Summary
Most plants grow and develop by taking up nutrients from the soil while continuously under threat from foraging animals. Carnivorous plants have turned the tables by capturing and consuming nutrient-rich animal prey, enabling them to thrive in nutrient-poor soil. To better understand the evolution of botanical carnivory, we compared the draft genome of the Venus flytrap (Dionaea muscipula) with that of its aquatic sister, the waterwheel plant Aldrovanda vesiculosa, and the sundew Drosera spatulata. We identified an early whole-genome duplication in the family as source for carnivory-associated genes. Recruitment of genes to the trap from the root especially was a major mechanism in the evolution of carnivory, supported by family-specific duplications. Still, these genomes belong to the gene poorest land plants sequenced thus far, suggesting reduction of selective pressure on different processes, including non-carnivorous nutrient acquisition. Our results show how non-carnivorous plants evolved into the most skillful green hunters on the planet.
Keywords: Droseraceae, WRKY transcription factors, tissue-specific genes, gene loss, whole-genome duplications, transposon classification, neofunctionalization, jasmonate signaling
Highlights
-
•
An early whole-genome duplication is the source of carnivory-associated genes
-
•
Trap-specific genes were recruited from the roots
-
•
Expansion of specific gene families enabled fine-tuning of hunting styles
-
•
Evolution of plant carnivory was paralleled by massive gene loss
Palfalvi et al. reconstruct the evolution of plant carnivory in the Droseraceae by comparative genome analysis. A common whole-genome duplication is the source for recruitment of genes to carnivory-related functions. Different hunting styles evolve by expansions of defined gene families. The analyzed genomes have massively lost genes.
Introduction
Carnivorous plants attract, capture, and digest small-animal prey with the consequent active uptake and usage of the prey-derived nutrients [1, 2]. Despite the high level of specialization, plant carnivory has evolved several times independently in flowering plants [3, 4]. This includes the evolution of a wide range of capture organs, with trapping mechanisms frequently including rapid movements: the touch-sensitive snap traps in Dionaea muscipula (Di. muscipula) and Aldrovanda vesiculosa (A. vesiculosa), the suction traps in Utricularia species, and the snap tentacles in certain Drosera species. Following the initial catch, ongoing mechanical activation [5] by the prey and chemical compounds from the prey [6] trigger the production of jasmonate (JA), which activates the carnivore’s digestive system. Specialized glands produce and secrete lytic enzymes that partially break down the insect’s chitin-based exoskeleton and degrade most of the prey’s internal soft tissues. The released nutrients are actively taken up by the plant cells from the digestive fluid through newly synthesized and membrane-placed transporters [7].
In recent years, the genomes of several carnivorous plants were sequenced, providing new insights into various aspects of the ecology and evolution of plant carnivory. Utricularia gibba [8] and Genlisea aurea [9], two members of the Lentibulariaceae family, were found to possess the smallest genomes of any known vascular plant. Studies on Drosera capensis revealed the rapid expansion of proteases, enzymes essential for the digestion of captured prey [10]. The analysis of specific adaptations in the pitcher plant Cephalotus follicularis and closely related non-carnivorous and carnivorous plants unveiled the origins of digestive enzymes, thus highlighting distinct trajectories for the evolution of this functional complex [11].
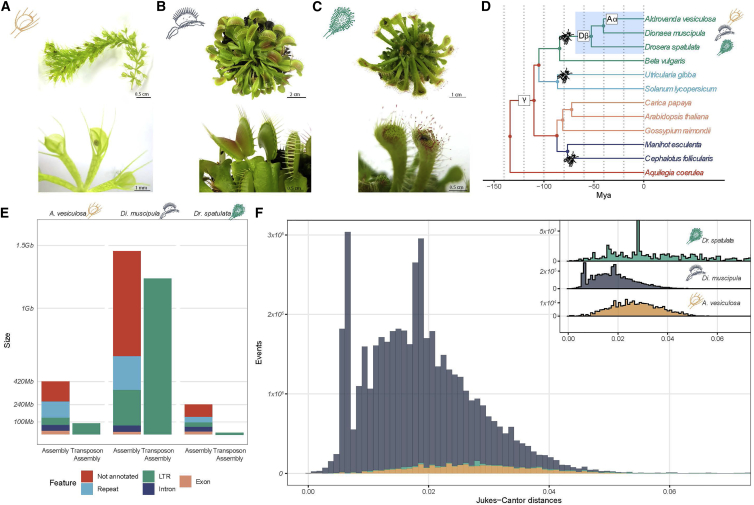
Here, we sequenced and compared the genomes of three related carnivorous species to reconstruct the evolutionary history of botanical carnivory and unravel clade- and species-specific adaptations. The analyzed species A. vesiculosa (Figure 1A), Di. muscipula (Figure 1B), and Drosera spatulata (Dr. spatulata) (Figure 1C) belong to one of the largest carnivorous families, the Droseraceae. Including members from all three genera of the Droseraceae enabled us to reconstruct early events in the emergence of carnivory.
Figure 1.
Genome Evolution
(A–C) Whole plants and traps of (A) A. vesiculosa, (B) Di. Muscipula, and (C) Dr. spatulata.
(D) Phylogenetic relationships of the three carnivorous Droseracaea and nine other species used in the study (Beta vulgaris, Utricularia gibba [carnivorous], Solanum lycopersicum, Carica papaya, Arabidopsis thaliana, Gossypium raimondii, Manihot esculenta, Cephalotus follicularis [carnivorous], and Aquilegia coerulea). Identified whole-genome duplications at the base of the Eudicots (γ), the base of the Droseraceae (Dβ), and in A. vesiculosa (Aα) as well as the independent emergences of carnivorous traits (fly) are indicated.
(E) Content of the genome and the transposon only assemblies.
(F) Age distribution of LTRs (long terminal repeats) indicated by number of substitutions as identified in transposon only assemblies. Upper right corner shows their relative distribution.
See also Figure S1 and Tables S1–S3.
Results and Discussion
A Whole-Genome Duplication Precedes Speciation
Di. muscipula lines that are commercially available for horticulture and for standardized physiological and molecular studies are produced by vegetative propagation over years to several decades. As this reduced selective pressure could affect genome metrics, we, in addition to cultured lines, collected leaf samples from native Di. muscipula in North Carolina and determined their genome size by flow cytometry (STAR Methods; Table S1). The genome size of both cultured and wild Di. muscipula is 3.18 Gbp and thus comparable in size with the human genome. In contrast, the genome sizes obtained for A. vesiculosa and Dr. spatulata are 509 Mbp (Table S1) and 323 Mbp [12], respectively. We sequenced (Data S1A), assembled, and annotated the genomes of these three species by integrating short- and long-read sequencing technologies (Table 1; STAR Methods). To unravel the evolutionary mechanisms leading to the large difference in genome size, we first searched the draft genome sequences for signs of whole-genome duplications (WGDs). The age distribution of paralogous genes revealed distinct peaks indicative of WGDs (STAR Methods; Figure S1). According to those data, a first WGD occurred at the base of the carnivorous Droseraceae family [13] and is thus shared by all three species considered here. Supported by copy numbers of syntenic sequences, A. vesiculosa appears to have undergone an additional genome triplication (Figure 1D; Table S2). This more recent event agrees with the increased fraction of duplicated genes compared to the other two carnivorous species (10.3% in A. vesiculosa but 3.1% in Dr. spatulata and Di. muscipula; Table 1). Despite its large genome size, no signs of additional WGD were identified in Di. muscipula.
Table 1.
