1080/21505594.2017.1397871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K., Reeves E. P. (2004). Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28 101–112. 10.1016/j.femsre.2003.09.002 [DOI] [PubMed] [Google Scholar]
- Kobayashi R., Kobayashi T., Sakai F., Hosoya T., Yamamoto M., Kurita-Ochiai T. (2017). Oral administration of Lactobacillus gasseri SBT2055 is effective in preventing Porphyromonas gingivalis-accelerated periodontal disease. Sci. Rep. 7:545. 10.1038/s41598-017-00623-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G. (2015). Probiotics research in Galleria mellonella. Virulence 6 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler G. A., Assefa S., Reid G. (2012). Probiotic interference of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 with the opportunistic fungal pathogen Candida albicans. Infect. Dis. Obstet. Gynecol. 2012:636474. 10.1155/2012/636474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõll-Klais P., Mändar R., Leibur E., Marcotte H., Hammarstrom L., Mikelsaar M. (2005). Oral Lactobacilli in chronic periodontitis and periodontal health: species composition and antimicrobial activity. Oral Microbiol. Immunol. 20 354–361. 10.1111/j.1399-302X.2005.00239.x [DOI] [PubMed] [Google Scholar]
- Krasse P., Carlsson B., Dahl C., Paulsson A., Nilsson A., Sinkiewicz G. (2006). Decreased gum bleeding and reduced gingivitis by the probiotic Lactobacillus reuteri. Swedish Dent. J. 30 55–60. [PubMed] [Google Scholar]
- Krisanaprakornkit S., Kimball J. R., Weinberg A., Darveau R. P., Bainbridge B. W., Dale B. A. (2000). Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68 2907–2915. 10.1128/iai.68.5.2907-2915.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont R. J., Hajishengallis G. (2015). Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21 172–183. 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyachi K., Ishihara K., Kimizuka R., Okuda K. (2007). Arg-gingipain A DNA vaccine prevents alveolar bone loss in mice. J. Dent. Res. 86 446–450. 10.1177/154405910708600511 [DOI] [PubMed] [Google Scholar]
- Miyata S., Casey M., Frank D. W., Ausubel F. M., Drenkard E. (2003). Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 71 2404–2413. 10.1128/iai.71.5.2404-2413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero E., Iniesta M., Rodrigo M., Marin M. J., Figuero E., Herrera D., et al. (2017). Clinical and microbiological effects of the adjunctive use of probiotics in the treatment of gingivitis: a randomized controlled clinical trial. J. Clin. Periodontol. 44 708–716. 10.1111/jcpe.12752 [DOI] [PubMed] [Google Scholar]
- Morales A., Gandolfo A., Bravo J., Carvajal P., Silva N., Godoy C., et al. (2018). Microbiological and clinical effects of probiotics and antibiotics on nonsurgical treatment of chronic periodontitis: a randomized placebo- controlled trial with 9-month follow-up. J. Appl. Oral Sci. 26:e20170075. 10.1590/1678-7757-2017-0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton D., Barnett R., Chadwick J. (1983). Structural alterations to Proteus mirabilis as a result of exposure to haemolymph from the larvae of Galleria mellonella. Microbios 39 177–185. [PubMed] [Google Scholar]
- Munoz-Quezada S., Bermudez-Brito M., Chenoll E., Genoves S., Gomez-Llorente C., Plaza-Diaz J., et al. (2013). Competitive inhibition of three novel bacteria isolated from faeces of breast milk-fed infants against selected enteropathogens. Br. J. Nutr. 109 (Suppl. 2), S63–S69. 10.1017/S0007114512005600 [DOI] [PubMed] [Google Scholar]
- Mylonakis E., Moreno R., El Khoury J. B., Idnurm A., Heitman J., Calderwood S. B., et al. (2005). Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73 3842–3850. 10.1128/IAI.73.7.3842-3850.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan S. (2014). New to Galleria mellonella: modeling an ExPEC infection. Virulence 5 371–374. 10.4161/viru.28338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisha S., Samyuktha G. S., Shashikumar P., Chandra S. (2017). Periodontal disease–Historical and contemporary hypothesis: a review. SRM J. Res. Dent. Sci. 8:121. [Google Scholar]
- Orsi C. F., Sabia C., Ardizzoni A., Colombari B., Neglia R. G., Peppoloni S., et al. (2014). Inhibitory effects of different lactobacilli on Candida albicans hyphal formation and biofilm development. J. Biol. Regul. Homeost. Agents 28 743–752. [PubMed] [Google Scholar]
- Prince T., Mcbain A. J., O’neill C. A. (2012). Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Appl. Environ. Microbiol. 78 5119–5126. 10.1128/AEM.00595-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L. G., Stevens E. J., Wolfort S. F., Shao J. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899. 10.1126/science.7604262 [DOI] [PubMed] [Google Scholar]
