Abstract
Lysophosphatidylserine (lyso-PS), a lysophospholipid derived from phosphatidylserine (PS), has emerged as a potent signaling lipid in mammalian physiology. In vivo, the metabolic serine hydrolases ABHD16A and ABHD12 are major lipases that biosynthesize and degrade lyso-PS respectively. Of biomedical relevance, deleterious mutations to ABHD12 causes accumulation of lyso-PS in the brain, and this deregulated lyso-PS metabolism leads to the human genetic neurological disorder PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract). While the roles of ABHD16A and ABHD12 in lyso-PS metabolism in the mammalian brain are well established, the anatomical and (sub)cellular localizations of both lipases, and the functional cross-talk between them towards regulating lyso-PS lipids remain under investigated. Here, using subcellular organelle fractionation, biochemical assays and immunofluorescence based high resolution microscopy, we show that the PS lipase ABHD16A is an endoplasmic reticulum (ER) localized enzyme, an organelle intricately regulating cellular PS levels. Further, leveraging immunohistochemical analysis using genetic ABHD16A and ABHD12 knockout mice as important controls, we map the anatomical distribution of both these lipases in tandem in the murine brain, and show for the first time, the distinct localization of these lipases to different regions and cells of the cerebellum. We complement the aforementioned immunohistochemical studies by quantitatively measuring lyso-PS concentrations in various brain regions using mass spectrometry, and find that the cerebellar lyso-PS levels are most affected by ABHD16A (decreased) or ABHD12 (increased) deletion. Taken together, our studies provide new insights into lyso-PS signaling in the cerebellum, the most atrophic brain region in human PHARC subjects.
Introduction
Understanding the molecular, biochemical and/or metabolic basis of several “orphan” human genetic neurological disorders, has only been possible because of the tremendous progress in whole genome sequencing technologies1. Following up on these genetic studies, complementary investigations have since revealed that deregulated lipid metabolism is central to a variety of human neurodegenerative disorders2–6. In 2010, a Norwegian family displaying Refsum-like neurological symptoms was clinically investigated, following which, their genomes were sequenced7. This genetic study found that this family harbored deleterious (null) mutations to the abhd12 gene on chromosome 20, that encodes the lipase α/β-hydrolase domain containing protein # 12 (ABHD12)7. Given the presentation of the symptoms clinically, this new autosomal recessive genetic neurological disorder was termed PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract) and deposited in the Online Mendelian Inheritance in Man (OMIM) database (ID: 612674)8. Since this study, several other genetic studies have mapped mutations (homozygous or compound heterozygous) in the abhd12 gene, all causing null full-length ABHD12 expression and in turn activity, leading to the symptoms of PHARC9–14. Most human PHARC subjects present with early onset (early teenage years) visual disturbances (cataract or retinal pigmentation) or hearing loss, with progressively worsening peripheral neuropathy and motor function (pes cavus), that eventually manifests into gross cerebellar atrophy8–14. At the time of this discovery, ABHD12 was tentatively annotated as a putative 2-arachidonoyl glycerol (2-AG) lipase, given its in vitro ability to hydrolyze the endocannabinoid 2-AG (along with monoacylglycerol lipase and ABHD6)15, and PHARC was thought to a genetic neurological disorder caused by defective endocannabinoid metabolism8.
In a seminal study to ascertain the biochemical and/or metabolic function of ABHD12, Cravatt and co-workers generated and rigorously characterized the ABHD12 knockout mouse, a murine model of PHARC16. Behaviorally, these mice mirror the age-dependent visual, auditory and motor defects seen in human PHARC subjects, and serve as an excellent pre-clinical model to investigate various aspects of this neurological disorder. The same study reported that the deletion of ABHD12 had no effect on the brain 2-AG levels16, and this meant two things: (i) in vivo, ABHD12 hydrolyzed some other lipid, and (ii) PHARC was most likely not a defect in endocannabinoid metabolism. To map the biological pathways regulated by ABHD12 in vivo, a comparative mass spectrometry based untargeted lipidomics experiment termed “discovery metabolite profiling”17 was performed on the brains of wild type and ABHD12 knockout mice. This study showed that deletion of ABHD12 resulted in a massive accumulation of the signaling lysophosphatidylserine (lyso-PS) lipids in the mouse brain. In the same study, biochemical substrate assays showed that ABHD12 efficiently hydrolyzed lyso-PS lipids in vitro, and together all of these studies conclusively annotated ABHD12 as a lyso-PS lipase16 (Figure 1). Based on the aforementioned findings, Cravatt and co-workers proposed a model for the disease progression, where as a function of age, the genetic deletion of ABHD12 results in sustained lyso-PS accumulation in the central nervous system (CNS), that in turn causes neuroinflammation from deregulated lyso-PS signaling, and eventually leads to progressively worsening neurobehavioral (auditory and motor) defects in ABHD12 null mice16.
Figure 1. The PS lipase and lyso-PS lipase activities catalyzed by ABHD16A and ABHD12 respectively.
Lyso-PS, a potent hormone-like signaling lysophospholipid, is present in all tissues, but is most abundant in the CNS (especially the brain) and in immune cells, where it regulates several physiologically important (immuno)biological processes like mast cell granulation18, macrophage activation and clearance of apoptotic cells19–21, facilitation of glucose metabolism in the brain and skeletal muscles22, 23, and chemotactic migration and stimulation of human cancer cells24–27. In the CNS, lyso-PS was thought to be biosynthesized from phosphatidylserine (PS) precursors via a PS lipase activity, and inhibitors to such a PS lipase might have tremendous therapeutic importance towards restoring the deregulated lyso-PS levels in the CNS, and in turn alleviating the pathophysiological effects seen in human PHARC subjects16. In search of such an enzyme, another previously unannotated lipase from the metabolic serine hydrolase family28, ABHD16A, was identified as a principal PS lipase in the CNS and in primary macrophages, where it biosynthesized lyso-PS lipids from PS precursors, and was found to function upstream of ABHD12 in regulating lyso-PS concentrations in vivo (Figure 1)29.
While the recent annotations of ABHD16A and ABHD12 as the principal PS lipase and lyso-PS lipase in the CNS respectively have significantly increased our understanding of lyso-PS metabolism29, very little is known of the anatomical, cellular and subcellular localization of these enzymes in the adult mammalian brain. We recently showed that ABHD12, at a subcellular level, is an integral membrane enzyme localized to the endoplasmic reticulum (ER) membrane in the mammalian brain30, where it regulates the secretion of very-long chain lyso-PS lipids from the ER16, 30. Following up on this finding, here, we report that the integral membrane PS lipase ABHD16A is also predominantly localized to the ER membrane in mammalian brain and cells, and we propose a model for the regulation of lyso-PS flux across the ER membrane from the interplay of these lipases. Further, using immunohistochemical analysis, we provide a detailed map of the cell specific anatomical localization of ABHD16A and ABHD12 in the mouse brain, using their respective knockout mice as important controls. Next, using substrate assays, and quantitative lipidomics, we measure PS/lyso-PS lipase activities and lyso-PS concentrations in different brain regions respectively, and correlate expression patterns to changes in lyso-PS levels following deletion of ABHD16A or ABHD12. Our findings taken together, provide new insights into the interplay of ABHD16A–ABHD12 and lyso-PS signaling in the adult mammalian brain, and a possible explanation as to why the cerebellum is the most affected brain anatomical region in human PHARC subjects.
Materials and Methods
Materials
Unless otherwise stated, all chemicals, buffers, solvents and reagents were purchased from Sigma-Aldrich, all lipids and lipid standards were purchased from Avanti Polar Lipids Inc., all tissue culture media, disposables and consumables were purchased from HiMedia, and all primary and secondary antibodies were purchased from Abcam.
Animal studies
All animal (mouse) studies described in the paper have received formal approval from the Institutional Animal Ethics Committee, IISER Pune (IAEC – IISER Pune), constituted as per the guidelines outlined by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India. All mice were maintained at the National Facility for Gene Function in Health and Disease (NFGFHD), IISER Pune, and had ad libitum access to water and food. All mice used in the study were generated by using a standard heterozygous x heterozygous breeding paradigm and the resulting litter were genotyped using previously reported protocols16, 29. This breeding paradigm afforded wild type mice with same-sex knockout littermates as controls for all experiments described in this paper.
Subcellular fractionation studies
The Neuro-2a cell line was purchased from ATCC and cultured in DMEM medium supplemented with 10% (v/v) Fetal Bovine Serum (FBS) and 1% (v/v) penicillin-streptomycin at 37°C in 5% CO2 (v/v) unless otherwise mentioned. The Neuro-2a cell line was routinely stained with 4’,6-diamidino-4-phenylindole (DAPI) using established protocols to ensure that it was devoid of any mycoplasma contamination31. Subcellular fractionation from tissues (mouse brain) and cells was performed to enrich the nuclear, mitochondrial, microsomal and cytosolic fractions using a protocol reported by us earlier30. The protein concentration of each subcellular fraction was estimated using Bradford reagent (Sigma-Aldrich), following which, the proteome from each fraction was denatured by boiling with 4X SDS loading dye at 95°C for 15 mins. This denatured proteome was separated over 10% SDS-PAGE gel and subsequently used for western blot analysis, to assess purity of enriched fractions, and detect the subcellular fraction to which ABHD16A enriches.