Assembly and Annotation Statistics
| Genome Size (Mbp) | Assembly Size (Mbp) | No. Contigs | Longest Contig (Mbp) | N50 (Kbp) | Completeness (BUSCO) | |
|---|---|---|---|---|---|---|
| A. vesiculosa | 509 | 420 | 2.408 | 3.4 | 314 | C: 86.9% [S: 76.6%, D: 10.3%] |
| Di. muscipula | 3.187 | 1.500 | 104.847 | 1 | 35 | C: 83.6% [S: 80.5%, D: 3.1%] |
| Dr. spatulata | 293 | 238 | 1.061 | 3.4 | 705 | C: 86.0% [S: 82.9%, D: 3.1%] |
| No. of Predicted Genes | Genes with Interpro Annotation | Average Intron Length (bp) | Average Exon Length (bp) | Completeness (BUSCO) | ||
| A. vesiculosa | 25.123 | 24.450 | 401 | 224 | C: 84.3% [S: 73.1%, D: 11.2%] | |
| Di. muscipula | 21.135 | 19.873 | 634 | 229 | C: 76.2% [S: 72.5%, D: 3.7%] | |
| Dr. spatulata | 18.111 | 17.645 | 400 | 222 | C: 83.6% [S: 80.1%, D: 3.5%] | |
Transposons Massively Bloated the Venus Flytrap Genome
Repetitive sequences, such as long transposable elements (LTRs), can make up a large part of plant genomes [14] but typically are poorly represented in draft assemblies [15]. As the draft assembly we obtained for Di. muscipula is, with 1.5 Gbp, substantially smaller than the size of 3.18 Gbp determined by flow cytometry, we used a genome-assembly-independent method for the characterization of the repeat landscapes of the three genomes [16] (Table S3). This method identifies reads originating from repetitive elements and assembles them de novo. According to this analysis, 1.236 Gbp of the Di. muscipula genome (38.78% of the measured genome size) consists of LTRs—largely exceeding the observations for A. vesiculosa (89 Mbp; 17.5%) and Dr. spatulata (17 Mbp; 5.7%; Figure 1E). Furthermore, the LTRs of Di. muscipula are highly self-similar (Figure 1F; STAR Methods), indicating that they arose in a recent expansion. For genes of Di. muscipula, the mean intron length is 1.5 times larger than for A. vesiculosa and Dr. spatulata (Table S1). Unlike the low amount of LTRs, Dr. spatulata has the highest count of tandem gene duplications observed (Figure S2). The presence of leucin-rich-repeat (LRR) and IQ domains in these genes suggests their role in the perception of prey via chemical cues. Taken together, the genomes of the three Droseraceae species were shaped by a common WGD followed by an additional whole-genome triplication in A. vesiculosa, extensive tandem gene duplications in Dr. spatulata, and a recent explosion of LTRs in Di. muscipula.
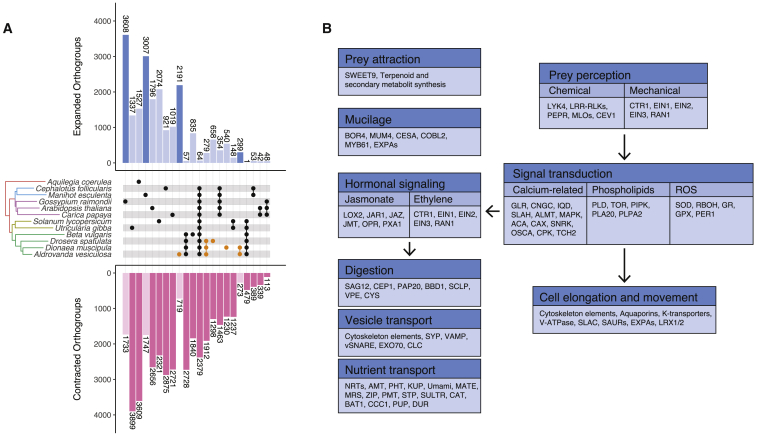
Evolution of Carnivory Is Associated with Massive Gene Loss
Despite the evolution of a new and complex trait, carnivory, the three Droseraceae considered here belong to the gene-poorest vascular plants sequenced to date (21,135 genes in Di. muscipula, 25,123 in A. vesiculosa, and 18,111 in Dr. spatulata; Figure S3). Still, we identified thirty orthogroups specific for the three Droseraceae, i.e., they are not shared with any of the nine outgroup species, including two carnivorous and seven non-carnivorous plants (Figure S4). Based on Gene Ontology (GO) terms, this gene set was significantly enriched in carboxypeptidases, hydrolases, and endopeptidase inhibitors, all of which can be directly associated with prey digestions (Data S1B). Additionally, genes involved in the regulation of transcription (GO: 0003700 and GO: 0000987) were enriched. To search for gene families that had significantly changed size in the carnivorous Droseraceae, we generated a birth-death-innovation model (STAR Methods) integrating the three carnivorous Droseraceae with two carnivorous and seven non-carnivorous angiosperm species (Figure 2A). In congruence with the observed overall reduction in gene content, 1,912 groups were contracted in the Droseraceae. These included genes involved in kinetochore formation [17, 18] (Figure S5), which have previously been shown to be associated with the occurrence of holocentric chromosomes [19, 20, 21]. Further losses affected the ubiquitin (UBQ) gene family often involved in stress responses (Figure S5) and genes related to root development (Figure S5). The last one is prominent in A. vesiculosa, where we could not identify key regulators of root development, such as WOX5, RHD6, LBD1, ANRs, and CASPs (Figure S5). This can be a consequence of the fact that its radicle is arrested early after germination and the lack of any root system in the adult plant [22].
Figure 2.
Gene Family Expansion and Contraction
(A) Numbers of expanded (blue) and contracted (pink) orthogroups shared among different lineages.
(B) Functional annotation of the 279 expanded gene families common to all three Droseraceae and their potential association with plant carnivory. Arrows indicate chronological order in the hunting cycle.
See also Figures S4 and S5.
Carnivory Is Associated with the Expansion of Distinct Gene Families
Contrasting the general trend of gene loss, we identified 279 expanded orthogroups (Figure 2A). Intriguingly, these are enriched in functions directly associated with the different steps of the hunting cycle—prey attraction, perception, digestion, and nutrient absorption (Figure 2B; Data S1C). The high number of enzymes involved in terpenoid and other secondary metabolite synthesis, as well as some sugar transporters, hint at their role in the attraction of prey. Expanded membrane receptor and signal transducer families appear to be adaptations related to the perception of prey, whereas peptidases, nucleases, and other hydrolases can be used for prey digestion. Finally, numerous transporters for nitrogen, phosphates, sulfate, amino acids, oligopeptides, sugars, and metallic ions were expanded, likely building the core of the nutrient uptake system. These are accompanied by expanded gene families related to vesicle transport, which were previously shown to play a role in nutrient uptake [23] as well as in digestive fluid release [24]. On the plant inner regulatory systems, GO terms related to JA metabolism and JA signaling were enriched. Because the expansion of the underlying gene families precedes the split of the three carnivores, our results suggest that this process likely evolved already in a common ancestor of the Droseraceae. Lastly, gene families that control structural features of the traps, namely the mucilage production and the ability to detect prey-induced trap movements, were over-represented. Although auxin was reported to be accumulated locally in Drosera traps following insect capture [25], further investigations have challenged the role of auxin in capture and leaf movement [26]. Interestingly, we found several small auxin upregulated RNA (SAUR) genes and related families, including V-ATPases, aquaporins, and expansins, are expanded in Droseraceae. Together, this supports the notion that gene duplications contributed to all four steps of plant carnivory (Figure 2B). At the base of the snap-trap-bearing species, we found 184 GO terms expanded (Data S1D). This includes leaf formation (GO:0010338), which suggests their contribution to the more complex leaf-trap morphology in this group. Further enriched terms cover “cell wall biogenesis” and “cell wall modification.” Indeed, the cell wall is crucial for the trap’s snapping mechanism. Additionally, we found terms possibly related to gland function (“fatty acid beta oxidation” and “Golgi vesicle transport”) as well as digestion (“metalloendopeptidase activity” and “cysteine-type endopeptidase inhibitors”).
