Abstract
Chimeric antigen receptor (CAR) T cell therapy relies on the ex vivo manipulation of patient T cells to create potent, cancer-targeting therapies, shown to be capable of inducing remission in patients with acute lymphoblastic leukemia and large B cell lymphoma. However, current CAR T cell engineering methods use viral delivery vectors, which induce permanent CAR expression and could lead to severe adverse effects. Messenger RNA (mRNA) has been explored as a promising strategy for inducing transient CAR expression in T cells to mitigate the adverse effects associated with viral vectors, but it most commonly requires electroporation for T cell mRNA delivery, which can be cytotoxic. Here, ionizable lipid nanoparticles (LNPs) were designed for ex vivo mRNA delivery to human T cells. A library of 24 ionizable lipids was synthesized, formulated into LNPs, and screened for luciferase mRNA delivery to Jurkat cells, revealing seven formulations capable of enhanced mRNA delivery over lipofectamine. The top-performing LNP formulation, C14—4, was selected for CAR mRNA delivery to primary human T cells. This platform induced CAR expression at levels equivalent to electroporation, with substantially reduced cytotoxicity. CAR T cells engineered via C14—4 LNP treatment were then compared to electroporated CAR T cells in a coculture assay with Nalm-6 acute lymphoblastic leukemia cells, and both CAR T cell engineering methods elicited potent cancer-killing activity. These results demonstrate the ability of LNPs to deliver mRNA to primary human T cells to induce functional protein expression, and indicate the potential of LNPs to enhance mRNA-based CAR T cell engineering methods.
Keywords: Lipid nanoparticles, mRNA delivery, CAR T, T cell engineering
Graphical Abstract
In 2017 the FDA approved CD19 chimeric antigen receptor (CAR) T cell therapy for the treatment of relapsed or refractory acute lymphoblastic leukemia (ALL), given its ability to induce high rates of remission.1,2 In the same year, the therapy was approved for the treatment of relapsed or refractory large B cell lymphoma, as it was also shown to induce remission in these patients.3,4 Based on these successes, CAR T cell therapy is now being explored for the treatment of several other cancers, including glioblastoma5 and refractory multiple myeloma,6 but have yet to receive FDA approval. The development of these autologous therapies relies on ex vivo cell engineering to produce CAR T cells. To produce this form of cancer immunotherapy, patient T cells are harvested, modified to express CD19-specific CAR, and reinfused into the patient. The transmembrane CAR construct allows T cells to target and bind cancerous B cells to induce apoptosis and, thus, eradicate the cancer using the patient’s own immune system.7
Though this process yields potent CAR T cells that induce durable remission,2–4 this therapy can have serious adverse effects that are attributed to patient immune response, as well as potential risks associated with viral transduction and production errors.3,8–12 Immediate reactions, which have been found to occur in nearly 70% of adult patients receiving the therapy,13 include macrophage activation syndrome, neurotoxicity, and cytokine release syndrome.3,9,14 While some of the initial adverse events may be mitigated with anti-IL-6 receptor antibodies,2 the long-term effects can be equally as severe. CD19-directed CAR T cells target both cancerous and normal B cells, often leading to the elimination of all CD19 positive cells, which results in B cell aplasia and hypogammaglobulinemia.9,15–17 Further, in the exploration of new CAR constructs to target biomarkers beyond CD19, several adverse events have been reported, which emphasizes the potential risks associated with uncontrolled CAR T cells and motivates the development of safer CAR T cells for early clinical investigations.18–21 In addition to the safety concerns associated with continuous targeting, CAR T cells with permanent CAR expression, including those that are FDA approved, are most commonly produced via viral transduction, which presents limitations for manufacturing and in vivo translation. Despite their costly and complex synthesis, viruses are limited in the amount of the genetic material they can carry and require elaborate protocols for CAR T cell manufacturing.22–24 Further, utilizing viruses to engineer T cells in vivo instead of ex vivo is restricted by the cost and immunogenicity of viral systems.23,24 In total, these adverse effects highlight the potential risks associated with this potent therapy, while the limitations of viral delivery motivate an investigation into improving CAR T cell production methods to generate safer, less expensive CAR T cells.
One potential solution to overcome these challenges associated with CAR T cell therapy is utilizing messenger RNA (mRNA) to induce CAR expression. mRNA allows for the transient expression of CAR, as it is translated without genomic integration.25 Furthermore, the customizable structure of in vitro transcribed (IVT) mRNA allows it to be engineered for potent transfection and translation.26,27 mRNA-based CAR T cell therapy has been validated in previous studies on a variety of cancers including ALL, melanoma, and Hodgkin’s lymphoma, and it has been shown to reduce the short-term disease burden as effectively as stably expressing CAR T cells.17,28–39 Given this potential, mRNA-based CAR T cell therapy is being evaluated in numerous ongoing clinical trials for cancers, including colorectal cancer and B cell lymphoma, among others.40 These previous investigations have confirmed that CAR expression typically persists for less than a week, which limits the ability of mRNA-based therapies to offer long-term therapeutic benefits without readministration.28,36,38 However, it also has the potential to cause fewer on-target, off-tumor effects and has been shown to offer lower toxicity.28,33,36–38 Additionally, the amount of mRNA delivered to T cells was shown to affect the level of CAR expression on T cells, indicating that mRNA-based CAR expression may offer a means to modulate the side effects, such as cytokine release syndrome, associated with CAR T cell therapy.28,33
However, because naked mRNA degrades rapidly and cannot readily cross the cell membrane, it requires delivery vectors and/or methods for uptake into T cells. Currently, electroporation (EP) is used clinically to deliver mRNA to a variety of cells, including T cells,27,28,41 but it has a number of disadvantages. The membrane disruption that occurs during EP requires specialized equipment, and risks the loss of cytoplasmic content and cytotoxicity, while failing to guarantee consistent membrane penetration across cells for delivery.41,42 This can lead to low viability, and altered gene and protein expression in the surviving cell population.41,43,44 Thus, further investigation into the long-term expression of transgenes and behavior in cells after EP is needed to understand the potential risks associated with this method of nucleic acid delivery.42,45
Nanoparticle (NP)-based delivery systems, composed of lipid- and polymer-based materials, offer a promising means to overcome the challenges faced using mechanical and viral cell engineering methods.41,46–48 NPs require no specialized equipment or elaborate protocols for cellular delivery, and they have numerous potential benefits including the ability to stabilize nucleic acid cargo, aid in intracellular delivery, and mitigate cytotoxicity.26,49–51 There have been numerous investigations into polymer-based NPs for mRNA delivery to cells with promising results, including reduced cytotoxicity compared to EP.47,52–55 Only a few of these investigations have specifically investigated the polymeric NP design for mRNA delivery to T cells, but they have successfully demonstrated the potential of NP platforms for T cell engineering.53,54 However, ionizable lipid nanoparticle (LNP) delivery systems are more clinically advanced than polymers in the context of RNA delivery, with the recent U.S. Food and Drug Administration’s (FDA) approval of Alnylam’s Onpattro.26,56 Additionally, LNPs contain an ionizable lipid core that remains neutral in a physiologically relevant pH but builds charge in acidic environments, such as the endosome, to ultimately aid in endosomal escape and enable potent intracellular nucleic acid delivery.46,57–59 Though the exact mechanisms governing uptake vary by cell type, LNPs enter cells mainly via membrane-derived endocytosis, making endosomal escape crucial for functional mRNA delivery.60,61 LNP delivery platforms have been validated across a variety of cell types, including immune cells, with minimal cytotoxicity, and previous work on lymphocyte delivery revealed that LNPs deliver mRNA more effectively than commercially available lipofectamine.46,47,50,57,58,62 Further, the easily modifiable composition of LNPs allows for the adjustment of their physicochemical properties to maximize their uptake into specific cell types, while their ionizable properties allow them to electrostatically complex with negatively charged nucleic acid cargo.46,47,50,57,62,63,64,65. These properties make LNPs a potentially promising platform for human CAR T cell engineering.
Here, a diverse library of LNPs was evaluated for the delivery of mRNA to T cells. Twenty-four distinct ionizable lipids were combined with set ratios of cholesterol, phospholipid, and lipid-anchored PEG and mixed via a microfluidic device with mRNA to form various LNP formulations (Figure 1A,B). These LNPs were first utilized to deliver luciferase mRNA to Jurkat cells, an immortalized T cell line. This screen identified seven LNP formulations that enhanced mRNA delivery compared to lipofectamine, and a top LNP formulation, C14—4, was identified for its potent transfection and low cytotoxicity. C14—4 LNPs were then optimized for the transfection of primary T cells, and it was shown that LNPs formulated with a purified saturated ionizable lipid led to improved mRNA delivery over LNPs formulated with a crude C14—4 lipid. Finally, to illustrate the translatability of the platform, the optimized C14—4 LNPs were used to encapsulate CAR mRNA to generate functional CAR T cells (Figure 1C). In comparison to EP-based mRNA delivery, C14—4 LNPs induced less T cell toxicity and resulted in similar amounts of CAR surface expression. In a coculture assay with ALL cells, LNP-engineered CAR T cells demonstrated the same potent cancer cell killing ability as both EP- and virally-engineered CAR T cells. Thus, LNPs were validated as a potentially promising alternative strategy for mRNA-based ex vivo engineering of CAR T cells.
Figure 1.
