Abstract
Recombinant adeno-associated virus (rAAV) vectors are increasingly popular gene delivery tools in biological systems. They are safe and lead to high-level, long-term transgene expression. rAAV are available in multiple serotypes, natural or engineered, which enable targeting to a wide array of tissues and cell types. In addition, rAAVs are relatively easily produced in a well-equipped lab or obtained from a viral vector core facility. Unfortunately, there is no standardization of quality control assays beyond titering and purity assessments. Next-generation sequencing (NGS) can be used to identify rAAV preparations. Because the rAAV genome is single stranded, previous studies have assumed that rAAV genomes must be converted to double strands before NGS. We demonstrate that rAAV DNA extracts exist primarily as double-stranded species. We hypothesize that these molecules form from the natural base pairing of complementary [+] and [−] strands after DNA extraction and show that rAAV DNA extracts are sufficient templates for downstream NGS without the labor-intensive double-stranding step. Here, we provide a detailed protocol for the simple and rapid NGS of rAAV genomes from DNA extracts. With this protocol, users can quickly confirm the identity of an rAAV preparation and detect the presence of contaminating rAAV DNA. In addition, we share custom Python scripts that allow users to accurately determine the serotype and detect Cre-independent DNA recombination events in rAAV containing Lox sites within minutes. We have used these scripts to analyze more than 100 rAAV preparations. Although we focused on the detection of cross-contaminating rAAV DNA and recombination events, our Python scripts can be customized to detect other sequences or events, such as reverse packaging of plasmid backbone or DNA from the packaging cell line. We find that the NGS of rAAV DNA extracts, termed viral genome sequencing, is a simple and powerful method for rAAV validation.
Keywords: AAV, rAAV, viral vectors, NGS, VGS, Addgene
Introduction
The advent of next-generation sequencing (NGS) has reduced the cost of obtaining whole-genome sequence data 50,000-fold since the days of the Human Genome Project.1 As the cost of NGS drops, the accessibility of this powerful method of data acquisition increases, and a method once considered too expensive for the average lab is becoming mainstream. One area that has been slow to adopt NGS as standard practice is in the quality control (QC) of research-grade recombinant adeno-associated virus (rAAV) vectors.
rAAVs are popular gene delivery tools used frequently in both basic research and drug development. rAAVs have several advantages over other viral gene delivery methods, including long-term expression of the transgene, reduced immunogenicity, and the ability to precisely target specific tissues and cell types by packaging in different serotypes. In addition, producing rAAV is fairly straightforward requiring a triple transfection of HEK293 cells with plasmids encoding the gene of interest, capsid and helper genes, and the isolation of the viral particles from cells, media, or both. Many research labs are able to produce rAAV on their own, and for those that cannot there are numerous viral vector cores that produce and assess the quality of rAAV.
Whether produced in house or by a vector core, rAAV QC tends to be sparse, usually consisting of titration by quantitative PCR (qPCR) and purity assessment by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The lack of comprehensive and standardized QC, especially among core facilities, is problematic for a variety of reasons.
First, scientists cannot be certain that the rAAV they are receiving is what they think it is. Most labs and core facilities titer by using primers targeting common features of the expression cassette such as inverted terminal repeats (ITRs), the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), or the SV40 polyA. Although this approach streamlines production by allowing multiple lots of viral vectors to be titered in parallel, there is no certainty that the correct transgene was packaged since primers do not target the gene of interest. In situations where multiple lots of viral vectors are being produced at the same time, this is especially troubling. In addition, qPCR and protein staining fail to distinguish serotypes; therefore, even if transgene specific primers are used for titration there is no confirmation that the correct capsid was used during packaging.
Second, the common rAAV QC assays fail to ascertain the level of non-cis plasmid DNA in vector preparations. Several studies have reported a range of common DNA contaminants in rAAV preparations, including plasmid backbone sequences such as antibiotic resistance markers or elements from the capsid and helper packaging plasmids, and genomic DNA from the packaging cells.2–11 Although the effect of contaminating DNA on downstream applications remains unclear, it has been demonstrated that it can persist in mammalian systems for months after the vector has been administered.3
Previous work using single molecule real-time sequencing (SMRT), ligation of thymine and adenine overhangs (TA-based ligation), and tagmentation-based library preparation methods have proved that NGS is an effective tool to confirm rAAV genome sequence and identify vector preparation anomalies such as truncated genomes, reverse packaging, and the presence of contaminating DNA.12–14 Unfortunately, at present, SMRT sequencing only works for self-complementary adeno-associated viral genomes precluding its widespread use.12 Further, both TA-based ligation and Fast-seq, a tagmentation-based method, included a laborious double-stranding step.13,14
Herein, we describe a rapid tagmentation-based library preparation method from viral DNA (vDNA) extracts termed viral genome sequencing (VGS). Using this approach, we are able to sequence to depths consistent with the Fast-seq method without a preliminary double-stranding step. Using these data, we are able to assess the identity and confirm the serotype of our rAAV preparations.
We show that VGS quickly and reliably confirms the identity of viral preparations, detects cross-contaminating rAAV DNA, and provides a simple platform for additional sequence-based analysis such as serotype confirmation, and detection of cre-independent recombination events.
In addition, herein we describe unique open-source Python scripts that confirm the serotype of our vector preparations and determine the rate of promiscuous recombination in Cre-dependent viral vectors. These scripts can be easily modified to detect other sequences or events, such as reverse packaging of plasmid backbone or the presence of specific DNA contaminants, and can analyze multiple samples within minutes. VGS is straightforward, requiring only a DNA extraction step before Illumina preparation and can be easily incorporated into the QC regimen of both independent laboratories and viral vector cores alike. Given the number of rAAV production services available and the widespread use of rAAV in both academia and industry, the standardization of AAV QC is a critical need for the research community.
Materials and Methods
AAV production
Vectors produced at Addgene were generated by using the helper-free triple plasmid transfection approach in adherent AAVPro-293T cells (Takara) in serum-containing conditions. Ninety-six hours after transfection, cells and media were harvested; cells were pelleted and resuspended in cold lysis buffer (50 mM Tris, 150 mM NaCl, 2 mM MgCl2) before sonication. After centrifugation, the clarified cell lysate was transferred to a clean tube. Viral particles present in the media were pelleted by addition of polyethylene glycol (PEG 8000) to a final concentration of 8%, stirred for 1 h, and incubated for 3 h at 4°C.
PEG-precipitated particles were pelleted by centrifugation, and the pellet was resuspended in cold lysis buffer. PEG-precipitated particles and clarified cell lysate were then combined and treated with Benzonase. The DNAse was added at a final concentration of 25 U/mL and incubated at 37°C for 45 min. Vectors were purified by iodixanol gradient ultracentrifugation at 350,000 g for 90 min. The purified viral particles were recovered from the 40% fraction, concentrated, and buffer exchanged in Amicon 100 kDa MWCO centrifugal filter units.
The final formulation buffer was phosphate-buffered saline supplemented with 150 mM NaCl and 0.001% Pluronic F68. Purified AAVs were aliquoted, and a QC aliquot was stored at 4°C for all QC assays whereas the remainder of aliquots were stored at −80°C long term. All additional AAV vectors were obtained from the University of Pennsylvania Vector Core. Complete reagent information is listed in Supplementary Table S5.
