Abstract
With the passage of time and more advanced societies, there is a greater emergence and incidence of disease and necessity for improved treatments. In this respect, nowadays, aptamers, with their better efficiency at diagnosing and treating diseases than antibodies, are at the center of attention. Here, in this review, we first investigate aptamer function in various fields (such as the detection and remedy of pathogens, modification of nanoparticles, antibiotic delivery and gene delivery). Then, we present aptamer-conjugated nanocomplexes as the main and efficient factor in gene delivery. Finally, we focus on the targeted co-delivery of genes and drugs by nanocomplexes, as a new exciting approach for cancer treatment in the decades ahead to meet our growing societal needs.
Keywords: aptamer, polyethyleneimine, hybrid nanocomplexes, antibiotic, targeting, gene delivery systems
Introduction
DNA and RNA are the most vital molecules that serve as the equivalent of an enormous hard disk to store information. They also can be used as a diagnostic gadget and therapeutic “doer”.1–4 The results from many types of research showed that the specific structural characteristics of these molecules have opened new research fields leading to the design of aptamers. Aptamers are oligonucleotides including ssRNA, ssDNA (single-stranded RNA, DNA) or peptides that bind to select targets with significant dependency and exclusivity due to their specific structural characteristics.5,6 Aptamers are known as an adequate alternative for antibodies due to their ability to overcome the many disadvantages of using antibodies.7–9 For example, it is well known that oligonucleotides are sturdier at higher temperatures than proteins, so that they can sustain their structures over frequent cycles of denaturation and renaturation. Therefore, the greatest benefit of aptamers based on oligonucleotides over antibodies based on proteins is their constant properties at high temperatures, even after a 95 °C denaturation.10–13
Aptamers are minimally toxic molecules with low immunogenicity because nucleic acids usually are not recognized as an external, foreign agent by the human immune system.14,15 Aptamers are derived from SELEX (Systematic Evolution of Ligands by Exponential Enrichment) or in vitro evolutionary processes, so toxins and other molecules that do not induce a strong immune response can be used as the object to develop aptamers. Therefore, based on the advantages described above, aptamers are more adaptable for different disease applications. They have been considered as a biological material and as a diagnostic and therapeutic tool they have been used in the expansion of novel drugs, drug delivery, gene delivery and antibiotic delivery systems, etc.16–18 They are actually considered for targeting an antibiotic and gene delivery system that controls the time and place of drug release or influence on gene delivery.19
Aptamers have been effectively established against metal ions, proteins, bacteria, viruses, and whole cells. High affinity and specificity are some of their characteristics comparable with antibodies. Unlike antibodies, they are capable of being selected under non-physiological conditions, such as very high or low temperatures or pH.20 These features make aptamers appropriate applicants for clinical use. A number of aptamers have been designated for various diseases, eg, types of cancers.21,22 Table 1 describes a list of the most common differences between an antibody and aptamer.
Table 1.
Different Properties Between Antibodies and Aptamers
| Aptamer | Antibody |
|---|---|
| Unlimited shelf life (several months) | Limited shelf life (few months) |
| High tissue penetration (small size, 3 nm) | Low tissue penetration (large size, 10–15 nm) |
| High specificity and affinity | High specificity and affinity |
| High stability in pH and temperature | Low stability in pH and temperature |
| Low immunogenicity | High immunogenicity |
| Variety of modification by diverse molecules | Hard to be modified |
| Cost of production of aptamer is less expensive | Cost of production of antibody is expensive |
| Presence of nuclease susceptibility (need for modification) | Absence of nuclease susceptibility |
Some published review articles have studied the role of aptamers in the diagnosis and treatment of cancer and disease.23–25 However, such work has been limited and does not emphasize the significant promise aptamers have to revolutionize medicine. This review was carried out to emphasize their importance in the future of antibiotic therapy, cancer diagnosis and treatment. Specifically, this review summarizes recent studies on the use of aptamers in disease diagnosis and treatment, antibiotic delivery and gene delivery, and also to review the use of aptamer-conjugated nanocomplexes in anti-cancer applications.26,27
Aptamers as Diagnostic and Therapeutic Agents
Pathogenic diseases are some of the most prevalent diseases that threaten the lives of many people all over the world, which have been created through various microorganisms. The inefficient transfer of antibiotics to the site of infection and side effects due to their great consumption by the immune system, and also the slow rate of developing new and improved anti-pathogenic drugs, are important global issues that need to be solved now. The development of targeted therapies and proprietary diagnostic methods for diseases caused by pathogens, infections and cancers is increasing. Targeted therapies have been developed based on aptamers in recent years to overcome these problems. Studies have been conducted on the use of aptamers in the field of diagnosis as well as targeted and early therapies.28,29
As just one example, Francisella tularensis, an aerobe bacterium, acts as a pathogenic agent and causes tularemia in men and animals. This pathogen is considered as one of the most significant biological threats that has a high degree of infectiousness, and a high capacity for disease and mortality (30%-60% were reported in the pre-antibiotic area). In fact, it is possible to point out the importance of the use of aptamers in the detection of the biological fight factor.30–32
An outstanding study generated aptamers (ssDNA) that bind to the F. tularensis subspecies. In this study, a set that contained 25 aptamers was isolated to specifically conjugate to the F. tularensis subspecies. Investigation into these aptamers under certain conditions indicated that the aptamer complex conjugated to the tularemia antigen specifically, which enabled it to be used as diagnostic tool for a biological fight factor like F. tularensis.33–35
Another prevalent bacterial infection is caused by Vibrio parahemolyticus and Staphylococcus aureus (S. aureus). V. parahaemolyticus is a common cause of intestinal illness after eating uncooked or raw seafood and as it is in environmental waters. Symptoms of V. parahaemolyticus are watery diarrhea, abdominal pain, vomiting, nausea, fever, headache, and bloody diarrhea. As a result, providing a method for its rapid and specific detection is of the utmost importance.36,37
The first whole-bacterium SELEX application to reconnoiter specific DNA aptamers was reported for V. parahemolyticus. This strategy was used for a set of FAM-labeled ssDNA molecules to identify the aptamers that were specifically attached to V. parahemolyticus. This identification was done by the use of flow cytometric analysis. According to the obtained results, the aptamer A3P indicated great binding affinity (76%) to V. parahemolyticus. This feature can be used for the detection of this bacterium even in food. Moreover, linking of this sequence to magnetic nanoparticles (NPs) and conjugating it to sensitive detection probes can help to control food safety.34,38
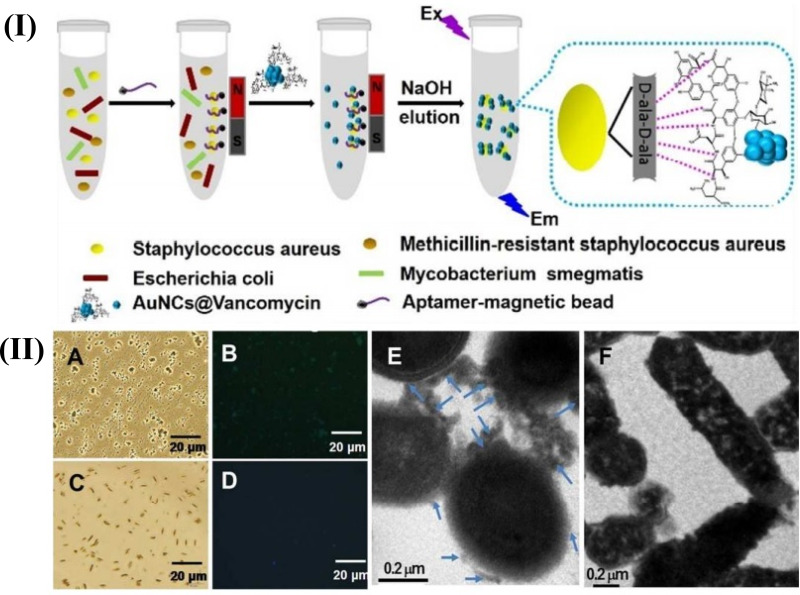
S. aureus is a round-shaped bacterium that can be pathogenic and a menace to human health.39 Evaluating S. aureus through a molecular recognition procedure with aptamer detectors is extremely effective.40 Moreover, enrichment and separation by magnetic particles and fluorescence detection by gold nanoclusters (AuNCs) are powerful tools for their evaluation because of their controllability and high sensitivity.41,42 In one study, Cheng et al indicated a new method for the detection of S. aureus in which recognition molecules, such as an aptamer, and antibiotic were combined together. In this study, S. aureus was quantified via the use of aptamer-coated magnetic beads (Apt-MB) and Vancomycin (Van)-functionalized fluorescent nanoclusters (AuNCs@Van, with 2.0±0.6 nm) in a mixture of other non-targeted bacteria and was also detected in authentic samples including milk and human serum.
After the synthesis of AuNCs@Van, its formation was confirmed by a transmission electron microscope (TEM) and X-ray photoelectron spectroscopy (XPS). Since the nanoprobes should be chemically and photochemically stable, the stability of AuNCs@Van nanoprobes was checked by UV illumination and the surrounding buffer. Observations showed the successful synthesis and stability of AuNCs@Van in various buffers and pH levels. Next, the attachment of AuNCs@Van to the bacteria was proved by fluorescence microscopy and TEM. The detection of S. aureus from complex samples and in authentic samples was shown in the range of 32–108 CFU/mL and the limit of detection (LOD) of 16 CFU/mL. Finally, it was confirmed that the magnetic beads coated by the aptamer and fluorescent AuNCs covered by the antibiotic could be used to quantify and detect bacteria from complex samples in a binary assessment process. Apt-MBs and AuNCs@Van dual recognition could detect S. aureus in milk and human serum as authentic samples with high efficiency (96.94% to 101.24%, respectively), so that it could be applied for the assessment of food contamination related to S. aureus bacterium and infection disease recognition43 (Figure 1).
Figure 1.
Schematic illustration of the recognition process of S. aureus using a AuNCs@Van and Apt-MB (I) dual recognition assay, and the characterization (II) (A–F) represents the optical, fluorescence and transmission electron microscopy characterization of this synthesized particle. The figure was reprinted with permission from Cheng D, Yu M, Fu F, et al. Dual recognition strategy for specific and sensitive detection of bacteria using aptamer-coated magnetic beads and antibiotic-capped gold nanoclusters. Analytical Chemistry. 2016;88(1):820–825. Copyright © 2016 American Chemical Society.43
This strategy has several advantages over previous studies, including it is easy-to-use and it is much cheaper to synthesize AuNCs @ Van than antibodies, and has a bacterial detection at concentrations as low as a ppm concentrations. However, from the limitations of this study, absent the pre-separation for targeting bacteria from other interfering components prior to fluorescence measurements, it is not likely to yield a real result.
Also in another study, Charoenphol et al applied a cell-specific aptamer-DNA nanostructure system for treatment purposes.44 DNA nanostructures have been vastly used in various applications because of their special characteristics such as programmability, uniform structures, biocompatibility and biodegradability. For example, DNA nanostructures have the ability to carry several therapeutic cargos (such as aptamers, small interfering RNA (siRNA), etc.) without any chemical correction. In this study, they merged AS1411 aptamers into DNA pyramids and observed selective inhibition in the growth of Hela cells (a kind of cancer cell). In addition, it was found that aptamer-displaying DNA pyramids were more stable to nucleus decadence than single-stranded aptamers. However, in this study, a single cargo type was indicated, but DNA can be used in a combination therapy.44 For example, a combination of chemotherapy and small RNA-based therapy (siRNA) is an example of these combination therapies. It is assumed that chemotherapy and siRNA can be delivered simultaneously to the same tumor cell to exert their synergistic effect, however, the development of these systems through systemic administration is still facing challenges.45
With the development of nanomedicine and aptamer selection methods, aptamer-functionalized NPs have opened a new direction to diagnostic and therapeutic applications. There have been many reports about the use of NP-aptamer conjugates for the delivery of NPs into cancer cells. Early diagnosis of cancer is a challenge for effective treatment and aptamers attached to NPs are considered as highly promising molecular detectors in this field worthy of more attention and clinical trials.46,47
Magnetic NPs associated with fluorescence moieties could detect and elicit exfoliated tumor cells. Recently, a study based on the application of aptamer-conjugated dual NPs with two features (including magnetism and fluorescence) to detect and elicit exfoliated tumor cells which are in the blood was conducted. Due to the low numbers of these cells that were surrounded by various other cells, an approach that created selective extraction and sensitive detection was required. For this purpose, the researchers used aptamer-conjugated NPs (ACNPs) with fluorescent NPs (FNPs) to extract and detect tumor cells. When the various parameters were combined, the most selective and sensitive ACNP system was specified. This approach can be adjusted for a variety of cancers to determine the possible ACNPs for the recognition and extraction of cancer cells.41 In another study, aptamers conjugated with magnetic NPs (ACMNPs) were utilized by Bamrungsap et al to diagnose cancer cells.48 The ability to detect at least 10 cancer cells in 250 µL of the cancer cell sample is a feature of this biosensor. ACMNPs utilize the features of aptamers to bind specifically to target cancer cells, and the broad surface of these NPs provides for the binding of several aptamers. In addition, a set of ACMNPs can be used to accurately detect molecules at the molecular and cellular level and detect them in fetal bovine serum, human plasma and whole blood.
