Abstract
The outbreak caused by the novel coronavirus SARS-CoV-2 has been declared a global health emergency. G-quadruplex structures in genomes have long been considered essential for regulating a number of biological processes in a plethora of organisms. We have analyzed and identified 25 four contiguous GG runs (G2NxG2NyG2NzG2) in the SARS-CoV-2 RNA genome, suggesting putative G-quadruplex-forming sequences (PQSs). Detailed analysis of SARS-CoV-2 PQSs revealed their locations in the open reading frames of ORF1 ab, spike (S), ORF3a, membrane (M) and nucleocapsid (N) genes. Identical PQSs were also found in the other members of the Coronaviridae family. The top-ranked PQSs at positions 13385 and 24268 were confirmed to form RNA G-quadruplex structures in vitro by multiple spectroscopic assays. Furthermore, their direct interactions with viral helicase (nsp13) were determined by microscale thermophoresis. Molecular docking model suggests that nsp13 distorts the G-quadruplex structure by allowing the guanine bases to be flipped away from the guanine quartet planes. Targeting viral helicase and G-quadruplex structure represents an attractive approach for potentially inhibiting the SARS-CoV-2 virus.
Keywords: SARS-CoV-2, G-quadruplex, PQSs, Coronaviridae, viral helicase nsp13, helicase inhibitor
Introduction
G-quadruplexes are guanine-rich nucleic acid sequences, which form a stable four-stranded helical structure by pairing of four guanine bases via Hoogsteen hydrogen bonds (Figure 1A). The conformation of a G-quadruplex is polymorphic, by changing its base sequence, loop connection and embedded cations [1]. G-quadruplexes are highly conserved secondary structures found in the genomes of various organisms. G-quadruplexes are formed by nucleic acid (DNA or RNA) containing the sequences G(≥2)NxG(≥2)NyG(≥2)NzG(≥2).x, y and z might be different. And it is ≥ because two layers are just about sufficient to form a G-quadruplex. This article focuses on G-quadruplexes of two layers. Studies of G-quadruplexes in the human genome have shown their regulatory roles in DNA replication, transcription and recombination [2]. Putative G-quadruplex-forming sequences (PQSs) have been found in different pathogens [3], including viruses [4], bacteria [5] and even protozoa [6].
Figure 1.
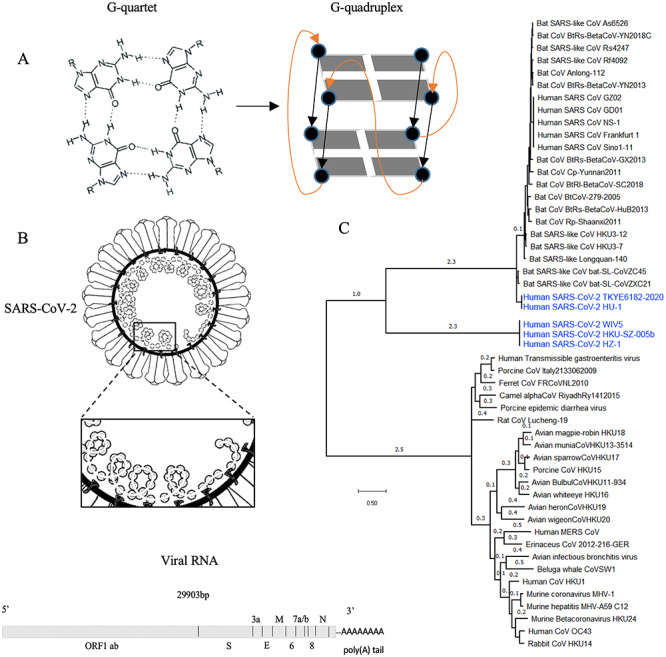
The formation of G-quadruplex structures in the SARS-CoV-2 genome. (A) a G-tetrad structure(left) and intramolecular folding of a parallel G-quadruplex structure (right). Four guanine bases form a G-tetrad, and two or more G-tetrads to build a G-quadruplex. (B) PQSs have been found in the SARS-CoV-2 genome. Close analysis of those PQSs revealed their location in the viral open reading frames of the ORF1 ab, spike (S), ORF3a, membrane (M) and nucleocapsid (N) genes. (C) Phylogenetic analysis of the whole genomes of SARS-CoV-2 (highlighted in blue) and other Coronaviridae.
Previous studies have shown that G-quadruplex structure participates in genome recognition, recombination, dimerization and packaging of human immunodeficiency virus-1 (HIV-1) [4]. Besides, G-quadruplexes also act as repressor elements in the transcriptional activation of HIV-1 [7]. Evidence suggests that G-quadruplexes play a role in the translational control and immune evasion of EBV [8]. Furthermore, G-rich regions in human papillomaviruses (HPVs) were shown to fold into G-quadruplex structures controlling the viral replication and transcription [9, 10]. PQSs were also identified in the Zika viral genome [11].
Previous studies showed that SARS-unique domain (SUD)-NM binds oligo(G)-nucleotides capable of forming G-quadruplexes. SUD-oligo(G) interaction is required for SARS-CoV genome replication. The SARS-CoV genome contains a number of conserved G-quadruplex stretches, which are the potential binding-partners for SUD [12]. The novel coronavirus (SARS-CoV-2), a SARS-like RNA virus causing pneumonia-like symptoms, has raised global health concern. Here, we show that the genetic sequence of SARS-CoV-2 contains a number of four contiguous GG runs (G2NxG2NyG2NzG2), suggesting the presence of PQSs (Figure 1B). We demonstrate that some of those PQSs are conserved in the Coronaviridae family. We confirm G-quadruplex structure forming in the top-ranked PQSs by multiple spectroscopic assays in vitro and characterize the cross-talk between G-quadruplexes and viral helicase by microscale thermophoresis (MST) and molecular docking. Our analysis of G-quadruplex-forming sequences in SARS-CoV-2 provides insights into the design of anti-viral treatment by targeting the viral helicase and G-quadruplex structures.