DNA extraction for sequencing and restriction digests
vDNA was extracted from 20 μL of purified AAV by using PureLink Viral RNA/DNA purification kit (Thermo Fisher) following the manufacturer's instructions. DNA was eluted in 30 μL of nuclease-free water and transferred into a 2D barcoded micronic tube. DNA concentration and purity were determined by using the Nanodrop spectrophotometer, and the samples were stored at −20°C. rAAV DNA extracts or ΦX174 DNA (New England Biolabs) were digested with SacII (New England Biolabs) and HaeIII (New England Biolabs) according to the manufacturer's instructions. Digested DNA was separated on 1% agarose gels alongside a 1 Kb control DNA ladder (New England Biolabs).
qPCR of SacII cleavage sites
Undigested and SacII digested DNA were cleaned by using QIAquick PCR Purification Kit (Qiagen) and eluted in water. DNA concentration and purity were determined by using the Nanodrop spectrophotometer, and the samples were stored at −20°C. Samples were thawed, and 0.2 ng of DNA was amplified with PowerUp SYBR Green Master Mix and 500 nM forward and reverse primers targeting the region surrounding the SacII cleavage site or the green fluorescent protein gene (gfp) as a control. SacII amplification was normalized to gfp, and the fold change of the undigested to SacII digested samples was calculated by using the delta Ct method. Complete reagent information is listed in Supplementary Table S5.
Primer sequences:
37825 amplification
SacII amplification forward primer: GTGGTTTGTCCAAACTCATC
SacII amplification reverse primer: CTGACAATTCCGTGGTGTTGTCGG
50465 amplification
SacII amplification forward primer: GTGGCAACTTCCAGGGCC
SacII amplification reverse primer: CTGACAATTCCGTGGTGTTGTCGG
gfp control amplification
gfp forward primer: GAACTCCAGCAGGACCATGT
gfp reverse primer: ACGACGGCAACTACAAGACC
Sample preparation for MiSeq and sequencing
vDNA was extracted from 20 μL of purified AAV by using PureLink Viral RNA/DNA purification kit (Thermo Fisher) following the manufacturer's instructions. DNA was eluted in 30 μL of nuclease-free water and transferred into a 2D barcoded micronic tube. DNA concentration and purity were determined by using the Nanodrop spectrophotometer, and the samples were stored at −20°C. Twenty microliters of vDNA extract was sent to Seqwell for library preparation by using a plexWell 96-well Library Preparation Kit and sequenced.
First, the average concentration of a 96-well plate was adjusted to 2.5 ng/μL by using a global dilution factor. Next, transposase complexes were added to tagment each sample in the 96-well plate with a unique P7 adaptor. After the first tagmentation step, the samples were pooled into a single tube and a second transposase was added to incorporate the p5 adaptor. Carrier DNA was included in the second tagmentation step to ensure a high enough DNA concentration of the pool to promote even incorporation of the adaptor. The prepared library was then sequenced on a MiSeq System in a 2 × 250 bp paired-end run.
Data analysis using Geneious
After sequencing, FASTQ files were imported into Geneious and paired as Paired End (inward pointing) with the insert size set at 500. The paired file was then trimmed by using BBDuk trimmer to remove poor-quality reads and adaptors. Adaptors were trimmed based on paired read overhangs with a minimum overlap set to 24. The minimum quality of reads was set at 20, and low-quality reads were trimmed from both ends. Reads under 50 bp were discarded.
Trimmed files were then aligned to a reference map of the cis plasmid used for transfection by using Geneious Mapper. Sensitivity was set to Medium Sensitivity/Fast and Fine Tuning was set to iterate up to five times. Cis plasmids were previously sequenced by using Seqwell's plexWell platform as described earlier to provide an accurate reference map. Alignments were then analyzed to ensure even ITR–ITR coverage, determine sequencing depth, and confirm consensus to the reference sequence. To identify non-cis plasmid sequences present in the viral prep, a Megablast search of the NCBI nucleotide database was performed on paired and unpaired unmapped reads.
Data analysis using custom Python scripts
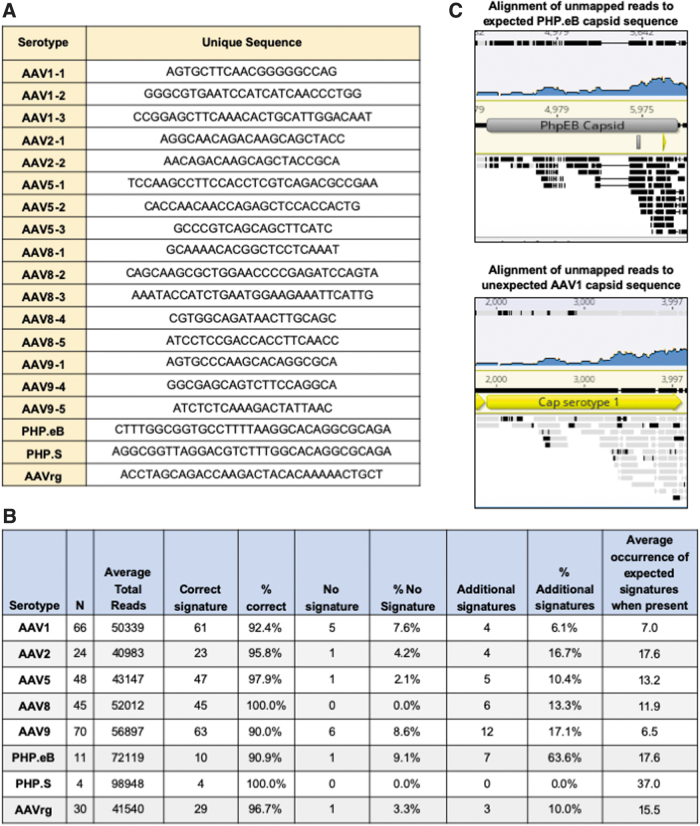
The serotype detection Python program was designed to analyze FASTQ sequencing files and determine serotype based on the presence of predefined signature sequences. The program takes, as configuration parameters, one or more signature sequences per serotype, as well as the directory where the FASTQ files containing the VGS data reside. When executed, the program loads each FASTQ file by using the BioPython library, extracts the sequence reads, and tallies the incidence of the different signature sequences by simply counting matches in both the reads and their reverse complement. The program then makes a serotype call based on the signature with the highest number of matches. The program outputs a spreadsheet summarizing the findings for all files in the input directory, as well as the detailed tallies for each file.
For the serotype determination script, unique signature sequences were identified by aligning the nucleotide sequences of the various Cap genes and manually identifying regions of divergence. Candidate sequences were interrogated against the VGS dataset and sequences that called an incorrect serotype were eliminated. Whenever possible, multiple signatures for a given serotype were used to increase the likelihood of a signature being present in the viral preparation.
The following signature sequences were used for these studies:
AAV1-1: AGTGCTTCAACGGGGGCCAG
AAV1-2: GGGCGTGAATCCATCATCAACCCTGG
AAV1-3: CCGGAGCTTCAAACACTGCATTGGACAAT
AAV2-1: AGGCAACAGACAAGCAGCTACC
AAV2-2: AACAGACAAGCAGCTACCGCA
AAV5-1: TCCAAGCCTTCCACCTCGTCAGACGCCGAA
AAV5-2: CACCAACAACCAGAGCTCCACCACTG
AAV5-3: GCCCGTCAGCAGCTTCATC
AAV8-1: GCAAAACACGGCTCCTCAAAT
AAV8-2: CAGCAAGCGCTGGAACCCCGAGATCCAGTA
AAV8-3: AAATACCATCTGAATGGAAGAAATTCATTG
AAV8-4: CGTGGCAGATAACTTGCAGC
AAV8-5: ATCCTCCGACCACCTTCAACC
AAV9-1: AGTGCCCAAGCACAGGCGCA
AAV9-4: GGCGAGCAGTCTTCCAGGCA
AAV9-5: ATCTCTCAAAGACTATTAAC
PHPS: AGGCGGTTAGGACGTCTTTGGCACAGGCGCAGA
PHPeB: CTTTGGCGGTGCCTTTTAAGGCACAGGCGCAGA
AAVrg: ACCTAGCAGACCAAGACTACACAAAAACTGCT
The recombination calculation Python program was designed to analyze FASTQ sequencing files and determine the percent of recombination at specific recombinase sites. The program takes, as configuration parameters, the expected sequence of the predefined recombinase site, the number of base pairs before and after the recombinase site to consider for determining recombination (the head and tail), and the directory where the FASTQ files containing the VGS data reside.