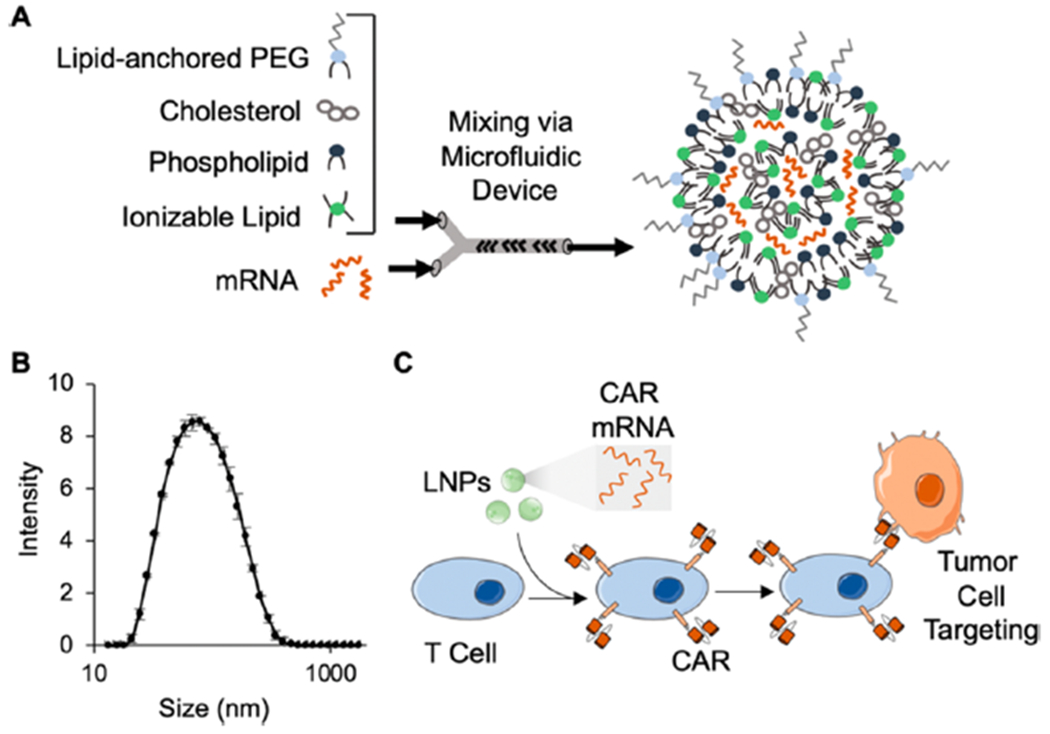
(A) Schematic of the components used to generate LNPs via microfluidic mixing and the expected structure of the resulting LNPs. (B) The size (z-average) distribution of a representative sample of C14—4 LNPs, revealing a diameter of approximately 70 nm using dynamic light scattering. Error bars represent the standard deviation across three samples. (C) Schematic of CAR mRNA loaded LNPs inducing CAR expressionin T cells, resulting in tumor cell targeting and killing.
CHARACTERIZATION OF LNP LIBRARY
In this study, ionizable lipid nanoparticles (LNPs) were investigated for mRNA delivery to T cells. LNPs were selected because they have been shown to deliver mRNA intracellularly with high potency and low cytotoxicity to a range of cell and tissue targets in vivo and ex vivo. Most recently, LNPs have been utilized for nucleic acid delivery to a range of immune cell types.52,61–63 Here, to investigate mRNA delivery specifically to T cells, a library of 24 different LNP formulations was generated by first synthesizing ionizable lipid materials via Michael addition chemistry, where polyamine cores were reacted with an excess of epoxide-terminated alkyl chains of varying lengths (Figure 2, Table S1). The specific ionizable lipids synthesized in this library are structural analogues of an ionizable lipid that was previously formulated into LNPs and shown to deliver siRNA and mRNA to immune cells.58,62,66 Here, LNPs were evaluated for mRNA delivery to T cells specifically. To formulate LNPs, ionizable lipids were combined in ethanol with three excipients: (i) cholesterol, to enhance LNP stability and enable membrane fusion, (ii) dioleoylphosphatidylethanolamine (DOPE), to fortify the bilayer structure of the LNP and promote endosomal escape, and (iii) lipid-anchored polyethylene glycol (C14-PEG), to reduce aggregation and nonspecific endocytosis.67–69 This ethanol phase was then mixed with aqueous phase mRNA in a microfluidic device (Figure 1A). These excipients and their molar ratios were chosen based off of previously optimized LNP formulations for mRNA delivery, which generally utilized (i) DOPE as the phospholipid component, (ii) a decreased molar percentage of ionizable lipid, and (iii) increased concentrations of cholesterol and lipid-anchored PEG.63,70 Given that alterations in the molar ratio of excipients impact the physicochemical properties and ultimately potent delivery of LNPs, the ratio of the components was held constant throughout these experiments.63,70,71
Figure 2.
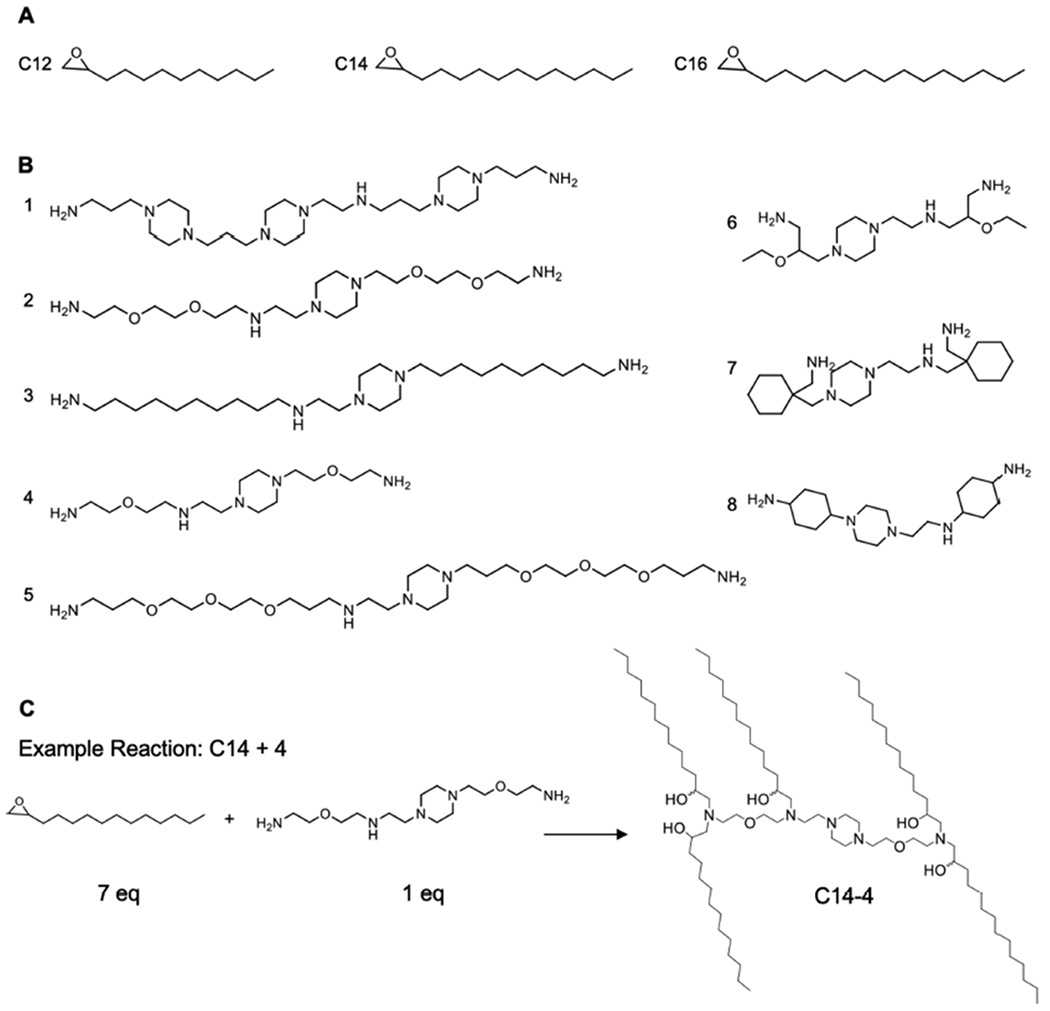
Structures of the alkyl chains (A) and polyamine cores (B) used to generate the ionizable lipid library and a reaction scheme (C) of the Michael addition chemistry used to synthesize the ionizable lipids by reacting an excess of alkyl chains with the polyamine cores. C14—4 is used here as a representative reaction.
The resulting LNPs were then characterized for size and mRNA concentration using dynamic light scattering (DLS) and A260 absorbance measurements. The diameter of the LNPs, reported as the z-average measurement, ranged from 51.05 to 97.01 nm with PDIs below 0.3 (Table S2). The mRNA concentration measured as A260 absorbance showed consistency across LNP formulations, ranging from 33.3 to 48.3 ng/ μL. Collectively, these results confirmed the formation of 24 different LNP formulations encapsulating mRNA to be used in this investigation for T cell delivery.
SCREENING OF LNPS FOR MRNA DELIVERY TO JURKAT CELLS
To evaluate LNPs for their ability to deliver mRNA, luciferase was chosen as the encoded reporter protein. After the addition of luciferin, only luciferase protein translated from the mRNA reacts to generate a luminescent signal, creating an easily detectible output that correlates with functional mRNA delivery.72 The luciferase mRNA used in these experiments utilized N1-Methyl-PseudoU and 5-Methyl-C modifications, which have been shown to enhance mRNA translation by both increasing ribosome density and enabling encapsulation within LNPs.30,73,74 These modifications may alter mRNA encapsulation in LNPs, delivery of the mRNA, and overall immunogenicity, and further investigation into the optimized modifications for these specific LNP delivery vehicles could be explored in future work30,50,75–78
Here, LNP-mediated delivery of luciferase mRNA was assessed in Jurkat cells, a line of immortalized human T cells commonly utilized to study T cell behavior.52,79,80 LNPs encapsulating luciferase mRNA were used to treat Jurkat cells at a concentration of 30 ng/60 000 cells. After 48 h, luciferase expression was assessed via luminescence measurements. The luminescence measurements from LNP formulations were normalized to an untreated cell group and compared to commercially available lipofectamine, a commonly used transfection reagent that is widely considered as the gold standard in vitro.81,82 The library screen identified seven LNP formulations that resulted in significantly higher luciferase expression than lipofectamine, indicating enhanced luciferase mRNA delivery to Jurkat cells (Figure 3A). Enhanced delivery did not correlate with LNP size, mRNA concentration, or ionizable lipid pKa (Figure S1). Of these seven top-performing LNPs, three formulations were comprised of ionizable lipids with C12 alkyl chains, three with C14 alkyl chains, and one with C16 alkyl chains. For the majority of the polyamine cores, C16 alkyl chains resulted in the lowest mRNA delivery. Polyamine cores 3, 6, and 7 did not enhance transfection compared to lipofectamine, regardless of the alkyl chain length. However, polyamine cores 2, 4, and 5, all with similar structures of only one ring and additional oxygens, were responsible for producing the five formulations with the highest resulting luciferase expression, C14—4, C14–2, C14–5, C16–2, and C12–4 LNPs. The two other polyamine cores with only one ring structure were 3 and 6, with polyamine core 3 missing additional oxygen groups and polyamine core 6 including branched features dissimilar in structure to the successful polyamine cores. Thus, it is possible that oxygen atoms in the polyamine core and unbranched chains, in addition to a single ring structure, play a role in ionizable lipid-based mRNA delivery to T cells, though further investigation would be necessary to better understand this potential structure-function relationship.