Breast cancer is a type of severe form of cancer and the second reason of death in women today. Investigations showed that relatively large numbers of circulating tumor cells (CTCs) were detected in the peripheral blood cells of people who suffer from breast cancer, so the diagnosis of blood CTCs is a hopeful method for the detection and determination of breast cancer cells. There are two cancer cells which cause breast cancer: human epidermal growth factor receptor 2 (HER2) and Mucin 1 (MUC1). HER2 and MUC1 are over-expressed in 15–20% and 90% of breast cancer patients, respectively. So, through the use of multi-target methods with HER2 and MUC1 probes, CTCs of breast cancer could be detected with high accuracy and precision. NPs have been used in various studies for the identification of breast cancer due to their advantages such as easy surface modification, etc.49–51
In a distinct study, a dual aptamer system including HER2 and MUC1 modified-SiNP (Dual-SiNP) for the diagnosis of breast cancer was considered. The dual aptamer approach provided the first and wide detection and diagnosis system for HER2 and MUC1 breast cancer cells. This synthesized probe was assessed by some techniques such as fluorescence spectroscopy, dynamic light scattering and UV-Vis spectroscopy. The application of this system was limited to in vitro systems because of several difficulties of in vivo systems including: inconsistency of nucleic acids in the blood, the decadence of fluorescence signals of the probes in blood, NPs with short half-life in blood flow, etc.52,53
Heavy metals are an important environmental contaminant. For example, the mercuric ion (Hg2+) is a toxic non-biodegradable heavy metal in which its accumulation in an organism will have irreparable effects even death. There are many analytical methods for measuring Hg2+ such as UV-Vis spectroscopy, atomic absorption/emission spectroscopy (AAS/AES) and X-ray absorption spectroscopy. These methods require expensive and complicated instruments, so there is an intense inclination to develop new methods with greater efficiency for Hg2+ ions detection. An effective method for the recognition of Hg2+ions requires the application of oligonucleotides. In order to form thymine- Hg2+-thymine complexes (T-Hg2+-T), Hg2+ ions interact with thymine bases and also could make an aptamer-thymine- Hg2+-thymine nanocomplex for biological applications.54–56
From another perspective, detecting Hg2+ ions has been considered as a strategic method by designing a mercury DNA (MSD) functionalized gold nanoparticle (AuNPs) for Hg2+ ion detection. The mechanism for this process was based on a T-Hg2+-T complex and suppressing the fluorescence feature of AuNPs. AuNPs-MSD probes were formed through conjugation of MSDs labeled with fluorescein (FAM) and AuNPs by Au-S bonds. Hg2+ detection was investigated by changing fluorescence signals. Also, this method showed high selectivity for Hg2+ between additional metal cations (such as Fe3+, Ca2+ and Mg2+, Cr3+, Mn2+, Cu2+, Ni2+, Pb2+ and Co2+). Further, this method could be used for Hg2+ detection in aqueous solutions. In addition, this method could be applied for detecting heavy metallic ions in body fluids by conjugating them with an aptamer and formation of aptamer nanocomplexes.57–59 Table 2 describes the types of aptamer-based nanostructures for applications in diagnostics with their advantages and limitations.
Table 2.
Types of Aptamer-Based Nanostructures in Diagnostics with Their Advantages and Limitations
| Aptamer-Based Nanostructures | Size of Nanostructures | Detection Methods | LOD | Advantages | Limitations | Ref |
|---|---|---|---|---|---|---|
| Au NPs/Graphene (Au NPs/Gr) | 500 nm | Colorimetric detection | 0.01–0.5 μM and 0.01–1.0 μg/L | Detection of various targets by this aptamer, rapid colorimetric assay, design of specific aptamer |
Investigation of other lengths of aptamers | 60 |
| Anti-IL-6 Aptamer-AuNPs | 15 and 50nm | Colorimetric detection | 1.95 μg/·mL | Successful detection of murine IL-6 (in the μg·mL−1 range) in buffers and a biological sample matrix | Low sensitiv-ity, more work is needed to increase the sensitivity to detect IL-6 in healthy serum/plasma | 61 |
| Graphene quantum dots (GQDs) -gold nanorods (AuNRs) |
2–3 nm size of GQDs, 2nm size of AuNRs | Electrochemical impedance spectroscopy (EIS), cyclic voltammetry, differential pulse voltammetry | 0.14 ng/mL | Detect PSA in human blood serum with high sensitivity and repeatability, stability, cost effectiveness |
Using the three analytical methods from this study to measure the signal at the same time, but can also be used for better measurement | 62 |
| Apt-Fe3O4@mTiO2 | 200 nm | - | 10 to 2000 CFU/mL | Rapid and sensitive detection of pathogenic bacteria, high selectivity and strong affinity of aptamers | Although both systems exhibit effective monitoring and high sensitivity, the expensive cost of detection limits their widespread use | 63 |
| MNPs-specific aptamer | 20nm size of Fe3O4 MNPs | Colorimetric detection | 7.5×105 CFU/mL | Use of specific aptamers, easy separation, low cost | Low LOD, need to enhance the LOD and to increase the cross-reactiv- ity to other pathogens | 64 |
Aptamers as Targeting Components for Antibiotic Delivery Systems
Antibiotic delivery aptamers have been illustrated in a number of reports.65–67 Aptamers were used for immobilizing antibiotic molecules instead of targeting them. For example, a nanocarrier for neomycin (aminoglycoside antibiotic) was developed by aptamers for delivery purposes, wherein aptamers immobilize antibiotic molecules via affinity binding. Mesoporous silica NPs (MSNs) could be an appropriate nanocarrier in stimuli-responsive drug delivery systems.68–70
In a study, an aptamer-gate mechanism was designed using a binding aptamer (AS1411). By converting aptamer sequences to the hairpin structure, a S. aureus (bacterium) specific molecular gate was gained. The interaction between S. aureus surface antigens and aptamer succession disrupted the aptamer structure. Increasing the rate of fluorescence depended on the presence of the objective. In this study, SA20hp was selected as a molecular gate aptamer because of its high fluorescence peak response. The destruction mechanism of the pathogenic agent was as follows: Van (antibiotic) was absorbed into the pores of MSNs and covered with the SA20hp molecule. When the NPs conjugated to surface of S. aureus cells through aptamer successions, the Van antibiotic was released to destroy the bacteria. These aptamer-gated silica NPs provided for a control of antibiotic dosage and unique internal release, and then, enabled the possibility of using stronger therapeutic compounds.71,72 In addition, several presitigous articles address different analytical methods and biosensors with capping different types of metal nanoparticles with biomolecules, as well as porous nanomaterials (Figure 2).73
Figure 2.
AuNPs-aptamer was capped on the MSA surface due to the binding reaction of ATP aptamer to the adenosine molecule. The delivery of the entrapped guest (fluorescein) was selectively triggered by an effective displacement reaction in the presence of the target molecule (ATP). Reprinted with permission from Zhu CL, Lu CH, Song XY, Yang HH, Wang XR. Bioresponsive controlled release using mesoporous silica nanoparticles capped with aptamer-based molecular gate. J Am Chem Soc. 2011;133(5):1278-1281. Copyright (2020) American Chemical Society.73
Aptamers as Targeting Components of Gene Delivery Systems
Nowadays, scientists are looking for new methods to treat and control diseases such as gene therapy. Gene therapy is the treatment process of nucleic acid transference into the patient’s cells by viral or non-viral carriers to repair faulty genes or provide further biological functions. In this approach, genes and nucleic acids including plasmid DNA (pDNA), anti-micro RNA (miRNA) oligonucleotides or siRNA are used. For nucleic acids, physical properties, such as negative charges, limit their binding to the cell surface and inactive diffusion through the cell membrane. Therefore, the delivery procedure is the single significant and difficult challenge in gene and nucleic acid therapy. Although gene delivery through viral vectors is well known, many drawbacks (such as irregular cytotoxicity, etc.) limit their clinical usage. In contrast, non-viral vectors provide many advantages such as low toxicity, no infective danger, flexibility and capability of production and modification. Non-viral vectors (including liposomes, cationic polymers, cationic lipids, peptides and dendrimers) are known for their applications in gene delivery. Additionally, due to the success of gene therapy dependent on the efficient delivery of genes to specific cells (targeted gene delivery), today, many targeting ligands (such as antibodies, nanobodies, peptides and aptamers) have been used for gene and drug transfer into particular cancer cells.74–78
Some cancer cells, such as melanoma (skin cancer), cannot be easily treated in the clinic today because of their resistance to radiotherapy, insignificant diagnosis and numerous speed of deformation (the BRAF gene mutation in many melanomas makes treatment difficult). Gene therapy is a sufficient treatment for melanoma versus common treatments (such as radiation therapy) because of its high specificity and low toxicity. Therefore, gene therapy using SiRNA (small RNAi) which targets the BRAF gene could be an effective treatment method for melanoma.79,80
A new strategy suggested by Liyu et al in which nucleolin-targeting liposomes were constructed to deliver SiBraf (anti-BRAF SiRNA) could be promising for therapeutic purposes (Figure 3). AS1411 is an aptamer with a specific binding ability to nucleolin. AS1411 was tagged to polyethylene glycosylated (PEGylated) (PEGylated) liposomes (ASLP) that acted as a targeting probe. AS1411ePEG-liposomes (ASLP) and SiRNA were conjugated to each other, electrostatically, to create a ASLP/siRNA complex. Observations indicated that the AS1411 aptamer conjugated liposomes, targeted specific cancer cells (A375) and showed considerable silencing activity in this cell line. So, the application of ASLP and AS411 aptamers to deliver siRNA showed high efficiency for melanoma therapy and should be further studied in clinical trials.80,81
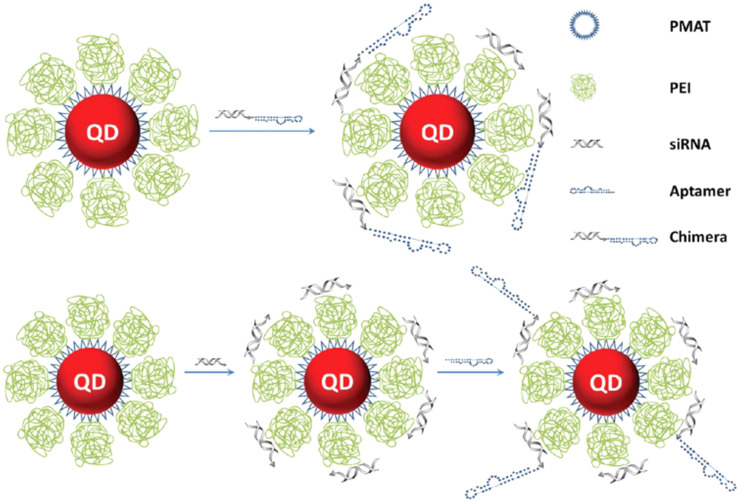
Figure 3.
