Abstract
Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 differentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in differentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream effector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer.
Subject terms: Cancer epigenetics, Oncogenes
Introduction
Activating KRAS mutations can be found in nearly 25 percent of all cancers1. Pancreatic and lung cancers, in particular, exhibit high rates of oncogenic KRAS mutation, at 95% and 30%, respectively2. In this respect, KRAS has been established as a crucial oncoprotein in the progression and maintenance of KRAS-mutant pancreatic and lung cancers3–8. The important role of oncogenic KRAS in cancer has been met with nearly four decades of effort to develop therapeutic strategies to target aberrant KRAS function for cancer treatment9,10. Recently, direct inhibitors of mutant KRAS have been developed10,11, and have entered clinical evaluation12. While the G12C mutation is prevalent in KRAS-mutant lung adenocarcinoma (~46%), this mutation is found in only 2% of PDAC13. Therefore, indirect approaches remain the best option for the majority of KRAS-mutant PDAC. Among indirect approaches, the inhibition of downstream effectors, the RAF-MEK-ERK MAPK cascade and the PI3K-AKT-mTOR pathways, remain the most promising direction14–18.
In addition to aberrant effector signaling, most cancer cells also undergo genome-scale epigenetic changes. The most widely studied biochemical modification governing epigenetics is DNA methylation of CpG dinucleotides19. DNA methylation in mammalian organisms occurs by the covalent addition of a methyl group to the C-5 position of cytosine base in a CpG sequence context. The human genome is CpG depleted, while nearly 70% of all CpGs are methylated, mostly in transposable elements and intergenic regions of the human genome. DNA methylation can impact proximal chromatin structure and regulate gene expression, playing critical roles in biological processes including embryonic development, X-chromosome inactivation, genomic imprinting, and chromosome stability19. Hence, determining the methylation status at a single base resolution in the genome is an important step in elucidating its role in regulating many cellular processes and its disruption in disease states. CpG methylation can be dynamically regulated and this process is reversible.
Global DNA hypomethylation and focal hypermethylation at CpG islands have become hallmarks of cancer20–23. Moreover, oncogenic KRAS expression has specifically been shown to induce aberrant DNA methylation, promoting hypomethylation across the genome while silencing key tumor suppressors through hypermethylation24–27. Gazin et al.24 reported an ordered pathway associated with RAS-induced epigenetic signaling. KRAS-associated differential DNA methylation could have a significant impact across the genome and lead to important oncogenic transcriptional changes. Discovering an essential and predictable epigenetic response to mutant KRAS expression either within one cancer type, across multiple cancer types, or specificity to a particular KRAS mutation (i.e. G12D), could reveal other potential anti-cancer targets. Interestingly, Xie et al.28. found that HRAS-transformed cells show methylation patterns diverging dramatically from reproducible methylation pattern of senescence. The authors suggest that cell transformation involves stochastic epigenetic patterns from which malignant cells may evolve. Ultimately, a better understanding of the DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches for KRAS-driven cancers and provide a platform for understanding the intrinsic heterogeneous nature of these cancers.
We have previously shown that mutant KRAS drives distinct molecular changes in pancreatic29 and lung30 cancer cells. However, it remains unclear whether these molecular changes are associated with epigenetic changes. Here we perform a genome-scale analysis using KRAS-mutant human pancreatic and lung cancer cell lines to investigate whether knock-down or overexpression of mutant KRAS as well as pharmacological inhibtion of ERK correlates with differential DNA methylation. We found that while KRAS-mediated DNA methylation changes were cell type specific, gene ontology analysis revealed that many of the genes were associated with development and differentiation. Furthermore, we found that ERK inhibition did not reverse the great majority of KRAS-mediated methylation changes, suggesting that ERK is not a main driver for KRAS-mediated DNA methylation changes.
Results
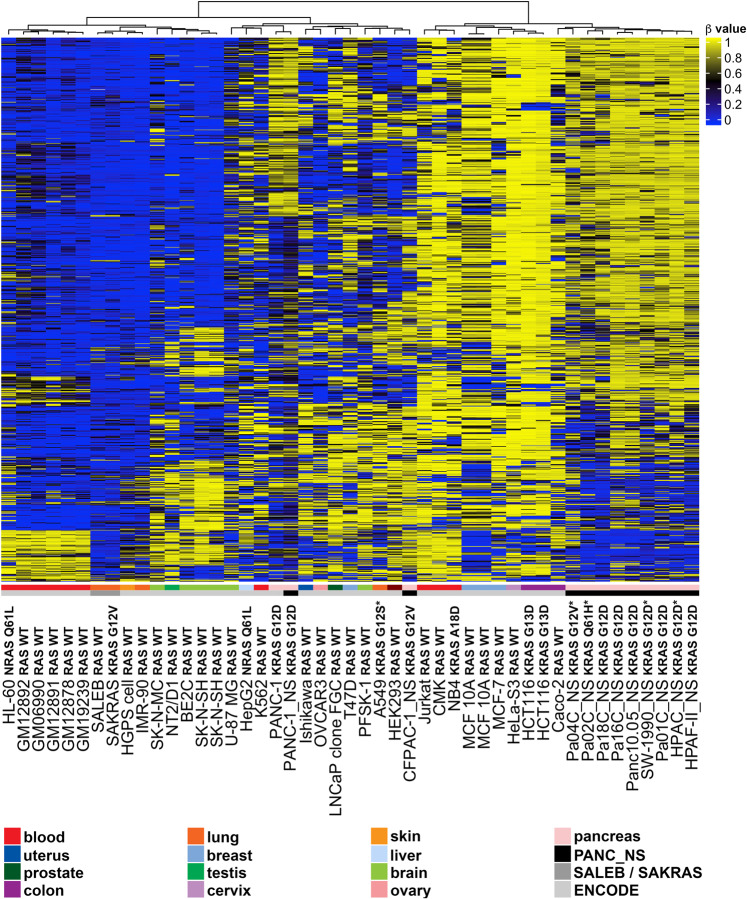
CpG methylation in a panel of 47 cell lines shows clustering of cell lines with similar tissue of origin independent of KRAS mutation status
Given the essential role of oncogenic KRAS in the great majority of pancreatic cancer15,29 (see cell line information, Supplementary Fig. S1), we investigated whether the presence of an activating KRAS mutation correlates with specific patterns of global DNA methylation. We first performed genome-wide DNA methylation profiling of 11 KRAS-dependent pancreatic cancer cell lines using the Infinium HumanMethylation450 BeadChip Array31. We also surveyed the CpG methylation patterns in low passage, immortalized lung epithelial cells transduced with KRAS G12V (SAKRAS cells) and non-transformed empty vector controls (SALEB cells). We compared the panel of 11 KRAS-mutant pancreatic cancer cell lines to DNA methylation data collected from SALEB and SAKRAS lung epithelial cells and published Infinium methylation data from ENCODE32 (Fig. 1). The published ENCODE data include three non-transformed human cell lines (HGPS and IMR-90 fibroblasts, and two different MCF 10 A breast epithelial cell lines) and 30 cell lines of varying cell types, genetic backgrounds, and tumorigenicity. As the pancreatic cancer cell lines were transduced with non-silencing (NS) shRNA, which could potentially affect the methylome of the transduced cells, we performed the same analysis while excluding these cells (Supplementary Fig. S2). After unsupervised hierachial clustering of the top 1,000 most variable CpG probes across all 47 cell lines, the pancreatic cancer cell lines formed a distinct cluster with the exception of CFPAC-1_NS and PANC-1_NS cells. These data suggest that the panel of KRAS-mutant pancreatic cancer cell lines contain similar overall basal DNA methylation patterns. Other KRAS mutant lines were clustered in the same branch of the dendrogram. However, in general, the cell lines formed clusters based on cell type with a few exceptions, and this was true regardless of the exclusion of the transduced pancreatic cancer cell lines. This suggests that even as KRAS may influence some key changes to the epigenome, DNA methylation patterns observed are more influenced by cell type.
Figure 1.
CpG methylation in a panel of 47 cell lines with varying KRAS status. Unsupervised hierarchical clustering analysis using the top 1000 most variable CpG probes across a panel of 47 cell lines is displayed above. Eleven human pancreatic cancer cell lines were transduced with non-silencing (NS) shRNA (black bar above). DNA methylation patterns in these pancreatic cells were compared to the DNA methylation in lung epithelial SALEB/SAKRAS cells and Infinium methylation data obtained from ENCODE (www.encodeproject.org). The β value for each probe is represented with a color scale as shown in the key. Values closer to 1 represent highly methylated CpGs, while values closer to zero represent least methylated CpGs.