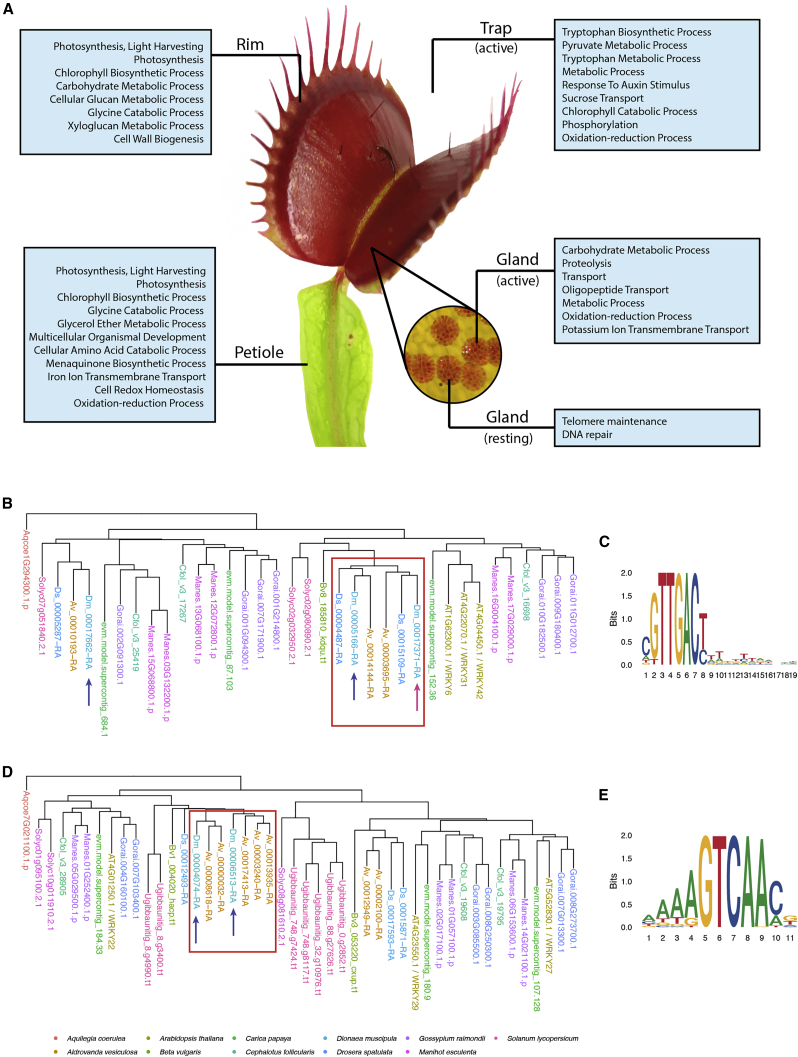
Recruitment of Genes to Carnivory
To understand the evolutionary origin of the genes involved in carnivory in the Droseraceae, we integrated the genomic data of Di. muscipula with the transcriptomes of ten different tissues, including resting and prey-processing traps and their glands [27]. For 15,964 predicted genes, we found an FPKM (fragments per kilobase of exon model per million reads mapped) >1 in at least one tissue. We classified the tissue specificity of each gene over all tissues and conditions using a Shannon entropy-derived method [28] (STAR Methods). This allowed us to functionally characterize different parts of the trap and their associated genes (Figure 3; Data S1M). We extracted all Arabidopsis thaliana orthologs of genes specific for activated glands in Di. muscipula. Surprisingly, these are significantly enriched in root-associated terms (Data S1E). This situation strongly suggests that genes used for prey-derived nutrient absorption in Di. muscipula were recruited from the root, the organ engaged with soil nutrient exploration and absorption in non-carnivorous plants.
Figure 3.
Function and Recruitment of Trap-Specific Genes in Di. muscipula
(A) Enriched biological function Gene Ontology terms from the tissue-specific expression list.
(B and D) Phylogenetic trees of WRKY transcription factor orthogroups predicted as potential regulators of carnivorous functions in Di. muscipula traps. (B) WRKY6 and (D) WRKY29.
(C and E) Enriched TF-binding motifs found in the upstream region of trap-specific genes, corresponding to the WRKY orthogroups. (C) WRKY6 and (E) WRKY29.
See also Table S4.
In Di. muscipula, the rim of the trap secretes volatiles to attract prey [29, 30]. Orthologs of rim-specific genes can be found in the nectaries of non-carnivorous plants [31]. This includes the sugar transporter SWEET9, which rewards pollinating insects [32] in floral nectaries, as well as genes responsible for volatile and secondary metabolite synthesis. Interestingly, this set of genes overlaps with the set of expanded gene families described in the previous section, further highlighting the importance of recruitment and expansion of genes with originally non-carnivorous functions to carnivory-related processes (Table S9).
Specific WRKY Transcription Factors Regulate Carnivory Genes
In the roots of non-carnivorous plants, nutrient transporters are constitutively expressed. In their glands, however, the expression of transporters is switched on only after nutrient-rich prey has been caught [27, 33]. This raises the more general questions whether and, if so, how carnivory-related genes came under the control of gland-specific promoters. To this end, we analyzed transcription factor binding motifs in the upstream regions of gland-associated genes in Di. muscipula (STAR Methods). In addition to insect-based stimulation, we also used coronatine (COR), a molecular analogon of JA-Ile, to mimic the presence of prey in the trap. COR induces the secretion of digestive enzymes and the expression of nutrient transporters in the glands [27]. We found that the upstream regions of genes activated upon COR stimulation or insect capture are enriched specifically in WRKY6 and WRKY29 binding sites, respectively (Figures 3B–3E; Table S4). Consistently, genes orthologous to WRKY6 and WRKY29 transcription factors are specifically expressed in the activated traps (Data S1G and S1H). Furthermore, the genomes of A. vesiculosa and Dr. spatulata also code for orthologs of these transcription factors (Data S1I). Phylogenetic reconstructions revealed a duplication common to all three Droseraceae for WRKY6 (Figure 3B) and one specific for the snap-trap plants for WRKY29 (Figure 3D). This might hint to a sub-functionalization following the duplication event. In non-carnivorous plants, orthologs of WRKY6 and WRKY29 are involved in responses to biotic and abiotic stresses [34, 35], including pathogens [36, 37] and herbivore attack [38]. This supports the notion that botanical carnivory originated from plant defense mechanisms [27, 39]. Taken together, our findings suggest that these transcription factors (TFs) could play a role in gland-specific, prey-induced gene expression. If this is the case, they could be of importance for the recruitment of genes to carnivory-related functions and thereby for the evolution of plant carnivory.
Parallel Evolution of Carnivory in Droseracaea and Nepenthaceae
The closest relative to Droseraceae is a clade containing the fully carnivorous Nepenthaceae and Drosophyllaceae, the partly carnivorous Dioncophyllaceae, and the non-carnivorous Ancistrocladaceae families. Members of this clade hunt with passive pitfall traps or mucilaginous glandular traps. Furthermore, stalked glands evolved independently in Droseraceae, Drosophyllaceae, and Dioncophyllaceae [40]. Together with the fundamentally different hunting strategies, this raises the question of whether carnivory evolved in the last common ancestor of these clades or independently in both. To address this, we compared the Droseraceae genomes with transcriptomic data for Nepenthes alata [41] and close non-carnivorous relatives in Caryophyllales and Arabidopsis thaliana. Orthogroups unique for all carnivorous species did not reveal functions related to plant carnivory. Furthermore, gene duplications before the split of Droseraceae and Nepenthaceae are enriched in 39 GO terms (Data S1J). Of those, only lipases and potassium transporters had a possible association with plant carnivory. By contrast, gene duplications at the base of the Droseraceae are enriched in carnivory-associated terms, including JA signaling, transport, and leaf formation (Data S1K). Thus, although first adaptations to plant carnivory might have evolved before the split of Droseraceae and Nepenthaceae [42], the present data suggest that the sophisticated mechanisms of carnivory evolved independently in these sister clades. Similar to the Droseraceae, the Nepenthaceae also underwent a WGD at the base of their clade [43]. It would be interesting to see whether this genomic event also was the basis for the evolution of plant carnivory in this sister clade.