Results
Identification of PQSs in the SARS-CoV-2 genome
The novel coronavirus SARS-CoV-2 is a positive and single-stranded RNA virus belonging to the family of enveloped coronaviruses (CoVs). We searched the 29.9 kb genome of SARS-CoV-2 stored in the China National Microbiological Data Center (accession number CNP0000881) for PQSs using QGRS-Mapper [13]. We set up the QGRS-mapper software to identify all PQSs with four repeats of at least two guanines interrupted by loops of a maximum length of 36 nucleotides. We performed this relaxed G-quadruplex definition as a number of recent studies have revealed G-quadruplex structure with unconventional features, such as two G-quartets and long loops [14–16]. This analysis revealed the presence of 25 four contiguous GG runs (G2NxG2NyG2NzG2) in the genome of SARS-CoV-2. x, y and z might be different (Table 1). Four contiguous GG runs represent two tetrad G-quadruplex motifs. But, PQSs of four contiguous GGG runs (three G-tetrads) or GGGG runs (four G-tetrads) were not found in the genome of SARS-CoV-2. PQSs of four contiguous GG runs (two G-tetrads) generally exhibit lower stability in folding into a G-quadruplex structure [17]. This is an average of one PQS per ~1 kb in the genome of SARS-CoV-2. However, there are 705 580 PQSs with at least three G-tetrads and 13 948 893 PQSs with two or more G-tetrads in human genome. The human genome has 3095.69 Mb [18]. The density of PQS = 2 × (genome size)/PQS number. The ratio of PQSs with at least three G-tetrads is 1 PQS per ~8.77 kb [19], and PQSs with at least two G-tetrads is 1 PQS per ~0.44 kb. Notably, PQS density of human genome was reported to be strongly enriched at gene promoters (1 kb upstream of TSS) and in 5′ untranslated regions [18].
Table 1.
PQSs found in the SARS-CoV-2 genome
| Position | Gene | Length | Putative G4-forming sequences | G-score |
|---|---|---|---|---|
| 353 | ORF1ab | 25 | GGCUUUGGAGACUCCGUGGAGGAGG | 16 |
| 644 | ORF1ab | 20 | GGUAAUAAAGGAGCUGGUGG | 15 |
| 1463 | ORF1ab | 26 | GGUGGUCGCACUAUUGCCUUUGGAGG | 6 |
| 1574 | ORF1ab | 26 | GGUGUUGUUGGAGAAGGUUCCGAAGG | 18 |
| 2714 | ORF1ab | 29 | GGCGGUGCACCAACAAAGGUUACUUUUGG | 10 |
| 3467 | ORF1ab | 17 | GGAGGAGGUGUUGCAGG | 15 |
| 4162 | ORF1ab | 27 | GGUUAUACCUACUAAAAAGGCUGGUGG | 6 |
| 4261 | ORF1ab | 29 | GGGUUUAAAUGGUUACACUGUAGAGGAGG | 10 |
| 8687 | ORF1ab | 23 | GGAUACAAGGCUAUUGAUGGUGG | 14 |
| 10261 | ORF1ab | 30 | GGCUGGUAAUGUUCAACUCAGGGUUAUUGG | 9 |
| 13385 | ORF1ab | 20 | GGUAUGUGGAAAGGUUAUGG | 19 |
| 14947 | ORF1ab | 28 | GGUUUUCCAUUUAAUAAAUGGGGUAAGG | 4 |
| 15208 | ORF1ab | 27 | GGAACAAGCAAAUUCUAUGGUGGUUGG | 6 |
| 15448 | ORF1ab | 29 | GGCGGUUCACUAUAUGUUAAACCAGGUGG | 3 |
| 18296 | ORF1ab | 23 | GGAUUGGCUUCGAUGUCGAGGGG | 9 |
| 22316 | S | 29 | GGUGAUUCUUCUUCAGGUUGGACAGCUGG | 10 |
| 24215 | S | 20 | GGUUGGACCUUUGGUGCAGG | 17 |
| 24268 | S | 24 | GGCUUAUAGGUUUAAUGGUAUUGG | 19 |
| 25197 | S | 22 | GGCCAUGGUACAUUUGGCUAGG | 17 |
| 25951 | ORF3a | 29 | GGUGGUUAUACUGAAAAAUGGGAAUCUGG | 8 |
| 26746 | M | 30 | GGAUCACCGGUGGAAUUGCUAUCGCAAUGG | 7 |
| 28781 | N | 29 | GGCUUCUACGCAGAAGGGAGCAGAGGCGG | 9 |
| 28903 | N | 15 | GGCUGGCAAUGGCGG | 18 |
| 29123 | N | 19 | GGAAAUUUUGGGGACCAGG | 14 |
| 29234 | N | 30 | GGCAUGGAAGUCACACCUUCGGGAACGUGG | 11 |
Note: G-score was used to evaluate the probability to form a stable G-quadruplex. Higher scoring sequences represent better candidates for G-quadruplexes. Guanine nucleotides predicted to participate in the formation of G-quadruplex structure are underlined.
The PQS positions were inspected with respect to their location in the SARS-CoV-2 genome. Previous analysis showed that the SARS-CoV-2 genome shares 79.5 and 96% sequence homology to SARS-CoV and bat coronavirus, respectively [20]. The G-quadruplex motifs were found located in the open reading frame regions of ORF1 ab, spike (S), ORF3a, membrane (M) and nucleocapsid (N) genes (Figure 1B, Table 1). ORF1ab of SARS-CoV-2, used for CoVs species classification, has 94.6% sequence homology to SARS-CoV. The sequence of the spike (S) gene, which binds to the human cell membrane receptor ACE2 and mediates the internalization of the virus into the host cell, is highly variable in CoVs [20]. M gene encodes membrane glycoprotein [21]. Function of ORF3a in SARS-CoV-2 is unknown; however, in SARS-CoV, ORF3a encodes transmembrane protein localized to the plasma membrane especially in the endoplasmic reticulum (ER)-Golgi region. It activates the PKR-like ER kinase (PERK) signaling pathway, which protects viral proteins against ER-associated degradation and apoptosis [22]. In human CoVs, ORF3a protein has been associated with virulence by controlling not only the expression of cytokines and chemokines but also inducing necrotic cell death [22]. Similar to the N gene of SARS-CoV, the N gene of SARS-CoV-2 is hypothesized to be involved in the viral RNA packaging and to play an essential role in viral RNA transcription and replication [23]. Previous studies suggested that the presence of the G-quadruplexes in the ORF region can halt the translation elongation in vivo [8]. Therefore, G-quadruplex motifs presenting in the SARS-CoV-2 ORFs might play a fundamental role in viral replication, assembly and immune response modulation.
Homology analysis of PQSs in the Coronaviridae family
The National Center for Biotechnology Information database (NCBI) harbors more than 50 sequenced genomes of CoVs, including bat CoVs, Middle East respiratory syndrome-related CoV (MERS-CoV) and SARS-CoV. Phylogenetic tree based on whole genomes of CoVs stored in NCBI using the Molecular Evolutionary Genetics Analysis (MEGA) shows that SARS-CoV-2 is closely related to CoVs found in bats (Figure 1C). To analyze the conservation of the PQSs identified in the SARS-CoV-2, we used the genomes of CoVs stored in NCBI for alignment to identify conserved PQSs throughout the Coronaviridae family (see Supplementary Information S1 available online at https://academic.oup.com/bib and Table 2). The PQSs at positions 353, 644, 2714, 3467, 8687, 10261, 13385, 14947, 15448, 24215, 24268, 26746, 28781 and 28903 are well conserved in a wide range of CoVs (see Supplementary Information S1 available online at https://academic.oup.com/bib and Table 2). Notably, PQSs at positions 14947 and 15448 share identical sequences with We have inserted/amended the running author. Please check and provide correct wording if necessary. a wide range of CoVs, including bat CoVs and SARS-CoV (see Supplementary Information S1 available online at https://academic.oup.com/bib and Table 2). However, those two PQSs have the lowest G-scores, indicating low possibility in folding into G-quadruplexes. Interestingly, PQSs at positions 13385 and 24268 with the highest G-scores indicating high probability to adopt G-quadruplex structures only share high sequence similarity to the bat CoVs (see Supplementary Information S1 available online at https://academic.oup.com/bib and Table 2). The sequence homology of PQSs can suggest conserved biological functions, where SARS-CoV-2 and bat coronavirus share an undiscovered common ancestor [20].