Unsupervised hierachical clustering shows cell line specific differential CpG methylation associated with KRAS suppression in pancreatic cancer cells
We have previously shown that silencing KRAS caused distinct molecular changes in pancreatic cancer cell lines29. Silencing of KRAS may therefore also lead to differential DNA methylation. To test this, we performed RNA-seq and genome-wide DNA methylation analysis using Illumina’s Infinium arrays to determine the effect of silencing of KRAS in the 11 KRAS-mutant and -dependent pancreatic cancer cells. Briefly, cells were harvested for RNA and genomic DNA 4 to 7 days following infection with lentivirus shRNA targeting KRAS. Despite being KRAS-dependent, KRAS knockdown was not sufficient to cause dramatic cell death in pancreatic cell lines. This has been observed previously, and these cells lines were shown to be able to activate compensatory pathways in response to KRAS suppression29. Reduced KRAS mRNA levels were observed in KRAS-depleted cells relative to NS controls as determined by RNA sequencing (Fig. 2A). We then performed GSEA to compare the KRAS-depleted cells to the NS controls, and found a reduction in KRAS signaling (Supplementary Fig. S3), as evident from decreased enrichment in genes which are upregulated by KRAS (HALLMARK_KRAS_UP) and increase enrichment in genes downregulated by KRAS (HALLMARK_KRAS_DN). There was also a decrease in both PI3K/AKT and mTORC signaling, which are pathways downstream of KRAS.
Figure 2.
Effects of KRAS inhibition on DNA methylation. (A) KRAS mRNA levels from 10 pancreatic cancer cell lines transduced with KRAS shRNA compared to non-silencing (NS) controls as measured by RNA sequencing. RNA was not collected for SW-1990 cells due to insufficient material. (B) Unsupervised hierarchical clustering analysis was performed using the top 1000 most variable CpG probes across the panel of 11 pancreatic cell lines transduced with NS shRNA or KRAS shRNA. The β value for each probe is represented with a color scale as shown in the key. (C) Bar graph showing the number of differentially methylated (DM) CpGs with Δβ values ≥0.2 or ≤−0.2 in cell lines transduced with KRAS shRNA (hypermethylated CpGs represented in yellow and hypomethylated CpGs represented in blue).
Unsupervised hierarchical clustering of genome-wide DNA methylation data using the top 1000 most variable CpG probes revealed co-clustering of isogenic cell line pairs in that all 11 KRAS-depleted cell lines and their isogenic controls appear more similar to each other than any other cell line (Fig. 2B). There was also no clear separation based on specific KRAS mutations (G12D vs G12V). There were two distinct branches separating the 11 isogeneic pairs (Fig. 2B), representing a group with a lower degree of methylation than the other. To identify common methylation changes between the pancreatic cell lines, we performed heirachical clustering using the union of differentially methylated probes (Δβ values ≥0.2 or ≤−0.2) appearing in at least 3 out of the 11 cell line pairs (a total of 204 CpG probes) and observed co-clustering of Pa16C, Pa01C, and Panc 10.05 (Supplementary Fig. S4A). From this list of 204 CpG probes, which represents the most frequently differentially methylated (DM) probes, we compiled the top 10 DM hypo or hypermethylated CpGs per cell line into a list (Supplementary Fig. S4B).
Next we examined the number of DM probes per cell line as a measure of the extent of DNA methylation response due to KRAS inhibition. The DNA methylation profiles of Pa16C, Pa01C, PANC-1, and Panc 10.05 cells showed the most robust response to KRAS suppression. These four responsive cell lines showed at least 5-fold more DM CpGs compared to the seven other pancreatic cell lines tested (Fig. 2C and Table 1). Although Pa16C cells are derived from Panc 10.05 cells33, Pa16C cells had more than 4-fold the number of DM CpGs (Fig. 2C and Table 1). The four responsive cell lines showed a significant number of DM CpGs located in the promoter region (200–1500 nt upstream of the transcription start site) of dozens of functionally important genes (Table 2). The methylation changes associated with KRAS suppression appeared to be cell line specific and were not generalizable within pancreatic cell lines. Although the methylation patterns in the NS shRNA-treated control cells were similar (Fig. 1), each cell line responded differently to the depletion of KRAS. Furthermore, two distinct groups emerged from the pancreatic cell lines, with seven lines displaying significantly less differential methylation compared to the four responsive cell lines (Pa16C, Pa01C, PANC-1, and Panc 10.05 cells). Taken together these results suggest that depleting oncogenic KRAS expression is cell line specific but also stochastic in nature. It is possible that whether a cell’s CpG methylation profile is responsive or refractory to KRAS suppression likely depends on its genetic background and other factors.
Table 1.
Mutation status of crucial genes and the total number of differentially methylated (DM) CpGs with Δβ value ≥0.2 or ≤−0.2 in KRAS-depleted pancreatic cancer cell lines.
| Cell Line | KRAS | CDKN2A | TP53 | SMAD4 | Hypermethylated Promoter CpGs/total CpGs | Hypomethylated Promoter CpGs/total CpGs | All CpGs with Δβ ≥0.2 or ≤−0.2 |
|---|---|---|---|---|---|---|---|
| Pa16C | G12D/WT | I255N* | 434/3275 | 764/5613 | 8888 | ||
| Pa01C | G12D/WT | T155P* | Del* | 175/1248 | 393/2998 | 4246 | |
| PANC-1 | G12D/WT | Del* | R273H* | 128/717 | 556/3136 | 3853 | |
| Panc 10.05 | G12D/WT | I255N/WT | 59/452 | 275/1508 | 1960 | ||
| Pa04C | G12V* | Del* | Del* | 13/172 | 25/200 | 372 | |
| Pa02C | Q61H* | Del* | L247P* | Del* | 28/185 | 28/184 | 369 |
| CFPAC-1 | G12V/WT | C242R* | Del* | 7/89 | 12/115 | 204 | |
| HPAC | G12D* | Stop/Stop | 16/104 | 15/92 | 196 | ||
| HPAF-II | G12D/WT | Del-FS* | P151S* | 14/96 | 5/68 | 164 | |
| SW-1990 | G12D* | Del* | 11/78 | 15/79 | 157 | ||
| Pa18C | G12D/WT | Del* | Del* | 6/72 | 6/72 | 144 |
The CpG methylation in Pa16C, Pa01C, PANC-1 and Panc 10.05 cells appears to be the most responsive to KRAS depletion. Homozygous mutations are represented with an asterisk.
Table 2.
Categorization of differentially methylated (DM) Promoter CpGs in KRAS-inhibited most responsive cell lines (Pa16C, Pa01C, PANC-1 and Panc 10.05 cells).
| Pa16C cells | Pa01C cells | PANC-1 cells | Panc 10.05 cells | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyper- methylated |
Hypo- methylated |
Hyper- methylated |
Hypo- methylated |
Hyper- methylated |
Hypo- methylated |
Hyper- methylated |
Hypo- methylated |
||||||
| Transcription Factors | |||||||||||||
| ASCL2 | PFDN5 | ARNT | HOXD8 | SMARCA5 | ALX1 | ALX4 | PAX7 | ID3 | AIRE | LSR | ASCL1 | BNC2 | LHX4 |
| CBX4 | RAX | ATOH7 | INSM2 | TAF3 | ALX3 | BARHL2 | PDX1 | KLF14 | CUX2 | MAF | BRF1 | C13orf15 | LIN28A |
| CRIP1 | RING1 | BACH2 | IRF7 | THRB | EZH1 | BHLHE22 | PHOX2A | MLLT6 | EBF4 | NEUROG1 | MKX | CASZ1 | LYL1 |
| E2F2 | RREB1 | BAZ2B | IRX1 | TMF1 | HMGB2 | BNC1 | PHOX2B | MSX2 | EGR3 | NFATC4 | NEUROG1 | CBFA2T3 | MSC |
| EBF4 | SALL4 | BRF1 | IRX2 | TRIM13 | HMX2 | BRF1 | PITX2 | NKX2-5 | EMX1 | NKX2-6 | ZFP30 | ESR2 | MSX1 |
| ELK3 | SIX3 | BTF3 | LIN28A | TRIM27 | HOXB1 | DBX1 | PLAGL1 | NKX3-1 | ETV7 | NKX6-2 | EVX2 | PAX7 | |
| EOMES | TCF7 | CECR6 | LMX1B | TSC22D2 | IRX1 | DBX2 | PRDM13 | PAX1 | FOXA2 | NRIP1 | FEZF2 | PCGF3 | |
| EYA2 | TLX2 | CNPY3 | MED24 | TSHZ3 | KDM3B | DLX1 | RNF2 | PRDM8 | FOXB1 | PBX4 | FOXE3 | RARG | |
| FOXC2 | TUB | CSRP1 | MIXL1 | TWIST1 | LHX8 | DMRT1 | RORB | TBX2 | FOXD3 | PER1 | FOXI1 | RUNX3 | |
| FOXE1 | TULP1 | CSRP2 | MKX | ULK2 | NEUROG1 | DMRTA2 | RUNX3 | TCF7L2 | FOXF1 | PHF11 | GBX2 | SALL1 | |