The program analyzes each FASTQ file and produces a report per file. It reads each file by using the BioPython library, extracts the reads and their reverse complements, and compiles a list of all the sequences containing the recombinase site plus a head and a tail. The two most frequent sequences are assumed to be the nonrecombined sequences. The program then analyzes the remaining sequences to determine whether they contain recombination (i.e., the head and tail of the nonrecombined sequence are flipped) and tallies them to compute the overall percentage of recombination for the sample.
Transduction
Seven thousand AAVpro cells were transduced with 1 μL of AAV9-AAV-FLEX-rev-ChR2(H134R)-mCherry in the presence or absence AAVrg-pAAV-Cre-GFP in a 96-well plate. mCherry expression was assessed 3–9 days post-transduction by direct fluorescence. Wells were examined until 9 days post-transduction for promiscuous expression of mCherry in the non-Cre-containing wells.
Plasmids
Plasmids for transfection were purified by using Qiagen's endotoxin-free HiSpeed Gigaprep kit (Qiagen) and quantified by spectrophotometry. The integrity of the cis plasmids and ITRs were confirmed by restriction digest and NGS. Complete reagent information is listed in Supplementary Table S5.
The following plasmids were used to prepare the rAAV described in this study; pAAV-EF1a-double floxed-hChR2(H134R)-EYFP-WPRE-HGHpA (Addgene 20298, a gift of Karl Deisseroth, unpublished) AAV-Cre-GFP (Addgene 68544, a gift of Eric Nestler), AAV-EF1a-DIO-GCaMP6s-P2A-nls-dTomato (Addgene 51082, a gift of Jonathan Ting), AAV-FLEX-rev-ChR2(H134R)-mCherry (Addgene 18916, a gift from Scott Sternson), rAAV2-retro helper (Addgene 81070, a gift of Alla Karpova), pAAV-hSyn-DIO-hM3D(Gq)-mCherry (Addgene 44361, a gift of Bryan Roth), pAAV-hSyn-DIO-hM4D(Gi)-mCherry (Addgene 44362, a gift of Bryan Roth), pAAV-hSyn-hM3D(Gq)-mCherry (Addgene 50474, a gift of Bryan Roth, unpublished), pAAV-hSyn-EGFP(Addgene 50465, a gift of Bryan Roth, unpublished), pAAV-CAG-GFP (Addgene 37825, a gift from Edward Boyden, unpublished), pAAV-CAG-tdTomato (Addgene 59462, a gift of Edward Boyden, unpublished), and pUCmini-iCAP-PHP.eB (Addgene 103005, a gift of Viviana Gradinaru).15–20
Software
FASTQ data were analyzed by using Geneious Prime. The custom viral serotype determination and recombination Python scripts can be accessed at GitHub in Addgene's Open Toolkit (https://addgene.github.io/openbio).
Results and Discussion
To prepare for VGS, DNA is extracted from 20 μL of DNAse-treated purified rAAV by using commercially available vDNA extraction kits. The titers of the rAAV preparations range from 2 × 1012 to 2 × 1013 genome copies (GC)/mL, therefore each 20 μL aliquot contains between 4 × 1010 and 4 × 1011 GC.
Purified DNA is then sent to Seqwell for library preparation and NGS. At Seqwell, libraries are prepared via a tagmentation reaction in which transposons individually tag samples with sample-specific barcoded adaptors. Samples are pooled and tagged with pool-specific barcoded adaptors and then amplified with universal primers. The tagmentation library preparation method has been shown to work for circular plasmid DNA and, in addition to VGS, is used to sequence verify the plasmids used for AAV production.21 After amplification, the library is normalized, sequenced and the data are provided to Addgene for analysis (Fig. 1).
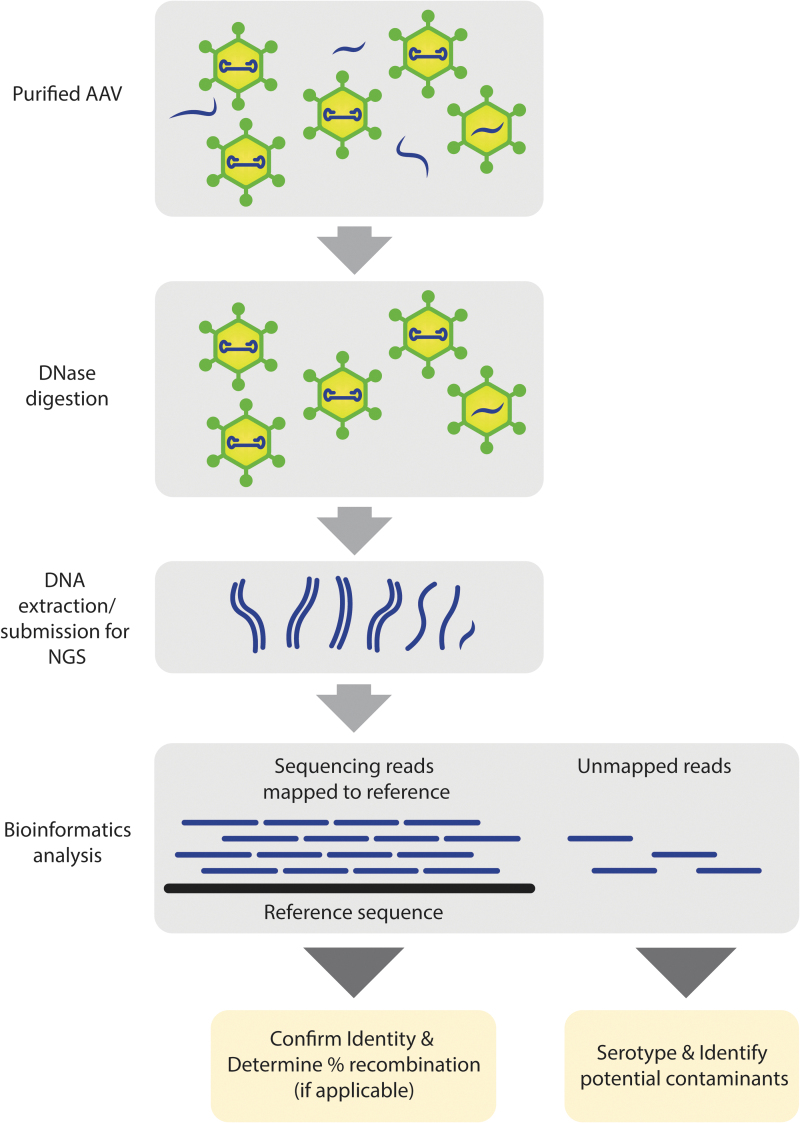
Figure 1.
