Our objective was to evaluate the diagnostic yield and accuracy of the BioFire FilmArray pneumonia panel (BFPP) for identification of pathogens in lower respiratory tract specimens (n = 200) from emergency department (ED) and intensive care unit (ICU) patients at a tertiary care academic medical center. Specimens were collected between January and November 2018, from patients ≥18 years of age, and culture was performed as part of standard-of-care testing.
KEYWORDS: BioFire, quantitative PCR, syndromic testing, pneumonia
ABSTRACT
Our objective was to evaluate the diagnostic yield and accuracy of the BioFire FilmArray pneumonia panel (BFPP) for identification of pathogens in lower respiratory tract specimens (n = 200) from emergency department (ED) and intensive care unit (ICU) patients at a tertiary care academic medical center. Specimens were collected between January and November 2018, from patients ≥18 years of age, and culture was performed as part of standard-of-care testing. The BFPP identified a viral or bacterial target in 117/200 (58.5%) samples, including Staphylococcus aureus in 22% of samples and Haemophilus influenzae in 14%, and both a viral and bacterial target in 4% of samples. The most common viruses detected by BFPP were rhinovirus/enterovirus (4.5%), influenza A virus (3%), and respiratory syncytial virus (RSV) (2%). Overall, there was strong correlation between BFPP and standard methods for detection of viruses (99.2%) and bacteria (96.8%). Most bacteria (60/61 [98.4%]) detected by standard methods were also identified by BFPP, and 92 additional bacteria were identified by BFPP alone, including 22/92 (23.9%) additional S. aureus isolates and 25/92 (27.2%) H. influenzae isolates, which were more frequently discordant when detected at low concentrations (S. aureus, P < 0.001; H. influenzae, P < 0.0001) and in sputum-type specimens (S. aureus, P < 0.05). A potential limitation of the BFPP assay is the absence of fungal targets and Stenotrophomonas maltophilia, which were detected in 26 and 4 of 200 specimens, respectively. Real-time specimen analysis with BFPP has the potential to identify bacterial pathogens and resistance markers 44.2 and 56.3 h faster than culture-based methods. The BFPP is a rapid and accurate method for detection of pathogens from lower respiratory tract infections.
INTRODUCTION
Pneumonia is one of the most common causes of hospitalization in the United States and is one of the leading causes of death worldwide (1; www.hcup-us.ahrq.gov/faststats/landing.jsp). For bacterial pneumonia, prompt treatment with antibiotics can improve outcomes, and delay in effective antimicrobial therapy is associated with increased mortality and length of hospital stay (2–4). Current guidelines recommend empirical use of antimicrobial therapy among patients with suspected pneumonia (5). While empirical antibiotics are critical for patients with pneumonia, the extensive use of antibiotics among patients with respiratory conditions threatens to erode the efficacy of these medications. Recent estimates suggest that half of antibiotic prescriptions for acute respiratory conditions are unnecessary, primarily in the setting where antibiotics are given for viral infections (6). Unnecessary antibiotic use has led to a rise in number of antibiotic-resistant infections, which now affect over 2.8 million people each year and result in over 35,000 deaths annually (7). Rapid diagnostic testing for pneumonia has the potential to guide clinical decisions and reduce the use of broad-spectrum antibiotics; however, these benefits are dependent upon the accuracy of these test methods.
The BioFire FilmArray pneumonia panel (BFPP) was recently FDA cleared (November 2018) for detection and identification of multiple respiratory viral and bacterial pathogens in addition to selected antimicrobial resistance genes from sputum or bronchial alveolar lavage (BAL)-like specimens from individuals with suspected lower respiratory tract (LRT) infections. This assay includes targets for 18 bacteria and 8 viruses that commonly cause pneumonia as well as 7 antibiotic resistance genes. The BFPP assay reports the presence or absence of 3 atypical bacteria and provides a semiquantitative organism abundance for the other 15 bacteria, analogous to the semiquantitative reporting provided with traditional culture-based testing. Although this is a novel and potentially useful feature for a PCR-based method, data on the correlation of this method with conventional cultures are scarce, and it is yet unknown how these results might be interpreted and acted upon clinically.
The objective of our investigation was to evaluate the performance characteristics and potential clinical impact of the BFPP in the context of a large academic medical center.
(This study was presented in part at the June 2019 ASM Microbe meeting in San Francisco, CA [presentation number AAR-725], and at the May 2019 Academy of Clinical Laboratory Physicians and Scientists annual meeting in Salt Lake City, UT.)
MATERIALS AND METHODS
Sample selection.
This study was conducted at Barnes Jewish Hospital, a 1,250-bed, tertiary care, academic medical center located in St. Louis, MO. Following approval from the Institutional Review Board (IRB) of Washington University in St. Louis, consecutively available samples were enrolled and frozen at –80°C based on the following inclusion criteria: (i) an aerobic lower respiratory bacterial culture was ordered and performed as part of standard-of-care (SOC) testing, (ii) the specimen was acceptable for culture based on specimen type Gram stain criteria, (iii) the sample was stored at refrigerator temperatures for <24 h prior to freezing, (iv) adequate residual volume (≥1.5 ml) was available from aerobic culture, and (v) a lower respiratory sample was collected in the emergency department (ED) or intensive care unit (ICU) or from a patient care unit with immunocompromised patients at Barnes-Jewish Hospital (Fig. 1). Patients were excluded from the study based on the following criteria: (i) having a previous sample included in the study within 14 days, (ii) the patient was <18 years of age, (iii) clinical information was not available for the patient (i.e., immediately transferred to different institution), and (iv) the patient did not meet eligibility criteria as outlined above. A total of 265 patients were screened for eligibility between January and November 2018; 200 samples from 198 patients were eligible. Specimen aliquots were stored at 4°C for no more than 1 day prior to being frozen at –80°C in preparation for testing. Aliquots were frozen for a range of 71 to 213 days before being thawed and analyzed by the BFPP. Aliquots were coded and assigned a study number by an honest broker. We use the term standard of care to refer to real-world clinical data from tests that are routinely ordered in our facility for the care of individuals with suspected lower respiratory illness. We acknowledge that the standard of care may vary by region and clinical setting and that this represents real-world clinical data from our specific setting.
FIG 1.
Study design. A total of 200 consecutively available lower respiratory tract samples were collected between January and November 2018. Samples were from adult patients with respiratory symptoms at a large tertiary care academic medical center. Remnant samples were stored at –80°C prior to being tested with the BFPP on the BioFire FilmArray 2.0. Antibiotic utilization and standard-of-care test results were obtained by retrospective chart review, and these data were used to determine turnaround time and test performance characteristics.
BioFire FilmArray pneumonia panel.
Frozen (–80°C) lower respiratory tract (LRT) specimens were analyzed on the BioFire FilmArray 2.0 using the BioFire FilmArray pneumonia panel. Testing was performed in accordance with the manufacturer’s instructions in a single research laboratory by qualified personnel. In brief, the BioFire pouch was removed from packaging, inserted into the pouch loading station, and hydrated with the manufacturer-supplied hydration solution. The BAL-like or sputum-like specimen was then mixed with sample buffer and was injected into the BioFire pouch. The pouch was scanned and loaded into the BioFire FilmArray 2.0, and the run was initiated. The BFPP includes two process controls in each pouch, which must both be positive for a run to pass. Runs that failed these internal process controls in our study were repeated with a new pouch. In the event of an invalid or indeterminate result, specimen testing was repeated once. BFPP results were for internal study purposes only and were not reported to members of the patient’s care team.
Bacterial culture.
Bacterial cultures were performed as part of SOC testing in the Clinical Microbiology Laboratory at Barnes-Jewish Hospital.
Sputum.