| GBX1 | UNCX | CUX2 | MSX2P1 | UTF1 | NKX2-2 | ESR1 | SALL3 | YAF2 | GATA5 | PITX3 | GSC | SALL3 | |
| GSX1 | VENTX | ELK4 | NCALD | VAX1 | PAX3 | EYA4 | TBX3 | ZNF213 | GFI1 | POU3F1 | HKR1 | T | |
| HAND1 | ZAR1 | EN1 | NEUROG1 | VSX1 | SOX8 | FEZF2 | TCF4 | ZNF222 | GSC2 | RARA | HOXA9 | TLX3 | |
| HAND2 | ZBTB16 | ERMP1 | NEUROG3 | YBX2 | TLX2 | FOXB1 | ULK2 | GSX1 | RAX | HOXB13 | TUB | ||
| HES2 | ZFP28 | ESR1 | NFYC | ZBTB22 | TWIST1 | GBX2 | ZIC1 | HAND1 | RORB | HOXB2 | VEZF1 | ||
| HNF1A | ZSCAN12 | ESRRG | OLIG1 | ZFP30 | ZNF213 | GCM2 | ZNF16 | HAND2 | SIM2 | HOXB4 | ZBTB16 | ||
| HOXB1 | EYA4 | PDLIM5 | ZIC1 | ZNF593 | GFI1 | ZNF18 | HES4 | SIX2 | HOXB8 | ZBTB17 | |||
| HOXC10 | FOXE3 | PHOX2A | ZMYND11 | HIC1 | ZNF256 | HEYL | TBX5 | IRX2 | ZFP28 | ||||
| HOXC4 | FOXG1 | PLAGL1 | ZNF124 | HOXA6 | ZNF331 | HLX | THRB | IRX3 | ZIC1 | ||||
| HOXD12 | GBX2 | POU3F2 | ZNF135 | HOXA9 | HMX3 | TOX | LEF1 | ZNF236 | |||||
| HOXD3 | GLI3 | POU4F1 | ZNF18 | HOXB13 | HNF1A | VENTX | |||||||
| HOXD4 | GRHL1 | PPARG | ZNF207 | HOXB2 | HNF1B | WT1 | |||||||
| HOXD9 | HIF3A | PRDM13 | ZNF211 | HOXC9 | HOXC13 | YBX2 | |||||||
| IRF4 | HMX3 | PRDM14 | ZNF219 | HOXD3 | HOXD1 | ZAR1 | |||||||
| KCNIP3 | HOXA5 | PRDM6 | ZNF232 | ID4 | HR | ZFP37 | |||||||
| MYCNOS | HOXA9 | RARG | ZNF268 | IRF7 | IRF6 | ZNF229 | |||||||
| NEUROG2 | HOXB13 | RBBP9 | ZNF295 | MKX | IRF8 | ZNF334 | |||||||
| NFIC | HOXB3 | RUNX3 | ZNF318 | MSC | ISL1 | ZNF701 | |||||||
| NKX6-1 | HOXB4 | SALL1 | ZNF532 | MSX1 | LEF1 | ZSCAN12 | |||||||
| NKX6-3 | HOXB8 | SALL3 | ZNF682 | NKX2-5 | |||||||||
| OSR1 | HOXC8 | SAMD4B | NKX6-2 | ||||||||||
| OTP | HOXD1 | SIM2 | PAX1 | ||||||||||
| Cytokines & Growth Factors | |||||||||||||
| BDNF | LEFTY1 | BMP3 | FGF2 | NRG3 | FGF20 | CALCA | GRP | CXCL5 | BMP2 | LTBP2 | CALCA | CMTM2 | SCT |
| CALCA | LTBP3 | CCK | FGF20 | PTHLH | GDF6 | CMTM2 | HAMP | KL | BMP7 | MDK | CCK | FGF2 | SEMA5A |
| CSF1 | PDGFRA | CMTM1 | FGF5 | SEMA6D | IL28B | EPO | NGF | BMP8A | NGF | GRP | SLIT1 | ||
| CXCL12 | PENK | CMTM3 | FGF9 | SLIT1 | NRG3 | FGF11 | PSPN | CXCL16 | NRG1 | NPY | |||
| FGF19 | PTH2 | DKK1 | GDNF | TNFSF13 | PDGFA | FGF12 | SEMA5A | EDN3 | NRG3 | ||||
| GDF7 | SCGB3A1 | EPO | GREM1 | SEMA6B | FGF2 | SLIT1 | FAM3B | RLN3 | |||||
| KL | SECTM1 | FGF11 | KITLG | GREM1 | SLIT2 | FGF22 | STC2 | ||||||
| FGF6 | TNFSF12 | ||||||||||||
| GDF10 | TYMP | ||||||||||||
| GDF7 | VEGFC | ||||||||||||
| GRP | |||||||||||||
| Homeodomain Proteins | |||||||||||||
| GBX1 | NKX6-1 | CUX2 | HOXC8 | POU3F2 | ALX1 | ALX4 | HOXD3 | MSX2 | CUX2 | ISL1 | MKX | EVX2 | HOXB8 |
| GSX1 | NKX6-3 | EN1 | HOXD1 | POU4F1 | ALX3 | BARHL2 | MKX | NKX2-5 | EMX1 | NKX2-6 | GBX2 | IRX2 | |
| HNF1A | OTP | GBX2 | HOXD8 | TSHZ3 | HMX2 | DBX1 | MSX1 | NKX3-1 | GSC2 | NKX6-2 | GSC | IRX3 | |
| HOXB1 | RAX | HMX3 | IRX1 | VAX1 | HOXB1 | DBX2 | NKX2-5 | GSX1 | PBX4 | HOXA9 | LHX4 | ||
| HOXC10 | SIX3 | HOXA5 | IRX2 | VSX1 | IRX1 | DLX1 | NKX6-2 | HLX | PITX3 | HOXB13 | MSX1 | ||
| HOXC4 | TLX2 | HOXA9 | LMX1B | LHX8 | GBX2 | PAX7 | HMX3 | POU3F1 | HOXB2 | PAX7 | |||
| HOXD12 | UNCX | HOXB13 | MIXL1 | NKX2-2 | HOXA6 | PDX1 | HNF1A | RAX | HOXB4 | TLX3 | |||
| HOXD3 | VENTX | HOXB3 | MKX | PAX3 | HOXA9 | PHOX2A | HNF1B | SIX2 | |||||
| HOXD4 | HOXB4 | MSX2P1 | TLX2 | HOXB13 | PHOX2B | HOXC13 | VENTX | ||||||
| HOXD9 | HOXB8 | PHOX2A | HOXB2 | PITX2 | HOXD1 | ||||||||
| HOXC9 | |||||||||||||
| Protein Kinases | |||||||||||||
| CAMK2B | PDGFRA | AATK | MAPK7 | PINK1 | CDK6 | CDKL3 | BRAF | ACVR1C | KDR | PRKAA2 | CDC42BPB | NEK3 | |
| FASTK | STK19 | BRAF | MYO3A | SGK1 | DDR1 | DCLK2 | CSNK1A1 | BCR | KSR2 | RIPK3 | FGFR1 | NTRK3 | |
| HUNK | TNK2 | CDC42BPB | NEK10 | SNRK | EIF2AK2 | NEK9 | CDKL2 | MAST4 | |||||
| KDR | CDKL3 | NEK3 | STYK1 | HIPK3 | PDK2 | CSNK1G2 | MST1R | ||||||
| LCK | FGFR1 | NRBP1 | ULK2 | MAP2K1 | PINK1 | DAPK1 | PBK | ||||||
| MAP3K6 | FGR | NTRK3 | MAPK4 | RIOK3 | DMPK | STK32A | |||||||
| MAPK12 | FYN | PBK | MATK | ULK2 | EPHA6 | STK32B | |||||||
| PRKD1 | FGFR1 | STK33 | |||||||||||
| RYK | FLT3 | SYK | |||||||||||
| HCK | TNK1 | ||||||||||||
| HUNK | WNK2 | ||||||||||||
| INSR | |||||||||||||
| Oncogenes | |||||||||||||
| CCND2 | ZBTB16 | ARNT | GAS7 | PPARG | CCND2 | HOXA9 | BRAF | BCR | KDR | CBFA2T3 | NTRK3 | ||
| IRF4 | BRAF | GNAS | TOP1 | CDK6 | PAX7 | CCND1 | CDH11 | MAF | FGFR1 | PAX7 | |||
| KDR | ELK4 | HOXA9 | TRIM27 | JAK2 | ZNF331 | MLLT6 | DDX6 | PER1 | HOXA9 | TLX3 | |||
| LCK | FGFR1 | HSP90AB1 | JAK3 | FGFR1 | RARA | LYL1 | ZBTB16 | ||||||
| PDGFRA | FIP1L1 | NTRK3 | PAX3 | FLT3 | SEPT9 | ||||||||
| TCL1A | HOXC13 | SYK | |||||||||||
| Cell Differentiation Markers | |||||||||||||
| CD40 | PROM1 | ADAM17 | IL17RA | TNFRSF10B | DDR1 | CD248 | PVRL2 | CDH1 | INSR | FGFR1 | TNFRSF1B | ||
| CD81 | FGFR1 | ITGB3 | TNFRSF8 | NCAM1 | CDH2 | CDH2 | KDR | IFITM1 | |||||
| IL10RA | FZD10 | MME | TNFSF13 | TNFRSF8 | FGFR1 | LAMP3 | |||||||
| KDR | IFITM1 | NGFR | FLT3 | MME | |||||||||
| PDGFRA | IGF2R | FZD10 | MST1R | ||||||||||
| GP1BB | THBD | ||||||||||||
| ICOSLG | |||||||||||||
| Tumor Suppressors | |||||||||||||
| EXT2 | BRCA1 | PHOX2B | BRCA1 | WT1 | FANCA | ||||||||
| HNF1A | CDH1 | XPA | |||||||||||
| HNF1A | |||||||||||||
Inhibitor treatment shows limited role for ERK in differential CpG methylation of Pa16C pancreatic cancer cells
Next, we investigated whether the methylation changes associated with KRAS suppression are dependent on ERK signaling, a major downstream effector of KRAS. We used Pa16C cells, the cell line with the greatest number of DM CpGs associated upon KRAS suppression (Fig. 2C), to test the effects of ERK inhibition on DNA methylation. Pa16C cells were treated with the ERK1/2-selective inhibitor, (ERKi, SCH772984)34 (Supplementary Fig. S5) and the cells were harvested for protein and genomic DNA 3 and 7 days after treatment. The dose response of SCH772984 on Pa16C cell growth was determined (Supplementary Fig. S5A). Based on this, Pa16C cells were treated with 0.25 μM (3.6 on log scale), which resulted in the highest inhibition of cell growth. ERKi treatment led to growth arrest as evidenced by the lower cell confluency compared to DMSO control (Supplementary Fig. S5B, Right) and also reduced total ERK protein and phosphorylated ERK protein as measured by western blot (Supplementary Fig. S5B, Left). Three and 7 days of ERKi treatment resulted in 29 and 37 DM probes, respectively. Only 1 CpG probe cg18988094 was hypomethylated in both 3- and 7-day ERKi-treated samples. This DM CpG is found near the gene STIP1, which has been reported to activate ERK signaling (Supplementary Fig. S5C). We compared DM CpG profiles of the ERKi-treated Pa16C cells to the Pa16C cells transduced with shKRAS. However, there were no overlapping DNA methylation changes between the ERKi-treated and the KRAS shRNA-transduced Pa16C cells despite the similar effects on cell growth observed in both conditions (Supplementary Fig. S5A, Right)15. Furthermore, KRAS shRNA induced >8000 DNA methylation changes compared to <40 DM CpGs after pharmacological ERK inhibition. These observations suggest that targeted ERK inhibition leads to Pa16C cell growth arrest similar to the growth arrest observed in KRAS shRNA transduced Pa16C cells. However, ERK does not appear to be consequential to the thousands of KRAS-associated DM CpGs present in the KRAS shRNA transduced Pa16C cells, at least after 7 days and suggests that KRAS suppression leads to sustained DM changes not affected by inhibition of downstream targets like ERK. However, it remains possible that ERK could still be responsible for KRAS-associated methylation changes that occur over a longer time frame.