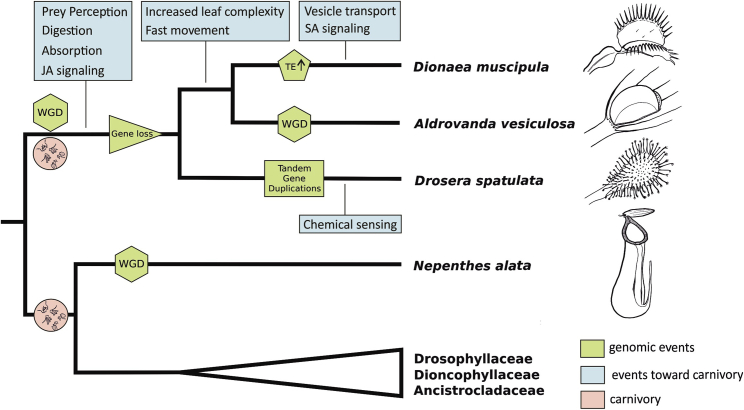
Conclusions
In summary, our study of the three carnivorous sister species Di. muscipula, A. vesiculosa, and Dr. spatulata suggests a three-step scenario in the evolution of plant carnivory in the Droseraceae (Figure 4). First, a whole-genome duplication in their last common ancestor provided gene material for diversification into carnivorous functions. This included the expression of ancestral root genes in leaf-derived traps hand in hand with a co-option for carnivory-specific processes. Second, the use of a new nutrient source reduced the selective pressure on genes involved in non-carnivorous nutrition. This led to massive gene losses, resulting in three of the gene-poorest plant genomes sequenced thus far. Finally, different species-specific mechanisms enabled the emergence of clade-specific, independent hunting styles. Obviously, these three steps can overlap and might not have happened independently.
Figure 4.
Reconstruction of Key Steps in the Evolution of Plant Carnivory in the Droseraceae
Green boxes refer to gained features for carnivory; blue boxes indicate events related to genome evolution. The red circle with a decomposed fly refers to the possible emergence points of carnivory.
See also Figures S2 and S3.
The genetic material underlying plant carnivory is present in most non-carnivorous plants, and whole-genome duplications happened frequently throughout the plant kingdom. Thus, the path to carnivory could have been open to most plants. To the relief of the animal kingdom, only a select few have evolved along this route and became green hunters.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Dionaea muscipula tissue | Own greenhouse or axenic culture | N/A |
| Aldrovanda vesiculosa | Own greenhouse or axenic culture | N/A |
| Drosera spatulata | Own greenhouse or axenic culture | N/A |
| Critical Commercial Assays | ||
| Genomic-tip 20/G | QIAGEN | 10223 |
| innuPREP Plant DNA Kit | Alalytik Jena | 845-KS-1060250 |
| Fruit-mate | TaKaRa Clontech | 9192 |
| NucleoSpin RNA Plant Kit | Macherey & Nagel | 740949.50 |
| PureLink Plant RNA Reagent | ThermoFisher | 12322012 |
| RNeasy Plant Mini Kit | QIAGEN | 74904 |
| Deposited Data | ||
| Dionaea muscipula sequence data | N/A | PRJEB35195 |
| Aldrovanda vesiculosa sequence data | N/A | PRJEB35196 |
| Drosera spatulata sequence data | N/A | PRJDB9009 |
| Experimental Models: Organisms/Strains | ||
| Dionaea muscipula | Own greenhouse or axenic culture | N/A |
| Aldrovanda vesiculosa | Own greenhouse or axenic culture | N/A |
| Drosera spatulata | Own greenhouse or axenic culture | N/A |
| Software and Algorithms | ||
| Canu v1.5 | [44] | https://github.com/marbl/canu/releases/tag/v1.5 |
| Bowtie2 v2.3.1 | [45] | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Pilon v1.22 | [46] | https://github.com/broadinstitute/pilon |
| ALLPATHS | [47] | http://software.broadinstitute.org/allpaths-lg/blog/ |
| Redundans | [48] | https://github.com/lpryszcz/redundans |
| PBJelly | [49] | https://sourceforge.net/p/pb-jelly/wiki/Home/ |
| BUSCO | [50] | https://busco.ezlab.org/ |
| RepeatMasker | [51] | http://www.repeatmasker.org |
| RepeatModeller | [52] | http://www.repeatmasker.org |
| Reper | [16] | https://github.com/nterhoeven/reper |
| Maker | [53] | https://yandell-lab.org/software/maker.html |
| Augustus | [54] | http://bioinf.uni-greifswald.de/augustus/ |
| STAR | [55] | https://github.com/alexdobin/STAR |
| Cufflinks | [56] | http://cole-trapnell-lab.github.io/cufflinks/ |
| InterProScan | [57] | https://www.ebi.ac.uk/interpro/download/ |
| Orthofinder | [58] | https://github.com/davidemms/OrthoFinder |
| MCScanX | [59] | http://chibba.pgml.uga.edu/mcscan2/ |
| Meme-suite | [60] | http://meme-suite.org/ |
| BadiRate | [61] | https://github.com/fgvieira/badirate |
| R 3.5.1 | N/A | https://www.R-project.org/ |
| Other | ||
| Assemblies, predicted transcripts and proteins | N/A | http://www.carnivorom.org/resources |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Jörg Schultz (Joerg.Schultz@uni-wuerzburg.de).
This study did not generate new unique reagents.
Experimental Model and Subject Details
Plant growth conditions
A. vesiculosa: Plants were axenically grown in a 16h light (40-55 μMol ∗ m2 ∗ s-1) /8h dark regime at 24°C/16°C. The medium was composed as follows: KNO3 - 5 mM; CaCl2 x 2 H2O - 500 μM; MgSO4 x 7 H2O - 500 μM; NaH2PO4 - 500 μM; (NH4)2SO4 - 500 μM; H3BO3- 100 μM; MnSO4 x 1 H2O - 100 μM; ZnSO4 x 7 H2O - 60 μM; KJ - 4 μM; Na2MoO4 x 2 H2O - 1,2 μM; CuSO4 x 5 H2O - 100 nM; CoCl2 x 6 H2O - 100 nM; Peptone - 250 mg/l; Inosit - 550 nM; Glycin - 27 nM; Nicotinic acid - 4 nM; Thiamin-HCl - 0,3 nM; Pyridoxin-HCl - 3 nM; FeNaEDTA - 100 nM; Sucrose - 20 g/l. pH was adjusted to 5.8.
Di. muscipula: Callus tissue was grown under sterile conditions on 1/3 MS Medium supplemented with 3% sucrose, 1% Agar, pH 5.6-5.8. 3 mg/ml 6-Benzylamonopurine and 0,5μg/ml 2,4-D (synthetic auxin) were added. Calli were grown at 23°C in the dark.
Dr. spatulata: Plants were grown in half strength MS medium supplemented with 30 g/L sucrose and Gamborg’s vitamins (pH 5.8) under continuous light and 25C.
Method Details
Flow cytometric determination of nuclear DNA contents
Absolute nuclear DNA contents (pg/2C) were determined on nuclei isolated from very young leaves/traps (about 1-1.5 cm long) of Dionaea muscipula plants grown in the greenhouse and shoot tips of in vitro grown Aldrovanda vesiculosa. Therefore, this plant material was co-chopped using a razor blade in 400 μL of the nuclei isolation buffer (Kit: CyStain PI Absolute P, Sysmex Deutschland GmbH, Norderstedt, Germany) with 0.5 cm2 young leaf tissue of the respective references chosen as internal standards, i.e., Pisum sativum cultivar ‘Viktoria, Kifejto Borso’ (9.07 pg/2C, genebank IPK Gatersleben acc. no. PIS 630) for D. muscipula and Solanum lycopersicum cultivar ‘Stupicke Polni Rane’ (1.96 pg/2C, genebank IPK Gatersleben acc. no. LYC 418). After 2 min, the suspension was filtered through 30 μm mesh (Celltric filters, Sysmex) and the filtrate was mixed with 1600 μL of the staining solution of the kit (CyStain PI Absolute P, Sysmex) containing propidium iodide, RNase and supplemented with 1% polyvinylpyrrolidone-10. After an incubation of at least 1 h in dark conditions and on ice, flow cytometric measurements were carried out using a CyFlow Ploidy Analyzer (Sysmex Partec GmbH, Münster, Germany). For each genotype, five measurements of independent samples were taken at different days, recording a minimum number of 632 nuclei in the main peak of the species of interest, with an average number of 2130 nuclei. Peaks were automatically detected by the flow cytometer software and nuclear DNA contents of the samples (pg/2C) were calculated by dividing the sample mean G0/G1 peak position by the reference mean G0/G1 peak position and multiplication by the reference DNA content (pg/2C).
