Abstract
Coronavirus disease 2019 (COVID-19) initiated in December 2019 in Wuhan, China and became pandemic causing high fatality and disrupted normal life calling world almost to a halt. Causative agent is a novel coronavirus called Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2/2019-nCoV). While new line of drug/vaccine development has been initiated world-wide, in the current scenario of high infected numbers, severity of the disease and high morbidity, repurposing of the existing drugs is heavily explored. Here, we used a homology-based structural model of transmembrane protease serine 2 (TMPRSS2), a cell surface receptor, required for entry of virus to the target host cell. Using the strengths of molecular docking and molecular dynamics simulations, we examined the binding potential of Withaferin-A (Wi-A), Withanone (Wi-N) and caffeic acid phenethyl ester to TPMRSS2 in comparison to its known inhibitor, Camostat mesylate. We found that both Wi-A and Wi-N could bind and stably interact at the catalytic site of TMPRSS2. Wi-N showed stronger interactions with TMPRSS2 catalytic residues than Wi-A and was also able to induce changes in its allosteric site. Furthermore, we investigated the effect of Wi-N on TMPRSS2 expression in MCF7 cells and found remarkable downregulation of TMPRSS2 mRNA in treated cells predicting dual action of Wi-N to block SARS-CoV-2 entry into the host cells. Since the natural compounds are easily available/affordable, they may even offer a timely therapeutic/preventive value for the management of SARS-CoV-2 pandemic. We also report that Wi-A/Wi-N content varies in different parts of Ashwagandha and warrants careful attention for their use.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, Ashwagandha, Withanone, Withaferin-A, honey bee, propolis, caffeic acid phenethyl ester, molecular docking, binding, transmembrane protease serine 2 (TMPRSS2), inhibition
1. Introduction
Coronaviruses (family Coronaviridae; order Nidovirales) are large, enveloped, positive-stranded RNA viruses. Their genome, largest among all RNA viruses, is packed inside a helical capsid constituted of nucleocapsid protein (N), surrounded by an envelope comprised of three structural proteins; (i) the membrane protein-M, (ii) the envelope protein-E and (iii) the spike protein-S. Whereas M and E proteins are involved in virus assembly, the S protein (forms large protrusions from the virus surface, giving it an appearance of crown) mediates virus entry into target host cells and serves as a critical determinant of viral host range (Boopathi et al., 2020; Li, 2016). Some coronaviruses also encode an envelope-associated hemagglutinin-esterase protein (HA). Among the four Coronaviruses genera (α, β, γ and δ), α and β coronaviruses infect mammals, γ infect avian species and δ infect both mammalian and avian species, and pose serious health threats (Li, 2016). Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) when transmitted from animals to humans, caused severe respiratory diseases (SARS and MERS in 2002 and 2012 with fatality rates of ∼10% and 36%, respectively). Porcine epidemic diarrhea coronavirus caused ∼100% fatality rate in piglets in 2013 in USA. A novel coronavirus, SARS-coronavirus 2 (SARS-CoV-2), closely related to SARS-CoV, has recently been causing infectious respiratory disease, coronavirus disease 19 (COVID-19). Originated and rapidly spreading (by human to human transmission) from Wuhan city of China to all over the world (Huang et al., 2020; Wang et al., 2020; Zhu et al., 2020), SARS-CoV-2 has infected 5,061,478 people with around 331,475 deaths as of 23 May 2020, and raised global health emergency (WHO Coronavirus Disease (COVID-19) Dashboard, 2020). There is an urgent need to understand the biology of the new coronavirus and develop prophylactic and therapeutic drugs.
Coronavirus infection is initiated by binding of viral surface glycoprotein (spike protein-S) to target host cell membrane receptor ACE-2 (Hasan et al., 2020). ACE-2 expression has also been reported to determine the severity of SARS-CoV-2 infection (Nejadi Babadaei, Hasan, Haj Bloukh et al., 2020). The viral protein is synthesized as a precursor protein. Its cleavage by host cell proteases including transmembrane protease serine 2 (TMPRSS2), endosomal cathepsin L protease or human airway trypsin-like protease (HAT) is an essential step for virus fusion to the host cell membrane, infectivity and propagation (Böttcher-Friebertshäuser et al., 2010; Yamaya et al., 2015). Based on these information, protease inhibitors are considered as drugs for treatment/prevention of viral infections. Indeed, overexpression of TMPRSS2 and HAT was shown to promote the growth of different subtypes of human and avian influenza viruses (Böttcher et al., 2009; Shen et al., 2017). On the other hand, serine protease inhibitors abrogated the virus proliferation (Böttcher et al., 2009; Böttcher-Friebertshäuser et al., 2011). Peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) that sterically blocked TMPRSS2 transcription caused significant suppression of viral titers (Böttcher-Friebertshäuser et al., 2011). Benzylsulfonyl-d-arginine-proline-4-amidinobenzylamide, a potent inhibitor of HAT and TMPRSS2 was also shown to cause strong suppression of virus propagation (Böttcher-Friebertshäuser et al., 2012, 2013). Camostat mesylate (an inhibitor of serine protease, clinically used to treat chronic pancreatitis) has also been suggested as a candidate antiviral drug to prevent or suppress TMPRSS2-dependent infection by SARS-CoV (Kawase et al., 2012). Cells constitutively expressing TMPRSS2 showed higher susceptibility and larger syncytia when infected with MERS-CoV. Camostat mesylate although completely inhibits the syncytium formation, could only partially block virus entry into cells. The simultaneous treatment with inhibitors of cathepsin L and TMPRSS2 was shown to completely block virus entry into Vero-TMPRSS2 cells (Shirato et al., 2013). Cysteine protease inhibitor K11777 (used for the treatment of tropical parasitic disease, American trypanosomiasis) and its closely related vinylsulfones have been shown to target cathepsin-mediated cell entry and act as broad-spectrum antivirals. It was shown to inhibit SARS-CoV and Ebola virus entry into the host cells. Activity of K11777, its derivative and Camostat mesylate were compared in pathogenic animal model of SARS-CoV infection. It was demonstrated that the viral pathogenesis of SARS-CoV is dependent on serine rather than cysteine proteases and hence Camostat mesylate was recommended as a potential therapeutic intervention for respiratory coronavirus infections (Yamaya et al., 2015; Zhou et al., 2015). TMPRSS2 was recently proven to be crucial for hemagglutinin cleavage of some human influenza viruses. Since the catalytic sites of the diverse serine proteases linked to influenza, parainfluenza and coronavirus activation are structurally similar, active site inhibitors of these airway proteases were predicted to possess broad therapeutic applicability against multiple respiratory viruses (Limburg et al., 2019). Alternatively, superior selectivity could be achieved with allosteric inhibitors of TMPRSS2 or another critical protease (Laporte & Naesens, 2017). Recently, SARS-CoV-2 was shown to use the ACE2 receptor for entry and the serine protease TMPRSS2 for S protein priming. Camostat mesylate blocked the entry of SARS-CoV-2 virus into the cells (Hoffmann et al., 2020).
Genomic characterization of the SARS-CoV-2, its variance, evolution, transmission dynamics (human to human transmission by droplets or direct contact) have also recently been reported (Benvenuto et al., 2020; Ceraolo & Giorgi, 2020; Li, Wang, et al., 2020; Li, Zai, et al., 2020; Lu et al., 2020; Paraskevis et al., 2020). It is an enveloped single-stranded RNA β-coronavirus similar to the SARS and MERS viruses and closely related to two bat-derived SARS-like coronaviruses (CoVZC45 and bat-SL-CoVZXC21; Lai et al., 2020). Lacking any specific treatment modalities for the new virus, the current treatment options are tailored from earlier SARS-CoV and MERS-CoV experiences and repurposing of existing drugs. These include broad-spectrum antibiotics, corticosteroids, interferons, ribavirin, lopinavir-ritonavir or mycophenolate mofetil and have not been subjected to clinical trials as yet. Shirato et al. (2017) examined the mechanism of cell entry used by a pseudo typed virus bearing the HCoV-229E spike (S) protein in the presence or absence of protease inhibitors. They showed that compared with a laboratory strain isolated in 1966, the recent clinical isolates of HCoV-229E were less likely to use endosomal protease cathepsin L due to two viral mutations (amino acid substitutions, R642M and N714K) that altered their sensitivity to a cathepsin L inhibitor. They suggested that the cell surface TMPRSS2 is a better target than endosomal cathepsin for therapeutic agents for SARS and MERS (Shirato et al., 2017). Explorations on repurposing of drugs continues worldwide to find immediate therapeutic strategies for deadly SARS-CoV-2. Computational approaches for quick screening of available compounds is gaining attention among the scientific community for the same. In a recent study, Indinavir and Remdesivir were investigated for their docking potential to SARS-CoV-2 protease. Though no binding was observed within the catalytic site of the protease, the active form (CHEMBL2016761) of Remdesivir could dock in the overlapping region of the NTP binding motif urging its validation through clinical trials for COVID-19 infection (Chang et al., 2020). Remdesivir has also been tested for its potential to bind to RNA dependent RNA polymerase of SARS-CoV-2 (Nejadi Babadaei, Hasan, Vahdani, et al., 2020). The U.S. Food and Drug Administration (FDA) has now issued an emergency use authorization for Remdesivir for the treatment of suspected or laboratory-confirmed adults and children hospitalized with severe symptoms of SARS-CoV-2 disease (FDA news release, 2020; Hendaus, 2020). In line with this, many research groups across the globe are exploring various drugs for potential repurposing to find an immediate and effective therapeutic candidate for COVID-19 pandemic, by mainly targeting SARS-CoV-2 main protease (Aanouz et al., 2020; Al-Khafaji et al., 2020; Das et al., 2020; Enmozhi et al., 2020; Gyebi et al., 2020; Islam et al., 2020; Joshi et al., 2020; Khan, Jha, et al., 2020; Khan, Zia, et al., 2020; Kumar, Kumari, et al., 2020; Lobo-Galo et al., 2020; Muralidharan et al., 2020; Pant et al., 2020; Umesh et al., 2020), E and N protein (Gupta et al., 2020; Sarma et al., 2020), RNA-dependent RNA polymerase (Elfiky, 2020), NSP15 and prefusion 2019-nCoV spike glycoprotein (Sinha et al., 2020) and cell surface receptors (ACE-2 and TMPRSS2) of host cells (Abdelli et al., 2020; Elmezayen et al., 2020; Thuy et al., 2020; Wahedi et al., 2020).