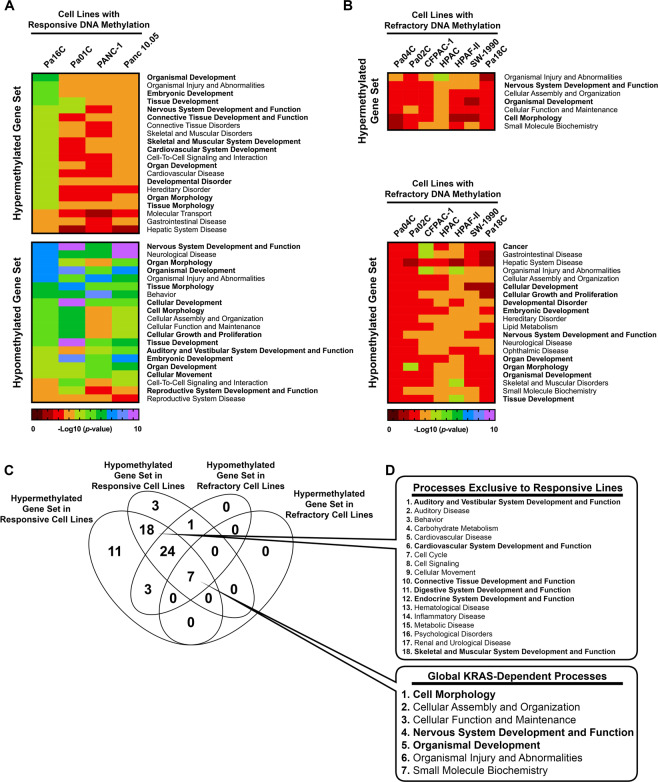
Gene ontology analysis of differentially methylated promoters in KRAS-depleted pancreatic cancer cell lines
Due to the limited number of overlapping DM CpGs (Supplementary Fig. S4), we attempted to isolate biological processes associated with KRAS knockdown that are common between KRAS-depleted cell lines. First, we grouped the KRAS-depleted cell lines into “responsive” cells (Pa16C, Pa01C, PANC-1, and Panc 10.05 cells) and “refractory” cells referring to the other seven pancreatic cell lines in our panel, based on the number of DM CpGs identified (Fig. 2C). To focus our analysis on genes with DM CpGs most likely to produce transcriptional effects, we isolated DM CpGs found within promoter regions, 200–1500 bases upstream of the transcription start site of a gene, and within 4 kb of a CpG island, including shores and shelves. We then kept only the gene promoters that had consistent CpG differential methylation, where all of the CpGs were either hypermethylated or hypomethylated. Genes encoding transcription factors, oncogenes, kinases, and growth factors showed differential DNA methylation at their promoters in KRAS-depleted cells (Table 2). Gene ontology analysis was performed using lists of promoters from each KRAS-depleted cell line that were hypermethylated or hypomethylated. The top ≤ 20 overlapping biological processes were compiled in a heat map (Fig. 3). Hypermethylated promoters in the responsive cells were enriched for genes involved in development and differentiation (Fig. 3A, bold); however, the number of hypermethylated promoters was significantly reduced in the refractory cells (Table 1), which limited the number of associated biological processes (Fig. 3B). The hypomethylated promoters in both the responsive cells and the refractory cells were enriched for genes involved in development and differentiation (Fig. 3A,B). Gene ontology analysis produced a significantly lower number of biological processes for the refractory cell lines compared to responsive cells due to the paucity of DM CpGs in the these cell lines (Fig. 3C). A total of 18 biological processes were found exclusively in the responsive lines with 6 of these related to development (Fig. 3D, Top, bold). In addition, our analysis showed 7 processes that were potentially affected by KRAS suppression in both responsive and refractory cell lines (Fig. 3D, Bottom). Together these results suggest that KRAS suppression leads to differential DNA methylation affecting genes involved in development and differentiation, especially in responsive cell lines, and corroborates previous gene ontology analyses of DM genes in HRAS-transformed fibroblasts, which also showed an enrichment for genes involved in development and differentiation28.
Figure 3.
Gene ontology analysis of differentially methylated (DM) promoters in KRAS- inhibited pancreatic cancer cells. (A,B) Gene Ontology analysis of DM genes in cells with (A) responsive or (B) refractory DNA methylation. Processes related to development and differentitation are in bold. (C) Venn diagram showing the number of biological processes associated with responsive or refractory promoter CpG methylation in KRAS-depleted cell lines. (D) (Top) List of affected biological processes exclusive to cell lines responsive to KRAS-depletion, or (Bottom) common among all of the KRAS-depleted cell lines.
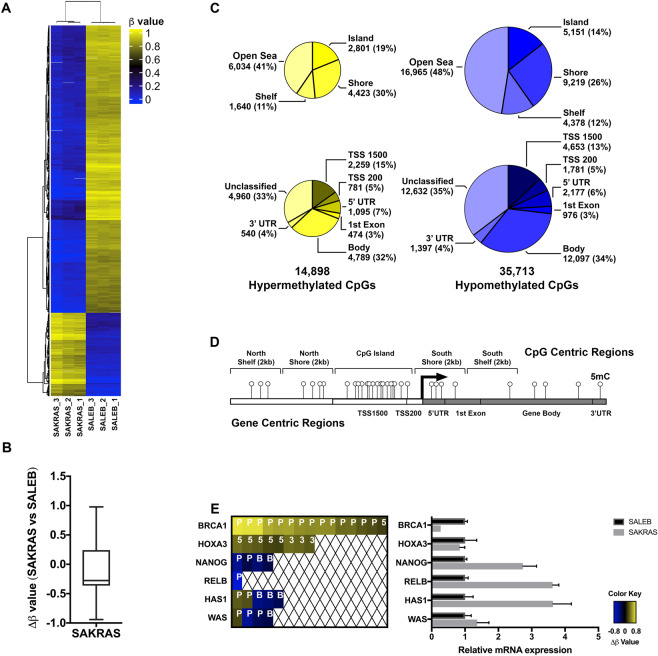
DNA methylation changes associated with mutant KRAS overexpression in lung cells
Since our results indicate that KRAS suppression is associated with CpG methylation changes in pancreatic cancer cell lines, we hypothesized that the overexpression of oncogenic KRAS would also lead to DNA methylation changes. To isolate the effects of oncogenic KRAS overexpression, we used an isogenic lung model for this experiment and performed the experiment in triplicate. KRAS is mutated in approximately 30% of all lung cancers35, making lung cells a relevant model to study the effects of activating KRAS mutations. We surveyed the CpG methylation patterns in low passage, immortalized lung epithelial cells stably expressing exogenous KRAS G12V (SAKRAS cells) and compared these cells to non-transformed empty vector controls (SALEB cells). Our analysis showed significantly greater DM CpGs in SAKRAS lung cells overexpressing KRAS G12V (50,611 DM CpGs) compared to Pa16C pancreatic cells with KRAS knockdown (8,888 DM CpGs). Compared to non-transformed SALEB cells, SAKRAS lung cells overexpressing KRAS G12V displayed significantly greater hypomethylated CpGs (Fig. 4A,B). Further categorization of the DM CpGs into “CpG centric” (Fig. 4C, top) and “gene centric” (Fig. 4C, bottom) regions reveal the postional and functional distribution of the methylation changes associated with KRAS G12V overexpression (Fig. 4C,D). The effects on mRNA expression corresponding to six genes of interest haboring DM CpGs was measured using qRT-PCR (Fig. 4E). Promoter hypermethylation correlated with reduced mRNA expression of BRCA1, and hypomethylation correlated with increased expression of NANOG and RELB. However, the relationship between promoter methylation and transcription was not directly correlated in other genes (Fig. 4E). Although changes at individually important CpGs may alter gene expression, alterations to an entire CpG region may be better correlated with changes in gene expression (Fig. 4E, BRCA1). Taken together, these data indicate that overexpression of oncogenic KRAS G12V is associated with significant CpG methylation changes in SALEB cells.