Sputum specimens were evaluated by unconcentrated conventional Gram stain prior to plating. Specimens with over 10 epithelial cells per low-power field on conventional Gram stain were considered to have excessive oral flora. These samples were rejected and culture was not performed, making these specimens ineligible for inclusion in this study. On average, 30% of sputum specimens submitted to the laboratory are rejected based on Gram stain characteristics. Acceptable sputum specimens were plated on Trypticase soy agar with 5% sheep blood (BAP), chocolate agar (CHOC), and MacConkey agar (MAC) from Remel (Lenexa, KS), were streaked for isolation using the quadrant streak method, and were incubated at 35°C in 5% CO2. All plates were examined for growth at 24 h of incubation. MAC plates without growth at 18 to 24 h were discarded. BAP and CHOC plates were reexamined at 48 h and were discarded at that time. Bacterial growth from sputum samples was reported qualitatively based on the following criteria: “rare,” 10 colonies or fewer in the first quadrant; “few,” greater than 10 colonies in the first quadrant; “moderate,” greater than 10 colonies and growth into the second quadrant; and “abundant,” heavy growth of colonies in the second quadrant, leading to growth into the third or fourth quadrant. Pathogens with a potential to be respiratory microbiota, such as Haemophilus influenzae and Streptococcus pneumoniae, were reported only if they were present in moderate to abundant quantities or if they were the predominant organism. Cultures were reported as “upper respiratory flora” if only normal upper respiratory microbiota were isolated in the absence of significant pathogens.
Tracheal aspirate.
Tracheal aspirate specimens were evaluated by unconcentrated conventional Gram stain prior to plating. Samples were inoculated onto BAP, CHOC, and MAC plates using a cross-streak pattern with a 0.001-ml calibrated loop and were incubated at 35°C in 5% CO2. All plates were examined for growth at 24 h of incubation. MAC plates without growth at 18 to 24 h were discarded. BAP and CHOC plates were reexamined at 48 h and were discarded at that time. Pathogens with a potential to be respiratory microbiota, such as H. influenzae and Streptococcus pneumoniae, were worked up for identification and susceptibility testing if isolated at or above 1 × 105 colonies (col)/ml. Growth of bacterial isolates above this threshold amount were reported as ≥100,000 col/ml. Isolates present at fewer than 1 × 105 col/ml were reported as “insignificant growth.” Cultures were reported as “upper respiratory flora” if only normal upper respiratory microbiota were isolated in the absence of significant pathogens.
Bronchial alveolar lavage and bronchial washing.
Acceptable BAL-type specimens (bronchial wash and bronchial alveolar lavage) were concentrated by Cytospin and then evaluated by conventional Gram stain. Specimens were inoculated onto BAP, CHOC, and MAC plates using a cross-streak pattern with a 0.001-ml calibrated loop and were incubated at 35°C in 5% CO2. All plates were examined for growth at 24 h of incubation. MAC plates without growth at 18 to 24 h were discarded. BAP and CHOC plates were reexamined at 48 h and were discarded at that time. Pathogens with a potential to be respiratory microbiota, such as H. influenzae and Streptococcus pneumoniae, were worked up for identity and susceptibility if isolated at ≥1 × 103 col/ml for a BAL specimen or ≥1 × 104 col/ml for a bronchial washing. Bacterial isolates quantified below these thresholds were reported as “insignificant growth.” Growth at concentrations above these thresholds was reported semiquantitatively as either ≥1,000 col/ml, 10,000 to 100,000 col/ml, or >100,000 col/ml for BAL samples or as 10,000 to 100,000 col/ml or >100,000 col/ml for bronchial washing samples. Cultures were reported as “upper respiratory flora” if only normal upper respiratory microbiota were isolated in the absence of a significant pathogen.
All specimen types.
Isolated bacterial colonies were identified with a combination of spot biochemical testing (e.g., catalase, oxidase) and matrix-assisted laser desorption/ionization–time of flight mass spectrometry (MALDI-TOF MS) using the Bruker Biotyper. Alpha-hemolytic colonies were screened with bile solubility reagent (sodium deoxycholate) or P-disk (optochin) to rule out Streptococcus pneumoniae. Susceptibility testing was performed by a combination of disk diffusion and gradient diffusion methodologies in accordance with CLSI standards, and results were interpreted using current CLSI interpretive criteria (8, 9). S. aureus isolates were classified as either methicillin-resistant (MRSA) or methicillin-susceptible (MSSA) based on the Alere PBP2a SA culture colony test (Abbott Laboratory, Chicago, IL) and disk diffusion testing with cefoxitin (8). The Cepheid (Sunnyvale, CA) Xpert Carba-R assay was used as part of SOC testing for detection and differentiation of blaKPC, blaNDM, blaVIM, blaOXA-48, and blaIMP from isolated colonies of Enterobacterales, which tested intermediate or resistant to any carbapenem tested and were positive by modified carbapenemase inactivation method (mCIM) testing (8). Other SOC test results were included in the study if the testing was ordered 48 h prior to or 48 h after ordering of the bacterial culture. SOC result reporting was performed in real time per the clinical laboratory’s standard operating procedure.
Viral detection.
SOC testing for influenza A virus, influenza B virus, and respiratory syncytial virus (RSV) was performed using the Cepheid Xpert Xpress Flu/RSV assay. Herpes simplex virus (HSV), cytomegalovirus (CMV), and adenovirus were detected by PCR performed on a Diasorin Liaison MDX (Cypress, CA). Biofire respiratory panel V2.0 (RP2) was performed on the BioFire FilmArray Torch with either nasopharyngeal (NP) samples (according to manufacturer’s guidelines) or LRT samples (performed off-label following verification within our laboratory). Each LRT sample was mixed with BioFire sample buffer and was injected into an RP2 pouch by means of the BioFire pouch loading station. The BioFire respiratory viral panel V2.0 includes targets for the following viruses and bacteria: adenovirus, coronavirus (HKU1, NL63, 229E, and OC43), human metapneumovirus, human rhinovirus/enterovirus, influenza viruses (A/H1, A/H3, A/H1-2009, and B), parainfluenza virus (1 to 4), RSV, Bordetella pertussis, Bordetella parapertussis, Chlamydia pneumoniae, and Mycoplasma pneumoniae. All SOC viral test results, including results from BioFire RP2, were reported to clinicians in real time and were collected retrospectively as part of this study.
Other testing.
Several other tests were ordered as part of SOC on a subset of the specimens, including fungal culture, acid-fast bacillus (AFB) culture, Legionella culture, Legionella urine antigen testing, and direct fluorescent antibody (DFA) staining for Pneumocystis. Legionella culture was performed by streaking specimen onto a buffered charcoal-yeast extract agar (BCYE) plate from BD (Franklin Lakes, NJ) and a buffered charcoal-yeast extract agar plate with polymyxin B, anisomycin, and vancomycin (BCYE/PAV) (Remel). Plates were incubated in air at 35○C and were examined for growth at 3, 5, and 7 days; presumptive Legionella isolates were submitted to the Missouri State Public Health Laboratory for confirmation. Legionella urine antigen testing was performed using the Binaxnow Legionella urinary antigen card from Abbott (Abbott Park, IL). DFA staining for Pneumocystis jirovecii was performed using an immunofluorescence test kit from Bio-Rad (Hercules, CA). Any positive blood culture results within 5 days of the BFPP testing were also recorded as part of the chart review.
Detection of microbial species within a microorganism complex.
We evaluated the potential of the BFPP to detect closely related species using prepared suspensions of bacteria, including Klebsiella variicola and Staphylococcus argenteus, by preparing a fresh subculture of clinical isolates. After 18 to 24 h of incubation at 37°C, isolates were inoculated into sterile saline to make a 0.5 McFarland suspension. The McFarland suspension was diluted 1:100 in saline and was run as a BAL specimen according to the package insert for the BFPP.
Chart review and statistics.
Study data, including antibiotic utilization and SOC test results, were collected by retrospective chart review using a REDCap (Research Electronic Data Capture) data collection instrument that was designed as part of this study (see Table S1 in the supplemental material); the RedCap database and tools were hosted at the Institute for Informatics at Washington University School of Medicine (10). Data were analyzed in GraphPad Prism version 8.2.1 (GraphPad, San Diego, CA) using parametric two-sided t test, chi-square test, and least-squares linear regression.
