Abstract
In early-to-mid March 2020, 20 of 46 (43%) COVID-19 cases at a tertiary care hospital in San Francisco, California were travel related. Cases were significantly associated with travel to either Europe (odds ratio, 6.1) or New York (odds ratio, 32.9). Viral genomes recovered from 9 of 12 (75%) cases co-clustered with lineages circulating in Europe.
Keywords: SARS coronavirus 2 (SARS-CoV-2), hospital epidemiology, pandemic, travel history, risk-based screening
INTRODUCTION
As of 4 April 2020, the coronavirus disease 2019 (COVID-19) pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], has infected more than 1.2 million people worldwide and the increase in cases has been exponential. In particular, New York cases in the United States quickly surged from 22 to more than 10 000 between 10 and 22 March [2]. By 4 April there were more than 150 000 cases in New York and nearby New Jersey, threatening to overwhelm hospitals and other regional healthcare systems in the city.
In San Francisco, we validated a quantitive reverse transcriptase–polymerase chain reaction (qRT-PCR) test to detect SARS-CoV-2 infection from nasopharyngeal swab samples based on the EUA (Emergency Use Authorization)-approved US Centers for Disease Control and Prevention assay [3]. Here we present travel-associated findings among the COVID-19 patients seen at University of California, San Francisco (UCSF) during the first 10 days since launch.
METHODS
The institutional review board (IRB) at UCSF approved the clinical and epidemiological association study (IRB #20-30538) and the phylogenetic study (IRB #10-01116, 11-05519). Non-identifying clinical, demographic, and laboratory data were extracted from clinical testing results and the electronic medical board by retrospective chart review. Informed consent was waived for this minimal risk study. Documented history was recorded by a physician or nurse practitioner and included sick contacts, health care worker status, and travel history.
For sequencing of SARS-CoV-2 genomes, RNA was extracted from nasopharyngeal (NP) swab samples on a Qiagen EZ1 Advanced XL instrument, followed by reverse transcription to cDNA, PCR amplification, and Illumina NextSeq sequencing using tiled primers spanning the genome as previously described [4, 5]. Viral genomes were aligned using MAFFT v7.427 [6] with 762 high-coverage viral genomes deposited in the GISAID (Global Initiative for Sharing All Influenza Data, recently adapted to include SARS-CoV-2 genomes) database [7, 8] as of March 20, 2020, in addition to the most recent viral genomes sequenced in California as of May 3, 2020 [9], for a total of 983 sequences. A maximum likelihood phylogenetic tree was constructed using IQTREE (version 2) using an HKY (Hasegawa-Kishino-Yano) substitution model [10].
Assembled SARS-CoV-2 genomes in this study were uploaded to the GISAID database [7, 8] (accession numbers EPI_ISL_417330, EPI_ISL_417331, EPI_ISL_429881, EPI_ISL_EPI_ISL450232 - EPI_ISL450240) and were also submitted to the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers MT419851, MT419852, MT419860, MT510718-726). Raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) database (BioProject accession number PRJNA629889 and umbrella BioProject accession number PRJNA171119). De-identified data from UCSF patients is available upon request.
RESULTS
We performed SARS-CoV-2 testing on 947 samples collected from 10 March through 20 March from patients with suspected SARS-CoV-2 infection admitted to UCSF hospitals or seen in outpatient clinics. We reviewed the electronic medical records from the first 46 consecutive SARS-CoV-2–positive cases. Data from these patients with COVID-19 were matched with 102 randomly selected negative controls who were patients who tested negative for SARS-CoV-2 over the same time period. Documented history was recorded by a physician or nurse practitioner and included sick contacts, healthcare worker status, and travel history. Among the 46 COVID-19–positive patients, the median age was 44 years, 46% were female, and 65% were outpatients (Supplementary Table 1).