Figure 4.
DNA methylation changes associated with mutant KRAS overexpression in SALEB lung cells. (A) Hierarchical clustering of the top 1000 differentially methylated probes for SALEB and SAKRAS cell lines. (B) Box plot showing overall delta β vales (median of −0.27664) in the SAKRAS cells compared to SALEB cells. (C) Annotation of hypermethylated (left; yellow) and hypomethylated (right; blue) CpGs to CpG islands (top) and gene functional regions (bottom). (D) Diagram showing examples of CpG centric and gene functional centric regions analyzed by the Infinium DNA methylation array. (E) Genes of interest with DM CpGs in SAKRAS cells. Each colored block represents one DM CpG at the respective region of the stated gene. P, promoter region, 5, 5’UTR; B, Body, gene body; 3, 3′UTR (left panel); The mRNA expression of these genes was measured using qRT-PCR (right panel).
Gene ontology analysis of DM CpGs reveals enrichment of genes involved in development and differentiation associated with changes in KRAS expression
To focus our analysis on genes with DM CpGs most likely to produce transcriptional effects in SAKRAS cells overexpressing KRAS G12V, we isolated promoter regions with consistently DM CpGs as previously described for the pancreatic cell lines (Fig. 3). Five hundred and forty-seven genes met these conditions, including 196 genes with hypermethylated promoters. Genes encoding transcription factors, oncogenes, kinases, and growth factors showed differential DNA methylation at their promoters in SAKRAS cells overexpressing KRAS G12V (Table 3). Gene ontology analysis using the list of 196 hypermethylated gene promoters, and 351 hypomethylated gene promoters in SAKRAS cells overexpressing KRAS G12V, showed an enrichment for genes involved in development and differentiation (Fig. 5), consistent with our previous mutant KRAS loss-of-function studies performed in pancreatic cancer cells (Fig. 3 and Supplementary Fig. S6D,E). Gene ontology analysis using the list of hypermethylated and hypomethylated gene promoters from both SAKRAS KRAS G12V expressing cells and Pa16C KRAS knockdown cells, showed the common enrichment for genes involved in differentiation and development (Supplementary Fig. S6D,E). It is noteworthy that while mutant KRAS knockdown and overexpression ultimately results in DM CpGs of genes involved in similar biological processes, the specific number of genes and location of DM affected are distinct and unique to each cell line.
Table 3.
Categorization of gene promoters with differentially methylated (DM) CpGs associated with KRAS overexpression in SAKRAS lung cell line.
| SAKRAS cells | |||
|---|---|---|---|
| Hyper- methylated |
Hypo- methylated |
||
| Transcription Factors | |||
| BARHL2 | HOXA5 | AFF2 | OTX1 |
| BARX2 | IRX1 | BRDT | PAX2 |
| C11orf9 | IRX3 | CITED1 | PAX9 |
| CDX1 | KEAP1 | CRIP1 | SALL1 |
| CDX2 | KLF11 | ELF4 | SIM2 |
| CTNNB1 | MYBL2 | EMX1 | SNAPC2 |
| ETV7 | NKX2-3 | FOXC2 | SOX1 |
| FEZF2 | NKX6-2 | FOXG1 | SOX11 |
| FHL2 | PAX7 | FOXO4 | SOX3 |
| FOXA2 | POU3F2 | GSC | TAF1 |
| GATA5 | PRDM2 | HEYL | TBX1 |
| GBX2 | SOX21 | HIC1 | TBX2 |
| HAND1 | TBX3 | HOXA9 | TBX4 |
| HES5 | ULK2 | HSF4 | TLX2 |
| HES6 | UNCX | ISL2 | TSC22D3 |
| HHEX | UTF1 | LHX2 | ZFP161 |
| HNF1B | VAX1 | LMX1A | ZIC3 |
| HOXA2 | ZIM2 | NFYB | ZNF132 |
| NKRF | ZNF318 | ||
| OLIG2 | ZNF630 | ||
| Cytokines & Growth Factors | |||
| APLN | ADM2 | NGF | |
| CMTM2 | EDN3 | NPY | |
| FGF22 | FGF13 | OXT | |
| NPPC | GAL | STC2 | |
| PYY | GDF7 | ||
| Homeodomain Proteins | |||
| BARHL2 | IRX1 | EMX1 | |
| BARX2 | IRX3 | GSC | |
| CDX1 | NKX2-3 | HOXA9 | |
| CDX2 | NKX6-2 | ISL2 | |
| GBX2 | PAX7 | LHX2 | |
| HHEX | POU3F2 | LMX1A | |
| HNF1B | UNCX | OTX1 | |
| HOXA2 | VAX1 | PAX2 | |
| HOXA5 | TLX2 | ||
| Protein Kinases | |||
| CSNK1D | BRDT | MST4 | |
| EPHA8 | CDKL5 | PDK3 | |
| FLT1 | FASTK | RPS6KA3 | |
| GUCY2D | IRAK3 | TAF1 | |
| STK32C | MAPK4 | ||
| ULK2 | |||
| Oncogenes | |||
| CDX2 | ELF4 | MSI2 | |
| CTNNB1 | FOXO4 | OLIG2 | |
| FEV | GNAS | SEPT9 | |
| PAX7 | HOXA9 | TCL1A | |
| Cell Differentiation Markers | |||
| FUT4 | CD151 | ||
| GP1BB | CD8A | ||
| IL13RA1 | |||
| PTPRJ | |||
| Tumor Suppressors | |||
| BRCA1 | FAM123B | ||
Figure 5.
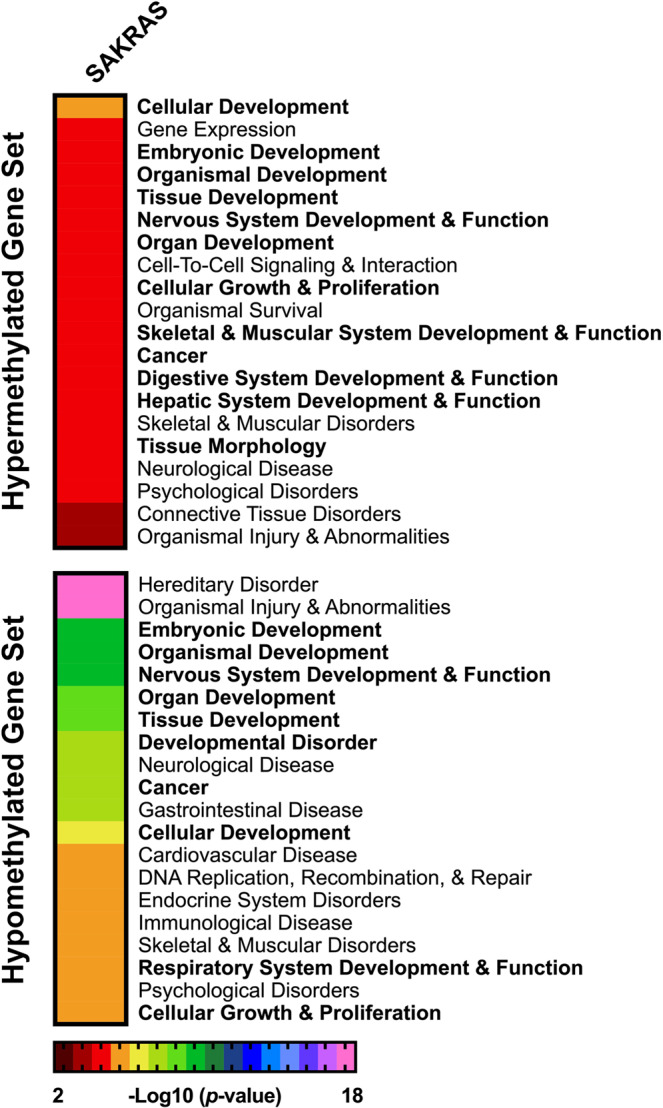
Gene ontology analysis of differentially methylated promoters associated with KRAS G12V overexpression in lung cancer cells. Gene ontology analysis of the hypermethylated (Top) and hypomethylated (Bottom) gene sets from the SAKRAS lung cell line are ranked using a negative log10 scale of the p-values. The top 20 biological processes are shown. Biological processes involved in cell development and differentiation shown in bold.