RESULTS
Patient population.
The mean age of study subjects was 57.7 years (range, 18 to 89 years); most subjects were men (62.8%) being treated on a critical care floor (93.5%) and presenting for medical care primarily during the winter (29%) and spring (54.5%) seasons. Nearly half of subjects (95/200 [47.5%]) had an onset of respiratory symptoms prior to or within 48 h of admission. Nearly two-thirds of subjects (129/200 [64.5%]) were on a ventilator for at least 24 h prior to aerobic culture collection, and one-third of subjects (66/200 [33.3%]) were reported as deceased within 30 days of sample collection (Table 1).
TABLE 1.
Population demographics
| Characteristic | Sample type |
|||||
|---|---|---|---|---|---|---|
| BAL type |
Sputum type |
Total |
||||
| No. | Category % | No. | Category % | No. | Category % | |
| Sexa | ||||||
| Female | 29 | 41.4 | 45 | 34.9 | 74 | 37.2 |
| Male | 41 | 58.6 | 84 | 65.1 | 125 | 62.8 |
| Age (yrs) | ||||||
| 18–34 | 9 | 12.9 | 14 | 10.8 | 23 | 11.5 |
| 35–65 | 36 | 51.4 | 81 | 62.3 | 117 | 58.5 |
| >65 | 25 | 35.7 | 35 | 26.9 | 60 | 30.0 |
| Setting | ||||||
| ICU/oncologyb | 69 | 98.6 | 118 | 90.8 | 187 | 93.5 |
| Emergency department | 1 | 1.4 | 12 | 9.2 | 13 | 6.5 |
| Season | ||||||
| Winter (December–February) | 16 | 22.9 | 42 | 32.3 | 58 | 29.0 |
| Spring (March–May) | 40 | 57.1 | 69 | 53.1 | 109 | 54.5 |
| Summer (June–August) | 2 | 2.9 | 12 | 9.2 | 14 | 7.0 |
| Fall (September–November) | 12 | 17.1 | 7 | 5.4 | 19 | 9.5 |
| Hosp onsetc | ||||||
| Yes | 36 | 51.4 | 69 | 53.1 | 105 | 52.5 |
| No | 34 | 48.6 | 61 | 46.9 | 95 | 47.5 |
| Ventd | ||||||
| Yes | 46 | 65.7 | 83 | 63.8 | 129 | 64.5 |
| No | 24 | 34.3 | 47 | 36.2 | 71 | 35.5 |
| 30-day mortalitye | ||||||
| Yes | 20 | 28.6 | 46 | 35.4 | 66 | 33.0 |
| No | 46 | 65.7 | 77 | 59.2 | 123 | 61.5 |
| Unknown | 4 | 5.7 | 7 | 5.4 | 11 | 5.5 |
| Row total | 70 | 35.0 | 130 | 65.0 | 200 | 100.0 |
Gender information was unavailable for one of the study participants.
Including 180 ICU patients and 7 oncology patients on an immunocompromised patient floor.
Onset of respiratory symptoms >48 h after admission to the hospital (Hosp).
Intubated with ventilator support for >24 h prior to aerobic culture.
Death within 30 days of aerobic culture specimen collection.
Overview of BFPP results by specimen type.
BFPP detected bacteria exclusively in 44.5% (89/200) of samples, viruses exclusively in 10% of samples (20/200), and co-occurring bacteria and viruses in 4% (8/200) of samples (Fig. 2). There was no significant difference (P = 0.36) in the number of viruses and bacteria detected by BFPP in sputum-type samples (sputum and tracheal aspirates; n = 130) compared to BAL-type specimens (BAL fluid and bronchial wash; n = 70). Nearly half of positive samples (50/117) contained more than one viral or bacterial target.
FIG 2.
BFPP detection. Panel a shows that BFPP detected a bacterium or virus in 117/200 (58.5%) of samples, with similar distributions between sample types (P = 0.36 [chi-square test] and degrees of freedom [df] = 3). Panel b demonstrates that more than one pathogen was detected by BFPP in 50/116 (43%) of positive samples.
Viral detection.
We next evaluated BFPP test performance characteristics by comparing results from BFPP to those obtained by SOC viral testing. We observed a high overall agreement (overall accuracy; mean, 95% confidence interval [CI]) (99.2% [98.4%, 99.6%]) between these two methods for viral detection; however, positive agreement between methods was lower than expected (82.6% [62.9%, 93.0%]) due to four cases in which a virus was detected by the SOC BioFire RP2 but not by BFPP. These discordant results included two for coronavirus HKU1, one for adenovirus, and one for influenza A virus (Table 2 and Fig. 3). In three cases, the same sample was used for both SOC and retrospective viral testing (Table 3), and a different sample was used in the fourth case in which a coronavirus was detected by SOC RP2 but not by BFPP. For three of these cases, it was possible to compare results with RP2 testing that had been undertaken earlier or later in the hospital course (date of aerobic culture collection ± 5 days). Only one of the SOC-positive, BFPP-negative results was positive at a second time point; this case was also positive for the same target (influenza A virus; cycle threshold [CT], 34.8) when tested retrospectively using the Cepheid Xpert Flu/RSV Xpress assay. The other two discrepant cases had negative results from RP2 testing 2 h earlier (tracheal aspirate negative, subsequent BAL positive for coronavirus) and 4 days earlier (initial BAL negative, subsequent BAL positive for coronavirus), respectively. The percent positive agreement (PPA) between the remaining five viral targets on the BFPP was 100% (Table 2).
TABLE 2.
BFPP performance summary for BAL-type and sputum-type specimens
| Analytec | PPA |
NPA |
BFPP-pos SOC-NTb |
BFPP-neg SOC-NT | ||||
|---|---|---|---|---|---|---|---|---|
| Prop.a | % | 95% CId (%) | Prop. | % | 95% CId (%) | |||
| Viruses | ||||||||
| Rhinovirus/enterovirus | 5/5 | 100 | 56.6–100 | 103/103 | 100 | 96.4–100 | 4 | 88 |
| Influenza A virus | 4/5 | 80 | 37.6–99.0 | 104/105 | 99.0 | 94.8–99.9 | 1 | 89 |
| Coronavirus | 1/3 | 33.3 | 1.7–88.2 | 106/107 | 99.1 | 94.9–99.9 | 1 | 91 |
| Influenza B virus | 3/3 | 100 | 43.9–100 | 107/107 | 100 | 96.5–100 | 1 | 89 |
| RSV | 4/4 | 100 | 51.0–100 | 106/106 | 100 | 96.5–100 | 0 | 90 |
| Metapneumovirus | 1/1 | 100 | 5.1–100 | 107/107 | 100 | 96.5–100 | 2 | 90 |
| Parainfluenza virus | 1/1 | 100 | 5.1–100 | 106/107 | 99.1 | 94.9–99.9 | 0 | 92 |
| Adenovirus | 0/1 | 0 | 0–94.9 | 107/107 | 100 | 96.5–100 | 0 | 92 |
| Total | 19/23 | 82.6 | 62.9–93.0 | 846/849 | 99.5 | 99.0–99.9 | 9 | 721 |
| Bacteria | ||||||||
| Staphylococcus aureus | 22/22 | 100 | 85.1–100 | 156/178 | 87.6 | 82.0–91.7 | 0 | 0 |
| Haemophilus influenzae | 3/3 | 100 | 43.9–100 | 172/197 | 87.3 | 81.9–91.3 | 0 | 0 |
| Pseudomonas aeruginosa | 8/8 | 100 | 67.6–100 | 185/192 | 96.4 | 92.7–98.2 | 0 | 0 |
| Enterobacter cloacae complex | 3/4 | 75 | 30.1–98.7 | 189/196 | 96.4 | 92.8–98.3 | 0 | 0 |
| Escherichia coli | 6/6 | 100 | 61.0–100 | 190/194 | 97.9 | 94.8–99.2 | 0 | 0 |
| Streptococcus pneumoniae | 2/2 | 100 | 17.8–100 | 191/198 | 96.5 | 92.9–98.3 | 0 | 0 |
| Klebsiella pneumoniae group | 4/4 | 100 | 51.0–100 | 192/196 | 98.0 | 94.9–99.2 | 0 | 0 |
| Streptococcus agalactiae | 2/2 | 100 | 17.8–100 | 193/198 | 97.5 | 94.2–98.9 | 0 | 0 |
| Klebsiella aerogenes | 2/2 | 100 | 17.8–100 | 196/198 | 99.0 | 96.4–99.8 | 0 | 0 |
| Klebsiella oxytoca | 3/3 | 100 | 43.9–100 | 196/197 | 99.5 | 97.2–99.9 | 0 | 0 |
| Moraxella catarrhalis | 1/1 | 100 | 5.1–100 | 196/199 | 98.5 | 95.7–99.6 | 0 | 0 |
| Proteus spp. | 0/0 | NAe | NA | 196/200 | 98.0 | 95.0–99.2 | 0 | 0 |
| Serratia marcescens | 3/3 | 100 | 43.9–100 | 197/197 | 100 | 98.1–100 | 0 | 0 |
| Acinetobacter baumannii–calcoaceticus complex | 0/0 | NA | NA | 199/200 | 99.5 | 97.2–99.9 | 0 | 0 |
| Legionella pneumophila | 0/0 | NA | NA | 44/44 | 100 | 92.0–100 | 1 | 156 |
| Mycoplasma pneumoniae | 1/1 | 100 | 5.1–100 | 86/86 | 100 | 95.7–100 | 0 | 80 |
| Total | 60/61 | 98.4 | 91.3–99.9 | 2778/2870 | 96.8 | 96.1–97.4 | 1 | 236 |
Prop., proportion for positive percent agreement (PPA) or negative percent agreement (NPA), calculated as follows: PPA = concordant positive/all positive results by SOC, and NPA = concordant negative/all negative results by SOC.