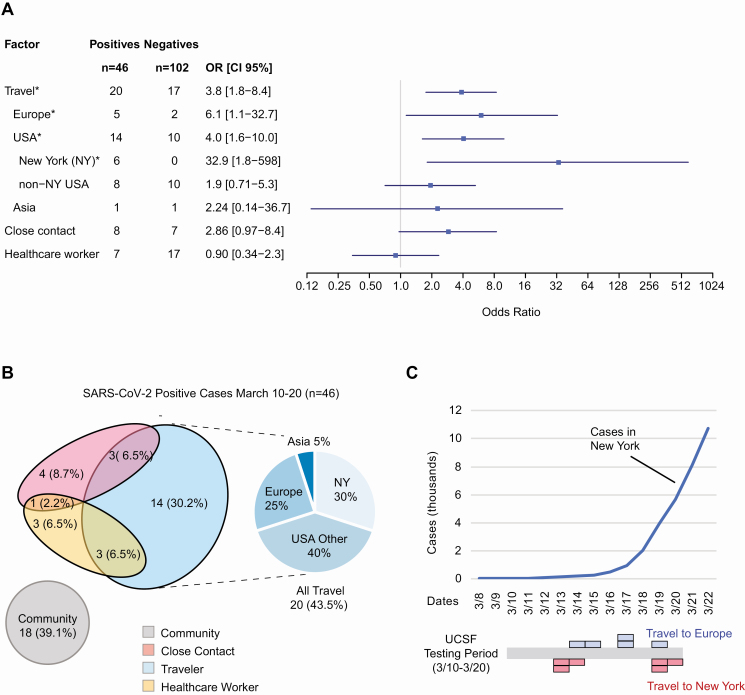
We noted that a travel history within 2 weeks of symptom onset (median date, 11 March 2020) was significantly associated with COVID-19 infection (odds ratio [OR], 3.8; 95% confidence interval [CI] 1.8–8.4), comprising 43% (20/46) of newly diagnosed cases (Figure 1A). Out of the 20 travelers with COVID-19 infection, there were significant associations for prior travel to Europe (5 travelers; OR, 6.1; 95% CI 1.1-32.7), travel outside of San Francisco to other cities within California or other states (United States) (14 travelers; OR: 4.0; 1.6–10.0), and specifically travel to New York (6 travelers; OR, 32.9; 95% CI 1.8-598) as compared with 17 travelers without infection (Figure 1B and Supplementary Tables 2 and 3). The association with travel may be due to direct exposure to SARS-CoV-2 while in high-prevalence regions (eg, New York) or exposure while traveling (close contact with fellow travelers or airport personnel). One cluster of 3 positive cases associated with COVID-19 infection in an airport worker was categorized as a case of community- rather than travel-associated transmission. No significant associations were found with regard to close contacts with persons with known COVID-19 infection or frontline healthcare workers. Those who did not have a recent travel history, a close contact who was COVID-19 positive, or were not a frontline healthcare worker were categorized as community transmission with an unknown source of infection and comprised 39% of cases.
Figure 1.
A, Associations with positive COVID-19 RT-PCR testing. ORs with 95% CIs are shown. Positives (n = 46) were consecutive cases from 10 to 20 March 2020. Negatives (n = 102) were randomized from the same time period. Significant risk factors (P < .05) are designated with “*” and were recent travel, including Europe, United States (domestic), and/or New York (P values are shown in Supplementary Table 2). B, Venn diagram of risk factors in positive SARS-CoV-2 cases. All positive cases and their associations are shown here categorized as those with a recent travel history, who had a close contact who was COVID-19 positive, a frontline healthcare worker, or a combination of the previous categories (left). Those who did not match one of those categories were uniformly categorized as a community case. The most common association with a positive case was a travel history immediately prior to symptoms. Travelers (n = 20) are subdivided by travel region: New York (NY), non-NY USA, Europe, or Asia (right). C, Timeline of cumulative COVID-19 cases diagnosed in New York (top), and UCSF positive cases found in San Francisco who recently traveled to New York or Europe over time (bottom). Each colored block represents a single patient. Abbreviations: CI, confidence interval; COVID-19, coronavirus disease 2019; GISAID, Global Initiative to Share All Influenza Data (recently adapted to include SARS-CoV-2 sequences); Neg, negative; OR, odds ratio; Pos, positive; RT-PCR, reverse transcriptase–polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; UCSF, University of California, San Francisco.