To directly assess the role of mutant KRAS in maintaining DNA methylation patterns in the isogenic lung cells, we identified differentially methylated (DM) CpGs from SAKRAS cells in which KRAS expression had been suppressed transiently with KRAS siRNA and compared these to cells transfected with control siRNA (Supplementary Fig. S6A,B). We observed 86 DM CpGs in SAKRAS cells following siRNA-mediated KRAS knockdown (Supplementary Fig. S6B). Interestingly, only two of these CpGs were also DM in the SAKRAS vs SALEB cell comparison. This included LRRC7 and the pluripotency transcription factor, NANOG, which were both hypomethylated in SAKRAS cells compared to SALEB cells, and then hypermethylated following KRAS depletion via siRNA (Supplementary Fig. S6B, Left). We identified 10 probes that were differentially methylated in opposite directions when comparing the Pa16C cells in which KRAS had been depleted with shRNA to the SAKRAS cells (Supplementary Fig. S6C). Taken together, the lists of DM genes affected by changes in KRAS expression while distinct between cell lines, showed an enrichment for genes involved in development and differentiaion.
Discussion
The RAS small GTPase is the most commonly mutated oncoprotein in cancer1. RAS and its downstream effectors control key aspects of cancer development. However, until recently, attempts to directly target oncogenic KRAS have been unsuccessful. In addition to aberrant signaling, the expression of mutant KRAS is correlated with global differential DNA methylation24,25. Therefore, epigenetic changes associated with oncogenic KRAS expression could be an avenue where the survival of KRAS-dependent cancer cells may be vulnerable. Here we demonstrated that the that cell type was more impactful than mutant KRAS on DNA methylation. KRAS-mutant PDAC cell lines were also classified based on the responsiveness of their methylome to KRAS depletion. Furthermore, a number of studies suggest that the majority of differential DNA methylation associated with cancer may be stochastic in nature - contributing to low levels of overlap and high heterogeneity between cell lines, even when they share the same genetic background and/or origin28,36–40. It is most likely due to this stochastic nature that we did not observe previously described methylation events driven by RAS, such as the silencing of proapoptotic FAS by HRAS in fibroblasts24, and the silencing of IRAK3 by mutant KRAS21. However, we did identify novel changes in genes related to development and differentiation after KRAS silencing, which was common to all our pancreatic cancer cell lines but was more pronounced in the KRAS-responsive lines. This suggests that while many DNA methylation changes could be stochastic in nature and simply “passenger” events, or a consequence of their cell state and cell lineage, KRAS is likely still able to influence key changes to the epigenome that are ultimately crucial for the cancer phenotype. More studies are needed to determine whether stratification, such as by cancer subtype, will reveal more consistent changes in methylation patterns.
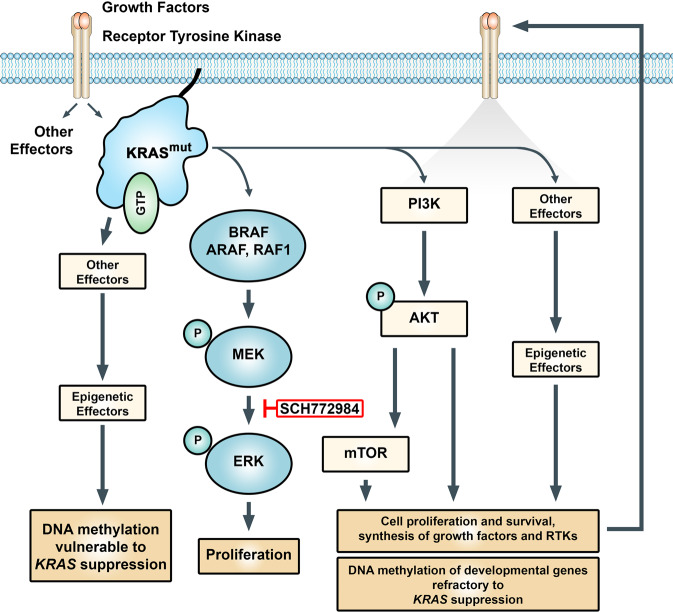
Another interesting observation is the variable number of DM CpGs associated with KRAS knockdown and/or KRAS overexpression. KRAS remains crucially linked to cell proliferation through RAF-MEK-ERK mitogen activated protein kinase (MAPK) cascade, its main effector pathway, and inhibition of this pathway reliably leads to growth arrest15. However, we showed that ERK was not responsible for changes in the methylome, at least over the time frame observed. While it is possible that ERK could play a role in methylation changes over a longer period of time, the question remains, if not ERK, which KRAS effectors are leading to short term DNA methylation changes. In mouse lung adenocarcinoma cells, YAP1 was able to rescue KRAS depleted cells, suggesting a relevant mechanism to bypass loss of KRAS signaling41. In the same study, KRAS also induced PI3K expression, and yet, the subsequent suppression of KRAS has no effect on the upregulated AKT activation. PI3K has been shown to compensate for KRAS suppression in pancreatic cancer cells and regulate epigenetic modifiers including DNMTs42. Cells in which KRAS levels have been genetically reduced display sensitivity to PI3K inhibitors and dual PI3K and MEK inhibitors have been found to be more effective than blocking the individual pathways alone43. PI3K/AKT signaling has been shown to be an epigenetic regulator in multiple cancers by modulating the activity of DNA methyltransferase I (DNMT1)44. It is possible that persistent PI3K-AKT activation, even after KRAS suppression, may be able maintain the majority of methylation changes induced by mutant KRAS. This kind of sustained activity by effector pathways could maintain the methylation status of the majority of the changes initially induced by mutant KRAS expression, but were not reversed upon KRAS knockdown (Fig. 6).
Figure 6.
Model showing epigenetic regulation of developmental genes by mutant KRAS. Activating KRAS mutations lead to persistant induction of effector pathways that drive the cancer phenotype including the differential DNA methylation of genes involved in development and differentiation. In some cell lines, effector pathways such as PI3K and others, are able to maintain their abberant activity independent of KRAS signaling. As a consequence of feed forward loops initiated by mutant KRAS, kinome reprogramming, or the establishment of stable epigenetic patterns, the majority of DNA methylation changes associated with mutant KRAS activity remains refractory to KRAS suppression. However, independent of the changes in DNA methylation, KRAS knockdown and ERK inhibition still both lead to growth arrest in KRAS driven cell lines. SCH772984, type I and type II ERK inhibitor.
DM CpGs associated with KRAS overexpression in our study have been localized to the promoters of important tumor suppressors, oncogenes, transcription factors, and regulators of differentiation, with gene ontology analysis revealing an enrichment for differentially methylated genes involved in differentiation and development. The regulation of pluripotency and lineage-specific genes requires the integration of multiple signaling pathways, epigenetic modifiers, and transcription factors45. In response to KRAS suppression, KRAS-driven cells may rely on compensatory survival pathways such as the PI3K pathway. PI3K-AKT has been shown to affect the expression of differentiation and stemness genes. In our pancreatic cells, particularly KRAS-responsive cells such as Pa16C, we identified many differentially methylated genes associated with stemness following KRAS knockdown, suggesting that KRAS could be involved in inducing stemness in cancer cells through PI3K/AKT. This includes promoter hypomethylation upstream regulators of AKT signaling, such as FGF946,47, and NRG3, a ligand that activates HER3, an EGFR member of receptor tyrosine kinase (RTK) signaling upstream of PI3K-AKT48. POU3F2 and OLIG2 were both hypomethylated - two out of the four genes, from a core set of neurodevelopmental transcription factors (POU3F2, SOX2, SALL2, and OLIG2) essential for GBM propagation. These transcription factors coordinately bind and activate regulatory elements sufficient to fully reprogram differentiated GBM cells into tumor propagating stem-like cells49. Another promoter hypomethylated upon KRAS knockdown is HOXA9, a major transcription factor that regulates stem cells during development. Aberrant expression of HOX genes occurs in various cancers, and HOXA9 transcriptomes are specifically associated with cancer stem cell features50. Hypomethylation was also found at the BMP3 promoter. BMPs are implicated in activation of signaling pathways that drive epithelial-mesenchymal transition (EMT), including WNT signaling, TGFB signaling and PI3K signaling, all important pathways in pancreatic cancer cells41,51,52. And finally, another promoter which appeared as hypomethylated was TWIST1, a canonical EMT transcription factor shown to promote cancer stem cell properties53. Overexpression of TWIST1 is reported to override Myc-induced apoptosis in tumor cells and along with the other changes, could be a compensatory response by the Pa16C KRAS-mutant pancreatic cells to survive KRAS suppression.