Simplified viral genome sequencing workflow. Packaged DNA is extracted from purified AAV and sequenced. Individual NGS reads are mapped to a reference sequence to confirm the identity of the viral genome. Unmapped reads are further analyzed to detect and identify potential contaminants. In addition to identifying confirmation and contaminant detection, the NGS data are used further to confirm serotype identity and determine relative recombination rate in Cre-dependent genomes (containing Lox sites). AAV, adeno-associated virus; NGS, next-generation sequencing. Color images are available online.
Previous work utilized TA-based ligation of adaptors for rAAV sequencing library preparation.13 In addition, a recent method termed Fast-Seq successfully used a Tn5 tagmentation-based approach for rAAV library preparation.14 A major drawback of these methods is that they require a laborious double-stranding step to generate a suitable ligation template. It has been demonstrated that rAAV package either a [+] or [−] strand DNA genome at equal rates.22
Here, we hypothesize that the following DNA extraction complementary [+] and [−] strands, present in approximately equal numbers, hybridize in silico to generate double-stranded (ds)DNA templates. These ds templates can serve as suitable ds substrates for library preparation protocols, including TA-based ligation and transposon-based tagmentation, and obviate the need for the labor-intensive and costly double-stranding step. As a tagmentation-based plasmid sequencing pipeline already existed for plasmid QC, we chose to focus on this method instead of TA-based adaptor ligation for rAAV genome sequencing.
To determine whether [+] and [−] rAAV genomic extracts exist as single-stranded (ss) or ds species, DNA extracts were treated with the restriction enzyme SacII and the enzyme's ability to cleave the DNA was assessed. SacII cleaves dsDNA at the palindromic sequence CCGCGG and should not be able to cleave the rAAV genomes if they exist as ss species. DNA extracts were run directly on an agarose gel after extraction (Ctrl), or they were incubated with (37°C + SacII) or without (37°C) SacII in the appropriate digest conditions before gel loading. To demonstrate that SacII is specific for dsDNA, DNA from bacteriophage ΦX174 was included as a negative control; the ΦX174 genome is [+], circular, and ss.
Unlike the rAAV extracts that contain equal proportions of [+] and [−] genomes, the ΦX174 genome is entirely [+] and therefore cannot hybridize. SacII was able to cleave the rAAV DNA extracts, efficiently yielding the expected digest pattern but was unable to cleave ΦX174 (Fig. 2A). To ensure that this result was due to the ss nature of the ΦX174 genome, ΦX174 DNA was digested with HaeIII, a restriction enzyme that has been demonstrated to cleave ssDNA at specific recognition sites.23 When the AAV extracts were incubated with SacII, there was a clear increase in intensity at the expected product sizes, indicating that the correct cleavage products were present. Of note, rAAV DNA extracts consistently ran as two distinct bands on an agarose gel (Fig. 2A, Ctrl lanes).
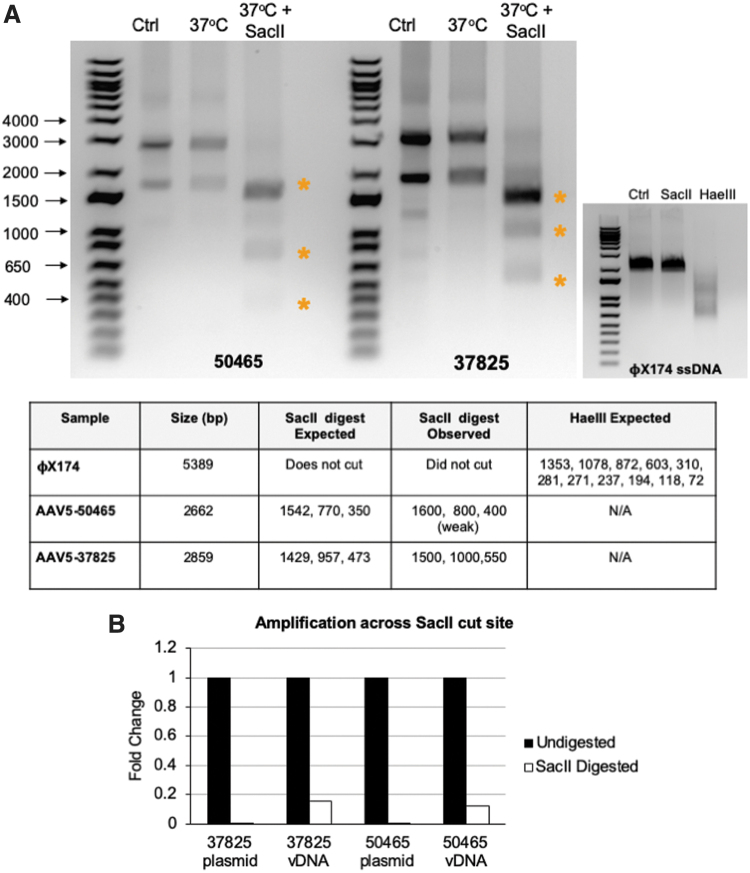
Figure 2.
Analysis of DNA conformation of rAAV vectors. (A) DNA extracted from the rAAV vectors pAAV-hSyn-EGFP (AAV-50465) and pAAV-CAG-GFP (AAV-37825) were left untreated (Ctrl) or incubated in the presence (37°C + SacII) or absence (37°C) of the restriction enzyme SacII and products separated by agarose gel electrophoresis. ΦX174 DNA was incubated in the presence of SacII, HaeIII or left untreated. The table lists the size of the viral genomes and the expected products after digestion. (B) Undigested and SacII digested DNA from plasmid DNA or extracted vDNA was amplified with primers targeting the region surrounding the SacII cleavage site or gfp as a control. SacII amplification was normalized to gfp amplification, and fold change of undigested to SacII digested samples was calculated and plotted. gfp, green fluorescent protein gene; rAAV, recombinant adeno-associated virus; vDNA, virus DNA. Color images are available online.
Initially, we hypothesized that the bands might arise from distinct ss and ds species that migrate differently through the gel. To address this, the bands were extracted from the agarose gel, purified, and submitted for NGS. Both the high-molecular-weight and low-molecular-weight bands were sequenced with 2,730 and 1,478 total reads, respectively (Supplementary Table S1 and Supplementary Fig. S1). The majority of reads, 90% and 76%, from the high- and low-molecular-weight bands, respectively, aligned to the reference sequence. Given that both bands can be sequenced and are completely digested by SacII, we now believe that they are likely different conformations of ds DNA species.
Although the data suggest that much of the DNA in the extract exists as a double-stranded species, it remains possible that the DNA extract is a heterogeneous mix of single- and double-stranded species. To address this, DNA extracts were left undigested or digested with SacII and primers were used to amplify across the SacII cleavage site. Primers targeting a gfp sequence without a SacII cut site were used as a control. Samples treated with SacII had a marked 6.4- and 8.3-fold reduction in SacII cleavage site amplification as compared with the undigested control (Fig. 2B).
Of note, amplification of the cleavage site was not completely lost. To determine whether intact SacII sites were due to incomplete digestion or the presence of ss species, we included the ds cis plasmids as a digest control. Amplification of the SacII sites in plasmid DNA was barely detectable, suggesting that intact SacII sites in the vDNA extracts were, indeed, arising from the presence of ss species. Taken together, these data suggest that the extracted rAAV DNA exists as a heterogeneous mix of ss and dsDNA with the majority associating as ds DNA species (Fig. 1).
To determine whether rAAV DNA extracts, shown to be present primarily as ds species, could serve as efficient tagmentation substrates without double stranding via random hexamer priming, DNA from several rAAV preparations was extracted and submitted directly to Seqwell for NGS. Seqwell was able to obtain tens of thousands of reads for the vast majority of rAAV samples (Supplementary Table S2).