Figure 3.
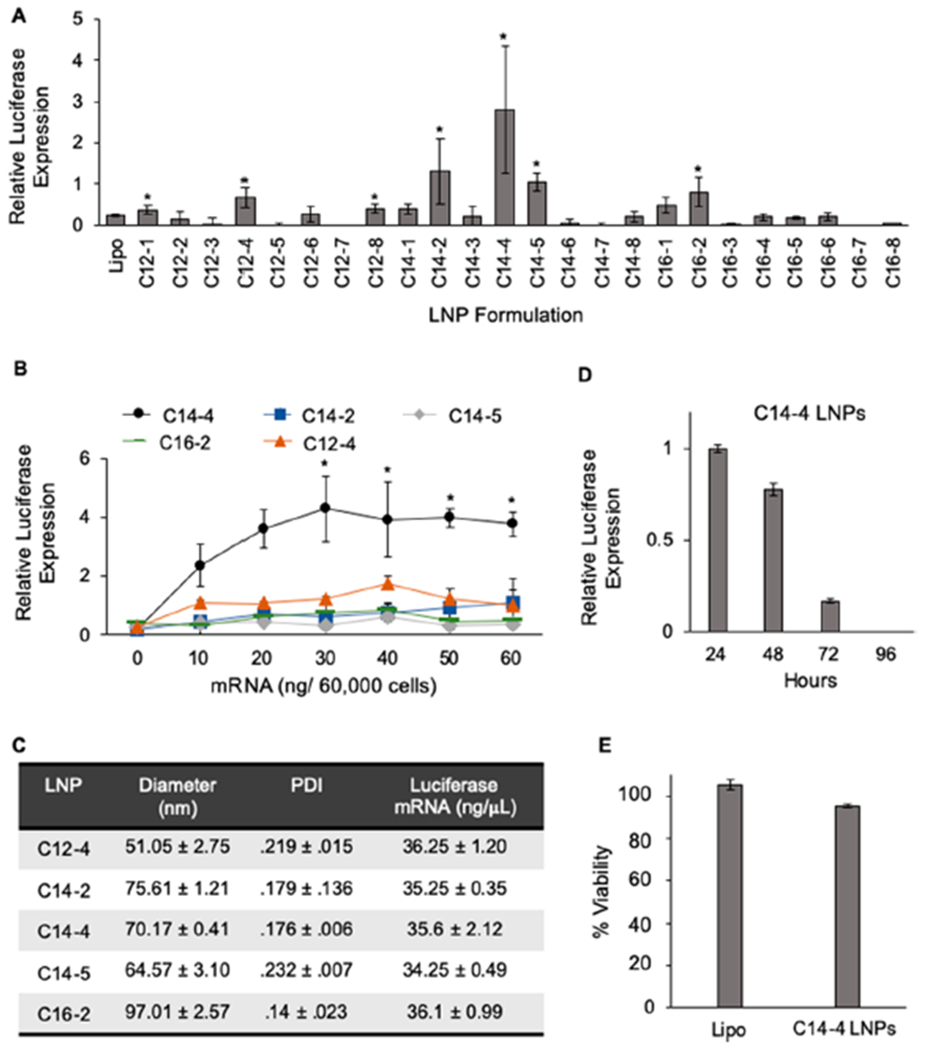
(A) Luciferase expression in Jurkat cells after treatment with the LNP library and lipofectamine for 48 h at a dose of 30 ng/60 000 cells identifies top-performing LNPs. Results were normalized to untreated cells, and the background luminescence was subtracted. *: p < 0.05 in paired student t test to lipofectamine. n = 4 biological replicates. (B) Luciferase expression of Jurkat cells treated with the top five performing LNP formulations to determine the top-performing LNP formulation. Results were normalized to untreated cells, and the averaged luminescent background was subtracted. *: p < 0.05 in Tukey’s multiple comparison test between C14—4 and each formulation. n = 3 biological replicates. (C) Table reporting the diameters (z-average), polydispersity index, and mRNA concentration (± standard deviation) of the top five performing LNP formulations. n = 3. (D) Luciferase expression over time in Jurkat cells treated with 30 ng/60 000 cells of C14—4 for 24 h confirms transient expression of the protein. Results normalized to expression at 24 h with the background subtracted. n = 3 biological replicates. (E) Viability of Jurkat cells treated with 30 ng mRNA/60 000 cells for 48 h using lipofectamine or C14—4, showing minimal cytotoxicity associated with C14—4 LNP treatment. Results normalized to untreated cells with the background subtracted. n = 3 biological replicates.
After the initial screen, the top five performing LNP formulations, C14—4, C14–2, C14–5, C16–2, and C12–4, were compared over a range of mRNA concentrations to determine both the top performing LNP formulation and the optimal LNP dose for Jurkat cell transfection. The results confirmed that C14—4, the top-performing LNP formulation from the original library screen, induced the highest luciferase expression out of the top five performing formulations (Figure 3B). The increase in luciferase expression was significant compared to all other LNP formulations at doses greater than 20 ng, indicating that the optimal dose for C14—4 LNPs in Jurkat cells was 30 ng. The enhanced performance of C14—4 LNPs does not reflect a difference in LNP size or mRNA concentration, as the LNP formulation has a diameter of 70.17 nm and a concentration of 35.6 ng/ μL (Figure 3C). Further, to verify the transient expression of mRNA delivered via C14—4 LNPs, luciferase expression in Jurkat cells treated with LNPs was observed over 96 h (Figure 3D). The results show a 23% decrease after 48 h, confirming transient luciferase expression and informing the use of 24 h time points for subsequent experiments. To determine the cytotoxicity of C14—4 LNPs, Jurkat cells treated with either C14—4 LNPs or lipofectamine for 48 h were compared to an untreated group, and no significant difference in viability was observed (Figure 3E). Collectively, these results allowed for the selection of C14—4 LNPs as the top performing formulation for mRNA delivery, and provided optimized transfection methods for C14—4 LNPs in vitro.
LIPID NANOPARTICLE-MEDIATED MRNA DELIVERY TO PRIMARY HUMAN T CELLS
Because CAR T cells used for clinical cancer immunotherapy are generated using harvested patient T cells, the top-performing C14—4 LNPs were evaluated for mRNA delivery to primary human T cells to demonstrate translatability beyond the Jurkat cell line. Limitations of the Jurkat cell line include that it is derived from only CD4+ T cells, whereas primary T cells also include CD8+ phenotypes.79 However, primary T cells require activation to achieve transfection.21,22 Dynabeads, widely utilized magnetic beads coated with stimulatory CD3 and CD28 antibodies, were used for the activation of T cells in a similar fashion as is employed for clinical CAR T cell production.72,83,84,85 CD4+ and CD8+ T cells were isolated from the peripheral blood of healthy human donors and combined at a 1:1 ratio.37 These cells were then treated with C14—4 LNPs, encapsulating luciferase mRNA, at a range of concentrations. After 24 h, luciferase expression and cell viability were quantified (Figure 4A). LNPs induced luciferase expression in T cells in an mRNA dose-dependent manner, indicating successful delivery of luciferase mRNA to the T cells. Further, minimal cytotoxicity was observed at only the highest doses, indicating minimal toxic effects of C14—4 LNPs on primary T cells.
Figure 4.
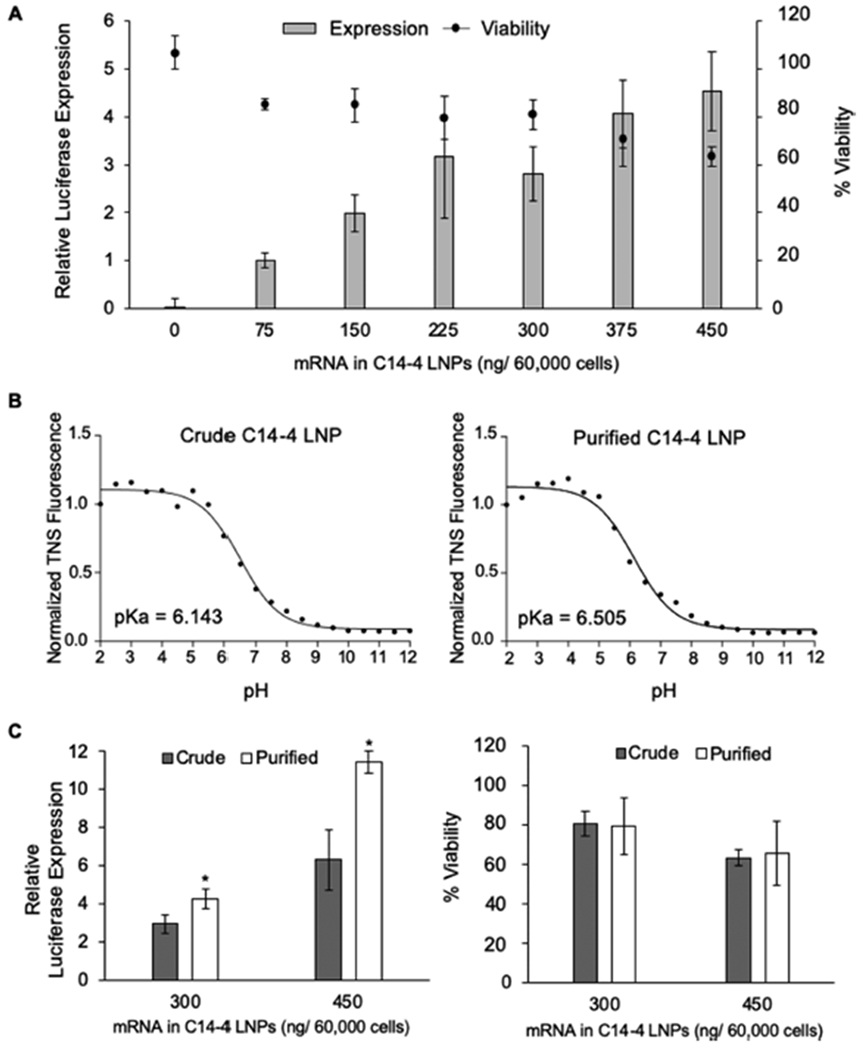
(A) Luciferase expression and viability of primary T cells treated with crude C14—4 LNPs for 24 h. n = 3 biological replicates. (B) Results of the TNS assay to determine LNP pKa for the crude and purified C14—4 LNPs encapsulating luciferase mRNA. pKa is calculated as the pH corresponding to half of the maximum TNS fluorescence value. (C) Luciferase expression and viability of primary T cells treated with either crude or purified C14—4, showing increased luciferase expression with no increase in cytotoxicity. *: p < 0.05 in paired student t test comparison of the crude and purified LNPs. n = 3 biological replicates. For panels A and C, luciferase expression was normalized to the lowest treatment (75 ng/60 000 cells), and the viability was normalized to no treatment with the background subtracted.
