Abstract
Background: Dendrimers are an attractive alternative to viral vectors due to the low cost of production, larger genetic insert-carrying capacity, and added control over immune- and genotoxic complications through versatile functionalization. However, their transfection rates pale in comparison to their viral counterparts, resulting in widespread research efforts in the attempt to improve transfection efficiency.
Materials and Methods: In this work, we designed a synthetic diblock nuclear-localization sequence peptide (NLS) (DDDDDDVKRKKKP) and complexed it with polyamidoamine (PAMAM) dendrimer G4 to form a duplex for gene delivery. We conducted transmission electron microscopy, gel mobility shift assay, and intracellular trafficking studies. We also assessed its transfection efficiency for the delivery of a green fluorescent protein-encoding plasmid (pGFP) to NIH3T3 cells.
Results: PAMAM dendrimer G4, NLS, and plasmid DNA can form a stable three-part polyplex and gain enhanced entry into the nucleus. We found transfection efficiency, in large part, depends on the ratio of G4:NLS:plasmid. The triplex prepared at the ratio of 1:60:1 for G4:NLS:pGFP has been shown to be more significantly efficient in transfecting cells than the control group (G4/pGFP, 0.5:1).
Conclusions: This new diblock NLS peptide can facilely complex with dendrimers to improve dendrimer-based gene transfection. It can also complex with other polycationic polymers to produce more potent nonviral duplex gene delivery vehicles.
Keywords: nuclear-localization sequence peptide, gene delivery, diaminobutane (DAB)-core dendrimer, fluorescence imaging
Introduction
The recent advances in gene therapy, especially since the discovery of CRISPR-Cas9, have expanded its use as a viable therapeutic option for the treatment of metabolic, ophthalmologic, and oncological diseases.1–3 Although this area becomes more sophisticated in its approach to producing corrected genes or removing harmful mutations, the need for effective delivery of the genetic material persists.4
Viral vectors remain the gold standard such as adenovirus (AV), adeno-associated virus (AAV), as well as lentivirus and retrovirus vectors employed for the incorporation of genetic transcripts into host genomes.5–10 These virus types vary in their duration of expression (transient or long term), double or single strandedness, insert-size capacity, and associated complications. AVs are double stranded, can carry gene inserts up to 7.5 kb, and potentially cause strong immunogenic reactions; whereas retroviruses are single stranded, produce long gene expression, and can cause random insertions into the host genome.11 Although improvements to viral vector design are being made for better control of expression and reducing immunogenic responses, insertional oncogenesis, and genotoxicity, the variability in the vectors can be difficult to navigate for specific treatment applications.12
Nonviral vectors represent an attractive alternative to viral gene delivery in their ability to reduce immunogenic and toxic effects while providing increased gene transcript-carrying capacity and produce long-term gene expression.13 Current commercially available vectors include Lipofectamine™, a cationic liposome formulation that delivers nucleic acids by fusing with the cell membrane, and Turbofect™, a cationic polymer that electrostatically complexes with nucleic acids.
Polymers are being widely explored as gene delivery vehicles for their efficient and low-cost production.14–16 The positive charge of these formulations has a multipronged effect on the delivery of genetic payloads into cells: (1) The positive surface charge allows for electrostatic self-assembly by condensation of nucleic acid via its negatively charge phosphate backbone;17 (2) maintaining an overall positive charge, the stable complex is drawn toward the negative charge lipid bilayer cell membrane;18 and (3) the positive charge for polymer specifically enables endosomal escape by the buffering capacity and associated osmotic swelling and rupture of the early endosome.19
Although the advantages of nonviral vectors are manifold, they are not without their drawbacks. Despite the afforded control and reduction of virus-associated disadvantages, the transfection efficiency can be orders of magnitude lower than that of viruses without causing material and concentration-associated cytotoxicity.20 Although the positive charge is useful for payload packaging and intracellular trafficking, high concentrations can destabilize the membranes and cause cell lysis. Many researchers address this by modifying polymer structures to reduce the charge-associated toxicity while maintaining the high payload-carrying capacity.21
The polymer utilized in this work is a generation 4 (G4) polyamidoamine (PAMAM) dendrimer with 64 surface primary amines. Dendrimers are a type of nanopolymer that are well defined, hyperbranched, and multivalent while maintaining a small hydrodynamic range relative to linear polymers of equal molecular weight.22 Although gene accumulation in the cytosol can be achieved with amine-terminated dendrimers and other polycationic polymers, the levels of nuclear translocation and transfection are limited due to the rate-limiting cytosolic dissociation of the polyplexes.23,24
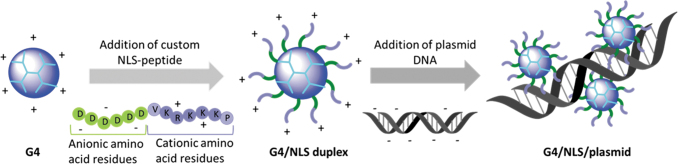
To increase the transfection efficiency of native G4 dendrimers at biocompatible concentrations, we propose an addition to the dendrimer/nucleic acid complex—the electrostatic interaction of a small peptide containing at one end a nuclear localization sequence (NLS), and at the other end a series of anionic amino acid residues for orientation-specific interaction.25,26 Work by Parelkar et al. showed that modified polymers with a reverse NLS peptide sequence accumulate in the nucleus more than plain NLS-sequence peptides.25,27 These findings led us to develop a facile approach to promote the nuclear translocation of the entire dendrimer/nucleic acid polyplex by electrostatic interaction with a small diblock peptide designed to interact with the dendrimer in a manner such that an NLS sequence would be exposed on the surface of the dendrimer in the reverse orientation (Fig. 1).
