Abstract
Three-dimensional (3D) organoid models derived from human pluripotent stem cells provide a platform for studying human development and understanding disease mechanisms. Most studies that examine biallelic inactivation of the cell cycle regulator Retinoblastoma 1 (RB1) and the link to retinoblastoma is in mice, however, less is known regarding the pathophysiological role of RB1 during human retinal development. To study the role of RB1 in early human retinal development and tumor formation, we generated retinal organoids from CRISPR/Cas9-derived RB1-null human embryonic stem cells (hESCs). We showed that RB is abundantly expressed in retinal progenitor cells in retinal organoids and loss of RB1 promotes S-phase entry. Furthermore, loss of RB1 resulted in widespread apoptosis and reduced the number of photoreceptor, ganglion, and bipolar cells. Interestingly, RB1 mutation in retinal organoids did not result in retinoblastoma formation in vitro or in the vitreous body of NOD/SCID immunodeficient mice. Together, our work identifies a crucial function for RB1 in human retinal development and suggests that RB1 deletion alone is not sufficient for tumor development, at least in human retinal organoids.
Keywords: retinoblastoma, human embryonic stem cells, organoid, retinal disease, CRISPR/Cas9, cell cycle
INTRODUCTION
Retinoblastoma 1 (RB1) regulates the G1/S transition during cell cycle progression by modulating the activity of E2F transcription factors [1]. Phosphorylation of RB by cyclin-dependent kinases lead to the dissociation of RB from E2F and release of RB-mediated repression [2]. In addition to controlling S-phase entry, RB has been implicated in other cellular processes including regulation of apoptosis, terminal differentiation, and tumor formation [3-6]. In humans, inheritance of a mutant allele of RB1 results in retinoblastoma [7]. In contrast to most human tumors, retinoblastomas usually arise during the first few years of life [6]. Retinoblastomas have also been found in the developing human fetus in utero [8]. Although many observations indicate that RB1 suppresses tumorigenesis during early retinal development, the retinal cell type and developmental stage in which RB1 functions has not been fully defined.
One of the reasons for the incomplete understanding of RB1’s pathophysiological role is the lack of reliable human disease models [9]. Pre-clinical research to delineate molecular mechanisms that drive cancer growth and progression is usually carried out in two-dimensional (2D) cell culture systems which are efficient and reliable but lack the appropriate cell-cell contact environment typically observed in vivo. Although genetically-modified animals are used as an alternative to overcome the limitation of 2D cell culture, they are costly and time-consuming experiments to conduct. Moreover, unlike the human situation, in mice, loss of Rb1 does not predispose to retinal tumors. Instead they develop a multiple endocrine neoplasia syndrome manifested by development of pituitary and thyroid tumors [10]. Only combined loss of RB1, and either of the related proteins p107 or p130, result in tumors with amacrine but not photoreceptor differentiation [11].
Advancement of novel technologies, such as growth of three-dimensional (3D) human retinal organoids and CRISPR/Cas9-mediated gene editing, opens up the possibility of recapitulating the process of human retinal development. This 3D organotypic model provides an alternative to both 2D culture and in vivo animal model systems, suggesting that they may be useful in disease modeling. We therefore chose 3D human retinal organoids as a platform to test the effect of biallelic RB1 inactivation in retinal development and disease.
MATERIALS AND METHODS
Generation of RB1−/− H9 hESC lines
The CRISPR design tool (https://chopchop.cbu.uib.no/) was used to design a guide RNA (gRNA) targeting the first exon of the RB1 gene. This guide sequence (CACCGCCGCCGCTGCCGCCG) was cloned into the lenti-CRISPR-Cas9-EGF plasmid (Addgene #57818) according to Addgene’s protocol. The Cas9-gRNA plasmid was transduced into H9 ESCs by electroporation. GFP+ cells were isolated by FACS sorting, expanded and subcloned by manually picking colonies. Clonal lines were screened to detect clones with indels by PCR amplification (Forward primer: TTGTAACGGGAGTCGGGAGA; Reverse primer: TCAAGTTGAAGCCGAGACCC). PCR products were cloned into TOPO-cloning vectors and sequenced by Sanger sequencing. Clonal hESC lines showed biallelic disruption of the RB1 gene. Loss of RB protein expression was verified by immunoblotting and immunocytochemistry.
Culture of wild-type and RB1−/− human ESCs
H9 ESCs were maintained in feeder-free conditions according to standard protocols from WiCell (www.wicell.org). Cells were cultured in mTeSR1 medium (Stemcell Technologies) on growth factor-reduced Matrigel (BD Biosciences) coated plates with medium changed every day.
hESC retinal organoidogenesis
Retinal differentiation of hESCs was performed by adapting a previously reported protocol [12, 13]. hESCs were dissociated to single cells by treatment with accutase and 10 μM Y-27632 for 5 min and quickly reaggregated using low-cell-adhesion 96-well plates (9,000 cells/well, 100 μl) with V bottomed conical wells with retinal differentiation medium (RDM) containing 20 μM Y-27632. The RDM was G-MEM supplemented with 20% KSR, 0.1 mM nonessential amino acids, 1 mM pyruvate, 0.1 mM 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin. IWR1e (Merck) was added to the culture to a final concentration of 3 μM from day 0 to day 12. Matrigel (BD Biosciences) was added from day 2 to day 18. IWR1e was removed from the culture by being washed with RDM medium with 10% FBS on day 12. From day 15 to day 18, 3 μM CHR99021 and 100 nM SAG were added to differentiation medium containing 10% FBS. On day 18, the culture was transferred to an NR culture medium (DMEM/F12-Glutamax medium containing N2 supplement). On day 20, the retinal epithelium was excised from the main body and divided into three evenly sized independent portions with fine forceps under a dissecting microscope and cultured in suspension under 40% O2/5% CO2 conditions in DMEM/F12-Glutamax medium (GIBCO) containing N2 supplement (Thermo Fisher Scientific), 10% FBS, 0.5 μM retinoic acid (Sigma-Aldrich), 0.25 μg/ml Fungizone (GIBCO), 100 U/ml penicillin, and 100 μg/ml streptomycin. The size of the retinal organoid was assessed by measuring the long-axis diameters (surface-to-surface) of 30 samples using the NIS-Elements software.