As a negative control, the single-stranded ΦX174 genome was submitted for sequencing. Approximately 800 reads were obtained from the ΦX174 sample. Of these reads, only 89 aligned to the reference ΦX174 genome and those that did had very low (<10 × ) coverage and poor consensus to the reference (Supplementary Fig. S1, bottom panel). Of note, one study has shown that the Tn5 transposase is able to bind to certain conformations of ss DNA.24 With this in mind, one cannot rule out the possibility that a low level of tagmentation may be occurring with ss DNA samples.
In the VGS analysis platform, rAAV NGS data are first analyzed by using Geneious Prime software to confirm identity, check packaging efficiency, and detect the presence of contamination. Briefly, using BBDuk (decontamination using kmers), low-quality reads and adaptors on paired read overhangs are trimmed and reads shorter than 50 base pairs are discarded. The reads are then aligned to the reference sequence of the cis plasmid used for transfection; cis plasmids undergo the same sequencing approach before transfection to provide an accurate reference map.
In a typical sample, the vast majority of reads, >90%, map to the reference sequence. The alignment is then checked to ensure an even distribution of reads over the entire expression cassette and consensus to the reference. The depth of coverage varies between samples but is typically between 500 and 1,000 × throughout most regions. This coverage is similar to the 1,400 × coverage observed by using the Fast-Seq method.14
Of note, certain regions such as ITRs and GC-rich sequences such as the CAG and synapsin promoters are notoriously difficult to sequence. In addition, tagmentation-based methods have known sequence biases toward GC rich sequences and structural biases toward synapses.25,26 Although a drop in coverage in these regions is expected and frequently observed, sequencing depths of >100 × are obtained in these regions, allowing for transgene identification and consistent with the Fast-Seq method.14
Once the identity of the sample is confirmed, the files are analyzed for contamination via a Megablast search on all unmapped reads and a manual review of the hits. As previously mentioned, it is common for non-rAAV sequences such as plasmid backbones, elements from the capsid, and helper packaging plasmids, and genomic DNA from the packaging cells to be packaged with the rAAV genome.2–9 In confirmation of these findings, these sequences were often present at low levels in the Megablast search.
In a typical clean sample, the number of hits to a given non-cis plasmid sequence is very small, usually fewer than 10. Therefore, to parse out the true contamination from the background noise, a threshold of >100 hits was established; any sample with >100 hits to an unexpected gene or vector undergoes further analysis. In these cases, the hits are assembled de novo whereby Geneious uses the overlapping sequences of the reads to assemble larger DNA contigs without using a reference map. The top five assemblies are blasted against the NCBI nucleotide collection database to determine their identity. Any sample with DNA >100 hits to DNA that has not arisen from the packaging, that is, plasmid elements, genes from packaging cells, is discarded.
To validate this approach, rAAV preparations were prepared in which 0.1–20% of sample 2 was spiked into sample 1 (Fig. 3A). After preparing the spiked samples, DNA was extracted and the samples were sent to Seqwell for VGS. Sequencing results were blinded and analyzed following the standard VGS procedure. In the Megablast analysis of the unmapped reads, WPRE and the EF1alpha promoter—elements unique to the spiked DNA—were identified in samples spiked with as little as 0.1% of sample 2 (Fig. 3A, B).
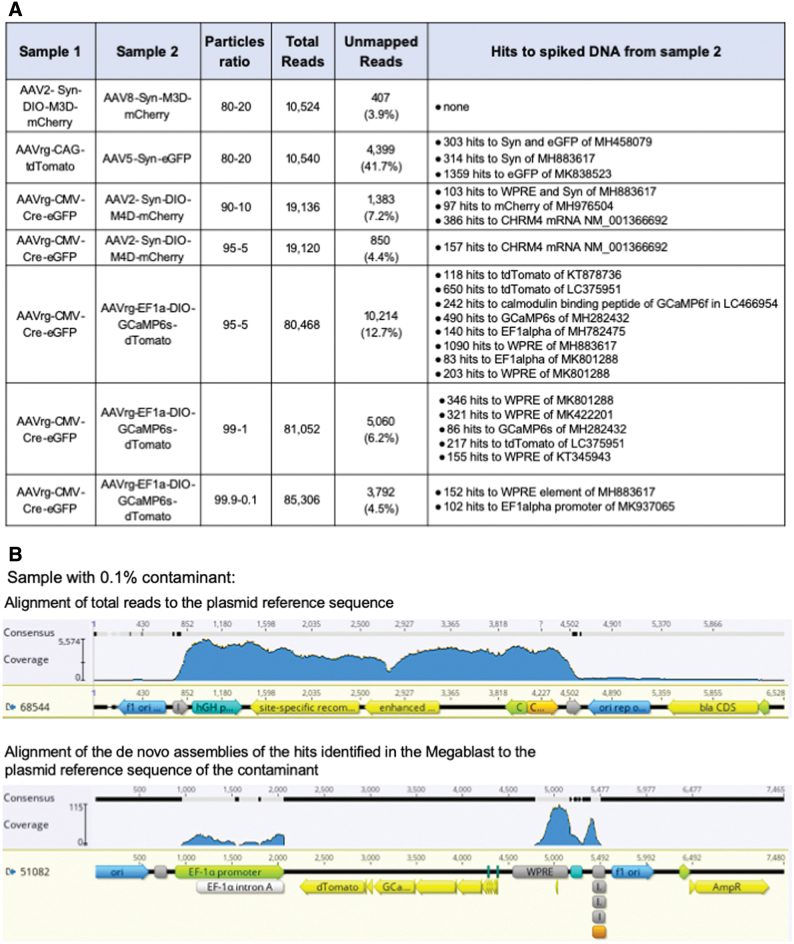
Figure 3.
Validation of VGS workflow and analysis. (A) To validate our assay, DNAse-treated purified viral particles from two different samples were mixed at a defined ratio before DNA was extracted. Samples were blinded before being sent to NGS and subsequently analyzed. (B) Top panel, reads obtained from AAV-Cre-GFP (AAV-68544) spiked with 0.1% of AAVrg-EF1a-DIO-GCaMP6s-dTomato (AAV-51082) were aligned to the reference plasmid sequence. (B) Bottom panel, contaminants identified as having >100 hits in the Megablast analysis were assembled de novo and contigs were aligned to the reference plasmid sequence of the spiked in sample AAV-51082. AAV, recombinant adeno-associated virus; NGS, next-generation sequencing; VGS, viral genome sequencing. Color images are available online.
The only sample in which the spiked DNA sequences were unable to be detected was an instance in which the sequences of vector and contaminant differed only by the absence of two 34 bp lox sites in the spiked sample. In this case, the difference between samples would likely only be observed as a drop in coverage at the lox sites. This specific type of contamination was not able to be detected at levels up to 20% of the sample. Of note, since the spiked DNA lacks the lox sites, this would not be identified in the Megablast analysis stage.
Sequencing depth will influence the ability to detect contaminants using this method. For these analyses, depths ranged from 10,540 to 85,306 total reads, a range that is consistent with the recently reported Fast-Seq method for research-grade rAAV identification.14 Overall, 19,120 reads were sufficient to detect a sample with 5% spiked DNA, and the sequencing depth was much higher for the sample in which 0.1% of spiked DNA was detected; in this case, of 85,306 reads only 102 and 150 hits to spiked DNA sequences were detected. Consequently, if adopting this method to identify low levels of contaminating DNA in research-grade rAAV, we would recommend sequencing to depths greater than 80,000 total reads.