FIG. 1.
Schematic illustration of preparation of G4:NLS: plasmid complexes. G4, generation 4; NLS, nuclear-localization sequence peptide.
We hypothesize that the addition of the NLS peptide to the polyplex will allow for its final movement into the nucleus, where dissociation can occur at the relevant site for transfection, thereby improving the transfection efficiency over a native dendrimer/nucleic acid polyplex.
Methods and Materials
Materials
PAMAM dendrimer (diaminobutane-core, generation 4) was purchased from NanoSynthons (Mt. Pleasant, MI). Lipofectamine 2000, phosphate-buffered saline (PBS), paraformaldehyde (PFA), Triton X-100, rhodamine phalloidin, and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Thermo Fisher Scientific (Waltham, MA). Dulbecco's modified Eagle medium (DMEM), trypsin-EDTA (0.25%), penicillin-streptomycin (10,000 U/mL), and ethidium bromide were purchased from Life Technologies (Carlsbad, CA). pMAX-green fluorescent protein (GFP) plasmid (pGFP) and cosmic calf serum (CS) were purchased from Lonza (Gaithersburg, MD). LabelIT® Cy3™ Plasmid Delivery Control (pCy3) (red) was purchased from Mirus Bio (Madison, WI). Cell proliferation reagent WST-1 and N,N-diisopropylethylamine (DIPEA) were purchased from Sigma-Aldrich (St. Louis, MO). Custom-designed peptide (DDDDDDVKRKKKP) with and without tetramethylrhodamine (TAMRA) C-terminal modification was synthesized by BioBasic (Amherst, NY). Dimethyl sulfoxide (DMSO) and fluorescein isothiocyanate (FITC) were purchased from Sigma Aldrich. Vectashield mounting medium was purchased from Vector Laboratories (Burlingame, CA). Roswell Park Memorial Institute (RPMI) 1640 Medium was purchased from ATCC (Gaithersburg, MD).
Synthesis of FITC-labeled G4 dendrimers
FITC dissolved in DMSO was added dropwise to G4 in the presence of DIPEA (with a molar ratio of DIPEA:FITC:G4 of 30:5:1). The reaction was stirred overnight in the dark. FITC-G4 conjugates were dialyzed against deionized water and lyophilized.
Complexation of dendrimer/NLS-peptide/plasmid
G4 dissolved in PBS was complexed first with NLS peptide pre-dissolved in PBS, based on previous literature.26 The mixture was allowed to equilibrate at room temperature for 30 min. Plasmid was then added to the solution, mixed by gentle pipetting, and finally allowed to equilibrate for an additional 30 min at room temperature (Fig. 1).
Gel mobility shift assay
G4/plasmid and G4/NLS/plasmid complexes at dendrimer:plasmid ratios of 0.5, 1, and 2 (corresponding to N/P charge ratios of 0.75, 1.5, and 3) were tested for complex charge stability via gel electrophoresis, keeping the plasmid constant at 1 μg. One percent agarose gels were prepared in tris-acetate-EDTA (TAE) buffer with ethidium bromide (0.5 μg/mL). After complexation, samples were loaded into wells and run at a constant voltage of 100 V for 30 min. DNA bands were visualized with a UV-transilluminator (BioRad ChemiDoc, Hercules, CA).
Dynamic light scattering
The zeta potential of G4 dendrimer, plasmid, G4/plasmid (1:1), G4/NLS (1:120), and G4/NLS/plasmid (1:120:1) was measured at room temperature by using a Malvern Zetasizer Nano ZS90 (Malvern Instruments, Worcestershire, UK).
Transmission electron microscopy
Particle and complex size and morphology were imaged by using a Jeol JEM-1400 series 120 kV transmission electron microscopy (TEM) equipped with an integrated sCMOS camera.