Western blot analysis
The proteins (typically 50 μg) were resolved on a 10% SDS-PAGE gel, transferred onto a methanol activated PVDF membrane (GE Healthcare) at 60V for 12 h at 4°C and processed post-transfer using a standard protocol reported by us earlier32. All primary and secondary antibody incubations described here were done at 1:1000 and 1:10,000 dilutions respectively. The blots were developed using the Thermo West Pico western blotting substrate (Thermo Fisher Scientific) and visualized on a Syngene G-Box Chemi-XRQ gel documentation system. The primary antibodies used in this study were: anti-ABHD16A (Rabbit, Abcam, 185549), anti-ABHD12 (Rabbit, Abcam, ab182011), anti-ATP5A (Mouse, Abcam, ab14748), anti-GM130 (Mouse, Abcam, ab169276), anti-Calnexin (Rabbit, Abcam, ab10286), anti-PMP70 (Mouse, Sigma-Aldrich, SAB4200181), anti-Lamin (Rabbit, Cloud Clone Corp, CAF548Hu01), anti-Tubulin (Rabbit, Cloud Clone Corp, CAB870Hu01), anti-GAPDH (Mouse, Cloud Clone Corp, CAB932Hu01) and anti-β-actin (Rabbit, Cloud Clone Corp, CAB340Hu01). The secondary antibodies used in this study were HRP-conjugated anti-Rabbit IgG (Goat, Thermo Fisher Scientific, 31460), and HRP-linked anti-Mouse IgG (Goat, Cloud Clone Corp, SAA544Mu19). All densitometric analysis of bands for quantitative western blot estimations were done using ImageJ 1.50i software (NIH, for Windows OS)33, 34.
Lipase substrate assays
All PS lipase and lyso-PS lipase substrate assays were performed using established liquid chromatography mass spectrometry (LC-MS) protocols reported earlier30. All lipase assays were done using 20 μg proteome on 100 μM lipid substrate (PS substrate: C18:0/18:1 PS, lyso-PS substrate: C18:1 lyso-PS) in 100 μL final volume for 30 mins. All free fatty acids released in these assays from the lipid substrates were detected using established high-resolution MS method on Sciex X500R QTOF instrument. The LC and MS parameters were identical to those reported earlier30. All MS data was acquired and analyzed using Sciex–OS software. All quantifications were done by calculating area under the curve for the desired fatty acid released and normalizing it to the area under the curve to internal standard (internal standard: C17:1 free fatty acid, 0.5 nmol per reaction) and the amount of protein used.
Cellular immunofluorescence studies
The Neuro-2a cell line was cultured as described earlier30, 32, and the live cells were counted using trypan blue method on a BTC20 automated cell counter (BioRad) as per as manufacturer’s instructions. 3 x 106 cells were seeded on an 18 mm coverslip in 35 mm tissue culture dish and processed thereafter using established protocols reported by us earlier30, 32. All confocal microscopy images were acquired on a Leica SP8 Confocal Laser Scanning Microscope fitted with 63X, 1.4 oil immersion objective. High resolution microscopy images were obtained on a Leica TCS STED super resolution microscope with HC PL APO CS2, 100X, 1.4 oil immersion objective using previously reported protocols30. All raw images were deconvoluted using Huygens Professional Software (Scientific Volume Imaging BV) and all microscopy images were analyzed using ImageJ 1.50i software (NIH, for Windows OS)33–35. The primary antibodies used in these studies were anti-ABHD16A (Rabbit, Abcam, ab1855409), anti-ABHD12 (Rabbit, Abcam, ab182011), anti-ATP5A (Mouse, Abcam, ab14748, mitochondrial marker), anti-PMP70 (Mouse, Sigma-Aldrich, SAB4200181, peroxisome marker), and anti-Sec63A (Mouse, Abcam, ab68550, endoplasmic reticulum marker). Goat anti-rabbit IgG (H&L) conjugated with DyLight 488 (Thermo Fisher Scientific, 35553) or goat anti-mouse IgG (H&L) conjugated with Alexa Fluor 568 (Thermo Fisher Scientific, A-11004) were used as secondary antibodies in this immunofluorescence study.
Harvesting thioglycollate elicited peritoneal macrophage
Peritoneal macrophages were elicited by injecting 3 mL of 4% (w/v) thioglycollate intraperitoneally using standard protocols reported earlier29, 31. Three days after injection, peritoneal macrophages were harvested in chilled Dulbecco’s phosphate buffered saline (DPBS) by a peritoneal lavage and the RBCs from the lavage were separated from the macrophages using RBC lysis buffer. Cells were pelleted at 200g for 3 mins and re-suspended in RPMI 1640 medium supplemented with 20% (v/v) FBS and 1% (v/v) penicillin-streptomycin (MP Biomedicals). Cells were cultured at 37°C, 5% CO2 (v/v) as described earlier, to yield approximately 15 x 106 cells per mouse. Lipopolysaccharide (LPS) treatment (5 μg/mL) of thioglycollate elicited peritoneal macrophages was done for 7 hours, and DPBS was used as a vehicle for these treatments. Cellular immunofluorescence and western blot analysis on primary macrophages were done as described in the sections above.
Immunohistochemical (IHC) analysis
Mice (10 – 12 weeks age) were deeply anaesthetized using isoflurane and subsequently perfused first with chilled phosphate buffer saline (PBS), followed by 4% (w/v) paraformaldehyde (PFA). Brains were dissected gently to maintain anatomical integrity, transferred 4% (w/v) PFA overnight (~ 16 hours) at 4°C and eventually transferred into 30% (w/v) sucrose solution in PBS until brains sank to bottom of the tubes (~ 3 days). Coronal sections (25 μm) were cut on a freezing microtome maintained at –30 °C, and sections were transferred to a reservoir containing ice-cold PBS. For DAB based brightfield IHC analysis, endogenous peroxidases were first inactivated with 3% (v/v) hydrogen peroxide in PBS for 15 min at 25°C in dark. Thereafter, the sections were washed twice with 1% (w/v) bovine serum albumin (BSA) in PBS and subsequently permeabilized with 0.1% (w/v) Triton X-100 and 0.5% (w/v) BSA in PBS for 45 mins at 25°C. The sections were then incubated with primary antibody in 1% (w/v) BSA overnight at 4°C, following which, the sections were washed three times with 0.5% (w/v) BSA in PBS and incubated with secondary antibody, biotinylated horse-anti rabbit [Vector labs, BP-1100] or biotinylated goat-anti mouse [Vector labs, BP-9200] in 0.5% (w/v) BSA in PBS for 1 hour at 25°C.
Following this treatment, the sections were washed twice in 0.5% (w/v) BSA in PBS, incubated with ABC Elite Vectastatin (Vector labs, PK-6100) for 1 hour at 25°C and subsequently washed twice with excess PBS. Finally, the sections were stained with ImmPACT DAB (Vector labs, SK-4105) for 3 min in dark, washed with PBS to remove excess DAB, and mounted using VectaMount (Vector labs, H-5000). Slides were imaged using an OptraSCAN Desktop or Precipoint M8 Digital Brightfield Microscope. For confocal laser scanning based IHC analysis, collected sections were directly permeabilized with 0.1% (v/v) Triton X-100 and 0.5% (w/v) BSA in PBS for 45 mins at 25°C and blocked with 1% (w/v) BSA in PBS for 1 hour at 25°C. Thereafter, sections were washed three times with 0.5% (w/v) BSA in PBS and incubated with primary antibody for 16 hours at 25°C. Sections were washed in 0.5% (w/v) BSA in PBS (3 times) and incubated with secondary antibody (1:1000 dilution) for 1 hour at 25°C. Sections were then washed with PBS and mounted with FluoromountTM aqueous mounting medium (Sigma-Aldrich). Slides were imaged using a Leica SP8 Confocal Laser Scanning Microscope. Primary antibodies used in the study were anti-ABHD16A [Rabbit, Abcam – ab185549, 1:100 dilution], anti-ABHD12 [Rabbit, Abcam – ab182011, 1:100 dilution], anti-NeuN [Mouse, Abcam – ab104224, 1:100 dilution], and anti-Calbindin [Mouse, Abcam – ab82812, 1:50 dilution]. Secondary antibodies used in the study were Goat anti-rabbit IgG (H&L) conjugated with DyLight 488 (Thermo Fisher Scientific, 35553) and goat anti-mouse IgG (H&L) conjugated with Alexa Fluor 568 (Thermo Fisher Scientific, A-11004).
Quantitative lipid measurements
Lipids (in particular PS and lyso-PS) were extracted with mass spectrometry grade organic solvents based the Folch extraction method with minor modifications and quantitatively estimated by LC-MS using a protocol previously reported by us31, 36. All lipids were quantified using an established multiple reaction monitoring high resolution (MRM-HR) method on Sciex X500R QTOF instrument36. Data was acquired and analyzed using Sciex–OS software. All quantifications were done by calculating the area under the curve for the desired analyte and normalizing it to the area for respective internal standard (lyso-PS and PS standards for absolute quantification, and free fatty acid standard for relative quantifications) and weight of the tissue or amount of protein as applicable.
Primary cerebellar neurons culture
Timed pregnancies were set up for wild type C57BL/6J mice, to afford pregnant female mice for harvesting embryos. To obtain an enriched pool of cerebellar neurons, the cerebellum was dissected from day 18 (E18) embryos and minced in DMEM medium supplemented with 10% (v/v) heat inactivated FBS and 1X antibiotic-antimycotic solution (Thermo Fisher Scientific). Minced tissue was digested with 0.25% (w/v) trypsin in PBS for 10 mins on ice, following which DNAse (0.1 mg/mL in PBS) was added for 5 mins on ice. Cells were pelleted down by spinning at 300g for 3 mins and washed twice with Hank’s Balanced Salt Solution. Cells were then re-suspended in Neurobasal Medium supplemented with 1X antibiotic-antimycotic solution (Thermo Fisher Scientific), 1X B-27 (Thermo Fisher Scientific), 1X glutamine (Thermo Fisher Scientific) at 37°C with 5% (v/v) CO2. Cells were cultured for 14 days, and media was changed every 2 days during this period. Purity of neurons was confirmed by cellular immunofluorescence and western blot analysis by probing for NeuN using established protocols reported in the sections above.