Sequencing
Genomic
A. vesiculosa: Genomic DNA was isolated from axenically grown Aldrovanda vesiculosa plants using a modified CTAB/Chloroform-Isoamylalcohol–based protocol. In brief, 1 g of frozen plant material was powdered in liquid nitrogen and thawed in 19ml of extraction buffer (2% CTAB, 2% PVP/MW 10,000, 100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA) and 120 mg PVPP. After incubation at 63°C for 1 h, cell debris was removed by centrifugation (30 min, RT, 3,200 x g). 2.5 mg RNase were added to the supernatant and incubated at 37°C for 1h. Following addition of 50 μl Proteinase K (20 mg/ml) and 45min incubation at 37°C, the sample was extracted with 1 volume of chloroform/isoamyl alcohol (24:1, v/v). Phases were separated by centrifugation (15 min, RT, 2700xg) and the DNA was precipitated from the aequeous phase by addition of 0.6 volumes of Isopropanol. After 1 h of incubation on ice, the gDNA was precipitated by centrifugation (30min, RT, 3,200 x g). After two washing steps with 70% ethanol, gDNA was dried at 37°C for 15 min, resuspended in TE-buffer and stored for subsequent use at 4°C. Quantity and quality of the resulting DNA were determined by capillary electrophoresis (Experion, Bio-Rad Laboratories), fluorometrically (Qubit, Thermo Fisher Scientific) or spectrophotometrically (NanoDrop 2000, Thermo Fisher Scientific).
Accession number: PRJEB35196
Dr. spatulata: Genomic DNA was isolated from shoots of aseptically grown plants of the “common” strain of diploid Dr. spatulata. Collected shoots were homogenized in liquid nitrogen using a mortar and a pestle. The homogenate was transferred into 2x CTAB extraction buffer (2% cetyltrimethylammonium bromide [CTAB], 1.4 M NaCl, 100 mM Tris-HCl [pH 8.0], 20 mM EDTA [pH 8.0], 1% PVP40) preheated to 80°C and supplied with 1/1,000 volume of 14.1 M β-mercaptoethanol, and was gently agitated at 65°C for 1 h. An equal volume of chloroform: isoamyl alcohol (25:1) was added, and agitated using a rotator at 30 rpm for 10 min at room temperature. After centrifugation at 10,000 rpm for 30 min at room temperature, supernatants were transferred to new tubes, and supplemented with 1/10 volume of 10% CTAB and an equal amount of chloroform:isoamyl alcohol (25:1). The tubes were shaken with a rotator at 30 rpm for 10 min. After centrifugation at 10,000 rpm for 30 min, supernatants were again transferred to new tubes and an equal volume of isopropanol was added. The tubes were centrifuged at 10,000 rpm for 30 min, and supernatants were discarded. The crude DNA pellet was rinsed twice with 5 mL of 70% EtOH and air-dried for 5 min. The pellet was dissolved in 100 μL of TE (pH 8.0) containing 0.1-1 μL 1 μg/μL RNase A, and gently agitated for 60 min at 37°C. A 1/20 volume of 20 mg/ml Proteinase K was added, and tubes were incubated at 56°C for 30 min. Subsequently, the DNA solution was further purified using Genomic-tip (QIAGEN) according to the manufacturer’s instruction. DNA concentration was determined using fluorometer Qubit 2.0 (Thermo Scientific).
Accession number: PRJDB9009
Di. muscipula: Genomic DNA from Dionaea muscipula was extracted from 500 mg of sterile callus tissue using the innuPREP Plant DNA Kit (analytikjena) according to the manufacturerś instructions. Quantity and quality of the resulting DNA were determined by capillary electrophoresis (Experion, Bio-Rad Laboratories), fluorometrically (Qubit, Thermo Fisher Scientific) or spectrophotometrically (NanoDrop 2000, Thermo Fisher Scientific).
Accession number: PRJEB35195
Transcriptomic
A. vesiculosa
Plant material of Aldrovanda vesiculosa was kindly provided by Lubomir Adamec from the collection in the Institute of Botany of the Czech Academy of Sciences at Trebon, Czech Republic, and cultivated in the Botanic Gardens Wuerzburg in a water tank [62].
Mature, healthy & non-flowering Aldrovanda vesiculosa plants were frozen in liquid nitrogen and ground to a fine powder. To remove polysaccharides and polyphenols, 200 mg of frozen plant powder were resuspended in 350μl Fruit-mate (TaKaRa Clontech). After cell debris was removed by centrifugation (10 min, 4°C, 11,000 x g), the supernatant was subjected to RNA purification using the NucleoSpin RNA Plant Kit (Macherey & Nagel) according to the manufacturerś instructions. Quantity and quality of the resulting RNA were determined by capillary electrophoresis (Experion, Bio-Rad Laboratories), fluorometrically (Qubit, Thermo Fisher Scientific) or spectrophotometrically (NanoDrop 2000, Thermo Fisher Scientific). One TruSeq RNA library was generated, and RNA sequencing was performed using an Illumina MiSeq sequencer at LGC Genomics.
Accession number: ERX3632913
Dr. spatulata
Total RNA was isolated from whole plant, mature leaves, and roots of aseptically grown plants, and inflorescences of plants grown on peat pots in three biological replicates. Plant materials were put in a 2 mL tube with a ϕ5 mm zirconia bead, frozen in liquid nitrogen, and ground using TissueLyser (QIAGEN) at 25 Hz for 2 min. Then, total RNA was extracted using PureLink Plant RNA Reagent (Thermo Scientific) according to the manufacture’s protocol. Subsequently, the DNA solution was further purified using RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s instruction. Total RNA quality was analyzed using 2100 Bioanalyzer (Agilent). Total RNA concentration was determined using Qubit and NanoDrop (Thermo Fisher Scientific).
Sequencing libraries for whole transcriptome analysis were produced from total RNA of Dr. spatulata using TruSeq Stranded mRNA HT Sample Prep Kit (Illumina) as the manufacturer’s protocol. Sequencing was performed with HiSeq1500 (Illumina) at the conditions of HO mode, v4 reagent, and paired-end sequencing of 2x126 bp.
Accession numbers: PRJDB9009
Assembly Strategies
The PacBio reads of Dr. spatulata and A. vesiculosa were assembled with canu [44] (version 1.5, default parameter settings, except for genomeSize). Following, Illumina reads were mapped to the assembly [45] (bowtie2 version 2.3.1; default parameter settings) and an error correction was performed with pilon [46] (version 1.22; default parameter).
For Di. muscipula, an assembly was generated using ALLPATHS [47] with a minimum contig size of 200 bp and haploidification enabled. As overlapping paired-end libraries, digitally normalized reads obtained through an improved normalization run with bbnorm.sh were used. For scaffolding, the mate-pair libraries Dm_GenIl_[001-018] plus the paired-end libraries with insert sizes of 500 bp and 800 bp (Dm_GenIl_[027-032]) as well as artificial mate-pair generated from corrected PacBio reads (dm-pb2il-[1235]000-01, dm-pb2il-15000-01) were supplied. Two additional post-processing steps were performed to further improve the assembly. First, Redundans [48] was used to identify and merge redundant scaffolds that resulted from the assembly of different heterozygous regions into separate contigs. Second, PBJelly [49] was applied to further increase the contiguity of the assembly by scaffolding and extending the available sequences and filling N containing gaps.