Cell culture
NIH3T3 mouse embryonic fibroblasts, HN12 head and neck squamous cell carcinoma were cultured in DMEM containing high glucose supplemented with L-glutamine, 10% CS, penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C in 95% air/5% CO2. Y79 cells in suspension were cultured in RPMI-1640 containing 20% CS at 37°C in 95% air/5% CO2.
Intracellular trafficking studies
FITC-labeled G4 dendrimer was complexed with either an unlabeled NLS and Cy3-labeled plasmid, or a TAMRA-labeled NLS and unlabeled plasmid following the procedure described earlier. NIH3T3 cells were seeded on coverslips in 12-well plates and allowed to attach overnight. Complexes were then added to the fresh cell culture medium for predetermined time points. Cells were then fixed with 4% PFA for 30 min at room temperature, permeated with 0.1% Triton X-100, and washed with 1 × PBS. Nuclei were then counterstained with DAPI and again washed with PBS. Coverslips were transferred to glass slides and then imaged by using a Zeiss LSM 710 confocal laser scanning microscope.
In vitro transfection
Unlabeled complexes were prepared as previously described and added to cell culture media of NIH3T3, HN12, and Y79 cells. Cells were incubated with transfection materials with combinations of G4/NLS/pGFP ratios (G4: 0.5, 1, 2; NLS: 0, 60, 120; pGFP: 1) for 4–6 h, followed by fresh media replacement. Cells were incubated for an additional 72 h before further analysis. Transfected cells were fixed and counterstained with DAPI (as previously described) and imaged by using a Nikon Eclipse Ti-U inverted fluorescence microscope using DAPI, GFP, and brightfield channels. The number of transfected cells was determined by cell counting in ImageJ.28
Statistical analysis
All data are expressed as means ± standard error of the mean (SEM). Transfection data (grouped by varying amounts of NLS-peptide) are evaluated for a two-way analysis of variance with replication. Differences between the means of G4:NLS:plasmid weight ratios are compared by using the Student's t-test for unpaired samples. A value of p < 0.05 was considered statistically significant.
Results
Preparation and characterization of G4/NLS/plasmid complexes
We designed a 13-amino-acid diblock peptide (DDDDDDVKRKKKP) to enhance entry of dendrimer-based gene complexes into the nucleus. This new NLS peptide includes a block of negatively charged amino acids at physiological pH at one end, and the neutral and cationic residues of the nuclear localization unit in reverse order on the other end. The sequence and orientation of the nuclear localization portion was based on previous literature.25 A series of six aspartic acid (Asp, D) residues was chosen for the negatively charged block due to its smaller side-chain size relative to the other anionic amino acid, glutamic acid (Glu, E). The inclusion of this block is to orient the peptide via electrostatic interaction with positively charged dendrimer surface while leaving the reverse NLS-peptide exposed.
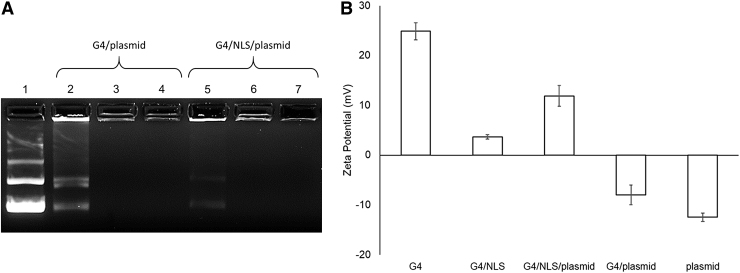
Gel mobility shift data (Fig. 2A) reveal stable G4/plasmid complexes formed at a ratio between 0.5:1 and 1:1 in the absence of NLS peptide. In the presence of NLS, G4/NLS/plasmid formed stable complexes also between 0.5:1, but can be assumed to be at a lower ratio of G4: plasmid, suggesting that the addition of the diblock NLS peptide contributes to the charge stabilization of the complex. This is based on the difference in band intensity between lanes 2 and 5. The weaker intensity of the G4/NLS/peptide at the 0.5:1 ratio (lane 5) suggests that the overall complex is more positively charged than the G4/peptide and therefore does not run down the lane as much due to the voltage differential across the gel (lane 2).
FIG. 2.
(A) Gel mobility shift assay of G4/plasmid complexes with or without the NLS peptide. Lane 1: pGFP; Lanes 2–4: 0.5:0:1, 1:0:1, 2:0:1 weight ratios G4/NLS/pGFP, respectively; Lanes 5–7: 0.5:120:1, 1:120:1, 2:120:1 weight ratios G4/NLS/pGFP, respectively. (B) Zeta potentials of individual components and their respective combinations at the weight ratio G4/NLS/plasmid ratio of 1:120:1. pGFP, plasmid green fluorescent protein.