Cryosectioning and immunostaining of organoids
Each organoid was fixed in 4% paraformaldehyde overnight at 4°C, dehydrated by 30% sucrose in PBS and embedded in OCT Compound (Thermo Fisher Scientific). Cryostat sections (14 μm) were cut and mounted onto slides (Thermo Fisher Scientific). Mounted sections were incubated for 1 h at room temperature with blocking solution (3% normal donkey serum+0.3% Triton X-100 in TBS) and incubated with primary antibodies (Table S1) diluted in blocking solution overnight at 4°C. After three washes with TBS, corresponding fluorophore-conjugated secondary antibodies (Table S2) diluted in the blocking solution were added for 2 h at room temperature and followed by DAPI staining. Finally, stained slides were rinsed with TBS three times, mounted and analyzed using a Nikon A1R confocal microscope. For Brdu labeling experiments, organoids were treated with BrdU (Sigma # B9285) at a final concentration of 30 μM every 2 hours and fixed 10 h after the first BrdU treatment [14]. BrdU-treated organoids were incubated with 2 N HCl for 30 min at RT and then with 0.1 M sodium borate (pH 8.5) for 10 min at room temperature. After rinsing with TBS three times, standard immunostaining was performed as described above. All quantitative analysis of immunostained sections were carried out using Nikon NIS-Elements AR and Image J software. To determine the type of cells that express RB, we counted all RB+ cells in two randomly selected fields (318.2 μm×318.2 μm each) in three immunostained slices per organoid and calculated the percentage of marker-positive cells out of the total RB+ cells. We used Image J to measure marker positive areas using the same thresholding in three immunostained slices per organoid and calculated the percentage of marker positive cells out of the total DAPI+ area. The number of organoids used for marker quantification is included in the figure legends.
Cell cycle analysis
Organoids or hESCs were dissociated to single cells by treatment with accutase for 10 min and fixed in 70% ethanol overnight. After washing with PBS, the samples were incubated for 30 min with RNase A (200 μg/ml), PI (20 μg/ml) and X-100 Triton (0.01%) in PBS and their DNA content was analyzed by flow cytometer (BD FACSAria) with 20,000 events per determination. Cell cycle profiles were generated using Flowjo software (Tree Star).
RNA isolation, RT-PCR and qRT-PCR
Organoid RNA was extracted according to the protocol supplied with TRIzol reagent (Thermo Fisher Scientific). The concentration and purity of the RNA samples were measured using Nanodrop (Thermo Fisher Scientific). The extracted RNA (400 ng) was reverse transcribed according to the protocol supplied with SuperScriptIII First-Strand Synthesis System for RT-PCR (catalog # 18080-051, Invitrogen Life Technologies). Quantitative real-time PCR (qRT-PCR) was carried out using ViiA (Applied Biosystems). Reactions were run in triplicate and expression of each gene was normalized to the geometric mean of GAPDH as a housekeeping gene and analyzed by using the ΔΔCT method. The primer sequences of each gene are listed in Table S3.
Soft agar colony formation assay
A base coat of 0.8% agarose was added into the wells of a 24-well plate and further covered with cell suspension (1000 cells/well in 0.48% agarose). Plates were incubated for 2 weeks and the resulting colonies were fixed with 3.7% paraformaldehyde. The images of the colonies were taken using a Nikon A1R confocal microscope.
Western blot
Proteins were extracted using RIPA buffer with protease inhibitors (Roche). After quantification, proteins were resolved on 4-12% SDS-PAGE gels and transferred to a PVDF membrane. The membrane was incubated with primary antibodies (Table S1) after blocking for 1 h with 5% BSA/TBS Tween. Immunoreactive bands were visualized by densitometry using HRP conjugated secondary antibodies.
Intravitreal injection
Animals were treated humanely and protocols were approved by the Institutional Animal Care and Use Committee of University of Texas Southwestern Medical Center at Dallas, USA. The animals were kept in individually ventilated cages (IVC) in our animal facility. The pupils were dilated with 1 to 2 drops of Medriaticum drops and a drop of topical anesthetic Novesine was applied. Methocel eye drops were used to avoid drying of the eyes. Injections were performed using a surgical microscope. Two microlitres (2uL, 10,000 cell/uL) of sterile phosphate buffered saline containing 2×10E4 cells dissociated from D30 or 90 organoids were injected into the vitreous of the right eye through the sclera using a Hamilton syringe with a 26-gauge cannula. All procedures were made using a binocular lens. Special care was taken to prevent lens damage or posterior retinal punctures. After the injection, the eyes were treated with antibiotic eye drops (e.g., tobramycine). The animals were examined 2, 12 and 24 h after surgery and then daily. Clinical findings regarding the presence of tumor was recorded.
In vivo imaging
Imaging of the right eye of the mice was carried out after intravitreal injection at 12 weeks. Because the precise area of tumor growth was not known in advance, the whole retina of the mouse was scanned during each imaging session. Pupils were dilated with a drop of tropicamide 1% (Mydriaticum; Laboratoire Thea, Clermont-Ferrand, France) and a drop of phenylephrin chlorhydrate 2.5% (Neosynephrine; Laboratoire Europhta, Monaco). Mice were then anesthetized by an intraperitoneal injection of the mixture of ketamine and xylazine. Both eyes were examined using the Micron IV rodent imaging system (Phoenix Research Labs, Pleasanton, CA, USA) after the instillation of moisturizing drops (Goniovisc; Hubs Pharmaceuticals, Rancho Cucamonga, CA, USA).
Histological analysis and immunohistochemistry
Animals were euthanized by cervical dislocation and enucleation of both eyes was performed. Eyes were fixed overnight by immersion in 4% paraformaldehyde before paraffin-embedding and sectioning. Eyes were sectioned using a microtome into 7 or 10 μm sections, which were stained with haematoxylin and eosin (H&E). Immunostainings were performed on Ventana discovery XT™ (Ventana, Roche Diagnostics, Vilvoorde, Belgium) using the DABMap detection system according to manufacturer's recommendations. The slides were examined by a light microscope.
Statistical analysis
An unpaired two-tailed Student’s t-test was used to compare the mean ± s.e.m. values from the wild-type and RB1−/− cells with Welch’s correction when the F-test indicated significant differences between the variances of both groups. All analyses were carried out with GraphPad Prism software (version 6.0) and values of P<0.05 were considered statistically significant.