Schematic representation of cationic nanoparticles for targeted delivery of siRNA-aptamer chimeras. Immobilization of preformed siRNA-aptamer chimeras onto positively charged QD-PMAT-PEI nanoparticles. The aptamer block collapsed on the carrier results in reduced binding activity. Two-step immobilization of chimeras on a cationic nanoparticle surface. siRNA molecules with a thiol-reactive terminal group are first adsorbed on the QD-PMAT-PEI surface to reduce the positive charge; subsequently aptamers with a single thiol group are brought in to form siRNA-aptamer chimeras on the nanoparticle surface. Adapted with permission from Bagalkot V, Gao X. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano. 2011;5(10):8131-8139.82,138 Copyright (2011) American Chemical Society.
Gene Delivery Through PEI-Based Vectors
Polyethyleneimine (PEI), a polycation molecule with the ability to compress DNA (through electrostatic interactions between anionic phosphate of nucleic acids and primary amines of PEI) into NPs and high transfection efficiency during in vitro and vivo conditions, is a very effective non-viral vector. PEI delivers NPs which contain genes into the cytoplasm before degradation by some enzymes (such as lysosomal enzymes) through a proton sponge (proton buffering) mechanism. For PEIs, molecular weight is so important because of its effect on transfection efficacy. The low molecular weight of PEIs represents low cell toxicity and transfection efficacy. For this matter, scientists designed a kind of ligand-conjugated PEIs-based vector with low toxicity and great transfection efficacy. Aptamers are the best option as functional targeting ligands for specific targeting.83–86
Targeted Gene Delivery by Aptamer-PEI Nanocomplexes
Non-specific binding of pDNA/PEI complexes to proteoglycans on cell membranes would lead to non-competence for gene transfer to specific cells. An outstanding research group developed some approaches in which pDNA/PEI complexes were covered with polyadenylic acid (PolyA) as a polynucleotide. Aptamer-based molecular assessments illustrated that these aptamer-coated complexes could enter cells via a cellular uptake process. In addition, studies showed that MUC1 would be adequate for tumor targeting. Therefore, Kurosaki et al synthesized pDNA/PEI/MUC1 complexes for the treatment of cancer cells through targeted gene delivery. In this research, several complexes at several weight ratios of pDNA/PEI/aptamer were constructed. This experiment was done in vivo and in vitro using a human and mice lung cancer cell line, A549 cells. The efficiency of in vivo and in vitro transfections for various weight ratios of aptamer complexes was investigated. The results indicated that for A549 cells with the MUC1 cell surface protein, the use of the pDNA-based aptamer complexes with weight ratios between pDNA, PEI and MUC1 of 1:1:0.25, 1:1:0.5, 1:1:1 and 1:1:2 increased gene expression. Also, large amounts of the MUC1 aptamer reduced aptamer-complex gene expression. It seems that robust anionic surface charges act on cellular membranes and the backlash might be higher than the binding strength of the MUC1 aptamer to MUC1.
In addition, results showed that the application of non-specific aptamers reduced transgene expression. Besides, studies have shown that the aptamer complex used for tumor specific gene delivery, by simply changing aptamers, this complex could be used for other specific gene delivery systems. The targeting and coating system may be pioneering knowledge for cell specific gene delivery.87–89
Lymphocytes are the main cellular species in the lymph modes. Special features of these cells (such as re-introduction into the body, mobility and distribution all over the body) provide for gene therapy for genetic and pathogenic diseases. Hamedani et al designed a novel complex in which the sgc-8c aptamer was non-covalently conjugated to the PEI polycation to raise transfection efficiency of the targeted gene delivery of pDNA into MOLT-4 cells. In general, the efficient transfection of pDNA via a vector will be obtained when there is an equilibrium between the condensation and fast diffusion of pDNA in the cytoplasm. For measuring the condensation of DNA and PEI-aptamer conjugation, ethidium bromide (EtBr) molecules were used. These molecules showed fluorescence until they were accommodated in double-stranded nucleic acids. By adding PEI to the pDNA/EtBr solution and condensation of pDNA by PEI, because of the release of some accommodated EtBrs molecules from pDNA and negligible interactions between them (because of the structural transformation of pDNA from a helix to a globular form), fluorescence intensity decreased.90–93
Moreover, the addition of the aptamer did not have any effect on the fluorescence and shape of the curves. Particle size and zeta potential were additional measurable features in this study. It is necessary to distinguish carriers according to these two factors because nanoparticle size and their surface charge affect gene delivery efficiency. By using small particles of ≤ 500 nm size, faster and easier cellular uptake as well as transfection efficacy occurred. Generally, in this study, the addition of a 10% w/w aptamer (with a negative charge) to a 10kDa PEI-pDNA polyplex caused an increase in particle size and decrease in zeta potential (reduced net positive surface charge). Positive surface charges of PEI interacted with cell membrane negative charges and caused PEI to become cytotoxic and instable in the cell membrane. In addition, the cytotoxicity of the pDNA-based aptamer complexes was determined for a protein tyrosine kinase 7 (PTK7)-positive and -negative, MOLT-4 (human, peripheral blood, leukemia, T cells) and U266 cell lines, respectively. All vectors represented low toxicity, however, by increasing PEI/pDNA proportions, toxicity increased and the presence of the aptamer did not have a significant effect on cytotoxicity. Researchers have used many strategies to treat lymphocytes, such as treating them with large amounts of genetic material but because these cells are resistant to most gene transfection systems, association of vectors increased cytotoxicity. One study used targeting aptamer PEI-based vectors to increase transfection efficacy. The evaluation of transfection efficiency and targeted delivery of PEI-aptamers to PTK7 receptors in MOLT-4 and U266 cell lines showed that in MOLT-4 cells with PTK7 receptors, the PEI/pDNA polyplexes and sgc-8c aptamer conjugation, increased transfection efficacy. Moreover, information showed that the conjugation of a nonspecific aptamer to PEI/pDNA polyplexes, prevented luciferase gene expression. Finally, the results obtained in this study showed that a cell specific sgc-8c aptamer which tagged the PEI vector through electrostatic interaction, could increase transfection efficacy. In addition, the sgc-8c aptamer kept its conformation after electrostatic binding to PEI and retained specificity and dependency of PTK7 to bind to MOLT-4 cells, selectively.94–97
Also in a recent study, a new EpCAM Aptamer -PEI-siRNA nanocomplex (PEI-EpApt-SiEP) was designed for EpCAM (epithelial cell adhesion molecule, with features such as overexpression in solid tumors and being a diagnostic marker for various cancer cells) targeting (Figure 4). In recent years, RNA and DNA aptamers instead of antibodies, were developed against EpCAM by the use of SELEX technologies. Moreover, siRNA (operating within RNA interference (RNAi)) was considered as an appropriate nucleic acid for targeted gene therapy especially in humans. In this study, to prepare an optimal PEI nanocore, sodium citrate was used for stabilizing its charge. For nanocomplex synthesis, at first, siRNA was added to the PEI nanocore and after, the aptamer was added to create a nanocomplex with EpCAM detection ability. Since the exact concentration of the aptamer (200 nM) and siRNA was capable of saturating the PEI-citrate nanosphere, the optimized concentration of SiEp and EpApt were mixed with PEI-citrate for constructing a novel PEI-EpApt-SiEp nanocomplex. As higher concentrations of PEI leads to higher cell toxicity (ie, 3 µg/mL), 0.3 µg/mL of PEI was used to functionalize the aptamer and siRNA. Zeta potential and particle size of the PEI nanocore and nanocomplex were investigated. To evaluate the application of this nanocomplex for in vivo systems, serum was shown to influence the size and charge of the nanocomplex as determined in RPMI.
Figure 4.
Schematic of the newly synthesized aptamer-PEI-siRNA nanocomplex (PEI-EpApt-SiEP) and its function to silence the target gene and decrease cell proliferation. The figure was reproduced from Subramanian N, Kanwar JR, Athalya P, et al. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J Biomed Sci. 2015;22(1):4. Copyright © Subramanian et al; licensee BioMed Central. 2015. Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0).98
The results indicated that there was a negligible difference in size and charge of nanocomplexes in the presence and absence of serum. Cellular uptake and cell binding of the complex (PEI-EpApt-SiEp) were investigated in breast cancer and WERI-Rb1 cell lines. Observations showed that the expression of EpCAM proteins and aptamer binding were higher in MCF-7 compared to WERI-Rb1 cells. Concerning cellular uptake for both of these cells, binding of cells to PEI-EpApt-SiEp caused a boost of the intensity of the fluorescent spectra in comparison with the EpApt alone. The greater binding of the PEI-EpApt-SiEp nanocomplex compared to EpApt and no connection of the ScrApt-nanocomplex or ScrApt to both cell lines, showed cell specificity of the EpCAM aptamer. The EpCAM silencing effect using the PEI-EpApt-SiEp nanocomplex was evaluated by checking the protein and mRNA amount by Western blotting and qPCR techniques, respectively. In treating MCF-7 and WERI-Rb1 with PEI-EpApt-SiEp, down regulation of the EpCAM gene was higher than the SiEp treatment (64%, 72% versus 56%, 62%, respectively). Moreover, PEI-ScrApt-SiEp and PEI alone treated cells, did not illustrate any EpCAM silencing. The EpCAM protein expression and mRNA levels did not change substantially compared to untreated cells. For MCF-7 and WERI-Rb1 treated with PEI-EpApt, EpCAM levels were reduced by 38% and 52%, respectively.98
Based on the previously obtained results, although EpCAM silencing in this method was less than by siRNA delivery via a targeted antibody, this nanocomplex demonstrated more functional activity and EpApt in this structure induced cytotoxicity with overexpressed levels of EpCAM. This novel nanocomplex included an aptamer and siRNA and effectively targeted EpCAM positive cells and suitable gene silencing activity that can be applied for in vivo systems.70,99
Targeted Gene Delivery Using Aptamer-Conjugated Copolymers
Bcl-xL (B-cell lymphoma-extra large) is a transmembrane molecule in the mitochondria. It is a kind of protein with anti-apoptosis properties which causes programmed cell death by preventing the release of mitochondrial content. In many cases, the overexpression of Bcl-xL protein leads to lung cancer. Gene and drug delivery are effective treatment methods for lung cancer due to some features of lung cells (such as low enzymatic activity and wide surface area). However, plenty of research has been conducted concerning PEI derivative applications for nucleic acid transference to the lung, but more attempts should be made for introducing new strategies based on cationic polymer vectors for more efficient gene delivery systems.100–102
Recently, a novel carrier based on a poly(L-lysine) (PLL)-alkyl-PEI copolymer was designed and the effect of aptamer conjugation on short hairpin RNA (shRNA) plasmid gene delivery efficiency was investigated for gene delivery of this modified vector to lung cancer cells. Beside PEI, PLL is a cationic polymer non-viral vector that can be used for most gene delivery purposes. Both of these polymers have disadvantages which limit their use as a vector for gene transference. In this research, a novel copolymer vector including PEI (with DNA condensation ability) and PLL (biodegradable) was designed to overcome deficiencies of these polymers. For the preparation of a copolymer, at the beginning, an alkylcarboxylate (6-bromohexanoic acid) was coupled to PLL at three molar ratios. Next, PEI was conjugated to PLL (via hexanoate linker) to produce PLL-hexanoate-50%-PEI (PLPE36%) copolymers and PLL-hexanoate-10%-PEI (PLPE8%). Vectors have many roles in gene delivery (such as carrying nucleic acid cargos and also their condensation and protection from extracellular degradation), so their optimal size and charge density are crucial for all medical applications.