We can determine the stability of the complexes by the lack of visible aggregates forming in solution as well as successful gel mobility shift data over the duration of the gel preparation and electrophoresis, as well as by other groups having reported varying long-term colloidal stability of polycationic polymer/DNA complexes near neutrality depending on concentration and complex hydrodynamic ranges.29–31 In the preparation of G4/NLS/plasmid, G4 was first complexed with NLS and then plasmid. The negatively charged block of the peptide interacts with the polycationic surface of the dendrimer, allowing the positively charged end of the peptide to be exposed on the surface. This G4/NLS complex, in effect, acts as a larger G4, thereby being able to condense more of the anionic plasmid. Therefore, less G4 is required to stably complex plasmid in the presence of this particular NLS-peptide.
Dynamic light scattering (DLS) data (Fig. 2B) provide zeta potential values for the individual G4 and plasmid components, as well as the 1:1 ratio of G4/plasmid, 1:120 ratio of G4/NLS, and the whole 1:120:1 ratio of G4/NLS/plasmid. As expected, plasmid is negatively charged at -12.5 mV ±0.8 mV whereas native G4 is positively charged at 24.9 mV ±1.7 mV. This combination of G4/plasmid remains slightly anionic, whereas the G4/NLS and G4/NLS/plasmid complexes are increasingly cationic but have a lower net charge than native G4.
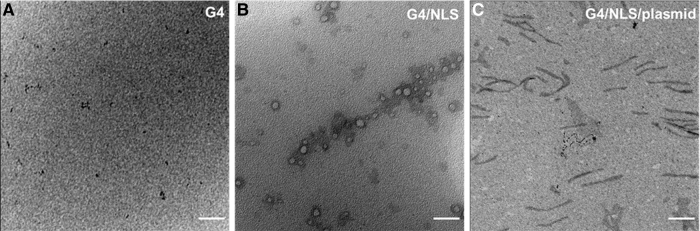
To confirm complexation, G4, G4/NLS, and G4/NLS/pGFP were imaged by using TEM for size and morphology analysis (Fig. 3). The reported size of 4 nm for a G4 PAMAM dendrimer was confirmed as well as the appropriate increases in size with the addition of each component.32 By complexing G4 with NLS peptide, the size of the complex increases to 6–12 nm. The variation in size is attributed to the nature of the physical interactions between these two components, as well as the possibility of the NLS peptide binding to itself. The plasmid is structurally much larger than either G4 or NLS. Once it is added, the complex changes shape from rounded structures to rods in the hundreds of nanometers range. These rod-like shapes are characteristic of condensed DNA with low-generation dendrimers.33
FIG. 3.
TEM images of G4 (A), G4/NLS duplex (1:120) (B), and G4/NLS/plasmid complex (1:120:1) (C). Scale bars: 100 nm (A), 50 nm (B), and 300 nm (C). TEM, transmission electron microscopy.
Cellular uptake of G4/NLS/plasmid complexes and intracellular trafficking
We have previously shown that G4 is minimally toxic at concentrations below 2 μM, as confirmed across multiple literature reports.34–36 The concentration of G4 chosen for all experiments was 0.7 μM based on the determined complexation ratios and the working volumes of 12-well tissue culture plates, which should produce virtually no toxicity to our cells.
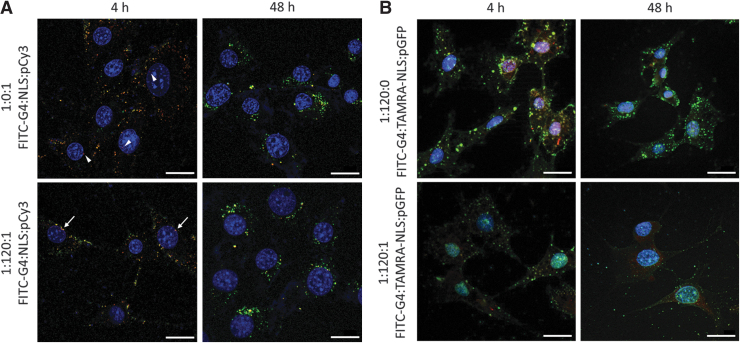
Cellular uptake was confirmed by complexing FITC-G4 with either unlabeled NLS and Cy3-labeled plasmid (Fig. 4A), or TAMRA-labeled NLS and unlabeled pGFP (Fig. 4B). By 4 h, the complexes have been taken into the cells, and depending on the formulation, they began to localize near or within the nucleus. In the FITC-G4/pCy3 sample, minimal red signal can be seen within the boundaries of the nucleus (white arrowheads). Strong electrostatic interactions between the dendrimer and plasmid components limit their dissociation. Without a nuclear chaperone signal such as the NLS, the entry of dendrimer into the nucleus is limited. By contrast, stronger red and yellow signals can be seen within the nucleus in the FITC-G4/NLS/pCy3 group.