In addition to confirming identity and detecting cross-contamination, VGS data can be used to examine the propensity of non-cis plasmid packaging. To assess this, the percentage of reads that did not align to the cis plasmid (unmapped reads) was first calculated and found to range from 1% to 12% (Fig. 4A). The unmapped reads were then aligned to the capsid plasmid and helper plasmid reference sequences. To focus on the viral elements of these plasmids, such as Rep, Cap, and the adenoviral helper genes, the plasmid backbone sequence was omitted from the alignments.
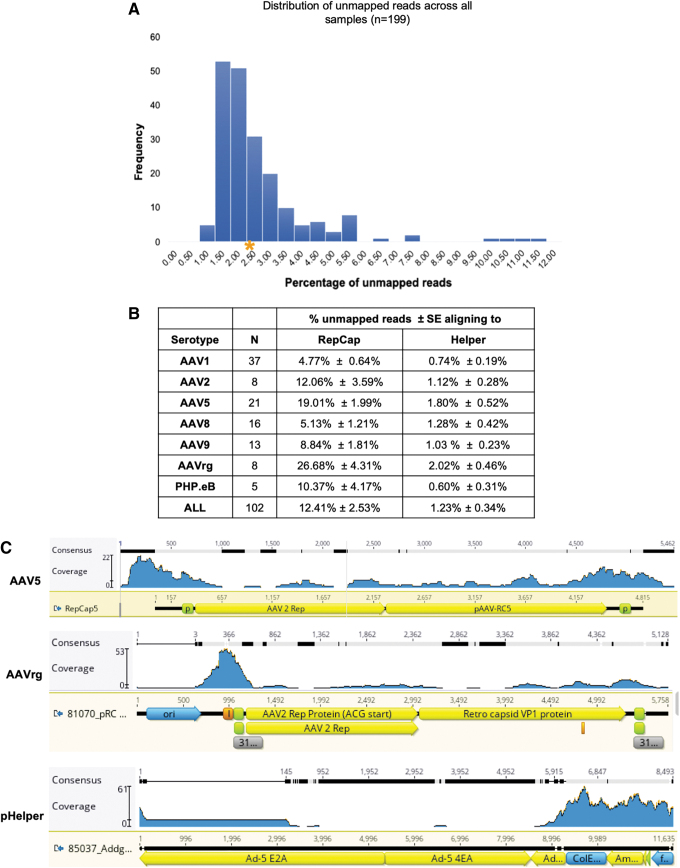
Figure 4.
Analysis of unmapped reads. (A) The percentage of unmapped reads for all samples was plotted, and the average (*) was calculated. (B) The average percentage of unmapped reads aligning to capsid plasmid or helper plasmid is listed. (C) Representative alignments of unmapped reads to an AAV5 capsid plasmid (top), AAVrg capsid plasmid (middle), or helper plasmid (bottom). AAV, recombinant adeno-associated virus. Color images are available online.
The percentage of packaged capsid sequences was significantly higher than that of the helper sequences (p-value ≤0.001); across serotypes, packaged capsid sequences ranged from 4.77% to 26.68% whereas those from the helper plasmid were only 0.74–2.02% (Fig. 4B). With an average of 26.7%, AAVrg had significantly higher levels of packaged capsid than all other serotypes tested, with the exception of AAV5 (p-value ≤0.05; Fig. 4B). To rule out differences due to the transgenes being expressed, pAAV-CAG-GFP and pAAV-hSyn-EGFP were packaged in each serotype and the percentage of unmapped reads aligning to the capsid was compared (Supplementary Tables S3 and S4).
For both viral vectors, AAV2, AAV5, and AAVrg had the highest levels of capsid packaging. Previous work examining rAAV packaging demonstrated that the p5 promoter elements present in the capsid plasmids are prone to packaging.12 Consistent with these findings, a clustering of reads around the p5 promoter region was observed in the samples (Fig. 4C, top panel). In addition, an adenoviral ITR element adjacent to one of the p5 promoters in the AAVrg capsid was frequently packaged (Fig. 4C, middle panel, peak at position 366). We speculate that this element may be structurally similar to the rAAV ITR elements, making it prone to packaging.
One concern, especially for those using AAV in the clinic, is the formation of a complete Rep gene if multiple AAV particles carrying different pieces of the Rep sequence infect the same cell. A fully functional Rep gene would render the virus replication competent and would be a serious safety concern.
We estimate that the DNA extracted for VGS contains between 4 × 1010 and 4 × 1011 GC. Despite the high number of genomes present, in the vast majority of cases, the Rep sequences present in a single sample do not span the entire gene. Given the low level of Rep coverage and the likelihood of the described coinfection event, the risk of generating a replication competent Rep is expected to be minimal. When reads that aligned to adenoviral helper genes were examined, the overall coverage was too low to identify any meaningful trends (Fig. 4C, bottom panel).
Although vector cores assess samples for titer and purity, verification of the serotype is not routinely done. Historically, serotype confirmation has required expensive and resource-intensive methods such as immunostaining and mass spectrophotometry precluding its incorporation into standard viral vector QC. Since DNA sequences from the capsid plasmids are often detected in the Megablast analysis stage, we hypothesized that we could use NGS reads to confirm the serotype of a sample. To address this, we developed a Python script that allows us to interrogate our VGS data for signature sequences that are unique to each serotype (Fig. 5A).
Figure 5.
Serotype determination from NGS reads. (A) Unique signature sequences from the capsid genes were identified for each serotype. (B) The percentage of correct serotype calls, percentage of calls returning no signature, and percentage of samples with additional calls in the serotype determination Python script are listed. (C) FASTQ files from a sample with a suspected mix-up were aligned to the PHP.eB, and AAV1 capsid sequences and the alignments were depicted. Black bars indicate a lack of consensus, whereas gray bars indicate good consensus with the reference sequence. AAV, recombinant adeno-associated virus; NGS, next-generation sequencing. Color images are available online.
The Python script analyzes multiple samples within minutes without the need for expensive reagents. The program tallies the incidence of the signature sequences and makes a serotype call based on the signature with the highest number of hits. One limitation of the program is that it is designed to search for exact matches to the signature and will not tally divergent signatures. In addition, for highly homologous capsids it may be difficult to identify unique signatures. To date, this script has been used to analyze more than 250 vectors. The program accurately detects greater than 90% of samples across all serotypes tested (Fig. 5B). In the cases where the program fails to call the expected serotype, it is not due to miscalling but instead is due to the absence of the signature sequence in the rAAV DNA extract.
The presence of a serotype's signature sequences in the NGS data depends on several factors such as the sequencing depth, rate of signature sequence packaging, and number of signatures being interrogated. For this dataset, the percentage of unmapped reads ranged from 1% to 12% (Fig. 4A). The percentage of these unmapped reads aligning to the capsid gene varied across serotype, with AAV1 having the lowest at 4.8% (Fig. 4B). At a sequencing depth of 10,000 reads, if the sample were at the low end of the distribution, only 1% or 100 reads would align to the capsid.
In the case of AAV1 where less capsid elements are packaged, only 4.8% or 4.8 of the unmapped reads would align to the capsid. Assuming equal packaging of the capsid sequence, 4.8 reads at a read length of 250 bp would span 1,200 bp and would not provide complete coverage of the ∼2,200 bp capsid gene. At 20,000 reads, coverage would span 2,400 and provide roughly 1 × coverage of the capsid gene. Consequently, we recommend 20,000 reads as a minimum for this Python script, especially if only one single signature sequence is used.
It should be noted that this recommendation is based on the assumption that all sequences of the capsid gene are packaged with the same efficiency. Given that previous work and these data have demonstrated that particular regions are more prone to packaging than others, this is likely not the case. Consequently, some signatures may be more likely to be present or absent than others. We recommend using as many unique signatures as possible to increase the likelihood of the program returning a serotype call.