To further explore the potential of C14—4 LNPs for mRNA delivery to T cells, the fully saturated ionizable lipid was purified via flash chromatography, and the purified product was utilized to formulate C14—4 LNPs. These purified C14—4 LNPs were compared with C14—4 LNPs made from the crude C14—4 ionizable lipid, to verify that this structure was essential for potent mRNA delivery. DLS and A260 absorbance characterization of the purified C14—4 LNPs revealed a diameter of 65.19 nm and mRNA concentration of 29.8 ng/ μL, which did not greatly differ from the LNPs made with crude C14—4 product (Table S3). Using a Ribogreen assay to evaluate mRNA encapsulation in each formulation, it was revealed that the crude and purified formulations had similar encapsulation efficiencies of 92.5% and 86.3%, respectively. Lastly in a TNS assay, the two LNP formulations were evaluated for their pKa, which is defined as the pH at which the LNPs are 50% protonated and is indicative of how pH affects the surface charge and stability of the LNP.72 Ionizable lipids have a pKa below 7, which enables them to become charged in acidic endosomal compartments, which can enable the release of encapsulated mRNA.72,86 Both the crude and purified C14—4 LNPs were shown to be ionizable, with the purified formulation having a slightly higher pKa value (Figure 4B).
The crude and purified C14—4 LNPs were then compared for their ability to deliver mRNA in primary T cells. The T cells were suspended at a 1:1 ratio of CD4+ to CD8+ and activated with Dynabeads before treatment with LNPs. Crude and purified C14—4 LNPs encapsulating luciferase mRNA were investigated at two concentrations for luciferase expression and viability (Figure 4C). At both concentrations, the purified C14—4 LNPs had significantly increased luciferase expression compared to the crude LNP formulation, and both formulations had minimal effects on cell viability. Though the exact mechanism by which the purified product out-performed the crude C14—4 LNPs is unknown, the crude C14—4 was characterized as containing mostly the fully saturated lipid with a small fraction of oversaturated lipid, indicated by its higher molecular weight measured via liquid chromatography—mass spectrometry (Figure S2). These oversaturated ionizable lipids may have an altered polyamine core structure to allow for the addition of a sixth alkyl chain, which could impact their delivery. Overall, the increase in luciferase expression without an increase in cytotoxicity suggests purified C14—4 LNPs as the top-performing formulation for primary T cell mRNA delivery.
LNPS DELIVER CAR MRNA AS EFFECTIVELY AS EP TO PRIMARY T CELLS WITH LOWER CYTOTOXICITY
After quantifying luciferase-encoded mRNA delivery to T cells via luminescence measurements, C14—4 LNPs were utilized for CAR mRNA delivery as a clinically relevant application of the delivery vehicle. CAR T cells generated with mRNA have been utilized in numerous clinical trials as initial investigations suggest that transient CAR expression may overcome obstacles associated with toxicity and off-target effects that result from permanent modification.28,33,40 However, in these investigations, mRNA was delivered to T cells via electroporation (EP), a commonly used, potent but toxic method of transfection that relies on electrically permeabilizing T cell membranes.27,28,31,41,43,44 Here, C14—4 LNPs encapsulating CD19 CAR mRNA were compared to EP to determine their ability to generate functional CAR T cells with minimal cytotoxicity.
LNPs were used to treat primary T cells at a 1:1 CD4+/CD8+ ratio in comparison to EP at identical 450 ng/μL mRNA concentrations. The resulting T cell populations were analyzed for CAR expression by flow cytometry, wherein surface CAR expression was quantified using mean fluorescence intensity (MFI) (Figure 5A). The highest resulting MFIs came from the T cells treated using EP and purified C14—4 LNPs, while crude C14—4 LNPs generated a more modest MFI. When these values were normalized to the untreated control group of T cells, EP showed a 10-fold increase in MFI over untreated cells, and purified C14—4 LNPs showed a 9.4-fold increase, while crude C14—4 LNPs only increased MFI by 4.5-fold. Thus, as observed with luciferase mRNA delivery, the crude C14—4 LNP formulation was less effective in inducing expression than its purified counterpart, despite no major differences in size or mRNA concentration (Figure 5B,C). The viability of T cells treated with LNP formulations in comparison to EP treatment was also quantified (Figure 5D), indicating significantly reduced cytotoxicity with LNP treatment compared to EP. As observed previously with luciferase mRNA delivery, cells treated with purified and crude LNPs have a similar, high viability of 76% and 78%, respectively. This was contrasted by the 31% viability observed in EP treated T cells, emphasizing the cytotoxic nature of this currently utilized mRNA delivery process. Overall, these results highlight the safety benefits of utilizing C14—4 LNPs over EP for mRNA delivery to T cells ex vivo.
Figure 5.
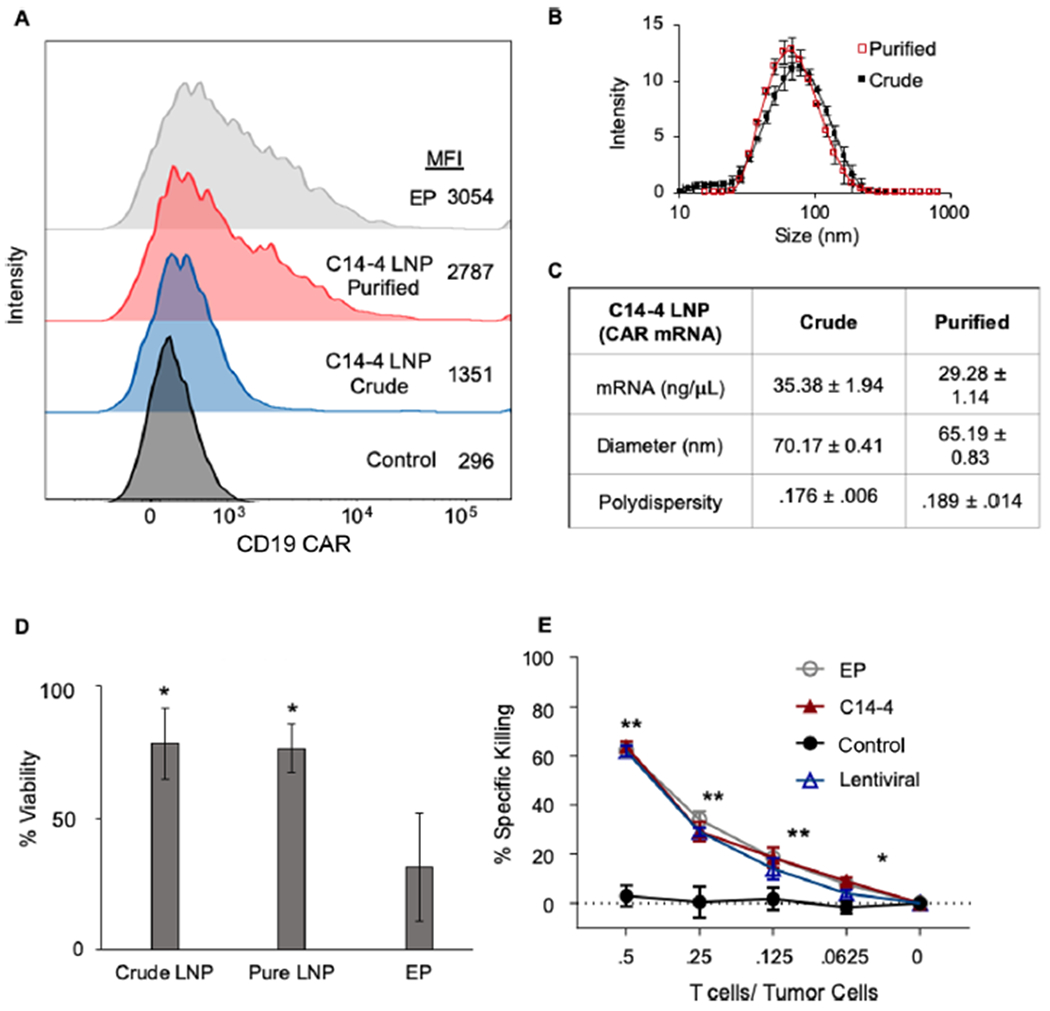
(A) Surface expression of CAR on primary T cells assessed using flow cytometry, indicating increased CAR surface expression, evaluated as the mean fluorescent intensity (MFI), in purified C14—4 LNP and EP treated groups compared to those treated with crude C14—4 LNPs. (B) DLS measurments of crude and purified C14—4 LNPs to indicate their respective sizes. Error bars represent the standard deviation across three samples. (C) Table reporting the mRNA concentration, diameters (z-average), and polydispersity index (±standard deviation) of the crude and purified C14—4 LNPs encapsulating mRNA. (D) Viability of primary T cells treated with each group normalized to treatment with the background subtracted, n = 3 biological replicates, *: p < 0.05 in paired t test to EP. (E) Results of Nalm6 and CAR T cell coplating at different effector-to-target ratios for 48 h normalized to Nalm6 cells co-plated with untreated T cells as the control group. n = 3 wells. *: p < 0.05, **: p < 0.01 in the paired t test to control.