Statistical analysis
All data was plotted and statistical analysis was performed using GraphPad Prism 7 for Mac OS X software. All bar data is represented as mean ± standard deviation (s. d.) for three or more independent biological replicates per experimental group
Results
Subcellular localization of ABHD16A
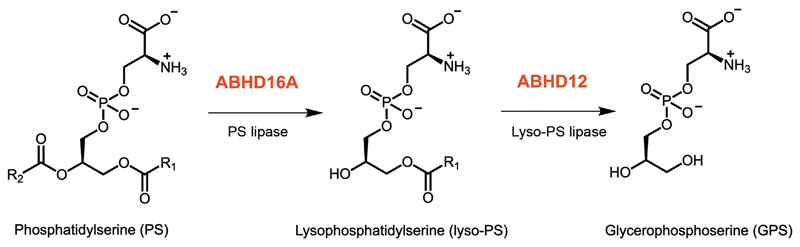
Previous protein deglycosylation studies and the detergent-independent sensitivity of ABHD16A to proteolytic degradation, have suggested that ABHD16A is an integral membrane lipase with a cytosolically oriented active site (α/β hydrolase domain)29 (Figure S1). To build on this finding, we first wanted to map the subcellular localization of ABHD16A. To address this, we used the biochemical organelle fractionation along with PS lipase substrate assays, followed by high resolution microscopy. Using established subcellular fractionation protocols30, we purified the cytoplasmic, microsomal, mitochondrial and nuclear fraction from mice brain (Figure 2A), and Neuro-2a cells, an immortalized mammalian neuronal cell line (Figure 2B). In accordance with literature precedence, we confirmed by western blot analysis, that these subcellular fractions were approximately 90% pure by probing for an organelle specific marker (ATPase5a for mitochondria, Lamin for nucleus, Calnexin for the ER, PMP70 for peroxisomes, GM130 for the Golgi and Actin for cytosol). Next, using western blot analysis, we probed for ABHD16A in all these subcellular fractions, and found in both the mouse brain (Figure 2A), and in Neuro-2a cells (Figure 2B), that ABHD16A was most enriched in the microsomal fraction. To corroborate the results from the western blot experiments, we performed PS lipase activity assay using C18:0/18:1 PS (stearoyl-oleayl phosphatidylserine, SOPS) as a substrate on the different subcellular organelle fractions obtained from the mouse brain and Neuro-2a cells. Consistent with the enrichment of ABHD16A to the microsomal fraction, we found that the specific PS lipase activity was highest in the microsomal fraction obtained from both the mouse brain (Figure 2C) and Neuro-2a cells (Figure 2D) as compared to any other subcellular fraction. The β-lactone KC01 is a potent inhibitor of mammalian ABHD16A, while the β-lactone KC02 serves as a structurally similar inactive control to assay for mammalian ABHD16A activity29, 37. When all subcellular fractions were pre-treated with KC01 or KC02 (both at 1 μM, 1 hour, 37°C), we found that the microsomal PS lipase activity in particular, was significantly inhibited by KC01, but not KC02, for both the mouse brain (Figure 2C) and Neuro-2a samples (Figure 2D). This result confirmed that the majority of the PS lipase activity from the purified microsomal fraction was indeed coming from ABHD16A, matching the microsomal enrichment profile for this lipase.
Figure 2. ABHD16A is enriched in the microsomal fraction.
Biochemical fractionation of various subcellular organelles from (A) mouse brains, and (B) Neuro-2a cells, shows that ABHD16A is enriched in the microsomal fraction. This experiment was repeated three times with reproducible results each time. Consistent with the localization studies, the PS lipase activity is also enriched in the microsomal fractions from (C) mouse brains, and (D) Neuro-2a cells. In both (C) and (D), the PS lipase activity is inhibited by KC01 but not KC02, consistent with this activity coming from ABHD16A. Data represents mean ± standard deviation for three biological replicates per group.
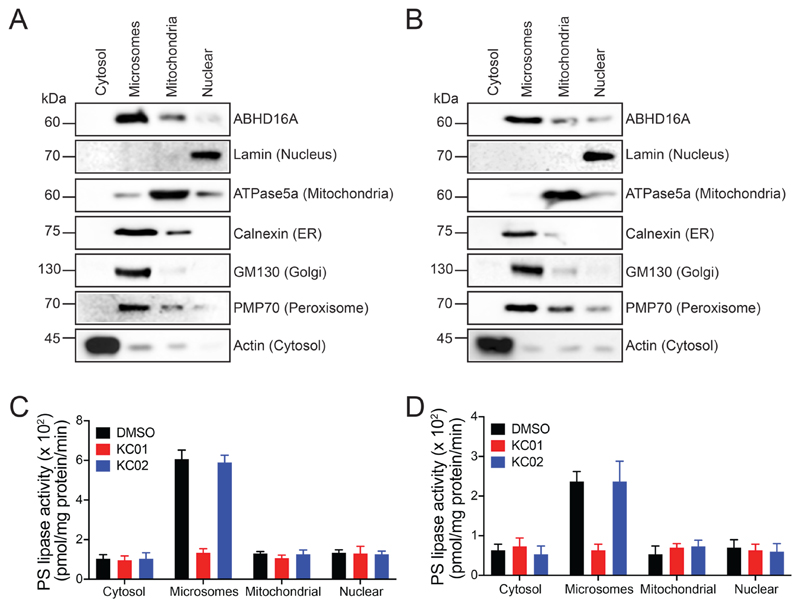
As reported by us earlier30, the microsomal fraction consists mostly of proteins from the ER, peroxisomes, Golgi and the plasma membranes. Given that most of the intracellular PS is localized to ER, peroxisomal and mitochondrial membrane38, we hypothesized, that ABHD16A would likely be localized to the ER or the peroxisomes. Using high resolution stimulated emission depletion (STED) microscopy, we found in Neuro-2a cells that ABHD16A co-localizes very well with Sec63a, an ER marker (Figure 3A). In the same cell line, we did not find any significant co-localization of ABHD16A with PMP70, a peroxisome marker or ATP5a, a mitochondrial marker, to rule out its presence of ER-mitochondrial contact sites (Figure S2). We have previously shown that mammalian ABHD12 is enriched at the ER membrane30, and here, we checked for the co-localization of ABHD16A with ABHD12 using STED microscopy. Indeed, we found in Neuro-2a cells, that ABHD16A co-localizes with ABHD12, further confirming that ABHD16A is enriched at the ER membrane (Figure S3). To ensure that the cellular localization studies were not an artefact of an immortalized cell line, we generated and cultured primary cerebellar neurons using standard protocols. Here, using STED microscopy, we found that ABHD16A co-localized well with Sec63a, thus confirming the subcellular localization of ABHD16A at the ER membrane even in primary neurons (Figure 3B).
Figure 3. ABHD16A is localized to the ER.
Cellular immunofluorescence studies in (A) Neuro-2a cells and (B) primary cerebellar neurons, show significant co-localization of ABHD16A with Sec63A (an ER marker). Bars on normal images are 5-μm, while bar on zoomed image is 2-μm. This experiment was repeated three times with reproducible results each time.
Mapping ABHD16A–ABHD12 localization in primary peritoneal macrophages upon an immune challenge
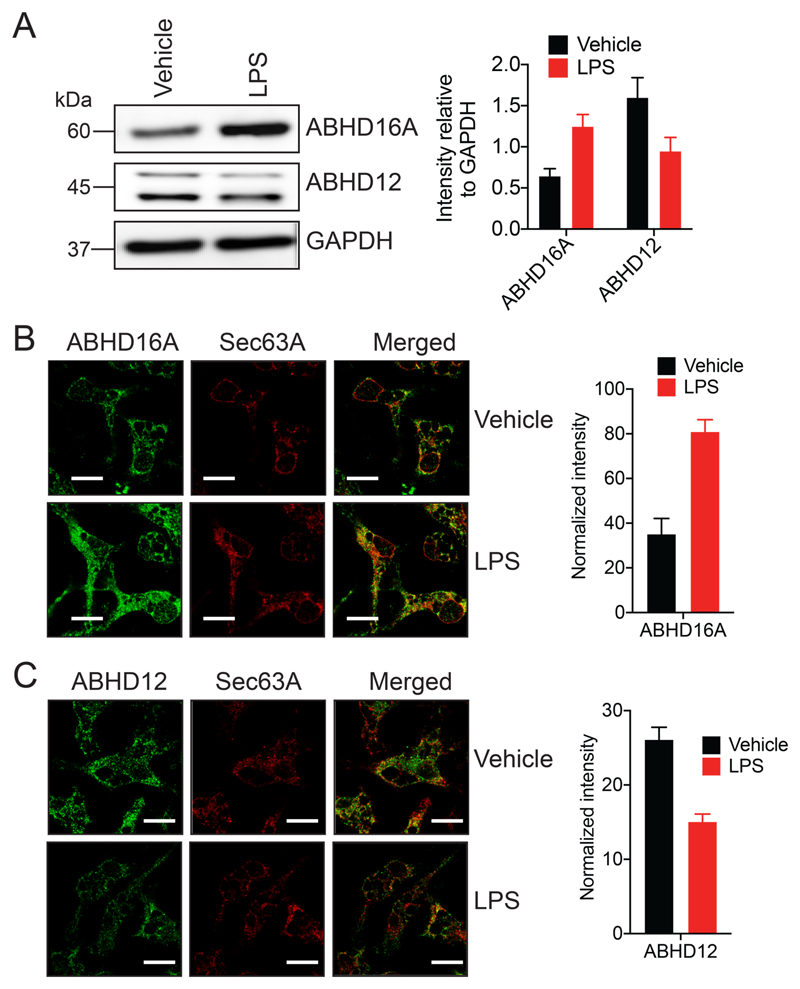
Previous large-scale gene expression studies39, 40 and biochemical characterization of the activities of ABHD16A and ABHD12 in primary peritoneal macrophages (PPMs) elicited using thioglycollate29, have shown that upon lipopolysaccharide (LPS) stimulation (1 μg/mL, 7 hours at 37°C), the activity of ABHD16A increases (~ 2.5 fold), while that of ABHD12 decreases (~ 2-fold), thereby increasing the secreted lyso-PS concentrations from PPM and enabling the resolution of an inflammatory stimuli29. However, it is unclear during an inflammatory stimulus, whether or not, the subcellular localization of ABHD16A and/or ABHD12 change, and we wanted to investigate this. Previously, we have reported using cellular immunofluorescence studies that ABHD12 in localized to the ER in PPM, using PPMs derived from ABHD12 knockout mice as a control for this assay30. Following up on this study, we first wanted to perform the same study on PPMs obtained from wild type (WT) or ABHD16A knockout mice, to determine the subcellular localization of ABHD16A and to ensure that our immunofluorescence readout of ABHD16A in PPM was accurate. Consistent with findings from the mouse brain and Neuro-2a cells, we found that WT PPM showed ER localization of ABHD16A, and this immunofluorescent signal was significantly diminished in PPM derived from ABHD16A knockout mice (Figure S4).