Indian Ayurvedic herb, Ashwagandha (Withania sominifera) and honey bee propolis have been heavily used in traditional home medicine systems. Both of these are trusted to boost the immune function, possess a variety of prophylactic and therapeutic activities (Latheef et al., 2017; Munagala et al., 2011). Several studies have provided evidence that honeybee propolis inhibits variety of viruses including herpes simplex virus, sindbis virus, parainfuenze-3 virus, human cytomegalovirus, dengue virus type-2, influenza virus A1 and rhinovirus (Amoros et al., 1994; Kwon et al., 2019; Munagala et al., 2011; Serkedjieva et al., 1992). Withaferin-A (Wi-A), a bioactive withanolide from Ashwagandha, was shown to possess inhibitory activity for HPV and influenza viruses (Cai et al., 2015). In light of these literature, we examined the potential of three natural compounds, i.e. Wi-A and Withanone (Wi-N) derived from Ashwagandha, and caffeic acid phenethyl ester (CAPE) from honey bee propolis for their ability to dock and inhibit TMPRSS2, while comparing with Camostat mesylate by molecular docking and molecular dynamics (MD) simulations analyses. We found that both Wi-A and Wi-N could bind and stably interact at the catalytic site of TMPRSS2. Wi-N showed stronger interactions with TMPRSS2 catalytic residues than Wi-A and was also able to induce changes in the allosteric site of TMPRSS2 similar to the ones reported for Camostat mesylate. We also examined if Wi-N could affect the endogenous expression of TMPRSS2 and found its transcriptional downregulation in Wi-N treated cells. The data predicts dual action of Wi-N to block SARS-CoV-2.
2. Materials and methods
2.1. Preparation of TMPRSS2 structure and ligands for the molecular docking
The 3D structure of TMPRSS2 is not yet available in the Protein Data Bank, however, a structure modeled using a homologous protein, serine protease hepsin (5ce1.1), is available in the Swiss model repository (O15393). The identity between the sequence of TMPRSS2 and hepsin is 33.82%, and the Q mean of the modeled structure was –1.62. Also, the catalytic domain in both the proteins was found to be well conserved with identical catalytic residues His296, Asp345 and Ser441 (Afar et al., 2001). This modeled structure was further prepared for docking using the protein preparation wizard of the maestro from Schrodinger suite (Schrödinger, 2020). The preparation steps mainly included the addition of missing disulfide and hydrogen bonds, filling of missing amino acids side chains and optimization of hydrogen bonds. Then OPLS3e forcefield was used for restrained minimization until the average root mean square deviation (RMSD) of the non-hydrogen atoms converged to 0.30 Å (Harder et al., 2016).
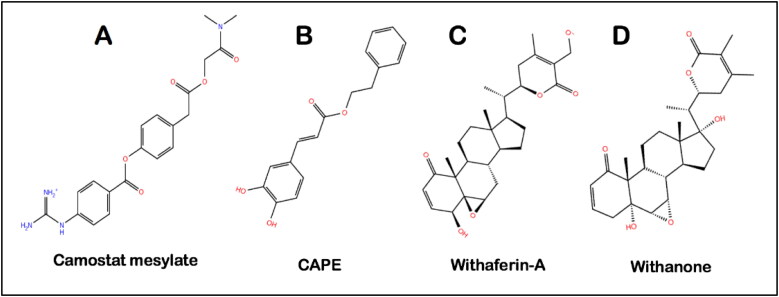
The structure of Camostat mesylate (CID 5284360), Wi-A (CID 265237), Wi-N (CID 21679027) and CAPE (CID 5281787) was downloaded from the PubChem database. The structure of all the ligands shown in Figure 1 was prepared using the LigPrep module of the Schrodinger suite (Schrödinger, 2020). For the ligand preparation, OPLS3e force field was used for energy minimization and a pH of 7.0 ± 2.0 was set to generate all the possible ionization states. Following this, the ligands were desalted, their tautomers were generated, while retaining the chiralities, possible stereoisomers were also generated (Sastry et al., 2013).
Figure 1.
Molecular structures of (A) Camostat mesylate, (B) CAPE, (C) Withaferin-A and (D) Withanone.
2.2. Molecular docking of ligands at the catalytic site of TMPRSS2
The grid was generated at the catalytic site of TMPRSS2, mainly covering His296, Asp345 and Ser441 residues for performing molecular docking studies. Extra precision flexible docking protocol was used for all the dockings (Friesner et al., 2006).
2.3. Molecular dynamics (MD) simulations in water
Desmond package with OPLS3e force field from Schrodinger was used to study the stability of the protein-ligand system in the presence of explicit water molecules (Schrödinger, 2020). Each protein-ligand complex was solvated with the TIP4P water model in an orthorhombic periodic boundary box. To prevent interaction of the protein complex with its own periodic image, the distance between the complex and the box wall was kept 10 Å. The system was then neutralized by the addition of ions. Energy of the prepared systems was minimized for 5000 steps using the steepest descent method or until a gradient threshold of 25 kcal/mol/Å was achieved. It was followed by L-BFGS (Low-memory Broyden-Fletcher-Goldfarb Shanno quasi-Newtonian minimizer) until a convergence threshold of 1 kcal/mol/Å was met (Harder et al., 2016). The equilibrated system was then subjected to 100 ns simulation in NPT ensemble with 300 K temperature, constant pressure of 1 atm and a time step of 2 fs.
2.4. Analysis of the MD simulations trajectories
The MD trajectories were analyzed post MD simulations using the Desmond Simulation event analysis tool. RMSD of conformations acquired by all protein-ligand complexes during the 100-ns simulation run with respect to their docked pose was analyzed and compared with the behavior of Camostat mesylate-TMPRSS2 complex as a function of time to investigate the stability of ligands within the active site pocket of TMPRSS2. Root mean square fluctuation (RMSF) of TMPRSS2 residues was also analyzed to investigate the flexibility of the protein region interacting with the ligands. The interaction pattern of all three ligands- Wi-A, Wi-N and CAPE within the catalytic domain of TMPRSS2 was studied and compared with the interactions of Camostat mesylate with TMPRSS2. Various residues residing in the catalytic domain of the protein were investigated for their time of contact with the different ligands to calculate their occupancy percentage for interaction during the simulation time. The radius of gyration and RMSD of ligands along the simulation trajectory was calculated and analyzed.
2.5. MMGBSA free binding calculations
A total of 100 frames representing the protein-ligand conformations spanned in between 40 and 100 ns of simulation run were extracted to calculate the MM/GBSA (molecular mechanics energies combined with the 906generalized Born and surface area continuum solvation) free binding energy using the Prime module of Schrodinger suite (Schrödinger, 2020).
The equation used for the calculation is:
ΔGcomplex, Greceptor and Gligand represent the free energies of the complex, receptor and ligand, respectively.