Table 2.
Homology analysis of SARS-CoV-2 PQSs to other CoVs
| Position | Genome with the highest conservation (E value, % coverage) |
|---|---|
| 353 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (2e-04, 100%) |
| 644 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (2e-04, 100%) |
| Bat SARS-like coronavirus isolate bat-SLCoVZC45 (2e-04, 100%) | |
| 2714 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (1e-04, 100%) |
| 3467 | Coronavirus BtRt-BetaCoV/GX2018 (1.6, 100%) |
| 8687 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (5e-06, 100%) |
| Bat SARS-like coronavirus isolate bat-SLCoVZC45 (5e-06, 100%) | |
| 10261 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (3e-05, 100%) |
| 13385 | Rhinolophus affinis coronavirus isolate LYRa11 (2e-04, 100%) |
| Bat coronavirus Cp/Yunnan2011 (2e-04, 100%) | |
| 14947 | Wide range of Coronaviridae |
| 15448 | Wide range of Coronaviridae |
| 24215 | Bat coronavirus Cp/Yunnan2011 (0.051, 100%) |
| 24268 | Bat SARS-like coronavirus isolate bat-SLCoVZC45 (3e-04, 100%) |
| 26746 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (0.008, 100%) |
| Bat SARS-like coronavirus isolate bat-SLCoVZC45 (0.008, 100%) | |
| 28781 | Wide range of Coronaviridae |
| 28903 | Bat SARS-like coronavirus isolate bat-SLCoVZXC21 (0.067, 100%) |
| Bat SARS-like coronavirus isolate bat-SLCoVZC45 (0.067, 100%) |
Note: SARS-CoV-2 PQSs can be found in other members of the Coronaviridae family.
G-quadruplex formation confirmed by multiple spectroscopic assays in vitro
G-quadruplex formation from the predicted PQSs cannot be safely assumed without experimental evidence, since it has been understood that not all PQS can fold into a G-quadruplex structure. In vitro PQSs of four contiguous GG runs (a.k.a. a two-quartet motif) in general also have lower propensity to fold into a G-quadruplex structure [17]. To validate G-quadruplex formation of top-ranked PQSs identified in our study, circular dichroism (CD) spectroscopy analysis, ultraviolet (UV) melting assay and fluorescence turn-on assay, which are commonly used to detect G-quadruplex formation [24], were conducted.
First, CD spectroscopy was used to determine the topology of RNA Please provide the significance for bold and underline present in Table 1. G-quadruplexes for PQSs at positions 13385 and 24268 (13385/PQS and 24268/PQS). The CD spectra recorded for both top-ranked PQSs in the presence of K+ or Li+ displayed only a positive band at ~265 nm and a negative band at 240 nm (Figure 2A and D). CD signal decreased, especially in 13385/PQS, upon substituting the K+ (G-quadruplex-stabilizing) with Li+ (G-quadruplex non-stabilizing) (Figure 2A). Consistent with literature [24], this CD profile and the monovalent ion-dependent feature are indicative of the formation of G-quadruplex with parallel topology. Second, UV melting data on the 13385/PQS and 24268/PQS exhibited a hyperchromic shift at 295 nm under K+ condition, which is a hallmark of G-quadruplex formation [25]. The melting temperature (Tm) of 13385/PQS and 24268/PQS were determined to be 31.5 and 30.5°C under 150 mM K+ conditions, close to the physiological temperature of 37°C (Figure 2B and E). Finally, RNA G-quadruplex structure-selective binding of N-methyl mesoporphyrin IX (NMM) was employed to confirm the G-quadruplex structures formation in 13385/PQS and 24268/PQS. NMM is a fluorescence light-up probe upon binding to the G-quadruplex structures [26, 27]. The much stronger fluorescence in K+ than Li+ condition results from the specifically binding of NMM to RNA G-quadruplex, which suggests the formation of G-quadruplex structures in PQSs at positions of 13385 and 24268 (Figure 2C and F). To further improve our understanding of RNA G-quadruplexes, investigating protein–RNA-binding events will be critical to completing an overall picture of their roles in SARS-CoV-2.
Figure 2.
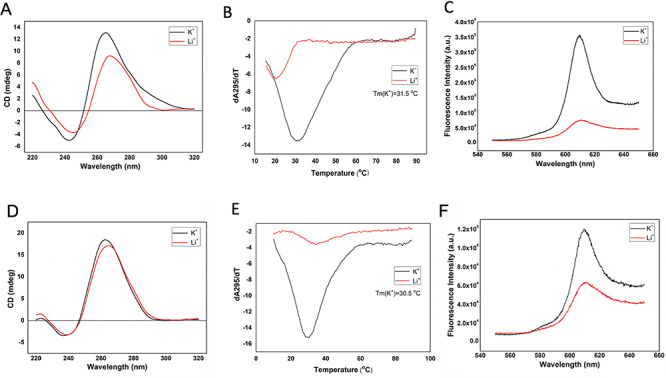
Biophysical characterization of G-quadruplex structure in 13385/PQS and 24268/PQS. (A) CD spectrum of 5 μM RNA 13385/PQS under 150 mM KCl and LiCl conditions. CD spectra suggest the formation of a parallel G-quadruplex structure in RNA 13385/PQS. (B) UV melting curve of 5 μM RNA 13385/PQS under 150 mM KCl and LiCl conditions. Hypochromic shift was observed and the melting temperature (Tm) was determined to be 31.5°C under K+ condition. (C) Fluorescence emission spectra of NMM with 1 μM RNA 13385/PQS under 150 mM KCl and LiCl conditions. The stronger fluorescence under K+ condition suggests the formation of G-quadruplex structure. (D) CD spectrum of 5 μM RNA 24268/PQS under 150 mM KCl and LiCl conditions. CD spectra suggest the formation of a parallel G-quadruplex structure in RNA 24268/PQS. (E) UV melting curve of 5 μM RNA 24268/PQS under 150 mM KCl and LiCl conditions. Hypochromic shift was observed and the melting temperature (Tm) was determined to be 30.5°C under K+ condition. (F) Fluorescence emission spectra of NMM with 1 μM RNA 24268/PQS under 150 mM KCl and LiCl conditions. The fluorescence enhancement between KCl and LiCl conditions suggests the G-quadruplex structure formation.