Together, our findings suggest that while oncogenic KRAS-associated DNA methylation changes may be stochastic in nature and superseded by cell type, the changes nevertheless converge on biological processes most notably involving pathways of development and differentiation. That ERK inhibition was not analogous to KRAS suppression in Pa16C cells suggests that KRAS-mediated DNA methylation are sustained independent of ERK. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream effector signaling. This may therefore represent a non-canonical mechanism for enhancing tumorigenic potential and possibly help explain the ineffectiveness of KRAS effector inhibition in the clinic. Exploring the KRAS-mediated methylation changes in these pathways may be a deserving direction toward identifying supplementary strategies to target KRAS-driven cancers.
Methods
Cell culture
PDAC cell lines were obtained from ATCC and were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal calf serum (FCS) (HPAC and PANC-1), in RPMI 1640 supplemented with 10% FCS (CFPAC-1, HPAF-II, Panc 10.05, and SW-1990). Low passage SALEB and SAKRAS cells were generous gifts from Dr. Scott H. Randell (UNC-Chapel Hill) and were grown as described previously54. The SALEB cells were generated by infecting small airway lung epithelial cells with an amphotropic retrovirus that transduces SV40 ER, which encodes both the LT and small t antigens, and a neomycin drug resistance marker. These cells were subsequently infected with a retrovirus vector that transduces the hTERT gene together with the hygromycin resistance marker. Expression of these genetic elements was sufficient to immortalize the SALEB* cells. Finally, SALEB* cells were infected with retrovirus that transduces (i) the puromycin resistance marker (SALEB) or (ii) mutant KRAS G12V oncogene together with the puromycin resistance marker (SAKRAS). All other cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; EMD Millipore). Cell lines were used for no longer than six months before being replaced. Stable cell lines were generated by selection in 2 μg/ml puromycin.
Western Blot reagents
Cells were lysed in 1% NP-40 lysis buffer (phosphatase and protease inhibitors from Sigma-Aldrich added fresh). Protein extracts were quantified by Bradford assay (Bio-Rad Laboratories) and analyzed by SDS-PAGE. Blot analyses were done with phospho-specific antibodies to ERK1/2 (T202/Y204) and antibodies recognizing total ERK1/2 to control for total protein expression. Antibody to KRAS4B was obtained from Calbiochem. Antibody for β-actin was used to verify equivalent loading of total cellular protein. Antibodies were purchased from Cell Signaling Techonology unless otherwise stated.
Small molecule inhibitors
The ERK1/2-selective inhibitor SCH772984 was provided by A. Samatar (Merck). Inhibitors for in vitro studies were dissolved in dimethyl sulfoxide (DMSO) to yield a 10 mM or 20 mM stock concentration and stored at −20 or −80 °C, respectively.
siRNA and shRNA transfection reagents
The following human siRNA (siGenome SMARTpool) was purchased from Dharmacon as a pool of four annealed dsRNA oligonucleotides: KRAS (L-005069–00) and non-targeting control #3 (D-001210-03). Dharmafect transfection reagent 1 was used to transfect 20–40 nM siRNA according to manufacturer’s instruction and cells were harvested 96 hours after transfection. The target sequence for the validated shRNA construct used to target KRAS was CAGTTGAGACCTTCTAATTGG. The lentivirus vector encoding shRNA targeting KRAS (TRCN0000010369) was provided by J. Settleman (Genentech). Target cells were transduced by combining viral particle-containing medium with complete media at a ratio of 1:4 in the presence of polybrene (8 μg/ml). Media were exchanged 8–10 h later and selection was initiated following 16 h incubation in complete media. Samples were collected 72–120 h after the initiation of selection.
DNA methylation analysis
Global DNA methylation was evaluated using the Infinium HumanMethylation450 BeadChip Array using more than ~450,000 Infinium CpG probes (Illumina, San Diego, CA). 1 μg of each DNA sample underwent bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer’s recommendation for the Illumina Infinium Assay. Bisulfite-treated DNA was then hybridized to arrays according to the manufacturer’s protocol. We used GenomeStudio V2011.1 (Illumina) for methylation data assembly and acquisition. Methylation levels for each CpG residue are presented as β values, estimating the ratio of the methylated signal intensity over the sum of the methylated and unmethylated intensities at each locus. The average β value reports a methylation signal ranging from 0 to 1, representing completely unmethylated to completely methylated values, respectively. Methylation data was preprocessed using the DMRcate package55. Data preprocessing included background correction, probe scaling to balance Infinium I and II probes, quantile normalization, and logit transformation. A logit transformation converts otherwise heteroscedastic beta values (bounded by 0 and 1) to M values following a Gaussian distribution. Additionally, detection p-values>0.05 in 25% of samples, probes on X and Y chromosomes, and probes situated within 10 bp of putative SNPs were removed. Differential methylation analysis on logit-transformed values was performed to compare samples in IMA. Wilcox rank test was conducted between experimental and control samples and p-values were corrected by calculating the false discovery rate by the Benjamini-Hochberg method. Probes with adjusted p-values <0.05, and delta β values ≥0.2 or ≤ −0.2 to 4 significant figures are considered statistically significant and differentially methylated. The methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119548. The ENCODE methylation data used in this publication were retrieved from the ENCODE Data Coordination Center and are accessible at https://www.encodeproject.org/ucsc-browser-composites/ENCSR037HRJ.
RNA sequencing and analysis
RNA sequencing was performed as described in Bryant et al.29. Briefly, a panel of human PDAC cell lines was infected with lentiviral vectors encoding shRNA targeting KRAS or a scrambled control construct for 8–10 h, and then selected for 48–96 h (depending on cell line). Following selection, cells were washed twice in ice cold phosphate-buffered saline (PBS), scraped in ice cold PBS, collected by centrifugation, and flash frozen. Total RNA (50 ng) for the pancreatic cell lines was used to generate whole transcriptome libraries for RNA sequencing using Illumina’s TruSeq RNA Sample Prep. Poly-A mRNA selection was performed using oligo(dT) magnetic beads, and libraries were enriched using the TruSeq PCR Master Mix and primer cocktail. Amplified products were cleaned and quantified using the Agilent Bioanalyzer and Invitrogen Qubit. The clustered flowcell was sequenced on the Illumina HiSeq. 2500 for paired 100-bp reads using Illumina’s TruSeq SBS Kit V3. Lane level fastq files were appended together if they were sequenced across multiple lanes. These fastq files were then aligned with STAR 2.3.1 to GRCh37.62 using ensembl.74.genes.gtf as GTF files. Transcript abundance was quantified and normalized using Salmon in the unit of transcripts per million (TPM). Clustering was performed using R heatmap.2 package with Euclidean Distance and McQuitty clustering method. Binary sequence alignment/map (BAM) files of RNA-seq data is available from the EMBL-EBI European Nucleotide Archive (ENA) database - http://www.ebi.ac.uk/ena/ with accession number PRJEB25797. The data are accessible at http://www.ebi.ac.uk/ena/data/view/PRJEB25797. The sample accession number is ERS2363485-ERS2363504.
Gene ontology analysis
The differentially methylated (DM) CpGs (i) in a promoter region (200–1500 bases upstream of the transcription start site of a gene) and (ii) within 4 kb of a CpG island (including CpGs at shores and shelves) are referred to as Promoter CpGs in this study. If a gene contains Promoter CpGs that did not all change in the same direction (all hypermethylated or all hypomethylated), that gene was excluded from analysis. Gene sets with hypermethylated or hypomethylated Promoter CpGs are loaded into Molecular Signature Database (MSigDB)56 (http://www.broad.mit.edu/gsea/) and members of each gene set are categorized by gene families. The gene ontology analyses were generated using IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis)57. The gene set of interest was uploaded into IPA (Ingenuity Systems, Redwood City, CA) and the Core Analysis workflow was run with default parameters. The Core Analysis provides an assessment of significantly altered pathways, molecular networks, and biological processes represented in the samples’ gene list. The relative ranking order of biological processes were determined using a negative log10 scale of their p-values. The most enriched (top 20) biological processes with p-value <0.01 were picked. The gene sets used for analysis either contained hypermethylated Promoter CpGs only or hypomethylated Promoter CpGs only. Individual promoters with both hypermethylated and hypomethylated Promoter CpGs were excluded from gene set enrichment analysis.
Supplementary information
Acknowledgements
Thanks to the members of the Baldwin lab for their friendship and support. Many thanks to Drs. Brian Strahl, Yue Xiong, William Kim and Whitney Henry for productive discussions. We would also like to acknowledge the ENCODE Consortium for providing cell line DNA methylation data. This work was supported in part by the NCI grant R35CA197684. This work was supported by the NCI grant R35CA197684 (A.S.B.), and the Department of Defense W81XWH-15-1-0611 (C.J.D.). K.L.B. was supported by NCI T32CA009156 and a grant from the Pancreatic Cancer Action Network/AACR (15-70-25-BRYA). T.K.H. was supported by NCI T32CA071341 and NCI F3180693.