With EP and purified C14—4 LNPs inducing similar CD19 CAR expression in T cells, the two mRNA delivery methods were evaluated for the CAR-driven effector function using a coplated cancer cell killing assay. By engineering Nalm6 ALL cells to express luciferase, cancer cell killing can be assessed by change in luminescence after the coculture with CAR T cells as compared to the signal from Nalm6 cells plated with T cells lacking CAR expression. Both this assay and cell line have been utilized routinely in previous studies to demonstrate the CAR T cell functionality and therapeutic potential.28,87–90 Here, CAR T cells engineered with mRNA delivered via purified C14—4 LNPs or EP were compared at a range of T cell to effector cell ratios. After 48 h, the EP and C14—4 LNP treatment groups performed nearly identically and resulted in significantly more cancer cell killing than the control group (Figure 5E). Thus, the purified C14—4 LNPs successfully generated mRNA-engineered CAR T cells that are comparable to those generated using EP, the current standard of delivery for mRNA CAR T cells. Collectively, these results validate purified C14—4 LNPs as a method for mRNA-based CAR T cell engineering with similar potency and reduced T cell cytotoxicity compared to EP.
In conclusion, this investigation utilized LNPs to achieve mRNA delivery, while lowering cytotoxicity compared to EP treatment of primary human T cells. Utilizing luciferase mRNA delivery to Jurkat cells to assess T cell transfection, a library of 24 distinct LNP formulations was screened, and C14—4 LNPs were identified as the top performer. This formulation was then used to transfect primary human T cells with luciferase-encoding mRNA and was further optimized by purifying the fully saturated C14—4 ionizable lipid for LNP formulation. This purified C14—4 LNP was then able to successfully deliver CD19-targeted CAR mRNA to primary human T cells while lowering the cytotoxicity compared to EP. Further, CAR T cells engineered with C14—4 LNPs, EP, or lentivirus induced the same potent cancer cell killing in a coplated assay using ALL cells. These results demonstrate the ability of C14—4 LNPs to deliver CAR mRNA to primary T cells and generate functional CAR T cells, validating the delivery platform as a new means to engineer CAR T cells that avoids the use of specialized EP equipment. Additionally, by demonstrating the delivery of both luciferase and CAR mRNA, the potential for C14—4 LNPs to be optimized and utilized for a broad range of mRNA-based T cell engineering applications is established.
MATERIALS AND METHODS
Ionizable Lipid Synthesis.
Ionizable lipids were synthesized by reacting epoxide-terminated alkyl chains (Avanti Polar Lipids) with polyamine cores (Enamine, Monmouth Jct, NJ) using Michael addition chemistry. The components were combined with a 7-fold excess of alkyl chains and mixed with a magnetic stir bar for 48 h at 80 °C. The crude product was then transferred to a Rotavapor R-300 (BUCHI, Newark, DE) for solvent evaporation, and the lipids were suspended in ethanol. Finally, to purify the top-performing lipid (C14—4), the lipid fractions were separated via a CombiFlash Nextgen 300+ chromatography system (Teledyne ISCO, Lincoln, NE), and the saturated lipid fraction was identified by molecular weight using liquid chromatography-mass spectrometry.
CAR mRNA Synthesis.
mRNA was produced using standard in vitro transcription methods, as previously described.44 Briefly, plasmid DNA encoding a second-generation lentiviral vector for CD19 targeting CAR bearing the CD3ζ and 4—1BB costimulatory domains was linearized overnight, followed by the production of mRNA using the T7 mMessage ULTRA kit (Thermo Fisher) as per manufacturer instructions. mRNA was then polyA tailed, capped, and purified using the RNeasy mini kit (Qiagen).
Lipid Nanoparticle (LNP) Formulation and Characterization.
To synthesize LNPs, an aqueous phase containing mRNA and an ethanol phase containing lipid and cholesterol components were mixed using a microfluidic device as previously described.91 Briefly, the aqueous phase was prepared using 10 mM citrate buffer and either luciferase mRNA with N1-Methyl-PseudoU and 5-Methyl-C substitutions (Trilink Biotechnologies, San Diego, CA) or CAR mRNA (synthesized as described above) at 1 mg/mL. To prepare the ethanol phase, ionizable lipid, 1,2-distearoyl-sn-glycero-3-phosphoe-thanolamine (DOPE) (Avanti Polar Lipids, Alabaster, AL), cholesterol (Sigma, St. Louis, MO), and lipid-anchored polyethylene glycol (PEG) (Avanti Polar Lipids) components were combined at a molar ratio of 35%, 16%, 46.5%, and 2.5%, respectively. Pump33DS syringe pumps (Harvard Apparatus, Holliston, MA) were used to mix the ethanol and aqueous phases at a 3:1 ratio in a microfluidic device.91 After mixing, LNPs were dialyzed against 1× PBS for 2 h before sterilization via 0.22 μm filters. Dynamic light scattering (DLS) performed on a Zetasizer Nano (Malvern Instruments, Malvern, U.K.) was then used to measure, in triplicate, the diameter (z-average) and polydispersity index (PDI) of the LNPs suspended in 1× PBS. A NanoDrop ND-1000 Spectrophotometer (ThermoFisher, Waltham, MA) was used to obtain the mRNA concentration of each LNP formulation.
Further analysis of top-performing LNP formulations included Quant-iT RiboGreen (ThermoFisher) and 6-(p-toluidinyl)naphthalene-2-sulfonic acid (TNS) assays to determine the encapsulation efficiency and pKa of the LNPs, respectively. The Quant-iT Ribogreen was performed as previously described.92 Briefly, equal concentrations of LNPs were treated with Triton X-100 (Sigma) to lyse the LNPs or left untreated, and after 10 min, the groups were plated in triplicate in 96-well plates alongside RNA standards. The fluorescent Ribogreen reagent was added per manufacturer instructions, and the resulting fluorescence was measured on a plate reader. A standard curve was used to quantify RNA content and calculate encapsulation efficiency. To determine either LNP or ionizable lipid pKa, a TNS assay was used to measure surface ionization as previously described.72 Buffered solutions of 150 mM sodium chloride, 20 mM sodium phosphate, 25 mM ammonium citrate, and 20 mM ammonium acetate were adjusted to reach pH values ranging from 2 to 12 in increments of 0.5. LNPs or ionizable lipids were added to each pH-adjusted solution in triplicate wells in a 96-well plate. TNS was then added to each well to reach a final TNS concentration of 6 μM, and the resulting fluorescence was read on a plate reader. The pKa was then calculated as the pH, at which the fluorescence intensity was 50% of its maximum value, reflective of 50% protonation.
mRNA Delivery to Jurkat Cells in Vitro.
Jurkat cells (ATCC no. TIB-152), an immortalized human T cell line,79 were cultured in RPMI-1640 with l-glutamine (ThermoFisher) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Cells were plated at 60 000 cells per well in 96-well plates in 60 μL of media and were immediately treated with 60 μL of LNPs diluted in PBS to varying concentrations. The lipofectamine MessengerMAX transfection reagent (ThermoFisher), used here as a positive control comparison, was combined with mRNA for 10 min per the manufacturer protocol and used to treat wells using the same mRNA concentration as the LNP groups. After 48 h of incubation, the cells were centrifuged at 300g for 4 min and resuspended in 50 μL of 1× lysis buffer (Promega, Madison, WI) and 100 μL of the luciferase assay substrate (Promega). The luminescence was then quantified using an Infinite M Plex plate reader (Tecan, Morrisville, NC). The luminescent signal from each group was normalized to either untreated cells or the lowest concentration treatment group, and the background, measured as wells with reagents but no cells, was subtracted. To assess the cytotoxicity, Jurkat cells were plated under the same conditions and treated with either C14—4 or lipofectamine at 30 ng mRNA per 60 000 cells. After 48 h, 60 μL of CellTiter-Glo (Promega) was added to each well, and luminescence corresponding to ATP production was quantified using a plate reader. The luminescent signal from each group was normalized to untreated cells, and the background was subtracted. The relative luminescent signal of each LNP was then graphed in comparison to size, mRNA concentration, and ionizable lipid pKa to determine if any correlation between these values and functional mRNA delivery existed (Figure S1).
mRNA Delivery to Primary T Cells ex Vivo.
Primary human T cells (CD3+) were collected from healthy volunteer donors and procured for these studies through the Human Immunology Core service. These cells were combined at an equal ratio of CD4+ and CD8+. Cells to be treated with LNPs were activated overnight with Human T-activator CD3/CD28 Dynabeads (ThermoFisher) at a 3:1 bead to cell ratio. After activation, the cells were plated at 60 000 cells per well in 96-well plates in 60 μL of media and treated with LNPs at varying mRNA concentrations. For electroporation, T cells were washed three times with serum-free media, resuspended at 108 cells/mL, and mixed with transcribed mRNA at a concentration of 100 μg mRNA per 106 T cells. The cells were then electroporated in a 2 mm cuvette using an ECM830 Electro Square Wave Porator (Harvard Apparatus BTX).