2.6. Cell culture and viability assays
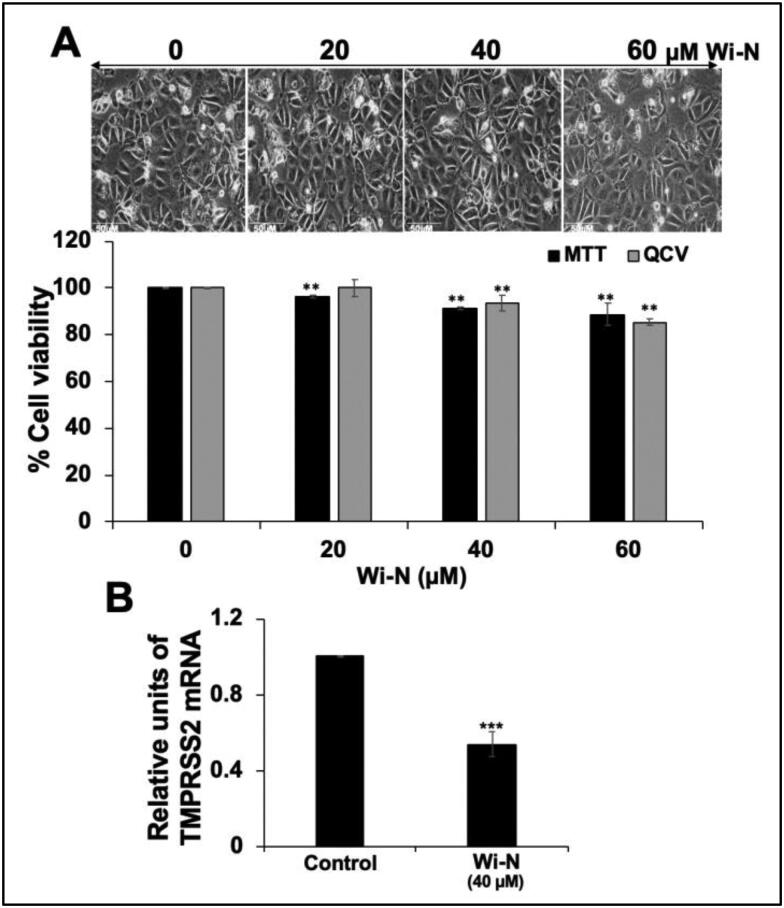
MCF7 (human breast carcinoma known to express TMPRSS2) cells were bought from The Japanese Collection of Research Bioresources Cell Bank (Tokyo, Japan) and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Invitrogen, Carlsbad, California, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a humidified incubator (37 °C and 5% CO2). Five thousand cells/well were plated in 96-well plate and allowed to settle overnight, followed by treatment with varying doses of Wi-N. Control cells were treated with dimethyl sulfoxide (DMSO). After 48 h, 10 μL of MTT {3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide} (Sigma-Aldrich, M2003-1G) in DMEM was added to each well and incubated for 4 h. The media and MTT from the wells were aspirated out and replaced with 100% DMSO, followed by measurement of optical density at 570 nm using Tecan infinite M200® Pro using a microplate reader (Infinite 200 PRO; Tecan Group Ltd., Mannedorf, Switzerland). For Quantitative Crystal Voilet (QCV) staining, after 48 h treatment, cells were fixed, stained with CV and incubated with de-staining solution (40% methanol:10% glacial acetic acid:50% water) for 5–10 mins. The optical density of de-staining solution was measured by Tecan infinite M200® Pro using a microplate reader at 570 nm. The data represents mean ± SD from three experiments. Statistical significance was calculated by Unpaired t test (GraphPad Prism, GraphPad Software, San Diego, CA; **represents p value < 0.05 for significant change). Phase contrast images of control and treated cells were captured on bright field microscopy (10X) Nikon ECLIPSE TE300 (Nikon Instruments Inc. Tokyo, Japan).
2.7. Real-time quantitative RT-PCR analysis
MCF7 cells were treated with non-toxic (determined by independent experiments) dose (20-40 μM) of Wi-N for 48 h. Control and Wi-N treated cells were harvested by trypsin-EDTA (Wako, Japan). Total RNA was extracted using TRIzol™ (Thermo Fisher Sci., Japan). cDNA synthesis was performed using QuantitTect Reverse Transcription Kit (Qiagen, Germany) followed by real-time RT-PCR on the ECOTM real time system (Illumina, San Diego, CA, USA) using SYBR® Green Select Master Mix (Applied Biosystem, Japan). The expression was normalized against 18S as an internal control. Single PCR product amplification was confirmed by melt-curve analysis 2–ΔΔCT. Primer sequences used in this study were – TMPRSS2 [forward] 5′-GAGGACGAGAATCGGTGTGT-3′ and [reverse] 5′-TCCAGTCGTCTTGGCACA-3′ (Foulds et al., 2010) and internal control 18S [forward] 5′-CAGGGTTCGATTCCGTAGAG-3′ and [reverse] 5′-CCTCCAGTGGATCCTCGTTA-3′. The data represents mean ± SD from three independent experiments. Statistical significance was calculated by unpaired t test (GraphPad Prism, GraphPad Software, San Diego, CA; p values *** < 0.001 represent highly significant change).
2.8. Analyses of Wi-N and Wi-A in the roots, stems and leaves of Ashwagandha
Dry powder of roots, stems and leaves from organically raised Ashwagandha were extracted in β-cyclodextrin-water and the amount of Wi-N and Wi-A was evaluated in each by HPLC, as described earlier (Kaul et al., 2016). The analyses were performed using Develosil (150 mm L × 4.6 mm i.d. 5 μm) C30-UG column (Nomura Chemical Co. Ltd., Japan) on Shimadzu HPLC CBM-20A/20Alite (Shimadzu Corporation, Tokyo, Japan) equipped with UV detector (SPD-20A/20AV) and Lab solutions Software. The separation was carried out using a linear gradient elution program with gradient method: methanol:water (50:50; 0–20 min) followed by (80:20; 21–30 min) at 237 nm at the flow rate of 1.0 μg mL−1 at column temperature maintained at 40 °C. Calibration curves of the standards Wi-A and Wi-N (5 mM) were prepared and diluted with DMSO to obtain 0.02 to 2.5 mM working solutions.
3. Results and discussion
3.1. TMPRSS2, an attractive therapeutic target for COVID-19
TMPRSS2, a single-pass type 2 membrane protein, has been reported as the activator of S protein of various strains of SARS-CoV (Belouzard et al., 2012; Bertram et al., 2011). A very recent report has confirmed that SARS-CoV-2 entry to human cells also depends on the S protein priming by TMPRSS2 (Hoffmann et al., 2020). It is also reported that SARS-CoV-2 can use the endosomal cysteine proteases CatB/L for S priming in TMPRSS2– cells, however, TMPRSS2 but not CatB/L is essential for the virus entry and its spread (Iwata-Yoshikawa et al., 2019). The main catalytic residues of TMPRSS2 involved in the proteolytic activity are His296, Asp345 and Ser441 (Paoloni-Giacobino et al., 1997; Wilson et al., 2005). Further, TMPRSS2 has been shown to be dispensable in mice knockout model that showed no significant effect on development, survival or normal organ structure or functions, suggesting its functional redundancy with other transmembrane serine proteases or its non-vital function in case of stressed or disease scenario (Kim et al., 2006). In these premises, TMPRSS2 has emerged as an attractive therapeutic target against COVID-19. Camostat mesylate (a serine protease inhibitor), well documented for the treatment of cancer, pancreatitis and liver fibrosis, has recently been shown to inhibit TMPRSS2 and entry of SARS-CoV-2 virus into the cells (Hoffmann et al., 2020). In earlier studies, it was shown to significantly reduce H1N1 influenza virus titers in the culture supernatant (Yamaya et al., 2015) and protect mice from H1N1 influenza virus infection (Bahgat et al., 2011; Lee et al., 1996). It was also shown to inhibit TMPRSS2 and yield 10-fold reduction in SARS-CoV virus titre (Shulla et al., 2011) and protected mice from the infection thus increasing the survival rate to 60% (Zhou et al., 2015). Its action against MERS-CoV has also been reported. Concentration of 10 μM could reduce the entry of virus in Vero-TMPRSS2 cells by 15-folds. Two hundred and seventy-fold decrease in the amount of viral RNA in the culture supernatant of Calu-3 cells was also observed three days post MERS-CoV infection with 100 μM dose of Camostat (Shirato et al., 2017). Camostat mesylate, similar to the other non-specific serine protease inhibitors, targets a highly conserved catalytic domain comprising of His-Asp-Ser triad. The protease catalytic domain of TMPRSS2 is constituted of amino acid residues His296, Asp345 and Ser441, corresponding to His57, Asp102 and Ser195 of chymotrypsinogen, another well-known serine protease (Lin et al., 1999; Park, 2011). Although the exact molecular mechanism of action of camostat remains elusive, to be effective against different serine proteases, it must target the active site harboring the conserved catalytic triad residues. Based on the anti-viral activity of Camostat mesylate, we decided to use it as a reference molecule. We first modelled its interactions with the catalytic residues of TMPRSS2 and then compared them with our small molecules; Wi-A, Wi-N and CAPE. The crystal structure of TMPRSS2 has not been reported yet. Therefore, we used a homology-based structural model from Swiss model repository (O15393). This structure was modeled using the structural coordinates of human hepsin as a template. Both TMPRSS2 and hepsin belong to the same family of the proteases i.e. type-II membrane-associated serine peptidase (Afar et al., 2001). Both Hepsin and TMPRSS2 are reported to share the same arrangement of the catalytic triad amino acids – His296, Asp345 and Ser441 (Afar et al., 2001). Based on this assessment of model confidence, we further processed this structure for carrying out molecular docking and MD simulations through restrained minimization. It allowed hydrogen atoms to be freely minimized. It also allowed sufficient movement of heavy atom to relax strained bonds, angles and clashes.