FIG. 4.
(A) Intracellular trafficking of FITC-G4/pCy3 with or without NLS in NIH3T3 cells at 4 and 48 h post-addition. (B) Intracellular trafficking of FITC-G4/TAMRA-NLS with or without pGFP in NIH3T3 cells at 4 and 48 h post-addition. Green: FITC-G4; Red: pCy3; Blue: DAPI nuclei. Scale bar: 20 μm. FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole; TAMRA, tetramethylrhodamine.
This observation suggests that the entire complex is transported into the nucleus with the aid of the NLS peptide. The red signal represents loosely associated pCy3 with FITC-G4, that is, released from the complex and moves into the nucleus, or pCy3 that has begun to dissociate from the rest of the complex inside the nucleus. Imaging was done at 48 h to determine the complex make-up and presence by the minimum time required to observe transfection. In both cases, no signal is seen within the nucleus, whereas mostly green and yellow signals remain in the cytosolic compartment of the cell. The loss of the red signal from pCy3 within the nucleus is presumably due to the degradation as a result of integration within the host genome. The FITC-G4 that localized within the cells was likely exported from the nucleus, accounting for the lack of green light overlapping with the nucleus.
To complement these data, the cellular localization was followed for FITC-G4/TAMRA-NLS and FITC-G4/TAMRA-NLS/plasmid samples to confirm the intended function of the NLS-peptide. In both samples at 4 h, a yellow signal can be seen within every cell's cytosol and nucleus. This proves that the NLS peptide assists in moving the entire complex into the nucleus. However, noticeably, the signal across the whole cell is stronger in the FITC-G4/TAMRA-NLS group relative to the FITC-G4/TAMRA-NLS/plasmid group. Because of competitive interaction, the added plasmid tends to lower the amount of NLS associating with dendrimer and may directly interact with NLS without dendrimer involvement, becoming more vulnerable to degradation during transport. This trend is seen at the 48 h time point as well.
However, at the later time point, a yellow signal is still seen within the nucleus of both groups as well as in the cytosol. This suggests that the NLS peptide remains associated with the dendrimer throughout the delivery and cellular residence. The FITC and TAMRA signals are not vanishing as with the pCy3 signal in 3A since there is negligible reduction in the signal from 4 h to 48 h. Its association with the NLS helps extend G4's localization within the nucleus.
Transfection evaluation
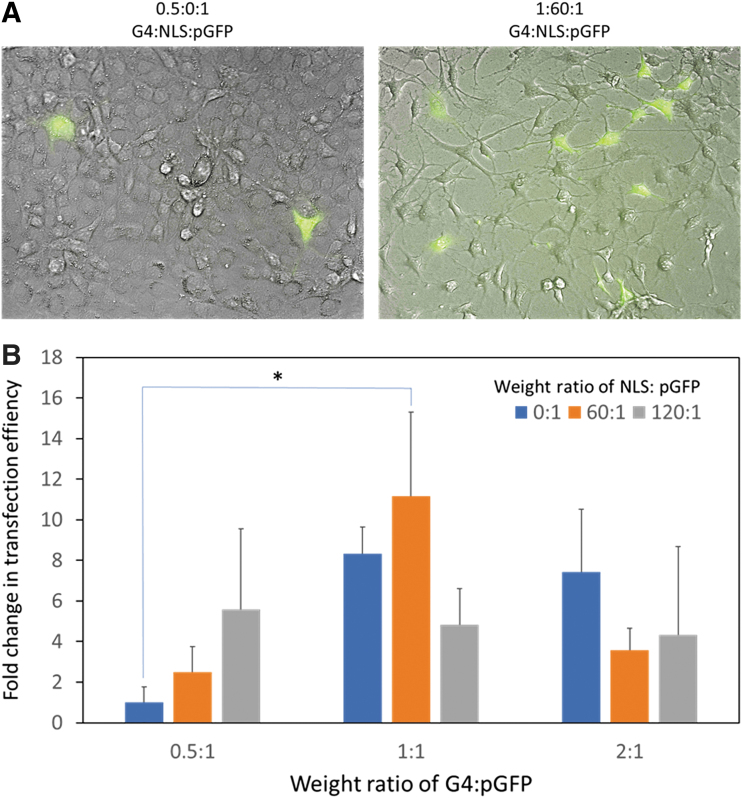
We chose pGFP as a model gene to assess transfection efficiency of G4/NLS duplex in terms of proportion of cells expressing GFP. We tested a series of G4:NLS: pGFP complexes prepared at varying weight ratios (Fig. 5). We used G4/pGFP (0.5:1) as a control and normalized transfection efficiency of other formulations to this control. We found that with increasing amounts of NLS in the G4:pGFP (0.5:1) complexes did not significantly improve transfection efficiency although an increasing transfection trend was observed.