Of note, for some samples the program detects additional signatures from unexpected serotypes. On closer examination, when this occurs, the hits tend to be from capsids that have a high level of homology and the rate of unexpected signatures is significantly lower than that of the expected capsid. For example, in one sample there were 57 hits to the expected AAV2 signatures and 2 hits to AAVrg, a derivative of AAV2.
Although most serotypes have a relatively low level of additional signature calls, the average for PHP.eB is quite high at 64% (Fig. 5B). This is due to the high degree of homology between PHP.eB and its parental capsid, AAV9. Despite designing a signature sequence in the area of divergence, AAV9 signatures are consistently observed. Thus far, the number of hits to the PHP.eB signature has been high enough to easily make the correct call; of 11 samples, the average occurrence per sample of the PHP.eB signature is 17.8 whereas that of the AAV9 is only 1.6. Given that most labs do not routinely use PHP.eB and the large disparity in the number of PHP.eB and AAV9 calls, we do not think this will be a problem for users choosing to adopt this analysis method.
During this analysis, one sample was identified in which the serotype determination software provided an unexpected serotype call. The sample was labeled as PHP.eB, yet the software identified 13 AAV1 signatures and no PHP.eB signatures. Although it is common to see additional AAV9 signatures in PHP.eB samples, AAV1 is completely unexpected, especially to such a high degree. To confirm the program's findings, the FASTQ files from the sample were first aligned to the reference cis plasmid and the unmapped reads were aligned to both the AAV1 and PHP.eB capsid sequences.
As indicated by black bars in Fig. 5C, there was no consensus between the unmapped reads and the expected PHP.eB capsid sequence. In contrast, although the overall coverage is low, there was very good consensus with the AAV1 capsid, especially at the 3′ end of the sequence (see gray bars in Fig. 5C). This confirmed that the serotype detection program was, indeed, correct and the sample had been inadvertently swapped with an AAV1 of the same cis plasmid. Importantly, this was missed in the manual FASTQ analysis and would have gone unnoticed had the data not been analyzed with the serotype detection software. This example serves to highlight the need for a serotype confirmation step in routine rAAV QC procedures and validates the serotype determination script as a robust detection method.
More recently, a thermostability-based approach termed AAV-ID was developed that can distinguish AAV serotypes based on melting temperature.27 AAV-ID can be done in a 96-well format and can yield results in as little as 6 h. Although AAV-ID is a marked improvement on previous methods, it does have some limitations. First, melting temperature will vary between different formulation buffers and can be affected by sample purity. In addition, some serotypes such as AAV6.2 and AAV9 have very similar melting temperatures despite limited homology in the VP3 protein and cannot be easily distinguished.
In cases such as this, where there is a high level of divergence of VP3 between serotypes, using a sequence-based approach for serotype identification would be beneficial. For viral cores producing large numbers of viral vectors, sequence-based identification and AAV-ID could be used as complementary methods of serotype confirmation. However, it is notable that if a core is already sequencing its rAAV preps, there is no additional cost to determine the serotype using the software, whereas additional lab work would be needed for the AAV-ID method.
The Cre-Lox system is a common tool used to control transgene expression. To obtain meaningful data when using the Cre-Lox system, it is critical that transgene expression is suppressed in the absence of the Cre recombinase. Consequently, scientists using Cre-dependent rAAV preparations are often concerned about promiscuous expression caused by random recombination events during plasmid propagation and viral packaging.
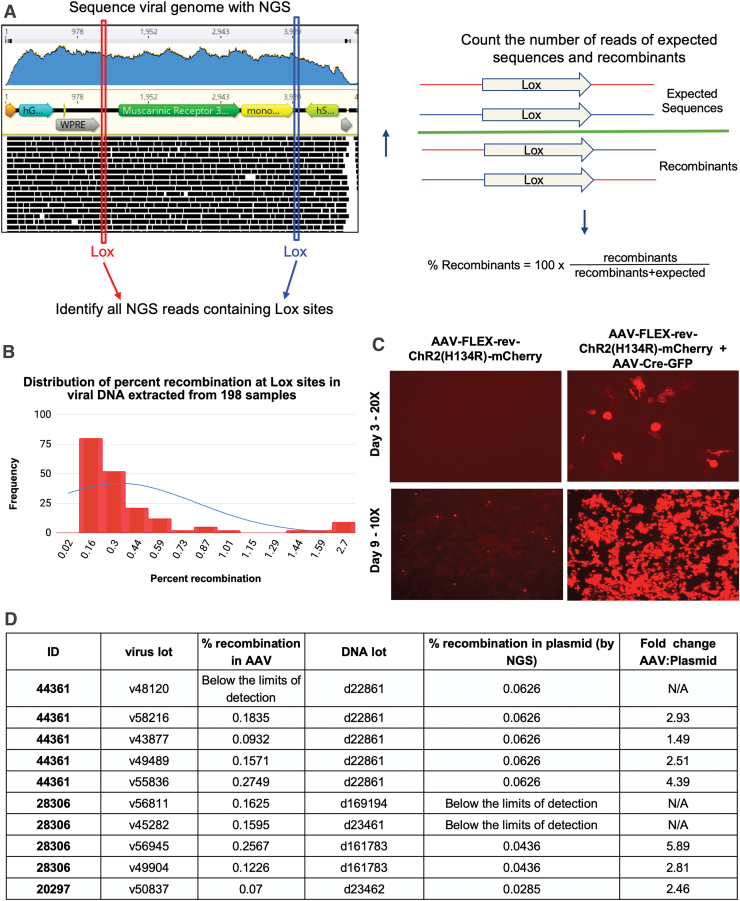
To address these concerns, we developed a Python script to interrogate VGS data for recombination events. Specifically, the program uses the Lox sequences as signatures and identifies which adjacent sequences are as expected and which could only exist if Cre-independent recombination occurred at those Lox sites. The color-coded cartoon in Fig. 6A shows the differences between the expected and recombined sequences. Using this program, we determined the relative recombination level in 198 rAAV preparations. Cre-independent recombination events range from 0% (below the limits of detection) to 2.70%, with most samples falling below a ∼0.2% recombination level (Fig. 6B).
Figure 6.
Analysis of promiscuous recombination at Lox sites in Cre-dependent viruses. (A) Schematic outlining the premise of the recombination detection Python script and the relative percentage of recombination at Lox sites across samples. (B) Frequency of the percentage of Cre-independent recombination at LOX sites from 198 samples. (C) Cells were infected with AAV-FLEX-rev-ChR2(H134R)-mCherry (AAV9-18916) in the presence and absence of Cre, and mCherry expression was assessed by direct fluorescence. (D) Comparison of the rates of Cre-independent recombination between plasmid DNA and its associated viral DNA extracts. Color images are available online.
Although the overall level of recombination across samples is quite low, there were some samples with a high percentage of recombinants. To determine whether recombinants could be detected in vitro, AAVpro cells were infected with AAV-FLEX-rev-ChR2(H134R)-mCherry at a high multiplicity of infection (MOI) of 2.6 × 106 GC/cell in the presence or absence of AAV-Cre-GFP.
mCherry expression was assessed by direct fluorescence up to 9 days after transduction. A high MOI was used as a worst-case scenario as to not limit the chances of observing Cre-independent expression. The lot of AAV-FLEX-rev-ChR2(H134R)-mCherry chosen had one of the highest recombination rates observed at 2.46%. Although mCherry expression was undetectable at 3 days, after incubating the sample for 9 days post-transduction, mCherry expression was present (Fig. 6C).