For luciferase mRNA experiments, the same protocols described above were used to assess luminescence after 48 h and cytotoxicity after 24 h. For CAR mRNA treatments, T cells were stained using an anti-idiotype antibody to the CD19 CAR (generously provided by Novartis Pharmaceuticals), followed by secondary staining with an antihuman IgG linked to R-phycoerythrin (Jackson Laboratories). T cells were then washed, and the surface CAR expression was evaluated on a BD LSRII Fortessa. Gating was performed as per the standard protocol with doublet exclusion. Cytotoxicity of the CAR mRNA treatments was assessed as described above, using a CellTiter Glo kit.
Lentiviral Vector Production and Primary T Cell Transduction.
293T human embryonic kidney cells were used to generate high-titer, replication-defective lentiviral vectors.93 Here, 107 cells were seeded in T150 tissue culture flasks for 24 h and then treated with 7 μg of pMDG.1, 18 μg of pMDLg/p.RRE packaging plasmids, and 15 μg of transfer plasmid with 96 μL of the Lipofectamine 2000 transfection reagent (Life Technologies, Grand Island, NY). The transfer plasmids containing CAR constructs utilized an EF-1α promoter.94 At 24 and 48 h post-transfection, the viral supernatant was collected and concentrated via ultracentrifugation overnight at 10 500g. Twenty-four h after undergoing Dynabead activation as described above, T cells were combined with lentiviral vectors at a concentration of 5–10 infectious particles per cell.
Functional Assays.
CAR T cells were coplated with luciferase-expressing Nalm-6 cells, a CD19+ pre-B ALL cell line, at varying effector-to-target ratios. Non-engineered, CAR negative T cells were also co-plated with the same cell line to provide a control. After 48 h in the coculture, D-luciferin potassium salt (PerkinElmer, Waltham, MA) was added to cell cultures to reach a final concentration of 15 μg/mL and incubated at 37 °C for 10 min. Luminescence was then detected using a Synergy H4 imager (BioTek, Winooski, VT), and the signal was analyzed using BioTek Gen5 software. The percentage of specific lysis was calculated using the control of target cells without effectors.
Supplementary Material
ACKNOWLEDGMENTS
M.J.M. acknowledges support from a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a US National Institutes of Health (NIH) Director’s New Innovator Award (DP2 TR002776), the National Institutes of Health(-NCI R01 CA241661 and NCI R37 CA244911), a grant from the American Cancer Society (129784-IRG-16-188-38-IRG), an Abramson Cancer Center (ACC)-School of Engineering and Applied Sciences (SEAS) Discovery Grant (P30 CA016520), and a 2018 AACR-Bayer Innovation and Discovery Grant (18-80-44-MITC). M.M.B. is supported by a Tau Beta Pi Graduate Research Fellowship.
ABBREVIATIONS
- LNP
lipid nanoparticle
- NP
nanoparticles
- IVT
in vitro transcribed
- EP
electroporation
- ALL
acute lymphoblastic leukemia
- DLS
dynamic light scattering
- MFI
mean fluorescent intensity
- DOPE
1,2-distearoyl-sn-glycero-3-phosphoe-thanolamine
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.9b04246.
Experimental details and supplementary figures showing the full nomenclature of polyamine core structures, the size and concentration characterizations of all LNP formulations in the library, the characteristics of crude and purified C14—4 LNPs, and the MFI and % specific killing data for crude C14—4 LNPs compared to EP in a coculture assay (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.nanolett.9b04246
Contributor Information
Margaret M. Billingsley, Department of Bioengineering, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
Nathan Singh, Division of Oncology, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States; Division of Oncology, Section of Stem Cell Biology, Department of Medicine, Washington University School of Medicine, St. Louis, Missouri 63110, United States.
Pranali Ravikumar, Division of Oncology, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Rui Zhang, Department of Bioengineering, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
Carl H. June, Abramson Cancer Center, Perelman School of Medicine and Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States
Michael J. Mitchell, Department of Bioengineering, Abramson Cancer Center, Perelman School of Medicine, Institute for Immunology, Perelman School of Medicine, Cardiovascular Institute, Perelman School of Medicine, and Institute for Regenerative Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States.
REFERENCES
- (1).Liu Y; Chen X; Han W; Zhang Y Tisagenlecleucel, an Approved Anti-CD19 Chimeric Antigen Receptor T-Cell Therapy for the Treatment of Leukemia. Drugs Today 2017, 53 (11), 597–608. [DOI] [PubMed] [Google Scholar]
- (2).Maude SL; Frey N; Shaw PA; Aplenc R; Barrett DM; Bunin NJ; Chew A; Gonzalez VE; Zheng Z; Lacey SF; et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. N. Engl. J. Med 2014, 371 (16), 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bouchkouj N; Kasamon YL; Claro RA de; George B; Lin X; Lee S; Blumenthal GM; Bryan W; McKee AE; Pazdur R FDA Approval Summary: Axicabtagene Ciloleucel for Relapsed or Refractory Large B-Cell Lymphoma. Clin. Cancer Res 2019, 25 (6), 1702–1708. [DOI] [PubMed] [Google Scholar]
- (4).Yip A; Webster RM The Market for Chimeric Antigen Receptor T Cell Therapies. Nat. Rev. Drug Discovery 2018, 17, 161. [DOI] [PubMed] [Google Scholar]
- (5).Brown CE; Alizadeh D; Starr R; Weng L; Wagner JR; Naranjo A; Ostberg JR; Blanchard MS; Kilpatrick J; Simpson J; et al. Regression of Glioblastoma after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med 2016, 375 (26), 2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Berdeja JG; Lin Y; Raje NS; Siegel SD; Munshi NC First-in-Human Multicenter Study of Bb2121 Anti-BCMA CAR T-Cell Therapy for Relapsed/Refractory Multiple Myeloma: Updated Results. J. Clin. Oncol 2017, 35 (15), 3010.28715249 [Google Scholar]
- (7).Benmebarek M; Karches CH; Cadilha BL; Lesch S; Endres S; Kobold S Killing Mechanisms of Chimeric Antigen Receptor (CAR) T Cells. Int. J. Mol. Sci 2019, 20, 1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).June CH; Riddell SR; Schumacher TN Adoptive Cellular Therapy: A Race to the Finish Line. Sci. Transl. Med 2015, 7 (280), 280ps7. [DOI] [PubMed] [Google Scholar]
- (9).June CH; Maus MV; Plesa G; Johnson LA; Zhao Y; Levine BL; Grupp SA; Porter DL Engineered T Cells for Cancer Therapy. Cancer Immunol. Immunother 2014, 63 (9), 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fesnak AD; June CH; Levine BL Engineered T Cells: The Promise and Challenges of Cancer Immunotherapy. Nat. Rev. Cancer 2016, 16, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Vormittag P; Gunn R; Ghorashian S; Veraitch FS A Guide to Manufacturing CAR T Cell Therapies. Curr. Opin. Biotechnol 2018, 53, 164–181. [DOI] [PubMed] [Google Scholar]
- (12).Ruella M; Xu J; Barrett DM; Fraietta JA; Tyler J; Ambrose DE; Klichinsky M; Shestova O; Patel PR; Nazimuddin F Induction of Resistance to Chimeric Antigen Receptor T Cell Therapy by Transduction of a Single Leukemic B Cell. Nat. Med 2018, 24 (10), 1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hartsell A Emerging Trends in Chimeric Antigen Receptor T-Cell Immunotherapy in Adults from the Vizient Clinical Database. Biol. Blood Marrow Transplant 2019, 25 (3), S336–S337. [Google Scholar]
- (14).Zheng PP; Kros JM; Wang G Elusive Neurotoxicity in T Cell-Boosting Anticancer Therapies. Trends Immunol 2019, 40 (4), 274–278. [DOI] [PubMed] [Google Scholar]
- (15).Porter DL; Levine BL; Kalos M; Bagg A; June CH Chimeric Antigen Receptor-Modified T Cells in Chronic Lymphoid Leukemia. N. Engl. J. Med 2011, 365 (8), 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Dotti G; Gottschalk S; Savoldo B; Brenner MK Design and Development of Therapies Using CAR Expressing T Cells. Immunol. Rev 2014, 257 (1), 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yang G; Sau C; Lai W; Cichon J; Li W The Principles of Engineering Immune Cells to Treat Cancer Wendell. Cell 2015, 344 (6188), 1173–1178. [Google Scholar]
- (18).Morgan RA; Yang JC; Kitano M; Dudley ME; Laurencot CM; Rosenberg SA Case Report of a Serious Adverse Event Following the Administration of T Cells Transduced with a Chimeric Antigen Receptor Recognizing ERBB2. Mol. Ther 2010, 18 (4), 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Brentjens R; Yeh R; Bernal Y; Riviere I; Sadelain M Treatment of Chronic Lymphocytic Leukemia with Genetically Targeted Autologous T Cells: Case Report of an Unforeseen Adverse Event in a Phase I Clinical Trial. Mol. Ther 2010, 18 (4), 666–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Heslop HE Safer CARS. Mol. Ther 2010, 18 (4), 661–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Büning H; Uckert W; Cichutek K; Hawkins RE; Abken H Do CARs Need a Driver’s License? Adoptive Cell Therapy with Chimeric Antigen Receptor-Redirected T Cells Has Caused Serious Adverse Events. Hum. Gene Ther 2010, 21 (9), 1039–1042. [DOI] [PubMed] [Google Scholar]