- Ramarao N., Nielsen-Leroux C., Lereclus D. (2012). The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. JoVE 70:e4392. 10.3791/4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro F., Barros P., Rossoni R. D., Junqueira J. C., Jorge A. O. C. (2017). Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 122 201–211. 10.1111/jam.13324 [DOI] [PubMed] [Google Scholar]
- Rossoni R. D., Dos Santos Velloso M., Figueiredo L. M. A., Martins C. P., Jorge A. O. C., Junqueira J. C. (2018). Clinical strains of Lactobacillus reduce the filamentation of Candida albicans and protect Galleria mellonella against experimental candidiasis. Folia Microbiol. 63 307–314. 10.1007/s12223-017-0569-9 [DOI] [PubMed] [Google Scholar]
- Rossoni R. D., Fuchs B. B., De Barros P. P., Velloso M. D., Jorge A. O., Junqueira J. C., et al. (2017). Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLoS One 12:e0173332. 10.1371/journal.pone.0173332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabia C., Anacarso I., Bergonzini A., Gargiulo R., Sarti M., Condo C., et al. (2014). Detection and partial characterization of a bacteriocin-like substance produced by Lactobacillus fermentum CS57 isolated from human vaginal secretions. Anaerobe 26 41–45. 10.1016/j.anaerobe.2014.01.004 [DOI] [PubMed] [Google Scholar]
- Sandros J., Papapanou P., Nannmark U., Dahlen G. (1994). Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Period. Res. 29 62–69. 10.1111/j.1600-0765.1994.tb01092.x [DOI] [PubMed] [Google Scholar]
- Scalfaro C., Iacobino A., Nardis C., Franciosa G. (2017). Galleria mellonella as an in vivo model for assessing the protective activity of probiotics against gastrointestinal bacterial pathogens. FEMS Microbiol. Lett. 364:fnx064 10.1093/femsle/fnx064 [DOI] [PubMed] [Google Scholar]
- Seed K. D., Dennis J. J. (2008). Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 76 1267–1275. 10.1128/IAI.01249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J. (2017). Periodontitis: facts, fallacies and the future. Periodontology 2000 7–23. 10.1111/prd.12221 [DOI] [PubMed] [Google Scholar]
- Slots J., Bragd L., Wikström M., Dahlén G. (1986). The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J. Clin. Periodontol. 13 570–577. 10.1111/j.1600-051x.1986.tb00849.x [DOI] [PubMed] [Google Scholar]
- Socransky S. S., Haffajee A. D., Cugini M. A., Smith C. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25 134–144. [DOI] [PubMed] [Google Scholar]
- Sookkhee S., Chulasiri M., Prachyabrued W. (2001). Lactic acid bacteria from healthy oral cavity of Thai volunteers: inhibition of oral pathogens. J. Appl. Microbiol. 90 172–179. 10.1046/j.1365-2672.2001.01229.x [DOI] [PubMed] [Google Scholar]
- Spurbeck R. R., Arvidson C. G. (2010). Lactobacillus jensenii surface-associated proteins inhibit Neisseria gonorrhoeae adherence to epithelial cells. Infect. Immun. 78 3103–3111. 10.1128/IAI.01200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M., Hammer B. (2000). Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54 567–613. 10.1146/annurev.micro.54.1.567 [DOI] [PubMed] [Google Scholar]
- Teughels W., Durukan A., Ozcelik O., Pauwels M., Quirynen M., Haytac M. C. (2013). Clinical and microbiological effects of Lactobacillus reuteri probiotics in the treatment of chronic periodontitis: a randomized placebo-controlled study. J. Clin. Periodontol. 40 1025–1035. 10.1111/jcpe.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshimitsu T., Ozaki S., Mochizuki J., Furuichi K., Asami Y. (2017). Effects of Lactobacillus plantarum strain OLL2712 culture conditions on the anti-inflammatory activities for murine immune cells and obese and type 2 diabetic mice. Appl. Environ. Microbiol. 83 e3001–e3016. 10.1128/AEM.03001-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. J., Loh J. M., Proft T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7 214–229. 10.1080/21505594.2015.1135289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela S. F., Barbosa J. O., Rossoni R. D., Santos J. D., Prata M. C., Anbinder A. L., et al. (2015). Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence 6 29–39. 10.4161/21505594.2014.981486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivekananda M. R., Vandana K. L., Bhat K. G. (2010). Effect of the probiotic Lactobacillus reuteri (Prodentis) in the management of periodontal disease: a preliminary randomized clinical trial. J. Oral Microbiol. 2:2. 10.3402/jom.v2i0.5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H. F., Rateitschak-Plüss E. M., Hassell T. M., Rateitschak K. H. (2005). Color Atlas of Periodontology. New York, NY: Thieme. [Google Scholar]
- Zhu L., Li H., Yang X., Xue L., Li X., Du J. (2019). Effects of Streptococcus salivarius K12 on experimental periodontitis and oral microbiota in mice. J. Biosci. Med. 7 95–111. [Google Scholar]
- Zhu Y., Xiao L., Shen D., Hao Y. (2010). Competition between yogurt probiotics and periodontal pathogens in vitro. Acta Odontol. Scand. 68 261–268. 10.3109/00016357.2010.492235 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.