To validate previous findings, we first assessed by quantitative western blot analysis, the levels of ABHD16A and ABHD12 following LPS treatment of WT PPM (1 μg/mL, 7 hours at 37°C), and found that upon LPS stimulation, the total protein levels (and in turn activity) of ABHD16A and ABHD12 were increased (~ 2.5 fold) and decreased (~ 1.8 fold) respectively relative to a GAPDH loading control (Figure 4A). Having confirmed these protein level changes, we next asked if the localization of ABHD16A and/or ABHD12 changed upon LPS stimulation using our established cellular immunofluorescence assays. We found from this assay, that the upon LPS treatment of WT PPM (1 μg/mL, 7 hours at 37°C), the total cellular protein levels of ABHD16A increased > 2-fold (Figure 4B), while that of ABHD12 decreased by ~ 2-fold (Figure 4C) consistent with all previous studies29. Interestingly however, we found that for vehicle or LPS treated WT PPMs, both ABHD16A and ABHD12 were still predominantly localized to the ER membrane (Figure 4B, 4C), suggesting negligible changes (if any) in cellular localization upon LPS stimulation.
Figure 4. ABHD16A and ABHD12 are ER localized in primary peritoneal macrophages (PPM) after an inflammatory stimulus.
(A) Representative blot and quantitation from western blot analysis of PPM, showing an increase and decrease in ABHD16A and ABHD12 levels respectively following LPS treatment. This experiment was done three times with reproducible result each time. (B, C) Representative images and quantitation from cellular immunofluorescence analysis of PPM, showing an increase and decrease in (B) ABHD16A and (C) ABHD12 levels respectively following LPS treatment. This experiment was done four times with reproducible result each time. Data represents mean ± standard deviation for three or four biological replicates per group. Bars on images in (B) and (C) are 5-μm.
Mapping the gross anatomical localization of ABHD16A, ABHD12 and lyso-PS in the adult mouse brain
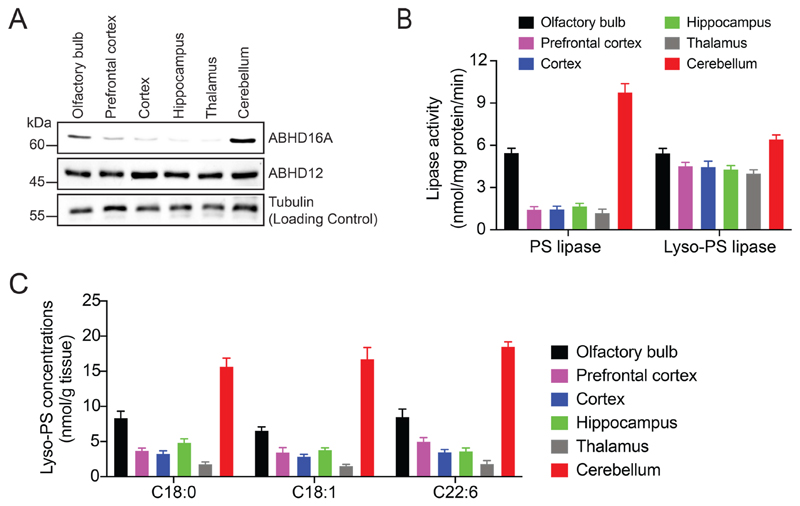
Having mapped the subcellular localization of ABHD16A and ABHD12 in the mouse brain and, in mammalian immortalized and primary cells, we next asked where in the adult mouse brain were both these lipases’ maximally enriched. To address this, we first micro-dissected the different anatomical regions of the brain namely the olfactory bulb, prefrontal cortex, cortex, hippocampus, thalamus, and cerebellum from adult WT mice (10 -12 weeks age). We prepared membrane proteomes of these brain regions using standard protocols30, and probed them for ABHD16A and ABHD12 expression using western blot analysis. We found relative to the tubulin protein loading control, that ABHD16A was maximally enriched in the cerebellum, and the olfactory bulb, while ABHD12 is ubiquitously expressed at near equal levels in all the aforementioned brain regions (Figure 5A). Next, we measured the specific PS lipase and lyso-PS lipase activities for the membrane proteomes of the different micro-dissected brain anatomical listed above using standard protocols30. We found that consistent with the ABHD16A and ABHD12 expression profiles, the specific PS lipase activity was highest in the cerebellum, and the olfactory bulb, while the specific lyso-PS lipase activity was nearly equivalent in all brain regions (Figure 5B). In the cerebellum, we confirmed by treatment with a selective inhibitor29, that ABHD16A was indeed contributing to the majority of the PS lipase activity (Figure S5).
Figure 5. Mapping the brain distribution of ABHD16A, ABHD12 and lyso-PS lipids.
(A) Western blot analysis on membrane lysates from different anatomical regions of the brain showing enrichment of ABHD16A to the cerebellum, while ABHD12 is more ubiquitously expressed in the brain. Tubulin was used as a loading control for this experiment. This western blot analysis was done three times with reproducible results each time. (B) Lipase activity profiles on membrane lysates from different anatomical regions of the brain showing enrichment of the PS lipase activity in the cerebellum, while lyso-PS lipase activity is comparable across all regions of the brain. Data represents mean ± standard deviation for three biological replicates per group. (C) Quantitative lyso-PS measurements on abundant lyso-PS species (C18:0, C18:1 and C22:6) on different anatomical regions of the brain showing enrichment of these lipids to the cerebellum. Data represents mean ± standard deviation for four biological replicates per group.
To determine if there was any correlation between the expressions of ABHD16A/ABHD12 and their respective lipase activities to lyso-PS concentrations in different brain regions, we quantitatively measured abundant lyso-PS lipids (C18:0, C18:1 and C22:6) in the aforementioned micro-dissected brain regions using established targeted LC-MS16, 29. Interestingly, we found that the cerebellum had the highest lyso-PS concentration, followed by the olfactory bulb (2-fold less that cerebellum), with all other brain regions having near equal lyso-PS concentrations, approximately 4 to 5-fold less than that of the cerebellum (Figure 5C). Consistent with ABHD16A being the major lyso-PS biosynthetic enzyme (PS lipase) in the mammalian brain29, its heightened cerebellar expression (and in turn activity) correlates very well with the cerebellum having the highest lyso-PS concentration in the adult mouse brain. Further, since ABHD12 expression (and in turn activity) is ubiquitous in the brain, it seems likely that the varying lyso-PS concentrations observed in the different anatomical regions of the WT adult mouse brain under normal conditions, are largely governed by the expression (and in turn activity) of ABHD16A.
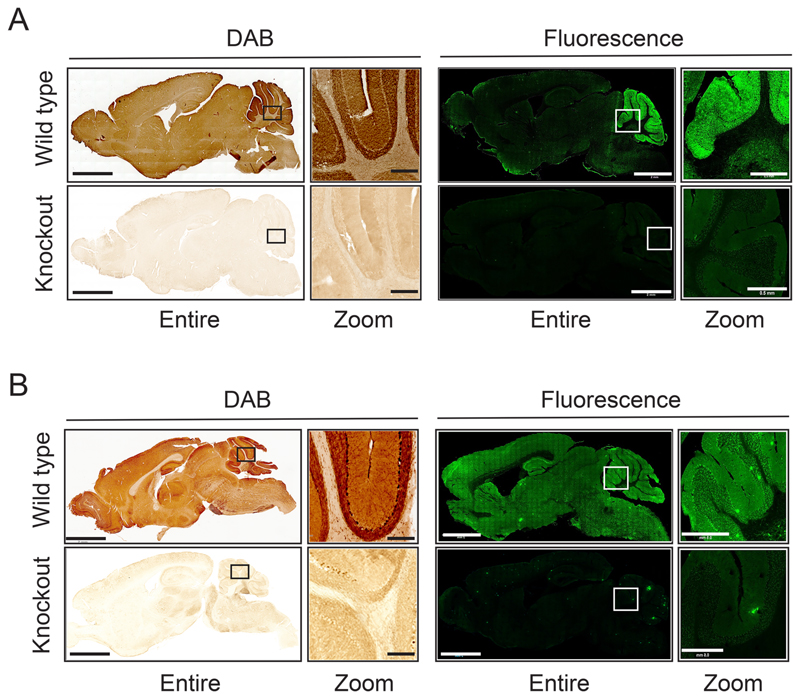
To complement these findings, we used two well established immunohistochemistry methods: a 3,3’-diaminobenzidine (DAB) based method and the fluorescence based confocal microscopy method to visualize the localization of ABHD16A and ABHD12 in whole brain sagittal sections. Previous studies on the brains of ABHD12 knockout mice have revealed no anatomical defects16 but such a validation study on the brains of ABHD16A knockout mice is currently lacking. To ensure that the loss of ABHD16A activity does not affect the gross brain anatomy, we performed Cresyl Violet (Nissl) and Hematoxylin and Eosin (H&E) staining on coronal sections on brains derived from WT and ABHD16A knockout mice, and found that the gross anatomy of the brain does not change upon ABHD16A deletion (Figure S6). From the immunohistochemical analysis using their respective knockout mice as controls, we found in sagittal brain sections by both DAB and immunofluorescence-based staining, that consistent with all previous findings, ABHD16A was significantly enriched in the cerebellum compared to any other brain region (Figure 6A), while ABHD12 was more ubiquitous, and found enriched in the cerebellum, hippocampus and cortex (Figure 6B). To ensure that the results from the sagittal sectioning were not artefacts, we also performed DAB staining on coronal sections from WT mice using ABHD16A or ABHD12 knockout mice as controls, and found similar results from this immunohistochemical study (Figure S7).