RESULTS
Efficient generation of human retinal organoids from hESCs
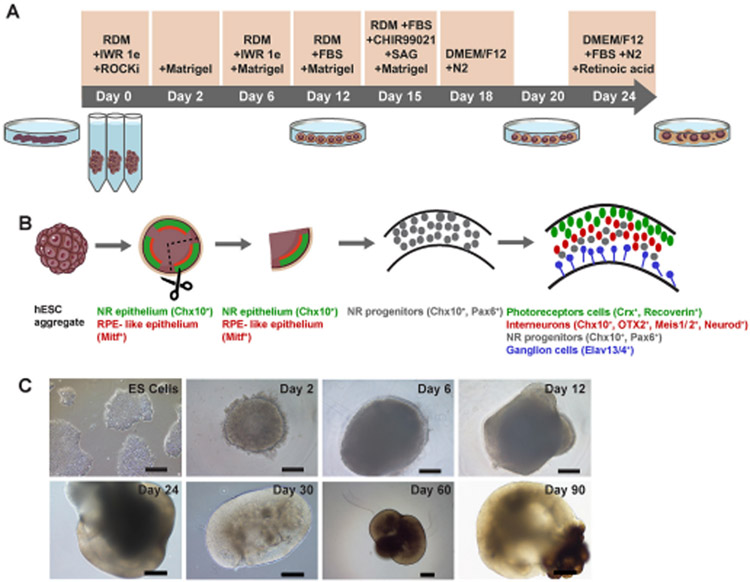
To generate human retinal organoids from hESCs, we took advantage of the original retinal organoid protocol as previously described [13, 15], but made modifications to increase the efficiency of the process. Dissociated hESCs were reaggregated in a V-bottomed 96-well plate (9000 cells/well) in 20% KSR-containing medium supplemented with IWRle, Y-27632 and Matrigel for the first 12 days. Fetal bovine serum (FBS, 10%) was added for optimized retinal differentiation after Day (D) 12. In addition, the Hedgehog signaling pathway smoothened agonist (SAG, 100nM) and the selective small molecule GSK3 inhibitor CHIR99021 to activate Wnt signaling, were added during D15-18 (Fig. 1A, B). Upon aggregation, hESCs formed continuous epithelial structures by D10-12 (Fig. 1C). The neural retina (NR) epithelia was isolated on D20 with fine forceps under a dissecting microscope and continuously grew under high O2 condition (40%) in retina maturation medium. During excision, without the help of a retinal epithelium reporter, we manually dissected organoids at the eye-field stage into three evenly sized independent portions (trisection step) according to a previous report [12]. Because these dissected organoids could all potentially contain eye-field-determined neuroepithelium, the growth of 3 retinal organoids per starting aggregate was possible (Fig. 1B). Following neuroepithelium trisection, the majority of organoids developed into large, continuous epithelial structures, resulting in efficient formation of high numbers of large, stratified retinal organoids (Fig. 1B, C). This trisection of the starting (mother) aggregate resulted in a 3-fold increase in organoids that continued to develop. This resulted in twice as many differentiated large stratified retinal organoids at D30 as starting aggregates; only a minority were non-retinal (22±3%, independent experiment [N]=3, total organoid number [n]=90), which had no compact neuroepithelium, suggesting non-retinal phenotypes. These non-retinal organoids were removed from the dish.
Figure 1. Generation of hESC-derived retinal organoids.
(A and B) Schematic overview and timeline of retinal organoid generation. (C) Representative phase contrast images of hESC-derived aggregates. Aggregates are shown before D2-12 and after D24-90 organoid trisection. RDM, retinal differentiation medium. hESC, human embryonic stem cell. NR, neural retina. RPE, retinal pigment epithelium. Scale bars, 200 μm.
Generation of RB1−/− human ESCs
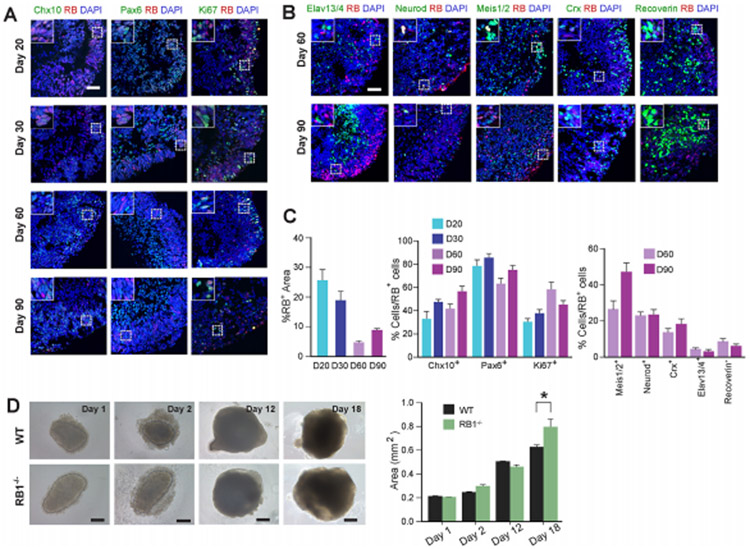
We used the CRISPR/Cas9 gene-editing approach to generate hESCs with homozygous knockout of RB1 (Fig. S1A, B) [14]. First, we analyzed the expression of RB in wild-type and RB1−/− cells by immunocytochemistry and western blot. Although the majority of wild-type hESCs expressed RB, we did not find any expression of RB in RB1−/− cells (Fig. S1C). Similar results were found by western blot (Fig. S1D). As RB was previously shown to be important for chromosomal stability, genetic integrity of the clone was verified (Fig. S1E). We also did not detect any RB+ cells in organoids derived from RB1−/− hESCs (Fig. S1F). However, in wild-type organoids, 4.5-25.3% of the cells expressed RB at 20, 30, 60 and 90 days in vitro (Fig. 2A, C). Since RB expression decreased over time, this suggested RB1 may play an important role at early stages of retinal organoid development. Next, using immunohistochemistry we analyzed the expression of RB in retinal ganglion (Elavl3/4), photoreceptor (Crx, Recoverin), bipolar (Chx10), interneuron (Chx10, Meis1/2, Neurod), NR progenitor (Chx10, Pax6) and proliferating cells (Ki67) in wild-type organoids (Fig. 2A, B and Fig. S2). In wild-type organoids, 31-86% of the RB+ cells were Chx10+, Pax6+ or Ki67+ cells. Most of the RB+ cells co-localized with Chx10+ and Pax6+ cells, suggesting that RB may be involved in retinal organoidogenesis. The average percentage of RB+ cells that overlapped with Meis1/2+, Neurod+, Crx+, Elavl3/4+ and Recoverin+ cells were 27.1%, 23.6%, 14.2%, 4.9%, and 9.1% at D60 and 47.8%, 24%, 18.9%, 3.6% and 6.6% at D90, respectively (Fig. 2C). These results support the conclusion that RB expression in retinal organoids is highest during progenitor cell stages and is downregulated in maturation stages.
Figure 2. RB is expressed in progenitor cells during retinal organoid development.
(A) Representative wild-type organoid at D20, 30, 60 and 90 sectioned and immunostained against Chx10 and RB; Pax and RB; Ki67 and RB, and counterstained with DAPI. Scale bar: 50 μm (B) Representative wild-type organoid at D60 and 90 sectioned and immunostained against Elavl3/4 and RB; Neurod and RB; Meis1/2 and RB, Crx and RB; Recoverin and RB; and counterstained with DAPI. Scale bar: 50 μm. (C) The graphs showed the percentage of RB+ cells in the organoid at D20, 30, 60 and 90, and the RB+ cells that colocalized with the aforementioned markers. (D) Representative organoids from wild-type and RB1−/− hESCs grown to D18 before organoid trisection. The graphs showed the mean area of wild-type and RB1−/− organoids. The results are mean±s.e.m. of 40 organoids from three independent experiments (*P<0.05, Student’s t-test). Scale bars, 200 μm.