These two properties were measured via laser Doppler velocimeter and DLS. The results showed that after this successful coupling (the exact conjugation of PEI to alkylcarboxylated-PLL), the positive surface charge and nanoparticle size increased. The buffering capacity of vectors is crucial in polycation mediated gene delivery. The investigations illustrated that unlike PLL with a weak buffering capacity, copolymers had suitable buffering capacity, almost the same as the PEI polycation. Moreover, fluorescence suppression analysis with the EtBr molecule indicated that the modified polymers could be capable of condensing DNA at the determined C/P ratio, above 0.5. The obtained information showed that electrostatic interactions which come from the attachment of the AS1411 aptamer to the copolymer that has been modified, could enhance transfection efficiency with an insignificant consequence on charge density and size. Finally, the aptamer-conjugated at the determined C/P was selected for shRNA delivery to silence human Bcl-xL gene expression in A549 cells. Protein expression and mRNA levels of Bcl-xL substantially decreased after shRNA plasmid transfection by aptamer-vector system against the Bcl-xL gene. So, this study suggested an efficient novel vector (apt-PLPE8%) for targeted non-viral gene delivery in lung cancer and pulmonary systems.77–79,103
Targeted Gene Delivery Using Aptamer Modified Carbon Nanotubes
As we know, shRNA and siRNA can be used for therapeutic purposes and there are two significant challenges in targeted gene delivery including low toxicity and great transfection efficiency of the synthesized vectors. PEI is a kind of polycation and an effective non-viral vector with buffering capacity. In reality, branched PEI consists of primary, secondary and tertiary amines, so it can be protonated and provide a buffer. Also, the molecular weight of PEI plays a major role in transfection efficiency of this molecule. As the PEI molecular weight increases, the growth of a positive surface charge and reduction of transfection efficiency will result. Alkylcarboxylation of PEI improves transfection efficiency.104–107
Single-walled carbon nanotubes (SWNTs) can be used in delivery systems due to its easy passage through the plasma membrane. Since SWNTs have poor solubility, they cannot be used as an efficient carrier. By inducing positive charges onto a SWNT surface, in addition to simplifying solubility in water, they can connect to molecules including DNA and siRNA with a negative charge. Generally, conjugation of polycations as PEI is an effective strategy to enhance DNA condensation of CNTs.108,109
As an example, a new system based on single-walled nanotubes functionalized with piperazine-PEI and an aptamer for siRNA transference into breast cancer cells proved that gene delivery could be used to diagnose cancer cell presence. EpCAM is a membrane protein which can be overexpressed in tumors and is recognized as a promising marker for cancer stem cells; it can be a cancer stem cell marker (CSCs). In the present study, PEI-piperazine was attached to functionalized SWNTs. Afterwards, SWNT-PEI-piperazine NPs tagged the EpCAM aptamer. For conjugation of SWNTs to PEI, surface carbons on SWNTs were oxidated to become COOH. So, this functional group could conjugate PEI amine groups via formation of amide bonds. To have an effective gene carrier, it is necessary to deliver therapeutic mediators to their specific cells, selectively. For this purpose, a targeting aptamer was used. Prior studies showed that conjugation between an aptamer and NPs increased delivery efficiency. So, a SWNT-PEI-piperazine/DNA complex was bound to the EpDT3 aptamer. Observations showed that transfection efficiency largely increased. As a result, in this study, the effect of the novel SWNT-PEI-piperazine NP to transfer BCL91 siRNA to EpCAM cells was assessed. This type of vector-aptamer derivative could increase DNA transfection efficiently. This strategy also could be used for breast cancer treatment by gene therapy.110–112
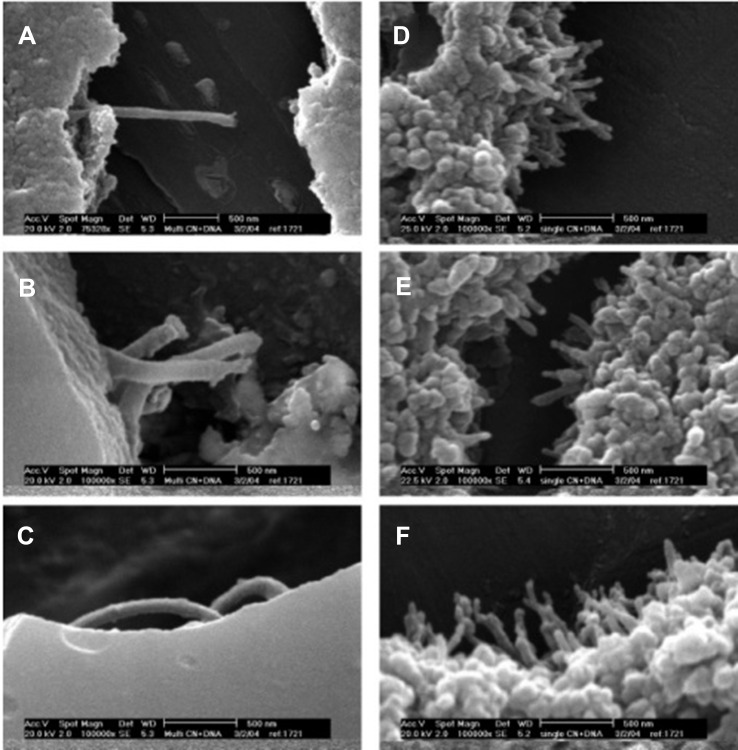
In a similar study, a novel targeted nanocomplex inclusive of PEI-CNT conjugated to the 5TR1 aptamer created for the specific delivery of Bcl-xL shRNA for breast cancer cell treatment.113 For the first step, functionalized single-walled SWCNs were synthesized. Because of negligible solubility of SWCNs in aqueous solutions, they were oxidized at the edge carboxylate groups via concentrated nitric acid solutions. Also, SWNTs and poly(ethylene glycol) (PEG)/PEI were conjugated covalently to prevent aggregation in biological environments and improve their DNA condensation ability. PEI (10kDa) was modified by various kinds of bromo-alkylcarboxylic acids. In reality, PEI primary amine alkylcarboxylation created PEI vectors with increased lipophilicity which caused a reduction in PEI toxicity and incremental increases in gene delivery efficacy. To conjugate PEI and modified-PEI to SWCNT-PEG, -COOH groups of PEG were covalently bound to primary amines of PEI. Formation of modified SWCNT and PEI was verified by IR spectroscopy. The existence of strong absorption bands at 1645.66 cm−1 and 1657 cm−1 verified the existence of amide bonds between SWCNT-COOH/PEG and amide conjugated PEI/SWCNT-COOH-PEG. To evaluate pDNA condensation, fluorescence intensity of the NPs were investigated in the vicinity of pDNA and EtBr. A reduction in fluorescence intensity proved the condensation of pDNA through SWCNT-PEG-PEI NPs. Evaluation of the results showed that SWNT-PEG-PEI compared with SWNT-PEG-n-x%-PEI (with a lower primary amine percent) and SWCNT-PEG modified PEI could condense pDNA to lower C/P ratios and in sizes lesser than 150 nm with zeta potentials between 6.3–30.8 mV. This method enables the study of the binding ability of different forms of genetic materials to the different conjugates of MWCNTs, and also the morphology of the synthesized MWCNTs is an important parameter in the binding ability process (Figure 5).114
Figure 5.
SEM images of carbon nanotube: DNA complexes formed at a 6:1 charge ratio: (A-C) MWNT-NH3 +:DNA; (D-F) SWNT-NH3 +:DNA. Reprinted with permission from Singh R, Pantarotto D, McCarthy D, et al. Binding and condensation of plasmid DNA onto functionalized carbon nanotubes: toward the construction of nanotube-based gene delivery vectors. J Am Chem Soc. 2005;127(12):4388-4396.114,139 Copyright (2020) American Chemical Society.
Since targeted gene delivery through aptamer-functionalized NPs improves the efficacy of the antitumor treatment, targeted gene delivery of this NP was done with the 5TR1 aptamer. Image analysis demonstrated that about half of the aptamers were covalently bound to the NPs and the remainder of aptamers were conjugated electrostatically onto the surface of NPs. Cytotoxicity evaluation of the polyplexes with and without aptamers in MCF-7 (MUC1+) and MDA-MB231 (MUC1-) via MTT colorimetric testing illustrated no toxicity in these cells. The evaluation of polyplex transfection efficiencies using Renilla luciferase showed SWCNT-PEG-10–10%-PEI with a maximum transfection efficacy at a C/P ratio of 6. Finally, a SWCNT-PEG-10-10% vector was used to transfect plasmid Bcl-xL shRNA to MCF-7 and MDA-MB231 cells (breast cancer cells) and assess the expression of the Bcl-xL protein. The analysis of the obtained data indicated that unlike MDA-MB231, transfection of MCF-7 cells with plasmid Bcl-Xl shRNA by using a SWCNT-based aptamer, decreased expression of Bcl-xL to 41%. So, this novel nanocomplex including Bcl-Xl shRNA and a SWCNT-based aptamer, could be used to provoke cell death in the MCF-7 cell line and, of course, MUC1 with low toxicity, high selectively and great efficiency.115–117
A Novel Approach: Tumor-Targeted Co-Delivery Systems via Aptamer Hybrid Nanocomplexes
Cancer is the most prevalent disease throughout the world. Many efforts have been used to treat it more effectively and faster with less side effects. Although chemotherapy is the most impressive and common method to control cancer, many features (such as low specificity, development of chemotherapeutic-resistant cancer cells, etc.) that cause healthy cell cytotoxicity and damaging side effects restrict its application. Furthermore, in most cases, drug resistance generated during the chemotherapy procedure leads to the eventual failure of chemotherapy. In addition to overcoming these difficulties, gene therapy can suppress oncogene activity, increase the expression of tumor inhibitors and cause immune responses against tumors.118,119
In recent years, many scholars have focused on a better approach to combine chemotherapeutic drugs and nucleic acids (such as siRNA and shRNA) and use their synergistic treatment effects to reduce the harmful side effects and drug resistance of anti-cancer drugs. Designing a secure and efficient delivery system is a significant challenge in combinational drug and gene delivery.120,121
A novel system based on a SWCNT-PEG-10-10%-PEI nanocomplex has been designed as an efficient tumor-targeted drug-gene delivery system by a AS1411 kind of aptamer-modified with SWCNT-PEG-10-10%-PEI NPs and the therapeutic efficacy of Bcl-xL shRNA and doxorubicin (Dox, a chemotherapy medication used to treat cancer) has been evaluated.122 Agarose gel electrophoresis proved Apt-NPs conformation. Moreover, the obtained results showed that 80% of the AS1411 aptamers were covalently conjugated to the nano particles and the remainder of aptamers were electrostatically adsorbed onto their surface. The size and surface charge of the NPs and NPs-aptamer were measured. The size of the modification segment of the AS1411 aptamer, to be exact SWCNT-PEG-10-10%-PEI, with pDNA was about 80±3 nm with a surface charge of 18.15±2.14mv. After attachment, particle sizes increased to 109±1.13 nm and positive charges reduced to 10.35±2.8 mv. Immunocytochemical analysis and investigation of colored spots proved the existence of nucleolin receptors on AGS cells (a human gastric adenocarcinoma cell-line) not L929 cells (NCTC clone 929 Clone of strain L). To demonstrate the main role of the AS1411 aptamer in targeted cargo delivery of Apt-NP complexes to cancer cells, AGS cells were saturated with increased amounts of AS1411 aptamer due to the linkage of the AS1411 aptamer with AGS cell nucleolin; this aptamer uptake was very low. So, the results showed that the aptamer had an efficient effect on the tumor cells against the modification segment and proved to be the main function of the aptamer in targeted drug/gene delivery.