Figure 6. Mapping the anatomical distribution of ABHD16A and ABHD12 using immunohistochemical (IHC) analysis.
Representative images from the IHC analysis on sagittal brain sections from (A) wild type and ABHD16A knockout mice and (B) wild type and ABHD12 knockout mice, using DAB staining or immunofluorescence. For both the DAB based staining and immunofluorescence experiments of the sagittal sections, the left image represents the entire sagittal section, while the right image represents a zoomed section of the cerebellum. Knockout controls for both IHC analysis show very little signal. Bars on entire sagittal image are 2 mm, while bars on zoomed cerebellar image are 0.5 mm. The IHC experiments were done four independent times with reproducible results each time.
Effect of ABHD16A or ABHD12 deletion on lyso-PS and PS concentrations in different anatomical regions of the brain
Previous quantitative lipidomic measurements have shown that the deletion of ABHD16A and ABHD12 result in decreased and increased bulk lyso-PS levels in the mammalian brain respectively16, 29. Interestingly, ABHD12 knockout mice brains, also show accumulation of arachidonoyl (C20:4) containing PS lipids16, recently shown to be through the activation of MBOAT5, a C20:4 specific acyltransferase, that converts excess lyso-PS back to PS41, while ABHD16A knockout mice brains show no changes in bulk PS levels29. So, having mapped the gross anatomical distribution of ABHD16A and ABHD12 in the adult mouse brain, we next asked, what was the effect of deletion of ABHD16A or ABHD12, on the lyso-PS and PS levels in specific anatomical regions of the adult brain. To address this, we again harvested the olfactory bulb, prefrontal cortex, cortex, hippocampus, thalamus, and cerebellum from age and gender matched: (i) WT and ABHD16A knockout mice, or (ii) WT and ABHD12 knockout mice (10 -12 weeks age); and measured their relative lyso-PS and PS content using established targeted LC-MS analysis16, 29, 31.
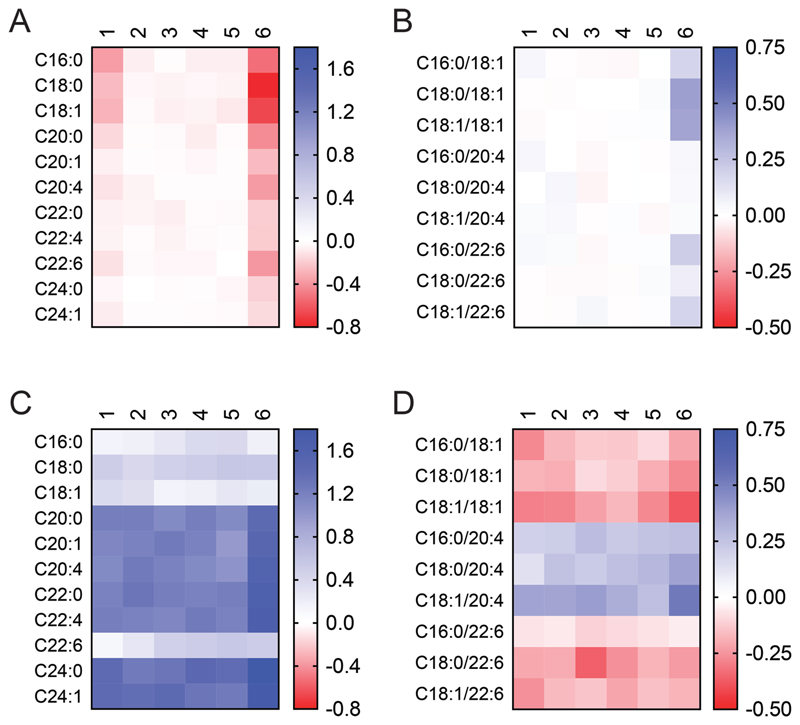
Consistent with the enriched cerebellar localization of ABHD16A, we found that its deletion resulted in a significant decrease of all lyso-PS species (especially long chain lyso-PSs) in this brain region (Figure 7A). We also found a modest decrease in long chain lyso-PSs in the olfactory bulb, where ABHD16A is also enriched, while all other brain regions showed negligible to no changes for the different lyso-PS lipids (Figure 7A). Interestingly, we also found that disruption of ABHD16A resulted in a modest accumulation of a handful PS lipid species esterified with long chain fatty acids only in the cerebellum, while all other brain regions including the olfactory bulb, showed little to no changes in brain PS profiles (Figure 7B). On the other hand, we found that the deletion of ABHD12 resulted in accumulation of lyso-PS lipids, in particular very long chain lyso-PSs, throughout the brain (Figure 7C). The accumulation of lyso-PSs in ABHD12 knockout mice brains was most profound in the cerebellum and the olfactory bulb (Figure 7C), consistent with ABHD16A expression, and its role as the principal lyso-PS biosynthetic enzyme in these brain regions. Interestingly, across the different brain regions of ABHD12 knockout mice, we found that several PS lipids were significantly decreased, while C20:4 containing PS lipids were elevated (Figure 7D), consistent with previously reported PS measurements from the whole brain16. Of note, the deregulation of PS lipids from ABHD12 knockout mice was most pronounced in the cerebellum (Figure 7D).
Figure 7. Relative lyso-PS and PS measurements in different brain regions following deletion of ABHD16A or ABHD12.
(A) Lyso-PS, and (B) PS concentrations in the different regions of the brain of ABHD16A knockout mice relative to corresponding regions from wild type mice. (C) Lyso-PS, and (D) PS concentrations in the different regions of the brain of ABHD12 knockout mice relative to corresponding regions from wild type mice. The relative data for each lyso-PS or PS species was obtained by taking the ratio of the knockout value to the wild type value, where the respective value was calculated as an average of independent 4 biological replicates per genotype. The scale adjoining the heat map plot is log10(ratio of knockout value/wild type value). The numbers denoted above each heat map plot correspond to different brain regions: 1 = olfactory bulb, 2 = prefrontal cortex, 3 = cortex, 4 = hippocampus, 5 = thalamus, and 6 = cerebellum. For comparison purposes, (A) and (C), and (B) and (D) are plotted on the same logarithmic scale.
Mapping the cell type distribution of ABHD16A and ABHD12 in the cerebellum
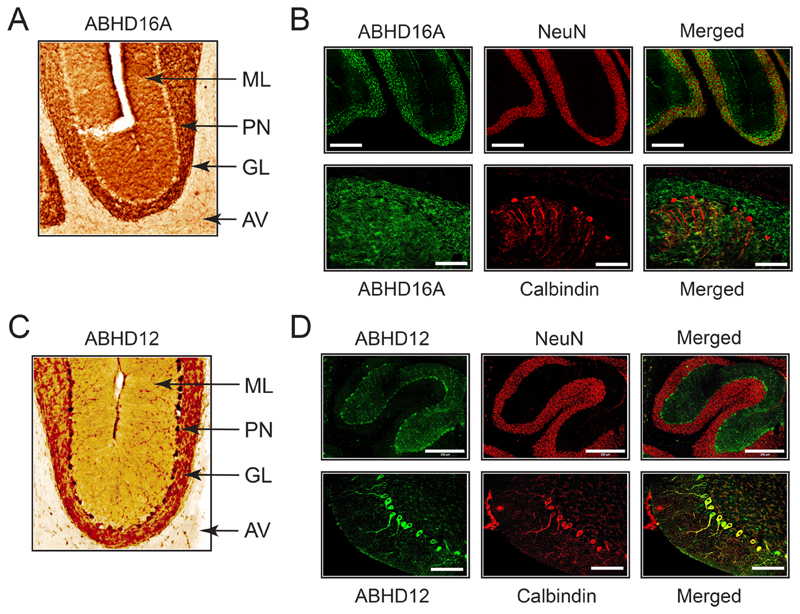
Since the cerebellum is most affected brain region in human PHARC subjects8, and because it shows the highest lyso-PS deregulation in both ABHD16A and ABHD12 knockout mice, we next wanted to understand the cerebellar localization of both ABHD16A and ABHD12. To address this, we obtained cerebellar (sagittal and coronal) sections from adult WT mice (10 - 12 weeks of age) using their respective knockout mice (ABHD16A or ABHD12) as age and gender matched controls, and assessed the localization of ABHD16A and ABHD12 using DAB and fluorescence based immunohistochemical analysis. In our fluorescence based immunohistochemical analysis, alongside the antibodies of our lipases’ of interest (ABHD16A and ABHD12), we used the established anti-NeuN42 and anti-Calbindin43 antibodies as specific markers for the granular layer neurons, and Purkinje neurons respectively, to assess the specific cerebellar localization of ABHD16A and ABHD12.