BFPP-pos, SOC-NT, analyte detected by BFPP but not tested by standard of care.
SOC testing also detected herpes simplex virus (n = 5), cytomegalovirus (n = 4), Stenotrophomonas maltophilia (n = 4), Acinetobacter spp. (n = 1), Citrobacter freundii complex (n = 1), Klebsiella variicola (n = 1), and Staphylococcus intermedius group (n = 1), which are not included in the BFPP panel. SOC testing detected yeast or mold in an additional 26 samples, including 21 yeasts, 2 Aspergillus spp., 1 Blastomyces dermatitidis/B. gilchristii isolate, 1 Paecilomyces species, and 1 Pneumocystis jirovecii isolate.
CI, confidence agreement for a binomial distribution, calculated by the Wilson/Brown method.
NA, not applicable.
FIG 3.
Concordance of BFPP with standard of care (SOC) for viral and bacterial identification. Panel a shows a high overall agreement (99%) of BFPP with SOC for detection of viruses. Panel b shows a predominance of S. aureus and H. influenzae among detected bacterial targets and displays 92 bacterial targets detected by BFPP and not by SOC testing.
TABLE 3.
Details on discordant viral test resultsa
| Sample | Viral target | Discordance | Specimen type |
Same specimen analyzed? | Difference in sample collection times (h) | |
|---|---|---|---|---|---|---|
| BFPP | SOC | |||||
| 1 | Coronavirus | SOC RP2+, BFPP− | BAL | BAL | No | 34:35 |
| 2 | Coronavirus | SOC RP2+, BFPP− | BAL | BAL | Yes | NA |
| 3 | Adenovirus | SOC RP2+, BFPP− | Tracheal aspirate | Tracheal Aspirate | Yes | NA |
| 4 | Influenza A virusb | SOC RP2+, BFPP− | Tracheal aspirate | Tracheal aspirate | Yes | NA |
| 5 | Influenza A virus | BFPP+, SOC RP2− | BAL | BAL | Yes | NA |
| 6 | Coronavirus | BFPP+, SOC RP2− | Sputum | BAL | No | 9:25 |
| 7 | Parainfluenza virus | BFPP+, SOC RP2− | Tracheal aspirate | Tracheal aspirate | Yes | NA |
The Biofire respiratory panel V2.0 (RP2) was performed for each of the lower respiratory tract (LRT) samples included as a laboratory-developed test.
Specimen retrospectively positive for influenza A virus by Xpert Flu/RSV Xpress assay (cycle threshold [CT], 34.8).
A virus was detected by BFPP but not by SOC in an additional three cases. These discordant viral results were for influenza A virus, coronavirus, and parainfluenza virus (Table 3). Of note, BFPP detected a viral target in an additional nine cases (4.5% of specimens) where there was no SOC testing for the virus within a 96-h period before or after aerobic culture. In contrast, SOC testing detected a virus that was not on the BioFire panel in nine cases (4.5%), including five cases with herpes simplex virus (HSV) and four cases with cytomegalovirus (CMV).
Bacterial detection.
Overall agreement between BFPP and SOC for detection and identification of bacteria was 96.8% (96.1%, 97.4%). Likewise, we observed a high PPA for bacterial detection, 98.4% (91.3%, 99.9%) (Table 2). An on-panel bacterium (Enterobacter cloacae complex) was detected by SOC but not by BFPP in only one case. An additional eight samples (4%) grew bacteria that were not part of the BFPP panel, including four Stenotrophomonas maltophilia isolates, one non-baumannii-complex Acinetobacter sp., one Citrobacter freundii complex isolate, one Staphylococcus intermedius group isolate, and one Klebsiella variicola isolate. Two bacteria from the BFPP panel, Chlamydia pneumoniae and Streptococcus pyogenes, were not detected in this cohort by either SOC or BFPP.
BFPP also showed a high negative percent agreement (NPA) with results from aerobic culture. NPA for bacterial identification was 96.8% (96.1%, 97.4%). Two bacteria, S. aureus and H. influenzae, had an NPA of less than 90%. These organisms accounted for over half of the 92 bacteria that were detected by BFPP alone, including 22 S. aureus and 25 H. influenzae isolates that were detected only by BFPP, compared to 22 S. aureus and 3 H. influenzae isolates that were detected concordantly. NPA was greater than 90% for the remaining bacteria; however, the number of discordant bacterial detections exceeded the number of concordant detections (noted as discordant: concordant in the following passage) for Enterobacter cloacae complex (7:3), Streptococcus pneumoniae (7:2), Streptococcus agalactiae (5:2), Proteus spp. (4:0), and Moraxella catarrhalis (3:1). SOC testing detected yeast or mold in an additional 26 samples, including 21 yeasts, 2 Aspergillus spp., 1 Blastomyces dermatitidis/Blastomyces gilchristii isolate, 1 Paecilomyces species, and 1 Pneumocystis jirovecii isolate.
Detection of S. aureus and determination of antimicrobial resistance.
Twenty-two additional S. aureus were detected by BFPP alone, including 15 MSSA and 7 MRSA isolates that were positive for mecA/C and MREJ (Table 4). For the majority of cases with discordant S. aureus results (15/22 [68.2%]), the subject was on effective antibiotics (based on hospital antibiogram and BFPP mecA/C status) at the time of aerobic culture, reflecting the widespread use of antibiotics in this population (149/200 [74.5%] of patients on ≥1 antibiotic at the time of aerobic culture). BFPP also identified MRSA in four cases where SOC testing reported MSSA, including one discordant result in BAL-type specimens and three discordant results among sputum-type specimens (Table 4), resulting in a moderately decreased NPA for both BAL-type (75% [30.1%, 98.7%]) and sputum-type (70% [39.7%, 89.2%]) specimens. These four additional MRSA detections by BFPP were from polymicrobial samples.
TABLE 4.