It should be stressed that this assay cannot be relied on as a detection method for leaky expression of Cre-dependent systems. To date, hundreds of samples have been analyzed and oftentimes, samples with high levels of recombinants are simply undetectable in vitro. Further, given the vast differences between viral transduction efficiency in vivo and in vitro, cell-based assays simply cannot be used to assume that a viral vector should or should not be used in vivo. Consequently, we strongly urge that all experiments with Cre-dependent constructs are designed with careful titration of doses and stringent controls and the optimal concentration is empirically determined.
Finally, the plasmid DNA used for viral production was compared with DNA extracts from the derived viral vector lots, to determine whether the Cre-independent recombinants were present in the cis plasmid used for transfection, or whether recombination occurred during AAV production in the mammalian packaging cells. The FASTQ data obtained therein were analyzed by using the recombination program, and the relative percentage of recombination was determined. Evidence of recombination was present in most of the plasmid stocks. In two cases, recombination in the DNA was not detected; of note, it cannot be assumed that recombination has not taken place, as the level of recombination might simply be below the threshold of detection.
In one lot of 44361 plasmid DNA, d22861, the level of recombination was measured at 0.06%. This lot of DNA was used to produce five rAAV lots in which the rate of recombination ranged from undetectable to 0.27%, which was 4.4-fold higher than that observed in the plasmid DNA (Fig. 6D). Of note, the number of reads for the sample in which recombination was not detected was only ∼16,000, limiting the likelihood of signature-containing reads. Therefore, it is recommended that the program only be used to analyze samples with >20,000 reads. The broad range of recombination across viral lots demonstrates that first, the rate of recombination in the viral preparation cannot be inferred from that of the plasmid and second, recombination is occurring during the packaging step in mammalian cells.
Herein, we describe a powerful tool to rapidly characterize the identity of purified AAV from rAAV DNA extracts that we term “VGS.” VGS does not require a double-stranding step before tagmentation, reducing both the time and cost of sample preparation. Without the double-stranding step, it is likely that the DNA extract is a heterogeneous mix of ss and ds species, a theory supported by the low level of intact SacII sites observed after enzyme digestion. Despite this, we achieve sequencing depth and coverage similar to that observed with the FAST-seq method.14
The VGS method combined with our open-source custom Python scripts allows scientists to quickly and reliably confirm the identity and serotype of their AAV preparations and detect low levels of rAAV cross-contamination and recombination events. For scientists handling or producing multiple viral vectors, this allows for validation similar to plasmid sequencing and, because samples can be batched together on a single lane for sequencing, prices can be kept low.
In addition to providing a rapid method of rAAV identification, VGS can be used to create sequence datasets for the study of rAAV genomics. In this study, we used the data to identify non-cis plasmid sequences present in viral preparations and examined the rates of promiscuous recombination in Cre-dependent viral vectors. By comparing the sequence of plasmids and then the viral genomes created from these plasmids, we can learn more about what happens during the packaging step and how factors such as differences in production facilities, reagents, and purification process affect packaging.
Supplementary Material
Acknowledgments
The authors thank Joanne Kamens, Lianna Swanson, Marcella Patrick, Will Arnold, and all other members of Addgene for their advice, helpful discussion, and support during the preparation of this article. The authors would also like to thank Benjie Chen for valuable advice on the Python scripts.
Author Disclosure
K.G., M.R., D.B., I.E., L.H., K.H.D., E.S., M.T., M.K., M.T., L.M.H., I.M., A.M., and M.F. are currently or have been employed by Addgene, a company that may be affected financially by the research reported in the enclosed article.
Funding Information
Addgene did not receive any funding for this work.
Supplementary Material
References
- 1. Wetterstrand KA. DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP). National Human Genome Research Institute; (updated January 15, 2016) [Google Scholar]
- 2. Allay JA, Sleep S, Long S, et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for hemophilia B clinical trial. Hum Gene Ther 2011;22:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chadeuf G, Ciron C, Moullier P, et al. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol Ther 2005;12:744–753 [DOI] [PubMed] [Google Scholar]
- 4. Wright JF, Zelenaia O. Vector characterization methods for quality control testing of recombinant adeno-associated viruses. Methods Mol Biol 2011;737:247–278 [DOI] [PubMed] [Google Scholar]
- 5. Ye GJ, Scotti MM, Liu J, et al. Clearance and characterization of residual HSV DNA in recombinant adeno-associated virus produced by an HSV complementation system. Gene Ther 2011;18:135–144 [DOI] [PubMed] [Google Scholar]
- 6. Noordman Y, Lubelski J, Bakker AC. Mutated rep encoding sequences for use in AAV production 2013. www.google.com/patents/US20130023034 (last accessed January10, 2020)
- 7. Nony P, Chadeuf G, Tessier J, et al. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J Virol 2003;77:776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hauck B, Murphy SL, Smith PH, et al. Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol Ther 2009;17:144–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu H, Qu G, Yang X, et al. Systemic elimination of de novo capsid protein synthesis from replication-competent AAV contamination in the liver. Hum Gene Ther 2011;22:625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kapranov P, Chen L, Dederich D, et al. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum Gene Ther 2012;23:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Ling C, Song L, et al. Limitations of encapsidation of recombinant self-complementary adeno-associated viral genomes in different serotype capsids and their quantitation. Hum Gene Ther Methods 2012;23:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tai PWL, Xie J, Fong K, et al. Adeno-associated virus genome population sequencing achieves full vector genome resolution and reveals human-vector chimeras. Mol Ther Methods Clin Dev 2018;9:130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lecomte E, Tournaire B, Cogné B, et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol Ther Nucleic Acids 2015;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maynard LH, Smith O, Tilmans NP, et al. Fast-Seq: a simple method for rapid and inexpensive validation of packaged single-stranded adeno-associated viral genomes in academic settings. Hum Gene Ther Methods 2019;30:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kennedy PJ, Feng J, Robinson AJ, et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci 2013;16:434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oh SW, Harris JA, Ng L, et al. A mesoscale connectome of the mouse brain. Nature 2014;508:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Atasoy D, Aponte Y, Su HH, et al. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci 2008;28:7025–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tervo DG, Hwang BY, Viswanathan S, et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 2016;92:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krashes MJ, Koda S, Ye C, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest 2011;121:1424–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan KY, Jang MJ, Yoo BB, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 2017;20:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suckling L, McFarlane C, Sawyer C, et al. Miniaturisation of high-throughput plasmid DNA library preparation for next-generation sequencing using multifactorial optimisation. Synth Syst Biotechnol 2019;4:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berns KI, Adler S. Separation of two types of adeno-associated virus particles containing complementary polynucleotide chains. J Virol 1972;9:394–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Horiuchi K, Zinder ND. Site-specific cleavage of single-stranded DNA by a Hemophilus restriction endonuclease. Proc Natl Acad Sci USA 1975;72:2555–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. York D, Reznikoff WS. DNA binding and phasing analyses of Tn5 transposase and a monomeric variant. Nucleic Acids Res 1997;25:2153–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ason B, Reznikoff WS. DNA sequence bias during Tn5 transposition. J Mol Biol 2004;335:1213–1225 [DOI] [PubMed] [Google Scholar]
- 26. Green B, Bouchier C, Fairhead C, et al. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob DNA 2012;3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pacouret S, Bouzelha M, Shelke R, et al. AAV-ID: a rapid and robust assay for batch-to-batch consistency evaluation of AAV preparations. Mol Ther 2017;25:1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.