From the DAB based immunohistochemical analysis, we found that ABHD16A is maximally enriched in the granular (and perhaps molecular) layers of the cerebellum but not in the cerebellar ganglionic layer (Figure 8A). We found from the fluorescence based immunohistochemical analysis that ABHD16A is co-localized with NeuN, but not with Calbindin, suggesting that ABHD16A is present in granular neurons, but not in Purkinje neurons (Figure 8B). Though not conclusive from our results, given its expression in the molecular layer from DAB IHC experiments, it is likely that ABHD16A may also be expressed in the parallel fibers of molecular layer which originate from the granular neurons, and synapse with Purkinje neurons. Interestingly, despite the ubiquitous expression of ABHD12 in the adult mouse brain, within the cerebellum, we found by DAB based immunohistochemical analysis, significant enrichment of ABHD12 in the ganglionic layer of the cerebellum (Figure 8C), suggesting that ABHD12 was likely present on Purkinje neurons. Indeed, fluorescence based immunohistochemical analysis also showed that ABHD12 co-localized with Calbindin, but not NeuN, and confirmed that this lipase was localized to Purkinje neurons (Figure 8D). To ensure that the deletion of ABHD16A or ABHD12 did not cause any structural abnormalities in Purkinje neurons, we also assessed cerebellar sections from the respective knockout mice, and found by DAB and fluorescence based immunohistochemical analysis, that morphology, number and integrity of Purkinje neurons from knockout mice was near identical to those from WT mice (Figure S8). Taken together, these studies show for the first time, the discrete localization of ABHD16A and ABHD12 within the cerebellum, and suggest for the first time, that cerebellar lyso-PS signaling might be paracrine in nature.
Figure 8. Mapping the anatomical localization of ABHD16A and ABHD12 in wild type mouse cerebellum.
(A) Representative image from DAB based IHC analysis for ABHD16A in a sagittal cerebellar section. (B) Representative image from fluorescence based IHC analysis of ABHD16A, in conjunction with NeuN and Calbindin. (C) Representative image from DAB based IHC analysis for ABHD12 in a sagittal cerebellar section. (D) Representative image from fluorescence based IHC of ABHD12, in conjunction with NeuN and Calbindin. Note: images represented in (A) and (C) are obtained from the sagittal sections of WT mice represented in Figure 6. For (A) and (C), ML = molecular layer, PN = Purkinje neurons, GL = granular layer, and AV = arbor vitae. For (B) and (D), bars for the NeuN IHC panel are 300 μm, while bars for the Calbindin panel are 100 μm.
Discussion
Deregulated lipid signaling pathways have been attributed as a primary cause to several human neurological diseases2–6, and understanding the biochemical basis for such disorders are important towards developing therapies for these. Recently, the early onset progressively debilitating neurodegenerative disease PHARC, caused by deleterious mutations to a gene that encodes the lipase ABHD12, was shown to involve a deregulated lipid signaling pathway, namely lyso-PS8, 9, 16. The integral membrane serine hydrolase enzyme ABHD12 has been annotated as a principal lyso-PS lipase in the mammalian brain, where it terminates lyso-PS signaling through its lipase activity (Figure 1)16, and its deletion results in gross accumulation of this lipid class in the CNS, where sustained deregulated signaling causes the pathophysiology of this neurological disease16. An upstream PS lipase, ABHD16A, was also recently identified as a principal lyso-PS biosynthetic enzyme in the CNS (Figure 1)29, and inhibitors to this enzyme, can in principle reverse the PHARC pathophysiology by preventing lyso-PS biosynthesis. While the biochemical activities of both these lipases are well understood, their (sub)anatomical and subcellular localizations, and the crosstalk between them, particularly in the brain remain relatively poorly understood. To understand this, in this paper, using biochemical assays, LC-MS based lipidomics, along with cellular immunofluorescence and tissue immunohistochemical analysis, here we set out to map the neuroanatomy of ABHD16A and ABHD12 and expand our current understanding as to how this localization likely influences brain lyso-PS distribution and signaling.
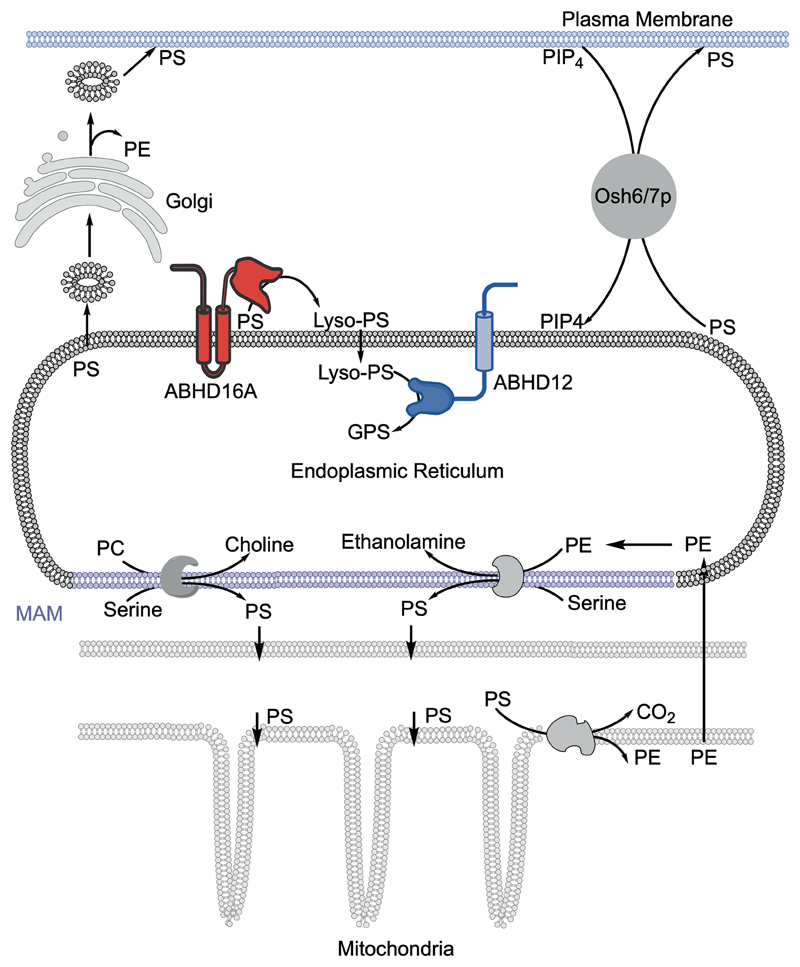
First, we show using biochemical subcellular fractionation (Figure 2) and cellular immunofluorescence assays (Figure 3) that the PS lipase ABHD16A, like ABHD1230, is an ER membrane localized lipase in the brain and neuronal cells. We also show in PPM, where upon LPS treatment the expression (and in turn activity) of ABHD16A increases and ABHD12 decreases29, that the ER localization of both these enzymes remains unchanged upon this inflammatory stimulus (Figure 4). PS regulates many facets of mammalian physiology, and several studies have shown in metabolism across various cellular components38, 44. PS is synthesized by phosphatidylserine synthase 1 (PSS1) and 2 (PSS2) at mitochondrial associated membranes (MAMs) from other phospholipid precursors, and is subsequently fluxed to mitochondria and metabolized to phosphatidylethanolamine (PE) or is directly transported from MAMs to plasma-membrane in exchange of phosphatidylinositol-4-phosphate (PIP4). PS is also transported to plasma membrane as vesicles through the Golgi network (Figure 9)38, 44. Quite interestingly, these lipid cell biological studies have also shown that PS is cytosolically oriented across the ER membrane38, 44. That ABHD16A is ER localized, and has an active site which is cytosolically oriented, enables it to access PS on ER membranes to biosynthesize lyso-PS lipids (Figure 9). We hypothesize that the while ABHD16A is continually making lyso-PS lipids, a small fraction of this is likely secreted from cells towards homeostatic signaling, while the remaining fraction in metabolized on the ER membrane by the lyso-PS lipase ABHD12 to recycle the head group for PS production (Figure 9). Thus, our mapping of ABHD16A here, along with previous reports of ABHD12’s ER localization30, expand our current understanding of PS metabolism in the ER (Figure 9).
Figure 9. Schematic representation of PS metabolism in the ER.
The mitochondrial associated membrane (MAM), where PS biosynthesis occurs is shown in purple. PC = phosphatidylcholine, PE = phosphatidylethanolamine, PIP4 = phosphatidylinositol-4-phosphate.
Second, we show using immunoblotting, biochemical assays, LC-MS based lipidomics measurements and immunohistochemical analysis, that PS lipase ABHD16A, and the brain’s specific PS lipase activity are enriched in the cerebellum, while lyso-PS lipase ABHD12 and the brain’s specific lyso-PS lipase activity are ubiquitous (Figure 5, 6). Given this disparity, lyso-PS lipids are enriched in the cerebellum, where the principal PS lipase ABHD16A resides (Figure 5). Under normal conditions ABHD16A seems to exert more influence on lyso-PS brain distribution, and we find that deletion of ABHD16A causes a decrease in lyso-PS lipids, and a modest corresponding increase in PS lipids, only in the cerebellum, while other brain regions mostly remain unaffected by this disruption (Figure 7). On the other hand, given its ubiquitous distribution, we find that deletion of ABHD12 has a more profound influence on lyso-PS and PS lipids in all regions of the brain, with the effects most evident in the cerebellum (Figure 7). The cerebellum is responsible for proper motor functions, posture and balance45–47, all of which are affected in human PHARC subjects8, 9, and here, we show that the cerebellum is most susceptible to lyso-PS deregulation upon disruption of ABHD16A and/or ABHD12 activity (Figure 7). Given all this, and the fact that the cerebellum undergoes maximum atrophy in human PHARC subjects8, 9, we wanted to investigate the (sub)anatomical localization of both ABHD16A and ABHD12 in the cerebellum. Interestingly, using IHC analysis, we found that ABHD16A and ABHD12 reside in spatially distinct regions of the cerebellum, i.e. the granular layer neurons and the Purkinje neurons respectively (Figure 8), that are known to communicate with one another towards regulating normal cerebellar functions45–47.