Loss of RB1 increases organoid size and S-phase entry
As RB is expressed in progenitor cells, we hypothesized that RB1 may play a role in regulating retinal organoid growth. The organoids derived from RB1−/− hESCs were slightly bigger than the organoids derived from wild-type hESCs at D18 (P<0.05, Fig. 2D), although we did not find any statistically significant difference in organoid size before D18. We also found RB1−/− retinal organoids readily formed cysts (data not shown).
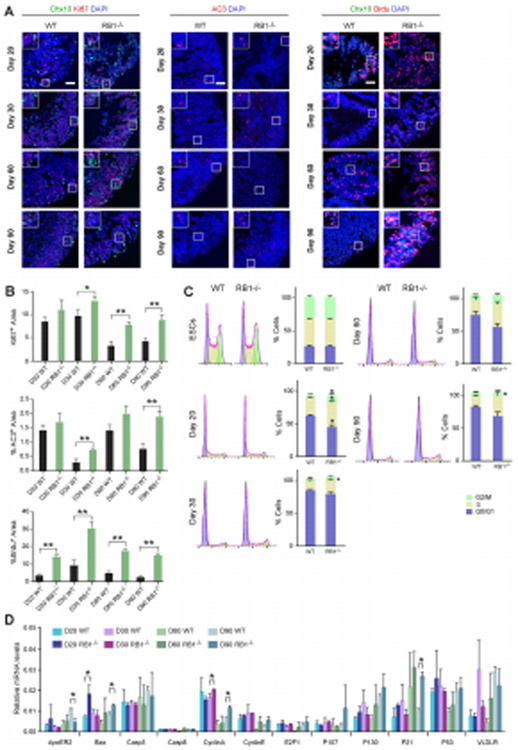
The increased size of RB1−/− organoids and the known role of RB1 in cell cycle regulation prompted us to analyze the effect of the lack of RB1 in cell proliferation during organoid development. Our analysis of the percentage of Ki67+ cells at different time points showed higher number of Ki67+ cells in RB1−/− organoids compared with wild-type at D30, 60 and 90 (Fig. 3A, B). To further analyze the role of RB1 in cell cycle progression, we dissociated the organoids into individual cells and stained with propidium iodide (PI) (Fig. 3C). hESCs have a shorter G1 phase of the cell cycle as compared with most other cell types after differentiation [16]. RB1−/− hESCs cells did not exhibit an altered cell cycle comparing with wild-type hESC cells (Fig. 3C). However, after starting differentiation, at D20, the percentage of cells in each phase of the cell cycle was different in organoids derived from wild-type compared to RB1−/− hESCs. There was increased percentage of cells in S phase in RB1−/− organoids at D20. At 30 and 90 DIV, although RB1−/− organoids did not have a significant change in the percentage of cells in G0/G1 (P>0.05, n=3) or G2/M (P>0.05, n=3), there was a significant increase in the percentage of cells in S phase compared with wild-type organoids (P<0.05, n=3; Fig. 3C). At D60, there was also slightly higher percentage of cells in S phase compared with wild-type organoids. To identify the cells in S phase in the organoids from RB1−/− organoids, we added a pulse of BrdU and immunostained the organoids. The overall percentage of BrdU+ cells were higher in RB1−/− organoids compared to wild-type at D20, 30, 60 and 90 (Fig. 3A, B). Our data suggest that RB1 deficiency produces an increase in organoid size by promoting S-phase entry.
Figure 3. Loss of RB increases S-phase entry.
(A) Representative wild-type and RB1−/− organoids at D20, 30, 60 and 90 sectioned and immunostained against Ki67 and Chx10, AC3, Chx10 and Brdu; and counterstained with DAPI. (B) The graphs show the percentage of Ki67+/DAPI+ area, AC3+/DAPI+ area and Brdu+/ DAPI+ area at different time points. The results are mean±s.e.m. of 9 organoids from three independent experiments. Relative mRNA levels of RB family genes and genes involved in cell cycle regulation and apoptosis as measured by RT-qPCR. The results are mean±s.e.m. of 3 organoids from three independent experiments (*P<0.05, **P<0.01, Student’s t-test). (C) Plots on the left show representative wild-type and RB1−/− organoid cell cycle profiles at D20, 30, 60 and 90. The graphs on the right show the percentage of cells in each cell cycle phase at different time points. The results are mean±s.e.m. of 3 organoids from three independent experiments (*P<0.05, Student’s t-test). (D). Relative mRNA levels of RB related genes measured by qRT-PCR. The results are mean±s.e.m. of 3 organoids from three independent experiments (*P<0.05, Student’s t-test). Scale bars: 50 μm.
Next, we analyzed whether the absence of RB1 resulted in cell death using a marker of apoptosis (cleaved caspase 3, AC3). At D30 and 90, RB1−/− organoids showed an increase in the number of AC3+ cells, respectively (P<0.01, Fig. 3A, B) compared with wild-type organoids. The organoids from RB1−/− hESCs revealed significant numbers of pyknotic nuclei indicative of apoptosis (data not shown). Combined with the data suggesting that RB1 is required for proper cell-cycle exit, the increased apoptosis in the organoids suggests that defects in the coordination of cell-cycle withdrawal and onset of differentiation may lead to increased cell death. To corroborate the role of RB1 in cell survival and proliferation and gain mechanistic insight into the nature of effector genes, we performed qRT-PCR to survey the expression of E2F targets involved in cell cycle regulation and apoptosis (Fig. 3D). We found an increase in the relative mRNA levels of cyclin A2 (CCNA2), BAX and p21 in RB1−/− organoids compared with wild-type. In addition, there was a decrease in the relative ApoER2 mRNA levels in RB1−/− organoids compared with wild-type, whereas we did not find significant differences in the expression of RB family member genes or of other genes related to the cell cycle and apoptosis.