Bcl-xL suppression assessment was done through Western blot analysis. Results indicated that in L929 cells and AGS which were treated by the modified segment of the aptamer, expression of the Bcl-xL protein was down regulated by Bcl-xL shRNA. However, aptamer addition to SWCNT-based shRNA in L929 cells did not have any effect on Bcl-xL expression due to the lack of a nucleolin receptor on L929 cells. So, all observations and results confirmed that functionalized-CNT-Apt polyplexes could induce apoptosis in cancer cells by down-regulating the Bcl-xL gene by the use of Bcl-xL shRNA. Loading of Dox onto the SWCNT-PEG-10-10%-PE-aptamer-I/pBcl-xL shRNA surface was studied by assessing fluorescence intensity. Dox and the polyplexes combination reduced fluorescence intensity of free Dox at 595nm. Lastly, the synergistic effects of Dox and Bcl-xL shRNA into AGS cells was investigated via MTT testing. The combination of Dox (100Nm)/SWCNT-based shRNA caused a decrease in cell viability and increased cell death compared to Dox only. So, this Dox/Bcl-xL shRNA co-delivery system with high targeting ability because of the nano-needle structure of SWCNTs and aptamer conjugation to the nucleolin receptor on specific cells, caused a significant anti-tumor effect.122–124
miRNA, a small RNA molecule which plays a main role in RNA silencing and gene expression regulation, can be applied as a significant therapeutic agent in tumor gene therapy. MiR-34a, as a miRNA, is capable of inhibiting the appearance and advancement of cancer by reducing the expression levels of protein-coding genes, such as Bcl-2, survivin, etc. Also, in many types of research, it has been proven that the co-delivery of chemotherapeutic agents and miRNA through nanosystems play an effective therapeutic role both in vivo and in vitro. Scholars have attempted to create stimuli-responsive delivery systems which cause agglomeration of nanocarriers and quickly release cargos at tumor sites and within tumor cells.125–127
A novel ATP-triggered nanosystem was developed based on the ATP-responsive aptamer duplex (the aptamer and its supplementary ss-DNA) designed with Dox and a miR-34a co-delivery capacity using a PEI 25kDa vector to prevent cell proliferation and migration. A hybridization process was completed to convert the ATP aptamer and its nucleic acid sequence to a duplex. After that, Dox was loaded onto the duplex to create a Dox-Duplex. This loading process was evaluated by fluorescence spectrometry. Moreover, the ATP-responsive Dox release was completed in the vicinity of ATP. Results indicated that at a high concentration of ATP, Dox was released from Dox-Duplex. Generally, it was proven that Dox can be encapsulated and released through the duplex which will be suitable for an ATP-responsive delivery system. A PEI 25kDa carrier was applied to condense the Dox-Duplex and miR-34a and to co-deliver these two cargos. The obtained optimal mass ratio for PEI/miR-34a and PEI/Dox-Duplex was 1.5 and 2, respectively, so the ternary nanocomplex with a mass ratio of 3:1.5:2 was used in the following studies. The size and zeta potential of this optimal nanocomplex were 262.73±2.32 nm and +33.10±1.25 mv, respectively, which enabled the nanocomplex for cellular uptake and anti-tumor efficacy. Assessment of fluorescence intensity confirmed intracellular miR-34a/Dox release through PEI/Dox-Duplex/miR-34a because of the high concentration of ATP in the cytosol. In addition, it was proved which Dox was placed more in the nuclei and that miR-34a was released in both the cytosol and nuclei.
Using MTT tests to assess cell proliferation inhibition revealed that among various samples, such as PEI, miR-34a, Dox-Duplex, the two PEI/Dox-Duplex/miR-34a nanocomplexes indicated significant inhibition of cell proliferation. Furthermore, live/dead tests were completed to investigate the anti-proliferative effect. Observations proved that the amount of dead cells in the PEI/Dox-Duplex/miR-34a structure was more than the PEI/Dox-Duplex and PEI/miR-34a. Actually, the synergistic effects of Dox/miR-34a (due to the control of cancer cell proliferation through RNA interference of miR-34a and DNA intercalation of Dox) and their fast release led to significant cell proliferation inhibition. Cell apoptosis assessment was studied by V-FITC/PI and PI staining. In this case, PEI/Dox-Duplex/miR-34a nanocomplexes with a mass ratio of 3:1.5:2 and 4:1.5:2 showed high cell apoptosis compared to single cargo structures. Also, cell cycle prevention was evaluated. Results indicated that PEI/Dox-Duplex/miR-34a compared to PEI/Dox-Duplex reduced the G2 phase. So, it could be concluded that the anti-proliferative effect of the PEI/Dox-Duplex/miR-34a nanocomplex is the result of cancer cell death and its cycle arrest process. The effect of PEI/Dox-Duplex/miR-34a on the expression level of specific proteins was investigated and the results showed that Dox/miR-34a co-delivery could prevent Bcl-2 expression. Also, observations showed that the ATP-responsive co-delivery system caused significant cell migration inhibition. As a result, the PEI/Dox-Duplex/miR-34a nanocomplex with a synergistic effect and ATP-trigged release of polyloads, could have effective anti-proliferative and anti-migration efficacy.128–130
In recent years, scholars have attempted to synthesize novel nanosystems with the ability to target drug delivery and control drug release. Polymers which are sensitive to acidic surroundings and the proton sponge effect of lysosomes, speed up drug release and siRNA in tumor cells.131 For instance, a new graft polymeric micelle was synthesized as a carrier with duplex pH/redox sensitivity and a AS1411 aptamer as a targeting ligand which was loaded by gene and anticancer drugs. A chitosan-ss-PEI-urocanic acid (CSO-ss-PEI-UA, (CPU)) graft polymer was synthesized by the acylation reaction between primary amines of CSO or PEI and carboxyl groups of UA. Evaluation by 1H NMR indicated that PEI was conjugated to CSO-ss and UA was introduced into CP. In the CPU copolymer, CSO was covalently attached to PEI through disulfide bonds to reduce the positive surface charge of PEI and UA as a hydrophobic agent and was added to a loading of Dox. Moreover, to diminish the cytotoxicity of CPU micelles and enhance its safety and efficiency as a vector, the AS1411 aptamer (with specific binding to nucleolin and high expression in the membrane of cancer cells such as A549 cells) was added to the CPU micelle as a targeting ligand. 1H NMR assessment showed that the CPU copolymer and AS1411 aptamer were covalently bound together. TEM images of the AS1411-chitosan-ss-polyethylenimine-urocanic acid (ACPU) micelle showed particle sizes of 124.6±1.068 nm and an average zeta potential 24.2±0.529 mV. Also, comparison of the CD spectra of the AS1411 aptamer and ACPU confirmed that the secondary structure of the AS1411 aptamer was preserved after attachment to a CPU micelle. In addition, the low critical micelle concentration (CMC) of the ACPU copolymer showed its self-assembly into a micelle which was stable in vitro.
The pH/redox sensitivity of the ACPU micelle was assayed by checking changes in micelle size during incubation in a weakly acidic phosphate-buffered saline (PBS) and PBS plus GSH. With PBS (pH 7.4)+GSH and PBS (pH 5.1), micelles inflated from 124 nm to 365.3 and 622.6 nm, respectively. GSH caused an enhance drug and gene release through destroying disulfide bonds. The Dox release from Dox micelles and siRNA release from micelleplexes were done in pH 5.3 PBS or with GSH 10Mm. These two factors including protonation of UA in an acidic environment and breaking of disulfide bonds in reductive conditions caused the rapid release of Dox and siRNA.
Cellular uptake experiments illustrated that Dox and siRNA were delivered to cells via micelles and then released into the nucleus and cytoplasm. So, these results proved the simultaneously delivery of drugs and Dox into cells via the Dox-siRNA ACPU micelle system. To specify transfection efficiency of the siRNA-ACPU micelle, the toll-like receptor 4 (TLR4) mRNA level and protein expression were evaluated by quantitative polymerase chain reaction (qPCR) and Western blot techniques. After treatment with siRNA-ACPU micelles, the mRNA level and protein expression of TLR4 reduced. The antitumor efficacy of the Dox-siRNA ACPU micelles was evaluated by a luc-A549 cell orthotopic tumor model. The results confirmed that the tumor inhibition from Dox-siRNA ACPU micelles was very significant. Moreover, Dox-siRNA ACPU micelles down-regulated TLR4 protein levels to prevent tumor invasion and improve tumor treatment efficacy. There is hope to use this novel co-deliver nanosystem in other relevant diseases.57–59,132–134
Table 3 describes the types of aptamer-based structures and their roles in gene delivery systems.
Table 3.
Types of Aptamer-Based Structures in Gene Delivery Systems
| Aptamer-Based Structure | Targeted Cells | Agents | Results | Ref |
|---|---|---|---|---|
| RNA aptamer EpDT3 | HepG2 cells | Tumor-suppressor gene of PTEN | Increase the antitumor effect on liver cancer, decrease disadvantages of naked Ad5-PTEN, selective and accurate target ability to EpCAM-positive HepG2 cells in vivo | 135 |
| PEI- sgc-8c aptamer | MOLT-4 cells and U266 cell lines | PDNA | A six- to eight-fold increase in transfection efficiency by pDNA/PEI/sgc-8c aptamer polyplexes than the pDNA/PEI polyplex, a higher transfection efficiency without significantly induced cytotoxicity due non-covalently conjugation to PEI by electrostatic interactions | 136 |
| PEI/ARAD | HeLa cells | PDNA (p53) | Lower cell toxicity, high gene transfection efficiency | 137 |
| PEI-PEG-Wy5a Aptamer | PC-3 prostate cancer cells, DU145 cells | PDNA | PEI-PEG-Wy5a appropriate for prostate cancer-specific gene delivery, Wy5a increase uptake of the complex into the tumors than an AS1411 aptamer, formation of stable complexes with pDNA | 138 |
| 5TR1- aptamer SWCNT-PEG-PEI | MCF-7 and MDA-MB231 cell lines | Bcl-xL shRNA | 8.5–10 fold improvement in transfection activity at C/P ratios of 6 than PEI 25 kDa, low cytotoxicity and high gene delivery efficiency | 113 |
| AS1411 aptamer- SWNT-PEG-PEI | AGS and L929 cells | pBcl-xL shRNA-DOX | This targeted delivery system inhibited the progress of nucleolin-abundant gastric cancer cells with high selectivity | 122 |
| Scorpion stingers | SK-BR-3 cells, MDA-MB-231 cells | DNA nano-scorpion nanostructure (termed AptDzy-DNS) | AptDzy-DNS shows high efficiency for targeted gene therapy with good biocompatibility and no side effects | 139 |
Conclusions and Future Outlook
The scientific society is looking for new techniques to control, diagnose and treat diseases with gene therapy. Gene therapy is the treatment process of nucleic acid transference into the patient’s cells by viral or non-viral carriers to remedy faulty genes or provide further biological functions. Pathogenic diseases are some of the most prevalent diseases that threaten many people all over the world which have been created through microorganisms. One of the most important goals of cancer research is to target cancer cells specifically, which increases the efficacy of treatment and reduces the side effects of drugs and genes. Since the aptamers appeared, we have seen significant progress over the past two decades. To date, various types of anticancer drugs (such as chemotherapy drugs and siRNAs) have been successfully delivered to cancer cells in vitro. However, the successful delivery of gene-based drugs to tumor tissue after systemic injection of aptamer-based compounds has been achieved only in a low number of studies. In fact, we need more work before achieving an aptamer-based agent, including clinical trials and eventually managing daily cancer patients. The AS1411 aptamer has been applied in several phases of a clinical trial and it represents a candidate for approval by the US Food and Drug Administration (FDA).
Along with advancements in this field, the inefficient transfer of antibiotics to infected cells and controlling the side effects and the releasing rate, are considered important issues which may be solved by aptamers. Targeted therapies for both gene therapy and pathogenic diseases have been developed based on aptamer hybrid nanocomplexes in recent years to overcome these problems.
Nanotechnology will also play an important role in the delivery of genes and therapeutic agents in the future. The creation of nanostructure complexes will enhance the efficiency of aptamer transfer and delivery. The large surface area of NPs can play an important role in the binding of multiple aptamers and targets simultaneously. However, there are many challenges to delivering NPs-based genes. Delivery and increased permeability can be effective in many cases, such as liposome-based drugs that have been FDA-approved. However, the half-life of the NPs is important because if too long, certain nanoparticle chemistries may cause toxicity in different organs, requiring more optimization and research.
Generally, aptamers can form a constructive interaction between different types of structures and lead to a positive selection against any given target. The chemical structure, geometry and relative reactivity with targeted molecules makes them a cost-effective and valuable choice for the diagnosis of many diseases, therapies and array-based applications. In addition, different attachment mechanisms of aptamers with antibiotic molecules provides for precious affinity ligands, making this field of research significantly worth it.
Research Involving Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Ethics and Consent Statement
All ethical standards were followed for the construction of this review.
Disclosure
All authors declare that they have no conflicts of interest.