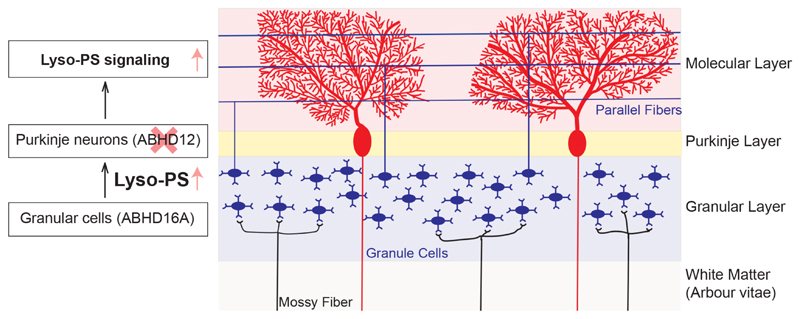
Although preliminary at this stage, based on the findings reported in this paper, we propose a paracrine signaling model for the role of ABHD16A and ABHD12, and lyso-PS lipids in regulating cerebellar function (Figure 10). We believe that lyso-PS lipids are biosynthesized by ABHD16A rich neuronal cells in granular layer, following which they are transported by as-of-yet unknown mechanism(s) to the Purkinje layer (and possibly other regions of the brain). Here, Purkinje neurons rich in ABHD12, tightly regulate lyso-PS concentrations, and maintain a basal threshold concentration necessary for homeostatic signaling and regulatory activities of the cerebellum (and perhaps other brain regions too). In the scenario of PHARC, where ABHD12 activity is absent, this homeostatic threshold is breached, and there is an excess of lyso-PS, constantly stimulating the Purkinje neurons, leading to deregulated cerebellar functioning, eventually manifesting into the motor defects and peripheral neuropathy seen in human PHARC subjects8, 9.
Figure 10. A preliminary paracrine signaling model for lyso-PS signaling in the cerebellum regulated by ABHD16A and ABHD12 activities.
Projecting ahead, the studies we report here open up several new exciting questions. First, given that the ER is a hub for PS metabolism, understanding the specific contribution of ABHD16A and its dynamic interlay with ABHD12 in regulating PS fluxes within the cellular organelle. Second, understanding and characterizing the paracrine signaling model for lyso-PS signaling we propose here, would provide new mechanistic insights into cerebellar functioning under the influence of this signaling lipid, and help map pathways downstream of lyso-PS signaling that are deregulated in PHARC. Recently, three G-Protein Coupled Receptors (GPR34, GPR174 and P2Y10)48 and the Toll-like Receptor 249, have putatively been identified as lyso-PS receptors, and understanding the localization of these receptors in the cerebellum, and characterizing their interplay with the ABHD16A-ABHD12 and lyso-PS network, will likely provide new therapeutic interventions in treating PHARC. Third, it would be important to determine in pre-clinical models, if disrupting ABHD16A activity can indeed rescue or reverse the neurological symptoms seen in PHARC. Fourth, it has been recently shown that ABHD12 also regulates levels of the pro-apoptotic oxidized phosphatidylserine lipids under oxidative stress conditions31, and that pharmacological disruption of ABHD12 activity leads to cell death ferroptosis in human cancer cells50–52. It would be interesting to understand whether ferroptosis if any, contributes to the PHARC pathophysiology, and how ABHD12 spatiotemporally regulates oxidized phosphatidylserine lipids in the brain under oxidative stress. Also, it would be useful to determine whether there are any changes in the brain anatomical distributions of ABHD16A and ABHD12 under an inflammatory-stimuli (e.g. systemic LPS treatment) and if and how that contributes to the neurological symptoms of PHARC. Fifth, we find from our profiling study here, that ABHD16A is also present in the olfactory bulb, and this expression corroborates the PS lipase activity profiles for the different brain regions. Given the involvement of visual disturbances, i.e. the early onset cataract and retinal pigmentation, in human PHARC subjects8, 9, understanding the role of lyso-PSs in the development of this pathophysiology should also be investigated. Finally, there is no structural information available for ABHD16A or ABHD12, given that they are integral membrane proteins, and have been difficult to purify and hence crystallize. With the advent of emerging structural biology techniques (e.g. cryo-electron microscopy), determination of the three-dimensional structures of ABHD16A and ABHD12 would enable a better mechanistic understanding of these lipases of biomedical importance.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: xxxx
Acknowledgement
Prof. Benjamin F. Cravatt, The Scripps Research Institute, is thanked for generously gifting ABHD12 and ABHD16A knockout mice breeders, and chemical probes (KC01 and KC02) used in this study. Prof. Vidita Vaidya and her lab members (TIFR Mumbai) are thanked for teaching us dissections of different anatomical regions of the mouse brain and for helpful discussions on this project. We thank Dr. Nagaraj Balasubramanian, and Dr. Richa Rikhy (both from IISER Pune) for providing antibodies used in this study. Dr. Mayurika Lahiri and her lab members are thanked for assistance with analyzing the immunohistochemical data. The NFGFHD IISER Pune, is thanked for maintaining and providing mice for this study. The IISER Pune – Leica Microscopy Facility is thanked for providing access to the microscopes for the immunofluorescence and immunohistochemical studies. Dr. Nishikant Subhedar (IISER Pune) and members of the Kamat lab are thanked for critical comments and helpful discussions on this study.
Funding
This work was supported by a DBT/Wellcome Trust India Alliance Fellowship [grant number IA/I/15/2/502058] awarded to S.S.K., and a Department of Science and Technology – Funds for Improvement of S&T Infrastructure Development (DST–FIST) grant (to Department of Biology, IISER Pune). S.S. is supported by a graduate student fellowship from IISER Pune.
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Author contributions: S.S. performed all the studies with assistance from A.J. and S.S.K.; S.S. and S.S.K. conceived the project, analyzed the data, and wrote the manuscript with inputs from A.J.; S.S.K. acquired funding for this project.
References
- [1].Galperin MY, Koonin EV. 'Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Aufschnaiter A, Kohler V, Diessl J, Peselj C, Carmona-Gutierrez D, Keller W, Buttner S. Mitochondrial lipids in neurodegeneration. Cell Tissue Res. 2017;367:125–140. doi: 10.1007/s00441-016-2463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Radak Z, Zhao Z, Goto S, Koltai E. Age-associated neurodegeneration and oxidative damage to lipids, proteins and DNA. Mol Aspects Med. 2011;32:305–315. doi: 10.1016/j.mam.2011.10.010. [DOI] [PubMed] [Google Scholar]
- [4].Yadav RS, Tiwari NK. Lipid integration in neurodegeneration: an overview of Alzheimer's disease. Molecular neurobiology. 2014;50:168–176. doi: 10.1007/s12035-014-8661-5. [DOI] [PubMed] [Google Scholar]
- [5].Darios F, Mochel F, Stevanin G. Lipids in the Physiopathology of Hereditary Spastic Paraplegias. Front Neurosci. 2020;14:74. doi: 10.3389/fnins.2020.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fiskerstrand T, Knappskog P, Majewski J, Wanders RJ, Boman H, Bindoff LA. A novel Refsum-like disorder that maps to chromosome 20. Neurology. 2009;72:20–27. doi: 10.1212/01.wnl.0000333664.90605.23. [DOI] [PubMed] [Google Scholar]
- [8].Fiskerstrand T, H'Mida-Ben Brahim D, Johansson S, M'Zahem A, Haukanes BI, Drouot N, Zimmermann J, Cole AJ, Vedeler C, Bredrup C, Assoum M, et al. Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism. Am J Hum Genet. 2010;87:410–417. doi: 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eisenberger T, Slim R, Mansour A, Nauck M, Nurnberg G, Nurnberg P, Decker C, Dafinger C, Ebermann I, Bergmann C, Bolz HJ. Targeted next-generation sequencing identifies a homozygous nonsense mutation in ABHD12, the gene underlying PHARC, in a family clinically diagnosed with Usher syndrome type 3. Orphanet journal of rare diseases. 2012;7:59. doi: 10.1186/1750-1172-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen DH, Naydenov A, Blankman JL, Mefford HC, Davis M, Sul Y, Barloon AS, Bonkowski E, Wolff J, Matsushita M, Smith C, et al. Two novel mutations in ABHD12: expansion of the mutation spectrum in PHARC and assessment of their functional effects. Hum Mutat. 2013;34:1672–1678. doi: 10.1002/humu.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yoshimura H, Hashimoto T, Murata T, Fukushima K, Sugaya A, Nishio SY, Usami S. Novel ABHD12 mutations in PHARC patients: the differential diagnosis of deaf-blindness. Ann Otol Rhinol Laryngol. 2015;124(Suppl 1):77S–83S. doi: 10.1177/0003489415574513. [DOI] [PubMed] [Google Scholar]
- [12].Tingaud-Sequeira A, Raldua D, Lavie J, Mathieu G, Bordier M, Knoll-Gellida A, Rambeau P, Coupry I, Andre M, Malm E, Moller C, et al. Functional validation of ABHD12 mutations in the neurodegenerative disease PHARC. Neurobiology of disease. 2017;98:36–51. doi: 10.1016/j.nbd.2016.11.008. [DOI] [PubMed] [Google Scholar]
- [13].Lerat J, Cintas P, Beauvais-Dzugan H, Magdelaine C, Sturtz F, Lia AS. A complex homozygous mutation in ABHD12 responsible for PHARC syndrome discovered with NGS and review of the literature. J Peripher Nerv Syst. 2017;22:77–84. doi: 10.1111/jns.12216. [DOI] [PubMed] [Google Scholar]