The completeness of all three assemblies was evaluated using BUSCO [50] version 3.0.1.
Assemblies and predicted genes are accessible at http://www.carnivorom.org/resources
Annotation
Repetitive elements
All assemblies were annotated with RepeatMasker [51] and species-specific repeat libraries generated with RepeatModeler [52]. Complementary, repetitive elements were annotated in an assembly free approach based on Illumina sequences using reper [16].
TE Age estimation
As result, reper reports cluster with one or many assembled retrotransposon sequences. For each cluster, the distance of each sequence to an exemplar (the longest sequence) is calculated according to the Jukes and Cantor model [35] as dAB = −3/4 ln(1 – 4/3 fAB) where dAB is the evolutionary distance between the sequences A and B and fAB is the fraction of observed differences between A and B. Additionally, read coverage for each sequence as well as average Jukes Cantor distance of all reads compared to the assembled sequence was considered. The number of occurrences of each sequence in the genome was calculated as: q = (nread /coverage) ∗ (lread /laseq) with the read-count (nread), genomic coverage, read-length (lread) and assembled-sequence-length (laseq). Finally, every distance between an exemplar and an individual sequence was counted once and the distance between an individual sequence and its reads (q − 1) times.
Gene Prediction
An iterative gene prediction was performed with Maker [53]. As evidence, all plant proteins in the swissprot database [57] (downloaded on 6. Dec 2017), predicted proteins of Amaranth (Amaranthus hypocondriacus) [63] and Quinoa (Chenopodium quinoa) [64] and an augustus model [54] of A. thaliana was used. Additionally, transcriptomic data for each plant were used. Therefore, transcriptomic reads were aligned to the respective genome assembly [55] and assembled [56]. The first iteration of maker was run using the assembled transcriptome as gff file, the A. thaliana augustus model, as well as the plant proteins mentioned above as evidence. The resulting annotation was then used to train the snap HMMs. These HMMs were then used as evidence for the second maker iteration. Then, a second snap training was conducted using the genes from this maker run. Finally, a third maker run was started using the second snap results.
Functional annotation
Domains and GeneOntology annotations were predicted with Interproscan Version 5.25-64.0 [65]. GO enrichments were calculated in R 3.5.1 environment using hypergeometric tests and Benjamini-Hochberg correction for multiple testing.
A. thaliana orthologs of Di. muscipula genes were used for Plant Ontology (PO)-term enrichment with “parent-child-union” and Benjamini-Hochberg correction for multiple testing [66].
Orthology prediction
Orthology prediction for the 12 species Eudicot and for the 11 species Caryophyllales dataset, was performed using Orthofinder 2.2.6 [58] with msa option using mafft aligner version 7.158b [67].
Species tree reconstruction
The Species tree was reconstructed using STRIDE in the Orthofinder suite [68]. Time estimation was performed with r8s setting the split of Aquilegia coerulea and Eudicots to 122-134 Mya and the Rosid and (Asterid-Caryophyllales) split to 110-124 Mya based on TimeTree estimates [69].
Genome Duplication
Fourfold degenerative transversion (4dTv) rates were calculated by aligning all orthologous or paralogous gene pairs using DECIPHER’s codon alignment suit (http://www2.decipher.codes) and then calculating 4dTv distances using the seqinr 3.1-5 R package (http://seqinr.r-forge.r-project.org). To assess the depth of duplications and investigate tandemly duplicated genes, we analyzed the 3 genomes using MCScanX [59].
TF binding site prediction
To identify candidate TF factor binding site in the tissue specific genes, sequences 950 bases upstream to 50 bases into the gene were extracted. JASPAR CORE [70] redundant database was used as reference for plant transcription factor motifs [33]. Enrichment of motifs was calculated by “Analysis of Motif Enrichment” (AME) [60]of the meme-suite ver. 5.0.1. Control sequences for a set of sequences specific to a single tissue were composed of the entirety of all tissue specific gene sequences except for the tissue in question. FIMO (“Find Individual Motif Occurrences”) [71], also found in the meme suite ver. 5.0.1, was used to scan tissue specific sequences for motifs of transcription factors identified by AME.
Quantification and Statistical Analysis
Birth-Death Innovation Model
BadiRate branch models
| model name | setup | replicates | parameters |
|---|---|---|---|
| gr | Global Rate | 3 | 2 |
| fr | Free rate | 3 | 44 |
| ar | Asteraceae + Caryophyllales + Background | 3 | 6 |
| db | Drosera + Background | 3 | 4 |
| vb | Dionaea + Background | 3 | 4 |
| ab | Aldrovanda + Background | 3 | 4 |
| av | Snap trap + Background | 3 | 4 |
Using the 12-species Eudicots orthogroups we calculated a Birth-Death-Innovation model for the family turnover rate using BadiRate [61] as described in [11]. We selected seven different branch models (Table 1). Analysis and results were calculated using the badirater R package (https://palfalvi.github.io/badirater/), which was developed as an R interface for large-scale studies using BadiRate. Branch models were considered significant if a weighted Akaike information criterion (wAIC) ratio was larger than 2.7.
Tissue Specificity
To identify tissue specific genes the Shannon entropy for every gene in every tissue was calculated. Therefore, all transcriptomic reads were mapped onto the reference genome using STAR [55]. Next, expression was quantified using Cuffquant [56] and normalized with Cuffnorm resulting in FPKM values for each gene and sample. For each tissue, the mean was taken over all samples. Following, R was used to calculate the Shannon Entropy [28]. First, the relative expression (pt|g) of each gene (g) in each tissue (t) based on the FPKM-value wg,t was calculated as pt|g = wg,t / Σwg,t. Next, the entropy of the gene’s expression distribution (Hg) was computed as Hg = Σ −pt|g∗log2(pt|g). Finally, the tissue specificity (Qg|t) could be derived as Qg|t = Hg − log2(pt|g). Here, a small value for Q-value indicates tissue specificity of gene g for tissue t. The Q-value cut-off to determine the 1 and 5% most specific genes was calculated by means of probability density: Q-values for each gene in each tissue were collectively pooled for calculation of the probability density curve. A Laplace-distribution was fitted to the probability density and the Q-value where the area under the curve accounted for 1% or 5% of the data respectively, was then used as a threshold for specificity across each tissue individually.
Data and Code Availability
All sequence information has been uploaded to the Short Read Archive (SRA). Genomic data: A. vesiculosa - PRJEB35196; Dr. spatulata - PRJDB9009; Di. muscipula - PRJEB35195. Transcriptomic data: A. vesiculosa - ERX3632913; Dr. spatulata - PRJDB9009.
Acknowledgments
This work was supported by the European Research Council (ERC) under the EU 7th Framework Program (FP/20010- 2015)/ERC grant agreement 250194 Carnivorom to R.H., by a DFG-funded Reinhart Koselleck project (HE 1640/42-1; project number 415282803) to R.H., by a JSPS KAKENHI grant (22128008 to T.N. and 22128001, 22128002, 16H06378, and 17H06390 to M.H.), and by a Researchers Supporting Project (NSRSP-2019), King Saud University, Riyadh, Saudi Arabia to K.A.-R. and R.H. C.K. was supported by the RA program of National Institute for Basic Biology. Dr. spatulata cultivation, genome sequence, and computer analyses were partly supported by MPRF-NIBB, DIAF-NIBB, and ROIS National Institute of Genetics. The ORCIDs for the authors are as follows: https://orcid.org/0000-0002-0838-7700 (J.S.), https://orcid.org/0000-0001-7425-8758 (M.H.), and https://orcid.org/0000-0003-3224-1362 (R.H.).