RB1 is required for photoreceptor, ganglion, and bipolar differentiation
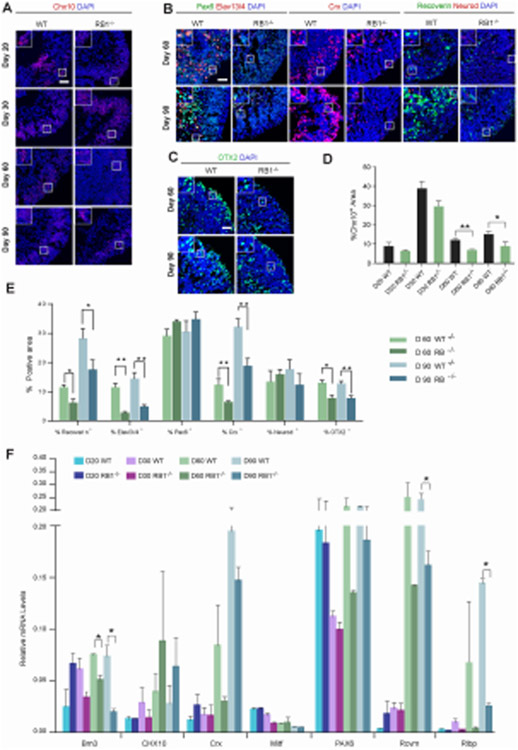
In addition to proliferation and cell survival, RB is also reported to play roles in retinal differentiation [11]. Immunohistochemistry analyses revealed Chx10+ cells were significantly decreased in RB1−/− organoids compared to wild-type organoids after D30 (p<0.05, Fig. 4A, D). In addition, we observed a dramatic reduction in the number of Recoverin+, Elavl3/4+ and Crx+ cells in D60 and D90 RB1−/− organoids compared with wild-type organoids (Fig. 4B, E). The bipolar marker, OTX2, was also reduced in RB1−/− organoids compared to wild-type organoids (Fig. 4C, E). These data indicated loss of RB1 led to reduced numbers of photoreceptor, ganglion and bipolar cells. Furthermore, we performed qRT-PCR analysis of select retinal differentiation genes to confirm the immunohistochemistry results (Fig. 4F). Among the retinal differentiation genes that showed significant changes between wild-type and RB1−/− organoids, genes related to ganglion cells (Brn3) and müller glia cells (Rlbp) were downregulated in RB1−/− organoids compared to wild-type. Moreover, a gene related to photoreceptor cells (Rcvrn) was also lower in RB1−/− organoids than wild-type. These results reinforce the role of RB1 in photoreceptor and ganglion cell differentiation. Since there was ongoing proliferation which did not indicate a decrease in a specific cell type(s), we performed co-labeling of Ki67 and Chx10, Recoverin, Elavl3/4, and Crx in wild-type and RB1−/− organoids. As shown in Fig. 3A and Fig. S3, most of the cells expressing subtype-specific markers were not Ki67+ in both wild-type and RB1−/− retinal organoids, consistent with RB1 playing a role in retinal differentiation in addition to its role in proliferation and cell survival.
Figure 4. Loss of RB1 reduces photoreceptor, ganglion and bipolar layers.
(A) Representative wild-type and RB1−/− organoids at D20, 30, 60 and 90 sectioned and immunostained against Chx10 and DAPI. (B) Representative wild-type and RB1−/− organoids at D60 and 90 sectioned and immunostained against Elavl3/4 and Pax6; Crx; Recoverin and Neurod; counterstained with DAPI. (C) Representative wild-type and RB1−/− organoids at D60 and 90 sectioned and immunostained against OTX2 and counterstained with DAPI. (D) The graphs show the percentage of Chx10+/DAPI+ area at different time points. (E) The percentage of Elavl3/4+, Pax6+, Crx+, Recoverin+, Neurod+ and OTX2+ cells out of the DAPI+ area at D60 and 90. The results are mean±s.e.m. of 9 organoids from three independent experiments (*P<0.05, **P<0.01, Student’s t-test). (F) Relative mRNA levels of retinal genes measured by qRT-PCR. The results are mean±s.e.m. of 3 organoids from three independent experiments (*P<0.05, **P<0.01, Student’s t-test). Scale bars: 50 μm.
Lack of tumorigenicity in RB1−/− organoids
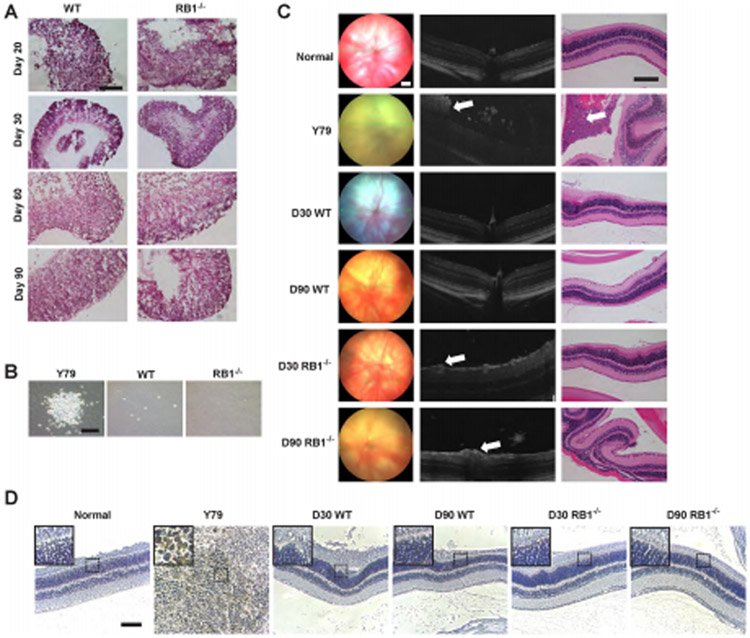
Since retinoblastoma is initiated by biallelic inactivation of the RB1 gene, we wanted to determine whether RB1−/− organoids would develop retinal tumors in a dish. H&E staining confirmed that no retinoblastoma cells, such as rosettes or calcifications found in human retinoblastoma, were detected in RB1−/− organoids (Fig. 5A). To evaluate tumorigenicity in vitro, we performed the colony formation assay which is the ability of tumor cells to form colonies in soft agar. As a positive control, the human retinoblastoma cell line Y79 formed large numbers of colonies in soft agar after 2 weeks of culture. However, cells from RB1−/− and wild-type organoids did not exhibit anchorage-independent growth and failed to form clones in soft agar (Fig. 5B).
Figure 5. Lack of tumorigenicity in RB1−/− organoids.
(A) Representative images of H&E staining of wild-type and RB1−/− organoids at D20, 30, 60 and 90. (B) Representative brightfield images of colonies using soft agar colony formation assay. (C) The fundus examination (left column), corresponding OCT scans (central column) and histology sections (right column) after intravitreal injection at 12 weeks. Suspicious positive sign (white arrow) of tumor in OCT scan. Verification of tumor (white arrow) in H&E staining. (D) Immunohistochemistry of Ku80, a human-specific nuclear marker. Scale bars: 100 μm.
In order to evaluate the in vivo tumorigenicity of RB1−/− organoids, we performed intravitreal injection of suspension cells from RB1−/− and wild-type organoids into the right eye of immunodeficient mice. Tumor growth was monitored by fundus and spectral-domain optical coherence tomography (SD-OCT) imaging and compared with histology (Fig. 5C). We included SLO/OCT analysis which is an imaging modality that could only be performed in eyes with early staged tumors or in eyes without a tumor because tumors at later stages will cover the fundus and obscure imaging. First, Y79 cells injected intravitreally caused eye swelling at 6 weeks post-injection, suggestive of tumor formation. According to the stages for tumor progression, the tumors from 7/11 cases of injected Y79 cells broke through the cornea (stage III). We found tumors in 72.7% of the analyzed eyes (8 of 11) 12 weeks post-injection of the Y79 cells. In the other 3 eyes, tumor growth could not be detected. Using OCT, we confirmed all tumor-bearing eyes in the Y79 cell-injected group 12 weeks after injection (Fig. 5C). Histologically, the tumors were composed of typical undifferentiated hyperchromatic cells with scanty cytoplasm having a rosette-like growth pattern, as described for the original tumors (Fig. 5C). All tumors showed a high mitotic and necrotic activity. The same fundus, OCT, and histological analysis was performed for the RB1−/− and wild-type cell-injected groups, however, we did not observe any eye abnormalities. For the D30 and D90 RB1−/− groups, although 1/11 and 2/13 showed a suspicious positive sign which can be seen on OCT image, respectively, tumors were not confirmed on the corresponding HE-stained slide (Fig. 5C). To confirm the presence of transplanted human cells in the Y79 group, the slides were stained using the human-specific nuclear biomarker Ku80 [17]. As expected, Ku80+ human cells were detected only after transplantation of Y79 cells but not after transplantation of wild-type or RB1−/− cells, consistent with the presence of tumors 12 weeks post-injection of the Y79 cells (Fig. 5D).