Virus–host cell interplays through G-quadruplexes and helicases
A number of studies showed that G-quadruplexes in RNA can be unfolded by helicase [28]. Viral helicase nsp13 plays a vital role in catalyzing the unwinding of double-stranded oligonucleotides into single strands [29] and might also catalyze the unwinding of G-quadruplex structures. The crystal structure of the full-length SARS-CoV nsp13 was reported recently [30]. Moreover, nsp13 is highly conserved in most CoVs. Sequence alignment between SARS-CoV and SARS-CoV-2 revealed almost 100% identity in nsp13 (Figure 3). Previous work employing yeast two-hybrid screening and co-immunoprecipitation showed that SARS-CoV helicase nsp13 interacts with cellular RNA helicase DDX5. SARS-CoV replication was significantly inhibited by knocking down the expression of DDX5 with small interfering RNA [31]. Asp-Glu-Ala-Asp (DEAD)-box polypeptide 5 (DDX5) is a prototypical member of the large ATP-dependent RNA helicase family and plays crucial roles in RNA biology, including translation, splicing, transcription regulation, ribosome biogenesis, mRNA nuclear export and micro RNA (miRNA) processing [32]. DDX5 was reported to be a G-quadruplex resolvase, which actively unfolds DNA and RNA G-quadruplex structures [33]. The DEAH-box helicases DHX9 and DHX36 (RHAU) were also reported to unwind both RNA and DNA G-quadruplexes [34]. The G-quadruplex resolvase DHX36 efficiently and specifically disrupts DNA G-quadruplexes via a translocation-based helicase mechanism [35]. The DEAD-box RNA helicases DDX21, DDX1 and DDX3X were reported to exhibit G-quadruplex unfolding only in RNA [36, 37]. Host DDX5 or other DEAD-box helicase may be hijacked by CoVs to enhance the transcription and proliferation of viral genome through G-quadruplex unfolding.
Figure 3.
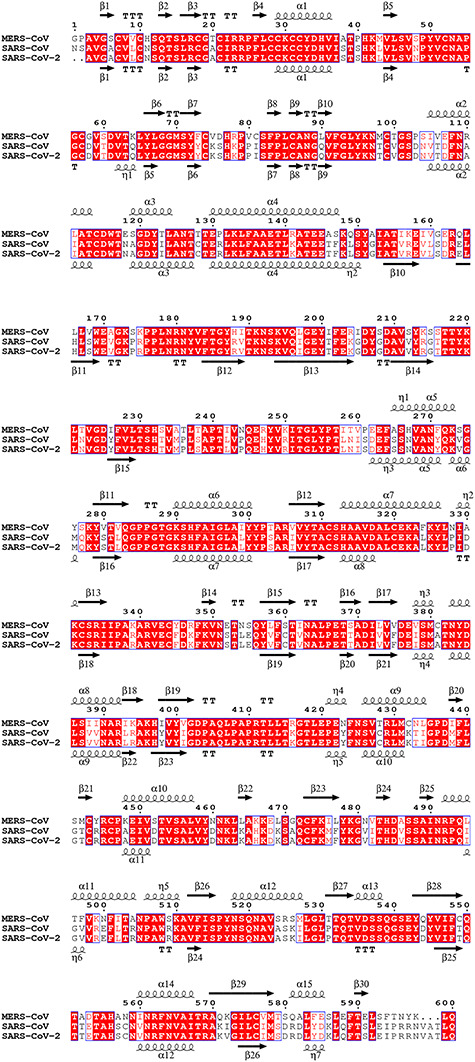
The multiple sequence alignment of helicase nsp13 from MERS-CoV, SARS-CoV and SARS-CoV-2. Invariant residues are highlighted with red background; conserved residues are in red. Secondary structure elements of MERS-CoV and SARS-CoV are aligned to the top and bottom of the sequences, respectively. Multiple sequence alignments were carried out using the program MUSCLE. The illustration of the result was generated by the program ESPript v3.0. The top line of secondary structure is MERS-CoV, and the bottom line is SARS-CoV. α-helices and 310-helices (η-helices) are displayed as medium and small squiggles. β-strands are rendered as arrows, and strict β-turns as TT letters.
It has been reported that SARS-CoV helicase (nsp13) has both RNA and DNA duplex unwinding activity [38]. Amino acid sequences of nsp13 proteins from different CoVs were compared, including SARS-CoV, MERS-CoV, mouse hepatitis virus, transmissible gastroenteritis virus, and avian infectious bronchitis virus. The protein alignment result showed that all helicases have the conserved helicase motifs of superfamily 1 (SF1) [39]. Moreover, the structural alignment showed some degree of similarity between SARS-CoV helicase (nsp13) and human Upf1 [30]. Both nsp13 and Upf1 belong to helicase SF1 [29, 40, 41]. Upf1 plays an important role in mRNA regulation [42]. The UPF1 helicase is also capable of unfolding G-quadruplex DNA [41]. Consequently, nsp13 of SARS-CoV-2 is very likely to be a G-quadruplex unwinding helicase. Since G-quadruplex structures are present in the transcriptome of the viral host (e.g. humans), the possibility that viral nsp13 interacts with host G-quadruplex structures cannot be excluded. The binding/unfolding events might be crucial to viral infection and host antiviral response.
Notably, RNA aptamers [43] and DNA aptamers [44] against the SARS-CoV helicase nsp13 have been reported previously. Aptamers are short, single-stranded DNA or RNA that developed in vitro by SELEX (systematic evolution of ligands by exponential enrichment) and bind to targets with high affinity and specificity by folding into tertiary structures [45]. Interestingly, most of the enriched aptamers against nsp13 are putative G-quadruplexes forming with guanine repeats in their sequences (Table 3). Among those, DNA aptamers (G5 and G8) were confirmed to form G-quadruplex structure by CD spectra [44]. Furthermore, the selected RNA aptamers were identified with a AG-rich conserved sequence of 10–11 nucleotides [AAAGGR(G)GAAG; R, purine base] and/or additional sequence of 5 nucleotides [GAAAG]. It has been predicted that the AG-rich conserved sequence and the [GAAAG] sequence mainly reside at the loop region in all the structures (Table 3). It has been suggested that the exposed [AAAGGR(G)GAAG] and [GAAAG] sequences in RNA aptamers might be important for interaction with the SARS-CoV helicase nsp13. In comparison, our G-quadruplex search across the genome of SARS-CoV-2 also identified a number of GG PQSs. PQS at position 13385 was confirmed to adopt G-quadruplex structures, which also contains a [GAAAG] sequence in the middle (Table 1). The naturally occurring [GAAAG] sequence in the PQSs RNA of SARS-CoV-2 implies the important interaction motif and potential cross-talk with nsp13. Therefore, we hypothesize that PQSs identified in our study might also bind to nsp13 helicase in SARS-CoV-2.