References
- 1.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 2.de Vries JW, Schnichels S, Hurst J, et al. DNA nanoparticles for ophthalmic drug delivery. Biomaterials. 2018;157:98–106. doi: 10.1016/j.biomaterials.2017.11.046 [DOI] [PubMed] [Google Scholar]
- 3.Rabiee N, Yaraki MT, Garakani SM, et al. Recent advances in porphyrin-based nanocomposites for effective targeted imaging and therapy. Biomaterials. 2019;119707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahrami S, Baheiraei N, Mohseni M, et al. Three-dimensional graphene foam as a conductive scaffold for cardiac tissue engineering. J Biomater Appl. 2019;34(1):74–85. doi: 10.1177/0885328219839037 [DOI] [PubMed] [Google Scholar]
- 5.Dollins CM, Nair S, Sullenger BA. Aptamers in immunotherapy. Hum Gene Ther. 2008;19(5):443–450. doi: 10.1089/hum.2008.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy M, Palliser D, Wengerter BC, Almo SC. Aptamer-Targetted Antigen Delivery. Google Patents; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayraç C, Eyidoğan F, Öktem HA. DNA aptamer-based colorimetric detection platform for Salmonella enteritidis. Biosens Bioelect. 2017;98:22–28. doi: 10.1016/j.bios.2017.06.029 [DOI] [PubMed] [Google Scholar]
- 8.Hajebi S, Rabiee N, Bagherzadeh M, et al. Stimulus-responsive polymeric nanogels as smart drug delivery systems. Acta Biomater. 2019;92:1–18. doi: 10.1016/j.actbio.2019.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmadi S, Rabiee N, Rabiee M. Application of aptamer-based hybrid molecules in early diagnosis and treatment of diabetes mellitus: from the concepts towards the future. Curr Diabetes Rev. 2019;15(4):309–313. doi: 10.2174/1573399814666180607075550 [DOI] [PubMed] [Google Scholar]
- 10.Gooch J, Daniel B, Parkin M, Frascione N. Developing aptasensors for forensic analysis. TrAC Trends Anal Chem. 2017;94:150–160. doi: 10.1016/j.trac.2017.07.019 [DOI] [Google Scholar]
- 11.Nussbaum O, Oz MB, Tilayov T, Atiya H, Dagan S. A signal amplification probe enhances sensitivity of antibodies and aptamers based Immuno-diagnostic assays. J Immunol Methods. 2017;448:85–90. doi: 10.1016/j.jim.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 12.Nour S, Baheiraei N, Imani R, et al. Bioactive materials: a comprehensive review on interactions with biological microenvironment based on the immune response. J Bionic Eng. 2019;16(4):563–581. doi: 10.1007/s42235-019-0046-z [DOI] [Google Scholar]
- 13.Nour S, Baheiraei N, Imani R, et al. A review of accelerated wound healing approaches: biomaterial-assisted tissue remodeling. J Mater Sci. 2019;30(10):120. [DOI] [PubMed] [Google Scholar]
- 14.Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O’toole MG, Trent JO. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: uses and mechanisms. Biochimica Et Biophysica Acta (BBA)-General Subj. 2017;1861(5):1414–1428. doi: 10.1016/j.bbagen.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 15.Howard PT. Investigating the Mechanism of Novel Anticancer Agent, AS1411: Does Metabolism to Guanine Play a Role? 2017. [Google Scholar]
- 16.Feng S, Chen C, Wang W, Que L. An aptamer nanopore-enabled microsensor for detection of theophylline. Biosens Bioelect. 2018;105:36–41. doi: 10.1016/j.bios.2018.01.016 [DOI] [PubMed] [Google Scholar]
- 17.Pützer BM, Solanki M, Herchenröder O. Advances in cancer stem cell targeting: how to strike the evil at its root. Adv Drug Deliv Rev. 2017;120:89–107. doi: 10.1016/j.addr.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 18.Tan KX, Danquah MK, Sidhu A, Lau SY, Ongkudon CM. Biophysical characterization of layer‐by‐layer synthesis of aptamer‐drug microparticles for enhanced cell targeting. Biotechnol Prog. 2017. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Ma T, Ma S, et al. Fluorometric determination of the antibiotic kanamycin by aptamer-induced FRET quenching and recovery between MoS 2 nanosheets and carbon dots. Microchimica Acta. 2017;184(1):203–210. doi: 10.1007/s00604-016-2011-4 [DOI] [Google Scholar]
- 20.Jeong S, Rhee Paeng I. Sensitivity and selectivity on aptamer-based assay: the determination of tetracycline residue in bovine milk. ScientificWorldJournal. 2012;159456(10):1. doi: 10.1100/2012/159456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hori SI, Herrera A, Rossi JJ, Zhou J. Current advances in aptamers for cancer diagnosis and therapy. Cancers. 2018;10:1. doi: 10.3390/cancers10010009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Wang Q, Zhang H, et al. Characterization of a DNA aptamer for ovarian cancer clinical tissue recognition and in vivo imaging. Cell Physiol Biochem. 2018;51(6):2564–2574. doi: 10.1159/000495925 [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Zhou S, Cai Y, Tang F. Nucleic acid aptamer application in diagnosis and therapy of colorectal cancer based on cell-SELEX technology. NPJ Prec Oncol. 2017;1(1):37. doi: 10.1038/s41698-017-0041-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Chen J, Wu M, Zhao JX. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics. 2015;5(4):322. doi: 10.7150/thno.10257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucl Acids. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Li Z, Hu D, Lin C-T, Li J, Lin Y. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc. 2010;132(27):9274–9276. doi: 10.1021/ja103169v [DOI] [PubMed] [Google Scholar]
- 27.Beals N, Thiagarajan PS, Soehnlen E, et al. Five-part pentameric nanocomplex shows improved efficacy of doxorubicin in CD44+ cancer cells. ACS Omega. 2017;2(11):7702–7713. doi: 10.1021/acsomega.7b01168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitsche A, Kurth A, Dunkhorst A, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007;7(1):48. doi: 10.1186/1472-6750-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv. 2015;33(6):1141–1161. doi: 10.1016/j.biotechadv.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 30.Han K, Liang Z, Zhou N. Design strategies for aptamer-based biosensors. Sensors. 2010;10(5):4541–4557. doi: 10.3390/s100504541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin B, Wang S, Lin M, et al. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasensitive bacteria detection. Biosens Bioelect. 2017;90:525–533. doi: 10.1016/j.bios.2016.10.029 [DOI] [PubMed] [Google Scholar]
- 32.Triebenbach AN, Vogl SJ, Leda Lotspeich-Cole DS, Sikes GM, Happ HK. Detection of Francisella tularensis in Alaskan Mosquitoes (Diptera: culicidae) and assessment of a laboratory model for transmission. J Med Entomol. 2010;47(4):639–648. doi: 10.1093/jmedent/47.4.639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vivekananda J, Kiel JL. Methods and Compositions for Aptamers Against Anthrax. Google Patents; 2003. [Google Scholar]
- 34.Duan N, Wu S, Chen X, Huang Y, Wang Z. Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J Agric Food Chem. 2012;60(16):4034–4038. doi: 10.1021/jf300395z [DOI] [PubMed] [Google Scholar]
- 35.Li P, Yu Q, Zhou L, et al. Probing and characterizing the high specific sequences of ssDNA aptamer against SGIV-infected cells. Virus Res. 2018. [DOI] [PubMed] [Google Scholar]
- 36.Kong C, Wang Y, Fodjo EK, Yang G-X, Han F, Shen X-S. Loop-mediated isothermal amplification for visual detection of Vibrio parahaemolyticus using gold nanoparticles. Microchimica Acta. 2018;185(1):35. doi: 10.1007/s00604-017-2594-4 [DOI] [PubMed] [Google Scholar]
- 37.Ramlal S, Mondal B, Lavu PS, Bhavanashri N, Kingston J. Capture and detection of Staphylococcus aureus with dual labeled aptamers to cell surface components. Int J Food Microbiol. 2018;265:74–83. doi: 10.1016/j.ijfoodmicro.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Gao T, Gao X, et al. An aptamer based sulfadimethoxine assay that uses magnetized upconversion nanoparticles. Microchimica Acta. 2017;184(9):3557–3563. doi: 10.1007/s00604-017-2378-x [DOI] [Google Scholar]
- 39.Luzzago C, Locatelli C, Franco A, et al. Clonal diversity, virulence-associated genes and antimicrobial resistance profile of Staphylococcus aureus isolates from nasal cavities and soft tissue infections in wild ruminants in Italian Alps. Vet Microbiol. 2014;170(1–2):157–161. doi: 10.1016/j.vetmic.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Duan N, Shi Z, Fang C, Wang Z. Simultaneous aptasensor for multiplex pathogenic bacteria detection based on multicolor upconversion nanoparticles labels. Anal Chem. 2014;86(6):3100–3107. doi: 10.1021/ac404205c [DOI] [PubMed] [Google Scholar]
- 41.Medley CD, Bamrungsap S, Tan W, Smith JE. Aptamer-conjugated nanoparticles for cancer cell detection. Anal Chem. 2011;83(3):727–734. doi: 10.1021/ac102263v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu W, Teng I-T, Chen X, Tan W, Veige AS. Aptamer-mediated selective delivery of a cytotoxic cationic NHC-Au (I) complex to cancer cells. Dalton Trans. 2018;47(1):120–126. doi: 10.1039/C7DT02616A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng D, Yu M, Fu F, et al. Dual recognition strategy for specific and sensitive detection of bacteria using aptamer-coated magnetic beads and antibiotic-capped gold nanoclusters. Anal Chem. 2015;88(1):820–825. doi: 10.1021/acs.analchem.5b03320 [DOI] [PubMed] [Google Scholar]
- 44.Charoenphol P, Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol Pharm. 2014;11(5):1721–1725. doi: 10.1021/mp500047b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun T-M, Du J-Z, Yao Y-D, et al. Simultaneous delivery of siRNA and paclitaxel via a “Two-in-one” micelleplex promotes synergistic tumor suppression. ACS Nano. 2011;5(2):1483–1494. doi: 10.1021/nn103349h [DOI] [PubMed] [Google Scholar]
- 46.Farokhzad OC, Jon S, Khademhosseini A, Tran T-NT, LaVan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–7672. doi: 10.1158/0008-5472.CAN-04-2550 [DOI] [PubMed] [Google Scholar]
- 47.Pang X, Cui C, Wan S, et al. Bioapplications of Cell-SELEX-generated aptamers in cancer diagnostics, therapeutics, theranostics and biomarker discovery: a comprehensive review. Cancers. 2018;10(2):47. doi: 10.3390/cancers10020047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bamrungsap S, Chen T, Shukoor MI, et al. Pattern recognition of cancer cells using aptamer-conjugated magnetic nanoparticles. ACS Nano. 2012;6(5):3974–3981. doi: 10.1021/nn3002328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bidard F-C, Fehm T, Ignatiadis M, et al. Clinical application of circulating tumor cells in breast cancer: overview of the current interventional trials. Cancer Metastasis Rev. 2013;32(1–2):179–188. doi: 10.1007/s10555-012-9398-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203 [DOI] [PubMed] [Google Scholar]