DISCUSSION
In this study, we found RB1 regulates S-phase entry, proliferation, and apoptosis in 3D retinal organoids, consistent with previous reports in mouse models and 2D cell cultures [18]. We also observed loss of RB1 caused retinal differentiation defects, including photoreceptor, ganglion, and bipolar cells. Despite the crucial role of RB1 in 3D retinal organoid development, there was no tumor formation in vitro or in immunodeficient mice. Thus, while RB1 is important for proliferation, differentiation, and apoptosis during human retinal development in 3D organoids, there are likely differences between the organoid system and the developing human retina that limit the development of retinoblastoma due to loss of RB1. A model depicting the role of RB1 in retinal organoids is shown in Fig. 6.
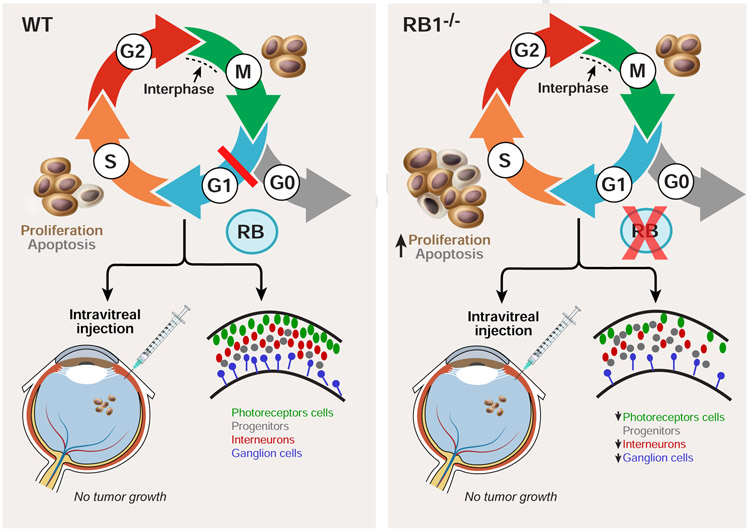
Figure 6. A model of the role of RB1 during human retinal development.
In wild-type cells, RB1 regulates entry into S-phase. However, loss of RB1 promotes an accumulation of cells in S-phase, increases apoptosis and reduces the number of photoreceptors, ganglion cells and bipolar cells. RB1 loss in retinal organoids did not result in retinoblastoma formation when injected into the vitreous body of NOD/SCID mice.
Our data demonstrate that RB1 deletion produces enlarged retinal organoids, possibly owing to an increase of cell proliferation and an accumulation of dead cells. The higher number of Ki67+ cells together with the increased percentage of cells in S phase in RB1−/− retinal organoids contribute to the increase in organoid size, which agrees with our previous work in cerebral organoids lacking RB1 [14]. We further found RB1−/− organoids showed an increase in the number of AC3+ cells. Combined with the data demonstrating that RB1 is required for proper cell-cycle exit, the increased apoptosis in the organoids suggest that defects in the coordination of cell-cycle withdrawal and onset of differentiation may lead to increased cell death. It is well known that RB’s role as a cell cycle regulator is correlated with its capability to negatively regulate cell cycle through its interaction with members of the E2F family of transcription factors [19]. Beyond its function as G1 checkpoint regulators, RB proteins are involved in many other cellular processes, such as preservation of chromosomal stability, induction and maintenance of senescence, regulation of cellular differentiation, angiogenesis, and apoptosis, which could all contribute to RB’s onco-suppressive activity. At the molecular level, RB is viewed as a platform for multiple protein interactions through which it regulates different cellular pathways [20, 21]. Our data is consistent with RB1 as a multifunctional protein, whose role in cell proliferation and apoptosis depends on both the cell type and the nature of the death inducers and whose function can be modulated by several post-translational modifications. Therefore, a careful evaluation of its functional status and context-dependent role in response to specific treatments in different tumors might be fundamental to guide therapeutic decisions.
In addition to RB1‘s role in regulating cell proliferation and cell survival, RB1 is dynamically expressed in multiple cell types during retinal development, suggesting it may play a key role in retinal cell differentiation [22]. While analyses of Rb1 deletion in mutant mice have led to a growing appreciation of the important role it plays in cell fate specification and differentiation [18, 23], less is known about the role of RB1 during retinal development in human tissue. We found RB is localized to differentiating photoreceptors as well as interneuron and ganglion cell types in retinal organoids. Furthermore, immunohistochemistry analyses of RB1−/− retinal organoids revealed that RB1 is required for photoreceptor, ganglion, and bipolar cell differentiation. It is still unclear whether RB promotes differentiation, either by enforcing cell cycle exit or by using additional mechanisms, to directly facilitate the execution of lineage-specific gene expression programs. Recent evidence suggests that RB-mediated mechanisms involved in cellular differentiation are separate from the ability of RB to restrain the cell cycle [24]. For example, Rb1 null mouse retinas show only a mild effect on retinal progenitor cell proliferation but a dramatic reduction in mature rod photoreceptors [25]. In addition, the role of RB may be distinct in different cell types. Studies from lineage and gene expression analysis suggest that the role of RB in rod photoreceptor differentiation is distinct from its role in retinal progenitor cell proliferation [26, 27]. Moreover, using a conditional RB1 deletion approach in mice, it was showed that multiple cells in the retina, including photoreceptor, bipolar and retinal ganglion cells respond to RB1 loss by undergoing apoptosis, whereas, amacrine cells and other cell types can tolerate RB1 mutation [18], a phenotype which is similar with our study. To begin to determine the underlying mechanism to account for RB’s effects in different cell types, a study profiled altered gene expression in RB1-deficient retinas [26], however it is not known which are direct targets of RB1. Therefore, additional studies are needed to address the mechanisms by which RB1 mediates distinct cellular effects on proliferation, cell survival, and lineage-specific differentiation.