Table 3.
PQSs found in the reported nsp13 aptamers
| Seq. name | Length | sequence | G-score | |
|---|---|---|---|---|
| RNA aptamers [43] | ES15–1 | 38 | GCAGAAAAGGGGGAAGAAGAGGGUGAUUCAGGCGAGAG | 14 |
| ES15–2 | 40 | CAGGGAGGAAAGGGGGAAGCGACUCAAGAACUGUAGAGGG | 19 | |
| ES15–3 | 41 | CCGGGCGGUCAAAGGAGAAGAAGAAAGAGAGAGCCCAGGGA | 0 | |
| ES15–4 | 40 | GGAGGGAAAAGGGGAAGCGGAAAGGUUAAGGAUGCGGAGG | 17/20 | |
| ES15–5 | 40 | GGUUAGGGGGAAAGGGGACCAGGUUCGCAGGAAAGCAGAG | 21 | |
| ES15–6 | 41 | GGAAGGGAGAGCGGGAACAAGGAGAAAGAGAAGGGGAAUCC | 18 | |
| DNA aptamers [44] | G1 | 29 | AACTTGGGGTGGGTGTGTTGTGTACGGGC | 9 |
| G2 | 30 | GGCTTGTGGGTGGTATCTGTGGTGTGTGC | 16 | |
| G3 | 29 | CGTCGGTGGTTGTGTCGGGGCGATGGGTT | 15 | |
| G4 | 30 | ATGGCATGTGTTTGGCGTGGATCGTGTGGC | 15 | |
| G5 | 30 | AGCGGGCATATGGTGGTGGGTGGTATGGTC | 20 | |
| G6 | 30 | AAGGGTGGAAAGTTGGGGGGTGTAGTGTTC | 17 | |
| G7 | 30 | ATGCCGCGTTGGGGAGTGGTGTTGGCTGGT | 19 | |
| G8 | 30 | AATGGAGTATGGATGGATTGCTAGTTCGGC | 12 | |
| G9 | 30 | GCTGGCCGGGATATGGTATGTTTGGCAGTT | 17 |
Note: Guanine nucleotides that participate in the formation of G-quadruplex structure were predicted by QGRS mapper and underlined. G-score was used to evaluate the probability to form a stable G-quadruplex. Higher scoring sequences represent better candidates for G-quadruplexes.
The interaction between nsp13 helicase protein and PQSs
To analyze the potential binding events, MST assay was performed to investigate the interaction between viral nsp13 protein and top-ranked PQSs. MST is a powerful tool to characterize molecular interactions in liquids including, but not limited to, protein, RNA, DNA, small molecules and ions [46]. The expression and purification of nsp13 protein from SARS-CoV were reported previously [39]. Moreover, the nsp13 protein of SARS-CoV is identical to SARS-CoV-2. As a result, SARS-CoV protein was expressed and purified as the target for binding (Figure 4A). We quantified the dissociation constants of 13385/PQS and 24268/PQS with nsp13 protein. We prepared a titration series of nsp13 protein (0.000153–5 μM) to mix with a constant concentration of 13385/PQS or 24268/PQS (30 nM). Binding affinity (Kd) is automatically calculated from the binding curves fitted by Nanotemper Analysis in MST. Kd values of 0.79 ± 0.14 and 0.37 ± 0.08 μM were obtained for 13385/PQS and 24268/PQS, respectively (Figure 4B). This confirms the physical interaction between SARS-CoV (or SARS-CoV-2) nsp13 protein and top-ranked PQSs in vitro.
Figure 4.
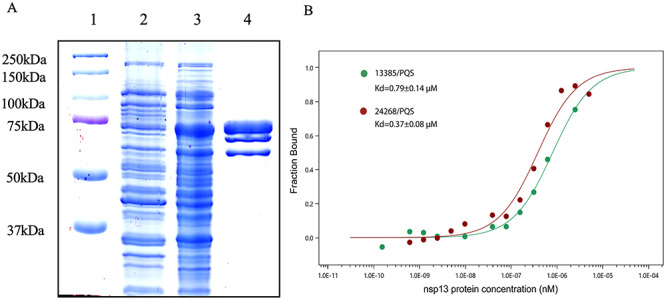
The interaction of nsp13 and 13385/PQS or 24268/PQS demonstrated by MST assay. (A) Expression and purification of nsp13. Lane 1, protein marker. Lane 2, BL21 cell transfected with nsp13 and extracted without induction. Lane 3, BL21 cell transfected with nsp13 and extracted after IPTG induction. Lane 4, helicase nsp13 purified by nickel affinity chromatography. (B) Dose–response curves for the binding interaction between the purified nsp13 protein and 13385/PQS or 24268/PQS RNAs.
Structural analysis of PQSs
As described above, top-ranked PQSs at positions of 13385 and 24268 were confirmed to form G-quadruplex structures. PQS at position 13385 contains a [GAAAG] motif found in the nsp13 binding aptamers. For PQSs at position 24268, two additional PQSs in the area upstream (position 24215) and downstream (position 25197) were identified in the S gene with similar high possibility (G-score = 17) to form G-quadruplexes (Table 1). Consequently, PQSs at positions of 13385 and 24268 are the most attractive candidates for investigation. To better understand the role of G-quadruplexes in SARS-CoV-2 and their mechanism of action, we performed the molecular docking of 13385/PQS or 24268/PQS with the SARS-CoV helicase nsp13 (PDB: 6JYT), respectively. Docking between PQS RNA molecules and nsp13 protein was performed using the docking server HDOCK (http://hdock.phys.hust.edu.cn/). HDOCK incorporates the binding interface information into the docking concept, which accepts both sequences and structures as input for the protein docking prediction [47]. According to the docking results, 13385/PQS was predicted to bind to the cleft between 1B domain and zinc-binding domain (ZBD), while 24268/PQS was predicted to bind to the cleft between 1B and 2A domains of SARS-CoV nsp13 (Figure 5A and C). It has been shown that 1B, 1A and 2A domains of SARS-CoV nsp13 are involved in the dsDNA binding [30]. Moreover, docking results suggested that the guanine bases deviated significantly from their expected placement in both cases. Those guanine bases are predicated to be flipped away from the G-quartets, thus disrupting their G-quadruplex structures (Figure 5B and D ). We hypothesize that the rearrangements of guanine bases induce G-quadruplex structure unfolding by the viral nsp13. In addition, the [GAAAG] motif found in both the nsp13-binding aptamers and 13385/PQS was predicted to closely interact with the cleft between 1B domain and ZBD (Figure 5B). The cleft at the base between the 1A and 2A domains represents a NTPase active pocket [30], which is farther away from the predicted G-quadruplex unwinding site. In summary, our bioinformatic docking analysis provides better understanding of the binding and unfolding of G-quadruplex oligonucleotides by SARS-CoV-2 nsp13 protein.