3.2. Wi-A, Wi-N and CAPE have the same binding affinity towards TMPRSS2 as Camostat mesylate
In these premises, we first investigated the docking score for Camostat mesylate, a known inhibitor, against the substrate-binding pocket of TMPRSS2. The docking score for the best pose of TMPRSS2/Camostat mesylate complex was found to be –5.90 Kcal/mol. Gly464 was mainly contributing to this by making a hydrogen bond with the ligand. Wi-A, Wi-N and CAPE were docked with TMPRSS2 using the same grid. All the ligands were binding at the catalytic site of the protein. For TMPRSS2/CAPE complex, the best pose had the binding energy of –6.20 Kcal/mol, and Gly464 and Ser436 were involved in the hydrogen bond interactions. Complex of TMPRSS2 with Wi-A had the docking score of –5.60 Kcal/mol. Here, Glu299 and Lys342 were mainly involved in the polar interactions. And in case of Wi-N, the docking score was –4.30 Kcal/mol and Gly462 of TMPRSS2 was involved in the polar interactions. The detailed list of residues involved in polar and non-polar interactions with all the four ligands – Camostat mesylate, Wi-A, Wi-N and CAPE is shown in Table 1. As molecular docking alone is not sufficient to give the real estimate of binding pose and binding affinity, the best-docked pose for each complex was further subjected to MD simulations and MM/GBSA free binding energy calculations for better understanding of the molecular basis of interactions.
Table 1.
Residues of TMPRSS2 interacting with the ligands after docking.
| Complex | Molecular docking (Kcal/mol) | Types of interactions and residues involved (Premolecular dynamic simulations) |
|
|---|---|---|---|
| H-bonds | Hydrophobic, polar and pi-pi stacking | ||
| TMPRSS2-Camostat mesylate | –5.90 | Gly464 | Val275, Gln276, Val278, Val 280, His296, Cys297, Leu302, Asp435, Ser436, Cys437, Gln438, Gly439, Ser441, Thr459, Trp461, Gly462, Cys465, Ala466, Gly472, Val473 |
| TMPRSS2-Wi-A | –5.60 | Glu299, Lys342 | His296, Tyr337, Glu389, Asp435, Asp435, Ser436, Cys437, Gln438, Asp440, Ser441, Thr459, Ser460, Trp461, Gly462, Ser463. Gly464, Cys465, Ala466, Gly472 |
| TMPRSS2-CAPE | –6.20 | Gly464, Ser436 | Cys281, Val280, His296, Cys297, Glu299, Leu302, Asp435, Cys437, Gln438, Gly439, Asp440, Ser441, Thr459, Ser460, Trp461, Gly462, Cys465 |
| TMPRSS2-Wi-N | –4.30 | Gly462 | His296, Glu299, Tyr337, Lys342, Glu389, Asp435, Ser436, Cys437, Gln438, Ser441, Thr459, Ser460, Trp461, Ser463, Gly464, Cys465, Gly472, Val473 |
3.3. Wi-N and Wi-A mimic the action of Camostat mesylate against TMPRSS2
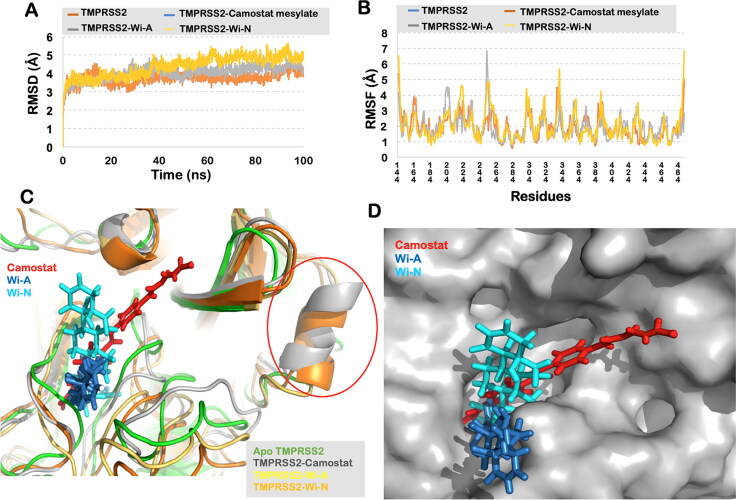
To investigate the dynamic behavior of each protein-ligand complex and account for the stability of the interactions we performed 100 ns long MD simulations in water solvated state. Visual analysis of the trajectory showed that TMPRSS2 could not retain CAPE in its substrate binding pocket for long. It lost its interaction and moved out within 37 ns of the simulation run. Wi-A and Wi-N, on the other hand, were found to be stably interacting at the docked site throughout the 100 ns of simulations and was very similar to Camostat mesylate. The overall conformation of TMPRSS2-Camostat mesylate, TMPRSS2-Wi-A and TMPRSS2-Wi-N were found stable and did not show much fluctuations (Figure 2(A)). The RMSF plot of the complexes was found to be aligned with TMPRSS2(Apo) residues, and no significant fluctuations were observed in the catalytic triad of TMPRSS2 (Figure 2(B)). To get further insights of the conformational changes, the simulated complexes were superimposed on TMPRSS2(Apo). The RMSD of TMPRSS2-Camostat mesylate with respect to TMPRSS2(Apo) was estimated to be 1.76 Å, for TMPRSS2-Wi-A it was 2.0 Å and for TMPRSS2-Wi-N it was 2.4 Å. In further scrutiny, it was found that the loop region from Gly245 to Gly265 had an outward conformation in the natural compound complexes, whereas it had an inward orientation in Camostat mesylate complex, when compared to TMPRSS2(Apo) structure. Further, it was found that Wi-N and Camostat mesylate were inducing the same conformation changes in TMPRSS2. Both these molecules forced the region from Arg316 to Tyr322 to acquire a helical secondary structure, while in case of Wi-A and TMPRSS2(Apo) this region was found to be a loop (Figure 2(C)). Furthermore, as all the ligands were binding within the same pocket of TMPRSS2, interacting with almost the same residues, suggested a similar mechanism of their action (Figure 2(D)). The stable binding of the ligands and the compactness of Wi-A and Wi-N were also seen through RMSD and radius of gyration calculations. All the calculations with the average values and standard deviation throughout the 100 ns of the simulations are shown in Table 2.
Figure 2.
(A) RMSD of the protein backbone along the simulation trajectory for the protein alone and all the docked complexes. The overall structure of TMPRSS2 did not change much after the binding of Wi-N or Wi-A when compared to Camostat mesylate. (B) RMSF of the amino acids comprising the interacting domain of TMRSS2. No abrupt fluctuations were observed in any region of the protein with or without the three ligands. (C) Superimposition of the three docked complexes with Apo-TMPRSS2. All the three small molecules – Camostat mesylate, Wi-N and Wi-A were bound in the same site suggesting their similar mechanism of action. Conformational change from loop to helix was observed in region Arg316 to Tyr322 in case of Camostat mesylate and Wi-N. (D) Surface representation of TMPRSS2 showing all the ligands embedded in its catalytic pocket.
Table 2.
Analysis of the MD trajectories in terms of RMSD, RMSF, H-bonds count, radius of gyration and solvent accessible surface area of the ligands for each protein-ligand complex.