One limitation of this study is that we did not have the Crx∷venus fluorescent reporter in our wild-type and RB1−/− organoids, which was used in the 3D retinal organoid method by Yoshiki Sasai and colleagues’ methods [13]. Therefore, we selected the epithelial structure based on morphology alone. This likely resulted in contaminating non-retinal tissue in our retinal organoids, thus affecting the quality of our differentiation and accounting for the low expression of Chx10 and Pax6. Consistent with the possibility of non-retinal tissue, we detected low levels of Emx1 and Foxg1 which are two forebrain markers (Fig. S4). While we observed some areas of disorganized and morphologically immature precursor cells, this is similarly observed by others [12, 28]. Compared with fetal retina, retinal organoids grown under static conditions exhibit low expression of precursor markers especially at early stages. Recently researchers have tried to culture retinal organoid under shaking conditions to obtain higher expression of precursor makers. However, this did not necessarily result in more mature retinal marker expression, suggesting that shaking cultures may enhance cellular proliferation but not overall maturation and static cultures may be a better representation of fetal retina tissue at later differentiation stages (21 weeks culture) [28]. Despite these retinal organoid culture methods, there are still limitations in retinal organoids in vitro such as the lack of mature retinal tissue found in vivo. Therefore in future experiments, more optimization is needed to produce retinal organoids that are structurally mature and resemble the in vivo situation.
Another limitation of this study is that we did not observe retinoblastoma in retinal organoids though RB1 inactivation. In humans, inheritance of a mutant allele of RB1 leads to retinoblastoma at nearly 90% frequency with loss of the remaining wild-type allele thought to be a rate-limiting step in tumorigenesis [29]. RB1 is also mutated in sporadic cancers of the brain, breast, bladder, lung and bone [30]. Although in our study we found an important role of RB1 during retinal development, RB1−/− cells from D30 or 90 retinal organoids did not result in retinoblastoma formation. It is likely that other genetic loci contribute to retinoblastoma. For example, p53 mutations cooperate with RB pathway inhibition and tumorigenesis by suppressing apoptosis [31]. Alternatively, the tumorigenic potential of RB1−/− retinal organoids may be different from the two-hit hypothesis process [32]. In hereditary retinoblastoma, patients have an inherited germline mutation (first “hit”) followed by an acquired second mutation (second “hit”) as their retinas develop [33]. This process may be challenging to recapitulate in RB1−/− retinal organoids. Furthermore, while biallelic loss of RB1 is required for retinoblastoma tumor development, additional mutational events (three or more “hits”) may be needed for tumor progression [33]. Lastly, there may be potential compensatory mechanisms for retinal RB1 loss, thus preventing tumor formation in retinal organoids. Additional RB family proteins p107 and p130 and their potential unique and overlapping functions as regulators of cell cycle progression and transcriptional modulation may compensate for RB function [34]. Future work to delete additional RB family proteins may prevent genetic compensation [35].
Another possibility to explain the lack of tumor formation in RB1-deficient retinal organoids may be due to the lack of RB1-sensitive cells. One recent study showed that human core precursors are uniquely sensitive to RB1 depletion [36]. We speculate because our 3D retinal organoids are derived from hESCs, and not directly from post-mitotic human cone precursors, we were unable to demonstrate that RB1 inactivation caused tumors in either retinal organoids or transplanted mice. These differences suggest that oncogenic deletion in organoids derived from specific sensitive cell types (hESCs vs. post-mitotic human cone precursors) may limit the development and efficacy of tumor modeling using organoid technology. Although we detected the photoreceptor markers Crx and Recoverin in our retinal organoids, cone-rich retinal organoids resembling the human macula/fovea have not been adequately demonstrated and characterized [37]. Rod and cone cells are photoreceptor cells with different distributions and functions in the human retina. Rods are dominant in the peripheral retina, whereas cones are enriched in the macula, which is responsible for central vision and visual acuity. Recently, Kim and colleagues reported the generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids differentiated from human embryonic stem cells using an improved retinal differentiation system [37]. In future experiments, it would be interesting to model the effects of RB1-depleted human embryonic stem cells differentiated into cone-rich human retinal organoids.
CONCLUSION
While we were unable to use RB1-mutant retinal organoids to model retinoblastoma, our work identifies crucial functions for RB1 in the retinal organoid model of human retinal development. Our work further highlights 3D self-organizing retinal structures derived from pluripotent stem cells can recapitulate many aspects of development and structural organization of their in vivo organ counterparts, thus holding great promise for biomedical research and translational applications.
Supplementary Material
Figure S1. Generation of a RB1−/− H9 hESC line using CRISPR/Cas9. (A) The DNA sequence of the first exon within the RB1 gene and the corresponding amino acid sequence in wild-type hESCs. The gRNA sequence is labeled in blue and the PAM sequence in purple. (B) Biallelic disruption of the RB1 gene in clonal hESCs. The red label shows insertion/deletion after gene editing and subsequent change(s) in the amino acid sequence. Asterisks represent the STOP codon. (C) Representative wild-type and RB1−/− hESCs immunostained against RB and counterstained with DAPI. (D) Western blot was performed to detect RB and GADPH proteins in samples from wild-type and RB1−/− hESCs. (E) Karyotype of wild-type and RB1−/− hESC lines showing normal 46+XY karyograms. (F) Representative wild-type and RB1−/− organoids at D20, 30, 60 and 90 sectioned and immunostained against RB and DAPI. Scale bars, 200 μm.
Figure S2. Representative wild-type organoids at D60 and 90 sectioned and immunostained against Chx10, Pax6, and Recoverin, and counterstained with DAPI. Scale bar: 100 μm
Figure S3. Representative wild-type and RB1−/− organoids at D60 and 90 sectioned and immunostained against Ki67 and OTX2, Ki67 and Elavl3/4, Ki67 and Crx, Ki67 and Recoverin, and counterstained with DAPI. Scale bar: 200 μm.
Figure S4. Relative mRNA levels of Emx1 and Foxg1 measured by qRT-PCR. The results are mean±s.e.m. of 3 organoids from three independent experiments (**P<0.01, Student’s t-test).
SIGNIFICANCE STATEMENT.
3D organoid models provide a platform for studying human development and understanding disease mechanisms. We developed a model to study the role of RB1 in early development and tumor formation by generating retinal organoids from RB1−/− human embryonic stem cells. While loss of RB1 did not result in retinoblastoma formation in retinal organoids, absence of RB1 promotes S-phase entry, increases widespread apoptosis and reduces the number of photoreceptor, ganglion, and bipolar cells, consistent with its essential role in proliferation and differentiation.
HIGHLIGHTS.
RB expression is highest in progenitor cells and downregulated during maturation stages.
RB1 is required for S-phase entry and cell survival in retinal organoids.
RB1 is necessary for photoreceptor, ganglion and bipolar cell differentiation.
Loss of RB1 is not sufficient for retinoblastoma formation in retinal organoids.
ACKNOWLEDGMENTS
We thank Bogale Aredo and Rafael L. Ufret-Vincenty (Department of Ophthalmology, UT Southwestern) for help with OCT and fundus examination. We also thank Jerry Niederkorn and Jessamee Mellon (Department of Ophthalmology, UT Southwestern) for help with vitreous injections. This work was supported by the National Institutes of Health (R01NS093992, R01NS089770 and R01NS113516 to J.H.). This work was also supported in part by funds from the San Antonio Life Sciences Institute, the Semmes Foundation, Inc., the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (to J.H.).