Figure 5.
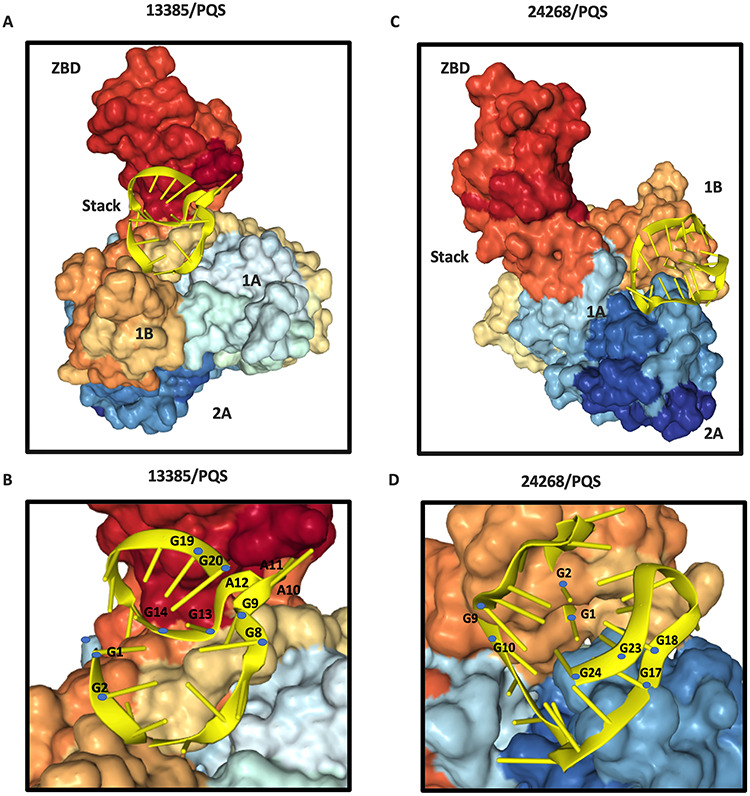
Molecular docking of 13385/PQS or 24268/PQS with the SARS-CoV helicase nsp13. Surface representation of SARS-CoV nsp13 (PDB ID: 6JYT) is composed of ZBD, stalk, 1B, 1A and 2A domains. The subdomains of the helicases are labeled. (A) Ribbon structure of 13385/PQS RNA molecule (gold color) bind to the 1B domain and ZBD of SARS-CoV nsp13. (B) Ribbon model of 13385/PQS RNA molecule (gold color). Guanine bases involved in the formation of G-quadruplex are colored with blue. The [G9A10A11A12G13] motif was predicted to closely contact with the cleft between 1B domain and ZBD. G-quadruplex structure was disrupted after binding. (C) Ribbon structure of 24268/PQS RNA molecule (gold color) bind to the 1B and 2A domains of SARS-CoV nsp13. (D) Ribbon model of 24268/PQS RNA molecule (gold color). Guanine bases involved in the formation of G-quadruplex are colored with blue. G-quadruplex structure was unfolded by the viral helicase, where the guanine bases were flipped out of the G-quartet after binding.
Potential therapeutic implications
The newly identified G-quadruplex-forming sequences might play important regulation roles in viral life cycle, serving as potential targets for antiviral treatment in SARS-CoV-2. As a result of their putative roles in binding and unwinding of the G-quadruplexes, helicases are also attractive antiviral targets for SARS-CoV-2. Several helicase inhibitors, such as bananins and 5-hydroxychromone derivatives, have been shown to exhibit antiviral activity against SARS-CoV by inhibiting ATPase activity [48–50]. Helicase inhibitors have the potential to be used as drugs against SARS-CoV-2. As discussed before, the human helicases share some degree of similarity or bind with SARS-CoV helicase (nsp13). SARS-CoV-2 might utilize those host helicases with similar conformation or function to bind with G-quadruplex molecules during infection. The inhibitory effects on cellular kinases and resulting potential cytotoxicity might limit clinical application of the helicase inhibitors.
Another class of helicase inhibitors that selectively inhibits the unwinding activity, but not the ATPase activity, was also shown to be an efficient antiviral agent, including aryl diketoacid compound in SARS-CoV [51] and SSYA10–001 in a broad range of CoVs [52]. Another approach to inhibit the G-quadruplex unwinding activity of viral helicase by stabilizing G-quadruplex structure can utilize small molecule ligands such as flavonoids [53], pyridostatin [54], GQC-05 [55] and CX-5461 [56]. Furthermore, nsp13 has been shown to interact with other SARS-CoV proteins, such as non-structural protein nsp12 (SARS-CoV polymerase), which leads to the enhanced helicase activity of nsp13 [30]. Consequently, targeting the nsp12-nsp13 complex could also be useful; however, this would require a more complicated docking of G-quadruplex RNAs with the nsp12-nsp13 complex.
Conclusions
Increasing evidence suggests that G-quadruplex structures play a regulatory role in gene transcription in humans and other organisms [2]. We identified 25 putative G-quadruplex folding sequences in the ORF regions of the SARS-CoV-2 genome. The identified top-ranked PQSs were confirmed to form G-quadruplex structures by multiple spectroscopic assays in vitro. Those G-quadruplex structures likely play a crucial role in the regulation of SARS-CoV-2 genes. Viral helicase (nsp13) is required for virus replication in the life cycle of SARS-CoV and MERS-CoV [57]. Our study showed that G-quadruplex-forming sequences interact with viral helicase nsp13. Helicase helps to catalyze the unfolding of G-quadruplexes [28] and facilitates the transcription and replication of the SARS-CoV-2. Given its biological importance, structural analysis of interaction between the G-quadruplex forming RNAs and the nsp13 helicase provides new insights into the RNA regulation of SARS-CoV-2. However, G-quadruplex nucleic acid structures are relatively difficult to probe in vivo, and only limited number of structures have been solved so far. As a result, molecular docking is an important tool to better understand the mechanism behind the G-quadruplex binding or unwinding in CoVs. Our docking analysis suggests rearrangement and flipping of the guanine bases drives G-quadruplex unfolding. A detailed understanding of the interactions between G-quadruplex forming RNAs with viral helicase is valuable for the rational design of new therapies. Notably, G-quadruplex RNA sequences might adopt various folding conformations, depending on flanking and loop sequences and environmental conditions. If the conformation is the most important feature shared by those PQSs found in SARS-CoV-2, we would expect the models built herein to be consistent with other RNAs sharing similar G-quadruplex structures. Consequently, even in the presence of polymorphism, this study can be used directly to complement experimental studies on the dynamics of G-quadruplex unwinding by SARS-CoV-2. However, the possibility of the predicted sequence adopting misfolded structures should also be taken into account in the future studies. G-quadruplex structure binding/unfolding capabilities of nsp13 protein need to be experimentally established in the future. The role of G-quadruplexes in the SARS-CoV-2 gene regulation, their structures and their use as potential drug targets will require further investigation.