Author Contributions
Y.H. and K.U. provided aseptic culture of Dr. spatulata. C.K. and K.F. maintained and collected Dr. spatulata samples and extracted DNA. T.F.S., T.N., S.S., and Y.T. performed genome sequencing of Dr. spatulata. L.A. cultured and provided A. vesiculosa plants. I.K. maintained aseptic cultures and extracted DNA for A. vesiculosa and Di. muscipula. T.W. established and provided aseptic cultures and determined genome size of Di. muscipula and A. vesiculosa. J.F. determined Di. muscipula genome size. G.P., T.F.S., T.N., and S.S. assembled and annotated Dr. spatulata genome. T.H., F.F., and M.A. developed and T.H. implemented the assembly strategy for the Di. muscipula genome. N.T. assembled A. vesiculosa genome, and G.P. and N.T. annotated all three species. G.P. identified WGDs, analyzed syntenic regions, identified and analyzed expanded and contracted gene families, designed figures, and performed comparison with N. alata. N.T. developed strategy for LTR identification, D.B. analyzed expanded protein families, F.S. performed tissue-specific gene analysis and performed orthogroup analyses, and M.F. analyzed composition and age of LTRs, performed TF binding site analysis, and analyzed A. thaliana orthologs of carnivory-specific genes. A.I. analyzed genome data and designed figures, I.S. analyzed genome data, and R.S. performed membrane protein classification. J.S. designed and directed computational analyses. G.P., J.S., M.H., R.H., and K.A.-R. wrote the paper with input from all authors. M.H. and R.H. devised the project. J.S., M.H., and R.H. are representatives of each group. G.P. and T.H. should be considered joint first authors.
Declaration of Interests
The authors declare no competing interests.
Published: May 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.04.051.
Contributor Information
Jörg Schultz, Email: joerg.schultz@uni-wuerzburg.de.
Mitsuyasu Hasebe, Email: mhasebe@nibb.ac.jp.
Rainer Hedrich, Email: hedrich@botanik.uni-wuerzburg.de.
Supplemental Information
A) Read statistics Illumina sequencing B) Read statistics PacBio sequencing C) GO Enrichment for the 30 carnivory specific orthogroups D) GO enrichment Droseraceae expanded orthogroups E) GO enrichment snap-trap expanded orthogroups (Di. muscipula and A. vesiculosa) F) Plant Ontology Enrichments G) GO enrichment for tissue specific genes H) Di. muscipula tissue specific genes (5%) I) Di. muscipula tissue specific genes (1%) J) Orthogroups of the 3 studied and 9 selected species K) GO enrichment for orthogroups expanded before the split of Droseraceae and Nepenthaceae L) GO enrichment of orthogroups expanded at the base of the Droseraceae, from Caryophyllales analysis M) Membrane protein classification N) Di. muscipula digestome
References
- 1.Charles Darwin . Second Edition. John Murray; 1875. Insectivorous Plants. [Google Scholar]
- 2.Givnish T.J., Burkhardt E.L., Happel R.E., Weintraub J.D. Carnivory in the bromeliad Brocchinia reducta, with a cost/benefit model for the general restriction of carnivorous plants to sunny, moist, nutrient-poor habitats. Am. Nat. 1984;124:479–497. [Google Scholar]
- 3.Albert V.A., Williams S.E., Chase M.W. Carnivorous plants: phylogeny and structural evolution. Science. 1992;257:1491–1495. doi: 10.1126/science.1523408. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann A., Schlauer J., Smith S.A., Givnish T.J. Evolution of carnivory in angiosperms. In: Ellis A.M., Adamec L., editors. Carnivorous Plants: Physiology, Ecology, and Evolution. Oxford University; 2018. pp. 22–42. [Google Scholar]
- 5.Böhm J., Scherzer S., Krol E., Kreuzer I., von Meyer K., Lorey C., Mueller T.D., Shabala L., Monte I., Solano R. The venus flytrap dionaea muscipula counts prey-induced action potentials to induce sodium uptake. Curr. Biol. 2016;26:286–295. doi: 10.1016/j.cub.2015.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mithöfer A., Reichelt M., Nakamura Y. Wound and insect-induced jasmonate accumulation in carnivorous Drosera capensis: two sides of the same coin. Plant Biol (Stuttg) 2014;16:982–987. doi: 10.1111/plb.12148. [DOI] [PubMed] [Google Scholar]
- 7.Hedrich R., Neher E. Venus flytrap: how an excitable, carnivorous plant works. Trends Plant Sci. 2018;23:220–234. doi: 10.1016/j.tplants.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Ibarra-Laclette E., Lyons E., Hernández-Guzmán G., Pérez-Torres C.A., Carretero-Paulet L., Chang T.H., Lan T., Welch A.J., Juárez M.J.A., Simpson J. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leushkin E.V., Sutormin R.A., Nabieva E.R., Penin A.A., Kondrashov A.S., Logacheva M.D. The miniature genome of a carnivorous plant Genlisea aurea contains a low number of genes and short non-coding sequences. BMC Genomics. 2013;14:476. doi: 10.1186/1471-2164-14-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butts C.T., Bierma J.C., Martin R.W. Novel proteases from the genome of the carnivorous plant Drosera capensis: structural prediction and comparative analysis. Proteins. 2016;84:1517–1533. doi: 10.1002/prot.25095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushima K., Fang X., Alvarez-Ponce D., Cai H., Carretero-Paulet L., Chen C., Chang T.H., Farr K.M., Fujita T., Hiwatashi Y. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat. Ecol. Evol. 2017;1:59. doi: 10.1038/s41559-016-0059. [DOI] [PubMed] [Google Scholar]
- 12.Hoshi Y., Azumatani M., Suyama C., Adamec L. Determination of ploidy level and nuclear DNA content in the Droseraceae by flow cytometry. Cytologia (Tokyo) 2017;82:321–327. [Google Scholar]
- 13.Yang Y., Moore M.J., Brockington S.F., Mikenas J., Olivieri J., Walker J.F., Smith S.A. Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytol. 2018;217:855–870. doi: 10.1111/nph.14812. [DOI] [PubMed] [Google Scholar]
- 14.Feschotte C., Jiang N., Wessler S.R. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 15.Tørresen O.K., Star B., Mier P., Andrade-Navarro M.A., Bateman A., Jarnot P., Gruca A., Grynberg M., Kajava A.V., Promponas V.J. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 2019;47:10994–11006. doi: 10.1093/nar/gkz841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terhoeven N., Schultz J., Hackl T. reper: Genome-wide identification, classification and quantification of repetitive elements without an assembled genome. J. Open Source Softw. 2018;3:527. [Google Scholar]
- 17.Sato H., Shibata F., Murata M. Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res. 2005;13:827–834. doi: 10.1007/s10577-005-1016-3. [DOI] [PubMed] [Google Scholar]
- 18.Komaki S., Schnittger A. The spindle assembly checkpoint in Arabidopsis is rapidly shut off during severe stress. Dev. Cell. 2017;43:172–185.e5. doi: 10.1016/j.devcel.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Kondo K., Nontachaiyapoom S. An evidence on diffused centromeres in Drosera chromosomes provided by scanning electron microscopy. Chromosom. Bot. 2008;3:79–81. [Google Scholar]
- 20.Kolodin P., Cempírková H., Bureš P., Horová L., Veleba A., Francová J., Adamec L., Zedek F. Holocentric chromosomes may be an apomorphy of Droseraceae. Plant Syst. Evol. 2018;304:1289–1296. [Google Scholar]
- 21.Drinnenberg I.A., deYoung D., Henikoff S., Malik H.S. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife. 2014;3:e03676. doi: 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross A. Redfern Natural History Productions; 2012. Aldrovanda: The Waterwheel Plant. [Google Scholar]
- 23.Adlassnig W., Koller-Peroutka M., Bauer S., Koshkin E., Lendl T., Lichtscheidl I.K. Endocytotic uptake of nutrients in carnivorous plants. Plant J. 2012;71:303–313. doi: 10.1111/j.1365-313X.2012.04997.x. [DOI] [PubMed] [Google Scholar]