FIG. 5.
(A) Representative fluorescence images of NIH3T3 cells transfected with the control G4/pGFP without NLS (0.5:0:1) and G4:NLS:pGFP (1:60:1). Blue: DAPI nuclei. Scale bar 20 μm. (B) Transfection efficiency (fold changes relative to the control) of G4:NLS:pGFP complexes prepared at the indicated weight ratios.
Increasing weight ratio of G4:pGFP from 0.5 to 1:1 and 2:1 generally augmented transfection efficiency. However, increasing weight ratio of NLS:pGFP did not necessarily exert additive effects. We identified 1:60:1 as the optimal ratio for G4:NLS:pGFP complexation as its transfection efficiency was significantly higher than the control (p = 0.03). These relationships within and between the grouping can be explained by the competitive interaction concept as well as the trend of increased transfection efficiency of a higher dose of polycationic polymers.37,38 As mentioned in the previous section, the higher the amounts of either NLS or pGFP relative to the cationic carrier means a decrease in the other component that can be delivered into the cell. In addition, higher generation dendrimers are more effective at cellular transfection (but suffer in their cytotoxicity).
The effect of increasing the amount of cationic groups (from weight ratio 0.5 to 1, and 1 to 2) mitigates the lack of an NLS peptide for transfection. Taken together, there seems to be an optimal ratio of NLS to pGFP that provides optimal nuclear localization and enough plasmid for transfection.
Discussion
Low-generation dendrimer gene delivery suffers from low transfection efficiency despite successful complexation and safe delivery into the cells, endosomal escape, and accumulation in the cytosol. Although the lack of dissociation is a factor in the low efficiency, the inability of the dendrimer to travel into the nucleus also contributes to this low transfection ability.
Here, we successfully developed a duplex of G4/custom NLS-sequence-containing peptide as a new gene delivery system. The combining of NLS peptide with G4 in the first step ensured that the NLS peptide would interact on the surface of the dendrimer with a specific orientation to allow the reverse NLS sequence to remain exposed. This interaction was confirmed and did not disrupt the ability of the G4/NLS complex to interact with plasmid constructs.
However, DLS data indicate that although G4/NLS (at 1:120) is a cationic complex, it has a lower positive charge than native G4 or the entire triplex (1:120:1). Although we expect the orientation of the negative end of the peptide to interact with the positively charged dendrimer surface, this interaction could be heterogenous and uneven with a potential peptide–peptide interaction. As such, the “layer” of NLS peptides may be shielding the dendrimer's surface charge, causing the DLS measurement to read a lower zeta potential compared with native G4.
Once the plasmid is added, peptide complexes may dissociate and form the correct orientation between the G4 and plasmid, accounting for the increased zeta potential of the final polyplex. Due to the positively charged and neutral amino acid residues at pH 7.4 in the NLS sequence itself, incorporating a block of negatively charged amino acid residues enabled its interaction with both the dendrimer and plasmid components. The design rationale was to promote the interaction between the dendrimer and the entire NLS peptide and to avoid only NLS/plasmid complexes. Any such complexes would be degraded by cellular nucleases and proteases during delivery. Results confirmed stable complexation at lower ratios than native G4/plasmid, as well as successful entry into the cells and nucleus in the presence of NLS peptide.
Competitive interactions between the three components affected the degree of nuclear translocation and transfection outcomes. The number of NLS peptides are reduced, as the number of available G4 surface groups is now split between NLS and plasmid. The weak electrostatic interactions cause dissociation and reassociation with natural variation in the complexation. This means that, at any given complex ratio, the nuclear translocation ability may be reduced due to the lower number of NLS peptides, or the covering up of NLS sequence by interaction with the plasmid. This was also apparent in the transfection data where low G4 ratios with high NLS-peptide ratios (0.5:120:1) and high G4 ratios and lower NLS-peptide ratios (1:60:1, 2:0:1) tend to transfect more cells.
Preliminary tests confirmed that suspension cells such as Y79 retinoblastoma and other types of adherence cells such as HN12 head and neck cancer cells can also be transfected with G4/NLS/GFP, although optimal formulations remain to be determined for those cell types (Supplementary Fig. S1). The strength of the association between plasmid and the G4/NLS component can be weakened to cause higher dissociation rates of the plasmid, another factor for improving transfection.