- (22).Nightingale SJ; Hollis RP; Pepper KA; Petersen D; Yu XJ; Yang C; Bahner I; Kohn DB Transient Gene Expression by Nonintegrating Lentiviral Vectors. Mol. Ther 2006, 13 (6), 1121–1132. [DOI] [PubMed] [Google Scholar]
- (23).Basarkar A; Singh J Nanoparticulate Systems for Polynucleotide Delivery. Int. J. Nanomedicine 2007, 2 (3), 353–360. [PMC free article] [PubMed] [Google Scholar]
- (24).Seow Y; Wood MJ Biological Gene Delivery Vehicles: Beyond Viral Vectors. Mol. Ther 2009, 17 (5), 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Riley RS; June CH; Langer R; Mitchell M Delivery Technologies for Cancer Immunotherapy. Nat. Rev. Drug Discovery 2019, 18 (March), 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pardi N; Hogan MJ; Porter FW; Weissman D MRNA Vaccines - a New Era in Vaccinology. Nat. Rev. Drug Discovery 2018, 17 (4), 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Smits E; Ponsaerts P; Lenjou M; Nijs G; Van Bockstaele DR; Berneman ZN; Van Tendeloo VFI RNA-Based Gene Transfer for Adult Stem Cells and T Cells. Leukemia 2004, 18 (11), 1898–1902. [DOI] [PubMed] [Google Scholar]
- (28).Barrett DM; Zhao Y; Liu X; Jiang S; Carpenito C; Kalos M; Carroll RG; June CH; Grupp SA Treatment of Advanced Leukemia in Mice with MRNA Engineered T Cells. Hum. Gene Ther 2011, 22 (12), 1575–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Harrer DC; Simon B; Fujii S.-i.; Shimizu K; Uslu U; Schuler G; Gerer KF; Hoyer S; Dorrie J; Schaft N RNA Transfection of γ/δ T Cells with a Chimeric Antigen Receptor or an α/β T-Cell Receptor: A Safer Alternative to Genetically Engineered α/β T Cells for the Immunotherapy of Melanoma. BMC Cancer 2017, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pardi N; Tuyishime S; Muramatsu H; Kariko K; Mui BL; Tam YK; Madden TD; Hope MJ; Weissman D Expression Kinetics of Nucleoside-Modified MRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Controlled Release 2015, 217, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Svoboda J; Rheingold SR; Gill SI; Grupp SA; Lacey SF; Kulikovskaya I; Suhoski MM; Melenhorst JJ; Loudon B; Mato AR; et al. Nonviral RNA Chimeric Antigen Receptor – Modi Fi Ed T Cells in Patients with Hodgkin Lymphoma. Blood 2018, 132 (10), 1022–1027. [DOI] [PubMed] [Google Scholar]
- (32).Rabinovich PM; Komarovskaya ME; Ye Z-J; Imai C; Campana D; Bahceci E; Weissman SM Synthetic Messenger RNA as a Tool for Gene Therapy. Hum. Gene Ther 2006, 17 (10), 060928063342003. [DOI] [PubMed] [Google Scholar]
- (33).Zhao Y; Moon E; Carpenito C; Paulos CM; Liu X; Brennan AL; Chew A; Carroll RG; Scholler J; Levine BL; et al. Multiple Injections of Electroporated Autologous T Cells Expressing a Chimeric Antigen Receptor Mediate Regression of Human Disseminated Tumor. Cancer Res 2010, 70 (22), 9053–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Singh N; Liu X; Hulitt J; Jiang S; June CH; Stephan A; Barrett DM; Zhao Y Differences in Control of Local versus Disseminated Neuroblastoma by Tansiently-Modified GD2 CAR T Cells Due to Variability in Tumor Penetration. Cancer Immunol. Res 2014, 2 (11), 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tasian SK; Kenderian SS; Shen F; Ruella M; Shestova O; Kozlowski M; Li Y; Schrank-Hacker A; Morrissette JJD; Carroll M; et al. Optimized Depletion of Chimeric Antigen Receptor T Cells in Murine Xenograft Models of Human Acute Myeloid Leukemia. Blood 2017, 129 (17), 2395–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Foster JB; Choudhari N; Perazzelli J; Storm J; Hofmann TJ; Jain P; Storm PB; Pardi N; Weissman D; Waanders AJ; et al. Purification of MRNA Encoding Chimeric Antigen Receptor Is Critical for Generation of a Robust T-Cell Response. Hum. Gene Ther 2019, 30 (2), 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Barrett DM; Singh N; Porter DL; Grupp SA; June CH Chimeric Antigen Receptor Therapy for Cancer. Annu. Rev. Med 2014, 65, 333–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Beatty GL; Haas AR; Maus MV; Torigian DA; Soulen MC; Plesa G; Chew A; Zhao Y; Levine BL; Albelda SM; et al. Mesothelin-Specific Chimeric Antigen Receptor MRNAEngineered T Cells Induce Anti-Tumor Activity in Solid Malignancies. Cancer Immunol. Res 2014, 2 (2), 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yoon SH; Lee JM; Cho HI; Kim EK; Kim HS; Park MY; Kim TG Adoptive Immunotherapy Using Human Peripheral Blood Lymphocytes Transferred with RNA Encoding Her-2neu-Specific Chimeric Immune Receptor in Ovarian Cancer Xenograft Model. Cancer Gene Ther 2009, 16 (6), 489–497. [DOI] [PubMed] [Google Scholar]
- (40).Foster JB; Barrett DM; Karikó K The Emerging Role of In Vitro -Transcribed MRNA in Adoptive T Cell Immunotherapy. Mol. Ther 2019, 27 (4), 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).DiTommaso T; Cole JM; Cassereau L; Buggé JA; Sikora Hanson JL; Bridgen DT; Stokes BD; Loughhead SM; Beutel BA; Gilbert JB Cell Engineering with Microfluidic Squeezing Preserves Functionality of Primary Immune Cells in Vivo. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (46), E10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Matthews KE; Dev SB; Toneguzzo F; Keating A Electroporation for Gene Therapy. Animal Cell Electroporation and Electrofusion Protocols. Methods in Molecular Biology 1995, 48, 273–280. [DOI] [PubMed] [Google Scholar]
- (43).Dullaers M; Breckpot K; Van Meirvenne S; Bonehill A; Tuyaerts S; Michiels A; Straetman L; Heirman C; De Greef C; Van Der Bruggen P; et al. Side-by-Side Comparison of Lentivirally Transduced and MRNA-Electroporated Dendritic Cells: Implications for Cancer Immunotherapy Protocols. Mol. Ther 2004, 10 (4), 768–779. [DOI] [PubMed] [Google Scholar]
- (44).Singh N; Liu X; Hulitt J; Jiang S; June CH; Grupp SA; Barrett DM; Zhao Y Nature of Tumor Control by Permanently and Transiently Modified GD2 Chimeric Antigen Receptor T Cells in Xenograft Models of Neuroblastoma. Cancer Immunol. Res 2014, 2 (11), 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lambricht L; Lopes A; Kos S; Sersa G; Préat V; Vandermeulen G Clinical Potential of Electroporation for Gene Therapy and DNA Vaccine Delivery. Expert Opin. Drug Delivery 2016, 13 (2), 295–310. [DOI] [PubMed] [Google Scholar]
- (46).Hajj KA; Whitehead KA Tools for Translation: Non-Viral Materials for Therapeutic MRNA Delivery. Nat. Rev. Mater 2017, 2 (10), 17056. [Google Scholar]
- (47).McKinlay CJ; Benner NL; Haabeth OA; Waymouth RM; Wender PA Enhanced MRNA Delivery into Lymphocytes Enabled by Lipid-Varied Libraries of Charge-Altering Releasable Transporters. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (26), E5859–E5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Mukalel AJ; Riley RS; Zhang R; Mitchell MJ Nanoparticles for Nucleic Acid Delivery: Applications in Cancer Immunotherapy. Cancer Lett 2019, 458 (April), 102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Fornaguera C; Guerra-Rebollo M; Ángel Lázaro M; Castells-Sala C; Meca-Cortés O; Ramos-Pérez V; Cascante A; Rubio N; Blanco J; Borrós S MRNA Delivery System for Targeting Antigen-Presenting Cells In Vivo. Adv. Healthcare Mater 2018, 7 (17), 1800335. [DOI] [PubMed] [Google Scholar]
- (50).Zhang R; Billingsley MM; Mitchell MJ Biomaterials for Vaccine-Based Cancer Immunotherapy. J. Controlled Release 2018, 292, 256–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Islam MA; Reesor EKG; Xu Y; Zope HR; Zetter BR; Shi J Biomaterials for MRNA Delivery. Biomater. Sci 2015, 3, 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Olden BR; Cheng Y; Yu JL; Pun SH Cationic Polymers for Non-Viral Gene Delivery to Human T Cells Brynn. J. Controlled Release 2018, 282 (March), 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Moffett HF; Coon ME; Radtke S; Stephan SB; McKnight L; Lambert A; Stoddard BL; Kiem HP; Stephan MT Hit-and-Run Programming of Therapeutic Cytoreagents Using MRNA Nanocarriers. Nat. Commun 2017, 8 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Démoulins T; Milona P; Englezou PC; Ebensen T; Schulze K; Suter R; Pichon C; Midoux P; Guzmán CA; Ruggli N; et al. Polyethylenimine-Based Polyplex Delivery of Self-Replicating RNA Vaccines. Nanomedicine 2016, 12, 711–722. [DOI] [PubMed] [Google Scholar]
- (55).Anderson DG; Lynn DM; Langer R Semi-Automated Synthesis and Screening of a Large Library of Degradable Cationic Polymers for Gene Delivery. Angew. Chem., Int. Ed 2003, 42, 3153–3158. [DOI] [PubMed] [Google Scholar]
- (56).Garber K Alnylam Launches Era of RNAi Drugs. Nat. Biotechnol 2018, 36, 777. [DOI] [PubMed] [Google Scholar]
- (57).Kauffman KJ; Webber MJ; Anderson DG Materials for Non-Viral Intracellular Delivery of Messenger RNA Therapeutics. J. Controlled Release 2016, 240, 227–234. [DOI] [PubMed] [Google Scholar]
- (58).Oberli MA; Reichmuth AM; Dorkin JR; Mitchell MJ; Fenton OS; Jaklenec A; Anderson DG; Langer R; Blankschtein D Lipid Nanoparticle Assisted MRNA Deliery for Potent Cancer Immunotherapy. Nano Lett 2017, 17 (3), 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Fan YN; Li M; Luo YL; Chen Q; Wang L; Zhang HB; Shen S; Gu Z; Wang J Cationic Lipid-Assisted Nanoparticles for Delivery of MRNA Cancer Vaccine. Biomater. Sci 2018, 6 (11), 3009–3018. [DOI] [PubMed] [Google Scholar]