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. [DOI] [PubMed] [Google Scholar]
- 2.Sheldon LA. Inhibition of E2F1 activity and cell cycle progression by arsenic via retinoblastoma protein. Cell Cycle. 2017:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sachdeva UM, O'Brien JM. Understanding pRb: toward the necessary development of targeted treatments for retinoblastoma. J Clin Invest. 2012;122:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang QE, Gwost I, Oatley MJ et al. Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol Reprod. 2013;89:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappas L, Xu XL, Abramson DH et al. Genomic instability and proliferation/survival pathways in RB1-deficient malignancies. Adv Biol Regul. 2017;64:20–32. [DOI] [PubMed] [Google Scholar]
- 6.Dimaras H, Kimani K, Dimba EAO et al. Retinoblastoma. The Lancet. 2012;379:1436–1446. [DOI] [PubMed] [Google Scholar]
- 7.Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu Rev Pathol. 2015;10:547–562. [DOI] [PubMed] [Google Scholar]
- 8.Soliman SE, Racher H, Zhang C et al. Genetics and Molecular Diagnostics in Retinoblastoma--An Update. Asia Pac J Ophthalmol (Phila). 2017;6:197–207. [DOI] [PubMed] [Google Scholar]
- 9.Weeber F, Ooft SN, Dijkstra KK et al. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem Biol. 2017;24:1092–1100. [DOI] [PubMed] [Google Scholar]
- 10.Naert T, Colpaert R, Van Nieuwenhuysen T et al. CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci Rep. 2016;6:35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee TC, Almeida D, Claros N et al. Cell cycle-specific and cell type-specific expression of Rb in the developing human retina. Invest Ophthalmol Vis Sci. 2006;47:5590–5598. [DOI] [PubMed] [Google Scholar]
- 12.Volkner M, Zschatzsch M, Rostovskaya M et al. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Reports. 2016;6:525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano T, Ando S, Takata N et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. [DOI] [PubMed] [Google Scholar]
- 14.Matsui T, Nieto-Estevez V, Kyrychenko S et al. Retinoblastoma protein controls growth, survival and neuronal migration in human cerebral organoids. Development. 2017;144:1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eiraku M, Takata N, Ishibashi H et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. [DOI] [PubMed] [Google Scholar]
- 16.Becker KA, Ghule PN, Therrien JA et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. Journal of Cellular Physiology. 2006;209:883–893. [DOI] [PubMed] [Google Scholar]
- 17.<Nodal signalling in the epiblast patterns the early mouse embryo.pdf>. [DOI] [PubMed] [Google Scholar]
- 18.MacPherson D, Sage J, Kim T et al. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18:1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol. 2013;14:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Indovina P, Pentimalli F, Casini N et al. RB1 dual role in proliferation and apoptosis: cell fate control and implications for cancer therapy. Oncotarget. 2015;6:17873–17890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yun J, Li Y, Xu CT et al. Epidemiology and Rb1 gene of retinoblastoma. Int J Ophthalmol. 2011;4:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer C, Pajovic S, Devlin H et al. Distinct patterns of expression of the RB gene family in mouse and human retina. Gene Expr Patterns. 2005;5:687–694. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich DW. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene. 2006;25:5233–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Wei R, Zhang L et al. Rb is required for retinal angiogenesis and lamination. Cell Death & Disease. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donovan SL, Dyer MA. Developmental defects in Rb-deficient retinae. Vision Res. 2004;44:3323–3333. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Gray J, Wu L et al. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DA, Donovan SL, Dyer MA. Mosaic deletion of Rb arrests rod differentiation and stimulates ectopic synaptogenesis in the mouse retina. J Comp Neurol. 2006;498:112–128. [DOI] [PubMed] [Google Scholar]
- 28.Mellough CB, Collin J, Queen R et al. Systematic Comparison of Retinal Organoid Differentiation from Human Pluripotent Stem Cells Reveals Stage Specific, Cell Line, and Methodological Differences. Stem Cells Transl Med. 2019;8:694–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter S, Vandezande K, Chen N et al. Sensitive and efficient detection of RB1 gene mutations enhances care for families with retinoblastoma. Am J Hum Genet. 2003;72:253–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macpherson D Insights from mouse models into human retinoblastoma. Cell Div. 2008;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacPherson D, Dyer MA. Retinoblastoma: from the two-hit hypothesis to targeted chemotherapy. Cancer Res. 2007;67:7547–7550. [DOI] [PubMed] [Google Scholar]
- 33.Mendoza PR, Grossniklaus HE. The Biology of Retinoblastoma. Prog Mol Biol Transl Sci. 2015;134:503–516. [DOI] [PubMed] [Google Scholar]
- 34.Costa C, Paramio JM, Santos M. Skin Tumors Rb(eing) Uncovered. Front Oncol. 2013;3:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McEvoy J, Flores-Otero J, Zhang J et al. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20:260–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu XL, Singh HP, Wang L et al. Rb suppresses human cone-precursor-derived retinoblastoma tumours. Nature. 2014;514:385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S, Lowe A, Dharmat R et al. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc Natl Acad Sci U S A. 2019;116:10824–10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Generation of a RB1−/− H9 hESC line using CRISPR/Cas9. (A) The DNA sequence of the first exon within the RB1 gene and the corresponding amino acid sequence in wild-type hESCs. The gRNA sequence is labeled in blue and the PAM sequence in purple. (B) Biallelic disruption of the RB1 gene in clonal hESCs. The red label shows insertion/deletion after gene editing and subsequent change(s) in the amino acid sequence. Asterisks represent the STOP codon. (C) Representative wild-type and RB1−/− hESCs immunostained against RB and counterstained with DAPI. (D) Western blot was performed to detect RB and GADPH proteins in samples from wild-type and RB1−/− hESCs. (E) Karyotype of wild-type and RB1−/− hESC lines showing normal 46+XY karyograms. (F) Representative wild-type and RB1−/− organoids at D20, 30, 60 and 90 sectioned and immunostained against RB and DAPI. Scale bars, 200 μm.
Figure S2. Representative wild-type organoids at D60 and 90 sectioned and immunostained against Chx10, Pax6, and Recoverin, and counterstained with DAPI. Scale bar: 100 μm
Figure S3. Representative wild-type and RB1−/− organoids at D60 and 90 sectioned and immunostained against Ki67 and OTX2, Ki67 and Elavl3/4, Ki67 and Crx, Ki67 and Recoverin, and counterstained with DAPI. Scale bar: 200 μm.
Figure S4. Relative mRNA levels of Emx1 and Foxg1 measured by qRT-PCR. The results are mean±s.e.m. of 3 organoids from three independent experiments (**P<0.01, Student’s t-test).