S. aureus identification and susceptibility by sample typea
| Sample type and BFPP result | SOC aerobic culture |
||
|---|---|---|---|
| MRSA | MSSA | S. aureus neg. | |
| Sputum typeb | |||
| MRSA | 3 | 3 | 7 |
| MSSA | 0 | 7 | 12 |
| S. aureus neg. | 0 | 0 | 98 |
| BAL typec | |||
| MRSA | 5 | 1 | 0 |
| MSSA | 0 | 3 | 3 |
| S. aureus neg. | 0 | 0 | 58 |
| Overall | |||
| MRSA | 8 | 4 | 7 |
| MSSA | 0 | 10 | 15 |
| S. aureus neg. | 0 | 0 | 156 |
MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; neg., negative.
Sputum-type specimens include sputum (n = 54) and tracheal aspirate (n = 76).
BAL-type specimens include bronchoalveolar lavage fluid (BAL) (n = 59) and bronchial washing (n = 11).
BFPP detected Gram-negative antimicrobial resistance (AMR) genes in eight samples (Table 5). Six of these samples were positive for only blaCTX-M, one sample was positive for both blaCTX-M and blaKPC, and an additional sample was positive for only blaKPC based on BFPP results. BFPP detected all four cases with AMR reported by SOC (overall PPA: 100%) as well as an additional three cases (three blaCTX-M) that were not evaluated for AMR by SOC because suspect bacteria were not present at threshold levels in culture, including one sample with Enterobacter cloacae complex (107 copies/ml by BFPP), one sample with Escherichia coli (106 copies/ml) and Klebsiella pneumoniae group (105 copies/ml), and one sample with Klebsiella pneumoniae group (104 copies/ml) reported by BFPP alone. Overall, 4/7 (57.1%) samples were from subjects with putative mecA/C and MREJ, who were on optimal antimicrobial regimens, and 1/3 (33.3%) of samples were from subjects with blaCTX-M who were receiving appropriate antibiotics at the time of culture.
TABLE 5.
BFPP performance characteristics for detection of AMR compared with SOC methods
| Analytea | PPA |
NPA |
Discordant bacterial ID |
|||||
|---|---|---|---|---|---|---|---|---|
| Prop.b | % | 95% CIh (%) | Prop.b | % | 95% CIh (%) | BFPP AMR-pos SOC AMR-NTc | SOC AMR-pos BFPP AMR-NTd | |
| mecA/C or MREJe | 8/8 | 100 | 67.6–100 | 10/14 | 71.4 | 45.3–88.3 | 7 | 0 |
| CTX-Mf | 4/4 | 100 | 51.0–100 | 24/24 | 100 | 86.2–100 | 3 | 0 |
| IMP | 0/0 | NA | NA | 29/29 | 100 | 88.3–100 | 0 | 0 |
| KPCg | 1/1 | 100 | 5.1–100 | 28/28 | 100 | 87.9–100 | 1 | 0 |
| NDM | 0/0 | NA | NA | 29/29 | 100 | 88.3–100 | 0 | 0 |
| VIM | 0/0 | NA | NA | 29/29 | 100 | 88.3–100 | 0 | 0 |
| OXA-48-like | 0/0 | NA | NA | 21/21 | 100 | 84.5–100 | 0 | 0 |
Disk diffusion testing was used as an initial SOC method for evaluating antimicrobial resistance (AMR).
Fractional proportion (Prop.) of PPA or NPA, expressed as follows: PPA = concordant positive/all positive results by SOC, and NPA = concordant negative/all negative results by SOC.
BFPP positive for AMR gene; SOC AMR was not tested (NT) because of discordant identification of bacteria by BFPP alone.
There were no cases where SOC AMR was positive and BFPP AMR was not tested.
mecA/C status was determined by disk diffusion testing (cefoxitin resistance) and positive results on Alere™ PBP2a SA culture colony test (Abbott).
Phenotypic evidence of extended-spectrum-beta-lactamase (ESBL) production on SOC disk diffusion testing was considered for the purposes of the table as concordant with CTX-M detection by BFPP.
KPC status was determined for SOC samples using Cepheid Xpert.
CI, confidence agreement for a binomial distribution, calculated by the Wilson/Brown method.
One of the two samples that were positive for blaKPC by BFPP was not evaluated for antimicrobial resistance by SOC because detected bacteria were below the threshold for workup and reporting for tracheal aspirates. In this case, aerobic culture reported yeast (≥100,000 col/ml) plus growth of clinically insignificant bacterial flora (<1 × 105 col/ml), whereas BFPP identified Enterobacter cloacae complex (104 copies/ml) and Pseudomonas aeruginosa (107 copies/ml). A carbapenem-susceptible Enterobacter cloacae complex was also reported on SOC blood cultures from the previous day, along with vancomycin-resistant Enterococcus faecium; blood cultures were negative when repeated the day after aerobic cultures were taken. This patient was discharged 8 days later in stable condition on meropenem and daptomycin but expired within the following 10 days due to unreported causes.
Factors related to discordant bacterial detection.
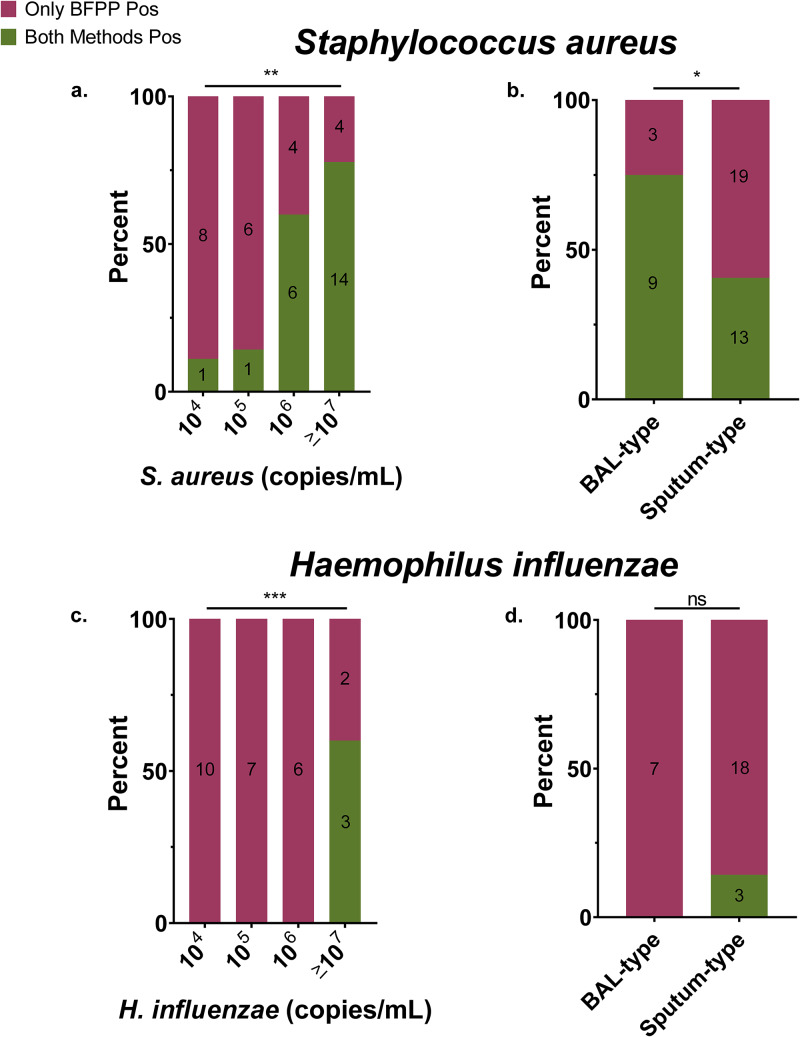
We examined factors that might predict S. aureus and H. influenzae discordance for SOC versus BFPP, including bacterial quantity from BFPP, rare or no polymorphonuclear leukocytes (PMNs) on Gram stain (marker for suboptimal specimens), abundant squamous epithelium on Gram stain (marker of oropharyngeal contamination), sputum or tracheal aspirate sample type, and empirical antibiotic coverage of detected bacteria (e.g., MRSA, MSSA, or H. influenzae). Least-squares regression was used to fit these covariates to a linear model. Multivariate analysis showed that low S. aureus quantity (P = 0.0019 and Pearson’s r = −0.49) and sputum/tracheal aspirate sample type (P = 0.03 and Pearson’s r = −0.13) were associated with discordant detection by BFPP (model fit: r-squared = 0.35 and degrees of freedom = 37) (Table S2 and Fig. S3). Gram stain PMNs (P = 0.63), squamous epithelium quantity (P = 0.63), and empirical coverage for S. aureus (P = 0.29) were not significantly associated with discordant detection of S. aureus. Similar to results from S. aureus, a low quantity of H. influenzae (P = 0.0001 and Pearson’s r = −0.74) organisms was associated with discordant detection of H. influenzae by BFPP (Fig. 4) (model fit: r-squared = 0.57 and degrees of freedom = 21), such that concordant results for H. influenzae were only detected at ≥107 copies/ml as measured by BFPP. Unlike with S. aureus, concordant detection of H. influenzae was not associated with specimen type. PMNs (P = 0.98), squamous epithelium quantity (P = 0.27), and empirical coverage for H. influenzae (P = 0.09) were not significantly associated with discordant detection of H. influenzae (Table S2 and Fig. S3).