- (60).Gilleron J; Querbes W; Zeigerer A; Borodovsky A; Marsico G; Schubert U; Manygoats K; Seifert S; Andree C; Stöter M; et al. Image-Based Analysis of Lipid Nanoparticle-Mediated SiRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol 2013, 31 (7), 638–646. [DOI] [PubMed] [Google Scholar]
- (61).Patel S; Ashwanikumar N; Robinson E; Duross A; Sun C; Murphy-Benenato KE; Mihai C; Almarsson O; Sahay G Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated MRNA. Nano Lett 2017, 17 (9), 5711–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Love KT; Mahon KP; Christopher G; Whitehead KA; Querbes W; Robert J; Qin J; Cantley W; Qin LL; Frank-Kamenetsky M; et al. Lipid-like Materials for Low-Dose, in Vivo Gene Silencing. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (5), 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kauffman KJ; Dorkin JR; Yang JH; Heartlein MW; Derosa F; Mir FF; Fenton OS; Anderson DG Optimization of Lipid Nanoparticle Formulations for MRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett 2015, 15 (11), 7300–7306. [DOI] [PubMed] [Google Scholar]
- (64).Midoux P; Pichon C Lipid-Based MRNA Vaccine Delivery Systems. Expert Rev. Vaccines 2015, 14 (2), 221–234. [DOI] [PubMed] [Google Scholar]
- (65).Lokugamage MP; Sago CD; Gan Z; Krupczak BR; Dahlman JE Constrained Nanoparticles Deliver SiRNA and SgRNA to T Cells In Vivo without Targeting Ligands. Adv. Mater 2019, 31, 1902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Leuschner F; Dutta P; Gorbatov R; Novobrantseva TI; Panizzi P; Lee WW; Iwamoto Y; Milstein S; Epstein H; Cantley W; et al. Therapeutic SiRNA Silencing in Inflammatory Monocytes Florian. Nat. Biotechnol 2011, 29 (11), 1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Granot Y; Peer D Delivering the Right Message: Challenges and Opportunities in Lipid Nanoparticles-Mediated Modified MRNA Therapeutics—An Innate Immune System Standpoint. Semin. Immunol 2017, 34, 68–77. [DOI] [PubMed] [Google Scholar]
- (68).Varkouhi AK; Scholte M; Storm G; Haisma HJ Endosomal Escape Pathways for Delivery of Biologicals. J. Controlled Release 2011, 151 (3), 220–228. [DOI] [PubMed] [Google Scholar]
- (69).Mui BL; Tam YK; Jayaraman M; Ansell SM; Du X; Tam YYC; Lin PJC; Chen S; Narayanannair JK; Rajeev KG; et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of SiRNA Lipid Nanoparticles. Mol. Ther.–Nucleic Acids 2013, 2 (139), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Ball RL; Hajj KA; Vizelman J; Bajaj P; Whitehead KA Lipid Nanoparticle Formulations for Enhanced Co-Delivery of SiRNA and MRNA. Nano Lett 2018, 18 (6), 3814–3822. [DOI] [PubMed] [Google Scholar]
- (71).Cheng Q; Wei T; Jia Y; Farbiak L; Zhou K; Zhang S; Wei Y; Zhu H; Siegwart DJ Dendrimer-Based Lipid Nanoparticles Deliver Therapeutic FAH MRNA to Normalize Liver function and Extend Survival in a Mouse Model of Hepatorenal Tyrosinemia Type I. Adv. Mater 2018, 30, 1805308. [DOI] [PubMed] [Google Scholar]
- (72).Hajj KA; Ball RL; Deluty SB; Singh SR; Strelkova D; Knapp CM; Whitehead KA Branched-Tail Lipid Nanoparticles Potently Deliver MRNA In Vivo Due to Enhanced Ionization at Endosomal PH. Small 2019, 15, 1805097. [DOI] [PubMed] [Google Scholar]
- (73).Svitkin YV; Cheng YM; Chakraborty T; Presnyak V; John M; Sonenberg N N1-Methyl-Pseudouridine in MRNA Enhances Translation through EIF2 -Dependent and Independent Mechanisms by Increasing Ribosome Density. Nucleic Acids Res 2017, 45 (10), 6023–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Trixl L; Lusser A The Dynamic RNA Modification 5-Methylcytosine and Its Emerging Role as an Epitranscriptomic Mark. Wiley Interdiscip. Rev.: RNA 2019, 10, e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Karikó K; Buckstein M; Ni H; Weissman D Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [DOI] [PubMed] [Google Scholar]
- (76).Li J; Wang W; He Y; Li Y; Yan EZ; Zhang K; Irvine DJ; Hammond PT Structurally Programmed Assembly of Translation Initiation Nanoplex for Superior MRNA Delivery. ACS Nano 2017, 11, 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Shen X; Corey DR Chemistry, Mechanism and Clinical Status of Antisense Oligonucleotides and Duplex RNAs. Nucleic Acids Res 2018, 46 (4), 1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Sahin U; Karikó K; Türeci Ö MRNA-Based Therapeutics — Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13 (10), 759–780. [DOI] [PubMed] [Google Scholar]
- (79).Abraham RT; Weiss A Jurkat T Cells and Development of the T-Cell Receptor Signalling Paradigm. Nat. Rev. Immunol 2004, 4 (April), 301. [DOI] [PubMed] [Google Scholar]
- (80).Jain R; Frederick JP; Huang EY; Burke KE; Mauger DM; Andrianova EA; Farlow SJ; Siddiqui S; Pimentel J; Cheung-Ong K; McKinney KM; Kohrer C; Moore MJ; Chakraborty T MicroRNAs Enable MRNA Therapeutics to Selectively. Nucleic Acid Ther 2018, 28 (5), 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Wang T; Larcher L; Ma L; Veedu R Systematic Screening of Commonly Used Commercial Transfection Reagents towards Efficient Transfection of Single-Stranded Oligonucleotides Tao. Molecules 2018, 23 (10), 2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Cardarelli F; Digiacomo L; Marchini C; Amici A; Salomone F; Fiume G; Rossetta A; Gratton E; Pozzi D; Caracciolo G The Intracellular Trafficking Mechanism of Lipofectamine-Based Transfection Reagents and Its Implication for Gene Delivery. Sci. Rep 2016, 6 (March), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Wang X; Rivière I Clinical Manufacturing of CAR T Cells: Foundation of a Promising Therapy. Mol. Ther. Oncolytics 2016, 3 (16015), 16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Hollyman D; Stefanski J; Przybylowski M; Bartido S; Taylor C; Yeh R; Capacio V; Hosey J; Sadelain M; Brentjens RJ Manufacturing Validation of Biologically Functional T Cells Targeted to CD19 Antigen for Autologous Adoptive Cell Therapy. J. Immunother 2009, 32 (2), 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Lee DW; Kochenderfer JN; Stetler-Stevenson M; Cui YK; Delbrook C; Feldman SA; Fry TJ; Orentas R; Wayne AS; Mackall CL T Cells Expressing CD19 Chimeric Antigen Receptors for Acute Lymphoblastic Leukaemia in Children and Young Adults: A Phase 1 Dose-Escalation Trial. Lancet 2015, 385 (9967), 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Zhang J; Fan H; Levorse DA; Crocker LS Interaction of Cholesterol-Conjugated Ionizable Amino Lipids with Biomembranes: Lipid Polymorphism, Structure Activity Relationship, and Implications for SiRNA Delivery. Langmuir 2011, 27, 9473–9483. [DOI] [PubMed] [Google Scholar]
- (87).Castella M; Boronat A; Martín-Ibáñez R; Rodríguez V; Suñé G; Caballero M; Marzal B; Pérez-amill L; Martín-antonio B; Castaño J; et al. Development of a Novel Anti-CD19 Chimeric Antigen Receptor: A Paradigm for an Affordable CAR T Cell Production at Academic Institutions. Mol. Ther.–Methods Clin. Dev 2019, 12 (March), 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).An N; Tao Z; Li S; Xing H; Tang K; Tian Z Construction of a New Anti-CD19 Chimeric Antigen Receptor and the Anti-Leukemia Function Study of the Transduced T Cells. Oncotarget 2016, 7 (9), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Li X; Daniyan AF; Spitzer MH; Brentjens RJ; Avanzi MP; Yeku O; Li X; Wijewarnasuriya DP; Leeuwen D. G. Van. Engineered Tumor-Targeted T Cells Mediate Enhanced Anti-Tumor Efficacy. Cell Rep 2018, 23 (7), 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Pan Y; Yoon S; Sun J; Huang Z; Lee C; Allen M; Wu Y; Chang Y-J; Sadelain M; Shung KK; Chien S; Wang Y Mechanogenetics for the Remote and Noninvasive Control of Cancer Immunotherapy. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (5), 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Chen D; Love K; Chen Y; Eltoukhy A; Kastrup C; Sahay G; Jeon A; Dong Y; Whitehead K; Anderson D Rapid Discovery of Potent SiRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc 2012, 134 (16), 6948–6951. [DOI] [PubMed] [Google Scholar]
- (92).Heyes J; Palmer L; Bremner K; Maclachlan I Cationic Lipid Saturation Influences Intracellular Delivery of Encapsulated Nucleic Acids. J. Controlled Release 2005, 107, 276–287. [DOI] [PubMed] [Google Scholar]
- (93).Parry RV; Rumbley CA; Vandenberghe LH; June CH; Riley JL CD28 and Inducible Costimulatory Protein Src Homology 2 Binding Domains Show Distinct Regulation of Phosphatidylinositol 3-Kinase, Bcl-XL, and IL-2 Expression in Primary Human CD4 T Lymphocytes. J. Immunol 2003, 171 (1), 166–174. [DOI] [PubMed] [Google Scholar]
- (94).Milone MC; Fish JD; Carpenito C; Carroll RG; Binder GK; Teachey D; Samanta M; Lakhal M; Gloss B; Danet-Desnoyers G; et al. Chimeric Receptors Containing CD137 Signal Transduction Domains Mediate Enhanced Survival of T Cells and Increased Antileukemic Efficacy in Vivo. Mol. Ther 2009, 17 (8), 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