| Properties | Complex |
Ligands |
|||||
|---|---|---|---|---|---|---|---|
| TMPRSS2-Camostat mesylate | TMPRSS2-Wi-A | TMPRSS2-Wi-N | Camostat mesylate | Wi-A | Wi-N | ||
| Root mean square deviation (Å) | 3.72 ± 0.31 | 4.03 ± 0.37 | 4.37 ± 0.56 | 1.28 ± 0.32 | 2.13 ± 0.26 | 2.10 ± 0.17 | |
| Root mean square fluctuation (Å) | 1.94 ± 0.80 | 1.88 ± 0.80 | 1.99 ± 0.98 | – | – | ||
| Hydrogen bonds count (protein-ligand complex) | 4.80 ± 1.00 | 1.00 ± 0.64 | 1.50 ± 1.12 | – | – | ||
| Radius of gyration (Å) | – | – | 6.21 ± 0.29 | 5.17 ± 0.06 | 4.72 ± 0.08 | ||
| Solvent accessible surface area (Å 2) | – | – | 191.15 ± 30.94 | 304.00 ± 44.00 | 245 ± 30.85 | ||
3.4. Wi-N binding to Ser441 of TMPRSS2 is likely to disrupt substrate binding at the catalytic site
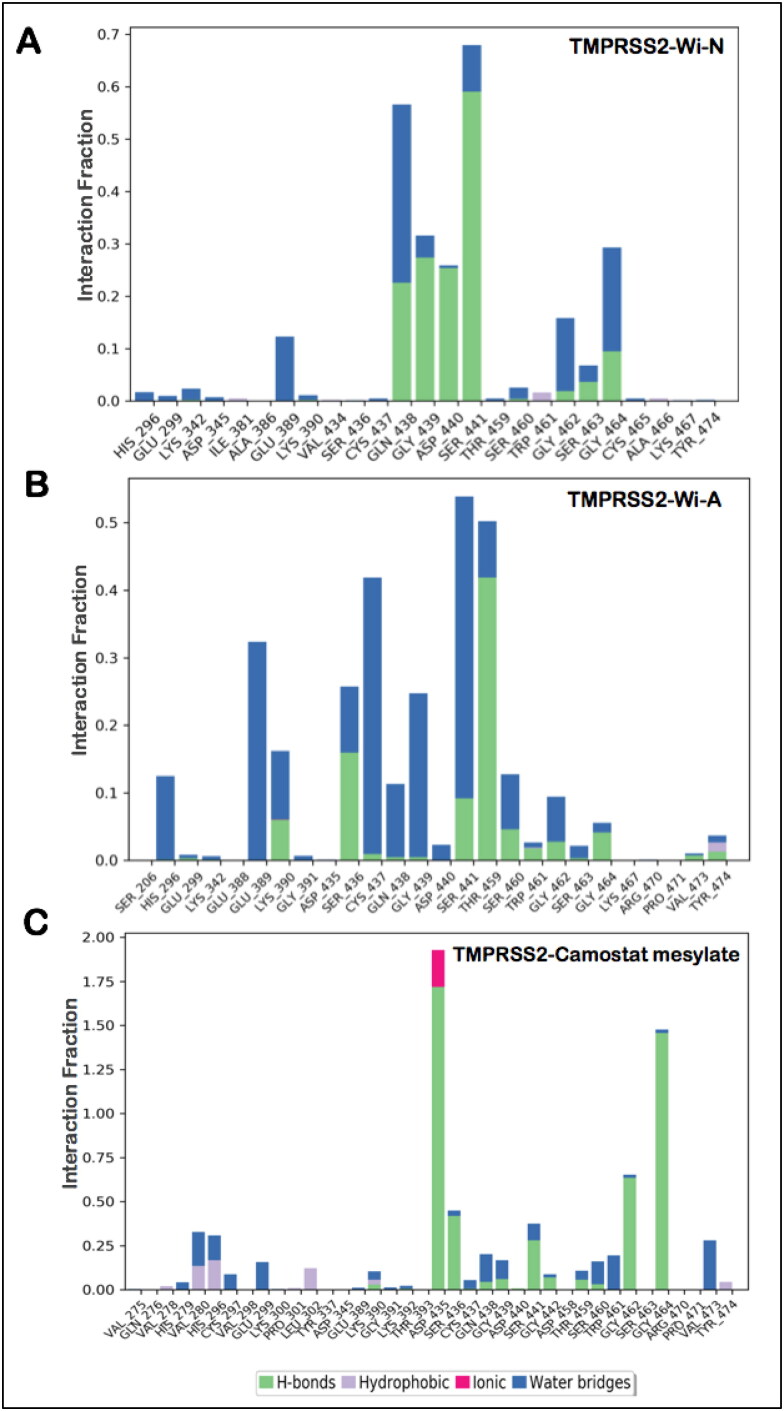
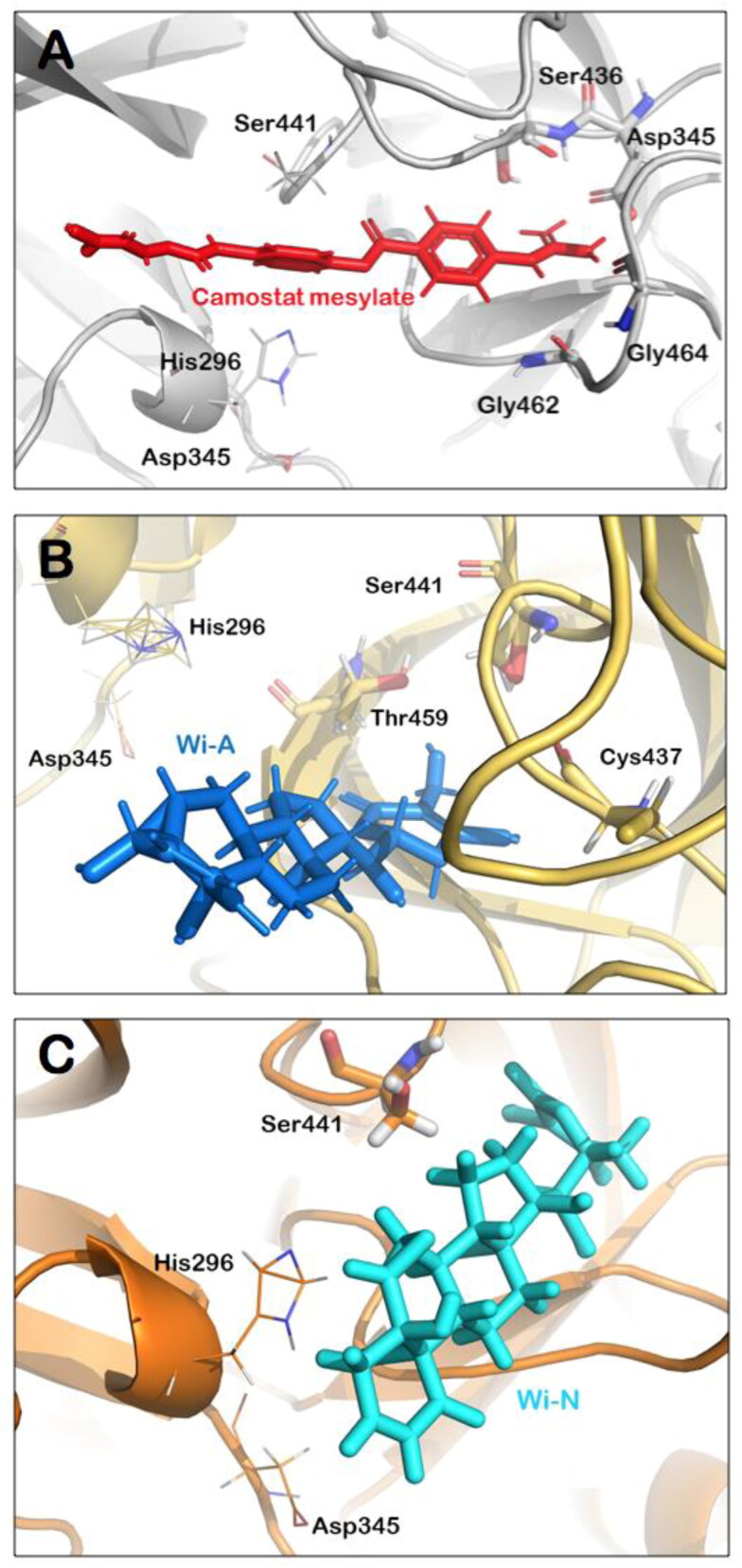
Hydrogen bonds predominantly contribute to the specificity of molecular recognition and are therefore exploited the most for the design of effective drug molecules. In view of this, we further investigated the hydrogen bond interactions made by the natural compounds during the simulation run. Hydrogen bonds were calculated taking the criteria that the distance between hydrogen bond donor and acceptor should be less than 2.5 Å, the donor angle should be less than or equal to 120 degrees between donor-hydrogen acceptor atoms and the acceptor angle should be less than or equal to 90 degrees between the hydrogen acceptor-bonded atoms. It was found that Wi-N was making significant polar contact (for more than 50% of the simulation time) with Ser441, a residue directly involved in catalytic activity of TMPRSS2, whereas Wi-A was making water-mediated contact with Ser 441 (Figure 3(A,B)). In both Wi-A and Wi-N, this interaction with the catalytic residues was maintained by the lactone ring. Though Camostat mesylate had numerous polar and non-polar contacts within the binding pocket of TMPRSS2, it was not making polar interaction with any of the catalytic triad residues (His296, Asp435, Ser441; Figure 3(C)). Overall analyses showed that at any fraction of time, all the ligands had a similar number of polar and non- polar interactions with TMPRSS2 while in terms of consistent interactions with catalytic residues of TMPRSS2, natural molecules were better. A detailed analysis of the interacting residues during the course of MD simulations has been shown in Table 3. The most significant interactions between the protein-ligand complexes are illustrated in Figure 4(A–C). This analysis taking Camostat mesylate as a reference molecule thus suggests that Wi-N and Wi-A have a good affinity towards the substrate-binding pocket of TMPRSS2 and could probably be a natural and readily available drug for the inhibition of S protein priming of SARS-Cov2.
Figure 3.
Hydrogen bond occupancy of various important residues of TMPRSS2 during the simulation run in case of binding with Wi-N (A), Wi-A (B) and Camostat mesylate (C).
Table 3.