The objective of this work was to elucidate whether the addition of an NLS peptide could enhance the transfection of low-generation and concentration dendrimer delivery vehicles by promoting its nuclear uptake in vitro. Future work will investigate the transfection efficiency in vivo, but it will require modifications to address the concerns of hemolysis, liver toxicity, and rapid drug clearance in systemic delivery administrations due to the polycationic membrane charge destabilization of blood cells and binding to blood serum albumins.39 Although the complexation of G4/NLS duplex with plasmid neutralizes the charge, other modifications to enhance the in vivo biocompatibility and half-life would be evaluated, such as dendrimer surface PEGylation or acetylation.40,41
Conclusions
In summary, we designed a 13-amino-acid diblock peptide (DDDDDDVKRKKKP) and used it to form a duplex with PAMAM dendrimer G4 for enhanced gene delivery. This peptide has been shown to enhance the nuclear translocation of G4/plasmid complexes into the nucleus. In addition, the competitive interactions among the three complexed components, that is, G4, NLS, and plasmid, destabilize the association between G4 and plasmid, allowing for dissociation to take place in the nucleus. An optimal ratio of 1:60:1 for G4:NLS:plasmid complexation has been identified for improved gene transfection. This new NLS peptide can also be applied to complex with higher generation dendrimers (e.g., G5) or other polycationic polymers to produce more potent nonviral duplex gene delivery vehicles.
Supplementary Material
Acknowledgment
The authors thank Mahmoud E. Moustafa for assistance in TEM imaging.
Author Confirmation Statement
R.C.C. and H.Y. conceptualized the project. R.C.C. conducted the experiments and data analysis. All co-authors have reviewed and approved of the article before submission. The article has been submitted solely to this journal and is not published, in press, or submitted elsewhere.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported, in part, by the National Institutes of Health (R01EY024072). Microscopy was performed at the VCU Microscopy Facility, supported, in part, by funding from NIH-NCI Cancer Center Support Grant P30CA016059.
Supplementary Material
References
- 1.Jarrett KE, Lee CM, Yeh Y-H, et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep 2017;7:44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltran WA, Cideciyan AV, Boye SE, et al. Optimization of retinal gene therapy for X-linked retinitis pigmentosa due to RPGR mutations. Mol Ther 2017;25:1866–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Gao P, Yuan S, et al. Oncolytic adenovirus complexes coated with lipids and calcium phosphate for cancer gene therapy. ACS Nano 2016;10:11548–11560 [DOI] [PubMed] [Google Scholar]
- 4.Mehier-Humbert S, Guy RH. Physical methods for gene transfer: Improving the kinetics of gene delivery into cells. Adv Drug Deliv Rev 2005;57:733–753 [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Kanasty RL, Eltoukhy AA, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet 2014;15:541–555 [DOI] [PubMed] [Google Scholar]
- 6.Iizuka S, Sakurai F, Tachibana M, et al. Neonatal gene therapy for hemophilia B by a novel adenovirus vector showing reduced leaky expression of viral genes. Mol Ther Methods Clin Dev 2017;6:183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Tai PWL, Gao GP. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Dis 2019;18:358–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurosaki F, Uchibori R, Mato N, et al. Optimization of adeno-associated virus vector-mediated gene transfer to the respiratory tract. Gene Ther 2017;24:290–297 [DOI] [PubMed] [Google Scholar]
- 9.Campochiaro PA, Lauer AK, Sohn EH, et al. Lentiviral vector gene transfer of endostatin/angiostatin for macular degeneration (GEM) study. Hum Gene Ther 2017;28:99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsner C, Bohne J. The retroviral vector family: Something for everyone. Virus Genes 2017;53:714–722 [DOI] [PubMed] [Google Scholar]
- 11.Lundstrom K. Viral vectors in gene therapy. Diseases 2018;6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David RM, Doherty AT. Viral vectors: The road to reducing genotoxicity. Toxicol Sci 2017;155:315–325 [DOI] [PubMed] [Google Scholar]
- 13.Ramamoorth M, Narvekar A. Non viral vectors in gene therapy—An overview. J Clin Diagn Res 2015;9:GE1–GE6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam HY, Nam K, Lee M, et al. Dendrimer type bio-reducible polymer for efficient gene delivery. J Control Release 2012;160:592–600 [DOI] [PubMed] [Google Scholar]
- 15.Dong RJ, Su Y, Yu SR, et al. A redox-responsive cationic supramolecular polymer constructed from small molecules as a promising gene vector. Chem Commun 2013;49:9845–9847 [DOI] [PubMed] [Google Scholar]