- 24.Scherzer S., Shabala L., Hedrich B., Fromm J., Bauer H., Munz E., Jakob P., Al-Rascheid K.A.S., Kreuzer I., Becker D. Insect haptoelectrical stimulation of Venus flytrap triggers exocytosis in gland cells. Proc. Natl. Acad. Sci. USA. 2017;114:4822–4827. doi: 10.1073/pnas.1701860114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bopp M., Weiler E. Leaf blade movement of Drosera and auxin distribution. Naturwissenschaften. 1985;72:434. [Google Scholar]
- 26.Nakamura Y., Reichelt M., Mayer V.E., Mithöfer A. Jasmonates trigger prey-induced formation of ‘outer stomach’ in carnivorous sundew plants. Proc. Biol. Sci. 2013;280:20130228. doi: 10.1098/rspb.2013.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bemm F., Becker D., Larisch C., Kreuzer I., Escalante-Perez M., Schulze W.X., Ankenbrand M., Van de Weyer A.L., Krol E., Al-Rasheid K.A. Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Res. 2016;26:812–825. doi: 10.1101/gr.202200.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schug J., Schuller W.P., Kappen C., Salbaum J.M., Bucan M., Stoeckert C.J., Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreuzwieser J., Scheerer U., Kruse J., Burzlaff T., Honsel A., Alfarraj S., Georgiev P., Schnitzler J.P., Ghirardo A., Kreuzer I. The Venus flytrap attracts insects by the release of volatile organic compounds. J. Exp. Bot. 2014;65:755–766. doi: 10.1093/jxb/ert455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jürgens A., El-Sayed A.M., Suckling D.M. Do carnivorous plants use volatiles for attracting prey insects? Funct. Ecol. 2009;23:875–887. [Google Scholar]
- 31.Escalante-Pérez M., Jaborsky M., Reinders J., Kurzai O., Hedrich R., Ache P. Poplar extrafloral nectar is protected against plant and human pathogenic fungus. Mol. Plant. 2012;5:1157–1159. doi: 10.1093/mp/sss072. [DOI] [PubMed] [Google Scholar]
- 32.Lin I.W., Sosso D., Chen L.Q., Gase K., Kim S.G., Kessler D., Klinkenberg P.M., Gorder M.K., Hou B.H., Qu X.Q. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature. 2014;508:546–549. doi: 10.1038/nature13082. [DOI] [PubMed] [Google Scholar]
- 33.Scherzer S., Krol E., Kreuzer I., Kruse J., Karl F., von Rüden M., Escalante-Perez M., Müller T., Rennenberg H., Al-Rasheid K.A.S. The Dionaea muscipula ammonium channel DmAMT1 provides NH4+ uptake associated with Venus flytrap’s prey digestion. Curr. Biol. 2013;23:1649–1657. doi: 10.1016/j.cub.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 34.Hsu F.-C., Chou M.-Y., Chou S.-J., Li Y.-R., Peng H.-P., Shih M.-C. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell. 2013;25:2699–2713. doi: 10.1105/tpc.113.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y.-F., Li L.-Q., Xu Q., Kong Y.-H., Wang H., Wu W.-H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21:3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 37.Robatzek S., Somssich I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–1149. doi: 10.1101/gad.222702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skibbe M., Qu N., Galis I., Baldwin I.T. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell. 2008;20:1984–2000. doi: 10.1105/tpc.108.058594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukushima K., Fang X., Alvarez-Ponce D., Cai H., Carretero-Paulet L., Chen C., Chang T.H., Farr K.M., Fujita T., Hiwatashi Y. Genome of the pitcher plant Cephalotus reveals genetic changes associated with carnivory. Nat. Ecol. Evol. 2017;1:59. doi: 10.1038/s41559-016-0059. [DOI] [PubMed] [Google Scholar]
- 40.Renner T., Specht C.D. A sticky situation: assessing adaptations for plant carnivory in the Caryophyllales by means of stochastic character mapping. Int. J. Plant Sci. 2011;172:889–901. [Google Scholar]
- 41.Leebens-Mack J.H., Barker M.S., Carpenter E.J., Deyholos M.K., Gitzendanner M.A., Graham S.W., Grosse I., Li Z., Melkonian M., Mirarab S., One Thousand Plant Transcriptomes Initiative One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heubl G., Bringmann G., Meimberg H. Molecular phylogeny and character evolution of carnivorous plant families in Caryophyllales--revisited. Plant Biol (Stuttg) 2006;8:821–830. doi: 10.1055/s-2006-924460. [DOI] [PubMed] [Google Scholar]
- 43.Smith S.A., Brown J.W., Yang Y., Bruenn R., Drummond C.P., Brockington S.F., Walker J.F., Last N., Douglas N.A., Moore M.J. Disparity, diversity, and duplications in the Caryophyllales. New Phytol. 2018;217:836–854. doi: 10.1111/nph.14772. [DOI] [PubMed] [Google Scholar]
- 44.Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., Earl A.M. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gnerre S., Maccallum I., Przybylski D., Ribeiro F.J., Burton J.N., Walker B.J., Sharpe T., Hall G., Shea T.P., Sykes S. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pryszcz L.P., Gabaldón T. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 2016;44:e113. doi: 10.1093/nar/gkw294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.English A.C., Richards S., Han Y., Wang M., Vee V., Qu J., Qin X., Muzny D.M., Reid J.G., Worley K.C., Gibbs R.A. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 51.Smit, A., Hubley, R., and Green, P. (2015). RepeatMasker Open-3.0. http://www.repeatmasker.org.
- 52.Smit, A., and Hubley, R. (2013-2015). RepeatMasker Open-4.0. http://www.repeatmasker.org.
- 53.Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanke M., Schöffmann O., Morgenstern B., Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLeay R.C., Bailey T.L. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Librado P., Vieira F.G., Rozas J. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics. 2012;28:279–281. doi: 10.1093/bioinformatics/btr623. [DOI] [PubMed] [Google Scholar]
- 62.Adamec L. How to grow Aldrovanda vesiculosa outdoors. Carniv. Plant Newsl. 1997;26:85–88. [Google Scholar]
- 63.Clouse J.W., Adhikary D., Page J.T., Ramaraj T., Deyholos M.K., Udall J.A., Fairbanks D.J., Jellen E.N., Maughan P.J. The amaranth genome: genome, transcriptome, and physical map assembly. Plant Genome. 2016;9:1–14. doi: 10.3835/plantgenome2015.07.0062. [DOI] [PubMed] [Google Scholar]
- 64.Zou C., Chen A., Xiao L., Muller H.M., Ache P., Haberer G., Zhang M., Jia W., Deng P., Huang R. A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value. Cell Res. 2017;27:1327–1340. doi: 10.1038/cr.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shameer K., Naika M.B., Mathew O.K., Sowdhamini R. POEAS: automated plant phenomic analysis using plant ontology. Bioinform. Biol. Insights. 2014;8:209–214. doi: 10.4137/BBI.S19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emms D.M., Kelly S. STRIDE: species tree root inference from gene duplication events. Mol. Biol. Evol. 2017;34:3267–3278. doi: 10.1093/molbev/msx259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 70.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R., Bessy A., Chèneby J., Kulkarni S.R., Tan G. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Read statistics Illumina sequencing B) Read statistics PacBio sequencing C) GO Enrichment for the 30 carnivory specific orthogroups D) GO enrichment Droseraceae expanded orthogroups E) GO enrichment snap-trap expanded orthogroups (Di. muscipula and A. vesiculosa) F) Plant Ontology Enrichments G) GO enrichment for tissue specific genes H) Di. muscipula tissue specific genes (5%) I) Di. muscipula tissue specific genes (1%) J) Orthogroups of the 3 studied and 9 selected species K) GO enrichment for orthogroups expanded before the split of Droseraceae and Nepenthaceae L) GO enrichment of orthogroups expanded at the base of the Droseraceae, from Caryophyllales analysis M) Membrane protein classification N) Di. muscipula digestome
Data Availability Statement
All sequence information has been uploaded to the Short Read Archive (SRA). Genomic data: A. vesiculosa - PRJEB35196; Dr. spatulata - PRJDB9009; Di. muscipula - PRJEB35195. Transcriptomic data: A. vesiculosa - ERX3632913; Dr. spatulata - PRJDB9009.