Author contributions
Conceptualization, J.K.D., K.L.B., C.J.D., A.S.B. and B.S. (Ideas; formulation or evolution of overarching research goals and aims); Methodology, J.K.D., K.L.B., T.K.H and B.S. (Development or design of methodology; creation of models); Software, B.T., D.N.B. and B.S. (Programming, software development; designing computer programs; implementation of the computer code and supporting algorithms; testing of existing code components); Validation, J.K.D., K.L.B. and T.K.H. (Verification, whether as a part of the activity or separate, of the overall replication/reproducibility of results/experiments and other research outputs); Formal Analysis, J.K.D, B.T., D.N.B, S.P., N.T. and B.S. (Application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data); Investigation, J.K.D., K.L.B., T.K.H, G.C.G., B.T. and B.S. (Conducting a research and investigation process, specifically performing the experiments, or data/evidence collection); Resources, C.J.D., A.S.B. and B.S. (Provision of study materials, reagents, materials, patients, laboratory samples, animals, instrumentation, computing resources, or other analysis tools); Data Curation, J.K.D., K.L.B., B.T. and B.S. (Management activities to annotate (produce metadata), scrub data and maintain research data (including software code, where it is necessary for interpreting the data itself) for initial use and later reuse); Writing – Original Draft, J.K.D. (Preparation, creation and/or presentation of the published work, specifically writing the initial draft (including substantive translation)); Writing – Review & Editing, J.K.D., B.T., A.S.B., C.J.D., G.C.G. and B.S. (Preparation, creation and/or presentation of the published work by those from the original research group, specifically critical review, commentary or revision – including pre- or postpublication stages); Visualization, J.K.D., K.L.B., T.K.H. and B.T. (Preparation, creation and/or presentation of the published work, specifically visualization/data presentation); Supervision, C.J.D., A.S.B. and B.S. (Oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team); Project Administration, J.K.D., C.J.D., A.S.B. and B.S. (Management and coordination responsibility for the research activity planning and execution); Funding Acquisition, C.J.D., A.S.B and B.S. (Acquisition of the financial support for the project leading to this publication).
Competing interests
C.J.D. is on the Scientific Advisory Board of Mirati Therapeutics. C.J.D. received funding support from Mirati Therapeutics and Deciphera Pharmaceuticals. C.J.D. has been a consultant with Deciphera Pharmaceuticals, Eli Lilly, Jazz Therapeutics and Ribometrix.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ben Yi Tew and Joel K. Durand.
Contributor Information
Albert S. Baldwin, Email: abaldwin@med.unc.edu
Bodour Salhia, Email: salhia@usc.edu.
Supplementary information
is available for this paper at 10.1038/s41598-020-66797-x.
References
- 1.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. Journal of Cell Science. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. British Journal of Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538:114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massarelli E, et al. KRAS Mutation Is an Important Predictor of Resistance to Therapy with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. Clinical Cancer Research. 2007;13:2890–2896. doi: 10.1158/1078-0432.ccr-06-3043. [DOI] [PubMed] [Google Scholar]
- 7.Riely GJ, Marks J, Pao W. KRAS Mutations in Non-Small Cell Lung Cancer. Proceedings of the American Thoracic Society. 2009;6:201–205. doi: 10.1513/pats.200809-107lc. [DOI] [PubMed] [Google Scholar]
- 8.Zeitouni D, Pylayeva-Gupta Y, Der C, Bryant K. KRAS Mutant Pancreatic Cancer: No Lone Path to an Effective Treatment. Cancers. 2016;8:45. doi: 10.3390/cancers8040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janes MR, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172(578–589):e517. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amgen. A Phase 1/2, Study Evaluating the Safety, Tolerability, PK, and Efficacy of AMG 510 in Subjects With Solid Tumors With a Specific KRAS Mutation, https://clinicaltrials.gov/show/NCT03600883 (2018).
- 13.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eser S, et al. Selective Requirement of PI3K/PDK1 Signaling for Kras Oncogene-Driven Pancreatic Cell Plasticity and Cancer. Cancer Cell. 2013;23:406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Hayes TK, et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell. 2016;29:75–89. doi: 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riquelme E, et al. Modulation of EZH2 Expression by MEK-ERK or PI3K-AKT Signaling in Lung Cancer Is Dictated by Different KRAS Oncogene Mutations. Cancer Research. 2015;76:675–685. doi: 10.1158/0008-5472.can-15-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wee S, et al. PI3K Pathway Activation Mediates Resistance to MEK Inhibitors in KRAS Mutant Cancers. Cancer Research. 2009;69:4286–4293. doi: 10.1158/0008-5472.can-08-4765. [DOI] [PubMed] [Google Scholar]
- 18.Wong K-K, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Current Opinion in Genetics & Development. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baylin, S. B. & Jones, P. A. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol8, 10.1101/cshperspect.a019505 (2016). [DOI] [PMC free article] [PubMed]
- 20.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nature Reviews Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 21.Carvalho D, et al. DNA Methylation Screening Identifies Driver Epigenetic Events of Cancer Cell Survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gazin C, Wajapeyee N, Gobeil S, Virbasius C-M, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serra, R. W., Fang, M., Park, S. M., Hutchinson, L. & Green, M. R. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. eLife3, 10.7554/elife.02313 (2014). [DOI] [PMC free article] [PubMed]
- 26.Su J, et al. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018;19:108. doi: 10.1186/s13059-018-1492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Tongelen A, Loriot A, De Smet C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017;396:130–137. doi: 10.1016/j.canlet.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 28.Xie W, et al. DNA Methylation Patterns Separate Senescence from Transformation Potential and Indicate Cancer Risk. Cancer Cell. 2018;33(309–321):e305. doi: 10.1016/j.ccell.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant KL, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25:628–640. doi: 10.1038/s41591-019-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basseres DS, Ebbs A, Cogswell PC, Baldwin AS. IKK is a therapeutic target in KRAS-Induced lung cancer with disrupted p53 activity. Genes Cancer. 2014;5:41–55. doi: 10.18632/genesandcancer.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bibikova M, et al. Genome-wide DNA methylation profiling using Infinium®assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 32.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones S, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 35.Tomasini P, Walia P, Labbe C, Jao K, Leighl NB. Targeting the KRAS Pathway in Non-Small Cell Lung Cancer. Oncologist. 2016;21:1450–1460. doi: 10.1634/theoncologist.2015-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480:250–253. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 37.Feinberg AP. Epigenetic stochasticity, nuclear structure and cancer: the implications for medicine. Journal of Internal Medicine. 2014;276:5–11. doi: 10.1111/joim.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landan G, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nature Genetics. 2012;44:1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 40.Raj A, van Oudenaarden A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao DD, et al. KRAS and YAP1 Converge to Regulate EMT and Tumor Survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fagnocchi L, Mazzoleni S, Zippo A. Integration of Signaling Pathways with the Epigenetic Machinery in the Maintenance of Stem Cells. Stem Cells International. 2016;2016:1–13. doi: 10.1155/2016/8652748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muzumdar, M. D. et al. Survival of pancreatic cancer cells lacking KRAS function. Nature Communications8, 10.1038/s41467-017-00942-5 (2017). [DOI] [PMC free article] [PubMed]
- 44.Spangle JM, Roberts TM, Zhao JJ. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim Biophys Acta Rev Cancer. 2017;1868:123–131. doi: 10.1016/j.bbcan.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang XQ, et al. CDK1-PDK1-PI3K/Akt signaling pathway regulates embryonic and induced pluripotency. Cell Death & Differentiation. 2016;24:38–48. doi: 10.1038/cdd.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai M-S, Cheng Y-S, Chen P-R, Tsai S-J, Huang B-M. Fibroblast Growth Factor 9 Activates Akt and MAPK Pathways to Stimulate Steroidogenesis in Mouse Leydig Cells. PLoS ONE. 2014;9:e90243. doi: 10.1371/journal.pone.0090243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, C. et al. FGF9 from cancer-associated fibroblasts is a possible mediator of invasion and anti-apoptosis of gastric cancer cells. BMC Cancer15, 10.1186/s12885-015-1353-3 (2015). [DOI] [PMC free article] [PubMed]
- 48.Mujoo, K., Choi, B.-K., Huang, Z., Zhang, N. & An, Z. Regulation of ERBB3/HER3 signaling in cancer. Oncotarget5, 10.18632/oncotarget.2655 (2014). [DOI] [PMC free article] [PubMed]
- 49.Suvà ML, et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seifert A, Werheid DF, Knapp SM, Tobiasch E. Role of Hox genes in stem cell differentiation. World J Stem Cells. 2015;7:583–595. doi: 10.4252/wjsc.v7.i3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCormack N, Molloy EL, O’Dea S. Bone morphogenetic proteins enhance an epithelial-mesenchymal transition in normal airway epithelial cells during restitution of a disrupted epithelium. Respiratory Research. 2013;14:36. doi: 10.1186/1465-9921-14-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zabkiewicz C, Resaul J, Hargest R, Jiang WG, Ye L. Bone morphogenetic proteins, breast cancer, and bone metastases: striking the right balance. Endocrine-Related Cancer. 2017;24:R349–R366. doi: 10.1530/erc-17-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mani SA, et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundberg AS, et al. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21:4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 55.Peters TJ, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.