- 16.Li J, He YZ, Li W, et al. A novel polymer-lipid hybrid nanoparticle for efficient nonviral gene delivery. Acta Pharmacol Sin 2010;31:509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang SB, Xu YM, Wang B, et al. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J Control Release 2004;100:165–180 [DOI] [PubMed] [Google Scholar]
- 18.Chen L, McCrate JM, Lee JCM, et al. The role of surface charge on the uptake and biocompatibility of hydroxyapatite nanoparticles with osteoblast cells. Nanotechnology 2011;22:105708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bus T, Traeger A, Schubert US. The great escape: How cationic polyplexes overcome the endosomal barrier. J Mater Chem B 2018;6:6904–6918 [DOI] [PubMed] [Google Scholar]
- 20.Dandekar P, Jain R, Keil M, et al. Cellular delivery of polynucleotides by cationic cyclodextrin polyrotaxanes. J Control Release 2012;164:387–393 [DOI] [PubMed] [Google Scholar]
- 21.Wen S, Zheng F, Shen M, Shi X. Surface modification and PEGylation of branched polyethyleneimine for improved biocompatibility. J Appl Polym Sci 2013;128:3807–3813 [Google Scholar]
- 22.Boas U, Heegaard PMH. Dendrimers in drug research. Chem Soc Rev 2004;33:43–63 [DOI] [PubMed] [Google Scholar]
- 23.Thibault M, Nimesh S, Lavertu M, et al. Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes. Mol Ther 2010;18:1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Won YW, Yoon SM, Lee KM, et al. Poly(oligo-D-arginine) with internal disulfide linkages as a cytoplasm-sensitive carrier for siRNA delivery. Mol Ther 2011;19:372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parelkar SS, Letteri R, Chan-Seng D, et al. Polymer-peptide delivery platforms: Effect of oligopeptide orientation on polymer-based DNA delivery. Biomacromolecules 2014;15:1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opanasopit P, Rojanarata T, Apirakaramwong A, et al. Nuclear localization signal peptides enhance transfection efficiency of chitosan/DNA complexes. Int J Pharm 2009;382:291–295 [DOI] [PubMed] [Google Scholar]
- 27.Branden LJ, Mohamed AJ, Smith CIE. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat Biotechnol 1999;17:784–787 [DOI] [PubMed] [Google Scholar]
- 28.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang MX, Szoka FC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther 1997;4:823–832 [DOI] [PubMed] [Google Scholar]
- 30.Lakshminarayanan A, Ravi VK, Tatineni R, et al. Efficient dendrimer-DNA complexation and gene delivery vector properties of nitrogen-core poly(propyl ether imine) dendrimer in mammalian cells. Bioconjug Chem 2013;24:1612–1623 [DOI] [PubMed] [Google Scholar]
- 31.Braun CS, Vetro JA, Tomalia DA, et al. Structure/function relationships of polyamidoamine/DNA dendrimers as gene delivery vehicles. J Pharm Sci 2005;94:423–436 [DOI] [PubMed] [Google Scholar]
- 32.Dobrovolskaia MA, Patri AK, Simak J, et al. Nanoparticle size and surface charge determine effects of PAMAM dendrimers on human platelets in vitro. Mol Pharm 2012;9:382–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ainalem ML, Nylander T. DNA condensation using cationic dendrimers-morphology and supramolecular structure of formed aggregates. Soft Matter 2011;7:4577–4594 [Google Scholar]
- 34.Yuan Q, Yeudall WA, Yang H. PEGylated polyamidoamine dendrimers with bis-aryl hydrazone linkages for enhanced gene delivery. Biomacromolecules 2010;11:1940–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jevprasesphant R, Penny J, Jalal R, et al. The influence of surface modification on the cytotoxicity of PAMAM dendrimers. Int J Pharm 2003;252:263–266 [DOI] [PubMed] [Google Scholar]
- 36.Kuo JHS, Jan MS, Chu HW. Mechanism of cell death induced by cationic dendrimers in RAW 264.7 murine macrophage-like cells. J Pharm Pharmacol 2005;57:489–495 [DOI] [PubMed] [Google Scholar]
- 37.Huth S, Hoffmann F, von Gersdorff K, et al. Interaction of polyamine gene vectors with RNA leads to the dissociation of plasmid DNA-carrier complexes. J Gene Med 2006;8:1416–1424 [DOI] [PubMed] [Google Scholar]
- 38.Haensler J, Szoka FC. Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug Chem 1993;4:372–379 [DOI] [PubMed] [Google Scholar]
- 39.Qi R, Gao Y, Tang Y, et al. PEG-conjugated PAMAM dendrimers mediate efficient intramuscular gene expression. AAPS J 2009;11:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luong D, Kesharwani P, Deshmukh R, et al. PEGylated PAMAM dendrimers: Enhancing efficacy and mitigating toxicity for effective anticancer drug and gene delivery. Acta Biomater 2016;43:14–29 [DOI] [PubMed] [Google Scholar]
- 41.Kolhatkar RB, Kitchens KM, Swaan PW, et al. Surface acetylation of polyamidoamine (PAMAM) dendrimers decreases cytotoxicity while maintaining membrane permeability. Bioconjug Chem 2007;18:2054–2060 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.