Residues of TMPRSS2 interacting with the ligands during the course of MD simulations along with the free binding energy of each protein-ligand complex.
| Complex | MM/GBSA free binding energy (Kcal/mol) | Types of interactions and residues involved during molecular dynamic simulations at any fraction of time |
|
|---|---|---|---|
| H-bonds | Hydrophobic and pi-pi stacking | ||
| TMPRSS2-Camostat mesylate | –65.20 ± 1.65 | Lys390, Asp345, Ser346, Gln438, Gly439, Ser441, Gly442, Gly462, Gly464 | Val275, Gln276, Val278, His279, Val280, Glu 299, Lys300, Pro301, Leu302, Tyr337, Asp345, Glu389, Lys390, Gly391, Lys392, Thr393, Asp435, Ser436, Cys437, Asp440, Asp458, Thr459, Ser460, Trp461, Ser463, Arg470, Pro471, Val473, Tyr474 |
| TMPRSS2-Wi-A | –37.80 ± 5.80 | Lys390, Ser436, Cys437, Ser441, Thr459, Ser460, Trp461, Gly462, Gly464, Tyr474 | Ser206, His296, Glu299, Lys342, Glu388, Glu389, Lys390, Gly391, Asp435, Ser436, Cys437, Gln438, Gly439, Asp440, Ser441, Thr459, Ser460, Trp461, Gly464, Lys467, Arg470, Pro471, Val473, Tyr474 |
| TMPRSS2-Wi-N | –46.80 ± 5.13 | Gln438, Gly439, Asp440, Ser441, Gly462, Ser463, Gly464 | His296, Glu299, Lys342, Asp435, Ile381, Ala386, Glu389, Lys390, Val434, Ser436, Cys437, Gln438, Gly439, Asp440, Ser441, Thr459, Ser460, Trp461, Gly462, Ser463, Gly464, Cys465, Ala466, Lys467, Tyr474 |
Figure 4.
Molecular interactions of Camostat mesylate (A), Wi-A (B) and Wi-N (C) within the catalytic site of TMPRSS2. Residues involved in hydrogen bond interactions are shown in sticks while the ones with non-polar interactions have been represented by lines. In all the three complexes, catalytic residues were participating in interaction with the ligands thereby blocking the site for priming of S protein, suggesting the therapeutic potential of Ashwagandha derived Wi-N and Wi-A.
3.5. Wi-N shows dual inhibitory activity towards TMPRSS2-inhibition of its catalytic activity and transcriptional downregulation
MM/GBSA is a method commonly used or estimating ligand-binding affinities in protein systems. We used 100 snapshots from the conformational space of each protein-ligand complex in between 40 to 100 ns for calculating the MMGBSA free binding energy. The average MMGBSA free binding energy of Camostat mesylate, Wi-N and Wi-A with TMPRSS2 was estimated to be –65.20 ± 1.64 Kcal/mol, −46.80 ± 5.13 Kcal/mol and –37.80 ± 5.80 Kcal/mol, respectively. Though the free binding energy of Camostat mesylate was more in comparison to Wi-N and Wi-A, the free binding energy of Wi-N was in range of –50 Kcal/mol clearly indicating equitable affinity of this natural molecule towards TMPRSS2. Camostat mesylate, an established inhibitor of TMPRSS2, has been shown to cause 30%–40% reduction in SARS infection (virus titer; Kawase et al., 2012). It was recently demonstrated that SARS-CoV-2 follows the mechanism similar to SARS for invasion into host cell and requires TMPRSS2 for its protein priming. When Vero cells expressing TMPRSS2 were treated with Camostat, the infection by SARS-CoV-2 was reduced to 40%–50% (Hoffmann et al., 2020). However, the mechanism of inhibition of TMPRSS2 by Camostat mesylate, per se, has not been clearly elucidated. Azouz et al. (2020) showed decrease in TMPRSS2 protein in Camostat mesylate-treated cells. On the other hand, Yamaya et al. (2015) reported that the expression levels of TMPRSS2 mRNA remained unaffected by Camostat mesylate.
Downregulation of endogenous TMPRSS2 expression clearly holds promise as an approach towards preventing transmission of coronavirus. Earlier, peptide-conjugated PPMO, single-stranded-DNA-like antisense agents that readily enter cells and sterically blocking cRNA, targeted to TMPRSS2 caused significant suppression of viral titers (Böttcher-Friebertshäuser et al., 2011). Most recently, Shen et al. (2020) reported an inhibition of influenza A virus propagation by benzoselenoxanthenes by downregulation of TMPRSS2 expression. We next considered and examined if Wi-N, in addition to the predicted inhibition of TMPRSS2, have such capability. We used human breast carcinoma (MCF7) cells, known to express TMPRSS2. Non-toxic doses of Wi-N were first determined by MTT and QCV assays. Expression of TMPRSS2 was then determined in control and Wi-N treated cells by real time RT-PCR using specific primers for TMPRSS2 and 18S (as an internal control). As shown in Figure 5, 40%–50% reduction in TMPRSS2 transcript was observed in cells treated with non-toxic doses of Wi-N. These data suggested that Wi-N may have dual impact on TMPRSS2, i.e. inhibition of its catalytic activity as predicted by molecular modelling data and transcriptional downregulation as shown in Figure 5. Both of these may collectively block the cleavage of viral protein and entry of the virus to host cells, effectively.
Figure 5.
Dose dependent cytotoxicity of Wi-N to MCF7 cells. Cell morphology and viability, as determined by MTT and QCV assays, are shown (A). TMPRSS2 mRNA expression was determined by real time quantitative PCR (RT-PCR) in control and Wi-N (40 μM) treated cells. The data represents mean ± SD from three experiments. Statistical significance was calculated by Unpaired t test (GraphPad Prism, GraphPad Software, San Diego, CA. ** and ***represent p value < 0.05 and < 0.001 for significant and very significant changes, respectively).
3.6. Source of Wi-N
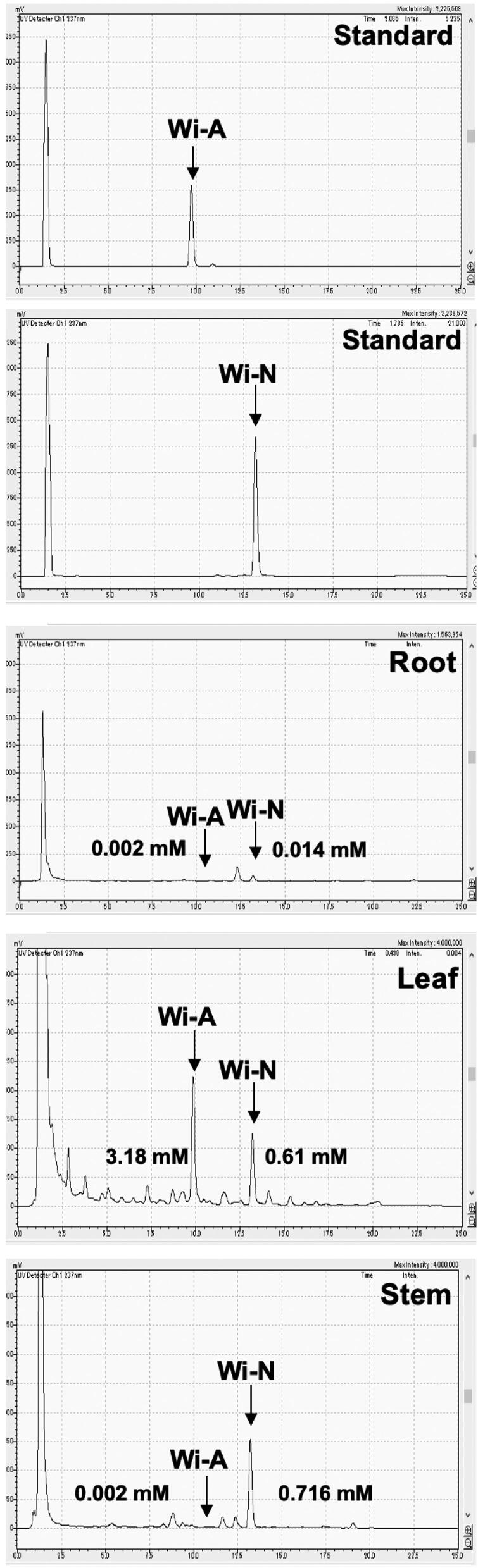
In light of the above findings, we considered to examine the best source of Wi-N. The alcoholic extract of Ashwagandha leaves and Wi-N have earlier been shown to possess anticancer potential, and safe for normal cells (Widodo et al., 2008). The normal cells were shown to be protected from oxidative, DNA damage, chemical and amnesia stresses in cell culture and mice models of study, respectively (Gao et al., 2014; Konar et al., 2011; Wadhwa et al., 2016; Widodo et al., 2009). It was earlier reported that Ashwagandha leaves possess higher amount of Wi-A and Wi-N, as compared to the roots (Kaul et al., 2016). Although there was a high ratio of Wi-N/Wi-A in roots, the total content was extremely (10 times) low as compared in leaves (Kaul et al., 2016). Methods of water-based extraction (using cyclodextrins) for these bioactive components, and hydroponic cultivation yielding leaves with enriched level of Wi-N have also been established (Kaul et al., 2016). The use of vermicompost as an organic fertilizer in Ashwagandha cultivation was seen to yield not only the higher leaf and root mass, but also higher level of Wi-N and Wi-A (Kaur et al., 2018). In view of the current findings that Wi-A and Wi-N may be particularly useful for the management of COVID-19 disease, we investigated their amount in the synchronous Ashwagandha sources and their extracts. Root, stem and leaves from the same Ashwagandha harvest were carefully separated, dried, powdered, extracted and accessed for Wi-A and Wi-N contents (Foulds et al., 2010). In multiple replicates, we found that β-CD assisted water extract from three parts of the plant contained both Wi-A and Wi-N. Interestingly, whereas the amount of Wi-A was highest in leaves, Wi-N was enriched in stem (Figure 6). Recently, Wi-N has also been predicted to possess the ability to interact with and inhibit the main protease of SARS-CoV-2 (Kumar, Dhanjal, et al., 2020), and may hence act as two-way sword, namely (i) inactivating the virus and (ii) blocking its entry to the target cells. Interestingly, an independent bioinformatics screening of thousands of phytochemicals selected Wi-N as a potential inhibition of ACE2, another cell surface receptor protein required for entry of virus to the target cells (Balkrishna et al., 2020). Taken together, these findings propose that (i) Wi-N could be a strong candidate for the management of COVID-19 and (ii) Ashwagandha stem and its Wi-N rich extract may provide preventive and therapeutic benefits in the current pandemic.