FIG 4.
Discordant detection of S. aureus and H. influenzae by bacterial quantity (left) and specimen type (right). Multivariate, least-squares, linear regression demonstrates that S. aureus (a and b) and H. influenzae (c and d) quantity as measured by BFPP was directly and independently associated with concordant detection by BFPP and SOC culture, compared to detection by BFPP alone (S. aureus model, r-squared = 0.35 and df = 37; H. influenzae model, r-squared = 0.57 and df = 21). P values from regression coefficients are represented as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05; and ns, nonsignificant. Values inside columns represent the number of samples per horizontal-axis category as grouped by test concordance.
Detection of microorganisms within an organism complex.
By running organism suspensions in pure cultures of Klebsiella variicola (member of the Klebsiella pneumoniae group) and S. argenteus (member of the S. aureus complex), we were able to determine the potential for BFPP to detect these clinically important species. Testing showed that 3/3 (100%) Klebsiella variicola isolates were identified as Klebsiella pneumoniae group. Likewise, 1/1 (100%) of Staphylococcus schweitzeri and 2/2 (100%) S. argenteus isolates were identified by the BFPP as the closely related species S. aureus.
Turnaround time comparison.
The BFPP has a turnaround time (TAT) and workflow considerations similar to those for the SOC BioFire RP2. Therefore, the TAT from RP2 testing that was performed as part of SOC for a subset of subjects in this study (n = 95) was used as a surrogate for turnaround time estimation for BFPP. Organism identification was, on average, 42.2 h faster with the BFPP surrogate than with SOC (P < 0.0001, 2-tailed, unpaired t test). Likewise, the turnaround time for detection of AMR markers by the BFPP surrogate was shorter (56.3-h reduction; P < 0.0001) than determination of antimicrobial susceptibility by standard phenotypic methods (Fig. 5).
FIG 5.
Projected time savings with BFPP compared to SOC aerobic culture. Turnaround time for the BioFire upper respiratory panel (surrogate for BFPP) was significantly shorter (****, P < 0.0001) than SOC culture for identification (ID) of bacteria (42.2-h reduction). Likewise, the turnaround time for detection of antimicrobial resistance (AMR) markers by BFPP was shorter (P < 0.0001) than determination of antimicrobial susceptibility by standard phenotypic methods (56.3-h reduction). Comparisons were made via two-tail, unpaired t tests.
Clinical outcomes.
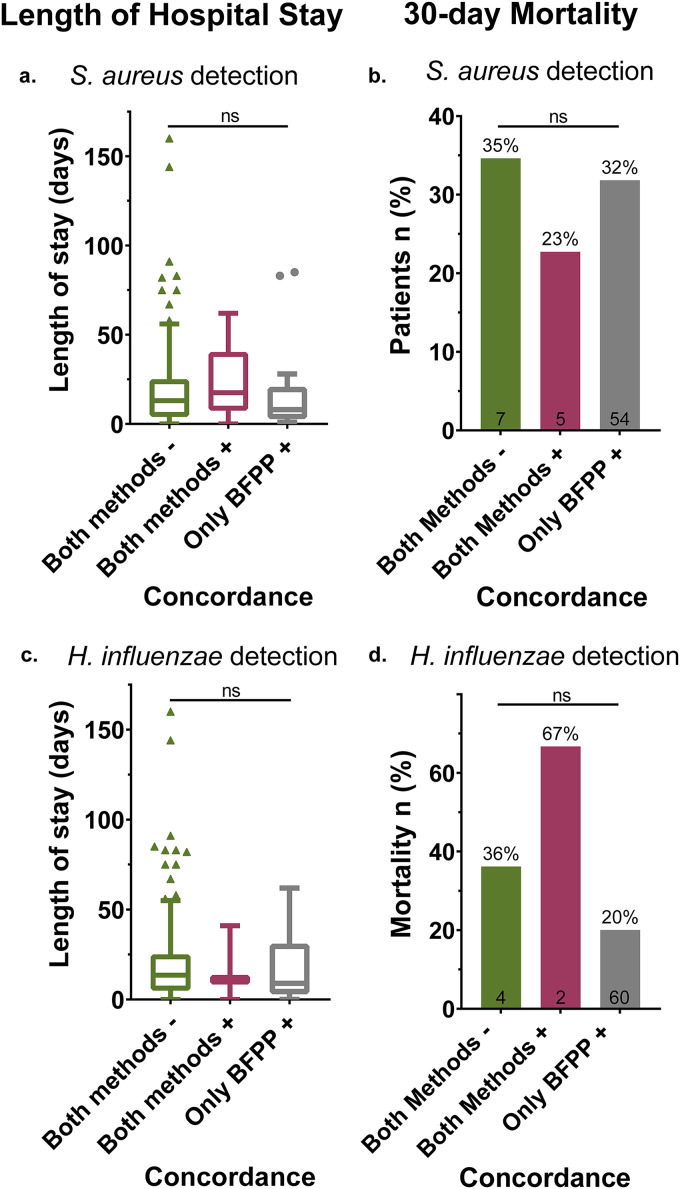
If BFPP were detecting clinically significant infections missed by SOC (true positive), one would expect worse clinical outcomes among these undiagnosed patients. Therefore, we used one-way analysis of variance (ANOVA) to test for a difference in length of hospital stay, and we applied a two-sided Fisher’s exact test to test for differences in 30-day mortality between these otherwise similar patient groups (Table S2). Analysis demonstrated that length of hospital stay and 30-day mortality, as measured from time of aerobic culture, were not significantly different (P > 0.05) for patients with concordant versus discordant S. aureus and H. influenzae detection (Fig. 6).
FIG 6.
Length of hospital stay and 30-day mortality grouped by test concordance between BFPP and SOC aerobic culture for detection of S. aureus and H. influenzae. One-way analysis of variance (ANOVA) demonstrates that length of hospital stay (left) was not significantly different for S. aureus [F(2,195) = 0.088, P = 0.92] (a) or H. influenzae [F(2,195) = 0.135, P = 0.87] (c) detected by both BFPP and SOC culture compared to detection by BFPP alone. Two-sided Fisher’s exact test demonstrated that 30-day mortality (right) was not significantly different for S. aureus (P = 0.74) (b) or H. influenzae (P = 0.15) (d) detected by both BFPP and SOC compared detection by BFPP alone. Values inside columns represent the number of patient deaths per x axis group that occurred within 30 days following aerobic culture testing.