- 51.Riethdorf S, O’Flaherty L, Hille C, Pantel K. Clinical applications of the CellSearch platform in cancer patients. Adv Drug Deliv Rev. 2018;125:102–121. doi: 10.1016/j.addr.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 52.Jo H, Her J, Ban C. Dual aptamer-functionalized silica nanoparticles for the highly sensitive detection of breast cancer. Biosens Bioelect. 2015;71:129–136. doi: 10.1016/j.bios.2015.04.030 [DOI] [PubMed] [Google Scholar]
- 53.Mittal S, Kaur H, Gautam N, Mantha AK. Biosensors for breast cancer diagnosis: a review of bioreceptors, biotransducers and signal amplification strategies. Biosens Bioelect. 2017;88:217–231. doi: 10.1016/j.bios.2016.08.028 [DOI] [PubMed] [Google Scholar]
- 54.Bernaus A, Gaona X, Esbrí JM, Higueras P, Falkenberg G, Valiente M. Microprobe techniques for speciation analysis and geochemical characterization of mine environments: the mercury district of Almadén in Spain. Environ Sci Technol. 2006;40(13):4090–4095. doi: 10.1021/es052392l [DOI] [PubMed] [Google Scholar]
- 55.Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kojima C, Ono A. 15N− 15N J-coupling across HgII: direct observation of HgII-mediated T− T base pairs in a DNA duplex. J Am Chem Soc. 2007;129(2):244–245. doi: 10.1021/ja065552h [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y, Xie X. A novel electrochemical aptamer biosensor based on DNAzyme decorated Au@ Ag core-shell nanoparticles for Hg2+ determination. J Braz Chem Soc. 2018;29(2):232–239. [Google Scholar]
- 57.Tan D, He Y, Xing X, Zhao Y, Tang H, Pang D. Aptamer functionalized gold nanoparticles based fluorescent probe for the detection of mercury (II) ion in aqueous solution. Talanta. 2013;113:26–30. doi: 10.1016/j.talanta.2013.03.055 [DOI] [PubMed] [Google Scholar]
- 58.Farzin L, Shamsipur M, Sheibani S. A review: aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta. 2017;174:619–627. doi: 10.1016/j.talanta.2017.06.066 [DOI] [PubMed] [Google Scholar]
- 59.Lan L, Yao Y, Ping J, Ying Y. Recent progress in nanomaterial-based optical aptamer assay for the detection of food chemical contaminants. ACS Appl Mater Interfaces. 2017;9(28):23287–23301. doi: 10.1021/acsami.7b03937 [DOI] [PubMed] [Google Scholar]
- 60.Tian J. Aptamer-based colorimetric detection of various targets based on catalytic Au NPs/Graphene nanohybrids. Sens Bio-Sensing Res. 2019;22:100258. doi: 10.1016/j.sbsr.2019.100258 [DOI] [Google Scholar]
- 61.Giorgi-Coll S, Marín MJ, Sule O, Hutchinson PJ, Carpenter KLH. Aptamer-modified gold nanoparticles for rapid aggregation-based detection of inflammation: an optical assay for interleukin-6. Microchimica Acta. 2019;187(1):13. doi: 10.1007/s00604-019-3975-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava M, Nirala NR, Srivastava SK, Prakash R. A comparative study of aptasensor vs immunosensor for label-free PSA cancer detection on GQDs-AuNRs modified screen-printed electrodes. Sci Rep. 2018;8(1):1923. doi: 10.1038/s41598-018-19733-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen H, Wang J, Liu H, et al. Rapid and selective detection of pathogenic bacteria in bloodstream infections with aptamer-based recognition. ACS Appl Mater Interfaces. 2016;8(30):19371–19378. doi: 10.1021/acsami.6b06671 [DOI] [PubMed] [Google Scholar]
- 64.Park JY, Jeong HY, Kim MI. Colorimetric detection system for salmonella typhimurium based on peroxidase-like activity of magnetic nanoparticles with DNA aptamers. J Nanomater. 2015;2015. [Google Scholar]
- 65.Ozyurt C, Bora B, Ugurlu O, Evran S. Chapter 18 - Pathogen-specific nucleic acid aptamers as targeting components of antibiotic and gene delivery systems In: Andronescu E, Grumezescu AM, editors. Nanostructures for Drug Delivery. Elsevier; 2017:551–577. [Google Scholar]
- 66.Lee B, Park J, Ryu M, et al. Antimicrobial peptide-loaded gold nanoparticle-DNA aptamer conjugates as highly effective antibacterial therapeutics against Vibrio vulnificus. Sci Rep. 2017;7(1):13572. doi: 10.1038/s41598-017-14127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu C-L, Wang X-W, Lin -Z-Z, Xie Z-H, Wang X-R. Cell microenvironment stimuli-responsive controlled-release delivery systems based on mesoporous silica nanoparticles. J Food Drug Anal. 2014;22(1):18–28. doi: 10.1016/j.jfda.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ladju RB, Pascut D, Massi MN, Tiribelli C, Sukowati CH. Aptamer: a potential oligonucleotide nanomedicine in the diagnosis and treatment of hepatocellular carcinoma. Oncotarget. 2018;9(2):2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shigdar S. What potential do aptamers hold in therapeutic delivery? Future Sci. 2017. [DOI] [PubMed] [Google Scholar]
- 71.Kavruk M, Celikbicak O, Ozalp V, et al. Antibiotic loaded nanocapsules functionalized with aptamer gates for targeted destruction of pathogens. Chem Commun. 2015;51(40):8492–8495. doi: 10.1039/C5CC01869B [DOI] [PubMed] [Google Scholar]
- 72.Sancenón F, Yu E, Aznar E, Marcos MD, Martínez-Máñez R. Gated porous materials for biomedical applications. Drug Del Syst. 2017;1:113. [Google Scholar]
- 73.Zhu C-L., Lu C-H., Song X-Y., Yang H-H., Wang X-R.. Bioresponsive controlled release using mesoporous silica nanoparticles capped with aptamer-based molecular gate. J Am Chem Soc. 2011;133(5):1278-1281. doi: 10.1021/ja110094g [DOI] [PubMed] [Google Scholar]
- 74.Elsabahy M, Nazarali A, Foldvari M. Non-viral nucleic acid delivery: key challenges and future directions. Curr Drug Deliv. 2011;8(3):235–244. doi: 10.2174/156720111795256174 [DOI] [PubMed] [Google Scholar]
- 75.Sadeqzadeh E, Rahbarizadeh F, Ahmadvand D, Rasaee MJ, Parhamifar L, Moghimi SM. Combined MUC1-specific nanobody-tagged PEG-polyethylenimine polyplex targeting and transcriptional targeting of tBid transgene for directed killing of MUC1 over-expressing tumour cells. J Controlled Release. 2011;156(1):85–91. doi: 10.1016/j.jconrel.2011.06.022 [DOI] [PubMed] [Google Scholar]
- 76.Khaleghi S, Rahbarizadeh F, Ahmadvand D, Hosseini HRM. Anti-HER2 VHH targeted magnetoliposome for intelligent magnetic resonance imaging of breast cancer cells. Cell Mol Bioeng. 2017;10(3):263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin L, Yuvienco C, Montclare JK. Protein based therapeutic delivery agents: contemporary developments and challenges. Biomaterials. 2017;134:91–116. doi: 10.1016/j.biomaterials.2017.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lam P, Steinmetz NF. Plant viral and bacteriophage delivery of nucleic acid therapeutics. Wiley Interdiscipl Rev. 2018;10:1. [DOI] [PubMed] [Google Scholar]
- 79.Pan J, Ruan W, Qin M, et al. Intradermal delivery of STAT3 siRNA to treat melanoma via dissolving microneedles. Sci Rep. 2018;8(1):1117. doi: 10.1038/s41598-018-19463-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li L, Hou J, Liu X, et al. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials. 2014;35(12):3840–3850. doi: 10.1016/j.biomaterials.2014.01.019 [DOI] [PubMed] [Google Scholar]
- 81.Wu S, Li J, Liang H, et al. Aptamer-based self-assembled supramolecular vesicles for pH-responsive targeted drug delivery. Sci China Chem. 2017;60(5):628–634. doi: 10.1007/s11426-016-0351-5 [DOI] [Google Scholar]
- 82.Bagalkot V, Gao X. siRNA-Aptamer Chimeras on Nanoparticles: Preserving Targeting Functionality for Effective Gene Silencing. ACS Nano. 2011;5(10):8131-8139. doi: 10.1021/nn202772p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19(7):1448–1455. doi: 10.1021/bc800065f [DOI] [PubMed] [Google Scholar]
- 84.Nimesh S, Aggarwal A, Kumar P, Singh Y, Gupta K, Chandra R. Influence of acyl chain length on transfection mediated by acylated PEI nanoparticles. Int J Pharm. 2007;337(1–2):265–274. doi: 10.1016/j.ijpharm.2006.12.032 [DOI] [PubMed] [Google Scholar]
- 85.Mazur J, Roy K, Kanwar JR. Recent advances in nanomedicine and survivin targeting in brain cancers. Nanomedicine. 2018;13(1):105–137. doi: 10.2217/nnm-2017-0286 [DOI] [PubMed] [Google Scholar]
- 86.Urmann K, Modrejewski J, Scheper T, Walter J-G. Aptamer-modified nanomaterials: principles and applications. BioNanoMaterials. 2017;18:1–2. [Google Scholar]
- 87.Kurosaki T, Higuchi N, Kawakami S, et al. Self-assemble gene delivery system for molecular targeting using nucleic acid aptamer. Gene. 2012;491(2):205–209. doi: 10.1016/j.gene.2011.09.021 [DOI] [PubMed] [Google Scholar]
- 88.Demeneix B, Behr JP. Polyethylenimine (PEI). Adv Genet. 2005;53:215–230. [DOI] [PubMed] [Google Scholar]
- 89.Qu J, Yu S, Zheng Y, Zheng Y, Yang H, Zhang J. Aptamer and its applications in neurodegenerative diseases. Cell Mol Life Sci. 2017;74(4):683–695. doi: 10.1007/s00018-016-2345-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Landi A, Babiuk LA, van Drunen Littel-van den Hurk S. High transfection efficiency, gene expression, and viability of monocyte-derived human dendritic cells after nonviral gene transfer. J Leukoc Biol. 2007;82(4):849–860. doi: 10.1189/jlb.0906561 [DOI] [PubMed] [Google Scholar]
- 91.Shahidi‐Hamedani N, Shier WT, Moghadam Ariaee F, Abnous K, Ramezani M. Targeted gene delivery with noncovalent electrostatic conjugates of sgc‐8c aptamer and polyethylenimine. J Gene Med. 2013;15(6–7):261–269. [DOI] [PubMed] [Google Scholar]
- 92.Askarian S, Abnous K, Ayatollahi S, Farzad SA, Oskuee RK, Ramezani M. PAMAM-pullulan conjugates as targeted gene carriers for liver cell. Carbohydr Polym. 2017;157:929–937. doi: 10.1016/j.carbpol.2016.10.030 [DOI] [PubMed] [Google Scholar]
- 93.Ozyurt C, Bora B, Ugurlu O, Evran S. Pathogen-specific nucleic acid aptamers as targeting components of antibiotic and gene delivery systems. Nanostruct Drug Del. 2017;551–577. [Google Scholar]
- 94.Cheng J, Teply BA, Sherifi I, et al. Formulation of functionalized PLGA–PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28(5):869–876. doi: 10.1016/j.biomaterials.2006.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elminger MW, Bell M, Schüett BS, Langkamp M, Kutoh E, Ranke MB. Transactivation of the IGFBP-2 promoter in human tumor cell lines. Mol Cell Endocrinol. 2001;175(1–2):211–218. doi: 10.1016/S0303-7207(00)00454-8 [DOI] [PubMed] [Google Scholar]
- 96.Reslan L, Mestas J-L, Herveau S, Béra J-C, Dumontet C. Transfection of cells in suspension by ultrasound cavitation. J Controlled Release. 2010;142(2):251–258. doi: 10.1016/j.jconrel.2009.10.029 [DOI] [PubMed] [Google Scholar]
- 97.Emami J, Maghzi P, Hasanzadeh F, Sadeghi H, Mirian M, Rostami M. PLGA-PEG-RA-based polymeric micelles for tumor targeted delivery of irinotecan. Pharm Dev Technol. 2018;23(1):41–54. doi: 10.1080/10837450.2017.1340950 [DOI] [PubMed] [Google Scholar]