Figure 6.
Content of Wi-A and Wi-N in root, stem and leaves of Ashwagandha as detected by high pressure liquid chromatography. Dry powders of each of the sample obtained from the same plant/harvest of Ashwagandha were extracted with β-CD water (Kaul et al., 2016). Content of Wi-A and Wi-N was determined by HPLC with respect to the standards shown on the top. X-axis and Y-axis show minutes and absorbance units, respectively.
4. Conclusion
The entry of SARS-CoV-2 into the host depends on the interaction with ACE-2 receptor of the host cell surface. After the successful attachment with ACE2 receptor, the S protein of SARS-CoV-2 needs to be cleaved and activated for its fusion into the host cell membrane, facilitated by the TMPRSS-2 enzyme of the host (Hoffmann et al., 2020). In the present study, the inhibitory potential of three natural compounds, Wi-A, Wi-N and CAPE for TMPRSS2 in comparison to its known inhibitor Camostat mesylate was investigated by molecular modeling tools. We demonstrate that both Wi-A and Wi-N could bind and stably interact at the catalytic site of TMPRSS2. The interactions of Wi-N with catalytic residues were stronger and significant than Wi-A. Wi-N also able to induce changes in the allosteric site of TMPRSS2 similar to the ones reported for Camostat mesylate. Furthermore, cells treated with Wi-N showed remarkable downregulation of TMPRSS2 suggesting dual potential of Wi-N to block TMPRSS2 and hence the entry of SARS-CoV-2 to host cells. While these preliminary data necessitate further in vitro and in vivo experimental evidence, we demonstrate that Wi-N/Wi-A (i) vary in their content in different parts of Ashwagandha plant and hence their careful recruitment may be helpful in the management of current outbreak of fatal SARS-CoV-2 and (ii) may provide a reliable direction to new drug development.
Funding Statement
This study was supported by the funds granted by AIST (Japan) and DBT (Government of India).
Acknowledgments
The computations were performed at the Bioinformatics Centre supported by the Department of Biotechnology (DBT) Govt. of India at IIT Delhi.
Author contributions
Conceptualization - V.K., D.S, R.W.; Bioinformatics work and formal analysis- V.K., J.K.D., D.S.; Experimental work - A.K., J.K.D, J.W., H.Z., P.B.; Funding acquisition - S.C.K., R.W., D.S.; Writing - review & editing V.K., J. K.D., S.C.K., R.W., D.S. All authors contributed to the development of this manuscript and read and approved the final version.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afar D. E., Vivanco I., Hubert R. S., Kuo J., Chen E., Saffran D. C., Raitano A. B., & Jakobovits A. (2001). Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Research, 61(4), 1686–1692. [PubMed] [Google Scholar]
- Al-Khafaji K., Al-DuhaidahawiL D., & Taskin Tok T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS -CoV- 2. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1764392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoros M., Lurton E., Boustie J., Girre L., Sauvager F., & Cormier M. (1994). Comparison of the anti-herpes simplex virus activities of propolis and 3-methyl-but-2-enyl caffeate. Journal of Natural Products, 57(5), 644–647. 10.1021/np50107a013 [DOI] [PubMed] [Google Scholar]
- Azouz N. P., Klingler A. M., & Rothenberg M. E. (2020). Alpha 1 antitrypsin is an inhibitor of the SARS-CoV2-priming protease TMPRSS2. bioRxiv, 1–16. 10.1101/2020.05.04.077826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahgat M. M., Błazejewska P., & Schughart K. (2011). Inhibition of lung serine proteases in mice: A potentially new approach to control influenza infection. Virology Journal, 8(1), 27 10.1186/1743-422X-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkrishna A., Pokhrel S., Singh J., & Varshney A. (2020). Withanone from Withania somnifera may inhibit novel coronavirus (COVID-19) entry by disrupting interactions between viral S-protein receptor binding domain and host ACE2 receptor. Virology Journal, (Preprint), 1–26. 10.21203/rs.3.rs-17806/v1 [DOI] [Google Scholar]
- Belouzard S., Millet J. K., Licitra B. N., & Whittaker G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., & Ciccozzi M. (2020). The 2019-new coronavirus epidemic: Evidence for virus evolution . Journal of Medical Virology, 92(4), 455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Müller M. A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E. J., Jahn O., Steffen I., & Pöhlmann S. (2011). Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. Journal of Virology, 85(24), 13363–13372. 10.1128/JVI.05300-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E., Freuer C., Steinmetzer T., Klenk H. D., & Garten W. (2009). MDCK cells that express proteases TMPRSS2 and HAT provide a cell system to propagate influenza viruses in the absence of trypsin and to study cleavage of HA and its inhibition. Vaccine, 27(45), 6324–6329. 10.1016/j.vaccine.2009.03.029 [DOI] [PubMed] [Google Scholar]
- Böttcher-Friebertshäuser E., Freuer C., Sielaff F., Schmidt S., Eickmann M., Uhlendorff J., Steinmetzer T., Klenk H. D., & Garten W. (2010). Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. Journal of Virology, 84(11), 5605–5614. 10.1128/JVI.00140-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshäuser E., Klenk H. D., & Garten W. (2013). Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathogens and Disease, 69(2), 87–100. 10.1111/2049-632X.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshäuser E., Lu Y., Meyer D., Sielaff F., Steinmetzer T., Klenk H. D., & Garten W. (2012). Hemagglutinin activating host cell proteases provide promising drug targets for the treatment of influenza A and B virus infections. Vaccine, 30(51), 7374–7380. 10.1016/j.vaccine.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Böttcher-Friebertshäuser E., Stein D. A., Klenk H. D., & Garten W. (2011). Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. Journal of Virology, 85(4), 1554–1562. 10.1128/JVI.01294-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Zhang G., Tang B., Liu Y., Fu X., & Zhang X. (2015). Promising anti-influenza properties of active constituent of Withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: Computational study. Cell Biochemistry and Biophysics, 72(3), 727–739. 10.1007/s12013-015-0524-9 [DOI] [PubMed] [Google Scholar]
- Ceraolo C., & Giorgi F. M. (2020). Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92(5), 522–528. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. C., Tung, Y. A., Lee, K. H., Chen, T. F., Hsiao, Y. C., Chang, H. C., Hsieh, T. T., Su, C. H., Wang, S. S., Yu, J. Y., & Shih, S. S. (2020). Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints. 10.20944/preprints202002.0242.v1 [DOI] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Singha Roy A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA News Release. (2020). Coronavirus (COVID-19) update: FDA Issues emergency use authorization for potential COVID-19 treatment Retrieved May 01, 2020, from https://www.fda.gov/news-events/pressannouncements/coronavirus-covid-19-update-fda-issues-emergencyuse-authorization-potential-covid-19-treatment [Google Scholar]
- Foulds C. E., Tsimelzon A., Long W., Le A., Tsai S. Y., Tsai M.-J., & O'Malley B. W. (2010). Research resource: Expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Molecular Endocrinology (Baltimore, MD), 24(5), 1090–1105. 10.1210/me.2009-0427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., & Mainz D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Gao R., Shah N., Lee J. S., Katiyar S. P., Li L., Oh E., Sundar D., Yun C. O., Wadhwa R., & Kaul S. C. (2014). Withanone-rich combination of Ashwagandha withanolides restricts metastasis and angiogenesis through hnRNP-K. Molecular Cancer Therapeutics, 13(12), 2930–2940. 10.1158/1535-7163.MCT-14-0324 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G. A., Ogunro O. B., Adegunloye A. P., Ogunyemi O. M., & Afolabi S. O. (2020). Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–19. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., Wang L., Lupyan D., Dahlgren M. K., Knight J. L., Kaus J. W., Cerutti D. S., Krilov G., Jorgensen W. L., Abel R., & Friesner R. A. (2016). OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., & Shahpasand K. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendaus M. A. (2020). Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1767691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., & Pöhlmann S. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell, 181(2), 271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., … Cao B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., & Nagata N. (2019). TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. Journal of Virology, 93(6), e01815–18. 10.1128/JVI.01815-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S. C., Ishida Y., Tamura K., Wada T., Iitsuka T., Garg S., Kim M., Gao R., Nakai S., Okamoto Y., Terao K., & Wadhwa R. (2016). Novel methods to generate active ingredients-enriched Ashwagandha leaves and extracts. PLoS One, 11(12), e0166945 10.1371/journal.pone.0166945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Singh B., Ohri P., Wang J., Wadhwa R., Kaul S. C., Pati P. K., & Kaur A. (2018). Organic cultivation of Ashwagandha with improved biomass and high content of active Withanolides: Use of Vermicompost. PLoS One, 13(4), e0194314 10.1371/journal.pone.0194314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., & Matsuyama S. (2012). Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. Journal of Virology, 86(12), 6537–6545. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G., Jain M., Singh E., Pathak A., Singh R. P., Muthukumaran J., & Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Zia K., Ashraf S., Uddin R., & Ul-Haq Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kim T. S., Heinlein C., Hackman R. C., & Nelson P. S. (2006). Phenotypic analysis of mice lacking the Tmprss2-encoded protease . Molecular and Cellular Biology, 26(3), 965–975. 10.1128/MCB.26.3.965-975.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konar A., Shah N., Singh R., Saxena N., Kaul S. C., Wadhwa R., & Thakur M. K. (2011). Protective role of Ashwagandha leaf extract and its component Withanone on scopolamine-induced changes in the brain and brain-derived cells. PLoS One, 6(11), e27265 10.1371/journal.pone.0027265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Jayaraj A., Kumar V., Kumar R. V., Dass S. K., Chandra R., & Singh P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Dhanjal J. K., Kaul S. C., Wadhwa R., & Sundar D. (2020). Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M. J., Shin H. M., Perumalsamy H., Wang X., & Ahn Y. J. (2019). Antiviral effects and possible mechanisms of action of constituents from Brazilian propolis and related compounds. Journal of Apicultural Research, 1–13. 10.1080/00218839.2019.1695715 [DOI] [Google Scholar]