- [14].Frasquet M, Lupo V, Chumillas MJ, Vazquez-Costa JF, Espinos C, Sevilla T. Phenotypical features of two patients diagnosed with PHARC syndrome and carriers of a new homozygous mutation in the ABHD12 gene. Journal of the neurological sciences. 2018;387:134–138. doi: 10.1016/j.jns.2018.02.021. [DOI] [PubMed] [Google Scholar]
- [15].Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc Natl Acad Sci U S A. 2013;110:1500–1505. doi: 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saghatelian A, Cravatt BF. Discovery metabolite profiling--forging functional connections between the proteome and metabolome. Life Sci. 2005;77:1759–1766. doi: 10.1016/j.lfs.2005.05.019. [DOI] [PubMed] [Google Scholar]
- [18].Iwashita M, Makide K, Nonomura T, Misumi Y, Otani Y, Ishida M, Taguchi R, Tsujimoto M, Aoki J, Arai H, Ohwada T. Synthesis and evaluation of lysophosphatidylserine analogues as inducers of mast cell degranulation. Potent activities of lysophosphatidylthreonine and its 2-deoxy derivative. J Med Chem. 2009;52:5837–5863. doi: 10.1021/jm900598m. [DOI] [PubMed] [Google Scholar]
- [19].Frasch SC, Berry KZ, Fernandez-Boyanapalli R, Jin HS, Leslie C, Henson PM, Murphy RC, Bratton DL. NADPH oxidase-dependent generation of lysophosphatidylserine enhances clearance of activated and dying neutrophils via G2A. J Biol Chem. 2008;283:33736–33749. doi: 10.1074/jbc.M807047200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Frasch SC, Bratton DL. Emerging roles for lysophosphatidylserine in resolution of inflammation. Prog Lipid Res. 2012;51:199–207. doi: 10.1016/j.plipres.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frasch SC, Fernandez-Boyanapalli RF, Berry KA, Murphy RC, Leslie CC, Nick JA, Henson PM, Bratton DL. Neutrophils regulate tissue Neutrophilia in inflammation via the oxidant-modified lipid lysophosphatidylserine. J Biol Chem. 2013;288:4583–4593. doi: 10.1074/jbc.M112.438507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chang HW, Inoue K, Bruni A, Boarato E, Toffano G. Stereoselective effects of lysophosphatidylserine in rodents. Br J Pharmacol. 1988;93:647–653. doi: 10.1111/j.1476-5381.1988.tb10322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yea K, Kim J, Lim S, Kwon T, Park HS, Park KS, Suh PG, Ryu SH. Lysophosphatidylserine regulates blood glucose by enhancing glucose transport in myotubes and adipocytes. Biochem Biophys Res Commun. 2009;378:783–788. doi: 10.1016/j.bbrc.2008.11.122. [DOI] [PubMed] [Google Scholar]
- [24].Park KS, Lee HY, Kim MK, Shin EH, Jo SH, Kim SD, Im DS, Bae YS. Lysophosphatidylserine stimulates L2071 mouse fibroblast chemotactic migration via a process involving pertussis toxin-sensitive trimeric G-proteins. Mol Pharmacol. 2006;69:1066–1073. doi: 10.1124/mol.105.018960. [DOI] [PubMed] [Google Scholar]
- [25].Lee SY, Lee HY, Kim SD, Jo SH, Shim JW, Lee HJ, Yun J, Bae YS. Lysophosphatidylserine stimulates chemotactic migration in U87 human glioma cells. Biochem Biophys Res Commun. 2008;374:147–151. doi: 10.1016/j.bbrc.2008.06.117. [DOI] [PubMed] [Google Scholar]
- [26].Iida Y, N HT, Kishikawa J, Kaneko K, Murono K, Kawai K, Ikeda T, Ishihara S, Yamaguchi H, Sunami E, Kitayama J, et al. Lysophosphatidylserine stimulates chemotactic migration of colorectal cancer cells through GPR34 and PI3K/Akt pathway. Anticancer Res. 2014;34:5465–5472. [PubMed] [Google Scholar]
- [27].Park KS, Lee HY, Kim MK, Shin EH, Bae YS. Lysophosphatidylserine stimulates leukemic cells but not normal leukocytes. Biochem Biophys Res Commun. 2005;333:353–358. doi: 10.1016/j.bbrc.2005.05.109. [DOI] [PubMed] [Google Scholar]
- [28].Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kamat SS, Camara K, Parsons WH, Chen DH, Dix MM, Bird TD, Howell R, Cravatt BF. Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay. Nat Chem Biol. 2015;11:164–171. doi: 10.1038/nchembio.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Joshi A, Shaikh M, Singh S, Rajendran A, Mhetre A, Kamat SS. Biochemical characterization of the PHARC-associated serine hydrolase ABHD12 reveals its preference for very-long-chain lipids. Journal of Biological Chemistry. 2018;293:16953–16963. doi: 10.1074/jbc.RA118.005640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kelkar DS, Ravikumar G, Mehendale N, Singh S, Joshi A, Sharma AK, Mhetre A, Rajendran A, Chakrapani H, Kamat SS. A chemical-genetic screen identifies ABHD12 as an oxidized-phosphatidylserine lipase. Nat Chem Biol. 2019;15:169–178. doi: 10.1038/s41589-018-0195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rajendran A, Vaidya K, Mendoza J, Bridwell-Rabb J, Kamat SS. Functional Annotation of ABHD14B, an Orphan Serine Hydrolase Enzyme. Biochemistry. 2020;59:183–196. doi: 10.1021/acs.biochem.9b00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol Reprod Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Arena ET, Rueden CT, Hiner MC, Wang S, Yuan M, Eliceiri KW. Quantitating the cell: turning images into numbers with ImageJ. Wiley Interdiscip Rev Dev Biol. 2017;6 doi: 10.1002/wdev.260. [DOI] [PubMed] [Google Scholar]
- [36].Pathak D, Mehendale N, Singh S, Mallik R, Kamat SS. Lipidomics Suggests a New Role for Ceramide Synthase in Phagocytosis. ACS Chem Biol. 2018;13:2280–2287. doi: 10.1021/acschembio.8b00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Camara K, Kamat SS, Lasota CC, Cravatt BF, Howell AR. Combining cross-metathesis and activity-based protein profiling: new beta-lactone motifs for targeting serine hydrolases. Bioorg Med Chem Lett. 2015;25:317–321. doi: 10.1016/j.bmcl.2014.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annual review of biophysics. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- [39].Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016;44:D313–316. doi: 10.1093/nar/gkv1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ichu TA, Reed A, Ogasawara D, Ulanovskaya O, Roberts A, Aguirre CA, Bar-Peled L, Gao J, Germain J, Barbas S, Masuda K, et al. ABHD12 and LPCAT3 Interplay Regulates a Lyso-phosphatidylserine-C20:4 Phosphatidylserine Lipid Network Implicated in Neurological Disease. Biochemistry. 2020;59:1793–1799. doi: 10.1021/acs.biochem.0c00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Weyer A, Schilling K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J Neurosci Res. 2003;73:400–409. doi: 10.1002/jnr.10655. [DOI] [PubMed] [Google Scholar]
- [43].Barski JJ, Hartmann J, Rose CR, Hoebeek F, Morl K, Noll-Hussong M, De Zeeuw CI, Konnerth A, Meyer M. Calbindin in cerebellar Purkinje cells is a critical determinant of the precision of motor coordination. J Neurosci. 2003;23:3469–3477. doi: 10.1523/JNEUROSCI.23-08-03469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kay JG, Grinstein S. Phosphatidylserine-mediated cellular signaling. Adv Exp Med Biol. 2013;991:177–193. doi: 10.1007/978-94-007-6331-9_10. [DOI] [PubMed] [Google Scholar]
- [45].Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- [46].Manto M, Bower JM, Conforto AB, Delgado-Garcia JM, da Guarda SN, Gerwig M, Habas C, Hagura N, Ivry RB, Marien P, Molinari M, et al. Consensus paper: roles of the cerebellum in motor control--the diversity of ideas on cerebellar involvement in movement. Cerebellum. 2012;11:457–487. doi: 10.1007/s12311-011-0331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013;80:807–815. doi: 10.1016/j.neuron.2013.10.044. [DOI] [PubMed] [Google Scholar]
- [48].Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, Higashiyama S, Ohwada T, Arai H, Makide K, Aoki J. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods. 2012;9:1021–1029. doi: 10.1038/nmeth.2172. [DOI] [PubMed] [Google Scholar]
- [49].van der Kleij D, Latz E, Brouwers JF, Kruize YC, Schmitz M, Kurt-Jones EA, Espevik T, de Jong EC, Kapsenberg ML, Golenbock DT, Tielens AG, et al. A novel host-parasite lipid cross-talk. Schistosomal lysophosphatidylserine activates toll-like receptor 2 and affects immune polarization. J Biol Chem. 2002;277:48122–48129. doi: 10.1074/jbc.M206941200. [DOI] [PubMed] [Google Scholar]
- [50].Kathman SG, Boshart J, Jing H, Cravatt BF. Blockade of the Lysophosphatidylserine Lipase ABHD12 Potentiates Ferroptosis in Cancer Cells. ACS Chem Biol. 2020;15:871–877. doi: 10.1021/acschembio.0c00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ogasawara D, Ichu TA, Vartabedian VF, Benthuysen J, Jing H, Reed A, Ulanovskaya OA, Hulce JJ, Roberts A, Brown S, Rosen H, et al. Selective blockade of the lyso-PS lipase ABHD12 stimulates immune responses in vivo. Nat Chem Biol. 2018;14:1099–1108. doi: 10.1038/s41589-018-0155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ogasawara D, Ichu TA, Jing H, Hulce JJ, Reed A, Ulanovskaya OA, Cravatt BF. Discovery and Optimization of Selective and in Vivo Active Inhibitors of the Lysophosphatidylserine Lipase alpha/beta-Hydrolase Domain-Containing 12 (ABHD12) J Med Chem. 2019;62:1643–1656. doi: 10.1021/acs.jmedchem.8b01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.