Materials and methods
Materials
RNA oligos (13385/PQS: GGUAUGUGGAAAGGUUAUGG; 24268/PQS: GGCUUAUAGGUUUAAUGGUAUUGG) were synthesized and purified by Genewiz Biotechnology Co., Ltd. (Suzhou, China). N-methyl-mesoporphyrin IX (NMM) was purchased from Frontier scientific (USA). All the stock and buffer were prepared using nuclease-free ultrapure distilled water (Thermo Fisher Scientific, USA).
Identification of the PQSs
RNA sequence of SARS-CoV-2 was obtained from the China National Microbiological Data Center (accession number CNP0000881). SARS-CoV-2 genome was analyzed using QGRS-Mapper (http://bioinformatics.ramapo.edu/QGRS/analyze.php) [13]. The parameters used were as follows: max length: 30; min G-group: 2; loop size: 0–36.
Sequence alignments
Alignments of PQSs within the Coronaviridae family were performed by using the BLAST algorithm adjusted for short input sequences from NCBI. Multiple sequence alignments of the PQSs in the members of the Coronaviridae were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Multiple sequence alignment on nsp13 protein was carried out using MUSCLE software (http://www.ebi.ac.uk/Tools/msa/muscle/). The illustration of sequence alignment was generated using ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/).
Phylogenetic analysis
Whole genome sequence alignment of SARS-CoV-2 genome with reference genomes stored in NCBI and phylogenetic analysis were done with the MEGA (version X), by the maximum likelihood method under the general time reversible nucleotide substitution model.
CD spectroscopy
CD spectra were detected on a Jasco CD J-150 spectrometer using 1 cm path length quartz cuvette. 13385/PQS or 24268/PQS RNAs were heated at 95°C for 3 min and cooled down on ice for 10 min before use. Samples were prepared in 10 mM LiCac buffer (pH 7.0) containing 5 μM 13385/PQS or 24268/PQS RNAs and 150 mM KCl/LiCl. Three scans from 220 to 320 nm at 1 nm interval were accumulated and averaged.
UV melting assay
UV melting assay was performed on a Cary 100 UV-vis spectrophotometer using 1 cm path length quartz cuvette. Samples were prepared as CD measurement above. The unfolding transitions were monitored at 295 nm. Spectra were collected over 0.5°C while heating over the temperature range from 5 to 95°C.
Fluorescence spectroscopy
Fluorescence detection was performed on a HORIBA FluoroMax-4 fluorescence spectrophotometer (Japan) using 1 cm path length quartz cuvette. 13385/PQS or 24268/PQS RNAs were heated at 95°C for 3 min and cooled down on ice for 10 min before use. 13385/PQS or 24268/PQS RNAs (1 μM) were mixed with 2 μM NMM in 10 mM lithium cacodylate (LiCac) buffer (pH 7.0) with 150 mM KCl or LiCl. The samples were incubated at room temperature for 30 min followed by fluorescence measurement. Spectra of NMM were excited at 399 nm and collected from 550 to 650 nm. Excitation and emission slits were set at 5 nm.
Expression and purification of nsp13 helicase protein
The SARS-CoV nsp13 expression plasmid was a kind gift from Dr Jiandong Huang, University of Hong Kong, China. The nsp13 protein was expressed in BL21 competent cells and purified as described previously [39].
Microscale thermophoresis
MST assays were carried out on a Monolith NT.115 instrument (NanoTemper) using the ‘nano-blue’ channel. Fluorescein amidite-labeled 13385/PQS and 24268/PQS RNAs were heated at 95°C for 3 min and cooled down on ice for 10 min before use. Different concentrations (0.000153–5 μM) of purified nsp13 protein were mixed with 13385/PQS or 24268/PQS RNAs (30 nM) in 25 mM Tris-HCl buffer (pH 7.5), 150 mM KCl and 5 mM MgCl2. The samples were incubated at 37°C for 30 min and subsequently applied to MST assay. Data were fitted by NanoTemper Analysis with a Kd model from the MST machine.
Docking analysis
Docking between PQS RNA molecules and nsp13 proteins were performed by HDOCK (http://hdock.phys.hust.edu.cn/).
Supplementary Material
Danyang Ji is a research student in Department of Chemistry, City University of Hong Kong, China. Her research interests include G-quadruplex and aptamer.
Mario Juhas worked as a research fellow at the universities of Oxford, Cambridge and Zurich. Currently, he works at the University of Fribourg. His work spans microbiology and synthetic biology.
Chi Man Tsang is a research assistant professor in Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong, China. Her research interests are mainly on the roles of Epstein-Barr virus (EBV) in the pathogenesis, progression of nasopharyngeal carcinoma (NPC) and aptamers for visualizing and targeting of NPC tumors.
Chun Kit Kwok is an assistant professor in Department of Chemistry, City University of Hong Kong, China, and Shenzhen Research Institute of City University of Hong Kong, Shenzhen,China. His research interests include RNA Biology, chemical biology, nucleic acids and G-quadruplex.
Yongshu Li is a PhD student in the Department of Anatomical and Cellular Pathology at The Chinese University of Hong Kong. He is working on the application of aptamer targeting NPC and EBV.
Yang Zhang is an assistant professor at the College of Science, Harbin Institute of Technology (Shenzhen). He finished PhD training in the Department of Pathology and Systems Biology Centre at the University of Cambridge. He is interested in developing computational and experimental methods for cancer, microbiology and nucleic acids research.
Key Points
Putative G-quadruplex-forming sequences (PQSs) were discovered in the RNA genome of SARS-CoV-2. PQSs were found located in the open reading frames of the ORF1 ab, spike (S), ORF3a, membrane (M) and nucleocapsid (N) genes.
The top-ranked PQSs at position 13385 and 24268 were confirmed to form G-quadruplex structures in vitro by circular dichroism spectroscopy, UV melting assay and fluorescence turn-on assay.
We demonstrated the direct interactions of nsp13 protein with PQSs at position 13385 or 24268 in vitro by microscale thermophoresis.
Molecular docking showed the guanine bases deviating significantly from their expected positions within guanine quartet planes, indicating that the G-quadruplex structure was unfolded by the viral helicase nsp13.
Targeting viral helicase nsp13 is an attractive approach for potentially inhibiting the SARS-CoV-2.
Funding
Shenzhen Science and Technology Innovation Commission (Shenzhen Basic Research Project No. JCYJ20180507181642811, JCYJ20180306172131515); The Research Grants Council, University Grants Committee (Project No. CityU 11101519, CityU 11100218, N_CityU110/17, CityU 21302317); The Croucher Foundation (Project No. 9500030, 9500039, and 9509003).