DISCUSSION
This study evaluated the BFPP in the setting of a large, academic medical center. Our results demonstrate that the BFPP is a rapid method for identifying pathogens and resistance markers in patients with suspected pneumonia. Our data show that a bacterial pathogen was detected in a high proportion of samples (48.5%) from our patient cohort, likely reflective of our inclusion criteria. Fourteen percent of samples were positive for viral targets, including 8 specimens (4%) with bacterial-viral codetection. Bacterial codetection was identified in more than a quarter (28.6%) of cases in which a virus was detected by BFPP. These results are comparable with the incidence of bacterial-viral coinfection reported elsewhere (11–13).
We report a high overall agreement between BFPP and SOC for detection of bacteria and viruses. However, we also note that BFPP detects additional bacteria, including S. aureus and H. influenzae, that are not reported by SOC. We show that these BFPP-only detections are more common among samples with a low concentration of bacteria as well as among sputum-type samples than among BAL-type specimens.
The BFPP demonstrated high negative percent agreement (NPA) compared to SOC viral detection, suggesting high clinical specificity. However, the average positive percent agreement (PPA) for BFPP viral testing was lower, at just over 80%, whereas the results from the 510(k) BFPP submission to the FDA reported an overall PPA of more than 90% for 1829 clinical samples (14). The majority of BFPP viral targets in our study were fully concordant with SOC; however, there were four discordant viral targets that reduced the overall test performance, including influenza A virus, coronavirus, adenovirus, and parainfluenza virus. Of note, only one out of three coronaviruses detected by SOC (detected by BioFire RP2) were also detected by BFPP. This level of discordance among coronaviruses significantly reduced the overall PPA for viral testing. These results are consistent with the BFPP 510(k) submission, which reported a PPA of less than 90% for detection of coronavirus. In contrast, the BioFire RP2 assay has been shown to perform very well for detection of coronavirus, with a PPA of 100% for coronavirus HKU1 (n = 43) (15).
It is notable that two of the discordant coronavirus results that were positive by BioFire RP2 and negative by BFPP were negative on prior RP2 testing performed 2 h before and 4 days before the positive SOC, suggesting that discordant results for coronavirus may be related less to the assay and more to preanalytical factors, such as sample quality, sample source, amount of target, or time of sampling. Sample-to-sample variability may have contributed to discordant coronavirus results; however, this was less of a factor in discordant detection of influenza A virus and adenovirus by SOC methods, given that both SOC and BFPP testing were performed on the same tracheal aspirate. A virus was detected by BFPP but not by SOC in an additional three cases. As was the case for discordant SOC viral detections, we did not identify a feature that clearly predicts viral test concordance. A direct comparison of BFPP with RP2 may be warranted and would be a useful benchmark for laboratories that are considering switching from a laboratory-developed RP2 assay for LRT specimens to the BFPP.
The BFPP assay showed high overall agreement with SOC culture for the detection of bacteria commonly associated with pneumonia. Average PPA and NPA were very similar to the values reported in the multisite 510(k) FDA submission (14). Only 1 of the 16 bacterial species detected by aerobic culture was negative when assayed by BFPP, namely, Enterobacter cloacae complex, which had a PPA of 75%. There were five off-panel bacterial species that were detected by SOC but not by BFPP, and most of these results were only obtained once in the study, suggesting that the panel includes most of the common causes of pneumonia. Only one such bacterium, Stenotrophomonas maltophilia, was detected in multiple samples (n = 4). The absence of Stenotrophomonas maltophilia from the BioFire pneumonia panel is a potential concern because this multidrug-resistant bacterium is commonly encountered among hospitalized patients and can cause significant morbidity and mortality among immunocompromised patients (16).
These results can be compared to those described in the Curetis Unyvero LRT application (Holzgerlingen, Germany), which received initial FDA clearance in 2018 for qualitative detection of 19 bacterial targets and 10 resistance markers in tracheal aspirates (17). The assay was recently expanded to include Pneumocystis jirovecii and was cleared for use on BAL specimens as well (18). A multicenter clinical trial of the Unyvero LRT in 603 fresh and 185 archived tracheal aspirate samples showed an overall weighted sensitivity of 92.5% and an overall specificity of 97.4% compared to those of culture for bacterial identification (19), and a recent retrospective comparison of the Unyvero LRT to routine bacterial culture of BAL fluids reported an overall PPA of 96.5% and an NPA of 99.6% (20). This study also reported a 77.8% (14/18) concordance between resistance targets and phenotypic susceptibility testing. The BFPP has notable differences from the Unyvero panel, including shorter turnaround time, quantitative reporting of bacteria, and inclusion of viral targets; however, the two assays have similar overall test performance characteristics.
Overall, our results suggest that BFPP is quite sensitive for the detection and identification of common bacterial causes of pneumonia. A potential implementation challenge of the assay is that it detects additional bacteria not reported by SOC culture. This was most pronounced for S. aureus and H. influenzae, which together accounted for more than half of bacteria detected by BFPP alone. These results are consistent with data from the FDA clearance study for BFPP, which reported an NPA of more than 90% for all the panel components except for S. aureus and H. influenzae, both of which had NPAs slightly less than 90%. It was initially unclear what factors best explained the large number of bacteria detected by BFPP alone. Our data showed that specimen type and bacterial quantity as determined by BFPP were significant predictors of detection by BFPP alone. The effect of bacterial quantity on test concordance is consistent with prior work showing that PCR-based methods generally detect both viable and nonviable bacteria as well as extracellular bacterial DNA (21, 22). This feature has been useful in determining the etiology of meningitis following antibiotics (15); however, it may be a liability in the setting of respiratory infection, where PCR would be expected to overestimate bacterial count, increase the number of clinically irrelevant detections, and drive excessive antimicrobial use. This may also have an impact on contact isolation, especially when MRSA is detected solely by BFPP. We noted an effect of specimen type on discordant detection of S. aureus, such that S. aureus was significantly more likely to be detected by BFPP alone in sputum and tracheal aspirate samples. This may be the result of criteria that are used in the clinical laboratory for reporting of results from respiratory culture, which tend to limit the reporting of potential upper respiratory flora, such as S. aureus and H. influenzae, when present at low quantities in sputum or tracheal aspirates or when found as part of normal flora in a polymicrobial culture or low-quality specimen as indicated by few leukocytes or numerous epithelial cells on Gram stain. While a reporting threshold is built into the BFPP, other criteria were not used to limit BFPP reporting. These differences in reporting may explain why nearly 100 additional bacteria were identified by BFPP alone.
BFPP showed complete agreement with SOC for detection of AMR (i.e., PPA of 100%), suggesting high assay sensitivity. NPA for mecA/C and MREJ detection was less than 90%, due to four cases in which BFPP identified MRSA but SOC testing reported MSSA. All of these discordant AMR results were detected in polymicrobial samples, suggesting one of the following: (i) SOC culture workup failed to detect a mixed specimen with co-occurring MRSA and MSSA, (ii) the detected S. aureus had an empty SCCmec cassette, or (iii) the MRSA was below the threshold for reporting.
There was complete agreement between methods for detection of Gram-negative AMR; however, this comparison was limited among samples with discordant bacterial identification, including four samples with AMR markers (3 blaCTX-M and 1 blaKPC) that were not fully characterized by SOC due to discordant bacterial identification. Results for AMR are similar to those reported as part of the 510(k) BFPP submission to the FDA, which likewise showed high positive agreement and modest (<90%) negative agreement with a comparator assay for mecA/C and MREJ detection (14). The FDA clearance study also reported a moderately decreased positive percent agreement (<90%) for blaCTX-M detection, which is not evident in the present study, but this may also vary depending on the prevalence, based on local epidemiology and resistance patterns. A comparison of results with the Unyvero LRT is limited by the modest sample size in the present study, but in general these two assays had similarly high PPA and NPA for detection of blaCTX-M and blaKPC. The Unyvero LRT assay is also reported to have an NPA over 90% for mecA detection, and PPA for the Unyvero LRT is slightly below 90% for mecA detection among tracheal aspirates (17). A potential benefit of the BFPP is detection of AMR among samples that might not otherwise be screened for resistance markers, such as the putative blaKPC-positive sample that was identified by BFPP but not by routine SOC testing or the 10 cases with blaCTX-M or mecA/C and MREJ on suboptimal therapeutic regiments for these putative ARM markers. This represents a potential opportunity for improved treatment, infection prevention, and monitoring of antimicrobial resistance.