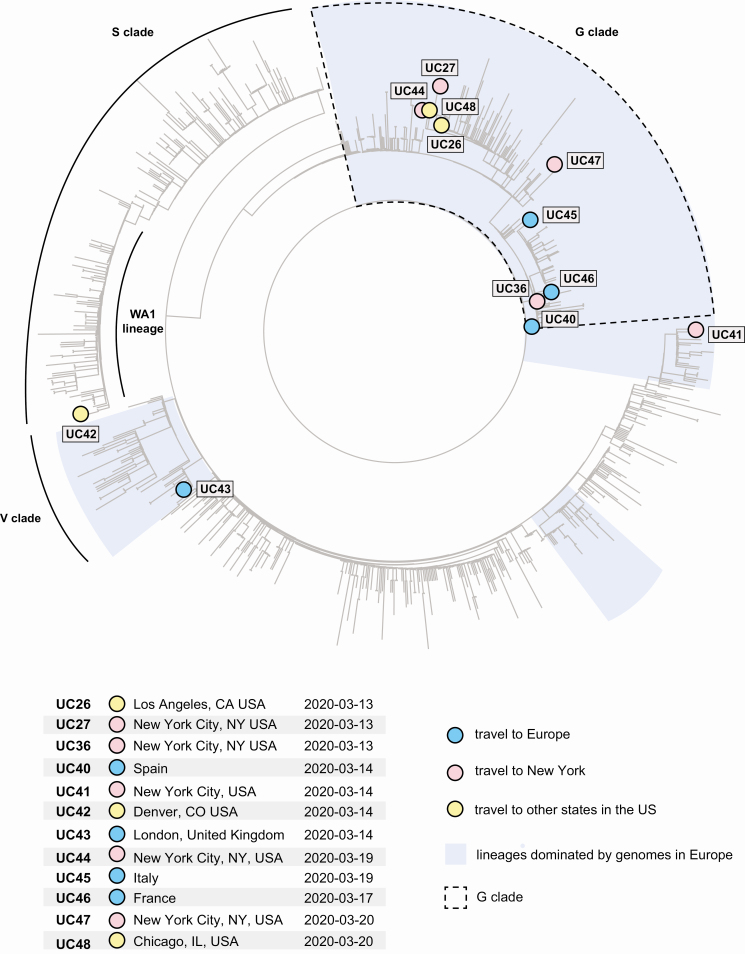
We conducted viral genomic sequencing and phylogenetic analysis of SARS-CoV-2 viruses from 12 of 20 travelers for whom the breadth of coverage of the viral genome was more than 90% (Figure 2).
Figure 2.
Phylogenetic analysis of SARS-CoV-2 viral genomes from domestic and international travelers. The 12 cases with sufficient viral genome coverage for phylogenetic analysis (≥90%) are highlighted by colored circles overlaying a global phylogenetic tree of 983 viruses, including 762 viruses in GISAID as of 20 March 2020 and the most recent viral genomes sequenced from California patients. The G, S, and V clades and the lineages dominated by genomes in Europe are highlighted. Abbreviations: GISAID, global initiative on sharing all influenza data; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
We defined genomic clades through the GISAID nomenclature found at that point in time on 20 March 2020 [7, 8]. The majority (9 of 12) of all travel cases clustered in the G clade as defined by the spike protein D614G variant marker (Figures 2Supplementary Figures 1 and 2), including 3 cases from Europe (UC40, UC45, UC46), 4 cases from New York (UC27, UC36, UC44, UC47), 1 case from Los Angeles (UC26), and 1 case from Chicago (UC48). Viruses in the G clade comprise most of the genomes sequenced from patients in Europe [7, 8], but notably have also been identified in the vast majority of cases associated with the New York SARS-CoV-2 outbreak in March to April of 2020, which occurred after the timeline of this study [11, 12]. The detection of G clade viral genomes in travelers to Los Angeles and Chicago suggests the possibility of dissemination of this clade to other states, either indirectly via New York or directly from Europe. Another case involving travel to Denver (UC42) was part of the WA1 lineage, which is associated with the first reported case of SARS-CoV-2 infection in the United States and is currently circulating in local communities in Washington State [13] and California [9]. Viruses from 2 additional travel-associated cases from Europe (UC43) and New York (UC41) were mapped to other clades circulating in Europe (Figure 2). The additional case from Europe was found to be part of the V clade, defined by a G251V mutation in the NS3 protein [7, 8].