References
- 1. Burge S, Parkinson GN, Hazel P, et al. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 2006;34:5402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 2015;43:8627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris LM, Merrick CJ. G-quadruplexes in pathogens: a common route to virulence control? PLoS Pathog 2015;11:e1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Metifiot M, Amrane S, Litvak S, et al. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res 2014;42:12352–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartas M, Cutova M, Brazda V, et al. The presence and localization of G-quadruplex forming sequences in the domain of bacteria. Molecules 2019;24:1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belmonte-Reche E, Martinez-Garcia M, Guedin A, et al. G-Quadruplex identification in the genome of protozoan parasites points to naphthalene diimide ligands as new antiparasitic agents. J Med Chem 2018;61:1231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karn J, Stoltzfus CM. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2012;2:a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murat P, Zhong J, Lekieffre L, et al. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat Chem Biol 2014;10:358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tluckova K, Marusic M, Tothova P, et al. Human papillomavirus G-quadruplexes. Biochemistry 2013;52:7207–16. [DOI] [PubMed] [Google Scholar]
- 10. Marusic M, Hosnjak L, Krafcikova P, et al. The effect of single nucleotide polymorphisms in G-rich regions of high-risk human papillomaviruses on structural diversity of DNA. Biochim Biophys Acta Gen Subj 1861;2017:1229–36. [DOI] [PubMed] [Google Scholar]
- 11. Fleming AM, Ding Y, Alenko A, et al. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the Flaviviridae family. ACS Infect Dis 2016;2:674–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson MA, Chatterjee A, Neuman BW, et al. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J Mol Biol 2010;400:724–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kikin O, D'Antonio L, Bagga PS. QGRS mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 2006;34:W676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwok CK, Marsico G, Sahakyan AB, et al. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat Methods 2016;13:841–4. [DOI] [PubMed] [Google Scholar]
- 15. Jodoin R, Bauer L, Garant JM, et al. The folding of 5'-UTR human G-quadruplexes possessing a long central loop. RNA 2014;20:1129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guédin A, Gros J, Alberti P, et al. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res 2010;38:7858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwok CK, Marsico G, Balasubramanian S. Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb Perspect Biol 2018;10:a032284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsico G, Chambers VS, Sahakyan AB, et al. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res 2019;47:3862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smargiasso N, Gabelica V, Damblon C, et al. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genomics 2009;10:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rehman SU, Shafique L, Ihsan A, et al. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens 2020;9:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yue Y, Nabar NR, Shi CS, et al. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis 2018;9:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Narayanan K, Kim KH, Makino S. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res 2003;98:131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kwok CK, Merrick CJ. G-quadruplexes: prediction, characterization, and biological application. Trends Biotechnol 2017;35:997–1013. [DOI] [PubMed] [Google Scholar]
- 25. Mergny JL, Lacroix L. UV melting of G-Quadruplexes. Curr Protoc Nucleic Acid Chem 2009; Chapter 17:Unit 17.11. [DOI] [PubMed] [Google Scholar]
- 26. Lyu K, Chen SB, Chan CY, et al. Structural analysis and cellular visualization of APP RNA G-quadruplex. Chem Sci 2019;10:11095–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Endoh T, Rode AB, Takahashi S, et al. Real-time monitoring of G-Quadruplex formation during transcription. Anal Chem 2016;88:1984–9. [DOI] [PubMed] [Google Scholar]
- 28. Mendoza O, Bourdoncle A, Boule JB, et al. G-quadruplexes and helicases. Nucleic Acids Res 2016;44:1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adedeji AO, Marchand B, Te Velthuis AJ, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One 2012;7:e36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia Z, Yan L, Ren Z, et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 2019;47:6538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen JY, Chen WN, Poon KM, et al. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol 2009;154:507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng W, Chen G, Jia H, et al. DDX5 RNA helicases: emerging roles in viral infection. Int J Mol Sci 2018;19:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu G, Xing Z, Tran EJ, et al. DDX5 helicase resolves G-quadruplex and is involved in. Proc Natl Acad Sci U S A 2019;116:20453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–65. [DOI] [PubMed] [Google Scholar]
- 35. Yangyuoru PM, Bradburn DA, Liu Z, et al. The G-quadruplex (G4) resolvase DHX36 efficiently and specifically disrupts DNA G4s via a translocation-based helicase mechanism. J Biol Chem 2018;293:1924–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McRae EKS, Booy EP, Moya-Torres A, et al. Human DDX21 binds and unwinds RNA guanine quadruplexes. Nucleic Acids Res 2017;45:6656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herdy B, Mayer C, Varshney D, et al. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res 2018;46:11592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee NR, Kwon HM, Park K, et al. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res 2010;38:7626–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adedeji AO, Lazarus H. Biochemical characterization of Middle East respiratory syndrome coronavirus helicase. mSphere 2016;1:e00235–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol 2010;20:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dehghani-Tafti S, Sanders CM. DNA substrate recognition and processing by the full-length human UPF1 helicase. Nucleic Acids Res 2017;45:7354–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fischer JW, Busa VF, Shao Y, et al. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jang KJ, Lee NR, Yeo WS, et al. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase. Biochem Biophys Res Commun 2008;366:738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shum KT, Tanner JA. Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem 2008;9:3037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Y, Lai BS, Juhas M. Recent advances in aptamer discovery and applications. Molecules 2019;24:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jerabek-Willemsen M, Wienken CJ, Braun D, et al. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 2011;9:342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan Y, Zhang D, Zhou P, et al. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017;45:W365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim MK, Yu MS, Park HR, et al. 2,6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV). Eur J Med Chem 2011;46:5698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanner JA, Zheng BJ, Zhou J, et al. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem Biol 2005;12:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adedeji AO, Sarafianos SG. Antiviral drugs specific for coronaviruses in preclinical development. Curr Opin Virol 2014;8:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Adedeji AO, Singh K, Calcaterra NE, et al. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother 2012;56:4718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Adedeji AO, Singh K, Kassim A, et al. Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother 2014;58:4894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tawani A, Mishra SK, Kumar A. Structural insight for the recognition of G-quadruplex structure at human c-myc promoter sequence by flavonoid Quercetin. Sci Rep 2017;7:3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moruno-Manchon JF, Koellhoffer EC, Gopakumar J, et al. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging (Albany NY) 2017;9:1957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Montoya JJ, Turnidge MA, Wai DH, et al. In vitro activity of a G-quadruplex-stabilizing small molecule that synergizes with Navitoclax to induce cytotoxicity in acute myeloid leukemia cells. BMC Cancer 2019;19:1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu H, Di Antonio M, McKinney S, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun 2017;8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pillaiyar T, Meenakshisundaram S, Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today 2020;25:668–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