The results of this study demonstrate that clinical outcomes were not significantly different regardless of whether S. aureus and H. influenzae were detected by both SOC and BFPP or by BFPP alone. Results suggest that either the effect size was too small to be detected in the current study, as may be the case for H. influenzae, that these patients were treated empirically, or that patients with positive results by BFPP alone for these bacteria do not have a true, clinically significant respiratory infection. Although BFPP results were not reported to clinicians in this study, putative false-positive results could lead to overtreatment if conveyed directly to clinicians. Our data do suggest that health care providers can predict detections by the BioFire alone based on semiquantitative bin value. Therefore, the reporting of bin values is critical for clinical interpretation of results. Given the necessity of and growing emphasis on antimicrobial stewardship, future research may explore the clinical impact of BFPP testing and evaluate methods for conveying these results to clinicians.
This retrospective study is one of the first to evaluate the performance of the BioFire FilmArray pneumonia panel among patients with suspected lower respiratory infection. The limitations of this work relate to its moderate sample size, retrospective format, and single-study-center design, which reduces generalizability. Another potential limitation to the study is that it did not evaluate BFPP test performance with low-quality sputum samples that were unsuitable for culture. Presumably the testing of such specimens would result in more frequent detection of clinically insignificant respiratory microbiota and a resultant decrease in clinical specificity. Though more data are needed regarding the negative effect of testing low-quality sputum specimens, laboratories should consider adopting strategies to avoid such practices. This is consistent with the manufacturer’s instructions suggesting that laboratories should adhere to their own established rules for the acceptance and rejection of sputum specimens (23). Strengths of this study include rigorous inclusion criteria, collection of detailed information on standard-of-care testing and clinical outcomes, representation of all four seasons during data collection, and assessment of both sputum-type and BAL-type specimens.
In conclusion, BFPP is a rapid and accurate method for detection of bacteria and viruses from lower respiratory tract infections.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by BioFire Diagnostics.
We thank Caitlin Johnson for coordinating sample enrollment for the study.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.WHO. 2019. World health statistics 2019: monitoring health for the SDGs, sustainable development goals. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Kuti EL, Patel AA, Coleman CI. 2008. Impact of inappropriate antibiotic therapy on mortality in patients with ventilator-associated pneumonia and blood stream infection: a meta-analysis. J Crit Care 23:91–100. doi: 10.1016/j.jcrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Meehan TP, Fine MJ, Krumholz HM, Scinto JD, Galusha DH, Mockalis JT, Weber GF, Petrillo MK, Houck PM, Fine JM. 1997. Quality of care, process, and outcomes in elderly patients with pneumonia. JAMA 278:2080–2084. doi: 10.1001/jama.1997.03550230056037. [DOI] [PubMed] [Google Scholar]
- 4.Houck PM, Bratzler DW, Nsa W, Ma A, Bartlett JG. 2004. Timing of antibiotic administration and outcomes for Medicare patients hospitalized with community-acquired pneumonia. Arch Intern Med 164:637–644. doi: 10.1001/archinte.164.6.637. [DOI] [PubMed] [Google Scholar]
- 5.Metlay JP, Waterer GW, Long AC, Anzueto A, Brozek J, Crothers K, Cooley LA, Dean NC, Fine MJ, Flanders SA, Griffin MR, Metersky ML, Musher DM, Restrepo MI, Whitney CG. 2019. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 200:e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance, London, United Kingdom. [Google Scholar]
- 7.CDC. 2019. Antibiotic resistance threats in the United States. US Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- 8.CLSI. 2018. M100 Performance standards for antimicrobial susceptibility testing. CLSI, Wayne, PA. [Google Scholar]
- 9.CLSI. 2015. M02-A12 Performance standards for antimicrobial disk susceptibility tests. Approved standard, 12th ed CLSI, Wayne, PA. [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. 2009. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah NS, Greenberg JA, McNulty MC, Gregg KS, Riddell J, Mangino JE, Weber DM, Hebert CL, Marzec NS, Barron MA, Chaparro-Rojas F, Restrepo A, Hemmige V, Prasidthrathsint K, Cobb S, Herwaldt L, Raabe V, Cannavino CR, Hines AG, Bares SH, Antiporta PB, Scardina T, Patel U, Reid G, Mohazabnia P, Kachhdiya S, Le B-M, Park CJ, Ostrowsky B, Robicsek A, Smith BA, Schied J, Bhatti MM, Mayer S, Sikka M, Murphy-Aguilu I, Patwari P, Abeles SR, Torriani FJ, Abbas Z, Toya S, Doktor K, Chakrabarti A, Doblecki-Lewis S, Looney DJ, David MZ. 2016. Bacterial and viral co-infections complicating severe influenza: incidence and impact among 507 US patients, 2013–14. J Clin Virol 80:12–19. doi: 10.1016/j.jcv.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice TW, Rubinson L, Uyeki TM, Vaughn FL, John BB, Miller Iii RR, Higgs E, Randolph AG, Smoot BE, Thompson BT. 2012. Critical illness from 2009 pandemic influenza A (H1N1) virus and bacterial co-infection in the United States. Crit Care Med 40:1487. doi: 10.1097/CCM.0b013e3182416f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings LC, Anderson TP, Beynon KA, Chua A, Laing RTR, Werno AM, Young SA, Chambers ST, Murdoch DR. 2008. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax 63:42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 14.BioFire Diagnostics LLC. 2018. 510(k) substantial equivalence determination summary K180966: FilmArray pneumonia panel. US Department of Health and Human Services, Food and Drug Administration, Washington, DC. [Google Scholar]
- 15.Leber AL, Everhart K, Balada-Llasat J-M, Cullison J, Daly J, Holt S, Lephart P, Salimnia H, Schreckenberger PC, DesJarlais S, Reed SL, Chapin KC, LeBlanc L, Johnson JK, Soliven NL, Carroll KC, Miller J-A, Dien Bard J, Mestas J, Bankowski M, Enomoto T, Hemmert AC, Bourzac KM. 2016. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol 54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Looney WJ, Narita M, Mühlemann K. 2009. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 17.Curetis GH. 2018. Evaluation of automatic class III designation for Unyvero LRT application and Unyvero system, DEN170047. US Department of Health and Human Services, Food and Drug Administration, Washington, DC. [Google Scholar]
- 18.Curetis GH. 2019. 510(k) substantial equivalence determination summary K191967: Unyvero LRT BAL application. US Department of Health and Human Services, Food and Drug Administration, Washington, DC. [Google Scholar]
- 19.Qi C, Wunderink RG, Sims M, Humphries RM, Kallstrom G, Carroll KC, Wu F, Dwight H, Butler-Wu S, Klein M, Schwarzer K, Patel R. 2018. Multicenter clinical trial of the Unyvero lower respiratory tract infection application (P0558). 28th European Congress of Clinical Microbiology and Infectious Diseases Meeting, Madrid, Spain. [Google Scholar]
- 20.Collins ME, Popowitch EB, Miller MB. 2020. Evaluation of a novel multiplex PCR panel compared to quantitative bacterial culture for the diagnosis of lower respiratory tract infections. J Clin Microbiol 58:e02013-19. doi: 10.1128/JCM.02013-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josephson KL, Gerba CP, Pepper IL. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol 59:3513–3515. doi: 10.1128/AEM.59.10.3513-3515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young G, Turner S, Davies JK, Sundqvist G, Figdor D. 2007. Bacterial DNA persists for extended periods after cell death. J Endod 33:1417–1420. doi: 10.1016/j.joen.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 23.BioFire Diagnostics. 2019. FilmArray pneumonia panel instructions for use. https://www.online-ifu.com/ITI0075.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.