DISCUSSION
Real-time dissemination of epidemiological survey data from positive COVID-19 cases is critical to support efforts to contain or reduce spread of viral infection in the community. Our evaluation of diagnosed COVID-19 cases in San Francisco in early March 2020 associates with travelers from New York prior to the recognized spike in New York cases in late March (Figure 1C). Travel from New York was underrecognized as a risk factor for COVID-19 infection in the United States in early March. Guidelines for COVID-19 testing have not included screening for domestic travel. Our findings in San Francisco here can be extrapolated across America as there are over 100 direct domestic destinations and more than 6 million domestic flights a month from JFK, Newark, and LaGuardia airports in the New York metropolitan area [14]. Similarly, travel by motor vehicle or train is also a plausible means of spread, especially if there are disproportionate numbers of cases between closely situated major population centers. Cryptogenic transmission of COVID-19 by individuals with mild illness or asymptomatic infection is a tremendous challenge to the containment of COVID-19 [15, 16]. As demonstrated here, stratifying the general population by their exposure risks, such as travel to specific hotspot regions, is one containment strategy that can be informed by real-time epidemiological and phylogenetic surveillance.
Limitations of our study include the use of epidemiological data from only the first 10 days of testing at a single institution. Nevertheless, in the setting of an emergent pandemic with shifting epidemiology, the results of our study reached statistical significance over 4 categories of travel (all travel, New York, USA, and Europe), and yielded data that may have presaged the exponential rise of New York cases and subsequent large-scale outbreak in the New York metropolitan area [11, 12].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Data Availability. Assembled SARS-CoV-2 genomes in this study were uploaded to global initiative on sharing all influenza data (GISAID) [8, 9] (accession numbers EPI_ISL_417330, EPI_ISL_417331, EPI_ISL_429881, EPI_ISL_EPI_ISL450232 - EPI_ISL450240) and were also submitted to the National Center for Biotechnology Information (NCBI) GenBank database (accession numbers MT419851, MT419852, MT419860, MT510718-726). Raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) database (BioProject accession number PRJNA629889 and umbrella BioProject accession number PRJNA171119). De-identified data from UCSF patients are available upon request.
The IRB at UCSF approved the clinical and epidemiological association study (IRB #20-30538) and the phylogenetic study (IRB #10-01116, 11-05519). Nonidentifying clinical, demographic, and laboratory data were extracted from clinical testing results and the electronic medical board by retrospective chart review. Informed consent was waived for this minimal risk study.
Financial support. This work was supported by the National Institutes of Health (grant number R33-AI129455; to C. Y. C.), by the National Institute of Allergy and Infectious Diseases (grant number K08-CA230156; to W. G.), the National Cancer Institute, the Charles and Helen Schwab Foundation (C. Y. C.), and the Burroughs-Wellcome Career Awards for Medical Scientists Award (W. G.).
Potential conflicts of interest. C. Y. C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Center and receives research support funding from Abbott Laboratories. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lu R, Zhao X, Li J, et al. . Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 2020; 395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. NYS-COVID19-Tracker [Internet] Available at: https://covid19tracker.health.ny.gov. Accessed 30 March 2020.
- 3. US Centers for Disease Control and Prevention. Real-Time RT-PCR Panel for Detection 2019-nCoV [Internet] 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-detection-instructions.html. Accessed 29 April 2020.
- 4. Deng X, Achari A, Federman S, et al. . Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat Microbiol 2020; 5:443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quick J, Grubaugh ND, Pullan ST, et al. . Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 2017; 12:1261–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002; 30:3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elbe S, Buckland‐Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Chall 2017; 1:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hadfield J, Megill C, Bell SM, et al. . Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng X, Gu W, Federman S, et al. . Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science 2020; 369:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 2015; 32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gonzalez-Reiche AS, Hernandez MM, Sullivan M, et al. . Introductions and early spread of SARS-CoV-2 in the New York City area. Science 2020; Available at: https://science.sciencemag.org/content/early/2020/05/28/science.abc1917. Accessed 29 May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maurano MT, Ramaswami S, Westby G, et al. . Sequencing identifies multiple, early introductions of SARS-CoV2 to New York City Region. medRxiv 2020; 2020.04.15.20064931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bedford T, Greninger AL, Roychoudhury P, et al. . Cryptic transmission of SARS-CoV-2 in Washington State. medRxiv 2020; 2020.04.02.20051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Port Authority of New York and New Jersey Airport Traffic Statistics [Internet] 2020. Available at: https://www.panynj.gov/airports/en/statistics-general-info.html
- 15. Li R, Pei S, Chen B, et al. . Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 2020. Available at: https://science.sciencemag.org/content/early/2020/03/13/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McMichael TM, Currie DW, Clark S, et al. . Epidemiology of Covid-19 in a Long-Term Care Facility in King County, Washington. N Engl J Med 2020; 382:2005–011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.