- 98.Subramanian N, Kanwar JR, Athalya P, et al. EpCAM aptamer mediated cancer cell specific delivery of EpCAM siRNA using polymeric nanocomplex. J Biomed Sci. 2015;22(1):4. doi: 10.1186/s12929-014-0108-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sarrach S, Huang Y, Niedermeyer S, et al. Spatiotemporal patterning of EpCAM is important for murine embryonic endo-and mesodermal differentiation. Sci Rep. 2018;8(1):1801. doi: 10.1038/s41598-018-20131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Castilla C, Congregado B, Chinchón D, Torrubia FJ, Japón MA, Sáez C. Bcl-xL is overexpressed in hormone-resistant prostate cancer and promotes survival of LNCaP cells via interaction with proapoptotic Bak. Endocrinology. 2006;147(10):4960–4967. doi: 10.1210/en.2006-0502 [DOI] [PubMed] [Google Scholar]
- 101.Beck-Broichsitter M, Merkel OM, Kissel T. Controlled pulmonary drug and gene delivery using polymeric nano-carriers. J Controlled Release. 2012;161(2):214–224. doi: 10.1016/j.jconrel.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Zhou Y, Wu M, et al. Functional nanocarrier for drug and gene delivery via local administration in mucosal tissues. Nanomedicine. 2018;13(1):69–88. doi: 10.2217/nnm-2017-0143 [DOI] [PubMed] [Google Scholar]
- 103.Yamazoe H. Multifunctional protein microparticles for medical applications. Biomaterials. 2018;155:1–12. doi: 10.1016/j.biomaterials.2017.10.045 [DOI] [PubMed] [Google Scholar]
- 104.Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. A mesoporous silica nanoparticle–PEI–fusogenic peptide system for siRNA delivery in cancer therapy. Biomaterials. 2013;34(4):1391–1401. doi: 10.1016/j.biomaterials.2012.10.072 [DOI] [PubMed] [Google Scholar]
- 105.Yin Q, Gao Y, Zhang Z, Zhang P, Li Y. Bioreducible poly (β-amino esters)/shRNA complex nanoparticles for efficient RNA delivery. J Controlled Release. 2011;151(1):35–44. doi: 10.1016/j.jconrel.2010.12.014 [DOI] [PubMed] [Google Scholar]
- 106.Parhiz H, Hashemi M, Hatefi A, Shier WT, Farzad SA, Ramezani M. Arginine-rich hydrophobic polyethylenimine: potent agent with simple components for nucleic acid delivery. Int J Biol Macromol. 2013;60:18–27. doi: 10.1016/j.ijbiomac.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 107.Nia AH, Behnam B, Taghavi S, et al. Evaluation of chemical modification effects on DNA plasmid transfection efficiency of single-walled carbon nanotube–succinate–polyethylenimine conjugates as non-viral gene carriers. MedChemComm. 2017;8(2):364–375. doi: 10.1039/C6MD00481D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kesharwani P, Gajbhiye V, Jain NK. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials. 2012;33(29):7138–7150. doi: 10.1016/j.biomaterials.2012.06.068 [DOI] [PubMed] [Google Scholar]
- 109.Moradian H, Fasehee H, Keshvari H, Faghihi S. Poly (ethyleneimine) functionalized carbon nanotubes as efficient nano-vector for transfecting mesenchymal stem cells. Colloids Surf B. 2014;122:115–125. doi: 10.1016/j.colsurfb.2014.06.056 [DOI] [PubMed] [Google Scholar]
- 110.Mohammadi M, Salmasi Z, Hashemi M, Mosaffa F, Abnous K, Ramezani M. Single-walled carbon nanotubes functionalized with aptamer and piperazine–polyethylenimine derivative for targeted siRNA delivery into breast cancer cells. Int J Pharm. 2015;485(1):50–60. doi: 10.1016/j.ijpharm.2015.02.031 [DOI] [PubMed] [Google Scholar]
- 111.Subramanian N, Raghunathan V, Kanwar JR, et al. Target-specific delivery of doxorubicin to retinoblastoma using epithelial cell adhesion molecule aptamer. Mol Vis. 2012;18:2783. [PMC free article] [PubMed] [Google Scholar]
- 112.Macdonald J, Henri J, Roy K, et al. EpCAM immunotherapy versus specific targeted delivery of drugs. Cancers. 2018;10(1):19. doi: 10.3390/cancers10010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taghavi S, HashemNia A, Mosaffa F, Askarian S, Abnous K, Ramezani M. Preparation and evaluation of polyethylenimine-functionalized carbon nanotubes tagged with 5TR1 aptamer for targeted delivery of Bcl-xL shRNA into breast cancer cells. Colloids Surf B. 2016;140:28–39. doi: 10.1016/j.colsurfb.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 114.Singh R, Pantarotto D, McCarthy D, et al. Binding and condensation of plasmid DNA onto functionalized carbon nanotubes: toward the construction of nanotube-based gene delivery vectors. J Am Chem Soc. 2005;127(12):4388–4396. [DOI] [PubMed] [Google Scholar]
- 115.Ferreira C, Matthews C, Missailidis S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biol. 2006;27(6):289–301. doi: 10.1159/000096085 [DOI] [PubMed] [Google Scholar]
- 116.Yang B, Chen B, He M, Yin X, Xu C, Hu B. An aptamer-based dual-functional probe for rapid and specific counting and imaging of MCF-7 cells. Anal Chem. 2018. [DOI] [PubMed] [Google Scholar]
- 117.Charbgoo F, Alibolandi M, Taghdisi SM, Abnous K, Soltani F, Ramezani M. MUC1 aptamer-targeted DNA micelles for dual tumor therapy using doxorubicin and KLA peptide. Nanomedicine. 2018;14(3):685–697. doi: 10.1016/j.nano.2017.12.010 [DOI] [PubMed] [Google Scholar]
- 118.Ebrahimian M, Taghavi S, Mokhtarzadeh A, Ramezani M, Hashemi M. Co-delivery of doxorubicin encapsulated PLGA nanoparticles and Bcl-xL shRNA using alkyl-modified PEI into breast cancer cells. Appl Biochem Biotechnol. 2017;183(1):126–136. doi: 10.1007/s12010-017-2434-3 [DOI] [PubMed] [Google Scholar]
- 119.Afsharzadeh M, Hashemi M, Mokhtarzadeh A, Abnous K, Ramezani M. Recent advances in co-delivery systems based on polymeric nanoparticle for cancer treatment. Artif Cells Nanomed Biotechnol. 2017;1–16. [DOI] [PubMed] [Google Scholar]
- 120.Zhang RX, Ahmed T, Li LY, Li J, Abbasi AZ, Wu XY. Design of nanocarriers for nanoscale drug delivery to enhance cancer treatment using hybrid polymer and lipid building blocks. Nanoscale. 2017;9(4):1334–1355. doi: 10.1039/C6NR08486A [DOI] [PubMed] [Google Scholar]
- 121.Ståhl S, Gräslund T, Karlström AE, Frejd FY, Nygren P-Å, Löfblom J. Affibody molecules in biotechnological and medical applications. Trends Biotechnol. 2017;35(8):691–712. doi: 10.1016/j.tibtech.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 122.Taghavi S, Nia AH, Abnous K, Ramezani M. Polyethylenimine-functionalized carbon nanotubes tagged with AS1411 aptamer for combination gene and drug delivery into human gastric cancer cells. Int J Pharm. 2017;516(1–2):301–312. doi: 10.1016/j.ijpharm.2016.11.027 [DOI] [PubMed] [Google Scholar]
- 123.Tyagi P, Subramony JA. Nanotherapeutics in oral and parenteral drug delivery: key learnings and future outlooks as we think small. J Controlled Release. 2018;272:159–168. doi: 10.1016/j.jconrel.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 124.Gou Y, Zhang Z, Li D, et al. HSA-based multi-target combination therapy: regulating drugs’ release from HSA and overcoming single drug resistance in a breast cancer model. Drug Deliv. 2018;25(1):321–329. doi: 10.1080/10717544.2018.1428245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.D’ANGELO B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36(11):5571–5575. doi: 10.21873/anticanres.11142 [DOI] [PubMed] [Google Scholar]
- 126.Duan Y, Xing Z, Yang J, et al. Chondroitin sulfate-functionalized polyamidoamine-mediated miR-34a delivery for inhibiting the proliferation and migration of pancreatic cancer. RSC Adv. 2016;6(75):70870–70876. doi: 10.1039/C6RA15716E [DOI] [Google Scholar]
- 127.Wu D, Wang C, Yang J, et al. Improving the intracellular drug concentration in lung cancer treatment through the codelivery of doxorubicin and miR-519c mediated by porous PLGA microparticle. Mol Pharm. 2016;13(11):3925–3933. doi: 10.1021/acs.molpharmaceut.6b00702 [DOI] [PubMed] [Google Scholar]
- 128.Wang Y, Chen J, Liang X, et al. An ATP-responsive codelivery system of doxorubicin and miR-34a to synergistically inhibit cell proliferation and migration. Mol Pharm. 2017;14(7):2323–2332. doi: 10.1021/acs.molpharmaceut.7b00184 [DOI] [PubMed] [Google Scholar]
- 129.Tang Q, Ma X, Zhang Y, Cai X, Xue W, Ma D. Self-sensibilized polymeric prodrug co-delivering MMP-9 shRNA plasmid for combined treatment of tumors. Acta Biomater. 2018;69:277–289. doi: 10.1016/j.actbio.2018.01.014 [DOI] [PubMed] [Google Scholar]
- 130.Yang Y, Xie X, Yang Y, et al. Polymer nanoparticles modified with photo-and pH-dual-responsive polypeptides for enhanced and targeted cancer therapy. Mol Pharm. 2016;13(5):1508–1519. doi: 10.1021/acs.molpharmaceut.5b00977 [DOI] [PubMed] [Google Scholar]
- 131.Yu S, He C, Lv Q, et al. pH and reduction-sensitive disulfide cross-linked polyurethane micelles for bio-triggered anti-tumor drug delivery. J Controlled Release. 2015;213:e99–e100. doi: 10.1016/j.jconrel.2015.05.166 [DOI] [PubMed] [Google Scholar]
- 132.Yang S, Ren Z, Chen M, et al. Nucleolin-targeting AS1411-aptamer-modified graft polymeric micelle with Dual pH/redox sensitivity designed to enhance tumor therapy through the codelivery of doxorubicin/TLR4 siRNA and suppression of invasion. Mol Pharm. 2017. [DOI] [PubMed] [Google Scholar]
- 133.Zhang C, Yang S, Zhu W, et al. Distinctive polymer micelle designed for siRNA delivery and reversal of MDR1 gene‐dependent multidrug resistance. J Biomed Mater Res Part B. 2017;105(7):2093–2106. doi: 10.1002/jbm.b.33748 [DOI] [PubMed] [Google Scholar]
- 134.Yi Y, Wang H, Wang X, et al. Photocontrollable drug release nanosystem for multifunctional synergistic cancer therapy. acs appl mater interfaces. 2017;9(7):5847–5854. doi: 10.1021/acsami.6b15414 [DOI] [PubMed] [Google Scholar]
- 135.Liu Z, Sun X, Xiao S, et al. Characterization of aptamer-mediated gene delivery system for liver cancer therapy. Oncotarget. 2017;9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shahidi-Hamedani N, Shier WT, Moghadam Ariaee F, Abnous K, Ramezani M. Targeted gene delivery with noncovalent electrostatic conjugates of sgc-8c aptamer and polyethylenimine. J Gene Med. 2013;15(6–7):261–269. [DOI] [PubMed] [Google Scholar]
- 137.Wang G-H, Huang G-L, Zhao Y, et al. ATP triggered drug release and DNA co-delivery systems based on ATP responsive aptamers and polyethylenimine complexes. J Mater Chem B. 2016;4(21):3832–3841. doi: 10.1039/C5TB02764K [DOI] [PubMed] [Google Scholar]
- 138.Lee J, Oh J, Lee E-S, Kim Y-P, Lee M. Conjugation of prostate cancer-specific aptamers to polyethylene glycol-grafted polyethylenimine for enhanced gene delivery to prostate cancer cells. J Ind Eng Chem. 2019;73:182–191. doi: 10.1016/j.jiec.2019.01.023 [DOI] [Google Scholar]
- 139.Wu D, Zhang Y, Xu X, et al. RGD/TAT-functionalized chitosan-graft-PEI-PEG gene nanovector for sustained delivery of NT-3 for potential application in neural regeneration. Acta Biomater. 2018;72:266–277. doi: 10.1016/j.actbio.2018.03.030 [DOI] [PubMed] [Google Scholar]