- Lai C. C., Shih T. P., Ko W. C., Tang H. J., & Hsueh P. R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges . Internatinal Journal of Antimicrobial Agents, 55(3), 105924 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte M., & Naesens L. (2017). Airway proteases: An emerging drug target for influenza and other respiratory virus infections. Current Opinion in Virology, 24, 16–24. 10.1016/j.coviro.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latheef S. K., Dhama K., Samad H. A., Wani M. Y., Kumar M. A., Palanivelu M., Malik Y. S., Singh S. D., & Singh R. (2017). Immunomodulatory and prophylactic efficacy of herbal extracts against experimentally induced chicken infectious anaemia in chicks: Assessing the viral load and cell mediated immunity. VirusDisease, 28(1), 115–120. 10.1007/s13337-016-0355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. G., Kim K. H., Park K. Y., & Kim J. S. (1996). Evaluation of anti-influenza effects of camostat in mice infected with non-adapted human influenza viruses. Archives of Virology, 141(10), 1979–1989. 10.1007/BF01718208 [DOI] [PubMed] [Google Scholar]
- Li F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3(1), 237–261. 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang W., Zhao X., Zai J., Zhao Q., Li Y., & Chaillon A. (2020). Transmission dynamics and evolutionary history of 2019-nCoV. Journal of Medical Virology, 92(5), 501–511. 10.1002/jmv.25701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Wang X., & Li Y. (2020). Potential of large “first generation” human-to-human transmission of 2019-nCoV. Journal of Medical Virology, 92(4), 448–454. 10.1002/jmv.25693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limburg H., Harbig A., Bestle D., Stein D. A., Moulton H. M., Jaeger J., Janga H., Hardes K., Koepke J., Schulte L., Koczulla A. R., Schmeck B., Klenk H.-D., & Böttcher-Friebertshäuser E. (2019). TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. Journal of Virology, 93(21), e00649–19. 10.1128/JVI.00649-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Ferguson C., White J. T., Wang S., Vessella R., True L. D., Hood L., & Nelson P. S. (1999). Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Research, 59(17), 4180–4184. [PubMed] [Google Scholar]
- Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., & Díaz-Sánchez Á. G. (2020). FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1764393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., … Tan W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munagala R., Kausar H., Munjal C., & Gupta R. C. (2011). Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis, 32(11), 1697–1705. 10.1093/carcin/bgr192 [DOI] [PubMed] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nejadi Babadaei M. M., Hasan A., Haj Bloukh S., Edis Z., Sharifi M., Kachooei E., & Falahati M. (2020). The expression level of angiotensin-converting enzyme 2 determine the severity of COVID-19: Lung and heart tissue as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1767211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejadi Babadaei M. M., Hasan A., Vahdani Y., Haj Bloukh S., Sharifi M., Kachooei E., Haghighat S., & Falahati M. (2020). Development of Remdesivir repositioning as a nucleotide analog against COVID-19 RNA dependent RNA polymerase. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1767210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoloni-Giacobino A., Chen H., Peitsch M. C., Rossier C., & Antonarakis S. E. (1997). Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics, 44(3), 309–320. 10.1006/geno.1997.4845 [DOI] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E. G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., & Tsiodras S. (2020). Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 79, 104212 10.1016/j.meegid.2020.104212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. (2011). TMPRSS2 (transmembrane protease, serine 2). Atlas of Genetics and Cytogenetics in Oncology and Haematology, 14(12), 1163–1165. 10.4267/2042/44922 [DOI] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., & Sherman W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments . Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schrödinger (2020). Schrödinger Release 2020-1: Maestro 019-3 SR, Glide, LigPrep, Protein Preparation Wizard, Prime, Desmond Molecular Dynamics System, Maestro-Desmond Interoperability Tools. Schrödinger, LLC.
- Serkedjieva J., Manolova N., & Bankova V. (1992). Anti-influenza virus effect of some propolis constituents and their analogues (esters of substituted cinnamic acids). Journal of Natural Products, 55(3), 294–302. 10.1021/np50081a003 [DOI] [PubMed] [Google Scholar]
- Shen L. W., Mao H. J., Wu Y. L., Tanaka Y., & Zhang W. (2017). TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie, 142, 1–10. 10.1016/j.biochi.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. W., Qian M. Q., Yu K., Narva S., Yu F., Wu Y. L., & Zhang W. (2020). Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Scientific Reports, 10(1), 7635 10.1038/s41598-020-64368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., & Matsuyama S. (2017). Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. Journal of Virology, 91(1), e01387–16. 10.1128/JVI.01387-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., & Matsuyama S. (2013). Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. Journal of Virology, 87(23), 12552–12561. 10.1128/JVI.01890-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., & Gallagher T. (2011). A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. Journal of Virology, 85(2), 873–882. 10.1128/JVI.02062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy B. T. P., My T. T. A., Hai N. T. T., Hieu L. T., Hoa T. T., Thi Phuong Loan H., Triet N. T., Anh T. T. V., Quy P. T., Tat P. V., Hue N. V., Quang D. T., Trung N. T., Tung V. T., Huynh L. K., & Nhung N. T. A. (2020). Investigation into SARS-CoV-2 Resistance of compounds in garlic essential oil. ACS Omega, 5(14), 8312–8320. 10.1021/acsomega.0c00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh D. K., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa R., Konar A., & Kaul S. C. (2016). Nootropic potential of Ashwagandha leaves: Beyond traditional root extracts. Neurochemistry International, 95, 109–118. 10.1016/j.neuint.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang Z., Chen X., Lu Y., Chen F., & Zhang W. (2020). Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Bioscience Trends, 14(1), 64–68. 10.5582/bst.2020.01030 [DOI] [PubMed] [Google Scholar]
- WHO Coronavirus Disease (COVID-19) Dashboard. (2020). Retrieved May 21, 2020, from https://covid19.who.int.
- Widodo N., Shah N., Priyandoko D., Ishii T., Kaul S. C., & Wadhwa R. (2009). Deceleration of senescence in normal human fibroblasts by Withanone extracted from ashwagandha leaves. Journal of Gerontology A Biological Sciences and Medical Sciences, 64(10), 1031–1038. 10.1093/gerona/glp088 [DOI] [PubMed] [Google Scholar]
- Widodo N., Takagi Y., Shrestha B. G., Ishii T., Kaul S. C., & Wadhwa R. (2008). Selective killing of cancer cells by leaf extract of Ashwagandha: Components, activity and pathway analyses. Cancer Letters, 262(1), 37–47. 10.1016/j.canlet.2007.11.037 [DOI] [PubMed] [Google Scholar]
- Wilson S., Greer B., Hooper J., Zijlstra A., Walker B., Quigley J., & Hawthorne S. (2005). The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. The Biochemical Journal, 388(Pt 3), 967–972. 10.1042/BJ20041066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya M., Shimotai Y., Hatachi Y., Lusamba Kalonji N., Tando Y., Kitajima Y., Matsuo K., Kubo H., Nagatomi R., Hongo S., Homma M., & Nishimura H. (2015). The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells. Pulmonary Pharmacology & Therapeutics, 33, 66–74. 10.1016/j.pupt.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R. Jr., Nunneley J. W., Barnard D., Pöhlmann S., McKerrow J. H., Renslo A. R., & Simmons G. (2015). Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Research, 116, 76–84. 10.1016/j.antiviral.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., & Tan W, & China Novel Coronavirus Investigating and Research Team (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.